Abstract
Polyhydroxyalkanoic acids (PHAs) are a class of polyesters stored in inclusion bodies and found in many bacteria and in some archaea. The terminal step in the synthesis of PHA is catalyzed by PHA synthase. Genes encoding this enzyme have been cloned, and the primary sequence of the protein, PhaC, is deduced from the nucleotide sequences of more than 30 organisms. PHA synthases are grouped into three classes based on substrate range, molecular mass, and whether or not there is a requirement for phaE in addition to the phaC gene product. Here we report the results of an analysis of a PHA synthase that does not fit any of the described classes. This novel PHA synthase from Bacillus megaterium required PhaC (PhaCBm) and PhaR (PhaRBm) for activity in vivo and in vitro. PhaCBm showed greatest similarity to the PhaCs of class III in both size and sequence. Unlike those in class III, the 40-kDa PhaE was not required, and furthermore, the 22-kDa PhaRBm had no obvious homology to PhaE. Previously we showed that PhaCBm, and here we show that PhaRBm, is localized to inclusion bodies in living cells. We show that two forms of PHA synthase exist, an active form in PHA-accumulating cells and an inactive form in nonaccumulating cells. PhaC was constitutively produced in both cell types but was more susceptible to protease degradation in the latter type. Our data show that the role of PhaR is posttranscriptional and that it functions directly or indirectly with PhaCBm to produce an active PHA synthase.
Polyhydroxyalkanoic acids (PHAs) are a class of aliphatic polyesters produced by many bacteria and archaea in response to various environmental conditions. PHAs are generally regarded as a carbon and energy reserve material (1, 10, 24). The accumulation of PHA increases in some bacteria when growth is limited by a nutrient other than carbon, while in other bacteria it readily accumulates during unrestricted growth (3, 17). These high-molecular-weight molecules, typically having molecular weights on the order of 2 × 105 to 3 × 106, are composed of a linear array of repeating 3-hydroxyacid monomeric units having the chemical structure -[O-CHR(CH2)xCO]- (10). The most common type of PHA found in natural isolates is polyhydroxybutyric acid, where the side chain R is a methyl group and x is equal to 1 (1, 26).
Much attention has been given in recent years to the synthesis of PHAs due to their perceived commercial potential (15, 28). PHA synthases catalyze the polymerization of hydroxyacyl thioesters (hydroxyacyl coenzyme A [HACoA]) into PHA with the release of CoA. This key enzyme is required regardless of the pathway used by the organism to generate HACoA substrates. Nucleotide sequences are available for more than 30 PHA synthases, and they are grouped into three classes based on the deduced amino acid sequences as well as data on the substrate ranges of some of the enzymes (20). Class I PHA synthases are encoded by phaC genes, are relatively large (≈64 kDa), and catalyze polymerization of short-chain-length (having a three- to five-carbon backbone) HACoAs. The prototype is that of Ralstonia eutropha (formally called Alcaligenes eutrophus). Class II PHA synthases are also encoded by phaC genes and are relatively large (≈63 kDa). They catalyze polymerization of medium-chain-length (having a 6- to 14-carbon backbone) HACoAs. The prototype is that of Pseudomonas oleovorans. Class III PHA synthases are heteromeric, requiring two subunits of approximately 40 kDa each (encoded by phaC and phaE) which show no homology to each other, and catalyze the polymerization of short-chain-length HACoAs. The prototype is that of Allochromatium vinosum (formally called Chromatium vinosum). There are two known exceptions to these three classes. One is PHA synthase of Thiocapsa pfennigii, which has two subunits, PhaC and PhaE, with approximately 85% similarity, respectively, to those of A. vinosum (class III), but unlike A. vinosum it can incorporate medium- as well as short-chain-length HACoAs into the polymer (11). The other exception is PHA synthase of Aeromonas caviae, which has approximately 45% similarity to that of R. eutropha (class I) and can incorporate 3-hydroxyhexanoyl thioester monomers into the polymer (5).
Genes for other proteins in PHA biosynthetic pathways, such as phaA, phaB, and phaZ, which encode a ketothiolase, a reductase, and a depolymerase, respectively, have been shown to occur in pha gene clusters (20). Also, genes that specify a heterogenous group of low-molecular-weight PHA inclusion body-associated proteins (≈14 to 24 kDa) occur in these clusters. They are designated granule-associated proteins or phasins, in analogy to oleosins found on the surface of oil bodies in plant seeds (8, 25). Phasins influence inclusion body size and yield of PHA but are not essential for PHA synthesis (29, 31).
Previously we cloned a cluster of genes, phaP, -Q, -R, -B, and -C, from Bacillus megaterium and ascribed functions to three of them, leaving the two small genes, phaR and -Q, with unknown functions (16). Here we focus on PhaC and the 22-kDa PhaR, encoded on the phaRBC operon. Using in vivo and in vitro methods, we show that PhaC and PhaR are essential for PHA synthase activity. We show two forms of PHA synthase, an active form in PHA-accumulating cells and an inactive form in nonaccumulating cells, where PhaC is more susceptible to degradation. Like PhaC, PhaR localized to PHA inclusion bodies in living cells, but unlike PhaC, it did not copurify with the inclusion bodies. These are the first PHA synthase genes to be cloned from the genus Bacillus. The enzymes are distinctly different from all known PHA synthases and may be the founding members of a new class.
MATERIALS AND METHODS
Bacterial strains and growth conditions.
The bacterial strains used in this study are listed in Table 1. Cultures were grown in Luria-Bertani (LB) broth medium at 250 rpm or on LB medium plus 1.5% agar (Sigma catalog no. A4550), unless otherwise stated. For plasmid selection in Escherichia coli, ampicillin at 200 μg/ml, erythromycin (ERY) at 200 μg/ml, or chloramphenicol (CHL) at 25 μg/ml was included in the medium. For plasmid selection in B. megaterium, CHL at 6 μg/ml or ERY at 1 μg/ml plus lincomycin at 25 μg/ml was used. E. coli was grown at 37°C and B. megaterium was grown at 30°C, unless otherwise stated.
TABLE 1.
Bacterial strains and plasmids used in this studya
| Strain or plasmid | Relevant characteristics | Reference or source |
|---|---|---|
| Strains | ||
| E. coli DH5α | deoR endA1 gyrA96 hsdR17(rK− mK+) recA1 relA1 supE44 thi-1 Δ(lacZYA-argFV169) φ80lacZΔM15 F−γ−; used for general cloning | Clontech |
| E. coli BLR DE3 | F−ompT hsdSB(rB− mB−) gal dcm recA λDE3 | Novagen |
| B. megaterium 11561 | Wild type | ATCC |
| B. megaterium PHA05 | PHA-negative mutant of 11561; Δ(phaP-phaC); Emr | This study |
| Plasmids | ||
| pBluescript II SK (pSKII) | colE1; Ampr | Stratagene |
| pET-21(+) | Transcriptional His tag fusion vector; colE1; Ampr | Novagen |
| pHPS9 | Bacillus-E. coli shuttle vector; colE1 and oriV of pTA1060; Emr Cmr | 7 |
| pLTV1 | Bacillus-E. coli shuttle vector; temperature-sensitive oriV from p194 | 32 |
| pGM29 | pSKII carrying the erm gene of pHPS9 in the EcoRI/EcoRV sites; Ampr Emr | This study |
| pGM10 | phaP, -Q, -R, -B, and -C in pSKII; Ampr | 16 |
| pGM10.2 | Derived from pGM10; the pha fragment's terminal EcoRI site was removed, leaving the EcoRI site in phaP intact; Ampr | This study |
| pGM23 | Derived from pGM10.2; a 1.5-kb fragment carrying the erm gene replaces the pha genes from the HindIII site in phaC to the EcoRI site in phaP; Ampr Emr | This study |
| pGM25 | Derived from pGM23; a 4-kb EcoRI fragment carrying the pE194 temperature-sensitive oriV cloned into the SmaI site; Ampr Emr | This study |
| pGM16.2 | pHPS9 carrying phaP::gfp phaQ phaR; Emr | 16 |
| pGM72b | Derived from pGM16.2; phaP (ΔC-terminal 167 aa)b::gfp phaQ phaR; Emr | This study |
| pGM92 | Derived from pGM72b; phaP (ΔC-terminal 167 aa)b::phaRc::gfp phaQ phaR; Emr | This study |
| pGM54 | Derived from pGM16.2; phaP (ΔC-terminal 81 aa) gfpd phaQ phaR; Emr | This study |
| pGM82 | Derived from pGM16.2; phaP::gfp phaQ; Emr | This study |
| pGM13 | pHPS9 carrying phaPe phaQ phaR phaB phaC::gfp; Emr | 16 |
| pGM61 | Derived from pGM13; phaR (aa, 1 to 200) was deleted; Emr | This study |
| pGM73S | Derived from pGM61; phaR (aa, 1 to 200) was restored in the sense orientation under control of the phaP promoter; Emr | This study |
| pGM73AS | Derived from pGM61; phaR (aa, 1 to 200) was restored in the antisense orientation under control of the phaP promoter; Emr | This study |
| pGM89 | Derived from pGM13; phaPe phaQ phaR ΔphaB phaC::gfp; Emr | This study |
| pGM78 | phaR cloned into the BamHI and HindIII sites of pET-21(+); Ampr | This study |
ATCC, American Type Culture Collection; aa, amino acids.
The first two codons of PhaP are fused out of frame to gfp.
Amino acids 33 to 200 (C terminus) of PhaR.
Out-of-frame fusion to GFP with alternative translation initiation of GFP.
C-terminal 29 amino acids deleted.
Analysis of PHA inclusion body-associated proteins.
PHA inclusion bodies were purified on sucrose gradients (17), and sodium dodecyl sulfate-polyacrylamide gel electrophoresis (SDS-PAGE) analysis of PHA inclusion body-associated proteins was carried out as previously described (16).
Construction of plasmids.
Plasmids (Table 1; Fig. 1) were constructed by standard procedures (2) and confirmed by restriction analysis and sequencing.
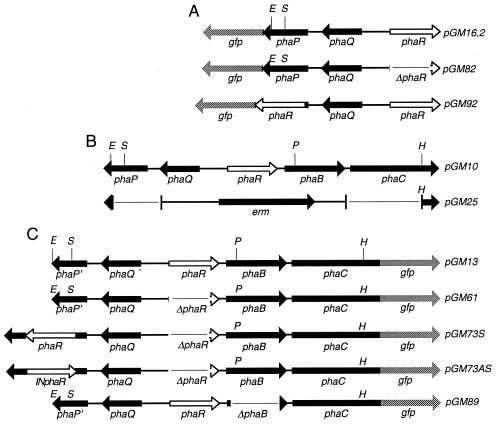
FIG. 1.
Maps of engineered plasmid inserts used in this study. Relevant restriction sites: E, EcoRI; H, HindIII; P, PstI; S, SnaBI. (A) Plasmids used to test the effect of PhaR on phaP expression (pGM82) and localization of PhaR (pGM92). (B) Plasmids used to construct PHA05. (C) Plasmids used to determine the effect of PhaR on PHA accumulation and PHA synthase activity.
pGM25, which was used to make the PHA-negative mutant strain PHA05, was constructed in three steps, starting with the modification of pGM10 to produce pGM10.2 followed by pGM23 and pGM25. pGM10.2 was constructed by removing the EcoRI site located in the vector (pHPS9) part of pGM10 through partial EcoRI digestion and treatment with T4 DNA polymerase, leaving intact the EcoRI site in the insert part of pGM10. To construct pGM23, the 1.5-kb HindIII-to-SmaI DNA fragment obtained from pGM29 harboring the erm marker was ligated to the 7.1-kb EcoRI (T4 DNA polymerase treated)-to-HindIII sites of pGM10.2. pGM25 was constructed by ligating a 4-kb EcoRI fragment (treated with T4 DNA polymerase) harboring a temperature-sensitive origin of replication from pLTV1 into the SmaI site of pGM23. pGM25 cannot replicate at 42°C in B. megaterium.
The plasmid pGM92, used to localize PhaR, was constructed in two steps, starting with pGM16.2 followed by pGM72b and then pGM92. In this plasmid, a translational fusion of two amino acids at the N terminus of PhaP was fused to residues 30 to 199 of PhaR, which was fused at its C terminus to green fluorescent protein (GFP). Transcriptional control of this chimera was under the direction of the phaP promoter. To construct pGM72b, an out-of-frame deletion of phaP in pGM16.2 was made using the ExSite PCR-based mutagenesis system (Stratagene) with primers 78 (5′-GATATCGGTGACAAAATGAGTAAAGG [the underline designates an EcoRV site]), located on the sense strand in the intergenic region spanning phaP and gfp, and 77 (5′-TGACATAATCAAATTCCTCC), located on the antisense strand at the 5′ end of phaP. A 512-bp DNA fragment spanning codons 30 through 199 of phaR was amplified with primers 85 (5′-GGGACAATATGAATCGTGAAG), located at the 5′ end of phaR, and 86 (5′-CTTGCGAGCCGGCTGCTCGA), located at the 3′ end of phaR, and cloned into the EcoRV site of pGM72b, resulting in the in-frame gene fusion phaP1–2::phaR30–199::gfp.
pGM82, which was used to test the effect of PhaR on phaP expression, was constructed by deleting phaR from pGM16.2. The ExSite PCR-based mutagenesis system was used with a pGM16.2 template and primers 33 (5′-CTCCATCTCCTTTCTTGTG), located on the antisense strand at the 5′ end of phaR, and 67 (5′-CGCAAGTAAGGTATCGGAG), located on the sense strand at the 3′ end of phaR. This resulted in a deletion of 594 bp spanning the 5′ end of phaR to codon 197.
The plasmids pGM13, pGM61, pGM73S, pGM73AS, and pGM89 were used to examine the influence of PhaR on PHA accumulation. To construct pGM61, a 594-bp deletion in phaR was made in pGM13 (16) using the ExSite PCR-based mutagenesis system with primers 33 (5′-CTCCATCTCCTTTCTTGTG), located on the antisense strand at the 5′ end of phaR, and 67 (5′-CGCAAGTAAGGTATCGGAG), located on the sense strand at the 3′ end of phaR. The resulting deletion spanned the 5′ end of phaR to codon 197. To construct pGM73S and pGM73AS, a 663-bp DNA fragment carrying phaR, including its putative translation signals, was cloned in frame into the SnaBI site in phaP. The 663-bp fragment was generated using PCR with primers 72 (5′-GTCCATATTGAAAGCCATAC [the underline indicates a stop codon]), located at the 5′ end of phaR on the sense strand, and 74 (5′-CTCCGATCCTTACTGCG [the underline indicates a stop anticodon]), located at the 3′ end of phaR on the antisense strand. pGM73S carries the insert in the same orientation as phaP, whereas it is in the opposite orientation in pGM73AS. To construct pGM89, a 635-bp phaB deletion was made in pGM13 using the ExSite PCR-based mutagenesis system along with primers 36 (5′ CCCCGATACCTTAGATCCGCC), located on the antisense strand at the 5′ end of phaB, and 79 (5′-GCGTACATTACAGGACAACAG), located on the sense strand at the 3′ end of phaB. The resulting deletion removed codons 20 through 231of phaB.
Plasmid pGM78 is a C-terminal His6-tagged fusion protein for the purification of PhaR. To construct pGM78, the template pGM16.2 was used along with primers 91 (5′-TAAGGATCCACAAGAAAAATGGAG [the underline designates a BamHI site]), located on the sense strand at the 5′ end of phaR, and 92 (5′-TTCAAGCTT/GCGAGCGGCTGCTC [the underline designates a HindIII site]), located on the antisense strand at the 3′ end of phaR, to amplify phaR, including its predicted coding region and translation signal. In pGM78, transcription of phaR was under the control of the vector-derived isopropyl-β-d-thiogalactopyranoside (IPTG)-inducible T7 promoter. Translation was dependent upon the native phaR translation signal. Also, PhaR formed an in-frame fusion to the N terminus of the vector-derived His6 sequence, where the PhaR stop codon was replaced by a multiple cloning site and His sequences from pET-21(+).
pGM54, which was used as a localization control, had the gfp gene transcriptionally, but not translationally, fused to a truncated phaP; hence, gfp expression was under the control of the phaP promoter and second-site translation initiation.
Construction of B. megaterium PHA05.
In PHA05, a PHA-negative mutant of strain 11561, the pha genes from the EcoRI site in phaP to the HindIII site in phaC were replaced with an ERY resistance gene using a double-crossover homologous recombination strategy (Fig. 1). Strain 11561(pGM25) was plated on ERY-containing medium following growth in LB medium at 30°C, the permissive temperature, or 42°C, the nonpermissive temperature for replication of the plasmid. Sectors of colonies with a less opaque appearance were examined for absence of PHA accumulation. Several independent isolates of PHA-negative mutants were subcultured. ERY marker substitution was confirmed by Southern blotting.
Transformations.
E. coli transformations were carried out using electroporation according to the manufacturer's instructions (Electroporator 2510; Eppendorf). B. megaterium was transformed based on previously published methods, with several modifications (27, 30). Twenty-milliliter cultures were grown from 1% inocula of fresh overnight cultures in 250-ml flasks at 35°C and 250 rpm to an optical density (OD) at 660 nm of 0.40. Protoplast preparation was at room temperature. Cells were harvested and resuspended in 2 ml of RHAF. (RHAF consists of [per liter] the following: NH4Cl, 1.0 g; Tris base, 12.0 g; KCl, 35 mg; NaCl, 58 mg; Na2SO4 · 10H2O, 300 mg; KH2PO4, 140 mg; MgCl2 · 5H2O, 4.26 g; yeast extract, 5 g; tryptone [Difco], 5 g; sucrose, 68.46 g; and 20% glucose, 10 ml. The pH was adjusted to 7.5 with HCl before addition of MgCl2 · 5H2O. Solid media contained 1% agar.) Protoplasts were formed by treatment with 600 μg of lysozyme per ml for 15 min. Protoplasts were pelleted at 1,000 × g for 5 min, washed gently in 2 ml of RHAF, pelleted again, and resuspended in 1 ml of RHAF. For transformation, 200 μl of protoplast suspension, up to 5 μg in 10 μl of plasmid DNA, and 200 μl of 35% polyethylene glycol (molecular weight, 8,000) prepared in RHAF were mixed gently and incubated at 37°C for 3 min, followed by immediate dilution with 3.0 ml of RHAF and pelleting at 1,000 × g for 5 min. For protoplast recovery, the pellet was resuspended in 1.0 ml of RHAF, incubated for 1.5 h at 37°C, plated on nonselective RHAF with 1.0% agar in 100-mm-diameter plates (350 μl per plate), and incubated overnight at 30°C. Recovered colonies were washed off the plates with 5 ml of LB medium and transferred to LB plates with appropriate selection.
Whole-cell extract preparations for activity assay and SDS-PAGE analysis.
A 200-ml culture in LB medium plus CHL (6 μg/ml) grown from a 1% inoculum (in similar medium) for 9 h at 35°C at 250 rpm was harvested at 9,000 × g at 4°C, washed in 100 ml of 20 mM Tris-HCl (pH 8.0), and finally resuspended in 20 ml of 20 mM Tris-HCl (pH 8.0). The 10× (relative to the original culture volume) cell suspension was subjected to a single French press passage at 12,000 lb/in2 at 4°C. The resulting 10× whole-cell extracts were used immediately for determination of PHA synthase activity and Western blotting (see below).
PHA synthase activity assay.
PHA synthase activity in 10× whole-cell extracts was determined with a discontinuous assay based on a previously published method that uses dithionitrobenzoic acid (DTNB) to monitor levels of free CoA (4, 13, 18). The reaction was carried out at 28°C in a final volume of 1 ml with the following components: 100 mM Tris-HCl, pH 8.0; 1 mM hydroxybutyryl-CoA substrate; and 40 μl of 10× whole-cell lysate. Fifty-microliter samples were removed at various time points and immediately quenched with 50 μl of 1.0% trichloroacetic acid in 100 mM Tris-HCl, pH 8.0. Samples were centrifuged for 1 min to remove particulate material, 95 μl of the supernatant was removed and combined with 505 μl of 100 mM Tris-HCl (pH 8.0) containing 1 mM DTNB, and absorbance at 412 nm was measured. The concentration of CoA was calculated using the extinction coefficient (412 nm) of 13,600 cm−1 M−1. One unit was defined as the amount of enzyme required to convert 1 μmol of substrate in 1 min.
Immunoblotting.
For each 10× whole-cell extract sample, approximately 30 μg of total cell protein was combined with 0.5 volume of 3× sample buffer (188 mM Tris-HCl [pH 6.8], 6% SDS, 30% glycerol, 0.03% bromphenol blue) and heated at 95°C for 5 min. Samples were briefly centrifuged, and the supernatant was loaded onto 12% polyacrylamide gels (Bio-Rad 161-0158; 37.5:1 acrylamide to bisacrylamide). Polypeptides were resolved at 15 V/cm for 2.5 h. Duplicate gels were prepared, one for Coomassie blue staining and the other for Western blotting, for which the protein was transferred to a polyvinylidene difluoride (PVDF) membrane (Qiagen). Immunoblotting was as follows. The PVDF membrane was washed for 5 min in phosphate-buffered saline (PBS) (pH 7.5), followed by incubation in 6% nonfat dairy milk for 2 h and three washes in PBS. The blot was incubated for 3 h in a 1/100 dilution of the primary antibody (anti-GFP; Clontech no. 8367-1) in PBS plus 0.5% bovine serum albumin (BSA) (A-7906; Sigma), rinsed five times in PBS–0.5% BSA, incubated for 1 h with a 1/1,000 dilution of the secondary antibody (goat anti-rabbit immunoglobulin G complexed with alkaline phosphatase) (catalog no. 111-055-045; Jackson Immuno Research), and rinsed five times in PBS–0.5% BSA followed by distilled water for 10 s. Bound antibody was detected using Sigma Fast 5-bromo-4-chloro-3-indolylphosphate–nitroblue tetrazolium alkaline phosphatase substrate tablets (B-5655) dissolved in distilled water. Between 1 and 2 min was sufficient for signal detection, which was quantified using a digital image of the blot and UN-SCAN-IT gel digitization software (Silk Scientific, Orem, Utah).
Expression, purification, and N-terminal identification of PhaR.
E. coli BLR DE3(pGM78) in log phase was induced with 1 mM IPTG for 3 h. The culture was harvested by centrifugation at 9,000 × g and resuspended in 1/10 volume of 20 mM Tris-HCl, pH 8.0. The cell suspension was subjected to two French press passes at 11,000 lb/in2. The whole-cell extracts were centrifuged at 17,000 × g to separate the soluble and insoluble proteins. SDS-PAGE analysis of the soluble and insoluble fractions indicated that the PhaR-His6 product was in the insoluble fraction, presumably as protein inclusions. As previously described, N-terminal sequences were identified by Edman degradation using at least 200 pmol of protein transferred to PVDF following separation on SDS–15% polyacrylamide gels (16).
Microscopy and image analysis.
PHA inclusion bodies were viewed either in phase contrast or stained with Nile Blue A (19). Phase-contrast microscopy was carried out on samples either prepared as wet mounts or embedded in 1.0% low-melting-point agarose at ×400 and ×1,000 magnifications. To view GFP, samples were prepared in 1.0% low-melting-point agarose at ×400 and ×1,000 magnifications under fluorescence (excitation, 390 to 450 nm; barrier filter, 480 to 520 nm; dichroic mirror, 470 nm). A Nikon Labophot-2 microscope with phase-contrast and fluorescence attachments was used. Images were acquired with a SPOT cooled color digital camera (Diagnostic Instruments, Inc.) and prepared using Adobe Photoshop version 5.0. Standardized exposure times were used for comparative data unless otherwise stated. Exposure times were in the range of 1.5 to 12 s.
RESULTS
PhaR.
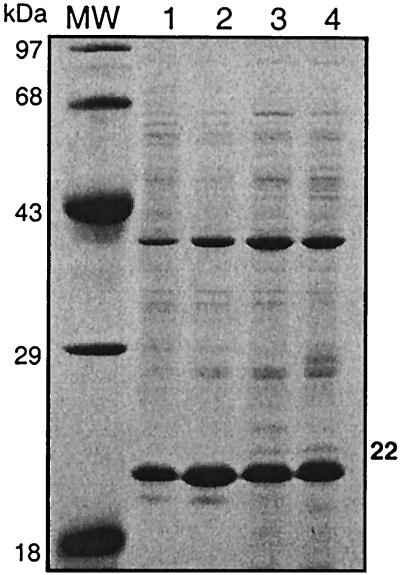
The phaR gene was originally identified in B. megaterium as the first open reading frame in the phaRBC operon, where the start of transcription was 182 bp upstream from the translational start of the putative PhaR. Also, a 22-kDa minor band (relative to PhaC) coinciding with the estimated size of PhaR was identified on SDS-polyacrylamide gels of proteins that copurified with PHA inclusion bodies (16). To further examine the putative PhaR, proteins stripped from PHA inclusion bodies, which were isolated from cells at different time points representing different growth phases, were run on SDS-polyacrylamide gels as previously described. The results showed that PhaC and PhaP were the two most abundant proteins that copurify with the PHA inclusion bodies at all time points. Several other proteins of much lower abundance were also present. These included a protein of about 22 kDa, the estimated size of PhaR, detected only in stationary-phase cells (Fig. 2). The abundance of PhaC relative to the 22-kDa protein on purified inclusion bodies suggests that they are not present in the cell in equimolar quantities. However, loss could have occurred during the purification process.
FIG. 2.
SDS-PAGE analysis of proteins that copurified with PHA inclusion bodies from cells harvested at different phases of growth. Strain 11561 was grown for 5 (OD at 600 nm, 0.5) (lane 1), 9 (OD, 1.0) (lane 2), 11 (OD, 2.0) (lane 3), and 24 (OD, 2.2) (lane 4) h under identical conditions in LB liquid medium, followed by purification of PHA inclusion bodies by density centrifugation using sucrose gradients. Inclusion bodies were stripped of protein using 2% SDS at 95°C, equivalent amounts (10 μg) of total protein from clarified soluble fractions were subjected to electrophoresis on a 12% polyacrylamide gel, and bands were visualized by staining with Coomassie blue. Molecular masses (MWs) of markers are indicated at the left of the gel, and the weak 22-kDa polypeptide band referred to in the text is indicated at the right of the gel.
N-terminal sequences of PhaR.
N-terminal sequences of PhaR are shown in Fig. 3. SDS-polyacrylamide gels of whole-cell extracts of E. coli(pGM78) showed two bands (data not shown) corresponding to the approximate size of PhaR: one at about 22 kDa and a smaller, less abundant band (about 50% abundance relative to the former band) at 19.5 kDa. Their N-terminal amino acid sequences were MEQQKVFDPF and MNREEFSQL, respectively. The larger protein is associated with a strongly conserved Shine-Dalgarno sequence and more optimal spacing than the smaller, less abundant protein, which starts at amino acid 32 of the larger protein and has a poorly conserved Shine-Dalgarno sequence. These data prove the existence of PhaR but do not indicate if one or both sizes occur in B. megaterium.
FIG. 3.
N-terminal and deduced amino acid sequences of PhaR. The black and boxed regions indicate the N-terminal amino acid sequences obtained for the 22- and 19.5-kDa PhaR-His tag products, respectively. The nucleotide sequence shown extends from −21 bp upstream of the TTG start codon of the 22-kDa PhaR-His6 product through codon 44 (GTG). Putative translation signals showing similarity to a Shine-Dalgarno sequence (AAGGAGG) are underlined upstream of the TTG (22-kDa protein) and ATG (19.5-kDa protein) start codons, beginning at nucleotides −9 and −6, respectively. The sequence is corrected for a frameshift error in the original published sequence, which resulted in a change in the first nine residues only. The corrected sequence is available under GenBank accession number AF109909.2.
PhaR localizes to PHA inclusion bodies in B. megaterium.
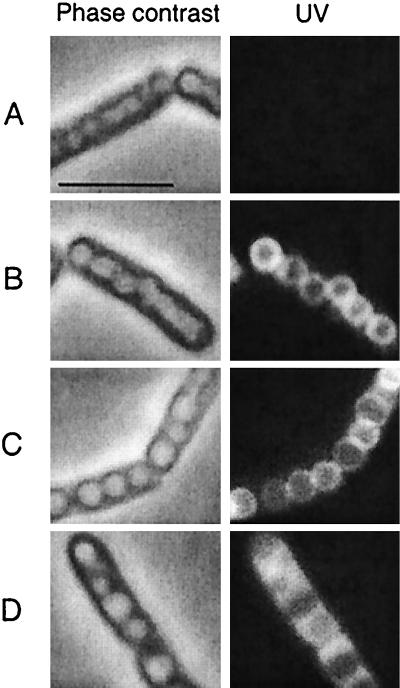
As a first step in identifying a function for PhaR, localization in living cells of B. megaterium was examined using a translational fusion of PhaR to GFP (pGM92). Cells from colonies and from all growth phases in liquid medium showed that PhaR-GFP localized as distinct rings of fluorescence that coincided with the perimeters of PHA inclusion bodies (Fig. 4). This localization pattern was similar to that previously reported for PhaP-GFP (16) and was distinctly different from the GFP nonlocalization control (pGM54). Fluorescence was not detected in the GFP-negative control, which was grown and examined under identical conditions. These results demonstrate that PhaR was localized to PHA inclusion bodies in living cells but was lost or mostly lost from purified inclusion bodies (see above). These data further strengthen the case for PhaR involvement in PHA accumulation.
FIG. 4.
Localization of PhaR-GFP to PHA inclusion bodies in plasmid-bearing 11561 strains. Cells were sampled from cultures grown on solid LB medium with CHL (6 μg/ml) for 16 h. The left and right columns show the same single cells in phase contrast and under fluorescence, respectively. The strains carry the following plasmids: pHPS9 (GFP negative control) (A), pGM16.2 (PhaP-GFP, GFP localization control) (B), pGM92 (PhaR-GFP) (C), and pGM54 (GFP nonlocalization control) (D). Bar, 5 μm.
PhaR is not required for expression of phaP or phaRBC.
One possible role for PhaR is that of a regulator of transcription of other genes in the pha cluster. To test the possible effect of PhaR on phaP expression, PhaP fused to the reporter GFP, with (pGM16.2) and without (pGM82) PhaR present, was examined in E. coli and B. megaterium strains. The results showed no visible difference in fluorescence in the presence or absence of PhaR, indicating that it is not a regulator of phaP expression. The effect of PhaR on phaQ expression was not separately tested, since phaQ is a transcriptional regulator of phaP expression (G. J. McCool and M. C. Cannon, unpublished data); therefore, an effect of PhaR on phaQ expression would have been observed as a phenocopy of phaP, and no effect was found. Using a similar strategy, the effect of PhaR on expression of phaRBC was tested. As was the case with PhaP expression, the results showed that PhaR was not required for expression of PhaC (see below for details).
PhaR is required for PHA accumulation.
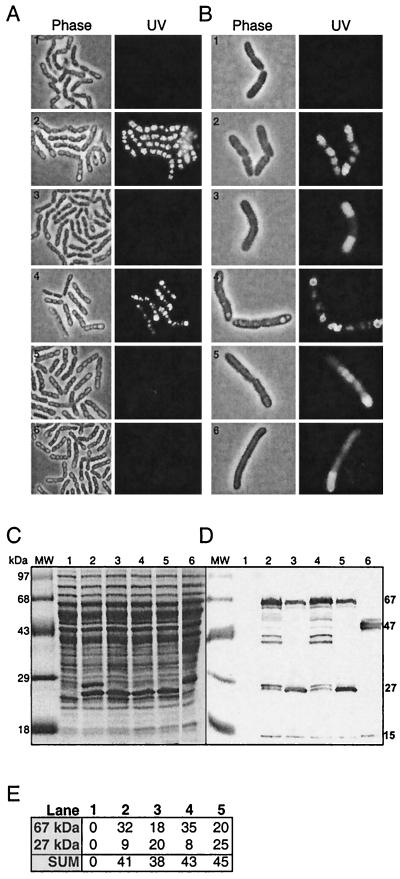
To examine a possible function of PhaR in both expression of phaC and activity of the gene product, a phaPQRBC deletion strain of B. megaterium, strain PHA05, was constructed for the purpose of reintroducing pha genes in desired combinations. Apart from a PHA-negative phenotype and resistance to ERY, PHA05 showed no apparent phenotypic difference and had the same growth rate as its progenitor, strain 11561 (data not shown). PHA05 carrying either pGM13 (phaPQRBC::gfp) or pGM61(phaPQBC::gfp) synthesized PhaC-GFP (Fig. 5B), indicating that PhaR is not required for expression of the phaRBC operon. In contrast to pGM13, however, pGM61 did not complement strain PHA05 with respect to PHA accumulation (Fig. 5A). No PHA was observed at any point during growth in batch cultures. This result shows that PhaC-GFP, although present, was unable to synthesize PHA in the absence of PhaR, which indicates that PhaR functions at the activity level of PHA synthase, either directly or indirectly. Since the absence of PHA accumulation could also be explained by a mutation in PhaC of pGM61, this finding was corroborated by purifying pGM61 from PHA05 and cloning the phaR gene, including the phaR coding region and translation signal, into the conveniently available SnaBI site in phaP. This generated a transcriptional fusion of phaP and phaR. The cloning was done in both sense (pGM73S) and antisense (pGM73AS) orientations to provide an additional negative control for the experiment. Following transformation of PHA05 with each of these plasmids, pGM73S was found to complement the PHA-negative B. megaterium strain PHA05, while pGM73AS did not complement, thus confirming the result that PhaR is required for PHA synthase activity. Western blotting confirmed the presence of PhaC-GFP in the strains tested (Fig. 5D). Interestingly, the PHA inclusion bodies in PHA05(pGM73S) were larger than those of both the wild type and PHA05(pGM13). This larger inclusion body size is in keeping with results obtained with phaP deletion strains (McCool and Cannon, unpublished results).
FIG. 5.
Effect of PhaR on PHA accumulation and phaC expression. PHA05 harboring various plasmids was grown and harvested under identical conditions. (A and B) Cells were examined for the presence of PHA inclusion bodies using phase-contrast and fluorescence microscopy (A) and for expression of PhaC-GFP and its localization using phase-contrast and fluorescence microscopy (B). Row 1, pHPS9; row 2, pGM13; row 3, pGM61; row 4, pGM73S; row 5, pGM73AS; row 6, pGM89. (C) Whole-cell extracts were prepared from all cultures under identical conditions, and 30-μg aliquots of total cell protein were loaded onto SDS–12% polyacrylamide gels. Bands were visualized with Coomassie blue stain. Molecular mass (MW) marker sizes are indicated on the left. (D) Western blotting was performed on a duplicate gel using anti-GFP antibody. Specific polypeptides detected by anti-GFP antibody as described in the text are indicated on the right in kilodaltons. For panels C and D: lane 1, pHPS9; lane 2, pGM13; lane 3, pGM61; lane 4, pGM73S; lane 5, pGM73AS; lane 6, pGM16.2. (E) Densitometry results for the 67- and 27-kDa bands in panel D, showing the total pixel values in lanes 1 to 5 and their sums (104).
PhaR is required for PHA synthase activity.
The results described above show that PhaR and PhaC are required for synthase activity. However, in vivo studies gave no indication as to the role played by PhaR. To gain further insights, PhaR and PhaC were tested for synthase activity in vitro. Extracts of the PHA-negative mutant PHA05 carrying pGM13, pGM61, or pGM73S were tested, along with appropriate controls (Table 2). The results were that extracts from PHA05 carrying pGM61, the PhaR-negative strain, showed no synthase activity, while PHA05 carrying pGM13 or pGM73S was positive. This is consistent with the in vivo results described above, where in the absence of PhaR, PhaC was synthesized but PHA did not accumulate. In vitro complementation assays were negative for PHA synthase activity when extracts of E. coli or strain PHA05 carrying pGM16.2 or pGM78 as a source of PhaR were used along with extracts of PHA05(pGM61). These results show that PhaC and PhaR had to be produced in the same cell to obtain synthase activity. We reasoned that the basis of this result could be that (i) PhaR and/or PhaC is in the active form only in PHA-accumulating cells and/or (ii) PhaR is instrumental in producing an unknown product required to activate PhaC or required along with PhaC for synthase activity. The hypothesis was tested by carrying out an in vitro assay of an extract from PHA05(pGM89). This plasmid had its phaB (acyl-CoA reductase) gene, an essential gene in the PHA biosynthetic pathway, deleted, and in its absence no PHA was produced (Fig. 5A). In vivo experiments that monitored PhaC-GFP (Fig. 5B) and Western blots of the extracts (data not shown) showed that PhaC was present in the cells, indicating expression of the phaRC operon. However, no PHA synthase activity was detected in the extracts. This result could also be explained by a mutation in phaC or phaR occurring during the plasmid construction process. This possibility was ruled out by confirming the sequence of pGM89, thus substantiating that PhaC and/or PhaR has synthase activity only in PHA-accumulating cells. This experiment gave no indication as to whether PhaC and/or PhaR is in an active form only in the metabolic milieu of cells accumulating PHA or whether the PHA inclusion bodies per se are a necessary requirement for PHA synthase activity. The latter is a less likely explanation, since the transfer of phaR, -B, and -C to E. coli results in PHA accumulation in the absence of PHA inclusion bodies (McCool and Cannon, unpublished data). The data in Fig. 5 and Table 2 demonstrate that PhaR is required either directly or indirectly through an unknown product along with PhaC for PHA synthase activity and that activity is tightly regulated.
TABLE 2.
PHA synthase activities in whole-cell extracts of plasmid-bearing PHA05 strains
| Whole-cell extract | Relevant phenotype
|
PHA accumulation | Total protein concn (mg/ml) | Total U | Sp act (U/mg) | |||
|---|---|---|---|---|---|---|---|---|
| PHA | PhaR | PhaB | PhaC | |||||
| pHPS9 | + | − | − | − | − | 2.4 | NDa | ND |
| pGM13 | + | + | + | + | + | 3.5 | 2,100 | 600 |
| pGM61 | − | − | + | + | − | 3.8 | ND | ND |
| pGM73S | + | + | + | + | + | 2.9 | 856 | 295 |
| pGM73AS | − | − | + | + | − | 2.6 | ND | ND |
| pGM89 | − | + | − | + | − | 3.2 | ND | ND |
| pGM16.2 | − | + | − | − | − | 4.4 | ND | ND |
| pGM61, pGM16.2b | −/− | −/+ | +/− | +/− | − | 4.1 | ND | ND |
| pGM61, pGM16.2bc | −/− | −/+ | +/− | +/− | − | 3.8 | ND | ND |
| pGM61, pGM78bd | −/− | −/+ | +/− | +/− | − | 3.7 | ND | ND |
ND, no detectable activity.
Equal volumes of each extract in mixture.
pGM16.2 cell extract is E. coli DH5α(pGM16.2).
pGM78 cell extract is E. coli BLR DE3(pGM78).
Regulation of PHA synthase activity.
We showed above that phaRBC was expressed in cells at different growth phases and in the presence and absence of PHA accumulation. However, in the absence of PHA accumulation PHA synthase showed no activity, indicating that it was in an inactive form in these cells. Western blotting of proteins in extracts of the different strains used, following separation by SDS-PAGE using an antibody to GFP, gave some insights into the states of PhaC in these strains. The Western blot (Fig. 5D) showed a different pattern of PhaC-GFP fusion product sizes in the presence (lanes 2 and 4) and absence (lanes 3 and 5) of PHA accumulation. A 15-kDa product occurred in all GFP fusion samples; this is likely to be an internal breakdown product of GFP and is not further considered here. PHA-accumulating cells (lanes 2 and 4) had many PhaC-GFP fusion products, the most abundant of which were approximately 27, 27.5, 41, 43, and 67 kDa in size, while non-PHA-accumulating cells had mostly products of approximately 27 and 67 kDa. GFP alone is 27 kDa, and the PhaC-GFP fusion is 67 kDa. There was significantly more of the 67-kDa protein and less of the 27-kDa protein in PHA-accumulating cells compared to nonaccumulating cells (Fig. 5D and E). Since the gels were loaded with equal quantities of total protein, the difference in quantities of the 27- and 67-kDa proteins is significant and was reproducible, thus allowing comparisons to be made between lanes as well as within lanes. The PhaP-GFP fusion product (47 kDa) in non-PHA-accumulating cells did not show smaller GFP fusion products (Fig. 5D, lane 6), except the 15-kDa product mentioned above, indicating that the multiple bands are not a general phenomenon. The cause of the partial digestion products is not understood at this time; however, it is notable that they occurred only when the phaR and -C genes were expressed. In summary, in PHA-accumulating cells the PhaC-GFP fusion was mostly intact but with many distinct breakdown products, and PHA synthase activity was detected in vitro, while in non-PHA-accumulating cells the PhaC-GFP fusion was more degraded to the 27-kDa product, which is the size of the GFP protein alone, and PHA synthase activity was not detected. These data are consistent with PhaC existing in an inactive form in the absence of PHA accumulation and as such being more susceptible to protease degradation, leading to the absence of partial digestion products and the greater quantity of the 27-kDa protein.
The PHA synthase of B. megaterium is novel.
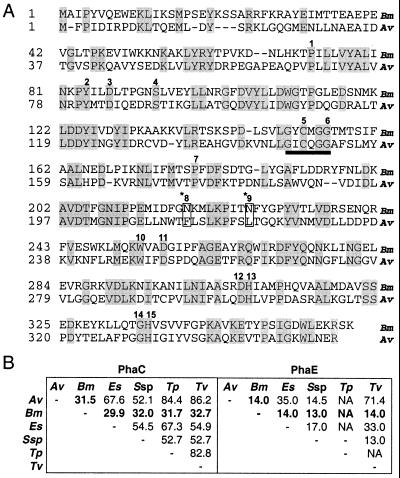
B. megaterium PhaC (PhaCBm) is 362 amino acids in length (41.5 kDa) and has the known salient features of PhaCs (Fig. 6A). It most closely resembles the PhaCs of class III PHA synthases in size and sequence, having identity to them in the range of 32.7 to 29.9% (Fig. 6B). The five known members of this class share a range of identity to each other of 82.6 to 52.1%, suggesting that PhaCBm is also distinctly different from them. PhaCBm has a putative lipase box as shown for other PhaCs (14). An alignment of 30 PhaC sequences, including the five known class III PhaC sequences, shows that there are 15 nonvariable amino acids (20). PhaCBm has 13 of these 15 amino acids. There is evidence to show that the active site in A. vinosum PhaC (PhaCAv) involved in the mechanism of polymerization includes three of these amino acids, a cysteine at position 149 (C149), a histidine at 331 (H331), and an aspartic acid at 302 (D302), which form a catalytic triad (9). In an alignment of class III PhaC sequences these amino acids correspond to C152, H336, and D307, with similar spacing, in PhaCBm (Fig. 6A). Class III PHA synthases are known to require the 40-kDa protein PhaE, as shown for A. vinosum (20). PHA synthase of B. megaterium does not require PhaE, as evidenced by the fact that the B. megaterium phaPQRBC gene cluster can synthesize PHA when transferred to E. coli and Pseudomonas putida, which are known not to have PhaE homologs (16). Furthermore, PhaC and PhaE copurify as PHA synthase in approximately equimolar quantities (18). They also copurify in equimolar quantities with PHA inclusion bodies and separate on SDS-polyacrylamide gels (12). Experiments with B. megaterium PHA synthase showed no such product present in addition to PhaC (Fig. 2). Pairwise alignments of the four available PhaE proteins with PhaR and with each other show that PhaR has an apparently insignificant homology (13 to 14% identity) to PhaEs overall, and alignments also show that they have no region of higher homology. However, Synechocystis PhaE also has very low homology (13 to 17% identity) to the other PhaEs, leaving open the possibility that PhaRBm is an orthologous replacement for PhaE.
FIG. 6.
Alignment of PhaCBm and PhaCAv and similarities between class III and B. megaterium PHA synthases. The Clustal method of Megalign (Lasergene; DNAStar) was used for all alignments. (A) Pairwise alignment of PhaCBm (Bm) (accession no. AAD05260) and PhaCAv (Av) (L01112). Black regions indicate identical residues. The 15 residues conserved among all PhaCs are indicated with a number above each residue. The asterisk and box at conserved residues 8 and 9 indicate a sequence difference in PhaCBm. Conserved residues 5, 12, and 15 form the proposed catalytic triad. (B) On the left are the results of pairwise alignments of all available class III PhaC sequences and PhaCBm expressed as percent identities; boldface highlights a comparison with PhaCBm. Av, A. vinosum (L01112); Bm, B. megaterium (AAD05260); Es, Ectothiorhodospora shaposhnikovii (AAG30259); Ssp, Synechocystis sp. (BAA17430); Tp, Thiocapsa phennegii (20); Tv, Thiocystis violacea (L01113). On the right are the results of pairwise alignments of all available PhaE sequences and PhaRBm expressed as percent identities; boldface highlights comparisons with PhaRBm. Protein accession numbers for PhaEs are as follows: Av, P45372; Bm, AAD05258; Es, AAG30260; Ssp, S77327; Tv, C48376. NA, not available.
DISCUSSION
This paper presents the results of an analysis of phaC and -R, carried on the phaRBC operon. The results showed that both PhaC and PhaR are essential for PHA accumulation and for PHA synthase activity, which is regulated by its ability to exist in active and inactive forms. PHA synthase was extracted in an active form from PHA-accumulating cells, while it was extracted in an inactive form from non-PHA-accumulating cells. Since cells have been shown to accumulate large quantities of PHA in the absence of inclusion body formation, it is reasonable to assume that the PHA synthase activity of B. megaterium responds to the metabolic milieu of the cell, which is the consequence of the cell's physiological status. This would be expected to differ in PHA-accumulating and nonaccumulating cells. PHA inclusion bodies not only are a storage system but also are considered to be a redox regulator within the cell (22). The ability of PHA synthase to exist in two different forms would allow it the capacity to respond rapidly to a change in its intracellular environment, which in turn could respond to the availability of nutrients and other environmental factors. Our results showing a requirement for PhaC and PhaR are in keeping with earlier results on PHA synthase in extracts of B. megaterium (6). Those authors found that following extraction of PHA inclusion bodies, both the supernatant and inclusion body fractions were required for synthase activity.
More recent studies on the PHA synthase of A. vinosum suggest that it may also exist in active and inactive forms. Liebergesell et al. (13) detected no activity after combining separate extracts of PhaCAv and PhaEAv derived from E. coli strains. However, the DTNB used in their continuous assay may have had an inhibitory effect (18). Using the discontinuous DTNB assay, Müh et al. (18) detected an extremely low level of activity (0.9 U/mg) with His-tagged PhaCAv purified from E. coli compared to that of the purified complex (150 U/mg). Complementing with a 10-fold excess of His-tagged PhaEAv gave about 50% of the activity of the purified complex. Since none of the data were obtained with enzymes extracted from A. vinosum, whose metabolic milieu may differ from that of E. coli, the data are consistent with PhaCAv and/or PhaEAv being in an inactive state in extracts from cells producing either one or the other protein. In addition, the inactivity or low activity of PhaCAv, or the excess of PhaE required for activity, is consistent with PhaEAv existing mostly in an inactive form. Taken together, these data are consistent with PhaE being a regulator of PhaC activity. In the B. megaterium system no activity was detected using the discontinuous DTNB assay when PhaCBm and PhaR were combined. PhaR localized to inclusion bodies, yet it did not copurify with inclusion bodies except perhaps to a very low level. These results are consistent with PhaR being an activator of PhaC, and as such a proportion of PhaR could be bound transiently to PhaC. Should PhaR-GFP be diffuse in the cytoplasm, it would be less detectable than protein concentrated at the surface of the inclusion body, and this may explain the localization results obtained with PhaR.
A mechanism of action has been proposed for PhaC of R. eutrophus (PhaE not required) (33) and A. vinosum (PhaE required) (9); however, a role for PhaE has not been described. Recently, class III PhaCs were reclassified with the αβ-hydrolase family of lipases, and a structure for PhaCAv was proposed (9). This is based on bacterial lipases for which open (active) and closed (inactive) structures have been determined (21). When we structurally aligned the two lipases (The Protein Data Bank accession nos. 5LIP and 1CVL) with each other using the combinatorial extension method (23), it was evident that there are two moving helices in the protein: helix 5 blocks the active site in the closed structure, and helix 6 (10 amino acids downstream) also moves. Interestingly, the two variant amino acids of PhaCBm occur in helix 6 of the structural model of PhaCAv, equivalent to the lipases' helix 6. In comparison to all known PhaCs, where there are 15 invariant amino acids with similar spacing in multiple alignments, PhaCBm has replaced two hydrophobic residues, F213 and L221 (PhaCAv numbering), with two positively charged residues, N218 and N226 (PhaCBm numbering). Since their positions are close together and are associated with helix 6 in the proposed structure of PhaCAv, it is reasonable to speculate that these two amino acids are instrumental in differentiating PhaCBm from other synthases, particularly in the requirement for PhaR.
The results of this paper are consistent with PhaR being an orthologous replacement for PhaE and with PhaR and/or PhaC having active and inactive forms. The results show that PhaC is more rapidly degraded in non-PHA-accumulating cells. The interactions of PhaC with PhaR or an unknown product that may be regulated by PhaR must now be examined to determine how one influences the other to achieve an active or inactive form of PHA synthase. A comparison of PhaCBm with other PhaCs has already been used in determining the proposed mechanism of action of PhaCAv (9), and it is very likely that the differences between other PHA synthases and that of B. megaterium will provide further insights into the mechanism of action and its regulation. The results of this study are consistent with PHA synthase of B. megaterium being significantly different from all known synthases, and since it is the first PHA synthase of the genus Bacillus to be studied at the molecular level, it is possibly the founding member of a new class of synthases.
ACKNOWLEDGMENTS
We thank Frank Cannon for valuable discussions and Shiming Zhang for help with the in vitro assays.
This research was supported by a grant from the National Science Foundation (MCB-9905419).
REFERENCES
- 1.Anderson A J, Dawes E A. Occurrence, metabolism, metabolic role, and industrial uses of bacterial polyhydroxyalkanoates. Microbiol Rev. 1990;54:450–472. doi: 10.1128/mr.54.4.450-472.1990. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Ausubel F M, Brent R E, Kingston D D, Moore J G, Seidman J G, Smith J G, Struhl K. Current protocols in molecular biology. New York, N.Y: John Wiley & Sons, Inc.; 1998. [Google Scholar]
- 3.Braunegg G, Lefebvre G, Genser F K. Polyhydroxyalkanoates: biopolyesters from renewable resources: physiological and engineering aspects. J Biotechnol. 1998;65:127–161. doi: 10.1016/s0168-1656(98)00126-6. [DOI] [PubMed] [Google Scholar]
- 4.Ellman G L. Tissue sulfhydryl groups. Arch Biochem Biophys. 1959;82:70–77. doi: 10.1016/0003-9861(59)90090-6. [DOI] [PubMed] [Google Scholar]
- 5.Fukui T, Doi Y. Cloning and analysis of the poly(3-hydroxybutyrate-co-3-hydroxyhexanoate) biosynthesis genes of Aeromonas caviae. J Bacteriol. 1997;179:4821–4830. doi: 10.1128/jb.179.15.4821-4830.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Griebel R, Merrick M. Metabolism of poly-β-hydroxybutyrate: effect of mild alkaline extraction on native poly-β-hydroxybutyrate granules. J Bacteriol. 1971;108:782–789. doi: 10.1128/jb.108.2.782-789.1971. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Haima P, van Sinderen D, Scholting H, Bron S, Venema G. Development of β-galactosidase α-complementation system for molecular cloning in Bacillus subtilis. Gene. 1990;86:63–69. doi: 10.1016/0378-1119(90)90114-7. [DOI] [PubMed] [Google Scholar]
- 8.Huang A H C. Oil bodies and oleosins in seeds. Annu Rev Plant Physiol Plant Mol Biol. 1992;43:177–200. [Google Scholar]
- 9.Jia Y, Kappock J T, Frick T, Sinskey A, Stubbe J. Lipases provide a new mechanistic model for polyhydroxybutyrate (PHB) synthases: characterization of the functional residues in Chromatium vinosumPHB synthase. Biochemistry. 2000;39:3927–3934. doi: 10.1021/bi9928086. [DOI] [PubMed] [Google Scholar]
- 10.Lee S Y. Bacterial polyhydroxyalkanoates. Biotechnol Eng. 1995;49:1–14. doi: 10.1002/(SICI)1097-0290(19960105)49:1<1::AID-BIT1>3.0.CO;2-P. [DOI] [PubMed] [Google Scholar]
- 11.Liebergesell M, Mayer J, Steinbüchel A. Analysis of polyhydroxyalkanoic acid-biosynthetic genes of anoxygenic phototrophic bacteria reveals synthesis of a polyester exhibiting an unusual composition. Appl Microbiol Biotechnol. 1993;40:292–300. [Google Scholar]
- 12.Liebergesell M, Schmidt B, Steinbüchel A. Isolation and identification of granule-associated proteins relevant for poly(hydroxyalkanoic acid) biosynthesis in Chromatium vinosumD. FEMS Microbiol Lett. 1992;99:227–232. doi: 10.1016/0378-1097(92)90031-i. [DOI] [PubMed] [Google Scholar]
- 13.Liebergesell M, Sonomoto K, Madkour M, Mayer F, Steinbüchel A. Purification and characterization of the poly(hydroxyalkanoic acid) synthase from Chromatium vinosumand localization of the enzyme at the surface poly(hydroxyalkanoic acid) granules. Eur J Biochem. 1994;226:71–80. doi: 10.1111/j.1432-1033.1994.tb20027.x. [DOI] [PubMed] [Google Scholar]
- 14.Liebergesell M, Steinbüchel A. Cloning and nucleotide sequences of genes relevant for biosynthesis of poly(3-hydroxybutyric acid) in Chromatium vinosum strainD. Eur J Biochem. 1992;209:135–150. doi: 10.1111/j.1432-1033.1992.tb17270.x. [DOI] [PubMed] [Google Scholar]
- 15.Madison L L, Huisman G W. Metabolic engineering of poly(3-hydroxyalkanoates): from DNA to plastic. Microbiol Mol Biol Rev. 1999;63:21–53. doi: 10.1128/mmbr.63.1.21-53.1999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.McCool G J, Cannon M C. Polyhydroxyalkanoate inclusion body-associated proteins and coding region in Bacillus megaterium. J Bacteriol. 1999;181:585–592. doi: 10.1128/jb.181.2.585-592.1999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.McCool G J, Fernandez T, Li N, Cannon M C. Polyhydroxyalkanoate inclusion-body growth and proliferation in Bacillus megaterium. FEMS Microbiol Lett. 1996;137:41–48. [Google Scholar]
- 18.Müh U, Sinskey A J, Kirby D P, Lane W S, Stubbe J. PHA synthase from Chromatium vinosum: cysteine 149 is involved in covalent catalysis. Biochemistry. 1999;38:826–837. doi: 10.1021/bi9818319. [DOI] [PubMed] [Google Scholar]
- 19.Ostle G A, Holt J G. Nile blue A as a fluorescent stain for poly-β-hydroxybutyrate. Appl Environ Microbiol. 1982;44:238–241. doi: 10.1128/aem.44.1.238-241.1982. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Rehm B H A, Steinbüchel A. Biochemical and genetic analysis of PHA synthases and other proteins required for PHA synthesis. Int J Biol Macromolecules. 1999;25:3–19. doi: 10.1016/s0141-8130(99)00010-0. [DOI] [PubMed] [Google Scholar]
- 21.Schrag J D, Li Y, Cygler M, Lang D, Burgdorf T, Hecht H J, Schmid R, Schomburg D, Ryder T J, Oliver J D, Strickland L C, Dunaway C M, Larson S B, Day J, McPherson A. The open conformation of a Pseudomonas lipase. Structure. 1997;5:187–190. doi: 10.1016/s0969-2126(97)00178-0. [DOI] [PubMed] [Google Scholar]
- 22.Senior P J, Dawes E A. Poly-β-hydroxybutyrate biosynthesis and the regulation of glucose metabolism in Azotobacter beijerinckii. Biochem J. 1971;125:55–66. doi: 10.1042/bj1250055. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Shindyalov I N, Bourne P E. Protein structure alignment by incremental combinatorial extension (CE) of the optimal path. Protein Eng. 1998;11:739–747. doi: 10.1093/protein/11.9.739. [DOI] [PubMed] [Google Scholar]
- 24.Steinbüchel A. Polyhydroxyalkanoic acids. In: Byrom D, editor. Biomaterials, novel materials from biological sources. Basingstoke, England: Macmillan Publishers Ltd.; 1991. pp. 123–213. [Google Scholar]
- 25.Steinbüchel A, Aerts K, Babel W, Follner C, Liebergesell M, Madkour M H, Mayer F, Pieper-Furst U, Pries A, Valentin H E, Wieczorek R. Considerations on the structure and biochemistry of bacterial polyhydroxyalkanoic acid inclusions. Can J Microbiol. 1995;41(Suppl. 1):94–105. doi: 10.1139/m95-175. [DOI] [PubMed] [Google Scholar]
- 26.Steinbüchel A, Valentin H E. Diversity of bacterial polyhydroxyalkanoic acids. FEMS Microbiol Lett. 1995;128:219–228. [Google Scholar]
- 27.Tao Y-J, Vary P S. Isolation and characterization of sporulation lacZ fusion mutants of Bacillus megaterium. J Gen Microbiol. 1991;137:797–806. [Google Scholar]
- 28.Valentin H E, Broyles D L, Casagrande L A, Colburn S M, Creely W L. PHA production, from bacteria to plants. Biol Macromolecules. 1999;25:303–306. doi: 10.1016/s0141-8130(99)00045-8. [DOI] [PubMed] [Google Scholar]
- 29.Valentin H E, Stuart E S, Fuller R C, Lenz R W, Dennis D. Investigation of the function of proteins associated to polyhydroxyalkanoate inclusions in Pseudomonas putidaBMO1. J Biotechnol. 1998;64:145–157. doi: 10.1016/s0168-1656(98)00097-2. [DOI] [PubMed] [Google Scholar]
- 30.Von Tersch M A, Carlton B C. Megacinogenic plasmids of Bacillus megaterium. J Bacteriol. 1983;155:872–877. doi: 10.1128/jb.155.2.872-877.1983. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Wieczorek R, Steinbüchel A, Schmidt B. Occurrence of polyhydroxyalkanoic acid granule-associated proteins related to the Alcaligenes eutrophusH16 GA24 protein in other bacteria. FEMS Microbiol Lett. 1996;135:23–30. doi: 10.1111/j.1574-6968.1996.tb07961.x. [DOI] [PubMed] [Google Scholar]
- 32.Youngman P, Poth H, Green B, York K, Olmedo G, Smith K. Methods for genetic manipulation, cloning, and functional analysis of sporulation genes in Bacillus subtilis. In: Smith I, Slepecky R, Setlow P, editors. Regulation of prokaryotic development. Washington, D.C.: American Society for Microbiology; 1989. pp. 65–87. [Google Scholar]
- 33.Zhang S, Yasuo T, Lenz R W, Goodwin S. Kinetic and mechanistic characterization of the polyhydroxybutyrate synthase from Ralstonia eutropha. Biomacromolecules. 2000;1:244–251. doi: 10.1021/bm005513c. [DOI] [PubMed] [Google Scholar]