Abstract
We identified an open reading frame, designated phcS, downstream of the transcriptional activator gene (phcR) for the expression of multicomponent phenol hydroxylase (mPH) in Comamonas testosteroni R5. The deduced product of phcS was homologous to AphS of C. testosteroni TA441, which belongs to the GntR family of transcriptional regulators. The transformation of Pseudomonas aeruginosa PAO1c (phenol negative, catechol positive) with pROR502 containing phcR and the mPH genes conferred the ability to grow on phenol, while transformation with pROR504 containing phcS, phcR, and mPH genes did not confer this ability. The disruption of phcS in strain R5 had no effect on its phenol-oxygenating activity in a chemostat culture with phenol. The phenol-oxygenating activity was not expressed in strain R5 grown in a chemostat with acetate. In contrast, the phenol-oxygenating activity in the strain with a knockout phcS gene when grown in a chemostat with acetate as the limiting growth factor was 66% of that obtained in phenol-grown cells of the strain with a knockout in the phcS gene. The disruption of phcS and/or phcR and the complementation in trans of these defects confirm that PhcS is a trans-acting repressor and that the unfavorable expression of mPH in the phcS knockout cells grown on acetate requires PhcR. These results show that the PhcS protein repressed the gratuitous expression of phenol-metabolizing enzymes in the absence of the genuine substrate and that strain R5 acted by an unknown mechanism in which the PhcS-mediated repression was overcome in the presence of the pathway substrate.
The expression of bacterial catabolic pathways for aromatic compounds is often controlled by one or more regulatory proteins, and the effectors of these regulatory proteins are usually either the initial substrates or catabolic intermediates of the pathways (35). The acquisition of an efficient transcriptional regulation system appears to be a major asset for a microorganism which enables it to thrive in an ever-changing environment, as the constitutively expressed pathway enzymes may impose an energy burden.
The expression of multicomponent phenol hydroxylase (mPH) (14, 19, 20, 31, 32, 49, 50) is thought to be controlled by a regulator of the XylR/DmpR subclass within the NtrC-type family of transcriptional regulators, resulting in the expression of phenol-metabolizing enzymes only in the presence of the pathway substrates or structural analogs (2, 20, 24, 30, 31, 36, 38, 42–45, 50). The regulators of this subclass are activated by direct interaction with an effector molecule which is normally the substrate for the catabolic pathway the regulators control (41). The physiological status of regulation is also thought to be overimposed on the system of the XylR/DmpR regulator–ς54-dependent promoter (7, 8, 13, 22, 23, 26, 47, 48). Another type of transcriptional regulator for the expression of mPH, belonging to the GntR family of transcriptional regulators, has been identified in Comamonas testosteroni TA441 (1). This regulator, named AphS, repressed the transcription of mPH genes and caused the inability of the bacterium to grow on phenol (1).
C. testosteroni R5 has been shown to exhibit high activity for both phenol oxygenation (55) and trichloroethylene degradation (18). We have cloned the DNA fragment encoding mPH (PhcKLMNOP) and its cognate transcriptional activator (PhcR) of the XylR/DmpR subclass from strain R5 (50). Our previous work (50) and sodium dodecyl sulfate-polyacrylamide gel electrophoretic analysis (unpublished data) indicated that the high activity of strain R5 was due to the high level of Phc mPH expression, leading us to investigate its transcriptional machinery. In this study, we found one open reading frame (phcS) downstream of phcR. The physiological role of PhcS on the expression of phenol-metabolizing enzymes in strain R5 was studied.
MATERIALS AND METHODS
Bacterial strains, plasmids, and oligonucleotides.
The bacterial strains, plasmids, and oligonucleotides used in this study are listed in Table 1.
TABLE 1.
Bacterial strains, plasmids, and oligonucleotides used in this study
| Strain, plasmid, or oligonucleotide | Relevant characteristic(s)a | Reference or source |
|---|---|---|
| Strains | ||
| C. testosteroni | ||
| R5 | Phl+, wild type, contains phc mPH genes | 55 |
| R5S | Phl+, ΔphcS::Tcr of R5 | This study |
| R5RS | Phl−, Δ(phcR-phcS)::Tcr of R5 | This study |
| TA441 | Phl−, wild type, contains aph mPH genes | 2 |
| P1 | Phl+, aphS mutant of TA441 | 1, 2 |
| P. putida CF600 | Phl+, wild type, contains dmp mPH genes | 40 |
| P. aeruginosa PAO1c | Phl− Cat+ | 21 |
| E. coli | ||
| DH5α | Host strain for DNA manipulation | Toyobo |
| S17-1 | Host strain for plasmid mobilization | 46 |
| Plasmids | ||
| pBluescript II KS(−) | Apr, cloning vector | Toyobo |
| pBSR502 | Apr, 7.8-kb SalI-Sse8387I fragment of pLAFRR501 cloned into SalI-PstI-cleaved pBluescript II KS(−) | 50 |
| pLAFRR501 | Tcr, 22-kb Sau3AI-digested (partially digested) DNA fragment from C. testosteroni R5 cloned into the BamHI site of pLAFR3 | 50 |
| pRO1614 | Apr/Cbr Tcr, cloning vector | 34 |
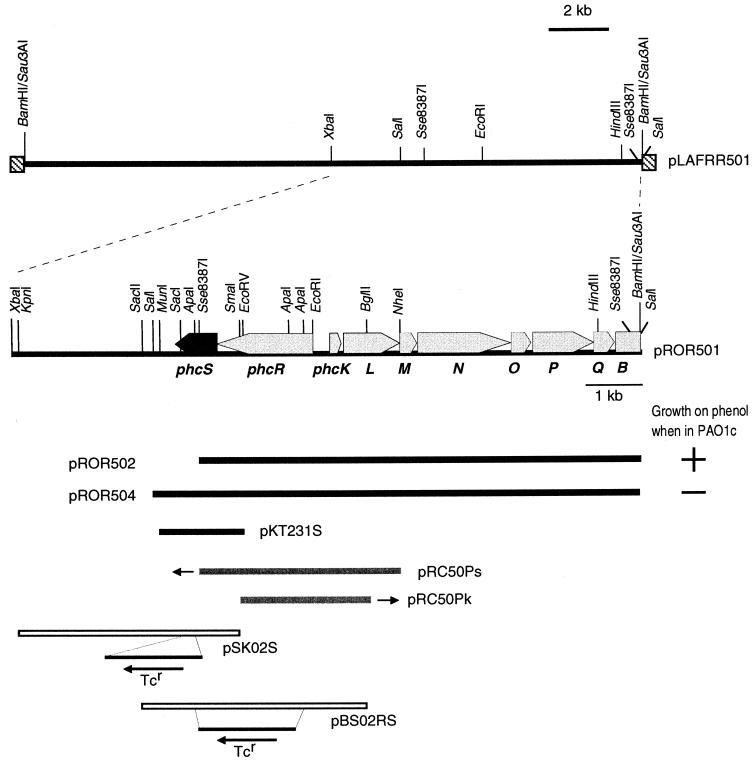
| pROR501 | Apr/Cbr, 11.1-kb XbaI-SalI fragment of pLAFRR501 cloned into NheI-SalI-cleaved pRO1614 | This study |
| pROR502 | Apr/Cbr, 7.8-kb BamHI-SalI fragment of pBSR502 cloned into pRO1614 | 50 |
| pROR504 | Apr/Cbr, 8.6-kb SalI fragment of pROR501 cloned into pRO1614 | This study |
| pMT5059 | Apr, pBR322 derivative carrying multiple cloning sites and an NotI site | 53 |
| pMT5056 | Apr Tcr, pBR322 derivative carrying a Tcr gene cartridge flanked by EcoRV and PvuII sites | 53 |
| pMT5071 | Cmr, plasmid containing an NotI-flanked mobilization cassette (the cassette contains the Cmr gene, sacB, and the Mob region) | 52 |
| pSK1 | Apr, 3.9-kb KpnI-SmaI fragment of pROR501 carrying the phcS gene cloned into pMT5059 | This study |
| pSK01S | Apr Tcr, 1.7-kb EcoRV fragment of pMT5056 carrying the Tcr gene cloned into a blunted ApaI-SacI site of pSK1 | This study |
| pSK02S | Apr/Cbr Tcr Cmr, NotI fragment carrying the mobilization cassette of pMT5071 cloned into pSK01S | This study |
| pBS2 | Apr, 4.0-kb Bgl/II-SacII (blunted) fragment of pROR501 carrying phcS and phcR genes cloned into pMT5059 | This study |
| pBS01RS | Apr Tcr, 1.7-kb EcoRV fragment of pMT5056 carrying the Tcr gene cloned into a blunted ApaI site of pBS2 | This study |
| pBS02RS | Apr/Cbr Tcr Cmr, NotI fragment carrying the mobilization cassette of pMT5071 cloned into pBS01RS | This study |
| pRW50 | Tcr, IncP, lacZ promoter-probe vector | 25 |
| pHSG398 | Cmr, cloning vector | Takara |
| pRC50 | Tcr Cmr, BstEII-cleaved PCR fragment carrying the Cmr gene amplified with Cm-BstEII-Fw and Cm-BstEII-Rv primers from pHSG398 cloned into a BstEII site of pRW50 | This study |
| pRC50Pk | Tcr Cmr, 2.2-kb EcoRV-BglII fragment of pROR501 and EcoRI-NotI-BamHI adapter (Takara) cloned into EcoRI-BamHI-cleaved pRC50; phcKL::lacZ transcriptional fusion | This study |
| pUC18 | Apr, cloning vector | Takara |
| pUC18Ps | Apr, 3.6-kb Sse8387I-NheI fragment of pROR501 cloned into a PstI-XbaI site of pUC18 | This study |
| pRC50Ps | Tcr Cmr, 3.6-kb HindIII-BamHI fragment of pUC18Ps carrying phcR cloned into pRC50, phcS::lacZ transcriptional fusion | This study |
| pBSNot | Apr, EcoRI-NotI-BamHI adapter (Takara) cloned into EcoRV-EcoRI-cleaved pBluescript II KS(−) | This study |
| pBSNotS | Apr, 1.5-kb MunI-EcoRV fragment carrying the phcS gene cloned into EcoRI-SmaI-cleaved pBSNot | This study |
| pKT231 | Kmr Smr, IncQ, broad-host-range cloning vector | 4 |
| pKT231S | Kmr, 1.5-kb NotI fragment of pBSNotS cloned into pKT231 | This study |
| pMMB67HE | Apr, IncQ, tac promoter | 17 |
| pHAM1102 | Apr, aphS in pMMB67HE | 1 |
| pET-29b(+) | Kmr, vector for protein expression | Novagen |
| pMMK67HE | Kmr, PacI-cleaved PCR fragment carrying the Kmr gene amplified with Km-Fw and Km-Rv primers from pET-29b(+) cloned into a PvuI site of pMMB67HE, Apr::Kmr | This study |
| pHAK1102 | Kmr, PacI-cleaved PCR fragment carrying the Kmr gene amplified with Km-Fw and Km-Rv primers from pET-29b(+) cloned into a PvuI site of pHAM1102, Apr::Kmr | This study |
| Oligonucleotides | ||
| Cm-BstEII-Fw | GGGGTTACCCGGAAGATCACTTCGC | |
| Cm-BstEII-Rv | GGGGTAACCGCACCAATAACTGCCT | |
| Km-Fw | CTTTAATTAAGGGATTTTGGTCATGAA | |
| Km-Rv | CATTAATTAATTCTTAGAAAAACTCATCG |
Abbreviations: Phl+, growth on phenol; Phl−, no growth on phenol; Cat+, growth on catechol; Apr, ampicillin resistant; Cbr, carbenicillin resistant; Apr/Cbr, resistant to both ampicillin and carbenicillin; Tcr, TET resistant; Cmr, CHL resistant; Kmr, KAN resistant; Smr, streptomycin resistant.
Media and growth conditions.
The culture media used in this study were Luria-Bertani (LB) medium (37), M9 medium (3), and an inorganic medium called MP (50). The Escherichia coli strains were grown at 37°C, while the C. testosteroni and Pseudomonas strains were grown at 30°C, unless stated otherwise. When required, the media were supplemented with the following antibiotics at the indicated concentrations: tetracycline (TET), 12 μg/ml; ampicillin, 100 μg/ml; carbenicillin, 500 μg/ml; kanamycin (KAN), 30 μg/ml (E. coli) or 400 μg/ml (C. testosteroni); and chloramphenicol (CHL), 20 μg/ml (E. coli) or 80 μg/ml (C. testosteroni).
Genetic techniques.
Plasmid isolation, restriction endonuclease digestion, and transformation of the E. coli strains were conducted by the methods of Sambrook et al. (37). The Pseudomonas aeruginosa and C. testosteroni strains were transformed by the method of Chakrabarty et al. (9).
Nucleotide sequencing and computer analysis.
To determine the nucleotide sequence of the 0.8-kb SalI-Sse8387I fragment downstream of phcR (Fig. 1), subfragments were cloned into the multicloning site of pBluescript II KS(−) (Toyobo). The nucleotide sequences of the subfragments were determined in both orientations by using M13 primers (Takara), a DNA sequencing kit (Dye Terminator Cycle Sequence; Perkin-Elmer), and a model 377 DNA sequencer (Perkin-Elmer) according to the manufacturers' instructions. The templates for the dideoxy chain-termination reaction mixtures were prepared by using Wizard minipreps (Promega). The DNA sequence data were aligned by using version 1.7 of CLUSTAL W (51).
FIG. 1.
Genetic organization of the regulatory and structural genes for mPH in C. testosteroni R5. phcS (in black) was identified in this study. The other genes were identified in our previous study (50). The arrows indicate the direction of transcription. The ability (+) or inability (−) of plasmids pROR502 and pROR504 to allow P. putida PAO1c to grow on phenol as the sole carbon source is indicated. pKT231S is a derivative of pKT231 and carries a MunI-EcoRV fragment which contains the phcS gene, while pRC50Ps is a derivative of pRC50 and carries an Sse8387I-NheI fragment which contains phcR. pRC50Pk is also a derivative of pRC50 and carries an EcoRV-BglII fragment which contains a phcK promoter region. The two small arrows indicate the direction of lacZ on plasmids pRC50Ps and pRC50Pk. pSK02S carries the phcS gene which was disrupted by the insertion of a tetracycline resistance gene (Tcr) cassette, while pBS02RS carries the phcS and phcR genes which were disrupted by the insertion of a Tcr cassette.
Construction of the phcS and phcRS knockouts.
An appropriate 3.9-kb KpnI-SmaI DNA fragment containing phcS on pROR501 (Fig. 1) was first subcloned into the multiple cloning site of pMT5059 (pSK1). The phcS gene was disrupted by inserting a 1.7-kb EcoRV fragment of pMT5056, which carried a Tcr gene, into the blunted ApaI-SacI site of pSK1 (pSK01S). A NotI fragment containing the mobilization cassette of pMT5071 was subsequently inserted into pSK01S. The plasmid thus constructed, pSK02S, was conjugally mobilized (10) from E. coli S17-1 to strain R5, and Tcr selection was done on an M9 agar plate containing 200 mg of phenol per liter, 5% (wt/vol) sucrose, and TET. The transconjugants were chosen for their sensitivity to carbenicillin, and their chromosomal DNAs were analyzed by PCR to confirm that gene replacement had occurred (data not shown).
The 4.0-kb BglII-SacII (blunted) DNA fragment containing phcS and phcR on pROR501 (Fig. 1) was first subcloned into the BglII-NruI (blunted) site of pMT5059 (pBS2). The phcS and phcR genes were disrupted by inserting a 1.7-kb PvuII fragment of pMT5056, which carried a Tcr gene, into the blunted ApaI site of pBS2 (pBS01RS). The NotI fragment of pMT5071 was subsequently inserted into pBS01RS. The plasmid thus constructed, pBS02RS, was conjugally mobilized from E. coli S17-1 to strain R5, and Tcr selection was done on an M9 agar plate containing 600 mg of sodium acetate per liter, 5% (wt/vol) sucrose, and TET. The transconjugants were chosen and analyzed as described above.
Continuous culture conditions.
The culture and sampling conditions were as described by Teramoto et al. (50), unless stated otherwise. The culture (working volume of 1 liter) was started at 25°C by inoculating cells grown in 100 ml of an LB medium into 1 liter of MP medium. The MP medium containing 1,000 mg of phenol per liter or 2,615 mg of sodium acetate per liter as the sole carbon source was then continuously supplied at the rate of 0.35 liter per day. For C. testosteroni P1 cells transformed with a pKT231 derivative, all media were supplemented with KAN.
Assay for phenol-oxygenating activity.
The phenol-oxygenating activity (oxygen uptake rate) was measured at 25°C with a Clark-type oxygen electrode (5/6 Oxygraph; Gilson) as described previously (50). The activity, which was measured in the presence of 10 mM potassium cyanide following the addition of phenol (final concentration of 10 μM), represents the amount of oxygen consumed equally by PH and catechol 2,3-dioxygenase (C23DOase). A previous study had indicated that the phenol-oxygenating activity was double the PH activity, showing that the activity of C23DOase is higher than that of PH (55). The cell weight (dry weight) was determined as described previously (50). Cells from a continuous culture were sampled immediately before the activity was measured.
Assay for C23DOase activity.
Cells (about 50 ml) from a continuous culture were assayed for C23DOase activity. They were collected by centrifugation and washed with 10 ml of ED buffer (10 mM ethylendiamine-H2SO4 [pH 7.5] and 10% isopropyl alcohol) before being stored at −80°C. They were then resuspended in 1 ml of the ED buffer and disrupted by sonication (Sonifier 250; Branson). The crude extract was centrifuged at 14,000 × g for 5 min at 4°C, and the resulting supernatant was used for the enzyme assay. This assay was performed by using a 100 mM potassium phosphate buffer (pH 7.4) at 25°C with 330 μM catechol as a substrate. The amount of the ring cleavage product from catechol (2-hydroxymuconic semialdehyde; the molar absorption coefficient at 375 nm is 33,000) was determined spectrophotometrically (33). The protein concentration was determined by the method of Bradford (5) with a protein assay kit (Bio-Rad), using bovine serum albumin as the standard.
Induction experiment.
The expression of mPH under batch culture conditions was examined next. C. testosteroni strains were grown in LB medium to the stationary phase, harvested, washed with MP medium, resuspended in the original culture volume of MP medium, and exposed to different substrates, phenol or acetate, at a final concentration of 2 mM at 30°C for the indicated periods of time while shaking at 100 rpm. Before the phenol-oxygenating activity was measured (see above), the culture was washed with MP medium and then resuspended in the same medium. C. testosteroni strains transformed with a pKT231 derivative, a pMMK67HE derivative, or a pRC50 derivative were grown in LB medium containing KAN (for the pKT231 and pMMK67HE derivatives) or CHL (for the pRC50 derivative) to the stationary phase and then washed and suspended in MP medium containing phenol or acetate as described above. After each experiment, maintenance of the plasmid was checked by using nonselective and selective plates (supplemented with KAN or CHL).
Other methods.
A growth test on P. aeruginosa PAO1c derivatives containing pROR502 or pROR504 was performed at 25°C with MP medium supplemented with 200 mg of phenol per liter as the sole carbon source, and the optical density at 660 nm of the culture was monitored by an automated recording system (TN2612; Advantec). The concentration of phenol in each culture medium was measured with a kit (color reagent tablets of Phenol-test Wako; Wako Pure Chemical Industries, Osaka, Japan) as described previously (50). The activity of β-galactosidase, the lacZ gene product, was determined by the protocol described by Miller (27).
Nucleotide sequence accession number.
The nucleotide sequence of the 0.8-kb SalI-Sse8387I region was deposited in the DDBJ/EMBL/GenBank database under accession no. AB050891.
RESULTS
Identification of the phcS gene.
We analyzed the DNA region downstream of phcR cloned in pLAFRR501 (50). The sequencing of the 0.8-kb SalI-Sse8387I region identified a 732-bp open reading frame, named phcS, preceded by a putative Shine-Dalgarno sequence (39) (Fig. 1). The phcS gene was immediately downstream of phcR, with only 22 bp between the two genes. The phcS gene encodes a protein of 244 amino acid residues with a predicted molecular mass of 27 kDa. The deduced product has 96% identity and 98% similarity to AphS, a transcriptional repressor for the expression of mPH in C. testosteroni TA441 (1). PhcS also shows 15% identity and 52% similarity to negative transcriptional regulator GntR of Bacillus subtilis (15, 16, 28). A possible helix-turn-helix (HTH) motif that is likely to be involved in DNA binding was thought to be located in the N-terminal region of the sequence (amino acid residues 43 to 64) (12). Members of the GntR family of transcriptional regulators have the putative HTH DNA-binding motif at their N terminus (11), and the residues conserved in the N-terminal domains of GntR-like proteins shown by Mouz et al. (29) were well conserved in the PhcS sequence, suggesting that the phcS gene product belongs to the GntR family of transcriptional regulators. Two AphS-binding sequences reported by Arai et al. (1) were identified in the corresponding regions in and around the phcR-phcK intervening sequence, indicating that PhcS may also bind to these two sites.
Analysis of phcS in PAO1c cells.
C. testosteroni TA441 has been reported to be unable to grow on phenol, as AphS represses transcription of the aph genes. In contrast, C. testosteroni R5 can grow on phenol and shows high phenol-oxygenating activity even in the presence of phcS. To investigate whether phcS encodes such a transcriptional repressor, the region encoding PhcS was deleted from the DNA fragment coding for mPH, and plasmids carrying the deleted and undeleted fragments (pROR502 and pROR504) were introduced into P. aeruginosa PAO1c (phenol-negative, catechol-positive) cells (Fig. 1). The PAO1c derivative with phcS was unable to grow on phenol. These data and the observations with AphS suggest that PhcS caused transcriptional repression of the phc mPH genes in the presence of phenol. Consequently, strain R5 should involve a mechanism to overcome the PhcS-mediated repression, as this strain expressed mPH in the presence of phenol even when a functional phcS gene was present.
Repression by PhcS of the gratuitous expression of phenol-metabolizing enzymes.
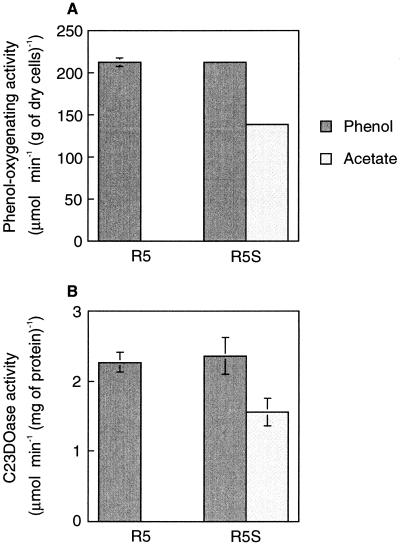
We constructed a phcS knockout of C. testosteroni strain R5 (R5S) to examine the physiological role of phcS. R5S and parental strain R5 were continuously cultured with phenol or acetate as the sole carbon source, and the phenol-oxygenating activity and C23DOase activity of the cultures were measured (Fig. 2). When the R5S strain was grown on phenol, its phenol-oxygenating activity was the same as that of strain R5. However, although acetate-grown R5 cells did not show any detectable phenol-oxygenating activity, acetate-grown R5S cells showed 66% of the activity of the phenol-grown cells.
FIG. 2.
Effect of disrupting phcS on expression of mPH in C. testosteroni strain R5 grown on phenol or acetate in a continuous culture. (A) Phenol-oxygenating activity of strains R5 and R5S. (B) C23DOase activity of strains R5 and R5S. Each value is the mean ± standard error from two or three independent cultures.
A C23DOase gene was found downstream of the phc mPH genes (50) and was thought to be transcribed in the same unit as mPH genes (40). Therefore, we measured C23DOase activity to monitor the transcriptional level of the mPH genes in C. testosteroni R5. The level of C23DOase activity was proportional to that of the phenol-oxygenating activity (Fig. 2). These results show that PhcS repressed the transcription of the phc phenol-metabolizing operon when strain R5 was grown on acetate and that no transcriptional repression occurred when it was grown on phenol.
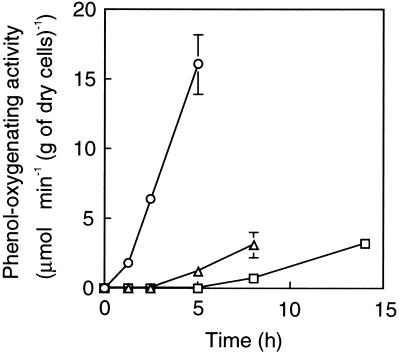
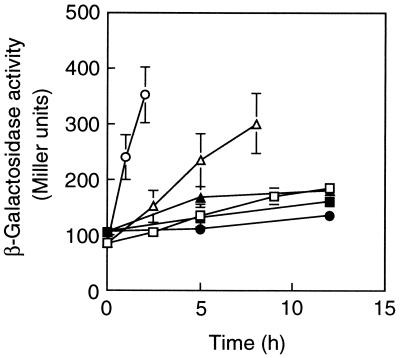
It has been reported that TbuT, which belongs to the DmpR/XylR subclass of transcriptional regulators from Burkholderia pickettii PKO1, activated the transcription of the toluene-3-monooxygenase operon not only in the presence of aromatic effectors but also in the presence of trichloroethylene (6). To test the possibility that acetate activates PhcR in C. testosteroni R5S, the patterns of expression of mPH in response to phenol and acetate in strain R5S were examined in batch cultures (Fig. 3). No phenol-oxygenating activity could be detected after R5S cells had been cultivated in the LB medium and then transferred to MP medium (0 h [Fig. 3]). The addition of phenol to the culture resulted in an immediate increase in the phenol-oxygenating activity. In contrast, no activity was apparent for the first 2.5 h, but activity was detected 5 h after the addition of acetate. The expression response to acetate was later than that to phenol, but the timing of expression was earlier than that in the culture incubated only in MP medium. In cells suspended in MP medium, the gratuitous expression which was observed in the absence of the genuine substrate would have been supported by cellular storage compounds. The transcriptional patterns of mPH genes in response to phenol and acetate in strain R5S were also examined in batch cultures (Fig. 4). The transcriptional response to acetate was slower than that to phenol, but the response was more rapid than that in the culture incubated in MP medium. The patterns of expression of mPH shown in Fig. 3 paralleled the transcriptional patterns of the mPH genes. These results do not indicate whether acetate was an inducer molecule for the transcription of the phc phenol-metabolizing operon. In contrast to strain R5S, strain R5 did not show any phenol-oxygenating activity in the absence of phenol, i.e., in MP medium with or without acetate (data not shown). Also, R5 transformed with pRC50Pk did not show any β-galactosidase activity in the absence of phenol, i.e., in MP medium with or without acetate (data not shown).
FIG. 3.
mPH expression of C. testosteroni strain R5S in response to the presence of phenol or acetate. Stationary-phase cultures grown on LB medium were exposed to MP medium with 2 mM phenol (circles) or acetate (triangles) or without growth substrate (squares). Each value is the mean ± standard error from three independent experiments.
FIG. 4.
Transcriptional activity of the phcK promoter in C. testosteroni strain R5S in response to the presence of phenol or acetate. The strain was transformed with pRC50Pk, which carries a transcriptional fusion of phcKL::lacZ (white symbols) or pRC50 (control vector [black symbols]). Stationary-phase cultures were transferred to MP medium with 2 mM phenol (circles) or acetate (triangles) or without growth substrate (squares). Each value is the mean ± standard error from two or three independent experiments.
To verify that phcS was a trans-acting factor in C. testosteroni R5, R5S was complemented with phcS in trans. The phenol-oxygenating activity of the resulting strain, R5S(pKT231S), was compared in batch cultures with that of R5S(pKT231) containing a control vector (Table 2). The expression after incubation with acetate was abolished in the phcS-complemented strain. It was thus confirmed that phcS was a trans-acting factor and that the PhcS protein repressed the gratuitous transcription of the phc phenol-metabolizing operon in minimal media (Fig. 2, 3, and 4 and Table 2).
TABLE 2.
Effects of PhcS, PhcR, and AphS on expression of mPH in strain R5 in the presence of acetatea
| Strain | Phenol-oxygenating activity (μmol min−1 g [dry wt] of cells−1 |
|---|---|
| R5 | NDb |
| R5S | 3.1 ± 0.9 |
| R5S(pKT231) | 4.1 ± 1.1 |
| R5S(pKT231S) | ND |
| R5RS(pRC50) | ND |
| R5RS(pRC50Ps) | 10.0 ± 0.2 |
| R5S(pMMK67HE) | 0.7 ± 0.2 |
| R5S(pHAK1102) | ND |
Stationary-phase cultures of R5 and its derivatives were assayed for their phenol-oxygenating activity 8 h after being exposed to 2 mM acetate. Each value is the mean ± standard error from two to four independent experiments.
ND, not detected.
Requirement of PhcR for the gratuitous expression.
To investigate the involvement of PhcR in the gratuitous expression of the Phc phenol-metabolizing enzymes in the phcS knockout, a phcRS knockout of C. testosteroni R5 (R5RS) and the knockout complemented by phcR in trans, R5RS(pRC50Ps), were both constructed (Table 2). Cells of R5RS(pRC50) containing a control vector did not express any phenol-oxygenating activity after being incubated with acetate, while cells of R5RS(pRC50Ps) incubated with acetate expressed a higher level of phenol-oxygenating activity than R5S did. This high level of activity is thought to be due to a copy number effect of phcR. These results show that the PhcR protein was essential for the gratuitous expression of the Phc phenol-metabolizing enzymes.
The R5 strain transformed with pRC50Ps also expressed phenol-oxygenating activity at a low level of 10 μmol min−1 g (dry weight) of cells−1 when grown in a chemostat culture on acetate. This result suggests that a high level of PhcR overcame the transcriptional repression caused by PhcS.
Phenol-oxygenating activity in a chemostat culture with acetate and with other phenol-degrading bacteria.
The phenol-oxygenating activity during growth on acetate was also examined with other phenol-degrading bacteria, Pseudomonas putida CF600 and C. testosteroni P1. Strain CF600 is a well-known phenol-degrading bacterium in which no gene homologous to phcS has been found. CF600 and P1 cells were continuously cultured in a chemostat of phenol or acetate as the sole carbon source, and the cultures were assayed. No phenol-oxygenating activity was detected in strain CF600 when it was grown on acetate, and the activity of the phenol-grown CF600 cells was 15 μmol min−1 g (dry weight) of cells−1. On the other hand, the phenol-oxygenating activity of strain P1 grown on acetate, 2 μmol min−1 g (dry weight) of cells−1, was 11% of that of the phenol-grown cells, 18 μmol min−1 g (dry weight) of cells−1.
Complementation of strain R5S with aphS and of strain P1 with phcS.
To investigate the functional equivalence of PhcS and AphS, C. testosteroni strains R5S and P1 were complemented with either aphS or phcS. Strains R5S and P1 were transformed with either a plasmid carrying aphS (pHAK1102) or its control plasmid (pMMK67HE), and the growth rates of P1(pHAK1102) and R5S(pHAK1102) on phenol were significantly slower than those of P1(pMMK67HE) and R5S(pMMK67HE), respectively (data not shown). Strains R5S and P1 were then transformed with either a plasmid carrying phcS (pKT231S) or its control plasmid (pKT231), and the growth rates of R5S(pKT231S) and P1(pKT231S) on phenol were not significantly different from those of R5S(pKT231) and P1(pKT231), respectively (data not shown). These results suggest that AphS may have had a strong repressive effect on the transcription of the phenol-metabolizing operon even in the presence of phenol and that PhcS may have had no apparent effect on this transcription in C. testosteroni strains growing on phenol.
C. testosteroni R5S(pMMK67HE) showed gratuitous expression of mPH with acetate, but R5S(pHAK1102) did not show such gratuitous expression (Table 2). Likewise, the phenol-oxygenating activity of P1(pKT231) continuously grown on acetate was 1.2 μmol min−1 g (dry weight) of cells−1, while no activity was observed in P1(pKT231S) continuously grown on acetate. These results show that PhcS and AphS may have had equal repressive effects on the gratuitous transcription of the phenol-metabolizing operon.
DISCUSSION
In this study we demonstrated that phcS encodes a functional transcriptional repressor of the GntR family for the phc phenol-metabolizing operon. Binding of GntR to the operator region of the gnt operon, which is responsible for gluconate metabolism, resulted in negative regulation, and this binding was specifically inhibited in vitro only by the substrates of the pathway, such as gluconate and glucono-δ-lactone (28). However, the activity of PhcS was not affected by phenol, as PAO1c(pROR504) was unable to grow on phenol. A comparison between the data with P. aeruginosa PAO1c and C. testosteroni R5 indicated that R5 incorporated an unknown mechanism by which the PhcS-mediated repression was overcome in the presence of the genuine substrate for Phc mPH. C. testosteroni TA441 described by Arai et al. (1) seems to have also incorporated such a mechanism, as C. testosteroni P1(pKT231S) was able to grow on phenol at a rate similar to the growth rate of P1(pKT231). However, this mechanism in TA441 may have been blocked by the presence of amino acid residues in AphS which are different from those present in PhcS, which may have resulted in the inability of TA441 to grow on phenol.
Our results show that the PhcS protein was important in repressing the gratuitous expression of the Phc phenol-metabolizing enzymes which occurs in minimal media. This type of expression did not occur in LB (rich) medium (at 0 h [Fig. 3]), showing that the expression was subject to regulation that was determined by the physiological status. It is possible that some amino acids contained in the LB medium may have triggered the repression, as has been seen in the inducer-activated XylR–ς54-dependent Pu and Ps promoter systems (26).
Regulators of the XylR/DmpR subclass have been thought to be activated by direct interaction with a substrate for the catabolic pathway they control, resulting in the expression of the pathway enzymes only in the presence of the substrate or a structural analog (36, 41–45). However, our data indicate that PhcR of the XylR/DmpR subclass caused the expression of the phenol-metabolizing operon in both the presence and absence of the genuine substrate and that the gratuitous expression which occurred in the absence of the genuine substrate was repressed by PhcS. C. testosteroni strain P1 which did not possess the PhcS-like protein (AphS) also demonstrated gratuitous expression. The regulation mechanisms for the expression of mPH in C. testosteroni R5 and P1 are therefore similar to each other but different from that in P. putida CF600 in terms of the gratuitous expression. The details of the mechanism for gratuitous expression and the reason why strain CF600 did not show such gratuitous expression are not known.
Acquiring phcS must have been crucial for C. testosteroni R5 to repress the highly unfavorable expression of phenol-metabolizing enzymes for survival in the natural environment. Strain R5 has in fact been found to be a major population in an oil refinery activated-sludge microbial community, its original habitat, where phenolic compounds were loaded intermittently as carbon and energy sources (54). Further investigation of the molecular mechanisms which regulate PhcS-mediated repression in strain R5 and control its high phenol-oxygenating activity will provide substantial information regarding bacterial survival strategies in the natural environment.
ACKNOWLEDGMENTS
We are grateful to Kouhei Ohnishi for helpful comments and technical advice. We thank Hiromi Awabuchi and Fusako Numazaki for technical assistance. We also thank Stephen Busby for providing pRW50, Hiroyuki Arai for providing strains P1 and TA441 and plasmid pHAM1102, Erich Lanka for providing pMMB67HE, Victoria Shingler for providing strain CF600, and Ronald H. Olsen for providing strain PAO1c.
This work was performed as a part of an industrial science and technology project, Technological Development of Biological Resources in Bioconsortia, supported by a grant from the New Energy and Industrial Technology Development Organization (NEDO).
REFERENCES
- 1.Arai H, Akahira S, Ohishi T, Kudo T. Adaptation of Comamonas testosteroni TA441 to utilization of phenol by spontaneous mutation of the gene for a trans-acting factor. Mol Microbiol. 1999;33:1132–1140. doi: 10.1046/j.1365-2958.1999.01554.x. [DOI] [PubMed] [Google Scholar]
- 2.Arai H, Akahira S, Ohishi T, Maeda M, Kudo T. Adaptation of Comamonas testosteroni TA441 to utilize phenol: organization and regulation of the genes involved in phenol degradation. Microbiology. 1998;144:2895–2903. doi: 10.1099/00221287-144-10-2895. [DOI] [PubMed] [Google Scholar]
- 3.Ausubel F M, Brent R, Kingston R E, Moore D D, Seidman J G, Smith J A, Struhl K, editors. Current protocols in molecular biology. New York, N.Y: John Wiley and Sons; 1994. [Google Scholar]
- 4.Bagdasarian M, Lurz R, Ruckert B, Franklin F C H, Bagdasarian M M, Frey J, Timmis K N. Specific-purpose plasmid cloning vectors. II. Broad host range, high copy number, RSF1010-derived vectors, and a host-vector system for gene cloning in Pseudomonas. Gene. 1981;16:237–247. doi: 10.1016/0378-1119(81)90080-9. [DOI] [PubMed] [Google Scholar]
- 5.Bradford M M. A rapid and sensitive method for the quantitation of microgram quantities of protein utilizing the principle of protein-dye binding. Anal Biochem. 1976;72:248–254. doi: 10.1006/abio.1976.9999. [DOI] [PubMed] [Google Scholar]
- 6.Byrne A M, Olsen R H. Cascade regulation of the toluene-3-monooxygenase operon (tbuA1UBVA2C) of Burkholderia pickettii PKO1: role of the tbuA1 promoter (PtbuA1) in the expression of its cognate activator, TbuT. J Bacteriol. 1996;178:6327–6337. doi: 10.1128/jb.178.21.6327-6337.1996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Carmona M, de Lorenzo V. Involvement of the FtsH (HflB) protease in the activity of ς54 promoters. Mol Microbiol. 1999;31:261–270. doi: 10.1046/j.1365-2958.1999.01169.x. [DOI] [PubMed] [Google Scholar]
- 8.Cases I, Perez-Martin J, de Lorenzo V. The IIANtr (PtsN) protein of Pseudomonas putida mediates the C source inhibition of the ς54-dependent Pu promoter of the TOL plasmid. J Biol Chem. 1999;274:15562–15568. doi: 10.1074/jbc.274.22.15562. [DOI] [PubMed] [Google Scholar]
- 9.Chakrabarty A M, Mylroie J R, Friello D A, Vacca J G. Transformation of Pseudomonas putida and Escherichia coli with plasmid-linked drug-resistance factor DNA. Proc Natl Acad Sci USA. 1975;72:3647–3651. doi: 10.1073/pnas.72.9.3647. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.de Lorenzo V, Timmis K N. Analysis and construction of stable phenotypes in gram-negative bacteria with Tn5- and Tn10-derived minitransposons. Methods Enzymol. 1994;235:386–405. doi: 10.1016/0076-6879(94)35157-0. [DOI] [PubMed] [Google Scholar]
- 11.Diaz E, Prieto M A. Bacterial promoters triggering biodegradation of aromatic pollutants. Curr Opin Biotechnol. 2000;11:467–475. doi: 10.1016/s0958-1669(00)00126-9. [DOI] [PubMed] [Google Scholar]
- 12.Dodd I B, Egan J B. Improved detection of helix-turn-helix DNA-binding motifs in protein sequences. Nucleic Acids Res. 1990;18:5019–5026. doi: 10.1093/nar/18.17.5019. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Duetz W A, Marqués S, de Jong C, Ramos J L, van Andel J G. Inducibility of the TOL catabolic pathway in Pseudomonas putida(pWW0) growing on succinate in continuous culture: evidence of carbon source repression control. J Bacteriol. 1994;176:2354–2361. doi: 10.1128/jb.176.8.2354-2361.1994. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Ehrt S, Schirmer F, Hillen W. Genetic organization, nucleotide sequence and regulation of expression of genes encoding phenol hydroxylase and catechol 1,2-dioxygenase in Acinetobacter calcoaceticus NCIB8250. Mol Microbiol. 1995;18:13–20. doi: 10.1111/j.1365-2958.1995.mmi_18010013.x. [DOI] [PubMed] [Google Scholar]
- 15.Fujita Y, Fujita T. The gluconate operon gnt of Bacillus subtilis encodes its own transcriptional negative regulator. Proc Natl Acad Sci USA. 1987;84:4524–4528. doi: 10.1073/pnas.84.13.4524. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Fujita Y, Fujita T, Miwa Y, Nihashi J, Aratani Y. Organization and transcription of the gluconate operon, gnt, of Bacillus subtilis. J Biol Chem. 1986;261:13744–13753. [PubMed] [Google Scholar]
- 17.Furste J, Pansegrau W, Frank R, Blocker H, Scholz P, Bagdasarian M, Lanka E. Molecular cloning of the plasmid RP4 primase region in a multi-host-range tacP expression vector. Gene. 1986;48:119–131. doi: 10.1016/0378-1119(86)90358-6. [DOI] [PubMed] [Google Scholar]
- 18.Futamata H, Watanabe K, Harayama S. Relationships between the trichloroethylene-degrading activities and the amino acid sequences of phenol hydroxylases in phenol-degrading bacteria. In: Wickramanayake G B, Hinchee R E, editors. Natural attenuation: chlorinated and recalcitrant compounds. Columbus, Ohio: Battelle Press; 1998. pp. 99–104. [Google Scholar]
- 19.Herrmann H, Muller C, Schmidt I, Mahnke J, Petruschka L, Hahnke K. Localization and organization of phenol degradation genes of Pseudomonas putida strain H. Mol Gen Genet. 1995;247:240–246. doi: 10.1007/BF00705655. [DOI] [PubMed] [Google Scholar]
- 20.Hino S, Watanabe K, Takahashi N. Phenol hydroxylase cloned from Ralstonia eutropha strain E2 exhibits novel kinetic properties. Microbiology. 1998;144:1765–1772. doi: 10.1099/00221287-144-7-1765. [DOI] [PubMed] [Google Scholar]
- 21.Holloway B W, Krishnapillai V, Morgan A F. Chromosomal genetics of Pseudomonas. Microbiol Rev. 1979;43:73–102. doi: 10.1128/mr.43.1.73-102.1979. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Holtel A, Marqués S, Möhler I, Jakubzik U, Timmis K N. Carbon source-dependent inhibition of xyl operon expression of the Pseudomonas putida TOL plasmid. J Bacteriol. 1994;176:1773–1776. doi: 10.1128/jb.176.6.1773-1776.1994. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Hugouvieux-Cotte-Pattat N, Köhler T, Rekik M, Harayama S. Growth-phase-dependent expression of the Pseudomonas putida TOL plasmid pWW0 catabolic genes. J Bacteriol. 1990;172:6651–6660. doi: 10.1128/jb.172.12.6651-6660.1990. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Jaspers M C M, Suske W A, Schmid A, Goslings D A M, Kohler H-P E, van de Meer J R. HbpR, a new member of the XylR/DmpR subclass within the NtrC family of bacterial transcriptional activators, regulates expression of 2-hydroxybiphenyl metabolism in Pseudomonas azelaica HBP1. J Bacteriol. 2000;182:405–417. doi: 10.1128/jb.182.2.405-417.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Lodge J, Fear J, Busby S, Gunasekaran P, Kamini N R. Broad host range plasmids carrying the Escherichia coli lactose and galactose operons. FEMS Microbiol Lett. 1992;95:271–276. doi: 10.1016/0378-1097(92)90441-p. [DOI] [PubMed] [Google Scholar]
- 26.Marqués S, Holtel A, Timmis K N, Ramos J L. Transcriptional induction kinetics from the promoters of the catabolic pathways of TOL plasmid pWW0 of Pseudomonas putida for metabolism of aromatics. J Bacteriol. 1994;176:2517–2524. doi: 10.1128/jb.176.9.2517-2524.1994. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Miller J H. A short course in bacterial genetics. A laboratory manual and handbook for Escherichia coli and related bacteria. Cold Spring Harbor, N.Y: Cold Spring Harbor Laboratory Press; 1992. [Google Scholar]
- 28.Miwa Y, Fujita Y. Purification and characterization of a repressor for the Bacillus subtilis gnt operon. J Biol Chem. 1988;263:13252–13257. [PubMed] [Google Scholar]
- 29.Mouz S, Merlin C, Springael D, Toussaint A. A GntR-like negative regulator of the biphenyl degradation genes of the transposon Tn4371. Mol Gen Genet. 1999;262:790–799. doi: 10.1007/s004380051142. [DOI] [PubMed] [Google Scholar]
- 30.Muller C, Petruschka L, Cuypers H, Burchhardt G, Herrmann H. Carbon catabolite repression of phenol degradation in Pseudomonas putida is mediated by the inhibition of the activator protein PhlR. J Bacteriol. 1996;178:2030–2036. doi: 10.1128/jb.178.7.2030-2036.1996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Ng L C, Shingler V, Sze C C, Poh C L. Cloning and sequences of the first eight genes of the chromosomally encoded (methyl) phenol degradation pathway from Pseudomonas putida P35X. Gene. 1994;151:29–36. doi: 10.1016/0378-1119(94)90629-7. [DOI] [PubMed] [Google Scholar]
- 32.Nordlund I, Powlowski J, Shingler V. Complete nucleotide sequence and polypeptide analysis of multicomponent phenol hydroxylase from Pseudomonas sp. strain CF600. J Bacteriol. 1990;172:6826–6833. doi: 10.1128/jb.172.12.6826-6833.1990. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Nozaki M, Kotani S, Ono K, Senoh S. Metapyrocatechase III. Substrate specificity and mode of ring fission. Biochim Biophys Acta. 1970;220:213–223. doi: 10.1016/0005-2744(70)90007-0. [DOI] [PubMed] [Google Scholar]
- 34.Olsen R H, DeBusscher G, McCombie W R. Development of broad-host-range vectors and gene banks: self-cloning of the Pseudomonas aeruginosa PAO chromosome. J Bacteriol. 1982;150:60–69. doi: 10.1128/jb.150.1.60-69.1982. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Parsek M R, McFall S M, Chakrabarty A M. Evolution of regulatory systems of biodegradative pathways. In: Nakazawa T, Furukawa K, Haas D, Silver S, editors. Molecular biology of pseudomonads. Washington, D.C.: American Society for Microbiology; 1996. pp. 135–152. [Google Scholar]
- 36.Pavel H, Forsman M, Shingler V. An aromatic effector specificity mutant of the transcriptional regulator DmpR overcomes the growth constraints of Pseudomonas sp. strain CF600 on para-substituted methylphenols. J Bacteriol. 1994;176:7550–7557. doi: 10.1128/jb.176.24.7550-7557.1994. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Sambrook J, Fritsch E F, Maniatis T. Molecular cloning: a laboratory manual. 2nd ed. Cold Spring Harbor, N.Y: Cold Spring Harbor Laboratory Press; 1989. [Google Scholar]
- 38.Schirmer F, Ehrt S, Hillen W. Expression, inducer spectrum, domain structure, and function of MopR, the regulator of phenol degradation in Acinetobacter calcoaceticus NCIB8250. J Bacteriol. 1997;179:1329–1336. doi: 10.1128/jb.179.4.1329-1336.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Shine J, Dalgarno L. Determination of cistron specificity in bacterial ribosomes. Nature. 1975;254:34–38. doi: 10.1038/254034a0. [DOI] [PubMed] [Google Scholar]
- 40.Shingler V. Metabolic and regulatory check points in phenol degradation by Pseudomonas sp. strain CF600. In: Nakazawa T, Furukawa K, Haas D, Silver S, editors. Molecular biology of pseudomonads. Washington, D.C.: American Society for Microbiology; 1996. pp. 153–164. [Google Scholar]
- 41.Shingler V. Signal sensing by ς54-dependent regulators: derepression as a control mechanism. Mol Microbiol. 1996;19:409–416. doi: 10.1046/j.1365-2958.1996.388920.x. [DOI] [PubMed] [Google Scholar]
- 42.Shingler V, Bartilson M, Moore T. Cloning and nucleotide sequence of the gene encoding the positive regulator (DmpR) of the phenol catabolic pathway encoded by pVI150 and identification of DmpR as a member of the NtrC family of transcriptional activators. J Bacteriol. 1993;175:1596–1604. doi: 10.1128/jb.175.6.1596-1604.1993. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Shingler V, Franklin C H, Tsuda M, Holroyd D, Bagdasarian M. Molecular analysis of a plasmid-encoded phenol hydroxylase from Pseudomonas CF600. J Gen Microbiol. 1989;135:1083–1092. doi: 10.1099/00221287-135-5-1083. [DOI] [PubMed] [Google Scholar]
- 44.Shingler V, Moore T. Sensing of aromatic compounds by the DmpR transcriptional activator of phenol-catabolizing Pseudomonas sp. strain CF600. J Bacteriol. 1994;176:1555–1560. doi: 10.1128/jb.176.6.1555-1560.1994. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Shingler V, Pavel H. Direct regulation of the ATPase activity of the transcriptional activator DmpR by aromatic compounds. Mol Microbiol. 1995;17:505–513. doi: 10.1111/j.1365-2958.1995.mmi_17030505.x. [DOI] [PubMed] [Google Scholar]
- 46.Simon R, Priefer U, Puhler A. A broad host range mobilization system for in vivo genetic engineering: transposon mutagenesis in gram negative bacteria. Bio/Technology. 1983;1:784–791. [Google Scholar]
- 47.Sze C C, Moore T, Shingler V. Growth phase-dependent transcription of the ς54-dependent Po promoter controlling the Pseudomonas-derived (methyl)phenol dmp operon of pVI150. J Bacteriol. 1996;178:3727–3735. doi: 10.1128/jb.178.13.3727-3735.1996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Sze C C, Shingler V. The alarmone (p)ppGpp mediates physiological-responsive control at the ς54-dependent Po promoter. Mol Microbiol. 1999;31:1217–1228. doi: 10.1046/j.1365-2958.1999.01264.x. [DOI] [PubMed] [Google Scholar]
- 49.Takeo M, Maeda Y, Okada H, Miyama K, Mori K, Ike M, Fujita M. Molecular cloning and sequencing of the phenol hydroxylase gene from Pseudomonas putida BH. J Ferment Bioeng. 1995;79:485–488. [Google Scholar]
- 50.Teramoto M, Futamata H, Harayama S, Watanabe K. Characterization of a high-affinity phenol hydroxylase from Comamonas testosteroni R5 by gene cloning and expression in Pseudomonas aeruginosa PAO1c. Mol Gen Genet. 1999;262:552–558. doi: 10.1007/s004380051117. [DOI] [PubMed] [Google Scholar]
- 51.Thompson J D, Higgins D G, Gibson T J. CLUSTAL W: improving the sensitivity of progressive multiple sequence alignment through sequence weighting, positions-specific gap penalties and weight matrix choice. Nucleic Acids Res. 1994;22:4673–4680. doi: 10.1093/nar/22.22.4673. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Tsuda M. Use of a transposon-encoded site-specific resolution system for construction of large and defined deletion mutation in bacterial chromosome. Gene. 1998;207:33–41. doi: 10.1016/s0378-1119(97)00601-x. [DOI] [PubMed] [Google Scholar]
- 53.Tsuda M, Miyazaki H, Nakazawa T. Genetic and physical mapping of genes involved in pyoverdin production in Pseudomonas aeruginosa PAO. J Bacteriol. 1995;177:423–431. doi: 10.1128/jb.177.2.423-431.1995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Watanabe K, Hino S. Identification of a functionally important population in phenol-digesting activated sludge with antisera raised against isolated bacterial strains. Appl Environ Microbiol. 1996;62:3901–3904. doi: 10.1128/aem.62.10.3901-3904.1996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Watanabe K, Hino S, Onodera K, Kajie S, Takahashi N. Diversity in kinetics of bacterial phenol-oxygenating activity. J Ferment Bioeng. 1996;81:560–563. [Google Scholar]