Abstract
In Shigella boydii 0-1392, genes encoding the synthesis and transport of the hydroxamate siderophore aerobactin are located within a 21-kb iron transport island between lysU and the pheU tRNA gene. DNA sequence analysis of the S. boydii 0-1392 island, designated SHI-3 for Shigella island 3, revealed a conserved aerobactin operon associated with a P4 prophage-like integrase gene and numerous insertion sequences (IS). SHI-3 is present at the pheU tRNA locus in some S. boydii isolates but not in others. The map locations of the aerobactin genes vary among closely related species. The association of the aerobactin operon with phage genes and mobile elements and its presence at different locations within the genomes of enteric pathogens suggest that these virulence-enhancing genes may have been acquired by bacteriophage integration or IS element-mediated transposition. An S. boydii aerobactin synthesis mutant, 0-1392 iucB, was constructed and was similar to the wild type in tissue culture assays of invasion and intercellular spread.
Iron is essential for the growth of most bacterial pathogens, and the ability to acquire iron is associated with bacterial virulence. To obtain iron, Shigella spp. can use host iron sources such as heme directly, and they have the ability to remove iron from host sources via siderophore-mediated uptake systems (22, 47). Siderophores are low-molecular-weight, high-affinity iron chelators synthesized and secreted into the environment. The iron-siderophore complex is transported back into the cell using specific receptors. Two different siderophore-mediated iron transport systems have been observed in Shigella spp. and clinical Escherichia coli isolates. The catechol siderophore enterobactin is produced by E. coli (39) and some, but not all, Shigella spp. (36, 38), while the hydroxamate siderophore aerobactin is synthesized by Shigella flexneri and Shigella boydii (20) and Shigella sonnei (34), as well as some E. coli clinical isolates (11, 34). The aerobactin operon encodes the IucABCD enzymes for aerobactin synthesis and Iut, the outer membrane receptor for aerobactin. Expression of the aerobactin operon is negatively regulated by the iron-binding repressor protein Fur (3). Under low-iron conditions, expression of the aerobactin operon is derepressed, and the siderophore synthesis proteins and receptor are produced to facilitate iron acquisition.
The aerobactin genes are found on the pColV and F1me plasmids in some strains of E. coli and Salmonella, respectively, and are found chromosomally in Shigella and other E. coli strains (11, 20, 22, 26, 46). While the aerobactin genes were shown to be located in the SHI-2 pathogenicity island downstream of the selC tRNA gene in S. flexneri and S. sonnei (30, 43), their location in S. boydii remained unknown. In this report, we show that the aerobactin operon is located in a 21-kb iron transport island between lysU and the pheU tRNA gene in S. boydii. While the sequence of the aerobactin genes is conserved, the sequences flanking the genes are distinct, and the aerobactin island in S. boydii has been designated SHI-3 for Shigella island 3.
MATERIALS AND METHODS
Bacterial strains, plasmids, and growth conditions.
Clinical isolates of S. boydii, S. dysenteriae, S. flexneri, and S. sonnei were obtained from the Texas Department of Health. Enteroinvasive E. coli (EIEC) strains were obtained from J. H. Crosa, Oregon Health Science University. E. coli 1017 (HB101 entF::Tn5) has been described previously (12). The iron chelator EDDA [ethylenediamine di(o-hydroxyphenylacetic acid)] was deferrated as described previously and used at a concentration of 300 μg/ml to induce iron starvation (19). Strains were grown in L broth or on L-agar plates with the addition of antibiotics at the following concentrations when necessary: 250 μg of carbenicillin/ml, 30 μg of chloramphenicol/ml, and 200 μg of streptomycin/ml.
Isolation of the S. boydii aerobactin genes.
A Sau3AI partial library of S. boydii 0-1392 was constructed in the cosmid vector pLAFR3 (13) and screened by colony hybridization using a probe to the S. flexneri iucA gene. Two overlapping cosmids, pGEP1 and pGEP2, were isolated, and their ability to confer aerobactin synthesis to E. coli 1017 was confirmed by the hydroxamate assay and siderophore bioassays (19, 35).
Construction of the iucB mutation.
An iucB mutation was constructed in S. boydii 0-1392 by allelic exchange using the suicide plasmid pGP704 (27). In S. boydii 0-1392 iucB, the wild-type allele is replaced with one containing a chloramphenicol resistance cassette inserted into the SmaI site of iucB.
Tissue culture, cell invasion, and plaque assays.
The ability of S. boydii 0-1392 and 0-1392 iucB to invade Henle cells was determined by the procedure of Hale and Formal (16). Plaque assays were performed as described by Oaks et al. (32).
Nucleotide sequence analysis.
DNA sequencing was performed using an ABI Prism 377 automatic sequencer. BamHI, HindIII, PstI, and EcoRI fragments of pGEP1 and pGEP2 were subcloned into either pBluescript SK(−) (Stratagene) or pWKS30 (44) plasmid vectors for sequencing. Routine sequence analysis was performed using MacVector software (33) (Oxford Molecular). Sequence homology to known genes and proteins was analyzed using the BlastN and BlastX algorithms, respectively, through the National Center for Biotechnology Information database (1, 2, 14).
PCR.
PCRs were performed in a GeneAmp PCR system 2400 (Perkin-Elmer). The primers used to amplify the SHI-3 int3 and int3-yjdC junction are as follows: primer 1 (5′-CGCTGGAGATGGTTGCTGAAC-3′), primer 2 (5′-GAATCAGGTTTGTGGTCC-3′), and primer 3 (5′-GGGTTATTACCTGCTCTC-3′). The primers used to amplify the SHI-2 int2 and int2-selC junction are as follows: primer 6 (5′-GCGGCGGTATGTATCTAC-3′) and primers 7 and 8, which correspond to primers 1 and 2, respectively, described by Vokes et al. (43). The SHI-3 int3 probe was generated by PCR using primer 2 and primer 3. The iucA probe was generated by PCR using primer 4 (5′-GGCAGCCCATACAGACAG-3′) and primer 5 (5′-CATCCCACGCTTCACTTC-3′). PCRs consisted of 30 cycles with an annealing temperature of 50°C and extension times of 30 s for primer pairs 2-3 and 6-8, 1 min for primer pair 4-5, and 2 min for primer pairs 1-3 and 7-8.
Southern hybridizations.
Genomic DNA was isolated using Qiagen Genomic-tip DNA isolation columns according to the manufacturer's instructions. Probe labeling, hybridization, standard stringency washes, and detection were performed as described in the ECL Direct Nucleic Acid Labeling Kit (Amersham Pharmacia).
Nucleotide sequence accession number.
The GenBank accession number for the sequence described here is AF335540.
RESULTS
Isolation of the S. boydii aerobactin genes.
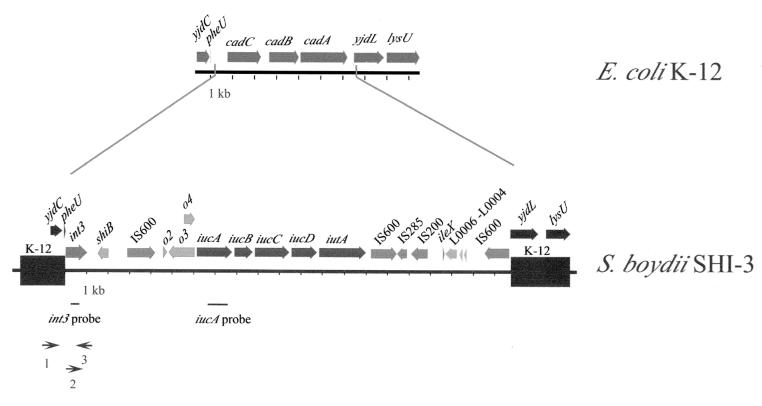
Strains of S. boydii, like those of S. flexneri, produce the siderophore aerobactin, but the S. boydii aerobactin synthesis genes do not map to the same location as those in S. flexneri (43). To map the location of the S. boydii 0-1392 aerobactin genes, cosmids containing the aerobactin genes were isolated from a library of strain 0-1392 by DNA hybridization using a probe to iucA. Two overlapping iucA-positive cosmids, pGEP1 and pGEP2, conferred the ability to synthesize aerobactin upon E. coli 1017 as shown by positive hydroxamate tests and siderophore bioassays (data not shown). The entire region containing the aerobactin genes was sequenced, revealing a 21-kb island between pheU and yjdL at min 93 to 94 of the E. coli K-12 map (Fig. 1). Aerobactin genes have not previously been mapped to this location in any species.
FIG. 1.
Map of the S. boydii 0-1392 aerobactin island, SHI-3, and the corresponding E. coli K-12 region between pheU and lysU at min 94. Sequences present in both E. coli K-12 and S. boydii are indicated by the black bars and are designated K-12 on the SHI-3 map. The ORFs, indicated as shiB and o2 to o4, were inferred from sequence analysis. The iucA probe and the int3 probe, as well as the approximate positions of primers used for PCR amplification of the junctions, are positioned below the SHI-3 map.
To confirm that the aerobactin genes at this locus are responsible for hydroxamate synthesis, an iucB mutation was constructed by inserting a chloramphenicol resistance cassette into the cloned iucB gene and transferring the mutation to 0-1392 by allelic exchange. 0-1392 iucB did not synthesize aerobactin, indicating that there is a single aerobactin operon in 0-1392. 0-1392 iucB invaded Henle cells at wild-type levels and produced plaques in a standard plaque assay (data not shown).
Structure of the S. boydii aerobactin island.
The S. boydii aerobactin island, which we have designated SHI-3, contains a functional aerobactin operon. SHI-3 is demarcated by a putative integrase gene inserted 200 bp downstream of the pheU tRNA at min 94 and by an IS600 interruption of yjdL, an uncharacterized open reading frame (ORF) adjacent to lysU (Fig. 1). Sequence scanning did not reveal direct repeats or any known sequence in the 84 bp between the conserved intergenic sequence downstream of pheU and the start of the integrase gene. The genes, ORFs, and insertion elements present in S. boydii SHI-3 are summarized in Table 1. The second ORF of the S. boydii aerobactin island shares 97% nucleotide identity with the S. flexneri M90T SHI-2 ORF of unknown function, shiB (30). The region between shiB and the aerobactin genes in S. boydii contains ORFs of unknown function with sequence similarity to ORFs upstream of aerobactin in S. flexneri SHI-2 and E. coli pColV-K30 (Table 1). There is 100% nucleotide identity between orf2 of 0-1392 and orf24 of the S. flexneri SA100 SHI-2 aerobactin island. The adjacent region is 99% identical to S. flexneri SA100 SHI-2 rorf25, an ORF transcribed off the minus strand, but the insertion of a thymine codon at base 5333 in the S. boydii SHI-3 island generates a stop codon, creating the defective rorf3 (43). The 398-bp orf4 is 99% homologous at the nucleotide and amino acid levels to SHI-2 orf27 and shares 93% nucleotide identity to sequence upstream of the aerobactin genes in E. coli pColV-K30. The SHI-3 iucA, -B, -C, and -D and iutA genes share 99% nucleotide identity with the SHI-2 aerobactin genes (30, 43) and 92 to 95% identity with the E. coli aerobactin genes (17, 18, 23). Thus, the aerobactin biosynthesis and transport genes and the immediate upstream region appear to be highly conserved among the different Shigella and E. coli strains that carry them.
TABLE 1.
ORFs within the S. boydii aerobactin island
| ORF, gene, or IS | Location | Length (bp) | Similar sequence (reference) | % Nucleotide (protein) identity |
|---|---|---|---|---|
| int3 | 84–1345 | 1,261a | P4 prophage integrase (37) | (63) |
| shiB | 1800–2249 | 449 | S. flexneri M90T shiB gene in SHI-2 (30) | 97 (94 over 143 of 153 aa)b |
| IS600 | 3198–4459 | 1,261 | S. sonnei IS600 (10, 24) | 98 |
| orf2 | 4815–5081 | 266 | S. flexneri SA100 orf24 in SHI-2 (43) | 100 (100) |
| rorf3c | 5037–6229 | 1,192 | S. flexneri SA100 rorf25 in SHI-2 (43) | 99 (89 for 297 of 397 aa)bd |
| orf4 | 5877–6305 | 428 | S. flexneri SA100 orf27 in SHI-2 (43) | 99 (99) |
| iucA, -B, -C, -D | 6308–12112 | 5,804 | S. flexneri M90T aerobactin synthesis genes in SHI-2 (30) | 99 (99) |
| iutA | 12115–14313 | 2,198 | S. flexneri M90T iutA in SHI-2 (30) | 99 (99) |
| IS600 | 14542–15809 | 1,267 | S. sonnei IS600 (10, 24) | 98 (93) |
| Partial IS285 | 15803–16159 | 356 | Y. pestis IS285 (21) | 86 (79 for 88 of 270 aa)b |
| Partial IS200 | 16374–17106 | 732 | E. coli IS200 (4) | 95 (87) |
| ileX | 17706–17778 | 72 | E. coli ileX tRNA gene (5) | 87 |
| L0006 | 17928–18590 | 662 | E. coli LEE pathogenicity island prophage gene L0006 (37) | 93 (95) |
| L0005 | 18604–18786 | 182 | E. coli LEE pathogenicity island prophage gene L0005 (37) | 92 (78) |
| Partial L0004 | 18799–19047 | 248 | E. coli LEE pathogenicity island prophage gene L0004 (37) | 90 (95 for 82 of 116 aa)b |
| IS600 | 19699–20961 | 1262 | S. sonnei IS600 (10, 24) | 98 (93) |
Three ORFs corresponding to the integrase-like sequence were corrected for apparent frameshifts and are presented here as a single ORF.
Amino acid identity for the partial ORF present in SHI-3.
rorf3 is an ORF transcribed off the minus stand.
Due to a premature stop codon, rorf3 is defective.
S. boydii SHI-3 contains genes and mobile elements that suggest either bacteriophage- or insertion sequence (IS) element-mediated horizontal transfer of the genes. The SHI-3 integrase, Int3, appears to be a member of the P4 prophage integrase family, which has been implicated in the integration of other iron transport and pathogenicity islands (30, 37, 43). Int3 shares 64% amino acid homology with the P4 prophage integrase of E. coli, 54% with the putative P4-like prophage integrase of the E. coli O157:H7 locus of enterocyte effacement (LEE) pathogenicity island, and 40% with the SHI-2 integrase, Int2 (8, 30, 37, 43). The SHI-3 integrase gene sequence has nucleotide insertions or deletions that generate frameshifts, preventing the translation of a functional integrase. In addition to the integrase gene, SHI-3 also contains the putative O157:H7 LEE prophage genes L0004, L0005, and L0006 (37) downstream of the aerobactin operon. L0004 and L0006 both resemble transposase genes, but the function of L0005 is unknown. While some prophage-like genes are present within SHI-3, this island does not encode an entire putative P4-like prophage.
IS elements, which potentially can mobilize intervening sequence as composite transposons when present in multiple copies, are also present in SHI-3. There are two intact copies of the S. sonnei IS600 downstream of iutA. A third IS600 is located upstream of the aerobactin operon, but it is unlikely to be functional, as the transposase gene contains premature stop codons. A partial copy of the E. coli IS200 transposase gene and a portion of the Yersinia pestis IS285 transposase gene are also present within SHI-3. No direct or inverted repeats are observed in the sequence flanking these IS elements. It is possible that IS elements may have been involved in the assembly of the S. boydii aerobactin island and that the present island evolved through several insertions or deletions mediated by IS elements. The presence of a region with 85% nucleotide identity to ileX tRNA between the putative prophage genes and the partial copies of IS200 and IS285 is indicative of the mosaic structure of the SHI-3 island. It is possible that the ileX tRNA gene was the target of a bacteriophage integration event and then was incorporated into SHI-3 via other bacteriophage- or IS element-mediated events.
Association between SHI-3 and loss of cadA.
In S. boydii 0-1392, there is a deletion of more than 6 kb relative to the E. coli K-12 sequence at the site occupied by SHI-3, including the lysine decarboxylase gene, cadA (Fig. 1). Cadavarine, produced by the decarboxylation of lysine, inhibits Shigella enterotoxin activity, and deletion of cadA has been shown to enhance the virulence of Shigella and EIEC (25). Thus, in S. boydii, the acquisition of SHI-3 may have resulted not only in the enhanced ability to scavenge iron from the host using the siderophore aerobactin but also in the loss of a gene whose absence is associated with an increase in virulence.
Distributions of SHI-3 int3 and SHI-2 int2 among enteric bacteria.
PCR was used to determine the distribution and location of the SHI-3 integrase gene, int3, among other Shigella strains, and hydroxamate tests were performed to assess the possible correlation of int3 presence with aerobactin production (Table 2). Primer pair 2-3 (Fig. 1) amplified a 395-bp product internal to int3 in several S. boydii, S. flexneri, S. sonnei, and S. dysenteriae serotypes, as well as EIEC strains, illustrating the distribution of this putative P4-like prophage integrase among Shigella spp. and E. coli. In those strains positive for int3, PCR to detect the yjdC-int3 junction was performed. Using primers 1 and 3, where primer 1 is in yjdC, the uncharacterized ORF upstream of pheU, and primer 3 is internal to int3, a 2.1-kb yjdC-int3 fragment was amplified in S. boydii 0-1392, 0-1393, and 224860, as well as in S. dysenteriae 1-130 (Fig. 1). All four of these strains produced aerobactin, as determined by hydroxamate assays and siderophore bioassays (data not shown). Therefore, it is possible that SHI-3, carrying the aerobactin genes, is located downstream of pheU in each of these strains. The presence of strains positive for int3 but lacking the yjdC-int3 junction, such as S. boydii 0-1591, suggests that another Int3-mediated bacteriophage integration event occurred at a different map location in these strains.
TABLE 2.
Presence of aerobactin and linkage of SHI-3 int to pheU in Shigella and E. coli strains
| Species and strain | Serotype | Aerobactin synthesis | int3a (linked to pheU)b | int2c (linked to selC)d |
|---|---|---|---|---|
| S. boydii | ||||
| 1-1660 | 1 | + | − | − |
| 1-1097 | 2 | + | − | − |
| 0-1392 | 5 | + | + (+) | − |
| 0-1393 | 5 | + | + (+) | − |
| 224860 | 7 | + | + (+) | − |
| 0-5353 | 9 | + | − | − |
| 0-1591 | 11 | + | + (−) | − |
| S. flexnerie | ||||
| SA100 | 2a | + | + (−) | + (+) |
| 5 strains | 1a | + | − | + (+) |
| 9 strains | 1b | + | − | + (+) |
| 20 strains | 2a | + | + (−) | + (+) |
| 9 strains | 2b | + | + (−) | + (+) |
| 10 strains | 3a | + | − | + (+) |
| 11 strains | 3b | + | − | + (+) |
| 8 strains | 3c | + | − | + (+) |
| 4 strains | 4a | + | − | + (+) |
| 9 strains | 4b | + | − | + (+) |
| 9 strains | 5 | + | + (−) | + (+) |
| 6 strains | 6 | + | − | − |
| S. dysenteriae | ||||
| 0-4576 | 1 | − | − | + (+) |
| 1-130 | 2 | + | + (+) | − |
| 0-4242 | 3 | + | − | − |
| 9-3485 | 4 | + | − | − |
| 217726 | 9 | + | − | − |
| S. sonnei | ||||
| 1-679 | + | + (−) | + (+) | |
| EN120 | + | + (−) | + (+) | |
| EIEC | ||||
| 930-78 | O124:H− | − | − | − |
| 550-3076 | + | + (−) | − | |
| 1107-81 | + | + (−) | − |
PCR with primer pair 2-3 amplifies a 395-bp product internal to SHI-3 int3.
Linkage determined by PCR with primer pair 1-3, where primer 1 is in yjdC and primer 3 is in SHI-3 int3.
PCR with primer pair 6-8 amplifies a 120-bp product internal to SHI-2 int2.
Linkage determined by PCR with primer pair 7-8, where primer 7 is in selC and primer 3 is in SHI-2 int2.
Each serotype pool contains the indicated number of clinical isolates from the Texas Department of Health.
The presence of int2, the related but distinct integrase gene found in SHI-2, among the Shigella and EIEC clinical isolates was also determined through PCR (Table 2). Although the two integrases are related, no cross-amplification of int3 and int2 genes occurs with primer pair 2-3 and primer pair 6-8. Using primers 6 and 8, a 120-bp fragment internal to int2 was amplified in all but one of the S. flexneri serotypes, in one of five S. dysenteriae strains, and in both S. sonnei strains. int2 was absent from the seven S. boydii strains and from the three EIEC strains tested (Table 2). In S. flexneri serotypes 2a, 2b, and 5, as well as S. sonnei strains, PCR amplification with both the int2 and int3 primer pairs suggested two independent bacteriophage integration events. The selC-int2 junction was present in the int2-positive strains as determined by an additional PCR with the primer pair 7-8, where primer 7 is located in selC and primer 8 is internal to int2. int2 is linked to selC in all tested strains; however, in S. dysenteriae the selC-int2 junction does not correlate with the presence of the SHI-2 aerobactin island, indicating that more than one pathogenicity island containing int2 exists (43). It is possible that the 3′ end of the selC tRNA gene was the target for a single Int2-mediated integration in Shigella and that different pathogenicity islands resulted from multiple horizontal gene transfers mediated by other mobile elements. Alternatively, other bacteriophage integration events mediated by int2 occurred at the same map location, introducing different pathogenicity islands into different lineages.
Distribution of SHI-3 among enteric bacteria.
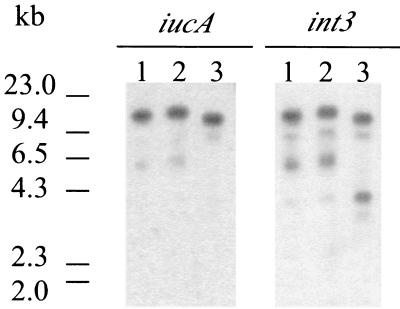
To determine whether the aerobactin and P4-like integrase genes are linked in S. boydii strains containing the yjdC-int3 junction, Southern hybridizations were performed using probes to iucA and int3 (Fig. 2). In S. boydii 0-1392, both probes hybridized to an approximately 12.9-kb PvuII fragment, the size predicted from the DNA sequence analysis. In 0-1392 and 224860, the sizes of the PvuII fragments were slightly different than in 0-1392, but in each strain the iucA and int3 probes both hybridized to fragments of the same size, suggesting linkage of the int3 and iuc genes. Thus, an SHI-3-like island may be found in multiple S. boydii serotypes.
FIG. 2.
The P4-like integrase and aerobactin genes are linked in several S. boydii serotypes. Genomic DNA was digested with PvuII, and the fragments were hybridized to a 1-kb iucA probe or a 395-bp int3 probe. Locations of the probes are shown in Fig. 1. Lane 1, S. boydii 0-1392; lane 2, S. boydii 0-1393; lane 3, S. boydii 224860.
DISCUSSION
SHI-3 is a 21-kb iron transport island carrying the aerobactin genes and is located downstream of the pheU tRNA gene in S. boydii 0-1392. SHI-3 has many characteristics of a pathogenicity island: it contains mobile elements, including a P4-like prophage integrase and IS elements; it is associated with a tRNA gene; and it may have been acquired via horizontal transfer. However, the role that this island plays in Shigella pathogenicity is unclear. The only potential virulence genes within this region encode enzymes for aerobactin synthesis and the outer membrane receptor Iut. Aerobactin is known to be important for bacterial survival in low-iron conditions and also may be important in the host. S. flexneri aerobactin mutants show reduced fluid accumulation in the rabbit ileal loop model of infection (31), yet iuc mutants are capable of wild-type invasion, form plaques in cultured epithelial cells, and are positive in the Serény test. In this study we have shown that S. boydii aerobactin mutants are also capable of wild-type invasion and plaque formation, most likely due to the presence of additional iron uptake systems. The presence of multiple functional iron transport systems in Shigella suggests that there is selection for the acquisition and maintenance of these genes. These iron acquisition systems may benefit Shigella in the different environments it encounters within the host, making the contribution of one iron transport system to virulence difficult to assess. The SHI-3 island also is associated with the absence of the gene encoding lysine decarboxylase activity, the loss of which contributes to Shigella pathogenicity (25). Thus, the aerobactin island may contribute to both the survival and the pathogenicity of these bacteria.
The acquisition of SHI-3 may have been bacteriophage mediated, as SHI-3 contains various phage genes, including that for an integrase similar to the LEE P4-like prophage integrase of E. coli. S. boydii 0-1392 SHI-3 appears to be stable (data not shown), although spontaneous deletions of the aerobactin genes in S. flexneri have been observed (19), suggesting the instability of the aerobactin genes in certain strains. The G+C content of the 21-kb SHI-3 island is similar to the observed base composition of the chromosome (51%). Either S. boydii acquired this island early in its evolution and the base composition has become similar to that of the chromosome, or SHI-3 was transferred from an organism with a base composition similar to that of Shigella.
Many pathogenicity islands are associated with tRNA genes or tRNA-like loci, and the 3′ ends of tRNA genes may act as sites for integration of foreign DNA (15). The S. boydii SHI-3 island is the second island to be associated with pheU in enteric pathogens. The LEE pathogenicity island is found at pheU in clinical isolates of enterohemorrhagic and enteropathogenic E. coli strains expressing β-intimin, while it is found immediately downstream of selC in strains expressing α- or γ-intimin (41, 45). The selC tRNA gene is also the site of the S. flexneri and S. sonnei SHI-2 aerobactin islands (30, 43).
SHI-3 is distinct from the previously described S. flexneri SHI-2, although the aerobactin genes are highly conserved. The G+C content of the 30-kb S. flexneri SHI-2 is slightly lower (46%) than that of the S. boydii SHI-3 (51%) and contains a colicin immunity gene (43). Additionally, SHI-3 contains three copies of IS600 and incomplete copies of IS200 and IS285, as well as the putative prophage genes L0004 to L0006, which are not present in SHI-2. The presence of different genes and IS elements and the difference between SHI-2 and SHI-3 in G+C content indicate that SHI-2 may have been acquired at a later time, or from another source, than SHI-3.
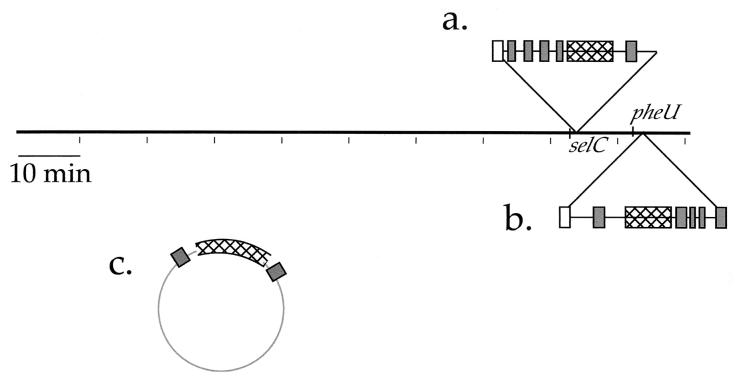
Horizontal gene transfer involves the introduction of genes into a single lineage via plasmid, bacteriophage, or IS elements, resulting in a scattered phylogenetic distribution among closely related species. Mapping of several iron transport loci among the Enterobacteriaceae suggests horizontal transfer of these genes. The various locations of the aerobactin genes, the distribution of the SHI-2 and SHI-3 aerobactin islands among enteric bacteria, and the association of aerobactin genes with bacteriophage or mobile elements suggest that genes for aerobactin synthesis and transport have been acquired through horizontal transfer (Fig. 3). Similarly, the presence of the shu heme transport locus in S. dysenteriae type 1 and various E. coli strains, but not in other S. dysenteriae serotypes or Shigella spp., suggests that the shu genes also spread via one of these transfer mechanisms (28, 29, 42, 47). Finally, the genes encoding the siderophore yersiniabactin and its receptor are present in the high-pathogenicity islands of Yersinia enterocolitica, Y. pestis, and Y. pseudotuberculosis (6, 7, 9). High-pathogenicity islands also have been found in the chromosomes of some pathogenic E. coli strains, suggesting horizontal transfer between these two species (40). The horizontal transfer of iron acquisition systems effectively alters the ecological and pathogenic characteristics of the recipients by allowing their survival in low-iron conditions. The aerobactin iron transport system may allow Shigella to compete for iron in certain iron-limited environments, including the host.
FIG. 3.
Map locations of the aerobactin genes among the Enterobacteriaceae. The islands containing aerobactin genes are shown as insertions relative to the E. coli K-12 chromosome. Hatched boxes indicate the aerobactin operon, white boxes indicate P4-like prophage integrase genes, and dark grey boxes indicate ISs. (a) The SHI-2 aerobactin pathogenicity island at selC in some S. flexneri strains and in S. sonnei. (b) The SHI-3 aerobactin island at pheU in S. boydii. (c) The aerobactin genes are encoded on the pColV and F1me plasmids in some strains of E. coli and Salmonella enterica, respectively. On pColV, the aerobactin genes are flanked by IS1 elements.
ACKNOWLEDGMENTS
This work was supported by grant AI16935 from the National Institutes of Health and contract DAAA21-93-C-0101 from the U.S. Department of the Army.
We thank Elizabeth Wyckoff, Stephanie Reeves, and Laura Runyen-Janecky for editorial guidance.
REFERENCES
- 1.Altschul S F, Gish W, Miller W, Myers E W, Lipman D J. Basic local alignment search tool. J Mol Biol. 1990;215:403–410. doi: 10.1016/S0022-2836(05)80360-2. [DOI] [PubMed] [Google Scholar]
- 2.Altschul S F, Madden T L, Schaffer A A, Zhang J, Zhang Z, Miller W, Lipman D J. Gapped BLAST and PSI-BLAST: a new generation of protein database search programs. Nucleic Acids Res. 1997;25:3389–3402. doi: 10.1093/nar/25.17.3389. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Bagg A, Neilands J B. Molecular mechanism of regulation of siderophore-mediated iron assimilation. Microbiol Rev. 1987;51:509–518. doi: 10.1128/mr.51.4.509-518.1987. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Bisercic M, Ochman H. The ancestry of insertion sequences common to Escherichia coli and Salmonella typhimurium. J Bacteriol. 1993;175:7863–7868. doi: 10.1128/jb.175.24.7863-7868.1993. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Blattner F R, Plunkett G, 3rd, Bloch C A, Perna N T, Burland V, Riley M, Collado-Vides J, Glasner J D, Rode C K, Mayhew G F, Gregor J, Davis N W, Kirkpatrick H A, Goeden M A, Rose D J, Mau B, Shao Y. The complete genome sequence of Escherichia coli K-12. Science. 1997;277:1453–1474. doi: 10.1126/science.277.5331.1453. [DOI] [PubMed] [Google Scholar]
- 6.Buchrieser C, Brosch R, Bach S, Guiyoule A, Carniel E. The high-pathogenicity island of Yersinia pseudotuberculosis can be inserted into any of the three chromosomal asn tRNA genes. Mol Microbiol. 1998;30:965–978. doi: 10.1046/j.1365-2958.1998.01124.x. [DOI] [PubMed] [Google Scholar]
- 7.Buchrieser C, Prentice M, Carniel E. The 102-kilobase unstable region of Yersinia pestis comprises a high-pathogenicity island linked to a pigmentation segment which undergoes internal rearrangement. J Bacteriol. 1998;180:2321–2329. doi: 10.1128/jb.180.9.2321-2329.1998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Burland V, Plunkett G R, Sofia H J, Daniels D L, Blattner F R. Analysis of the Escherichia coli genome. VI. DNA sequence of the region from 92.8 through 100 minutes. Nucleic Acids Res. 1995;23:2105–2119. doi: 10.1093/nar/23.12.2105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Carniel E, Guilvout I, Prentice M. Characterization of a large chromosomal “high-pathogenicity island” in biotype 1B Yersinia enterocolitica. J Bacteriol. 1996;178:6743–6751. doi: 10.1128/jb.178.23.6743-6751.1996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Chandler M, Fayet O. Translational frameshifting in the control of transposition in bacteria. Mol Microbiol. 1993;7:497–503. doi: 10.1111/j.1365-2958.1993.tb01140.x. [DOI] [PubMed] [Google Scholar]
- 11.Colonna B, Nicoletti M, Visca P, Casalino M, Valenti P, Maimone F. Composite IS1 elements encoding hydroxamate-mediated iron uptake in FIme plasmids from epidemic Salmonella spp. J Bacteriol. 1985;162:307–316. doi: 10.1128/jb.162.1.307-316.1985. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Daskaleros P A, Stoebner J A, Payne S M. Iron uptake in Plesiomonas shigelloides: cloning of the genes for the heme-iron uptake system. Infect Immun. 1991;59:2706–2711. doi: 10.1128/iai.59.8.2706-2711.1991. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Friedman A M, Long S R, Brown S E, Buikema W J, Ausubel F M. Construction of a broad host range cosmid cloning vector and its use in the genetic analysis of Rhizobium mutants. Gene. 1982;18:289–296. doi: 10.1016/0378-1119(82)90167-6. [DOI] [PubMed] [Google Scholar]
- 14.Gish W, States D J. Identification of protein coding regions by database similarity search. Nat Genet. 1993;3:266–272. doi: 10.1038/ng0393-266. [DOI] [PubMed] [Google Scholar]
- 15.Hacker J, Blum-Oehler G, Mühldorfer I, Tschäpe H. Pathogenicity islands of virulent bacteria: structure, function and impact on microbial evolution. Mol Microbiol. 1997;23:1089–1097. doi: 10.1046/j.1365-2958.1997.3101672.x. [DOI] [PubMed] [Google Scholar]
- 16.Hale T L, Formal S B. Protein synthesis in HeLa or Henle 407 cells infected with Shigella dysenteriae 1, Shigella flexneri 2a, or Salmonella typhimurium W118. Infect Immun. 1981;32:137–144. doi: 10.1128/iai.32.1.137-144.1981. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Herrero M, de Lorenzo V, Neilands J B. Nucleotide sequence of the iucD gene of the pColV-K30 aerobactin operon and topology of its product studied with phoA and lacZ gene fusions. J Bacteriol. 1988;170:56–64. doi: 10.1128/jb.170.1.56-64.1988. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Krone W J A, Stegehuis F, Koningstein G, van Doorn C, Roosendaal B, de Graaf F K, Oudega B. Characterization of the pColV-K30 encoded cloacin DF13/aerobactin outer membrane receptor protein of Escherichia coli; isolation and purification of the protein and analysis of its nucleotide sequence and primary structure. FEMS Microbiol Lett. 1987;26:153–161. [Google Scholar]
- 19.Lawlor K M, Daskaleros P A, Robinson R E, Payne S M. Virulence of iron transport mutants of Shigella flexneri and utilization of host iron compounds. Infect Immun. 1987;55:594–599. doi: 10.1128/iai.55.3.594-599.1987. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Lawlor K M, Payne S M. Aerobactin genes in Shigella spp. J Bacteriol. 1984;160:266–272. doi: 10.1128/jb.160.1.266-272.1984. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Lindler L E, Plano G V, Burland V, Mayhew G F, Blattner F R. Complete DNA sequence and detailed analysis of the Yersinia pestis KIM5 plasmid encoding murine toxin and capsular antigen. Infect Immun. 1998;66:5731–5742. doi: 10.1128/iai.66.12.5731-5742.1998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Marolda C L, Valvano M A, Lawlor K M, Payne S M, Crosa J H. Flanking and internal regions of chromosomal genes mediating aerobactin iron uptake systems in enteroinvasive Escherichia coli and Shigella flexneri. J Gen Microbiol. 1987;133:2269–2278. doi: 10.1099/00221287-133-8-2269. [DOI] [PubMed] [Google Scholar]
- 23.Martinez J L, Herrero M, de Lorenzo V. The organization of intercistronic regions of the aerobactin operon of pColV-K30 may account for the differential expression of the iucABCD iutA genes. J Mol Biol. 1994;238:288–293. doi: 10.1006/jmbi.1994.1290. [DOI] [PubMed] [Google Scholar]
- 24.Matsutani S, Ohtsubo H, Maeda Y, Ohtsubo E. Isolation and characterization of IS elements repeated in the bacterial chromosome. J Mol Biol. 1987;196:445–455. doi: 10.1016/0022-2836(87)90023-4. [DOI] [PubMed] [Google Scholar]
- 25.Maurelli A T, Fernandez R E, Bloch C A, Rode C K, Fasano A. “Black holes” and bacterial pathogenicity: a large genomic deletion that enhances the virulence of Shigella spp. and enteroinvasive Escherichia coli. Proc Natl Acad Sci USA. 1998;95:3943–3948. doi: 10.1073/pnas.95.7.3943. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.McDougall S, Neilands J B. Plasmid- and chromosome-coded aerobactin synthesis in enteric bacteria: insertion sequences flank operon in plasmid-mediated systems. J Bacteriol. 1984;159:300–305. doi: 10.1128/jb.159.1.300-305.1984. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Miller V L, Mekalanos J J. A novel suicide vector and its use in construction of insertion mutations: osmoregulation of outer membrane proteins and virulence determinants in Vibrio cholerae requires toxR. J Bacteriol. 1988;170:2575–2583. doi: 10.1128/jb.170.6.2575-2583.1988. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Mills M, Payne S M. Genetics and regulation of heme iron transport in Shigella dysenteriae and detection of an analogous system in Escherichia coli O157:H7. J Bacteriol. 1995;177:3004–3009. doi: 10.1128/jb.177.11.3004-3009.1995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Mills M, Payne S M. Identification of shuA, the gene encoding the heme receptor of Shigella dysenteriae, and analysis of invasion and intracellular multiplication of a shuA mutant. Infect Immun. 1997;65:5358–5363. doi: 10.1128/iai.65.12.5358-5363.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Moss J E, Cardozo T J, Zychlinsky A, Groisman E A. The selC-associated SHI-2 pathogenicity island of Shigella flexneri. Mol Microbiol. 1999;33:74–83. doi: 10.1046/j.1365-2958.1999.01449.x. [DOI] [PubMed] [Google Scholar]
- 31.Nassif X, Mazert M C, Mounier J, Sansonetti P J. Evaluation with an iuc::Tn10 mutant of the role of aerobactin production in the virulence of Shigella flexneri. Infect Immun. 1987;55:1963–1969. doi: 10.1128/iai.55.9.1963-1969.1987. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Oaks E V, Wingfield M E, Formal S B. Plaque formation by virulent Shigella flexneri. Infect Immun. 1985;48:124–129. doi: 10.1128/iai.48.1.124-129.1985. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Olson S A. MacVector: an integrated sequence analysis program for the Macintosh. Methods Mol Biol. 1994;25:195–201. doi: 10.1385/0-89603-276-0:195. [DOI] [PubMed] [Google Scholar]
- 34.Payne S M. Iron and virulence in the family Enterobacteriaceae. Crit Rev Microbiol. 1988;16:81–111. doi: 10.3109/10408418809104468. [DOI] [PubMed] [Google Scholar]
- 35.Payne S M. Synthesis and utilization of siderophores by Shigella flexneri. J Bacteriol. 1980;143:1420–1424. doi: 10.1128/jb.143.3.1420-1424.1980. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Payne S M, Niesel D W, Peixotto S S, Lawlor K M. Expression of hydroxamate and phenolate siderophores by Shigella flexneri. J Bacteriol. 1983;155:949–955. doi: 10.1128/jb.155.3.949-955.1983. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Perna N T, Mayhew G F, Posfai G, Elliott S, Donnenberg M S, Kaper J B, Blattner F R. Molecular evolution of a pathogenicity island from enterohemorrhagic Escherichia coli O157:H7. Infect Immun. 1998;66:3810–3817. doi: 10.1128/iai.66.8.3810-3817.1998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Perry R D, San Clemente C L. Siderophore synthesis in Klebsiella pneumoniae and Shigella sonnei during iron deficiency. J Bacteriol. 1979;140:1129–1132. doi: 10.1128/jb.140.3.1129-1132.1979. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Rogers H J. Iron-binding catechols and virulence in Escherichia coli. Infect Immun. 1973;7:438–444. doi: 10.1128/iai.7.3.438-444.1973. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Schubert S, Rakin A, Karch H, Carniel E, Heesemann J. Prevalence of the “high-pathogenicity island” of Yersinia species among Escherichia coli strains that are pathogenic to humans. Infect Immun. 1998;66:480–485. doi: 10.1128/iai.66.2.480-485.1998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Sperandio V, Kaper J B, Bortolini M R, Neves B C, Keller R, Trabulsi L R. Characterization of the locus of enterocyte effacement (LEE) in different enteropathogenic Escherichia coli (EPEC) and Shiga-toxin producing Escherichia coli (STEC) serotypes. FEMS Microbiol Lett. 1998;164:133–139. doi: 10.1111/j.1574-6968.1998.tb13078.x. [DOI] [PubMed] [Google Scholar]
- 42.Torres A G, Payne S M. Haem iron-transport system in enterohaemorrhagic Escherichia coli O157:H7. Mol Microbiol. 1997;23:825–833. doi: 10.1046/j.1365-2958.1997.2641628.x. [DOI] [PubMed] [Google Scholar]
- 43.Vokes S A, Reeves S A, Torres A G, Payne S M. The aerobactin iron transport system genes in Shigella flexneri are present within a pathogenicity island. Mol Microbiol. 1999;33:63–73. doi: 10.1046/j.1365-2958.1999.01448.x. [DOI] [PubMed] [Google Scholar]
- 44.Wang R F, Kushner S R. Construction of versatile low-copy-number vectors for cloning, sequencing and gene expression in Escherichia coli. Gene. 1991;100:195–199. [PubMed] [Google Scholar]
- 45.Wieler L H, McDaniel T K, Whittam T S, Kaper J B. Insertion site of the locus of enterocyte effacement in enteropathogenic and enterohemorrhagic Escherichia coli differs in relation to the clonal phylogeny of the strains. FEMS Microbiol Lett. 1997;156:49–53. doi: 10.1111/j.1574-6968.1997.tb12704.x. [DOI] [PubMed] [Google Scholar]
- 46.Williams P H. Novel iron uptake system specified by ColV plasmids: an important component in the virulence of invasive strains of Escherichia coli. Infect Immun. 1979;26:925–932. doi: 10.1128/iai.26.3.925-932.1979. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Wyckoff E E, Duncan D, Torres A G, Mills M, Maase K, Payne S M. Structure of the Shigella dysenteriae haem transport locus and its phylogenetic distribution in enteric bacteria. Mol Microbiol. 1998;28:1139–1152. doi: 10.1046/j.1365-2958.1998.00873.x. [DOI] [PubMed] [Google Scholar]