Abstract
Background
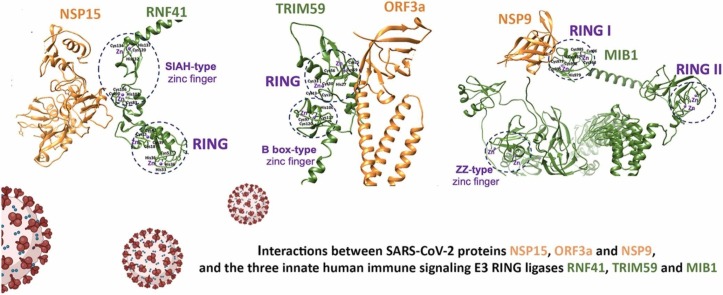
The ubiquitin system is a modification process with many different cellular functions including immune signaling and antiviral functions. E3 ubiquitin ligases are enzymes that recruit an E2 ubiquitin-conjugating enzyme bound to ubiquitin in order to catalyze the transfer of ubiquitin from the E2 to a protein substrate. The RING E3s, the most abundant type of ubiquitin ligases, are characterized by a zinc (II)-binding domain called RING (Really Interesting New Gene). Viral replication requires modifying and hijacking key cellular pathways within host cells such as cellular ubiquitination. There are well-established examples where a viral proteins bind to RING E3s, redirecting them to degrade otherwise long-lived host proteins or inhibiting E3’s ubiquitination activity. Recently, three binary interactions between SARS-CoV-2 proteins and innate human immune signaling Ε3 RING ligases: NSP15-RNF41, ORF3a-TRIM59 and NSP9-MIB1 have been experimentally established.
Methods
In this work, we have investigated the mode of the previous experimentally supported NSP15-RNF41, ORF3a,-TRIM59 and NSP9-MIB1 binary interactions by in silico methodologies intending to provide structural insights of E3-virus interplay that can help identify potential inhibitors that could block SARS-CoV-2 infection of immune cells.
Conclusion
In silico methodologies have shown that the above human E3 ligases interact with viral partners through their Zn(II) binding domains. This RING mediated formation of stable SARS-CoV-2-E3 complexes indicates a critical structural role of RING domains in immune system disruption by SARS-CoV-2-infection.
Data Availability
The data used to support the findings of this research are included within the article and are labeled with references.
Keywords: E3 RING ligases, SARS-CoV-2, Zinc binding RING domains, Haddock
Graphical Abstract
1. Introduction
The innate immune system is the human first line of defense against viral infection. Thus, viruses have evolved sophisticated mechanisms to inhibit antiviral innate immunity. Simultaneously, cytokine storm and subsequent multi-organ failure caused by unbalanced human immune responses are considered the major lethal contributors of SARS-CoV-2 infection. Viral proteins encoded by the SARS-CoV-2 genome are deeply involved in regulating innate immune signaling activation. One representative example is that NSP1, NSP12, ORF6, and M proteins of SARS-CoV-2 inhibited MDA 5 dependent type I interferon induction [1]. The human angiotensin-converting enzyme 2 (hACE2) has been identified as the main receptor for Spike protein-mediated SARS-CoV-2 infection [2]. However, the relatively low ACE2 expression in SARS-CoV-2 infected myeloid cells indicates the existence of an alternative infection mediator in these immune cells [3]. Therefore, the disruption of the innate immune system by SARS-CoV-2 infection still remains to be thoroughly investigated.
E3 ubiquitin ligases catalyze protein degradation by transferring ubiquitin from the E2 ubiquitin-conjugating enzyme to the protein substrate and are key enzymes in innate and adaptive immunity [4]. RING E3s domains bind two Zn(II) ions in a unique "cross-brace" arrangement through a defined motif of cysteine and histidine residues [5]. Usually, the ubiquitin transfer and the specificity of E3 RING ligases for substrate recognition are mediated by their RING domain [6], [7], [8].
Viruses promote viral replication and pathogenesis by interfering with host ubiquitin-dependent signaling pathways or by hijacking the cellular ubiquitination machinery [9], [10], [11]. There are well-established examples where the viral proteins bind to RING E3s. Many viruses hijack members of the Cullin-RING E3 Ligase (CRL) family, which is the largest family of E3 ligases. Viruses interact in many ways with CRLs to impact their ligase activity or redirect CRL complexes to degrade host targets that are otherwise not degraded within host cells [11]. Recently, three direct interactions between SARS-CoV-2 proteins and human innate immune signaling Ε3 ligases (NSP15-RNF41, ORF3a,-TRIM59 and NSP9-MIB1) have been identified using affinity–purification mass spectroscopy [12] and their in silico SARS-CoV-2-human interactome has been reported [13]. RNF41 has 317 aa length and regulates Toll-like receptor (TLR) responses in different ways: (i) inhibition of the production of proinflammatory cytokines (ii) increase of interferon-b production in Toll-like receptor–triggered macrophages by suppressing adapter MyD88–dependent activation of transcription factors NF-jB and AP-1, and iii) activation of the kinase TBK1 and transcription factor IRF3 [14]. TRIM59 has 403 aa length and can regulate the innate immune response through the Nuclear Factor Kappa-light-chain-enhancer of activated B cells (NF-κB) and interferon regulatory factor (IRF-3/7) -mediated signal pathways by interacting with evolutionarily conserved signaling intermediates in the Toll pathway (ECSIT) [15]. In addition, TRIM59 significantly suppresses lipopolysaccharide (LPS)-induced macrophage activation, whereas siRNA-mediated knockdown of TRIM59 enhances LPS-induced macrophage activation [16]. As activated macrophages play an important role in many inflammatory diseases, it is evident that TRIM59 is an important player in innate immunity and antiviral defense. The ubiquitin ligases-mind bomb 1 (MIB1) has a critical role in the regulation of the antiviral signaling pathway TANK-binding kinase 1 (TBK1)/ IκB kinase (IKKε) [17]. MIB1 is a long protein with 1006 residues, activates NF-kB, and suppresses RNA virus dissemination through its interaction with TBK1 and Nucleosome assembly protein (NAP1). In contrast, gene targeting of MIB1 enhances viral replication and inhibits interferon (IFN) production upon RNA virus infection [18].
In this work, we have investigated the mode of the previous experimentally supported NSP15-RNF41, ORF3a,-TRIM59 and NSP9-MIB1 binary interactions by in silico methodologies intending to provide structural insights of E3-virus interplay that can help identify potential inhibitors that could block SARS-CoV-2 infection of immune cells.
2. Experimental
2.1. Retrieval of protein structures
The structures of E3 ligases RNF41, TRIM59, and MIB1 have yet to be determined experimentally. Zinc binding amino acid motifs of RNF41, TRIM59 and MIB1 were characterized using the strategy previously applied to identify putative metal-binding sites in various proteomes [19], [20], [21], [22], [23]. The list of known zinc binding domains (such as: SIAH-type Zn(II) finger, B-box type Zn (II) finger, ZZ-type Zn(II) finger and zinc domains of metalloproteases [24], has been extracted from the Pfam library (http://pfam.xfam.org/) [25], and their 3D structures were retrieved from the Protein Data Bank (PDB; http://www.rcsb.org/pdb/) [26] and MetalPDB databases (https://metalpdb.cerm.unifi.it/) [27]. The three E3 ligases were analyzed for zinc-binding motifs with the search tool HMMER (http://hmmer.org/) [28]. The structures of the zinc binding RING and finger domains of E3 ligases were modeled via structural modeling using MODELLER v9.19 [29] and compared with the predicted models mined by the AlphaFold Protein Structure Database [30]. The full-length structures of E3 ligases RNF41, TRIM59, and MIB1 were mined from the AlphaFold Protein Structure Database (IDs: AF-Q9H4P4-F1, AF-Q8IWR1-F1 and AF-Q86YT6-F1, respectively). The X-ray structures of the viral proteins NSP15, ORF3, and NSP9 were mined from the protein data bank (PDB ids: 6VWW, 6XDC and 6W4B, respectively) [26].
2.2. Modeling of protein complexes
To perform blind docking between protein pairs, the interfacial contacts between the interacting proteins were predicted by two approaches: i) CPORT algorithm that predicts protein-protein interface residues [31] and ii) the RaptorX ultra deep learning model trained from single-chain proteins to predict contacts for a protein pair [32]. Among the contacts with higher probability (probability score value> 0,8 *average value of the scores of all contacts resulted per protein pair) those that were retrieved from both protocols (listed in the Supplementary Material: Tables S1-S3), were used as restrains in the HADDOCK (High Ambiguity Driven protein-protein DOCKing) platform [33]. Information from the identified protein interfaces was encoded in ambiguous interaction restraints (AIRs) to drive the docking process applying three successive steps: (i) rigid-body energy minimization, (ii) semi-flexible refinement in torsion angle space and (iii) final refinement in explicit solvent. The selection of the final 3D models of protein complexes has been based on a weighted sum of electrostatics, desolvation, van der Waals energy terms and the energetic contribution of the restraints used to drive the docking.
2.3. Molecular dynamics
MD simulations were run with YASARA [34]. The setup included an optimization of the hydrogen bonding network [35] to increase the solute stability, and a pKa prediction to fine-tune the protonation states of protein residues at the chosen pH of 7.4 [36]. All the structures were placed in the center of a dodecahedron box with a distance of 1 nm between the proteins and the wall of the box on all sides and the box was solvated using TIP3P water. NaCl was added with a physiological concentration of 0.9 %, with an excess of either Na+ or Cl- for cell neutrality. After steepest descent and simulated annealing minimizations to remove clashes, temperature equilibration for 100 ps at 298 K using the Berendsen thermostat was performed. In the next step, pressure was equilibrated to 1 atm in 1 ns using the Berendsen barostat. During both the equilibrations, all the heavy atoms of the proteins were position restrained with a force constant of 1000 kJ mol-1 nm-2. The simulation was run for up to 140 ns using the AMBER14 force field [37] for solute, Zinc AMBER Force Field (ZAFF) [38] for special parametrization of Zn2+ interactions and TIP3P for water. The cutoff was 8 Å for van der Waals forces (the default used by AMBER [39], and long range electrostatics were obtained by smooth Particle-mesh Ewald (SPME) algorithm [40]). The equations of motion were integrated with a multiple timestep of 2.5 fs for bonded intermolecular interactions and 5.0 fs for non-bonded interactions at temperature of 298 K and a pressure of 1 atm (NPT ensemble) using algorithms described in detail previously [41].
3. Results and discussion
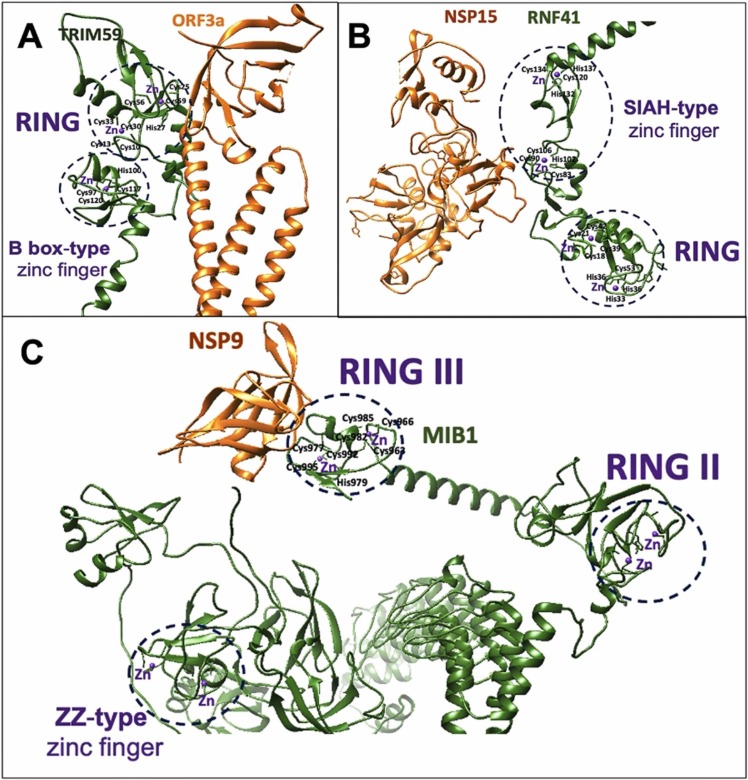
Structural modeling and in silico identification of metal-binding domains suggested that each of the three E3 ligases consists of multiple Zn(II) binding domains. RNF41 has a RING domain (Cys18-Arg57) and SIAH-type Zn(II) finger (Lys78-Leu138), TRIM59 has a RING domain (Cys10-Arg60), and B-box type Zn(II) finger (Pro92-Leu134) and MIB1 has three RING domains (Cys819-Lys854, Cys866-Arg901 and Cys963-Arg996) and ZZ-type Zn(II) finger (His80-Ser132) ( Fig. 1).
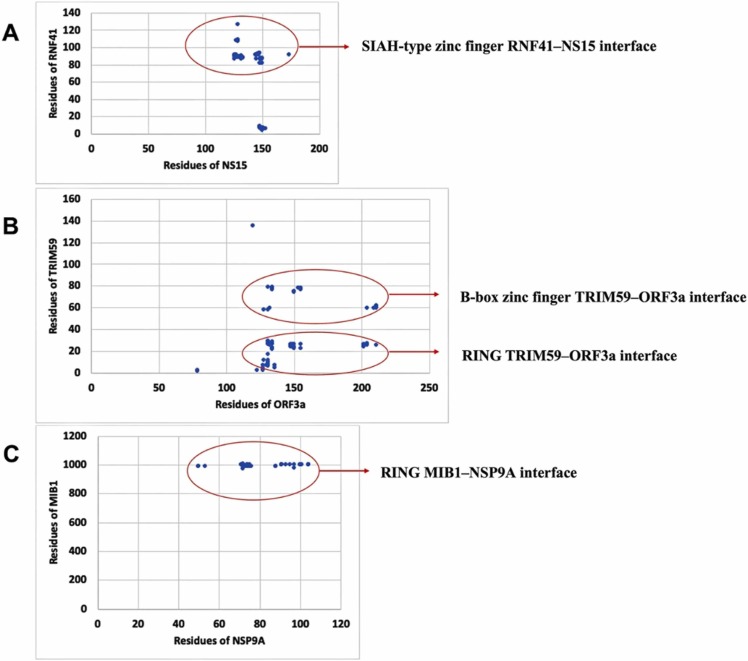
Fig. 1.
: The cluster of intermolecular contacts at the interface (within the threshold distance of 5.5 Å) for the complexes RNF41-NSP15 (A), TRIM59-ORF3a (B) and MIB1-NSP9 (C).
The RING domains of RNF41, TRIM59, and MIB1 showed the typical globular conformation of RING domains [7], [8], characterized by a central alpha-helix and variable-length loops separated by small beta-strands (Fig. 1).
Interestingly, all E3 ligases interacted with SARS-CoV-2 proteins through their Zn binding domains: TRIM59 and MIB1 through RING domain and RNF41 utilizes SIAH-type zinc finger ( Fig. 1, Fig. 2).
Fig. 2.
Zn(II) binding domains of TRIM59, RNF41 and MIB1, and their docking interfaces with ORF3a (A), NSP15 (B) and NSP9 (C), respectively.
Docking results of the protein assemblies, including HADDOCK scores, van der Waals and electrostatic energies, buried surface areas, and dissociation constants, are listed in Table 1.
Table 1.
Haddock results of E3-SARS-CoV-2 complexes.
| E3-SARS-CoV-2 complexes | RNF41-NSP15 | TRIM59-ORF3a | MIB1-NSP9 |
|---|---|---|---|
| HADDOCK score | -76.1 + /- 2.0 | -132.8 + /- 1.7 | -95.4 + /- 3.0 |
| Kd(M) at 25.0 ℃ | 3.8E-07 | 1.8E-08 | 3.5E-07 |
| ΔG (kcal mol-1) | -8.7 | -10.5 | -8.8 |
| Buried surface area (Å2) | 1249.8 + /- 93.7 | 1669.2 + /- 135.6 | 1545.5 + /- 99.2 |
| Van der Waals energy (kcal mol-1) | -49.2 + /- 2.6 | -55.7 + /- 7.5 | -45.1 + /- 3.3 |
| Electrostatic energy (kcal mol-1) | -137.1 + /- 31.3 | -147.2 + /- 26.2 | -342.8 + /- 50.7 |
The interface between RING and viral protein has the highest surface for TRIM59-ORF3a complex (1669.2 Å2), followed by MIB1-NSP9 (1545.5 Å2) and RNF41-NSP15 (1249.8 Å2). Accordingly, the TRIM59-ORF3a complex has the highest binding affinity (lowest Kd and ΔG) compared to the others.
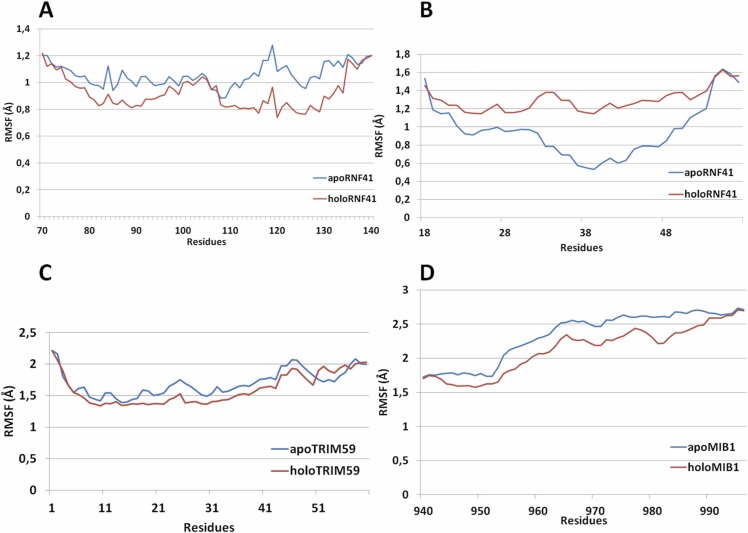
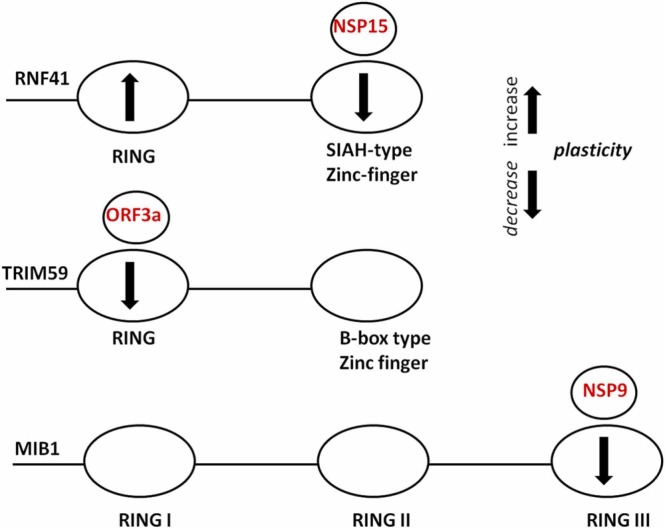
Comparison of 140 ns of molecular dynamics (MDs) simulation between apo and the complexed form of E3s showed that zinc-binding E3 regions located in the interfaces in the protein complexes exhibited decreased mobility (a decrease of root-mean-square fluctuation (RMSF) measures, Fig. 3).
Fig. 3.
Structural fluctuations (RMSF of Ca atoms per residue) for the polypeptide region (Lys78-Leu138) of RNF41 simulations (blue) and in complex with NSP15 (red) (A); for the RING domain (Cys18-Arg57) of RNF41 simulations (blue) and in complex with NSP15 (red) (B); for the RING domain (Cys10-Arg60) of TRIM59 simulations (blue) and in complex with ORF3a (red) (C); for the RING domain (Cys963-Arg996) of MIB1 simulations (blue) and in complex with NSP9 (red) (D).
Specifically, the overall plasticity of the polypeptide regions: (Lys78-Leu138 for RNF41, Cys10-Arg60 for TRIM59 and Cys943-Arg996 for MIB1) has decreased (Fig. 3A, C and D respectively). In addition, a remarkable increase in the plasticity of RING domain (Cys18-Arg57) that is not in RNF41-NSP15 interface was observed (Fig. 3B).
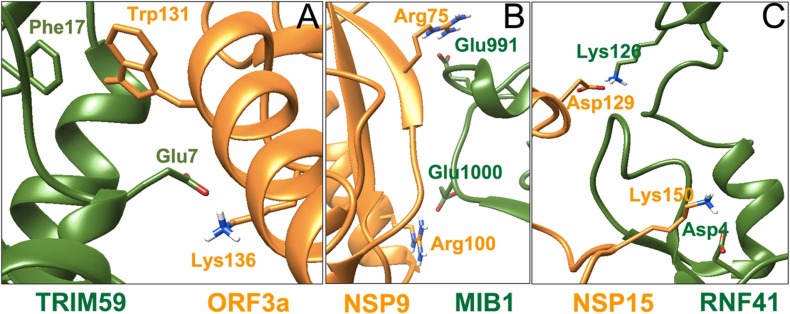
Molecular dynamics (MDs) simulation of protein ensembles showed that all Zn(II)-binding RING domains and viral proteins are linked through critical interactions. Specifically, the intermolecular binding of the complexes is strengthened by salt bridges between positively charged groups of lysines (amino) and arginines (guanidino) with negatively charged groups of glutamic and aspartic carboxylates. This type of interfacial contact has been identified for the following amino acid pairs: Lys136-Glu7 for TRIM59- ORF3 and Arg100-Glu1000, Arg75-Glu991, Glu71-Arg1001, Lys37-Asp291, Lys53-Asp987 and Lys37-Glu290 for the MIB1-NSP9 complex, and Asp4-Lys150 and Lys126-Asp129 for the RNF41- NSP15 complex ( Fig. 4).
Fig. 4.
Intermolecular contacts between (A) TRIM59 and ORF3a (π-π stacking and salt bridges, (B) MIB1 and NSP9 (salt bridges) and (C) RNF41 and NSP15 (salt bridges).
In addition, MDs reveal an intermolecular π-π interaction in the complex TRIM59- ORF3 between Phe17 and Trp131 respectively (Fig. 4), which may be a key force that strengthens the binding affinity of the complex compared to the others.
The zinc-binding domains of the three E3 ligases that, based on our results, mediate E3-SARS-CoV-2 interactions and show alterations in conformational plasticity upon complexation ( Fig. 5), have been previously proposed as structurally critical components for their ligase activity.
Fig. 5.
Alterations in conformational plasticity of zinc binding domains upon complexation between RNF41-NSP15 (upper panel), TRIM59-ORF3a (medium panel) and MIB1-NSP9 (lower panel).
The RING domain (Cys10-Arg60) of TRIM59, that interacts with the ORF3a protein of SARS-CoV-2, has been proposed to play a critical role in TRIM59’s ligase activity. TRIM59 is mediated osteosarcoma progression, via regulation and ubiquitin-mediated degradation of P53 in osteosarcoma cells though its RING domain [42], [43]. Furthermore, the binding of RING domain (Val963-Arg996) of MIB1 to NSP9 protein of SARS-CoV-2 causes allosterically decreased rigidity (decreased RMSF measures, Fig. 3D) in the region of Val943-Cys963 preceding the interacting RING domain. This fragment is located within a helical region separating two RING domains of MIB1. It was proposed that site mutations in this region revealed a potentially deleterious effect and inactivation of MIB1 ubiquitin ligase activity. MIB1 ubiquitin ligase promotes endocytosis of the NOTCH ligands DELTA and JAGGED and V943F mutant showed that causes left ventricular noncompaction cardiomyopathy, reduced ventricular Notch1 activity and abnormal expression of cardiac development and disease genes [44], [45]. Interestingly, the binding of SIAH-type Zn(II) finger (Lys78-Leu138) of RNF41 causes allosterically increased mobility (increased RMSF measures, Fig. 3B) to the RING domain (Cys18-Arg57). It was reported that RNF41 RING is responsible for E2-E3 interaction and the RNF41’s catalytic activity and biological experiments indicated that substitutions of zinc ligands in RING domain (C34S and H36Q) caused inhibition of its enzymatic activity [46], [47]. Thus, an increase in RING conformational plasticity induced by RNF41- NSP15 interaction may have an impact on the E3- E2 interplay of RNF41.
Previously, it was reported that the molecular surface of the RING E3 domain between the two metal-binding sites constitutes the interaction interface with E2 enzymes. Moreover, a strong association between destabilization of their RING core, enchantment of the RING’s backbone mobility, stretching of the Zn2+–Zn2 + distance and E3 ligase inactivation was observed in RING E3 mutant ubiquitination studies [8]. A remarkable similarity of our data is observed with the observations of the previous study: MDs of the RNF41- NSP15 protein complex showed that the increased conformational plasticity of the RING domain (Cys18-Arg57) of RNF41 was accompanied by increase in the distance between the Zn2+ ions (15,1 ± 0,3 Å and 16,2 ± 0,6 Å in apo and holo forms respectively).
4. Conclusions
In this work, the structural role of RING E3 domains has been investigated for the SARS-CoV-2 disruption of cellular ubiquitination pathways as an anti-innate immunity viral strategy. Since the RING or Zn(II) finger domains are responsible for the E3-E2 interactions, further detailed experimental investigations are needed to study the proposed RING-dependent host-virus interplay. Possible viral distribution of the formation of E2-E3 complexes, which are essential for controlling the ubiquitination pathway and the human immune system, should be further investigated. The analysis presented here of the RING-binding mode of SARS-CoV-2 protein interactions could provide insights into the future identification of inhibitors (drugs) that block SARS-CoV-2 infection of immune cells and emphasizes the crucial biological role of Zn(II).
Funding
M.P. acknowledges Università degli Studi di Sassari (UNISS) for the financial support received within the program “Fondo di Ateneo per la ricerca 2020, FAR 2020”.
CRediT authorship contribution statement
Christos T. Chasapis: Conceptualization, Data curation, Formal analysis, Writing – original draft preparation, Software, Visualization, Writing – review & editing, Supervision. Spyros P. Perlepes: Conceptualization, Writing – original draft preparation, Writing – review & editing. Geir Bjørklund: Conceptualization, Writing – review & editing. Massimiliano Peana: Conceptualization, Data curation, Funding acquisition, Investigation, Visualization, Writing – review & editing, Supervision.
CRediT authorship contribution statement
All authors confirmed they have contributed to the intellectual content of this paper and have met the following three requirements: (a) significant contribution to the conception and design, acquisition of data, or analysis and interpretation of data; (b) drafting or revising the article for intellectual content; (c) final approval of the published article.
Conflict of interest
The authors declare no potential conflicts of interest.
Footnotes
Supplementary data associated with this article can be found in the online version at doi:10.1016/j.jtemb.2022.127089.
Appendix A. Supplementary material
Supplementary material
.
References
- 1.Shemesh M., Aktepe T.E., Deerain J.M., McAuley J.L., Audsley M.D., David C.T., Purcell D.F.J., Urin V., Hartmann R., Moseley G.W., Mackenzie J.M., Schreiber G., Harari D. SARS-CoV-2 suppresses IFNbeta production mediated by NSP1, 5, 6, 15, ORF6 and ORF7b but does not suppress the effects of added interferon. PLOS Pathog. 2021;17(8) doi: 10.1371/journal.ppat.1009800. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Lan J., Ge J., Yu J., Shan S., Zhou H., Fan S., Zhang Q., Shi X., Wang Q., Zhang L., Wang X. Structure of the SARS-CoV-2 spike receptor-binding domain bound to the ACE2 receptor. Nature. 2020;581(7807):215–220. doi: 10.1038/s41586-020-2180-5. [DOI] [PubMed] [Google Scholar]
- 3.Lu Q., Liu J., Zhao S., Gomez Castro M.F., Laurent-Rolle M., Dong J., Ran X., Damani-Yokota P., Tang H., Karakousi T., Son J., Kaczmarek M.E., Zhang Z., Yeung S.T., McCune B.T., Chen R.E., Tang F., Ren X., Chen X., Hsu J.C.C., Teplova M., Huang B., Deng H., Long Z., Mudianto T., Jin S., Lin P., Du J., Zang R., Su T.T., Herrera A., Zhou M., Yan R., Cui J., Zhu J., Zhou Q., Wang T., Ma J., Koralov S.B., Aifantis I., Segal L.N., Diamond M.S., Khanna K.M., Stapleford K.A., Cresswell P., Liu Y., Ding S., Xie Q., Wang J. SARS-CoV-2 exacerbates proinflammatory responses in myeloid cells through C-type lectin receptors and Tweety family member 2. Immunity. 2021;54(6):1304–1319. doi: 10.1016/j.immuni.2021.05.006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Chasapis C.T., Spyroulias G.A. RING finger E(3) ubiquitin ligases: structure and drug discovery. Curr. Pharm. Des. 2009;15(31):3716–3731. doi: 10.2174/138161209789271825. [DOI] [PubMed] [Google Scholar]
- 5.Chasapis C.T., Loutsidou A.K., Orkoula M.G., Spyroulias G.A. Zinc binding properties of engineered RING finger domain of arkadia E3 ubiquitin ligase. Bioinorg. Chem. Appl. 2010 doi: 10.1155/2010/323152. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Kandias N.G., Chasapis C.T., Bentrop D., Episkopou V., Spyroulias G.A. High yield expression and NMR characterization of Arkadia E3 ubiquitin ligase RING-H2 finger domain. Biochem. Biophys. Res. Commun. 2009;378(3):498–502. doi: 10.1016/j.bbrc.2008.11.055. [DOI] [PubMed] [Google Scholar]
- 7.Chasapis C.T., Kandias N.G., Episkopou V., Bentrop D., Spyroulias G.A. NMR-based insights into the conformational and interaction properties of Arkadia RING-H2 E3 Ub ligase. Proteins. 2012;80(5):1484–1489. doi: 10.1002/prot.24048. [DOI] [PubMed] [Google Scholar]
- 8.Birkou M., Chasapis C.T., Marousis K.D., Loutsidou A.K., Bentrop D., Lelli M., Herrmann T., Carthy J.M., Episkopou V., Spyroulias G.A. A residue specific insight into the Arkadia E3 ubiquitin ligase activity and conformational plasticity. J. Mol. Biol. 2017;429(15):2373–2386. doi: 10.1016/j.jmb.2017.06.012. [DOI] [PubMed] [Google Scholar]
- 9.Valerdi K.M., Hage A., van Tol S., Rajsbaum R., Giraldo M.I. The role of the host ubiquitin system in promoting replication of emergent viruses. Viruses. 2021;13(3) doi: 10.3390/v13030369. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Wang L., Ning S. TRIMming type I interferon-mediated innate immune response in antiviral and antitumor defense. Viruses. 2021;13(2) doi: 10.3390/v13020279. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Mahon C., Krogan N.J., Craik C.S., Pick E. Cullin E3 ligases and their rewiring by viral factors. Biomolecules. 2014;4(4):897–930. doi: 10.3390/biom4040897. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Gordon D.E., Jang G.M., Bouhaddou M., Xu J., Obernier K., White K.M., O'Meara M.J., Rezelj V.V., Guo J.Z., Swaney D.L., Tummino T.A., Huttenhain R., Kaake R.M., Richards A.L., Tutuncuoglu B., Foussard H., Batra J., Haas K., Modak M., Kim M., Haas P., Polacco B.J., Braberg H., Fabius J.M., Eckhardt M., Soucheray M., Bennett M.J., Cakir M., McGregor M.J., Li Q., Meyer B., Roesch F., Vallet T., Mac Kain A., Miorin L., Moreno E., Naing Z.Z.C., Zhou Y., Peng S., Shi Y., Zhang Z., Shen W., Kirby I.T., Melnyk J.E., Chorba J.S., Lou K., Dai S.A., Barrio-Hernandez I., Memon D., Hernandez-Armenta C., Lyu J., Mathy C.J.P., Perica T., Pilla K.B., Ganesan S.J., Saltzberg D.J., Rakesh R., Liu X., Rosenthal S.B., Calviello L., Venkataramanan S., Liboy-Lugo J., Lin Y., Huang X.P., Liu Y., Wankowicz S.A., Bohn M., Safari M., Ugur F.S., Koh C., Savar N.S., Tran Q.D., Shengjuler D., Fletcher S.J., O'Neal M.C., Cai Y., Chang J.C.J., Broadhurst D.J., Klippsten S., Sharp P.P., Wenzell N.A., Kuzuoglu-Ozturk D., Wang H.Y., Trenker R., Young J.M., Cavero D.A., Hiatt J., Roth T.L., Rathore U., Subramanian A., Noack J., Hubert M., Stroud R.M., Frankel A.D., Rosenberg O.S., Verba K.A., Agard D.A., Ott M., Emerman M., Jura N., von Zastrow M., Verdin E., Ashworth A., Schwartz O., d'Enfert C., Mukherjee S., Jacobson M., Malik H.S., Fujimori D.G., Ideker T., Craik C.S., Floor S.N., Fraser J.S., Gross J.D., Sali A., Roth B.L., Ruggero D., Taunton J., Kortemme T., Beltrao P., Vignuzzi M., Garcia-Sastre A., Shokat K.M., Shoichet B.K., Krogan N.J. A SARS-CoV-2 protein interaction map reveals targets for drug repurposing. Nature. 2020;583(7816):459–468. doi: 10.1038/s41586-020-2286-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Chasapis C.T., Georgiopoulou A.K., Perlepes S.P., Bjørklund G., Peana M. A SARS-CoV-2 -human metalloproteome interaction map. J. Inorg. Biochem. 2021;219 doi: 10.1016/j.jinorgbio.2021.111423. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Wang C., Chen T., Zhang J., Yang M., Li N., Xu X., Cao X. The E3 ubiquitin ligase Nrdp1 'preferentially' promotes TLR-mediated production of type I interferon. Nat. Immunol. 2009;10(7):744–752. doi: 10.1038/ni.1742. [DOI] [PubMed] [Google Scholar]
- 15.Kondo T., Watanabe M., Hatakeyama S. TRIM59 interacts with ECSIT and negatively regulates NF-kappaB and IRF-3/7-mediated signal pathways. Biochem. Biophys. Res. Commun. 2012;422(3):501–507. doi: 10.1016/j.bbrc.2012.05.028. [DOI] [PubMed] [Google Scholar]
- 16.Jiang M.X., Hong X., Liao B.B., Shi S.Z., Lai X.F., Zheng H.Y., Xie L., Wang Y., Wang X.L., Xin H.B., Fu M., Deng K.Y. Expression profiling of TRIM protein family in THP1-derived macrophages following TLR stimulation. Sci. Rep. 2017;7 doi: 10.1038/srep42781. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Hu H., Sun S.C. Ubiquitin signaling in immune responses. Cell Res. 2016;26(4):457–483. doi: 10.1038/cr.2016.40. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Kumagai Y., Akira S. Mind bomb proteins in the antiviral arsenal. Immunity. 2011;35(3):320–322. doi: 10.1016/j.immuni.2011.09.003. [DOI] [PubMed] [Google Scholar]
- 19.Chasapis C.T., Andreini C., Georgiopolou A.K., Stefanidou M.E., Vlamis-Gardikas A. Identification of the zinc, copper and cadmium metalloproteome of the protozoon tetrahymena thermophila by systematic bioinformatics. Arch. Microbiol. 2017;199(8):1141–1149. doi: 10.1007/s00203-017-1385-y. [DOI] [PubMed] [Google Scholar]
- 20.Stefanidou M., Loutsidou A.C., Chasapis C.T., Spiliopoulou C.A. Immunotoxicity of cocaine and crack. Curr. Drug Abus. Rev. 2011;4(2):95–97. doi: 10.2174/1874473711104020095. [DOI] [PubMed] [Google Scholar]
- 21.Peana M., Chasapis C.T., Simula G., Medici S., Zoroddu M.A. A model for manganese interaction with deinococcus radiodurans proteome network involved in ROS response and defense. J. Trace Elem. Med. Biol. 2018;50:465–473. doi: 10.1016/j.jtemb.2018.02.001. [DOI] [PubMed] [Google Scholar]
- 22.Chasapis C.T. Interactions between metal binding viral proteins and human targets as revealed by network-based bioinformatics. J. Inorg. Biochem. 2018;186:157–161. doi: 10.1016/j.jinorgbio.2018.06.012. [DOI] [PubMed] [Google Scholar]
- 23.Chasapis C.T., Peana M., Bekiari V. Structural identification of metalloproteomes in marine diatoms, an efficient algae model in toxic metals bioremediation. Molecules. 2022;27(2) doi: 10.3390/molecules27020378. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Dalkas G.A., Chasapis C.T., Gkazonis P.V., Bentrop D., Spyroulias G.A. Conformational dynamics of the anthrax lethal factor catalytic center. Biochemistry. 2010;49(51):10767–10769. doi: 10.1021/bi1017792. [DOI] [PubMed] [Google Scholar]
- 25.Mistry J., Chuguransky S., Williams L., Qureshi M., Salazar G.A., Sonnhammer E.L.L., Tosatto S.C.E., Paladin L., Raj S., Richardson L.J., Finn R.D., Bateman A. Pfam: the protein families database in 2021. Nucleic Acids Res. 2021;49(D1):D412–D419. doi: 10.1093/nar/gkaa913. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Berman H.M., Westbrook J., Feng Z., Gilliland G., Bhat T.N., Weissig H., Shindyalov I.N., Bourne P.E. The protein data bank. Nucleic Acids Res. 2000;28(1):235–242. doi: 10.1093/nar/28.1.235. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Putignano V., Rosato A., Banci L., Andreini C. MetalPDB in 2018: a database of metal sites in biological macromolecular structures. Nucleic Acids Res. 2018;46(D1):D459–D464. doi: 10.1093/nar/gkx989. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Potter S.C., Luciani A., Eddy S.R., Park Y., Lopez R., Finn R.D. HMMER web server: 2018 update. Nucleic Acids Res. 2018;46(W1):W200–W204. doi: 10.1093/nar/gky448. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Sali A., Blundell T.L. Comparative protein modelling by satisfaction of spatial restraints. J. Mol. Biol. 1993;234(3):779–815. doi: 10.1006/jmbi.1993.1626. [DOI] [PubMed] [Google Scholar]
- 30.Jumper J., Evans R., Pritzel A., Green T., Figurnov M., Ronneberger O., Tunyasuvunakool K., Bates R., Zidek A., Potapenko A., Bridgland A., Meyer C., Kohl S.A.A., Ballard A.J., Cowie A., Romera-Paredes B., Nikolov S., Jain R., Adler J., Back T., Petersen S., Reiman D., Clancy E., Zielinski M., Steinegger M., Pacholska M., Berghammer T., Bodenstein S., Silver D., Vinyals O., Senior A.W., Kavukcuoglu K., Kohli P., Hassabis D. Highly accurate protein structure prediction with AlphaFold. Nature. 2021;596(7873):583–589. doi: 10.1038/s41586-021-03819-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.de Vries S.J., Bonvin A.M. CPORT: a consensus interface predictor and its performance in prediction-driven docking with HADDOCK. PLOS One. 2011;6(3) doi: 10.1371/journal.pone.0017695. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Zeng H., Wang S., Zhou T., Zhao F., Li X., Wu Q., Xu J. ComplexContact: a web server for inter-protein contact prediction using deep learning. Nucleic Acids Res. 2018;46(W1):W432–W437. doi: 10.1093/nar/gky420. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.van Zundert G.C.P., Rodrigues J., Trellet M., Schmitz C., Kastritis P.L., Karaca E., Melquiond A.S.J., van Dijk M., de Vries S.J., Bonvin A. The HADDOCK2.2 web server: user-friendly integrative modeling of biomolecular complexes. J. Mol. Biol. 2016;428(4):720–725. doi: 10.1016/j.jmb.2015.09.014. [DOI] [PubMed] [Google Scholar]
- 34.Krieger E., Vriend G. YASARA view – molecular graphics for all devices – from smartphones to workstations. Bioinformatics. 2014;30(20):2981–2982. doi: 10.1093/bioinformatics/btu426. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Krieger E., Dunbrack R.L., Jr., Hooft R.W., Krieger B. Assignment of protonation states in proteins and ligands: combining pKa prediction with hydrogen bonding network optimization. Methods Mol. Biol. 2012;819:405–421. doi: 10.1007/978-1-61779-465-0_25. [DOI] [PubMed] [Google Scholar]
- 36.Krieger E., Nielsen J.E., Spronk C.A., Vriend G. Fast empirical pKa prediction by Ewald summation. J. Mol. Graph Model. 2006;25(4):481–486. doi: 10.1016/j.jmgm.2006.02.009. [DOI] [PubMed] [Google Scholar]
- 37.Maier J.A., Martinez C., Kasavajhala K., Wickstrom L., Hauser K.E., Simmerling C. ff14SB: improving the accuracy of protein side chain and backbone parameters from ff99SB. J. Chem. Theory Comput. 2015;11(8):3696–3713. doi: 10.1021/acs.jctc.5b00255. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Peters M.B., Yang Y., Wang B., Fusti-Molnar L., Weaver M.N., Merz K.M., Jr. Structural survey of zinc containing proteins and the development of the Zinc AMBER force field (ZAFF) J. Chem. Theory Comput. 2010;6(9):2935–2947. doi: 10.1021/ct1002626. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Hornak V., Abel R., Okur A., Strockbine B., Roitberg A., Simmerling C. Comparison of multiple Amber force fields and development of improved protein backbone parameters. Proteins. 2006;65(3):712–725. doi: 10.1002/prot.21123. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Essmann U., Perera L., Berkowitz M.L., Darden T., Lee H., Pedersen L.G. A smooth particle mesh Ewald method. J. Chem. Phys. 1995;103(19):8577–8593. doi: 10.1063/1.470117. [DOI] [Google Scholar]
- 41.Krieger E., Vriend G. New ways to boost molecular dynamics simulations. J. Comput. Chem. 2015;36(13):996–1007. doi: 10.1002/jcc.23899. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Liang J., Xing D., Li Z., Shen J., Zhao H., Li S. TRIM59 is upregulated and promotes cell proliferation and migration in human osteosarcoma. Mol. Med. Rep. 2016;13(6):5200–5206. doi: 10.3892/mmr.2016.5183. [DOI] [PubMed] [Google Scholar]
- 43.Sharma A., Khan H., Singh T.G., Grewal A.K., Najda A., Kawecka-Radomska M., Kamel M., Altyar A.E., Abdel-Daim M.M. Pharmacological modulation of ubiquitin-proteasome pathways in oncogenic signaling. Int. J. Mol. Sci. 2021;22(21) doi: 10.3390/ijms222111971. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Luxan G., Casanova J.C., Martinez-Poveda B., Prados B., D'Amato G., MacGrogan D., Gonzalez-Rajal A., Dobarro D., Torroja C., Martinez F., Izquierdo-Garcia J.L., Fernandez-Friera L., Sabater-Molina M., Kong Y.Y., Pizarro G., Ibanez B., Medrano C., Garcia-Pavia P., Gimeno J.R., Monserrat L., Jimenez-Borreguero L.J., de la Pompa J.L. Mutations in the NOTCH pathway regulator MIB1 cause left ventricular noncompaction cardiomyopathy. Nat. Med. 2013;19(2):193–201. doi: 10.1038/nm.3046. [DOI] [PubMed] [Google Scholar]
- 45.Barsi J.C., Rajendra R., Wu J.I., Artzt K. Mind bomb1 is a ubiquitin ligase essential for mouse embryonic development and Notch signaling. Mech. Dev. 2005;122(10):1106–1117. doi: 10.1016/j.mod.2005.06.005. [DOI] [PubMed] [Google Scholar]
- 46.Qiu X.B., Goldberg A.L. Nrdp1/FLRF is a ubiquitin ligase promoting ubiquitination and degradation of the epidermal growth factor receptor family member, ErbB3. Proc. Natl. Acad. Sci. USA. 2002;99(23):14843–14848. doi: 10.1073/pnas.232580999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Wu X., Yen L., Irwin L., Sweeney C., Carraway K.L., 3rd Stabilization of the E3 ubiquitin ligase Nrdp1 by the deubiquitinating enzyme USP8. Mol. Cell. Biol. 2004;24(17):7748–7757. doi: 10.1128/MCB.24.17.7748-7757.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Supplementary material
Data Availability Statement
The data used to support the findings of this research are included within the article and are labeled with references.