Abstract
The regulation of cellular energy metabolism is central to most physiological and pathophysiological processes. However, most current methods have limited ability to functionally probe metabolic pathways in individual cells. Here, we describe SPICE‐Met (Single‐cell Profiling and Imaging of Cell Energy Metabolism), a method for profiling energy metabolism in single cells using flow cytometry or imaging. We generated a transgenic mouse expressing PercevalHR, a fluorescent reporter for cellular ATP:ADP ratio. Modulation of PercevalHR fluorescence with metabolic inhibitors was used to infer the dependence of energy metabolism on oxidative phosphorylation and glycolysis in defined cell populations identified by flow cytometry. We applied SPICE‐Met to analyze T‐cell memory development during vaccination. Finally, we used SPICE‐Met in combination with real‐time imaging to dissect the heterogeneity and plasticity of energy metabolism in single macrophages ex vivo and identify three distinct metabolic patterns. Functional probing of energy metabolism with single‐cell resolution should greatly facilitate the study of immunometabolism at a steady state, during disease pathogenesis or in response to therapy.
Keywords: energy, glycolysis, imaging, immunometabolism, OXPHOS
Subject Categories: Immunology, Metabolism
A new transgenic mouse model allows analysis of energy metabolism at single‐cell resolution in living cells and is applied here to T cells and perinatal macrophages.
Introduction
Mammalian cells use oxidative phosphorylation (OXPHOS) and glycolysis for energy production. Immune cells undergo metabolic changes during activation and within specific tissue microenvironments during infection, inflammation, or cancer development(Buck et al, 2017). For example, T cells or macrophages can switch from OXPHOS to glycolysis during activation (O'Neill et al, 2016; O'Neill & Pearce, 2016). These changes are associated with profound modifications of cellular functions highlighting that immunometabolism is a major determinant of immunological processes. Various methodologies are available to characterize metabolic pathways engaged in energy production (Artyomov & Van den Bossche, 2020). Extracellular flux analysis (SeaHorse technology) is a technique of choice that measures oxygen consumption rate (OCR) and extracellular acidification rate (ECAR) in the presence of specific metabolic inhibitors to estimate the contribution of respiration and glycolysis in a bulk of cells (van der Windt et al, 2016). Mass spectrometry‐based techniques provide quantitative measurements for hundreds of distinct metabolites in a given sample (Wang et al, 2019). In addition, several approaches have been developed to collect information regarding immunometabolism at the single‐cell level. Metabolic genes can be analyzed by single‐cell transcriptomics to provide information regarding energetic pathways in individual cells (Xiao et al, 2019). Single‐cell immunometabolism can also be inferred from protein levels of key enzymes using high‐dimensional flow cytometry (Ahl et al, 2020; Hartmann et al, 2021). SCENITH is an elegant flow cytometry‐based assay that has been described recently and uses the rate of protein synthesis and treatment with metabolic drugs to estimate the contribution of energy metabolic pathways in complex cell populations (Arguello et al, 2020). Fluorescent biosensors can also be exploited to collect metabolic information at the single‐cell level (Tantama et al, 2013; Mendelsohn et al, 2018; Postat et al, 2018; Lobas et al, 2019; Kondo et al, 2021). Finally, spatially resolved mass spectrometry (Kertesz & Cahill, 2021) and in situ evaluation of enzymatic activities (Miller et al, 2017) are paving the way for spatial immunometabolomics.
These powerful methodologies provide complementary and important biological information. Most of these techniques, however, either do not offer direct functional measurement of energy production or lack the single‐cell resolution. Here, we aimed to develop a simple and versatile methodology for ex vivo profiling energy metabolism in complex cellular populations. Our method, named SPICE‐Met (Single‐cell Profiling and Imaging of Cell Energy Metabolism), relies on a new transgenic mouse (termed SPICY) expressing the fluorescent sensor PercevalHR for intracellular measurements of ATP:ADP ratio (Tantama et al, 2013) and on interferences with metabolic drugs. We show that SPICE‐Met documents the respective dependencies on OXPHOS and glycolysis in a variety of immune cell types at a steady state or during activation. As a proof of concept, we characterized the kinetics of metabolic changes during memory T‐cell development and uncovered the heterogeneity and plasticity of energy metabolism in macrophages ex vivo.
Results and Discussion
Probing energy metabolism using a reporter mouse for ATP:ADP ratio
To devise a strategy for single‐cell measurement of energy metabolism, we sought to rely on a fluorescent biosensor, named PercevalHR (Tantama et al, 2013). PercevalHR is composed of a mutated version of the ATP‐binding bacterial protein GlnK1 and the circular permuted monomeric Venus fluorescent protein. ATP but not ADP binding to the PercevalHR causes a ratiometric shift in the probe fluorescence excitation spectrum, providing a read‐out of ATP:ADP intracellular ratio (Tantama et al, 2013). We envisioned that interfering with specific metabolic pathways will be reflected by changes in ATP:ADP ratio depending on the cellular energy metabolism (Fig 1A). For example, we have previously observed a rapid drop in ATP:ADP ratio in bone marrow–derived macrophages (BMDM) retrovirally transduced with PercevalHR (Tantama et al, 2013) and treated with oligomycin (an inhibitor of the ATP synthase) to block OXPHOS (Postat et al, 2018). To test whether this approach is applicable to complex cell populations, we generated a transgenic mouse line expressing a floxed version of the PercevalHR sensor under the control of the CAG promoter (SPICY mice).
Figure 1. Probing energy metabolism using SPICE‐Met.
-
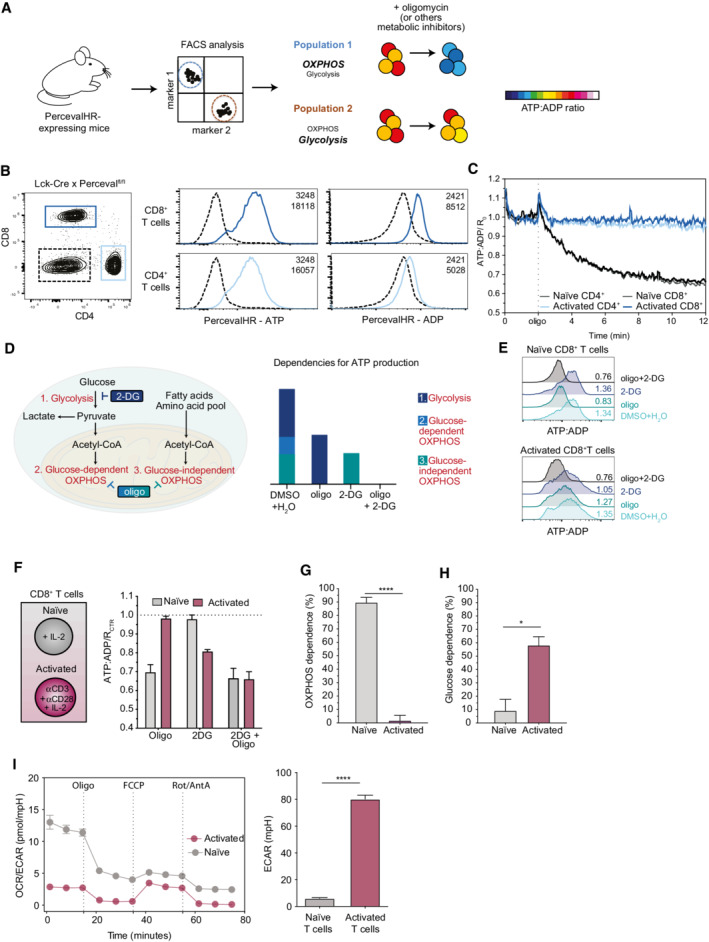
AScheme of the SPICE‐Met method. Cells from PercevalHR‐expressing mice can be stained with a desired antibody cocktail and treated with metabolic inhibitors such as oligomycin or other metabolic inhibitors. Changes in PercevalHR fluorescence are used to deduce the dependence of energy metabolism on OXPHOS and glycolysis in a specific cell population.
-
BPercevalfl/fl mice were crossed with Lck‐Cre mice, and the expression of PercevalHR was evaluated in both CD4+ and CD8+ T cells. Dotted line represents PercevalHR expression in double negative cells.
-
CNaïve or in vitro activated CD4+ and CD8+ T cells from Lck‐Cre x Percevalfl/fl mice were subjected to time‐resolved flow cytometry. After 2 min, oligomycin was added and the acquisition was continued for an additional period of 10 min. Relative ATP:ADP ratio values were calculated using PercevalHR fluorescence excited using a blue (488 nm) and violet (405 nm) laser to quantify ATP and ADP, respectively. The graph shows the geometric mean of the cellular ATP:ADP ratio normalized to the average value measured during the first 2 minutes of acquisition time (R 0). One representative experiment of three is shown.
-
DScheme illustrating the main energetic pathways used for ATP production and the expected impact of metabolic inhibitors.
-
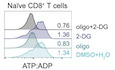
ERepresentative histograms of ATP:ADP ratio in naïve and activated CD8+ T cells upon 40 min treatment with DMSO+H2O as control, oligo, 2‐DG, or oligo+2‐DG.
-
FATP:ADP ratio values were normalized to the DMSO+H2O treatment ratio (RCTR) to visualize the relative contributions of the different metabolic inhibitors.
-
G, HOXPHOS and glucose dependencies were calculated as ((RDMSO+H2O − Roligo) / (RDMSO+H2O − Roligo+2‐DG)) × 100 and ((RDMSO+H2O − R2‐DG) / (RDMSO+H2O − Roligo+2‐DG)) × 100, respectively. Results were evaluated using an unpaired Student's t‐test, *P < 0.05, ****P < 0.0001.
-
ICD8+ T cells were isolated from a WT mouse and activated with anti‐CD3 and anti‐CD28 or kept in the presence of recombinant murine IL‐2. OCR and ECAR were measured using the SeaHorse technology during sequential treatments with oligomycin, FCCP, and Rot/AntA on resting and activated CD8+ T cells isolated from WT mice. OCR/ECAR ratios and basal ECAR are graphed for the indicated populations. Representative of 2 independent experiments (with 3 to 8 biological replicates for each condition). Results were evaluated using a two‐tailed unpaired Student's t‐test, *P < 0.05.
Data information: Results in (E–H) are representative of 2–3 independent experiments. Data are shown as mean ± SEM.
As a first proof of concept, we initially thought to compare naïve and activated T cells that are known to rely on distinct metabolic pathways (OXPHOS and glycolysis, respectively) for energy production (O'Neill et al, 2016). To this end, SPICY (Percevalfl/fl) mice were crossed with Lck‐Cre mice for T‐cell‐specific expression of the biosensor (Fig 1B). CD4+ and CD8+ T cells were activated anti‐CD3/CD28 monoclonal antibodies supplemented with IL‐2 or simply cultured with IL‐2 for 2 days. To analyze the energy metabolism of these T‐cell populations, we analyzed their ATP:ADP ratio by time‐resolved flow cytometry. After acquiring baseline values for 2 min, oligomycin was added and recording was continued for 10 min. As shown in Fig 1C, the ATP:ADP ratio rapidly dropped in naïve CD4+ and CD8+ T cells upon treatment with oligomycin. This drop corresponded to a decrease in the PercevalHR‐ATP signal and a moderate increase in the PercevalHR‐ADP signal (Appendix Fig S1B). By contrast, the ATP:ADP ratio in activated T cells was not affected by the presence of the inhibitor. These results suggest that our approach can readily estimate the dependencies on OXPHOS and glycolysis to cell energy metabolism. To further characterize energy metabolic pathways, we treated cells with 2‐DG that is expected to block glycolysis and glucose‐dependent OXPHOS, or the combination of 2‐DG and oligomycin to block both glycolysis and OXPHOS (Fig 1D) as previously done to evaluate metabolic pathways contributing to protein synthesis (Arguello et al, 2020). We analyzed the impact of these inhibitors after 40 min when ATP:ADP ratios were found to reach stable levels (Appendix Fig S1A) and chose this time point for all subsequent analyses. We deduced the OXPHOS dependence on the overall energy metabolism by relating the drop in ATP:ADP induced by oligomycin to the value obtained in untreated cells (maximum value) and that in cells treated with a mix of oligomycin and 2‐DG (minimum value) (Fig 1E and F; Appendix Fig S1B). As expected, OXPHOS dependence was high (84–89%) in naïve CD8+ and CD4+ T cells but low (2%) in activated CD8+ and CD4+ T cells (Fig 1G; Appendix Fig S1C). Similarly, glucose dependence was estimated by measuring the impact of 2‐DG treatment on cellular ATP:ADP and was found to be substantially higher in activated compared with naïve T cells (Fig 1H; Appendix Fig S1D). We confirmed that similar profiles were obtained for these two populations in CD8+ T cells using the SeaHorse technology, a standard approach for metabolic measurements in bulk cells (Fig 1I), and similar data were observed in human T cells using SCENITH (Arguello et al, 2020). Of note, in the presence of 2‐DG, activated T cells became sensitive to oligomycin treatment (Fig 1E and F), most likely reflecting plasticity and adaptation for energy production. Because our methodology is profiling energy metabolism at the single‐cell level, we termed this approach SPICE‐Met.
Profiling of energy metabolism in defined T‐cell populations ex vivo
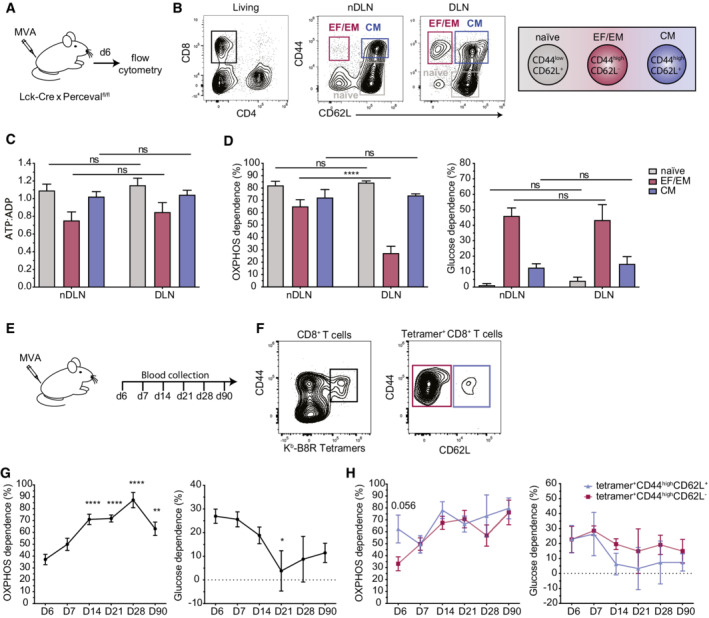
One fundamental feature of our methodology is the possibility to evaluate energy metabolism in complex cell populations identified by specific markers using flow cytometry. Taking advantage of this strategy, we explored the metabolic changes undergone by various T‐cell populations during the development of a T‐cell response in the lymph node upon footpad immunization with a Modified Vaccinia Ankara (MVA) candidate vaccine. CD8+ T‐cell populations were distinguished based on the surface expression of CD44 and CD62L (Fig 2A and B). To test for local changes in energy metabolism induced by MVA administration, we applied SPICE‐Met to draining and nondraining lymph nodes (as a control). For each population, the baseline ATP:ADP ratio was minimally affected by MVA administration (Fig 2C). We observed that CD8+ T cells residing in nondraining lymph nodes largely relied on OXPHOS with the strongest dependence for CD44lowCD62L+ naïve T cells (82%) followed by CD44highCD62L+ central memory T cells (72%) and CD44highCD62L− effector/effector memory T cells (65%) (Fig 2D). In the draining lymph node, CD44highCD62L− T cells exhibited a drop in OXPHOS dependence (37%) (Fig 2D), likely reflecting a burst of glycolytic effector T cells responding to MVA. Similar trends were obtained when the cells were analyzed immediately after harvesting or after 90 min of culture in complete media at 37°C, suggesting the measurements are relatively stable within this time window (Appendix Fig S2). Focusing on MVA‐specific T cells using pMHC tetramers (Fig 2E and F), we characterized the evolution of energy metabolism during memory T‐cell formation. At the peak of the response (d6), OXPHOS dependence in MVA‐specific T cells was minimal but then progressively increased over the next 3 weeks being maximal at day 28 postimmunization (Fig 2G). These changes were detected in both central (CD44highCD62L+) and effector memory (CD44highCD62L−) antigen‐specific T‐cell subsets (Fig 2H). Thus, the use of SPICE‐Met during immunization provided a precise kinetic of metabolic changes associated with the development of memory T cells.
Figure 2. Ex vivo profiling of metabolic activities on phenotypically defined T‐cell populations.
-
A–DLck‐Cre x Percevalfl/fl mice were injected with 2x106 PFU of MVA‐HIVB. Lymph node T cells were stained with the indicated markers and subjected to SPICE‐Met. (A) Experimental setup. (B) Representative contour plot showing CD8+ T cells in nondraining (nDLN) and draining lymph nodes (DLN) 6 days postinfection. Effector and effector memory (EF/EM), central memory (CM), and naïve T cells were defined by their expression of CD44 and CD62L. (C) ATP:ADP basal ratio was measured in EF/EM, CM, and naïve T‐cell populations. (D) OXPHOS and glucose dependencies were calculated as detailed in the Methods section. Data are pooled from n = 9 mice and were obtained in 3 independent experiments. Statistical significance was evaluated using a two‐way ANOVA. ns, significant, **P < 0.01, ****P < 0.0001. Data are represented as mean ± SEM.
-
E–HLck‐Cre x Percevalfl/fl mice were injected with 2x106 PFU of MVA‐HIVB, and blood was collected every week for the first month and at day 90. (E) Experimental setup. (F) Gating strategy. CD8+ T cells specific for the immunodominant epitope of MVA (Kb‐B8R) were identified by MHC–peptide tetramers and further distinguished based on their expression of CD62L. (G, H) OXPHOS and Glucose dependencies were measured by SPICE‐Met in (G) total MVA‐specific CD8+ T cells or (H) in CD62L+ and CD62L− MVA‐specific CD8+ T‐cell compartments. Results are pooled from three independent experiments corresponding to a total of n = 12–13 mice. Statistical significance was evaluated using a multiple unpaired t‐test. *P < 0.5, **P < 0.01, ****P < 0.0001.
Simultaneous analysis of energy metabolism in multiple immune cell populations
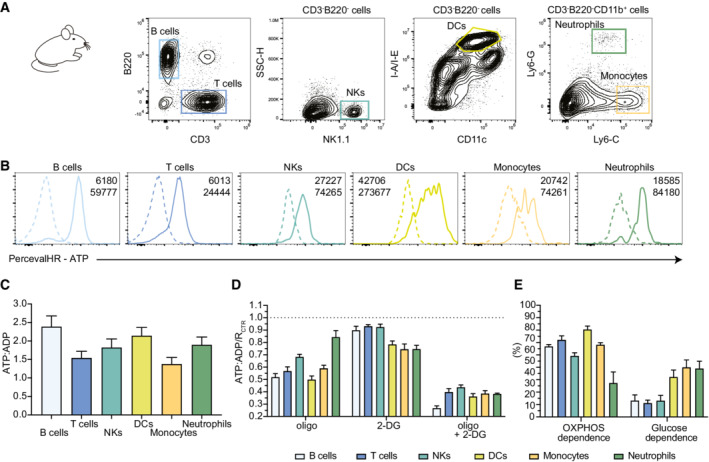
To generate a more versatile mouse model, we crossed the SPICY (Percevalfl/fl) animals to Vav‐iCre mice, which promote recombination in the vast majority of hematopoietic cells (Georgiades et al, 2002). As shown in Fig 3A and B, the reporter was expressed in the various cell types tested including T cells, B cells, NK cells, monocytes, and neutrophils. While the expression level of the probe varied between cell types (most likely due to the differential activity of the promoter), the range of ATP:ADP ratio at a steady state only showed mild variability among cell types (Fig 3C). Applying SPICE‐Met on immune cells from lymph nodes at a steady state, we could determine the OXPHOS and glucose dependencies in multiple cell types simultaneously (Fig 3D and E). Lymph node T cells (CD3+), B cells (B220+), dendritic cells (DCs) (CD3−B220−CD11c+MHCII+), and monocytes (CD3−B220−CD11b+Ly6G−Ly6C+) largely relied on OXPHOS for energy production. OXPHOS only accounted for half of the ATP production in NK cells (CD3−B220−NK1.1+) and around 30% in neutrophils (CD3−B220−CD11b+Ly6G+Ly6Cint). Glucose had the largest contribution to DCs, monocytes, and neutrophils (Fig 3D and E). SPICE‐Met using the Vav‐iCre x Percevalfl/fl mice therefore provides the opportunity to readily profile energy metabolism in multiple cellular populations in a single experiment.
Figure 3. A mouse model for the simultaneous measurement of energy metabolism in defined immune cell populations.
-
AGating strategy used from lymph node cells.
-
BReporter expression (as measured by the ATP signal) is detected in all immune cell populations analyzed. Negative dotted lines correspond to the value measured in the C57Bl/6 wild‐type mouse.
-
CATP:ADP ratios measured at steady state in B cells, T cells, NKs, DCs, monocytes, and neutrophils.
-
D, ESPICE‐Met was performed on various lymph node immune cell types simultaneously. (D) ATP:ADP ratios were normalized to the untreated condition and shown for different populations upon treatment with DMSO+H2O as control, oligo, 2‐DG, or oligo+2‐DG. (E) OXPHOS and glucose dependencies were calculated for B cells, T cells, NKs, DCs, monocytes, and neutrophils.
Data information: Results are pooled from three independent experiments (n = 6). Data are represented as mean ± SEM.
Dissecting energy metabolism in single macrophages using SPICE‐Met imaging
So far, we relied on flow cytometry to dissect energy metabolism in phenotypically defined cell populations. Imaging at the single‐cell level could offer the additional possibility to characterize cellular heterogeneity within a defined cell population. We therefore adapted our technique to real‐time imaging by tracking changes in PercevalHR fluorescent signals upon treatment with oligomycin. We applied this approach to the well‐characterized metabolic shift associated with macrophage activation (O'Neill & Pearce, 2016). Indeed, while nonactivated macrophages are known to largely rely on OXPHOS for energy metabolism, classically activated (but not alternatively activated) macrophages shift their metabolism toward glycolysis. BMDMs generated from Vav‐iCre x Percevalfl/fl mice were either left untreated, treated with IL‐4 (alternative activation), or activated with IFNγ and LPS (classical activation) (Fig 4A). ATP:ADP ratio using two‐photon imaging through sequential excitation at 830 and 1,040 nm following treatment with oligomycin. As expected, the majority of nonactivated or alternatively activated macrophages were sensitive to oligomycin treatment, whereas classically activated macrophages were not (Fig 4B and C; Appendix Fig S3). However, we identified a substantial degree of cellular heterogeneity in these different culture conditions, including variability in basal ATP:ADP levels and in response to oligomycin (Fig 4C and D; Movie EV1). Our imaging data were largely consistent with flow cytometry–based SPICE‐Met (Appendix Fig S4) that we also used to characterize the kinetics of macrophage metabolic switch (Appendix Fig S5A and B) and the role of iNOS in blocking OXPHOS during classical activation (Appendix Fig S5C–E).
Figure 4. SPICE‐Met imaging uncovers energy metabolism in single macrophages in vitro and ex vivo .
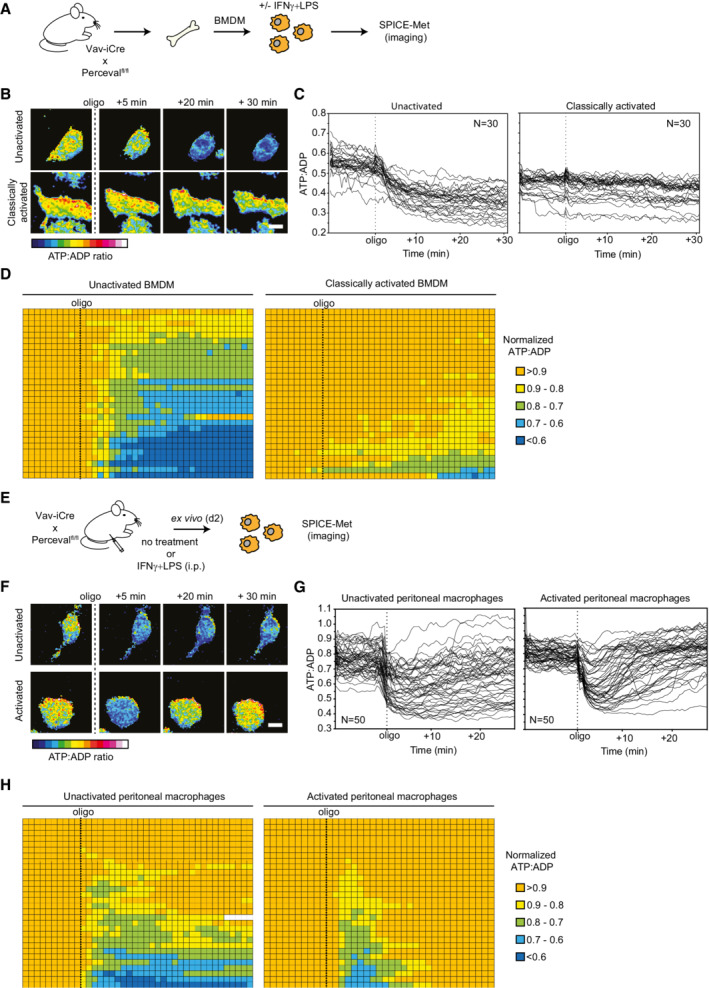
-
A–DBMDMs of Vav‐iCre × Percevalfl/fl mice were activated 24 h with LPS + IFNγ or left untreated. (A–D) Live imaging of ATP:ADP ratio in unactivated or activated BMDMs. Macrophages were exposed to oligomycin (1 μM) during image acquisition. (A) Experimental setup. (B) Representative time‐lapse images showing the ATP:ADP ratio in unactivated and activated macrophages in response to oligomycin. Scale bar, 10 μm. (C) Kinetic analysis of ATP:ADP ratios for single macrophages (n = 30 cells for each condition). (D) Table representing the ATP:ADP variations in individual macrophages treated with oligomycin. Ratios were normalized to the steady‐state (initial) values and were color‐coded as indicated. Each line represents one cell, and each square corresponds to 1 min. Results are representative of three independent experiments (n = 30 cells for each condition).
-
E–HVav‐iCre × Percevalfl/fl mice were injected i.p. with LPS + IFNγ or left untreated. Peritoneal macrophages were isolated 48 h later and immediately subjected to SPICE‐Met imaging. (E) Experimental setup. (F) Representative time‐lapse images showing the ATP:ADP ratio in ex vivo isolated macrophages in response to oligomycin. Scale bar, 10 μm. (G) Kinetic analysis of ATP:ADP ratios for single macrophages (n = 50 cells shown for each condition). (H) Table representing the ATP:ADP variations in individual macrophages (n = 30 cells are shown for each condition) treated with oligomycin. Ratios were normalized to the steady‐state values and were color‐coded as indicated. Each line represents one cell, and each square corresponds to 1 min. Results are representative of two independent experiments.
We extended our imaging approach to peritoneal macrophages activated in vivo (Fig 4E). SPICE‐Met imaging of ex vivo isolated macrophages revealed that most activated macrophages were transiently responsive to oligomycin but rapidly restored their ATP:ADP levels. By contrast, the effects of oligomycin on nonactivated macrophages were sustained (Fig 4F–H; Movie EV2). These results suggest that in vivo, activated macrophages have maintained their capacity to use OXPHOS but could rapidly rewire their energy metabolism toward glycolysis upon blockage of mitochondrial respiration.
To confirm that these distinct patterns all corresponded to bona fide macrophages, we performed SPICE‐Met in the presence of a fluorescent F4/80 antibody to control for cell identity. For this experiment, we recovered peritoneal macrophages following the classical thioglycolate stimulation (Fig 5). SPICE‐Met analyses performed on F4/80+ cells suggested that at least 3 patterns of macrophage metabolism can coexist (Fig 5A). In a first subset (subset O), energic resources are highly dependent on OXPHOS with ATP:ADP ratio dropping irreversibly upon oligomycin treatment. A second subset (subset G) of macrophage is irresponsive to oligomycin and therefore is hardwired toward glycolysis. Finally, a third subset (subset OG) of macrophage exhibits strong plasticity in energy production and rapidly restores energic resources after a transient drop induced by oligomycin (Fig 5B and C; Movie EV3). Of note, G and OG profiles (but not O profile) were lost in the absence of pyruvate and glucose in the medium suggesting that pyruvate is dispensable for respiration but that glucose is, as expected, essential for G and OG profiles (Appendix Fig S6).
Figure 5. Coexistence of macrophages with distinct energy metabolic profiles in vivo .
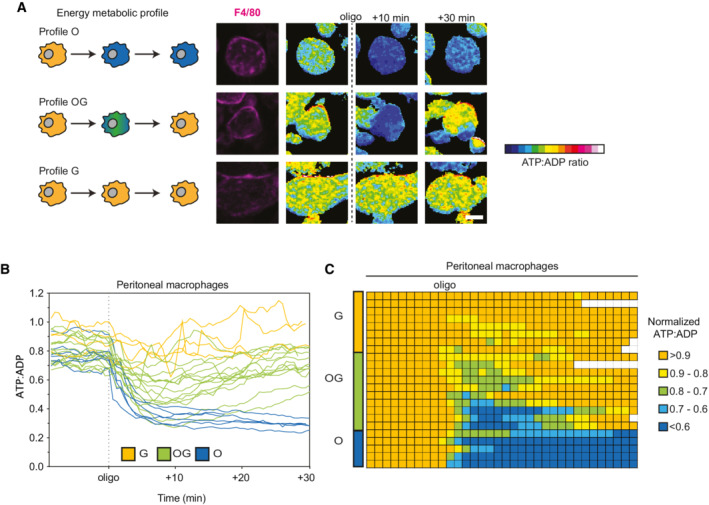
Vav‐iCre x Percevalfl/fl mice were injected i.p. with thioglycolate. Peritoneal macrophages were isolated 3 days later and immediately subjected to SPICE‐Met imaging in the presence of Alexa 594‐conjugated F4/80 antibody.
-
ARepresentative time‐lapse images showing the 3 patterns of ATP:ADP changes in response to oligomycin in peritoneal macrophages. Scale bar, 10 μm.
-
BKinetic analysis of ATP:ADP ratios for single macrophages (n = 25 cells).
-
CTable representing the ATP:ADP variations in individual macrophages (n = 25 cells are shown) treated with oligomycin. Ratios were normalized to the steady‐state values and were color‐coded as indicated. Each line represents one cell, and each square corresponds to 1 min. Results are representative of two independent experiments.
Thus, the capacity to visualize energetic resources in real‐time provides valuable information on metabolic plasticity and heterogeneity and reveals substantial differences between in vitro and in vivo activated macrophages. More generally, combining SPICE‐Met and imaging represents a valuable addition to profile cell–cell variability in energy metabolism within defined culture conditions or within ex vivo isolated cells.
Conclusions
In sum, we described here a new methodology to monitor the dependencies on OXPHOS and glycolysis in complex cell populations ex vivo. Like the recently described SCENITH technique (Arguello et al, 2020), SPICE‐Met is compatible with flow cytometry and can be combined with phenotypic markers and achieve energy metabolic profiling in multiple cell types simultaneously. A potential limitation of our technique lies in the composition of the culture media that may not necessarily match nutrient availability in vivo. In our assay, we choose to use complete media to maximize energic resources and to reveal how a given cell generate energy in the absence of constraints. Interestingly, by modifying the media (i.e., removing the glucose), we can also reveal plasticity in energy metabolism. For example, we observe that in the absence of glucose, activated T cells are able to reverse to OXPHOS (Appendix Fig S7).
Importantly, our technique can also be used for in vitro or ex vivo imaging at the single‐cell level to characterize heterogeneity in bioenergetic pathways within a given population. Future imaging applications may integrate microfluidic devices for high‐throughput single‐cell metabolic profiling, combination with single‐cell transcriptomics, and adaptation of the method for in vivo imaging. The transgenic mouse line expressing a floxed PercevalHR reporter should be suited for many applications in the study of immune and non‐immune cells. SPICE‐Met is a rapid and versatile assay and should represent a valuable tool for energy metabolic profiling in mouse models of infection, inflammation, and cancer.
Materials and Methods
Reagents and Tools table
| Reagent/Resource | Reference or Source | Identifier or Catalog Number |
|---|---|---|
| Experimental Models | ||
| C57BL/6J (M. musculus) | Envigo | |
| PercevalHR floxed transgenic mice (Percevalfl/fl or SPICY mice) | Produced by the Bousso Laboratory and Institut Pasteur | |
| Lck‐Cre | Stock center | |
| Vav‐iCre | Jackson | B6.Cg‐Commd10Tg(Vav1‐icre)A2Kio/J |
| Recombinant DNA | ||
| pSF‐CAG‐amp plasmid | Oxford Genetics | |
| loxNeoStoplox cassette (2,185 bp) | Genewiz | |
| pSF‑CAG‑loxStoplox‑PercevalHR | Subcloning the PercevalHR coding sequence into the loxNeoStoplox cassette and digested for transgenesis using PacI/AsiSI enzymes | |
| Antibodies | ||
| purified rat anti‑mouse CD3(17.A2) | BioLegend | 100202 |
| purified Syrian hamster anti‑mouse CD28(37.51) | BioLegend | 102102 |
| Alexa Fluor 594 anti‐F4/80 antibody (BM8) | BioLegend | 123140 |
| BV605 rat anti‑mouse/human CD44 (IM7) | BioLegend | 103047 |
| PerCpCy5.5 rat anti‑mouse CD11b (M1/70) | BioLegend | 101228 |
| PECy7 mouse anti‑mouse NK1.1 (PK136) | BioLegend | 108714 |
| BV605 rat anti‐mouse Ly6C (HK1.4) | BioLegend | 128036 |
| AlexaFluor 647 rat anti‑mouse Ly6G (1A8) | BioLegend | 127610 |
| APC‑fire750 rat anti‐mouse I‐A/I‐E (M5/114.15.2) | BioLegend | 107652 |
| BV785 rat anti‑mouse B220 (RA3‑6B2) | BioLegend | 103246 |
| BUV737 armenian hamster anti‑mouse CD11c (HL3) | BD | 612797 |
| BUV395 rat anti mouse CD4 (GK1.5) | BD | 565974 |
| BUV395 armenian hamster anti‐mouse CD3 (145‑2C11) | BD | 563565 |
| BUV395 rat anti‐mouse CD11b (M1/70) | BD | 563553 |
| BUV737 rat anti‐mouse CD11b (M1/70) | BD | 612801 |
| BV786 rat anti‑mouse CD8 (53‑6.7) | BD | 563332 |
| APC rat anti‑mouse CD62L (MEL‑14) | eBioscience | 17‐0621‐82 |
| pMHC tetramers (PE‐conjugated H2‑Kb‑TSYKFESV tetramers) | NIH Tetramer Core Facility | |
| Oligonucleotides and sequence‐based reagents | ||
| PercevalHR | Tantama et al (2013) | |
| Chemicals, enzymes and other reagents | ||
| recombinant human IL‑2 | Merk | 10799068001 |
| recombinant mouse IFNγ | Peprotech | 315‐05 |
| recombinant mouse IL‐4 | Peprotech | 214‐14 |
| LPS | Sigma | 82857‐67‐8 |
| EDTA | Gibco | 15575020 |
| oligomycin | Sigma | 1404‐19‐9 |
| 2‑deoxy‑D‑glucose(2‑DG) | Sigma | D8375 |
| Liberase TL | Sigma | 5401020001 |
| RBC lysis buffer | eBioscience | 00‐4333‐57 |
| Software | ||
| FlowJo software version 10.6 | Tree Star | |
| Prism version 9.2.0 | GraphPad | |
| Fiji | ImageJ | |
| Imaris | BitPlane | |
| Other | ||
| SeaHorse XF Cell Mito Stress Test Kit (Agilent Technologies) | Agilent Technologies | |
| FVMPE‑RS upright microscope (25X/1.05 NA objective) | Olympus | |
| Cytoflex LX | Beckman Culture | |
Methods and Protocols
Mice
Six‐ to eight‐week‐old C57BL/6J mice were purchased from ENVIGO. PercevalHR floxed transgenic mice (Percevalfl/fl), also referred to as SPICY mice, were generated by the Centre d'Ingénierie Génétique Murine at Institut Pasteur and performed in accordance with the European Community guidelines (2010/63/UE). Briefly, a conditional loxNeoStoplox cassette (2,185 bp) was synthetized at Genewiz and cloned into the pSF‐CAG‐amp plasmid (Oxford Genetics). The final construct pSF‐CAG‐loxStoplox‐PercevalHR was obtained by subcloning the PercevalHR coding sequence into this cassette and digested for transgenesis using PacI/AsiSI enzymes. Nine pups on the C57Bl/6 background had integrated the transgene with one pup showing germ‐line transmission and protein expression of the transgene upon Cre‐mediated recombination. The generated mice expressing a floxed version of PercevalHR were bred with Lck‐Cre or Vav‐iCre mice for expression of the biosensor in T cells and hematopoietic cells, respectively. All animal studies were approved by the Institut Pasteur Safety Committee in accordance with French and European guidelines (CETEA 190148).
In vitro cell stimulation
T cells were obtained from lymph nodes and spleens of Lck‐Cre × Percevalfl/fl mice. When indicated, T cells were activated in vitro in plates coated with 2.5 μg/ml purified rat anti‐mouse CD3 (17.A2, BioLegend) in the presence of 2.5 μg/ml soluble purified Syrian hamster anti‐mouse CD28 (37.51, BioLegend) and/or supplemented with 10 ng/ml recombinant human IL‐2 (Roche) for the naïve resting condition. Murine BMDMs were generated from the bone marrow of Vav‐iCre x Percevalfl/fl mice as previously described (Postat et al, 2018). On day 7, adherent cells were detached using 5 mM EDTA (Gibco) diluted in PBS and seeded into tissue culture‐treated dishes (TPP, Trasadingen, Switzerland) at a concentration of 2 × 106 cells/ml in RPMI medium 1640 ‐ GlutaMAX™ containing 10% heat‐inactivated fetal bovine serum (FCS), 100 U/ml penicillin, 100 ng/ml streptomycin, 1 mM sodium pyruvate, 10 mM HEPES and 5 μM 2β‐mercaptoethanol (complete RPMI) without phenol, supplemented or not with 1 μg/ml lipopolysaccharide (LPS) (Sigma) and 0.1 ng/ml IFNγ (Peprotech) or 10 ng/ml IL‐4 (Peprotech) for 24 h.
In vivo stimulations and cell isolation
Recombinant MVA‐HIVB (expressing full‐length HIV Gag, fused to three Pol and two Nef fragments)(Sagoo et al, 2016) was provided by the Agence Nationale de Recherche sur le Sida (ANRS). Lck‐Cre × Percevalfl/fl mice were injected into the footpad with 2 × 106 p.f.u. of MVA‐HIVB. To evaluate primary T‐cell responses against MVA, draining lymph nodes and nondraining lymph nodes were collected 6 days postinfection. Blood was collected and treated with RBC lysis buffer (eBioscience) every week for the first month and then at day 90.
For the imaging experiment, Vav‐iCre × Percevalfl/fl mice were injected intraperitoneally with a mix of 25 μg of IFNγ and 25 μg of LPS in a total volume of 200 μl PBS or left untreated or received a 1 ml intraperitoneal injection of 3% Brewer thioglycollate broth (Sigma BD Horizon). After 48 h or 3 days, macrophages were harvested as previously reported (Zhang et al, 2008). Cells were centrifuged and resuspended in a complete RPMI medium without phenol and plated in tissue culture‐treated dishes for 4 h to adhere. Nonadherent cells were washed away, and adherent macrophages were left in RPMI medium without phenol. In some experiments, macrophages were stained with a conjugated Alexa Fluor 594 anti‐F4/80 antibody (BM8, Biolegend) for 30 min at 37°C in complete RPMI.
Flow cytometry
Single‐cell suspensions were stained for 10 min at 37°C in complete RPMI without phenol with a combination of the following antibodies: BV786 rat anti‐mouse CD8 (53–6.7, BD Horizon), BUV395 rat anti‐mouse CD4 (GK1.5, BD Horizon), APC rat anti‐mouse CD62L (MEL‐14, eBioscience), BV605 rat anti‐mouse/human CD44 (IM7, BioLegend), PerCpCy5.5 rat anti‐mouse CD11b (M1/70, BioLegend), PECy7 mouse anti‐mouse NK1.1 (PK136, BioLegend), BV605 rat anti‐mouse Ly6C (HK1.4, BioLegend), AlexaFluor 647 rat anti‐mouse Ly6G (1A8, BioLegend), APC‐fire750 rat anti‐mouse I‐A/I‐E (M5/114.15.2, Biolegend), BUV737 Armenian hamster anti‐mouse CD11c (HL3, BD Horizon), BV785 rat anti‐mouse B220 (RA3‐6B2, Biolegend), and BUV395 Armenian hamster anti‐mouse CD3 (145‐2C11, BD). When indicated, cells were stained with pMHC tetramers (PE‐conjugated H2‐Kb‐TSYKFESV tetramers were obtained through the NIH Tetramer Core Facility) corresponding to the immunodominant B8R 20–27 epitope. For flow cytometry analyses of BMDM, we mixed differently labeled unactivated, classically activated, or alternatively activated macrophages to perform SPICE‐Met simultaneously. Labeling was performed using BUV737, BUV395, and PerCpCy5.5‐coupled anti‐CD11b antibodies (M1/70, BioLegend). Samples were acquired using a Cytoflex LX (Beckman Culture) and analyzed using FlowJo software version 10.6 (Tree Star).
Extracellular flux assay
Activated or resting T cells were collected, washed with XF Base Medium Minimal DMEM supplemented with 2% Sodium Pyruvate, 10 mM glucose, and 2% Glutamax (Gibco), and seeded into CELL‐TAK™ (Corning)‐coated 96‐well cell‐culture microplates (Agilent Seahorse XF96) (0.5 × 106 cells/ well) for OCR and ECAR measurements using SeaHorse XF Cell Mito Stress Test Kit (Agilent Technologies).
SPICE‐Met by flow cytometry
Following staining with the indicated antibodies, cells expressing PercevalHR were analyzed by flow cytometry. ATP:ADP ratios in individual cells were calculated from two fluorescence signals. In brief, excitation with a violet laser (405 nm) and signal detection with a 525 band‐pass filter was used to measure ADP contribution while ATP levels were estimated by excitation with a blue laser (488 nm) and signal detection with a 525 band‐pass filter. For time‐resolved (kinetic) flow cytometry, cells were recorded for 2 min and then treated with 1 mM oligomycin. Acquisition was continued for an additional 10 min to monitor change in ATP:ADP ratio. When indicated, the ATP:ADP ratio was normalized to the average value acquired during the first 2 min (R0). For analyses at a defined time point, cells were treated with either 1µM oligomycin, 100 mM 2‐deoxy‐D‐glucose (2‐DG), the combination of the two, or H2O and DMSO as control, and kept at 37°C for 40 min. Samples were then recorded for 1 min. A Cytoflex LX (Beckman Coulter) was used for the acquisition, and data analysis was performed using FlowJo 10.2 software. OXPHOS and glucose dependencies were calculated as ((RDMSO+H2O − Roligo) / (RDMSO+H2O − Roligo+2‐DG)) × 100 and ((RDMSO+H2O − R2‐DG) / (RDMSO+H2O − Roligo+2‐DG)) × 100, respectively. In experiments where the medium composition was tested, we used RPMI medium 1640 ‐ GlutaMAX™ without glucose supplemented with 1% FCS, 100 U/ml penicillin, 100 ng/ml streptomycin, and 1 mM sodium pyruvate. When indicated, 10 mM D‐glucose was added.
SPICE‐Met by imaging
For in vitro and ex vivo analysis of Perceval‐expressing macrophages, two‐photon imaging was performed using a 25X/1.05 NA objective (Olympus) installed into an FVMPE‐RS upright microscope (Olympus) equipped with an Insight deep see dual laser (Spectra physics) and a resonant scanner. PercevalHR fluorescence corresponding to ADP and ATP levels were detected using 830 nm and 1,040 nm excitation, respectively. Emitted fluorescences were collected sequentially using dichroic mirrors (Semrock), a 542/27 band‐pass filter (PercevalHR signal), and a 624/40 band‐pass filter (Alexa Fluor 594 anti‐mouse F4/80 antibody signal) before collection with GaAsP detectors. Images from 3 to 5 z‐planes were collected in every 20–30 s. Image acquisition was performed for 10 min in the absence of oligomycin and an additional 30 min after the addition of 1 μM oligomycin. Data were processed and analyzed using Fiji (ImageJ) and Imaris (Bitplane) software.
Statistical analysis
Data are expressed as mean ± SEM. The unpaired Student's t‐test, one‐ and two‐way analysis of variance (ANOVA) were used as indicated in individual figure legends. Significance was defined by a P‐value < 0.05. All statistical tests were performed using Prism v.9.2.0 (GraphPad). P‐values were reported as stars: *P < 0.05; **P < 0.01; ***P < 0.005, ****P < 0.0001; ns is nonsignificant.
Author contributions
Erica Russo: Conceptualization; formal analysis; investigation; methodology; writing – original draft; writing – review and editing. Fabrice Lemaitre: Data curation; investigation; methodology. Beatrice Corre: Investigation; methodology. Aleksandra S Chikina: Investigation; methodology. Francina Langa‐Vives: Resources; methodology. Philippe Bousso: Conceptualization; formal analysis; supervision; funding acquisition; investigation; writing – original draft; writing – review and editing.
In addition to the CRediT author contributions listed above, the contributions in detail are:
ER, FL, BC, AC, and FLV conducted the experiments. ER and PB designed the experiments. ER, FL, BC, AC, and PB analyzed the data, and ER and PB wrote the manuscript.
Disclosure and competing interests statement
The authors declare that they have no conflict of interest.
Supporting information
Appendix
Movie EV1
Movie EV2
Movie EV3
Acknowledgements
We thank members of the Bousso laboratory for the critical review of the manuscript. We thank CB_UTechS at Institut Pasteur for support in conducting the present study. The work was supported by Institut Pasteur, INSERM, the Vaccine Research Institute in Créteil, France, and an advanced grant (ENLIGHTEN) from the European Research Council. E.R. was supported by fellowships from the Swiss National Science Foundation and the Vaccine Research Institute.
The EMBO Journal (2022) 41: e111528
Data availability
This study includes no data deposited in external repositories.
References
- Ahl PJ, Hopkins RA, Xiang WW, Au B, Kaliaperumal N, Fairhurst AM, Connolly JE (2020) Met‐Flow, a strategy for single‐cell metabolic analysis highlights dynamic changes in immune subpopulations. Commun Biol 3: 305 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Arguello RJ, Combes AJ, Char R, Gigan JP, Baaziz AI, Bousiquot E, Camosseto V, Samad B, Tsui J, Yan P et al (2020) SCENITH: a flow cytometry‐based method to functionally profile energy metabolism with single‐cell resolution. Cell Metab 32: 1063–1075.e7 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Artyomov MN, Van den Bossche J (2020) Immunometabolism in the single‐cell era. Cell Metab 32: 710–725 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Buck MD, Sowell RT, Kaech SM, Pearce EL (2017) Metabolic instruction of immunity. Cell 169: 570–586 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Georgiades P, Ogilvy S, Duval H, Licence DR, Charnock‐Jones DS, Smith SK, Print CG (2002) VavCre transgenic mice: a tool for mutagenesis in hematopoietic and endothelial lineages. Genesis 34: 251–256 [DOI] [PubMed] [Google Scholar]
- Hartmann FJ, Mrdjen D, McCaffrey E, Glass DR, Greenwald NF, Bharadwaj A, Khair Z, Verberk SGS, Baranski A, Baskar R et al (2021) Single‐cell metabolic profiling of human cytotoxic T cells. Nat Biotechnol 39: 186–197 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kertesz V, Cahill JF (2021) Spatially resolved absolute quantitation in thin tissue by mass spectrometry. Anal Bioanal Chem 413: 2619–2636 [DOI] [PubMed] [Google Scholar]
- Kondo H, Ratcliffe CDH, Hooper S, Ellis J, MacRae JI, Hennequart M, Dunsby CW, Anderson KI, Sahai E (2021) Single‐cell resolved imaging reveals intra‐tumor heterogeneity in glycolysis, transitions between metabolic states, and their regulatory mechanisms. Cell Rep 34: 108750 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lobas MA, Tao R, Nagai J, Kronschlager MT, Borden PM, Marvin JS, Looger LL, Khakh BS (2019) A genetically encoded single‐wavelength sensor for imaging cytosolic and cell surface ATP. Nat Commun 10: 711 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mendelsohn BA, Bennett NK, Darch MA, Yu K, Nguyen MK, Pucciarelli D, Nelson M, Horlbeck MA, Gilbert LA, Hyun W et al (2018) A high‐throughput screen of real‐time ATP levels in individual cells reveals mechanisms of energy failure. PLoS Biol 16: e2004624 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Miller A, Nagy C, Knapp B, Laengle J, Ponweiser E, Groeger M, Starkl P, Bergmann M, Wagner O, Haschemi A (2017) Exploring metabolic configurations of single cells within complex tissue microenvironments. Cell Metab 26: 788–800.e6 [DOI] [PubMed] [Google Scholar]
- O'Neill LA, Kishton RJ, Rathmell J (2016) A guide to immunometabolism for immunologists. Nat Rev Immunol 16: 553–565 [DOI] [PMC free article] [PubMed] [Google Scholar]
- O'Neill LA, Pearce EJ (2016) Immunometabolism governs dendritic cell and macrophage function. J Exp Med 213: 15–23 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Postat J, Olekhhnovitch R, Lemaïtre F, Bousso P (2018) A metabolism‐based quorum sensing mechanism contributes to termination of inflammatory responses. Immunity 49: 654–665 [DOI] [PubMed] [Google Scholar]
- Sagoo P, Garcia Z, Breart B, Lemaitre F, Michonneau D, Albert ML, Levy Y, Bousso P (2016) In vivo imaging of inflammasome activation reveals a subcapsular macrophage burst response that mobilizes innate and adaptive immunity. Nat Med 22: 64–71 [DOI] [PubMed] [Google Scholar]
- Tantama M, Martinez‐Francois JR, Mongeon R, Yellen G (2013) Imaging energy status in live cells with a fluorescent biosensor of the intracellular ATP‐to‐ADP ratio. Nat Commun 4: 2550 [DOI] [PMC free article] [PubMed] [Google Scholar]
- van der Windt GJW, Chang CH, Pearce EL (2016) Measuring bioenergetics in T cells using a seahorse extracellular flux analyzer. Curr Protoc Immunol 113: 3.16B.11–13.16B.14 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang S, Blair IA, Mesaros C (2019) Analytical methods for mass spectrometry‐based metabolomics studies. Adv Exp Med Biol 1140: 635–647 [DOI] [PubMed] [Google Scholar]
- Xiao Z, Dai Z, Locasale JW (2019) Metabolic landscape of the tumor microenvironment at single cell resolution. Nat Commun 10: 3763 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang X, Goncalves R, Mosser DM (2008) The isolation and characterization of murine macrophages. Curr Protoc Immunol Chapter 14: Unit 14.1 [DOI] [PMC free article] [PubMed] [Google Scholar]


