Abstract
Polyhydroxyalkanoates (PHAs) are polyoxoesters that are produced by diverse bacteria and that accumulate as intracellular granules. Phasins are granule-associated proteins that accumulate to high levels in strains that are producing PHAs. The accumulation of phasins has been proposed to be dependent on PHA production, a model which is now rigorously tested for the phasin PhaP of Ralstonia eutropha. R. eutropha phaC PHA synthase and phaP phasin gene replacement strains were constructed. The strains were engineered to express heterologous and/or mutant PHA synthase alleles and a phaP-gfp translational fusion in place of the wild-type alleles of phaC and phaP. The strains were analyzed with respect to production of polyhydroxybutyrate (PHB), accumulation of PhaP, and expression of the phaP-gfp fusion. The results suggest that accumulation of PhaP is strictly dependent on the genetic capacity of strains to produce PHB, that PhaP accumulation is regulated at the level of both PhaP synthesis and PhaP degradation, and that, within mixed populations of cells, PhaP accumulation within cells of a given strain is not influenced by PHB production in cells of other strains. Interestingly, either the synthesis of PHB or the presence of relatively large amounts of PHB in cells (>50% of cell dry weight) is sufficient to enable PhaP synthesis. The results suggest that R. eutropha has evolved a regulatory mechanism that can detect the synthesis and presence of PHB in cells and that PhaP expression can be used as a marker for the production of PHB in individual cells.
Polyhydroxyalkanoates (PHAs) are polyoxoesters that are produced by diverse bacteria as intracellular storage compounds and that can be used to make biodegradable plastics (7, 14, 16, 21, 26). PHA synthases and phasins are proteins that play important roles in PHA production. PHA synthases play the central catalytic role in PHA synthesis and granule formation by catalyzing the polymerization of hydroxyacyl coenzyme A substrates to yield PHAs (9), which in turn associate to form PHA granules (8, 16). Studies on the PHA synthases of Ralstonia eutropha (PhaCRe) and Chromatium vinosum (PhaECCv) have yielded important insights on the mechanism of PHA synthesis, namely, that a cysteine residue conserved among these two proteins (PhaCRe C319 and PhaCCv C149) and among all known PHA synthases is involved in covalent catalysis (9, 20, 36) and that the synthases share structural and functional similarities with lipases (11, 12, 15). Phasins, on the other hand, play a poorly understood role in PHA synthesis and granule formation (32, 37). Phasins are low-molecular-weight proteins, designated PhaP, that share no sequence homology and have been identified from many bacterial strains based on their accumulation to high levels in cells producing PHAs and their association with PHA granules (17, 19, 23, 35). Phasins from several bacterial strains have been shown to increase production of PHAs and to promote the accumulation of PHAs as numerous small granules in cells (17, 35, 37). These two effects are likely related, but the precise role played by phasins remains to be determined. Efforts to understand the regulation and function of phasins are the major focus of the study reported here.
Several lines of evidence suggest that regulation of phasin accumulation is important for phasin function and that accumulation of phasins is tightly coupled to PHA synthesis. Studies with an R. eutropha phaP deletion strain and several strains expressing low levels of PhaP indicate that these strains exhibit a 50% decrease in PHA production relative to the wild-type (wt) strain (37), suggesting that phasin must accumulate to high levels in order to promote PHA synthesis. Studies of phaC::Tn5 and spontaneous PHA-null mutants of R. eutropha suggest that PhaP accumulation requires PHA synthesis (35), and studies of several spontaneous PHA-leaky mutants of Rhodococcus ruber suggest that phasin levels generally match PHA levels (23). These latter two studies, however, leave open the possibility that factors such as the physical absence of PhaCRe from cells, effects on expression of genes downstream of phaC, or defects in cell growth, rather than defects in PHA production, are actually responsible for defects in phasin accumulation. These studies are also ambiguous with regard to whether PhaP accumulation is regulated at the level of PhaP synthesis and/or degradation and whether PhaP accumulation is regulated at the level of individual cells or populations of cells. Recent studies of the PhaF protein of Pseudomonas oleovorans and the PhaR protein of Paracoccus denitrificans provide useful insights into how the expression of proteins involved in PHA synthesis, including phasins, may be negatively regulated in the absence of PHA (17, 24). Specifically, PhaF has been proposed to function as a negative regulator of transcription that can be titrated from DNA by PHA (24), and PhaR may function similarly (17). The generality of PhaF and PhaR-mediated regulation in PHA synthesis remains to be determined.
We are interested in developing a model for regulation of phasin accumulation. Recent advances in understanding of the mechanism of PhaCRe (9), combined with the fact that R. eutropha is readily amenable to genetic manipulation (22, 28, 31) and produces poly-[(R)-3-hydroxybutyrate] (PHB) under many standard cultivation conditions (16, 34), make R. eutropha an excellent organism in which to address this goal. Thus, we have constructed a set of R. eutropha phaC deletion and gene replacement strains and have analyzed these strains with respect to growth, PHB production, and PhaC and PhaP accumulation. An R. eutropha strain carrying a phaP-gfp translational fusion in place of the wt allele of PhaP was also constructed and analyzed with respect to expression of green fluorescent protein (GFP). The results suggest that accumulation of PhaP is strictly dependent on the genetic capacity of strains to produce PHB, that R. eutropha has evolved a regulatory mechanism that can detect the synthesis and presence of PHB in cells, and that PhaP expression can be used as a marker for the production of PHB in individual cells.
MATERIALS AND METHODS
Strains, plasmids, and oligonucleotides.
The strains and plasmids used in this study are listed in Table 1. The oligonucleotides used in this study are listed in Table 2.
TABLE 1.
Strains and plasmids used in this studya
| Strain or plasmid | Descriptionb | Reference or source |
|---|---|---|
| R. eutropha strains | ||
| AeH16 | wt, Gm resistant, also termed DSM 428 and ATCC 17699 | ATCC 17699 |
| Re1000 | phaC3::Tn5, blocked in PHB synthesis due to Tn5 insertion in phaC ORF | 22 |
| Re1001 | phaP-gfp translational-fusion gene replacement strain, derived from Ae H16/pGY15 | This study |
| Re1007 | phaP-gfp translational-fusion gene replacement strain, derived from phaC3::Tn5/pGY19 | This study |
| Re1017 | phaC partial-deletion gene replacement strain, derived from Ae H16/pGY47 | This study |
| Re1022 | phaCRe C319A gene replacement strain, derived from Re1017/pGY31 | This study |
| Re1031 | phaECCv gene replacement strain, derived from Re1034/pGY53 | This study |
| Re1034 | phaC precise-deletion gene replacement strain, derived from Ae H16/pGY46 | This study |
| Re1036 | phaCCv gene replacement strain, derived from Re1034/pGY52 | This study |
| Re1052 | phaP precise-deletion gene replacement strain, derived from Ae H16/pGY63 | 37 |
| Re1058 | phaECCv C149A gene replacement strain, derived from Ae H16/pGY67 | This study |
| E. coli strains | ||
| DH5α | Strain for ligation/cloning experiments | NEBc |
| S17-1 | Strain for conjugative transfer of plasmids into R. eutropha | 30 |
| Plasmids | ||
| pBluescript II KS | High-copy-number plasmid used for cloning, confers Ap resistance | Stratagene |
| pGY1a+ | phaP-gfp translational-fusion cloned into pSW213, useful for expression of GFP in E. coli | This study |
| pGY15 | phaP-gfp translational-fusion gene replacement plasmid, confers Km resistance | This study |
| pGY19 | phaP-gfp translational-fusion gene replacement plasmid, confers Sm/Sp resistance | This study |
| pGY31 | phaCRe C319A gene replacement plasmid, confers Km resistance | This study |
| pGY46 | phaC precise-deletion gene replacement plasmid, confers Km resistance | This study |
| pGY47 | phaC partial-deletion gene replacement plasmid, confers Km resistance | This study |
| pGY52 | phaCCv gene replacement plasmid, confers Km resistance | This study |
| pGY53 | phaECCv gene replacement plasmid, confers Km resistance | This study |
| pGY63 | phaP precise-deletion gene replacement plasmid, confers Km resistance | 37 |
| pGY67 | phaECCv C149A gene replacement plasmid, confers Km resistance | This study |
| pJQ200mp18 | Gene replacement vector; encodes sacB, oriV, oriT, traJ; confers Gm resistance | 25 |
| pJQ200mp18Km | Derivative of pJQ200mp18, Gm resistance gene disrupted, confers Km resistance | 37 |
| pJQ200mp18SmSp | Derivative of pJQ200mp18, Gm resistance gene disrupted, confers Sm/Sp resistance | This study |
| pKAS4-C319A | phaCRe C319A allele cloned into plasmid pKAS4 | 9 |
| pKENgfpmut2 | gfpmut2 allele cloned into multicloning site of pKEN1, confers Ap resistance | 4, 6 |
| pSW213 | Low-copy-number plasmid, confers Tc resistance | 3 |
| pUC19 | High-copy-number plasmid used for cloning, confers Ap resistance | NEB |
| pUM4-C149A | phaECCv C149A allele cloned into plasmid pUM4 | 20 |
| pUT-miniTn5-Km | Source of Km resistance gene for pGY15 | 5 |
| pUT-miniTn5-SmSp | Source of Sm/Sp resistance gene for pJQ200mp18SmSp | 5 |
Plasmids constructed in this study that were used only as intermediates for construction of other plasmids are described only in Materials and Methods.
Abbreviations: Ap, ampicillin; Gm, gentamicin; Km, kanamycin; Sm, streptomycin; Sp, spectinomycin; Tc, tetracycline.
NEB, New England Biolabs.
TABLE 2.
Oligonucleotides used in this study
| Oligonucleotide | Sequencea | Location and orientationb |
|---|---|---|
| gfp1 | AATTCCTCGAGATGAGTAAAGGAGAAGAACTTTTC | 5′ end of gfp ORF (+) |
| gfp2 | AGCTTGGATCCGCATGCCTGCAGGTCTGG | 3′ end of gfp ORF (−) |
| phaC2 | AGCTTGGATCCGATGCGAGCGCTGCATACC | 5′ end of region upstream of phaC ORF (+) |
| phaC3 | P–GATTTGATTGTCTCTCTGCCG | 3′ end of region upstream of phaC ORF (−) |
| phaC4 | P–CGCTTGCATGAGTGCCGGCG | 5′ end of region downstream of phaC ORF (+) |
| phaC5 | AGCTTGGATCCGGCGCTCATGTTTTCCTGG | 3′ end of region downstream of phaC ORF (−) |
| phaC6 | P–ATTGAGCAGGTAGAACGCGG | Internal to phaC ORF (−) |
| phaC7 | P–ATCGCCGGTGTGATCAACCC | Internal to phaC ORF (+) |
| phaCcv1 | P–ATGTTCCCCATCGACATCCG | 5′ end of phaCCv ORF (+) |
| phaCcv2 | P–TTATCGCTCGTTGAGCCACTT | 3′ end of phaCCv ORF (−) |
| phaEcv1 | P–ATGAGCAACACTAATTTCTTCAATG | 5′ end of phaECCv ORF (+) |
| phaP2c | CCGAGGATCCATCGCCGGACAAGGCAGC | Region downstream of phaP ORF (−) |
| phaP4 | CTAGCGAATTCGGATCCGCAATCGCGCATCGTTG | 5′ end of region upstream of phaP ORF (+) |
| phaP5 | CTAGTGCGTCGACCATTGCTGGTCTCCAGTGGTG | 3′ end of region upstream of phaP ORF (−) |
Restriction sites engineered into sequences are indicated in boldface. P– indicates presence of 5′ phosphate group.
Forward (+) or reverse (−) orientation relative to ORF is indicated.
Residue underlined in phaP2 corresponds to G in correct sequence of phaP but was designed as C based on an error in original published sequence. Despite the error, phaP2 is functional in PCR amplification of the phaP gene.
Growth media.
R. eutropha strains were cultivated on one of the following media, depending on the particular application: Luria-Bertani (LB) medium (18), tryptic soy broth dextrose-free (TSB) medium (Becton Dickinson Microbiology Systems, Cockeysville, Md.), PHA(no carbon), PHA(med), PHA(high), or PHB production medium. PHA(no carbon), PHA(med), and PHA(high) are based on a minimal medium (22) supplemented with fructose (0, 0.5, and 1%, respectively) and ammonium chloride (0.5, 0.1, and 0.01%, respectively). PHB production medium is identical to PHA(high) except that it contains 40% less of the following components: fructose, ammonium chloride, and trace salts. Escherichia coli strains were cultivated on LB medium.
Antibiotics.
Antibiotics were added to growth media to the following final concentrations: for R. eutropha, gentamicin (10 μg/ml), kanamycin (270 μg/ml), and spectinomycin (250 μg/ml); for E. coli, ampicillin (100 μg/ml), gentamicin (10 μg/ml), kanamycin (25 μg/ml), spectinomycin (100 μg/ml), and tetracycline (10 μg/ml).
Cultivation conditions.
R. eutropha and E. coli strains were cultivated with aeration at 30 and 37°C, respectively. For the preparation of genomic DNA or selection for resistance to antibiotics, R. eutropha strains were cultivated in liquid TSB medium or solid LB agar (1.2%). For PHB production analyses, R. eutropha strains were cultivated in 4 ml of TSB in test tubes to saturation (24 to 30 h). Aliquots (1 ml) were transferred into 50 ml of TSB in 250-ml baffled flasks and cultivated for 12 h. Aliquots of washed cells were transferred into 200 ml of PHB production medium to yield cultures with an initial optical density at 600 nm (OD600) of 1.0 and were cultivated for 72 h. For PHB utilization analyses, aliquots (50 ml) of washed cells were resuspended in 200 ml of PHA(no carbon) and were cultivated for an additional 72 h. For immunoblot analyses, R. eutropha strains were cultivated in 5 ml of TSB to saturation (approximately 36 h). Aliquots (100 μl) of culture were transferred into 5 ml of TSB and were cultivated to saturation (approximately 24 h). Aliquots of washed cells were transferred into 5 ml of TSB, PHA(med), and/or PHA(high) to yield cultures with an initial OD600 of 1.0 (∼1 × 109 CFU/ml) and were cultivated for 48 h. For cocultivation experiments, R. eutropha strains were cultivated as described for immunoblot analyses, except that final cultures were inoculated with two strains, each added to an initial OD600 of 0.5 (∼5 × 108 CFU of each strain/ml).
DNA preparation and manipulation.
Standard approaches were used for preparation and manipulation of DNA and for the PCR (1). Genomic DNA was prepared from R. eutropha strains by the hexadecyltrimethyl ammonium bromide method (1) with one important modification: DNA was prepared from 250 μl of culture (rather than 1.5 ml of culture) without proportional adjustment of reagents. All constructs containing PCR products were confirmed by sequencing at the Massachusetts Institute of Technology Biopolymer Lab.
Construction of phaC precise-deletion (and phaC partial-deletion) gene replacement plasmid pGY46 (and pGY47).
A 0.41-kb (or 0.78-kb) fragment of R. eutropha DNA, corresponding to the region immediately upstream of the phaCRe open reading frame (ORF) (or this region plus part of the phaCRe ORF), was amplified by PCR with the oligonucleotides phaC2 and phaC3 (or phaC2 and phaC6) such that a BamHI site was introduced at the upstream end of the PCR product. A 0.45-kb (or 0.69-kb) fragment of R. eutropha DNA, corresponding to the region immediately downstream of the phaCRe ORF (or this region plus part of the phaCRe ORF), was amplified by PCR with the oligonucleotides phaC4 and phaC5 (or phaC7 and phaC5) such that a BamHI site was introduced at the downstream end of the PCR product. A three-way ligation was conducted between the two PCR products and the vector pBluescript II KS, each of which had been digested with BamHI and gel purified. The product, designated pGY26 (or pGY27), contains a 0.86-kb (or 1.47-kb) BamHI fragment corresponding to a fusion of the regions upstream and downstream of phaCRe (or these regions plus part of the phaCRe ORF), cloned into the BamHI site of pBluescript II KS. The 0.86-kb (or 1.47-kb) BamHI fragment from pGY26 (or pGY27) was cloned into the BamHI site of pJQ200mp18Km to yield pGY46 (or pGY47). Note that the phaC partial deletion gene replacement plasmid pGY47 and the resulting R. eutropha strain Re1017 were used as intermediates for construction of the phaCRe C319A mutant strain and were not studied further.
Construction of phaCCv (and phaECCv) gene replacement plasmid pGY52 (and pGY53).
A 1.1-kb (or 2.2-kb) fragment of C. vinosum DNA, corresponding to the phaCCv ORF (or phaECCv ORFs), was amplified by PCR with oligonucleotides phaCcv1 and phaCcv2 (or phaECcv1 and phaCcv2). The 1.1-kb phaCCv (or 2.2-kb phaECCv) PCR fragment was digested with AatII to yield a 0.30-kb (or 1.43-kb) N-terminal fragment and a 0.77-kb C-terminal fragment. A 0.41-kb fragment of R. eutropha DNA, corresponding to the region immediately upstream of the phaCRe ORF, was amplified by PCR with oligonucleotides phaC2 and phaC3 such that a BamHI site was introduced at the upstream end of the PCR product. This fragment was digested with BamHI. A 0.45-kb fragment of R. eutropha DNA, corresponding to the region immediately downstream of the phaCRe ORF, was amplified by PCR with oligonucleotides phaC4 and phaC5 such that a BamHI site was introduced at the downstream end of the PCR product. This fragment was digested with BamHI. A three-way ligation was conducted with the 0.41-kb BamHI-blunt upstream fragment, the 0.30-kb (or 1.43-kb) blunt-AatII N-terminal fragment, and the 2.2-kb BamHI-AatII fragment of pUC19, to yield pGY49 (or pGY36), in which the region upstream of the phaCRe ORF is cloned immediately upstream of, and in the same orientation as, the 0.30-kb (or 1.43-kb) N-terminal end of the phaCCv ORF (or phaECCv ORFs). A three-way ligation was conducted with the 0.77-kb AatII-blunt C-terminal fragment, the 0.45-kb blunt-BamHI downstream fragment, and the 2.2-kb BamHI-AatII fragment of pUC19, to yield pGY48, in which the region downstream of the phaCRe ORF is cloned immediately downstream of, and in the same orientation as, the 0.77-kb C-terminal end of the phaCCv ORF. A contiguous fragment of DNA (phaCRe upstream-phaCCv ORF [or phaECCv ORFs]-phaCRe downstream) was constructed by three-way ligation of pBluescript II KS (digested with BamHI), the 0.7-kb (or 1.84-kb) BamHI/AatII fragment of pGY49 (or pGY36), and the 1.2 kb AatII/BamHI fragment of pGY48, to yield pGY50 (or pGY51). The 1.9-kb (or 3.0-kb) BamHI fragment of pGY50 (or pGY51) was cloned into the BamHI site of pJQ200mp18Km to yield pGY52 (or pGY53).
Construction of phaCRe C319A gene replacement plasmid pGY31.
A 1.8-kb EcoRI/BamHI fragment corresponding to the entire phaCRe C319A ORF was isolated from pKAS4-C319A, treated with Klenow to generate blunt ends, and cloned into the SmaI site in the multicloning site of pJQ200mp18Km to yield pGY31.
Construction of phaECCv C149A gene replacement plasmid pGY67.
A 0.72-kb AscI-DraIII fragment of pUM4-C149A, including the phaCCv C149A mutation, was used to replace the corresponding fragment of pGY53 in order to yield pGY67.
Construction of gene replacement vector pJQ200mp18SmSp.
pJQ200mp18SmSp was constructed by cloning the 2-kb BamHI fragment encoding streptomycin/spectinomycin resistance from pUT-miniTn5-SmSp into the BglII site within the gentamicin resistance gene of pJQ200mp18. pJQ200mp18SmSp can be used to select for maintenance of plasmids in R. eutropha strains carrying Tn5 insertions, given that it encodes spectinomycin resistance.
Construction of phaP-gfp translational-fusion gene replacement plasmids pGY15 and pGY19 and GFP expression plasmid pGY1a+.
A 0.77-kb fragment of R. eutropha DNA, corresponding to the region immediately upstream of the phaP ORF, was amplified by PCR (oligonucleotides, phaP4 and phaP5) such that a BamHI site and a SalI site were introduced at the upstream and downstream ends, respectively. The PCR product was cloned into the EcoRV site of pBluescript II KS, yielding pphaP2. A 0.75-kb fragment of pKENgfpmut2, corresponding to the gfp ORF, was amplified by PCR (oligonucleotides, gfp1 and gfp2) such that an XhoI site and a BamHI site were introduced at the upstream and downstream ends, respectively. The PCR product was cloned into the EcoRV site of pBluescript II KS, yielding pgfp2. The 0.77-kb BamHI-SalI fragment of pphaP2 and the 0.75-kb XhoI-SalI fragment of pgfp2 were excised from their respective plasmids, ligated, and treated with BamHI, SalI, and XhoI. The resulting 1.5-kb ligation product was cloned into the BamHI site of pSW213, yielding pGY1a+, and was also cloned into the BamHI site of pBluescript II KS, yielding pKSphaPgfp7. A 1.5-kb fragment of R. eutropha DNA, corresponding to the region upstream of the phaP gene and the entire phaP ORF, was amplified by PCR (oligonucleotides, phaP4 and phaP2) such that BamHI sites were introduced at both ends of the PCR product. The PCR product was digested with BamHI and cloned into the BamHI site of the vector pSW213 to yield pGY4+. A 1.6-kb HindIII-PstI (partial digest) fragment of pKSphaPgfp7 was cloned into pGY4+, which had been digested by HindIII and PstI. The resulting plasmid, pGY11a, contains the phaP promoter-gfp ORF fusion adjacent to a truncated version of the phaP ORF. The 2.1-kb BamHI fragment of pGY11a was cloned into the BamHI site of pJQ200mp18 to yield pGY12. pGY15 was constructed by cloning the 2.0-kb BamHI fragment encoding kanamycin resistance from pUT-miniTn5-Km into the BglII site within the gentamicin resistance gene of pGY12. pGY19 was constructed by cloning the 2.14-kb BamHI fragment of pGY11a into the BamHI site of pJQ200mp18SmSp.
Construction of phaC and phaP gene replacement strains.
Gene replacement was accomplished by adaptation of standard protocols (25, 31). The combinations of starting strains and plasmids used for construction of each gene replacement strain are indicated in Table 1. Each gene replacement construction was designed and carried out such that a successful gene replacement strain could be distinguished from the starting strain based on the size of the phaC or phaP allele in the chromosome, as determined by PCR. Successful gene replacement strains were identified and confirmed based on PCR analyses and Southern blot analyses.
Quantitation of PHB in R. eutropha cells.
PHB was quantitated by the sulfuric acid/high-pressure liquid chromatography method of Karr et al. (13) (column, Aminex HPX 87H [Bio-Rad, Hercules, Calif.]; column temperature, 50°C; gradient, isocratic; mobile phase, 0.014 N sulfuric acid; flow rate, 0.7 ml/min; detection system, UV detector, 210 nm). Samples corresponded to cells from 5 or 10 ml of culture that had been dried and weighed.
Preparation and quantitation of PhaCRe, PhaECCv, and PhaP proteins.
PhaCRe (expressed as an N-terminal histidine tag fusion protein), PhaECCv, and PhaP were purified as previously described (12, 20, 37). Experimentally determined extinction coefficients were used for quantitation of each protein (11, 12, 37).
Polyclonal antibodies against PhaCRe, PhaECCv, PhaP, and GFP.
Rabbit polyclonal antibodies against PhaCRe and PhaP were generated by use of standard protocols (10) at Covance Antisera Services (Denver, Pa.). Preparation of antibodies against PhaECCv (20) has been described previously. Rabbit serum was filtered (0.45-μm-pore-size filter) prior to use. Antibodies against PhaP were further purified by binding and elution from Hi-Trap NHS (N-hydroxysuccinimide)-activated resin (Amersham Pharmacia Biotech, Piscataway, N.J.) to which PhaP had been cross-linked and by binding and elution from Hi-Trap protein G resin (Amersham Pharmacia Biotech), in both cases according to the manufacturer's instructions. Anti-GFP antibodies were obtained from Clontech (Palo Alto, Calif.).
Immunoblot analyses.
Whole bacterial cells or purified protein samples were separated by sodium dodecyl sulfate–10 or 15% polyacrylamide gel electrophoresis (SDS–10 or 15% PAGE) (1). Bacterial cell samples corresponded to 10-μl aliquots of cells resuspended to an OD600 of 1.0. Proteins were transferred to Immobilon P polyvinyl difluoride membrane (Millipore, Bedford, Mass.) by electroblotting at 100 V for 1.5 h at ∼4°C. Protein detection was accomplished by use of the Western-Light chemiluminescent detection system kit (Tropix, Bedford, Mass.) according to the manufacturer's instructions. Antibodies were used at 1/500 to 1/1,500 dilutions. The chemiluminescent signal was captured by exposure of blots to film.
For quantitative immunoblot analyses, 2.5-μl aliquots of cultures and five standards of PhaP (55, 27.5, 13.75, 6.88, and 3.44 ng) were included for each SDS-PAGE gel. A Hewlett-Packard ScanJet 4C desktop scanner and Deskscan 2.0 and Adobe Photoshop 5.0 software were used to convert signal on film to TIFF files. Automatic contrast adjustment was turned off during scanning. Quantitation of signal was performed on a Macintosh computer using the public domain NIH Image 1.60 program (developed at the National Institutes of Health and available on the Internet at http://rsb.info.nih.gov/nih-image/).
RESULTS
Construction of phaC gene replacement strains.
To test whether PhaP accumulation is dependent on PHB production in R. eutropha cells, we constructed a set of five phaC gene replacement plasmids and the corresponding R. eutropha gene replacement strains (Table 1). In the first strain, Re1017, the phaCRe ORF has been precisely deleted. In the second and third strains, Re1031 and Re1036, the phaCRe ORF has been replaced with the phaECCv and phaCCv alleles, both of which encode active PHA synthase (20). In the fourth and fifth strains, Re1022 and Re1058, the phaCRe ORF has been replaced with the phaCRe C319A and phaECCv C149A alleles, both of which encode inactive PHA synthase (9, 20). These strains were generated to determine how specific changes in the PHA synthase genes affect PhaP accumulation. The phaC precise-deletion strain was generated to test whether a nonpolar null mutation of phaC would be sufficient to block PhaP accumulation. The phaECCv and phaCCv strains were generated to test whether expression of heterologous, active PHA synthases is sufficient to promote PhaP accumulation. Finally, the phaCRe C319A and phaECCv C149A strains were generated to establish whether expression of inactive PHA synthases could promote PhaP accumulation.
PHB production requires active PHA synthases.
As the first step toward testing whether PhaP accumulation depends on PHB production in R. eutropha, we determined the amount of PHB produced by each strain. For these analyses, strains were cultivated in PHB production medium and were harvested after 72 h of cultivation, and the PHB present in cells was quantitated by the sulfuric acid/high-pressure liquid chromatography method (13). The wt strain was analyzed in parallel for comparison. Results for culture OD600, cell dry weight (cdw), and PHB quantitation are shown in Table 3. The results indicate that the strains expressing active PHA synthase produce detectable amounts of PHB, whereas those expressing inactive or no PHA synthase produce no detectable PHB. The observation that the phaECCv strain produces high amounts of PHB (91% cdw) like the wt strain (80% cdw) indicates that the heterologous PhaECCv PHA synthase can functionally replace PhaCRe in R. eutropha. The observation that the phaCCv strain produces much less PHB (1.7% cdw) is consistent with the report of Müh et al. (20) that PhaCCv in vitro exhibits 1/150 the activity of PhaECCv. Thus, the PhaECv cosynthase is also required for production of PHB to high levels by the C. vinosum PHA synthase during expression in R. eutropha.
TABLE 3.
Analysis of PHB production in R. eutropha wt and phaC gene replacement strainsa
| Strain | Culture OD600 | cdw (mg/ml of culture) | PHB (mg/ml of culture) | PHB (% cdw) |
|---|---|---|---|---|
| wt | 10.2 ± 0.79 | 1.82 ± 0.15 | 1.45 ± 0.12 | 80 |
| phaC precise deletion | 0.94 ± 0.28 | 0.29 ± 0.01 | <0.0003 | <0.1b |
| phaECCv | 11.0 ± 0.21 | 1.84 ± 0.01 | 1.67 ± 0.02 | 91 |
| phaCCv | 1.36 ± 0.04 | 0.38 ± 0.11 | 0.0065 ± 0.0026 | 1.7 |
| phaCRe C319A | 1.10 ± 0.06 | 0.22 ± 0.06 | <0.0003 | <0.1 |
| phaECCv C149A | 1.17 ± 0.07 | 0.41 ± 0.01 | <0.0003 | <0.1 |
Data correspond to analyses of strains inoculated in PHB production medium at an initial OD600 of 1.0 and cultivated for 72 h. Data for wt represent six independent cultures. Data for all other strains represent two independent cultures. The limit of detection is 0.0003 mg of PHB/ml of culture.
Lower limit of detection for PHB = 0.1% cdw.
Interestingly, the results also indicate that strains that lack highly active PHA synthase exhibit little or no growth during cultivation under conditions that promote the production of PHB to high levels (Table 3; see OD600 and cdw). Measurements of CFU in cultures in these and additional experiments indicate that the phaC deletion, phaCRe C319A, phaECCv C149A, and phaCCv strains typically remain viable over the course of these types of cultivations (data not shown) but exhibit little or no increase in biomass. This observation raises the possibility that defects in PHB production might affect PhaP accumulation indirectly, by negatively affecting cell growth.
Active and inactive PHA synthases accumulate in the R. eutropha strains.
As the second step toward testing whether PhaP accumulation depends on PHB production in R. eutropha cultures, the extent to which PHA synthases accumulate in each of the strains was determined by immunoblotting. We were interested in determining the stability of inactive PHA synthases under different growth conditions. We focused on two growth media, TSB and PHA(high), in which the wt strain accumulates PHB to low and high levels, respectively (37). Cells were harvested after 48 h of cultivation, and the presence of PHA synthase was detected by immunoblot analyses with anti-PhaCRe and anti-PhaECCv antibodies. The wt strain was analyzed in parallel for comparison. The results are shown in Fig. 1.
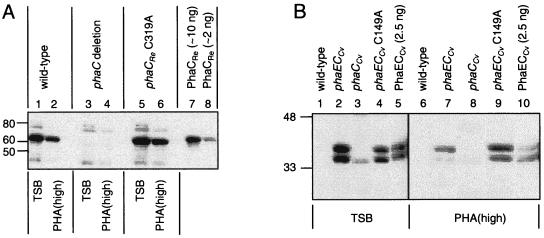
FIG. 1.
(A) Accumulation of PhaCRe in R. eutropha wt, phaC deletion, and phaCRe C319A strains, as detected by anti-PhaCRe antibody. Proteins were separated by SDS–10% PAGE and were subjected to immunoblot analysis. Molecular-mass standards are indicated in kilodaltons. Cells from R. eutropha cultures were harvested after cultivation for 48 h in TSB or PHA(high). Bacterial samples correspond to cells from 10 μl of culture diluted to an OD600 of 1.0. Purified PhaCRe was included as a positive control. (B) Accumulation of PhaECCv in R. eutropha wt, phaECCv, phaCCv, and phaECCv C149A strains, as detected by anti-PhaECCv antibody. Samples were analyzed as described above. Purified PhaECCv was included as a positive control. Note that the data correspond to two blots (first blot, lanes 1 to 5; second blot, lanes 6 to 10).
PHA synthases are detectable in strains cultivated in TSB or PHA(high), regardless of their activity. The anti-PhaCRe antibody detects a 64-kDa protein consistent with PhaCRe in the wt (Fig. 1A, lanes 1 and 2) and phaCRe C319A strains (Fig. 1A, lanes 5 and 6) but not in the phaC deletion strain (Fig. 1A, lanes 3 and 4). The anti-PhaECCv antibody detects a 39-kDa protein, consistent with PhaCCv, that is present in the phaECCv, phaCCv, and phaECCv C149A strains (Fig. 1B, lanes 2 to 4, 7, and 9). The detection of the 39-kDa protein in the phaCCv strain cultivated in PHA(high) requires prolonged exposure of immunoblots to film (data not shown). The anti-PhaECCv antibody also detects a 41-kDa protein, consistent with PhaECv, that is present in the phaECCv and phaECCv C149A strains (Fig. 1B, lanes 2, 4, 7, and 9) but not in the phaCCv strain (Fig. 1B, lanes 3 and 8). The results indicate some variability in amounts of the different synthases in R. eutropha. The basis for this variability has not been determined. Importantly, PhaCRe C319A and PhaECCv C149A can both be expressed detectably in R. eutropha; thus, the corresponding gene replacement strains can be used to test whether the physical presence of PHA synthase might be sufficient to enable PhaP accumulation.
PhaP accumulation is dependent on expression of active PHA synthase.
We proceeded to test whether expression of heterologous and/or inactive synthases is sufficient to promote PhaP accumulation. We also tested whether PhaP accumulation is altered in particular strains due to altered cell growth or an inability to produce PHB. Strains were cultivated in TSB, PHA(med), and PHA(high), and cultures were analyzed after 48 h. In parallel, the wt strain and the phaC3::Tn5 and phaP deletion strains were analyzed as positive and negative controls, respectively. Results for culture OD600 and immunoblot analyses are shown in Fig. 2.
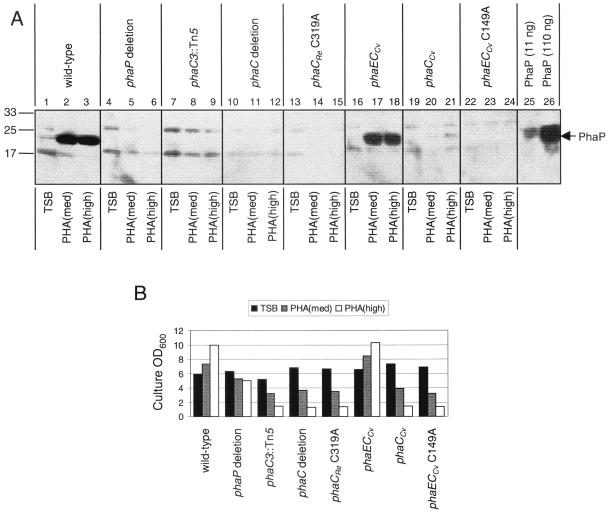
FIG. 2.
(A) Accumulation of PhaP in R. eutropha wt, phaP deletion, phaC3::Tn5, phaC deletion, phaCRe C319A, phaECCv, phaCCv, and phaECCv C149A strains. Proteins were separated by SDS–15% PAGE and were subjected to immunoblot analysis for detection of PhaP. Molecular-mass standards are indicated in kilodaltons. Cells from R. eutropha cultures were harvested after cultivation for 48 h in TSB, PHA(med), and PHA(high). Bacterial samples correspond to cells from 10 μl of culture diluted to an OD600 of 1.0. Data correspond to three blots (first blot, lanes 1 to 3; second blot, lanes 4 to 15; third blot, lanes 16 to 26). Purified PhaP was included as a positive control on each blot. Blots were exposed to film for 5, 10, and 30 min. Data correspond to 10-min exposures for the first and second blots and a 30-min exposure for the third blot. (B) Measurements of OD600 for R. eutropha strains after cultivation for 48 h in TSB, PHA(med), and PHA(high). Data were extrapolated from 10-fold dilutions of cultures.
The results indicate that PhaP accumulation is dependent on expression of active PHA synthase. Anti-PhaP antibody detects a 24-kDa protein, consistent with PhaP, in the wt strain (Fig. 2A, lanes 1 to 3), the phaECCv strain (Fig. 2A, lanes 16 to 18), and, to a lesser extent, in the phaCCv strain (Fig. 2A, lane 21). This 24-kDa protein is not detected in the phaC deletion, phaCRe C319A, and phaECCv C149A strains nor in the phaC3::Tn5 and phaP deletion strains (Fig. 2A, lanes 4 to 15 and 22 to 24). The observation that PhaP accumulates in the phaECCv and the phaCCv strains suggests that expression of any active PHA synthase is sufficient to enable PhaP accumulation. The observation that PhaP accumulates to low levels or fails to accumulate in the phaECCv and the phaCCv strains during cultivation in TSB is consistent with our previous report that PhaP accumulates only transiently in the wt strain in TSB (37). The observation that PhaP is not detectable in strains that are genetically blocked for PHB synthesis, independent of the precise nature of the genetic block, suggests that PhaP accumulation is strictly dependent on the ability of cells to express active PHA synthase. The observation that PhaP fails to accumulate in strains that are genetically blocked in PHB synthesis, even when the strains are cultivated in PHA(med) and thus are exhibiting substantial growth (Fig. 2B), argues against the possibility that phaC mutations block PhaP accumulation indirectly due to effects on growth. Taken together, these observations strongly suggest that PhaP accumulation in a given strain is specifically dependent on PHB production in that strain.
PhaP accumulation is regulated at the level of PhaP synthesis.
Our studies are consistent with the possibilities that PhaP accumulation is regulated at the level of PhaP synthesis, PhaP degradation, or both. To test for regulation of PhaP accumulation at the level of PhaP synthesis, we constructed a phaP-gfp translational fusion and tested the effects of growth conditions and mutations in phaC on expression of this fusion in R. eutropha. We reasoned that a phaP-gfp translational fusion could serve as a useful reporter for PhaP synthesis, based on the assumption that GFP would not be subjected to degradation by any mechanisms which may exist to specifically degrade PhaP. We designed the phaP-gfp fusion such that it includes transcriptional and translational start signals of phaP and thus corresponds to a translational fusion (29) and such that it encodes a variant of GFPmut2 (N-Met-Val-Glu-GFPmut2-C) in place of PhaP. R. eutropha strains in which the phaP gene has been replaced by the phaP-gfp translational fusion were constructed in the wt and phaC3::Tn5 backgrounds to yield a phaP-gfp strain, designated Re1001, and a phaP-gfp phaC3::Tn5 strain, designated Re1007.
To test regulation of expression of the phaP-gfp fusion, the phaP-gfp and phaP-gfp phaC3::Tn5 strains were cultivated in TSB, PHA(med), and PHA(high); cells were harvested after 48 h of cultivation; and the presence of GFP in cells was detected by immunoblot analysis with anti-GFP antibodies. An E. coli strain carrying the phaP-gfp fusion on a plasmid and a strain carrying the corresponding vector without the phaP-gfp fusion were included as positive and negative controls, respectively. The R. eutropha wt and phaC3::Tn5 strains were also analyzed in parallel as negative controls. Results for culture OD600 and immunoblot analyses are shown in Fig. 3. These results parallel those of PhaP immunoblot analyses (Fig. 2). Specifically, the anti-GFP antibody recognizes a 27-kDa protein, consistent with GFP, that accumulates to much higher levels in the phaP-gfp strain (Fig. 3A, lanes 7 to 9) than in the phaP-gfp phaC3::Tn5 strain (Fig. 3A, lanes 10 to 12). Unfortunately, the anti-GFP antibodies cross-react with a 27-kDa R. eutropha protein (Fig. 3A, lanes 1 to 6), obscuring a lower limit of detection for GFP. Nonetheless, the results indicate that PhaP accumulation is regulated at least in part at the level of PhaP synthesis. In addition, the observation that the phaP-gfp phaC3::Tn5 strain exhibits substantial increases in OD600 during cultivation in TSB and PHA(med) (Fig. 3B) suggests that the strain fails to express the phaP-gfp fusion not due to lack of growth but rather due to lack of PHB production.
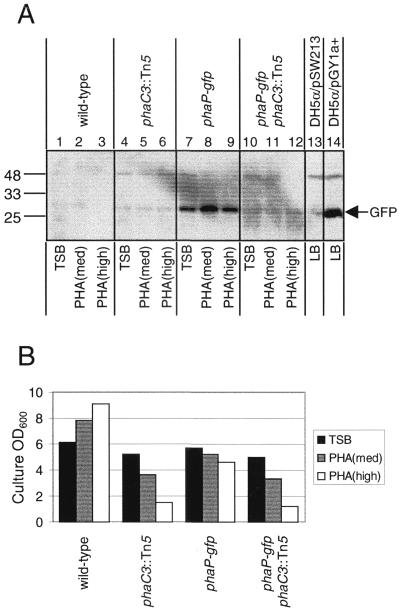
FIG. 3.
(A) Accumulation of GFP in wt, phaC3::Tn5, phaP-gfp, and phaP-gfp phaC3::Tn5 R. eutropha strains. Proteins were separated by SDS–15% PAGE and were subjected to immunoblot analysis for detection of GFP. Molecular-mass standards are indicated in kilodaltons. Cells from R. eutropha cultures were harvested after cultivation for 48 h in TSB, PHA(med), and PHA(high). Bacterial samples correspond to cells from 10 μl of culture diluted to an OD600 of 1.0. The E. coli strains DH5α/pGY1a+, which carries the phaP-gfp fusion on a plasmid, and DH5α/pSW213, which carries the corresponding vector lacking the phaP-gfp fusion, were included as positive and negative controls, respectively. (B) Measurements of OD600 for R. eutropha strains after cultivation for 48 h in TSB, PHA(med), and PHA(high). Data were extrapolated from 10-fold dilutions of cultures.
We recently became aware of a study, reported by Steinbüchel's group in a conference paper (33), which supports regulation at the level of PhaP synthesis. This study indicates that expression of the phaP gene is regulated at the level of transcription in a manner dependent on PHB production in cells, based on S1 nuclease analyses. Our results are consistent with those reported in this study, except in one important respect. The observation of Steinbüchel's group that the wt strain produces little or no phaP transcript during cultivation in rich medium (33), along with their earlier report that the wt strain produces no PHB and no PhaP protein during cultivation in rich medium (35), contradicts recent observations of our group and others that the wt strain produces both PHB (27, 37) and PhaP (37) during cultivation in rich medium.
PhaP stability is dependent on the presence of PHB in cells.
The observation that PhaP accumulation is regulated at the level of PhaP synthesis does not rule out the possibility that PhaP accumulation may also be regulated at the level of PhaP degradation. In fact, we recently reported that PhaP levels rise and fall with PHB levels in the wt strain cultivated in TSB (37), which suggests that PhaP accumulation is also regulated at the level of PhaP degradation. The basis for this degradation is not known. One possibility is that net PHB utilization is sufficient to trigger PhaP degradation. Alternatively, intracellular PHB may need to decrease below some minimal threshold level in order to trigger PhaP degradation.
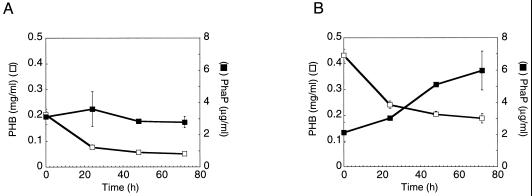
To distinguish between these possibilities, the cells of the wt strain were first cultivated in PHA(med) or PHA(high) for 72 h to allow the accumulation of PHB and PhaP and were then washed, diluted fourfold, and cultivated in PHA(no carbon) for an additional 72 h to trigger partial utilization of intracellular PHB. PHB and PhaP were quantitated over time. The results are shown in Fig. 4. PhaP levels remain constant for cells that were previously cultivated in PHA(med) (Fig. 4A) and, somewhat surprisingly, increase for cells that were previously cultivated in PHA(high) (Fig. 4B). This is the case even though PHB levels decrease during these cultivations (Fig. 4): PHA(med), 51 to 21% cdw; PHA(high), 92 to 48% cdw. Taken together with results from our previous analyses of the wt strain cultivated in TSB (37), these results tend to rule out the possibility that net PHB utilization is sufficient to trigger PhaP degradation. Instead, the results suggest that net PhaP degradation begins only after levels of PHB have decreased below a certain minimal level in cells (PHB level < ∼20% cdw, for example). Furthermore, these results indicate that net PhaP synthesis can occur in cells simultaneously with net PHB utilization. This observation contradicts models whereby PhaP accumulation is strictly dependent on PHB synthesis and suggests rather that the presence of a certain minimal amount of PHB in cells (PHB level > ∼50% cdw, for example) is sufficient to trigger PhaP synthesis.
FIG. 4.
Comparison of levels of PhaP versus PHB for wt strain cultivated under PHB utilization conditions (PHA[no carbon]) for 72 h. Cells were cultivated in PHA(med) (A) or PHA(high) (B) (200 ml) for 72 h, washed, diluted fourfold, and were then cultivated in PHA(no carbon) for 72 h. Time zero corresponds to start of cultivation in PHA(no carbon). All data points represent average value for two cultures (error bars represent standard deviations).
PhaP accumulation is regulated at the level of individual cells.
Our studies thus far do not distinguish between the possibilities that PhaP accumulation is regulated at the level of individual cells or populations of cells. If PhaP accumulation is regulated by a mechanism involving direct detection of PHB, then PhaP accumulation in a given cell will strictly depend on the production of PHB in that cell. In contrast, if PhaP accumulation is regulated by a mechanism involving indirect detection of PHB—for example, based on the presence of particular metabolites or extracellular signals in culture supernatants—then PhaP accumulation in a given cell may depend only on the production of PHB in a subset of cells in the surrounding population. We were particularly interested in distinguishing between these two possibilities, given the suggestion of Campos-García et al. (2) that in Pseudomonas aeruginosa the expression of a ketoacyl reductase implicated in PHA synthesis may be regulated by quorum sensing and given that the potential usefulness of PhaP expression as a marker for PHB production in cells would depend on knowing the basis for regulation of PhaP accumulation.
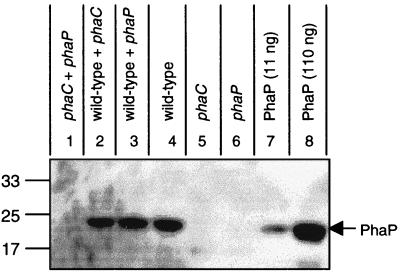
To distinguish between regulation of PhaP accumulation at the level of individual cells versus populations of cells, we tested whether the presence of cells of the phaP deletion strain, which produce PHB (37), could cause the accumulation of PhaP in cells of the phaC deletion strain, which normally do not express PhaP. We reasoned that if PhaP accumulation is regulated at the level of individual cells, then PHB production by the phaP deletion strain would not be sufficient to cause PhaP accumulation in the phaC deletion strain. In contrast, if PhaP accumulation is regulated at the level of populations of cells, then PHB production by the phaP deletion strain would be sufficient to cause PhaP accumulation in the phaC deletion strain. For this experiment, equal amounts of the phaP and phaC deletion strains (based on culture OD600) were combined, cultivated together in PHA(med) for 48 h, and subjected to PhaP immunoblot analysis. PHA(med) was used as the growth medium because it is sufficient to trigger the production of substantial amounts of PHB by the phaP deletion strain (37) and to promote substantial growth of the phaC deletion strain (Fig. 3). Cocultivation of the wt strain with the phaP or phaC deletion strain and cultivation of each strain alone were conducted in parallel for comparison. Results for the immunoblot analyses are shown in Fig. 5.
FIG. 5.
Accumulation of PhaP in cultures of wt, phaC deletion, and phaP deletion strains, cultivated alone or cocultivated in pairs. Proteins were separated by SDS–15% PAGE and were subjected to immunoblot analysis for detection of PhaP. Molecular-mass standards are indicated in kilodaltons. Cells from R. eutropha cultures were harvested after cultivation for 48 h in PHA(med). Bacterial samples correspond to cells from 10 μl of culture diluted to an OD600 of 1.0. Purified PhaP was included as a positive control.
No signal consistent with PhaP is observed for cocultivation of the phaP deletion strain and phaC deletion strains (Fig. 5, lane 1), indicating that the presence of cells that can produce PHB in a culture is not sufficient to trigger PhaP accumulation in other cells in the same culture. The observation that PhaP accumulates to detectable levels in cultures of the wt strain cultivated with either the phaP deletion strain (Fig. 5, lane 3) or the phaC deletion strain (Fig. 5, lane 2) tends to rule out the possibility that the presence of the phaP or phaC deletion strains interferes with PhaP expression or PHB production in other cells in the same cultures. The same results were observed in an independent experiment in which coinoculations of cells of the phaP and phaC deletion strains were conducted in ratios ranging from 1:9 to 9:1 (data not shown). The results suggest that accumulation of PhaP in a given cell depends on the production and/or accumulation of PHB in that cell.
DISCUSSION
Based on our results we propose a model for regulation of PhaP phasin accumulation in R. eutropha. According to our model, net PhaP synthesis is triggered by either of two conditions, net synthesis of PHB or the presence of relatively high amounts of intracellular PHB (>50% cdw). Our results suggest that R. eutropha has evolved a regulatory mechanism that can detect either of these conditions. Net PhaP degradation is triggered by the combination of two conditions, net utilization of PHB and the presence of relatively low amounts of PHB in cells. Our results are consistent with the possibilities that the combination of these conditions triggers expression of a mechanism for degradation of PhaP in cells or that such a mechanism is expressed constitutively but that PhaP is susceptible to degradation only when the combination of conditions applies.
How might PhaP synthesis be regulated? One possibility is that cells express a negative regulator of phasin expression and that this negative regulator is titrated by binding to intracellular PHB. Prior to PHB synthesis, the negative regulator would prevent phasin expression. During net synthesis of PHB, the negative regulator would bind the newly synthesized PHB, resulting in derepression of phasin expression. During net utilization of PHB, the negative regulator would remain bound to the PHB until the PHB had decreased below a certain level (∼50% cdw). At this point the negative regulator would begin to be released and would again repress phasin expression. Such a model could explain how net PHB synthesis or the presence of large amounts of PHB in cells could trigger PhaP accumulation.
This model for negative regulation of phasin accumulation in R. eutropha seems particularly attractive, given that a regulatory system of this type has been proposed previously by Prieto et al. (24) for PhaF-mediated regulation of PHA synthase expression in P. oleovorans and has been anticipated by Maehara et al. (17) for PhaR-mediated regulation of phasin accumulation in P. denitrificans. Genetic evidence suggests that PhaF and PhaR may be transcriptional repressors that are titrated from DNA by intracellular PHA (17, 24). Given the observation of Maehara et al. (17) that homologs of PhaR occur in many PHA-producing strains, including R. eutropha, it seems likely that this type of regulatory mechanism will prove to be a general feature of PHA synthesis.
How might PhaP degradation be regulated? One possibility is that PhaP is protected from proteolytic degradation as long as it is bound to PHB granules and that once PHB decreases below a certain level (∼20% cdw), PhaP protein is released into the cytosol and is degraded. This idea seems particularly attractive, given the report of Wieczorek et al. (35) that PhaP in R. eutropha cells is detected only associated with PHB granules and not in the cytosol.
Regulation of phasin accumulation is important for PHA production. Improved understanding of this regulation may be useful in efforts aimed at precisely manipulating the timing and levels of phasin expression in PHA-producing strains, which in turn should be useful in determining the precise nature of the role of phasins. The observation that phasin accumulation is regulated at the level of individual cells is particularly important because it suggests that a cell expresses phasin not as a general response to growth conditions or even to PHA production within other cells in the same population but strictly as a response to the presence of PHA within the given cell. Thus, expression of PhaP or a surrogate marker such as the phaP-gfp translational fusion could serve as an indicator of production of PHB, not just in cultures but in individual cells in a mixed population. This point could prove useful in efforts to optimize PHA synthases through genetic engineering.
ACKNOWLEDGMENTS
We thank Ute Müh, JoonHo Choi, and Wei Yuan for useful discussions and Jimmy Jia and Jiamin Tian for supplying purified PhaCRe and PhaECCv protein.
G.M.Y. is a DOE-Energy Biosciences Research Fellow of the Life Sciences Research Foundation. This work was supported by NIH Grant GM 49171 to A.J.S. and J.S.
REFERENCES
- 1.Ausubel F M, Brent R, Kingston R E, Moore D D, Seidman J G, Smith J A, Struhl K. Current protocols in molecular biology. New York, N.Y: John Wiley & Sons, Inc.; 1994. [Google Scholar]
- 2.Campos-García J, Caro A D, Nájera R, Miller-Maier R M, Al-Tahhan R A, Soberón-Chávez G. The Pseudomonas aeruginosa rhlG gene encodes an NADPH-dependent β-ketoacyl reductase which is specifically involved in rhamnolipid synthesis. J Bacteriol. 1998;180:4442–4451. doi: 10.1128/jb.180.17.4442-4451.1998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Chen C Y, Winans S C. Controlled expression of the transcriptional activator gene virG in Agrobacterium tumefaciens by using the Escherichia coli lac promoter. J Bacteriol. 1991;173:1139–1144. doi: 10.1128/jb.173.3.1139-1144.1991. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Cormack B P, Valdivia R H, Falkow S. FACS-optimized mutants of the green fluorescent protein (GFP) Gene. 1996;173:33–38. doi: 10.1016/0378-1119(95)00685-0. [DOI] [PubMed] [Google Scholar]
- 5.de Lorenzo V, Herrero M, Jakubzik U, Timmis K N. Mini-Tn5 transposon derivatives for insertion mutagenesis, promoter probing, and chromosomal insertion of cloned DNA in gram-negative eubacteria. J Bacteriol. 1990;172:6568–6572. doi: 10.1128/jb.172.11.6568-6572.1990. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Ezaz-Nikpay K, Uchino K, Lerner R E, Verdine G L. Construction of an overproduction vector containing the novel srp (sterically repressed) promoter. Protein Sci. 1994;3:132–138. doi: 10.1002/pro.5560030117. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.García B, Olivera E R, Miñambres B, Fernández-Valverde M, Cañedo L M, Prieto M A, García J L, Martínez M, Luengo J M. Novel biodegradable aromatic plastics from a bacterial source: genetic and biochemical studies on a route of the phenylacetyl-CoA catabolon. J Biol Chem. 1999;274:29228–29241. doi: 10.1074/jbc.274.41.29228. [DOI] [PubMed] [Google Scholar]
- 8.Gerngross T U, Martin D P. Enzyme-catalyzed synthesis of poly[(R)-(-)-3-hydroxybutyrate]: formation of macroscopic granules in vitro. Proc Natl Acad Sci USA. 1995;92:6279–6283. doi: 10.1073/pnas.92.14.6279. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Gerngross T U, Snell K D, Peoples O P, Sinskey A J, Csuhai E, Masamune S, Stubbe J. Overexpression and purification of the soluble polyhydroxyalkanoate synthase from Alcaligenes eutrophus: evidence for a required posttranslational modification for catalytic activity. Biochemistry. 1994;33:9311–9320. doi: 10.1021/bi00197a035. [DOI] [PubMed] [Google Scholar]
- 10.Harlow E, Lane D. Antibodies: a laboratory manual. Cold Spring Harbor, N.Y: Cold Spring Harbor Laboratory; 1988. [Google Scholar]
- 11.Jia Y, Kappock T J, Frick T, Sinskey A J, Stubbe J. Lipases provide a new mechanistic model for polyhydroxybutyrate (PHB) synthases: characterization of the functional residues in Chromatium vinosum PHB synthase. Biochemistry. 2000;39:3927–3936. doi: 10.1021/bi9928086. [DOI] [PubMed] [Google Scholar]
- 12.Jia Y, Yuan W, Wodzinska J, Park C, Sinskey A J, Stubbe J. Mechanistic studies on class I polyhydroxybutyrate (PHB) synthase from Ralstonia eutropha: class I and class III synthases share a similar catalytic mechanism. Biochemistry. 2001;40:1011–1019. doi: 10.1021/bi002219w. [DOI] [PubMed] [Google Scholar]
- 13.Karr D B, Waters J K, Emerich D W. Analysis of poly-β-hydroxybutyrate in Rhizobium japonicum bacteroids by ion-exclusion high-pressure liquid chromatography and UV detection. Appl Environ Microbiol. 1983;46:1339–1344. doi: 10.1128/aem.46.6.1339-1344.1983. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Kellerhals M B, Kessler B, Witholt B, Tchouboukov A, Brandl H. Renewable long-chain fatty acids for production of biodegradable medium-chain-length polyhydroxyalkanoates (mcl-PHAs) at laboratory and pilot plant scales. Macromolecules. 2000;33:4690–4698. [Google Scholar]
- 15.Liebergesell M, Steinbüchel A. Cloning and nucleotide sequences of genes relevant for biosynthesis of poly(3-hydroxybutyric acid) in Chromatium vinosum strain D. Eur J Biochem. 1992;209:135–150. doi: 10.1111/j.1432-1033.1992.tb17270.x. [DOI] [PubMed] [Google Scholar]
- 16.Madison L L, Huisman G W. Metabolic engineering of poly(3-hydroxyalkanoates): from DNA to plastic. Microbiol Mol Biol Rev. 1999;63:21–53. doi: 10.1128/mmbr.63.1.21-53.1999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Maehara A, Ueda S, Nakano H, Yamane T. Analyses of a polyhydroxyalkanoic acid granule-associated 16-kilodalton protein and its putative regulator in the pha locus of Paracoccus denitrificans. J Bacteriol. 1999;181:2914–2921. doi: 10.1128/jb.181.9.2914-2921.1999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Maniatis T, Fritsch E F, Sambrook J. Molecular cloning: a laboratory manual. Cold Spring Harbor, N.Y: Cold Spring Harbor Laboratory Press; 1982. [Google Scholar]
- 19.McCool G J, Cannon M C. Polyhydroxyalkanoate inclusion body-associated proteins and coding region in Bacillus megaterium. J Bacteriol. 1999;181:585–592. doi: 10.1128/jb.181.2.585-592.1999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Müh U, Sinskey A J, Kirby D P, Lane W S, Stubbe J. PHA synthase from Chromatium vinosum: cysteine 149 is involved in covalent catalysis. Biochemistry. 1999;38:826–837. doi: 10.1021/bi9818319. [DOI] [PubMed] [Google Scholar]
- 21.Olivera E R, Carnicero D, García B, Miñambres B, Moreno M A, Cañedo L, DiRusso C C, Naharro G, Luengo J M. Two different pathways are involved in the β-oxidation of n-alkanoic and n-phenylalkanoic acids in Pseudomonas putida U: genetic studies and biotechnological applications. Mol Microbiol. 2001;39:863–874. doi: 10.1046/j.1365-2958.2001.02296.x. [DOI] [PubMed] [Google Scholar]
- 22.Peoples O P, Sinskey A J. Poly-beta-hydroxybutyrate (PHB) biosynthesis in Alcaligenes eutrophus H16. Identification and characterization of the PHB polymerase gene (phbC) J Biol Chem. 1989;264:15298–15303. [PubMed] [Google Scholar]
- 23.Pieper-Fürst U, Madkour M H, Mayer F, Steinbüchel A. Purification and characterization of a 14-kilodalton protein that is bound to the surface of polyhydroxyalkanoic acid granules in Rhodococcus ruber. J Bacteriol. 1994;176:4328–4337. doi: 10.1128/jb.176.14.4328-4337.1994. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Prieto M A, Bühler B, Jung K, Witholt B, Kessler B. PhaF, a polyhydroxyalkanoate-granule-associated protein of Pseudomonas oleovorans GPo1 involved in the regulatory expression system for pha genes. J Bacteriol. 1999;181:858–868. doi: 10.1128/jb.181.3.858-868.1999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Quandt J, Hynes M F. Versatile suicide vectors which allow direct selection for gene replacement in gram-negative bacteria. Gene. 1993;127:15–21. doi: 10.1016/0378-1119(93)90611-6. [DOI] [PubMed] [Google Scholar]
- 26.Rehm B H, Steinbüchel A. Biochemical and genetic analysis of PHA synthases and other proteins required for PHA synthesis. Int J Biol Macromol. 1999;25:3–19. doi: 10.1016/s0141-8130(99)00010-0. [DOI] [PubMed] [Google Scholar]
- 27.Saegusa H, Shiraki M, Kanai C, Saito T. Cloning of an intracellular poly[d(−)3-hydroxybutyrate] depolymerase gene from Ralstonia eutropha H16 and characterization of the gene product. J Bacteriol. 2001;183:94–100. doi: 10.1128/JB.183.1.94-100.2001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Schubert P, Steinbüchel A, Schlegel H G. Cloning of the Alcaligenes eutrophus genes for synthesis of poly-β-hydroxybutyric acid (PHB) and synthesis of PHB in Escherichia coli. J Bacteriol. 1988;170:5837–5847. doi: 10.1128/jb.170.12.5837-5847.1988. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Silhavy T J. Gene fusions. J Bacteriol. 2000;182:5935–5938. doi: 10.1128/jb.182.21.5935-5938.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Simon S, Priefer T, Pühler A. A broad host range mobilization system for in vivo genetic engineering: transposon mutagensis in Gram negative bacteria. Bio/Technology. 1983;1:784–791. [Google Scholar]
- 31.Slater S, Houmiel K L, Tran M, Mitsky T A, Taylor N B, Padgette S R, Gruys K J. Multiple β-ketothiolases mediate poly(β-hydroxyalkanoate) copolymer synthesis in Ralstonia eutropha. J Bacteriol. 1998;180:1979–1987. doi: 10.1128/jb.180.8.1979-1987.1998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Steinbüchel A, Aerts K, Babel W, Föllner C, Liebergesell M, Madkour M H, Mayer F, Pieper-Fürst U, Pries A, Valentin H E, Wieczorek R. Considerations on the structure and biochemistry of bacterial polyhydroxyalkanoic acid inclusions. Can J Microbiol. 1995;41:94–105. doi: 10.1139/m95-175. [DOI] [PubMed] [Google Scholar]
- 33.Steinbüchel A, Wieczorek R, Krüger N. PHA biosynthesis, its regulation and application of C1-utilizing microorganisms for polyester production. In: Lidstrom M E, Tabita F R, editors. Microbial growth on C1 compounds. Dordrecht, The Netherlands: Kluwer Academic Publishers; 1996. pp. 237–244. [Google Scholar]
- 34.Taidi B, Anderson A J, Dawes E A, Byrom D. Effect of carbon source and concentration on the molecular mass of poly(3-hydroxybutyrate) produced by Methylobacterium extorquens and Alcaligenes eutrophus. Appl Microbiol Biotechnol. 1994;40:786–790. [Google Scholar]
- 35.Wieczorek R, Pries A, Steinbüchel A, Mayer F. Analysis of a 24-kilodalton protein associated with the polyhydroxyalkanoic acid granules in Alcaligenes eutrophus. J Bacteriol. 1995;177:2425–2435. doi: 10.1128/jb.177.9.2425-2435.1995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Wodzinska J, Snell K D, Rhomberg A, Sinskey A J, Biemann K, Stubbe J. Polyhydroxybutyrate synthase: evidence for covalent catalysis. J Am Chem Soc. 1996;118:6319–6320. [Google Scholar]
- 37.York G M, Stubbe J, Sinskey A J. New insight into the role of the PhaP phasin of Ralstonia eutropha in promoting the synthesis of polyhydroxybutyrate. J Bacteriol. 2001;183:2394–2397. doi: 10.1128/JB.183.7.2394-2397.2001. [DOI] [PMC free article] [PubMed] [Google Scholar]