Abstract
Background
DNA methylation is an effective epigenetic process that is frequently linked to changes in gene expression. Zinc is a vital micronutrient that plays a crucial role in DNA methylation. Therefore, abnormal zinc levels may cause aberrant DNA methylation and other diseases.
Objectives
To investigate the influence of zinc on gene-specific and global DNA methylation in humans and rodents, their tissues and their cells.
Method
Systematic literature searches were conducted using Medline, Scopus, Google Scholar, and Web of Science databases. Studies that met the inclusion criteria and were published in English language were included. Data including the first author, sample size, subjects, targeted genes, tissue types or cells analysed, zinc level, molecular techniques, DNA methylation outcomes, and consequences were extracted.
Results
From a total of 2360 articles screened by title and abstract, 15 met the inclusion criteria. Qualitative analysis indicates that there are associations between zinc deficiency and gene-specific hypomethylation in humans and between zinc deficiency and hypermethylation in rodents. Zinc did not influence LINE-1 methylation in humans. Depending on cell type, zinc could have a positive or negative effect on global methylation in humans and rodents. As predicted, in general, gene expression was elevated by DNA hypomethylation and the corresponding protein levels were also upregulated. However, some studies showed that zinc deficiency led to reduced gene expression or no alteration in mRNA levels and corresponding protein levels.
Conclusion
Our study shows links between zinc levels and DNA methylation. However, greater significance may be achieved if more than one independent investigator analyses the same set of genes in the same cell type. Therefore, gene-cell and animal-specific investigations are recommended to reduce variability and allow comparisons across studies.
Keywords: zinc, Gene-specific DNA methylation, Global DNA methylation, Methylation consequences
Gene-specific DNA methylation; Global DNA methylation; Methylation consequences.
1. Introduction
Environmental factors, including micronutrients, have been proven to impact epigenetics. Zinc is an essential ubiquitous micronutrient required for normal metabolic reactions, growth, and development of all organisms [1]. Its deficiency is still a challenge in developing and developed nations, leading to high rates of morbidity and mortality, particularly in low-income populations [2]. Due to the importance of zinc, its altered level will affect various processes in the body [3]. Through epigenetic changes, a gene's expression and function can be influenced by epigenetic changes without affecting a gene's sequence. Epigenetic alterations, such as DNA methylation and histone modifications, are mediated by several proteins containing zinc-binding domains. These zinc metalloproteins are divided into two classes: zinc metalloproteins and zinc finger proteins. Zinc-binding DNA methyltransferases (DNMTs) carried out the process of DNA methylation [4]. However, these flexible epigenetic signals can be triggered by environmental events such as diet, hormones, stress, drugs and chemicals [5].
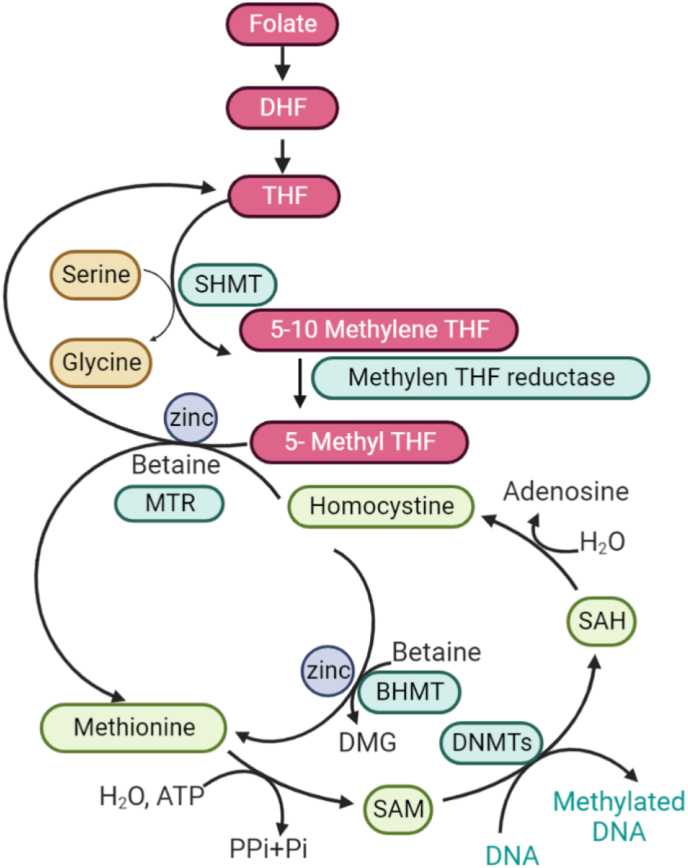
Dietary methyl groups are necessary for DNA methylation. The methyl-group donors engaged in the one-carbon cycle are methionine, folate and choline. Co-factors required for DNA methylation are zinc and the vitamins B2, B6, and B12. Several studies revealed the importance of zinc in DNA methylation [6, 7, 8]. In both folate-dependent and folate-independent cycles, methionine is produced from homocysteine by the enzymes 5-methyltetrahydrofolate-homocysteine methyltransferase (MTR) and betaine homocysteine methyltransferase (BHMT) in a zinc-dependent process, respectively. Methionine is converted into the methyl donor S-adenosylmethionine, the substrate of DNMT [4, 9] (Figure 1). Methyl groups from dietary methionine, folate, and choline are ultimately added to the 5’ carbon of cytosine within CpG (cytosine phosphate guanine) dinucleotides' found in DNA. In the promoter regions of many housekeeping or developmentally regulated genes, CpG islands are mostly unmethylated in mammals. When CpG islands are methylated, chromatin condensation is induced, and gene expression is silenced. Since most transcription factors binding sites within DNA contain CpG dinucleotides, methylation of these areas might prevent transcription factors from accessing the DNA, therefore suppressing gene transcription [10].
Figure 1.
The role of zinc in DNA methylation. The mechanisms involving one-carbon metabolism and DNA methylation are illustrated in this diagram. BHMT = Betaine-homocysteine S-methyltransferase; DHF = Dihydrofolate; DMG = Dimethylglycine; DNMTs = DNA methyltransferases; MTR = 5-methyltetrahydrofolate-homocysteine methyltransferase; SAH = S- Adenosylhomocysteine; SAM = S-Adenosylmethionine; SHMT = Serine hydroxymethyltransferase; THF = Tetrahydrofolate.
Knowledge of epigenome alterations, namely alterations in gene methylation and the specific genomic areas where they occur, may be critical to human health. In addition, since epigenetic alterations are reversible, a better knowledge of the particular changes caused by dietary variables such as zinc might provide new targets for intervention.
This review aims to determine if there is an association between zinc level and methylation of specific genes and global DNA and the physiological consequences in primary literature on human and rodent studies. We report a wide range of effects of zinc on DNA methylation and their consequences. To our knowledge, this is the first such systematic review exploring zinc level and its possible association with DNA methylation.
2. Methods
This review is registered with the PROSPERO database (registration number: CRD42021278212) and has been reported in compliance with the guideline of the Preferred Reporting Items for Systematic Reviews and Meta-Analyses (PRISMA) Statement, revision 2020 [11].
2.1. Search strategy
A comprehensive search of the literature was undertaken using four biomedical electronic databases Scopus, Web of Science, Google Scholar, and MEDLINE as sources for this study. The search aimed to identify relevant articles published in peer-reviewed journals written in English, with the assumption that most of the important findings will be reported in English regardless of country of origin. The Boolean search was performed on each database as follows:
SCOPUS: TITLE-ABS-KEY (((zinc) AND ("DNA methylation" OR epigenetic) AND NOT ("zinc finger binding protein") AND NOT ("zinc finger protein") AND NOT ("zinc-finger"))) AND (LIMIT-TO (DOCTYPE, "ar")) AND (LIMIT-TO (LANGUAGE, "English")) AND (LIMIT-TO (SRCTYPE, "j")). Web of Science: ((zinc) AND ("DNA methylation" OR epigenetic) NOT ("zinc finger binding protein") NOT ("zinc finger protein") NOT ("zinc-finger")). MEDLINE: AB ("Zinc") AND ("DNA methylation" OR "methylation"). Google Scholar: allintitle: zinc "DNA methylation". All searches were concluded up through August 2021.
2.2. Inclusion and exclusion criteria
The inclusion criteria include studies on humans and rodents, used DNA methylation techniques, original data to show the relationship between zinc level and DNA methylation and the establishment of the outcome of changes in gene-specific DNA methylation or changes in global DNA methylation, effects of zinc on DNA methylation altered by other elements or molecules.
The exclusion criteria include studies that did not specify the zinc level and with no clear outcome, studies other than the English language, review articles and studies of nanoparticles and environmental factors including particulate matter.
2.3. Selection criteria and study selection
After completing the search, articles were then organized into Mendeley Software. Duplication articles were removed by Zotero software. One reviewer performed this task via the “Find and Remove Duplicate References” function, followed by manual screening, as some of the same articles were entered slightly differently into different databases. After removing duplicates, articles were assessed for eligibility independently by two reviewers in two stages. First, the title and abstract of the database were screened and assessed for relevance. Next, the full text of potentially relevant publications was retrieved and reviewed for inclusion. Both stages of the study selection were performed independently by two reviewers and cross-validated to assess for disagreements. For any disagreement, consensus was sought where possible, and in cases where that was not possible, a third reviewer was assigned. Only full-text articles were included in the review to enable quality assessments. Forward and backward citations search was not attempted due to resource limitations.
2.4. Data extraction
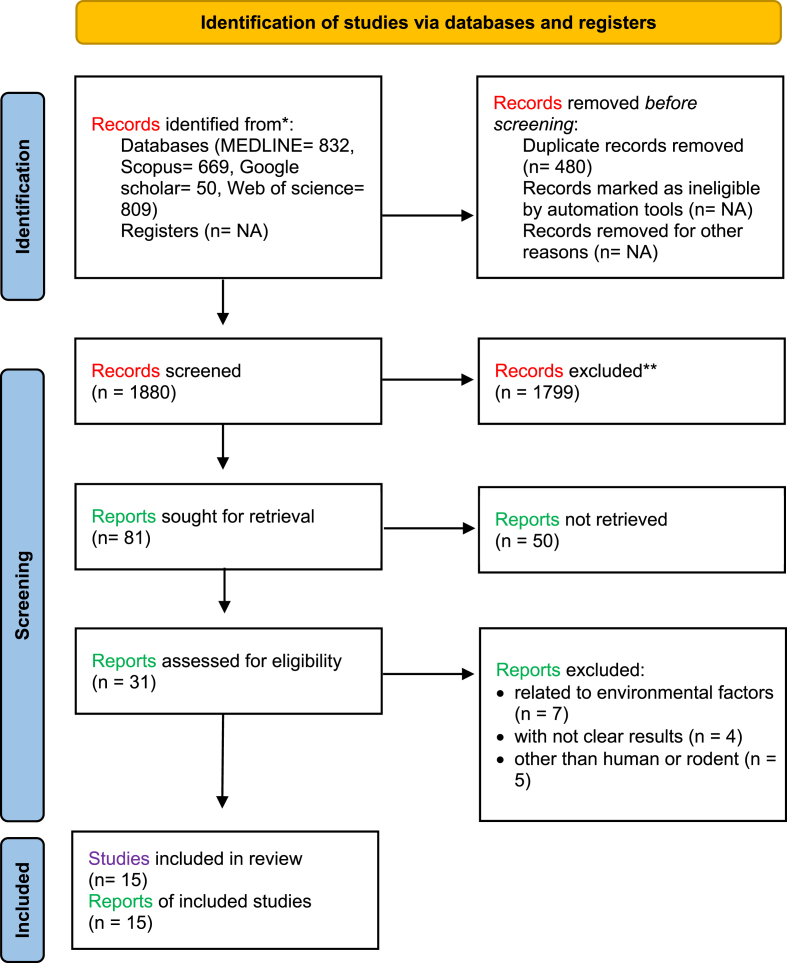
Two independent researchers performed the data extractions to establish inter-rater reliability and avoid data entry errors. Reported findings were extracted onto a data extraction form. For each of the included studies, data included author(s), year of publication, sample size, type of samples, experimental methods, type of tissues, candidate genes, and significant findings. The process of article searches and selection performed in this study is shown in Figure 2.
Figure 2.
Flow diagram of the study selection process.
3. Results
A total of 2360 records were identified by searching four databases (Web of Science, MEDLINE Complete, Google Scholar, SCOPUS). After removing the duplicates, titles and abstracts were screened, and 81 records were selected for full-text assessment. Among them 66 records that did not report zinc levels and DNA methylation, reported research on organisms other than humans and rodents and reported environmental were removed, leaving 15 articles that met the eligibility criteria for our qualitative analysis. An overview of these selected studies in research characteristics created in the PRISMA chart is shown in Figure 2.
The data extracted from the 15 articles are summarized in Table 1. Seven of these studies were on humans, seven on rodents (rats or mice), and one study was on both humans and mice. The articles were published between 2012 and 2020.
Table 1.
Data extracted from 15 studies investigating the effect of zinc level on gene-specific or global DNA methylation in humans and rodents.
| Autor and Reference | Country | Subjects/Sample size | DNA Methylation Method | Biological samples | Targeted genes | Zinc measurement method/related tissues | Zinc level | Methylation status | Consequences |
|---|---|---|---|---|---|---|---|---|---|
| Kumar, (2015) [12] | Australia | Two breastfed human neonates (1. premature at 36 weeks, 2. premature at 37 weeks) | Pyrosequencing | Fibroblast and lymphoblast cells | SLC30A5, SLC30A6 | N.A/Venous blood and milk | Zinc deficiency | Methylation of SLC30A5 was reduced in lymphoblasts but increased in fibroblasts | Reduced levels of SLC30A5 mRNA and protein in lymphoblasts were associated with a reduction of zinc in breast milk |
| Perng, (2014) [20] | USA | 987 adults aged 45–84 years | Pyrosequencing | Blood | LINE-1 and Alu | FFQ/N.A | Zinc supplementation | Increase Alu methylation but not effect on LINE-1 | Elevation of LINE-1 methylation found in subjects with higher BMI, positive correlation between height and Alu methylation |
| Iqbal, (2019) [21] | Bangladesh | 324 children aged 2–3 years | 5-mC DNA ELISA | Blood | Global DNA | Semiquantitative FFQ/N.A | Low level of zinc in the diet | Significantly increase in global DNA methylation | Positive correlation between lower zinc intake by children and the prevalence of stunting |
| Vidal, (2015) [13] | USA | 319 infant-mother pairs | Pyrosequencing | Venous and umbilical cord blood | PEG3, PLAGL1, DMRs and MEG3 DMRs. | Solution-based ICP-MS/Venous blood | Low and higher zinc concentrations | Lower zinc and a high level of Cd reduced PLAGL1 methylation, higher zinc increased PEG3 methylation but no association with MEG3 methylation in offspring | Maternal Cd exposure in early pregnancy alters DNA methylation at multiple DMRs in offspring with sex and possibly race/ethnic-specific effects, and Zn may mitigate these effects. |
| Perng, (2012) [22] | Colombia | 568 children, 5–12 years | Pyrosequencing | Blood | LINE-1 | Atomic absorption technique/Venous blood | serum zinc level | No zinc showed no effect on LINE-1 methylation in total, but the LINE-1 methylation level of girls was lower compared to boys | DNA methylation was not related to erythrocyte folate, serum zinc, plasma vitamin B12 or ferritin. |
| Rodenkirchen, (2020) [14] | Germany | Healthy adults | Methylation-specific qPCR | Blood PBMC | IFN-γ and IRF-4. | Used zinc deficient, zinc adequate and zinc excessive/In-vitro | Zinc supplementation, zinc deficiency | Neither zinc-deficient nor zinc-supplemented PBMC showed a methylation pattern of the IFN-γ promoter region differing from the control | Significant increase in the cytokine’s mRNA and a reduction of IFN-γ protein level compared with zinc-supplemented cells. |
| Farahzadi, (2017) [15] | Iran | Six normal women, 45–58 years old) | Methylation-specific qPCR | Mesenchymal stem cells from adipose tissue | Human telomerase reverse transcriptase (hTERT) | Zinc deficient, zinc adequate/In-vitro | Higher and lower levels of ZnSO4 | Among the 45 sites of CpG sites, 5 sites were changed to methylated, 9 sites were changed to unmethylated and 7 unmethylated sites and 10 methylated sites remained unchanged. | Increased telomere length, the hTERT gene expression, and the telomerase activity |
| Tian, (2013) [23] | USA | Female CD1 mice | Immuno-fluorescence | Oocytes | Global DNA | Fed zinc deficient and zinc control diet/in vivo | Zinc deficiency | Decrease in global DNA methylation in zinc deficient oocytes | Increase Lap, Line-1, Sineb1 and Sineb2 transcription and decrease of Gdf9, Zp3 and Figla transcription, a significant reduction for the mature eggs to reach divided two cells |
| Finke, (2020) [24] | Germany | 18, Male C57BL/6J mice | High Performance Liquid Chromatography-Tandem Mass Spectrometry | Jejunum, liver, cortex, and kidney | Global DNA | Fed zinc deficient and zinc control diet/in vivo | Zinc deficiency | No effect on the global DNA methylation and the hydroxymethylation | Splenomegaly and genomic instability in the liver of mice |
| Davison, (2010) [25] | UK | Pregnant mice and their pups | Luminometric methylation assay (LUMA) assay using a pyrosequencer | Pups liver | Global DNA | Zn-restricted, Zn-adequate or Zn-supplemented/in-vivo | Zinc restriction, zinc supplementation | Significantly higher methylation in zinc deficient and zinc supplementation | Significant increase of DNMT1 and DNMT3a expression in higher zinc |
| Jiang, (2020) [16] | China | 32 pregnant rats | Quantitative real-time PCR. Methylation-specific PCR |
Hippocampus of offspring | BDNF promoter | Fed zinc deficient and zinc control diet/in vivo | Zinc deficiency and zinc supplementation | Reduction of BDNF methylation after zinc supplementation to offspring from zinc-deficient mother | Supplementation of zinc at age 21 to 60, improved BDNF expression but the MECP2 and GADD45b expression were still significantly lower |
| Kurita, (2012) [17] | Japan | Pregnant mice | Bisulfite genomic sequencing | Livers (approx. 0.1 g) and blood (approx. 0.2 g) specimens | Metallothionein 2 (MT) promoter | Fed zinc deficient and zinc control diet/in vivo | A significant increase in DNA methylation at the -820 CpG site, | Zn deficiency in utero induces fetal epigenetic alterations and these changes are stored as an epigenetic memory until adulthood | |
| Khadivi, (2020) [26] | Iran | 40 male rats | Immunofluorescence staining | Rats’ sperm | Global DNA | Fed zinc deficient/in vivo | Zinc supplementation against hypomethylation effect of chemotherapeutic agents. | Global DNA methylation increased compared to (bleomycin, etoposide, and cisplatin) group, yet no significant difference compared to the control group. | Improved protamination but lower than the control group. Zinc reduced DNA fragmentation compared to chemotherapeutic agents treated and exhibited recovery of body and testis weight with an increase in spermatogonia and Leydig cells |
| Hu, (2016) [18] | China | Rats and 0∼2 months offspring | Methylation-specific quantitative real-time PCR | Rats’ hippocampus | BDNF | Fed zinc deficient and zinc control diet/in vivo | zinc deficiency | DNA methylation of the BDNF exon IX was significantly increased in the zinc-deficient pups | Reduction of BDNF protein level, BDNF mRNA and DNMT3A mRNAs expression, but increase in the DNMT1 mRNA |
| Wong, (2015) [19] | USA | 10 old (92–101) and 10 young (20–28) years human, and mice | Pyrosequencing | Human lymphoblastoid cell line THP-1 cells and female C57Bl/6 mouse spleens | Human IL-6 promoter Mouse IL-6 | Zinc deficient, zinc adequate/In-vitro | Zinc deficiency | Reduced IL-6 promoter methylation in zinc deficient THP1 cells, in aged mice and in human lymphoblastoid cell lines derived from aged individuals. No significant differences in total DNA methylation levels between zinc deficiency and control groups |
Increased IL-6 protein production |
In three human studies, the association between zinc levels and DNA methylation was studied in venous blood, umbilical cord blood, fibroblasts, and lymphoblast cells. Two studies used solution-based inductively coupled plasma mass spectrometry and atomic absorption techniques to quantify zinc levels in blood samples. In contrast, the third study did not specify the zinc measuring method. In three other studies, human peripheral blood mononuclear cells (PBMC), mesenchymal stem cells (MSCs) from adipose tissue, and human lymphoblastic cell lines were cultured in vitro using zinc-restricted and zinc-adequate media. Two human studies used food frequency questionnaires to estimate zinc levels and venous blood and relate that information to DNA methylation. In animal studies, rodents such as rats and mice were used. Female CD1 mice, male c57BL/6J mice, pregnant mice, male rats, and neonate rats were used.
In the majority of animal studies, zinc-altered diets were used to assess the relationship between nutritional zinc and DNA methylation in cells and tissues, including blood, oocytes, Jejunum, liver, cortex, kidney, liver, hippocampus, and sperm. One animal study assessed zinc levels in the plasma and hippocampus of animals fed zinc-deficient and zinc-adequate diets. Another examined DNA methylation levels in spleens and liver cells cultured in zinc-deficient and zinc-adequate media.
DNA methylation targets are included in a range of genomic locations. Eight studies examined gene-specific DNA methylation (SLC30A5, SLC30A6, PEG3, PLAGL1, MEG3, IFN-γ, IRF-4, hTERT, BDNF, MT2, IL-6) [12–19], and seven studies examined global DNA methylation using long interfering nuclear element-1 (LINE-1) and Arthrobacter luteus (Alu) genes as proxies for global DNA methylation [20, 21, 22, 23, 24, 25, 26]. DNA methylation analysis utilized six different methods, bisulfite pyrosequencing (n = 5) [12,13,20,22,27] and methylation-specific qPCR (n = 4) [[14], [15], [16],18], immune fluorescence (n = 2) [23,26], enzyme-linked immunosorbent assay (ELISA) (n = 1) [21], high-performance liquid chromatography-tandem mass spectrometry (HPLC TMS) (n = 1) [24], luminometric methylation assay (LUMA) (n = 1) [25], and bisulfite genomic sequencing (n = 1) [17].
3.1. Zinc level and DNA methylation with its consequences
3.1.1. Gene-specific DNA methylation
Results from human studies indicate that zinc has a heterogeneous effect on gene-specific DNA methylation, transcript levels and protein levels. In one study, of cells from two zinc-deficient breastfeeding mothers, DNA methylation level at CpG sites in zinc transporter genes (solute carrier 30, SLC30As genes) was dependent on cell type. Zinc deficiency exhibited a positive association with SLC30A5 methylation in the lymphoblasts of both subjects and a negative association in the fibroblasts of one subject [12]. Zinc deficiency had no association with the methylation of other SLC30As genes in the same subject. Changes in the pattern of DNA methylation were associated with a significant reduction in the transcript level of SLC30A5 and SLC30A6. SLC30A5 and SLC30A6 protein levels were significantly reduced in lymphoblasts in both individuals, whereas SLC30A6 protein levels were unaltered in fibroblasts. However, SLC30A1, SLC30A3, SLC30A4, SLC30A7, SLC30A9, and SLC30A10 mRNA expression levels in fibroblasts and lymphoblasts were not different from matching controls. These changes were associated with a reduction of zinc in the breast milk of subjects [12]. In another human study, dietary cadmium (Cd) was associated with hypomethylation of specific genes paternally expressed 3 (PEG3) and maternally expressed 3 (MEG3). Dietary zinc supplementation reverted the methylation level in these genes. In addition, supplemented zinc in pregnant women was associated with a higher birth weight of offspring [13].
In another study, treatment of human mesenchymal stem cells from adipose tissue with high and low levels of zinc in the culture medium showed different effects on telomerase reverse transcriptase (TERT) DNA methylation [15]. However, at high zinc levels, the majority of CpG sites were changed from unmethylated to methylated and the minority of CpG sites were changed from methylated to unmethylated. This change was associated with an elevation in TERT gene expression, increased telomerase activity and telomere length [15]. In cultured human peripheral blood mononuclear cells, another study found no change in the methylation levels of Interferon-gamma (IFN-γ) and interferon regulatory factor 4 (IRF4) in zinc supplemented, zinc deficient, or control cells. IRF-4 activates cytokine mRNA transcription by binding to the major histocompatibility complex I promoter. In comparison to higher zinc levels, lower zinc levels exhibited significantly less IFN- γ protein and much higher cytokine mRNA levels [14].
In another study, low zinc concentration had a heterogeneous effect on the methylation of interleukin 6 (IL-6) in lymphoblastoid cell lines and the human monocytic cell line THP-1 from both aged and young subjects [19]. In this study, cells from elderly subjects in zinc-deficient culture media exhibited significantly lower IL-6 promoter methylation. Interestingly cells from younger subjects exhibited hypermethylation on the IL-6 promoter. Global DNA methylation status in both groups remained unchanged.
Similar to human studies, the effect of zinc on gene-specific methylation was also heterogeneous in rodents. In rodents, zinc deficiency is often associated with an increase in the methylation status of specific genes. Two studies found a strong association between maternal zinc-deficient diet (<1 mg zinc/kg) and increased brain-derived neurotrophic factor (BDNF) methylation in the hippocampus of offspring. Methylation was associated with low levels of BDNF and DNMT3A mRNA, but higher levels of DNMT1A mRNA [16, 18]. Furthermore, low protein levels of BDNF, DNMT1, and growth arrest and DNA damage-inducible beta (GADD45b) were reported. However, a zinc adequate diet (30 mg zinc/kg) for pups from zinc-deficient mothers reduced the BDNF methylation and increased methyl CpG binding protein 2 (MeCP2) and GADD45b expression. This change was associated with an improvement in cognitive ability in rat pups [16]. Another study showed that feeding a low-zinc diet to pregnant mice increased methylation of the metallothionein (MT2) gene in the lower distal area of the offspring's liver, but did not affect methylation near responsive elements of this gene. The decline in MT2 mRNA expression was associated with this methylation alteration. A single dose of Cd was given to the pups as a hypomethylation-inducing agent. The Cd treatment reversed MT2 methylation and enhanced gene expression [17]. In another study, the spleens of older mice fed a zinc-deficient diet revealed lower IL-6 promoter methylation, which was associated with an increase in IL-6 and intercellular adhesion molecule 1 (ICAM1) expression, but these changes were not observed in younger mice [19]. This effect of zinc-deficient diet on IL-6 expression in young and old rodent cells contrasts with the effect seen in young and old human cells.
Generally, zinc deficiency in human subjects showed diverse effects with hypomethylation, hypermethylation or no change in specific gene DNA methylation status. In human studies, zinc addition mitigated the hypomethylation effect of Cd. In rodent studies, Cd mitigated the hypermethylation response of zinc deficiency and reduced the methylation status in specific genes. However, in rodents, zinc deficiency was associated with an increase in gene methylation and reduction of gene expression. Only one study found a decrease in gene methylation in response to zinc deficiency which was associated with overexpression of gene [19]. In the majority of cases, elevation in gene methylation was associated with a reduction of gene expression.
3.1.2. Global DNA methylation
In human studies, zinc level has shown an association with LINE-1 methylation, as a global DNA marker, but only one study showed a positive effect of zinc supplementation on Alu methylation. One study reported hypermethylation in global DNA within zinc-deficient children. Compared to a control group, zinc supplementation in adults did not affect LINE-1 methylation status. However, in this study, there was an elevation in the LINE-1 methylation in the population with a higher BMI than that found in the population with an average normal BMI. Zinc supplementation increased the Alu methylation with a positive association between height and Alu methylation [20]. A study with children aged 5–12 years found no association of serum zinc level with LINE-1 methylation [22]. Another study on 2–3 years old boys and girls showed a higher global DNA methylation in zinc-deficient subjects, which had a positive correlation between the elevation of global DNA methylation and the prevalence of stunting [21].
In cell culture studies from rodents, zinc deficiency reduced global DNA methylation in oocytes which was associated with an increase in transcription of some repetitive elements such as Lap, Line-1, Sineb1 and Sineb2 and a decreased transcription of Gdf9, Zp3 and Figla genes. However, depending on the severity of zinc deficiency there was a significant reduction in the ability of mature eggs to undergo cell division [23]. In mice, increased zinc levels and zinc deficiency in female mice had an impact on global DNA methylation in the livers of pups, which was associated with a considerable increase in the expression of DNMT1 and DNMT3 when compared to control mice [25]. However, zinc treatment showed to mitigate the effect of demethylation agents. A study reported the methylation restoring effect of zinc on rat sperm pre-treated with the combination of chemotherapeutic agents bleomycin, etoposide, and cisplatin that reduced global DNA methylation [26]. Although, zinc treatment improved protamination, which was still lower than the control group. Zinc treatment of chemotherapeutic agent pre-treated cells also showed a reduced DNA fragmentation compared to cells not pre-treated. Zinc treatment partially restored body and testis weight with numbers of spermatogonia and Leydig cells [26]. Another rodent study found that neither lower nor higher zinc supplements affected global DNA methylation in the liver, jejunum, kidney and cortex of male mice. But, splenomegaly and genomic instability in the liver were observed [24].
4. Discussion
This review explored the association of extracellular zinc treatment with DNA methylation in humans and rodents. Our review includes an analysis of specific gene and global DNA methylation studies conducted on whole organisms and cells. A total of 15 articles were included in this review. In both human and rodent studies, SLC30A5, SLC30A6, PLAGL1, PEG3, MEG3, IFN-γ, IRF-4, hTERT, BDNF, MT2, IL-6, LINE-1, and Alu DNA are presented. The zinc levels were measured or estimated in venous blood, plasma, food frequency questionnaire (FFQs), diet and cell culture media. Some in vivo studies in humans estimate zinc levels in the blood using solution-based ICP-MS methods, atomic absorption technique, and flame atomic absorption spectrophotometry. Zinc levels in most studies were estimated typically in the blood and the DNA methylation was measured in other cell types, however, it is reported that the methylation level of a gene in the blood can reflect its status in other cells [28]. The methylation of genes in human studies was examined in fibroblast, lymphoblast, venous blood, umbilical cord blood, blood PBMC, lymphoblastoid cell lines and human monocytic cell line THP-1 and mesenchymal stem cells from adipose tissue. However, the cells used in animal studies were oocytes, Jejunum, cortex, kidney, liver, hippocampus, blood, sperm and spleens.
To assess DNA methylation at a specific locus, three major experimental approaches are used: methods based on bisulfite conversion, restriction enzyme-based procedures, and affinity enrichment-based approaches [29]. The four human studies in this review employed pyrosequencing to determine the methylation status of specific genes. Bisulfite-converted genomic DNA sequencing is frequently used, whereas pyrosequencing is considered as a gold standard. Two studies used methylation-specific PCR to determine gene-specific DNA methylation and one study used a 5-mC DNA ELISA kit. Other methods used were Immuno-fluorescence, methylation-specific PCR, High-Performance Liquid Chromatography-Tandem Mass Spectrometry, and bisulfite genomic sequencing.
Hypermethylation of genes was frequently associated with lower mRNA levels and hypomethylation with an increase in gene expression and, subsequently, downregulation and upregulation of related proteins, respectively. However, some studies showed no correlation between change in gene methylation and its expression level though the protein level was altered, which suggests a post-translational effect of zinc.
4.1. Gene-specific studies
In candidate gene studies, higher and lower zinc levels in both humans and rodents showed diverse associations with gene-specific DNA methylation. Most of the studies also examined the consequences of specific-gene methylation of target genes in response to zinc levels and its effect on DNA methylation, including gene expression and protein levels. Only one study did not examine the consequence of DNA methylation on gene expression or protein level [13]. While it is commonly accepted that hypermethylation of DNA at the gene promoter represses transcription, there are several documented exceptions, DNA methylation can either suppress or enhance gene expression depending on the cell type [30]. In zinc-induced changes in gene methylation, the majority of studies reviewed here show that gene expression levels were downregulated in hypermethylated genes. Only one study reported high levels of protein expressed after its cognate gene showed increased gene methylation [14].
The effect of zinc level on DNA methylation was quite variable, depending on cell type and tissue type. All human studies reported a direct association of zinc level with gene-specific DNA methylation. Zinc levels in in-vivo studies were assumed using dietary questionnaires or by measuring blood zinc levels. In in-vitro studies, lower and higher concentrations of zinc were used. Hypomethylations in SLC30A5 [12] and IL-6 genes (in aged subjects) [19] were observed in two studies in zinc-deficient adults and children. Similarly, another study reported hypermethylation in higher zinc levels in TERT [15]. However, higher zinc concentration mitigated the hypomethylation of PEG3 gene [13], caused by Cd, and elevated DNA methylation. Contrary to the above results, one study did not find any association between either low or high zinc levels with IFN-γ gene methylation levels.
Though, human studies showed specific effects of zinc on specific genes in specific tissues. Zinc deficiency showed a reduction in SLC30A5 methylation in lymphoblasts compared to fibroblasts in the same subject. However, in this study in both tissues the methylation status of another gene, SLC30A6, was not changed in both cells [12]. Nevertheless, the methylation of IL-6 promoter in zinc-deficient human lymphoblastoid and monocytic cell line THP-1 in aged but not in younger subjects showed a significant reduction, though, the total DNA methylation in both cell lines was not affected in both subjects [19]. This result showed an age-specific effect of zinc on gene-specific methylation. Correspondingly, age-related zinc deficiency has been suggested as a possible contributor to immunological dysfunction and chronic inflammation during the ageing process [31]. The reduction in the methylation of IL-6 had a positive association with IL-6 protein production. Moreover, the cell activation markers such as ICAM1, MHC class II and CD86 were also upregulated in THP1 cells [19].
Mothers with a lower level of zinc and a high level of Cd represented a reduction in DNA methylation in the pleiomorphic adenoma-like protein 1 (PLAGL1) gene in offspring. Elevation of zinc increased PEG3 (parentally expressed 3) gene methylation but had no association with the methylation of MEG3 (maternally expressed 3). In this study, subjects with a higher concentration of Cd and lower zinc showed an association with lower birth weight compared with higher zinc and higher Cd concentrations. PEG3 is a paternal gene that encodes a zinc-finger protein with tumour suppressive action, that helps to improve apoptosis through p53/c-myc [32]. PEG3 is controlled by allele-specific DNA methylation. The PEG3 regulatory differentially methylated region (DMR), which is considered to impede the proapoptotic activity of this gene, results in a reduction in PEG3 transcription. Aberrant methylation in this DMR has been related to reduced PEG3 expression, which was found in 18 cancer types, including ovarian and endometrial cancers [32, 33]. Zinc finger protein PLAGL1 is a paternally expressed gene that is methylated in the female germline and is important for regulating several genes required for the growth and development of the mice foetus or placenta. PLAGL1 biallelic expression has been linked to transitory newborn hyperglycaemia and severe intrauterine growth retardation in humans [34]. Unlike PEG3, zinc deficiency did not affect the methylation level of the human MEG3 gene which is found in the paraxial mesoderm, the developing central nervous system, and the epithelia of the salivary glands, kidneys, and pancreas throughout development [35].
IFN-γ is a proinflammatory cytokine that stimulates effector immune cells and improves antigen presentation in vivo. Natural killer cells release IFN-γ as part of the innate immune response, while CD4+ Th1 cells and CD8+ cytotoxic T lymphocytes secrete IFN-γ after adaptive immunity has been initiated [36]. IFN-γ stimulates cytotoxic T cells and promotes cell-mediated cytotoxicity, and, IFN-γ gene expression is linked to the methylation of the IFN-γ promoter. Methylation of the IFN- γ -54 CpG in PBMC has been shown to reduce promoter activity by 60–80% [37]. In contrast to the above studies, zinc deficiency and zinc supplementation in cultured PBMC did not affect the methylation level of the human IFN-γ promoter region compared to zinc adequate cells. Surprisingly, no change in the methylation status of the IFN-γ gene was associated with a significant increase in the cytokine’s mRNA and a reduction of IFN-γ protein level compared with zinc supplemented cells and suggesting that this effect probably be related to the post-transcription effect of zinc on IFN-γ [14].
Changes in gene methylation status have been linked to changes in gene expression. The expression of SLC30A5 and SLC30A6 was downregulated in lymphoblasts due to differences in their methylation state. However, there were no changes in the methylation and the expression of other zinc transporter genes in response to zinc deficiency. SLC30A5 and SCL30A6 are enterocyte transporters of zinc [38]. SLC30A5 gene coding zinc transporter ZnT5 has an important role in absorbing dietary zinc, especially for the foetus. On the other hand, SLC30A6 coding zinc transporter ZnT6 is expressed at low levels in the intestine and is essential for zinc transportation from cells to the body [38, 39, 40]. Alteration in the expression of zinc transporters in the lymphoblasts of zinc-deficient lactating women was shown to be associated with a reduction of zinc concentration in breast milk, which caused the offspring to be zinc-deficient [12].
On the other hand, in response to higher zinc concentration, most unmethylated CpG sites in the TERT gene were methylated but fewer methylated sites changed to unmethylated, and there was no change in some other CpG sites [15]. Despite these findings, the result showed a positive effect of zinc on the methylation of human TERT in cultured mesenchymal stem cells. The positive effect of zinc on TERT mRNA expression was associated with an increase in concentration-dependent telomerase activity. In line with this, the expression of ALP, OCN, PPAR-α, and PPAR-γ was also increased. Elevation in the expression of the TERT gene raised telomerase activity which resulted from an increase in telomere length. TERT is the main protein component of telomerase, which is responsible for the elongation of telomeres. In somatic cells, TERT gene expression is significantly associated with telomerase activity. Telomere length is a measure of cellular replication ability. Telomere dysfunction is caused by very short telomeres [41]. TERT promoter methylation and mutation were found in 53% and 31% of TERT-expressing cancer cell lines, respectively, and suggested that epigenetic changes are linked to TERT dysregulation and cellular transformation [42].
Zinc levels were manipulated by feeding zinc deficient, zinc adequate or higher zinc content diets to rodents. An inverse association of zinc level with gene-specific DNA methylation was found in these studies.
Zinc deficiency in rodents showed a significant increase in DNA methylation. Zinc deficiency significantly increased DNA methylation in the BDNF IX in the pup’s hippocampus [25]. Similarly, zinc supplementation to pups of zinc-deficient mothers, from the age of 21 days–60 days was found to reduce the DNA methylation of BDNF exon IX. It is suggested that zinc supplementation may be beneficial for early life zinc deficiency impairment [16]. BDNF is a major growth factor in the neurotrophins family [43]. In addition to neurons and astrocytes, serotonergic and noradrenergic systems, BDNF is extensively expressed in the CNS, the intestines and other tissues. BDNF is also expressed in endothelium and mononuclear blood cells [43, 44]. BDNF regulates learning and memory, as well as synaptic plasticity. Growing data suggest that DNA methyltransferase and methyl-binding proteins may influence BDNF gene expression. DNA methylation and other epigenetic changes are important in the activity-dependent regulation of the BDNF gene [18]. Zinc supplementation has been shown to regulate BDNF expression [45]. Hypermethylation of BNDF promoter in zinc-deficient pups was associated with a reduction in the expression of BDNF mRNA and DNMT3A mRNAs, whereas the DNMT1 mRNA was increased [25]. The protein level of BDNF, DNMT1 and GADD455b in neonates were reduced. Feeding a zinc-adequate diet at age 21 to 60, improved BDNF expression but the MECP2 and GADD45b expression were still significantly lower compared to the zinc-adequate group. In agreement with this study, another study also found the regulatory effect of zinc on BDNF expression [45]. Similarly, the reduction of zinc in cultured hippocampal neurons significantly reduced the DNMT3a, DNMT1, BDNF, and GADD45b protein levels whereas the MeCP2 was increased. In this study, the expression of DNMT3a and DNMT1 showed a reduction, while the expression of MeCP2, GADD45b and BDNF were upregulated [46].
Likewise, the liver of offspring from zinc-deficient pregnant mice showed hypermethylation in the -820 CpG region of the MT2 gene but no methylation in the -480 to +140 region of this gene which contains metal responsive elements (MREs) [17]. MT has been shown to engage in heavy metal ions, such as zinc and copper transport, metabolism and homeostasis in tissue and cells [47]. MT methylation intensity was shown to be linked with the downregulation of gene expression [48]. The demethylation of CpG sites near MREs in the MT gene was associated with higher expression of MT in mouse lymphosarcoma cells [49]. However, a change in the methylation level of the MT2 gene was associated with a reduction in its mRNA expression, where Cd influenced the expression of MT1 but had no effect on the expression of MT2 17.
In contrast, lower zinc levels in aged animals had a reduction in the methylation of IL-6 promoter in a lymphoblastoid cell line. But, in young animals, there was an increase in methylation of IL-6 promoter by 8 to 10-fold. In this study, no significant change in total DNA methylation level has been found in both groups [19]. IL-6 is generated quickly and transiently to help protect the host by stimulating acute phase response, haematopoiesis, and immunological responses to infection and tissue damage [50]. Reduction in the methylation of IL-6 had a positive association with IL-6 protein production. Moreover, the cell activation markers such as ICAM1, MHC class II and CD86 were also upregulated in THP1 cells [19].
Overall, the results of both human and rodent studies showed a heterogeneous association between zinc level and gene-specific DNA methylation. However, most human gene-specific studies showed a positive association of zinc with elevation in DNA methylation, while, zinc deficiencies were correlated with hypomethylation. The effect of zinc was typically cell-specific and in some cases, zinc deficiency or zinc supplementation had a diverse effect on different genes, while, zinc did not affect the methylation level of some other genes. Contrariwise, most rodent studies showed a negative association between zinc in gene-specific DNA methylation. In rodents studies, zinc deficiency resulted in hypermethylation, whereas zinc supplementation resulted in hypomethylation in particular genes. Fewer rodent studies showed the elevation of DNA methylation in genes with a lower level of zinc compared with control, whereas the effect was age-dependent. Generally, in most studies, we found a reduction in specific-gene expression in response to the elevation in DNA methylation.
4.2. Global DNA methylation
From the total of seven studies (three on humans and four on rodents) that examined the effect of zinc on global DNA methylation, five used the term global DNA methylation with no specification of gene region, one study analysed LINE-1, and one analysed both LINE-1 and Alu elements. Approximately 17% and 11% of total DNA is composed of repeating elements LINE-1 and Alu in the human genome, respectively [51]. Approximately 80% of CpG islands occur in LINEs and SINEs (short interspersed nuclear elements). The degree of global DNA methylation can be linked to functional implications, such as increased mutational events, genomic instability, or changed gene expression. Global hypomethylation within repetitive regions throughout the genome can lead to gene overexpression and transposable element activation, both of which can contribute to illness [52].
To determine if there is an association of zinc with global DNA methylation in adults and children, zinc levels were estimated using food frequency questionnaires. No association was found between zinc level and LINE-1 methylation in peripheral leukocytes of adult male and female individuals but displayed an elevation in the methylation of Alu element [20]. Similarly, no association was found between serum zinc level and LINE-1 methylation in male and female school-age children. However, generally LINE-1 methylation level of girls was lower compared to the boys [22]. The lower level of zinc intake for stunted and higher of zinc for no-stunted children showed a seemingly higher but statistically not significant zinc level and low global DNA methylation. There was a significant increase in global DNA methylation in children with lower zinc intake showed a positive correlation with the prevalence of stunting [21]. Elevation in the LINE-1 methylation in the adult population was associated with higher BMI. A zinc supplementation diet increased the Alu methylation and there is a positive correlation has been shown between height and Alu methylation [20].
Overall, there was no association between zinc deficiency and LINE-1 methylation level in human studies. However, one study showed elevation in the DNA methylation of Alu element in zinc-deficient subjects [20] and one study reported increased global DNA methylation in children with low zinc intake [21].
To find the association of zinc level with global DNA methylation in rodents studies, animals were fed zinc deficient, zinc adequate or higher zinc diets. A significant reduction in global DNA methylation was observed in zinc-deficient female CD1 mice oocytes fed less than 1 mg zinc/kg compared to control mice oocytes fed 29 mg zinc/kg [23]. Changes in global DNA methylation were associated with an increase in transcription of some repetitive elements such as Iap, Line-1, Sineb1 and Sineb2 and a decrease in transcription of Gdf9, Zp3 and Figla. However, depending on the intensity of zinc deficiency there was a significant reduction for the mature eggs to reach to divided into two cells [23]. Contrariwise, the liver of foetal mice of mothers fed zinc supplemented (150 μg/g) and zinc restricted (15 μg/g) diet during pregnancy for 17 days, showed significantly elevated levels of global DNA methylation compared to the foetus of mothers fed zinc adequate (50 μg/g) diets. An elevated level of zinc was associated with a significant increase in DNMT1 and DNMT3 expression compared to control [25]. Likewise, male Wister rats were treated with a combination of chemotherapeutic agents (bleomycin, etoposide, and cisplatin) and chemotherapeutic agents + zinc compared to zinc or saline-treated animals showed an increase in global DNA methylation in their sperm. Zinc treatment improved protamination on rat sperm which was still lower than the control group. Zinc treatment showed a reduced DNA fragmentation compared to chemotherapeutic agents treated group and also exhibited recovery of body and testis weight with an increase in spermatogonia and Leydig cells [26]. In contrast, a low zinc diet containing 4.7 mg/kg of zinc in male mice showed no effect on the global DNA methylation and the hydroxymethylation level in the liver and other tissues. But, splenomegaly and genomic instability in the liver of mice were reported [24].
Overall, the effect of zinc in rodent studies was mostly cell-dependent. Decreased global DNA methylation was associated with rodents fed a zinc-deficient diet. Both higher and lower levels of zinc correlated with an increase in global DNA methylation. Zinc supplementation in cultured rodent cells restored global DNA methylation that was lost upon pre-treatment with chemotherapeutic agents. One study showed no effect of lower zinc levels on global DNA methylation in the liver and other tissues of male mice.
4.3. Strengths and limitations
This review summarizes studies reporting an association between zinc and DNA methylation in humans and rodents. Study population, targeted gene, types of tissues, methylation assessment technique, the consequence of DNA methylation, and consideration of downstream effects, were evaluated. However, there are some limitations to be aware of. First and foremost, the heterogeneity of research parameters and the low member of studies in the literature prevented us from performing a meta-analysis. Second, variability of tissue type, research design, measured outcomes, comparison groups, analytical methods, and sample sizes may result in inconsistent associations between zinc level and DNA methylation. Third, only English databases were searched, which may have limited the number of qualified articles.
Overall, we found that in most cases zinc was associated with either hyper- or hypomethylation of nuclear DNA in specific genes and global DNA. Future studies should investigate the zinc-mediated shift in methylation levels using longitudinal prospective study methodologies. Gene expression and protein level measurements should be incorporated into zinc studies to more deeply understand the consequences of gene methylation alteration by zinc.
Declarations
Author contribution statement
Ziauddin Azimi: Conceived and designed the experiments; Performed the experiment; Analyzed and interpreted the data; Contributed reagents, materials, analysis tools or data; Wrote the paper.
Mohamad Rodi Isa, Zaliha Ismail: Analyzed and interpreted the data; Contributed reagents, materials, analysis tools or data; Wrote the paper.
Jesmine Khan, Seok Mui Wang: Analyzed and interpreted the data; Wrote the paper.
Funding statement
This research did not receive any specific grant from funding agencies in the public, commercial, or not-for-profit sectors.
Data availability statement
Data included in article/supp. material/referenced in article.
Declaration of interests statement
The authors declare no conflict of interest.
Additional information
No additional information is available for this paper.
Acknowledgements
The authors would like to thank Jamil Momand for his thorough review of the article, as well as BioRender for allowing us to employ their tool to create the graphical model.
Appendix A. Supplementary data
The following is the supplementary data related to this article:
References
- 1.Ho E., Beaver L.M., Williams D.E., Dashwood R.H. Dietary factors and epigenetic regulation for prostate cancer prevention. Adv. Nutr. 2011;2:497–510. doi: 10.3945/an.111.001032. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Ogawa Y., Kawamura T., Shimada S. Zinc and skin biology. Arch. Biochem. Biophys. 2016;611:113–119. doi: 10.1016/j.abb.2016.06.003. [DOI] [PubMed] [Google Scholar]
- 3.Sanusi K.O., et al. Effect of maternal zinc deficiency on offspring health: the epigenetic impact. J. Trace Elem. Med. Biol. 2021;65 doi: 10.1016/j.jtemb.2021.126731. [DOI] [PubMed] [Google Scholar]
- 4.Yusuf A.P., et al. Zinc metalloproteins in epigenetics and their crosstalk. Life. 2021;11:186. doi: 10.3390/life11030186. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Keil K.P., Lein P.J. DNA methylation: a mechanism linking environmental chemical exposures to risk of autism spectrum disorders? Environ. Epigenetics. 2016;2:1–15. doi: 10.1093/eep/dvv012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Mandaviya P.R., et al. Association of dietary folate and Vitamin B-12 intake with genome-wide DNA methylation in blood: a large-scale epigenome-wide association analysis in 5841 individuals. Am. J. Clin. Nutr. 2019;110:437–450. doi: 10.1093/ajcn/nqz031. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.McGee M., Bainbridge S., Fontaine-Bisson B. A crucial role for maternal dietary methyl donor intake in epigenetic programming and fetal growth outcomes. Nutr. Rev. 2018;76:469–478. doi: 10.1093/nutrit/nuy006. [DOI] [PubMed] [Google Scholar]
- 8.Łoboś P., Regulska-Ilow B. Link between methyl nutrients and the DNA methylation process in the course of selected diseases in adults. Rocz. Państwowego Zakładu Hig. 2021;72:123–136. doi: 10.32394/rpzh.2021.0157. [DOI] [PubMed] [Google Scholar]
- 9.O’Neill R.J., Vrana P.B., Rosenfeld C.S. Maternal methyl supplemented diets and effects on offspring health. Front. Genet. 2014;5 doi: 10.3389/fgene.2014.00289. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Bogdanović O., Veenstra G.J.C. DNA methylation and methyl-CpG binding proteins: developmental requirements and function. Chromosoma. 2009;118:549–565. doi: 10.1007/s00412-009-0221-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Page M.J., et al. The PRISMA 2020 statement: an updated guideline for reporting systematic reviews. BMJ. 2021;n71 doi: 10.1136/bmj.n71. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Kumar L., et al. Altered expression of two zinc transporters, SLC30A5 and SLC30A6, underlies a mammary gland disorder of reduced zinc secretion into milk. Genes Nutr. 2015;10:38. doi: 10.1007/s12263-015-0487-x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Vidal A.C., et al. Maternal cadmium, iron and zinc levels, DNA methylation and birth weight. BMC Pharmacol. Toxicol. 2015;16:20. doi: 10.1186/s40360-015-0020-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Rodenkirchen V., Schettgen T., Rink L. Zinc deficiency impairs interferon-γ production on post-transcriptional level. J. Trace Elem. Med. Biol. 2020;62 doi: 10.1016/j.jtemb.2020.126598. [DOI] [PubMed] [Google Scholar]
- 15.Farahzadi R., Fathi E., Mesbah-Namin S.A., Zarghami N. Zinc sulfate contributes to promote telomere length extension via increasing telomerase gene expression, telomerase activity and change in the TERT gene promoter CpG island methylation status of human adipose-derived mesenchymal stem cells. PLoS One. 2017;12 doi: 10.1371/journal.pone.0188052. [DOI] [PMC free article] [PubMed] [Google Scholar] [Retracted]
- 16.Jiang Y.-G.Y.G., Wang Y., hui Y.-H.Y., Zhang H., Wang Z.-Y.Z.-Y.Y., Liu Y.-Q.Y.-Q.Q. Effects of early-life zinc deficiency on learning and memory in offspring and the changes in DNA methylation patterns. Nutr. Neurosci. 2020:1–10. doi: 10.1080/1028415X.2020.1831259. [DOI] [PubMed] [Google Scholar]
- 17.Kurita H., Ohsako S., Hashimoto S., Yoshinaga J., Tohyama C. Prenatal zinc deficiency-dependent epigenetic alterations of mouse metallothionein-2 gene. J. Nutr. Biochem. 2013;24:256–266. doi: 10.1016/j.jnutbio.2012.05.013. [DOI] [PubMed] [Google Scholar]
- 18.Hu Y.D.Y.-D., et al. The cognitive impairment induced by zinc deficiency in rats aged 0∼2 months related to BDNF DNA methylation changes in the hippocampus. Nutr. Neurosci. 2017;20:519–525. doi: 10.1080/1028415X.2016.1194554. [DOI] [PubMed] [Google Scholar]
- 19.Wong C.P., Rinaldi N.A., Ho E. Zinc deficiency enhanced inflammatory response by increasing immune cell activation and inducing IL6 promoter demethylation. Mol. Nutr. Food Res. 2015;59:991–999. doi: 10.1002/mnfr.201400761. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Perng W., et al. Dietary intake, plasma homocysteine, and repetitive element DNA methylation in the Multi-Ethnic Study of Atherosclerosis (MESA) Nutr. Metabol. Cardiovasc. Dis. 2014;24:614–622. doi: 10.1016/j.numecd.2013.11.011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Iqbal M.S., et al. Lower intakes of protein, carbohydrate, and energy are associated with increased global DNA methylation in 2- to 3-year-old urban slum children in Bangladesh. Matern. Child Nutr. 2019;15:1–9. doi: 10.1111/mcn.12815. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Perng W., et al. Micronutrient status and global DNA methylation in school-age children. Epigenetics. 2012;7:1133–1141. doi: 10.4161/epi.21915. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Tian X., Diaz F.J. Acute dietary zinc deficiency before conception compromises oocyte epigenetic programming and disrupts embryonic development. Dev. Biol. 2013;376:51–61. doi: 10.1016/j.ydbio.2013.01.015. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Finke H., et al. Effects of a cumulative, suboptimal supply of multiple trace elements in mice: trace element status, genomic stability, inflammation, and epigenetics. Mol. Nutr. Food Res. 2020;64 doi: 10.1002/mnfr.202000325. [DOI] [PubMed] [Google Scholar]
- 25.Davison G., Mathers J.C., Ford D., Valentine R.A. Effects of dietary zinc supply during pregnancy on global DNA methylation. Proc. Nutr. Soc. 2010;69:E17. [Google Scholar]
- 26.Khadivi F., Razavi S., Hashemi F. Protective effects of zinc on rat sperm chromatin integrity involvement: DNA methylation, DNA fragmentation, ubiquitination and protamination after bleomycin etoposide and cis-platin treatment. Theriogenology. 2020;142:177–183. doi: 10.1016/j.theriogenology.2019.09.039. [DOI] [PubMed] [Google Scholar]
- 27.Wong C.P., Rinaldi N.A., Ho E. 2015. Zinc Deficiency Enhanced Inflammatory Response by Increasing Immune Cell Activation and Inducing IL6 Promoter Demethylation. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Kundakovic M., et al. DNA methylation of BDNF as a biomarker of early-life adversity. Proc. Natl. Acad. Sci. U.S.A. 2015;112:6807–6813. doi: 10.1073/pnas.1408355111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Pajares M.J., et al. Methods for analysis of specific DNA methylation status. Methods. 2021;187:3–12. doi: 10.1016/j.ymeth.2020.06.021. [DOI] [PubMed] [Google Scholar]
- 30.Ions L.J., et al. Effects of Sirt1 on DNA methylation and expression of genes affected by dietary restriction. Age (Omaha). 2013;35:1835–1849. doi: 10.1007/s11357-012-9485-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Wong C.P., Ho E. Zinc and its role in age-related inflammation and immune dysfunction. Mol. Nutr. Food Res. 2012;56:77–87. doi: 10.1002/mnfr.201100511. [DOI] [PubMed] [Google Scholar]
- 32.Zhang M., Zhang J. PEG3 mutation is associated with elevated tumor mutation burden and poor prognosis in breast cancer. Biosci. Rep. 2020;40:1–9. doi: 10.1042/BSR20201648. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Nye M.D., et al. Associations between methylation of paternally expressed gene 3 (PEG3), cervical intraepithelial neoplasia and invasive cervical cancer. PLoS One. 2013;8 doi: 10.1371/journal.pone.0056325. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Vincent R.N., Gooding L.D., Louie K., Chan Wong E., Ma S. Altered DNA methylation and expression of PLAGL1 in cord blood from assisted reproductive technology pregnancies compared with natural conceptions. Fertil. Steril. 2016;106:739–748. doi: 10.1016/j.fertnstert.2016.04.036. [DOI] [PubMed] [Google Scholar]
- 35.Hsieh P.F., Yu C.C., Chu P.M., Hsieh P.L. Long non-coding rna meg3 in cellular stemness. Int. J. Mol. Sci. 2021;22:1–11. doi: 10.3390/ijms22105348. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Merli P., Quintarelli C., Strocchio L., Locatelli F. The role of interferon-gamma and its signaling pathway in pediatric hematological disorders. Pediatr. Blood Cancer. 2021;68:1–11. doi: 10.1002/pbc.28900. [DOI] [PubMed] [Google Scholar]
- 37.Hashimoto H., et al. Association of IFNG gene methylation in peripheral blood cells with the development and prognosis of autoimmune thyroid diseases. Cytokine. 2019;123 doi: 10.1016/j.cyto.2019.154770. [DOI] [PubMed] [Google Scholar]
- 38.Ford D. Intestinal and placental zinc transport pathways. Proc. Nutr. Soc. 2004;63:21–29. doi: 10.1079/PNS2003320. [DOI] [PubMed] [Google Scholar]
- 39.Adelino J., et al. A genetic variant within SLC30A6 has a protective role in the severity of rheumatoid arthritis. Scand. J. Rheumatol. 2017;46:326–327. doi: 10.1080/03009742.2016.1209551. [DOI] [PubMed] [Google Scholar]
- 40.Jackson K.A., Valentine R.A., Coneyworth L.J., Mathers J.C., Ford D. Mechanisms of mammalian zinc-regulated gene expression. Biochem. Soc. Trans. 2008;36:1262–1266. doi: 10.1042/BST0361262. [DOI] [PubMed] [Google Scholar]
- 41.Denham J., Sellami M. Exercise training increases telomerase reverse transcriptase gene expression and telomerase activity: a systematic review and meta-analysis. Ageing Res. Rev. 2021;70 doi: 10.1016/j.arr.2021.101411. [DOI] [PubMed] [Google Scholar]
- 42.Dogan F., Forsyth N.R. Telomerase regulation: a role for epigenetics. Cancers. 2021;13:1–24. doi: 10.3390/cancers13061213. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Rahmani M., Rahmani F., Rezaei N. The brain-derived neurotrophic factor: missing link between sleep deprivation, insomnia, and depression. Neurochem. Res. 2020;45:221–231. doi: 10.1007/s11064-019-02914-1. [DOI] [PubMed] [Google Scholar]
- 44.Bathina S., Das U.N. Brain-derived neurotrophic factor and its clinical Implications. Arch. Med. Sci. 2015;11:1164–1178. doi: 10.5114/aoms.2015.56342. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Yu X.D., Ren T.H., Yu X.G. Disruption of calmodulin-dependent protein kinase II α/brain-derived neurotrophic factor (α-CaMKII/BDNF) signalling is associated with zinc deficiency-induced impairments in cognitive and synaptic plasticity. Br. J. Nutr. 2013;110:2194–2200. doi: 10.1017/S0007114513001657. [DOI] [PubMed] [Google Scholar]
- 46.He C., cong C.-C.C., et al. DNA methylation mechanism of intracellular zinc deficiency-induced injury in primary hippocampal neurons in the rat brain. Nutr. Neurosci. 2018;21:478–486. doi: 10.1080/1028415X.2017.1312090. [DOI] [PubMed] [Google Scholar]
- 47.Yang L., et al. Cardiac-specific overexpression of metallothionein attenuates L-NAME-induced myocardial contractile anomalies and apoptosis. J. Cell Mol. Med. 2019;23:4640–4652. doi: 10.1111/jcmm.14375. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Maleckaite R., et al. DNA methylation of metallothionein genes is associated with the clinical features of renal cell carcinoma. Oncol. Rep. 2019;41:3535–3544. doi: 10.3892/or.2019.7109. [DOI] [PubMed] [Google Scholar]
- 49.Ogushi S., Yoshida Y., Nakanishi T., Kimura T. CPG site-specific regulation of metallothionein-1 gene expression. Int. J. Mol. Sci. 2020;21:1–9. doi: 10.3390/ijms21175946. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Tanaka T., Narazaki M., Kishimoto T. IL-6 in inflammation, immunity, and disease. Cold Spring Harbor Perspect. Biol. 2014;6:a016295. doi: 10.1101/cshperspect.a016295. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Lisanti S., et al. Comparison of methods for quantification of global DNA methylation in human cells and tissues. PLoS One. 2013;8 doi: 10.1371/journal.pone.0079044. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Stanković A. DNA Methylation Mechanism. IntechOpen; 2020. Global DNA methylation as a potential underlying mechanism of congenital disease development. [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Data Availability Statement
Data included in article/supp. material/referenced in article.