Abstract
Urothelial carcinoma (UC) is an umbrella term for bladder cancers (BCa) and upper‐tract urothelial carcinoma (UTUC), with BCa and UTUC sometimes detected concomitantly. The methods of detection for UC are often inaccurate or highly invasive, and, therefore, are thought to be unsatisfactory. Previously, we reported seven serum miRNAs as diagnostic markers for BCa. Here, we re‐evaluated potential diagnostic miRNAs in different institutions. We prospectively analyzed serum samples obtained from 126 UC patients (BCa: 106 samples; UTUC: 14 samples; UTUC with BCa: six samples) and 50 noncancer controls by microarray analysis. We randomly assigned these samples into a training or a validation set. Biomarker candidate miRNAs were selected based on cross‐validation scores in the training set of samples, with diagnostic power confirmed in the validation set. Among the diagnostic miRNAs identified in this way, miR‐1343‐5p and miR‐6087 had been identified as potential diagnostic miRNAs in our previous study. In addition, we evaluated the association between the serum levels of identified miRNAs and the presence of UC risk conditions. The expression levels of several miRNAs correlate with the risk factors in participants without UC, which may be explained by the presence of a microscopic tumor or a precancerous lesion. In conclusion, we identified two robust miRNA diagnostic markers for UC detection. Further functional analysis is required to elucidate the mechanism by which alterations in the expression of these miRNAs occur.
Keywords: diagnostic, liquid biopsy, miRNA, urothelial carcinoma
The methods of detection for urothelial carcinoma are often inaccurate or highly invasive, and, therefore, are thought to be unsatisfactory. In the present study, we prospectively evaluated the expression of circulating miRNAs in urothelial carcinoma patients, identifying miR‐1343 and miR‐6087, as reliable diagnostic markers for urothelial carcinoma.

Abbreviations
- BCa
bladder cancer
- DM
diabetes mellitus
- miRNA
microRNAs
- UC
urothelial carcinoma
- UTUC
upper‐tract urothelial carcinoma
1. INTRODUCTION
Urothelial carcinoma (UC) is the fourth most common malignancy worldwide. 1 While BCa accounts for 90% of UC, UTUC is less common. 2 However, the incidence of concomitant BCa at the time of primary UTUC is about 20%. 3 Currently, the methods used in the detection of UC are urinary cytology, imaging, and endoscopic examinations, such as cystoscopy or ureteroscopy; however, these methods are unsatisfactory. as they are often inaccurate and can be highly invasive. 4 , 5 Thus, the development of effective and unintrusive diagnostic markers is urgently required.
miRNAs are noncoding, single‐chain RNAs of approximately 18–24 base pairs, which act as post‐transcriptional regulators of gene expression. 6 Recently, differential miRNA expression has emerged as a hallmark of some human diseases, including cancers, cardiovascular disease, and DM. 7 Moreover, miRNAs are stable in the blood stream and their expression has been shown to be reproducible. 8 Therefore, circulating miRNAs have garnered a great deal of attention as novel targets for noninvasive diagnostic biomarkers in liquid biopsy.
In a previous study, we investigated circulating miRNAs as noninvasive biomarkers for BCa. 9 We established a promising diagnostic model of circulating miRNAs with high accuracy [sensitivity, 95%; specificity, 87%; area under the curve (AUC), 0.97]; however, most of the samples were stored in the National Cancer Center biobank for several years before analysis; thus, further analysis is required. 9
In the present study, we prospectively collected serum samples from different institutions and conducted comprehensive miRNA microarray analyses, independently extracting biomarker miRNAs. We used this new dataset to investigate the consistency of our previous study. 9 We additionally evaluated the association between serum level of identified miRNAs and the presence of risk conditions for UC, such as age, male, smoking history, and DM.
2. MATERIALS AND METHODS
2.1. Patient selection
Consecutive serum samples were obtained from 128 UC patients (BCa: 108 samples; UTUC: 14 samples; UTUC with BCa: six samples) admitted or referred to the Kanagawa Cancer Center or National Hospital Organization Yokohama Medical Center between October 2016 and December 2018. Patients previously diagnosed with cancers other than UC were excluded, with two BCa patients excluded on this basis. Fifty control samples were collected from the National Hospital Organization Yokohama Medical Center: 20 were obtained from DM patients hospitalized for DM management education, and the remaining 30 were obtained from participants receiving a medical checkup. For inclusion as a control sample, the participant must have had no history of cancer. Serum was immediately prepared from whole blood and stored at −80°C. Briefly, whole blood was collected in sterilized vacuum tubes for serum separation, gently inverted five times, and incubated at room temperature for 30 min to allow complete coagulation. The clot was removed by centrifugation at 2500 g for 10 min in a refrigerated centrifuge, and the resulting supernatant was designated serum.
2.2. Analysis of miRNA expression by microarray profiling
Total RNA was extracted from 300 μl serum using 3D‐Gene RNA extraction reagent from a liquid kit (Toray Industries) and purified with the RNeasy 96 QIAcube HT kit (Qiagen). Comprehensive miRNA expression analysis was performed using a 3D‐Gene miRNA labeling kit and 3D‐Gene Human miRNA Oligo Chip (Toray Industries), which was designed to detect 2565 miRNA sequences registered in miRBase release 21, 10 with three miRNAs (miR‐149‐3p, miR‐2861, and miR‐4463) selected as internal normalization controls. 11 Microarray analysis was performed within 5 months of sample collection, and all data are publicly available through the GEO database (GSE201359).
2.3. Evaluation of the diagnostic power of miRNAs
Samples were randomly assigned to the training or validation set. As a quality control, we selected miRNAs with a signal >26 in more than 50% of UC or noncancer control samples in the training set. In the training set, the best miRNAs to detect UC were investigated using Fisher linear discriminant analysis with leave‐one‐out cross‐validation. miRNAs with a cross‐validation score of >0.75 were selected as candidate biomarkers. Cutoffs to evaluate diagnostic performance in the validation set were selected using Youden index in the training set.
2.4. Statistics
χ 2 or unpaired t test was used to compare categorical or continuous variables in the two clinical groups, respectively. Linear discriminant analysis was performed using R version 3.4.3 (R Foundation for Statistical Computing, http://www.R‐project.org), compute.es package version 0.2‐4, hash package version 2.2.6.1, MASS package version 7.3‐51.3, mutoss package version 0.1‐12, and pROC package version 1.14.0. Heat maps were created using the online tool Morpheus (https://software.broadinstitute.org/morpheus). Other statistical analyses were performed using STATA version 14 (StataCorp.). A two‐sided p value <0.05 was considered statistically significant and, where appropriate, significance at a Bonferroni adjusted p value is also indicated.
3. RESULTS
3.1. Patient characteristics
The discovery and validation sets consisted of 63 UC patients and 25 noncancer controls, respectively (Figure 1A). No significant difference in participant characteristics was observed between the training and validation set (Table 1). Characteristic differences between UC patients and noncancer controls are summarized in Table S1. In UC patients, the median age was 76 years (range 32–92). Ninety‐six (76.2%) were male, and 25 (19.8%) were diagnosed with DM. The median Brinkman index was 663 (range 0–3060). In noncancer controls, the median age was 76 years (range 32–92). Twenty‐three participants (46.0%) were male, and 21 (42.0%) were diagnosed with DM. The median Brinkman index was 0 (range 0–2280). These factors were significantly different between UC patients and noncancer controls.
FIGURE 1.
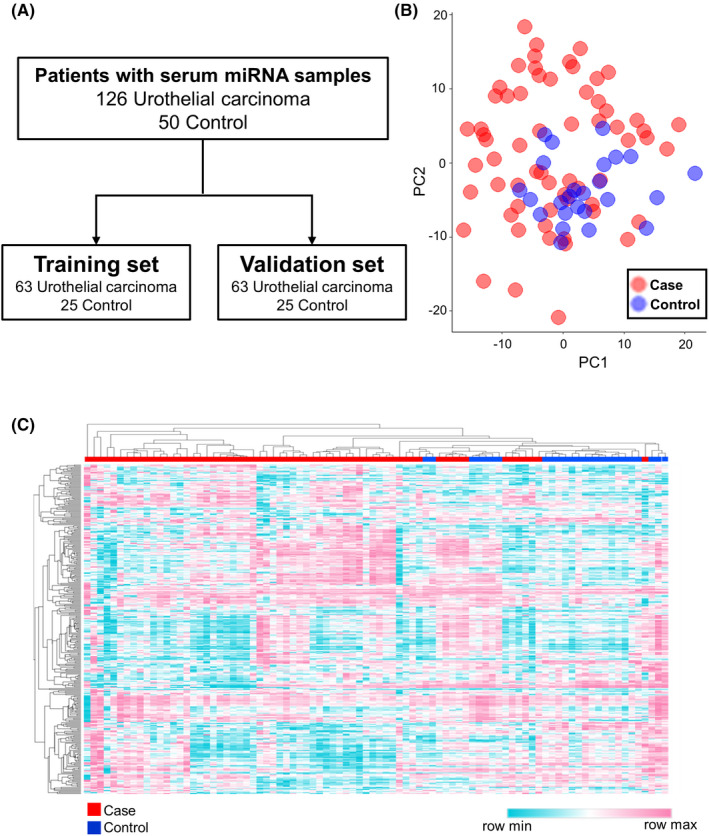
Candidate miRNA selection strategy in urothelial carcinoma. (A) Sample selection and inclusion in the training and validation sets. (B) Principal component analysis map of miRNA profiles from the 320 miRNAs in the training set. (C) Heat map showing differences in miRNA expression between patients with urothelial carcinoma and noncancer controls.
TABLE 1.
Patient characteristics
| Characteristics | Discovery set | Validation set | p Value |
|---|---|---|---|
| Cancer | 63 | 63 | |
| BCa, N (%) | 53 (84.1) | 53 (84.1) | 1.00 |
| UTUC, N (%) | 7 (11.1) | 7 (11.1) | |
| UTUC with BCa | 3 (4.8) | 3 (4.8) | |
| Age (median [range]) | 74 (32–92) | 76 (51–91) | 0.17 |
| Sex, N (%) | |||
| Male | 51 (81.0) | 45 (71.4) | 0.21 |
| Female | 12 (19.0) | 18 (28.6) | |
| DM, N (%) | 13 (20.6) | 12 (19.0) | |
| Tumor stage, N (%) | |||
| <pT2 | 41 (65.0) | 48 (76.2) | 0.12 |
| ≥pT2 | 22 (35.0) | 14 (23.8) | |
| Metastasis, N (%) | 2 (3.2) | 3 (4.8) | 0.65 |
| Urothelial carcinoma, N (%) | 59 (93.7) | 60 (95.2) | 0.68 |
| Control | 25 | 25 | |
| Age (median [range]) | 58 (46–81) | 57 (46–80) | 0.58 |
| Sex, N (%) | |||
| Male | 9 (36.0) | 14 (56.0) | 0.16 |
| Female | 16 (64.0) | 11 (44.0) | |
| DM, N (%) | 11 (44.0) | 10 (40.0) | 0.77 |
Abbreviations: BCa, bladder cancer; DM, diabetes mellitus; UTUC, upper‐tract urothelial carcinoma.
3.2. Investigation of potential diagnostic miRNAs in UC
A total of 320 miRNAs passed the quality control criteria, with principal component analysis mapping suggesting a separation between UC patients and noncancer controls (Figure 1B). Unsupervised hierarchical clustering using a heat map of the 320 selected miRNAs also suggested a segregation of UC from noncancer controls (Figure 1C). Fisher linear discriminant analysis was performed to select miRNAs for the diagnosis of UC. Twelve miRNAs were selected using cross‐validation scores (Table 2). Among these, miR‐1343‐5p and miR‐6087 had been selected as diagnostic miRNAs in our previous study. 9 The diagnostic performance of each miRNA was confirmed in the validation set with high accuracy (Figure 2). The highest AUC value was obtained from miR‐1273g‐3p (AUC, 0.89; sensitivity, 0.81; specificity, 0.96), and miR‐1343‐5p and miR‐6087 were validated for their performance (miR‐1343‐5p: AUC, 0.87; sensitivity, 0.83; specificity, 0.68; and miR‐6087: AUC, 0.76; sensitivity, 0.59; specificity, 0.84, respectively). Additionally, we investigated the diagnostic performance of each miRNA for BCa and UTUC, separately. As shown in Figures S1 and S2, each miRNA had favorable accuracy for BCa and UTUC. Furthermore, the signal intensity individually plotted on the graph showed that BCa and UTUC had the same trends against controls (Figure S3).
TABLE 2.
Discriminant analysis for urothelial carcinoma in the training set
| Model | miRNA | Sensitivity | Specificity | Accuracy | AUC | Cross‐validated accuracy | Cutoff |
|---|---|---|---|---|---|---|---|
| Model 1 | miR‐1273 g‐3p | 0.87 | 0.92 | 0.89 | 0.92 | 0.83 | 7.48 |
| Model 2 | miR‐1343‐5p | 0.81 | 0.88 | 0.83 | 0.89 | 0.80 | 10.21 |
| Model 3 | miR‐6778‐5p | 0.84 | 0.84 | 0.84 | 0.89 | 0.80 | 7.93 |
| Model 4 | miR‐619‐5p | 0.86 | 0.84 | 0.85 | 0.90 | 0.80 | 6.89 |
| Model 5 | miR‐4732‐5p | 0.78 | 0.88 | 0.81 | 0.89 | 0.79 | 5.63 |
| Model 6 | miR‐6087 | 0.60 | 1.00 | 0.72 | 0.85 | 0.78 | 11.65 |
| Model 7 | miR‐8071 | 0.59 | 0.96 | 0.69 | 0.82 | 0.77 | 6.75 |
| Model 8 | miR‐6768‐5p | 0.76 | 0.92 | 0.81 | 0.85 | 0.77 | 8.93 |
| Model 9 | miR‐4433a‐3p | 0.56 | 0.96 | 0.67 | 0.79 | 0.75 | 7.23 |
| Model 10 | miR‐7641 | 0.59 | 0.92 | 0.68 | 0.78 | 0.75 | 7.05 |
| Model 11 | miR‐4454 | 0.68 | 0.84 | 0.73 | 0.81 | 0.75 | 9.53 |
| Model 12 | miR‐1260a | 0.70 | 0.84 | 0.74 | 0.80 | 0.75 | 6.26 |
FIGURE 2.
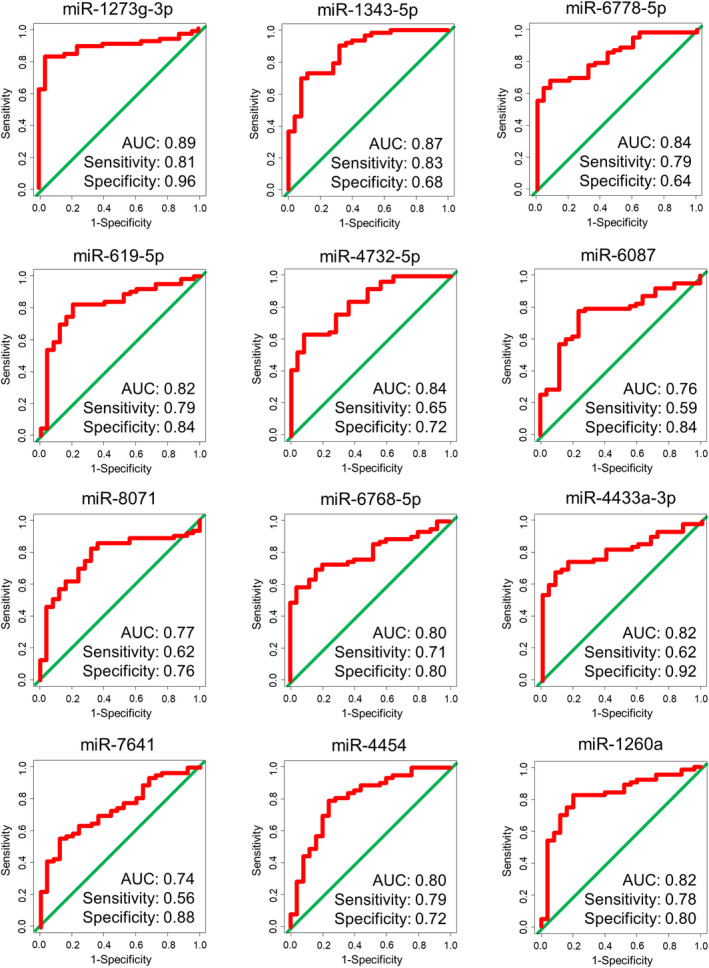
ROC curve analysis of the selected miRNAs in the validation set.
3.3. The influence of risk factors on miRNA expression
It has previously been suggested that DM and smoking history are associated with increased incidence and poor prognosis in UC, 12 , 13 while age and gender are also accepted risk factors. 14 , 15 Therefore, we investigated the relationship between miRNA expression and these risk factors.
Expression of miR‐1343‐5p and miR‐6768‐5p was significantly higher in noncancer control participants with DM than in those without DM. Conversely, miR‐4433a‐3p and miR‐7641 expression was significantly lower in noncancer controls with DM than in those without DM. Interestingly, these trends were not observed in UC patients (Table 3). In addition, expression of miR‐1343‐5p and miR‐6768 increased significantly with age in noncancer controls but not in UC patients (Table 4). There was no relationship between miRNA expression and gender or smoking history (Tables 5 and 6).
TABLE 3.
The differences in miRNA expression profiles between patients with and without DM
| Tumor (+) | Tumor (−) | |||||
|---|---|---|---|---|---|---|
| DM (+) | DM (−) | p‐Value | DM (+) | DM (−) | p Value | |
| miR‐1273g‐3p | 7.09 (0.55) | 6.97 (0.67) | 0.36 | 7.82 (0.46) | 8.03 (0.43) | 0.10 |
| miR‐1343‐5p | 10.49 (0.21) | 10.42 (0.24) | 0.2 | 10.16 (0.25) | 9.89 (0.19) | 0.0001* |
| miR‐6778‐5p | 7.42 (0.57) | 7.51 (0.63) | 0.51 | 8.30 (0.40) | 8.39 (0.46) | 0.45 |
| miR‐619‐5p | 6.39 (0.91) | 6.31 (0.76) | 0.67 | 7.30 (0.68) | 7.50 (0.68) | 0.31 |
| miR‐4732‐5p | 4.81 (1.80) | 4.97 (1.53) | 0.65 | 6.20 (0.47) | 6.29 (0.53) | 0.54 |
| miR‐6087 | 11.86 (0.21) | 11.83 (0.32) | 0.65 | 11.59 (0.28) | 11.49 (0.16) | 0.14 |
| miR‐8071 | 7.04 (0.82) | 7.03 (0.59) | 0.95 | 6.45 (0.37) | 6.46 (0.34) | 0.96 |
| miR‐6768‐5p | 9.28 (0.47) | 9.15 (0.45) | 0.19 | 8.83 (0.16) | 8.61 (0.16) | <0.0001* |
| miR‐4433a‐3p | 7.05 (0.29) | 7.06 (0.30) | 0.91 | 7.46 (0.18) | 7.30 (0.11) | 0.0003* |
| miR‐7641 | 6.59 (1.98) | 6.60 (0.97) | 0.97 | 7.90 (0.61) | 7.11 (0.29) | <0.0001* |
| miR‐4454 | 8.97 (0.53) | 9.11 (0.70) | 0.33 | 10.13 (0.70) | 9.78 (0.70) | 0.086 |
| miR‐1260a | 5.98 (0.44) | 5.92 (0.51) | 0.55 | 6.65 (0.49) | 6.46 (0.48) | 0.17 |
Note: Mean (SD).
Abbreviation: DM, diabetes mellitus.
Significant at a Bonferroni adjusted p value of 0.002.
TABLE 4.
The relationship between miRNA expression profiles and age
| Age | ||||
|---|---|---|---|---|
| Tumor (+) | Tumor (−) | |||
| R | p Value | R | p Value | |
| miR‐1273g‐3p | 0.39 | 0.67 | −0.40 | 0.016 |
| miR‐1343‐5p | −0.083 | 0.36 | 0.49 | 0.0003* |
| miR‐6778‐5p | −0.16 | 0.86 | −0.28 | 0.046 |
| miR‐619‐5p | 0.018 | 0.84 | −0.24 | 0.098 |
| miR‐4732‐5p | −0.013 | 0.89 | −0.25 | 0.085 |
| miR‐6087 | −0.037 | 0.68 | 0.28 | 0.049 |
| miR‐8071 | 0.10 | 0.26 | −0.0073 | 0.96 |
| miR‐6768‐5p | −0.0071 | 0.94 | 0.52 | 0.0001* |
| miR‐4433a‐3p | −0.13 | 0.16 | 0.22 | 0.12 |
| miR‐7641 | −0.036 | 0.69 | 0.41 | 0.003 |
| miR‐4454 | −0.041 | 0.65 | 0.076 | 0.60 |
| miR‐1260a | 0.0003 | 1.00 | 0.0049 | 0.97 |
Significant at a Bonferroni adjusted p value of 0.002.
TABLE 5.
The difference in miRNA expression profiles from male and female patients
| Tumor (+) | Tumor (−) | |||||
|---|---|---|---|---|---|---|
| Male | Female | p Value | Male | Female | p Value | |
| miR‐1273g‐3p | 7.00 (0.55) | 7.01 (0.68) | 0.93 | 7.92 (0.40) | 7.96 (0.49) | 0.78 |
| miR‐1343‐5p | 10.43 (0.23) | 10.43 (0.24) | 0.91 | 10.08 (0.23) | 9.94 (0.25) | 0.055 |
| miR‐6778‐5p | 7.43 (0.61) | 7.70 (0.62) | 0.037 | 8.37 (0.42) | 8.34 (0.45) | 0.81 |
| miR‐619‐5p | 6.31 (0.78) | 6.35 (0.84) | 0.82 | 7.38 (0.62) | 7.44 (0.75) | 0.77 |
| miR‐4732‐5p | 4.82 (1.70) | 5.32 (1.05) | 0.13 | 6.25 (0.43) | 6.25 (0.56) | 0.98 |
| miR‐6087 | 11.85 (0.29) | 11.82 (0.32) | 0.68 | 11.55 (0.26) | 11.52 (0.19) | 0.69 |
| miR‐8071 | 7.03 (0.64) | 7.05 (0.63) | 0.84 | 6.47 (0.36) | 6.44 (0.35) | 0.76 |
| miR‐6768‐5p | 9.17 (0.41) | 9.17 (0.57) | 1.00 | 8.77 (0.20) | 8.65 (0.17) | 0.026 |
| miR‐4433a‐3p | 7.05 (0.28) | 7.09 (0.34) | 0.55 | 7.36 (0.16) | 7.37 (0.18) | 0.93 |
| miR‐7641 | 6.55 (1.33) | 6.75 (0.82) | 0.45 | 7.53 (0.60) | 7.36 (0.59) | 0.32 |
| miR‐4454 | 9.03 (0.65) | 9.28 (0.72) | 0.073 | 9.75 (0.77) | 10.07 (0.64) | 0.12 |
| miR‐1260a | 5.88 (0.46) | 6.08 (0.57) | 0.065 | 6.42 (0.54) | 6.64 (0.42) | 0.11 |
Note: Mean (SD).
TABLE 6.
The relationship between miRNA expression profiles and Brinkman index
| Brinkman index | ||||
|---|---|---|---|---|
| Tumor (+) | Tumor (−) | |||
| R | p Value | R | p Value | |
| miR‐1273g‐3p | 0.032 | 0.72 | −0.073 | 0.62 |
| miR‐1343‐5p | 0.11 | 0.21 | 0.32 | 0.023 |
| miR‐6778‐5p | −0.042 | 0.64 | −0.018 | 0.90 |
| miR‐619‐5p | 0.055 | 0.54 | −0.089 | 0.54 |
| miR‐4732‐5p | −0.10 | 0.27 | 0.042 | 0.77 |
| miR‐6087 | 0.10 | 0.27 | 0.07 | 0.63 |
| miR‐8071 | −0.071 | 0.43 | 0.17 | 0.24 |
| miR‐6768‐5p | 0.056 | 0.53 | 0.22 | 0.13 |
| miR‐4433a‐3p | −0.040 | 0.66 | 0.20 | 0.16 |
| miR‐7641 | −0.0023 | 0.98 | 0.28 | 0.049 |
| miR‐4454 | −0.22 | 0.014 | 0.018 | 0.90 |
| miR‐1260a | −0.18 | 0.049 | 0.043 | 0.77 |
Finally, as the risk factors were not matched between participants with or without UC, we performed multivariate logistic regression analysis. After adjusting for these risk factors, miRNA expression was still a significant indicator for UC, suggesting that these miRNAs are independently associated with the presence of UC (Table S2).
4. DISCUSSION
In our previous study, we comprehensively analyzed miRNA profiles from 392 BCa patients, 100 noncancer controls, and 480 patients with other cancers. Using the Fisher linear discriminant analysis, we identified several miRNAs that can be used to discriminate BCa from noncancer and other cancers with high accuracy. 9 However, samples were collected from different institutions and stored under different conditions, which might have had an impact on data quality. In the present study, therefore, we evaluated the association between circulating miRNA profiles and UC from samples stored under unified conditions. Samples were collected and stored at −80°C for no more than 5 months before microarray analysis. In addition, we collected detailed patient information to enable us to control for the presence of UC risk factors in our patient samples. This is the first study to evaluate circulating miRNA profiles in relation to UC risk factors. Furthermore, we investigated their diagnostic power for UC, which is composed of BCa and UTUC. Most UC, whether BCa or UTUC, originate in the transitional epithelium. 16 The detection methods for these subpopulations of UC are very similar, and BCa and UTUC are sometimes detected concurrently. 3 , 17 Establishing diagnostic markers that can detect both BCa and UTUC is, therefore, clinically important.
miR‐6087 and miR‐1343‐5p were identified as potential diagnostic miRNAs in both our current and previous study. Especially, miR‐6087 was previously reported to be a highly sensitive and selective marker (AUC of 0.89, sensitivity of 93%, and specificity of 77%). 9 Although the interinstitutional bias could critically influence the miRNA expression, the identification of the two suggests their stability and versatility. On the contrary, the other miRNAs were not selected in our previous study. In the present study, we managed the sample conditions more strictly than in our previous study, in which we used preserved samples from the several biobanks. The differences of sample management methods may affect the result of the miRNA selection. Additionally, DM patients who were in hospital for the purpose of education of DM were included in the present study. The difference in characteristics of control samples may also influence the selected miRNAs.
We further examined the effect of the risk factors (DM, age, sex, and smoking history) on circulating miRNA expression in UC, which revealed that the expression of miR‐1343‐3p and miR‐6768 is upregulated in UC patients with DM. In noncancer controls, these miRNAs were similarly upregulated in DM patients compared with non‐DM participants. As DM is a risk factor for UC, 12 this upregulation may reflect the presence of a precancerous lesion. Similar trends for these two miRNAs were observed when we focused on patient age; miR‐1343‐3p and miR‐6768 were upregulated in older noncancer controls. By contrast, miR‐4433a‐3p and miR‐7641 expression was downregulated in UC patients compared with noncancer controls, and upregulated in noncancer controls from DM patients compared with non‐DM participants. Although the role of these miRNAs in the tumor microenvironment has not been elucidated, these trends may reflect a reactive response of tumor environment.
This study has several limitations. Although we prospectively collected samples, the sample size was relatively small; therefore, further external validation is still required.
In conclusion, we prospectively evaluated the expression of circulating miRNAs in UC patients, identifying miR‐1343 and miR‐6087, among others, as reliable diagnostic markers for UC. The expression of several miRNAs appears to be influenced by UC risk factors. While further study is required, these data suggest circulating miRNAs are attractive biomarkers for the diagnosis of UC.
AUTHOR CONTRIBUTIONS
KH and TO conceived this study. FU, JM, KH, and TO designed the methodology. FU and KH conducted data analysis with support from JM and TO. FU drafted the manuscript, conducted data interpretation with JM, FT, TK, TO, and KH. All authors have contributed to the manuscript drafting.
FUNDING INFORMATION
This study was supported by the “Japan Agency for Medical Research and Development, Development of Diagnostic Technology for Detection of miRNA in Body Fluids” and “Takeda Research Support (TKDS20190430007)”.
DISCLOSURE
The authors declare no competing interests.
ETHICAL APPROVAL
Approval of the research protocol by an Institutional Review Board: The present study involving human subjects was approved by the Ethical Committee of the Independent Administrative Institution National Hospital Organization Yokohama Medical Center (28‐7); the Ethics and Conflict of Interest Committee of the Kanagawa Cancer Center Hospital (28‐43); and the Human Tissue Samples Ethics Committee for R&D, Toray Industries.
Informed consent: Written informed consent was obtained from each subject. This study was conducted in accordance with the ethical guideline of the “Declaration of Helsinki.”
Registry and the registration no. of the study/trial: N/A.
Animal studies: N/A.
Supporting information
Figure S1.
Figure S2.
Figure S3.
Table S1.
Table S2.
ACKNOWLEDGEMENTS
The authors thank Yutaka Koga for the management of serum samples, Dr. Makoto Ujihara and Dr. Kazuye Okubo for the recruitment of DM patients, Kamakura Techno‐Science for performing the microarray assays, and Junpei Kawauchi and Satoko Takizawa for technical support. Some of the samples were obtained from KCCH. The authors also thank Akiko Sekiyama and Tomoe Tajima for the management of serum samples and Kumusu Tokiwa for the management of the participant information.
Urabe F, Matsuzaki J, Takeshita F, Kishida T, Ochiya T, Hirai K. Independent verification of circulating miRNA as diagnostic biomarkers for urothelial carcinoma. Cancer Sci. 2022;113:3510‐3517. doi: 10.1111/cas.15496
REFERENCES
- 1. Rouprêt M, Babjuk M, Burger M, et al. European Association of Urology guidelines on upper urinary tract urothelial carcinoma: 2020 update. Eur Urol. 2021;79(1):62‐79. doi: 10.1016/j.eururo.2020.05.042 [DOI] [PubMed] [Google Scholar]
- 2. Leow JJ, Chong KT, Chang SL, Bellmunt J. Upper tract urothelial carcinoma: a different disease entity in terms of management. ESMO Open. 2017;1(6):e000126. doi: 10.1136/esmoopen-2016-000126 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3. Cosentino M, Palou J, Gaya JM, Breda A, Rodriguez‐Faba O, Villavicencio‐Mavrich H. Upper urinary tract urothelial cell carcinoma: location as a predictive factor for concomitant bladder carcinoma. World J Urol. 2013;31(1):141‐145. doi: 10.1007/s00345-012-0877-2 [DOI] [PubMed] [Google Scholar]
- 4. Yafi FA, Brimo F, Steinberg J, Aprikian AG, Tanguay S, Kassouf W. Prospective analysis of sensitivity and specificity of urinary cytology and other urinary biomarkers for bladder cancer. Urol Oncol. 2015;33(2):66.e25‐31. doi: 10.1016/j.urolonc.2014.06.008 [DOI] [PubMed] [Google Scholar]
- 5. Almallah YZ, Rennie CD, Stone J, Lancashire M. Urinary tract infection and patient satisfaction after flexible cystoscopy and urodynamic evaluation. Urology. 2000;56(1):37‐39. doi: 10.1016/s0090-4295(00)00555-0 [DOI] [PubMed] [Google Scholar]
- 6. Lin S, Gregory RI. MicroRNA biogenesis pathways in cancer. Nat Rev Cancer. 2015;15(6):321‐333. doi: 10.1038/nrc3932 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7. Rupaimoole R, Slack FJ. MicroRNA therapeutics: towards a new era for the management of cancer and other diseases. Nat Rev Drug Discov. 2017;16(3):203‐222. doi: 10.1038/nrd.2016.246 [DOI] [PubMed] [Google Scholar]
- 8. Matsuzaki J, Ochiya T. Circulating microRNAs and extracellular vesicles as potential cancer biomarkers: a systematic review. Int J Clin Oncol. 2017;22(3):413‐420. doi: 10.1007/s10147-017-1104-3 [DOI] [PubMed] [Google Scholar]
- 9. Usuba W, Urabe F, Yamamoto Y, et al. Circulating miRNA panels for specific and early detection in bladder cancer. Cancer Sci. 2019;110(1):408‐419. doi: 10.1111/cas.13856 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10. Kozomara A, Griffiths‐Jones S. miRBase: annotating high confidence microRNAs using deep sequencing data. Nucleic Acids Res. 2014;42:D68‐D73. doi: 10.1093/nar/gkt1181 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11. Shimomura A, Shiino S, Kawauchi J, et al. Novel combination of serum microRNA for detecting breast cancer in the early stage. Cancer Sci. 2016;107(3):326‐334. doi: 10.1111/cas.12880 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12. Xu Y, Huo R, Chen X, Yu X. Diabetes mellitus and the risk of bladder cancer: a PRISMA‐compliant meta‐analysis of cohort studies. Medicine. 2017;96(46):e8588. doi: 10.1097/MD.0000000000008588 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13. Cumberbatch MGK, Jubber I, Black PC, et al. Epidemiology of bladder cancer: a systematic review and contemporary update of risk factors in 2018. Eur Urol. 2018;74(6):784‐795. doi: 10.1016/j.eururo.2018.09.001 [DOI] [PubMed] [Google Scholar]
- 14. Miyazaki J, Nishiyama H. Epidemiology of urothelial carcinoma. Int J Urol. 2017;24(10):730‐734. doi: 10.1111/iju.13376 [DOI] [PubMed] [Google Scholar]
- 15. Bianchi M, Roghmann F, Becker A, et al. Age‐stratified distribution of metastatic sites in bladder cancer: a population‐based analysis. Can Urol Assoc J. 2014;8(3–4):E148‐E158. doi: 10.5489/cuaj.787 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16. Birtle A, Johnson M, Chester J, et al. Adjuvant chemotherapy in upper tract urothelial carcinoma (the POUT trial): a phase 3, open‐label, randomised controlled trial. Lancet. 2020;395(10232):1268‐1277. doi: 10.1016/S0140-6736(20)30415-3 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17. Miyake M, Owari T, Hori S, Nakai Y, Fujimoto K. Emerging biomarkers for the diagnosis and monitoring of urothelial carcinoma. Res Rep Urol. 2018;10:251‐261. doi: 10.2147/RRU.S173027 [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Figure S1.
Figure S2.
Figure S3.
Table S1.
Table S2.


