Abstract
Patients with bladder cancer often have a poor prognosis due to the highly invasive and metastatic characteristics of bladder cancer cells. Epithelial-to-mesenchymal transition (EMT) has been causally linked to bladder cancer invasion. The E3 ubiquitin ligase, tumor necrosis factor receptor–associated factor 4 (TRAF4) has been implicated as a tumor promoter in a wide range of cancers. In contrast, here we show that low TRAF4 expression is associated with poor overall survival in patients with bladder cancer. We show that the TRAF4 gene is epigenetically silenced and that ERK mediates TRAF4 phosphorylation, resulting in lower TRAF4 protein levels in bladder cancer cells. In addition, we demonstrate that TRAF4 is inversely correlated with an EMT gene signature/protein marker expression. Functionally, by manipulating TRAF4 expression, we show that TRAF4 regulates EMT genes and epithelial and invasive properties in bladder cancer cells. Transcriptomic analysis of dysregulated TRAF4 expression in bladder cancer cell lines revealed that high TRAF4 expression enhances the bone morphogenetic protein (BMP)/SMAD and inhibits the NF-κB signaling pathway. Mechanistically, we show that TRAF4 targets the E3 ubiquitin ligase SMURF1, a negative regulator of BMP/SMAD signaling, for proteasomal degradation in bladder cancer cells. This was corroborated in patient samples where TRAF4 positively correlates with phospho-SMAD1/5, and negatively correlates with phospho-NFκb-p65. Lastly, we show that genetic and pharmacologic inhibition of SMURF1 inhibits the migration of aggressive mesenchymal bladder cancer cells.
Implications:
Our findings identify E3 ubiquitin ligase TRAF4 as a potential therapeutic target or biomarker for bladder cancer progression.
Introduction
Bladder cancer is a highly prevalent cancer with poor clinical outcomes, especially in advanced stages of progression when the cancer starts invading the bladder muscle (1). Epithelial-to-mesenchymal transition (EMT) has been implicated in bladder cancer progression and metastasis. EMT is a dynamic process in which epithelial cells lose their cell–cell contacts and apical–basal polarity and gain mesenchymal traits with increased migration and invasion abilities (2, 3). Cells lose the expression of epithelial markers such as E-cadherin and gain the expression of mesenchymal markers such as N-cadherin (4). This process is orchestrated by EMT-inducing transcription factors, including SNAIL and SLUG (5, 6). TGFβ signaling pathway is known to stimulate EMT (7).
Subsets of patients with (mesenchymal) bladder cancer were found to have an overly active mitogen-activated protein (MAP) kinase pathway; patients with advanced and/or muscle-invasive bladder cancer were found to have gain-of-function mutations in upstream activators of ERK i.e., in FGF or amplification of RAF1 kinase (8, 9). Active ERK MAP kinase may cooperate with other signaling pathways to promote EMT of bladder and other cancer cells (10, 11).
Tumor necrosis factor receptor–associated factor 4 (TRAF4) encodes a ring domain containing E3 ubiquitin ligase that belongs to the TRAF protein family. Most TRAF proteins control immune and inflammation processes by mediating signaling via tumor necrosis factor receptors (TNFR) and IL1/Toll-like receptors (12). TRAF4, however, was found to be mainly involved in embryogenesis and morphogenesis (13). In breast cancer, TRAF4 was identified as a promoter of invasion and metastasis, and functions by targeting E3 homologous to the E6-AP carboxyl terminus (HECT) domain containing ubiquitin ligase SMURF2 for proteasomal degradation (14). SMURF2 is recruited to TGFβ type I receptor (TβRI) to mediate TβRI polyubiquitination and proteasomal degradation. By targeting the negative regulator SMURF2, TRAF4 promoted TGFβ signaling, EMT, and metastasis in breast cancer cells (14). Moreover, TRAF4 was found to be a critical factor driving breast, prostate, lung, and glioma tumor progression (15–18). The role of TRAF4 in bladder cancer has not been investigated.
Here, we report that in contrast to its role in other cancer subtypes, TRAF expression positively associated with good prognosis in bladder cancer. We uncovered how TRAF4 expression becomes compromised in aggressive bladder cancer cells and elucidated how this low TRAF4 expression influences EMT and may trigger bladder cancer progression. Moreover, using transcriptional profiling, as well as genetic and pharmacologic intervention approaches, we elucidated the contribution of the NF-κb and bone morphogenetic protein (BMP) pathways that are affected upon TRAF4 dysregulation. Moreover, we confirmed these correlations using material from patients with bladder cancer. Our findings may, therefore, be of importance for the treatment of patients with bladder cancer with low TRAF4 expression.
Materials and Methods
Cell culture conditions
Bladder cancer cell lines and 293T were purchased from ATCC. Cells were grown in DMEM supplemented with 10% FBS and penicillin/streptomycin. Cells were regularly tested for the absence of Mycoplasma contamination and were genotyped and authenticated. Cells were grown in 5% CO2 atmosphere incubator at 37°C. Where appropriate, cells were treated with BMP6 (50 ng/mL), TNFα (10 ng/mL), MG132 (2 μmol/L), LDN193189 (120 nmol/L), SMURF1i-A01 (5 or 10 μmol/L), MEK inhibitor (PD0325901, 2 μmol/L), 5′-Azacytidine (5 μmol/L), and Cycloheximide (10 μg/mL) for the indicated hours.
Transient transfection
293T cells were transfected with the indicated plasmids using calcium chloride and HEPES buffered saline (pH 6.95). After an overnight incubation, cells were washed twice with 1X PBS solution and replenished with fresh serum containing media. The following vectors and its derivates were used: pcDNA3.1 (6xMyc) TRAF4, pFLAG-CMV SMURF1, GFP-ERK1 was a gift from Rony Seger (Addgene plasmid # 14747; ref. 19) and GFP-TRAF4 was a gift from Ying Zhang (Addgene plasmid # 58318; ref. 20).
Stable transfection
Stable knockdown of TRAF4 in RT4, HT1376 or overexpression of TRAF4 in T24 cells were performed through lentiviral transduction. 293T cells were transfected with pLKO.1 puro vectors (Sigma Mission shRNAs) or pLV-IRES Lenti Puro TRAF4 along with lentiviral packaging plasmids (pCMV-VSVG, pMDLg-RRE, and pRSV-REV). The media containing viral particles were collected 48 hours later and passed through 0.45 μM filter. The supernatant with Polybrene (0.01%) was used to transduce bladder cancer cells. The cells were further selected with Puromycin (1 μg/mL) containing medium. The list of short hairpins used for knockdown are provided in Supplementary Table S5.
Immunofluorescence
Labelling of plasma membrane was achieved with CellMask Orange plasma membrane stain (Thermo Fisher) solution. RT4 cells were treated with the solution according to the manufacturer's instructions and images were captured using Leica fluorescence microscope.
In vivo phosphorylation experiment
Transfected 293T cells were lysed in ELB buffer (250 mmol/L NaCl, 0.5% Nonidet P-40, HEPES 50 mmol/L, pH 7.3), supplemented with protease inhibitors and serine/threonine phosphatase inhibitors: 50 mmol/L sodium fluoride and 10 mmol/L β-glycerophosphate. Protein concentration was estimated on the lysates using a Bicinchoninic Acid Protein Assay Kit (5000111, Bio-Rad). Equal protein concentrations for each of the samples were incubated with Myc antibodies overnight, followed by incubation with Protein G beads for 1 hour at 4°C. After several washing steps with 1X lysis buffer, beads were boiled in 2X sample buffer. The resulting supernatants were processed for immunoblotting.
Culturing of cell spheroids
Cell spheroids were generated using RT4 cells. 1.5% agarose was boiled until it dissolved in 1XPBS, then added onto a sterile 96-well plate (100 μL each well) and it was let to solidify. After one hour RT4 cells were trypsinized, counted, and diluted in media. 200 μL of media containing the appropriate amount of cells were added onto the agarose beds formed on the 96-well plate. The plate was spun down at 1,000 RPM for 2 minutes and incubated at 37°C CO2 incubator overnight. The following day, cell spheroids were checked under a Leica microscope. Spheroids were assessed for circularity using ImageJ software.
Assessment of ubiquitination
Cells were washed in ice cold 1XPBS (twice) and then lysed with RIPA buffer (25 mmol/L Tris HCl, pH 7.4, 150 mmol/L NaCl, 1%Nonidet P-40, 1%SDS, 0.5%sodium deoxycholate), supplemented with protease inhibitors and 10 mmol/L N-ethylmaleimide. Lysates were sonicated, boiled at 95°C for 5 minutes and diluted with RIPA buffer containing 0.1% SDS. Lysates were centrifuged at 4°C for 15 minutes. Thereafter, protein estimation was performed and equal amounts of lysates were incubated with Myc antibodies overnight, followed by incubation with Protein G beads for 1 hour at 4°C. After several washing steps with 1X lysis buffer, beads were boiled in 2Xsample buffer. The resulting supernatant was processed for immunoblotting.
Transcriptomics, gene signatures, pathway analysis, and enrichment scores
TRAF4 was knocked down using shRNA in HT1376 using lentiviral transduction. Cells with empty vector (pLKO) was used as control. Four independent experimental replicates were used for each condition. T24 cells stably overexpressing Myc-TRAF4 or empty vector (Myc-tag) were generated. Again, four independent experimental replicates were used. The cells were processed for RNA extraction and sent to BGI Tech (Hong Kong) for further processing. RNA sequencing (RNA-seq) files were processed using the opensource BIOWDL RNA-seq pipeline version 3.0.0 (https://zenodo.org/record/3713261#.X4GpD2MzYck) developed at the LUMC. The pipeline performs FASTQ preprocessing (including quality control, quality trimming, and adapter clipping), RNA-seq alignment, read quantification, and optionally transcript assembly. FastQC was used for checking raw read QC. Adapter clipping was performed using Cutadapt (v2.8) with default settings. RNA-seq reads’ alignment was performed using STAR (v2.7.3a) on GRCh38 reference genome. The gene read quantification was performed using HTSeq-count (v0.11.2). The gene annotation used for quantification was Ensembl version 99. Using the gene read count matrix, CPM was calculated per sample on all annotated genes. EdgeR (v3.28.1) with TMM normalization was used to perform differential gene expression analysis. Benjamini and Hochberg FDR was computed to adjust P values obtained for differentially expressed genes. For pathway analysis, gene signatures were obtained from previous studies (ref. 21, 22; Supplementary Table S3). Thereafter, changes in gene expression were compared with gene signatures and enrichment scores were obtained (Supplementary Table S4). The enrichment scores of gene signatures were estimated using R GSVA v1.36.2 (23).
Quantitative real-time PCR
Total RNA from cells was isolated using the NucleoSpin RNA II kit (740955, BIOKÉ) using the manufacturer's instructions. Thereafter, 1 μg of RNA from each sample was used to perform cDNA synthesis using the RevertAid First Strand cDNA Synthesis Kit (K1621, Thermo Fisher Scientific). Real time PCR was performed with GoTaq qPCR Master Mix (A6001, Promega) using CFX Connect Detection System (1855201, Bio-Rad). GAPDH was used as internal control for normalization. Experiments are performed as technical triplicates. A list of primers that were used are provided in Supplementary Table S5.
MTS cell viability/proliferation assay
To measure the proliferative capacities, cells were seeded on 96-well plates with 100 μL of media. The following and subsequent days, 20 μL of MTS solution was added per well and incubated in CO2 incubator for 1.5 hours. Thereafter, absorbance was measured on a luminometer at 490 nm.
Luciferase reporter assay
Luciferase reporter assays were performed using Dual luciferase reporter system (Promega). BRE-luciferase or NF-κB reporter plasmid was transfected with either control empty vector or TRAF4 and CMV-Renilla in 293T cells seeded on a 24-well plate. About 72 hours post-transfection, cells were stimulated overnight with BMP6 (50 ng/mL) and/or TNFα (10 ng/mL) in serum-free media. The following day, cells were lysed in Passive Lysis Buffer (Promega) and relative luciferase units and Renilla values were measured using a Luminometer.
Transwell invasion assays
HT1376, T24, or MBT-2 cells were grown in 10-cm dishes and serum starved overnight. The following day, cells were trypsinized and resuspended in 0.5% serum-containing media; 50,000 were seeded onto the upper chambers. The lower chambers (wells) were filled with 2% serum-containing media and incubated overnight. The following day, cells were fixed in ice-cold methanol for 10 minutes and stained with crystal violet solution. The inner side of the chambers were wiped clean using cotton swabs dipped in 1XPBS to remove remaining cells. Migrated cells were visualized through brightfield microscope and images were captured at four random sites and quantified.
Site directed mutagenesis
PCR reactions were performed using QuikChange XL Kit by Agilent Technologies (catalog no. 200517-4) according to the manufacturer's instructions. The presence of mutants was confirmed by sequencing. The list of primers that were used is provided in Supplementary Table S5.
Immunoblotting
Cells were lysed in RIPA buffer (150 mmol/L NaCl, 1% Nonidet P-40, 0.5% sodium deoxycholate, 0.1% sodium dodecyl sulfate, 50 mmol/L Tris pH 8.0), supplemented with protease inhibitors and phosphatase inhibitors, 50 mmol/L sodium fluoride, 100 mmol/L β-glycerophosphate, and 1 mmol/L sodium orthovanadate. Protein estimation was performed on the lysates and equal amounts of protein lysates were boiled in 2X sample buffer. Thereafter, samples were loaded onto 10% SDS-PAGE gels and transferred onto 0.45 μM PVDF membranes (Millipore). The membranes were blocked in 5% milk and probed with specific antibodies overnight at 4°C. For visualization of protein signals, blots were incubated with secondary antibodies which were HRP-linked and detected using chemiluminescence. The following antibodies were used: TRAF4 1:2,000 (D1N3A, CST); E-cadherin 1:1,000 (Catalog no. 610181, BD); N-cadherin 1:1,000 (Catalog no. 610920, BD); Vimentin 1:5,000 (CST); SLUG 1:1,000 (C19G7, CST); SNAIL 1:1,000 (C15D3, CST); Flag 1:5,000 (M2, Sigma-Aldrich); phospho-Serine 1:1,000 (612546, BD); SMURF1 1:1,000 (45-K, Santa Cruz Biotechnology); Myc 1:5,000 (9E10, Santa Cruz Biotechnology); HA 1:5,000 (Y11, Santa Cruz Biotechnology); GFP 1:5,000 (FL, Santa Cruz Biotechnology); and GAPDH 1:10,000 (MAB374, Millipore).
Wound-healing assays on Incucyte
T24 cells were trypsinized and counted, then 25,000 cells were seeded on each well of a 96-well plate (Essen ImageLock) and let to attach in the CO2 incubator for 5 hours. Thereafter, media containing serum was removed and replaced with serum-free media and cultured overnight. The following day, a Woundmaker Tool (4563, Essen) was used to produce wounds on the 96-well plate. After washing 2 times with 1XPBS, cells were replenished with 100 μL of 0.5% serum-containing media with the indicated treatments. The plate was then placed into the Incucyte Systems for Live-Cell Imaging and Analysis. Real-time images of (migrating) cells were captured every 1 hour and wound closure was analyzed. Ten to 12 well replicates were used for each condition to produce statistical error and significance.
IHC staining
Tissue microarrays containing bladder cancer samples of Stages 1, 2, and 3, as well as adjacent normal tissue and healthy bladder tissue were purchased from Biomax (BL802b, Biomax). Sections were heated at 60°C for 30 minutes prior to staining. Sections were deparaffinized and rehydrated, followed by heat induced antigen retrieval for 20 minutes. For the NFkb p65 and phospho-SMAD1/5/8 antibodies, antigen retrieval was performed in 0.01 mol/L sodium citrate/0.05% Tween (pH 6). For the TRAF4 antibody, this was performed in 10 mmol/L TRIS/1 mmol/L EDTA/0.05% Tween (pH 9). Sections were blocked for 30 minutes with 1% BSA and 0.1% Tween, followed by overnight incubation with primary antibodies at 4°C. Primary antibodies used were TRAF4 (1:50, HPA052377, Atlas, Bromma Sweden), NF-κB-p65 (phospho-Ser311; 1:100, #11260, Signalway antibody, Uithoorn, the Netherlands) and phospho-SMAD1/5/8 (1:50, #9511, Cell Signaling Technology, Leiden, the Netherlands). Sections were incubated with secondary antibody Alexa Fluor 488 donkey anti-rabbit (1:250, A-21206, Invitrogen, Landsmeer, the Netherlands) for 2 hours at room temperature, followed by 10 minutes of DAPI staining. Slides were digitalized using Pannoramic 250 Flash III slide scanner (3DHISTECH, Budapest, Hungary) and staining for all antibodies were scored by two independent observers and their average scores were considered. TRAF4, NF-κB-p65 (phospho-Ser311), and phospho-SMAD1/5/8s staining were scored combining the staining intensity (0, no staining; 1, low staining; 2, medium staining; 3, high staining) and percentage of positive tumor cells (TRAF4) or percentage of tumor cells with nuclear staining (NF-κB-p65 (phospho-Ser311) and phospho-SMAD1/5/8 (0, 0%; 1, 1%–5%; 2, 6%–25%; 3, 26%–50%; 4, 51%–75%; 5, 76%–100%). Staining from normal bladder tissue samples (n = 8) were not considered for analysis for pSMAD1/5/8 and p-p65 samples. Representative photos of TRAF4 staining on grade 1, 2, and 3, as well as low and high NF-κB-phospho-p65 and phospho-SMAD1/5/8 staining were generated using the CaseViewer software version 2.0 (3DHISTECH, Budapest, Hungary).
Statistical analyses
Bar graphs show mean SD or SEM as indicated in the figure legends. Student t test, one-way ANOVA or two-way ANOVA, as indicated in the figure legends, were used for the analysis of significance and P value. Kaplan–Meier graph was plotted using survival curve (GraphPad Prism). For regression plots, Pearson r was used to analyze correlation. All tests were two-tailed.
Data availability
The data generated in this study are available within the article and its Supplementary Data files. Publicly available expression data analyzed in this study, The Cancer Genome Atlas Urothelial Bladder Carcinoma (TCGA BLCA), was obtained from the Human Protein Atlas www.proteinatlas.org/about/download, version 21).
Results
TRAF4 expression negatively correlates with bladder cancer progression
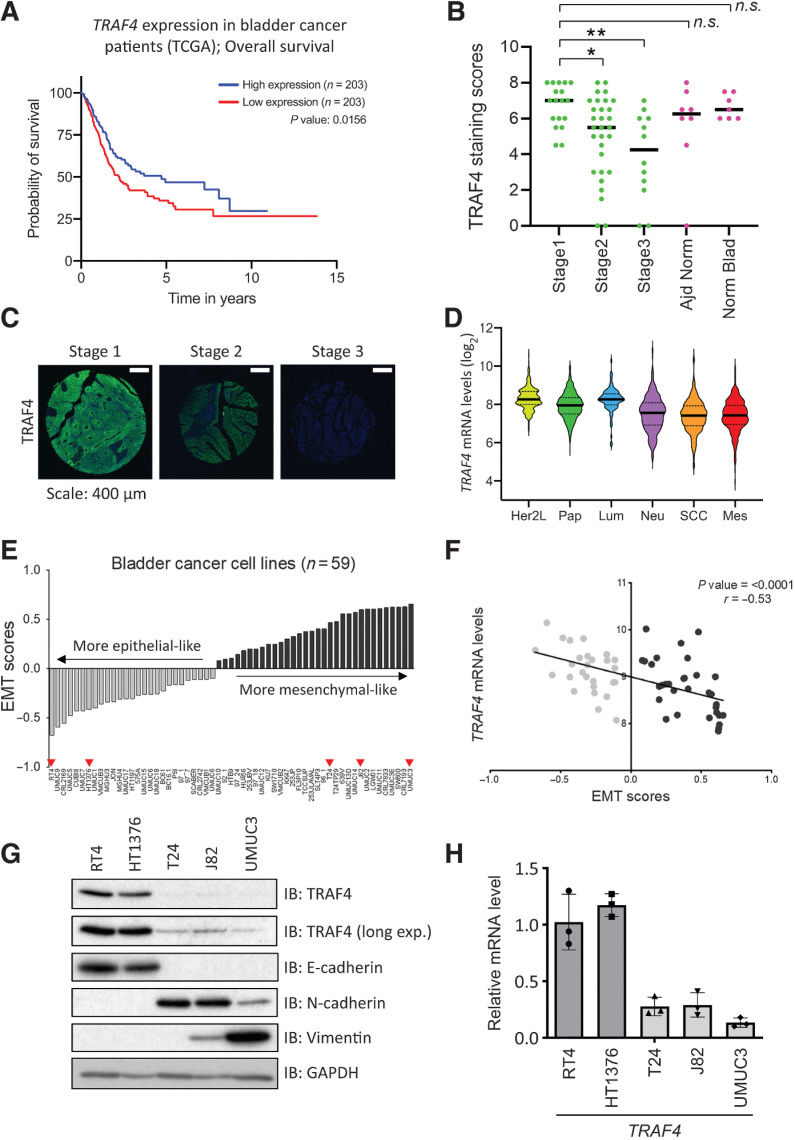
We investigated the correlation between TRAF4 mRNA expression and overall survival in patients with bladder cancer across all stages. Kaplan–Meier analysis of publicly available data from TCGA BLCA dataset (24), which was obtained from the Human Protein Atlas (www.proteinatlas.org/about/download, version 21; ref. 25), revealed that patients with bladder cancer with a lower level of TRAF4 expression had a significantly lower survival probability than those with a higher TRAF4 expression level (Fig. 1A). To further confirm these observations, we performed IHC analysis of TRAF4 protein expression using tissue microarray samples obtained from Biomax U.S. (BL802b). Our data revealed significant differences in TRAF4 protein expression between stage 1 and stage 2/3 tumor samples, but the expression differences between stage 1 and adjacent normal tissue or normal bladder tissue samples were nonsignificant (Fig. 1B and C). To further corroborate our initial observations, we cross-checked TRAF4 expression in a recently compiled meta-cohort study of 2411 sets of bladder tumor data (26). The classification comprised of six distinct molecular subtypes: Her2-like, papillary, neural, luminal, squamous cell carcinoma (SCC), and mesenchymal. We observed that TRAF4 expression was lowest in the SCC and mesenchymal subtypes, which had the poorest survival outcomes (Fig. 1D). EMT scoring can be performed using a specific EMT gene signature (27). It is noteworthy that the SCC and mesenchymal subtypes had the highest EMT scores, meaning that cells of these tumors are most likely to be mesenchymal-like in phenotype (26). Furthermore, we extended our EMT scoring to bladder cancer cell lines. We calculated the EMT scores using publicly available data obtained in 59 (human) bladder cancer cell lines (ref. 28; Fig. 1E; Supplementary Table S1). We defined cell lines with a negative EMT score as more epithelial-like and cell lines with a positive EMT score as more mesenchymal-like. Consistent with the results obtained in patient biopsies, significant negative correlations were found between TRAF4 expression and the EMT score in these 59 bladder cancer cell lines (Fig. 1F). We selected 5 bladder cancer cell lines, i.e., RT4 and HT1376, with negative EMT scores, and T24, J82 and UMUC3, with positive EMT scores, for further consideration in our study. We next examined whether TRAF4 expression correlates with the ‘EMT status’ of these cell lines at the protein level by Western blot analysis. As shown in Fig. 1G, TRAF4 expression was higher in epithelial cell lines expressing higher levels of E-cadherin and lower levels of the mesenchymal markers N-cadherin and Vimentin (5). In contrast, TRAF4 expression was lower in mesenchymal cell lines with lower E-cadherin levels but higher N-cadherin and Vimentin levels. Importantly, the epithelial cell lines also had higher TRAF4 mRNA expression levels than the mesenchymal cell lines (Fig. 1H). Collectively, our results indicate that TRAF4 expression is higher in epithelial bladder cancer cells than in more mesenchymal bladder cancer cells.
Figure 1.
TRAF4 is downregulated in aggressive bladder tumors and mesenchymal bladder cancer cell lines. A, Kaplan–Meier plot showing the overall survival of patients with bladder cancer stratified by TRAF4 expression. Data were obtained and reproduced from TCGA (ref. 24; obtained from Human Protein Atlas; ref. 25), and the median Fragments Per Kilobase of transcript per Million mapped reads (FKPM) value was taken as the TRAF4 expression cutoff, n = 203. B, Graph showing TRAF4 expression through scores obtained from IHC analysis of a tissue microarray; *, P ≤ 0.05 and **, P ≤ 0.01 calculated using one-way ANOVA; n.s. indicates a nonsignificant P value. C, Representative IHC images of TRAF4 expression (green) in the tissue microarray from stage 1–3 bladder tumors are shown. Scale bar, 400 μm. D, Violin plot showing the TRAF4 expression level (and distribution) in different subtypes of bladder cancer; Her2L: Her2-like (n = 253), Pap: papillary (n = 674), Lum: luminal (n = 107), Neu: neural (n = 448), SCC (n = 333) and Mes: mesenchymal (n = 308). The black bars in the middle of the distribution indicate the medians. The subtypes are arranged according to their EMT scores (26, 27). E, Plot showing the EMT scores in 59 bladder cancer cell lines; the light grey bars indicate cell lines with a negative EMT score, the dark grey bars indicate cell lines with a positive EMT score, and the red arrowheads indicate the cell lines that were used for further investigation (28). F, Regression plot of TRAF4 expression levels vs. EMT scores in 59 bladder cancer cell lines. G, Immunoblot analysis showing the expression of TRAF4 and other EMT marker proteins. GAPDH, loading control. Representative results of 2 independent biological replicates. H, Real-time PCR data showing TRAF4 mRNA expression in cell lines. The error bars indicate ± SD. Epithelial cell lines (dark grey bars) had significantly higher TRAF4 expression than mesenchymal cell lines (light grey bars). Performed in 3 technical repeats.
TRAF4 is epigenetically repressed and ERK mediates its phosphorylation at serine 334
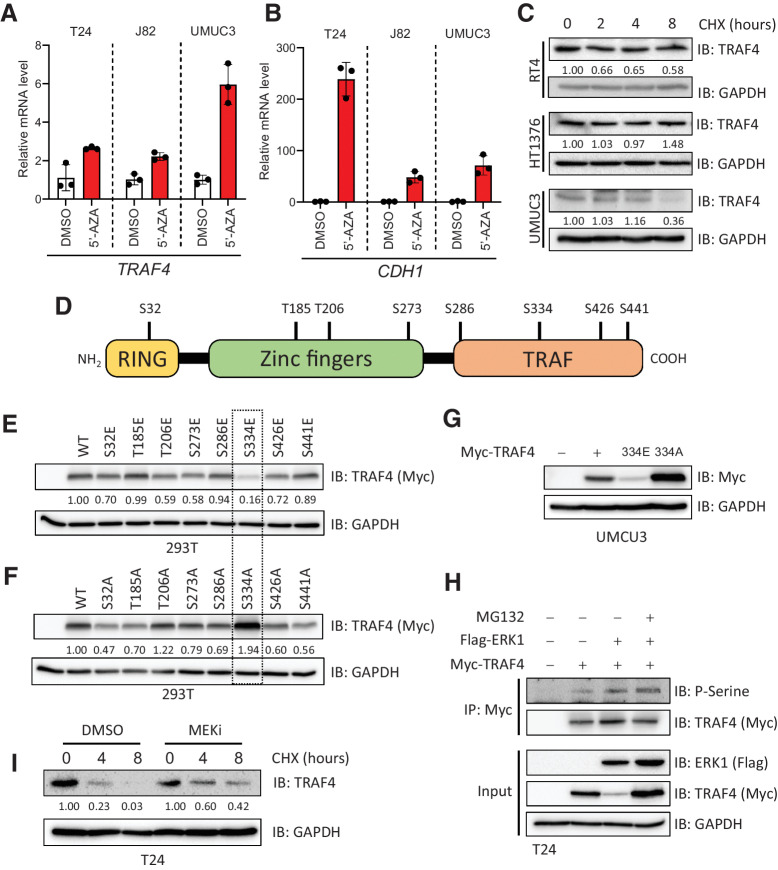
We then sought to determine the reasons for the low expression of TRAF4 in mesenchymal cells. We subjected three mesenchymal cell lines to 5-azacitidine (5-AZA, a compound that blocks DNA methylation) treatment for a week. As shown in Fig. 2A, TRAF4 expression was rescued upon treatment in these cell lines, suggesting that TRAF4 is epigenetically repressed. As a positive control, we measured the expression of CDH1 (encoding E-cadherin), which is known to be epigenetically repressed in many mesenchymal cancer cells; its expression was also increased upon 5-AZA treatment (Fig. 2B). Surprisingly, however, the treated cells did not show consistent upregulation of TRAF4 protein expression, although E-cadherin expression was upregulated in 2 of the 3 cell lines (Supplementary Fig. S1A). This suggested the existence of additional mechanisms that control TRAF4 protein levels. We performed a cycloheximide pulse-chase experiment to analyze the protein stability of TRAF4 in different cell lines. The TRAF4 protein was less stable in UMUC3 cells than in RT4 and HT1376 cells (Fig. 2C). To determine the existence of specific posttranslational modifications that regulate TRAF4 protein stability, we performed mass spectrometric analysis after overexpression of epitope Flag tagged-TRAF4 in 293T cells (Supplementary Fig. S2). We observed that TRAF4 undergoes several phosphorylation events at serine and threonine residues scattered through its length (Fig. 2D; Supplementary Fig. S2).
Figure 2.
TRAF4 is repressed in mesenchymal (bladder cancer) cell lines at the epigenetic and proteomic levels. A, Real-time PCR results showing changes in the TRAF4 mRNA level in mesenchymal cell lines after treatment with 5-AZA, performed in 3 technical repeats. B, Real-time PCR results showing changes in the CDH1 mRNA level in cell lines after treatment with 5-AZA; the error bars indicate ± SD, performed in 3 technical repeats. C, Immunoblot results showing the endogenous TRAF4 levels in the indicated cell lines after treatment with cycloheximide (CHX). GAPDH, loading control. The numbers indicate the relative quantitative TRAF4 levels with respect to the loading control GAPDH, n = 1 D, Schematic representation of TRAF4 showing the distinct domain structures and the candidate phosphorylated serine and threonine residues that were identified using mass spectrometric analysis. E, Immunoblot results from 293T cells transfected with expression constructs for either TRAF4 or the TRAF4 glutamic acid (E) mutant. GAPDH, loading control. The numbers indicate the relative quantitative TRAF4 levels with respect to the loading control GAPDH. F, Immunoblot results from 293T cells transfected with expression constructs for either TRAF4 or the TRAF4 alanine (A) mutant. GAPDH, loading control. The numbers indicate the relative quantitative TRAF4 levels with respect to the loading control GAPDH. G, Western blot analysis of ectopic Myc-TRAF4 WT and the S334E and S334A mutants in the UMUC3 cell line. GAPDH, loading control. H, Immunoprecipitation of Myc-TRAF4 with or without Flag-ERK1 overexpression, GAPDH, loading control. I, Western blot analysis of TRAF4 expression in T24 cells treated with CHX at the indicated times, in the presence of DMSO (control) or MEKi. GAPDH, loading control. The numbers indicate the relative quantitative TRAF4 levels for both DMSO and MEKi (PD0325901) treatment separately, with respect to the loading control GAPDH.
To determine whether phosphorylation at these sites affects the TRAF4 expression level, we performed site-directed mutagenesis to mutate the candidate serine/threonine residues to glutamic acid (E) residues in order to mimic phosphorylation, or alanine (A) residues in order to block phosphorylation. We ectopically expressed these TRAF4 mutants in 293T cells and assessed their expression levels. As shown in Fig. 2E and F, compared with modifications at the other sites, mutation of serine 334 to glutamic acid significantly reduced the TRAF4 expression level, and reciprocally, mutation of serine 334 to alanine increased its expression level. Serine 334 in TRAF4 appears to be highly conserved in the other species examined (Supplementary Fig. S1B). Ectopic expression of Myc-TRAF4 mutants in UMUC3 cells confirmed the results obtained in 293T cells (Fig. 2G). A cycloheximide pulse-chase assay revealed that the S334E mutant was indeed less stable than wild-type (WT) TRAF4 or the S334A mutant (Supplementary Fig. S1C and S1D). Moreover, addition of the proteasome inhibitor MG132 rescued the low expression level of the S334E mutant, suggesting that its low expression is due to proteasomal-mediated degradation (Supplementary Fig. S1E). A phosphorylation prediction tool (Human Protein Reference Database, PhosphoMotif Finder) revealed that serine 334 could be a putative ERK-MAP kinase phosphorylation site. Moreover, the sequence KRRLLCPLC that resembles a MAPK docking site (D-site) consensus sequence ((R/K)2–3-X2–6-ΦA-X-ΦB (Φ is any hydrophobic residue) is present in TRAF4 (29). When we overexpressed ERK1 along with TRAF4 in 293T cells, the TRAF4 expression level was significantly reduced (Supplementary Fig. S1F). Importantly, ERK1 was not able to influence the expression level of the S334A mutant (Supplementary Fig. S1F). MG132 was also able to rescue the ERK1-mediated decrease in the expression of TRAF4, suggesting that ERK1-mediated phosphorylation of TRAF4 induces its proteasomal degradation (Supplementary Fig. S1G). To determine whether ERK1 can convey the phosphorylation of TRAF4 and whether this effect can be enhanced by MG132, we assessed the levels of TRAF4 pSerine upon ERK1 expression. As shown in Fig. 2H, co-transfection of ERK1 with TRAF4 indeed increased TRAF4 phosphorylation, and this effect was further enhanced upon treatment with MG132. Moreover a selective small molecule MEK (an upstream kinase of ERK) inhibitor (MEKi, PD0325901) prolonged TRAF4 stability T24 cells overexpressing TRAF4 (Fig. 2I) and slightly enhanced steady-state TRAF4 level in 293T cells expressing Myc-TRAF4 (Supplementary Fig. S1H). This observation was consistent in the UMUC3 and T24 cell lines, where the endogenous TRAF4 level was slightly increased by MEKi treatment, while TRAF4 mRNA level remained relatively unaffected (Supplementary Fig. S1I–1L). Taken together, our results suggest that phosphorylation of TRAF4 at serine 334 mediated by ERK leads to a decrease in its expression via proteasome-mediated degradation.
Knockdown of TRAF4 in epithelial cell lines leads to loss of epithelial integrity and gain of mesenchymal markers
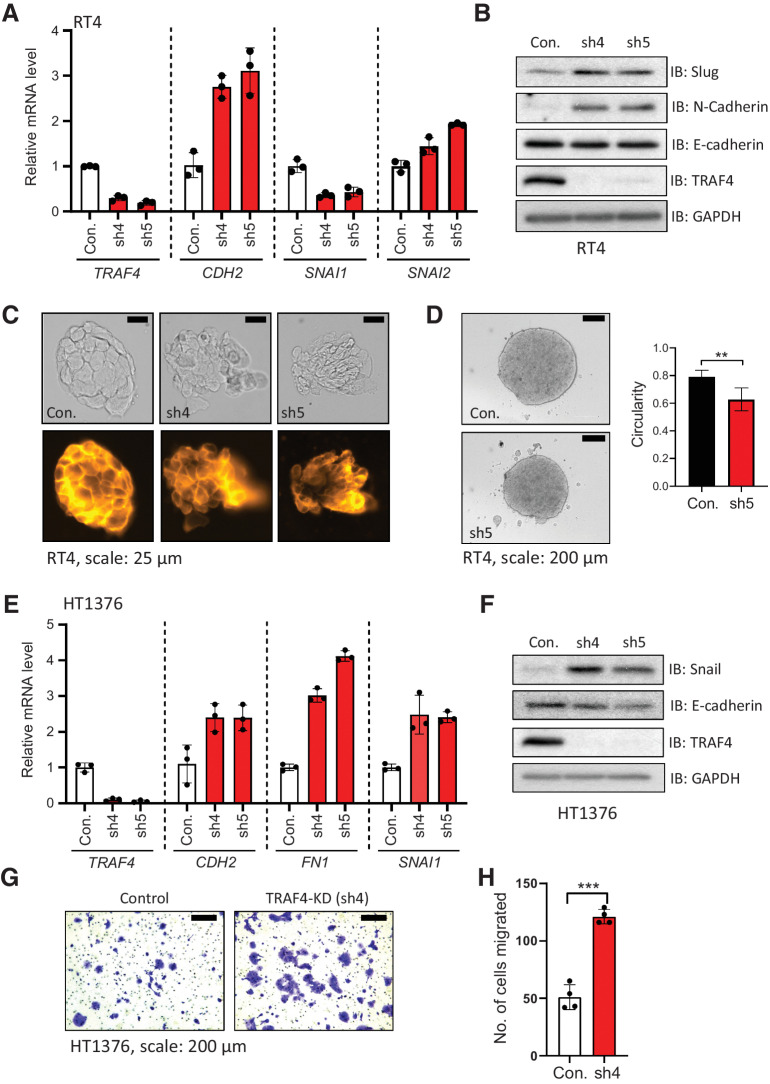
Since we observed that TRAF4 expression is reduced in mesenchymal cells compared with epithelial bladder cancer cells, we next investigated the consequences of TRAF4 knockdown in the epithelial cancer cell lines RT4 and HT1376. Depletion of TRAF4 in RT4 cells using the two independent short hairpin RNAs (#4 and #5) with the highest knockdown efficiency (Supplementary Fig. S3A) led to increases in the mRNA expression levels of the mesenchymal markers CDH2 (encoding N-cadherin) and SNAI2 (encoding SLUG; Fig. 3A). SLUG is a well-studied EMT transcription factor that has been described to play roles in cadherin switching and malignancy in bladder cancer progression (30). Notably, another major EMT-inducing transcription factor (TF), SNAI1 (encoding SNAIL), was consistently downregulated upon TRAF4 knockdown. The increases in SLUG and N-cadherin (but not E-cadherin) expression were also observed at the protein level (Fig. 3B). Besides changes in gene expression of EMT markers, we observed phenotypic changes in the integrity and architecture of cell colonies upon TRAF4 knockdown (Fig. 3C). Staining with a membrane dye revealed that the borders of cells within RT4 cell colonies became disordered and that loosely attached cells appeared upon TRAF4 knockdown. The membrane staining of colonies formed by control (pLKO vector) cells resembled that of colonies formed by WT RT4 cells (Supplementary Fig. S3B).
Figure 3.
Knockdown of TRAF4 in epithelial (bladder cancer) cell lines leads to loss of epithelial integrity and changes in EMT marker expression. A, Real-time PCR results from RT4 cells showing the mRNA expression levels of the indicated genes; the error bars indicate ± SD, performed in 3 technical repeats. B, Immunoblot results showing changes in EMT marker protein expression in RT4 cells upon TRAF4 knockdown. GAPDH, loading control. Representative result of 2 biological replicates. C, RT4 cell colonies visualized by brightfield imaging (top) or after staining with CellMask Orange Plasma membrane stain (bottom); scale bar: 25 μm. Representative of n = 5. D, Images showing RT4 spheroids formed from control (empty pLKO vector) and TRAF4 knockdown (sh5) cells; scale bar: 200 μm. The graph shows circularities calculated from five independent spheroids of different sizes (n = 5). The error bars indicate ± SD; **, P ≤ 0.01 calculated using two-tailed Student t test. E, Real-time PCR results from HT1376 cells showing the mRNA expression levels of the indicated genes; the error bars indicate ± SD performed in 3 technical repeats. F, Immunoblot results showing EMT marker protein expression levels in HT1376 cells with or without TRAF4 knockdown. GAPDH, loading control. Representative results of 2 biological repeats. G, Representative images of Transwell assays performed on HT1376 cells are shown. Cells were stained with crystal violet; scale bar: 200 μm. H, Quantification of the number of migrated cells in four random fields; the error bars indicate ± SD; ***, P ≤ 0.001 calculated using two-tailed Student t test, 4 technical repeats.
To examine whether TRAF4 knockdown affects the 3-dimensional structural architecture of cells, RT4 cell spheroids were generated. Spheroids recapitulate tumor cell clusters and can be considered, in many ways, a model more representative of in vivo conditions than 2-dimensional–cultured cells. Fig. 3D shows spheroids formed from control RT4 (empty pLKO vector) and TRAF4 knockdown cells. We found that upon TRAF4 knockdown, several clumps of cells within the spheroids were dissociated or excluded from the main bodies. Moreover, the spheroids in the knockdown group were more irregular in shape compared with those in the control group, as determined by measurement of their circularity (Fig. 3D). This cell exclusion phenotype has been observed in previous studies and is reflective of the loss of certain tight junction components in epithelial cells (31, 32). We further demonstrated the effects of TRAF4 knockdown using the epithelial cell line, HT1376. TRAF4 knockdown using 2 different shRNAs (Supplementary Fig. S3C) led to a significant increase in the mRNA expression levels of the mesenchymal markers FN1 (encoding Fibronectin) and SNAI1 (Fig. 3E), and an increasing trend of CDH2, encoding N-cadherin. Although we saw a concomitant increase in the level of the EMT-inducing TF SNAIL (Fig. 3F), we could not observe whether the N-cadherin level changed as it remained below the limit of detection. Moreover, the E-cadherin level remained at best unchanged upon TRAF4 knockdown in these cells. We hypothesized that because EMT has been linked to migratory and invasive properties, knockdown of TRAF4 would lead to enhanced invasive behavior. Knockdown of TRAF4 indeed increased the number of invaded cells, as determined by transwell invasion assays (Fig. 3G and H). Thus, TRAF4 knockdown in epithelial cells disrupts their epithelial architecture and organization. Importantly, TRAF4 knockdown leads to an increase in gene expression of EMT markers, disruption in epithelial architecture and organization, and importantly, an increase in their invasive capacities. This suggests that upon knockdown of TRAF4, epithelial bladder cancer cells become more mesenchymal.
Stable overexpression of TRAF4 diminishes the migratory and invasive properties of mesenchymal cells
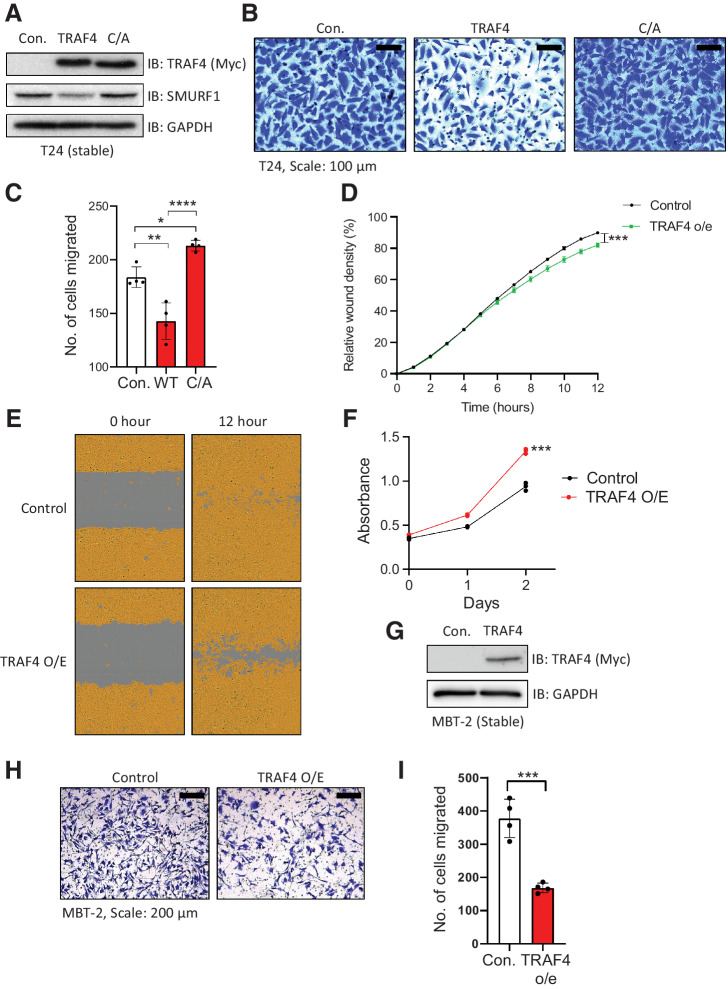
We next assessed whether ectopic expression of TRAF4 in mesenchymal cell lines affects their functional properties. To this end, we stably overexpressed empty vector (Myc epitope tagged), TRAF4, or the TRAF4 (C/A) mutant, as shown in Fig. 4A. Ectopic expression of TRAF4 slightly but significantly decreased T24 cell invasion (Fig. 4B and C) and migration (Fig. 4D and E) in a catalytically dependent manner, while proliferation was slightly increased upon ectopic expression of TRAF4 (Fig. 4F). Next, we used the highly invasive mouse cell line MBT-2, which has a low TRAF4 level (Supplementary Fig. S4A). Stable overexpression of TRAF4 in MBT-2 cells (Fig. 4G) decreased their invasive ability (Fig. 4H and I). Upon closer examination, morphologically, TRAF4-overexpressing cells tended to cluster more closely together than control cells, especially when seeded at a low density (Supplementary Fig. S4B). Collectively, these findings demonstrate that TRAF4 overexpression diminishes some of the aggressive characteristics of mesenchymal cell lines.
Figure 4.
Ectopic expression of TRAF4 in mesenchymal cells inhibits their migration and invasion. A, Immunoblot showing T24 cells stably expressing either control vector (Myc-tag), TRAF4 or the catalytically inactive TRAF4 mutant (C/A: cysteine substituted with alanine at residue C18). GAPDH, loading control. B, Representative images of Transwell assays performed on T24 cells stably expressing TRAF4 or the catalytically inactive TRAF4 mutant (C/A) are shown. Cells were stained with crystal violet; scale bar: 100 μm. C, Quantification of the number of migrated cells in four random fields. The error bars indicate ± SD; **, P ≤ 0.01 and ****, P ≤ 0.0001 calculated using one-way ANOVA, 4 technical repeats. D, Graph showing the relative wound widths as determined with an IncuCyte system. Representative results from ten independent experiments are shown; the error bars indicate ± SEM; ***, P ≤ 0.001 calculated using two-tailed Student t test (n = 10). E, Representative images related to the graph shown in D; the brown area represents the cell coverage, and the gray area indicates the initial wound produced and the remaining wound after 12 hours. F, MTS cell viability/proliferation assay performed with either control T24 cells or T24 cells stably expressing TRAF4. The absorbance was measured at the indicated time points; the error bars indicate ± SD from three sample replicates; ***, P ≤ 0.001 calculated using two-tailed Student t test (n = 3). G, Immunoblot results for MBT-2 cells stably expressing either control vector (empty vector with a Myc-tag) or Myc-TRAF4. GAPDH, loading control. H, Representative images of Transwell assays performed on control and TRAF4-overexpressing MBT-2 cells stained with crystal violet; scale bar: 200 μm. I, Quantification of the number of migrated cells in four random fields. The error bars indicate ± SD; ***, P ≤ 0.001 calculated using two-tailed Student t test, 4 technical repeats.
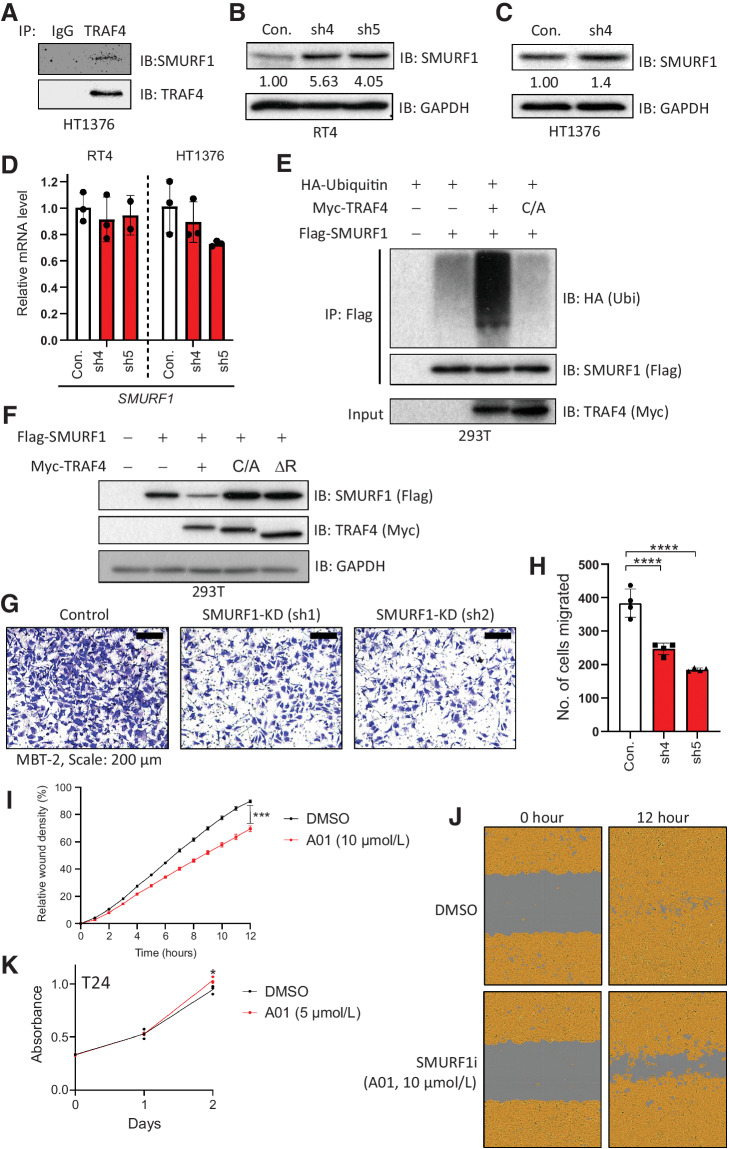
TRAF4 targets SMURF1 for polyubiquitination and degradation
Previously, TRAF4 was shown to interact with the E3 ligase SMURF1, a negative regulator of BMP/SMAD signaling and positive regulator of EMT progression (14, 33, 34). We hypothesized that TRAF4 targets SMURF1 for proteasomal degradation and that this event may explain the inhibitory effect of TRAF4 on EMT in bladder cancer cells. We found that TRAF4 interacts with SMURF1 in HT1376 bladder cancer cells (Fig. 5A; ref. 13). Consistent with the hypothesis that TRAF4 targets SMURF1 for degradation, we observed that in RT4 cells, the SMURF1 level was decreased by overexpression of WT TRAF4 but not by its catalytically inactive mutant TRAF4 (C/A; Fig. 4A). Moreover, upon TRAF4 knockdown in RT4 cells, the level of SMURF1 increased compared with that in control cells (Fig. 5B). This change was also observed in HT1376 cells (Fig. 5C). The SMURF1 mRNA level did not change upon TRAF4 knockdown in both of these cell lines (Fig. 5D). Next, we tested whether TRAF4 can induce SMURF1 polyubiquitination and decrease its expression level. Co-expression of TRAF4, but not the inactive TRAF4 (C/A) mutant or the RING deletion mutant, increased the polyubiquitination of SMURF1 (Fig. 5E) and decreased the steady-state level of SMURF1 (Fig. 5F).
Figure 5.
TRAF4 targets SMURF1 for ubiquitination and degradation. A, Immunoprecipitation of SMURF1 followed by Western blot analysis of TRAF4 in HT1376 cells. B, Immunoblot results in control (empty pLKO vector) and TRAF4 knockdown (sh4 and sh5) RT4 cells probed with the indicated antibodies. GAPDH, loading control. The numbers indicate the relative quantitative SMURF1 levels with respect to the loading control GAPDH. C, Immunoblot results in control (empty pLKO vector) and TRAF4 knockdown (sh4) HT1376 cells probed with the indicated antibodies. The numbers indicate the relative quantitative SMURF1 levels with respect to the loading control GAPDH. D, Real-time PCR results showing SMURF1 mRNA expression levels in RT4 and HT1376 (control and TRAF4 knockdown) cells; the error bars indicate ± SD, performed in 3 technical repeats. E, A ubiquitination assay was performed with anti-Flag antibodies in 293T cells overexpressing the indicated plasmids. Cells were treated with MG132 (2 μmol/L) overnight prior to lysis. Representative results from three independent experiments are shown. F, Immunoblot results in 293T cells transfected with the indicated plasmids. GAPDH, loading control. G, Representative images of Transwell assays performed on control and Smurf1 knockdown MBT-2 cells stained with crystal violet; scale bar: 200 μm. H, Quantification of the number of migrated cells in four random fields. The error bars indicate ± SD; ***, P ≤ 0.001 and ****, P ≤ 0.0001 calculated using one-way ANOVA, 4 technical repeats. I, Graph showing relative wound widths as determined with an IncuCyte system. Images were acquired every hour after wounding. T24 cells were treated with either DMSO or the SMURF1 inhibitor A01 (10 μmol/L); the error bars indicate ± SEM; ***, P ≤ 0.001 calculated using two-tailed Student t test. Representative results from ten independent experiments are shown (n = 10). J, Representative images related to the graph in I. The brown area represents the cell coverage, and the gray area indicates the wound initially produced and remaining after 12 hours. K, An MTS assay was performed on T24 cells treated with either control (DMSO) or the SMURF1i A01 at 5 μmol/L. The absorbance was measured at the indicated time points; the error bars indicate ± SD from three sample replicates; *, P ≤ 0.05 calculated using two-tailed Student t test (n = 3).
The role of SMURF1 in promoting cancer cell invasion is well documented (34). To examine whether SMURF1 plays a similar role in mesenchymal bladder cancer cells, we used the MBT-2 cell line. We observed that upon SMURF1 knockdown (using two independent shRNAs, Supplementary Fig. S4C and S4D), the invasion of MBT-1 cells was significantly decreased (Fig. 5G and H). These results were confirmed in T24 cells using a commercially available SMURF1 inhibitor, A01 (35). Of note, this SMURF1 inhibitor does not target E3 ligase activity but targets the ability of SMURF1 to bind to BMP pathway effector protein SMAD1 and SMAD5 and subsequently induce their proteasomal degradation (36). Treatment of T24 cells with A01 significantly reduced the wound healing ability compared with those in vehicle control (DMSO-treated) cells (Fig. 5I and J), while having a slightly enhanced effect on proliferation (Fig. 5K). Taken together, our results demonstrate that TRAF4 can reduce the SMURF1 protein level in bladder cancer cells and that SMURF1 enhances the migration and invasion of these mesenchymal bladder cancer cells.
Dysregulated expression of TRAF4 in bladder cancer cell lines affects the NF-κB and BMP signaling pathways
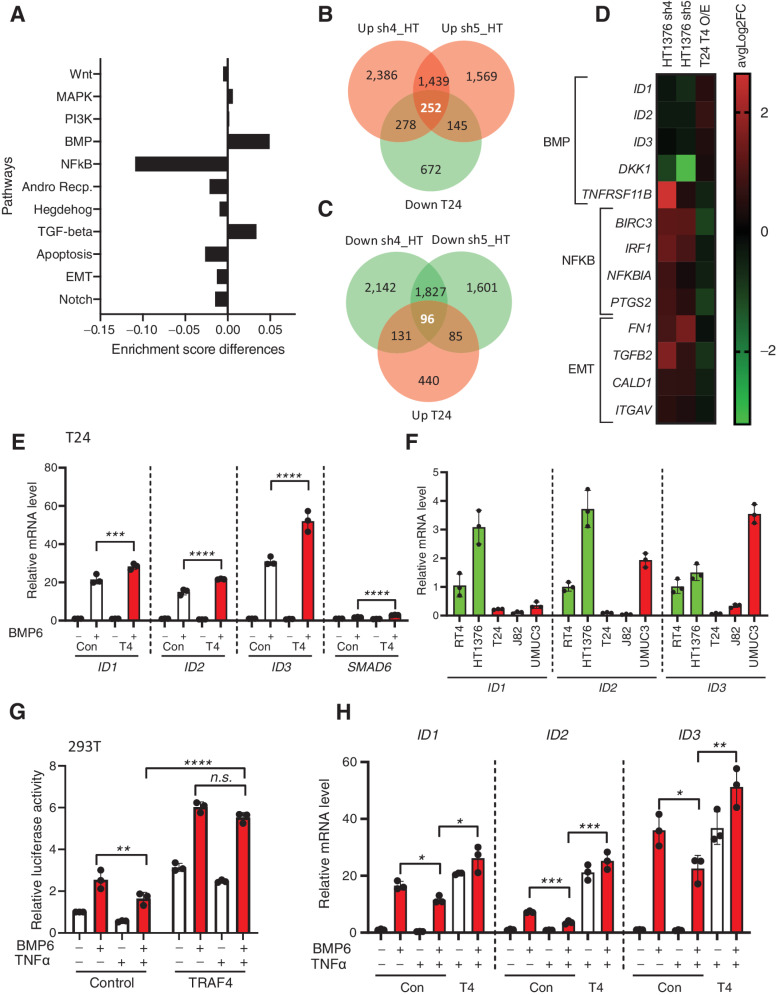
Next, to obtain insights into the mechanisms by which TRAF4 affects bladder cancer cell behavior, we examined the effect of TRAF4-mediated dysregulation of signaling pathways. To this end, we stably overexpressed TRAF4 in T24 cells (Supplementary Fig. S4E), performed transcriptomic (RNA-seq) analysis and looked for changes in the gene response signatures of the eleven most commonly studied oncogenic signaling pathways. Volcano plot of T24 cells expressing empty vehicle versus TRAF4 is shown in Supplementary Fig. S4F. Nine of the eleven signaling pathways showed varying degrees of changes in the enrichment score. The two most prominently changes in pathways were the downregulation of the NF-κB signaling pathway and the upregulation of the BMP/SMAD signaling pathway (Fig. 6A). These findings are consistent with previous reports indicating that TRAF4 can inhibit NF-κB signaling and promote BMP signaling (13, 36).
Figure 6.
Dysregulated expression of TRAF4 in bladder cancer cell lines affects BMP/SMAD- and NF-κb-responsive genes. A, Graph showing differences in enrichment scores when TRAF4 was overexpressed in T24 cells. Gene signatures of eleven major cancer-associated signaling pathways were considered for analysis. B, Venn diagram showing the numbers of genes that were upregulated in HT1376 cells transfected with two independent shRNAs targeting TRAF4 (pink) and downregulated in T24 cells with stable overexpression of TRAF4 (green) compared with the corresponding control cells. The 252 genes in the middle are the reciprocally affected common genes. The results were obtained from four independent replicates for each sample (n = 4). C, Venn diagram showing the numbers of genes that were downregulated in HT1376 cells transfected with two independent shRNAs targeting TRAF4 (green) and upregulated in T24 cells with stable overexpression of TRAF4 (pink) compared with the corresponding control cells. The 96 genes in the middle represent the reciprocally affected common genes. The results were obtained from four independent replicates for each sample (n = 4). D, Heatmap showing the common dysregulated genes in the BMP,NF-κB and EMT gene signatures in both cell lines. E, Real-time PCR results showing the mRNA expression levels of the indicated genes in control (empty vector with a Myc tag) vs. TRAF4-overexpressing T24 cells upon stimulation with BMP6 (50 ng/mL) for 1 hour. The error bars indicate ± SD, performed in 3 technical repeats. F, Real-time PCR results showing the mRNA expression levels of ID1, ID2, and ID3 in the indicated cell lines, performed in 3 technical repeats. G, A luciferase reporter assay was conducted in 293T cells transfected with the BRE-luciferase reporter, SV40 Renilla and either empty vector control or TRAF4. Transfected cells were stimulated overnight with BMP6 (50 ng/mL) and/or TNFα (10 ng/mL) where indicated. The error bars indicate ± SD; **, P ≤ 0.01 and ****, P ≤ 0.0001 calculated using two-way ANOVA; n.s. indicates a nonsignificant P value. Representative results from three independent experiments are shown (n = 3). H, Real-time PCR results showing the mRNA expression levels of the indicated genes in control (empty vector with a Myc tag) vs. TRAF4-overexpressing T24 cells upon stimulation with BMP6 (50 ng/mL) and/or TNFα (10 ng/mL) as indicated for 1 hour. The error bars indicate ± SD, performed in 3 technical repeats.
We also examined the changes in gene expression upon TRAF4 depletion in HT1376 cells using two independent shRNAs. Volcano plots of HT1376 cells expressing empty vehicle versus TRAF4 sh4 and sh5 are shown in Supplementary Fig. S4G and S4H respectively. As shown in Fig. 6B, 252 genes were upregulated upon TRAF4 knockdown in HT1376 cells (common to both shRNAs) and reciprocally downregulated in T24 cells upon TRAF4 overexpression (Supplementary Table S2). Similarly, we detected 96 genes that were upregulated in T24 cells and downregulated in HT1376 cells compared with the corresponding control cells (Fig. 6C; Supplementary Table S2). Five BMP/SMAD target genes, i.e., ID1, ID2, ID3, DKK1, and TNFRSF11B (37), were reciprocally regulated in the two cell lines upon TRAF4 dysregulation. These genes were downregulated upon TRAF4 knockdown in HT1376 cells and upregulated upon TRAF4 overexpression in T24 cells (Fig. 6D). Upon examining the NF-κB gene signature within the set of reciprocally regulated genes, we found four common genes showing downregulation upon TRAF4 overexpression in T24 cells and upregulation upon TRAF4 knockdown in HT1376 cells (Fig. 6D). We found that TRAF4 inversely correlated with the EMT status of bladder cancer cells (Fig. 1F); therefore, we examined EMT gene markers within the set of reciprocally regulated genes between the two cell lines and found four genes, i.e., FN1 (encoding Fibronectin), TGFB2 (encoding TGFβ2), CALD1 (encoding Caldesmon 1), and ITGAV (encoding Integrin subunit α-V) that are inversely linked to TRAF4 expression (Fig. 6D). Our transcriptomic analysis thus shows that dysregulated expression of TRAF4 affects NF-κb and BMP signaling pathways, as well as EMT-related genes.
TRAF4 promotes BMP/SMAD signaling in bladder cancer cells and antagonizes the inhibitory effect of TNFα signaling on BMP/SMAD signaling
As TRAF4 expression was positively associated with BMP signaling, we next determined the consequences of BMP stimulation on mesenchymal cells. Interestingly, we observed that canonical BMP pathway target genes (ID1, ID2, ID3, and SMAD6 were expressed at slightly higher levels upon BMP6 stimulation in T24 cells stably expressing TRAF4 (Fig. 6E; ref. 37, 38). This effect was also observed in MBT-2 cells (Supplementary Fig. S5A). Next, a BMP/SMAD response element (BRE)-luciferase transcriptional reporter assay was used to measure downstream BMP/SMAD signaling activity in 293T cells. BMP6 stimulation led to significantly higher luciferase activity in 293T cells transfected with TRAF4 than in the non-transfected control cells (Supplementary Fig. S5B). We then hypothesized that perhaps these three ID genes are expressed at higher levels in epithelial cell lines due to the difference in TRAF4 expression. Indeed, we observed that ID gene expression in the epithelial cell lines RT4 and HT1376 was generally higher than that in mesenchymal cell lines (Fig. 6F). Moreover, we observed that a selective BMP type I receptor kinase inhibitor (LDN193189) rescued the inhibitory effects of TRAF4 on the wound healing ability (Supplementary Fig. S5C and S5D). Likewise, BMP6 stimulation inhibited T24 cell migration (Supplementary Fig. S5E and S5F), while having minimal effects on proliferation (Supplementary Fig. S5G). Taken together, our results suggest that TRAF4 inhibits bladder cancer cell migration by promoting BMP/SMAD signaling.
Consistent with previous observations, overexpression of TRAF4 negatively affected NF-κB reporter activity (Supplementary Fig. S5H). As we observed that TRAF4 promotes BMP/SMAD signaling in bladder cancer cells, we next explored the possibility of cross-talk between BMP and NF-κB signaling and whether TRAF4 has a role therein. TNFα stimulation inhibited BMP/SMAD signaling, as determined by measuring BRE-luc transcriptional reporter activity (Fig. 6G). Interestingly, overexpression of TRAF4 mitigated the negative effect of TNFα on BMP/SMAD signaling. Similar effects were observed when T24 cells were stimulated with a combination of BMP and TNFα and the expression of ID genes was analyzed (Fig. 6H). Thus, the increased NF-κB signaling associated with a low TRAF4 level in mesenchymal cells may lead to an decrease in BMP/SMAD signaling. Thus, TRAF4 can promote BMP/SMAD indirectly by antagonizing the inhibitory effect of TNFα/NF-κB signaling on BMP/SMAD signaling.
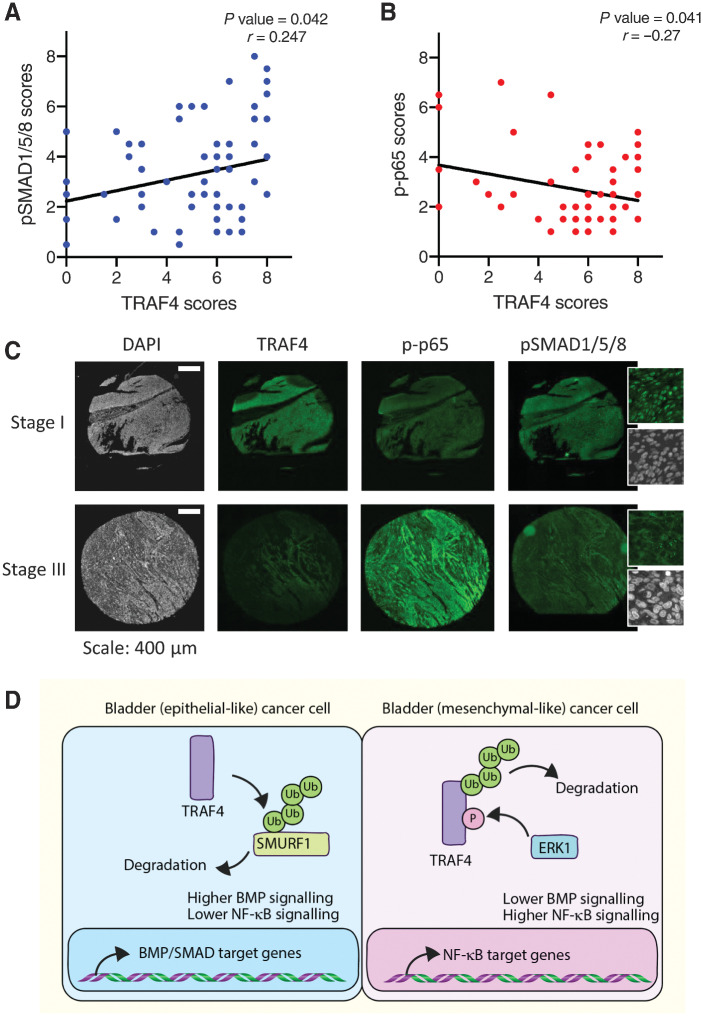
TRAF4 expression positively correlates with pSMAD1/5/8 levels and negatively correlates with the p-p65 level in bladder cancer patient samples
To further extend our observations described above regarding the differential regulatory effects of TRAF4 on BMP/SMAD and NF-κB signaling and whether these findings can be extended to biopsy material from bladder cancer patients, we performed IHC using anti-TRAF4 and phospho-specific antibodies on tissue microarray samples obtained from Biomax U.S. (BL802b). To determine the activation status of the BMP/SMAD and NF-κB signaling pathways, we used validated anti-pSMAD1/5/8 antibodies and anti–p-p65 antibodies, respectively. We observed significant positive and negative correlations between the TRAF4 and pSMAD1/5/8 levels (Fig. 7A and C; r = 0.247) and the TRAF4 and p-p65 levels (Fig. 7B and C; r = –0.27), respectively. Taken together, our findings confirmed that high TRAF4 expression is associated with low NF-κB activity and increased BMP activity and that low TRAF4 expression has the inverse associations in bladder cancer patient biopsies.
Figure 7.
TRAF4 expression correlates positively with pSMAD1/5/8 levels and negatively with the p-p65 level in bladder tumors. A, Regression analysis showing the correlations between the TRAF4 expression level (score) and phospho (p)SMAD1/5/8 scores in patients with bladder cancer. Pearson χ2 test was used to determine the correlations between the TRAF4 and pSMAD1/5/8 scores. B, Regression analysis showing the correlations between the TRAF4 expression level (score) and phospho (p)-p65 score in bladder cancer patients. Pearson χ2 test was used to determine the correlations between the TRAF4 and p-p65 scores. C, Representative images of continuous sections of tissue microarray samples probed with the indicated antibodies using fluorescent IHC. The magnified insets for pSMAD1/5/8 show nuclear staining. Scale bar: 400 μm. D, Schematic representation of TRAF4 signaling dynamics in epithelial-like and mesenchymal-like bladder cancer cells. Ub denotes ubiquitin and P stands for phosphorylation of serine 334.
Discussion
In this study, we investigated the role of TRAF4 during bladder cancer progression and in contrast to other cancer types, observed strong positive correlations between its expression and increased overall patient survival. Mining of publicly available data and our own IHC analysis of patient samples revealed that TRAF4 expression gradually decreases as bladder cancer progresses. Pathologically, stage 2 and 3 bladder tumors are more aggressive than stage 1 tumors and have muscle invasiveness, and consistent with these characteristics, we observed higher TRAF4 levels in stage 1 tumors. Furthermore, in both cultured cells and material from bladder cancer patients, we observed strong links between TRAF4 expression and increased BMP/SMAD and decreased NF-κB pathway signaling. Mechanistically, we showed that TRAF4 ubiquitinates and degrades SMURF1, a pro-EMT and oncogenic protein. TRAF4 directly and indirectly promotes BMP/SMAD signaling. Our results give credence to the claim that the interplay between TRAF4 and SMURF1 expression levels and activity functions as an important functional node in the interactions that enhance BMP/SMAD and NF-κB signaling cross-talk during bladder cancer progression (Fig. 7D).
EMT is a prominent event during bladder cancer metastasis, especially in bladder carcinoma, where (epithelial) cancer cells usually have to gain mesenchymal properties to penetrate through the bladder muscle wall. Mesenchymal markers FN1 and ITGAV have been well documented as potential biomarkers or targets for bladder carcinoma (39, 40). Interestingly, we found a strong correlation between the EMT status of bladder cancer cells and TRAF4 expression. Knocking down TRAF4 in the epithelial cell line RT4 led to loose attachment of cells to colonies, suggesting possible loss of epithelial tight junction components. Physiologically, bladder urothelial cells have high expression of TRAF4, which perhaps enables the bladder to maintain a strong barrier against leakage of stored urine. Changes in the expression of the EMT transcription factors SNAIL and SLUG were observed upon TRAF4 knockdown. For example, in RT4 cells, there was an increase in SLUG and a decrease in SNAIL, and vice versa in HT1376 cells. This mutually exclusive expression pattern was observed across 5 bladder cancer cell lines used in this study, suggesting a certain level of functional redundancy or compensation. Indeed, such reciprocal effects of SNAIL and SLUG expression have been previously documented (41).
We investigated why the steady-state TRAF4 level is lower in mesenchymal bladder cells than in epithelial bladder cancer cells. Epigenetic repression of genes is commonly seen during cancer progression and is mediated by DNA methylation enzymes that methylate certain regions in promoters to diminish their transcriptional activity (42). Treatment of mesenchymal bladder cell lines with 5-AZA, a compound that blocks DNA methylation, rescued TRAF4 expression. This suggests that TRAF4 is (directly or indirectly) epigenetically repressed. Our results also showed that the TRAF4 protein is less stable in more aggressive mesenchymal bladder cancer cells. We found that ERK mediated the phosphorylation of TRAF4 that is linked to a decrease in the steady-state TRAF4 protein level and TRAF4 stability. The presence of ERK and MAPK docking consensus sites suggests that TRAF4 is a direct ERK substrate. However, for formal proof of this additional biochemical experiments with purified proteins are needed. We therefore do not exclude the possibility that ERK activation may induce TRAF4 Ser334 phosphorylation in an indirect manner. Many of these aggressive (bladder) cancer cells have mutations in components of MAPK pathways, such as Raf or Ras, that increase the activity of downstream ERK signaling (10, 11).
In this study, we observed that TRAF4 affected the SMURF1 protein level in bladder cancer cells; TRAF4 maintained an appropriate SMURF1 level, and as the TRAF4 level decreased, the steady-state SMURF1 level increased. We previously reported that TRAF4 is able to ubiquitinate SMURF2, thereby potentiating the TGFβ signaling and promoting breast cancer metastasis (14). SMURF1 has high sequence similarity to SMURF2 and belongs to the same E3 ubiquitin ligase subfamily (HECT domain, NEDD4 subgroup). TRAF4 has been reported to promote BMP signaling neural crest development and neural plate morphogenesis through SMURF1 inhibition (13). BMP is a family member of TGFβ, which inhibits TGFβ-induced EMT and promotes mesenchymal-to-epithelial transition. In addition, there have been other important studies reflecting the dynamic interplay between TRAF4 and SMURF1 (13, 20). Our observations reveal that upregulation of SMURF1 due to reduced TRAF4 expression in later stages of bladder cancer progression could indeed potentially dampen the BMP signaling output.
Whereas TRAF4 knockdown in epithelial bladder cancer cells promotes migration and induces loss of epithelial integrity, ectopic expression of TRAF4 in mesenchymal cells inhibits migration and invasion. Consistent with these observations, we found that exogenous addition of BMP ligand or a SMURF1 inhibitor (A01) to mesenchymal cells inhibited their migration. The latter compound inhibits the degradation of the BMP signaling pathway components SMAD1 and SMAD5 by SMURF1. These data are in line with the hypothesis that TRAF4, by inhibiting SMURF1, potentiates BMP/SMAD signaling and thereby inhibits bladder cancer cell migration.
We performed unbiased transcriptomic and pathway analyses, which underlined our findings that TRAF4 promotes BMP signaling, and revealed that TRAF4 inhibits NF-κB signaling pathway activity in bladder cells. In contrast to other TRAF family members that mediate NF-κB signaling, TRAF4 has been shown to counteract other TRAF members and to antagonize NF-κB signaling. This is in line with our findings. NF-κB signaling was found to promote EMT and to play a role in bladder cancer progression (43). The correlation of TRAF4 with BMP and NF-κB signaling pathways were confirmed in material from patients with bladder cancer; Phosphorylated SMAD1/5 levels, indicative of active BMP receptor signaling, were found to be positively associated with high TRAF4 expression, while higher levels of phosphorylated NF-κB-p65 were associated with lower TRAF4 expression. This is consistent with our finding that TRAF4 targets SMURF1, for proteasomal degradation (44–46). The negative correlation of TRAF4 with NF-κB gene response signature and NF-κB-p-p65 in patient samples is in line with our in vitro findings. Moreover, we observed that TNFα, an upstream activator of the NF-κB pathway, can diminish the BMP signaling output, and that this effect that can be reduced by TRAF4. The link of SMURF1 and TRAF4-induced inhibition of NF-κB have been identified previously (47). Taken together, these and our studies demonstrate the intimate cross-talk between TRAF4 and SMURF1 in regulating BMP/SMAD and NK-κb signaling.
In summary, we identified TRAF4 expression level as a key determinant in the progression of bladder cancer. We uncovered that TRAF4 has a negative role in this process by enhancing BMP/SMAD and inhibiting NF-κB signaling. Low TRAF4 expression may be useful as a biomarker to detect aggressive types of bladder cancer. In future studies, it may be interesting to explore therapeutic potential of SMURF1 inhibitors or other BMP agonists in bladder cancer treatment.
Supplementary Material
Acknowledgments
P.V. Iyengar was supported by European Union's Horizon 2020 research and innovation programme under the Marie Skłodowska-Curie Individual Fellowship. P. ten Dijke and L. Ritsma are supported by Cancer Genomics Centre, Netherlands. P. ten Dijke is also supported by Dutch Cancer Society (KWF) grant (BUIT 2015-7526) and L. Ritsma is also supported by a Veni grant (NWO, 016.176.081) and a LUMC Gisela Thier grant. We are grateful to Slobodan Vukicevics (University of Zagreb) for providing BMP6. We thank Midory Thorikay and Gerard van der Zon for technical assistance and Martijn Rabelink for the shRNA constructs. The MBT-2 cell line was a kind gift from Simon Dovedi. RNA-seq was performed by BGI Tech (Hong Kong). Karien Wiesmeijer and Annelies Boonzaier-van der Laan helped with scanning the tissue microarray slides. We also thank Kees Fluiter for providing advice on the pathway studies.
The costs of publication of this article were defrayed in part by the payment of page charges. This article must therefore be hereby marked advertisement in accordance with 18 U.S.C. Section 1734 solely to indicate this fact.
This article is featured in Highlights of This Issue, p. 1469
Footnotes
Note: Supplementary data for this article are available at Molecular Cancer Research Online (http://mcr.aacrjournals.org/).
Authors' Disclosures
C.S. Verma reports grants from research funding from MSD via A*STAR, A*STAR and NMRC grants; and grants from BDT outside the submitted work; and is cofounder and nonexecutive director of two spinoffs - Sinopsee Therapeutics and Aplomex; there is no conflict with this work. P. ten Dijke reports grants from Centre for Cancer Genomics Netherlands; and grants from Dutch Cancer Society during the conduct of the study. No disclosures were reported by the other authors.
Authors' Contributions
P.V. Iyengar: Conceptualization, resources, data curation, formal analysis, supervision, funding acquisition, validation, investigation, visualization, methodology, writing–original draft, project administration, writing–review and editing. D.L. Marvin: Validation, investigation, writing–review and editing. D. Lama: Formal analysis, investigation, writing–review and editing. T.Z. Tan: Investigation, writing–review and editing. S. Suriyamurthy: Investigation, writing–review and editing. F. Xie: Investigation, writing–review and editing. M. van Dinther: Investigation, writing–review and editing. H. Mei: Formal analysis, investigation, writing–review and editing. C.S. Verma: Writing–review and editing. L. Zhang: Investigation, writing–review and editing. L. Ritsma: Formal analysis, investigation, writing–review and editing. P. ten Dijke: Conceptualization, supervision, writing–review and editing.
References
- 1. Siegel RL, Miller KD, Jemal A. Cancer statistics, 2020. CA Cancer J Clin 2020;70:7–30. [DOI] [PubMed] [Google Scholar]
- 2. Lim J, Thiery JP. Epithelial–mesenchymal transitions: insights from development. Development 2012;139:3471–86. [DOI] [PubMed] [Google Scholar]
- 3. Polyak K, Weinberg RA. Transitions between epithelial and mesenchymal states: acquisition of malignant and stem cell traits. Nat Rev Cancer 2009;9:265–73. [DOI] [PubMed] [Google Scholar]
- 4. Wheelock MJ, Shintani Y, Maeda M, Fukumoto Y, Johnson KR. Cadherin switching. J Cell Sci 2008;121:727–35. [DOI] [PubMed] [Google Scholar]
- 5. Nieto MA, Huang RY-J, Jackson RA, Thiery JP. EMT: 2016. Cell 2016;166:21–45. [DOI] [PubMed] [Google Scholar]
- 6. Yang J, Antin P, Berx G, Blanpain C, Brabletz T, Bronner M, et al. Guidelines and definitions for research on epithelial–mesenchymal transition. Nat Rev Mol Cell Biol 2020;21:341–52. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7. Hao Y, Baker D, Ten Dijke P. TGFβ-mediated epithelial–mesenchymal transition and cancer metastasis. Int J Mol Sci 2019;20:2767. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8. Glaser AP, Fantini D, Shilatifard A, Schaeffer EM, Meeks JJ. The evolving genomic landscape of urothelial carcinoma. Nat Rev Urol 2017;14:215–29. [DOI] [PubMed] [Google Scholar]
- 9. Bekele RT, Samant AS, Nassar AH, So J, Garcia EP, Curran CR, et al. RAF1 amplification drives a subset of bladder tumors and confers sensitivity to MAPK-directed therapeutics. J Clin Invest 2021;131:e147849. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10. Sim WJ, Iyengar PV, Lama D, Lui SKL, Ng HC, Haviv-Shapira L, et al. c-Met activation leads to the establishment of a TGFβ-receptor regulatory network in bladder cancer progression. Nat Commun 2019;10:4349. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11. Janda E, Lehmann K, Killisch I, Jechlinger M, Herzig M, Downward J, et al. Ras and TGF[beta] cooperatively regulate epithelial cell plasticity and metastasis: dissection of Ras signaling pathways. J Cell Biol 2002;156:299–313. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12. Park HH. Structure of TRAF family: current understanding of receptor recognition. Front Immunol 2018;9:1999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13. Kalkan T, Iwasaki Y, Park CY, Thomsen GH. Tumor necrosis factor receptor—associated factor 4 is a positive regulator of transforming growth factor-beta signaling that affects neural crest formation. Mol Biol Cell 2009;20:3436–50. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14. Zhang L, Zhou F, García De Vinuesa A, De Kruijf EM, Mesker WE, Hui L, et al. TRAF4 promotes TGF-beta receptor signaling and drives breast cancer metastasis. Mol Cell 2013;51:559–72. [DOI] [PubMed] [Google Scholar]
- 15. Singh R, Karri D, Shen H, Shao J, Dasgupta S, Huang S, et al. TRAF4-mediated ubiquitination of NGF receptor TrkA regulates prostate cancer metastasis. J Clin Invest 2018;128:3129–43. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16. Kim E, Kim W, Lee S, Chun J, Kang J, Park G, et al. TRAF4 promotes lung cancer aggressiveness by modulating tumor microenvironment in normal fibroblasts. Sci Rep 2017;7:8923. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17. Li W, Peng C, Lee M-H, Lim D, Zhu F, Fu Y, et al. TRAF4 is a critical molecule for Akt activation in lung cancer. Cancer Res 2013;73:6938–50. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18. Liu Y, Duan N, Duan S. MiR-29a inhibits glioma tumorigenesis through a negative feedback loop of TRAF4/Akt signaling. Biomed Res Int 2018;2018:2461363. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19. Yung Y, Yao Z, Aebersold DM, Hanoch T, Seger R. Altered regulation of ERK1b by MEK1 and PTP-SL and modified Elk1 phosphorylation by ERK1b are caused by abrogation of the regulatory C-terminal sequence of ERKs. J Biol Chem 2017;292:8852. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20. Wang X, Jin C, Tang Y, Tang LY, Zhang YE. Ubiquitination of tumor necrosis factor receptor—associated factor 4 (TRAF4) by Smad ubiquitination regulatory factor 1 (Smurf1) regulates motility of breast epithelial and cancer cells. J Biol Chem 2013;288:21784–92. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21. Irshad S, Bansal M, Guarnieri P, Davis H, Al Haj Zen A, Baran B, et al. Bone morphogenetic protein and Notch signaling cross-talk in poor-prognosis, mesenchymal-subtype colorectal cancer. J Pathol 2017;242:178–92. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22. Van De Stolpe A, Holtzer L, Van Ooijen H, Inda MAD, Verhaegh W. Enabling precision medicine by unravelling disease pathophysiology: quantifying signal transduction pathway activity across cell and tissue types. Sci Rep 2019;9:1603. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23. Jones G, Willett P, Glen RC, Leach AR, Taylor R. Development and validation of a genetic algorithm for flexible docking. J Mol Biol 1997;267:727–48. [DOI] [PubMed] [Google Scholar]
- 24. Robertson AG, Kim J, Al-Ahmadie H, Bellmunt J, Guo G, Cherniack AD, et al. Comprehensive molecular characterization of muscle-invasive bladder cancer. Cell 2017;171:540–56. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25. Uhlen M, Zhang C, Lee S, Sjöstedt E, Fagerberg L, Bidkhori G, et al. A pathology atlas of the human cancer transcriptome. Science 2017;357:eaan2507. [DOI] [PubMed] [Google Scholar]
- 26. Tan TZ, Rouanne M, Tan KT, Huang RY-J, Thiery JP. Molecular subtypes of urothelial bladder cancer: results from a meta-cohort analysis of 2411 tumors. Eur Urol 2019;75:423–32. [DOI] [PubMed] [Google Scholar]
- 27. Tan TZ, Miow QH, Miki Y, Noda T, Mori S, Huang RY-J, et al. Epithelial–mesenchymal transition spectrum quantification and its efficacy in deciphering survival and drug responses of cancer patients. EMBO Mol Med 2014;6:1279–93. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28. Earl J, Rico D, Carrillo-De-Santa-Pau E, Rodríguez-Santiago B, Méndez-Pertuz M, Auer H, et al. The UBC-40 Urothelial Bladder Cancer cell line index: a genomic resource for functional studies. BMC Genomics 2015;16:403. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29. Peti W, Page R. Molecular basis of MAP kinase regulation. Protein Sci 2013;22:1698. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30. Wu K, Zeng J, Zhou J, Fan J, Chen Y, Wang Z, et al. Slug contributes to cadherin switch and malignant progression in muscle-invasive bladder cancer development. Urol Oncol 2013;31:1751–60. [DOI] [PubMed] [Google Scholar]
- 31. Stadler M, Scherzer M, Walter S, Holzner S, Pudelko K, Riedl A, et al. Exclusion from spheroid formation identifies loss of essential cell–cell adhesion molecules in colon cancer cells. Sci Rep 2018;8:1151. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32. Smyrek I, Mathew B, Fischer SC, Lissek SM, Becker S, Stelzer EHK. E-cadherin, actin, microtubules and FAK dominate different spheroid formation phases and important elements of tissue integrity. Biol Open 2019;8:bio037051. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33. Wang HR. Degradation of RhoA by Smurf1 ubiquitin ligase. Methods Enzym 2006;406:437–47. [DOI] [PubMed] [Google Scholar]
- 34. Sahai E, Garcia-Medina R, Pouysségur J, Vial E. Smurf1 regulates tumor cell plasticity and motility through degradation of RhoA leading to localized inhibition of contractility. J Cell Biol 2007;176:35–42. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35. Cao Y, Wang C, Zhang X, Xing G, Lu K, Gu Y, et al. Selective small molecule compounds increase BMP-2 responsiveness by inhibiting Smurf1-mediated Smad1/5 degradation. Sci Rep 2014;4:4965. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36. Takeshita F, Ishii K J, Kobiyama K, Kojima Y, Coban C, Sasaki S, et al. TRAF4 acts as a silencer in TLR-mediated signaling through the association with TRAF6 and TRIF. Eur J Immunol 2005;35:2477–85. [DOI] [PubMed] [Google Scholar]
- 37. Hollnagel A, Oehlmann V, Heymer J, Rüther U, Nordheim A. Id genes are direct targets of bone morphogenetic protein induction in embryonic stem cells. J Biol Chem 1999;274:19838–45. [DOI] [PubMed] [Google Scholar]
- 38. Takase M, Imamura T, Sampath TK, Takeda K, Ichijo H, Miyazono K, et al. Induction of Smad6 mRNA by bone morphogenetic proteins. Biochem Biophys Res Commun 1998;244:26–29. [DOI] [PubMed] [Google Scholar]
- 39. Yang X, Huang H, Zeng Z, Zhao L, Hu P, He D, et al. Diagnostic value of bladder tumor fibronectin in patients with bladder tumor: a systematic review with meta-analysis. Clin Biochem 2013;46:1377–82. [DOI] [PubMed] [Google Scholar]
- 40. Van Der Horst G, Bos L, Van Der Mark M, Cheung H, Heckmann B, Clément-Lacroix P, et al. Targeting of alpha-v integrins reduces malignancy of bladder carcinoma. PLoS One 2014;9:e108464. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41. Nakamura R, Ishii H, Endo K, Hotta A, Fujii E, Miyazawa K, et al. Reciprocal expression of Slug and Snail in human oral cancer cells. PLoS One 2018;13:e0199442. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42. Suriyamurthy S, Baker D, Ten Dijke P, Iyengar PV. Epigenetic reprogramming of TGFβ signaling in breast cancer. Cancers 2019;11:726. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43. Wu Q, Zhou X, Li P, Ding M, You S, Xu Z, et al. ROC1 promotes the malignant progression of bladder cancer by regulating p-IκBα/NF-κB signaling. J Exp Clin Cancer Res 2021;40:1–13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44. Zhu H, Kavsak P, Abdollah S, Wrana JL, Thomsen GH. A SMAD ubiquitin ligase targets the BMP pathway and affects embryonic pattern formation. Nature 1999;400:687–93. [DOI] [PubMed] [Google Scholar]
- 45. Ying SX, Hussain ZJ, Zhang YE. Smurf1 facilitates myogenic differentiation and antagonizes the bone morphogenetic protein-2—induced osteoblast conversion by targeting Smad5 for degradation. J Biol Chem 2003;278:39029–36. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46. Murakami G, Watabe T, Takaoka K, Miyazono K, Imamura T. Cooperative inhibition of bone morphogenetic protein signaling by Smurf1 and inhibitory Smads. Mol Biol Cell 2003;14:2809–17. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47. Li S, Lu K, Wang J, An L, Yang G, Chen H, et al. Ubiquitin ligase Smurf1 targets TRAF family proteins for ubiquitination and degradation. Mol Cell Biochem 2010;338:11–17. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Data Availability Statement
The data generated in this study are available within the article and its Supplementary Data files. Publicly available expression data analyzed in this study, The Cancer Genome Atlas Urothelial Bladder Carcinoma (TCGA BLCA), was obtained from the Human Protein Atlas www.proteinatlas.org/about/download, version 21).