Abstract
Forsythia suspensa is a famous ornamental and medicinal plant in Oleaceae. CCD family is involved in the synthesis of pigments, volatiles, strigolactones, and abscisic acid (ABA) in plants. In this study, the CCD family in F. suspensa was analyzed at the genome level. A total of 16 members of the CCD family were identified, which included 11 members of the carotenoid cleavage dioxygenases (CCD) subfamily and 5 members of the 9-cis epoxycarotenoid dioxygenases (NCED) subfamily. The expression analysis of different tissues demonstrated that three FsCCD1 genes might be involved in the synthesis of pigments and volatiles in flowers and fruits. Three CCD4 genes were effectively expressed in flowers, while only FsCCD4-3 was effectively expressed in fruits. Comparison of CCD4 between Osmanthus fragrans and F. suspensa showed that the structure of FsCCD4-1 is was comparable that of OfCCD4-1 protein, indicating that the protein might be performing, especially in catalyzing the synthesis of β-ionone. However, further comparison of the upstream promoter regions showed that the proteins have major differences in the composition of cis-elements, which might be responsible for differences in β-ionone content. On the other hand, four NCED genes were significantly up-regulated under cold stress while two were up-regulated in drought stress. The data showed that these genes might be involved in the synthesis of ABA. Taken together, our data improves understanding of the CCD family and provides key candidate genes associated with cold and drought stresses in F. suspensa.
Keywords: cold stress, drought stress, CCD family, gene expression, Forsythia suspensa
Introduction
Ornamental plants play an important role in garden and landscape design and improvement of the environment (Zheng et al., 2021). Due to the wide varieties in ornamental plants, there is no single variety that occupy a large share in the market. Therefore, it is difficult to achieve high economic benefits from planting ornamental plant species. Thus, planting the ornamental plants with both edible and ornamental value, or medicinal and ornamental value, presents an important opportunity for high economic benefit. Forsythia suspensa (Thunb.) Vahl. is a famous medicinal and ornamental plant, which belongs to Oleaceae family. F. suspensa blooms in early spring, with flowers first and leaves later. F. suspensa trees are golden during flowering period, which confers the plants with excellent ornamental effects (Fu et al., 2014). On the other hand, F. suspensa fruits contain phillyrin, phillyrin A, α-pinene, β-pinene, terpinen-4-ol, and other volatile components, and it is widely used as a Chinese patent medicine for treatment of colds (Xiang et al., 2021). In fact, recent studies have demonstrated that F. suspensa can reduce covid-19 symptoms (Hu et al., 2021). F. suspensa is widely cultivated as a medicinal crop in Hebei, Henan, Shanxi, and Shaanxi provinces in China (Li et al., 2022). Because of the important ornamental and medicinal values of F. suspensa, its basic and applied research is on the rise (Qiao et al., 2020).
Carotenoid cleavage dioxygenase (CCD) family is a relatively small gene family in plants, which include CCD and 9-cis-epoxy carotenoid dioxygenase (NCED) subfamilies (Ohmiya, 2009). This family catalyzes the cleavage of carotenoids with the conjugated double bonds to form various apocarotenoids and their derivatives (Tian et al., 2021). Four members of the CCD subfamily were identified in Arabidopsis, and included CCD1, CCD4, CCD7, and CCD8 (Auldridge et al., 2006). Previous data has shown that CCD1 and CCD4 are involved in the synthesis of pigments and volatiles (such as α-ionone, β-ionone) in flowers and fruits of many plants (Simkin et al., 2004; Phadungsawat et al., 2020). CCD7 and CCD8 are two key genes involved in the synthesis pathway of strigolactones (Umehara et al., 2008), which regulates in the regulation of senescence, root growth, branching and tillering and flower development (Liu et al., 2019). On the other hand, five members of NCED subfamily were identified in Arabidopsis, which included NCED2, NCED3, NCED5, NCED6, and NCED9 (Auldridge et al., 2006). The NCED genes are involved in the synthesis of abscisic acid (ABA) (Frey et al., 2012; Hamzah et al., 2020; Truong et al., 2021). ABA is an important plant hormone that plays major roles in seed development and dormancy, and mediates plant responses to various environmental stresses (Seo and Koshiba, 2002). The CCD family has been identified in the genome of many crops, vegetables, and flowers, such as Brassica napus (Zhou et al., 2020), Populus trichocarpa (Wei et al., 2022), Gossypium species (Zhang et al., 2021), Cucurbitaceae species (Cheng et al., 2022), and Rosaceae species (Zhang et al., 2021). However, data on the whole genome characterization and expression analysis of the CCD family in F. suspensa remains scant.
In this study, we identified the CCD family members based on the published F. suspensa genome (Li et al., 2022). We then analyzed the expression patterns of the CCD genes in fruit, stem, leaf and flower tissues as well as the expression responses to cold and drought stresses. The data showed that unlike F. suspensa, Osmanthus fragrans, a plant from the same family, has a strong floral fragrance. We further analyzed differences in the CCD4 gene, a gene associated with the synthesis of β-ionone, between the F. suspensa and O. fragrans. Therefore, this study provides in-depth data on the number and classification, gene structure, and expression of the CCD gene family at the genome level. Besides, our study provides key candidate genes associated with cold and drought stresses in F. suspensa.
Materials and methods
Data sources and sequence searches
The genome of F. suspensa was obtained from the National Center for Biotechnology Information (NCBI, accession no. JAHHPY000000000; Li et al., 2022). The keywords “CCD” and “NCED” were used to search for the CCD genes in the annotation file, and then the candidate genes were blasted in NCBI (Altschul et al., 1990)1 to identify the REP65 or PLN02258 domain. The genes with conserved REP65 or PLN02258 domains were considered the true CCD genes. Physicochemical properties of the CCD protein in F. suspensa, such as molecular weight, isoelectric point, amino acid number, fat index, instability index, and hydrophobicity were predicted using the ExPASy online tool (Artimo et al., 2012).2 Subcellular localization of the CCD genes was predicted by Plant-mPLoc online software (Chou and Shen, 2010),3 while the secondary structure was predicted by the online software SOPMA (Geourjon and Deleage, 1995).4
According to the IDs of the identified CCD genes and the F. suspensa genome sequence, the CCD genes were mapped on the chromosomes of F. suspensa. The chromosome position of the CCD genes was visualized using TBtools software (Chen et al., 2020). The genome and protein sequence data of O. fragrans were obtained from NCBI (accession no. PRJNA529305; Yang et al., 2018), while the genome and protein sequence data of Arabidopsis thaliana were from the Arabidopsis information resource.5
On the other hand, the genome and protein sequence data of Oryza sativa were obtained from the Rice Genome Annotation Project (Kawahara et al., 2013).6
Phylogenetic relationship and gene structure
Maximum likelihood (ML) tree (Felsenstein, 1996) was constructed to elucidate the phylogenetic relationship of the CCD genes based their amino acid sequences. The ML tree was constructed using MEGA 7.0 (Kumar et al., 2016) with the Jones-Taylor-Thornton model (Jones et al., 1992), pairwise deletion option, and 1,000 bootstrap resampling times. The phylogenetic tree was drawn using FigTree v1.4.4 (Rambaut, 2009), while the introns and exons of all the F. suspensa CCD genes were visualized using TBtools (Chen et al., 2020). In addition, the protein domains and conserved motifs of all the F. suspensa CCD genes were analyzed by the MEME online tool (Bailey et al., 2009).7 The protein domains of the CCD gene family of F. suspensa were visualized by TBtools (Chen et al., 2020), and the analysis value of conserved motifs was set to 10. The upstream 2,000 bp sequences of all the CCD genes in F. suspensa were extracted using the TBtools (Chen et al., 2020), and the potential cis-acting elements of the CCD genes were predicted by PlantCARE online software (Thijs et al., 2002).8 The predicted results were visualized by the TBtools (Chen et al., 2020). Amino acid sequences of the CCD4 genes in O. fragrans and F. suspensa were compared using DNAMAN 6.0 (Lynnon Crop., Quebec, Canada), while conservative domain analysis of the CCD4 genes of O. fragrans and F. suspensa was performed using the online software NCBI Conserved Domain Search (Lu et al., 2020).9 The possible cis-acting elements of in the upstream 2,000 bp sequences in the CCD4 genes of O. fragrans were predicted using the PlantCARE (see text footnote 8; Thijs et al., 2002).
Expression profile of the carotenoid cleavage dioxygenases genes in different tissues and under cold and drought stresses
Expression patterns of the CCD genes in different tissues of F. suspensa was extracted from the RNA-seq data in NCBI. The patterns included data from fruits, stems, leaves (accession no. SRR17386487-SRR17386495), and flowers (accession no. SRX11342985, SRX11342993, and SRX11342994). Fresh fruits, stems, leaves from the F. suspensa fruits in the harvest season (July) were sampled from three individuals (Li et al., 2022). Flowers at the budding stage (March) were also sampled from three individuals. All the samples were treated with liquid nitrogen, and then kept in the ultra-low temperature refrigerator at −80°C until extraction of RNA. Leaves are often the most sensitive to drought and cold treatments. Therefore, the gene expression data in leaves was used for analysis in our study. Previous studies (Li et al., 2021a,b) showed that Wuzhishan population has the highest cold and drought tolerance when compared with the other three populations. Thus, the gene expression data of the Wuzhishan population was used as a representative. Expression patterns of the CCD genes in F. suspensa under drought stress were from Wuzhishan population under 80 and 20% soil water content (accession no. SRX7503009, SRX7503010, SRX7503012-SRX7503015; Li et al., 2021a). Expression patterns of the CCD genes in F. suspensa under cold stress were from Wuzhishan populations at 25 and 4°C (accession no. SRX7440183-SRX7440188; Li et al., 2021b).
The RNA-seq data from F. suspensa were further processed. Low-quality reads with more than 50% of bases possessing a value Q ≤ 10 and more than 10% anonymous nucleotides (N) were eliminated from original sequencing data. Fragments per kilobase of transcript per million fragments mapped (FPKM) was used to profile the gene expression in these samples using StringTie (Pertea et al., 2015). The expression patterns of the F. suspensa CCD genes in different tissues and in response to drought and cold stresses were visualized using the R package Heatmap. Significantly expressed CCD genes in different tissues of F. suspensa were analyzed, and a Log2FPKM ≥ 1 was used as the threshold. FC ≥ 2 and FDR ≤ 0.05 were used as thresholds to screen for the CCD genes involved in drought and cold stress responses.
Quantitative real-time transcription PCR validation of carotenoid cleavage dioxygenases genes under cold and drought stresses
To verify the expression patterns of 16 CCD genes from transcriptomic data under cold and drought stresses, quantitative real-time transcription PCR (qRT-PCR) was performed. The primers (Supplementary Table 1) of 16 CCD genes for qRT-PCR were designed using primer premier 5.0 (Lalitha, 2000). qRT-PCR reaction was performed using the TB Green Premix Ex Taq II (TaKaRa, Beijing, China) on the ABI QuantStudio®3 Real-Time System (Applied Biosystems, CA, USA). The amplification procedure was as described in Li et al. (2021b). α elongation factor (Rosati et al., 1999) was used as an internal control, and all these reactions with three repeats. The expression levels of the CCD genes were calculated by using the 2–△△Ct method (Livak and Schmittgen, 2001).
Results
Gene identification and sequence characteristics of the carotenoid cleavage dioxygenases gene family
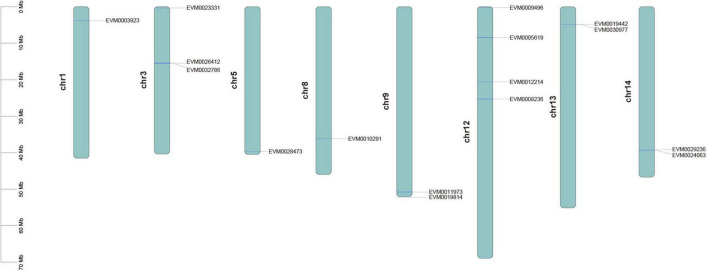
Our search of the genome annotation files identified a total of 16 possible CCD genes from the F. suspensa genome. Domain analysis showed that all the genes had the REP65 or PLN02258 conserved domains. Thus, the 16 genes were considered as the real CCD genes. CCD proteins in F. suspensa demonstrated great variation; where their amino acid length ranged from 123 (FsCCD4-2) to 602 aa (FsCCD7), protein molecular weight ranged from 16.651 (FsCCD4-2) to 67.671 KDa (FsCCD7), while the isoelectric point ranged from 5.11 (FsCCD4-3) to 8.88 (FsCCD4-4) (Supplementary Table 2). Four CCD proteins were localized in the mitochondrion, 4 were in the peroxisome, 2 in the cytoplasm, while the remaining 6 were in the chloroplast (Supplementary Table 2). All the 16 CCD proteins were hydrophilic proteins (Supplementary Table 2). Among the 16 CCD proteins, 5 (FsCCD4-5, FsNCED1-1, FsNCED5-1, FsNCED5-2, and FsNCED6), whose instability index was higher than 40, were predicted to be unstable proteins, while the rest whose value under 40 were stable proteins. The CCD genes were randomly distributed on eight chromosomes of F. suspensa. Chr12 bore most of the CCD genes (25%), while Chr1, Chr5, and Chr8 each accounted for the least (6.25%) of the CCD genes (Figure 1).
FIGURE 1.
Distribution location of CCD gene family in the Forsythia suspensa genome.
Phylogenetic relationship of the carotenoid cleavage dioxygenases proteins in F. suspensa
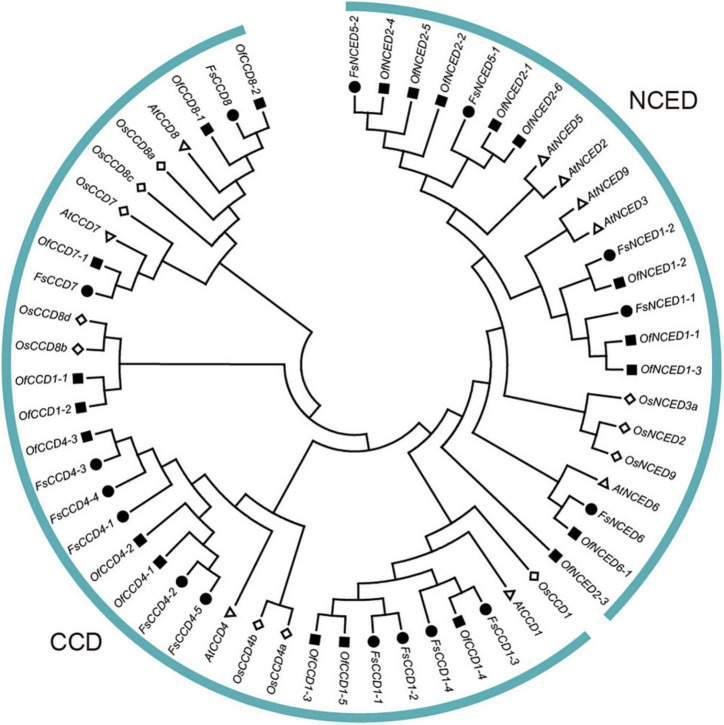
To demonstrate the phylogenetic relationship in the CCD proteins of F. suspensa, a phylogenetic tree involving 9 CCD proteins in A. thaliana, 11 CCD proteins in O. sativa, 16 CCD proteins in F. suspensa, and 21 CCD proteins in O. fragrans was constructed using the ML method. The phylogenetic tree showed that the members of CCD protein family in F. suspensa and O. fragrans were grouped into two clades, i.e., NCED and CCD clades (Figure 2). NCED proteins were clustered into three subclades, which included FsNCED6, FsNCED1, and FsNCED5. On the other hand, CCD proteins were clustered into three subclades, which included FsCCD1, FsCCD4, and FsCCD7 and FsCCD8. Due to the lack of the NCED5 protein in O. fragrans, the two FsNCED5 proteins in F. suspensa were clustered together with the four OfNCED2 proteins in O. fragrans. Our clustering results showed a close relationship between FsCCD7 and FsCCD8 or FsNCED1 and FsNCED5.
FIGURE 2.
Phylogenetic tree of CCD proteins in Forsythia suspensa, Osmanthus fragrans, and Arabidopsis thaliana.
Gene and protein structure of the carotenoid cleavage dioxygenases gene family
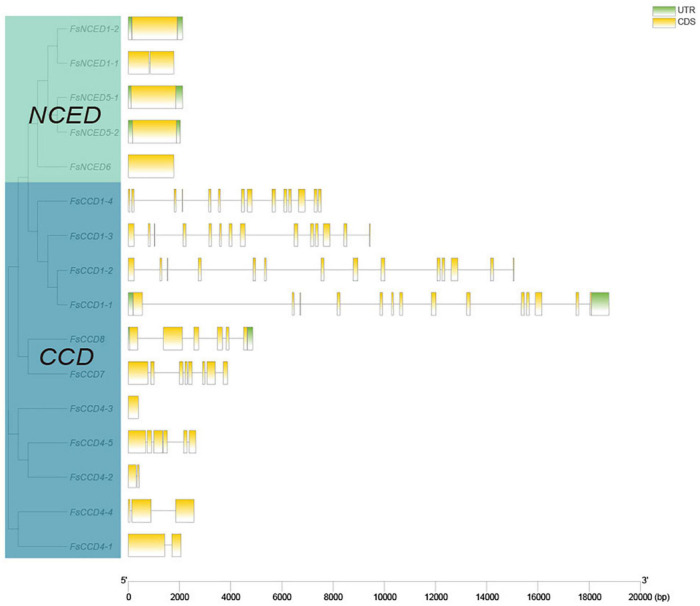
The gene intron and exon structure usually reflect the evolutionary relatedness of the members of a gene family. Here, we analyzed the CCD gene sequences of F. suspensa and visualized the gene structure using TBtools (Figure 3). The average gene length of the NCED subfamily showed minor changes, where four of them had no introns, and only FsNCED1-1 had a shorter intron. In contrast, there were more changes in the average gene length of the CCD subfamily. Four FsCCD1 genes were significantly longer than the other CCD genes. However, they all consisted of 14 exons and 13 introns (Figure 3). Five FsCCD4 genes were shorter than other CCD genes and their number of exons ranged from 1 to 6. FsCCD7 had 8 exons and 7 introns, while FsCCD8 had 6 exons and 5 introns.
FIGURE 3.
Gene structure analysis of CCD genes of Forsythia suspensa.
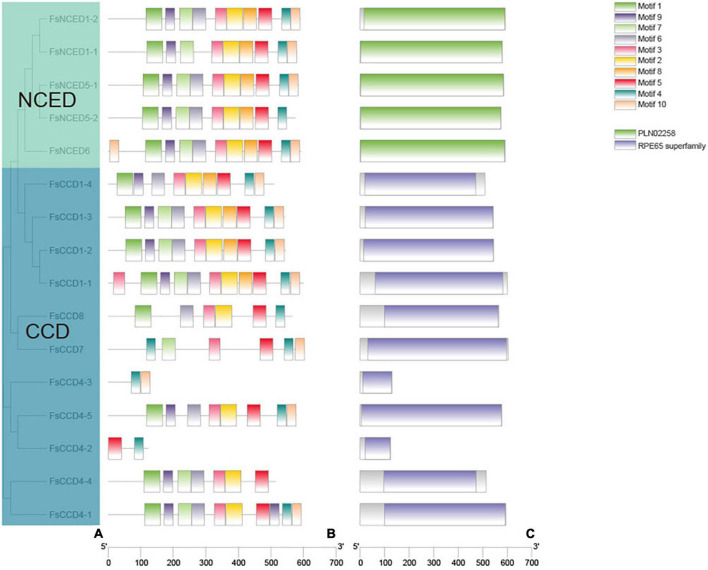
To further analyze the structure and function of the CCD protein in F. suspensa, we further identified the conserved domain and motif (Figure 4). Conservative domain analysis showed that the 5 members of NCED subfamily contained the PLN02258 domain, while the CCD subfamily members contained the REP65 domain. Conservative motif analysis showed that the PLN02258 domain was composed of two motif combinations (motif1, motif9, motif7, motif6, motif3, motif2, motif8, motif5, motif4 or motif1, motif9, motif7, motif6, motif3, motif2, motif8, motif5, motif4, motif10). On the other hand, the REP65 domain was composed of multiple combinations. The first combination was motif1, motif9, motif7, motif6, motif3, motif2, motif8, motif5, motif4, motif10, and included FsCCD1-1, FsCCD1-2, FsCCD1-3. FsCCD1-4 lacked motif 7 relative to the first one. FsCCD4-1 contained motif1, motif9, motif7, motif6, motif3, motif2, motif5, motif9, motif4, and motif10. FsCCD4-5 lacked motif 7 and motif 8 relative to the first one. FsCCD4-4 lacked motif 8, motif4, and motif10 relative to the first one. FsCCD7 and FsCCD8 contained six motifs, while FsCCD4-2 and FsCCD4-3 contained two motifs.
FIGURE 4.
The protein motifs and protein domain analysis of CCD genes of Forsythia suspensa. (A) Phylogenetic tree of CCD genes; (B) protein motif of CCD genes; (C) protein domain of WRKY genes.
Prediction of protein secondary structure of the CCD family showed that the proportion of alpha helices ranged from 7.75% (FsCCD4-3) to 18.15% (FsNCED5-1), while beta turns ranged from 4.33% (FsCCD4-5) to 9.76% (FsCCD4-2). Extended strands ranged from 20.76% (FsCCD7) to 34.88% (FsCCD4-3), while random coils ranged from 47.97% (FsCCD4-2) to 57.50% (FsCCD4-1) (Supplementary Table 3). The results showed that the secondary structure of the CCD protein in F. suspensa was mainly composed of extended strands and random coils.
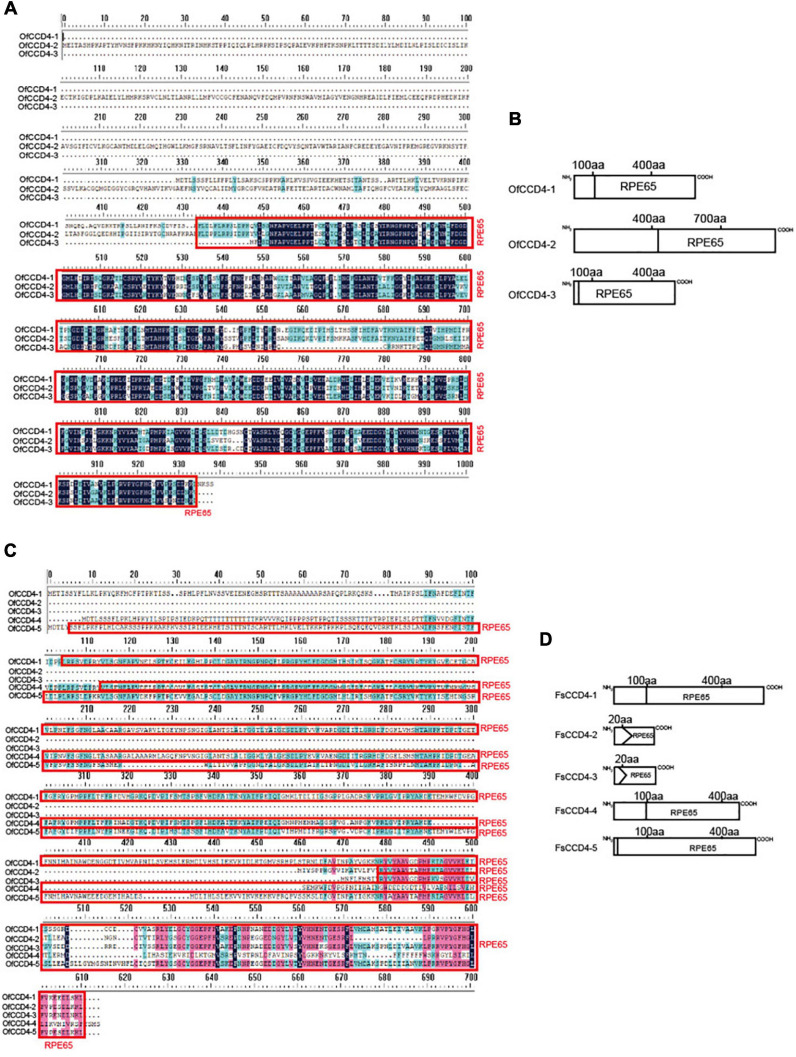
A previous study showed that the OfCCD4 protein of O. fragrans cleaved β-carotene to produce β-ionone (Zhang et al., 2016). However, F. suspensa is not an aromatic plant. Thus, we compared the structural differences in the CCD4 proteins between the F. suspensa and O. fragrans. Three CCD4 genes were found in O. fragrans and five CCD4 genes were in F. suspensa. From the protein domain, FsCCD4-1 in F. suspensa resembled OfCCD4-1 in O. fragrans, while FsCCD4-5 in F. suspensa was similar to OfCCD4-3 in O. fragrans. However, none of the proteins was similar to the OfCCD4-2 in O. fragrans (Figure 5).
FIGURE 5.
Amino acid sequence and conserved domain of CCD4 genes of Forsythia suspensa and Osmanthus fragrans. (A) Amino acid sequence of CCD4 genes in Osmanthus fragrans; (B) conserved domain of CCD4 genes in Osmanthus fragrans; (C) amino acid sequence of CCD4 genes in Forsythia suspensa; (D) conserved domain of CCD4 genes in Forsythia suspensa.
Cis-acting elements of the carotenoid cleavage dioxygenases gene family
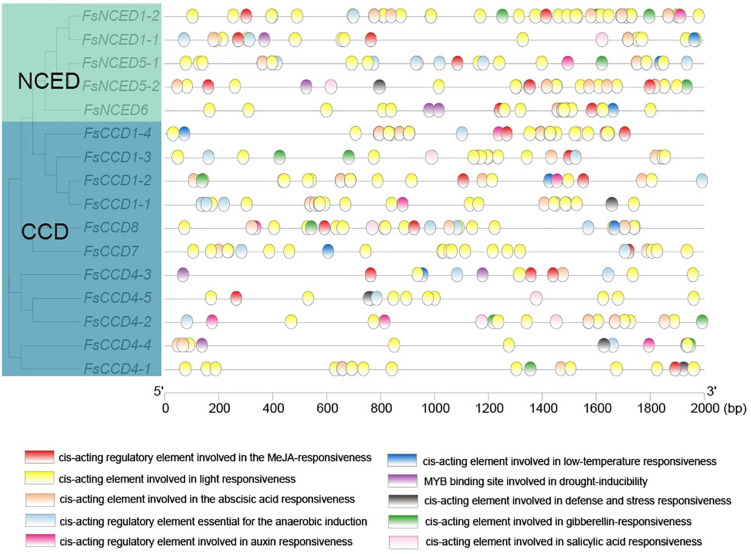
A total of 417 possible cis-acting regulatory elements were identified in the upstream 2,000 bp range of 16 CCD genes (Supplementary Table 4 and Figure 6). The results showed that there were many cis-acting elements in the promoter region of the CCD gene in F. suspensa. In addition to many light-responsive elements, the cis-acting elements were associated with plant hormones, such as methyl jasmonate (MeJA), ABA, gibberellin (GA), auxin, salicylic acid (SA), and cis-acting elements related to stress, such as low temperature, drought, anaerobic environment, and defense and stress, were also found in the CCD genes of F. suspensa.
FIGURE 6.
Cis-acting elements analysis of the promoters of CCD genes of Forsythia suspensa.
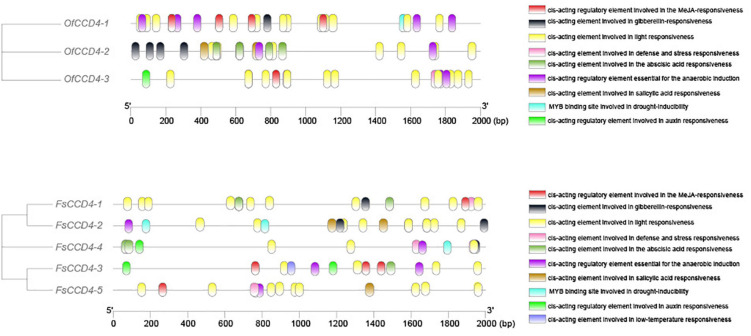
We then compared the cis-acting elements upstream of the CCD4 gene between O. fragrans and F. suspensa. The data showed that between the similar FsCCD4-1 and OfCCD4-1, OfCCD4-1 had more anaerobic induction and MeJA-responsiveness cis-components compared to FsCCD4-1, but had fewer ABA responsiveness cis-components than FsCCD4-1 (Figure 7). Between the similar FsCCD4-5 and OfCCD4-3, OfCCD4-3 had more auxin responsiveness cis-components than FsCCD4-5, but had fewer salicylic acid responsiveness cis-components than FsCCD4-5 (Figure 7). The difference in the cis-elements upstream of the CCD4 genes might lead to the difference of β-ionone between O. fragrans and F. suspensa.
FIGURE 7.
Cis-acting elements analysis of the promoters of CCD4 genes of Forsythia suspensa and Osmanthus fragrans.
Expression patterns in the carotenoid cleavage dioxygenases gene in different tissues, cold and drought stresses
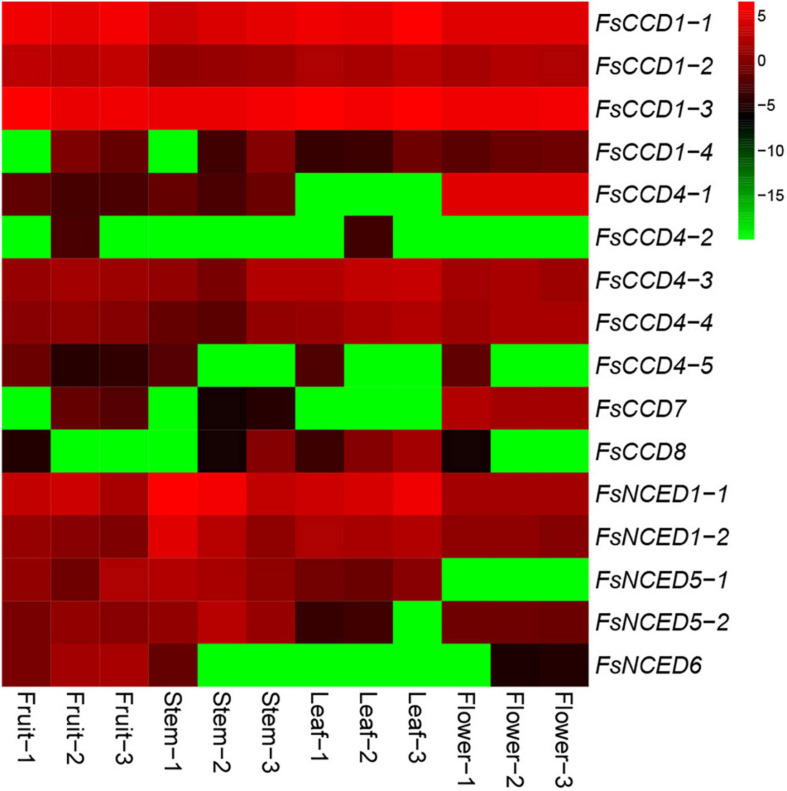
We investigated the expression patterns of the CCD genes in fruits, stems, leaves and flowers of F. suspensa. The results showed that 7 CCD genes were expressed in fruits, 8 in stems, 7 in leaves, and 8 in flowers (Supplementary Table 5 and Figure 8). Our results indicated that about a half of the CCD genes might be involved in the development and morphogenesis of fruit, stem, leaf, and flower tissues in F. suspensa. In addition, four FsCCD4 genes were expressed in flowers, and the expression level of FsCCD4-1 was highest among the four FsCCD4 genes. Meanwhile, FsCCD4-1 had specific expression in flowers.
FIGURE 8.
Heat map of CCD gene expression (Log2FPKM) in different tissues of Forsythia suspensa. Fruit-1 to Fruit-3, Stem-1 to Stem-3, Leaf-1 to Leaf-3, and Flower-1 to Flower-3 indicate the three biological replicates from each tissue.
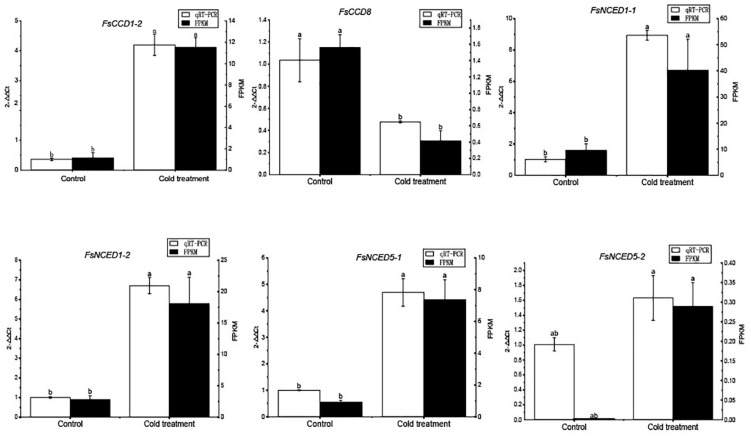
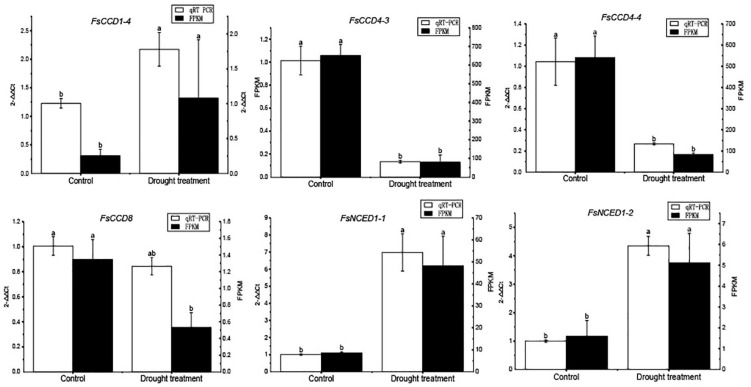
We further profiled the expression patterns of the CCD genes in leaves under cold and drought stresses. Transcriptomic data showed that six CCD genes responded to cold stress (Supplementary Table 6 and Supplementary Figure 1), and qRT-PCR confirmed these genes responded to cold stress (Figure 9). Of which, 4 (FsCCD1-2, FsCCD8, FsNCED1-1, and FsNCED5-1) had cis-elements related to low temperature stress. Six CCD genes were significantly differentially expressed under drought stress (Supplementary Table 7 and Supplementary Figure 2), and qRT-PCR confirmed all of them responded to drought stress, while FsCCD8 was only slightly decreased when drought stress (Figure 10). All of them had cis-elements with ABA responsiveness (FsCCD1-4, FsCCD4-3, FsCCD4-4, FsCCD8, FsNCED1-1, and FsNCED1-2), where 3 of the genes had drought-related cis-elements involved in MYB transcription factors (FsCCD4-3, FsCCD4-4, and FsNCED1-1), and three of them (FsCCD4-3, FsCCD4-4, and FsNCED1-1) had the two kinds of cis-elements.
FIGURE 9.
qRT-PCR verification of the CCD genes of Forsythia suspensa under cold stress. Comparison of qRT-PCR (white bar) with RNA-seq data (black bar). The relative changes were calculated with 2–△△ Ct. The relative qRT-PCR expression level is shown on the left y-axis. The FPKM from the RNA-Seq data are indicated on the right y-axis. The letters above the bars indicates the significance among different samples.
FIGURE 10.
qRT-PCR verification of the CCD genes of Forsythia suspensa under drought stress. Comparison of qRT-PCR (white bar) with RNA-seq data (black bar). The relative changes were calculated with 2–△△Ct. The relative qRT-PCR expression level is shown on the left y-axis. The FPKM from the RNA-Seq data are indicated on the right y-axis. The letters above the bars indicates the significance among different samples.
Discussion
In this study, we systematically analyzed CCD genes in F. suspensa at the genome level. We profiled the CCD genes in F. suspensa and O. fragrans, and the clustering results supported the division of CCD and NCED subfamilies of F. suspensa. A total of 16 CCD genes were identified in F. suspensa, which included 11 members in the CCD subfamily and 5 members in the NCED subfamily., They were more than the number of CCD genes (10, 9, 9, 13, 8, 8) identified in six Cucurbitaceae species (Cheng et al., 2022), Pyrus bretschneideri (12), Fragaria vesca (11), Prunus mume (8), and Prunus persica (10) (Zhao et al., 2021) and G. raimondi (15) (Zhang et al., 2021), but less than the 21 members identified in O. fragrans, the 23 members in Populus trichocarpa (Wei et al., 2022), 33 and 31 members in G. hirsutum and G. barbadense (Zhang et al., 2021), 20 members in Malus domestica (Zhao et al., 2021), as well as 30 members in B. napus (Zhou et al., 2020).
Current studies suggest that the CCD family has obvious functional differentiation (Cheng et al., 2022). CCD1 and CCD4 are associated with the synthesis of pigments and aromatic substances synthesis in plant flowers and fruits (Xi et al., 2020). The four FsCCD1 genes were relatively conservative, with little differences in protein length and motif composition. Three CCD1 genes, FsCCD1-1, FsCCD1-2, and FsCCD1-3, were effectively expressed in fruits and flowers of F. suspensa, and might be involved in the synthesis of pigments and volatiles in fruits and flowers. There was also major changes in the CCD4 gene in F. suspensa, with most differences observed in protein length and motif composition. Three CCD4 genes; FsCCD4-1, FsCCD4-3, and FsCCD4-4, were effectively expressed in F. suspensa flowers, while only FsCCD4-3, with two motifs, was expressed in fruits.
In O. fragrans, OfCCD4-1, a famous aromatic plant in the Oleaceae family, which is similar to F. suspensa FsCCD4-1 protein domain, has been shown to be involved in the synthesis of β-ionone (Zhang et al., 2016). Therefore, FsCCD4-1 might be having similar functions, while the other two genes, FsCCD4-3 and FsCCD4-4, might have undergone functional differentiation. Although FsCCD4-1 was similar to OfCCD4-1 protein domain, they had great differences in the cis-elements in the promoter region. OfCCD4-1 had cis-components with more anaerobic induction and MeJA-responsiveness, but had fewer ABA responsiveness cis-components than that of FsCCD4-1 (Figure 7). In a recent study of O. fragrans, the cultivated variety “Zaohuang” (Albus group) had an ethylene response factor binding cis-element, which was absent in “Chenghong Dangui” (Aurantiacus group), resulting in a higher content of β-ionone in “Zaohuang” than that in “Chenghong Dangui” (Han et al., 2022). The cis-element differences in the upstream promoter region of FsCCD4-1 and OfCCD4-1 might have resulted in the difference in the β-ionone content between F. suspensa and O. fragrans.
CCD7 and CCD8 are involved in the synthesis of strigolactones (Umehara et al., 2008), which participate in regulation of aging, root growth, branching, and tillering as well as flower development (Liu et al., 2019). However, our data showed that only FsCCD7 was effectively expressed in flowers, CCD7 and CCD8 were not expressed in stems, leaves and fruits of F. suspensa. Here, FsCCD7 was demonstrated to be involved in the development of F. suspensa flowers.
Members of the NCED subfamily are involved in the synthesis of ABA (Truong et al., 2021), which is involved in seed development (Seo and Koshiba, 2002). Three NCED genes, FsNCED1-1, FsNCED5-1, and FsNCED6, were found to be expressed in F. suspensa fruits (Figure 8 and Supplementary Table 5), and were considered to be the candidate genes involved in the development of F. suspensa seeds. In addition, ABA was also shown to confer resistance to adverse environment (Seo and Koshiba, 2002). FsNCED1-1, FsNCED1-2, FsNCED5-1, and FsNCED5-2 in F. suspensa were significantly up-regulated under cold stress (Figure 9). Similarly, FsNCED1-1 and FsNCED1-2 genes in F. suspensa were significantly up-regulated under drought stress (Figure 10). These genes might be involved in ABA synthesis in F. suspensa under cold and drought stress environment, which enhances the ability of F. suspensa to withstand cold and drought. The up-regulated expression of the genes in the NCED subfamily might be one of the reasons underlying the high cold and drought resistance of F. suspensa.
Conclusion
In this study, a total of 16 members of the CCD family were identified, including 11 members of the CCD subfamily and 5 members of the NCED subfamily. The expression analysis of different tissues showed that three FsCCD1 genes might be involved into the synthesis of pigments and volatiles in flowers and fruits. Three CCD4 genes were effectively expressed in flowers, and only one FsCCD4-3 with two motifs was effectively expressed in fruits. Comparison of the CCD4 in Osmanthus fragrans and F. suspensa showed that the structure of FsCCD4-1 was similar to that of OfCCD4-1 protein, indicating that it might have similar functions, especially in catalyzing the synthesis of β-ionone. However, further analysis of the upstream promoter regions showed that they had great differences in the composition of cis-elements, which might be associated with differences in the β-ionone content in F. suspensa and O. fragrans. In addition, four and two NCED genes were significantly up-regulated under cold and drought stresses, respectively. These genes might be involved into the synthesis of ABA, and could be used as candidate genes in improving the cold and drought resistance in F. suspensa. Taken together, this study improves our understanding of the CCD gene family and provides key candidate genes associated with cold and drought stresses in F. suspensa.
Data availability statement
The datasets presented in this study can be found in online repositories. The names of the repository/repositories and accession number(s) can be found in the article/Supplementary material.
Author contributions
X-LZ and YL coordinated execution of this study. X-LZ performed the RNA-seq analysis. Y-LY performed the gene family analysis. H-XX performed the qPCR experiment. YL wrote the manuscript. All authors have read and agreed to the submission of the manuscript.
Acknowledgments
We thank Qian Li for her valuable help in carrying out gene family analysis.
Footnotes
Funding
This work was supported by the Open Fund of State Key Laboratory of Tree Genetics and Breeding (Chinese Academy of Forestry) (Grant No. TGB2021004) and the Key Scientific Research Project of Higher Education Institutions of Henan Province (Grant No. 14A180035).
Conflict of interest
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
Publisher’s note
All claims expressed in this article are solely those of the authors and do not necessarily represent those of their affiliated organizations, or those of the publisher, the editors and the reviewers. Any product that may be evaluated in this article, or claim that may be made by its manufacturer, is not guaranteed or endorsed by the publisher.
Supplementary material
The Supplementary Material for this article can be found online at: https://www.frontiersin.org/articles/10.3389/fpls.2022.998911/full#supplementary-material
References
- Altschul S. F., Gish W., Miller W., Myers E. W., Lipman D. J. (1990). Basic local alignment search tool. J. Mol. Biol. 215, 403–410. [DOI] [PubMed] [Google Scholar]
- Artimo P., Jonnalagedda M., Arnold K., Baratin D., Csardi G., de Castro E., et al. (2012). Expasy: Sib bioinformatics resource portal. Nucleic Acids Res. 40 W597–W603. 10.1093/nar/gks400 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Auldridge M. E., Block A., Vogel J. T., Dabney-Smith C., Mila I., Bouzayen M., et al. (2006). Characterization of three members of the Arabidopsis carotenoid cleavage dioxygenase family demonstrates the divergent roles of this multifunctional enzyme family. Plant J. 45 982–993. 10.1111/j.1365-313X.2006.02666.x [DOI] [PubMed] [Google Scholar]
- Bailey T., Bodén M., Buske F., Frith M., Grant C., Clementi L., et al. (2009). MEME SUITE: Tools for motif discovery and searching. Nucleic Acids Res. 37 W202–W208. 10.1093/nar/gkp335 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chen C., Chen H., Zhang Y., Thomas H. R., Frank M. H., He Y., et al. (2020). TBtools: An integrative toolkit developed for interactive analyses of big biological data. Mol. Plant 13 1194–1202. 10.1016/j.molp.2020.06.009 [DOI] [PubMed] [Google Scholar]
- Cheng D. H., Wang Z. Y., Li S. Y., Zhao J., Wei C. H., Zhang Y. (2022). Genome-wide identification of CCD gene family in six Cucurbitaceae species and its expression profiles in melon. Genes 13:262. 10.3390/genes13020262 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chou K. C., Shen H. B. (2010). Plant-mPploc: A top-down strategy to augment the power for predicting plant protein subcellular localization. PLoS One 5:e11335. 10.1371/journal.pone.0011335 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Felsenstein J. (1996). Inferring phylogenies from protein sequences by parsimony, distance, and likelihood methods. Meth. Enzym. 266 419–427. 10.1016/S0076-6879(96)66026-1 [DOI] [PubMed] [Google Scholar]
- Frey A., Effroy D., Lefebvre V., Seo M., Perreau F., Berger A., et al. (2012). Epoxycarotenoid cleavage by NCED5 fine-tunes ABA accumulation and affects seed dormancy and drought tolerance with other NCED family members. Plant J. 70 501–512. 10.1111/j.1365-313X.2011.04887.x [DOI] [PubMed] [Google Scholar]
- Fu Z. Z., Lei Y. K., Peng D. D., Li Y. (2014). Population genetics of the widespread shrub Forsythia suspensa (Oleaceae) in warm-temperate China using microsatellite loci: Implication for conservation. Plant Syst. Evol. 302 1–9. 10.1007/s00606-015-1241-y [DOI] [Google Scholar]
- Geourjon C., Deleage G. (1995). SOPMA: Significant improvements in protein secondary structure prediction by consensus prediction from multiple alignments. Comput. Appl. Biosci. 11 681–684. 10.1093/bioinformatics/11.6.681 [DOI] [PubMed] [Google Scholar]
- Hamzah M. A., Kasim N. A. M., Shamsuddin A., Mustafa N., Rusli N. I. M., Teh C. Y. (2020). Nucleotide variations of 9-cis-epoxycarotenoid dioxygenase 2 (NCED2) and pericarp coloration genes (Rc and Rd) from upland rice varieties. 3 Biotech 10:105. 10.1007/s13205-020-2092-y [DOI] [PMC free article] [PubMed] [Google Scholar]
- Han Y., Lu M., Yue S., Li K., Dong M., Liu L., et al. (2022). Comparative methylomics and chromatin accessibility analysis in Osmanthus fragrans uncovers regulation of genic transcription and mechanisms of key floral scent production. Hortic. Res. 9:uhac096. 10.1093/hr/uhac096 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hu K., Guan W. J., Bi Y., Zhang W., Li L. J., Zhang B. L., et al. (2021). Efficacy and safety of lianhuaqingwen capsules, a repurposed Chinese herb, in patients with coronavirus disease 2019: A multicenter, prospective, randomized controlled trial. Phytomedicine 85:153242. 10.1016/j.phymed.2020.153242 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jones D., Taylor W., Thornton J. (1992). The rapid generation of mutation data matrices from protein sequences. Comput. Appl. Biosci. 8 275–282. 10.1093/bioinformatics/8.3.275 [DOI] [PubMed] [Google Scholar]
- Kawahara Y., de la Bastide M., Hamilton J. P., Kanamori H., McCombie W. R., Ouyang S., et al. (2013). Improvement of the Oryza sativa Nipponbare reference genome using next generation sequence and optical map data. Rice 6:4. 10.1186/1939-8433-6-4 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kumar S., Stecher G., Tamura K. (2016). MEGA7: Molecular evolutionary genetics analysis version 7.0 for bigger datasets. Mol. Biol. Evol. 33 1870–1874. 10.1093/molbev/msw054 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lalitha S. (2000). Primer premier 5.0. Biotechnol. Softw. Int. Rep. 1 270–272. 10.1089/152791600459894 [DOI] [Google Scholar]
- Li Y., Shi L. C., Pei N. C., Cushman S. A., Si Y. T. (2021a). Transcriptomic responses to drought stress among natural populations provide insights into local adaptation of weeping forsythia. BMC Plant Biol. 21:273. 10.1186/s12870-021-03075-6 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li Y., Shi L. C., Cushman S. A. (2021b). Transcriptomic responses and physiological changes to cold stress among natural populations provide insights into local adaptation of weeping forsythia. Plant Physiol. Biochem. 165 94–103. 10.1016/j.plaphy.2021.05.020 [DOI] [PubMed] [Google Scholar]
- Li Y., Wang F., Pei N. C., Li Q., Liu H. L., Yuan W. J., et al. (2022). The updated weeping forsythia genome reveals the genomic basis for the evolution and the forsythin and forsythoside A biosynthesis. Hortic. Plant J. 10.1016/j.hpj.2022 [Epub ahead of print]. [DOI] [Google Scholar]
- Liu Y. C., Zhang C., Dong B., Zhao H. B. (2019). Advances of CCD subfamily in higher plants. J. Agr. Biotech. 27 720–734. [Google Scholar]
- Livak K. J., Schmittgen T. D. (2001). Analysis of relative gene expression data using real-time quantitative PCR and the 2-△△CT method. Methods 25 402–408. 10.1006/meth.2001.1262 [DOI] [PubMed] [Google Scholar]
- Lu S., Wang J., Chitsaz F., Derbyshire M. K., Geer R. C., Gonzales N. R., et al. (2020). CDD/SPARCLE: The conserved domain database in 2020. Nucleic Acids Res. 48 D265–D268. 10.1093/nar/gkz991 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ohmiya A. (2009). Carotenoid cleavage dioxygenases and their apocarotenoid products in plants. Plant Biotechnol. 26 351–358. 10.5511/plantbiotechnology.26.351 [DOI] [Google Scholar]
- Pertea M., Pertea G. M., Antonescu C. M., Chang T. C., Mendell J. T., Salzberg S. L. (2015). Stringtie enables improved reconstruction of a transcriptome from RNA-seq reads. Nat. Biotechnol. 33 290–295. 10.1038/nbt.3122 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Phadungsawat B., Watanabe K., Mizuno S., Kanekatsu M., Suzuki S. (2020). Expression of CCD4 gene involved in carotenoid degradation in yellow-flowered Petunia x hybrida. Sci. Hortic. 261:108916. 10.1016/j.scienta.2019.108916 [DOI] [Google Scholar]
- Qiao Y., Cao Y., Jia M., Wang Y., He J., Zhang X., et al. (2020). Research on flower buds growth development and pollination habits of Forsythia suspensa heterostyly. Acta Hortic. Sin. 47 699–707. [Google Scholar]
- Rambaut A. (2009). FigTree, a graphical viewer of phylogenetic trees. Edinburgh: Institute of Evolutionary Biology University of Edinburgh. [Google Scholar]
- Rosati C., Cadic A., Duron M., Ingouff M., Simoneau P. (1999). Molecular characterization of the anthocyanidin synthase gene in Forsythia×intermedia reveals organ-specific expression during flower development. Plant Sci. 149 73–79. 10.1016/S0168-9452(99)00146-6 [DOI] [Google Scholar]
- Seo M., Koshiba T. (2002). Complex regulation of ABA biosynthesis in plants. Trends Plant Sci. 7 41–48. 10.1016/S1360-1385(01)02187-2 [DOI] [PubMed] [Google Scholar]
- Simkin A. J., Schwartz S. H., Auldridge M., Taylor M. G., Klee H. J. (2004). The tomato carotenoid cleavage dioxygenase 1 genes contribute to the formation of the flavor volatiles beta-ionone, pseudoionone, and geranylacetone. Plant J. 40 882–892. 10.1111/j.1365-313X.2004.02263.x [DOI] [PubMed] [Google Scholar]
- Thijs G., Marchal K., Lescot M., Rombauts S., De Moor B., Rouze P., et al. (2002). A gibbs sampling method to detect overrepresented motifs in the upstream regions of coexpressed genes. J. Comput. Biol. 9 447–464. 10.1089/10665270252935566 [DOI] [PubMed] [Google Scholar]
- Tian S. H., Cheng H., Zhang Y., Liu C., Xia D. A., Wei Z. G. (2021). Genome-wide identification and expressional analysis of carotenoid cleavage dioxygenases (CCD) gene family in Populus trichocarpa under drought and salt stress. Bull. Bot. Res. 41 993–1005. [Google Scholar]
- Truong H. A., Lee S., Trinh C. S., Lee W. J., Chung E. H., Hong S. W., et al. (2021). Overexpression of the HDA15 gene confers resistance to salt stress by the induction of NCED3, an ABA biosynthesis enzyme. Front. Plant Sci. 12:640443. 10.3389/fpls.2021.640443 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Umehara M., Hanada A., Yoshida S., Akiyama K., Arite T., Takeda-Kamiya N., et al. (2008). Inhibition of shoot branching by new terpenoid plant hormones. Nature 455 195–200. 10.1038/nature07272 [DOI] [PubMed] [Google Scholar]
- Wei H., Movahedi A., Liu G., Li Y., Liu S., Yu C., et al. (2022). Comprehensive analysis of carotenoid cleavage dioxygenases gene family and its expression in response to abiotic stress in poplar. Int. J. Mol. Sci. 23:1418. 10.3390/ijms23031418 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Xi W. P., Zhang L. A., Liu S. Y., Zhao G. H. (2020). The genes of CYP, ZEP, and CCD1/4 play an important role in controlling carotenoid and aroma volatile apocarotenoid accumulation of apricot fruit. Front. Plant Sci. 11:607715. 10.3389/fpls.2020.607715 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Xiang M. F., Jin C. T., Sun L. H., Zhang Z. H., Yao J. J., Li L. C. (2021). Efficacy and potential mechanisms of Chinese herbal compounds in coronavirus disease 2019: Advances of laboratory and clinical studies. Chin. Med. 16:130. 10.1186/s13020-021-00542-y [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yang X., Yue Y., Li H., Ding W., Chen G., Shi T., et al. (2018). The chromosome-level quality genome provides insights into the evolution of the biosynthesis genes for aroma compounds of Osmanthus fragrans. Hortic. Res. 5:72. 10.1038/s41438-018-0108-0 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang S. L., Guo Y. T., Zhang Y. Q., Guo J. G., Li K., Fu W. W., et al. (2021). Genome-wide identification, characterization and expression profiles of the CCD gene family in Gossypium species. 3 Biotech. 11:249. 10.1007/s13205-021-02805-9 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang X., Pei J., Zhao L., Tang F., Fang X., Xie J. (2016). Overexpression and characterization of CCD4 from Osmanthus fragrans and β-ionone biosynthesis from β-carotene in vitro. J. Mol. Catal. B Enzy. 134 105–114. 10.1016/j.molcatb.2016.10.003 [DOI] [Google Scholar]
- Zhao J. R., Li J. Y., Zhang J., Chen D., Zhang H. P., Liu C. Y., et al. (2021). Genome-wide identification and expression analysis of the carotenoid cleavage oxygenase gene family in five Rosaceae species. Plant Mol. Biol. Rep. 39 739–751. 10.1007/s11105-021-01284-9 [DOI] [Google Scholar]
- Zheng T. C., Li P., Li L. L., Zhang Q. X. (2021). Research advances in and prospects of ornamental plant genomics. Hortic. Res. 8:65. 10.1038/s41438-021-00499-x [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhou X. T., Jia L. D., Duan M. Z., Chen X., Qiao C. L., Ma J. Q., et al. (2020). Genome-wide identification and expression profiling of the carotenoid cleavage dioxygenase (CCD) gene family in Brassica napus L. PLoS One 15:e0238179. 10.1371/journal.pone.0238179 [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Data Availability Statement
The datasets presented in this study can be found in online repositories. The names of the repository/repositories and accession number(s) can be found in the article/Supplementary material.