Abstract
Recently, we reported that TMEM163 is a zinc efflux transporter that likely belongs to the mammalian solute carrier 30 (Slc30/ZnT) subfamily of the cation diffusion facilitator (CDF) protein superfamily. We hypothesized that human TMEM163 forms functional heterodimers with certain ZNT proteins based on their overlapping subcellular localization with TMEM163 and previous reports that some ZNT monomers interact with each other. In this study, we heterologously expressed individual constructs with a unique peptide tag containing TMEM163, ZNT1, ZNT2, ZNT3, and ZNT4 (negative control) or co-expressed TMEM163 with each ZNT in cultured cells for co-immunoprecipitation (co-IP) experiments. We also co-expressed TMEM163 with two different peptide tags as a positive co-IP control. Western blot analyses revealed that TMEM163 dimerizes with itself but that it also heterodimerizes with ZNT1, ZNT2, ZNT3, and ZNT4 proteins. Confocal microscopy revealed that TMEM163 and ZNT proteins partially co-localize in cells, suggesting that they exist as homodimers and heterodimers in their respective subcellular sites. Functional zinc flux assays using Fluozin-3 and Newport Green dyes show that TMEM163/ZNT heterodimers exhibit similar efflux function as TMEM163 homodimers. Cell surface biotinylation revealed that the plasma membrane localization of TMEM163 is not markedly influenced by ZNT co-expression. Overall, our results show that the interaction between TMEM163 and distinct ZNT proteins is physiologically relevant and that their heterodimerization may serve to increase the functional diversity of zinc effluxers within specific tissues or cell types.
Keywords: SV31, ZnT1, ZnT2, ZnT3, ZnT4, Zinc
Graphical abstract
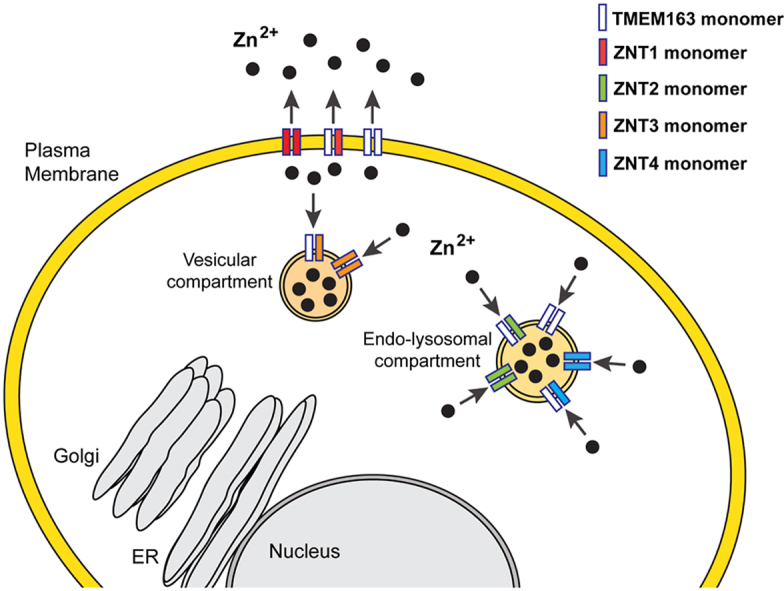
Highlights
-
•
TMEM163 protein heterodimerizes with ZNT1, ZNT2, ZNT3 and ZNT4 zinc efflux transporters.
-
•
Partial co-localization of TMEM163 and ZNT proteins in cells suggests distinct roles as homodimers and heterodimers.
-
•
Zinc efflux activity of TMEM163 or ZNT protein homodimers did not differ from their TMEM163/ZNT heterodimer counterparts.
-
•
TMEM163/ZNT heterodimerization attests to the role of TMEM163 as a bona fide SLC30 protein family member.
1. Introduction
Rodent Transmembrane 163 (Tmem163) protein was initially coined as synaptic vesicle 31 (Sv31), based on its identification from rat brain synaptosomes using proteomics and calculated molecular weight [1]. Human TMEM163 and mouse Tmem163 transcripts are detected in many tissues, notably in the brain, lung, pancreas, kidney, ovary, and testis [[2], [3], [4]]. Heterologous expression of TMEM163 in cultured HEK-293 cells exhibits localization within plasma membrane (PM), lysosomes, and vesicular compartments [[2], [3], [4]]. Similarly, immunocytochemistry and subcellular fractionation studies of cultured PC-12 cells stably expressing Tmem163 show that it is detected in the PM, lysosomes, early endosomes, and synaptic vesicles [5]. It was also reported that Tmem163 localizes within platelet dense granules (DG) and insulin granules, suggesting an important role in DG biogenesis [6] and insulin packaging or secretion [4]. Rodent Tmem163 has been shown to bind and transport zinc as a dimer in a proton-dependent manner [1,5,7] and we recently reported that human TMEM163 is a zinc efflux transporter [3]. Based on its function and subcellular localization, we proposed that TMEM163 should be classified as a member of the mammalian ZnT (Slc30, solute carrier 30) subfamily of the cation diffusion facilitator (CDF) protein superfamily [8]. The human ZNT (SLC30) family consists ten highly conserved, proton-dependent zinc transport proteins, namely, ZNT1 to ZNT10 [[9], [10], [11]]. The ZNT proteins efflux zinc from the cytoplasm into the extracellular space and into the lumen of membrane-bound compartments. ZNTs have predicted six transmembrane domains (TMD), but the structure of one of its members, ZNT8, was recently solved using cryo-electron microscopy, confirming six TMD with intracellular N- and C-termini regions [12].
Our interests in understanding the pathology of Mucolipidosis type IV (MLIV) disease, which is caused by the loss of Mucolipin-1 (TRPML1) protein function [[13], [14], [15]], led us to discover that zinc dyshomeostasis is present in MLIV patient cells, as well as cell culture and mouse models of MLIV disease [16]. We subsequently identified human TMEM163 protein as an interaction partner of the TRPML1 ion channel [2]. We proposed that the abnormal zinc metabolism in MLIV disease is possibly due to disrupted TMEM163 and TRPML1 interaction since TMEM163 is a zinc efflux transporter [3] that shuttles zinc into lumenal compartments while TRPML1 is permeable to zinc [17] that may serve as a lysosomal zinc release channel [18] especially in neurons [19]. The loss of TMEM163 and TRPML1 interaction could explain intracellular zinc accumulation in MLIV cells, particularly that RNA interference (RNAi)-induced knockdown (KD) of TMEM163 and TRPML1 in HEK-293 cells shows cytoplasmic zinc elevation, while over-expression of both proteins exhibits an opposite effect [2]. Further, KD or knockout (KO) of mouse Tmem163 in MIN6 cells and platelets, respectively, also produces intracellular zinc accumulation following zinc exposure [4,6]. It is interesting to note that KD of TRPML1 in cultured HeLa cells also results in anomalous lysosomal zinc accumulation when these cells are exposed to zinc [20]. This phenomenon is mediated by the SLC30A4 (ZNT4) zinc efflux protein [20]. Taken together, ZNT4 and TMEM163 appear to influence zinc transport into the lysosomes [20], and the roles of each protein could explain the cellular zinc imbalance linked to TRPML1 dysfunction [18]. We hypothesized that TMEM163 and ZNT proteins like ZNT4 form heterodimers based on several fronts. First, TMEM163 proteins localize where certain ZNT protein are also detected [2,20]. Second, TMEM163 shares an evolutionary relationship with ZNT proteins being all members of the CDF zinc efflux protein family [3,8]. Finally, although most CDF proteins exist as homodimers, several ZNT monomers heterodimerize with each other. For example, ZNT5 and ZNT6 proteins are only functional as heterodimers [21,22]. Other heterodimeric combinations that were reported include ZNT3/ZNT4, ZNT3/ZNT10 and ZNT2/ZNT10 proteins [[23], [24], [25]]. Similarly, ZNT1, ZNT2, ZNT3 and ZNT4 also form heterodimers with each other [26,27]. Interestingly, the subcellular localization of ZNT1 homodimer, which is typically observed in the PM, changes mostly into vesicular or compartmental sites upon heterodimerization with ZNT3 and ZNT4, but is only partially altered as ZNT1/ZNT2 heterodimers [27]. The respective heterodimers were reported to be functional using a Zinquin fluorescence assay that detects changes in cytoplasmic zinc levels [27]. In the current study, we report that human TMEM163 protein heterodimerizes with ZNT1, ZNT2, ZNT3, and ZNT4 efflux transporters.
2. Materials and methods
2.1. Materials
We purchased co-immunoprecipitation kits (Pierce), control agarose resin (Pierce), T-Per tissue lysis buffer (Thermo Scientific), n-Dodecyl-β-D-maltoside (DDM) (Thermo Scientific, Waltham, MA), poly-d-lysine (PDL, Sigma-Aldrich), Sulfo-NHS-LC-LC-Biotin (Pierce) and Neutravidin agarose beads (Pierce) from Fisher Scientific (Waltham, MA). Culture media (Dulbecco's Modified Eagle's Media [DMEM] with 4.50 g/L glucose, 0.58 g/L l-glutamine, 0.11 g/L sodium pyruvate; Corning) and supplements (fetal bovine serum [FBS]; 100 mM sodium pyruvate; 100X Glutamax™; Gibco) were also obtained from Fisher Scientific. Primary antibodies were purchased from the following sources: anti-HA rabbit polyclonal antibody (pAb; #NB600-345, Novus Biologicals, Englewood, CO; #F3165, Millipore-Sigma, St. Louis, MO), anti-DDK (M2 Flag) mouse monoclonal antibody (mAb; NBP2-43570, Novus Biologicals; #H6908, Millipore-Sigma), anti-TMEM163 rabbit pAb (#NBP1-06608, Novus Biologicals), anti-ZNT1 rabbit pAb (#PA5-77768, Thermo Scientific), anti-ZNT2 rabbit pAb (#PA5-99761, Thermo Scientific), anti-ZNT3 mouse mAb (#TA501498, Origene Technologies, Rockville, MD), and anti-ZNT4 rabbit pAb (#PA5-80028, Thermo Scientific). Secondary anti-mouse and anti-rabbit antibodies conjugated with infrared (IR)-Dye™ 800CW were purchased from LI-COR Biosciences (Lincoln, NE), while those antibodies conjugated with Alexa Fluor-488 and Alexa Fluor-568, respectively, were purchased from Fisher Scientific. Fluorescent zinc dyes, Fluozin-3 tetra-potassium salt (membrane impermeant, #F24194, Thermo Scientific) and Newport Green DCF acetate (membrane permeant, #N7791, Thermo Scientific) were purchased from Fisher Scientific.
2.2. Cell culture
Human embryonic kidney (HEK)-293T and HeLa cells were purchased from American Type Culture Collection (ATCC; Manassas, VA). The cells were cultured without antibiotics in DMEM supplemented with 10% fetal bovine serum (FBS; Thermo Scientific, Waltham, MA). The cells were maintained in a standard humidified 37 °C incubator supplied with 5% CO2. For functional zinc flux assays, we added 1 mM sodium pyruvate and 1X Glutamax™ as additional supplements in the media or buffer to improve cell viability during the experimental trials. We used HEK-293T cells for over-expression and co-immunoprecipitation experiments because these cells are highly amenable for transfection, but can easily lift off the culture plate surface when subjected to a series of washes. We used HeLa cells for functional zinc flux and confocal microscopy studies due to their strong attachment on culture plate surface, thereby minimizing or preventing cell detachment during a series of washes that are necessary to perform the techniques.
2.3. Recombinant DNA constructs
We used the In-Fusion™ (IF) HD cloning system online primer design website (Takara Bio USA, Mountain View, CA) to create primer sets (Table S1) for cloning and/or subcloning of open-reading frame (ORF) of TMEM163 (Origene Technologies), SLC30A1/ZNT1 and Slc30a1/Znt1 (Origene Technologies), SLC30A2/ZNT2 variant 1 and Slc30a2/Znt2 variant 2 (human variant 2, a kind gift from Dr. Shannon Kelleher, University of Massachusetts, Lowell and Origene Technologies, respectively), SLC30A3/ZNT3 and Slc30a3/Znt3 (a kind gift from Dr. Robert Palmiter, University of Washington, Seattle, and Origene Technologies, respectively), and SLC30A4/ZNT4 and Slc30a4/Znt4 (Origene Technologies). We used IF cloning for the pBI dual expression vector containing a minimal CMV promoter (Takara Bio USA) (Fig. S1). Specifically, we created two sets of pBI clones – one group consisting of pBI constructs with single ORF of TMEM163, ZNT1, ZNT2, ZNT3, and ZNT4; and another group that contains two cDNAs consisting of TMEM163 and one of each ZNT. For single expression of pBI constructs, the TMEM163 coding sequence was cloned at the second multiple cloning site (MCS2) leaving MCS1 empty, while the ZNT proteins were placed at the other site (MCS1) leaving MCS2 empty. For dual expression of the pBI constructs, each ZNT ORF was cloned in the MCS1 of the pBI vector containing TMEM163 in its MCS2. The use of pBI dual expression vector minimizes confounding variables arising from low transfection efficiency or biased expression for one of the two plasmids transfected, and potential cytotoxic effects of two-plasmid over-expression with CMV promoters. For protein-protein interaction experiments using the pCMV6 vector with HA or Myc-DDK peptide tag at the 3′-end (Origene Technologies), we used the same IF cloning system to subclone cDNAs of our genes-of-interests (GOIs) (Table S1). If the IF cloning approach failed, we cloned the GOIs into the pCMV6 vector using restriction enzymes, Sgf I at the 5′ end and Mlu I at the 3′ end, since we designed the IF cloning primers containing these restriction sites. For TMEM163 and ZNT4, we subcloned their ORFs into pCMV6 vector with HA or DDK tag directly from a commercially purchased pCMV6-GFP and pCMV6-HA expression clones (Origene Technologies), respectively, using Sgf I and Mlu I restriction enzymes. The sequence integrity of all expression constructs used in the study was verified by Sanger sequencing (Retrogen, San Diego, CA) and analyzed using Lasergene SeqMan version 17 (DNAStar, Madison, WI).
2.4. Co-immunoprecipitation (Co-IP) and Western blot (WB)
We heterologously expressed pCMV6 constructs of each TMEM163 and ZNT protein using HEK-293T cells. The cells were lysed the next day and processed for co-IP using the Pierce Co-IP kit according to the manufacturer's protocol (see Appendix A Supporting Information for details). All immunoblots were imaged using the Odyssey SA™ IR or Odyssey DLX™ imaging system (LI-COR Biosciences). WB images were processed using Adobe Photoshop 2022 to crop unwanted edges or empty lanes, equalize contrast levels, and straighten vertical position. We used Adobe Illustrator 2022 to compile and label images.
2.5. Immunocytochemistry (ICC) and Confocal microscopy analyses
To avoid potential confounding effects of a bulky fluorescence protein tag inducing mis-localization of the over-expressed protein, we used ICC technique on cells expressing TMEM163 and ZNT proteins with a peptide tag (see Appendix A Supporting Information for details). Confocal monochromatic images were processed using Adobe Photoshop 2022 to change grayscale images to green for Alexa-488 fluorescence, red for Alexa-568 fluorescence and blue for DAPI fluorescence. All images were cropped and auto-contrasted before the three channels were merged as a single image using Photoshop. We used Adobe Illustrator to compile and label images.
For protein colocalization study, we used the Colocalization Analysis pipeline of Cell Profiler v. 2.4.2 software (https://cellprofiler.org) to measure the correlation between confocal images of TMEM163 (green channel) and ZNT proteins (red channel). The Colocalization Analysis pipeline allowed us to perform both pixel-based and object-based correlation methods to determine the degree of overlap between two fluorescent images and spatial coincidence, respectively, in which the resulting data are indicative of protein-protein interaction. The Pearson correlation coefficient was calculated using the pixel-based technique, while the percentage of protein (object) colocalization was obtained using the object-based approach. The use of both correlation methods has been shown to produce reliable data [28]. The percentage of colocalized objects was represented as bar graph using the Graphpad Prism version 9 software (La Jolla, CA).
2.6. Cell surface biotinylation (CSB)
HEK-293T cells were plated at 1.5 × 106 cells in PDL-coated 10-cm petri dishes. The cells were transfected with 3 μg of DNA for single plasmids and 3 μg each plasmid for dual expression of pCMV6 constructs containing TMEM163-DDK, ZNT1-HA, ZNT2-HA, ZNT3-HA, and ZNT4-HA. The cells were then used for CSB as described previously [2]. The images were processed using Adobe Photoshop 2022 and the figure was created using Adobe Illustrator 2022.
To quantify WB band intensities, we used the Gel Analysis tool of the NIH ImageJ v. 1.53 software (https://imagej.nih.gov/ij/) to obtain the area under the curve (AUC) values. The AUC values were measured from three independent trials, averaged, and plotted as a bar graph using the Graphpad Prism version 9 software.
2.7. Spectrofluorometric zinc flux assay
To determine zinc flux in cells transfected with pBI constructs, we seeded HeLa cells at 10,000 cells per well on a 96-well culture plate treated with PDL. Untransfected cells served as negative control. We used cell membrane impermeant Fluozin-3 (FZ3, Kd ∼15 nM; ex = 494 nm, em = 516 nm) and cell membrane permeant Newport Green (NG, Kd ∼1 μM; ex = 505 nm, em = 535 nm) dyes to assay changes in extracellular and intracellular zinc levels, respectively, as described in our protocol paper [29]. For both assays, the cells were pre-loaded with zinc-pyrithione. To analyze zinc flux, we used the high affinity membrane impermeant FZ3 to ensure that cytoplasmic zinc ions that were extruded by the cells remain chelated by FZ3 in the extracellular milieu, which prevents extracellular zinc from being transported back into the cells as it accumulates outside of the cells over time. Meanwhile, we used a low affinity membrane permeant NG to allow NG-zinc chelates to readily dissociate in the cytoplasm, which would allow zinc ions to be transported out of the cells by zinc efflux transporters in response to its relatively higher intracellular levels over time. The data were plotted using Graphpad Prism version 9.
2.8. Statistical analysis
Spectrofluorometric data were evaluated for statistical significance using GraphPad Prism version 9 software. Relative fluorescent unit (RFU) values from Fluozin-3 trials were analyzed using one-way analysis of variance (ANOVA) with repeated measures followed by Tukey's post-hoc multiple comparisons test (control versus treatment). RFU values from Newport Green trials were analyzed using the Mixed-effects model with multiple comparisons test (control versus treatment) and Student's t-test. The significance level was set at p < 0.05.
3. Results
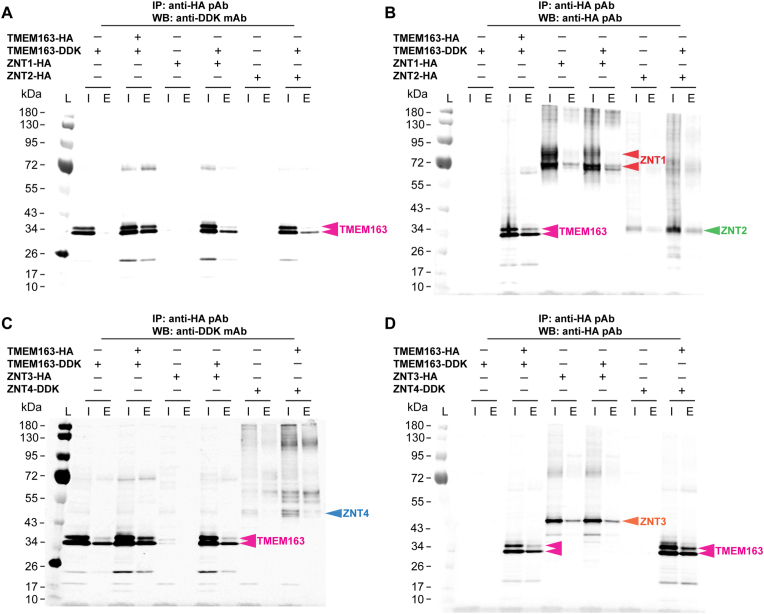
We initially performed co-IP experiments using the sample wash buffer from the Pierce co-IP kit, which is a standard buffer. However, to ensure that the co-IP results were specific, we subsequently used a highly stringent wash buffer in other experiments to show the extent of protein-protein interactions between TMEM163 dimer and TMEM163/ZNT heterodimers. As expected, co-IP results showed that human TMEM163 formed a dimer with itself as evidenced by TMEM163-DDK co-eluting with TMEM163-HA (Fig. 1). TMEM163 thus served as a positive control for all other co-IP experiments co-expressing TMEM163 with ZNT proteins. We found that ZNT1-HA or ZNT2-HA co-expressed with TMEM163-DDK resulted in co-elution of TMEM163-DDK when ZNT1-HA or ZNT2-HA was immunoprecipitated (Fig. 1A–B). Co-expression of ZNT3-HA with TMEM163-DDK also showed interaction between the two proteins as shown by the co-eluted TMEM163-DDK band upon immunoprecipitation of ZNT3-HA (Fig. 1C–D). Likewise, co-expression of ZNT4-DDK with TMEM163-HA resulted in co-elution of ZNT4-DDK after TMEM163-HA was immunoprecipitated (Fig. 1C–D). Interestingly, the highly stringent washing of the co-IP samples revealed that the heterodimeric binding between TMEM163 and ZNT proteins was not as physically strong as the homodimer formation between TMEM163 monomers (e.g., compare the WB band elution profiles of all TMEM163 co-IP with itself versus those WB elution bands of TMEM163 co-IP with specific ZNT proteins).
Fig. 1.
Co-immunoprecipitation assays of heterologously expressed TMEM163, ZNT1, ZNT2, ZNT3, and ZNT4 proteins. Co-expressions of TMEM163-DDK with ZNT1-HA, ZNT2-HA, and TMEM163-HA show heterodimerization as indicated by their respective eluted bands upon immunoblotting with A) anti-DDK monoclonal antibody (mAb) and B) anti-HA polyclonal antibody (pAb). Co-expressions of TMEM163-DDK with ZNT2-HA, and TMEM163-HA with ZNT4-DDK both show heterodimerization as indicated by their respective eluted bands upon immunoblotting with C) anti-DDK mAb and D) anti-HA pAb. Expression of TMEM163-HA or TMEM163-DDK in all trials serves as a positive (dimerization) control with itself, while single expressions of each respective protein serve as negative controls. L, protein ladder; I, input/cell lysate; E, elution; +, expressed; –, not expressed. Predicted molecular weight (MW) of human proteins: TMEM163 = 31.5 kDa, ZNT1 = 55.3–63.0 kDa, ZNT2 variant 2 = 35.0 kDa, ZNT3 = 41.8 kDa, and ZNT4 = 47.4 kDa. The images are representative of N ≥ 6 independent trials.
In a separate set of experiments, we performed co-IP of TMEM163 with each of the ZNT protein that included control agarose beads to further confirm the specificity of the co-IP results. We did not observe non-specific binding in the absence of the primary antibody (Fig. S2). To further corroborate our findings, we performed native co-IP assays using mouse tissues. We had a priori knowledge that Tmem163 is expressed in the pancreas and testis, and we then checked the tissue expression of mouse Znt1, Znt2, Znt3 and Znt4 proteins using the Human Protein Atlas database (www.proteinatlas.org). We found that Znt2 and Znt4 are present in the pancreas, while Znt1 and Znt3 are present in testis. We used commercial antibodies against human TMEM163 [2,3], ZNT1, ZNT2, ZNT3, and ZNT4 proteins that the suppliers indicated on their product datasheet to cross-react with their mouse protein counterparts. Our initial native co-IP data suggested that endogenous mouse Znt proteins co-eluted upon immunoprecipitation of mouse Tmem163. However, Znt1 and Znt2 protein bands appeared inconsistent with their predicted size relative to the positive control (not shown). We therefore performed anti-ZNT antibody validation to determine if the ZNT antibodies do cross-react with their respective mouse Znt proteins. We have previously validated the anti-TMEM163 antibody as cross-reactive to mouse Tmem163 proteins [2]. To accomplish the antibody validation, we transfected plasmids of mouse Znt and human ZNT proteins in HEK-293T cells and used their corresponding over-expression (OE) lysates for WB analysis. We found that antibodies against ZNT1, ZNT2, ZNT3, and ZNT4 detected human proteins (Figs. S3A–S3D); however, ZNT1 and ZNT2 antibodies were barely or completely unable to detect the mouse proteins (i.e., compare WB protein bands of mouse Znt proteins detected by anti-ZNT1 and anti-ZNT2 antibodies versus anti-DDK antibody) (Fig. S3). Notwithstanding, we found that endogenous mouse proteins of Tmem163, Znt3, and Znt4 co-eluted following native co-IP from tissue homogenates using anti-TMEM163 antibody (Fig. S4).
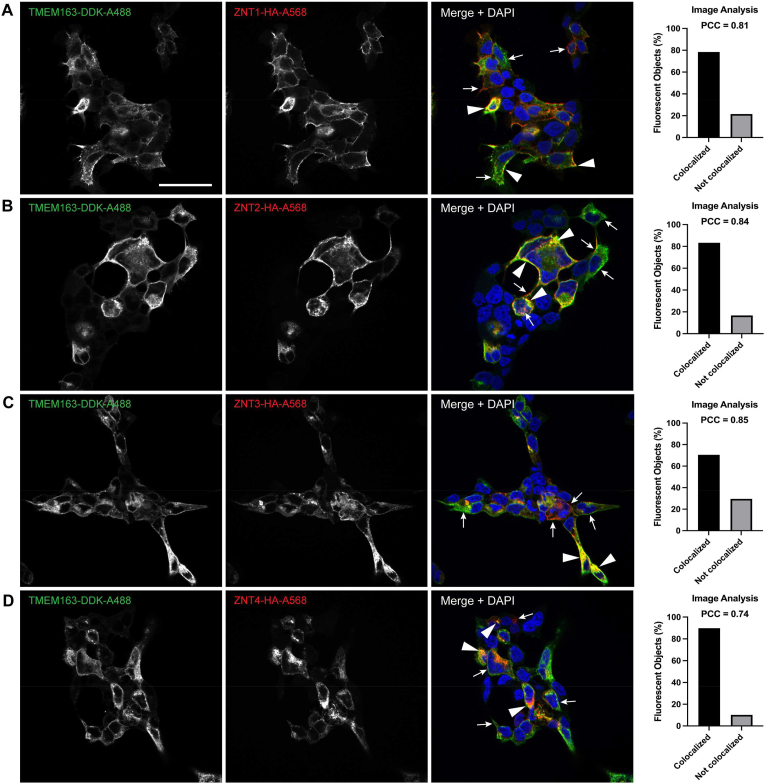
If TMEM163 and ZNT proteins form heterodimers, we expect them to co-localize together within specific subcellular sites. To this end, we performed ICC experiments upon co-expression of TMEM163 with ZNT proteins to further corroborate protein-protein interaction in cultured cells. We heterologously expressed TMEM163, ZNT1, ZNT2, ZNT3, and ZNT4 proteins individually to visually compare their subcellular localization with co-expressed TMEM163/ZNT heterodimers. Confocal microscopy showed partial co-localization between TMEM163 with each of the ZNT protein examined (Fig. 2). Semi-quantitative analysis of protein co-localization revealed a high percentage of fluorescent objects overlapped with TMEM163 and ZNT protein levels, while the Pearson correlation coefficient (PCC) values were indicative of a relatively high degree of fluorescence overlap between TMEM163 and ZNT proteins (Fig. 2). Meanwhile, single expression of TMEM163 protein showed that it is localized within the cell periphery and intracellular compartments (Fig. S5), which is consistent with our previous observations [2,3]. ZNT1 proteins exhibited a similar localization pattern as TMEM163, while ZNT2, ZNT3, and ZNT4 proteins showed mostly punctate distribution patterns in cells (Fig. S5). These findings are consistent with previous observations for each of the ZNT protein studied [23,27,[30], [31], [32]].
Fig. 2.
Immunocytochemistry of TMEM163 heterologously co-expressed with ZNT proteins. Representative confocal images of cells co-transfected with TMEM163-DDK and: A) ZNT1-HA, B) ZNT2-HA, C) ZNT3-HA, and D) ZNT4-HA. The DDK peptide was detected by anti-DDK mAb and visualized with anti-mouse secondary antibody conjugated with Alexa-488 (green). The HA peptide was detected by anti-HA pAb and visualized with anti-rabbit secondary antibody conjugated with Alexa-568 (red). DAPI stains the nuclei blue. Arrow indicates co-localization of TMEM163 and ZNT protein heterodimers within their respective cellular sites. Arrowhead indicates non-overlapping subcellular localization of protein homodimers. Protein co-localization was semi-quantitatively determined using the pixel-based and object-based analyses pipeline of the Cell Profiler software. Pearson correlation coefficient (PCC) shows the degree of overlapping fluorescence between the two channels. The fluorescent objects (in percent) provide the number of overlapping objects between the two channels. Scale bar: 100 μm. (For interpretation of the references to colour in this figure legend, the reader is referred to the Web version of this article.)
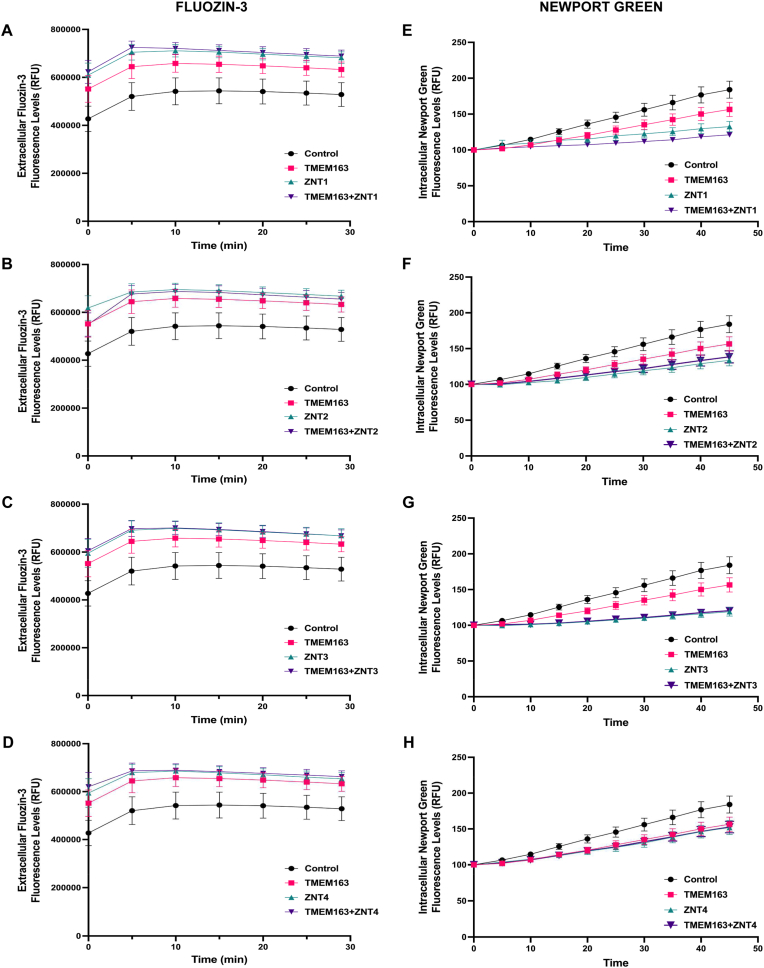
To determine the functional significance of TMEM163 interaction with each of the four ZNT proteins, we performed a standard zinc flux assay using Fluozin-3 and Newport Green fluorescent dyes [29]. We heterologously expressed pBI constructs with a single cDNA (TMEM163, ZNT1, ZNT2, ZNT3, and ZNT4) and pBI constructs with dual cDNAs (TMEM163+ZNT1, TMEM163+ZNT2, TMEM163+ZNT3, and TMEM163+ZNT4) in cultured cells for zinc flux analyses. Extracellular and intracellular zinc flux data confirmed our previous report that TMEM163 is actually a zinc effluxer. Specifically, cellular expression of TMEM163 significantly reduced intracellular zinc levels when compared to untransfected control cells (FZ3, p < 0.0001, ANOVA, Tukey's multiple comparisons test; NG, p < 0.001, Student's t-test) (Fig. 3). We also observed that the zinc efflux activity of TMEM163 homodimers appeared to be slightly different than TMEM163/ZNT heterodimers, but overall not significant in magnitude.
Fig. 3.
TMEM163 and ZNT proteins efflux zinc. Zinc flux assays using the cell membrane impermeant Fluozin-3 dye (A-D) and cell membrane permeant Newport Green dye (E-H) upon single expression and co-expression of TMEM163, ZNT1, ZNT2, ZNT3, and ZNT4. Heterologous expression of TMEM163 and ZNTx (where x denotes 1, 2, 3 or 4) show significant zinc extrusion when compared to untransfected control (FZ3, p < 0.0001, Tukey's multiple comparisons test; NG, p < 0.001, Student's t-test). Data shown are 5-min intervals and represented as mean ± SEM (N ≥ 4 independent trials). RFU, relative fluorescence unit. (For interpretation of the references to colour in this figure legend, the reader is referred to the Web version of this article.)
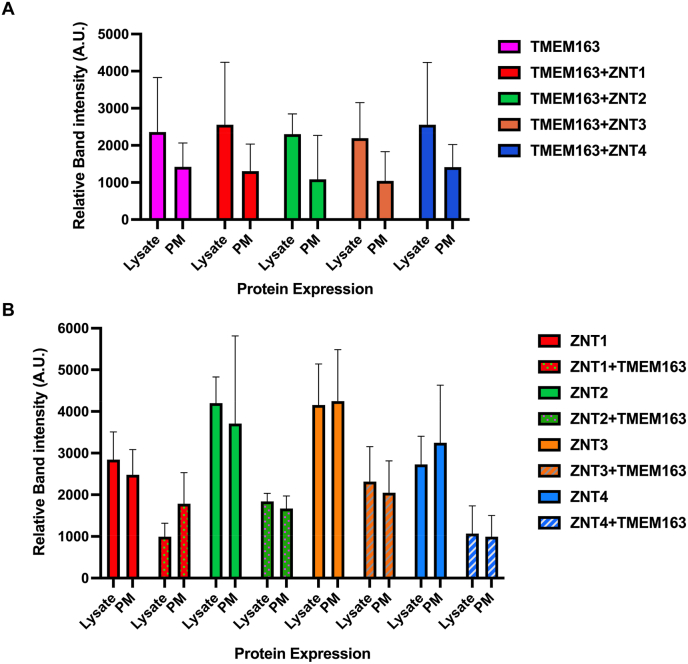
In 2015, Golan and colleagues previously reported that ZNT1, ZNT2, ZNT3 and ZNT4 monomers form heterodimers [27]. The group found that ZNT1/ZNT3 heterodimers were mostly localized within intracellular vesicles, which effectively reduced the PM localization of ZNT1 protein, while heterodimerization of ZNT2 or ZNT4 with ZNT1 increased the PM localization of both ZNT2 and ZNT4 [27]. The group's observations suggest that certain ZNT monomers influence their membrane localization. To investigate whether the interaction of TMEM163 with ZNT proteins also impacts their subcellular localization and vice versa, we performed cell surface biotinylation experiments of individually expressed protein and dual expression of TMEM163 and ZNT proteins. For this study, we biotinylated heterologously expressed TMEM163-DDK, ZNT1-HA, ZNT2-HA, ZNT3-HA and ZNT4-HA proteins and examined their cell surface expression using the WB technique (Fig. S6). Semi-quantitative analysis of the protein band intensities showed that the PM localization of TMEM163 did not change significantly when it was co-expressed with the ZNT proteins (Fig. 4A). Similarly, the expression of ZNT proteins was unaffected by the presence of TMEM163 (Fig. 4B). Note, however, that the vesicular proteins, ZNT2, ZNT3, and ZNT4 were detected on the cell surface with or without TMEM163 co-expression. This observation was likely an artifact of the heterologous over-expression system, since ZNT1 is only expected to be detected in the PM [32]. Despite such non-specific effects produced by protein over-expression, the results from the biotinylation studies appeared to have not altered the overall outcome of the biotinylation data.
Fig. 4.
Semi-quantitative analysis of cell surface expression of TMEM163 and ZNT proteins. WB of TMEM163-DDK co-expressed with each of ZNT1-HA, ZNT2-HA, ZNT3-HA, and ZNT4-HA proteins probed with anti-DDK to detect TMEM163 (A) and anti-HA peptide to detect each ZNT protein (B). The WB images associated with the analysis are shown in Fig. S6. Protein band intensities were analyzed using Image J. The numerical values were semi-quantitatively measured using the area under the curve (AUC) tool. The AUC values from proteins that appear as doublet were averaged. Lysate: Input; PM: plasma membrane; A.U. = arbitrary unit.
ZNT1 has been reported to undergo post-translational modification (PTM) such as N-glycosylation, which makes its WB protein band profile appears as double band [32]. The N-glycosylated isoform of ZNT1 (upper band) has an apparent molecular weight (MW) of 75 kDa, while its non-glycosylated isoform (lower band) is around 63 kDa even though the protein has a theoretical MW of 55.3 kDa [32]. At the qualitative level, we observed that the N-glycosylated isoform of ZNT1 protein (upper band) that was detected on the cell surface appeared to be relatively darker in the presence of TMEM163 compared to its non-glycosylated isoform (lower band) (Fig. S6). This observation is similar to the observed WB band profile of TMEM163 that was detected on the cell surface, which appeared to be a non-PTM isoform since its lower band was consistently darker than the higher band (i.e., see Fig. 1, Fig. S4, and Fig. S6). We attributed the upper band of TMEM163 protein as PTM based on our in silico analysis using the Phosphosite online software (www.phosphosite.org) that showed six predicted phosphorylation sites (five Serine residues and one Threonine residue) within its N-terminus region (data not shown). Overall, our observations support previous reports that certain protomers of the ZNT protein family form functional heterodimers with each other [23,25,27] but that this unique group also now includes TMEM163 protein. In addition, the predicted secondary structure of TMEM163 and ZNT proteins (Fig. S7) suggests that similarities in their structural features and function make it possible for their protomer to interact with each other and form heterodimers, giving additional credence to the general findings of this study.
4. Discussion
TMEM163 is a zinc transporter that shares evolutionary conservation with certain members of the CDF protein superfamily [3,8]. Our current study further confirms that TMEM163 extrudes intracellular zinc as a dimer or a heterodimer with distinct ZNT proteins. Thus, TMEM163 is a likely bona fide member of the SLC30/ZNT zinc efflux transporters as evidenced by its functional interaction with other ZNT proteins. The highly stringent wash employed in our co-IP experiments showed that the physical binding between TMEM163 and ZNT monomers is relatively weak compared to TMEM163 homodimer. This finding supports the report by Golan and co-workers that homodimers exhibit relatively strong binding than heterodimers [27]. Such difference suggests that the interaction domain between TMEM163 and ZNT proteins may not be the same as the dimerization domain of TMEM163. This conjecture is potentially bolstered by the previous observation that the dimerization of ZNT3 protomer is mediated by covalently linked dityrosine amino acid residues [23], but that the same tyrosine residues responsible for ZNT3 dimerization are not critical for ZNT3/ZNT4 or ZNT3/ZNT10 heterodimerization [25]. Future work should address the possibility that the dimerization and heterodimerization domains of TMEM163 correspond to that of the ZNT3 protein. Meanwhile, our attempt to corroborate our heterologous expression co-IP using native co-IP of mouse tissues is successful for two of the four mouse Znt proteins despite the presence of some non-specific bands due to the polyclonal nature of the antibodies. Inclusion of positive cell lysate over-expression controls, the use of IgG heavy/light chains as landmarks, and the use of molecular weight marker helped pinpoint the endogenous mouse Znt3 and Znt4 proteins co-eluting with the mouse Tmem163 protein. We are not aware of published reports on ZNT protomer interactions that have ever used native co-IP from post-mortem tissues of a rodent, and there are only two well-known studies that have used cultured cells to show endogenous ZNT protein interaction or co-localization using co-IP, subcellular fractionation, and microscopy [25,33]. The limitation of this particular experiment is that we were only able to verify endogenous interactions between mouse Tmem163 and Znt3, as well as Tmem163 and Znt4 due to the lack of commercially available antibodies that specifically detect mouse Znt1 and Znt2 proteins. Although the human antibodies we purchased supposedly cross-reacted with rodent epitopes, our antibody validation data indicate that species cross-reactivity should always be experimentally confirmed especially when performing native co-IP experiments. Nevertheless, our confocal imaging and co-localization analyses provided additional evidence that heterodimers of TMEM163 and ZNT proteins exist in their respective cellular sites albeit they only partially co-localize. Semi-quantitative analysis showed co-localization of the proteins examined. Note, however, that the restricted protein co-localization implies that TMEM163 and ZNT homodimers may function differently than their equivalent heterodimers. Although the functional zinc flux assays suggest this possibility, it appears that the interaction between TMEM163 and ZNT proteins do not significantly influence their inherent subcellular localization as shown by cell surface biotinylation results. The apparent mis-localization of vesicular ZNT2, ZNT3, and ZNT4 proteins in the PM is likely caused by the strong CMV promoter contained within their respective expression plasmid. It is worth noting that the pBI expression construct that we used for the functional zinc assays contained a minimal CMV promoter, which is designed to reduce the potential negative impact of the heterologous over-expression system. Overall, diversifying the protein composition of zinc effluxers in cells may be an effective way for them to eliminate excess cytoplasmic zinc when the need arises. This phenomenon may also explain why many cell types have redundant expressions of influx and efflux zinc transporters.
TMEM163 protein typically exhibits a double band on WB assays. We surmised that the top band is a product of PTM for two reasons. First, our in silico analysis revealed that TMEM163 has six predicted phosphorylation sites within its N-terminus (NT) region. Second, we previously showed that deletion of at least 42 amino acid residues in its NT region eliminates one of the double bands [2]. Third, a TMEM163 protein variant with a Serine to Arginine substitution at position 61 (S61R) resolves slightly faster and appears lower than the wildtype double bands on WB [3]. Finally, another relevant evidence to a possible PTM of TMEM163 is the observation that its WB elution profiles upon co-IP (Fig. 1) and cell surface biotinylation with or without co-expressed ZNT protomers (Fig. S6), seem to be mostly non-PTM protein isoforms (darker lower band relative to its upper band). In contrast, the WB elution profile of TMEM163 as a homodimer shows comparatively similar intensities between the upper and lower bands, suggesting that its dimerization domain may be subject to PTM (e.g., phosphorylation). These findings also indicate that the interaction between TMEM163 and ZNT monomers may be occurring before TMEM163 undergoes PTM or it may be that PTM favors the formation of TMEM163 homodimers. The former reasoning is supported by a similar observation that non-glycosylated ZNT1 isoform (lower band of approximately 63 kDa on WB) mainly co-eluted with TMEM163 protein (see Fig. 1 and Fig. S2). Taken together, it would be interesting to determine in future investigations whether or not the PTM of TMEM163 (predicted phosphorylation status) at the NT region influences its homodimerization, its heterodimerization with certain ZNT protomers, and its overall zinc efflux function.
5. Conclusion
In this study, we discovered that TMEM163 is a promiscuous protein that interacts not only with TRPML1 and P2X3/P2X4 receptor ion channels [2,34] and BLOC-1 cargo trafficking protein [6], but also functionally heterodimerizes with related zinc efflux transporters ZNT1, ZNT2, ZNT3 and ZNT4. The apparent differences in physical binding of TMEM163 with itself as homodimers versus TMEM163/ZNT heterodimers suggest that their respective interaction domains are unique and may be influenced by PTM. Further, TMEM163 and ZNT proteins partially co-localize within distinct subcellular location but they do not affect each other's subcellular distribution. In sum, the physiological significance of the interaction between TMEM163 and ZNT proteins not only provides a diverse kind of zinc transporters, but also adds an extra layer of redundancy to tightly regulate zinc levels in specific cells or tissues as a protective mechanism from the effects of intracellular zinc overload.
Funding
This work was supported by the National Institutes of Health (NIH), National Institute of Neurological Diseases and Stroke AREA grant 2R15 NS101594-02 and partly funded by the 1R03 NS123728-01 grant to MPC. The content of this paper is solely the responsibility of the authors and does not necessarily represent the official views of the National Institutes of Health.
Author contributions
Conceptualization, Funding acquisition (MPC); Methodology, Data analysis (AE, DJS, SA, MPC); Writing, Revision (AE, DJS, SA, MPC). All authors reviewed and approved the manuscript.
Declaration of competing interest
The authors declare that they have no known competing financial interests or personal relationships that could have appeared to influence the work reported in this paper.
Acknowledgments
We are grateful to Nimrah Ashfaq and Steve Karl for their technical support. We also thank Dr. Shannon Kelleher (University of Massachusetts, Lowell) for providing the ZnT2 clone, and Dr. Robert Palmiter (University of Washington, Seattle) for providing the ZnT3 clone.
Footnotes
Supplementary data to this article can be found online at https://doi.org/10.1016/j.bbrep.2022.101362.
Appendix A. Supplementary data
The following is the Supplementary data to this article:
References
- 1.Burre J., Zimmermann H., Volknandt W. Identification and characterization of SV31, a novel synaptic vesicle membrane protein and potential transporter. J. Neurochem. 2007;103:276–287. doi: 10.1111/j.1471-4159.2007.04758.x. [DOI] [PubMed] [Google Scholar]
- 2.Cuajungco M.P., Basilio L.C., Silva J., Hart T., Tringali J., Chen C.C., Biel M., Grimm C. Cellular zinc levels are modulated by TRPML1-TMEM163 interaction. Traffic. 2014;15:1247–1265. doi: 10.1111/tra.12205. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Sanchez V.B., Ali S., Escobar A., Cuajungco M.P. Transmembrane 163 (TMEM163) protein effluxes zinc. Arch. Biochem. Biophys. 2019;677 doi: 10.1016/j.abb.2019.108166. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Chakraborty S., Vellarikkal S.K., Sivasubbu S., Roy S.S., Tandon N., Bharadwaj D. Role of Tmem163 in zinc-regulated insulin storage of MIN6 cells: functional exploration of an Indian type 2 diabetes GWAS associated gene. Biochem. Biophys. Res. Commun. 2020;522:1022–1029. doi: 10.1016/j.bbrc.2019.11.117. [DOI] [PubMed] [Google Scholar]
- 5.Barth J., Zimmermann H., Volknandt W. SV31 is a Zn2+-binding synaptic vesicle protein. J. Neurochem. 2011;118:558–570. doi: 10.1111/j.1471-4159.2011.07344.x. [DOI] [PubMed] [Google Scholar]
- 6.Yuan Y., Liu T., Huang X., Chen Y., Zhang W., Li T., Yang L., Chen Q., Wang Y., Wei A., Li W. A zinc transporter, transmembrane protein 163, is critical for the biogenesis of platelet dense granules. Blood. 2021;137:1804–1817. doi: 10.1182/blood.2020007389. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Waberer L., Henrich E., Peetz O., Morgner N., Dotsch V., Bernhard F., Volknandt W. The synaptic vesicle protein SV31 assembles into a dimer and transports Zn(2) J. Neurochem. 2017;140:280–293. doi: 10.1111/jnc.13886. [DOI] [PubMed] [Google Scholar]
- 8.Styrpejko D.J., Cuajungco M.P. Transmembrane 163 (TMEM163) protein: a new member of the zinc efflux transporter family. Biomedicines. 2021;9 doi: 10.3390/biomedicines9020220. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Colvin R.A., Holmes W.R., Fontaine C.P., Maret W. Cytosolic zinc buffering and muffling: their role in intracellular zinc homeostasis. Metallomics. 2010;2:306–317. doi: 10.1039/b926662c. [DOI] [PubMed] [Google Scholar]
- 10.Eide D.J. Zinc transporters and the cellular trafficking of zinc. Biochim. Biophys. Acta. 2006;1763:711–722. doi: 10.1016/j.bbamcr.2006.03.005. [DOI] [PubMed] [Google Scholar]
- 11.Kambe T., Hashimoto A., Fujimoto S. Current understanding of ZIP and ZnT zinc transporters in human health and diseases. Cell. Mol. Life Sci. 2014;71:3281–3295. doi: 10.1007/s00018-014-1617-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Xue J., Xie T., Zeng W., Jiang Y., Bai X.C. Cryo-EM structures of human ZnT8 in both outward- and inward-facing conformations. Elife. 2020;9 doi: 10.7554/eLife.58823. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Bargal R., Avidan N., Ben-Asher E., Olender Z., Zeigler M., Frumkin A., Raas-Rothschild A., Glusman G., Lancet D., Bach G. Identification of the gene causing mucolipidosis type IV. Nat. Genet. 2000;26:118–123. doi: 10.1038/79095. [DOI] [PubMed] [Google Scholar]
- 14.Bassi M.T., Manzoni M., Monti E., Pizzo M.T., Ballabio A., Borsani G. Cloning of the gene encoding a novel integral membrane protein, mucolipidin-and identification of the two major founder mutations causing mucolipidosis type IV. Am. J. Hum. Genet. 2000;67:1110–1120. doi: 10.1016/S0002-9297(07)62941-3. S0002-9297(07)62941-3 [pii] [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Sun M., Goldin E., Stahl S., Falardeau J.L., Kennedy J.C., Acierno J.S., Jr., Bove C., Kaneski C.R., Nagle J., Bromley M.C., Colman M., Schiffmann R., Slaugenhaupt S.A. Mucolipidosis type IV is caused by mutations in a gene encoding a novel transient receptor potential channel. Hum. Mol. Genet. 2000;9:2471–2478. doi: 10.1093/hmg/9.17.2471. [DOI] [PubMed] [Google Scholar]
- 16.Eichelsdoerfer J.L., Evans J.A., Slaugenhaupt S.A., Cuajungco M.P. Zinc dyshomeostasis is linked with the loss of mucolipidosis IV-associated TRPML1 ion channel. J. Biol. Chem. 2010;285:34304–34308. doi: 10.1074/jbc.C110.165480. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Dong X.P., Cheng X., Mills E., Delling M., Wang F., Kurz T., Xu H. The type IV mucolipidosis-associated protein TRPML1 is an endolysosomal iron release channel. Nature. 2008;455:992–996. doi: 10.1038/nature07311. nature07311 [pii] 10.1038/nature07311. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Cuajungco M.P., Kiselyov K. The mucolipin-1 (TRPML1) ion channel, transmembrane-163 (TMEM163) protein, and lysosomal zinc handling. Front Biosci (Landmark Ed) 2017;22:1330–1343. doi: 10.2741/4546. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Minckley T.F., Zhang C., Fudge D.H., Dischler A.M., LeJeune K.D., Xu H., Qin Y. Sub-nanomolar sensitive GZnP3 reveals TRPML1-mediated neuronal Zn(2+) signals. Nat. Commun. 2019;10:4806. doi: 10.1038/s41467-019-12761-x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Kukic I., Lee J.K., Coblentz J., Kelleher S.L., Kiselyov K. Zinc-dependent lysosomal enlargement in TRPML1-deficient cells involves MTF-1 transcription factor and ZnT4 (Slc30a4) transporter. Biochem. J. 2013;451:155–163. doi: 10.1042/BJ20121506. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Suzuki T., Ishihara K., Migaki H., Ishihara K., Nagao M., Yamaguchi-Iwai Y., Kambe T. Two different zinc transport complexes of cation diffusion facilitator proteins localized in the secretory pathway operate to activate alkaline phosphatases in vertebrate cells. J. Biol. Chem. 2005;280:30956–30962. doi: 10.1074/jbc.M506902200. [DOI] [PubMed] [Google Scholar]
- 22.Fukunaka A., Suzuki T., Kurokawa Y., Yamazaki T., Fujiwara N., Ishihara K., Migaki H., Okumura K., Masuda S., Yamaguchi-Iwai Y., Nagao M., Kambe T. Demonstration and characterization of the heterodimerization of ZnT5 and ZnT6 in the early secretory pathway. J. Biol. Chem. 2009;284:30798–30806. doi: 10.1074/jbc.M109.026435. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Salazar G., Falcon-Perez J.M., Harrison R., Faundez V. SLC30A3 (ZnT3) oligomerization by dityrosine bonds regulates its subcellular localization and metal transport capacity. PLoS One. 2009;4 doi: 10.1371/journal.pone.0005896. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Patrushev N., Seidel-Rogol B., Salazar G. Angiotensin II requires zinc and downregulation of the zinc transporters ZnT3 and ZnT10 to induce senescence of vascular smooth muscle cells. PLoS One. 2012;7 doi: 10.1371/journal.pone.0033211. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Zhao Y., Feresin R.G., Falcon-Perez J.M., Salazar G. Differential targeting of SLC30A10/ZnT10 heterodimers to endolysosomal compartments modulates EGF-induced MEK/ERK1/2 activity. Traffic. 2016;17:267–288. doi: 10.1111/tra.12371. [DOI] [PubMed] [Google Scholar]
- 26.Lasry I., Golan Y., Berman B., Amram N., Glaser F., Assaraf Y.G. In situ dimerization of multiple wild type and mutant zinc transporters in live cells using bimolecular fluorescence complementation. J. Biol. Chem. 2014;289:7275–7292. doi: 10.1074/jbc.M113.533786. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Golan Y., Berman B., Assaraf Y.G. Heterodimerization, altered subcellular localization, and function of multiple zinc transporters in viable cells using bimolecular fluorescence complementation. J. Biol. Chem. 2015;290:9050–9063. doi: 10.1074/jbc.M114.617332. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Moser B., Hochreiter B., Herbst R., Schmid J.A. Fluorescence colocalization microscopy analysis can be improved by combining object-recognition with pixel-intensity-correlation. Biotechnol. J. 2017;12 doi: 10.1002/biot.201600332. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Ali S., Cuajungco M.P. Protocol for quantifying zinc flux in cultured cells using fluorescent indicators. STAR Protoc. 2020;1 doi: 10.1016/j.xpro.2020.100050. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Lopez V., Kelleher S.L. Zinc transporter-2 (ZnT2) variants are localized to distinct subcellular compartments and functionally transport zinc. Biochem. J. 2009;422:43–52. doi: 10.1042/BJ20081189. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.McCormick N.H., Kelleher S.L. ZnT4 provides zinc to zinc-dependent proteins in the trans-Golgi network critical for cell function and Zn export in mammary epithelial cells. Am. J. Physiol. Cell Physiol. 2012;303:C291–C297. doi: 10.1152/ajpcell.00443.2011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Nishito Y., Kambe T. Zinc transporter 1 (ZNT1) expression on the cell surface is elaborately controlled by cellular zinc levels. J. Biol. Chem. 2019;294:15686–15697. doi: 10.1074/jbc.RA119.010227. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Salazar G., Love R., Werner E., Doucette M.M., Cheng S., Levey A., Faundez V. The zinc transporter ZnT3 interacts with AP-3 and it is preferentially targeted to a distinct synaptic vesicle subpopulation. Mol. Biol. Cell. 2004;15:575–587. doi: 10.1091/mbc.E03-06-0401. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Salm E.J., Dunn P.J., Shan L., Yamasaki M., Malewicz N.M., Miyazaki T., Park J., Sumioka A., Hamer R.R.L., He W.W., Morimoto-Tomita M., LaMotte R.H., Tomita S. TMEM163 regulates ATP-gated P2X receptor and behavior. Cell Rep. 2020;31 doi: 10.1016/j.celrep.2020.107704. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.