Abstract
Lung cancer is by far the leading cause of cancer death in the US. Recent studies have demonstrated the effectiveness of screening using low dose CT (LDCT) in reducing lung cancer related mortality. While lung nodules are detected with a high rate of sensitivity, this exam has a low specificity rate and it is still difficult to separate benign and malignant lesions. The ISBI 2018 Lung Nodule Malignancy Prediction Challenge, developed by a team from the Quantitative Imaging Network of the National Cancer Institute, was focused on the prediction of lung nodule malignancy from two sequential LDCT screening exams using automated (non-manual) algorithms. We curated a cohort of 100 subjects who participated in the National Lung Screening Trial and had established pathological diagnoses. Data from 30 subjects were randomly selected for training and the remaining was used for testing. Participants were evaluated based on the area under the receiver operating characteristic curve (AUC) of nodule-wise malignancy scores generated by their algorithms on the test set. The challenge had 17 participants, with 11 teams submitting reports with method description, mandated by the challenge rules. Participants used quantitative methods, resulting in a reporting test AUC ranging from 0.698 to 0.913. The top five contestants used deep learning approaches, reporting an AUC between 0.87 - 0.91. The team’s predictor did not achieve significant differences from each other nor from a volume change estimate (p=.05 with Bonferroni-Holm’s correction).
Keywords: Lung cancer, nodules challenge, ISBI 2018, indeterminate pulmonary nodules, cancer detection in longitudinal CT, NLST, computed tomography
I. Introduction
Lung cancer is the leading cause of cancer related deaths in the US [1]. Despite the advancements in lung cancer treatment strategies, including the use of targeted therapies coupled with improved treatment regimens, the disease shows poor prognosis [2, 3]. Recent studies [4–6] demonstrated that screening high risk subjects with low dose computed tomography (LDCT) results in a reduction in lung cancer specific mortality. Specifically, the National Lung Screening Trial (NLST) reported a higher cancer detection rate for the LDCT arm of the trial (24.2%) compared to the conventional chest X-ray arm (6.9%). Recently, the United States Preventive Task Force (USPSTF) recommended use of LDCT for screening high risk individuals between 55 to 80 years with a smoking history of at least 30 pack years [7]. While lung nodules are detected with a high rate of sensitivity, lung cancer screening with CT has a low specificity rate and it is still difficult to separate benign lesions from malignant ones. False positive screening rates in a recent study were reported to be high in both LDCT and chest X-ray(96.4% and 94.5% respectively) [8]. Radiological determination of malignant nodules is challenging due to a wide variety of appearances coupled with varied solidity of these nodules. Nodule size metrics have been the only clinically-accepted metric to quantify and characterize abnormalities in lung cancer screening [9–11]. Even so, there has been well-documented literature outlining the limitations in the use of size based dimensions [12, 13]. A significant effort has been made to study the usefulness of nodule characteristics, including the relationship of volume/doubling time and life style/tobacco use to identification of a malignant nodule [14–19]. A number of methods have been proposed in the literature to relate malignancy status to nodule size measurements [20, 21], radiological or physiological aspects [19, 22, 23], quantitative characteristics (radiomics) [24–29] including deeper sub-voxel level nodules characterization (deep learning) [30–33], all with a goal to find surrogate digital markers to identify malignancy. The promise of longitudinal nodule assessment established in these multi-institutional studies, paired with the uncertainty around the reproducibility of nodule segmentations, motivated the creation of a common data set with an open international challenge that fosters novel imaging biomarker development. A few previous community efforts attempted to create consensus-based community data sets. Most notably, these include the Lung Image Database Consortium (LIDC) study, single time point, with radiologists opinions as truth; the LUNGx, single time point diagnostic scans (diagnostic CT), with pathological assessment; and the Data Science Bowl (DSB 2017), single time point with pathological assessment [34–36] challenges. A few other community challenges that have been focused on lung physiology (like nodules location, attachment status) with implication to the disease progression, such as nodule detection, vessel segmentation and vessel tree extraction [37–39]. This challenge, however, goes beyond size-based metrics and is the first challenge to provide two time points matched patient scans with nodule segmentations that are pathologically verified to the competition participants with a goal to assess nodule malignancy using both time point scans. This moves the field beyond segmentation related constraints and with a desire to redirects the focus towards finding clinically actionable decisions. This challenge explicitly encouraged the use of two sequential time point scans. This challenge was organized by members of the National Cancer Institute’s Quantitative Imaging Network (QIN) (co-authors: YB, AB, SN, MMG, LH, DG, JKC, KF and support from PP) through the 2018 Institute of Electrical and Electronics Engineers’ (IEEE) multi-society (EMBS and SPS) organized medical imaging conference, the International Symposium on Biomedical Imaging (ISBI), April 4-8, 2018 in Washington, DC, USA, and was conducted with no restriction on the participant’s affiliation or group size. The final team ranking and top winners were announced at the society’s annual conference, which had wide national and international participation. The teams could use any available datasets for training but were required to fully disclose the data sources with detailed description of the methods used in a report submitted to complete their participation. In this summary report, we present the design, implementation, and results of the challenge, and discuss commonalities between different participant methods, strength of ranking, methodological preferences in the community, and lessons learned in organizing this challenge, including problem mitigation.
II. Materials and methods
Dataset:
We curated data from 100 NLST subjects with an equal number of malignant and benign cases. Each subject had two LDCT scans, a baseline (T0) scan and a follow-up (T1) scan taken within a year. Nodule of interest (one per case) was located by a radiologist based on NLST’s clinical information. Table 1 shows the patient demographics and nodule size distribution for the study cohort used in this challenge. For each case, the organizers released image data in DICOM (Digital Imaging and Communications in Medicine) format, as well as the three-dimensional nodule segmentations in DICOM-RT (radiation therapy) and NIfTI (neuroimaging informatics technology initiative) formats. A nodule location file was also provided by the organizing team that showed a screen capture of the nodule for a selected slice with the slice number at sequential scan intervals. Teams could use the scans in any fashion (whole or partial) and with or without anatomical structures, but in a quantitative, non-manual methods needed to build their models. Due to the limited public access to NLST data, the organizers removed the identifying DICOM tag information, which was embedded in the DICOM images, that carried reference to the original NLST distribution. The image data was released to the participants through the challenge web portal maintained specifically for the challenge (http://isbichallenges.cloudapp.net/competitions/15). The data was open to the challenge participants for a period of ten weeks, eight of which were allocated for algorithms training and two of which were intended for algorithm evaluation. The NLST DTA agreement restricted open access to data beyond the challenge period. Fig. 1 shows a sample subject chest LDCT scan (2D slice) with a lung nodule across two time points.
TABLE I.
Patient clinical and demographics at baseline (t0) and follow-up scan (T1).
| A) Demographics | ||||
|---|---|---|---|---|
| Training (N=30) | Testing (N=70) | |||
| Cancer (N=15) | Benign (N=15) | Cancer (N=35) | Benign (N=35) | |
| Age (median/mean/st.dev) | 62/64.3/5.1 | 66/64.2/5.3 | 66/65.6/4.92 | 66/65.6/5.22 |
| Gender (M/F) | 8/7 | 14/1 | 18/17 | 15/15 |
| Race (White/African-American/Latino/Mixed) | 15/0/0/0 | 15/0/0/0 | 34/1 | 34/1 |
| Clinical Stage & Grade (# in: Well/Moderate/Poor/Undifferentiated/Unspecified) | ||||
| 1A | 10 (2/4/3/0/1) | 0 | 17 (3/9/3/1/1) | 0 |
| 1B | 0 | 0 | 7 (1/3/2/1/0) | 0 |
| IIA | 2 (0/1/1/0/0) | 0 | 2 (0/2/0/0/0) | 0 |
| IIB | 0 | 0 | 2 (0/1/1/0/0) | 0 |
| IIIA | 1 (0/0/0/0/1) | 0 | 0 | 0 |
| IIIB | 1 (0/0/0/0/1) | 0 | 3 (0/0/0/0/3) | 0 |
| IV | 1 (0/0/0/0/1) | 0 | 4 (0/0/0/1/2) | 0 |
| N/A | 0 | 15 | 0 | 35 |
Fig. 1.
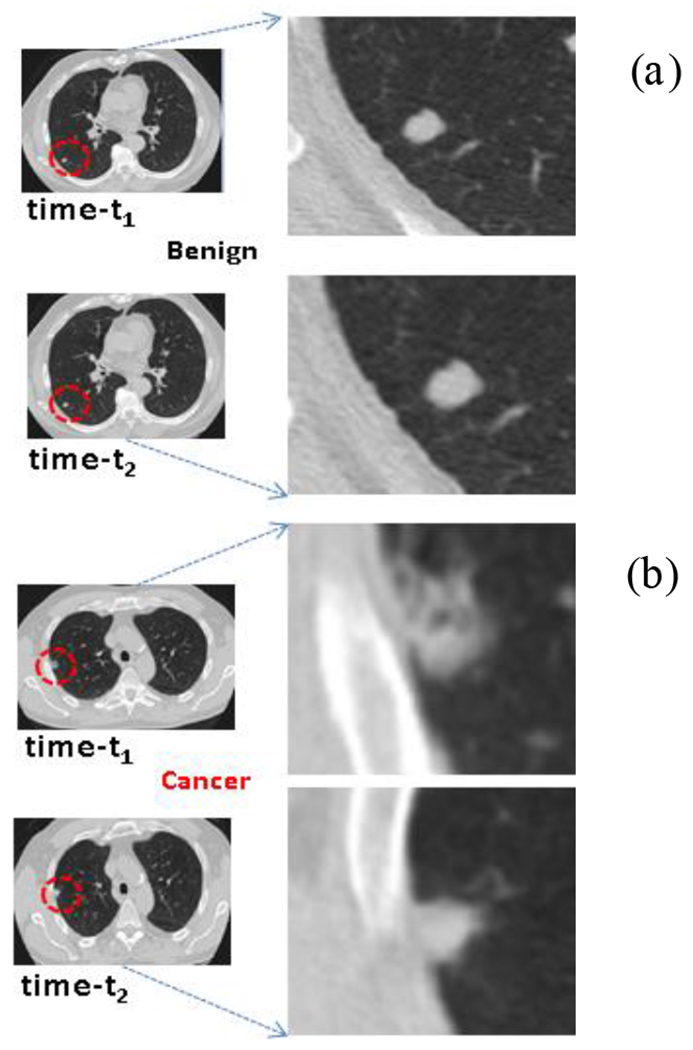
Representative patient cases showing 2D CT slice across two time points (t1 & t2) with a) Benign and b) Malignant diagnosis.
We randomly selected 30 cases with sequential time point scans, with an equal number of malignant and benign nodules that were released as a training (or calibration) dataset. Pathological diagnosis for the training dataset was provided to the competition participants. The participants were encouraged to calibrate their algorithms with the training cases. The test dataset contained the remaining 70 subjects with blinded clinical outcome, upon which competition participants were evaluated. Teams could use any external datasets to train their methods, but participants were asked to disclose the methods with details of such datasets in their summary report. The ‘Participant Methods’ section summarizes the methods used by the participants and additional detail is presented in the supplemental section.
The patient’s cancer status along with nodule pathology results were obtained from the NLST’s clinical diagnosis tables. The locations for malignant nodules were obtained from the NLST’s radiology report and verified in the sequential CT scans by a resident radiologist. The locations for benign nodules were selected based on a radiological consensus read using subject sequential LDCT scans, as patients were classified as non-cancerous by the NLST. Detail on nodule identification is described in prior publications [23, 40].
A. Evaluation Criteria
The challenge organizers used subjects’ nodule malignancy information along with investigated nodule location, obtained from the NLST abstracted clinical tables [4, 41] (as described earlier), that was used as ground truth. The organizing team used area under the receiver operating characteristic curve (AUC) [42, 43] based on participants’ submitted nodule malignancy scores to assess participant predictor performance. Participants were asked to submit an estimated malignancy score (ranging from [0,1]) for each case, one nodule per case). There was no limitation placed on participating teams’ methodology or algorithms, except that the proposed methods be non-manual (ideally, fully automated). Some extent of manual intervention, or correction was allowed, such as re-segmenting with additional (manual) seed points or altering the regions of interest. We placed no restriction on the teams during the training phase; they could adjust any functional (kernel) parameters or re-configure their networks. Participants were ranked based on their best AUC evaluated from the three allowed test phase submissions (during the initial days of the challenge the system allowed six entries, which was rectified to the stated limit). Full consideration upon challenge completion was limited to those teams that submitted their malignancy scores for each case in the test set, a mandatory abstract describing their proposed methodologies at the end of the eight-week training phase, and a final report describing details of their method with best performing model setting, by the end of the testing phase.
B. Challenge Execution
The Lung Nodule Malignancy Prediction challenge was implemented on a customized version of the CodaLab open source platform [44]. The challenge platform allowed prospective participants to register for the challenge and learn about the goals and the rules of the challenge. Once registered, participants had to download the training set (30 subjects) which had CT scans from two consecutive visits as well as diagnosis data. At the testing phase, participants could download a 70-case test set, which had CT scans from two consecutive visits without any diagnostic data. Using their respective trained models, participants submitted estimated malignancy scores (ranging between [0,1]) for the test cases. There were instances of participants creating duplicate user accounts and initiating additional submissions over the maximum allowed number of entries. The challenge organizers took additional effort to manually authenticate the users and compiled their submissions. This process resulted in removing or consolidating some of the user accounts. Teams without final report submissions were not considered for final validated teams results.
The competition participants were requested to submit their results via the challenge’s web-portal. Teams could submit their model predictions as many times as necessary during the training phase and a leader board displayed evaluation metrics for all team submissions. The teams were required to disclose the specific methodology behind their best performing submission in the final report. Individual submissions were manually verified and linked to the teams and the final leaderboard was updated accordingly. Top-performing teams were initially invited to present their findings, but the invitation was later extended to all willing participants to present their methods in a special challenge session convened during the 2018 ISBI Conference.
C. Nodule Segmentation and Prior Volume Estimate
It is widely believed that the lack of growth or slow growth over time is indicative of benign nodules. There have been a number of studies that showed that the nodule growth rate pattern had the ability to predict malignancy [21, 45, 46]. Lack of consensus in nodule segmentations as well as discrepancies in nodule size measurement practices (3D) are possible reasons that have impeded the use of these metrics in regular clinical practice. The effect of different segmentations on radiomic features derived using different software packages was studied and it was found that 68% of the 830 computed features showed a Concordance correlation coefficient (CCC) above 0.75 across different segmentations on the same set of nodules [47].
In a recent effort, several institutions from the National Cancer Institute (NCI) funded Quantitative Imaging Network (QIN) evaluated the variability of pulmonary nodule volume change estimated in a sequential scan using semi-automated methods. The study assessed each participating institutions’ volume change estimates for their ability to predict individual nodule malignancy and reported an AUC range of 0.65 to 0.89 [48] (see Suppl. S.F. 1). Notably, methods for calculating volume change between institutions had a range of CCC of [0.56 to 0.95], indicating that nodule segmentations and volumes varied considerably between participating institutions’ segmentation methods. This result bolstered findings that the group had previously found in a study to evaluate the differences in single time-point semi-automatic segmentation methods across institutions on a diverse set of lung lesions (including phantoms). They reported significant differences (p<0.05) in spatial overlap of segmented regions between different institutions, and even within the same institution but across different initializations within a given software package [20].
D. Volume Change as Baseline Predictor
We developed a predictive model using change in lesion volume to serve as a baseline comparator. We obtained volume estimates based on organizers delineation masks provided for the challenge. The nodules were semi-automatically delineated using a segmentation region growing method and the resulting boundary overread, corrected by a trained radiologist (Moffitt Team), reported in earlier work [48]. In the post challenge analysis, we compared the performance of nodule volume estimated by the participating teams to the organizer’s measurement (see Suppl. Table ST1, Suppl. Fig. SF2 & Table ST2). It is to be noted that participating teams were free to alter the segmentation boundaries as appropriate for their planned methods. Additionally, the volume estimate was used to predict the nodules malignancy status and form a baseline predictor.
E. Participant Methods
In this section, we summarize approaches used by participating teams of the ISBI 2018 Lung Nodule Malignancy Prediction challenge. There was no restriction on the participant team size, participant place of origin, or institutional affiliation. This section contains a summary of the methods used by the 11 participating teams that provided a complete description of their approaches; a supplemental document contains methodological details, model description, figures and supporting tables. Four other participating teams (T4, T7, T11, T15) had an incomplete method description and did not respond to our request for details. In addition, two teams (T13 and T17) decided to withdraw post-challenge. Following the challenge rules, abstracts for participants with missing reports have not been included.
Team 1:
The team used a deep learning network architecture that consisted of a nodule detector trained on the LIDC-IDRI dataset followed by a cancer predictor trained on the DSB 2017 dataset [34], totaling about 1593 LDCTs. The final evaluation was conducted on the ISBI 2018 Lung Nodule Malignancy Prediction test set. The team trained a multi-path network of five paths combined in a fully convolutional layer (each with 3D CNN architecture). After initial network training using DSB 2017, challenge data sets with segmentation were used to fine tune the network. To obtain change information, a subtraction was done between the nodules across the sequential scans. A linear combination of the probabilities of these network values provided the final malignancy score. A more detailed description is provided in the Suppl. Sub.T1.
Team 2:
This team used an ensemble of deep and machine learning methods and trained their system using two public external datasets LIDC-IDIR and DSB 2017 [34, 36], reporting to have used a total of 2481 patients with over 5149 nodules. The proposed method is composed of two deep convolutional neural networks, inspired by the VGG [36] and RESNET [49] architectures. The first network was trained to predict nodule characteristics including subtlety, internal structure, sphericity, calcification, lobulation, spiculation, texture, and malignancy likelihood based on radiologists’ annotations of the LIDC-IDRI cohort. The second network was trained on the DSB 2017 dataset to predict the patient cancer probability using the risk dominant nodule in a single scan. The features from the first system with addition of diameter and location of nodule were combined with the image representations of the nodule to predict the cancer likelihood in the second system. Finally, a logistic regression model was trained on the ISBI challenge dataset that included the growth of nodules in the provided time points. In the three submissions, the Ensemble method that used a geometric mean of deep network and growth model achieved the highest AUC on the test set. Further details are provided in the suppl. Sub.T2.
Team 3:
This team used a 3D convolutional neural network (CNN) model along with volume change information to estimate the malignancy scores (pseudo probability) of lung nodules. This team’s 3D CNN model consists of two networks, the feature extraction network and the malignancy prediction network. The first network is designed to extract image features from a nodule in an image. The malignancy prediction network is designed to reduce the number of the concatenated image features. The team used tumor doubling time (DT) and percentage volume change (PVC) to incorporate volume change information. An additional 40 patient image scans with two time points (80 LDCTs) obtained independently by the team from the NLST were used for training. Further details are provided in supplemental section Sub.T3.
Team 5:
This team used deep learning (DL) with clinically relevant hand-crafted features to assess the malignancy status of the nodule. The approach used by the team was designed to fuse global patient information with spatiotemporal evolution of the main nodule like changes in shape, position. They used two additional datasets, namely the Lung Nodule Analysis 2016 (LUNA16) [50] and DSW2017 [34], with a total of 3499 LDCTs. The radiomic features were extracted from nodule regions and combined with the normalized axial position that was fed into a deep network (ResNet) to obtain a malignancy score. The temporal information was encoded in this model through a linear combination of the malignancy scores of each CT scan. Further details are provided in Supplemental section Sub.T5.
Team 6:
This team’s method combines three independent methodologies to predict the degree of lung nodule malignancy that uses information from two sequential CT scans. Each model individually predicts the malignancy using the information provided in a single scan or from two sequential scans. The Borda Count algorithm [51] fuses the predictions obtained by each model and provides the final malignancy score from the three independent methods. The team used a mixed source of external data (Lung Nodule Analysis 2016 or LUNA16, Lung Image Database Consortium or LIDC and local), with about 1260 CT’s used for model training. The algorithm and modeling approaches are described in more details in Suppl. Sub.T6.
Team 8:
This team combined the latent features extracted from a stacked auto-encoder with quantitative features including nodule size, shape, intensity, and a few others. These metrics came both from a single scan and the changes between the two sequential scans. These features were used to train a logistic regression model to obtain nodule malignancy prediction. The team reported to have used challenges data set for their training, Further details are provided in Suppl. Sub.T8.
Team 9:
This team presented a fusion system based on deep learning and radiomics to analyze longitudinal CT scans that used growth of lung nodules between the time points to assess the risk of malignancy. The deep learning system was used to transform 3D patches around the nodules into high-dimensional feature vectors. These high-dimensional features were then concatenated with radiomic features that were extracted in the vicinity of nodules. A 436-length feature vector was extracted for each patient scan and these metrics were used to train a logistic regression model to assess the pathological malignancy status. A 3-fold cross validation was used on the training set. They used a mixed source of external data set (DSB2017, LUNA16), totaling 2481 LDCT’s for their model training. Further details are provided in Suppl. Sub.T9.
Team 10:
This team’s method was based on combining visual and temporal features extracted from the pulmonary nodules of interest. The patient malignancy score (pseudo probability) was computed by a simple averaging of these scores. The visual features were extracted by a 3D CNN that was first pre-trained for a separate task and then fine-tuned for malignancy classification. In this approach, a combination of three datasets was used for training: LIDC, challenge’s training data, and team’s institutional data. The change in volume and longest diameter of each nodule across time points were determined for a nodule using the segmentation masks. The first set of visual features was extracted by a deep 3D neural network similar to the VGG-16 [52], but contained six convolutional layers with 3D filters. The initial model was created by pre-training on the LUNA16 challenge [50]. A simple fusion method was performed by averaging the two percent differences based on volume and diameter change along with the neural network confidence score. The combination of these features boosted the accuracy of the overall malignancy score. They used data from LIDC and LUNA16 for model training, totaling to 1936 LDCTs. Further details are provided in suppl. Sub.T10.
Team 12:
This team used a feature-based approach, specifically focused on computed 3D-Grey Level Co-Occurrence Matrix (GLCM) features on the nodules, independently across the time points. The final malignancy status was obtained by averaging the score obtained by linear discriminant functions at these two time points. The team used the LIDC16 data set (524 LDCTs) along with challenge’s training data to build the model. The data was divided into training and testing with k-fold cross validation and a linear discriminant classifier model. The best-trained model was applied on the test data provided in the test phase of the challenge. Further details are provided in Suppl. section Sub.T12.
Team 14:
The team followed a radiomics approach to extract features from the nodules of interest and proposed an ensemble learning model based on a selected subset of features to obtain a malignancy score. The team used the training/calibration data provided and extracted radiomic features using the pyra diomics package[53] applied to the segmentation masks. The team used an ensemble of two step algorithms to select features of interest. The first step involved a gradient tree boosting and XGBoost as a feature selection approach [54]. Based on this step ten features were automatically selected from the pool of 297 available features. The choice of keeping ten features was converged based on a cross-validation approach on the training set. The second step was to use classification on the selected features. This step involved an ensemble of a bagging classifier, XGBoost, random forest [4], and multi-layer perceptron. The final prediction was made using a soft majority voting of predictions from all the models. Further details are provided in Supplemental section Sub.T14.
Team 16:
The team used a deep learning model to predict the malignancy score of lung nodules in CT. The CT images were first interpolated to a common resolution (0.72x0.72x2 mm) and used a network model motivated by a prior study [55]. They used external data sets from mixed sources (LIDC, DSB2017), totaling 1593 LDCT for their model training The team designed a model that combines ResNeXt [56] with the DenseNet [57] deep network architecture. The implemented architecture implemented consists of two identical parallel paths each with four general denseNext blocks (B) with five ResNext layers each, with a cardinality (K) of four and an increasing growth rate (G) within the blocks. The network model provided three probability maps (background, benign nodule, and malignant nodule) that were reported as nodules’ malignancy score. Further details are provided in Suppl. Sub.T13.
III. RESULTS
The challenge attracted wide international participation with over 120 registered participants, grouped into 17 teams that submitted the abstract, of those, 11 teams provided complete reports and were part of this challenge paper. Broadly the participant affiliation could be categorized into universities (6), clinical research laboratories (2), and industry (3). Authors were represented from four continents: Asia (2), Europe (1), North America (6) and South America (2). The participating teams reported the malignancy score for the annotated nodules in patients with two sequential time scans, following the approaches stated in their individual reports. Several teams used external datasets for training their methods. Most common datasets the participants used were the LIDC database, LUNA16, and DSB 2017 (see Table 2).
TABLE 2.
Overview of challenge participant methods and their best AUC estimate obtained on the test data. Volume change estimate was used as a comparator (post challenge), See Supplemental Tables S1, S2 for details.
| Teams | Reported Method | Training Data Source | Training Data-CT** | Sensitivity/Specificity | AUC, CI |
|---|---|---|---|---|---|
| T1 | Deep learning (3D CNN) | LIDC-IDRI, DSB 2017 | 2394 | 0.867/0.886 | 0.913 [0.844, 0.982] |
| T2 | Deep learning, ensemble-approach (volume change) | Selected-LIDC, DSB 2017 | 2481 | 0.714/0.971 | 0.897 [0.822, 0.972] |
| T3 | Deep learning, volume change | NLST cohort | 80 | 0.714/0.943 | 0.879 [0.797, 0.962] |
| T5 | Deep learning and hand crafted radiomics. | LIDC, DSB 2017, LUNA16. | 3499 | 0.714/0.971 | 0.868 [0.778, 0.958] |
| T6 | Deep learning and nodule growth | LUNA16, LIDC, Local data | 1078 | 0.714/1 | 0.867 [0.778, 0.956] |
| T8 | Fusion approach. Deep learning – Stacked auto-encoder (6features including size). | Training data (ISBI 2018) | 60 (2*30) | 0.686/0.943 | 0.855 [0.761, 0.949] |
| T9 | Deep learning (CNN) and Radiomics | DSB 2017, LUNA16. | 2481 | 0.571/1 | 0.854 [0.764, 0.943] |
| T10 | Deep Learning (CNN) | LIDC, 30 CT from local institution. LUNA16 pre-trained. | 1936 | 0.686/0.914 | 0.848 [0.756, 0.941] |
| T12 | GLCM with varying ROI and discriminant functions | Selected LIDC | 524 | 0.743/0.0886 | 0.809 [0.697, 0.92] |
| T14 | Radiomics | Not reported. | Training CTs | 0.629/0.914 | 0.789 [0.678, 0.901] |
| T16 | Deep learning | LIDC-IDRI and DSB 2017. | 1593 | 0.57/0.486 | 0.698 [0.572, 0.823] |
| Volume Change | Training data | 60 (2*30) | 0.743/0.943 | 0.866 [0.773, 0.96] |
Unless stated by the authors, the number of CT scans reported in the table was obtained from the respected studies, namely: DSB 2017 (Data Science Bowl 2017) had 1593 CTs, LUNA 16(Lung Image Analysis 2016) had 888 CTs, and LIDC (Lung Image Database Consortium) had1018 CT scans. ISBI 2018-challenge provided 30 sequential CTs: as training (or calibration set).
#: Teams’ method description was removed from assessment due to incomplete reports (T4, T7, T11, T15) or voluntary withdrawal (T13 and T17).
Participating teams’ predictive performance was measured by the AUC values. The Challenge participants with complete reports achieved AUCs ranging between 0.913 and 0.698, with nine out of the eleven participating teams achieving AUCs above 0.8. These measures are broadly grouped into quartiles for comparison purposes. The top quartile was composed of three teams (T1, T2, T3), reporting an AUC of 0.913 to 0.879. The second quartile group comprised three teams (T5, T6, T8) that reported an AUC of 0.868 to 0.855. The third quartile group consisted of two teams (T9, T10) that reported an AUC of 0.854 to 0.848. The last quartile group comprised three teams (T12, T14, T16) that reported an AUC of 0.809 to 0.698.
Notably, the difference in AUC was not statistically significant after multiple testing corrections (p≤0.05) between the first-ranked team and the last quartile group, which included teams greater than index-twelfth (T12, T14, T16). Furthermore, there was no statistical difference between the volume change for any other team. Based on the statistical difference based on their AUC comparisons (Delong’s test, un-corrected p-values), we could group the participant results into two broad groups. The first group was comprised of eight teams (T1, T2, T3, T5, T6, T8, T9, T10) and the second group comprised of three teams (T12, T14, T16) and six additional teams were removed due to incomplete report or withdrawal post-challenge. Figure. 2 illustrates the AUC values achieved by the participating teams (the best of each team’s three submitted algorithms). We find nine teams (T1,T2,T3,T5,T6,T8,T9,T10) using deep learning methods with an AUC range of [0.698, 0.913]. Two teams (T12, T14) reported to have used radiomic based methods with an AUC range of [0.789, 0.809]. It is interesting to see the distribution of the malignancy score as reported by the teams on a limited set of test patient data for benign and malignant cases (see Suppl. Figure SF4). To have a baseline comparison to the team’s estimate, we trained the volume change predictor on the training cohort (30 patients) using a logistic regression model and applied it on the test patients to estimate malignancy score; we obtained an AUC of 0.866 [0.773, 0.96]. Using volume at baseline and follow up time point, individually, we had an AUC of 0.672 and 0.839, respectively. In comparison, using the nodule’s size change (longest diameter change) as a predictor, it had an AUC of 0.76. Using nodules diameter/size information at baseline and follow-up, each gave an AUC of 0.694, 0.824 respectively (see Suppl. Table ST1). We also find that volume change shows better performance (AUC) compared to size (diameter) based measurement; a similar observation has been reported by other studies [58]. We believe the baseline comparator is useful to assess the ability of non-size-volumetric based predictors.
Fig. 2.
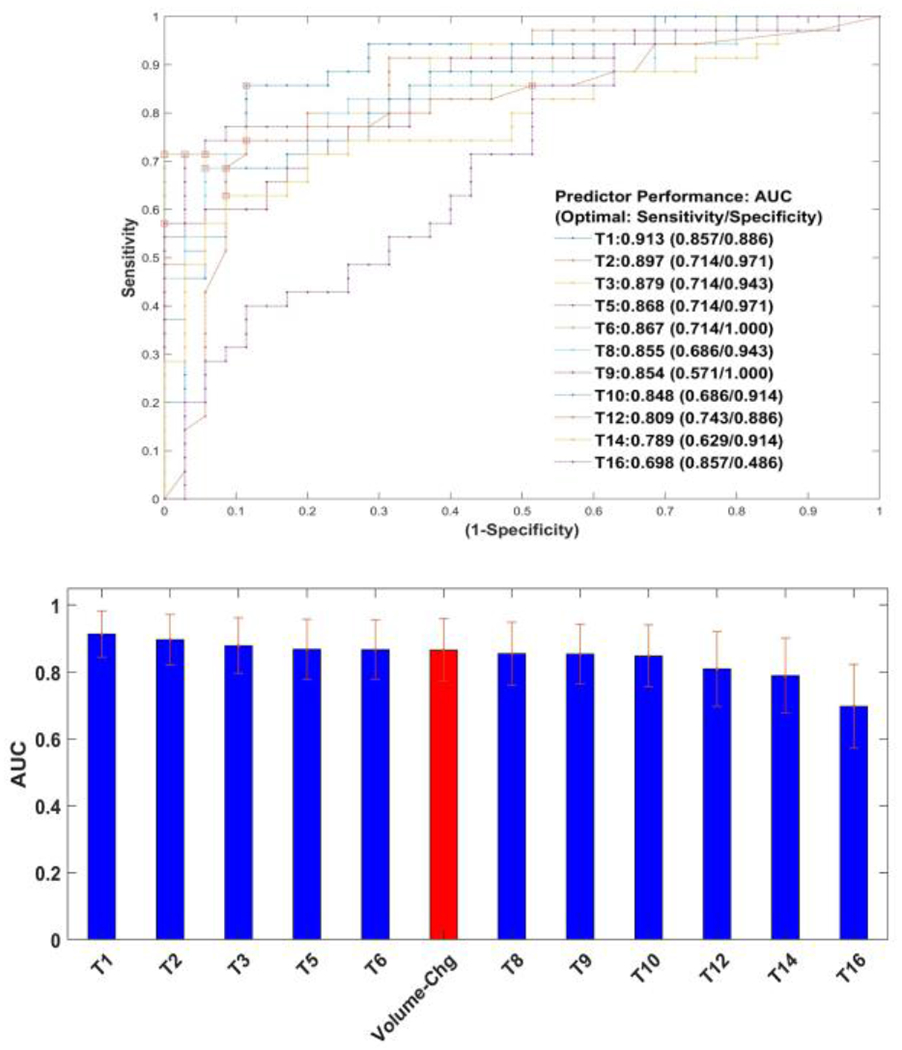
Challenge participant’s predictor performance. A) Receiver operator characteristic curve for participant predictors (11 teams). B) Bar graph showing the highest AUC values with 95% confidence limits computed by bootstrap resampling. Volume change predictor is shown for baseline comparison.
We compared the participating team’s predictor performance to assess their statistical significance (see Suppl. Table ST3 C) using De-long’s test [59]. We find no significant difference between any of the participant predictors nor with the volume change estimate, after applying multiple test corrections using the Bonferroni-Holm method to control family wise error rate [60]. To have an unbiased comparison True Positive Rate (TPR or sensitivity) and False Positive Rate (FPR or 1-specificity) were computed at different ranges, by fixing one metric and computing the other across the teams (see Table 3). It is interesting to note that the teams T2, T5, T6 showed best sensitivity (TPR) of 71.4% (all three) for an FPR of 3 to 5%. While teams T2, T1 show lowest FPR of 51.4%, 54.3% respectively, for a sensitivity (TPR) of 95 to 97%. Similar comparisons can be derived using partial AUCs. Teams T6, T9, T5 show the highest pAUC of 2.04%, 1.63%, 1.55% respectively for an FPR (1-specificity) interval of 0-5%. Teams T1, T2, T10 show highest pAUC of 1.19%, 1.5%, 1.18% respectively for a TPR (sensitivity) interval of 95-100% (see Suppl. Table ST4).
TABLE 3.
Performance comparison of participants predictor for, a) range of False positive rate (FPR or 1-specificity) and b) range of True positive rates (TPR or sensitivity). Highlighted top and bottom 5%.
| FPR | A. TPR (sensitivity) across teams | |||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|
| T1 | T2 | T3 | T5 | T6 | T8 | T9 | T10 | T12 | T14 | T16 | Vol. Chg | |
| 0.01 | 0.371 | 0.486 | 0.457 | 0.543 | 0.714 | 0.457 | 0.571 | 0.2 | 0.02 | 0.286 | 0 | 0.143 |
| 0.03 | 0.514 | 0.714 | 0.571 | 0.714 | 0.714 | 0.543 | 0.571 | 0.457 | 0.144 | 0.486 | 0.2 | 0.514 |
| 0.05 | 0.514 | 0.714 | 0.571 | 0.714 | 0.714 | 0.543 | 0.571 | 0.457 | 0.164 | 0.486 | 0.2 | 0.514 |
| 0.1 | 0.771 | 0.743 | 0.714 | 0.771 | 0.714 | 0.714 | 0.6 | 0.686 | 0.7 | 0.629 | 0.314 | 0.743 |
| 0.15 | 0.857 | 0.771 | 0.771 | 0.771 | 0.743 | 0.743 | 0.657 | 0.686 | 0.743 | 0.629 | 0.4 | 0.8 |
| 0.2 | 0.857 | 0.8 | 0.771 | 0.771 | 0.743 | 0.771 | 0.714 | 0.743 | 0.771 | 0.714 | 0.429 | 0.829 |
| 0.25 | 0.886 | 0.8 | 0.8 | 0.771 | 0.743 | 0.8 | 0.743 | 0.743 | 0.771 | 0.714 | 0.429 | 0.829 |
| 0.3 | 0.943 | 0.829 | 0.8 | 0.771 | 0.8 | 0.829 | 0.771 | 0.8 | 0.786 | 0.743 | 0.486 | 0.829 |
| 0.35 | 0.943 | 0.914 | 0.857 | 0.829 | 0.8 | 0.829 | 0.857 | 0.857 | 0.8 | 0.743 | 0.543 | 0.857 |
| 0.4 | 0.943 | 0.914 | 0.914 | 0.857 | 0.829 | 0.857 | 0.914 | 0.886 | 0.829 | 0.743 | 0.629 | 0.886 |
| 0.45 | 0.943 | 0.943 | 0.943 | 0.857 | 0.829 | 0.886 | 0.914 | 0.886 | 0.829 | 0.743 | 0.714 | 0.886 |
| 0.5 | 0.943 | 0.943 | 0.943 | 0.857 | 0.857 | 0.886 | 0.914 | 0.943 | 0.85 | 0.8 | 0.714 | 0.886 |
| 0.55 | 0.971 | 0.971 | 0.943 | 0.914 | 0.914 | 0.886 | 0.914 | 0.943 | 0.857 | 0.829 | 0.857 | 0.914 |
| 0.6 | 0.971 | 0.971 | 0.943 | 0.943 | 0.943 | 0.886 | 0.914 | 0.943 | 0.871 | 0.857 | 0.857 | 0.914 |
| TPR | B. FPR (1-specificity) across teams | |||||||||||
| T1 | T2 | T3 | T5 | T6 | T8 | T9 | T10 | T12 | T14 | T16 | Vol.Chg | |
| 0.4 | 0.029 | 0 | 0 | 0 | 0 | 0 | 0 | 0.029 | 0.057 | 0.029 | 0.171 | 0.029 |
| 0.45 | 0.029 | 0 | 0 | 0 | 0 | 0 | 0 | 0.029 | 0.064 | 0.029 | 0.257 | 0.029 |
| 0.5 | 0.029 | 0.029 | 0.029 | 0 | 0 | 0.029 | 0 | 0.057 | 0.081 | 0.057 | 0.314 | 0.029 |
| 0.55 | 0.057 | 0.029 | 0.029 | 0.029 | 0 | 0.057 | 0 | 0.086 | 0.086 | 0.057 | 0.371 | 0.057 |
| 0.6 | 0.057 | 0.029 | 0.057 | 0.029 | 0 | 0.057 | 0.143 | 0.086 | 0.086 | 0.086 | 0.4 | 0.057 |
| 0.65 | 0.057 | 0.029 | 0.057 | 0.029 | 0 | 0.057 | 0.143 | 0.086 | 0.086 | 0.171 | 0.429 | 0.057 |
| 0.7 | 0.057 | 0.029 | 0.057 | 0.029 | 0 | 0.086 | 0.2 | 0.171 | 0.1 | 0.2 | 0.429 | 0.057 |
| 0.75 | 0.086 | 0.114 | 0.114 | 0.086 | 0.257 | 0.2 | 0.257 | 0.286 | 0.2 | 0.486 | 0.514 | 0.143 |
| 0.8 | 0.114 | 0.286 | 0.314 | 0.343 | 0.371 | 0.257 | 0.343 | 0.314 | 0.371 | 0.514 | 0.514 | 0.2 |
| 0.85 | 0.114 | 0.314 | 0.343 | 0.371 | 0.457 | 0.371 | 0.343 | 0.343 | 0.5 | 0.6 | 0.514 | 0.343 |
| 0.9 | 0.286 | 0.314 | 0.371 | 0.543 | 0.543 | 0.686 | 0.4 | 0.486 | 0.664 | 0.743 | 0.629 | 0.514 |
| 0.95 | 0.543 | 0.514 | 0.686 | 0.657 | 0.771 | 0.8 | 0.657 | 0.714 | 0.786 | 0.857 | 0.686 | 0.771 |
| 0.97 | 0.543 | 0.514 | 0.686 | 0.657 | 0.771 | 0.8 | 0.657 | 0.714 | 0.906 | 0.857 | 0.686 | 0.771 |
| 0.99 | 0.686 | 0.829 | 0.857 | 0.943 | 0.771 | 0.857 | 0.857 | 0.8 | 0.97 | 0.857 | 0.857 | 0.971 |
IV. DISCUSSION
Challenges offer the opportunity to bring out the collective talents of scientific communities that would not normally subject their algorithms for performance comparison using a blinded-independent reference dataset. For this to occur, the challenge needs to be accessible and open. Ideally, it fosters research towards a common goal to address the most critical needs, which in our case is to improve clinical diagnosis in lung cancer using sequential screening LDCT. The ISBI 2018 Lung Malignancy Prediction Challenge provided the platform to organize this community wide challenge that was made possible by the availability of NLST patient data. The underlying mandate of this exercise was to encourage method development and to bring the best worldwide talents in the scientific community to propose new approaches, particularly, in the early diagnosis of lung cancer and to evaluate the validity of these methods on a common set of test cases.
There have been few prior pulmonary nodule challenges in the past. One example is the LUNGx challenge, whose goal was to identify malignant nodules in single time point diagnostic scans. This was organized by a team led by Armato et.al through a joint effort of International Society for Optics and Photonics (SPIE), along with the American Association of Physicists in Medicine (AAPM) and Quantitative Imaging Network (QIN) of the National Cancer Institute (NCI) [35, 61]. The imaging data for the challenge came from a single institution with 15 samples used for model calibration, and 73 (37 benign, 36 malignant, diagnostic scans) used for model evaluation. Participating teams achieved an AUC for this task in the range of 0.55 to 0.71, with only three out of 11 participant methods performing better than random guesses. Competition organizers also sought malignancy assessments for the evaluation datasets from six thoracic radiologists from two different institutions. The AUC performance of clinical opinion on the test data was in the range of 0.70 to 0.85, outperforming almost every method submitted by challenge participants. Recently, the NCI partnered with industry to conduct a prized challenge (Booz Allen Hamilton’s sponsored Data Science Bowl challenge or DSB 2017) [34], which used a larger cohort with over 1593 cases for the training phase and 200 patient scans used for testing. The challenge had a prize of $1 million USD provided by Laura and John Arnold Foundation[62], which attracted over 400 international participants. They used a log-loss metric to score participant methods, and the top ten teams had a score ranging from 0.399 to 0.444. In a recent review of past international challenges [63], several recommendations have been made, which include a needed focus on reproducibility and interpretability of results for proposed methods and robustness of ranking. The report documents factors that could affect the result, which could be due to choices of data sets or ensemble methods used. In our post challenge analysis, we reanalyzed participant-reported malignancy probabilities and evaluated the strength of their predictors, by computing TPR (True Positive Rate) and FPR (False Positive Rate) at different cut points, by fixing one and computing the other across the participants (see Table 3). One could choose an operating interval based on required sensitivity (TPR)/specificity (1-FPR). At a range of 3 to 5% FPR we find teams T2, T5 and T6’s predictor performs the best with a TPR of 71.4%. At a 95% TPR, Team T2 and T1’s predictor shows best results with an FPR of 51.4% and 54.3%, respectively. In comparison, volume change has a TPR (sensitivity) of 51.4% for an FPR of 3% or 5%, certainly lower compared to the two best-performing teams. For a TPR (sensitivity) of 95% to 97%, the volume change estimate has an FPR of 77.1%, higher than the top four (T1, T2, T3, T5) performing teams. Based on choices of the cut points (TPR, FPR), one can observe a possible shift in the participant ranking order. Our current comparative analysis provides unbiased means to compare participants’ predictors. It should be noted that small test sample size limits generalizability of these findings; nevertheless, it provides the possibility for a new discovery or to contrast and improve approaches. There needs to be some caution while cross comparing methods (see supplemental tables, figures for additional information). As reported in the prior study [63], the authors have shown that a change in summary statistics could alter the ranking of participants in a challenge. In our re-analysis of the participants’ predictors with finer threshold levels based on TPR/FPR, we could derive similar inference (see section B. ‘Strength of participant Ranking’ and supplemental Table SF4).
The current clinical convention in lung cancer screening is to use univariate maximal diameter to screen for detecting abnormal (possibly malignant) nodules; the NLST used a fixed diameter of 4mm as a cutoff for positive findings. Most oncologists follow consensus criteria derived either from the National Comprehensive Cancer Network (NCCN), the Fleischner Society recommendations, or American College of Radiology’s Lung RADS. The NCCN and Lung RADS criteria provide a range of 6 to 8mm for nodule diameter (size), and allow for additional consideration of nodule densities before malignancy classification [9, 64, 65]. The Lung RADS score is particularly relevant to our challenges, as it allows for the consideration of the patient’s previous scans in the malignancy classification, when available. In a prior study [66], authors used Lung RADS to predict the malignancy status of NLST data at initial screening and with a one-year follow-up and two-year follow-up. They reported a test AUC in the range of [0.4 to 0.72] at baseline and a range of [0.76, 0.973] at follow-up diagnostic time points. In the current challenge, participating team methods yielded an AUC range of 0.698 to 0.913 (based on the 11 teams with complete reports). The deep neural network methods report an AUC of 0.698 to 0.913, while the radiomic methods report an AUC of 0.789 to 0.809. The participants’ reported AUC values were higher than those reported in prior single time-point LUNGx challenge [35], and a controlled case-control Lung Rads study [66], while the former study used a single institutional data set with diagnostic scans and later used a subset of NLST scans (high resolution, low dose CTs) for their evaluation.
The challenge results provide a means to contrast different quantitative/AI methods against a common dataset and contrast it to a clinical variable (volume change) performance. The deep neural networks and fusion type approaches used by some participants (T2, T5, T6) showed improvement in sensitivity (TPR) compared to the current clinical metric (volume change) for a lower level of false positives (FPR of 3-5%). At a higher sensitivity (TPR) range of 95 to 97%, six participant methods (T2,T1,T5,T9,T3,T10) show lower FPR compared to volume change predictor. A conventional clinical support system is expected to perform in a variety of patients operating on a fixed (locked) model. The nature of adaptive learning in deep learning approaches is recognized by the Food and Drug Administration (FDA) [67] with no clear directions for clinical use. A survey of recently approved AI devices in medicine does not show extensive prospective trials, nor standardized metrics for detection or diagnosis [68, 69]. We acknowledge the variety of training datasets used by the participants, limited test data set with sequential time scans, where the organizers provided volumetric nodule segmentations carried out by a single institutional radiologist. Nevertheless, the segmentation mask provides a ‘common’ reference boundary for radiomic, size/volume-based approaches. We believe they provide accurate locational information for deep learning methods. The segmentation mask allows teams to compare (or build on) a clinically useful measurement, Suppl. Table T2 and Suppl. Figure F2 shows the performance of volume estimates across teams. Clinically, malignant nodules are often characterized as nodules that grow over multiple scanning visits, while benign nodules are considered to show no growth or a reduction, typically measured by its diameter/size [12]. The ability to use multiple time scans of the patient nodule from two sequential visits, instead of single time observation of the nodule may be responsible for the increased biomarker success in this challenge relative to past efforts.
The past decade has seen enormous growth in the development of quantitative imaging methods for radiological images, which has been fueled by availability of publicly available data (like the NLST, LIDC). In this challenge, the use of external training datasets played a role in training the network-based models. Eight out of the 11 participating teams used external datasets. Four teams used the DSB 2017 dataset, three used the LIDC dataset, three teams used data from the LUNA16 challenge and one team used scans obtained from the NLST study. There have been a number of malignancy prediction models [19, 70, 71] that use clinical characteristics, nodule size and subtle nodule characteristics. Notably, the study by McWilliams [19] reported an impressive AUC for the clinical model with nodule diameter which was in the range of 0.894 to 0.907 for Pan-Canadian Early Detection of Lung Cancer Study (PanCan) and British Columbia Cancer Agency (BCCA) data sets. In our challenge the clinical characteristics for the patients selected in our cohort (broadly age, smoking pack-years) were controlled, reported earlier [72]. Patient level information was not released to the participants as the focus of the challenge was to develop imaging-based biomarkers. A recent study that used over 6,716 LDCT’s from the NLST and a large validation cohort of 1,139 cases reported an AUC of 0.944 [33].
The challenge participants’ predictors are broadly illustrated by a bar plot in Fig. 3. Which broadly shows cases that were most often classified incorrectly by participant algorithms. Nodules, whose malignancy status is seemingly unrelated to changes in nodule volume, as defined by the provided segmentations, are not classified correctly; benign nodules that appear to grow are classified as malignant, while malignant nodules that shrink or stay the same are classified as benign. Sub-solid nodules are also found difficult to correctly classify by all participants. It has been well documented that clinical diagnosis of smaller size or ground glass opacity type nodules are difficult and still remains a challenge [73, 74], and automatic methods seem to offer no clear discrimination advantage on these nodules. It is expected that non-size based texture or deep learning methods would provide additional information to bolster size/volume-based predictors.
Fig. 3.
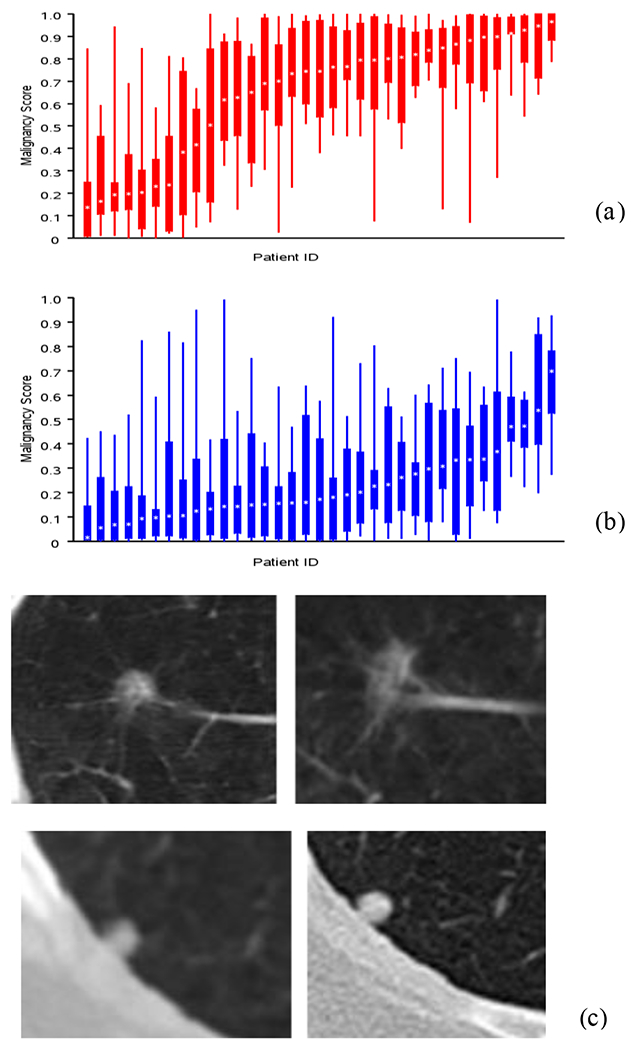
Boxplot show distribution of scores for cases and control (x-axis is the patient index, y-axis is the malignancy score) that helps to identify cases where most participants incorrectly reported the diagnostic status, a) Benign (see higher scores), b) Malignant (see lower scores) diagnosis. Representative patient cases showing 2D CT slice across two time points with most teams reporting incorrect malignancy scores. c) Benign diagnosis (average reported score 0.67) and d) Malignant diagnosis. (average reported score 0.26).
A. Organizing Teams Challenges:
The availability of open source data from the larger lung cancer clinical trials such as the NLST, LIDC and others has been a tremendous resource for the research community [4, 36], but poses a challenge to challenge organizers. In this case, some portions of the NLST dataset in an anonymized form have been used by other challenges and have been open to the public for a certain time period, similar to other challenges in the field, while the NLST data set can be obtained by anyone with a project proposal made through the Cancer Data Access System [64]. In this challenge, the organizers took an extra level of de-identification steps to re-anonymize the data set and removed identifiers that may indicate the specific patient details or origin of the data, especially any NLST reference tags.
Nevertheless, after the challenge’s completion, one of the competition participants reported an overlap between the evaluation dataset in this challenge and the dataset released in the DSB 2017. Four of the five top-performing teams made use of the DSB 2017 dataset, which used single time scans, but it is difficult to discern whether their methods performed better due to approach or due to the supplemental training data sets. Further analysis showed that post-hoc removal of the overlapping cases (20/70 of the cases) in the evaluation dataset would not create significant differences in participant AUCs, would result in similar performance changes for both teams that used the DSB 2017 dataset and teams that did not, and would maintain the high ranking of the top-performing teams. While unlikely to have affected the outcome of the challenge, this event displays the importance of knowing the exact provenance of a chosen dataset when organizing a challenge.
The open use of external datasets themselves can be a difficulty for any challenge organizer. In this challenge, participants could use external datasets and were required to disclose details on the data used. This led to two teams supplementing their algorithm training with private institutional datasets not available to other teams. While most teams performed very similarly in this challenge, the allowance of such datasets could in future challenges provide an advantage to larger institutions or hospitals with sole access to large, private datasets. Most daunting difficulty in organizing challenges in machine learning competitions is the problem of data leakage. There may be a factor independent of any relevant variable that can help competition participants correctly classify datasets. In this challenge, it was noticed by a participant early in the training stage that patients with benign nodules often had multiple nodules, identified using provided segmentation masks, while patients with malignant nodules always (most often) had a single nodule segmented. This discrepancy was an unintentional artifact of the clinical annotation process. Challenge organizers took care to fix these unexpected errors in the training and evaluation datasets and re-released the cohort after randomly re-ordering the patient identifiers with no effect on the results of the competition. Many participants had needed assistance interacting with data provided in the competition, as they were often not familiar with the software packages necessary to manipulate the data in its originally released file format. Others, particularly international participants, had difficulty downloading the large LDCT imaging datasets without connection interruptions. This was remedied by organizers splitting the data into smaller downloadable packets. Future challenge organizers should take care to provide clear tutorials on downloading, converting, and manipulating data provided in their challenges, and provide guidance towards software that correctly processes such data.
B. Strength of Participant Ranking:
We contrasted the team’s predictor by computing True Positive Rate (TPR or sensitivity) and false positive rate (FPR or 1-specificity) for a range of values, by fixing a level for one of the metrics and computing the other across the teams (see Table 3). The first part of the Table 3A, shows participant’s predictors sensitivity or TPR performance for a set level of FPR varied over a range, while Table 3B shows the predictors specificity (1-FPR) for a set level of TPR varied over a range. Based on these values, we re-ranked the team’s performance for a range of sensitivity (or TPR) and 1- specificity (or FPR) (see Suppl. Table ST4).
At 1% False positive rate (1-specificity), we find the mid-ranked team (T6) is placed at the top, yielding the highest TPR (sensitivity of 71.4%) and continues to remain at the top with same TPR, along with two other contenders (T6, T5, T2) for a FPR of 3-5%. Looking at their approaches, these two teams (T6, T2) used volume change along with deep learning models and T5 used radiomics along with deep learning models to predict malignancy, which seems to have aided their performance. On the contrary, at a fixed sensitivity (TPR) of 99%, teams T1 followed by T6 and T10 showed lowest FPR (at 68.6, 77.1, 80% respectively). For a sensitivity (TPR) of 95-97%, the top two team’s (T2, T1) along with T5’s predictors show lowest FPR of 51.4%, 54.3% and 65.7% respectively. But at a sensitivity (TPR) range of 85-90%, top ranked team’s (T1, T2, T3) predictors showed the lowest FPR(1-specificity). At sensitivity (TPR) of 80%, volume change is placed second with an FPR of 20%. It is evident that deep learning models along with volume or handcrafted features seem to help the performances of the predictors (especially T2, T3, T5, T6). Pure deep learning models (as used by T1) are ranked as the top contenders using sensitivity (TPR) as an evaluation criterion but does not hold up its place for lower levels of FPR (1-specificity). These additional analyses reveal improved predictor performance using deep learning methods with conventional volume or radiomics features to achieve lower FPR. Deep learning models seem to be the top contenders for high sensitivity (TPR).
It is apparent that using AUC as a metric to contrast the team’s performances may not have the ability to discern specific details of the predictors. We also compute the partial area under the receiver operating characteristic curve (pAUC) for different intervals of TPR and FPR (see Supplemental Table ST5). In a clinical setting, most often a practitioner finds an acceptable operating cutoff point based on required detection rate (sensitivity/specificity). Additional inferences could be derived to assess participants’ performances and shift in ranking at different levels of cut-points.
C. Need for Better Models:
It is well recognized that advancement in artificial intelligence applications is fast evolving with the prolific use of deep neural architectures in medicine and many other fields. These technologies poses many challenges that include models’ accuracy in prediction tasks, utility compared to current standard of care, reliability, and ethical use [75–78]. In this challenge, over 80% of the teams (9 of 11) proposed deep models to estimate malignancy status. Though the participant model’s performance was not statistically significant compared to the volume estimate. They may have several usages in subtasks such as segmentation and region of interest identification. This may hold promise to improve the current clinical workflow and reduce inter reader human expert (radiologist) variability[79–83].
V. Lessons Learned and limitations
We would like to summarize lessons learned through this community wide challenge and provide recommendations to future events. We acknowledge the need for improved infrastructure for better international data distribution, and software support for users interacting with unfamiliar data formats. The provenance of datasets provided in challenges should be thoroughly investigated, to make sure that they have not been published in a previous venue. Rules regarding external dataset usage should be explicit for competition organizers. Users can be asked to submit source code for their algorithms in addition to their results on the evaluation set, which may lead to better clarity in participant methods. This will encourage third party validation (team that is not related to the developers) and error identification. This will also improve reproducibility and interpretability of the proposed methods. Otherwise, researchers must rely on method descriptions submitted by competition entrants, which may be cursory or lacking significant implementation details. In this challenge it was voluntary for the teams to release their code.
Organizers should limit the number of submissions and timing of the submission during the final testing period. This process will avoid undue guess work and reduce the possibility of overfitting to the evaluation set. In this challenge, submissions were limited to the best three for each team and participant reports were required to identify the submission differences.
Organizers are entrusted to validate participant affiliation and registered duplicate user accounts, both of which can be challenging. Due to the presence of multiple registrations or incomplete affiliation information, validation can become time consuming. Explicit rules regarding the minimum user information required should be determined before challenge operation, to reduce this work load on the organizers.
A. Limitations.
It is recognized that challenge evaluation datasets may not be large enough to show significant differences between challenge participants. Relatively small training datasets also limit the generalizability for methods development. We understand that required time between follow up scans was subjected to clinical decisions and most often did not fully adhere to the prescribed one-year period. We mitigated the training part of the challenge by encouraging the teams to use any available datasets. Nevertheless, the time series curated data cohort with accompanying 3D segmentation mask was the first of its kind that provided a basis for teams to compare their methods. It is acknowledged that nodule size and clinical staging distribution between the train/calibration and test sets were not consistent, which is due to random patient selection based on epidemiological cohort match.
Ideally, challenges should be evaluated on a diverse cohort when possible. A recent article reports several practices to follow and provides comparison of prior competitions [63].
B. Post challenge methods development.
There have been few large population studies published post ISBI 2018 challenge. One notable commercially lead study claims to have trained their deep learning model on 6,716 NLST trial CT scans (single time point) and validated their findings on a set of 1,139 cases, reporting an AUC of 0.944 [33]. The study claims to have reduced false positives by 11% and false negatives by 5% compared to a group of clinical radiologists. Another study that used public data sets (DSB 2017 and LUNA16) to train a deep network (CNN), reports a sensitivity of 84.4% and specificity of 83% in detecting malignant nodules [84]. In a radiomics-based study using curated NLST CT scans of 479 patients and a randomized case-control cohort based approach, the best models averaged an AUC of 0.85[76]. In an interesting use of fusion based methods, a recent study claims to use deep networks with radiomics on baseline CT scans of 498 NLST patients to detect cancer occurrences in following time points (years 1 or 2), and reports an accuracy of 90.29% with an AUC of 0.96 [85]. Other studies used a different set of curated NLST scans from 857 patients [86, 87]. In [87], a 2-D convolutional attention based network that allows for use of pre-trained feature extractors using 1,2 or 3 time points in a Siamese structure achieved AUCs in the range of 0.858-0.882. In a deep learning-based model [88], claims a recall rate of 99.6% using DSB 2017 cohort.
VI. Conclusion
The ISBI 2018 Lung Malignancy Prediction Challenge was the first lung nodule-based malignancy prediction challenge that used a longitudinal dataset. It was a successful, community-wide effort that highlighted challenges in diagnosing malignant lung nodules with sequential LDCT scans. The effort helped to improve a wareness and fostered methods development that attracted over 120 registered participants, with 11 teams that submitted evaluation results and detailed method descriptions. Despite a relatively small evaluation dataset, the challenge offered an avenue to benchmark some of the proposed innovative approaches to analyze LDCT lung screening data. Participating teams proposed a suite of quantitative methods including radiomics, deep learning, and fusions between the two. The participants presented promising approaches to improve malignancy prediction, little over the change in volume method. But none of the methods showed statistical significance compared to the volume change estimate. Most methods still struggled to perform well across a variety of nodule densities.
Supplementary Material
Acknowledgment
We would like to acknowledge the invaluable contribution rendered by the following individuals. Their effort had immensely helped to organize the challenge and contributed to the advancement of imaging methods in the field of quantitative medical imaging. Robert Gillies PhD (Cancer Physiology), Mathew Schabath PhD (Epidemiology), Alberto Garcia BS (Cancer Physiology), Mahmoud Abdalah PhD (IRAT Core), H. Lee. Moffitt Cancer Center (MCC), Tampa, FL. Dmitry Cherezov, University of South Florida (USF). Ying Liu, MD., Qian Li, MD., Tianjin Medical Hospital and Cancer Center, Tianjin, China. Justin Kirby, National Cancer Institute. Program Managers and Staff at Quantitative Imaging Network, NCI and Data Managers at The Cancer Data Access System (CDAS). We are thankful to Carolyn Klinger, Fredrick National Laboratory for Cancer Research, for her valuable comments and proofreading the manuscript. We would like to acknowledge the team members who participated in the challenge and acknowledge their contributions to the individual teams method development. Jae-hun Kim, Ph.D., Department of Radiology, Samsung Medical Center, Sungkyunkwan University School of Medicine, Seoul 06351, Korea. Jung Won Moon, M.D, Human Medical Imaging & Intervention Center, Seoul 06524, Korea. Chin A Yi, M.D, Ph.D., Sungkyunkwan University School of Medicine, Seoul 06524, Korea. Nova F. Smedley, Edgar A. Rios Piedra, Medical Imaging Informatics, University of California, Los Angeles. Adrià Barja Romero, Alberto Montes Gómez, Àlex Martín Alay, Carlos González Rotger, Daniel Rodrigo, Enric Cosp Arqué, Eric Valls, Ferran Vidal-Codina, Ivan Parrot Martinez, Javier Maroto Morales, Jon Liberal Huarte, Josep Marc Mingot Hidalgo, Juan Jose Garau Luis, Manuel Sarmiento Calder, Miguel Cidras Senra, Miguel Pérez Sanchis, Pau Batlle Franch, and Sergio Escosa Rodríguez, Alumni Team from the Universitat Politècnica de Catalunya, Barcelona, Spain. Peter O’Halloran, MD, Department of Radiology, Massachusetts General Hospital.
Footnotes
Supplemental files. Available on the journal’s website.
REFERENCES
- [1].Siegel RL, Miller KD, and Jemal A, “Cancer statistics, 2018,” CA Cancer J Clin, vol. 68, no. 1, pp. 7–30, Jan, 2018. [DOI] [PubMed] [Google Scholar]
- [2].Hirsch FR, Scagliotti GV, Mulshine JL et al. , “Lung cancer: current therapies and new targeted treatments,” Lancet, vol. 389, no. 10066, pp. 299–311, Jan 21, 2017. [DOI] [PubMed] [Google Scholar]
- [3].Govindan R, “Overcoming resistance to targeted therapy for lung cancer,” N Engl J Med, vol. 372, no. 18, pp. 1760–1, Apr 30, 2015. [DOI] [PubMed] [Google Scholar]
- [4].Aberle DR, Berg CD, Black WC et al. , “The National Lung Screening Trial: overview and study design,” Radiology, vol. 258, no. 1, pp. 243–53, Jan, 2011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [5].Walter JE, Heuvelmans MA, de Jong PA et al. , “Occurrence and lung cancer probability of new solid nodules at incidence screening with low-dose CT: analysis of data from the randomised, controlled NELSON trial,” Lancet Oncol, vol. 17, no. 7, pp. 907–916, Jul, 2016. [DOI] [PubMed] [Google Scholar]
- [6].de Koning HJ, van der Aalst CM, de Jong PA et al. , “Reduced Lung-Cancer Mortality with Volume CT Screening in a Randomized Trial,” N Engl J Med, vol. 382, no. 6, pp. 503–513, Feb 6, 2020. [DOI] [PubMed] [Google Scholar]
- [7].Moyer VA, “Screening for lung cancer: U.S. Preventive Services Task Force recommendation statement,” Ann Intern Med, vol. 160, no. 5, pp. 330–8, Mar 4, 2014. [DOI] [PubMed] [Google Scholar]
- [8].Aberle DR, Adams AM, Berg CD et al. , “Reduced lung-cancer mortality with low-dose computed tomographic screening,” N Engl J Med, vol. 365, no. 5, pp. 395–409, Aug 4, 2011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [9].Wood DE, Kazerooni E, Baum SL et al. , “Lung cancer screening, version 1.2015: featured updates to the NCCN guidelines,” J Natl Compr Canc Netw, vol. 13, no. 1, pp. 23–34; quiz 34, Jan, 2015. [DOI] [PubMed] [Google Scholar]
- [10].Mets OM, de Jong PA, Chung K et al. , “Fleischner recommendations for the management of subsolid pulmonary nodules: high awareness but limited conformance - a survey study,” European radiology, vol. 26, no. 11, pp. 3840–3849, 2016. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [11].Eisenhauer EA, Therasse P, Bogaerts J et al. , “New response evaluation criteria in solid tumours: revised RECIST guideline (version 1.1),” Eur J Cancer, vol. 45, no. 2, pp. 228–47, Jan, 2009. [DOI] [PubMed] [Google Scholar]
- [12].Larici AR, Farchione A, Franchi P et al. , “Lung nodules: size still matters,” European Respiratory Review, vol. 26, no. 146, 2017. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [13].Petkovska I, Brown MS, Goldin JG et al. , “The effect of lung volume on nodule size on CT,” Academic radiology, vol. 14, no. 4, pp. 476–485, 2007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [14].Yankelevitz DF, Yip R, Smith JP et al. , “CT Screening for Lung Cancer: Nonsolid Nodules in Baseline and Annual Repeat Rounds,” Radiology, vol. 277, no. 2, pp. 555–64, Nov, 2015. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [15].Henschke CI, Yankelevitz DF, Yip R et al. , “Lung cancers diagnosed at annual CT screening: volume doubling times,” Radiology, vol. 263, no. 2, pp. 578–83, May, 2012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [16].Yankelevitz DF, Reeves AP, Kostis WJ et al. , “Small pulmonary nodules: volumetrically determined growth rates based on CT evaluation,” Radiology, vol. 217, no. 1, pp. 251–6, Oct, 2000. [DOI] [PubMed] [Google Scholar]
- [17].Schultz EM, Sanders GD, Trotter PR et al. , “Validation of two models to estimate the probability of malignancy in patients with solitary pulmonary nodules,” Thorax, vol. 63, no. 4, pp. 335–41, Apr, 2008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [18].Gould MK, Donington J, Lynch WR et al. , “Evaluation of Individuals With Pulmonary Nodules: When Is It Lung Cancer?: Diagnosis and Management of Lung Cancer, 3rd ed: American College of Chest Physicians Evidence-Based Clinical Practice Guidelines,” Chest, vol. 143, no. 5 Suppl, pp. e93S–e120S, May, 2013. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [19].McWilliams A, Tammemagi MC, Mayo JR et al. , “Probability of cancer in pulmonary nodules detected on first screening CT,” N Engl J Med, vol. 369, no. 10, pp. 910–9, Sep 5, 2013. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [20].Kalpathy-Cramer J, Zhao B, Goldgof D et al. , “A Comparison of Lung Nodule Segmentation Algorithms: Methods and Results from a Multi-institutional Study,” Journal of digital imaging, pp. 1–12, 2016. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [21].Zhao YR, van Ooijen PM, Dorrius MD et al. , “Comparison of three software systems for semi-automatic volumetry of pulmonary nodules on baseline and follow-up CT examinations,” Acta Radiol, vol. 55, no. 6, pp. 691–8, Jul, 2014. [DOI] [PubMed] [Google Scholar]
- [22].Liu Y, Balagurunathan Y, Atwater T et al. , “Radiological Image Traits Predictive of Cancer Status in Pulmonary Nodules,” Clin Cancer Res, vol. 23, no. 6, pp. 1442–1449, Mar 15, 2017. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [23].Liu Y, Wang H, Li Q et al. , “Radiologic Features of Small Pulmonary Nodules and Lung Cancer Risk in the National Lung Screening Trial: A Nested Case-Control Study,” Radiology, vol. 286, no. 1, pp. 298–306, Jan, 2018. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [24].Aerts HJ, Velazquez ER, Leijenaar RT et al. , “Decoding tumour phenotype by noninvasive imaging using a quantitative radiomics approach,” Nat Commun, vol. 5, pp. 4006, Jun 3, 2014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [25].Balagurunathan Y, Schabath MB, Wang H et al. , “Quantitative Imaging features Improve Discrimination of Malignancy in Pulmonary nodules,” Sci Rep, vol. 9, no. 1, pp. 8528, Jun 12, 2019. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [26].Alilou M, Beig N, Orooji M et al. , “An integrated segmentation and shape-based classification scheme for distinguishing adenocarcinomas from granulomas on lung CT,” Med Phys, vol. 44, no. 7, pp. 3556–3569, Jul, 2017. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [27].Beig N, Khorrami M, Alilou M et al. , “Perinodular and Intranodular Radiomic Features on Lung CT Images Distinguish Adenocarcinomas from Granulomas,” Radiology, vol. 290, no. 3, pp. 783–792, Mar, 2019. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [28].Gu Q, Feng Z, Liang Q et al. , “Machine learning-based radiomics strategy for prediction of cell proliferation in non-small cell lung cancer,” Eur J Radiol, vol. 118, pp. 32–37, Sep, 2019. [DOI] [PubMed] [Google Scholar]
- [29].Coroller TP, Agrawal V, Huynh E et al. , “Radiomic-Based Pathological Response Prediction from Primary Tumors and Lymph Nodes in NSCLC,” J Thorac Oncol, vol. 12, no. 3, pp. 467–476, Mar, 2017. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [30].Hosny A, Parmar C, Quackenbush J et al. , “Artificial intelligence in radiology,” Nature reviews. Cancer, vol. 18, no. 8, pp. 500–510, 2018. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [31].Paul R, Hawkins SH, Balagurunathan Y et al. , “Deep Feature Transfer Learning in Combination with Traditional Features Predicts Survival Among Patients with Lung Adenocarcinoma,” Tomography (Ann Arbor, Mich.), vol. 2, no. 4, pp. 388–395, 2016. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [32].Hosny A, Parmar C, Coroller TP et al. , “Deep learning for lung cancer prognostication: A retrospective multi-cohort radiomics study,” PLoS medicine, vol. 15, no. 11, pp. e1002711–e1002711, 2018. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [33].Ardila D, Kiraly AP, Bharadwaj S et al. , “End-to-end lung cancer screening with three-dimensional deep learning on low-dose chest computed tomography,” Nat Med, vol. 25, no. 6, pp. 954–961, Jun, 2019. [DOI] [PubMed] [Google Scholar]
- [34].DSB-2017. “Data Science Bowl,” https://www.kaggle.com/c/data-science-bowl-2017/leaderboard.
- [35].Armato SG 3rd, Hadjiiski L, Tourassi GD et al. , “LUNGx Challenge for computerized lung nodule classification: reflections and lessons learned,” J Med Imaging (Bellingham), vol. 2, no. 2, pp. 020103, Apr, 2015. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [36].Armato SG 3rd, McLennan G, Bidaut L et al. , “The Lung Image Database Consortium (LIDC) and Image Database Resource Initiative (IDRI): a completed reference database of lung nodules on CT scans,” Med Phys, vol. 38, no. 2, pp. 915–31, Feb, 2011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [37].van Ginneken B, Armato SG 3rd, de Hoop B et al. , “Comparing and combining algorithms for computer-aided detection of pulmonary nodules in computed tomography scans: The ANODE09 study,” Med Image Anal, vol. 14, no. 6, pp. 707–22, Dec, 2010. [DOI] [PubMed] [Google Scholar]
- [38].Rudyanto RD, Kerkstra S, van Rikxoort EM et al. , “Comparing algorithms for automated vessel segmentation in computed tomography scans of the lung: the VESSEL12 study,” Med Image Anal, vol. 18, no. 7, pp. 1217–32, Oct, 2014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [39].Lo P, van Ginneken B, Reinhardt JM et al. , “Extraction of airways from CT (EXACT’09),” IEEE Trans Med Imaging, vol. 31, no. 11, pp. 2093–107, Nov, 2012. [DOI] [PubMed] [Google Scholar]
- [40].Balagurunathan ABY, Cramer JK, McNitt-Gray M, Hadjiiski L, Zhao B, Zhu J, Yang H, Yip SSF, Aerts HJWL, Napel S, Cherezov D, Cha K, Chan H, Flores C, Garcia A, Gillies R, Goldgof D., “Semi-Automated Pulmonary Nodule Interval Segmentation using the NLST data,” Medical Physics, vol. 45, no. 3, pp. 1093–1107, 2018. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [41].Marcus PM, Gareen IF, Miller AB et al. , “The National Lung Screening Trial’s Endpoint Verification Process: determining the cause of death,” Contemp Clin Trials, vol. 32, no. 6, pp. 834–40, Nov, 2011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [42].Metz CE, “Receiver operating characteristic analysis: a tool for the quantitative evaluation of observer performance and imaging systems,” J Am Coll Radiol, vol. 3, no. 6, pp. 413–22, Jun, 2006. [DOI] [PubMed] [Google Scholar]
- [43].Powers D, “Evaluation: From Precision, Recall and F-Score to ROC, Informedness, Markedness & Correlation,” Journal of Machine Learning Technologies, vol. 2, no. 1, pp. 37–63, 2011. [Google Scholar]
- [44].CodaLab. “Open source framework for running competitions,” 01/2021, 2021; https://github.com/codalab/codalab-competitions/wiki/Project_About_CodaLab.
- [45].Han D, Heuvelmans MA, and Oudkerk M, “Volume versus diameter assessment of small pulmonary nodules in CT lung cancer screening,” Transl Lung Cancer Res, vol. 6, no. 1, pp. 52–61, Feb, 2017. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [46].Liang M, Yip R, Tang W et al. , “Variation in Screening CT-Detected Nodule Volumetry as a Function of Size,” AJR Am J Roentgenol, pp. 1–5, Jun 01, 2017. [DOI] [PubMed] [Google Scholar]
- [47].Kalpathy-Cramer J, Mamomov A, Zhao B et al. , “Radiomics of Lung Nodules: A Multi-Institutional Study of Robustness and Agreement of Quantitative Imaging Features,” Tomography : a journal for imaging research, vol. 2, no. 4, pp. 430–437, 2016. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [48].Balagurunathan Y, Beers A, Kalpathy-Cramer J et al. , “Semi-automated pulmonary nodule interval segmentation using the NLST data,” Med Phys, vol. 45, no. 3, pp. 1093–1107, Mar, 2018. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [49].He K, Zhang X, Ren S et al. “Deep residual learning for image recognition,” 11/2018; arXiv:1512.03385. [Google Scholar]
- [50].Setio AAA, Traverso A, de Bel T et al. , “Validation, comparison, and combination of algorithms for automatic detection of pulmonary nodules in computed tomography images: The LUNA16 challenge,” Med Image Anal, vol. 42, pp. 1–13, Dec, 2017. [DOI] [PubMed] [Google Scholar]
- [51].Borda J. C. d., On elections by ballot., p.^pp. 83–89, 1995. [Google Scholar]
- [52].Simonyan Karen, and Zisserman A, “VERY DEEP CONVOLUTIONAL NETWORKS FOR LARGE-SCALE IMAGE RECOGNITION,” in ICLR, San Diego, 2015. [Google Scholar]
- [53].van Griethuysen JJM, Fedorov A, Parmar C et al. , “Computational Radiomics System to Decode the Radiographic Phenotype,” Cancer Research, vol. 77, no. 21, pp. e104–e107, 2017. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [54].Hsieh C, Chen Y, Beh W et al. , “Feature Selection Framework for XGBoost Based on Electrodermal Activity in Stress Detection.” pp. 330–335. [Google Scholar]
- [55].Castillo LS, Daza LA, Rivera LC et al. , “Brain Tumor Segmentation and Parsing on MRIs Using Multi resolution Neural Networks,” Brainlesion: Glioma, Multiple Sclerosis, Stroke and Traumatic Brain Injuries. pp. 332–343. [Google Scholar]
- [56].Xie Saining, Girshick Ross, Dollár Piotr et al. , “Aggregated residual transformations for deep neural networks,” in Computer Vision and Pattern Recognition (CVPR 2017), 2016. [Google Scholar]
- [57].Huang G, Liu Z., Weinberger KQ, “Densely connected convolutional networks,” in Computer Vision and Pattern Recognition (CVPR 2017), 2016. [Google Scholar]
- [58].Devaraj A, van Ginneken B, Nair A et al. , “Use of Volumetry for Lung Nodule Management: Theory and Practice,” Radiology, vol. 284, no. 3, pp. 630–644, Sep, 2017. [DOI] [PubMed] [Google Scholar]
- [59].DeLong ER, DeLong DM, and Clarke-Pearson DL, “Comparing the areas under two or more correlated receiver operating characteristic curves: a nonparametric approach,” Biometrics, vol. 44, no. 3, pp. 837–45, Sep, 1988. [PubMed] [Google Scholar]
- [60].Strassburger K, and Bretz F, “Compatible simultaneous lower confidence bounds for the Holm procedure and other Bonferroni-based closed tests,” Statistics in medicine, vol. 27, no. 24, pp. 4914–4927, 2008. [DOI] [PubMed] [Google Scholar]
- [61].Armato SG 3rd, Drukker K, Li F et al. , “LUNGx Challenge for computerized lung nodule classification,” J Med Imaging (Bellingham), vol. 3, no. 4, pp. 044506, Oct, 2016. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [62].Laura, and Arnold. https://www.arnoldventures.org/.
- [63].Maier-Hein L, Eisenmann M, Reinke A et al. “Is the winner really the best? A critical analysis of common research practice in biomedical image analysis competitions,” https://arxiv.org/ftp/arxiv/papers/1806/1806.02051.pdf.
- [64].CDAS-NLST. “National Lung Screening Trial,” https://biometry.nci.nih.gov/cdas/studies/nlst/.
- [65].A. C. o. Radiology. “Lung CT Screening Reporting and Data System (Lung-RADS),” http://www.acr.org/Quality-Safety/Resources/LungRADS.
- [66].Li Q, Balagurunathan Y, Liu Y et al. , “Comparison Between Radiological Semantic Features and Lung-RADS in Predicting Malignancy of Screen-Detected Lung Nodules in the National Lung Screening Trial,” Clinical lung cancer, vol. 19, no. 2, pp. 148–156.e3, 2018. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [67].F. d. Guidelines. “Clinical Decision Support Software,” 19 May 2021, 2021; https://www.fda.gov/media/109618/download.
- [68].Benjamens S, Dhunnoo P, and Meskó B, “The state of artificial intelligence-based FDA-approved medical devices and algorithms: an online database,” npj Digital Medicine, vol. 3, no. 1, pp. 118, 2020/09/11, 2020. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [69].Wu E, Wu K, Daneshjou R et al. , “How medical AI devices are evaluated: limitations and recommendations from an analysis of FDA approvals,” Nature Medicine, vol. 27, no. 4, pp. 582–584, 2021/04/01, 2021. [DOI] [PubMed] [Google Scholar]
- [70].Jaffee EM, Dang CV, Agus DB et al. , “Future cancer research priorities in the USA: a Lancet Oncology Commission,” Lancet Oncol, vol. 18, no. 11, pp. e653–e706, Nov, 2017. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [71].Strauss GM, Gleason RE, and Sugarbaker DJ, “Screening for lung cancer re-examined. A reinterpretation of the Mayo Lung Project randomized trial on lung cancer screening,” Chest, vol. 103, no. 4 Suppl, pp. 337s–341s, Apr, 1993. [PubMed] [Google Scholar]
- [72].Schabath MB, Massion PP, Thompson ZJ et al. , “Differences in Patient Outcomes of Prevalence, Interval, and Screen-Detected Lung Cancers in the CT Arm of the National Lung Screening Trial,” PLoS One, vol. 11, no. 8, pp. e0159880, 2016. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [73].Sanchez M, Benegas M, and Vollmer I, “Management of incidental lung nodules <8 mm in diameter,” J Thorac Dis, vol. 10, no. Suppl 22, pp. S2611–s2627, Aug, 2018. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [74].Kim H, Park CM, Koh JM et al. , “Pulmonary subsolid nodules: what radiologists need to know about the imaging features and management strategy,” Diagn Interv Radiol, vol. 20, no. 1, pp. 47–57, Jan-Feb, 2014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [75].Miller DD, and Brown EW, “Artificial Intelligence in Medical Practice: The Question to the Answer?,” Am J Med, vol. 131, no. 2, pp. 129–133, Feb, 2018. [DOI] [PubMed] [Google Scholar]
- [76].Balagurunathan Y, Mitchell R, and El Naqa I, “Requirements and reliability of AI in the medical context,” Phys Med, vol. 83, pp. 72–78, Mar 13, 2021. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [77].Jaremko JL, Azar M, Bromwich R et al. , “Canadian Association of Radiologists White Paper on Ethical and Legal Issues Related to Artificial Intelligence in Radiology,” Can Assoc Radiol J, vol. 70, no. 2, pp. 107–118, May, 2019. [DOI] [PubMed] [Google Scholar]
- [78].COMEST, Preliminary study on the ethics of Artificial Intelligence, UNESCO USA, 2019. [Google Scholar]
- [79].Park A, Chute C, Rajpurkar P et al. , “Deep Learning-Assisted Diagnosis of Cerebral Aneurysms Using the HeadXNet Model,” JAMA Netw Open, vol. 2, no. 6, pp. e195600, Jun 5, 2019. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [80].Sreekumari A, Shanbhag D, Yeo D et al. , “A Deep Learning-Based Approach to Reduce Rescan and Recall Rates in Clinical MRI Examinations,” AJNR Am J Neuroradiol, vol. 40, no. 2, pp. 217–223, Feb, 2019. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [81].Nagpal K, Foote D, Tan F et al. , “Development and Validation of a Deep Learning Algorithm for Gleason Grading of Prostate Cancer From Biopsy Specimens,” JAMA Oncol, vol. 6, no. 9, pp. 1372–1380, Sep 1, 2020. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [82].Hesamian MH, Jia W, He X et al. , “Deep Learning Techniques for Medical Image Segmentation: Achievements and Challenges,” J Digit Imaging, vol. 32, no. 4, pp. 582–596, Aug, 2019. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [83].Bernard O, Lalande A, Zotti C et al. , “Deep Learning Techniques for Automatic MRI Cardiac Multi-Structures Segmentation and Diagnosis: Is the Problem Solved?,” IEEE Transactions on Medical Imaging, vol. 37, no. 11, pp. 2514–2525, 2018. [DOI] [PubMed] [Google Scholar]
- [84].Zhang C, Sun X, Dang K et al. , “Toward an Expert Level of Lung Cancer Detection and Classification Using a Deep Convolutional Neural Network,” Oncologist, vol. 24, no. 9, pp. 1159–1165, Sep, 2019. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [85].Paul R, Schabath M, Gillies R et al. , “Convolutional Neural Network ensembles for accurate lung nodule malignancy prediction 2 years in the future,” Computers in Biology and Medicine, vol. 122, pp. 103882, 2020/07/01/, 2020. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [86].Veasey B, Farhangi MM, Frigui H et al. , “Lung Nodule Malignancy Classification Based ON NLSTx Data.” 2020 IEEE 17th Int. Symp. on Biomedical Img., pp. 1870–1874, 2020. [Google Scholar]
- [87].Veasey BP, Broadhead J, Dahle M et al. , “Lung Nodule Malignancy Prediction From Longitudinal CT Scans With Siamese Convolutional Attention Networks,” IEEE Open Journal of Engineering in Medicine and Biology, vol. 1, pp. 257–264, 2020. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [88].Perez G, and Arbelaez P, “Automated lung cancer diagnosis using three-dimensional convolutional neural networks,” Med Biol Eng Comput, vol. 58, no. 8, pp. 1803–1815, Aug, 2020. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.


