Abstract
Respiration in cyanobacterial thylakoid membranes is interwoven with photosynthetic processes. We have constructed a range of mutants that are impaired in several combinations of respiratory and photosynthetic electron transport complexes and have examined the relative effects on the redox state of the plastoquinone (PQ) pool by using a quinone electrode. Succinate dehydrogenase has a major effect on the PQ redox poise, as mutants lacking this enzyme showed a much more oxidized PQ pool. Mutants lacking type I and II NAD(P)H dehydrogenases also had more oxidized PQ pools. However, in the mutant lacking type I NADPH dehydrogenase, succinate was essentially absent and effective respiratory electron donation to the PQ pool could be established after addition of 1 mM succinate. Therefore, lack of the type I NADPH dehydrogenase had an indirect effect on the PQ pool redox state. The electron donation capacity of succinate dehydrogenase was found to be an order of magnitude larger than that of type I and II NAD(P)H dehydrogenases. The reason for the oxidized PQ pool upon inactivation of type II NADH dehydrogenase may be related to the facts that the NAD pool in the cell is much smaller than that of NADP and that the NAD pool is fully reduced in the mutant without type II NADH dehydrogenase, thus causing regulatory inhibition. The results indicate that succinate dehydrogenase is the main respiratory electron transfer pathway into the PQ pool and that type I and II NAD(P)H dehydrogenases regulate the reduction level of NADP and NAD, which, in turn, affects respiratory electron flow through succinate dehydrogenase.
Synechocystis sp. strain PCC 6803, a unicellular cyanobacterium, contains a respiratory electron transport chain on both the cytoplasmic and thylakoid membranes (11, 23). The cytoplasmic membrane forms the inner boundary of the periplasmic space and is known to contain proteins typically associated with respiratory electron transport, such as NAD(P)H dehydrogenase, cytochrome b6f, and terminal oxidases (presumably predominantly Cyd, a putative quinol oxidase) (7, 11, 23). Two types of NAD(P)H dehydrogenase have been found in cyanobacteria. One is a NADPH-preferring type I dehydrogenase (NDH-1) that is encoded by ndh genes, consists of about a dozen subunits, and contributes to a proton gradient across the membrane (2). The second type of dehydrogenase is a NADH-oxidizing type II dehydrogenase (NDH-2) consisting of a single subunit and presumably not contributing to a proton gradient across the membrane. Three genes for NDH-2 (ndbA, ndbB, and ndbC) are found in Synechocystis sp. strain PCC 6803.
The thylakoid membrane contains both a photosynthetic electron transport chain that includes photosystem I (PSI) and PSII and a respiratory electron transport chain containing, among others, NDH-1, succinate dehydrogenase (SDH), and a cytochrome aa3-type terminal oxidase (CtaI) (5, 19, 24). Genes for a third terminal oxidase (CtaII) are present in the genome, but no functional evidence for CtaII has been obtained thus far. The respiratory and photosynthetic electron transport chains in the thylakoids have electron carriers in common, including the cytochrome b6f complex, the plastoquinone (PQ) pool, and soluble redox-active proteins (21, 27).
Electron transport into and out of the PQ pool in cyanobacterial thylakoids is complex in that many different pathways exist and the relative rate at which electrons are transported via each pathway depends on the capacity of the pathway and the availability of oxidized substances to accept electrons (or reduced compounds to donate them), etc. Three intersecting pathways traditionally have been viewed as dominant in Synechocystis sp. strain PCC 6803 thylakoids. The three pathways are linear photosynthetic electron transport, respiratory transport from NADPH and succinate to cytochrome oxidase, and cyclic electron transport around PSI (electrons at the acceptor side of PSI returning to the PQ pool). However, electrons can easily cross from one pathway to another at the level of the PQ pool, cytochrome b6f, and/or soluble carriers. For example, in the absence of PSI, PSII-generated electrons are fed into cytochrome oxidase (25), and in darkness, respiratory electrons are used to reduce the acceptor side of PSII if terminal oxidases are blocked (8).
Even though a wealth of data has accumulated over the years regarding individual pathways, very little is known regarding the relative importance of the various electron transfer pathways in the organism in vivo. However, this information is critical to an understanding of the metabolism and its regulation in the organism. As cyanobacteria are thought to be related to the ancestor of chloroplasts, understanding of photosynthetic and respiratory electron transport interaction and regulation is likely to also impact our understanding of such processes in photosynthetic eukaryotes. Regulatory processes with respect to energy requirement (ATP production) (17, 22–24), metabolic cofactor reduction-oxidation (such as NADP+ reduction at PSI, allowing CO2 fixation via the Calvin cycle) (18), and redox sensing-poising of the PQ pool for gene regulation (8, 16) are well established phenomenologically, but the primary signals and mechanisms for such regulation remain unclear.
To aid in providing a comprehensive overview of photosynthesis and respiration in vivo, we determined the PQ pool redox state, the reduction level of NAD-NADP, and the concentration of organic acids such as succinate in a range of strains lacking one electron flow pathway or multiple electron flow pathways. Our findings indicate that succinate levels, the PQ pool redox state, and the NADP reduction state are interrelated, with the succinate concentration and SDH activity being much more important factors than has been realized thus far.
MATERIALS AND METHODS
Growth conditions.
Wild-type and mutant cultures of Synechocystis sp. strain PCC 6803 were grown in liquid BG-11 medium (20) at 30°C. Where indicated, 5 mM glucose was added for photoheterotrophic growth. Cultures were grown at a light intensity of 40 to 50 μmol of photons m−2 s−1, except when the cultures were grown at a low light intensity (3 to 5 μmol of photons m−2 s−1). Cells were grown in ambient air or, if indicated, in air enriched with CO2 (bubbling with air containing 3% CO2). Cells used for organic acid analysis, quinone (Q) redox state determination, and [NAD] or [NADP] quantifications were harvested at an optical density at 730 nm (OD730) of 0.5, as determined with a Shimadzu UV 160 spectophotometer. This corresponds to mid-exponential phase.
Deletion mutant construction and segregation.
The NDH-1 deficient strain, a gift from T. Ogawa, carries an insertional mutation of the ndhB gene. The two sdhB genes (sll1625 and sll0823) were deleted from the wild-type, NDH-2-deficient, and Cyd-deficient/CtaII-deficient strains by using plasmids pΔsll1625 (Cmr) and pΔsll0823 (Kmr), which were described previously (5). For deletion of the two sdhB genes from the PSI-deficient and CtaI-deficient strains, the antibiotic resistance cassette of the pΔsll1625 and pΔsll0823 plasmids, respectively, was replaced with the PvuII-PvuII fragment from pZeol (8), which contains the zeocin resistance (Zor) cassette. To create a PSII-deficient/SDH-deficient strain, the SDH-deficient strain (Δsll0823 Δsll1625) was used as the background strain for deletion of PSII by transformation with the pΔDICEmr plasmid, in which an erythromycin resistance cassette replaces part of the psbDIC operon.
Segregation analysis was performed by PCR with primers specific for the sequence of the flanking regions of the gene being deleted. In addition, PCR was performed by using primers recognizing sequences inside the wild-type sequence that was deleted in the mutant. The exclusive presence of a band corresponding to the inactivated gene in the first PCR and the absence of a product in the second PCR were taken as evidence of full segregation.
Organic acid isolation and analysis.
Organic acids were isolated, purified, derivatized, and analyzed by gas chromatography-mass spectrometry as previously described (5). One liter of cells to be harvested for organic acid analysis was grown to an OD730 of 0.5. The levels of malate and isocitrate were determined as described for succinate and fumarate (5): derivatized standards were run, and mass spectral fragmentation patterns coupled with retention time were used to identify and quantitate the derivatized organic acid of interest.
NADP-NADPH and NAD-NADH extraction.
Extraction and determination of NADP/NADPH and NAD/NADH ratios and concentrations were carried out by two separate methods. In the first method, extracts to be used for enzymatic cycling reactions were prepared by a modification of the procedure used by Wagner and Scott (26) for erythrocytes. One liter of cells was grown to an OD730 of ∼0.5 and harvested by centrifugation. The pelleted cells were resuspended in approximately 0.8 ml of breakage buffer (20 mM nicotinamide, 20 mM NaHCO3, 100 mM Na2CO3) precooled to 4°C. The resuspended cells were frozen in liquid N2 and thawed quickly in a water bath at 20°C. Glass beads (70 to 100-μm diameter; approximately one-third of the volume of the cell suspension) were added, and the cells were broken by using a mini-beadbeater (BioSpec Products) (4 × 30 s). Cells were incubated on ice for 1 min between breakage cycles. Complete breakage was achieved when the chlorophyll contents (measured by determining OD663) when extracted with 80% acetone and with methanol were nearly identical; 80% acetone does not efficiently extract chlorophyll from intact cells. Following breakage, all subsequent steps were carried out in darkness to avoid photodegradation of the pyridine nucleotides. The cell debris was spun at 14,000 rpm in an Eppendorf 5415 microcentrifuge, and the supernantant was removed to a new tube. To determine [NADP]total, 1 μl of the supernatant was added to a glass test tube containing 0.9 ml of ice-cold NADP cycling buffer (100 mM Tris-HCl [pH 8.0], 0.5 mM thiazolium blue [MTT], 2 mM phenazine ethosulfate [PES], 5 mM Na4EDTA, 1.3 IU of glucose-6-phosphate dehydrogenase [G-6-PDH] per ml) and incubated in the dark at 37°C for 10 min. Following incubation, 100 μl of glucose-6-phosphate was added and the spectrophotometric changes at 570 nm were monitored for 100 s. The reaction is termed a cycling reaction since NADP+ is reduced by G-6-PDH to NADPH, which is then oxidized by PES, which, in turn, is reoxidized by MTT, yielding a color change reaction (12). None of the samples remained in cycling buffer for more than 15 min to avoid degradation.
To determine the [NAD]total in the extracts, 1 μl of the extract was added to 0.9 ml of NAD cycling buffer (100 mM Tris-HCl [pH 8.0], 0.5 mM MTT, 1 mM PES, 0.2 mg of alcohol dehydrogenase per ml, 1% [wt/vol] bovine serum albumin). The principle of the cycling reaction for NAD determination is the same as for NADP, except that NAD-specific alcohol dehydrogenase is used rather than NADP-specific G-6-PDH. To initiate the reaction, 100 μl of 30% ethanol was added and the spectrophotometric changes at 570 nm were monitored for 100 s.
An aliquot of the cell extract was heated for 30 min at 60°C in a glass test tube in the dark. One microliter of this heated sample was then assayed for NAD and NADP as described above. The heating step denatures the oxidized, but not the reduced, form of the pyridine nucleotides, and the rates observed in the cycling reaction represent the NADH and NADPH concentrations. [NAD+] and [NADP+] were calculated by subtracting the reduced value from the [NAD]total and [NADP]total.
A second method by which to extract and detect NADP-NADPH and NAD-NADH involved fluorescence-based high-pressure liquid chromatography (HPLC) using a method derived from that of Klaidman et al. (14). First, 1 liter of cells (OD730 = 0.5) was pelleted and resuspended to 1 ml in a mixture containing 0.06 mM KOH, 0.2 mM KCN, and 1 mM bathophenanthrolinedisulfonic acid. In this solution, the oxidized forms of NAD and NADP will be derivatized with CN, making the oxidized form visible by fluorescence (emission at 460 nm upon excitation at 330 nm) at an efficiency nearly equivalent to that of the reduced form (14). Glass beads were added to a total volume of 1.5 ml, and the cells were broken as described above. Samples were spun at 14,000 rpm in an Eppendorf 5415 microcentrifuge to remove the insoluble matter, and the supernatant was further rinsed with 0.5 volume of chloroform to ensure the removal of all lipid material. Samples were spun through a 0.45-μm-pore-size microcentrifuge spin filter.
Concentrations and ratios of the oxidized and reduced forms of NAD and NADP were confirmed by fluorescence-based HPLC analysis as described by Klaidman et al. (14) using an HP1100 LC with an Agilent 1100 fluorescence detector and a Waters Spherisorb 5 μm ODS1 column (4.6- by 250-mm analytical C18 column).
Isocitrate dehydrogenase and malate dyhydrogenase coenzyme specificities.
An extract of soluble enzymes was prepared from 10 liters of wild-type Synechocystis sp. strain PCC 6803: Cells were harvested by centrifugation and resuspended in a buffer similar to that described for thylakoid isolation (5), with the exception that the MgCl2 and CaCl2 concentrations were increased to 25 mM MgCl2 and 50 mM CaCl2. Cells were broken with glass beads as described earlier, and the cells debris was removed by centrifugation. Solid ammonium sulfate was added in small aliquots while stirring at 0 to 4°C until the solution was 85% saturated (≅516 mg of ammonium sulfate per ml). The solution was centrifuged at 48,000 × g for 30 min, and the supernatant was discarded. The wet pellet was resuspended in 50 μl of 80 mM potassium phosphate buffer (pH 7.0). Assays were carried out by addition of 2 μl of resuspended protein to one of the following solutions (kept at 37°C).
For isocitrate dehydrogenase activity measurements, the cuvette contained 1 ml of the reaction solution (10 mM MnCl2, isocitrate [0 or 10 mM], either NADP [0 or 0.1 mM] or NAD [0 or 0.1 mM], and 80 mM potassium phosphate [pH 7.0]). Assays were conducted on an HP8452 diode array spectrophotometer by monitoring changes in A340 over time after addition of the protein extract.
Malate dehydrogenase activity measurements were carried out similarly, with the exception that malate replaced isocitrate as the enzyme substrate in the reaction mixtures.
Respiration measurements.
Oxygen uptake was measured as described previously (9), with the exception that for 2 min prior to the measurement, samples were exposed to saturating white light in the presence of DCMU [(3,3-dichlorophenyl)-1,1-dimethylurea] to ensure complete PQ pool oxidation at the beginning of the measurement. Without PSII activity, which is inhibited by DCMU, and in the presence of PSI activity at high light intensity, the PQ pool becomes oxidized very quickly. All cells were grown under photoheterotrophic conditions.
Q electrode.
The Q electrode apparatus was assembled and operated as described previously (4, 6, 28). Cells were harvested in mid-exponential phase (OD730 = 0.5) and resuspended to an OD730 of 4.0 in 10 mM HEPES-NaOH buffer (pH 7.4)–50 mM KCl. A 10-ml volume of the cell suspension was placed in a cuvette that was kept at 28°C and that contained a glassy carbon working electrode poised at +360 mV with respect to an Ag-AgCl reference electrode, as well as a platinum auxiliary electrode. Following initial electrode equilibration, exogenous ubiquinone-1 (UQ-1) was added to the cuvette to a final concentration of 0.2 μM. This Q acts as a redox mediator between the PQ pool in the cells and the electrode surface. The current created by the oxidation of the Q at the electrode surface was measured as a function of time using a CV-50W voltammeter (Bioanalytical Systems Inc., West Lafayette Ind.). The instrument's sensitivity was set at 100 nA V−1. Illumination was provided by a halogen lamp, and light was passed through a filter that kept wavelengths of less than 430 nm from reaching the sample. The light intensity was modulated with a combination of reflectance and wire mesh filters.
Cells were incubated in darkness for 5 min before traces were recorded. The light intensity was increased in a stepwise manner. To obtain full reduction of the PQ pool, cells were incubated with 0.2 mM KCN in darkness, which fully blocks oxidation of the PQ pool by terminal oxidases.
Chlorophyll fluorescence measurements.
The relative chlorophyll fluorescence yield during incubation in darkness was monitored by using a Walz fluorometer (PAM 101, 102) in order to help determine the kinetics of PQ pool reduction indirectly. The intensity of the measuring light was kept minimal (<0.01 μmol of photons m−2 s−1). The time constant of the instrument response was 960 ms. Cells were harvested in mid-exponential growth phase (OD730 = 0.5), spun down, and resuspended in 10 mM HEPES-NaOH (pH 7.0) buffer at a chlorophyll concentration of 10 μg ml−1. The fluorescence level (F0) was measured for 15 s with the measuring light on, the measuring light was turned off, and KCN was injected to a final concentration of 1 mM. The measuring light (<0.01 μmol of photons m−2 s−1) was turned on again for 5 s at 10- to 35-s intervals for the duration of the measurement. The measuring light itself did not have a noticeable actinic effect.
RESULTS
PQ pool redox state measurements.
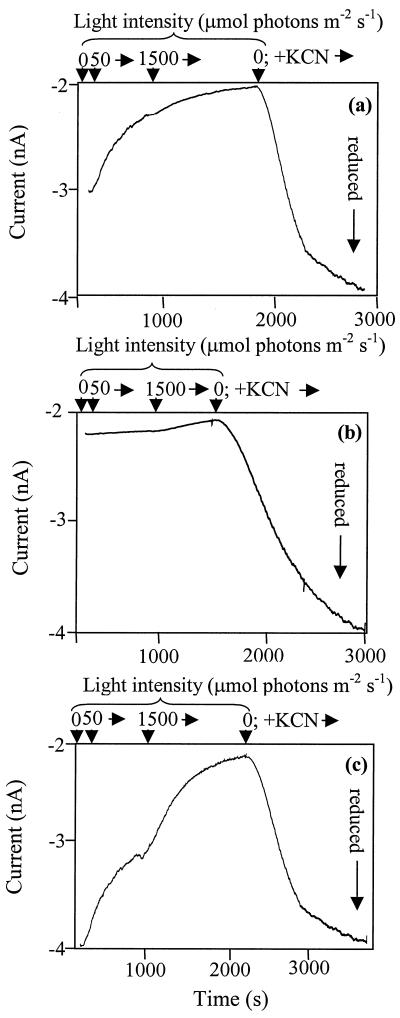
To determine the effects of individual redox-active complexes on the redox state of the PQ pool in the thylakoid membrane of Synechocystis sp. strain PCC 6803, we have utilized a set of mutant strains that lack one or more of the enzymes catalyzing electron transport into or out of the PQ pool in the thylakoid membrane. These mutants were analyzed for the relative redox state of the thylakoid PQ pool by means of a Q electrode. This apparatus has been used with plant mitochondria (6), pea chloroplasts (4), Rhodobacter membranes (28), and most recently with intact cells of Synechocystis sp. strain PCC 6803 (10). In PSI-containing strains, the PQ pool was fairly reduced in darkness and became increasingly oxidized at increasing light intensities (Table 1). The reason for this is that PSI is very abundant in Synechocystis sp. strain PCC 6803 and is a highly efficient and high-capacity electron acceptor in the light. In contrast, PSI-deficient strains became very reduced at increasing light levels (Table 1). The other major electron acceptor is cytochrome oxidase (CtaI), and indeed, in darkness the PQ pool was fully reduced when CtaI was absent (Table 1; Fig. 1).
TABLE 1.
Relative reduction state of UQ-1,a reflecting the redox state of the PQ pool, in different strains at various light intensities as measured with a Q electrode
| Light intensity (μmol photons m−2 s−1) | Relative UQ-1 reduction state
|
|||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|
| WTb | SDH Defc | NDH-2 Def | SDH Def/ NDH-2 Def | Cyd Def/ CtaII Def | SDH Def/ Cyd Def/ CtaII Def | PSII Def | SDH Def/ PSII Def | CtaI Def | SDH Def/ CtaI Def | NDH-1 Def | PSI Def | |
| 0 | 0.57 | 0.26 | 0.15 | 0.03 | 0.41 | 0.20 | 0.55 | 0.38 | 1.00 | 1.00 | 0.16 | 0.46 |
| 4 | 0.54 | 0.13 | 0.16 | 0.05 | 0.39 | 0.15 | 0.50 | 0.37 | 0.63 | 0.43 | 0.15 | 0.48 |
| 8 | 0.51 | 0.12 | 0.16 | 0.02 | 0.35 | 0.12 | 0.45 | 0.35 | 0.58 | 0.16 | 0.15 | 0.55 |
| 15 | 0.48 | 0.08 | 0.12 | 0.02 | 0.32 | 0.07 | 0.40 | 0.17 | 0.46 | 0.10 | 0.14 | 0.80 |
| 30 | 0.40 | 0.07 | 0.03 | 0.03 | 0.28 | 0.05 | 0.23 | 0.12 | 0.32 | 0.04 | 0.12 | 0.98 |
| 70 | 0.15 | 0.02 | 0.01 | 0.01 | 0.18 | 0.02 | 0.18 | 0.10 | 0.25 | 0.01 | 0.09 | 1.00 |
| 350 | 0.08 | 0.01 | 0.01 | 0.00 | 0.04 | 0.01 | 0.05 | 0.03 | 0.15 | 0.00 | 0.03 | 1.00 |
| >1,000 | 0.02 | 0.00 | 0.00 | 0.00 | 0.01 | 0.00 | 0.00 | 0.00 | 0.15 | 0.00 | 0.00 | 1.00 |
The relative reduction state of UQ-1 was calculated as follows: [UQH2]/([UQ] + [UQH2]).
WT, wild type.
Def, deficient.
FIG. 1.
Sample trace acquired with the Q electrode by using wild-type (a), NDH-2/SDH-deficient (b), and CtaI-deficient (c) strains upon illumination and upon addition of KCN. Light level changes (upper) and total time (lower) are indicated on the horizontal axes. The vertical axes show the current in nanoamperes. Under our experimental conditions, a fully reduced UQ-1 pool yields a current of about −4 nA; a fully oxidized pool corresponds to about −2 nA.
SDH, NDH-2, and NDH-1 are potential respiratory donors to the PQ pool. Deletion of any of these three donors led to a much more oxidized PQ pool in darkness (Table 1), whereas an even more pronounced effect was seen upon deletion of both SDH and NDH-2 (Fig. 1 and Table 1). In all strains, a fully reduced PQ pool was observed when KCN was added and the light was turned off, just as was observed in the CtaI-deficient strain in darkness without KCN {[UQH2]/([UQ]+[UQH2]) = 1.00} (Table 1). However, note that in a mutant lacking SDH and NDH-2, this KCN-induced reduction in darkness was much slower than in a strain retaining these dehydrogenases (Fig. 2). Not surprisingly, the quinol oxidase-deficient strain (Cyd/CtaII deficient) did not behave significantly differently from the wild type (Table 1): the quinol oxidases have not been shown to be active in the thylakoid membrane of Synechocystis sp. strain PCC 6803 under the growth conditions used here (9).
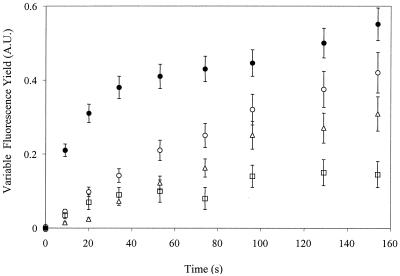
FIG. 2.
Changes in chlorophyll fluorescence yield in darkness after addition of 1 mM KCN. Variable fluorescence (arbitrary units [A.U.]) in intact cells of the wild-type (●), SDH-deficient (○), NDH-1-deficient (□), and NDH-2/SDH-deficient (Δ) strains was measured by using very weak illumination that had no actinic effect. KCN was added at time zero. Curves were normalized so that a value of 1 on the y axis corresponds to the maximal variable fluorescence yield obtained upon illumination in the presence of DCMU. Error bars are derived from three replicates each on separately grown cultures.
Upon removal of SDH in all background strains, the PQ pool became more oxidized than that in the parent strains, consistent with significant SDH activity occurring under essentially all conditions. However, a fully reduced PQ pool was observed in darkness in the CtaI-deficient strain even if SDH was absent, indicating that in the absence of CtaI, sufficient electron donation to the PQ pool can occur via other pathways (such as NDH-1 and NDH-2) to provide a fully reduced PQ pool (Table 1).
Physiological and metabolic effects of removal of respiratory complexes.
Removal of SDH, NDH-1, or NDH-2 led to a more oxidized PQ pool in darkness. If all three pathways were to feed in electrons at comparable rates, the effects of single deletions on the PQ redox state might be expected to be less pronounced, as two pathways remain. Therefore, it is likely that in some cases, the oxidized state of the PQ pool is a secondary effect. To check this, we examined the pool size of key organic acids in central metabolism (succinate, fumarate, isocitrate, and malate) via gas chromatography-mass spectrometry (5). Interestingly, the succinate levels in these mutants were decreased (Table 2), with the level in the NDH-1-deficient strain being so low that the decreased respiratory electron flow in the NDH-1-deficient mutant could be the consequence of low succinate levels rather than the primary lack of NDH-1 activity. We will return to this later. However, the succinate level in the NDH-2-deficient strain was higher and should be sufficient to not fully impair SDH activity. Indeed, in the fluorescence analysis of NDH-2-deficient mutants, the fast phase in fluorescence induction upon addition of KCN now interpreted to be a consequence of SDH activity (5) was still present (8). To further study the effect of NDH-1 on succinate levels, three strains lacking two ndhD genes each (18) were analyzed. The ΔndhD1 ΔndhD2 mutant exhibited decreased respiratory rates but normal CO2 uptake levels (18). In contrast, the ΔndhD3 ΔndhD4 mutant was impaired in CO2 uptake and the ΔndhD5 ΔndhD6 mutant is indistinguishable from the wild type (18). In two of the three strains lacking two NdhD copies (ΔndhD1 ΔndhD2 and ΔndhD3 ΔndhD4), the succinate levels were very low as well, even though they were higher than in the NDH-1-deficient strain.
TABLE 2.
Organic acid levels from various photoautotrophically grown strains
| Strain | Succinatea | Fumaratea | Malateac | Isocitratea |
|---|---|---|---|---|
| WTd | 2.60 | 2.55 | 1.6 ± 0.6 | 2.63 |
| SDH Defe | 0.45 | 0.03 | 0.5 ± 0.2 | 1.28 |
| NDH-1 Defb | 0.05 | 0.02 | 0.2 ± 0.1 | 3.40 |
| NDH-2 Def | 0.22 | 0.08 | 0.8 ± 0.2 | 2.24 |
| NdhD1/D2 Def | 0.11 | 0.10 | 0.3 ± 0.2 | 1.44 |
| NdhD3/D4 Defb | 0.11 | 0.03 | 0.6 ± 0.4 | 3.87 |
| NdhD5/D6 Def | 1.56 | 1.22 | 1.1 ± 0.6 | 6.62 |
Values are expressed as micromoles of organic acid per milligram of chlorophyll.
Bubbled with >3% CO2 balanced with air.
Malate concentrations were adjusted for the efficiency of the derivatization reaction (≅10%). Standard deviations for all samples were ≤8% of the average values presented, except for malate.
WT, wild type.
Def, deficient.
Analysis of fumarate, malate, and isocitrate levels was also informative regarding possible impairments in the various mutants. As previously observed (5), the fumarate concentration in the SDH-deficient strain was much more reduced than that of succinate, leading us to conclude that the enzyme functions as an SDH and not a fumarate reductase (Table 2). Indeed, in the NDH-1-deficient mutant, the NDH-2-deficient mutant, and two of the NdhD mutants, the fumarate level was also very low, which is suggestive of limited SDH activity. The level of malate in the cells was qualitatively consistent with that of fumarate in all strains except the NDH-2-deficient strain. This suggests that this strain is impaired in the conversion of malate. We will come back to this later. Isocitrate levels did not change by more than a factor of 3 in the various strains relative to the wild type (Table 2), but it is of note that most strains carrying NDH-1 mutations had high isocitrate levels, which is suggestive of an impairment of isocitrate conversion in these strains.
The conversions of isocitrate to 2-oxoglutarate and malate to oxaloacetate are both dependent on the oxidized form of NAD(P). The redox cofactor specificity of the two dehydrogenases depends on the organism. To examine whether the malate buildup in the strain lacking NDH-2 and the isocitrate accumulation in the strain lacking NDH-1 may have been due to a lack of the oxidized cofactor, we determined the pool size and redox state of NADP and NAD in the strains lacking SDH, NDH-1, and NDH-2 in comparison with the wild type. Whereas the wild-type and SDH-deficient strains contained significant levels of NADP, NADPH, NAD, and NADH (Table 3), in the NDH-1-deficient and NDH-2-deficient strains, the oxidized form of either NAD or NADP was virtually depleted. The NDH-1-deficient strain essentially lacked the oxidized form of NADP, whereas the NAD/NADH ratio in this strain was normal (Table 3). In contrast, in the NDH-2-deficient strain, virtually all NAD was reduced whereas ample oxidized NADP was present (Table 3). This confirms the NADPH specificity of NDH-1 and the NADH specificity of NDH-2 and suggests that NAD and NADP are not in redox equilibrium with each other.
TABLE 3.
NADP and NAD concentrations and their redox state in cellular extractsa
| Strain | Treatment | NADPtotal (μmol of NADP/mg of Chlb) | % NADP reduced | NADtotal (μmol of NAD/mg of Chl) | % NAD reduced |
|---|---|---|---|---|---|
| WT | Air | 6.4 ± 0.1 (5.0 ± 0.3) | 75 ± 10 (70 ± 5) | 0.6 ± 0.2 (0.9 ± 0.1) | 65 ± 15 (55 ± 6) |
| SDH Defd | Air | 2.5 ± 0.1 (2.1 ± 0.4) | 50 ± 15 (55 ± 6) | 0.1 ± 0.3 (0.4 ± 0.1) | 50 ± 21 (50 ± 5) |
| NDH-2 Def | Air | 8.7 ± 0.1 (6.6 ± 0.1) | 35 ± 20 (25 ± 6) | 0.8 ± 0.2 (0.6 ± 0.1) | 100 ± 5 (100 ± 3) |
| WTc | Air + 3% CO2 | 2.9 ± 0.1 (4.0 ± 0.4) | 50 ± 10 (50 ± 10) | 0.3 ± 0.4 (0.3 ± 0.1) | 70 ± 20 (45 ± 5) |
| NDH-1 Def | Air + 3% CO2 | 0.5 ± 0.1 (0.8 ± 0.1) | 100 ± 4 (100 ± 3) | 0.02 ± 0.3 (0.1 ± 0.1) | 70 ± 20 (39 ± 9) |
The values reported are averages of three separate determinations using cells grown at 45 μmol of photons m−2 s−1. The standard deviations are indicated. Concentrations were determined by NAD and NADP cycling reactions and fluorescence-based HPLC (values in parentheses) as described in the text.
Chl, chlorophyll.
WT, wild type.
Def, deficient.
The results presented in Table 3 suggest a reason for the isocitrate and malate accumulation in the NDH-1-deficient and NDH-2-deficient strains, respectively, if the corresponding dehydrogenases are specific for NADP and NAD, respectively. Indeed, in Synechocystis sp. strain PCC 6803, the nucleotide-binding motif in isocitrate dehydrogenase suggests NADP specificity because the negative charge (Glu or Asp) at the end of the second β sheet in this motif is missing (1). To test the cofactor specificity of isocitrate dehydrogenase, the activity of this enzyme was determined in a crude protein extract from wild-type Synechocystis sp. strain PCC 6803 with either NAD or NADP as a cofactor, and the NADP specificity of the enzyme was experimentally confirmed (Table 4). As NADP levels are very low in the NDH-1-deficient strain (Table 3), the isocitrate dehydrogenase activity may be limited, thus causing isocitrate accumulation and a drastic decrease in the level of succinate in the cells (Table 2).
TABLE 4.
Isocitrate and malate dehydrogenase activities in crude protein extracts from wild-type Synechocystis sp. strain PCC 6803 as a function of cofactor identity and availability
| Substrate | Pyridine nucleotide | [NAD(P)] (mM) | NAD(P)H formed, μmol (mg of chlorophyll)−1 min−1 |
|---|---|---|---|
| Isocitrate | NADP | 0 | −0.02 ± 0.04 |
| 0.1 | 1.25 ± 0.06 | ||
| Isocitrate | NAD | 0 | 0.04 ± 0.05 |
| 0.1 | −0.01 ± 0.04 | ||
| Malate | NADP | 0 | 0.02 ± 0.03 |
| 0.1 | 0.07 ± 0.05 | ||
| Malate | NAD | 0 | 0.01 ± 0.04 |
| 0.1 | 0.40 ± 0.05 |
In contrast, the primary sequence of malate dehydrogenase in Synechocystis sp. strain PCC 6803 contains a motif consistent with NAD-binding specificity (a negative charge is present 19 residues from the third Gly of the GXGXXG nucleotide binding motif). Indeed, a specificity of the enzyme for NAD was observed when assaying for malate dehydrogenase activity in a crude protein extract in the presence of either NADP or NAD (Table 4). In the NDH-2-deficient strain, the NAD level is very low and, indeed, malate accumulation is observed (Table 2).
Respiratory rate.
The results presented thus far suggest an important role of succinate and SDH in respiratory electron transfer. To determine whether this is reflected in electron transport rates, respiration was monitored as O2 consumption in darkness by the wild-type and SDH-deficient strains grown photoheterotrophically before the measurement. Wild-type cells respired at a rate of 34 ± 8 μmol of O2 mg of chlorophyll−1 h−1 in darkness. This rate was inhibited greater than 90% by addition of 1 mM KCN and was about 50% inhibited upon addition of 5 mM malonate, an inhibitor of SDH. In the SDH-deficient strain, the respiratory rate in darkness was 9 ± 5 μmol of O2 mg of chlorophyll−1 h−1, reflecting 76% inhibition relative to the wild type.
Chlorophyll fluorescence as a probe of PQ reduction rates following KCN addition.
Chlorophyll fluorescence yield measurements can be used as a tool with which to monitor (over)reduction of the PQ pool indirectly. When the PQ pool becomes fully reduced, some QA− (the reduced form of the first quinone-type electron acceptor in PSII) is formed due to reverse electron flow (3). The midpoint potential of QA/QA− is about 80 mV more negative than that of PQ/PQH2 (15), and therefore, the redox equilibrium constant between QA and PQ is 20 to 30. This formation of QA− when the PQ pool becomes fully reduced can be visualized as an increase in the chlorophyll fluorescence yield of PSII when excited by a weak (nonactinic) measuring beam. Upon addition of KCN to the cell suspension in the dark, the pathway of electrons out of the PQ pool to the terminal oxidases is blocked and the donation of electrons to the PQ pool via respiratory pathways can be measured by observing the increase in fluorescence yield as a function of time. The SDH-deficient strains lack the fast phase of QA− formation following KCN addition in the dark, which is a prominent phase in the wild type (2). After the fast phase of QA reduction, a slow phase appears that is independent of SDH and that was attributed to PQ reduction by a strong reductant such as NADPH or NADH (2). As the midpoint potential difference between succinate and QA is not in favor of QA reduction by succinate, only partial QA− formation via the fast (SDH) pathway can occur. To determine the pathway(s) of electron flow that is responsible for the slow phase, we examined the rise in chlorophyll fluorescence yield in various strains lacking one or more of these pathways. The initial rate of the rise in the SDH- and NDH-2-deficient strain was about half of that seen in the SDH-deficient strain and approximately 1/10 of the initial rise observed in wild-type cells (Fig. 2). In a first approximation, the difference between the rates in the SDH-deficient strain and the SDH- and NDH-2-deficient strain may be attributed to NDH-2 activity, and the activity remaining in the SDH- and NDH-2-deficient strain may be attributed to NDH-1 activity. However, in the NDH-1-deficient strain, not only was the fast phase of the rise essentially absent (consistent with the absence of succinate in this strain) but also the rate of the slower phase was reduced.
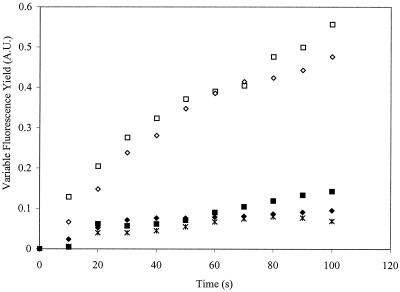
To test whether the absence of the fast phase in the NDH-1-deficient strain is due to a lack of succinate, cells were incubated with 1 mM succinate for different times prior to measurement. A fast phase of chlorophyll fluorescence yield increase was recovered (Fig. 3), indicating that the rapid loss of electron donation to the PQ pool in darkness in this mutant results from the lack of succinate. A further confirmation of this assessment was provided when malonate was added, in addition to succinate, to the NDH-1-deficient cells; no KCN-induced variable fluorescence was obtained under these conditions (Fig. 3).
FIG. 3.
Effect of succinate addition on chlorophyll fluorescence yield as a function of time after the addition of KCN (at time zero) in the NDH-1-deficient strain. Variable fluorescence (arbitrary units [A.U.]) was monitored in samples without (closed symbols) and with (open symbols) addition of 1 mM succinate or with both 1 mM succinate and 5 mM malonate (∗) 5 min prior to KCN addition. The NDH-1-deficient culture was incubated with or without succinate for 2 (□, ■) or 10 (◊, ⧫) min while bubbling with 3% CO2.
DISCUSSION
The PQ pool redox state is affected by several respiratory complexes.
Data collected by using the Q electrode are enlightening with respect to the steady-state redox state of the PQ pool under various irradiance conditions. The PQ pool of Synechocystis sp. strain PCC 6803 is much more oxidized in the light than in darkness. This differs from chloroplasts but is consistent with the relative overabundance of PSI in the cyanobacterial thylakoid membrane. Furthermore, when PSII is functionally deleted, the steady-state PQ pool redox state is not dramatically different from that in the wild type at any of the light intensities studied, which is consistent with a low ratio of PSII relative to PSI (23) in Synechocystis under our growth conditions.
The effect of respiratory complexes on the redox state of the PQ pool is best seen in darkness, as under this condition PSI does not draw electrons out of the PQ pool. A relatively surprising result presented in this paper (Table 1) is that deletion of any one of the three respiratory pathways feeding electrons into the PQ pool (SDH, NDH-1, or NDH-2) resulted in a much more oxidized PQ pool in darkness. However, according to reference 5 and results presented in Fig. 2, the capacity of SDH to feed electrons into the PQ pool is much larger than that of either of the NDH complexes. Therefore, the effects of NDH-1 and NDH-2 deletion on the redox state of the PQ pool are unlikely to be primary effects.
Organic acid and NAD(P) reduction levels.
The effects of deletion of SDH, NDH-1, and NDH-2 on organic acid accumulation levels are extensive. The mutant lacking NDH-2 accumulates NADH and malate, suggesting that the malate-to-oxaloacetate step of the tricarboxylic acid (TCA) cycle may have been blocked by NAD+ limitation. Indeed, malate dehydrogenase from Synechocystis sp. strain PCC 6803 was found to be an NAD-specific enzyme (Table 4). Interestingly, the succinate concentration in the NDH-2-deficient strain has been reduced about 10-fold compared to that in the control, suggesting a feedback inhibition of enzymes earlier in the pathway or a lack of flux through the TCA cycle. Sufficient succinate remains (Table 2) for SDH to impact the redox state of the PQ pool in the short term (8), but the steady-state SDH flux is low in the NDH-2-deficient mutant, as the fumarate level in this strain is low. This provides a reason why the NDH-2-deficient strain has an oxidized PQ pool in darkness.
The NDH-1-deficient mutant accumulated isocitrate and essentially lacked acids that are downstream of isocitrate in the TCA cycle. This suggests an inhibition of isocitrate dehydrogenase. A plausible reason for this apparent inhibition is a lack of an electron acceptor, as the NDH-1-deficient strain essentially lacked oxidized NADP+ and isocitrate dehydrogenase appears to be a NADP-dependent enzyme in Synechocystis sp. strain PCC 6803. The results presented in Fig. 3 and Table 2 present a plausible explanation for the oxidized PQ pool in darkness in the absence of NDH-1 activity: in the absence of NDH-1, the succinate level in the cell is very low and the SDH-mediated reduction of the PQ pool is inhibited due to a lack of substrate. Indeed, reduction of the PQ pool resumes upon addition of succinate (Fig. 3).
It is interesting that in Synechocystis sp. strain PCC 6803, the redox state of the NAD and NADP pools is able to fluctuate independently, as evidenced by the lack of oxidized NADP in the NDH-1-deficient strain and of oxidized NAD in the NDH-2-deficient strain, while the redox state of the other pyridine dinucleotide is fairly close to that in the wild type. This implies that there is no significant transhydrogenase activity in this cyanobacterium, although open reading frames have been identified in the Synechocystis sp. strain PCC 6803 genome sequence that appear to code for the A and B subunits of pyridine nucleotide transhydrogenase (slr1239 and slr1434) (13).
Another interesting finding is that the total level of NAD(H) and NADP(H) was variable, depending on the strain studied and the conditions under which it was grown. The reason for this, presumably, is that these compounds are produced via pathways starting at TCA cycle intermediates. NAD(P) is produced from the cyclization of glycerol and aspartate to form quinolinate. Aspartate is synthesized from oxaloacetate via aspartate aminotransferase or possibly from fumarate via fumarate dehydratase. Therefore, reduced levels of fumarate and oxaloacetate are likely to lead to lower levels of NAD(P).
A final observation about the cellular levels of NAD and NADP is that there is an order of magnitude more NADP than NAD, as determined by two independent methods. This difference in concentration probably is related to the fact that many important cellular reactions in the cyanobacterium are NADP specific rather than NAD specific and that NADP is used for storage of reductant generated by photosynthesis.
Relative activities of the respiratory complexes in the thylakoid membranes.
The reduction kinetics of the PQ pool in darkness upon addition of KCN were much slower in SDH-deficient strains (Fig. 1 and 2). The fast phase of the increase in the chlorophyll fluorescence yield was completely absent in the SDH-deficient strain (Fig. 2). From the estimated redox midpoint potential of QA/QA− versus PQ/PQH2, the amount of PQ, the amount of chlorophyll, and the rate of fluorescence yield increase upon KCN addition, the rate of reduction of the PQ pool in the wild type upon KCN addition was calculated to be 80 to 100 μmol of electrons (mg of chlorophyll)−1 h−1 (5), with a considerable potential underestimation because of the time it takes for KCN to diffuse in and to act. The observed respiratory rate (O2 uptake) in wild-type Synechocystis in the dark is about 120 to 200 μmol of electrons (mg of chlorophyll)−1 h−1 [30 to 50 μmol of O2 consumed (mg of chlorophyll)−1 h−1] (9). The rate of PQ reduction in the SDH-deficient strain was calculated to be 10 to 30 μmol of electrons (mg of chlorophyll)−1 h−1 (5), and therefore, the SDH complex accounts for the majority of the respiratory electrons donated to the PQ pool upon dark respiration. This implies that the remaining complexes (NDH-1 and NDH-2) account for less than half, and perhaps only a small fraction, of the respiratory electrons reaching the oxidases.
In line with this, initial results indicated that in the NDH-2-deficient strain, the rate of KCN-induced filling of the PQ pool was similar to that in the control (8). Indeed, according to the rate of KCN-induced QA reduction in darkness in the SDH-deficient strain relative to that in the SDH- and NDH-2-deficient strain (Fig. 2), the rate of PQ pool reduction by NDH-2 complexes is about 10 μmol of electrons (mg of chlorophyll)−1 h−1 and therefore accounts for only 5 to 10% of the total respiratory electrons donated to the PQ pool in darkness. This approximation assumes that NDH-1 activity remains the same in the presence and absence of NDH-2.
The observation that the rate of PQ pool reduction in the dark is about half in the SDH- and NDH-2-deficient strain compared to that in the SDH-deficient strain (Fig. 2) also implies that the rate of electron donation by NDH-1 (plus any unknown electron donors that may be present) to the PQ pool is similar to that of NDH-2. Therefore, the NDH-1 contribution to respiratory electron transport is relatively minimal (5 to 10% of the total respiratory electron transfer) in the thylakoid membrane of Synechocystis sp. strain PCC 6803.
The large contribution of SDH in providing electrons to the PQ pool in darkness is evidenced further by comparing the respiratory rates of the wild-type and SDH-deficient strains. The SDH-deficient strain grown photoheterotrophically showed very little KCN-sensitive oxygen consumption; the rate of oxygen uptake in this strain in darkness was 9 ± 5 μmol of O2 (mg of chlorophyll)−1 h−1, whereas in the presence of KCN, about 5 μmol of O2 (mg of chlorophyll)−1 h−1 remained (the latter rate of KCN-insensitive oxygen uptake is found in all of the Synechocystis sp. strain PCC 6803 isolates we have analyzed, and this oxygen uptake appears to be unrelated to respiration). This again illustrates that the contribution of other respiratory donors, NDH-1 and NDH-2, is minimal compared to that of SDH activity.
In summary, the SDH complex has a major role in respiratory electron flux and thereby in redox poising of the PQ pool. On the other hand, the direct role of the NDH-1 complex in electron donation to the PQ pool is minor, and changes in respiratory rates upon NDH-1 deletion need to be reinterpreted in terms of succinate depletion. Finally, as shown by evidence presented here and previously (8), NDH-2 also has a role in thylakoid PQ pool reduction that seems to be quantitatively similar to that of NDH-1.
ACKNOWLEDGMENTS
We thank Teruo Ogawa for the gift of the NdhB-deficient (M55) and NdhD-deficient strains, Mark Hayes and Karl Booksh (Department of Chemistry and Biochemistry, Arizona State University) for the use of the CV-50W analyzer, Lokesh Joshi (Department of Plant Biology, Arizona State University) for the use of the HPLC apparatus with fluorescence detection, and Crispin Howitt (currently at the Division of Plant Industry, Commonwealth Scientific and Industrial Research Organisation) for help with some of the Q electrode experiments. UQ-1 was a gift from David A. Ward (Department of Chemistry, University of Adelaide).
Support for this research was provided by grants from the Human Frontiers Science Program (RG 0051/19997M) and the U.S. Department of Agriculture National Research Initiative Competitive Program (97-35306-4881). Jason Cooley was supported by a Graduate Research Training grant from the National Science Foundation (DGE-9553456).
REFERENCES
- 1.Bellamacina C R. The nicotinamide dinucleotide binding motif: a comparison of nucleotide binding proteins. FASEB J. 1996;10:1257–1269. doi: 10.1096/fasebj.10.11.8836039. [DOI] [PubMed] [Google Scholar]
- 2.Berger S, Ellersiek U, Steinmüller K. Cyanobacteria contain a mitochondrial complex-I homologous NADH-dehydrogenase. FEBS Lett. 1991;286:129–132. doi: 10.1016/0014-5793(91)80957-5. [DOI] [PubMed] [Google Scholar]
- 3.Bouges-Boquet B. Electron transfer between the two photosystems in spinach chloroplasts. Biochim Biophys Acta. 1973;314:250–259. doi: 10.1016/0005-2728(73)90140-0. [DOI] [PubMed] [Google Scholar]
- 4.Cleland R E. Voltammetric measurement of the plastoquinone redox state in isolated thylakoids. Photosynth Res. 1998;58:183–192. [Google Scholar]
- 5.Cooley J W, Howitt C A, Vermaas W F J. Succinate:quinol oxidoreductase in the cyanobacterium Synechocystis sp. strain PCC 6803: presence and function in metabolism and electron transport. J Bacteriol. 2000;182:714–722. doi: 10.1128/jb.182.3.714-722.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Dry I, Moore A, Day D, Wiskich J. Regulation of alternative pathway activity in plant mitochondria: nonlinear relationship between electron flux and the redox poise of the quinone pool. Arch Biochem Biophys. 1989;273:148–157. doi: 10.1016/0003-9861(89)90173-2. [DOI] [PubMed] [Google Scholar]
- 7.Howitt C A, Smith G D, Day D A. Cyanide-insensitive oxygen uptake and pyridine nucleotide dehydrogenases in the cyanobacterium Anabaena PCC 7120. Biochim Biophys Acta. 1993;1141:313–320. [Google Scholar]
- 8.Howitt C A, Udall P K, Vermaas W F J. Type 2 NADH dehydrogenases in the cyanobacterium Synechocystis sp. strain PCC 6803 are involved in regulation rather than respiration. J Bacteriol. 1999;181:3994–4003. doi: 10.1128/jb.181.13.3994-4003.1999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Howitt C A, Vermaas W F J. Quinol and cytochrome oxidases in the cyanobacterium Synechocystis sp. PCC 6803. Biochemistry. 1998;37:17944–17951. doi: 10.1021/bi981486n. [DOI] [PubMed] [Google Scholar]
- 10.Howitt, C. A., J. W. Cooley, J. T. Wiskich, and W. F. J. Vermaas. A strain of Synechocystis sp. PCC 6803 without photosynthetic oxygen evolution and respiratory oxygen consumption: implications for the study of cyclic photosynthetic electron transport. Planta in press. [DOI] [PubMed]
- 11.Jeanjean R, Bédu S, Havaux M, Matthijs H C P, Joset F. Salt-induced photosystem I cyclic electron transfer restores growth on low inorganic carbon in a type 1 NAD(P)H dehydrogenase deficient mutant of Synechocystis PCC6803. FEMS Microbiol Lett. 1998;167:131–137. [Google Scholar]
- 12.Jorgenson B O, Rasmussen H J. Recycling analysis of nicotinamide-adenine dinucleotide phosphates (NADP and NADPH) Anal Biochem. 1979;99:297–303. doi: 10.1016/s0003-2697(79)80010-x. [DOI] [PubMed] [Google Scholar]
- 13.Kaneko T, Sato S, Kotani H, Tanaka A, Asamizu E, Nakamura Y, Miyajima N, Hirosawa M, Sugiura M, Sasamoto S, Kimura T, Hosouchi T, Matsuno A, Muraki A, Nakazaki N, Naruo K, Okumura S, Shimpo S, Takeuchi C, Wada T, Watanabe A, Yamada M, Yasuda M, Tabata S. Sequence analysis of the genome of the unicellular cyanobacterium Synechocystis sp. strain PCC6803. II. Sequence determination of the entire genome and assignment of potential protein-coding regions. DNA Res. 1996;3:109–136. doi: 10.1093/dnares/3.3.109. , 185–209. [DOI] [PubMed] [Google Scholar]
- 14.Klaidman L K, Leung A C, Adams J D. High-performance liquid chromatography analysis of oxidized and reduced pyridine dinucleotides in specific brain regions. Anal Biochem. 1995;228:312–317. doi: 10.1006/abio.1995.1356. [DOI] [PubMed] [Google Scholar]
- 15.Krieger A, Rutherford A W, Johnson G N. On the determination of redox midpoint potential of the primary quinone electron acceptor, QA, in photosystem II. Biochim Biophys Acta. 1995;1229:193–201. [Google Scholar]
- 16.Mourney N J, Kaplan S. Redox-dependent gene regulation in Rhodobacter sphaeroides 2.4.1(T): effects on dimethyl sulfoxide reductase (dor) gene expression. J Bacteriol. 1998;180:5612–5618. doi: 10.1128/jb.180.21.5612-5618.1998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Ogawa T. NAD(P)H dehydrogenase: a component of PS-I cyclic electron flow driving inorganic carbon transport in cyanobacteria. In: Murata N, editor. Research in photosynthesis III. Dordrecht, The Netherlands: Kluwer Academic Publishers; 1992. pp. 763–770. [Google Scholar]
- 18.Ohkawa H, Pakrasi H B, Ogawa T. Two types of functionally distinct NAD(P)H dehydrogenases in Synechocystis sp. strain PCC 6803. J Biol Chem. 2000;275:31630–31634. doi: 10.1074/jbc.M003706200. [DOI] [PubMed] [Google Scholar]
- 19.Pearce J, Leach C K, Carr N G. The incomplete citric acid cycle in the blue-green alga Anabaena variabilis. J Gen Microbiol. 1969;49:301–313. doi: 10.1099/00221287-55-3-371. [DOI] [PubMed] [Google Scholar]
- 20.Rippka R, Deruelles J, Waterbury J B, Herdman M, Stanier R Y. Generic assignments, strain histories and properties of pure culture of cynaobacteria. J Gen Microbiol. 1979;181:1875–1882. [Google Scholar]
- 21.Schmetterer G. Cyanobacterial respiration. In: Bryant D A, editor. The molecular biology of cyanobacteria. Dordrecht, The Netherlands: Kluwer Academic Publishers; 1994. pp. 409–435. [Google Scholar]
- 22.Shen J R, Vermaas W, Inoue Y. The role of cytochrome c-550 as studied through reverse genetics and mutant characterization in Synechocystis sp. PCC 6803. J Biol Chem. 1995;270:6901–6907. doi: 10.1074/jbc.270.12.6901. [DOI] [PubMed] [Google Scholar]
- 23.Sturzl E, Scherer S, Böger P. Interaction of respiratory and photosynthetic electron transport and evidence for membrane-bound pyridine-nucleotide dehydrogenase in Anabaena variabilis. Physiol Plant. 1984;60:479–483. [Google Scholar]
- 24.Tanaka Y, Katada S, Ishikawa H, Ogawa T, Takabe T. Electron flow from NAD(P)H dehydrogenase to photosystem I is required for adaptation to salt shock in the cyanobacterium Synechocystis sp. PCC6803. Plant Cell Physiol. 1997;38:1311–1318. [Google Scholar]
- 25.Vermaas W F J, Shen G, Styring S. Electrons generated by photosystem II are utilized by an oxidase in the absence of photosystem I in the cyanobacterium Synechocystis sp. PCC 6803. FEBS Lett. 1994;337:103–108. doi: 10.1016/0014-5793(94)80638-1. [DOI] [PubMed] [Google Scholar]
- 26.Wagner C T, Scott M D. Single extraction method for the spectrophotometric quantification of oxidized and reduced pyridine nucleotides in erythrocytes. Anal Biochem. 1994;222:417–426. doi: 10.1006/abio.1994.1511. [DOI] [PubMed] [Google Scholar]
- 27.Willeford K O, Gombos Z, Gibbs M. Evidence for chloroplastic succinate-dehydrogenase participating in the chloroplastic respiratory and photosynthetic electron transport chains of Chlamydomonas reinhardtii. Plant Physiol. 1989;90:1084–1087. doi: 10.1104/pp.90.3.1084. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Zannoni D, Moore A L. Measurement of the redox state of the ubiquinone pool in Rhodobacter capsulatus membrane fragments. FEBS Lett. 1990;271:123–127. doi: 10.1016/0014-5793(90)80387-x. [DOI] [PubMed] [Google Scholar]