Abstract
Women who are heterozygous for deleterious BRCA1 germline mutations harbor a high risk of hereditary breast cancer. Previous Brca1‐heterozygous animal models do not recapitulate the breast cancer phenotype, and thus all currently used knockout models adopt conditional, mammary‐specific homozygous Brca1 loss or addition of Trp53 deficiency. Herein, we report the creation and characterization of a novel Brca1 mutant rat model harboring the germline L63X mutation, which mimics a founder mutation in Japan, through CRISPR‐Cas9–based genome editing. Homozygotes (Brca1 L63X/L63X) were embryonic lethal, whereas heterozygotes (Brca1 L63X/+) showed apparently normal development. Without carcinogen exposure, heterozygotes developed mammary carcinoma at a comparable incidence rate with their wild‐type (WT) littermates during their lifetime. Intraperitoneal injection of 1‐methyl‐1‐nitrosourea (25 or 50 mg/kg) at 7 weeks of age induced mammary carcinogenesis at comparable levels among the heterozygotes and their littermates. After exposure to ionizing radiation (0.1–2 Gy) at 7 weeks of age, the heterozygotes, but not WT littermates, displayed dose‐dependent mammary carcinogenesis with 0.8 Gy−1 excess in hazard ratio during their middle age; the relative susceptibility of the heterozygotes was more prominent when rats were irradiated at 3 weeks of age. The heterozygotes had tumors with a lower estrogen receptor α immunopositivity and no evidence of somatic mutations of the WT allele. The Brca1 L63X/+ rats thus offer the first single‐mutation, heterozygous model of BRCA1‐associated breast cancer, especially with exposure to a DNA break‐inducing carcinogen. This implies that such carcinogens are causative and a key to breast cancer prevention in individuals who carry high‐risk BRCA1 mutations.
Keywords: animal model, breast cancer, genome editing, hereditary breast and ovarian cancer syndrome, radiation carcinogenesis
Carrying one copy of pathogenic variants of the DNA repair gene BRCA1 is a common cause of hereditary breast cancer but, as previous mouse models with one deficient copy do not mimic the disease, researchers had to use mice with a two‐copy deficiency or one‐copy deficiency with additional mutations, which may not mimic the very early steps of the disease. Here the authors used the rat, a less common but good model of breast cancer, to produce a Brca1 knockout that affects one copy and mimics a human variant common in Japan. The new model reproduces, especially under a DNA‐breaking genotoxic stress, aspects of the disease and can contribute to studies related to prevention.

Abbreviations
- CI
confidence interval
- ER
estrogen receptor
- HR
hazard ratio
- MNU
1‐methyl‐1‐nitrosourea
- qPCR
quantitative PCR
- QST
National Institutes for Quantum Science and Technology
- WT
wild type
1. INTRODUCTION
Breast cancer is experienced by 10% or more of women in developed countries, and pathogenic germline mutations have been estimated to cause 5%–10% of female breast cancers. 1 , 2 BRCA1 and BRCA2 are involved in homologous recombination repair of DNA double strand breaks, and one copy of pathogenic germline mutations of BRCA genes substantially increases the lifetime risk of breast cancer. 3 , 4 The risk of carcinogen exposure in people with defects in such DNA damage repair systems has hence been a matter of concern. Recent studies suggest that exposure at a young age to low‐dose radiation used in diagnostic procedures or cigarette smoke increases breast cancer risk in women harboring such germline mutations, albeit with possible influence of biases and necessity for further research. 5 , 6 , 7 Breast cancers associated with BRCA1 mutations are frequently negative for ovarian hormone receptors, whereas many sporadic breast cancers express those receptors. 4
Development of preventive measure and basic understanding of key mechanisms of BRCA‐related breast carcinogenesis are thus urgent, and good animal models will facilitate relevant research and development. More than 10 animal models of Brca1 mutations have thus far been developed. 8 , 9 , 10 Notably, none of the heterozygous Brca1 mouse mutants display increase in mammary tumorigenesis. 8 , 9 , 10 As mice with homozygous Brca1 mutations do not survive, several lines of conditional knockouts have been developed in which homozygous deletion was driven by mammary‐specific promoters. 8 , 9 , 10 Alternatively, Brca1 heterozygous mice require a heterozygous loss of Trp53 to achieve increased mammary tumorigenesis. 8 , 9 , 10 These models have contributed to establishing novel therapeutic concepts. 11 In contrast, it is difficult to assume that these models recapitulate the very early phases of breast carcinogenesis of heterozygous women carrying BRCA1 mutations. We supposed that this issue is attributed to the low spontaneous incidence of mammary cancer in most unmodified mouse strains. Rats have higher spontaneous mammary cancer incidence, although their use has been less widespread. 12 , 13 A random‐mutagenesis study has produced the first Brca1 knockout rat, but this strain did not show an increased incidence of mammary cancer. 14
BRCA1 variants have been found in ~1.4% of Japanese patients with breast cancer, and the BRCA1 L63X variant (a nonsense mutation of codon 63, which encodes leucine) is the most prevalent therein. 15 , 16 , 17 We thus introduced a point mutation mimicking L63X in the rat Brca1 gene. We further characterized the normal development of the rats, as well as their mammary cancer development, with or without treatment with carcinogens including ionizing radiation (as a DNA break‐inducing agent) and MNU (as a DNA alkylating agent). The present study is the first report that a single heterozygous germline mutation of Brca1 can impose breast cancer susceptibility in a laboratory rodent when exposed to ionizing radiation, thus suggesting the use of the Brca1 L63X/+ rat as a new model recapitulating the features of the human disease.
2. MATERIALS AND METHODS
2.1. Animal experiments
All procedures below were approved by the Institutional Animal Care and Use Committee of either Kyoto University or QST and carried out essentially as described. 18 , 19 , 20 In brief, fertilized eggs of Jcl:SD rats (Clea Japan) were injected with guide RNA, single‐stranded oligodeoxynucleotides (Figure 1A and Table 1) and Cas9 mRNA and transferred to a pseudopregnant foster mother. Genotyping was undertaken as described below. The strain (named Jcl:SD‐Brca1 em1kyo) was maintained by mating mutants with purchased closed‐colony Jcl:SD rats. The mutant rats and WT littermates of the second to seventh generations relative to the founder were injected intraperitoneally with MNU (Toronto Research Chemicals Inc.), whole‐body irradiated with 137Cs γ rays (dose rate, 0.5 Gy/min), or left untreated. The rats were palpated weekly, and tumors reaching ~2 cm in diameter were biopsied and diagnosed on H&E‐stained sections. When rats were identified as having mammary carcinoma or showed general deterioration, they were euthanized and autopsied. A portion of each tumor was fixed in 10% phosphate‐buffered formalin and histological diagnosis was confirmed.
FIGURE 1.
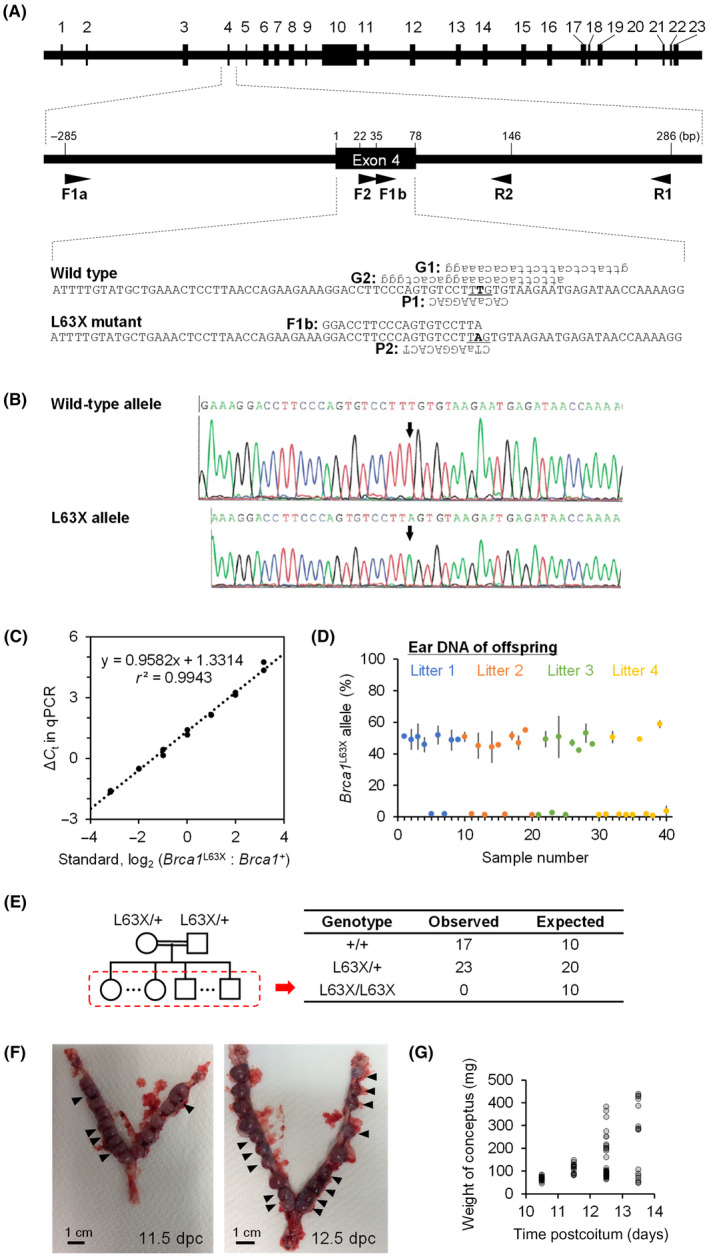
Generation and heredity of the Brca1 L63X allele. (A) Structure of rat Brca1 and oligonucleotides used. Top, exon–intron structure. Middle, a magnified view. Bottom, nucleotide sequences. Arrowheads, primers; G1 and G2, guide RNAs; P1 and P2, probes. Uppercase, deoxyribonucleotides; lowercase, ribonucleotides; bold and underlined, location of the c.188T>A (p.L63X) mutation. (B) Sequence of cloned PCR products from the founder rat Brca1 exon 4. Arrows, induced mutation. (C–E) Heredity of the Brca1 L63X allele. (C) Standard curve of the allele‐specific quantitative PCR. ΔC t, difference in the threshold cycles for the two molecular species. (D) Content of the Brca1 L63X allele in the genomic DNA of offspring (n = 40) of Brca1 L63X/+ heterozygotes. Mean and SD (duplicate measurements). (E) Contingency table of observed and expected numbers of offspring with each genotype. (F) Representative photographs of uteri of pregnant dams at designated days postcoitum (dpc). Arrowheads, maldeveloping conceptuses. (G) Weight of individual conceptuses
TABLE 1.
Oligonucleotides
| Use | ID | Type | Sequence (5′→3′) |
|---|---|---|---|
| Genome editing | G1 | Guide RNA | GTTATCTCATTCTTACACAAAGG |
| G2 | Guide RNA | ATTCTTACACAAAGGACACTGGG | |
| O1 | ssODN | ACTCCTTAACCAGAAGAAAGGACCTTCCCAGTGTCCTT[A]GTGTAAGAATGAGATAACCAAAAGGTAAATAACATGTGTAA | |
| Genotyping | F1a | Forward primer | TGCAGGTAAGTGTAATTTTCATAGG |
| F1b | Forward primer | GGACCTTCCCAGTGTCCTT[A] | |
| R1 | Reverse primer | CCGATGTGCATGGTACTGTC | |
| qPCR | F2 | Forward primer | TAACCAGAAGAAAGGAC |
| R2 | Reverse primer | CTACACATCAATTTCTACTT | |
| P1 | Probe, FAM | CAC[a]AAGGAC | |
| P2 | Probe, HEX | C[T]aAGGACACT | |
| S1 | Standard | CCACACAGTGCGACCACATATTTTGCAAATTTTGTATGCTGAAACTCCTTAACCAGAAGAAAGGACCTTCCCAGTGTCCTT[T]GTGTAAGAATGAGATAACCAAAAGGAGCCTACAAGGAAGTGCAAGGTTTAGTCAACTTGTTGAAGAGCTGCTGAAAAT | |
| S2 | Standard | CCACACAGTGCGACCACATATTTTGCAAATTTTGTATGCTGAAACTCCTTAACCAGAAGAAAGGACCTTCCCAGTGTCCTT[A]GTGTAAGAATGAGATAACCAAAAGGAGCCTACAAGGAAGTGCAAGGTTTAGTCAACTTGTTGAAGAGCTGCTGAAAAT | |
| Sequencing | F3 | Forward primer | CACTGCATAGGGAAACTGGC |
| R3 | Reverse primer | GGGACTGATCTAGGGGTGAC | |
| F4 | Forward primer | AACCCTACCCAGAAAGCTCC | |
| R4 | Reverse primer | ACCTGCAGCTGTCTTGAGAT | |
| F5 | Forward primer | GACCCCTTCATTGTCCTCCA | |
| R5 | Reverse primer | TCCCCTAGCTCCCTCATGAT | |
| F6 | Forward primer | AGTCAGTCCACCATGTCAGT | |
| R6 | Reverse primer | GCAGAGCAGACCCATCGATA | |
| F7 | Forward primer | GCACACCACATCTCTCCTCT | |
| R7 | Reverse primer | GCCAAACAACATGCATGACA | |
| F8 | Forward primer | GTTTCTGGTAAGCAAGCCCC | |
| R8 | Reverse primer | GATCAACTTCTGGGCCATGC | |
| F9 | Forward primer | TGCCCCTTCCCATTATCACA | |
| R9 | Reverse primer | ATCCATGGCAAAGGGAGACA | |
| F10 | Forward primer | CCTGTAACTCCAGCTCCCAA | |
| R10 | Reverse primer | GAAGGCAGGAGGTGAAGTCT | |
| F11 | Forward primer | GGCAGCCTCGTTACACAATC | |
| R11 | Reverse primer | GTGCCCCTCAGACTCTTCAT |
Note: Nucleotides in bracket, location of introduced T‐to‐A mutation. Allele‐specific quantitative PCR (qPCR) was carried out by using the cycleave PCR technique. This technique uses a chimeric DNA–RNA–DNA probe labeled with a fluorescent dye and a quencher at its ends; if the probe generates a perfect hybrid with the PCR product, it is digested with RNaseH at the RNA–DNA heteroduplex, leading to separation of the quencher and dye and increased fluorescence; any mismatch at or near the heteroduplex does not lead to RNaseH digestion. Lowercase letter, ribonucleotide; uppercase letter, deoxyribonucleotide. FAM, 6‐carboxyfluorescein‐labeled; HEX, hexachlorofluorescein‐labeled; ssODN, single‐stranded oligodeoxynucleotide.
2.2. Genotyping
DNA was extracted from ear punches as described 21 and used for PCR (94°C for 3 min, followed by 36 cycles of 94, 58, and 72°C each for 30 s and 72°C for 5 min; primers F1a, F1b, and R1 in Table 1). The PCR products were routinely analyzed by agarose gel electrophoresis. On occasions, PCR products were cloned by using TOPO TA Cloning Kit (Life Technologies Inc.) and sequenced by using BigDye Terminator version 3.1 Cycle Sequencing Kit (Thermo Fisher Scientific). Potential off‐target sequences of guide RNAs were searched by using the CRISPR Design webtool (http://crispr.mit.edu/ as of September 14, 2015) and analyzed using PCR primers F3–F11 and R3–R11 (Table 1).
2.3. Allele‐specific qPCR
Allele‐specific qPCR (95°C for 10 s, followed by 45 cycles of 95°C for 5 s, 55°C for 10 s, and 72°C for 20 s; primers F2 and R2 and probes P1 and P2 in Table 1) was run by using CycleavePCR Reaction Mix (Takara Bio) on a LightCycler 96 thermal cycler (Roche Diagnostics), 22 according to the manufacturer’s instructions. Mixtures of WT and mutant synthesized DNA (S1 and S2 in Table 1, respectively; 1000 copies in total; Eurofins Genomics Inc.) were used as standards.
2.4. Whole‐mount tissue preparation
Mammary tissue was extended on glass slides, fixed in 10% phosphate‐buffered formalin for 1–3 days, and stored in 70% ethanol at 4°C. Tissue was subsequently defatted in acetone for 3 days and stained with hematoxylin overnight at room temperature. Staining was adjusted by decolorization in 1% hydrochloric acid/70% ethanol (up to 1 h). The tissue was then dehydrated/defatted in acetone for 2–3 h, cleared in d‐limonene for 2 days and stored in mineral oil in heat‐sealed plastic bags.
2.5. Immunohistochemistry
Formalin‐fixed paraffin‐embedded sections were immunostained with an antibody against ERα (clone 6F11; Leica Biosystems), photographed, and analyzed as described. 19 The percentage of antigen‐positive epithelial tumor cells was determined by automated counting using Patholoscope software (Mitani Corporation).
2.6. Tumor genome analyses
Genomic DNA for the loss of heterozygosity (LOH) analysis was obtained from cryosections (1–3 mm2) of tumors and normal kidney tissues of Brca1 L63X/+ rats with a laser microdissection system (MMI CellCut; Molecular Machines & Industries) and QIAamp DNA Micro Kits (Qiagen) and used for allele‐specific qPCR as mentioned above. Genomic DNA for target sequencing was extracted from mammary carcinomas developed in the Brca1 L63X/+ rats in the untreated (n = 10), irradiated (3 weeks of age, n = 11), and MNU groups (dose 25 mg/kg, n = 3; 50 mg/kg, n = 8) or from normal mammary glands using Qiagen AllPrep DNA/RNA Micro Kits (Qiagen). Extracted DNA was then treated with ribonuclease A and purified using QIAmp DNA Micro Kits (Qiagen). DNA was quantified using a Qubit fluorometer (Life Technologies) and submitted to Azenta Japan for target sequencing with the DNBSEQ platform (MGI Tech). Capture probes were designed to target the rat Brca1 gene (chr10:86,418,468–86,477,304 on the mRatBN7.2/rn7 assembly; Twist Biosciences). Somatic single‐nucleotide variants and insertion/deletions in tumors were called with the VarScan 2 software (version 2.4.4). 23 A false‐positive filter was then applied to remove sequencing‐ and alignment‐related artifacts. Variants were annotated and the effect on coding sequences predicted using the SnpEff software (version 4.3). 24
2.7. Statistical analysis
All tests were two‐sided, p < 0.05 considered significant, and the open‐source free software R used. 25 Fisher’s exact probability tests, F tests, Student’s and Welch’s t‐tests (selected based upon the result of F tests), and two‐way ANOVA were run on R. Kruskal–Wallis tests and Wilcoxon rank sum tests were undertaken by using the “coin” package in R. 26 Log–rank tests and simple Cox regression analyses were carried out by using the “survival” package in R. 27 Cox regression fitting to dose–response models was carried out by using the “epifit” package in R. 28 Therein, the dose–response of HR was modeled as 1 + β 0 I L + (β L I L + β W I W)D, where β 0 is the excess HR for untreated Brca1 L63X/+ rats, I L and I W are dummy variables for Brca1 L63X/+ and Brca1 +/+ rats, β L and β W are the excess HR per Gy for Brca1 L63X/+ and Brca1 +/+ rats, and D is radiation dose in Gy. All R codes are provided in Document S1.
3. RESULTS
3.1. Brca1 L63X /+ rats are generated by gene editing
We injected guide RNA, single‐stranded oligodeoxynucleotides (Figure 1A, Table 1), and Cas9 mRNA into fertilized eggs of Jcl:SD rats (a Sprague–Dawley strain) and transferred these eggs to a pseudopregnant foster mother to produce knockout rats. One male rat having a c.188T>A (p.L63X) mutation of Brca1 (designated herein as the Brca1 L63X allele) was identified and used as a founder to establish the strain (named Jcl:SD‐Brca1 em1kyo). The genotype of the founder was confirmed by sequencing the cloned PCR products (primers F1a and R1; Figure 1A, Table 1) from the Brca1 alleles, indicating successful introduction of c.188T>A (Figure 1B). We compared 10 potential off‐target sequences of the guide RNAs between the founder and three purchased Jcl:SD rats and confirmed their intactness (Table 2).
TABLE 2.
Potential off‐target sequences in the founder Brca1 L63X/+ rat
| Guide RNA a | Location b | Reference sequence (5′→3′) c | Primers a | Result |
|---|---|---|---|---|
| G1 | Target (Brca1) | GTTATCTCATTCTTACACAAAGG | N/A | N/A |
| Chr 6: +73,719,173 | GGAATCTTATTCTTACACAATGG | F3/R3 | Intact | |
| Chr 13: −20,706,840 | CTGAACTCATTCTTACACAAAAG | F4/R4 | Intact | |
| Chr 14: +11,065,816 | CTTCTCCCATTCTTACACAAAAG | F5/R5 | Intact | |
| Chr 14: +11,066,038 | CTTCTCCCATTCTTACACAAAAG | F5/R5 | Absent d | |
| Chr 15: −77,797,047 | TTTATGTTATTCTTACACAAAGG | F6/R6 | Intact | |
| Chr 2: −229,084,162 | TTTATCTAATTATTACACAATAG | F7/R7 | Intact | |
| Chr 1: −280,828,323 | AGTGACTCATTCTTACACAAAAG | F8/R8 | Intact | |
| G2 | Target (Brca1) | ATTCTTACACAAAGGACACTGGG | N/A | N/A |
| Chr 7: +111,332,798 | TTCCTTACCCAAAGGACACTTAG | F9/R9 | Intact e | |
| Chr 1: −157,626,260 | TGTTTTAAACAAAGGACACTAAG | F10/R10 | Intact | |
| Chr 20: +10,695,088 | GTTTATACAGAAAGGACACTAAG | F11/R11 | Intact |
Guide RNA and primer sequences are shown in Table 1.
Location of the 5′‐end in the Rnor 6.0 assembly. Plus/minus signs indicate sense/antisense strands.
Sequence for the BN strain. Underlined bases indicate mismatches relative to the guide RNA sequence.
A 222 bp sequence (chromosome [Chr] 14: 11,065,964–11,066,185), present in the reference sequence of the BN strain, was absent in the founder as well as in three untreated Jcl:SD rats.
Single nucleotide polymorphism g.111332800C>T was present in the founder as well as in three untreated Jcl:SD rats. F, forward; N/A, not applicable; R, reverse.
The Significance of Bold values indicates guide RNA sequences.
3.2. Brca1 L63X homozygotes are embryonic lethal and heterozygotes are apparently normal
Using the strain established from this founder, we examined the heredity of the Brca1 L63X allele in offspring of four Brca1 L63X/+ female–male pairs. A well‐characterized allele‐specific qPCR analysis of genomic DNA (Figure 1C) indicated the absence of Brca1 L63X/L63X homozygotes, a departure from Mendelian inheritance (Figure 1D,E; p = 0.001 by Fisher’s exact probability test). Examination of uteri during pregnancy indicated many small conceptuses by 11.5 days postcoitum (Figure 1F,G). Evidence thus suggested that Brca1 L63X/L63X homozygotes were embryonic lethal. The Brca1 L63X/+ heterozygotes showed normal postnatal growth as seen in their body weight, the weights of several organs, and the whole‐mount tissue preparations of mammary gland (Figure 2).
FIGURE 2.
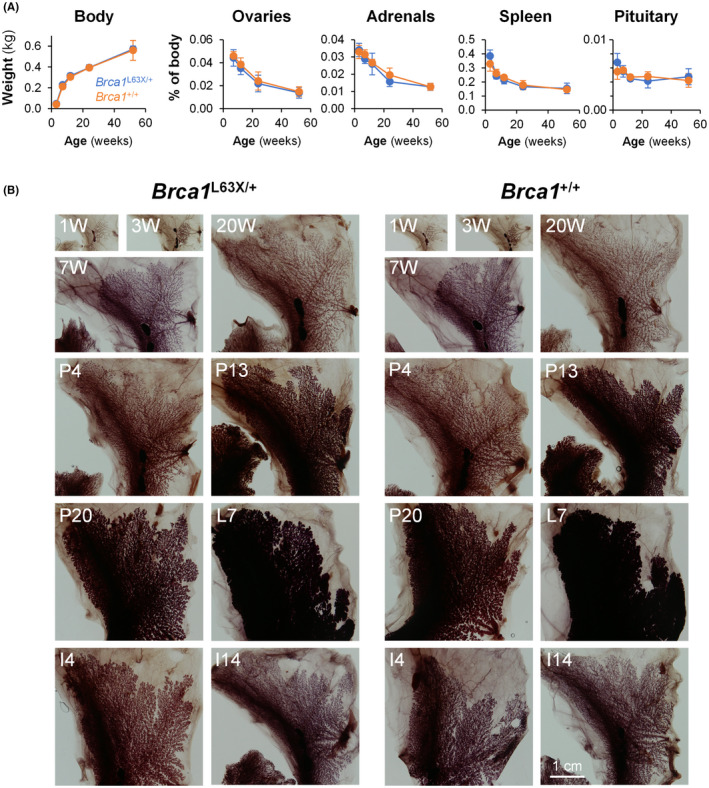
Normal development of Brca1 L63X/+ and Brca1 +/+ rats. (A) Body and organ weights of Brca1 L63X/+ rats (L63X/+, blue) and WT littermates (+/+, orange). Data are shown as mean ± SD (n = 4–6). No significant difference suggested between genotypes or interaction between genotypes and age, whereas age‐related differences were significant for all measures (p < 0.001), by two‐way ANOVA. (B) Mammary gland development. Whole mounts of inguinal mammary glands are shown. I, day of postlactational involution; L, day of lactation; P, day of pregnancy; W, weeks of age. For pregnancy and lactation, rats were mated at 11 or 12 weeks of age. Scale bar in the bottom right panel applies to all
3.3. Brca1 L63X /+ rats showed apparently unchanged susceptibility to spontaneous and MNU‐induced mammary carcinogenesis
We next set up untreated and carcinogen‐treated groups of Brca1 L63X/+ and Brca1 +/+ rats to study characteristics of both spontaneous and induced mammary carcinogenesis. We chose ionizing radiation and MNU as homologous recombination‐relevant and ‐irrelevant carcinogens, respectively, as radiation induces DNA double‐strand breaks whereas MNU induces methylation of DNA bases. The treatment regimens were chosen with reference to past experiments using WT Jcl:SD rats. 19 , 20
The crude analysis is summarized in Table 3. Ovarian cancers of epithelial origin, as described in humans, 4 were not observed. Comparison of untreated Brca1 L63X/+ and Brca1 +/+ rats illustrated no clear difference in mammary cancer incidence (Table 3, Figure 3A). Treatment with MNU increased the incidence rate of mammary cancer in a dose‐dependent manner (Table 3, Figure 3B). The age at autopsy was significantly younger, and the number of rats with carcinoma and the number of carcinomas developing per unit time were slightly higher, in Brca1 L63X/+ than Brca1 +/+ rats treated with 25 mg/kg MNU (Table 3); however, the slight increase in the mammary cancer incidence did not reach statistical significance (Figure 3B).
TABLE 3.
Mammary tumor incidence in Brca1 L63X/+ rats and their littermate Brca1 +/+ rats
| Genotype | Treatment | Age at treatment (weeks) | Dose | n a | Age at autopsy (weeks) b | Carcinoma | Benign tumors | ||
|---|---|---|---|---|---|---|---|---|---|
| Rats (%) | Tumors (10−2/week) c | Rats (%) | Tumors (10−2/week) c | ||||||
| Brca1 L63X/+ | None | – | – | 30 | 82.5 ± 26.0 | 11 (37) | 0.53 ± 0.15 | 15 (50) | 1.12 ± 0.28 |
| MNU | 7 | 25 mg/kg | 12 | 52.8 ± 18.3***, †† | 7 (58) | 1.66 ± 0.51* | 4 (33) | 1.42 ± 0.68 | |
| MNU | 7 | 50 mg/kg | 12 | 34.3 ± 10.4*** | 10 (83)* | 6.38 ± 1.33*** | 3 (25) | 0.58 ± 0.30 | |
| γ‐rays | 7 | 0.1 Gy | 30 | 78.6 ± 26.3 | 12 (40) | 0.73 ± 0.19 | 14 (47) | 0.98 ± 0.26 | |
| γ‐rays | 7 | 0.5 Gy | 30 | 77.8 ± 22.7 | 9 (30) | 0.50 ± 0.16 | 15 (50) | 1.29 ± 0.29 | |
| γ‐rays | 7 | 1 Gy | 30 | 63.8 ± 20.5** | 13 (43) | 1.16 ± 0.31 | 21 (70) | 1.72 ± 0.29 | |
| γ‐rays | 7 | 2 Gy | 30 | 60.3 ± 18.6*** | 17 (57) | 1.46 ± 0.30* | 22 (73) | 2.52 ± 0.40** | |
| γ‐rays | 3 | 2 Gy | 25 | 66.9 ± 21.2* | 11 (44) † | 0.93 ± 0.25 † | 20 (80)* | 2.78 ± 0.49** | |
| Brca1 +/+ | None | – | – | 30 | 76.6 ± 24.1 | 11 (37) | 0.81 ± 0.23 | 10 (33) | 0.69 ± 0.21 |
| MNU | 7 | 25 mg/kg | 12 | 67.8 ± 13.2 | 6 (50) | 1.34 ± 0.54 | 7 (58) | 2.46 ± 0.75* | |
| MNU | 7 | 50 mg/kg | 9 | 33.1 ± 6.2*** | 9 (100)** | 8.24 ± 0.88*** | 0 (0) | 0.00 ± 0.00 | |
| γ‐rays | 7 | 0.1 Gy | 30 | 88.1 ± 22.8* | 11 (37) | 0.81 ± 0.24 | 19 (63)* | 1.73 ± 0.34* | |
| γ‐rays | 7 | 0.5 Gy | 30 | 74.9 ± 22.7 | 12 (40) | 0.66 ± 0.16 | 18 (60) | 1.45 ± 0.26* | |
| γ‐rays | 7 | 1 Gy | 30 | 65.5 ± 24.8 | 13 (43) | 0.92 ± 0.23 | 21 (70)** | 1.94 ± 0.31** | |
| γ‐rays | 7 | 2 Gy | 30 | 56.3 ± 17.1** | 13 (43) | 1.25 ± 0.33 | 19 (63)* | 1.72 ± 0.27** | |
| γ‐rays | 3 | 2 Gy | 26 | 71.2 ± 19.5 | 4 (15) | 0.26 ± 0.13 | 22 (85)*** | 2.36 ± 0.37*** | |
Number of rats.
Mean ± SD.
Mean ± SEM of the number of tumors, divided by the number of weeks observed, in each rat.
p < 0.05.
p < 0.01.
p < 0.001 vs. the corresponding untreated group.
p < 0.05.
p < 0.01 vs. the corresponding Brca1 +/+ group.
The Kruskal–Wallis test was used to compare ages and tumor numbers among groups, followed by pairwise Wilcoxon rank sum tests to compare with the untreated group or between genotypes. Fisher’s exact probability test was used to compare the number of animals with tumors. MNU, 1‐methyl‐1‐nitrosourea.
FIGURE 3.
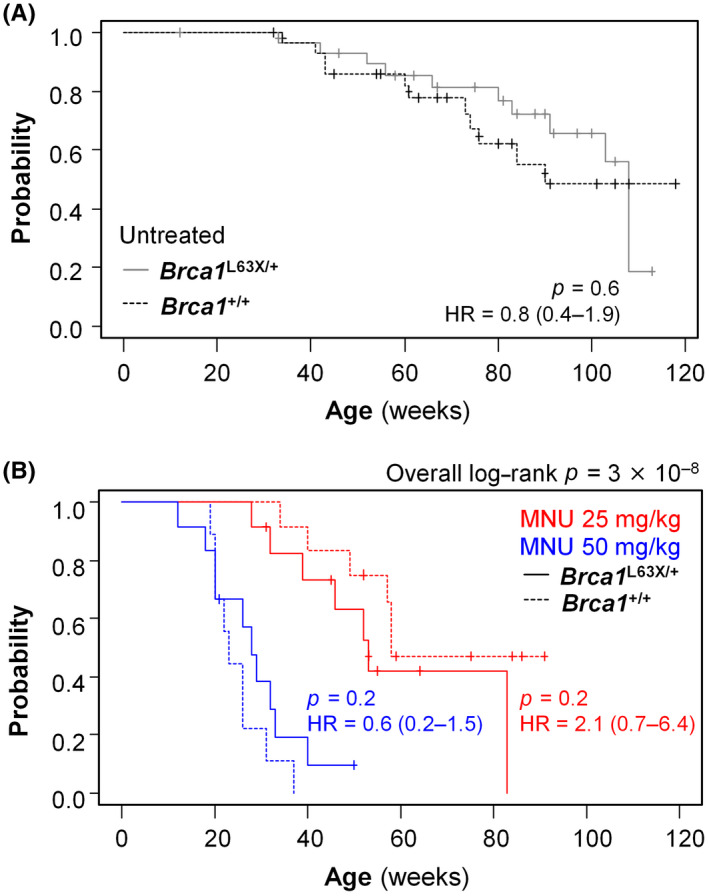
Spontaneous and 1‐methyl‐1‐nitrosourea (MNU)‐induced mammary carcinogenesis in Brca1 L63X/+ and Brca1 +/+ rats. (A) Kaplan–Meier plots depicting the incidence of palpable mammary carcinoma in the untreated groups. (B) Kaplan–Meier plots depicting the incidence of palpable mammary carcinoma in MNU‐treated Brca1 L63X/+ and Brca1 +/+ rats. p values, log–rank test. HR, hazard ratio (95% confidence interval)
3.4. Brca1 L63X /+ rats are more susceptible to mammary carcinogenesis induced by radiation than WT littermates
The incidence of mammary carcinoma displayed a tendency of increase related to radiation dose over the whole observation period (Figure 4A,B), and the slopes of the dose response of HR (with unirradiated Brca1 +/+ as reference) were positive in both Brca1 L63X/+ (p = 0.015) and Brca1 +/+ (p = 0.07) rats (Figure 4C). The HR of early‐onset mammary cancer (i.e., <40 weeks of age) showed a tendency toward dose dependence independently of the Brca1 genotype (Figure 4D), whereas HR during middle age (40–80 weeks) was significantly dose dependent only in Brca1 L63X/+ rats (p = 0.045; in Brca1 +/+, p = 0.82; Figure 4E); no clear dose response was observed later (Figure 4F). These results suggest that the early‐onset mammary cancer before 40 weeks is intrinsic to Jcl:SD rats irradiated at 7 weeks and independent of the Brca1 genotype, whereas mammary carcinogenesis during middle age was prominent in irradiated Brca1 L63X/+ rats.
FIGURE 4.
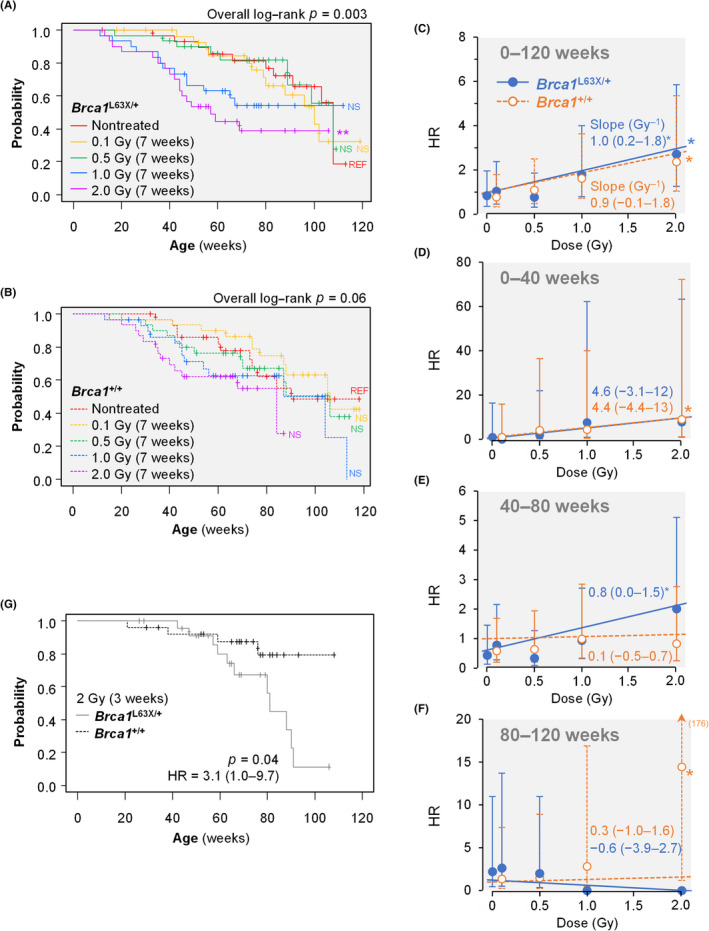
Radiation‐induced mammary carcinogenesis in Brca1 L63X/+ and Brca1 +/+ rats. (A,B) Kaplan–Meier plots depicting the incidence of palpable mammary carcinoma in (A) Brca1 L63X/+ and (B) Brca1 +/+ rats irradiated with 0.1–2 Gy at 7 weeks of age. Data for untreated groups (REF) are reproduced from Figure 3A. **p < 0.01 (compared with untreated, log–rank test). NS, not significant. (C–F) Cox regression–estimated hazard ratios (HR; dots) with nonirradiated Brca1 +/+ rats as the reference, their 95% confidence interval (CI; error bars), and the fitted linear dose–response models (lines) during designated time periods. Numbers are linear coefficients with (95% CIs); number in parentheses in (F) indicates the upper limit of the 95% CI; *p < 0.05 (Cox regression). (G) Kaplan–Meier plot depicting the incidence of palpable mammary carcinoma in rats irradiated with 2 Gy at 3 weeks of age
We previously showed that early‐onset mammary carcinogenesis is suppressed, whereas that during middle age is moderately increased, in Jcl:SD rats irradiated at 3 weeks of age. 19 In Brca1 L63X/+ and Brca1 +/+ rats irradiated at this age, as expected, the early‐onset component of carcinogenesis was diminished, and a clear and significant difference in mammary cancer incidence was indicated between the two genotypes (Figure 4G). Thus, the results hitherto indicate that the Brca1 L63X/+ rats are more susceptible to mammary carcinogenesis initiated by radiation, but not MNU, than their WT littermates.
3.5. Mammary carcinomas of Brca1 L63X /+ rats show low ERα positivity
Histology of mammary cancer showed no clear difference between Brca1 L63X/+ and Brca1 +/+ rats (Figure 5A,B). We evaluated the ERα‐positive rate of induced mammary carcinomas. Herein, all palpable carcinomas that developed for the first time in individual animals were immunohistochemically analyzed (Figure 5C,D). Of note, carcinomas that developed spontaneously in Brca1 L63X/+ rats had a significantly smaller fraction of ERα‐positive cells than those in Brca1 +/+ rats (Figure 5E). Overall, carcinomas developing in Brca1 L63X/+ and Brca1 +/+ rats irradiated at 7 weeks of age displayed similar rates of ERα positivity (Figure 5E). However, if the analysis was confined to carcinomas that developed between 40 and 80 weeks (i.e., the period when a significant dose dependence was observed, as mentioned above), we noted a significantly reduced ERα‐positive rate in Brca1 L63X/+ rats compared with Brca1 +/+ rats (Figure 5F). We previously showed that irradiation with 2 Gy at 3 weeks of age leads to premature ovarian failure and suppresses development of mammary carcinoma with high ERα positivity. 29 Indeed, carcinomas that developed after irradiation at 3 weeks showed low ERα‐positive rates regardless of the genotype (Figure 5E), concordant with the idea that this irradiation regimen revealed the distinct susceptibility of the Brca1 L63X/+ and Brca1 +/+ rats to estrogen‐independent mammary carcinogenesis through suppression of estrogen‐dependent mechanisms.
FIGURE 5.
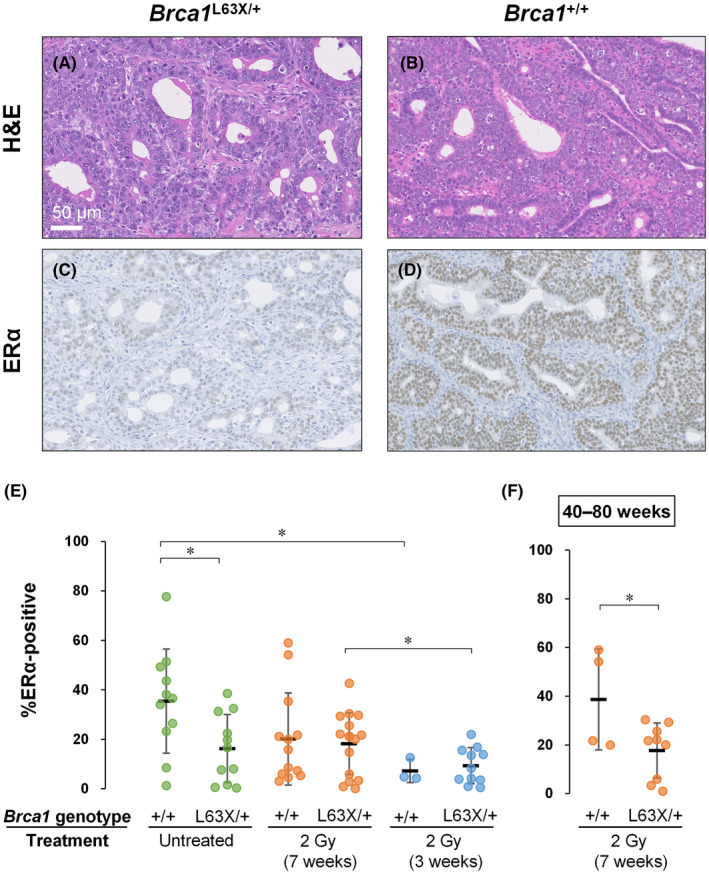
Estrogen receptor α (ERα)‐positive rate of mammary carcinomas in Brca1 L63X/+ and Brca1 +/+ rats. (A, B) H&E stained tissue sections. (C, D) Immunostained sections for ERα. Brown staining indicates immunoreactivity. Carcinomas that developed in untreated Brca1 L63X/+ (A,C) and Brca1 +/+ (B,D) rats. Scale bar in (A) applies to (A–D). (E,F) ERα‐positive rate of individual mammary carcinomas. Dots, individual data points; black horizontal line, mean; error bars, SD. *p < 0.05, t‐test
3.6. Mammary carcinoma of Brca1 L63X /+ rats retains a genetically intact Brca1 + allele
To obtain insights into the mechanism of tumor development in the Brca1 L63X/+rats, we quantified the ratio of Brca1 L63X and Brca1 + alleles in the tumors using allele‐specific qPCR (Figure 1C). This analysis revealed that the Brca1 + allele occupied nearly 50% (Figure 6A), indicating that LOH of the Brca1 locus is not involved. Thus, we then searched for de novo somatic mutations of 32 tumors in the Brca1 + allele by targeted sequencing. This analysis identified two exonic mutations that were either in the upstream sequence or synonymous (Figure 6B), with no evidence of loss of the Brca1 + allele. These results suggest that the tumors developed through nonmutational mechanisms in the Brca1 L63X/+rats.
FIGURE 6.
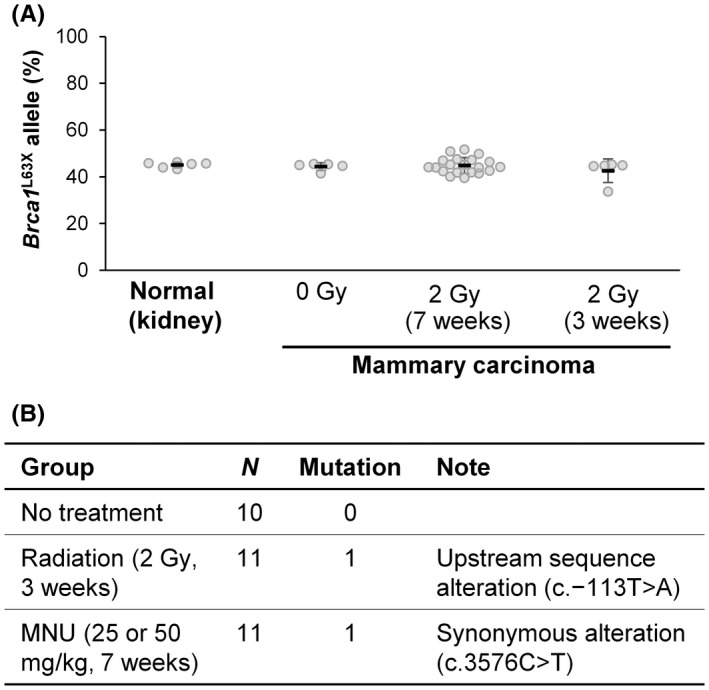
Analysis of somatic mutations of the Brca1 locus in the mammary carcinoma of the Brca1 L63X/+ rats. (A) Retention of heterozygosity as revealed by allele‐specific quantitative PCR. The occupancy of the Brca1 L63X allele was ~50% in normal kidney tissues and mammary carcinomas from rats that were unexposed (0 Gy) and irradiated (2 Gy) at 3 or 7 weeks of age. (B) Summarized results of the target sequencing of the Brca1 locus. MNU, 1‐methyl‐1‐nitrosourea; N, number of tumors analyzed. See Table S1 for detail
4. DISCUSSION
The present study reports phenotypes of a novel rat model harboring the Brca1 L63X allele. Whereas homozygotes were embryonic lethal, heterozygotes showed normal development, except that they developed mammary carcinomas with lower ERα positivity relative to WT littermates. Heterozygotes also displayed elevated susceptibility to radiation‐induced, but not MNU‐induced, mammary carcinogenesis.
Below, we compare the above phenotypes with those of previous Brca1 knockouts and humans to delineate the features of the present model. First, lethality of homozygotes is observed in humans, 30 as well as Brca1‐deficient mice between embryonic days 7.5 and 13.5, 8 consistent with the present observation. Second, many mouse strains with mammary‐specific homozygous Brca1 deficiency, as in humans, develop ERα‐low mammary cancer, 10 which is recapitulated by the present heterozygous rat model. Third, Brca1 L63X/+ rats did not display increased incidence of mammary cancer in the absence of carcinogenic exposure, a feature common to all previously reported Brca1 heterozygous mouse/rat models. 10 , 14 Fourth, susceptibility to radiation‐induced mammary carcinogenesis was elevated in the present model, a feature not reported on any other simple heterozygous Brca1 models 10 ; only those with double heterozygous deficiency (Brca1 and Trp53) and some homozygous models show susceptibility to radiation (but not MNU) compared with WT mice. 31 , 32 , 33 The radiation‐susceptible phenotype of the present model implies that the high spontaneous incidence of breast cancer in human carriers results from elevated susceptibility to continuous exposures to exogenous and/or endogenous carcinogens; the shorter life of rats might have hindered the susceptibility without external carcinogens. Thus, the present finding suggests that reduction of exposure to such carcinogens is key to prevention in human carriers. Finally, ovarian cancers of epithelial origin were not observed herein, unlike in human carriers. This is an expected result because epithelial ovarian cancers are rare in spontaneous and carcinogen‐induced animal models. 34 Thus, the Brca1 L63X/+ rat model, with a single‐gene, heterozygous mutation and an associated susceptibility to radiation‐induced mammary cancer, recapitulates important aspects of human BRCA1 deficiency that was not observed in previous models.
The susceptibility of carriers of BRCA mutations to radiation‐induced carcinogenesis is a matter of concern. The use of radiation for the treatment of primary breast cancer is not associated with a risk of second cancer in the contralateral breast in carriers of BRCA mutations, whereas increased breast cancer risk has been suggested in carriers receiving low‐dose diagnostic radiation at younger ages, albeit with a possible strong influence of biases. 35 The variability between the radiotherapy and diagnostic radiation studies could reflect difference in ages at exposure, as a recent Japanese atomic bomb survivor study indicates that the susceptibility of breast tissue to radiation‐induced carcinogenesis is particularly high during the peripubertal period. 36 If so, is the possible radiation‐related risk in humans quantitatively consistent with our observations in the rat? Pijpe et al. 6 reported that exposure to diagnostic radiation at an average level of ~0.01 Gy is associated with a breast cancer HR of 1.62 (95% CI, 1.02–2.58) in carriers exposed before 20 years of age. The rat age of 7 weeks is a postpubertal stage and can be compared to 15–20 years of age in humans. 37 The present observation did not suggest an irregularly high risk of radiation‐induced cancer at low doses, at least down to 0.1 Gy. Assuming a simple linear dose response, the observed radiation‐related excess HR of 0.8 Gy−1 (95% CI, 0.0–1.5) in Brca1 L63X/+ rats predicts a very low HR of ~1.008 at 0.01 Gy (i.e., 0.008 in excess), advocating a need for careful interpretation of the studies of diagnostic radiation. The increase in the HR was manifested during middle age (40–80 weeks) of Brca1 L63X/+ rats irradiated at 7 weeks of age. Mammary cancer incidence before age 40 weeks is characteristic of rats irradiated after, but not before, puberty, 19 implying a specific puberty‐related mechanism of tumor development before 40 weeks 29 ; the present result suggests that this mechanism is independent of the Brca1 genotype.
The BRCA1 protein suppresses the effects of estrogen and progesterone by binding to their receptors; hormone responsiveness is thus enhanced in cells with homozygous Brca1 deficiency and knockdown. 38 , 39 Estrogen and progesterone induce the secretion of cytokines by ERα‐expressing mature luminal cells, and the secreted cytokines in turn stimulate proliferation of luminal progenitor cells, which do not express ERα. 40 Thus, Brca1 deficiency can support proliferation of ERα‐negative luminal progenitors and lead to low ERα expression. Whether this mechanism holds in the situation of Brca1 haploinsufficiency remains an open question and could be answered by use of the present model.
We did not find evidence of LOH or other somatic mutations of the Brca1 locus in the tumors of Brca1 L63X/+ rats, which is surprising given that LOH is a major mechanism of biallelic inactivation of BRCA1. 41 Importantly, breast cancer with monoallelic BRCA1 inactivation is more frequently observed among carriers in the Japanese population than the TCGA cohort (http://cancergenome.nih.gov). 42 It is not well understood how tumors retaining the WT allele develop. A recent systematic review indicates that hypermethylation of the promoter region of BRCA1 is rare (i.e., ~4% of cases), although this might be an underestimation given that most studies have assessed limited methylation sites. 43 Noncoding variants are another possible explanation because functional assays provide partial evidence indicating that mutations of the promoter, upstream, intron, and 3′‐untranscribed sequences can affect the expression levels of BRCA1. 44 Thus, the Brca1 L63X/+ rat model can provide a tool to identify the nonmutational mechanisms of inactivating the WT Brca1 allele and to explore therapy strategies for BRCA1‐related tumors without LOH or other somatic mutations. A recent publication on renal carcinogenesis also supports this idea. 45
Taken together, our findings characterize the Brca1 L63X/+ rat mammary cancer model, the first single‐gene, heterozygous model mimicking aspects of BRCA1 deficiency in humans. This model is expected to provide a useful platform for studying the early phases of breast carcinogenesis and thereby preventive measures.
DISCLOSURE
Dr. Mitsue Saito supervises a researcher who receives a scholarship from Eisai Co. Ltd. The other authors have no conflict of interest.
APPROVAL OF THE RESEARCH PROTOCOL BY AN INSTITUTIONAL REVIEW BOARD
N/A.
INFORMED CONSENT
N/A.
REGISTRY AND REGISTRATION NO. OF THE STUDY/TRIAL
N/A.
ANIMAL STUDIES
Protocols were approved by the Institutional Animal Care and Use Committee.
Supporting information
Doc. S1 R codes for statistical analysis.
Table S1 Datasets.
ACKNOWLEDGMENTS
We thank Masami Ootawara, Mari Ogawa, Ken‐ichi Kudo, Hitomi Seo, Takahiro Hamoya, Misuzu Fujita, Hitomi Moriyama, staff at the Department of Radiation Effects Research and the Laboratory Animal and Genome Sciences Section, and the J‐SHARE project 46 of QST, for technical support. The study was funded in part by the Japan Society for the Promotion of Science, Grant Numbers JP26550036, JP15H02824, and JP21H03601 to TI.
Nakamura Y, Kubota J, Nishimura Y, et al. Brca1 L63X /+ rat is a novel model of human BRCA1 deficiency displaying susceptibility to radiation‐induced mammary cancer. Cancer Sci. 2022;113:3362‐3375. doi: 10.1111/cas.15485
Yuzuki Nakamura and Jo Kubota contributed equally to this work.
DATA AVAILABILITY STATEMENT
Data were generated by the authors and are provided with the article (Table S1).
REFERENCES
- 1. Heer E, Harper A, Escandor N, Sung H, McCormack V, Fidler‐Benaoudia MM. Global burden and trends in premenopausal and postmenopausal breast cancer: a population‐based study. Lancet Glob Health. 2020;8:e1027‐e1037. [DOI] [PubMed] [Google Scholar]
- 2. Casaubon JT, Kashyap S, Regan JP. BRCA 1 and 2. StatPearls; 2021. [Google Scholar]
- 3. Yoshida K, Miki Y. Role of BRCA1 and BRCA2 as regulators of DNA repair, transcription, and cell cycle in response to DNA damage. Cancer Sci. 2004;95:866‐871. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4. Kuchenbaecker KB, Hopper JL, Barnes DR, et al. Risks of breast, ovarian, and contralateral breast cancer for BRCA1 and BRCA2 mutation carriers. JAMA. 2017;317:2402‐2416. [DOI] [PubMed] [Google Scholar]
- 5. Ribeiro Guerra M, Coignard J, Eon‐Marchais S, et al. Diagnostic chest X‐rays and breast cancer risk among women with a hereditary predisposition to breast cancer unexplained by a BRCA1 or BRCA2 mutation. Breast Cancer Res. 2021;23:79. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6. Pijpe A, Andrieu N, Easton DF, et al. Exposure to diagnostic radiation and risk of breast cancer among carriers of BRCA1/2 mutations: retrospective cohort study (GENE‐RAD‐RISK). BMJ. 2012;345:e5660. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7. Li H, Terry MB, Antoniou AC, et al. Alcohol consumption, cigarette smoking, and risk of breast cancer for BRCA1 and BRCA2 mutation carriers: results from the BRCA1 and BRCA2 cohort consortium. Cancer Epidemiol Biomarkers Prev. 2020;29:368‐378. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8. Evers B, Jonkers J. Mouse models of BRCA1 and BRCA2 deficiency: past lessons, current understanding and future prospects. Oncogene. 2006;25:5885‐5897. [DOI] [PubMed] [Google Scholar]
- 9. Drost RM, Jonkers J. Preclinical mouse models for BRCA1‐associated breast cancer. Br J Cancer. 2009;101:1651‐1657. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10. Diaz‐Cruz ES, Cabrera MC, Nakles R, Rutstein BH, Furth PA. BRCA1 deficient mouse models to study pathogenesis and therapy of triple negative breast cancer. Breast Dis. 2010;32:85‐97. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11. Bouwman P, Jonkers J. Mouse models for BRCA1 associated tumorigenesis: from fundamental insights to preclinical utility. Cell Cycle. 2008;7:2647‐2653. [DOI] [PubMed] [Google Scholar]
- 12. Nandi S, Guzman RC, Yang J. Hormones and mammary carcinogenesis in mice, rats, and humans: a unifying hypothesis. Proc Natl Acad Sci U S A. 1995;92:3650‐3657. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13. Gould MN. Rodent models for the study of etiology, prevention and treatment of breast cancer. Semin Cancer Biol. 1995;6:147‐152. [DOI] [PubMed] [Google Scholar]
- 14. Smits BM, Cotroneo MS, Haag JD, Gould MN. Genetically engineered rat models for breast cancer. Breast Dis. 2007;28:53‐61. [DOI] [PubMed] [Google Scholar]
- 15. Yoshida R, Watanabe C, Yokoyama S, et al. Analysis of clinical characteristics of breast cancer patients with the Japanese founder mutation BRCA1 L63X. Oncotarget. 2019;10:3276‐3284. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16. Nakamura S, Takahashi M, Tozaki M, et al. Prevalence and differentiation of hereditary breast and ovarian cancers in Japan. Breast Cancer. 2015;22:462‐468. [DOI] [PubMed] [Google Scholar]
- 17. Momozawa Y, Iwasaki Y, Parsons MT, et al. Germline pathogenic variants of 11 breast cancer genes in 7,051 Japanese patients and 11,241 controls. Nat Commun. 2018;9:4083. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18. Yoshimi K, Kaneko T, Voigt B, Mashimo T. Allele‐specific genome editing and correction of disease‐associated phenotypes in rats using the CRISPR‐Cas platform. Nat Commun. 2014;5:4240. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19. Takabatake M, Daino K, Imaoka T, et al. Differential effect of parity on rat mammary carcinogenesis after pre‐ or post‐pubertal exposure to radiation. Sci Rep. 2018;8:14325. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20. Imaoka T, Nishimura M, Doi K, et al. Molecular characterization of cancer reveals interactions between ionizing radiation and chemicals on rat mammary carcinogenesis. Int J Cancer. 2014;134:1529‐1538. [DOI] [PubMed] [Google Scholar]
- 21. Truett GE, Heeger P, Mynatt RL, Truett AA, Walker JA, Warman ML. Preparation of PCR‐quality mouse genomic DNA with hot sodium hydroxide and tris (HotSHOT). Biotechniques. 2000;29(52):54. [DOI] [PubMed] [Google Scholar]
- 22. Yatabe Y, Hida T, Horio Y, Kosaka T, Takahashi T, Mitsudomi T. A rapid, sensitive assay to detect EGFR mutation in small biopsy specimens from lung cancer. J Mol Diagn. 2006;8:335‐341. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23. Koboldt DC, Zhang Q, Larson DE, et al. VarScan 2: somatic mutation and copy number alteration discovery in cancer by exome sequencing. Genome Res. 2012;22:568‐576. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24. Cingolani P, Platts A, Wang le L, et al. A program for annotating and predicting the effects of single nucleotide polymorphisms, SnpEff: SNPs in the genome of Drosophila melanogaster strain w1118; iso‐2; iso‐3. Fly (Austin). 2012;6:80‐92. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25. R Core Team . R: A language and environment for statistical computing. R Foundation for Statistical Computing; 2020. [Google Scholar]
- 26. Hothorn T, Hornik K, van de Wiel MA, Zeileis A. Implementing a class of permutation tests: the coin package. J Stat Softw. 2008;28:1‐23.27774042 [Google Scholar]
- 27. Therneau TM, Grambsch PM. Modeling survival data: extending the Cox model. Springer; 2020. [Google Scholar]
- 28. Doi K, Sakabe K, Taruri M. epifit: Flexible Modelling Functions for Epidemiological Data Analysis. R package version 0.1.2.2017.
- 29. Imaoka T, Nishimura M, Iizuka D, Nishimura Y, Ohmachi Y, Shimada Y. Pre‐ and postpubertal irradiation induces mammary cancers with distinct expression of hormone receptors, ErbB ligands, and developmental genes in rats. Mol Carcinog. 2011;50:539‐552. [DOI] [PubMed] [Google Scholar]
- 30. Arason A, Agnarsson BA, Johannesdottir G, et al. The BRCA1 c.4096+3A>G variant displays classical characteristics of pathogenic BRCA1 mutations in hereditary breast and ovarian cancers, but still allows homozygous viability. Genes (Basel). 2019;10:882. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31. Cressman VL, Backlund DC, Hicks EM, Gowen LC, Godfrey V, Koller BH. Mammary tumor formation in p53‐ and BRCA1‐deficient mice. Cell Growth Differ. 1999;10:1‐10. [PubMed] [Google Scholar]
- 32. Kim SS, Cao L, Li C, et al. Uterus hyperplasia and increased carcinogen‐induced tumorigenesis in mice carrying a targeted mutation of the Chk2 phosphorylation site in Brca1. Mol Cell Biol. 2004;24:9498‐9507. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33. Kim SS, Cao L, Baek HJ, et al. Impaired skin and mammary gland development and increased gamma‐irradiation‐induced tumorigenesis in mice carrying a mutation of S1152‐ATM phosphorylation site in Brca1. Cancer Res. 2009;69:9291‐9300. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34. Shan W, Liu J. Epithelial ovarian cancer: focus on genetics and animal models. Cell Cycle. 2009;8:731‐735. [DOI] [PubMed] [Google Scholar]
- 35. Drooger JC, Hooning MJ, Seynaeve CM, et al. Diagnostic and therapeutic ionizing radiation and the risk of a first and second primary breast cancer, with special attention for BRCA1 and BRCA2 mutation carriers: a critical review of the literature. Cancer Treat Rev. 2015;41:187‐196. [DOI] [PubMed] [Google Scholar]
- 36. Brenner AV, Preston DL, Sakata R, et al. Incidence of breast cancer in the life span study of atomic bomb survivors: 1958–2009. Radiat Res. 2018;190:433‐444. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37. Sengupta P. The laboratory rat: relating its age with Human's. Int J Prev Med. 2013;4:624‐630. [PMC free article] [PubMed] [Google Scholar]
- 38. Ma Y, Katiyar P, Jones LP, et al. The breast cancer susceptibility gene BRCA1 regulates progesterone receptor signaling in mammary epithelial cells. Mol Endocrinol. 2006;20:14‐34. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39. Rosen EM, Fan S, Isaacs C. BRCA1 in hormonal carcinogenesis: basic and clinical research. Endocr Relat Cancer. 2005;12:533‐548. [DOI] [PubMed] [Google Scholar]
- 40. Tharmapalan P, Mahendralingam M, Berman HK, Khokha R. Mammary stem cells and progenitors: targeting the roots of breast cancer for prevention. EMBO J. 2019;38:e100852. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41. Tung N, Miron A, Schnitt SJ, et al. Prevalence and predictors of loss of wild type BRCA1 in estrogen receptor positive and negative BRCA1‐associated breast cancers. Breast Cancer Res. 2010;12:R95. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42. Inagaki‐Kawata Y, Yoshida K, Kawaguchi‐Sakita N, et al. Genetic and clinical landscape of breast cancers with germline BRCA1/2 variants. Commun Biol. 2020;3:578. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43. Vos S, van Diest PJ, Moelans CB. A systematic review on the frequency of BRCA promoter methylation in breast and ovarian carcinomas of BRCA germline mutation carriers: mutually exclusive, or not? Crit Rev Oncol Hematol. 2018;127:29‐41. [DOI] [PubMed] [Google Scholar]
- 44. Santana Dos Santos E, Lallemand F, Burke L, et al. Non‐coding variants in BRCA1 and BRCA2 genes: potential impact on breast and ovarian cancer predisposition. Cancers. 2018;10:453. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45. Kong Y, Akatsuka S, Motooka Y, et al. BRCA1 haploinsufficiency promotes chromosomal amplification under Fenton reaction‐based carcinogenesis through ferroptosis‐resistance. Redox Biology. 2022;54:102356. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46. Morioka T, Blyth BJ, Imaoka T, et al. Establishing the Japan‐store house of animal radiobiology experiments (J‐SHARE), a large‐scale necropsy and histopathology archive providing international access to important radiobiology data. Int J Radiat Biol. 2019;95:1372‐1377. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Doc. S1 R codes for statistical analysis.
Table S1 Datasets.
Data Availability Statement
Data were generated by the authors and are provided with the article (Table S1).


