Abstract
Background: The PITX gene family, comprising PITX1, PITX2, and PITX3, is critical in organogenesis and has been evolutionary conserved in animals. PITX genes are associated with the advanced progression and poor prognosis of multiple cancers. However, the relationship between the PITX genes and head and neck squamous cell carcinoma (HNSC) has not been reported.
Methods: We used data from The Cancer Genome Atlas (TCGA) to analyze the association between PITX mRNA expression and clinicopathological parameters of patients with HNSC. The prognostic value of PITX genes was evaluated using the Kaplan-Meier plotter. Multivariate Cox analysis was used to screen out prognosis-associated genes to identify better prognostic indicators. The potential roles of PITX1 and PITX2 in HNSC prognosis were investigated using the protein-protein interaction (PPI) network, Gene Ontology (GO) analysis, and the Kyoto Encyclopedia of Genes and Genomes (KEGG) analysis. The correlation between PITX1 and PITX2 expression or methylation and immune cell infiltration was evaluated using the tumor-immune system interaction database (TISIDB). MethSurv was used to identify DNA methylation and its effect on HNSC prognosis.
Results: PITX genes expression was correlated with different cancers. PITX1 and PITX2 expression was lower in the patients with HNSC. In HNSC, PITX1 expression was significantly related to the clinical stage, histologic grade, and N stage, while PITX2 expression was only significantly related to the histologic grade. The high expression of PITX3 was significantly related to the histologic grade, T stage, and N stage. Survival analysis revealed that PITX genes had prognostic value in HNSC, which was supported by multivariate Cox analysis. PPI network and enrichment analysis showed that the genes interacting with PITX1 and PITX2 belonged predominantly to signaling pathways associated with DNA binding and transcription. Of the CpG DNA methylation sites in PITX1 and PITX2, 28 and 22 were related to the prognosis of HNSC, respectively. Additionally, PITX1 and PITX2 expression and methylation was associated with tumor-infiltrating lymphocytes (TILs).
Conclusion: The PITX genes were differentially expressed in patients with HNSC, highlighting their essential role in DNA methylation and tumor-infiltrating immune cell regulation, as well as overall prognostic value in HNSC.
Keywords: PITX gene family, HNSC, DNA Methylation, prognosis, Bioinformatic analysis
Introduction
Head and neck squamous cell carcinoma (HNSC) is the most common cancer of the head and neck. It originates from the squamous epithelium of the oral cavity, oropharynx, larynx, and hypopharynx (Solomon et al., 2018). It is the sixth most common cancer worldwide, with an incidence of ∼600,000 new cases each year, which is predicted to rise to 1.08 million new cases per year by 2030 (an increase of 30%) (Bray et al., 2018; Ferlay et al., 2019; Jiang et al., 2022). The early symptoms of HNSC are not obvious, meaning that the majority of patients are diagnosed at later stages of the disease, which is a serious threat to human health. At present, HNSC is primarily treated using resection, radiation, and systemic therapy; however, these strategies may lead to complications and poor long-term outcomes (Solomon et al., 2018). Accordingly, the 5-years survival rate for patients with HNSC remains less than 50%. Abnormal gene expression may be involved in tumorigenesis and is associated with the prognosis of HNSC patients (Zhang and Gao, 2021). In the past decade, elucidation of the molecular genetic landscape of HNSC has revealed new opportunities for therapeutic intervention, with a particular focus on immunotherapy. For instance, the immune checkpoint inhibitor pembrolizumab has been trialed as a first-line systemic treatment of HNSC (Saâda-Bouzid et al., 2017). Besides, detailed evaluation of the molecular characteristics of HNSC and immune profiling suggest that the inclusion of prognostic and predictive biomarkers in clinical studies may overcome the obstacles of targeted therapy and prolong the survival time of patients with HNSC (Oliva et al., 2019; Johnson et al., 2020). Hence, effective prognostic indicators for HNSC are urgently needed.
The PITX gene family belongs to the group of homeobox genes, which are highly conserved in all animals through evolution. To date, three PITX paralogs have been identified in mammalian cells: PITX1, PITX2, and PITX3 (Tran and Kioussi, 2021). Due to the early expression of PITX genes during embryonic development, their role has been mainly studied in the context of tissue and organ formation (Angotzi et al., 2008). PITX1 plays a role in pituitary development and hindlimb tissue configuration (Lanctôt et al., 1999). Meanwhile, PITX2 regulates the development of the pituitary, the face, teeth, skeletal muscle, the heart, and the intestine (Shih et al., 2007). Finally, PITX3 monitors dopaminergic neuronal function in the substantia nigra and during lens development. Recently, the PITX genes were shown to be involved in tumorigenesis. The expression of PITX activators is deregulated in some human malignancies, including hepatocellular carcinoma (Tai et al., 2016), lung cancer (Chen et al., 2007), prostate cancer (Poos et al., 2022), and cutaneous malignant melanoma (Osaki et al., 2013). PITX1 has been reported as a candidate tumor suppressor gene and a possible biomarker for predicting the chemical sensitivity of HNSC in humans (Takenobu et al., 2016). A recent study of PITX2 and an adjacent long non-coding RNA (lncRNA) methylation site, showed that hypermethylation was associated with improved survival in an HNSC cohort (Sailer et al., 2016). Moreover, PITX3 DNA methylation has proved to be an independent prognostic biomarker for overall survival (OS) in patients with HNSC and could potentially assist with risk-group assignment and subsequent treatment stratification (Sailer et al., 2017b). However, the role of PITX gene expression and methylation in the infiltration of immune cells into the tumor and the prognosis of HNSC has not been described.
Here, we used a bioinformatics approach to elucidate the relationship between PITX genes and the oncologic characteristics of patients with HNSC as well as assess their ability to predict the prognosis of HNSC. We used The Cancer Genome Atlas (TCGA) to analyze the expression of different PITX transcription factors in patients with HNSC, in order to determine their expression pattern, potential mechanism of action, DNA methylation status, and their relationship with immune infiltration and the prognosis of HNSC.
Materials and methods
Raw data acquisition and processing
TCGA has profiled and analyzed a large collection of clinical and molecular data from over 10,000 tumor patients across 33 different tumor types (Weinstein et al., 2013; Colaprico et al., 2016). Transcriptome RNA sequencing (RNA-seq) data were extracted from TCGA (https://portal.gdc.cancer.gov/) for 33 cancers, which included: adrenocortical carcinoma (ACC); bladder urothelial carcinoma (BLCA); breast invasive carcinoma (BRCA); cervical squamous cell carcinoma and endocervical adenocarcinoma (CESC); cholangiocarcinoma (CHOL); colon adenocarcinoma (COAD); lymphoid neoplasm diffuse large B-cell lymphoma (DLBC); esophageal carcinoma (ESCA); glioblastoma multiforme (GBM); head and neck squamous cell carcinoma (HNSC); kidney chromophobe (KICH); kidney renal clear cell carcinoma (KIRC); kidney renal papillary cell carcinoma (KIRP); acute myeloid leukemia (LAML); brain lower grade glioma (LGG); liver hepatocellular carcinoma (LIHC); lung adenocarcinoma (LUAD); lung squamous cell carcinoma (LUSC); mesothelioma (MESO); ovarian serous cystadenocarcinoma (OV); pancreatic adenocarcinoma (PAAD); pheochromocytoma and paraganglioma (PCPG); prostate adenocarcinoma (PRAD); rectum adenocarcinoma (READ); sarcoma (SARC); skin cutaneous melanoma (SKCM); stomach adenocarcinoma (STAD); testicular germ cell tumors (TGCT); thyroid carcinoma (THCA); thymoma (THYM); uterine corpus endometrial carcinoma (UCEC); uterine carcinosarcoma (UCS) and uveal melanoma (UVM). RNA-seq data from 502 HNSC tissues and 44 normal tissues were downloaded from TCGA. The RNA-seq data and the corresponding patient clinical information (Workflow Type: HTSeq-FPKM) were acquired using the UCSC Xena tool (https://xenabrowser.net/datapages/). The level 3 HTSeq-FPKM data were transformed to log2 for the following analysis.
Survival analysis and PITX genes expression
The Kaplan-Meier plotter (http://kmplot.com/analysis/index.php?p=service) is an online drawing tool used to evaluate the influence of 54,685 genes on survival by mining information from 10,471 samples. In this study, we used the Kaplan-Meier plotter to determine the prognostic value of PITX gene expression levels in predicting OS and relapse free survival (RFS) of patients with HNSC. A difference was considered statistically significant when the p-value was < 0.05. Univariate Cox proportional hazard regression analysis was conducted to screen the differentially expressed PITX genes significantly associated with OS of HNSC. Then, the differentially expressed PITX genes with a p-value of less than 0.1 were further identified by multivariate Cox proportional hazard regression. Genes with a p-value of less than 0.05 (according to the multivariate Cox proportional hazard regression analysis) were identified as prognosis-related genes.
Analysis of the biological function of PITX genes
GeneMANIA is a flexible web interface used to construct protein-protein interaction (PPI) networks, generate hypotheses on gene function, and explore gene lists, while prioritizing genes (Warde-Farley et al., 2010). In this study, we used GeneMANIA to visualize the gene network and predict the functions of the co-expressed PITX1 and PITX2 genes. Gene Ontology (GO) enrichment and Kyoto Encyclopedia of Genes and Genomes (KEGG) pathway analysis of co-expressed genes were performed using the “ClusterProfiler” package (V 3.14.3) and visualized using the “ggplot2” package (V 3.3.3) (Xu et al., 2012).
DNA methylation of prognosis-associated PITX1 and PITX2 genes
The MethSurv database (https://biit.cs.ut.ee/methsurv/) was used to analyze the DNA methylation sites of PITX1 and PITX2 in TCGA. Moreover, the prognostic value of PITX1 and PITX2 CpG methylation sites was evaluated using OS as the survival outcome measure.
Tumor-immune estimation resource (TIMER) database analysis
TIMER 2.0 (http://timer.comp-genomics.org/) is a web server used for the analysis of infiltrating immune cells and their clinical impact (Li et al., 2020). In this study, the “gene” module was used to evaluate the relationship between PITX gene expression and immune cell infiltration. We chose PITX1 or PITX2 as input, while B cells, CD8+ T cells, CD4+ T cells, neutrophils, macrophages, and dendritic cells (DCs), were selected as the immune cell types (Li et al., 2016).
Tumor-immune system interaction database (TISIDB) analysis
TISIDB (http://cis.Hku.hk/TISIDB/) is an online web integrated repository portal for tumor-immune system interactions (Ru et al., 2019). In this study, we used the TISIDB to determine the relationships between the abundance of tumor-infiltrating lymphocytes (TILs) and the expression or methylation of PITX1 and PITX2 across human cancers. Based on the gene expression profile, the relative abundance of TILs was inferred using gene set variation analysis. The correlations between the expression and methylation of PITX1 and PITX2 genes and TILs were measured using Spearman’s test.
Statistical analysis
All statistical analysis were performed using R software (V 4.2.0) (https://www.r-project.org/) and the R package ggplot2 was used to visualize gene expression differences. The Wilcoxon rank-sum test was used to determine the differences between tumor tissues and adjacent normal tissues. The associations between PITX gene expression and clinicopathological parameters were measured using the Student’s t-test.
Results
The expression of PITX gene family members in different cancers
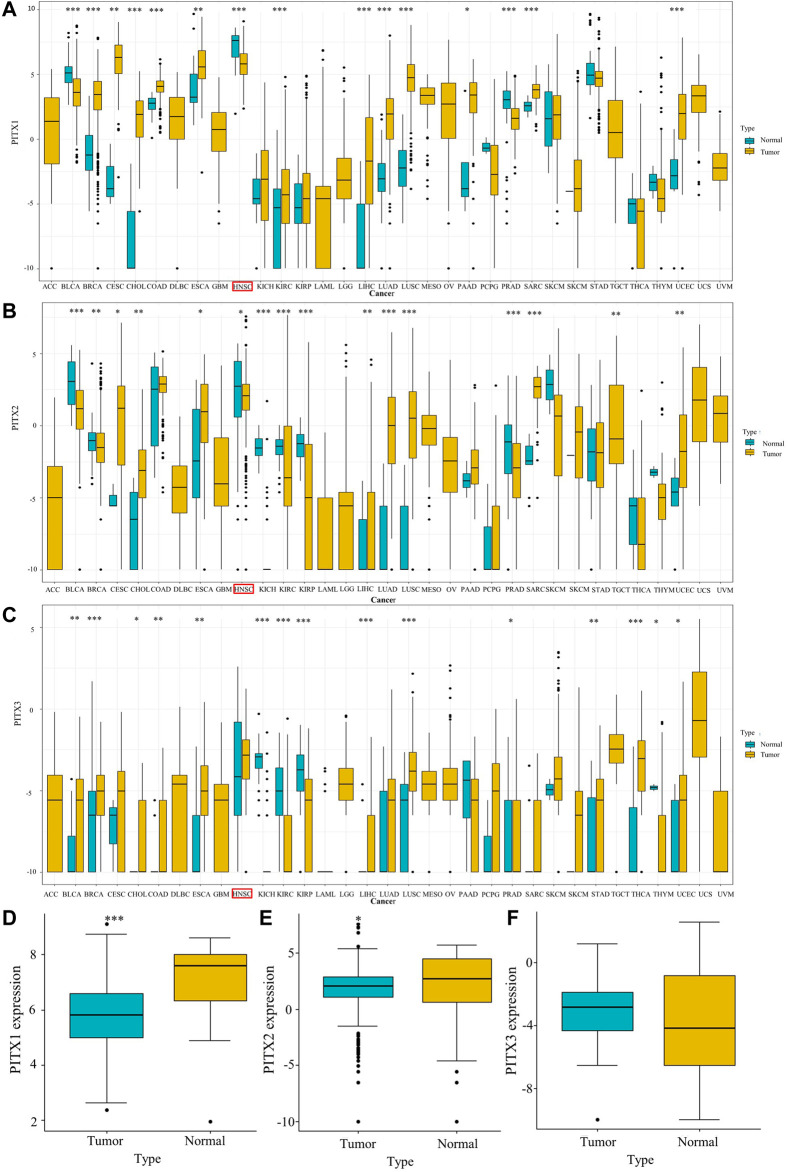
To understand whether the expression of PITX genes correlates with cancer, we evaluated PITX1, PITX2, and PITX3 mRNA expression in different human tumors (33 cancer types) and adjacent normal tissues using TCGA data. The results showed that PITX1 mRNA expression was associated with BLCA, BRCA, CESC, CHOL, COAD, ESCA, HNSC, KIRC, LIHC, LUAD, LUSC, PAAD, PRAD, READ, and UCEC (Figure 1A). The expression of PITX2 was linked to BLCA, BRCA, CESC, CHOL, ESCA, HNSC, KICH, KIRC, KIRP, LIHC, LUAD, LUSC, PRAD, READ, THCA, and UCEC (Figure 1B). The expression of PITX3 was related to BLCA, BRCA, CHOL, COAD, ESCA, KICH, KIRC, KIRP, LIHC, LUSC, PRAD, STAD, THCA, THYM, and UCEC (Figure 1C). Low expression of PITX1 (p < 0.001, Figure 1D) and PITX2 (p < 0.05, Figure 1E) was observed in HNSC tumor tissues compared with normal tissues in unpaired specimens. Meanwhile, the expression of PITX3 was higher in HNSC than in normal samples, but this difference was not statistically significant (p > 0.05, Figure 1F).
FIGURE 1.
Expression of PITX genes in HNSC and normal tissue. (A) PITX1, (B) PITX2, and (C) PITX3 expression levels in different tumor types from TCGA. (D) PITX1, (E) PITX2, and (F) PITX3 expression difference in unpaired HNSC samples. p ≥ 0.05; *, p < 0.05; **, p < 0.01; ***, p < 0.001. ACC, adrenocortical carcinoma; BLCA, bladder urothelial carcinoma; BRCA, breast invasive carcinoma; CESC, cervical squamous cell carcinoma and endocervical adenocarcinoma; CHOL, cholangiocarcinoma; COAD, colon adenocarcinoma; DLBC, lymphoid neoplasm diffuse large B-cell lymphoma; ESCA, esophageal carcinoma; GBM, glioblastoma multiforme; HNSC, head and neck squamous cell carcinoma; KICH, kidney chromophobe; KIRC, kidney renal clear cell carcinoma; KIRP, kidney renal papillary cell carcinoma; LAML, acute myeloid leukemia; LGG, brain lower grade glioma; LIHC, liver hepatocellular carcinoma; LUAD, lung adenocarcinoma; LUSC, lung squamous cell carcinoma; MESO, mesothelioma; OV, ovarian serous cystadenocarcinoma; PAAD, pancreatic adenocarcinoma; PCPG, pheochromocytoma and paraganglioma; PRAD, prostate adenocarcinoma; READ, rectum adenocarcinoma; SARC, sarcoma; SKCM, skin cutaneous melanoma; STAD, stomach adenocarcinoma; TGCT, testicular germ cell tumors; THCA, thyroid carcinoma; THYM, thymoma; UCEC, uterine corpus endometrial carcinoma; UCS, uterine carcinosarcoma; UVM, uveal melanoma.
Expression of PITX genes and clinicopathological characteristics of HNSC patients
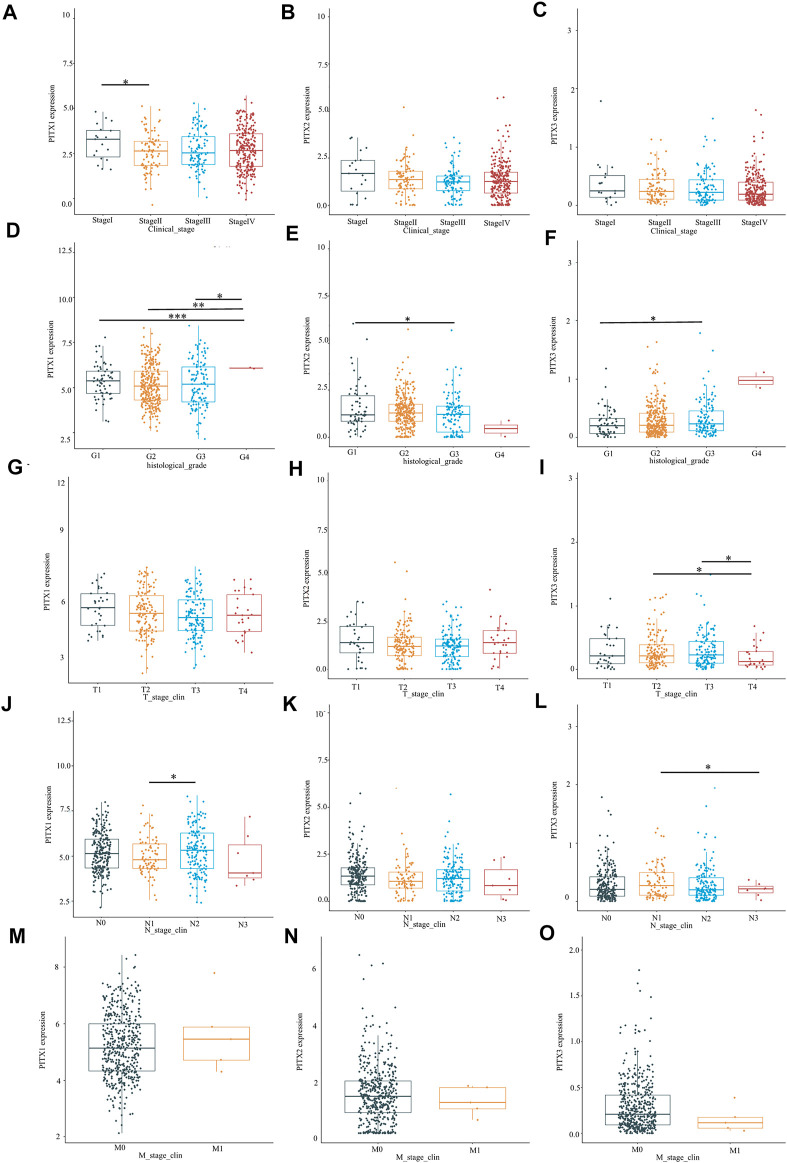
The clinicopathological characteristics of 502 patients with HNSC are summarized in Table 1. Roughly half of the HNSC patients were over 60 years old (51.1%) and had stage IV cancer (55.7%). The PITX1 expression was significantly related to the clinical stage (stage I vs. stage II, p < 0.05, Figure 2A), histologic grade (G1 vs. G4, G2 vs. G4, G3 vs. G4, p < 0.05, Figure 2D), and N stage (N1 vs. N2, p < 0.05, Figure 2J) of HNSC. However, no relationship was detected between PITX1 expression and HNSC T stage (p > 0.05, Figure 2G) or M stage (p > 0.05, Figure 2M). The PITX2 expression was only significantly related to the histologic grade of HNSC (G1 vs. G3, p < 0.05, Figure 2E). No correlations were detected between PITX2 expression and HNSC clinical stage (p > 0.05, Figure 2B), T stage (p > 0.05, Figure 2H), N stage (p > 0.05, Figure 2K), or M stage (p > 0.05, Figure 2N). The PITX3 mRNA expression was significantly related to the histologic grade (G1 vs. G3, p < 0.05, Figure 2F), T stage (T2 vs. T4, T3 vs. T4, p < 0.05, Figure 2I), and N stage (N1 vs. N3, p < 0.05, Figure 2L) of HNSC. Meanwhile, no correlations were detected between PITX3 expression and the HNSC clinical stage (p > 0.05, Figure 2C) or M stage (p > 0.05, Figure 2O).
TABLE 1.
Clinical characteristics of the HNSC patients (from TCGA).
| Characteristic | Levels | Overall |
|---|---|---|
| N | 502 | |
| T stage, n (%) | T1 | 33 (6.8%) |
| T2 | 144 (29.6%) | |
| T3 | 131 (26.9%) | |
| T4 | 179 (36.8%) | |
| N stage, n (%) | N0 | 239 (49.8%) |
| N1 | 80 (16.7%) | |
| N2 | 154 (32.1%) | |
| N3 | 7 (1.5%) | |
| M stage, n (%) | M0 | 472 (99%) |
| M1 | 5 (1%) | |
| Clinical stage, n (%) | Stage I | 19 (3.9%) |
| Stage II | 95 (19.5%) | |
| Stage III | 102 (20.9%) | |
| Stage IV | 272 (55.7%) | |
| Gender, n (%) | Female | 134 (26.7%) |
| Male | 368 (73.3%) | |
| Race, n (%) | Asian | 10 (2.1%) |
| Black or African American | 47 (9.7%) | |
| White | 428 (88.2%) | |
| Age, n (%) | <=60 | 245 (48.9%) |
| >60 | 256 (51.1%) | |
| Histologic grade, n (%) | G1 | 62 (12.8%) |
| G2 | 300 (62.1%) | |
| G3 | 119 (24.6%) | |
| G4 | 2 (0.4%) | |
| Smoker, n (%) | No | 111 (22.6%) |
| Yes | 381 (77.4%) | |
| Lymphovascular invasion, n (%) | No | 219 (64.2%) |
| Yes | 122 (35.8%) | |
| Lymph node neck dissection, n (%) | No | 90 (18%) |
| Yes | 409 (82%) | |
| OS event, n (%) | Alive | 284 (56.6%) |
| Dead | 218 (43.4%) | |
| DSS event, n (%) | Alive | 347 (72.7%) |
| Dead | 130 (27.3%) | |
| PFI event, n (%) | Alive | 308 (61.4%) |
| Dead | 194 (38.6%) |
DSS, disease-specific survival; PFI, progression-free interval.
FIGURE 2.
Relationship between PITX genes and clinicopathological parameters in patients with HNSC. The expression of PITX1, PITX2, and PITX3 markedly correlated with (A–C) clinical stage, (D–F) histologic grade, (G–I) T stage, (J–L) N stage, and (M–O) M stage. p ≥ 0.05; *, p < 0.05; **, p < 0.01; ***, p < 0.001.
Prognostic value of PITX mRNA expression in HNSC patients
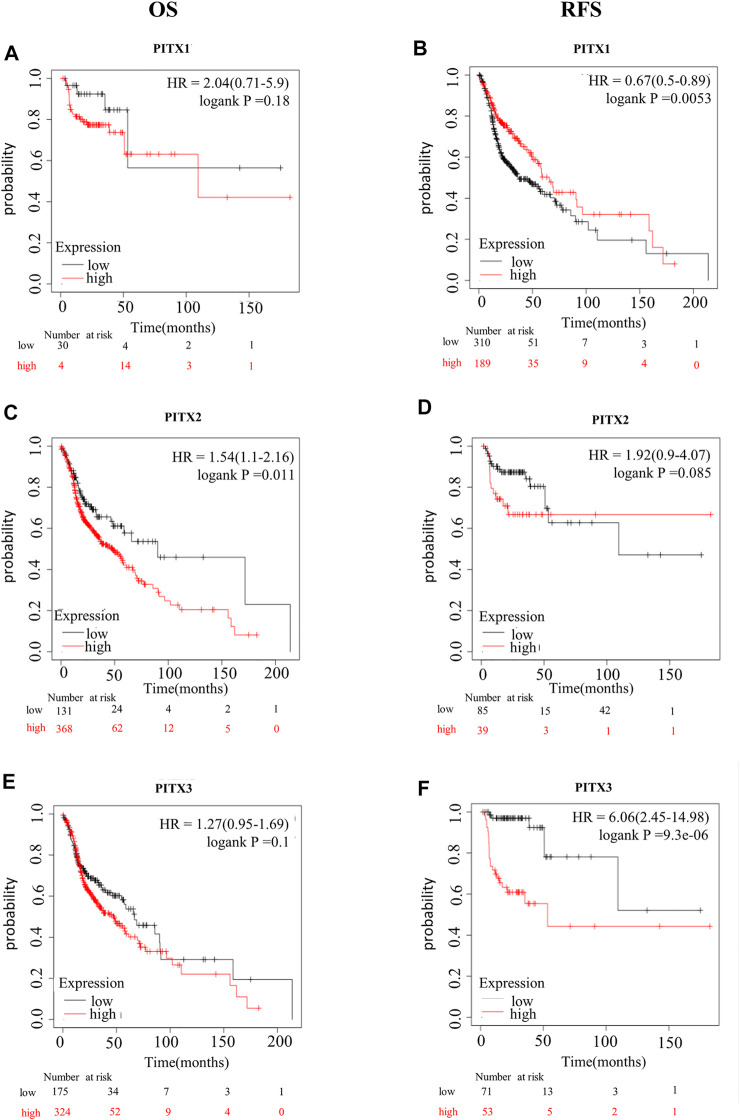
Next, we assessed the prognostic value of differentially expressed PITX genes in the context of HNSC. The correlations between different PITX genes and clinical outcomes were analyzed using the Kaplan-Meier plotter (Figure 3). We found that mRNA levels of PITX1 were significantly correlated with RFS but not with OS in all HNSC patients. HNSC patients with higher PITX1 mRNA levels PITX1 had better RFS [hazard ratio (HR) = 0.67 (0.5–0.89), p = 0.0053]. PITX2 expression was, however, significantly correlated with OS [HR = 1.54 (1.1–2.16), p = 0.011] but not with RFS. The expression of PITX3 was associated with RFS but not with OS. In contrast to PITX1, higher PITX3 mRNA levels were associated with poorer RFS in patients with HNSC [HR = 6.06 (2.45–14.98), p = 9.3e−06].
FIGURE 3.
Kaplan-Meier survival curves for HNSC prognostic PITX genes. Kaplan-Meier survival curves for PITX1 showing (A) OS and (B) RFS; Kaplan-Meier survival curves for PITX2 showing (C) OS and (D) RFS; Kaplan-Meier survival curves for PITX3 showing (E) OS and (F) RFS.
Next, univariate Cox regression analysis revealed that PITX1 and PITX2 co-expression was significantly correlated with OS. Subsequently, multivariate Cox regression analysis indicated that PITX1 (HR = 0.832, p < 0.05) and PITX2 (HR = 1.149, p < 0.05) exhibited independent prognostic values for HNSC (Table 2).
TABLE 2.
Univariate and multivariate Cox analysis of PITX gene expression for determining HNSC prognosis.
| Gene name | Total (N) | Univariate analysis | Multivariate analysis | ||
|---|---|---|---|---|---|
| Hazard ratio (95% CI) | p-value | Hazard ratio (95% CI) | p-value | ||
| PITX1 | 501 | 0.846 (0.750–0.954) | 0.006 | 0.832 (0.736–0.940) | 0.003 |
| PITX2 | 501 | 1.118 (0.989–1.263) | 0.073 | 1.149 (1.012–1.304) | 0.032 |
| PITX3 | 501 | 1.216 (0.753–1.962) | 0.424 | ||
Bold value means statistically significant.
PITX1 and PITX2 PPI network and enrichment analysis
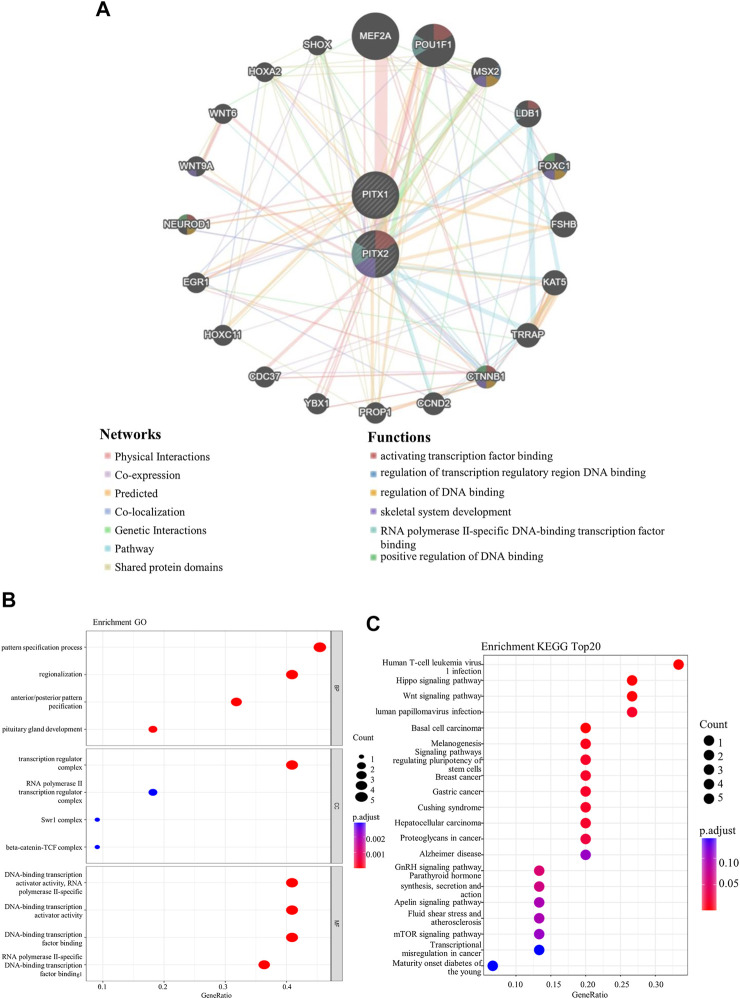
Using GeneMANIA, we conducted a PPI network analysis of PITX1 and PITX2 to explore the potential interactions. We found that the PITX genes interacted with genes involved in the binding of activating transcription factors, the regulation of transcription regulatory region DNA binding, the regulation of DNA binding, skeletal system development, RNA polymerase II-specific DNA-binding transcription factor binding, and positive regulation of DNA binding (Figure 4A).
FIGURE 4.
The network and functional enrichment analysis of PITX1 and PITX2. (A) The protein-protein interaction (PPI) network for PITX1 and PITX2 (generated using GeneMANIA). The functions of PITX1, PITX2, and co-expressed genes were predicted by performing (B) GO and (C) KEGG analysis.
The functions of PITX1, PITX2, and the associated co-expressed genes were predicted by performing GO and KEGG analysis. GO enrichment analysis predicts the functions of target host genes based on biological processes, cell components, and molecular functions. We found that the pattern specification process, regionalization, anterior/posterior pattern specification, transcription regulator complex, DNA-binding transcription activator activity RNA polymerase II-specific, DNA-binding transcription activator activity, DNA-binding transcription factor binding, and RNA polymerase II-specific DNA-binding transcription factor binding were markedly associated with PITX1 and PITX2 alterations in HNSC (Figure 4B and Supplementary Table S1). The corresponding genes are known to be associated with DNA-binding transcription.
KEGG analysis can define the pathways related to the functions of altered PITX1 and PITX2 genes, and the associated co-expressed genes. Twelve pathways related to the functions of PITX1 and PITX2 alterations in HNSC were found (Figure 4C and Supplementary Table S2), including Human T-cell leukemia virus1 infection, Hippo signaling pathway, Wnt signaling pathway, Human papillomavirus infection, and so on.
DNA methylation in promoter regions of PITX1 and PITX2
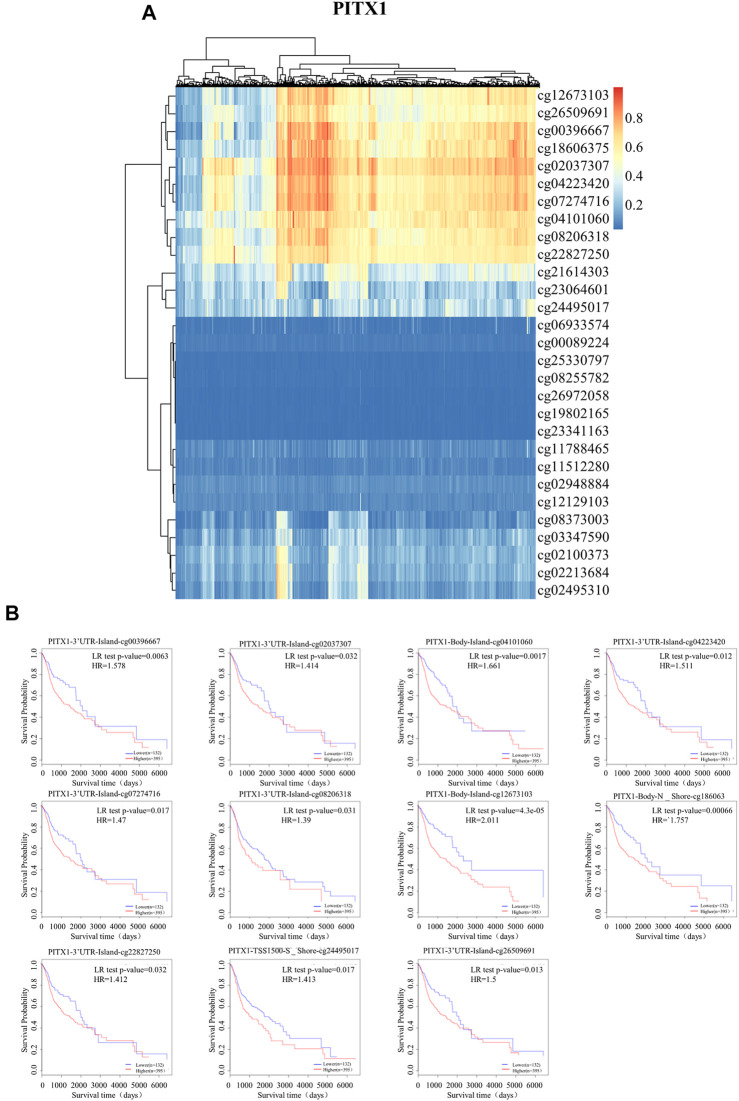
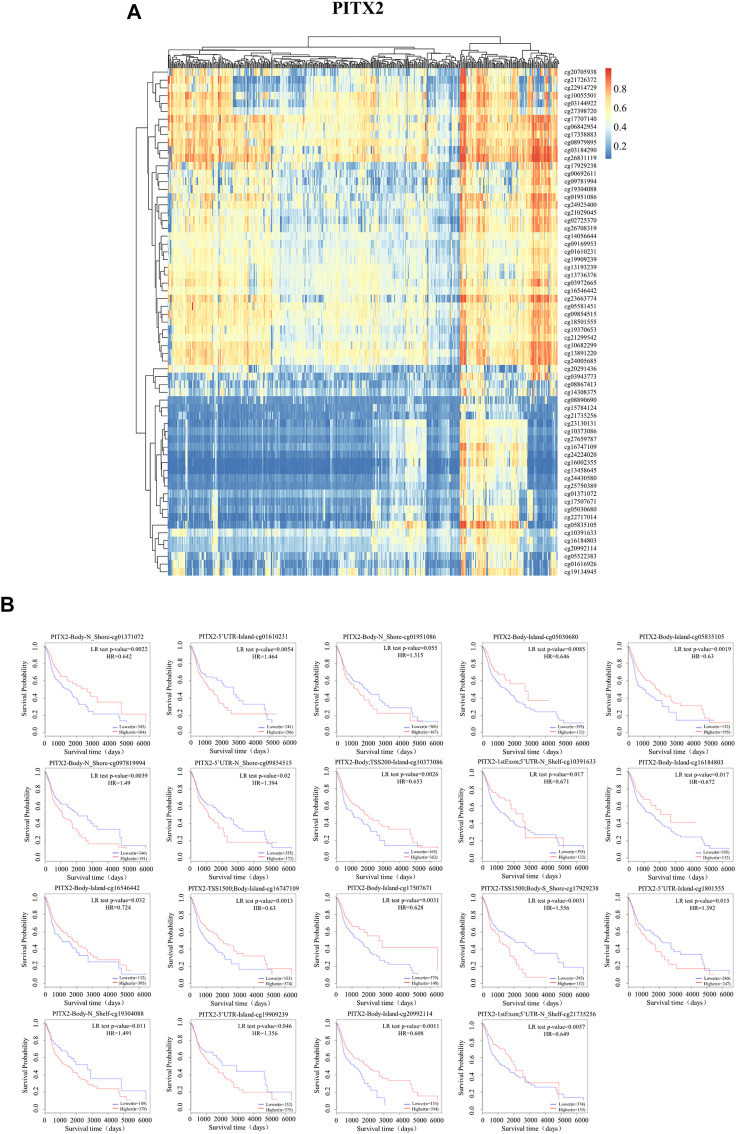
The results of GO and KEGG analysis showed that PITX1, PITX2, and the associated genes were mainly enriched for DNA transcription-related functions. Therefore, the promoter methylation level of PITX1 (Figure 5) and PITX2 (Figure 6) were analyzed by MethSurv; the prognostic value of methylation at each CpG site was assessed individually. We found that cg02037307 of PITX1 had the highest level of DNA methylation (Figure 5A). In addition, among the 29 CpGs in the PITX1 promoter region, 11 had significant prognostic value for patients with HNSC, based on Kaplan-Meier survival analysis (Figure 5B); data for the remaining 18 CpGs are shown in Supplementary Figure S1. PITX2 has 65 CpGs in the promoter region, of which cg26831119 had the highest level of methylation (Figure 6A). Nineteen of these CpGs had a significant prognostic value for HNSC, according to the Kaplan-Meier survival analysis (Figure 6B); data for the remaining 46 CpGs are shown in Supplementary Figure S2.
FIGURE 5.
DNA methylation of PITX1 and HNSC prognosis. (A) The DNA methylation of PITX1 was determined using MethSurv; (B) The prognostic values of CpGs in PITX1, based on Kaplan-Meier survival analysis. Red to blue: high expression to low expression.
FIGURE 6.
DNA methylation of PITX2. (A) The DNA methylation of PITX2 was determined using MethSurv; (B) The prognostic values of CpGs in PITX2, based on Kaplan-Meier survival analysis. Red to blue: high expression to low expression.
The expression and methylation levels of PITX1 and PITX2 correlate with immune infiltration in HNSC
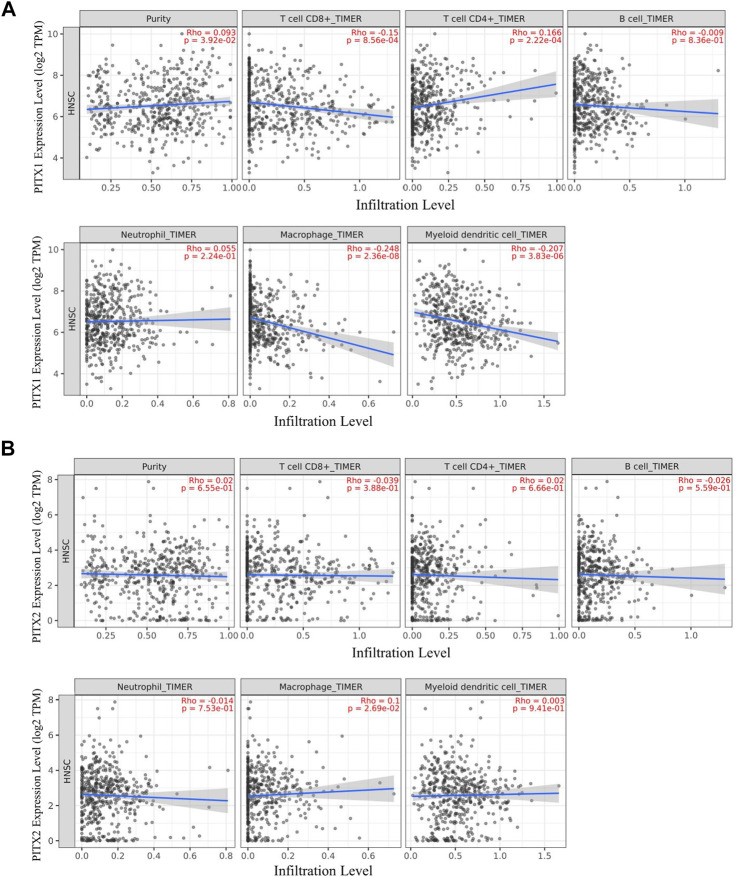
TIMER 2.0 is a tool for analyzing the associated between specific genes and immune infiltration. We used TIMER 2.0 to investigate the correlation between the expression of PITX1 and PITX2 and the infiltration levels of immune cells in HNSC. The expression of PITX1 was significantly correlation with tumor-infiltrating CD8+ T cells (Rho = −0.15, p = 8.56e−04), CD4+ T cells (Rho = 0.166, p = 2.22e−04), macrophages (Rho = −0.248, p = 2.36e−08), and DCs (Rho = −0.207, p = 3.83e−06) (Figure 7A). Additionally, a correlation between PITX2 expression and macrophage infiltrations (Rho = 0.1, p = 2.69e−02) was observed (Figure 7B).
FIGURE 7.
TIMER analysis of correlations between PITX1 and PITX2 expression and immune cell infiltration in HNSC. (A) Correlations between PITX1 expression and immune cell infiltration were evaluated using TIMER. (B) Correlation between PITX2 expression and immune cell infiltration levels using TIMER.
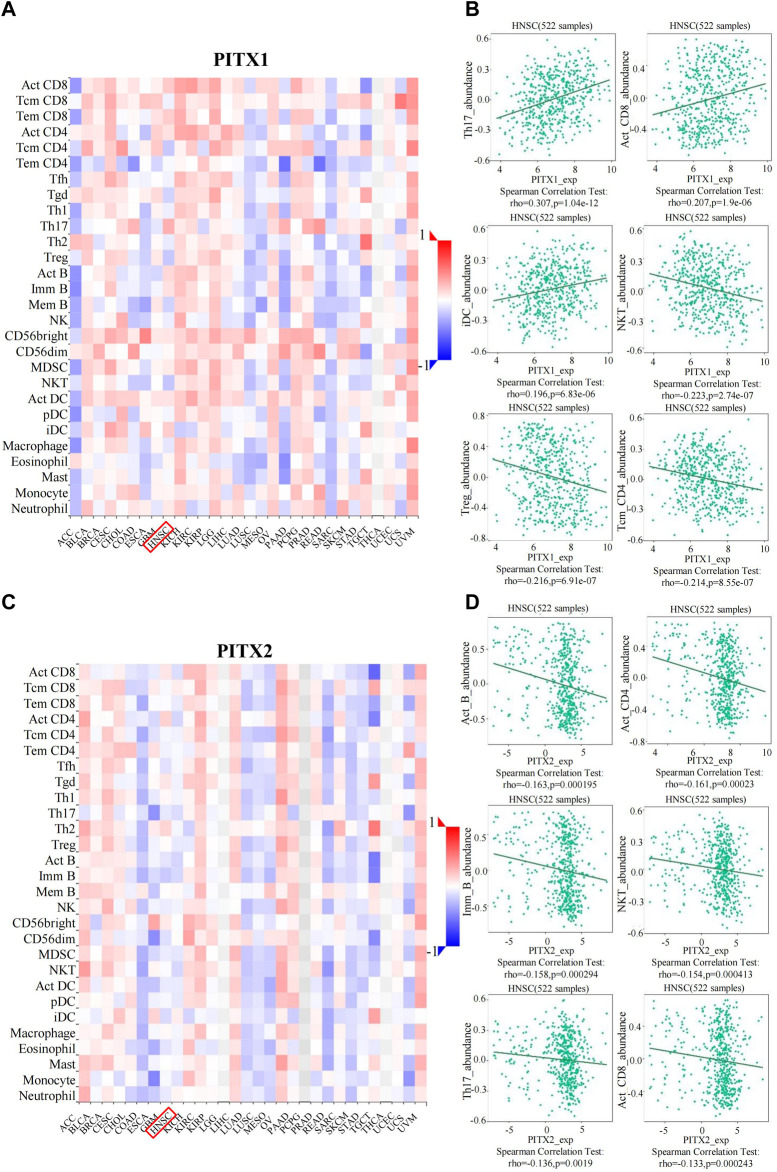
TILs can independently be used to predict sentinel lymph node positivity and cancer prognosis (Warde-Farley et al., 2010). Therefore, we used TISIDB to further explore the relationship between TIL numbers and the expression or methylation of PITX1 and PITX2. We found that the expression of PITX1 was associated with 16 immune cell subtypes in HNSC (Figure 8A and Table 3). The strongest associations with PITX1 expression involved T helper (Th)17 cells (r = 0.307, p = 1.04E−12), activated CD8+ T cells (Act_CD8, r = 0.207, p = 1.9E−06), immature (i) DCs (r = 0.196, p = 6.83E-06), natural killer T (NKT) cells (r = −0.223, p = 2.74E−07), regulatory T cells (Tregs) (r = −0.216, p = 6.91E−07), and central memory CD4+ T cells (Tcm_CD4, r = −0.214, p = 8.55E−07) (Figure 8B). The expression of PITX2 was associated with nine immune cell subtypes in HNSC (Figure 8C and Table 3). Among these, activated B cells (Act_B, r = −0.163, p = 0.000195), activated CD4+ T cells (Act_CD4, r = −0.161, p = 0.00023), immature B cells (Imm_B, r = −0.158, p = 0.000294), NKT cells (r = −0.154, p = 0.000413), Th17 cells (r = −0.136, p = 0.0019), and Act_CD8 (r = −0.133, p = 0.00243) displayed the strongest correlations with PITX2 expression (Figure 8D).
FIGURE 8.
TISIDB analysis of correlations between PITX1 and PITX2 expression and immune cell infiltration across human cancers. (A) The relationship between PITX1 expression and 28 types of TILs across various human cancers. (B) The top six TILs displaying the strongest Spearman’s correlation with PITX1 expression in HNSC. (C) The relationship between PITX2 expression and 28 types of TILs across various human cancers. (D) The top six TILs displaying the strongest Spearman’s correlation with PITX2 expression in HNSC. Act_CD8, activated CD8+ T cells; Tcm_CD8, central memory CD8+ T cells; Tem_CD8, effector memory CD8+ T cells; Act_CD4, activated CD4+ T cells; Tcm_CD4, central memory CD4+ T cells; Tem_CD4, effector memory CD4+ T cells; Tfh, T follicular helper cells; Tgd, gamma delta T cells; Th1, type 1 T helper cells; Th17, type 17 T helper cells; Th2, type 2 T helper cells; Treg, regulatory T cells; Act B, activated B cells; Imm_B, immature B cells; Mem_B, memory B cells; NK, natural killer cells; CD56bright, CD56bright NK cells; CD56dim, CD56dim NK cells; MDSC, myeloid derived suppressor cells; NKT, natural killer T cells; Act DC, activated dendritic cells; pDC, plasmacytoid DCs; iDC, immature DCs; Mast, mast cell.
TABLE 3.
The expression and methylation levels of PITX1 and PITX2 correlate with ITLs.
| PITX1 expression | PITX2 expression | PITX1 methylation | PITX2 methylation | |||||
|---|---|---|---|---|---|---|---|---|
| r | p | r | p | r | p | r | p | |
| Activated CD8+ T cell (act_CD8) | 0.207 | 1.9e−06 | −0.133 | 0.00243 | 0.346 | 5.28e−16 | 0.357 | <2.20e−16 |
| Central memory CD8+ T cell (Tcm_CD8) | −0.06 | 0.174 | 0.076 | 0.0818 | 0.375 | <2.20e−16 | −0.065 | 0.139 |
| Effector memory CD8+ T cell (Tem_CD8) | 0.195 | 7.22e−06 | −0.078 | 0.0744 | 0.365 | 9.4e−19 | 0.343 | 1.02e−15 |
| Activated CD4+ T cell (Act CD4) | 0.117 | 0.00742 | −0.161 | 0.00023 | 0.12 | 0.00629 | 0.368 | <2.20e−16 |
| Central memory CD4+ T cell (Tcm_CD4) | −0.214 | 8.55e−07 | −0.051 | 0.248 | 0.393 | <2.20e−16 | −0.062 | 0.16 |
| Effector memory CD4+ T cell (Tem_CD4) | −0.029 | 0.503 | −0.028 | 0.523 | 0.2 | 4.26e−06 | 0.218 | 5.14e−07 |
| T follicular helper cell (Tfh) | −0.047 | 0.283 | −0.033 | 0.446 | 0.478 | <2.20e−16 | 0.203 | 3.22e−06 |
| Gamma delta T cell (Tgd) | −0.173 | 7.26e−05 | −0.05 | 0.255 | 0.442 | <2.20e−16 | 0.006 | 0.885 |
| Type 1 T helper cell (Th1) | −0.068 | 0.123 | −0.042 | 0.341 | 0.437 | <2.20e−16 | 0.27 | 4.4e−10 |
| Type 17 T helper cell (Th17) | 0.307 | 1.04e−12 | −0.136 | 0.0019 | 0.091 | 0.0375 | 0.371 | <2.20e−16 |
| Type 2 T helper cell (Th2) | −0.077 | 0.0789 | −0.034 | 0.434 | 0.2 | 4.52e−06 | 0.146 | 0.000843 |
| Regulatory T cell (Treg) | −0.216 | 6.91e−07 | −0.082 | 0.0617 | 0.475 | <2.20e−16 | 0.134 | 0.00225 |
| Activated B cell (act B) | 0.148 | 0.000717 | −0.163 | 0.000195 | 0.259 | <2.20e−16 | 0.477 | <2.20e−16 |
| Immature B cell (Imm_B) | 0.118 | 0.00718 | −0.158 | 0.000294 | 0.371 | <2.20e−16 | 0.431 | <2.20e−16 |
| Memory B cell (Mem_B) | −0.178 | 4.37e−05 | 0.019 | 0.673 | 0.176 | 5.36e−05 | 0.187 | 1.76e−05 |
| Natural killer cell (NK) | −0.069 | 0.113 | −0.049 | 0.26 | 0.318 | 1.25e−13 | 0.225 | 2.27e−07 |
| CD56bright natural killer cell (CD56bright) | 0.102 | 0.0193 | −0.005 | 0.904 | 0.274 | 2.28e−10 | −0.071 | 0.107 |
| CD56dim natural killer cell (CD56dim) | 0.029 | 0.51 | 0.012 | 0.793 | 0.113 | 0.00974 | 0.007 | 0.877 |
| Myeloid derived suppressor cell (MDSC) | 0.082 | 0.0626 | −0.098 | 0.025 | 0.458 | <2.20e−16 | 0.273 | 2.83e−10 |
| Natural killer T cell (NKT) | −0.223 | 2.74e−07 | −0.154 | 0.000413 | 0.391 | <2.20e−16 | 0.184 | 2.38e−05 |
| Activated dendritic cell (Act_DC) | 0.108 | 0.0135 | −0.08 | 0.0664 | 0.336 | 3.66e−15 | 0.223 | 2.68e−07 |
| Plasmacytoid dendritic cell (pDC) | −0.041 | 0.353 | −0.092 | 0.0363 | 0.276 | 1.72e−10 | 0.091 | 0.0376 |
| Immature dendritic cell (iDC) | 0.196 | 6.83e−06 | 0.072 | 0.101 | 0.158 | 0.000293 | 0.126 | 0.00383 |
| Macrophage (macrophage) | −0.1 | 0.023 | −0.041 | 0.35 | 0.425 | <2.20e−16 | 0.188 | 1.59e−05 |
| Eosinophil (eosinophil) | 0.041 | <2.20e−16 | −0.056 | 0.203 | 0.242 | 2.39e−08 | 0.249 | 9.54e−09 |
| Mast cell (mast) | −0.028 | 0.525 | −0.022 | 0.614 | 0.227 | 1.37e−10 | 0.186 | 2e−05 |
| Monocyte (monocyte) | 0.038 | 0.389 | −0.11 | 0.0118 | 0.396 | <2.20e−16 | 0.154 | 0.000438 |
| Neutrophil (neutrophil) | 0.091 | 0.0371 | 0.041 | 0.346 | 0.239 | 3.41e−08 | −0.021 | 0.637 |
Bold value means statistically significant.
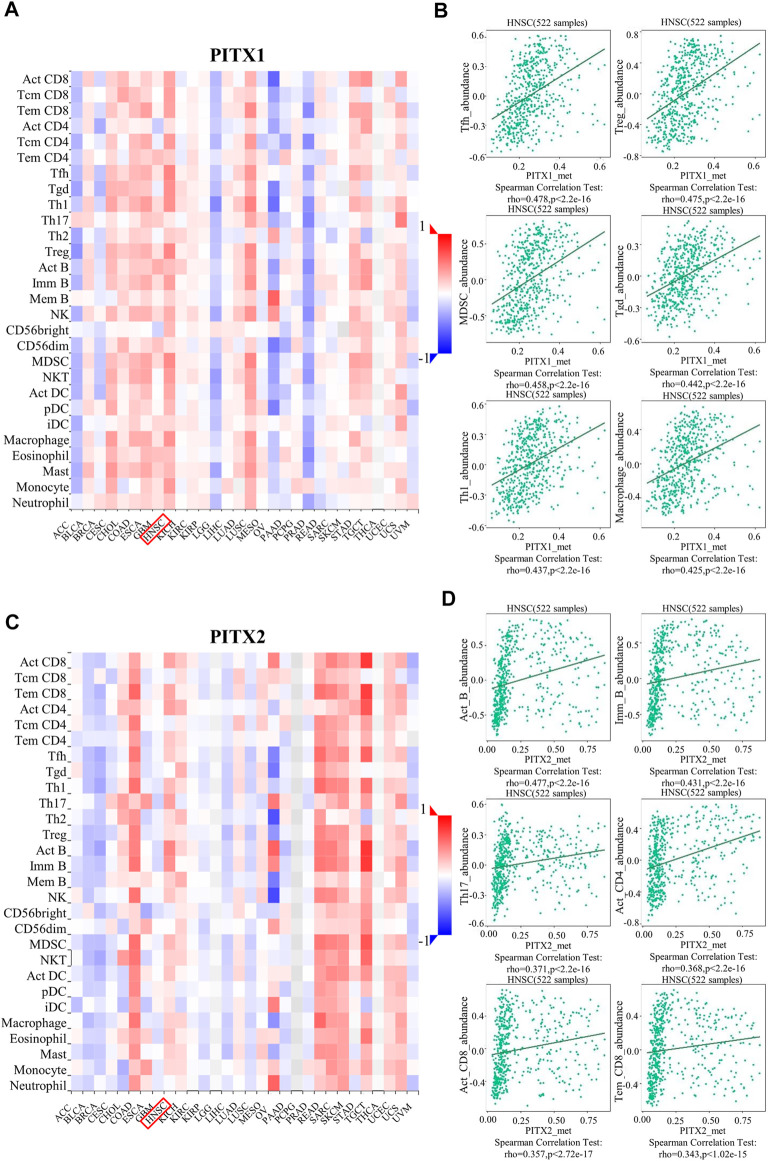
In addition, the methylation level of PITX1 and PITX2 had a significant impact on the prognosis of HNSC. We analyzed the correlation between TILs and the methylation level of PITX1 and PITX2 using the TISIDB. Our results showed that the methylation level of PITX1 was associated with 28 types of TILs in HNSC (Figure 9A and Table 3). Follicular helper T (Tfh) cells (r = 0.478, p < 2.2E−16), Tregs (r = 0.475, p < 2.2E−16), myeloid-derived suppressor cells (MDSCs) (r = 0.458, p < 2.2E−16), NKT cells (r = 0.442, p < 2.2E−16), Th1 cells (r = 0.437, p < 2.2E-16), and macrophages (r = 0.425, p < 2.2E-16) were most strongly correlated (r > 0.3) with PITX1 methylation (Figure 9B). PITX2 methylation was associated with 22 immune cell subtypes in HNSC (Figure 9C and Table 3). Among these TILs, Act_B (r = 0.477, p < 2.2E−16), Imm_B (r = 0.431, p < 2E−16), Th17 cells (r = 0.371, p < 2.2E−16), Act_CD4 (r = 0.368, p < 2.2E-16), Act_CD8 (r = 0.357, p < 2.2E−16), and effector memory CD8+ T cells (Tem_CD8, r = 0.343, p = 1.02E−15) were most strongly correlated with PITX2 methylation (Figure 9D). These results imply that the methylation level of PITX1 and PITX2 could serve as a regulator of immune cell infiltration into HNSC tumors.
FIGURE 9.
TISIDB analysis of correlations between PITX1 and PITX2 methylation and immune cells infiltration across human cancers. (A) The relationship between PITX1 methylation and 28 types of TILs across various human cancers. (B) The top six TILs displaying the strongest Spearman’s correlation with PITX1 methylation in HNSC. (C) The relationship between PITX2 methylation and 28 types of TILs across various human cancers. (D) The top six TILs displaying the strongest Spearman’s correlation with PITX2 methylation in HNSC. Act_CD8, activated CD8+ T cells; Tcm_CD8, central memory CD8+ T cells; Tem_CD8, effector memory CD8+ T cells; Act_CD4, activated CD4+ T cells; Tcm_CD4, central memory CD4+ T cells; Tem_CD4, effector memory CD4+ T cells; Tfh, T follicular helper cells; Tgd, gamma delta T cells; Th1, type 1 T helper cells; Th17, type 17 T helper cells; Th2, type 2 T helper cells; Treg, regulatory T cells; Act B, activated B cells; Imm_B, immature B cells; Mem_B, memory B cells; NK, natural killer cells; CD56bright, CD56bright NK cells; CD56dim, CD56dim NK cells; MDSC, myeloid derived suppressor cells; NKT, natural killer T cells; Act DC, activated dendritic cells; pDC, plasmacytoid DCs; iDC, immature DCs; Mast, mast cells.
Discussion
The dysregulation of the PITX gene family has been reported in various types of cancer (Tran and Kioussi, 2021). Although the role of PITX genes in the initiation and prognosis of certain cancers has been partially demonstrated (Zhang et al., 2021), further bioinformatic analysis of their involvement in HNSC need to be performed. To the best of our knowledge, this study was the first attempt to investigate the oncological characteristics and prognostic value of different members of the PITX gene family in HNSC.
In the present study, we demonstrated that the expression of the PITX gene family in HNSC is related to some clinicopathological features, including pathologic types and histologic grade (according to TCGA). We also found that the expression of PITX genes correlated with the prognosis of patients with HNSC. OS and RFS are commonly used as primary end points to assess prognostic value of a particular marker (Jabbour et al., 2018). Our bioinformatic analysis showed that PITX1 expression was decreased in patients with HNSC, which correlated with relatively poor RFS. Moreover, the Student’s t-test values indicated that PITX1 was significantly associated with tumor stage, histologic grade, and N stage, which are related to cancer progression. Univariate/multivariate Cox regression analysis results also indicated that the expression of PITX1 could be an independent prognostic factor for HNSC. Collectively, these results revealed that PITX1 may perform well as a prognostic predictor for HNSC.
PITX1 acts as a tumor suppressor gene in various human cancers (Kolfschoten et al., 2005). It was reported that the downregulation of PITX1 expression might contribute to the progression of cutaneous malignant melanoma by promoting cell proliferation (Osaki et al., 2013). In lung cancer, PITX1 expression was also decreased, with 62% of lung cancer patients displaying no evidence of PITX1 expression. Moreover, the lack of PITX1 mRNA expression was associated with a higher tumor grade (Chen et al., 2007). The low PITX1 expression in HNSC may therefore be correlated with a worse prognosis. In our study, the expression of PITX2 in patients with HNSC was lower than that in normal tissues; however, its expression was not correlated with tumor histologic stage, T stage, N stage, M stage, or RFS. In addition, PITX3 was proven to be associated with HNSC and serve as an independent prognostic biomarker (Sailer et al., 2017b). Consistently, we found that PITX3 may be positively correlated with the occurrence and development of HNSC. The higher expression of PITX3 in HNSC was associated with higher G and T stages and poorer RFS (Holmes et al., 2016). The high expression of PITX3 in HNSC may therefore also lead to poor prognosis. PPI network and functional enrichment analysis showed that the high expression of PITX1 and PITX2 was mainly associated with DNA binding, regulation and transcription, transcription factor activation, and RNA polymerase II-specific processes. Dysregulation or mutation of DNA-binding proteins has been implicated in the development and progression of various types of diseases, including cancer (Shiroma et al., 2020). We also found that PITX1 and PITX2 co-expression was strongly associated with the myocyte enhancer factor 2A (MEF2A), while PITX2 expression alone was closely functionally related to pituitary-specific positive transcription factor 1 (POU1F1). A promoter- and cell-specific functional interaction between PITX2 and MEF2A was previously reported, which is involved in the regulation of oral epithelial cells (Toro et al., 2004). MEF2A may also promote the transcriptional activity of other factors, thus promoting stem-like properties of oral squamous cell carcinoma and playing an important role in the development of HNSC (Wang et al., 2022). Moreover, PITX2 plays a crucial role in embryogenesis, ontogenesis, growth, and development via the Wnt/beta-catenin and POU1F1 pathways (Zhang et al., 2018).
DNA methylation is the most common epigenetic marker that usually acts as a transcriptional repressor and contributes to tumor progression (Meng et al., 2015; Zhou et al., 2020). DNA methylation can directly affect the interaction between DNA-binding proteins with their cognate sites (Zhu et al., 2016). It has been reported that DNA methylation plays an important role in the etiology, pathogenesis, and prognosis of HNSC (Zhou et al., 2018). Thus, we were interested in investigating the interaction between DNA methylation and DNA-binding proteins in the context of HNSC. Our MethSurv database analysis revealed that PITX1 and PITX2 expression was strongly correlated with DNA methylation. Our study found that the expression of PITX1 and PITX2 was significantly lower in HNSC tumors, compared to normal tissues, which was related to PITX1 and PITX2 hypermethylation. A recent bioinformatics analysis showed that high PITX1 expression was related to DNA methylation and poor prognosis in lung adenocarcinoma (Song et al., 2018). The DNA methylation level of PITX2 was also associated with the risk of occurrence and progression of lung cancer (Dietrich et al., 2012). PITX2 DNA methylation was proven to serve as a prognostic biomarker for patients with HNSC (Sailer et al., 2017a).
Cancers are inextricably linked with the immune response. The immune system acts as an external tumor suppressor but conversely can also promote cancer occurrence, development, and progression (Vesely et al., 2011). We therefore explored the potential relationship between PITX genes and immunity, focusing on the tumor microenvironment, tumor immune infiltration, and immune cell DNA methylation. As for tumor immune infiltration, PITX1 and PITX2 expression was positively correlated with the infiltration of Th17 cells and Act_CD8+ T cells, but negatively correlated with NKT cell infiltration. A recent clinical trial has shown that tumor immune cell infiltration is correlated with the sensitivity to immunotherapy and HNSC prognosis (Zhang et al., 2020). In tumors of the oral cavity, outcomes appear to improve with an increase in tumoral or stromal CD8+ T cell numbers (Oguejiofor et al., 2015). Individuals with more genetic defects affecting NKT cells had a higher risk of developing cancer and these inherited defects were associated with tumor immune microenvironment subtypes, recruitment of TILs, and clinical outcomes (Xu et al., 2019). Besides, we found that the methylation level of PITX1 and PITX2 was associated with various TILs in HNSC. DNA methylation plays a key role in TIL differentiation and plasticity, which can be associated with favorable or poor prognoses in many types of cancer (Zeiner et al., 2018; Merino et al., 2019; Renaude et al., 2020). Therefore, we speculate that the correlations that we have observed may help reveal the mechanism by which PITX1 and PITX2 regulate the function of immune cells in HNSC.
Acknowledgments
The authors are grateful to the patients whose data was used in this study.
Data availability statement
The original contributions presented in the study are included in the article/Supplementary Material, further inquiries can be directed to the corresponding author.
Author contributions
YZ and JZ designed, performed, and supervised experiments, analyzed data, and co-wrote the manuscript. MZ and QZ performed experiments and collected data. FY and YF designed and supervised experiments and analyzed data. YG provided conceptual input and edited the manuscript. All authors took part in the drafting, revising, or critically reviewing the manuscript, and approved the final draft version submitted for publication.
Funding
This study was supported by the National Natural Science Foundation of China (81800788 and 81773339), Science and Technology Department of Hunan Province, China (2017WK2041 and 2018SK52511), Scientific Research Project of Hunan Provincial Health Commission (202208043514), Natural Science Foundation of Changsha City (kq2202403 and kq2202412), Fund for the Xiangya Clinical Medicine Database of Central South University (2014-ZDYZ-1-16), Education and Teaching Reform Research Project of Central South University (2020jy165-3), Research Project on Postgraduate Education and Teaching Reform of Central South University (2021JGB072), Open Sharing Fund for the Large-scale Instruments and Equipment of Central South University and the Fundamental Research Funds for the Central Universities of Central South University.
Conflict of interest
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
Publisher’s note
All claims expressed in this article are solely those of the authors and do not necessarily represent those of their affiliated organizations, or those of the publisher, the editors and the reviewers. Any product that may be evaluated in this article, or claim that may be made by its manufacturer, is not guaranteed or endorsed by the publisher.
Supplementary material
The Supplementary Material for this article can be found online at: https://www.frontiersin.org/articles/10.3389/fgene.2022.982241/full#supplementary-material
References
- Angotzi A. R., Ersland K. M., Mungpakdee S., Stefansson S., Chourrout D. (2008). Independent and dynamic reallocation of pitx gene expression during vertebrate evolution, with emphasis on fish pituitary development. Gene 417, 19–26. 10.1016/j.gene.2008.03.013 [DOI] [PubMed] [Google Scholar]
- Bray F., Ferlay J., Soerjomataram I., Siegel R. L., Torre L. A., Jemal A. (2018). Global cancer statistics 2018: GLOBOCAN estimates of incidence and mortality worldwide for 36 cancers in 185 countries. Ca. Cancer J. Clin. 68, 394–424. 10.3322/caac.21492 [DOI] [PubMed] [Google Scholar]
- Chen Y., KnöSEL T., Ye F., Pacyna-Gengelbach M., Deutschmann N., Petersen I. (2007). Decreased PITX1 homeobox gene expression in human lung cancer. Lung Cancer 55, 287–294. 10.1016/j.lungcan.2006.11.001 [DOI] [PubMed] [Google Scholar]
- Colaprico A., Silva T. C., Olsen C., Garofano L., Cava C., Garolini D., et al. (2016). TCGAbiolinks: An R/bioconductor package for integrative analysis of TCGA data. Nucleic Acids Res. 44, e71. 10.1093/nar/gkv1507 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dietrich D., Hasinger O., Liebenberg V., Field J. K., Kristiansen G., Soltermann A. (2012). DNA methylation of the homeobox genes PITX2 and SHOX2 predicts outcome in non-small-cell lung cancer patients. Diagn. Mol. Pathol. 21, 93–104. 10.1097/PDM.0b013e318240503b [DOI] [PubMed] [Google Scholar]
- Ferlay J., Colombet M., Soerjomataram I., Mathers C., Parkin D. M., PiñEROS M., et al. (2019). Estimating the global cancer incidence and mortality in 2018: GLOBOCAN sources and methods. Int. J. Cancer 144, 1941–1953. 10.1002/ijc.31937 [DOI] [PubMed] [Google Scholar]
- Holmes E. E., Goltz D., Sailer V., Jung M., Meller S., Uhl B., et al. (2016). PITX3 promoter methylation is a prognostic biomarker for biochemical recurrence-free survival in prostate cancer patients after radical prostatectomy. Clin. Epigenetics 8, 104. 10.1186/s13148-016-0270-x [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jabbour E., Ravandi F., Kebriaei P., Huang X., Short N. J., Thomas D., et al. (2018). Salvage chemoimmunotherapy with inotuzumab ozogamicin combined with mini-hyper-CVD for patients with relapsed or refractory philadelphia chromosome-negative acute lymphoblastic leukemia: A phase 2 clinical trial. JAMA Oncol. 4, 230–234. 10.1001/jamaoncol.2017.2380 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jiang Y., Guo H., Tong T., Xie F., Qin X., Wang X., et al. (2022). lncRNA lnc-POP1-1 upregulated by VN1R5 promotes cisplatin resistance in head and neck squamous cell carcinoma through interaction with MCM5. Mol. Ther. 30, 448–467. 10.1016/j.ymthe.2021.06.006 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Johnson D. E., Burtness B., Leemans C. R., Lui V. W. Y., Bauman J. E., Grandis J. R. (2020). Head and neck squamous cell carcinoma. Nat. Rev. Dis. Prim. 6, 92. 10.1038/s41572-020-00224-3 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kolfschoten I. G., Van Leeuwen B., Berns K., Mullenders J., Beijersbergen R. L., Bernards R., et al. (2005). A genetic screen identifies PITX1 as a suppressor of RAS activity and tumorigenicity. Cell 121, 849–858. 10.1016/j.cell.2005.04.017 [DOI] [PubMed] [Google Scholar]
- Lanctôt C., Moreau A., Chamberland M., Tremblay M. L., Drouin J. (1999). Hindlimb patterning and mandible development require the Ptx1 gene. Development 126, 1805–1810. 10.1242/dev.126.9.1805 [DOI] [PubMed] [Google Scholar]
- Li B., Severson E., Pignon J. C., Zhao H., Li T., Novak J., et al. (2016). Comprehensive analyses of tumor immunity: Implications for cancer immunotherapy. Genome Biol. 17, 174. 10.1186/s13059-016-1028-7 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li T., Fu J., Zeng Z., Cohen D., Li J., Chen Q., et al. (2020). TIMER2.0 for analysis of tumor-infiltrating immune cells. Nucleic Acids Res. 48, W509–w514. 10.1093/nar/gkaa407 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Meng H., Cao Y., Qin J., Song X., Zhang Q., Shi Y., et al. (2015). DNA methylation, its mediators and genome integrity. Int. J. Biol. Sci. 11, 604–617. 10.7150/ijbs.11218 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Merino A., Zhang B., Dougherty P., Luo X., Wang J., Blazar B. R., et al. (2019). Chronic stimulation drives human NK cell dysfunction and epigenetic reprograming. J. Clin. Invest. 129, 3770–3785. 10.1172/JCI125916 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Oguejiofor K., Hall J., Slater C., Betts G., Hall G., Slevin N., et al. (2015). Stromal infiltration of CD8 T cells is associated with improved clinical outcome in HPV-positive oropharyngeal squamous carcinoma. Br. J. Cancer 113, 886–893. 10.1038/bjc.2015.277 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Oliva M., Spreafico A., Taberna M., Alemany L., Coburn B., Mesia R., et al. (2019). Immune biomarkers of response to immune-checkpoint inhibitors in head and neck squamous cell carcinoma. Ann. Oncol. 30, 57–67. 10.1093/annonc/mdy507 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Osaki M., Chinen H., Yoshida Y., Ohhira T., Sunamura N., Yamamoto O., et al. (2013). Decreased PITX1 gene expression in human cutaneous malignant melanoma and its clinicopathological significance. Eur. J. Dermatol. 23, 344–349. 10.1684/ejd.2013.2021 [DOI] [PubMed] [Google Scholar]
- Poos A. M., Schroeder C., Jaishankar N., RöLL D., Oswald M., Meiners J., et al. (2022). PITX1 is a regulator of TERT expression in prostate cancer with prognostic power. Cancers (Basel) 14, 1267. 10.3390/cancers14051267 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Renaude E., Kroemer M., Loyon R., Binda D., Borg C., Guittaut M., et al. (2020). The fate of Th17 cells is shaped by epigenetic modifications and remodeled by the tumor microenvironment. Int. J. Mol. Sci. 21, E1673. 10.3390/ijms21051673 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ru B., Wong C. N., Tong Y., Zhong J. Y., Zhong S. S. W., Wu W. C., et al. (2019). Tisidb: An integrated repository portal for tumor-immune system interactions. Bioinformatics 35, 4200–4202. 10.1093/bioinformatics/btz210 [DOI] [PubMed] [Google Scholar]
- Saâda-Bouzid E., Defaucheux C., Karabajakian A., Coloma V. P., Servois V., Paoletti X., et al. (2017). Hyperprogression during anti-PD-1/PD-L1 therapy in patients with recurrent and/or metastatic head and neck squamous cell carcinoma. Ann. Oncol. 28, 1605–1611. 10.1093/annonc/mdx178 [DOI] [PubMed] [Google Scholar]
- Sailer V., Holmes E. E., Gevensleben H., Goltz D., DröGE F., De Vos L., et al. (2016). PITX2 and PANCR DNA methylation predicts overall survival in patients with head and neck squamous cell carcinoma. Oncotarget 7, 75827–75838. 10.18632/oncotarget.12417 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sailer V., Gevensleben H., Dietrich J., Goltz D., Kristiansen G., Bootz F., et al. (2017a). Clinical performance validation of PITX2 DNA methylation as prognostic biomarker in patients with head and neck squamous cell carcinoma. PLoS One 12, e0179412. 10.1371/journal.pone.0179412 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sailer V., Holmes E. E., Gevensleben H., Goltz D., DröGE F., Franzen A., et al. (2017b). PITX3 DNA methylation is an independent predictor of overall survival in patients with head and neck squamous cell carcinoma. Clin. Epigenetics 9, 12. 10.1186/s13148-017-0317-7 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shih H. P., Gross M. K., Kioussi C. (2007). Expression pattern of the homeodomain transcription factor Pitx2 during muscle development. Gene Expr. Patterns 7, 441–451. 10.1016/j.modgep.2006.11.004 [DOI] [PubMed] [Google Scholar]
- Shiroma Y., Takahashi R. U., Yamamoto Y., Tahara H. (2020). Targeting DNA binding proteins for cancer therapy. Cancer Sci. 111, 1058–1064. 10.1111/cas.14355 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Solomon B., Young R. J., Rischin D. (2018). Head and neck squamous cell carcinoma: Genomics and emerging biomarkers for immunomodulatory cancer treatments. Semin. Cancer Biol. 52, 228–240. 10.1016/j.semcancer.2018.01.008 [DOI] [PubMed] [Google Scholar]
- Song X., Zhao C., Jiang L., Lin S., Bi J., Wei Q., et al. (2018). High PITX1 expression in lung adenocarcinoma patients is associated with DNA methylation and poor prognosis. Pathol. Res. Pract. 214, 2046–2053. 10.1016/j.prp.2018.09.025 [DOI] [PubMed] [Google Scholar]
- Tai W. T., Chen Y. L., Chu P. Y., Chen L. J., Hung M. H., Shiau C. W., et al. (2016). Protein tyrosine phosphatase 1B dephosphorylates PITX1 and regulates p120RasGAP in hepatocellular carcinoma. Hepatology 63, 1528–1543. 10.1002/hep.28478 [DOI] [PubMed] [Google Scholar]
- Takenobu M., Osaki M., Fujiwara K., Fukuhara T., Kitano H., Kugoh H., et al. (2016). PITX1 is a novel predictor of the response to chemotherapy in head and neck squamous cell carcinoma. Mol. Clin. Oncol. 5, 89–94. 10.3892/mco.2016.880 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Toro R., Saadi I., Kuburas A., Nemer M., Russo A. F. (2004). Cell-specific activation of the atrial natriuretic factor promoter by PITX2 and MEF2A. J. Biol. Chem. 279, 52087–52094. 10.1074/jbc.M404802200 [DOI] [PubMed] [Google Scholar]
- Tran T. Q., Kioussi C. (2021). Pitx genes in development and disease. Cell. Mol. Life Sci. 78, 4921–4938. 10.1007/s00018-021-03833-7 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vesely M. D., Kershaw M. H., Schreiber R. D., Smyth M. J. (2011). Natural innate and adaptive immunity to cancer. Annu. Rev. Immunol. 29, 235–271. 10.1146/annurev-immunol-031210-101324 [DOI] [PubMed] [Google Scholar]
- Wang X., Li R., Wu L., Chen Y., Liu S., Zhao H., et al. (2022). Histone methyltransferase KMT2D cooperates with MEF2A to promote the stem-like properties of oral squamous cell carcinoma. Cell Biosci. 12, 49. 10.1186/s13578-022-00785-8 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Warde-Farley D., Donaldson S. L., Comes O., Zuberi K., Badrawi R., Chao P., et al. (2010). The GeneMANIA prediction server: Biological network integration for gene prioritization and predicting gene function. Nucleic Acids Res. 38, W214–W220. 10.1093/nar/gkq537 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Weinstein J. N., Collisson E. A., Mills G. B., Shaw K. R., Ozenberger B. A., Ellrott K., et al. (2013). The cancer genome Atlas pan-cancer analysis project. Nat. Genet. 45, 1113–1120. 10.1038/ng.2764 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Xu X., Guo J., Vorster P., Wu Y. (2012). Involvement of LIM kinase 1 in actin polarization in human CD4 T cells. Commun. Integr. Biol. 5, 381–383. 10.4161/cib.20165 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Xu X., Li J., Zou J., Feng X., Zhang C., Zheng R., et al. (2019). Association of germline variants in natural killer cells with tumor immune microenvironment subtypes, tumor-infiltrating lymphocytes, immunotherapy response, clinical outcomes, and cancer risk. JAMA Netw. Open 2, e199292. 10.1001/jamanetworkopen.2019.9292 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zeiner P. S., Zinke J., Kowalewski D. J., Bernatz S., Tichy J., Ronellenfitsch M. W., et al. (2018). CD74 regulates complexity of tumor cell HLA class II peptidome in brain metastasis and is a positive prognostic marker for patient survival. Acta Neuropathol. Commun. 6, 18. 10.1186/s40478-018-0521-5 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang R., Gao Y. (2021). Identification of NUTF2 as a candidate diagnostic and prognostic biomarker associated with immune infiltration in head and neck squamous cell carcinoma. Onco. Targets. Ther. 14, 5455–5467. 10.2147/OTT.S337469 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang S., Xu H., Kang Z., Cai H., Dang R., Lei C., et al. (2018). Bovine pituitary homeobox 2 (PITX2): mRNA expression profiles of different alternatively spliced variants and association analyses with growth traits. Gene 669, 1–7. 10.1016/j.gene.2018.05.083 [DOI] [PubMed] [Google Scholar]
- Zhang X., Shi M., Chen T., Zhang B. (2020). Characterization of the immune cell infiltration landscape in head and neck squamous cell carcinoma to aid immunotherapy. Mol. Ther. Nucleic Acids 22, 298–309. 10.1016/j.omtn.2020.08.030 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang C., Chen X., Chen Y., Cao M., Tang J., Zhong B., et al. (2021). The PITX gene family as potential biomarkers and therapeutic targets in lung adenocarcinoma. Med. Baltim. 100, e23936. 10.1097/MD.0000000000023936 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhou C., Ye M., Ni S., Li Q., Ye D., Li J., et al. (2018). DNA methylation biomarkers for head and neck squamous cell carcinoma. Epigenetics 13, 398–409. 10.1080/15592294.2018.1465790 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhou F., Shen D., Xiong Y., Cheng S., Xu H., Wang G., et al. (2020). CTHRC1 is a prognostic biomarker and correlated with immune infiltrates in kidney renal papillary cell carcinoma and kidney renal clear cell carcinoma. Front. Oncol. 10, 570819. 10.3389/fonc.2020.570819 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhu H., Wang G., Qian J. (2016). Transcription factors as readers and effectors of DNA methylation. Nat. Rev. Genet. 17, 551–565. 10.1038/nrg.2016.83 [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Data Availability Statement
The original contributions presented in the study are included in the article/Supplementary Material, further inquiries can be directed to the corresponding author.