Abstract
Specific DNA methylation at CpG and non-CpG sites is essential for chromatin regulation. The DNA methyltransferase DNMT3A interacts with target sites surrounded by variable DNA sequences with its TRD and RD loops, but the functional necessity of these interactions is unclear. We investigated CpG and non-CpG methylation in a randomized sequence context using WT DNMT3A and several DNMT3A variants containing mutations at DNA-interacting residues. Our data revealed that the flanking sequence of target sites between the −2 and up to the +8 position modulates methylation rates >100-fold. Non-CpG methylation flanking preferences were even stronger and favor C(+1). R836 and N838 in concert mediate recognition of the CpG guanine. R836 changes its conformation in a flanking sequence-dependent manner and either contacts the CpG guanine or the +1/+2 flank, thereby coupling the interaction with both sequence elements. R836 suppresses activity at CNT sites but supports methylation of CAC substrates, the preferred target for non-CpG methylation of DNMT3A in cells. N838 helps to balance this effect and prevent the preference for C(+1) from becoming too strong. Surprisingly, we found L883 reduces DNMT3A activity despite being highly conserved in evolution. However, mutations at L883 disrupt the DNMT3A-specific DNA interactions of the RD loop, leading to altered flanking sequence preferences. Similar effects occur after the R882H mutation in cancer cells. Our data reveal that DNMT3A forms flexible and interdependent interaction networks with the CpG guanine and flanking residues that ensure recognition of the CpG and efficient methylation of the cytosine in contexts of variable flanking sequences.
Keywords: DNMT3A, DNA methylation, enzyme mechanism, enzyme specificity, protein-DNA interaction
Abbreviations: MBP, maltose binding protein; NGS, next generation sequencing; TRD, target recognition domain; WT, wildtype
In human cells, DNA is methylated at the cytosine-C5 position in about 70 to 80% of all CpG but also at non-CpG sites (1, 2, 3). DNA methylation has important roles in the regulation of protein–DNA interactions and it is involved in gene expression, X-chromosome inactivation, genomic stability, cell differentiation, and mammalian development (1, 4, 5). The DNA methyltransferases DNMT3A and DNMT3B are so-called de novo DNA methyltransferases (6), which set up DNA methylation patterns during gametogenesis and postimplantation development (5, 7). The DNMT3A enzyme is essential in mammalian development, but it also has important roles in carcinogenesis (8, 9) and in the brain (10, 11). The catalytically inactive DNMT3-like protein (DNMT3L) has an important regulatory role in gametogenesis by acting as a stimulator of DNMT3A (6).
The catalytically active C-terminal domain of DNMT3A (DNMT3AC) forms a linear heterotetrameric complex with the C-terminal domain of DNMT3L (DNMT3LC) in a 3L-3A-3A-3L arrangement (Fig. 1A) (12). Biochemical studies showed that the DNMT3L subunits in the DNMT3A/3L heterotetramer can be replaced by two additional subunits of DNMT3AC (13, 14), yielding a DNMT3A homotetramer as the smallest catalytically active form of DNMT3A. The central DNMT3AC subunits of such homo- or hetero-tetramers interact via the so-called RD interface and form the DNA-binding site of the complex (12). Structures of the DNMT3A/3L complex bound to DNA showed that these subunits interact with two CpG sites in 12 base pair distance and methylate them in opposite DNA strands (15, 16) (Fig. 1A), but biochemical data showed that other arrangements of comethylation of CpG sites are also possible (17).
Figure 1.
Structure and activity of DNMT3A complexes.A, structure of the DNMT3A/3L heterotetramer (15). DNMT3L is shown in light and dark gray, and DNMT3A in orange and light orange. The DNA is colored blue. The flipped cytosine and the CpG guanine are colored cyan, and the +1 to +3 flank base pairs are colored turquoise. The mutated residues are indicated in red in the light orange DNMT3A subunit. B, structural snapshots of the TRD loop regions in DNMT3A (CGT complex 6BRR (15) and CGA complex 6W8B (16)) and DNMT3B (CGT complex 6U8W (18)). The flipped cytosine is shown in cyan and the CpG guanine in dark blue. W, water. C, structural snapshots of the RD loop regions in DNMT3A and DNMT3B. The flipped cytosine is shown in cyan and the CpG guanine in dark blue. D, multiple sequence alignment of DNMT3A and DNMT3B in representative vertebrate species. Sea urchin and amphioxus were included as examples of nonvertebrates. E, activity analysis of the mutants investigated in this study with radioactive assays or the Deep Enzymology NGS assay. NGS activity of R882A is based on the MBP-cleaved data, and WT refers to the activity of MBP-cleaved WT taken from Mack et al., 2022 (24). NGS activity of R882H is based on the data from Emperle et al. (2019) (22). TRD, target recognition domain.
Structural studies showed that the DNA interaction of DNMT3A and DNMT3B is mediated by three protein regions (15, 18): (1) a loop directly following the active center (residues G707–K721 in DNMT3A), (2) a loop from the target recognition domain (TRD-loop, residues R831–F848), and (3) the RD interface (RD-loop, residues S881-R887), which together create a continuous DNA-binding surface. Among them, residues in the TRD and RD loops form an interconnected network of contacts with the DNA which mediates recognition of the CpG guanine and of the flanking sequence in the 3′ direction (Fig. 1A). DNMT3A and DNMT3B interact with the CpG guanine with residues from the TRD loop and they prefer methylation of DNA within CpG sites, but they also introduce methylation at lower level at non-CpG sites (19). Non-CpG methylation has been connected with gene regulatory and chromatin modulating functions and to X-chromosome inactivation mainly in the nervous system (2, 20). Previous Deep Enzymology studies showed that DNMT3A and DNMT3B methylate CpG and non-CpG sites in strong dependence on their flanking sequences (19). Differences in flanking sequence preferences of DNMT3A and DNMT3B could be connected to sequence and structural differences in the RD loops of both enzymes (18). In addition, the somatic R882H mutation in the RD loop of DNMT3A occurs in a large fraction of acute myeloid leukemia tumors (8, 9), and it was shown to alter the flanking sequence preferences of DNMT3A strongly, making them more similar to those of DNMT3B (21, 22, 23, 24). Moreover, R882H was shown to increase the stability of the RD interface which explains its dominant cellular phenotype (24).
Structural studies have documented the involvement of several amino acid residues from the TRD and RD loops in the DNA interaction of DNMT3A (Fig. 1B). In a CGT structure (15), the R836 side-chain from the TRD loop forms an H-bond with the O6 of the CpG guanine plus one additional water-mediated contact to the guanine N7. However, in a CGA structure (16), the R836 side chain moved up and it formed a side-chain H-bond to the N6 atom of the adenine in the +1 flanking position, A(+1), and a second H-bond to the +2 flank site, indicating flexible and combined readout of CpG specificity and the +1/+2 flank. In the CGA structure, the side chain of N838 contacts the O6 of the CpG guanine, while in the CGT structure, this residue is flipped up and it forms an H-bond to the phosphodiester group connecting the +2 and +3 flanking site and a direct contact to the +2′ base (where ‘ refers to the non-target DNA strand). Interestingly, the CpG recognition pattern in the CGA structure of DNMT3A resembles the structure of DNMT3B, where N779 (corresponding to N838 of DNMT3A) mediates the CpG guanine recognition. In both DNMT3A structures, the N7 atom of the CpG guanine is bound with a water-mediated H-bond to T834, a corresponding contact is also observed in DNMT3B. Moreover, S837 from the TRD loop forms a side chain H-bond to the N7 of G(+3’) in both DNMT3A structures, thereby directly contacting the +3 flank. The RD loop residues of DNMT3A are engaged in several contacts to the flanking residues at the 3′ side of the CpG (Fig. 1C). S881 and R887 form side chain H-bonds to the phosphodiester group between the flanking bases at +5′ and +6’. R882 contacts the phosphodiester group between the flanking bases at +3′ and +4’. L883 is sandwiched between S837 and the DNA backbone at the +4′ and +5′ flanking bases with multiple van-der-Waals contacts and a backbone H-bond to the phosphodiester group at this site. The TRD and RD loops are directly connected by the positioning of R836, which pushes R882 away from the DNA in its uplifted conformation (Fig. 1C). Moreover, the R882 side chain contacts the side chain of S837 in both structures.
While structural studies document the dependence of the dynamic DNA interaction networks of DNMT3A on the flanking sequences of the CpG site, the functional relevance of this context-dependent DNA recognition has not been investigated. It was the aim of this study to unravel the functional consequences of the interconnected recognition of the CpG guanine, the +1 to +3 flanking sequences and flanking sequences further away from the CpG site and determine their effects in CpG and non-CpG methylation. To this end, several mutants of critical residues in DNMT3A were prepared and their activity, CpG specificity, and flanking preference in CpG and non-CpG context were determined in Deep Enzymology experiments. Our data provide a comprehensive view on the dynamic interaction of DNMT3A with the CpG sites and the 3′ flanking sequences, which defines the roles of individual amino acids residues in this contact network and their interdependence.
Results
Selection of residues for mutagenesis, mutant generation and purification, and initial activity assessment
To investigate the interaction of the catalytic domain of DNMT3A with the target CpG guanine and the 3′ flanking DNA, seven DNMT3A residues in the TRD and RD loop were selected from the available structures (15, 16) and mutated to alanine, viz. R836, S837, N838, S881, R882, L883, and R887. The cancer mutant R882H was investigated as well because of its medical relevance. A multiple sequence alignment of DNMT3A and DNMT3B enzymes from several vertebrate species and DNMT3 from amphioxus and sea urchin as representatives of nonvertebrates (Fig. 1D) showed that all these residues are fully conserved in DNMT3A enzymes, which is matching their putative highly important mechanistic role. However, there are strong differences in these residues between DNMT3A and DNMT3B. In the TRD loop, R836 is replaced by K777 (using human DNMT3B numbering) in most of the DNMT3B sequences, and the RD loop is one of the most divergent regions of DNMT3A and DNMT3B, where the DNMT3A residues S881-R882-L883-…-R887 are replaced by G822-R823-G824 and K828 in DNMT3B. DNMT3A catalytic domain mutations were generated by site-directed mutagenesis, overexpressed, and purified as maltose binding protein (MBP)-tagged proteins. All mutants were obtained at comparable purity (Fig. S1). Protein concentrations were determined by A280nm and validated on Coomassie BB-stained polyacrylamide gels. Methylation activity was initially determined using radioactively-labeled AdoMet and a biotinylated double-stranded 30-mer oligonucleotide substrate with a single CpG site that has been employed as reference substrate in several previous studies (24, 25). Initial reaction rates were calculated from time courses of the methylation reactions and averaged over several (3–6) independent experimental repeats (blue bars in Fig. 1E). The results revealed a striking five-fold enhancement of activity for L883A, slight reductions in activity of R836A and R882H, and a stronger reduction of activity in the case of R882A. The catalytic activities of N838A, S837A, S881A, and R887A were similar to the DNMT3A wildtype (WT) activity.
Analysis of the flanking preferences of DNMT3A mutants in CpG context
To explore the flanking sequence preferences of CpG methylation by DNMT3A WT and the mutants, we next conducted Deep Enzymology experiments (19). We first investigated the methylation of a pool of DNA substrates, in which a target CpG site was flanked by 10 random nucleotides on either side. The substrate pool was methylated by DNMT3A WT and mutants, followed by hairpin ligation, bisulfite conversion, PCR amplification, and next generation sequencing (NGS) analysis as described previously (18, 22, 26). In each case, two independent experiments were conducted as detailed in Table S1. R882H CpG methylation data were used from our previous publication (22). To exclude effects of the fixed sequences outside of the randomized region, the following analyses were restricted to the −8 to +8 region, which then still have two random nucleotides on both sides. Using these data sets, we determined the average methylation levels of all substrates containing a particular base at one of the −8 to +8 flank sites to identify bases favorable or unfavorable for methylation activity. The data were expressed in observed/expected ratios of the corresponding base in the methylated DNA strands. We first compared the results of the experimental repeats conducted with each mutant, which always showed high correlation of the derived profiles, which was also the case for the WT data determined here and previously (18) (Table S2). In general, the average methylation activities determined in the Deep Enzymology experiments were similar to the radioactive methylation rates of the respective mutant (orange bars in Fig. 1E). Most strikingly, the rate enhancement with L883A seen in the radioactive kinetics was observed in the NGS assay as well.
Comparison of the effects of flanking sites on CpG activity
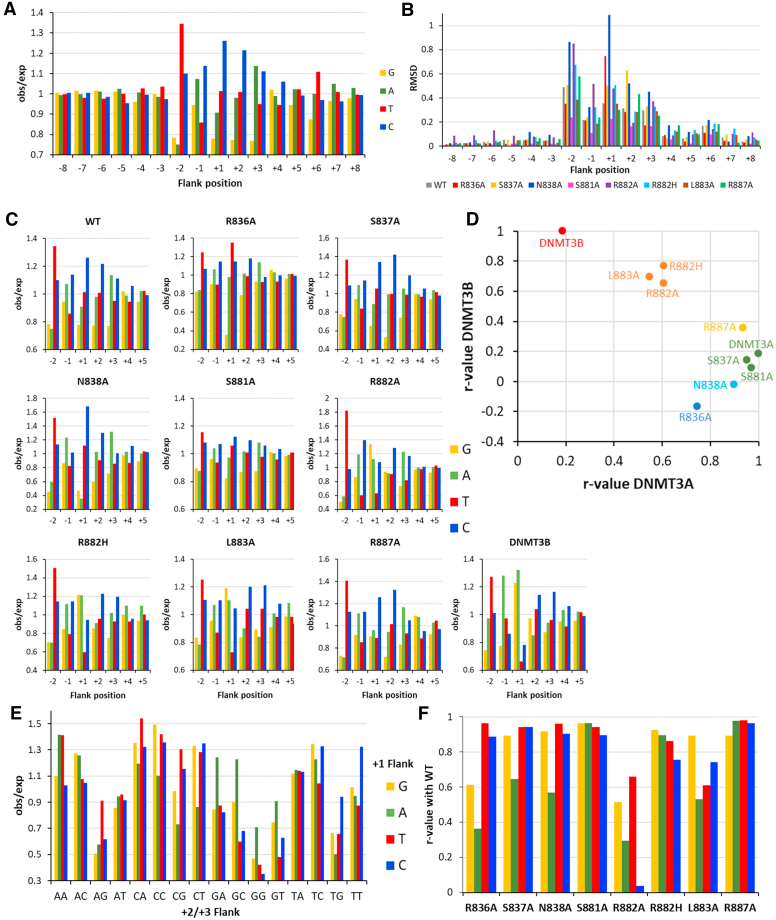
Detailed analysis of the flanking sequence preferences revealed strong position and mutant specific effects. DNMT3A WT flanking effects were detectable between the −2 and (mainly) +6 positions (Fig. 2A). At the −2 site, T was strongly preferred, while G and A were disfavored. C and A were preferred at −1, where T was most disfavored. At +1 to +3, C was strongly preferred (in addition to A at +3) and G was disfavored. Weaker effects were detectable at +4 (preference for C) and +6 (preference for T and disfavor for G). Due to the limited number of reads, average methylation levels could only be determined for all NNCGNNN flanks. The numbers varied between 95% methylation for TCCGCCC and no detectable methylation for 11 NNCGNNN sequence motifs that were all very G-rich, illustrating the strong effect of the flanking sequences on DNMT3A activity in agreement with previous data (18, 22, 24).
Figure 2.
Analysis of flanking sequence effects in CpG methylation.A, flanking sequence preferences of WT DNMT3A for the −8 to +8 region. B, compilation of RSMD values of flanking sequence effects of WT DNMT3A and mutants at each flank position. C, flanking sequence preferences of WT and DNMT3A mutants in the −2 to +3 flank region. D, compilation of the r-values of the correlation of mutant flanking sequence preferences with WT DNMT3A and DNMT3B. E, +2/+3 flanking preferences in dependence of the base at the +1 flank site of WT DNMT3A. Mutant profiles are shown in Fig. S4. F, correlation of mutant +2/+3 flank preferences with WT DNMT3A for different bases at the +1 position.
To identify the flanking base pairs most relevant for the catalytic activity, the RMSD value for the deviation of the occurrence of each base from the expected value (obs/exp = 1.0) was determined for each position. The RMSD values of WT and mutant profiles showed effects between the −2 and +7 flanking sites (Fig. 2B). Strong overall flanking sequence preferences were observed with R836A, N838A, R882A, and R882H, while effects at S837A, S881A, L883A, and R887A were smaller. Largest effects were observed at the −2 and +1 sites, the largest difference between DNMT3A and DNMT3B profiles (18) were observed at −1 and +1 sites. Detailed analyses of the mutant preferences were conducted for the −2 to +5 positions and compared with WT (Fig. 2C). R836A showed a strong change of preferences at the +1 site, where the preferences changed from C>T>A>G (WT) to T>C>A>>G, indicating that the disfavor for G was even more enhanced and the preferences for T and C swapped. In addition, the disfavor for G(+2) and G(+3) was reduced for R836A. In case of S837A, no big effects were observed, but at the +1 and +2 sites, the preferences of WT were enhanced and the preference for A(+3) was reduced. N838A led to strong changes in the flanking sequence preferences at the +1 position, where the preferences changed from C>T>A>G (WT) to C>T>>G∼A. In addition, the preference for C(+3) was reduced. S881A did not show big effects. R882A showed a strong elevation of WT effects at −1 and a strong shift of preferences at +1 toward G>A∼C>>T, indicating that the preference for G increased while T and C dropped. Moreover, the preference for G(+2) was increased as well. A similar change in preferences at the +1 and +2 sites as R882A was observed for R882H. In addition, the preference for A(+3) dropped. L883A also caused the same change in preferences at +1 as R882A and R882H and the drop of the A(+3) preference as observed for R882H. For R887A, a mild change of preferences at +4 was observed, where G and A were more preferred than by WT.
In summary, these data reveal strong >100-fold flanking sequence preferences of DNMT3A that were heavily influenced by R836, N838, R882, and L883. To further compile the data and compare the deviations of the preferences of the DNMT3A mutants from the preference profiles of DNMT3A and DNMT3B (taken from (18)), the R-values of the correlation of −2 to +5 flanking profiles of mutants with DNMT3A and DNMT3B WT were determined and plotted, revealing several distinct groups of mutants (Fig. 2D). The R882A, R882H, and L883A mutants caused large and similar changes of the profile making them similar to DNMT3B. In contrast, R836A caused an equally large but different shift of preferences that does move it toward DNMT3B. R887A and N838A also caused distinct changes of flanking sequence preferences. Finally, S837A and S881A caused only weak effects and cluster together with WT DNMT3A.
Combined readout of the +1 to +3 flank sites
Next, we were interested to determine the combined readout of the base pairs at the +1 to +3 flank sites. To this end, the relative preferences of all 16 +2/+3 flank dinucleotides were determined for each given base at the +1 site, revealing several interesting combined effects with WT DNMT3A (Fig. 2E). For example: (1) T(+1) is favored in the +2/+3 context of AG, (2) A(+1) is favored in the +2/+3 context GA, GC, and GG but disfavored in combination with CG, and (3) C(+1) is favored in the TG and TT context. The corresponding patterns were determined for all mutants (Fig. S2) and the correlations of mutant dinucleotide preferences with the WT were determined (Fig. 2F). This analysis revealed that R882A by far showed the weakest correlation, which underscores the importance of R882 for the +1 to +3 flank interaction. The more conservative R882H exchange led to much smaller changes. Interestingly, the mutations in the TRD loop led to reduction of the correlation of +2/+3 flank preferences in the presence of A(+1), and in the case of R836A, also for G(+1). Finally, L883A showed reduced correlation with all bases at the +1 site, except G. Based on this, one can conclude that R836 forms an essential interaction with G(+1), the TRD loop mediates interactions with A(+1), and L883A is required for the +1 to +3 flank interaction, if there is no G at the +1 site. Among the detailed results, the strong changes of +2/+3 site effects of G(+1) with R836A, strong preferences for AAT, CAA, and CAT with N838A, and TCC with R882H, R882A, and L883A are the strongest effects (Fig. S2).
In summary, these data show a combined readout of DNMT3A with the +1 and +2/3 flanking sites that is mediated by R836 and the RD loop residues, mainly R882.
Effects of the outer flanks
In the previous experiments shown in Figure 2, A and B, flanking effects on CpG methylation were detectable also at the +4 to +7 positions. Related to this, in a previous study, we showed strong effects of these sites on the comethylation of CpG sites in a distance of 12 base pairs and found that A/T bases were preferred (17). We suspected that effects of these regions, which we now call “outer flanks”, might have been partially masked by the stronger effects of the inner −3 to +3 flanks. Hence, we studied outer flank effects in five different substrates with fixed inner flanks, which were chosen to cover the spectrum from highly preferred to strongly disfavored inner flanks. The substrates were prepared with the different inner flanks surrounded by randomized −10 to −4 and +4 to +10 regions, methylated by DNMT3A WT and the mutants in two experimental repeats, and analyzed as described above for the inner flank experiments (Table S3). The individual repeats showed very good correlation in most cases (Table S4) and the data were merged for further analysis.
As shown in Figure 3A, clear and distinct outer flank effects were observed with WT mainly at the −4 and +4 to +8 sites. To estimate the overall effect size, average methylation levels were determined for substrates with each combination of N-NNN bases at the 4, +4, +5, and +6 positions showing that these values differed by up to 8-fold. In general, our data indicate that only small sequence preferences and effects on activity were observed in the context of a highly preferred inner flank (GTACGTCA in Fig. 3A), but the influence of the outer flanks on activity increased with more and more disfavored inner flanks. In general, the effect of 3’ flanks was stronger than of the 5’ flanks, and often A/T bases were preferred at these places in agreement with our previous data (17). Interestingly, strong 5′ outer flank effects were only observed in the TGCCGTTG substrate which is missing the highly favored T(−2) residue. Corresponding outer flank profiles were also determined for the mutants (Fig. S3). For comparison, the RMSD deviation of the base distributions averaged for all positions were compiled for all five substrates (Fig. 3B). The data revealed very small outer flank effects on the most preferred substrate for WT and all mutants. Interestingly, on the more disfavored substrates, the WT showed more pronounced flanking effects than the mutants, which is also clearly visible when comparing the individual profiles (Fig. S3).
Figure 3.
Outer flank effect on DNMT3A activity.A, effects of the −8 to +8 flanks on the methylation of five substrates with fixed −3 to +3 inner flank with DNMT3A WT. Rank refers to the preference of the corresponding inner flank where small numbers indicate high preference. Methylation range refers to the combined sequences of the −4, +4, +5, and +6 flank sites. B, RMSD values of the deviation of the flanking profiles averaged for all −8 to −4 and +4 to +8 sites. C, RMSD values of the deviation of the flanking profiles of WT and mutants averaged for all five substrates at the different flanking sites. D, +4 to +8 outer flank sequence preferences for WT and mutants averaged for all five substrates.
Next, the RMSD values of the differences between the outer flanking sequence preferences of the mutants and WT were calculated for each position and averaged for the five substrates (Fig. 3C). The data revealed strong differences between WT and mutant preferences at the 3′ side (+4 to +8), which is in agreement with the fact that the mutants were selected for their potential interaction with this part of the DNA. Strongest effects were observed at +4, where many mutants showed strong deviations, like a strong disfavor for G(+4) of L883A in the ATT-ATG and TGC-TTG substrates or a strong preference for G(+4) of R836A and S837A in the GTC-CGA substrate. It is noticeable that strong outer flank effects were observed with R836A, R887A, S881A, and R882A/H even at the +6 to +8 sites. To compare the data at a combined level, +4 to +8 outer flanking sequence preferences were averaged for all five substrates for WT and all mutants (Fig. 3D). The data clearly indicate the preference of WT for A or T at all these sites, which has been lost or strongly reduced with all mutants with the exception that the T(+6) preference is still observed, though reduced with R887 and R882 mutants.
In summary, these data show that outer flank sequences influence the activity of DNMT3A and largest outer flank effects were observed with unfavored inner flanks.
Analysis of the CpG recognition of DNMT3A WT and mutants
To study the activity and flanking sequence preferences of non-CpG methylation by DNMT3A and its mutants, Deep Enzymology experiments were also conducted with a substrate containing a CpN target site in a randomized sequence context and the data were split into CpG, CpA, CpT, and CpC methylation. Read counts and methylation levels of the individual experimental repeats are listed in Table S5, and correlations of the flanking profiles of individual repeats are shown in Table S6. In general, correlations of the CpG and CpA data were good, which is related to the fact that all enzymes were most active in these sequence contexts. In addition, the CpG flank preferences determined with the CpN substrate were compared with the preferences determined with the CpG substrate and found to be highly correlated, despite the fact that CpG methylation levels in this CN methylation analysis were intentionally higher, to make non-CpG methylation more detectable (Table S6). Correlations of the CpT data were lower due to the low overall methylation activity leading to small numbers of methylated reads in some cases. The CpC correlation was only satisfying in case of the WT and the most active mutants. Hence, the CpT and CpC flanking profiles of some mutants could not be analyzed in detail.
First, the average methylation levels of C in each CpN context were determined and the data fitted to an exponential reaction progress curve to determine the relative methylation rates in the four sequence contexts (CpG, CpA, CpT and CpC) (Figs. 4A and S4). For the most accurate measurement of the relative methylation rates, the time points describing the individual reaction progress in each methylation experiment were included in the fitting. The relative activity of WT DNMT3A at CpA, CpT, and CpC sites in random flanking sequence context was found to be 4.9 %, 2.1 %, and 0.4 %, respectively, of the CpG methylation rate. The largest change in CpG recognition was observed with R836A showing a more than 40-fold increase in CpC methylation, combined with a 3- to 4-fold increase in CpA and CpT methylation. S837A and S881A showed similar relative CpA and CpT methylation as WT together with a mild increase in relative CpC methylation. N838A, R882A, R882H, L883A, and R887A showed reduced relative non-CpG methylation. Our results demonstrate that R836A is a very critical residue for CpG recognition as already observed before with individual test substrates (15). We show that CpC methylation (which is very low with WT DNMT3A) becomes elevated most prominently, followed by CpA and CpT. Actually, methylation activity of R836A at CpC sites was about 4-fold higher than at CpT sites, while it is 5-fold lower in case of WT. These results are in very good agreement with published cellular non-CpG methylation levels of WT and R836A where a strong increase in CpC methylation was observed as well (15). Of note, mutation of N838 led to a decrease in non-CpG methylation indicating a higher CpG specificity although this residue forms direct contacts to the CpG guanine. This paradoxical finding will be discussed later.
Figure 4.
Non-CpG methylation of WT and mutants.A, relative non-CpG methylation rates of DNMT3A WT and mutants. B, flanking sequence preferences of WT DNMT3A for non-CpG methylation. Relative methylation rates of 0.05 correspond to the limit of detection. C, combined readout of the CpN base and flank position +1. Gray shaded experiments could not be analyzed due to the low methylation levels.
Analysis of the flanking preferences of DNMT3A WT and mutants in non-CpG context
As described above for the CpG flanking sequence profiles, we determined the average methylation levels of all substrates containing a particular base at one of the −8 to +8 flank sites to identify bases favorable or unfavorable for activity. In Figure 4B, the profiles of WT are shown for the −4 to +4 region, revealing strong and CpN-specific effects. At the +1 site, which is directly adjacent to the CpN site, the most variable and CpN specific preferences were observed. In case of CpG, a trend of +1 preferences was observed with C>T>A>G. For CpA methylation, the preferences for C(+1) and G(+1) were elevated and a C>G>T>A profile was observed, indicating a relative increase in the preference of CAG methylation. For CpT methylation, a very strong C(+1) preference was seen and CTC by far was the most preferred methylation motif (C>>G>A∼T). In case of CpC methylation (C>T∼A>>G), the disfavor for G(+1) was elevated, which may be related to the fact that a CCG site generates a new CpG dinucleotide, which represents the preferred methylation target. Hence, the second and not the first cytosine in CCG sites will be methylated preferably. No noticeable changes of preferences for non-CpG methylation were observed at the +2 to +4 flanking positions except the general observation that effects were elevated. At the −1 site, C was slightly preferred over A for CpG methylation. In contrast, for all non-CpG activities, only A is preferred at this position indicating that CCG methylation is particularly favored. At the −2 site, the highly characteristic preference profile T>C>A, G was observed for CpG and non-CpG methylation, though effects were even more pronounced in the case of non-CpG activity. No noticeable effects were observed at the −3 and −4 sites.
The −3 to +3 flanking sequence preferences of DNMT3A WT, DNMT3A mutants, and DNMT3B WT (taken from Dukatz et al., 2020 (27)) are compiled in Figure 5. Comparison of all data reveals that WT DNMT3A and most mutants recognize the CpN site and +1 flank in a combined manner. This is clearly illustrated by the relative methylation of dinucleotides comprising the NX part of the CNX sequences (Fig. 4C). By comparing these CNX preference patterns, several groups of mutants can be distinguished:
-
(1)
WT and some mutants showed small changes in preferences (S837A, S881A, and R887A).
-
(2)
R836A showed a preference for T(+1) in all methylation contexts, in particular at non-CpG sites.
-
(3)
N838A showed low methylation of CGA and CGG.
-
(4)
R882A, R882H, and L883A showed very similar patterns that partially overlap with DNMT3B. These profiles are characterized by a disfavor for C(+1) in all sequence contexts and the fact that CAG is strongly preferred.
Figure 5.
Flanking sequence preferences of non-CpG methylation of WT and mutants. The −3 to +3 flanking preference profiles are shown. Some experiments could not be analyzed due to the low methylation levels (n.d.).
More complex, CpN and mutant-specific preference profiles are visible in the individual +3 to −3 flanking sequence preferences of the mutants shown in Figure 5, but here, only the strongest effects will be highlighted. In case of R836A, the preference for C(+1) observed with WT was lost and replaced by an increased preference for T(+1), in particular in non-CpG methylation. At the same time, the CAG preference observed for WT was lost as well. Moreover, A(+2) was more preferred for CpT methylation. N838A showed a specific drop in CGA and (weaker) CGG methylation, corresponding to a loss of the preference for A(+1) and G(+1) in CpG methylation, but its +1 flanking preferences are similar to WT for non-CpG methylation. With R882A, R882H, and L883A, G(+1) was strongly preferred in CpA methylation. Moreover, the strong C(+1) preference for WT in CpA and CpT methylation has been lost in these mutants and T(+1) is disfavored.
In summary, these data reveal strong flanking sequence preferences of DNMT3A in non-CpG methylation activity with a prominent preference for C(+1), which in RD loop mutants is altered to a G(+1) preference similarly as observed in DNMT3B.
Discussion
The DNA interaction of DNMT3A faces two critical challenges. First, target CpG sites must be identified and methylation should be mainly introduced at them. Of note, non-CpG methylation exists in the human genome and it has been connected with important functions, but it must be introduced in a controlled manner and aberrant methylation at arbitrary non-CpG sites must be prevented. One reason for this is that the C5-methyl group of thymine is a critical specificity determinant in protein–DNA interaction used by sequence-specific DNA-binding proteins to discriminate thymine from cytidine (28, 29, 30). Hence, aberrant cytosine methylation in a CpG or non-CpG context has the ability to disturb the DNA interaction of transcription factors and MBD proteins at enhancers and promoters leading to altered gene activity patterns. While CpG methylation has been demonstrated to have strong effects on DNA binding of these proteins (31, 32, 33, 34, 35), the potential role of non-CpG methylation has not been studied systematically for most DNA-binding proteins.
The second challenge for DNMT3A is that it interacts with a very small two-base pair CpG “recognition sequence” that is embedded in a very variable sequence context in genomic DNA. It is known that the static and dynamic conformational properties of DNA are heavily modulated by the DNA sequence (29, 36, 37). For structural reasons, DNMTs interact with larger regions of the DNA leading to the challenge to establish an accurate positioning of the target CpG in the active site of the enzyme although the structural properties of different DNA substrate sequences vary. Previous work has shown that different DNMTs and also TET enzyme act on target sites embedded in different sequence contexts with different efficiency and strong preferences for some flanking contexts over other ones were reported (18, 26, 27, 38). Flanking sequence preferences have been documented to affect cellular 5 mC and 5hmC patterns and they could be connected with divergent biological roles of DNMT3A and DNMT3B and the pathogenicity of the R882H cancer mutation in DNMT3A (19). In this work, we have focused on the combined recognition and interaction of DNMT3A with the CpG guanine residue, the adjacent −3 to +3 inner flanking region, and the −8 to +8 outer flanking sequence. We have mutated several DNA-interacting residues and analyzed DNA methylation of WT DNMT3A and its mutants using libraries of substrates containing CpG and non-CpG sites in randomized flanking sequence context using the Deep Enzymology approach (19). Our data illustrate strong (>100 fold) flanking sequence effects and combined readout of the CpG guanine with the +1 to +3 flank sites, which is further connected with readout of outer flanks (+4 to +8). In some cases, these effects were mutant specific, allowing us to identify the roles of individual amino acid residues in the formation of the contact networks between DNMT3A and its target DNA.
CpG guanine recognition
In structural studies, R836 or N838 were found to contact the CpG guanine in a manner depending on the +1 flanking sequence (15, 16). In addition, a water-mediated contact to the N7-atom of the guanine is mediated by T834. In contrast, in DNMT3B, only the N779- and T775-mediated contacts were observed (corresponding to DNMT3A T834 and N838), while DNMT3B K777 (corresponding to R836) adopts an orientation pointing away from the CpG guanine (18). Based on our activity measurements in all possible flanking contexts, both enzymes qualitatively exhibit similar preferences for CpN sites (CpG>>CpA>CpT>CpC). The preference for a CpA as second-best target site could be due to the highly conserved water-mediated contact between T834/T775 and the N7 atom of the CpG guanine that could be equally formed with adenine. This interpretation is supported by the finding that a T775A mutation in DNMT3B caused an almost complete loss of activity (27).
However, the relative non-CpG activity of DNMT3B was stronger than that of DNMT3A, which may be related to the lack of the possibility to form an arginine-guanine contact in DNMT3B. An important role of R836 for CpG guanine recognition by DNMT3A is further supported by our finding that mutation of R836 led to a pronounced reduction of CpG specificity, while mutation of N838 even led to an increase in CpG recognition. Our observation that the DNMT3A R836A mutant retains a considerable CpG specificity suggests that N838 can partially take over the role of R836 in CpG recognition, when R836 is mutated. The finding of an increased CpG specificity of N838A can be explained by a similar mechanism, proposing that R836 may use the space emptied by the N838A mutation, and this might strengthen its contact to the CpG guanine and thereby improve CpG recognition. The reduced non-CpG activities of R882A, R882H, L883A, and R887A can be understood, because weakening of the DNA contacts by the RD loop through these mutations might increase the requirements for a correct positioning of the TRD loop, leading to more stringent readout of the CpG guanine.
Flanking sequence preferences in CpG and non-CpG methylation
Our data show that DNMT3A has pronounced preferences for inner flanking sequences, which lead to >100-fold changes in methylation rates, because in the same library, several NNCGNNN flanks showed zero methylation when the most preferred sequence context was already 95% methylated. One general result of the non-CpG flanking sequence analyses of DNMT3A WT and mutants was that flanking effects are elevated on disfavored CpN sites, again indicating that difficult substrates require a supportive flanking context for efficient methylation. One interesting observation with WT DNMT3A was that the C(−1) preference was high for CpG methylation, while in case of all other methylation contexts, A(−1) was preferred. This effect cannot be further interpreted as the mutational study conducted here did not cover the amino acid residues putatively involved in −1 flank interaction. Another very strong effect was that CpT methylation had a very pronounced preference for C(+1) meaning that CTC methylation is highly preferred.
DNA interaction of TRD loop residues R836 and N838
R836 is a key residue in CpG guanine and flanking interaction of DNMT3A. Structural studies detected a striking movement of R836, which contacts the CpG guanine in a CGT structure, but the +1 and +2 flank in a CGA structure. Our data revealed several biochemical effects that are connected to R836. (1) R836A showed a strong change of flanking sequence preferences at the +1 side, where the preference for C(+1) is lost, which is highly characteristic for DNMT3A non-CpG methylation and also has been observed in cells (39). DNA methylation in CpA context has been observed in neurons and it has been shown to have very important biological roles (10, 11). Hence, R836 may contact C(+1) in particular on non-CpG target sites where the CpG guanine interaction partner for R836 is absent. (2) R836A showed a strong decrease in G(+1) preference of CpA methylation indicating that this residue stimulates methylation of CAG sites. These effects may be related to the contact of R836 to the N7 atom of A(+1) that is observed in the CGA structure and may be similarly possible with CAG. (3) R836A shows a strong preference for T(+1) in particular in non-CpG methylation which is specific for this mutant. This effect might indicate that the mutated A836 could engage in a van-der-Waals contact with the methyl group of T(+1) in non-CpG sites. (4) In CpG methylation, R836A showed a specific effect on the +2/+3 flank recognition in the context of G(+1), suggesting that R836 adopts a specific conformation on CGG complexes, which may be related to the very low preference for this sequence. This effect may be related to the ability of R836 to contact G(+1) and also the +2 base by hydrogen bonds, thereby affecting the +2 site interaction on CGG sites. (5) The influence of the folded back conformation of R836 on the +2 and +3 flank interaction of the entire RD loop is also visible in the altered CpG +2/+3 dinucleotide recognition pattern of all TRD loop mutants in the context of A(+1). Based on this, one can conclude that the TRD loop mediates interactions with A(+1) that depend on the correct conformation of R836, S837, and N838.
N838 is the second most important residue for CpG recognition and flank interactions of DNMT3A. In the DNMT3A CGA structure, the side chain of N838 occupies the place of R836 and it contacts the CpG guanine, while in the CGT structure, it forms an H-bond to the +2/+3 phosphodiester group and the +2′ base. The N838A exchange led to strong changes in the flanking sequence preferences at the +1 position, which were distinct from R836A. In particular, the N383A mutant showed a very strong and unique drop of A(+1) and G(+1) preference in CpG (but not in non-CpG) methylation. This effect can be explained by the combined contact of N838 to the CpG guanine and the A(+1) N7 atom seen in the CGA structure. This contact could equally be formed with G(+1), suggesting that N838 is necessary for efficient CGA and CGG methylation. Hence, N838 functions to buffer the effect of R836 preventing an “overshooting” of the C(+1) preference. In summary, R836 and N838 interact with the CpG guanine and flanking sites in a flexible manner ensuring high activity and specificity of DNMT3A in diverse flanking sequence contexts.
Roles of RD loop residues L883 and R882
One of the most striking observations of this study was the very strong rate enhancement caused by the L883A exchange. This finding can be explained in the context of the DNMT3A structure, showing that the hydrophobic L883 side chain is placed at an exposed and hydrophilic position pointing toward the DNA. Nevertheless, this residue is conserved in all DNMT3A enzymes, indicating that it must have an important biological role. Our data show that L883 is required to stabilize the DNMT3A-specific conformation of the RD loop, because the L883A mutation caused similar changes in flanking preferences as R882A and R882H. The +1 to +3 flanking sequence preferences of R882A, R882H, and L883A in CpG and non-CpG methylation showed similarity to DNMT3B, including the preference for G(+1) and A(+1) in CpG methylation, G(+1) in non-CpG methylation, and absence of the DNMT3A-characteristic C(+1) and C(+2) preference in CpG, and even more in non-CpG methylation. These changes convert the DNMT3A type preference profile for the +1 site (C>T>A>G) into a DNMT3B-like profile (G>A>C>T). Our data indicate that the RD loop residues are needed to stabilize its DNMT3A-specific conformation, allowing for several DNMT3A-specific DNA contacts to the flanking sequence. This function apparently is so important and dependent on the presence of L883 that a reduction of the activity of DNMT3A by this residue was tolerated in evolution.
Disruption of the DNMT3A-specific DNA contacts of the RD loop leads to flanking sequence preference profiles that are DNMT3B-like, and this process is one of the important carcinogenic mechanisms of R882H (21, 22, 23). However, the prominent and specific role of the R882H cancer mutation is also due to two more unrelated effects. (1) H882 in the R882H mutant has been shown to form new interface contacts leading to a preferred formation of mutant/mutant RD interfaces, which finally causes a dominant effect of the R882H mutation (24). (2) The R882H mutation arises from the deamination of a methylated cytosine in a CpG site, leading to a high mutational pressure at this site.
Effects of the outer flanks and DNA bending
We demonstrate here that the outer flank sequences also showed up to 8-fold and inner flank–dependent effects on methylation rates indicating that long range effects connect DNA flanking sequence and the active sites of DNMT3A. Overall, DNMT3A showed a preference for A/T bases in the +4 to +8 outer flank region. This finding supports results of a previous work, where we showed that A and T bases in this region are preferable for comethylation of CpG sites on a two-site substrate (17). It was structurally interpreted in the context of the DNA bending required for the interaction of two CpG sites with the two active centers at the RD-interface of the DNMT3A tetramer leading to comethylation of both CpG sites. Our data with the single CpG site substrates presented here reveal similar preferences, which might suggest that these substrates also adopt a bent conformation when being bound to DNMT3A, although they are lacking a second CpG site. Surprisingly, this effect was lost or reduced with almost all mutants. This may be due to the fact that the mutations weaken the DNA interaction, making the combined DNA interaction of both active subunit with the DNA less favorable. Instead, in case of the mutants, only one subunit of DNMT3A might contact the DNA at any time. This would remove the requirement for DNA bending and hence reduce the need to have A/T bases in the +4 to +8 flanking region. However, there was one exception to this rule, because T(+6) was preferred by all mutants, suggesting a more specific role of this base or its A(+6') partner base.
Conclusion and outlook
Our experimental data document the combined and interdependent readout of the CpG site and flanking residues up to eight base pairs away from it. By this, DNMT3A generates networks of interactions that are necessary for the efficient methylation of CpG sites embedded into different flanking sites. In general, we observed that a substrate containing an unfavorable feature (e.g., a non-CpG target site or an unfavored inner flank) shows elevated flanking preferences for the remaining part of the substrate. Mutational analyses allowed us to associate the CpG guanine and +1 to +8 flank DNA-interaction mainly with R836 and N838 from the TRD loop and R882 and L883 from the RD loop.
DNMT3A and DNMT3B arose in the whole genome duplication that occured before the origin of vertebrates estimated at 500 myo (40). Appearance of two DNMT3 paralogs apparently has allowed for an improvement of the DNA methylation machinery by specialization of the two DNMT3 genes that led to the preservation of both paralogs in further evolution. Specialization of DNMT3A and DNMT3B include their regulation and targeting by interaction with distinct sets of transcription factors (41) and divergence of their chromatin interaction, for example, providing the PWWP domain of DNMT3A with a preference for H3K36me2, while the PWWP domain of DNMT3B prefers H3K36me3 (42, 43). Diverging flanking sequence preferences of DNMT3A and DNMT3B represent another factor of evolutionary specialization of DNMT3 enzymes. Different flanking sequence preferences of DNMT3A and DNMT3B have been shown to explain the specific role of DNMT3B for methylation of SATII sequences (18), leading to the connection of DNMT3B with the ICF syndrome (44, 45).
Based on structural studies, the RD loop of both proteins was proposed to contribute to these effects (18). Here, we show experimentally the strong influence of the RD loop conformation on DNMT3A-specific flanking sequence preferences. Our observation that DNMT3A mutants with different mutations in the RD loop exhibit flanking sequence preferences similar to DNMT3B illustrates the key role of this structural element for the DNMT3A/DNMT3B divergence and extends earlier observations for R882H (22, 23). In addition, our data illustrate an important role of the TRD loop in the flanking sequence interactions as well, in particular of R836 and N838. This provides another source of mechanistic divergence between DNMT3A and DNMT3B, because R836 is replaced by K777 in DNMT3B which has a different contact potential than arginine and, for example, has been shown to be responsible for the DNMT3B-specific preference for G(+1) in non-CpG methylation (18, 27), while we show here that R836 mediates the C(+1) preference in non-CpG methylation of DNMT3A. One role of N838 is to balance this effect of R836 by supporting CGA and CGG methylation, to avoid a too strong preference for C(+1).
We consider it very plausible that flanking sequence preferences would be observed with other DNA interaction enzymes and DNA-binding proteins, once experimental studies tailored to observe such effects had been conducted. Unfortunately, due to the large number of different enzyme-DNA complexes that need to be considered, a structural analysis of the detailed effects discovered here is far beyond the current state of structural analysis or molecular dynamics (MD) simulations. Hence, our data illustrate the clear demand for improved methods to allow a mechanistic understanding of the basic processes determining the interaction of DNMT3A with its DNA substrate. Improved methods should allow the parallel investigation of several protein–DNA complexes for example by automated MD simulations using force fields that correctly describe the structural parameters of the DNA and the energetics of its protein interaction. At the same time, simulation times must be long enough to capture the specific conformational changes that accompany complex formation.
Experimental procedures
Mutagenesis and protein expression
The catalytic, C-terminal domain of DNMT3A (residues 612–912 of Q9Y6K1) and its mutants were cloned as MBP-tagged fusion proteins. Protein expression and purification was conducted essentially as described (24, 25). Protein overexpression was carried out in Escherichia coli BL21 (DE3) Codon Plus RIL cells transformed with the corresponding plasmids. If needed, cleavage of the MBP tag was performed as described (24). Mutagenesis was conducted as described (27). The purity of the preparations was estimated to be >95% from Coomassie-stained SDS gels. The concentrations of the proteins were determined by UV spectrophotometry and confirmed by densitometric analysis of Coomassie BB–stained SDS–polyacrylamide gels.
Radioactive DNA methylation kinetics
Methylation activities of DNMT3A WT and mutants were determined using an avidin-biotin methylation plate assay using a biotinylated double-stranded 30-mer oligonucleotide with a single CpG site (GAA GCT GGG ACT TCCGGG AGG AGA GTG CAA) basically as described (24, 27). Methylation reactions with DNMT3A were conducted at 37 °C with 2 μM enzyme in methylation buffer (20 mM Hepes pH 7.5, 1 mM EDTA, 50 mM KCl, 0.25 mg/ml bovine serum albumin) in the presence of 1 μM of the biotinylated substrate. The reactions were started by adding 0.76 μM radioactively labeled AdoMet (PerkinElmer). The initial slope of the enzymatic reaction was determined by linear regression.
Flanking sequence preference analysis with a randomized substrate and bioinformatics analysis
The preparation of the substrate with CpN site in a 10 base pair randomized sequence context, methylation, bisulfite conversion, and library preparation was conducted as described (18, 26). In addition, the effect of the outer flanks was tested using five substrates with fixed −3 to +3 inner flanks and randomized −10 to −4 and +4 to +10 outer flanks, which were selected to cover the entire range of preferred (low ranks) to disfavored inner flanks (high ranks):
-
•
PB955: inner flank GTACGTCA, rank 57
-
•
MD221: inner flank CTACGGCA, rank 112
-
•
MD222: inner flank GTCCGCGA, rank 564
-
•
PB954: inner flank ATTCGATG, rank 3814
-
•
PB956: inner flank TGCCGTTG, rank 3883
Library methylation was conducted at 37 °C as described (18, 26) using 5 ng/μl of the library in buffer containing 20 mM Hepes pH 7.5, 1 mM EDTA, 50 mM KCl, 100 μg/ml bovine serum albumin, and 0.8 mM AdoMet (Sigma). Enzyme concentrations (0.5–5 μM) and incubation times (30–120 min) were as indicated. The methylation reactions were stopped by shock freezing in liquid nitrogen, then treated with proteinase K (NEB) for 2 h at 42 °C, and purified by PCR Clean-up kit (Macherey-Nagel). Afterward, bisulfite conversion and NGS library preparation were conducted as described (18, 24, 26). Different sets of barcodes were introduced in the PCR steps to distinguish different samples and experiments. NGS data analysis was conducted basically as described (27).
Data availability
NGS kinetic raw data will be available at DaRUS at https://doi.org/10.18419/darus-2993
Biochemical data are provided with this article.
Supporting information
This article contains supporting information.
Conflict of interest
The authors declare no competing interests.
Acknowledgments
This work has been supported by the Deutsche Forschungsgemeinschaft (JE 252/10 and JE252/36).
Author contributions
A. J. and Mi. D. devised the study. Mi. D. performed the experimental work with support from Ma. D. and E. S. Mi. D., S. A. and P. B. conducted the NGS analyses. Mi. D., S. A., P. B. and A. J. were involved in NGS data analysis and bioinformatic analyses. A. M. prepared the MSA. All authors were involved in final data analysis and interpretation and writing of the manuscript. All authors approved the final version of the manuscript. Mi. D. and A. J. conceptualization; Mi. D., Ma. D., E. S., and A. J. formal analysis; Mi. D., Ma. D., E. S., S. A., A. d. M., and P. B. investigation; Mi. D., A. d. M., and A. J. visualization; Mi. D. and A. J. writing–original draft; Mi. D., Ma. D., E. S., S. A., A. d. M., P. B., and A. J. writing–review and editing; S. A. and P. B. methodology; P. B. software; A. J. project administration; A. J. funding acquisition.
Edited by Joseph Jez
Supporting information
References
- 1.Schubeler D. Function and information content of DNA methylation. Nature. 2015;517:321–326. doi: 10.1038/nature14192. [DOI] [PubMed] [Google Scholar]
- 2.He Y., Ecker J.R. Non-CG methylation in the human genome. Annu. Rev. Genomics Hum. Genet. 2015;16:55–77. doi: 10.1146/annurev-genom-090413-025437. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Jeltsch A., Broche J., Bashtrykov P. Molecular processes connecting DNA methylation patterns with DNA methyltransferases and histone modifications in mammalian genomes. Genes. 2019;10:388. doi: 10.3390/genes10050388. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Bergman Y., Cedar H. DNA methylation dynamics in health and disease. Nat. Struct. Mol. Biol. 2013;20:274–281. doi: 10.1038/nsmb.2518. [DOI] [PubMed] [Google Scholar]
- 5.Zeng Y., Chen T. DNA methylation reprogramming during mammalian development. Genes. 2019;10:257. doi: 10.3390/genes10040257. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Gowher H., Jeltsch A. Mammalian DNA methyltransferases: new discoveries and open questions. Biochem. Soc. Trans. 2018;46:1191–1202. doi: 10.1042/BST20170574. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Chen Z., Zhang Y. Role of mammalian DNA methyltransferases in development. Annu. Rev. Biochem. 2019;89:135–158. doi: 10.1146/annurev-biochem-103019-102815. [DOI] [PubMed] [Google Scholar]
- 8.Yang L., Rau R., Goodell M.A. DNMT3A in haematological malignancies. Nat. Rev. Cancer. 2015;15:152–165. doi: 10.1038/nrc3895. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Hamidi T., Singh A.K., Chen T. Genetic alterations of DNA methylation machinery in human diseases. Epigenomics. 2015;7:247–265. doi: 10.2217/epi.14.80. [DOI] [PubMed] [Google Scholar]
- 10.Kinde B., Gabel H.W., Gilbert C.S., Griffith E.C., Greenberg M.E. Reading the unique DNA methylation landscape of the brain: non-CpG methylation, hydroxymethylation, and MeCP2. Proc. Natl. Acad. Sci. U. S. A. 2015;112:6800–6806. doi: 10.1073/pnas.1411269112. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Christian D.L., Wu D.Y., Martin J.R., Moore J.R., Liu Y.R., Clemens A.W., et al. DNMT3A haploinsufficiency results in behavioral deficits and global epigenomic dysregulation shared across neurodevelopmental disorders. Cell Rep. 2020;33 doi: 10.1016/j.celrep.2020.108416. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Jia D., Jurkowska R.Z., Zhang X., Jeltsch A., Cheng X. Structure of Dnmt3a bound to Dnmt3L suggests a model for de novo DNA methylation. Nature. 2007;449:248–251. doi: 10.1038/nature06146. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Jurkowska R.Z., Anspach N., Urbanke C., Jia D., Reinhardt R., Nellen W., et al. Formation of nucleoprotein filaments by mammalian DNA methyltransferase Dnmt3a in complex with regulator Dnmt3L. Nucl. Acids Res. 2008;36:6656–6663. doi: 10.1093/nar/gkn747. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Jurkowska R.Z., Rajavelu A., Anspach N., Urbanke C., Jankevicius G., Ragozin S., et al. Oligomerization and binding of the Dnmt3a DNA methyltransferase to parallel DNA molecules: heterochromatic localization and role of Dnmt3L. J. Biol. Chem. 2011;286:24200–24207. doi: 10.1074/jbc.M111.254987. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Zhang Z.M., Lu R., Wang P., Yu Y., Chen D., Gao L., et al. Structural basis for DNMT3A-mediated de novo DNA methylation. Nature. 2018;554:387–391. doi: 10.1038/nature25477. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Anteneh H., Fang J., Song J. Structural basis for impairment of DNA methylation by the DNMT3A R882H mutation. Nat. Commun. 2020;11:2294. doi: 10.1038/s41467-020-16213-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Emperle M., Bangalore D.M., Adam S., Kunert S., Heil H.S., Heinze K.G., et al. Structural and biochemical insight into the mechanism of dual CpG site binding and methylation by the DNMT3A DNA methyltransferase. Nucl. Acids Res. 2021;49:8294–8308. doi: 10.1093/nar/gkab600. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Gao L., Emperle M., Guo Y., Grimm S.A., Ren W., Adam S., et al. Comprehensive structure-function characterization of DNMT3B and DNMT3A reveals distinctive de novo DNA methylation mechanisms. Nat. Commun. 2020;11:3355. doi: 10.1038/s41467-020-17109-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Jeltsch A., Adam S., Dukatz M., Emperle M., Bashtrykov P. Deep enzymology studies on DNA methyltransferases reveal novel connections between flanking sequences and enzyme activity. J. Mol. Biol. 2021;433 doi: 10.1016/j.jmb.2021.167186. [DOI] [PubMed] [Google Scholar]
- 20.Guo J.U., Su Y., Shin J.H., Shin J., Li H., Xie B., et al. Distribution, recognition and regulation of non-CpG methylation in the adult mammalian brain. Nat. Neurosci. 2014;17:215–222. doi: 10.1038/nn.3607. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Emperle M., Rajavelu A., Kunert S., Arimondo P.B., Reinhardt R., Jurkowska R.Z., et al. The DNMT3A R882H mutant displays altered flanking sequence preferences. Nucl. Acids Res. 2018;46:3130–3139. doi: 10.1093/nar/gky168. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Emperle M., Adam S., Kunert S., Dukatz M., Baude A., Plass C., et al. Mutations of R882 change flanking sequence preferences of the DNA methyltransferase DNMT3A and cellular methylation patterns. Nucl. Acids Res. 2019;47:11355–11367. doi: 10.1093/nar/gkz911. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Norvil A.B., AlAbdi L., Liu B., Tu Y.H., Forstoffer N.E., Michie A.R., et al. The acute myeloid leukemia variant DNMT3A Arg882His is a DNMT3B-like enzyme. Nucl. Acids Res. 2020;48:3761–3775. doi: 10.1093/nar/gkaa139. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Mack A., Emperle M., Schnee P., Adam S., Pleiss J., Bashtrykov P., et al. Preferential self-interaction of DNA methyltransferase DNMT3A subunits containing the R882H cancer mutation leads to dominant changes of flanking sequence preferences. J. Mol. Biol. 2022;434 doi: 10.1016/j.jmb.2022.167482. [DOI] [PubMed] [Google Scholar]
- 25.Emperle M., Dukatz M., Kunert S., Holzer K., Rajavelu A., Jurkowska R.Z., et al. The DNMT3A R882H mutation does not cause dominant negative effects in purified mixed DNMT3A/R882H complexes. Sci. Rep. 2018;8 doi: 10.1038/s41598-018-31635-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Adam S., Anteneh H., Hornisch M., Wagner V., Lu J., Radde N.E., et al. DNA sequence-dependent activity and base flipping mechanisms of DNMT1 regulate genome-wide DNA methylation. Nat. Commun. 2020;11:3723. doi: 10.1038/s41467-020-17531-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Dukatz M., Adam S., Biswal M., Song J., Bashtrykov P., Jeltsch A. Complex DNA sequence readout mechanisms of the DNMT3B DNA methyltransferase. Nucl. Acids Res. 2020;48:11495–11509. doi: 10.1093/nar/gkaa938. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Garvie C.W., Wolberger C. Recognition of specific DNA sequences. Mol. Cel. 2001;8:937–946. doi: 10.1016/s1097-2765(01)00392-6. [DOI] [PubMed] [Google Scholar]
- 29.Rohs R., Jin X., West S.M., Joshi R., Honig B., Mann R.S. Origins of specificity in protein-DNA recognition. Annu. Rev. Biochem. 2010;79:233–269. doi: 10.1146/annurev-biochem-060408-091030. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Slattery M., Zhou T., Yang L., Dantas Machado A.C., Gordan R., Rohs R. Absence of a simple code: how transcription factors read the genome. Trends Biochem. Sci. 2014;39:381–399. doi: 10.1016/j.tibs.2014.07.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Maurano M.T., Wang H., John S., Shafer A., Canfield T., Lee K., et al. Role of DNA methylation in modulating transcription factor occupancy. Cell Rep. 2015;12:1184–1195. doi: 10.1016/j.celrep.2015.07.024. [DOI] [PubMed] [Google Scholar]
- 32.Yin Y., Morgunova E., Jolma A., Kaasinen E., Sahu B., Khund-Sayeed S., et al. Impact of cytosine methylation on DNA binding specificities of human transcription factors. Science. 2017;356 doi: 10.1126/science.aaj2239. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Kribelbauer J.F., Laptenko O., Chen S., Martini G.D., Freed-Pastor W.A., Prives C., et al. Quantitative analysis of the DNA methylation sensitivity of transcription factor complexes. Cell Rep. 2017;19:2383–2395. doi: 10.1016/j.celrep.2017.05.069. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Shimbo T., Wade P.A. Proteins that read DNA methylation. Adv. Exp. Med. Biol. 2016;945:303–320. doi: 10.1007/978-3-319-43624-1_13. [DOI] [PubMed] [Google Scholar]
- 35.Liu K., Xu C., Lei M., Yang A., Loppnau P., Hughes T.R., et al. Structural basis for the ability of MBD domains to bind methyl-CG and TG sites in DNA. J. Biol. Chem. 2018;293:7344–7354. doi: 10.1074/jbc.RA118.001785. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Rohs R., West S.M., Sosinsky A., Liu P., Mann R.S., Honig B. The role of DNA shape in protein-DNA recognition. Nature. 2009;461:1248–1253. doi: 10.1038/nature08473. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Abe N., Dror I., Yang L., Slattery M., Zhou T., Bussemaker H.J., et al. Deconvolving the recognition of DNA shape from sequence. Cell. 2015;161:307–318. doi: 10.1016/j.cell.2015.02.008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Adam S., Bracker J., Klingel V., Osteresch B., Radde N.E., Brockmeyer J., et al. Flanking sequences influence the activity of TET1 and TET2 methylcytosine dioxygenases and affect genomic 5hmC patterns. Commun. Biol. 2022;5:92. doi: 10.1038/s42003-022-03033-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Lee J.H., Park S.J., Nakai K. Differential landscape of non-CpG methylation in embryonic stem cells and neurons caused by DNMT3s. Sci. Rep. 2017;7 doi: 10.1038/s41598-017-11800-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.de Mendoza A., Poppe D., Buckberry S., Pflueger J., Albertin C.B., Daish T., et al. The emergence of the brain non-CpG methylation system in vertebrates. Nat. Ecol. Evol. 2021;5:369–378. doi: 10.1038/s41559-020-01371-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Jeltsch A., Jurkowska R.Z. Allosteric control of mammalian DNA methyltransferases - a new regulatory paradigm. Nucl. Acids Res. 2016;44:8556–8575. doi: 10.1093/nar/gkw723. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Baubec T., Colombo D.F., Wirbelauer C., Schmidt J., Burger L., Krebs A.R., et al. Genomic profiling of DNA methyltransferases reveals a role for DNMT3B in genic methylation. Nature. 2015;520:243–247. doi: 10.1038/nature14176. [DOI] [PubMed] [Google Scholar]
- 43.Weinberg D.N., Papillon-Cavanagh S., Chen H., Yue Y., Chen X., Rajagopalan K.N., et al. The histone mark H3K36me2 recruits DNMT3A and shapes the intergenic DNA methylation landscape. Nature. 2019;573:281–286. doi: 10.1038/s41586-019-1534-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Okano M., Bell D.W., Haber D.A., Li E. DNA methyltransferases Dnmt3a and Dnmt3b are essential for de novo methylation and mammalian development. Cell. 1999;99:247–257. doi: 10.1016/s0092-8674(00)81656-6. [DOI] [PubMed] [Google Scholar]
- 45.Xu G.L., Bestor T.H., Bourc'his D., Hsieh C.L., Tommerup N., Bugge M., et al. Chromosome instability and immunodeficiency syndrome caused by mutations in a DNA methyltransferase gene. Nature. 1999;402:187–191. doi: 10.1038/46052. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Data Availability Statement
NGS kinetic raw data will be available at DaRUS at https://doi.org/10.18419/darus-2993
Biochemical data are provided with this article.