Abstract
Binding of the neurotransmitter acetylcholine to its receptors on muscle fibers depolarizes the membrane and thereby triggers muscle contraction. We sought to understand at the level of 3D structure how agonists and antagonists alter nicotinic acetylcholine receptor conformation. We used the muscle-type receptor from the Torpedo ray to first define the structure of the receptor in a resting, activatable state. We then determined the receptor structure bound to the agonist carbachol, which stabilizes an asymmetric, closed channel desensitized state. We find conformational changes in a peripheral membrane helix are tied to recovery from desensitization. To probe mechanisms of antagonism, we obtained receptor structures with the active component of curare, a poison arrow toxin and precursor to modern muscle relaxants. Intriguingly, d-tubocurarine stabilizes the receptor in a desensitized-like state in the presence and absence of agonist. These findings define the transitions between resting and desensitized states and reveal divergent means by which antagonists block channel activity of the muscle-type nicotinic receptor.
Editor summary:
Here the authors reveal the structural basis of how the nicotinic acetylcholine receptor type found on skeletal muscle and in fish electric organs desensitizes in response to agonist, and how the arrow poison curare antagonizes the channel by stabilizing a desensitized state.
Introduction
The Cys-loop superfamily of cell surface receptors mediates chemical neurotransmission in the brain, spinal cord, and periphery. Nicotinic receptors are the founding members of this superfamily and comprise a wide range of receptor subtypes. In contrast to neuronal subtypes, the nicotinic receptor found postsynaptically at the neuromuscular junction is strictly defined in subunit composition1,2. This muscle-type receptor is a pentamer comprising four types of subunits, with two non-equivalent neurotransmitter-binding sites localized to interfaces in the extracellular domain3. Binding of the neurotransmitter acetylcholine shifts the conformational equilibrium from a resting state with a closed ion channel to a short-lived activated state with an open ion channel permeable to cations. The receptor rapidly closes in the sustained presence of agonist, adopting an agonist-bound, closed pore conformation termed the desensitized state4. The conformational changes in the muscle-type nicotinic receptor that underlie state transitions are poorly understood at the 3D structural level.
Neuromuscular blockers are routinely used during anesthesia to relax skeletal muscle; the first such drug was curare. Curare is a mixture of plant toxins first identified in the 16th century as an arrow poison used by South American indigenous peoples for hunting5. The experiments of Bernard (1844) and Langley (1906) demonstrated that curare induces neuromuscular block by antagonizing nicotinic receptors on skeletal muscle5,6. In 1935, King isolated and determined the chemical structure of the most active component of curare, the alkaloid d-tubocurarine (d-tubo)5,7,8. Curare has been leveraged to define the nature of the muscle nicotinic receptor, its architecture, how it signals, and was foundational in the development of modern muscle relaxants3,5. While this natural product has been used for over a century to study this archetypal ligand-gated ion channel, our understanding of where it binds and how it alters the channel structure to inhibit neuromuscular transmission is limited by a paucity of direct structural information.
Here we used the muscle-type nicotinic receptor found abundantly in the electric organ of the Torpedo ray to structurally define the resting and desensitized channel states and relate the process of desensitization to how d-tubo antagonizes the muscle-type receptor. We present five high-resolution cryo-EM structures of the Torpedo nicotinic receptor in the ligand free resting state with and without supplemental cholesterol, in the canonical desensitized state in complex with the agonist carbachol, in a d-tubo bound state, and in a combined d-tubo and carbachol bound state. These structures address fundamental questions in channel gating. First, we define the location and nature of high affinity cholesterol binding sites important for channel function, and thereby establish a robust reference for the resting state of this receptor. Second, we uncover differences between the resting and desensitized states that involve asymmetric conformational changes and distinguish the muscle-type receptor from the neuronal nicotinic receptors. Mutagenesis and electrophysiology measurements allow us to connect specific changes in the membrane domain to the rate of recovery from desensitization. Third, we show d-tubo stabilizes a desensitized-like structure by occupying the two orthosteric sites and two allosteric sites, distinct from how another antagonist, α-bungarotoxin, acts9. Lastly, the combined complex of d-tubo and carbachol provides insight into receptor desensitization by agonist in the presence of d-tubo, relevant to neuromuscular block, and reveals the distinct affinity determinants in the two non-equivalent orthosteric sites. Together, these results define a structural pathway from resting to desensitization and resolve how classical antagonists act through surprisingly diverse ways to block neuromuscular transmission.
Results and Discussion
Biochemistry and architecture
We purified the native muscle-type nicotinic acetylcholine receptor from the electric organ of Torpedo californica 10 and reconstituted receptors into soy lipid nanodiscs for structure determination by cryo-EM. We collected a cryo-EM dataset on this apo sample, as well as an apo sample supplemented with cholesterol, and reconstructed the receptor structures at 2.5 Å and 2.7 Å overall resolutions, respectively, to define lipid interactions (Table 1, Fig. 1a, and Extended Data Figs. 1, 2). The map quality was sufficient in both cases to model all ordered components of the receptor. We further obtained structures with bound carbachol to examine the desensitized state (2.8 Å), and with d-tubo (3.2 Å), and d-tubo plus carbachol (2.6 Å), to explore mechanisms of antagonism (Table 1 and Extended Data Figs. 1, 2).
Table 1.
Cryo-EM data collection, refinement and validation statistics
| Apo (EMD- 25202) (PDB 7SMM) |
Apo + cholesterol (EMD- 25205) (PDB 7SMQ) |
Carbachol (EMD- 25202) (PDB 7SMM) |
d-Tubo (EMD- 25207) (PDB 7SMS) |
d-Tubo + carbachol (EMD- 25208) (PDB 7SMT) |
|
|---|---|---|---|---|---|
| Data collection and processing | |||||
| Magnification | 81,000 | 81,000 | 81,000 | 81,000 | 81,000 |
| Voltage (kV) | 300 | 300 | 300 | 300 | 300 |
| Electron exposure (e–/Å2) | 50 | 50 | 50 | 50 | 50 |
| Defocus range (μm) | 0.5 - 2.5 | 0.5 - 2.5 | 1.0 – 3.0 | 0.8 – 2.5 | 0.5 – 2.5 |
| Pixel size (Å) | 1.0694 | 1.0694 | 1.0700 | 1.0694 | 1.0694 |
| Symmetry imposed | C1 | C1 | C1 | C1 | C1 |
| Initial particle images (no.) | 1,247,954 | 868,740 | 1,987,523 | 807,922 | 1,228,929 |
| Final particle images (no.) | 295,228 | 140,634 | 204,581 | 320,279 | 318,847 |
| Map resolution (Å) | 2.51 | 2.74 | 2.77 | 3.18 | 2.56 |
| FSC threshold | 0.143 | 0.143 | 0.143 | 0.143 | 0.143 |
| Map resolution range (Å) | 2.3 – 3.3 | 2.5 – 3.5 | 2.5 – 3.5 | 3.0 – 4.0 | 2.4 – 3.4 |
| Refinement | |||||
| Initial model used (PDB code) | 6UWZ | 6UWZ | 6UWZ | 6UWZ | 6UWZ |
| Model resolution (Å) | 2.57 | 2.82 | 2.83 | 3.18 | 2.61 |
| FSC threshold | |||||
| Model resolution range (Å) | |||||
| Map sharpening B factor (Å2) | −20 | −15 | −40 | −70 | −20 |
| Model composition | |||||
| Non-hydrogen atoms | 17,256 | 17,427 | 17,037 | 17,051 | 17,016 |
| Protein residues | 2,030 | 2,030 | 2,022 | 2,015 | 2,022 |
| Ligands | 0 | 0 | 2 | 4 | 2 |
| B factors (Å2) | |||||
| Protein | 67.96 | 67.29 | 65.41 | 74.03 | 71.60 |
| Ligand | - | - | 49.79 | 82.13 | 65.79 |
| R.m.s. deviations | |||||
| Bond lengths (Å) | 0.004 | 0.005 | 0.005 | 0.005 | 0.005 |
| Bond angles (°) | 0.538 | 0.592 | 0.536 | 0.663 | 0.586 |
| Validation | |||||
| MolProbity score | 1.42 | 1.47 | 1.36 | 1.42 | 1.38 |
| Clashscore | 3.39 | 4.84 | 4.25 | 5.16 | 5.16 |
| Poor rotamers (%) | 0 | 0 | 0 | 0 | 0 |
| Ramachandran plot | |||||
| Favored (%) | 95.87 | 96.57 | 97.15 | 97.24 | 97.50 |
| Allowed (%) | 4.13 | 3.43 | 2.85 | 2.76 | 2.50 |
| Disallowed (%) | 0 | 0 | 0 | 0 | 0 |
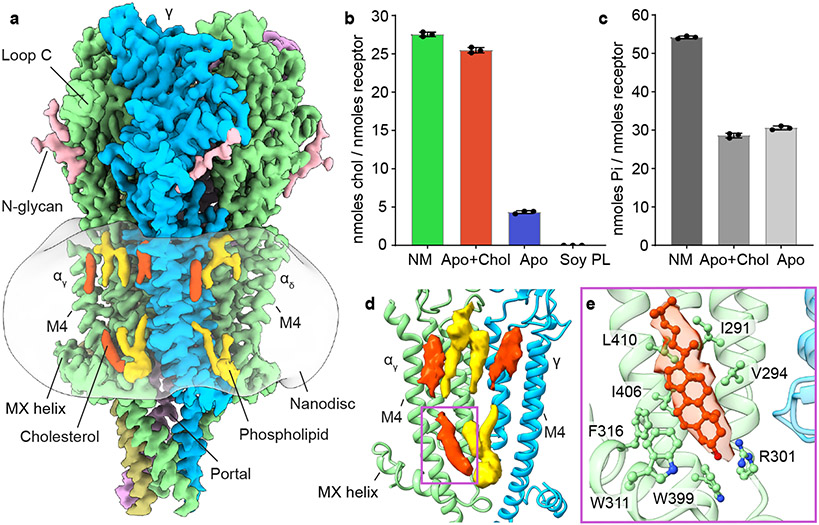
Fig. 1: Resting state structure and cholesterol interactions.
a, Side view of apo/resting state map; α subunit - green, β subunit - khaki, γ subunit - blue, δ subunit - violet, cholesterols - red, phospholipids - yellow and N-glycans - pink; lipid nanodisc is semi-transparent surface. b, c, Cholesterol and phospholipid quantification assays: native membrane (NM); apo + cholesterol (Apo+Chol); apo (Apo) and soy polar lipid (Soy PL). Data are presented as the mean +/− SD of 3 separate measurements from each sample. Data for graphs b in c are available as source data. d, Lipid densities at a representative interface, αγ-γ. e, Cholesterol interaction with receptor. Corresponding cholesterol density is shown as semi-transparent surface.
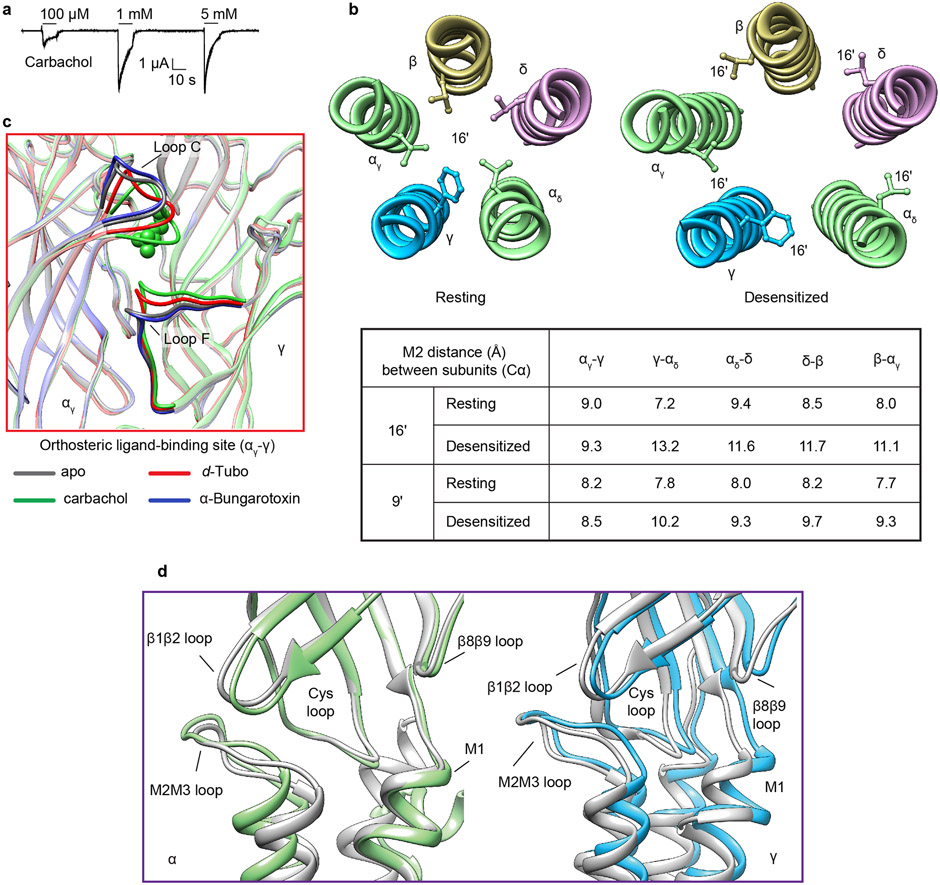
The Torpedo nicotinic receptor consists of four homologous subunit types arranged α-γ-α-δ-β counterclockwise around a pseudo-five-fold symmetry axis (Figs. 1, 2) 3,11,12. Two non-equivalent orthosteric binding sites are found at the α-γ and α-δ subunit interfaces in the extracellular domain (ECD). In both, an α-subunit forms the “principal” side and either a γ or δ subunit forms the “complementary” side of the interface3. These two α-subunits are termed αγ and αδ based on their complementary neighbors. The transmembrane domain (TMD) of each subunit comprises four helices, M1-M4, with M2 lining the ion channel and M4 being most peripheral. The intracellular domain (ICD) of each subunit is formed by a partially ordered loop between the M3 and M4 helices: an amphipathic MX helix following M3, and a long helix called MA that leads into and is continuous with M4. The MA helices form a bundle at their N-termini and frame lateral portals for ion flux13. The N- and C-termini of each subunit are extracellular, with long ordered structures in the C-termini of the δ and γ subunits9.
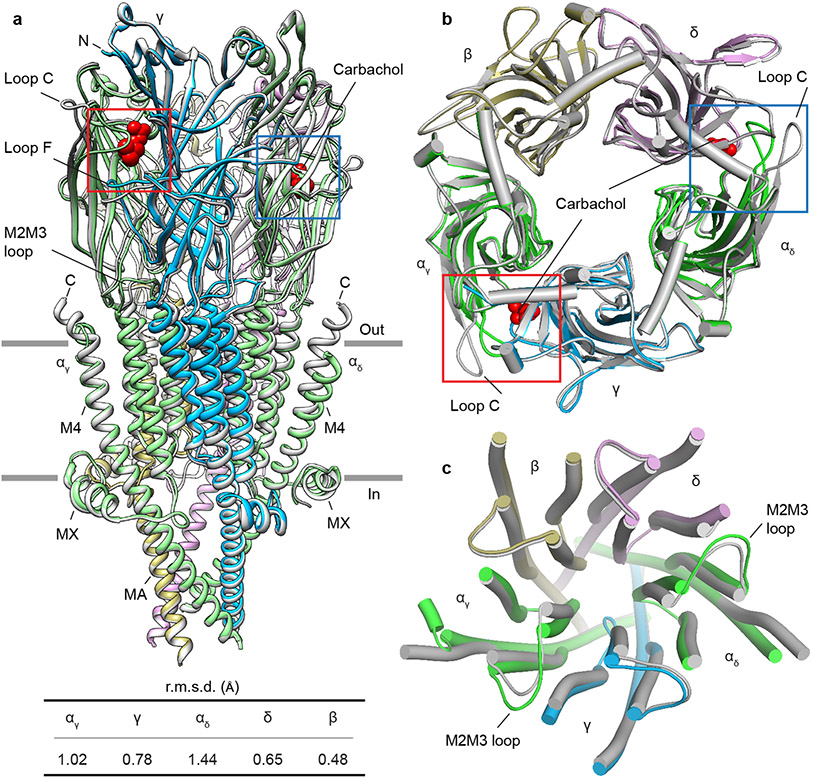
Fig. 2: Asymmetric conformational transition between resting and desensitized states.
a, Superposition of resting and desensitized states; desensitized model is colored (α- green, β- khaki, γ- blue, δ- violet) and resting is in gray. Carbachols are shown as red spheres. Table contains the r.m.s.d. values of subunits between resting and desensitized states, where the αδ subunit showed the largest conformational change and the β- subunit the least. b, c, Top views of superimposed ECD and TMD, respectively.
Cholesterol sites
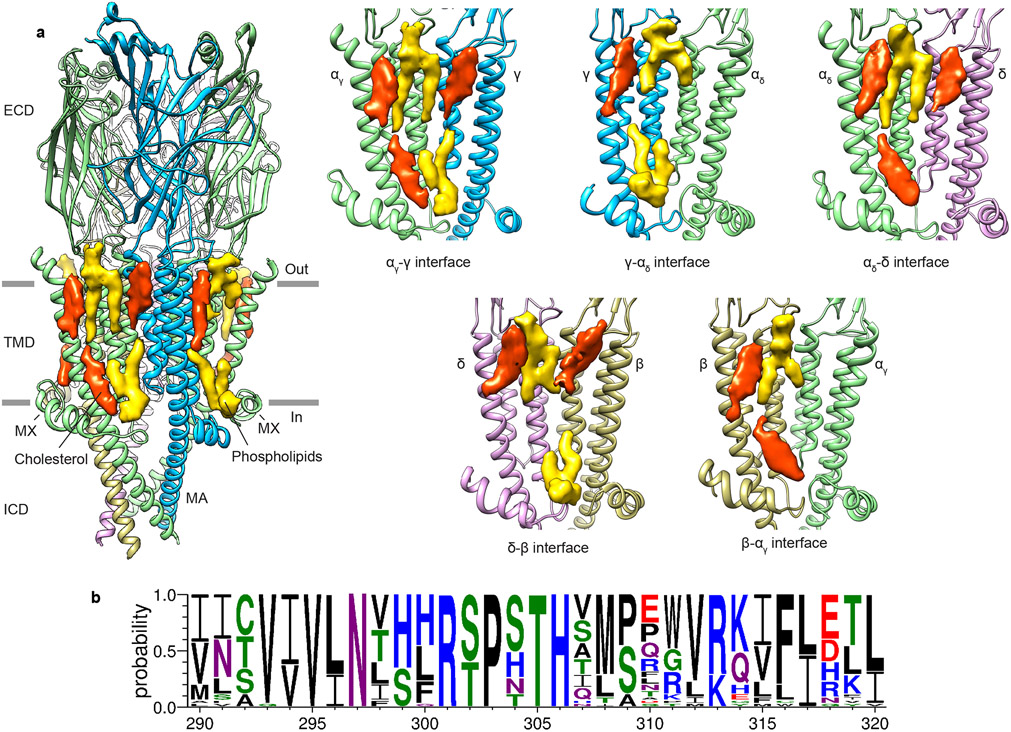
We aimed to first establish a reference structure for the resting state receptor before investigating gating transitions and antagonism. Lipids are known to modulate the ability of agonists to gate the acetylcholine receptor channel 14-16; both anionic phospholipids and cholesterol potentiate channel activity 16,17. Cholesterol quantification estimated that 4-5 cholesterol molecules per receptor remained bound through purification in the presence of soy lipids alone (Fig. 1b), a condition that yields functional protein9. The Torpedo native membrane is comparatively rich in cholesterol, containing 27-28 cholesterol molecules per receptor, in a molar ratio of 1:2 with phospholipids (Figs. 1b, c). The sample purified in soy lipids supplemented with cholesterol contains 25-26 molecules of cholesterol per receptor, nearly identical to the native membrane ratio (Fig. 1b). The resting state receptor structures, with and without added cholesterol, allowed us to better define high and low affinity cholesterol binding sites, to differentiate cholesterol-associated density from phospholipids, and to map cholesterol binding interactions. Structural comparison between the cholesterol poor and cholesterol rich preparations revealed minimal conformational differences in the receptor (Cα r.m.s.d. 0.16 Å). Lipid densities were observed near the MX helices and in the outer membrane leaflet (Figs. 1a, d), consistent with lower resolution studies of the receptor in native-like tubular membranes18,19. The clearest cholesterol densities observed in both resting-state structures are present in the inner leaflet near the MX-helices and represent a presumed high-affinity site (Figs. 1a, d and Extended Data Fig. 3a). Among the interacting residues in the high-affinity cholesterol binding sites, R301 (M3-MX loop) and F316 (MX) are conserved across subunits and species (Fig. 1e and Extended Data Fig. 3b). The cholesterol densities that became stronger after adding cholesterol define the low affinity binding sites and are present in the outer membrane leaflet (Figs. 1a, d and Extended Data Fig. 3a). Together, these findings define sites of cholesterol interaction and establish a robust structural standard for the apo receptor in its activatable resting state.
Structural transition between resting and desensitized states
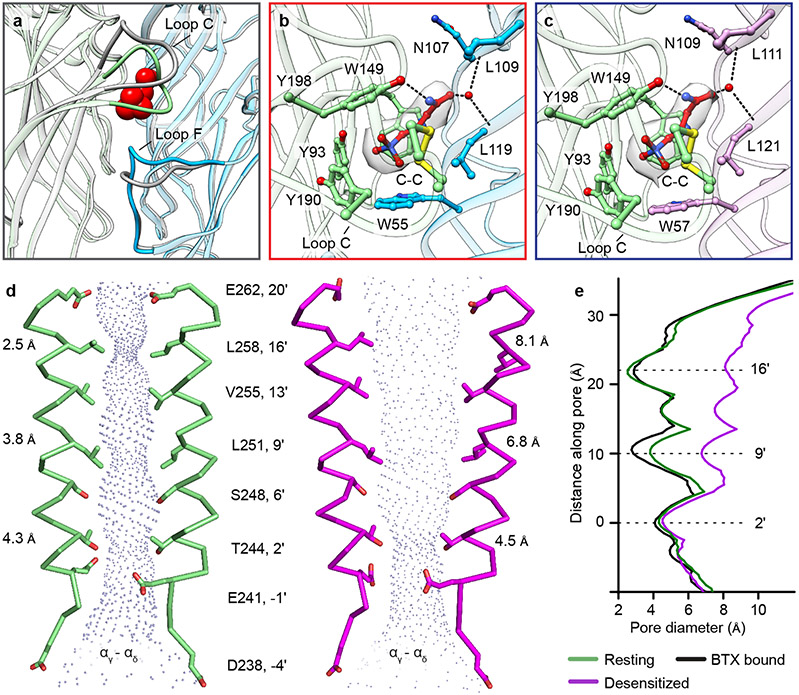
Most ligand-gated ion channels desensitize in the sustained presence of agonist, a phenomenon first described in studies of the muscle nicotinic receptor4. The carbachol-bound Torpedo receptor structure represents this desensitized state (Fig. 2, and Extended Data Fig. 4a). Structural comparison of the resting and desensitized states reveals asymmetry in this conformational change that begins in the agonist site (Fig. 2 and Extended Data Fig. 4b). Carbachol, a stable acetylcholine analog, binds at α-γ and α-δ interfaces in the ECD (Figs. 2a, b). Among the six interfacial loops at the neurotransmitter-binding interfaces, loop C from principal α-subunits and loop F from complementary γ and δ subunits undergo local hinge motions that make the agonist site more compact (Figs. 2a, b and 3a). Bound carbachol is thereby stabilized in similar orientations in the two binding pockets, largely inaccessible to solvent (Figs. 3b, c). Its choline quaternary ammonium group orients toward the membrane, packing against the faces of four aromatic residues contributed by both subunits, with αW149 contributing to a cation-π interaction20. The agonist carbamoyl group orients away from the membrane to form electrostatic interactions with loop C and with an ordered water molecule deep in the pocket (Figs. 3b, c). The atomic interactions of carbachol with the Torpedo receptor are superficially consistent with those in the soluble acetylcholine binding protein21; in the latter, stabilization of the carbamoyl group through electrostatic interactions with loop C and ordered water were not observed.
Fig. 3: Agonist binding site and ion pore profile.
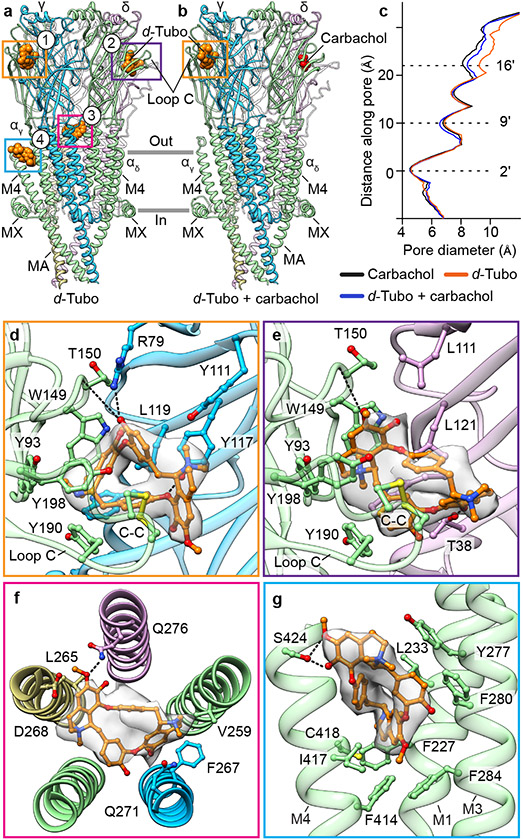
a, Conformational changes in loop C of an α-subunit and Loop F of a complementary subunit (γ) after carbachol binding. Desensitized model is colored (α- green, γ- blue) and resting is in gray; carbachol is shown as red spheres. b, c, Carbachol at α/γ interface and α/δ interface, respectively; α- green, γ- blue, δ- violet. Carbachols are shown as red sticks and corresponding density is shown as semi-transparent surface; water as red spheres. d, Pore profile of resting state (green) and desensitized state (magenta) with M2 helices and their pore-lining residues shown for the two α subunits. e, Pore diameter comparison between resting, desensitized and α-bungarotoxin (BTX) bound structures (PDB ID: 6UWZ).
Agonist binding brings the extracellular domains of the principal and complementary subunits closer together (Figs. 2a, b, 3a and Extended Data Fig. 4c), a mechanism conserved in pentameric ligand-gated ion channels22,23. The conformational change between resting to desensitized in the Torpedo receptor can be described as a rotation, upon agonist binding, of the ECD half of the each of the principal and complementary subunits, counterclockwise about an axis approximately perpendicular to the membrane and through the center of mass of each subunit, in combination with local translations (Fig. 2b and Movies S1-2). In contrast to the α, γ and δ subunits, the β subunit behaves as a rigid scaffold, with neither substantial local hinge motions nor larger domain movements between the resting and desensitized states (Figs. 2 a-c and Movies S1-2). The conformational changes initiated by agonist binding in the ECD are transmitted to the TMD through the coupling region (Extended Data Fig. 4d and Movies S1-2). Here, the Cys-loop and β1β2 loop from each subunit impinge on its own M2M3 loop, and as the α subunits move the most in the ECD to accommodate agonist binding, the TMDs of the two α-subunits undergo the largest conformational changes (Fig. 2c), consistent with functional studies in mammalian muscle nicotinic receptors and low-resolution reconstructions of the receptor from Torpedo24-28. The Cys-loop and β1β2 loop interactions with the M2M3 loop drive the extracellular ends of the M2 and M3 helices away from the channel axis. This transition occurs in all subunits but is subtle in the rigid β subunit and most pronounced in the αδ subunit, likely because the latter is sandwiched between two complementary subunits that, unlike the β subunit, participate directly in agonist binding (Figs. 2a-c and Movies S1-2). Accordingly, among the complementary subunits, the conformational change in the γ subunit, positioned between two principal subunits, are larger than in δ, which packs against the rigid β subunit (Figs. 2a-c and Movies S1-2). The ICD bundle of MA helices does not undergo a substantial conformational change (Fig. 2a), consistent with it acting as an anchor in the gating cycle29.
Activation and desensitization channel gates are distinct
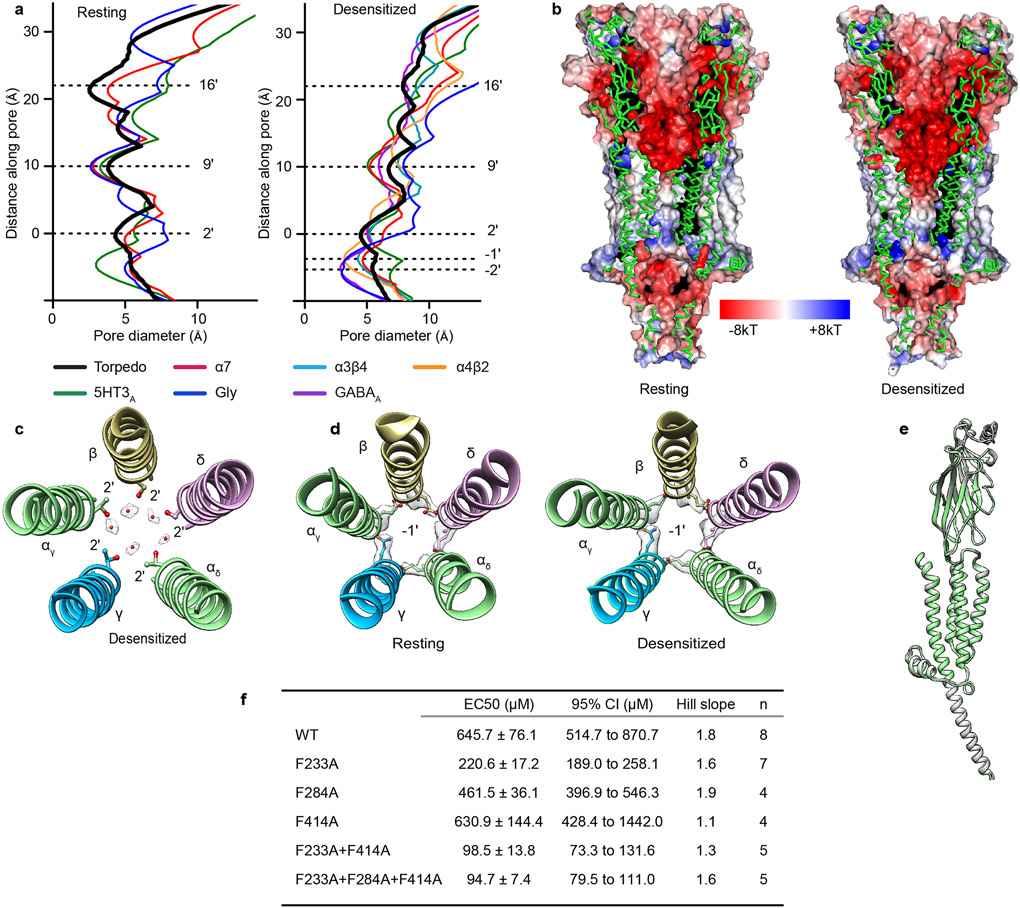
The physiologically critical consequence of these structural changes is their effect on ion channel conformation and conductance. The Torpedo receptor resting state structure reveals a tightly closed transmembrane pore with two distinct hydrophobic pinch points in its extracellular half. The main constrictions are defined by side chains of 9′L and 16′ (L/ α, β, δ; F/ γ) that result in minimal diameters of 3.8 and 2.5 Å respectively (Figs. 3d, e). This pair of hydrophobic constrictions presents an extensive barrier to hydrated cations, the permeant species 30, and defines the activation gate. That this gate comprises more than the conserved 9′L is consistent with prior studies wherein channels with polar substitutions still open and close31,32. The pore conformations in the resting state and in the complex with α-bungarotoxin 9 are similar but not identical (Fig. 3e), where the latter shifts the tightest constriction from the 16′ position to the 9′ position. Comparison with other Cys-loop receptors reveals conservation of the 9′ residue as a component of the activation gate (Extended Data Fig. 5a)23,33-36. The Torpedo resting structure therefore comprises a closed channel with two constrictions (9′ and 16′) impermeable to hydrated ions, as opposed to a simple 9′L gate.
The pore conformation in the desensitized state is funnel shaped and distinct from that in the resting state37 (Figs. 3d, e). The main constriction and hydrophobic gate of the resting state (16′ position) becomes wider and electronegative in the desensitized state (diameter increases from 2.5 to 8.1 Å; Figs. 3d, e, and Extended Data Fig. 5b). The 9′ constriction also widens in the desensitized state (from 3.8 to 6.8 Å), coincident with a ring of five water molecules appearing at the 2′ level (Extended Data Figs. 5 b, c). The main constriction (4.5 Å) is near the cytoplasmic mouth at the 2′ position, which contains either threonine or serine in each subunit (Figs. 3d, e). In contrast, neuronal nicotinic receptors have their principal desensitized state constriction at the −1′ position, delimited by side chains of glutamate residues29,38-40 (Extended Data Fig. 5a). Intriguingly, these −1′ side chains in the Torpedo receptor, four glutamates and one asparagine, do not orient toward the pore axis, but instead cap the N-termini of M2 helices in adjacent subunits (Extended Data Fig. 5d). The result is a more extracellular desensitization gate in the Torpedo nicotinic receptor than in neuronal nicotinic receptors 1. Comparison with other Cys-loop receptors reveals diversity in the desensitization gate position; GABAA and glycine receptor structures share −2′ gates while a desensitized-like 5-HT3A receptor structure had a 2′ constriction, akin to what we observe in the Torpedo receptor23,36,41-44 (Extended Data Fig. 5a). Together, the Torpedo receptor structures reveal an activation gate spanning much of the extracellular half of the pore that widens dramatically in the desensitized state, and a desensitization gate at the cytosolic mouth of the pore that is equivalently closed in both the resting and desensitized states.
Desensitization involves asymmetric uncoupling of M4
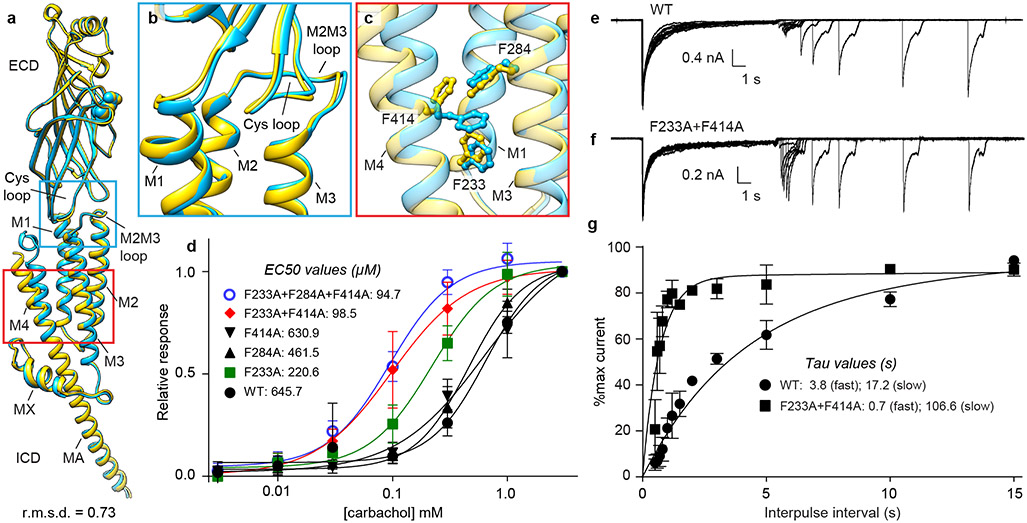
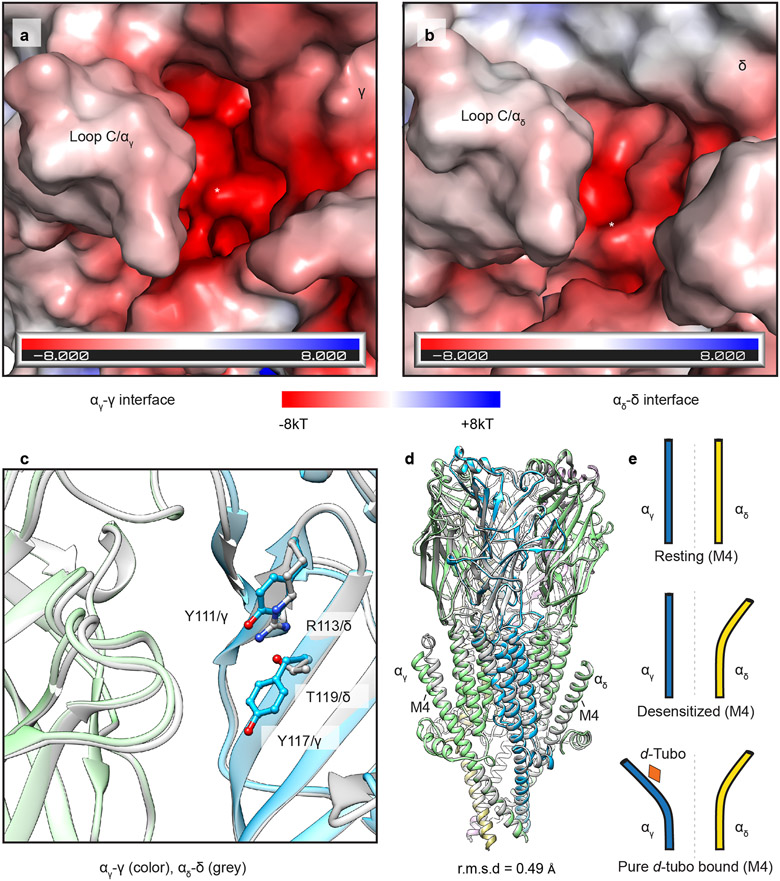
The two α-subunits adopt nearly identical conformations in the more symmetric resting state and differ from each other in the carbachol-bound desensitized state (Fig. 4a and Extended Data Fig. 5e). The most striking difference occurs in the extracellular end of the M4 helix of the αδ subunit, which is no longer in direct contact with the core of the receptor, rendering it conformationally disordered compared to the αγ subunit (Fig. 4a). This conformational difference in M4 occurs in concert with a smaller movement of M2 and M3 relative to M1 in the αδ TMD bundle (Figs. 4b, c). The bundle movement alters interactions among three phenylalanine residues (F233, F284 and F414) at the M1, M3 and M4 helical junction, causing a rotameric flip in F414 on M4; clashes in the resting-state conformation of these side chains detach M4 at its extracellular end from the coupling region (Fig. 4c).
Fig. 4: M4 uncoupling in the desensitized state.
a, Superposition of two α-subunits; αγ in blue, αδ in yellow, agonist carbachol as spheres. Conformational differences in b, coupling region and c, TMD. d, Dose response by TEVC of WT and mutants. Confidence intervals are in Extended Data Fig. 5f. e, f, Repeated two-pulse agonist application was used to measure apparent desensitization recovery kinetics for WT and mutant, shown for single representative cells. g, The rate of recovery was fitted with a sum of two exponentials. For d, WT (n=8), F233A (n=7), F284A (n=4), F414A (n=4), F233A-F414A (n=5), F233A-F284A-F414A (n=5) and g, n=5 independent cells, and data are presented as the mean +/− SD. Data for graphs in d-g are available as source data.
To functionally probe the role of these three residues in Torpedo receptor function, we made single, double, and triple mutants to alanine, and performed dose-response experiments using two-electrode voltage-clamp (TEVC) electrophysiology (Fig. 4d and Extended Data Fig. 5f). Among the three residues, F233A increased agonist potency ~3-fold and the F233A + F414A double mutant 7-fold, consistent with an alanine scanning study that identified both F233 and F414 in the human muscle receptor as gain of function mutants 45. F284A did not cause a measurable change in agonist potency for Torpedo. We hypothesized that the gain of function could stem from a decrease in stability of the desensitized state. We tested this hypothesis by measuring the apparent rate of recovery from desensitization. As the Torpedo muscle-type receptor does not express well in HEK cells, and solution exchange in oocytes is slow, we conducted these experiments by patching the αβγδ mouse muscle nicotinic receptor, WT and the mutant F233A + F414A, expressed in HEK cells (Figs. 4e-g). We found that the mutant recovers ~6-fold faster from desensitization than WT (Fig. 4g). These data suggest that the bulky phenylalanine side chains may act like a doorstop to stabilize the desensitized state; when they are pruned to alanine, the desensitized state is less stable and channel activity recovers faster.
The conformational detachment of M4, coincident with altered interactions among the phenylalanines, is not observed in the αγ subunit. The αγ M1 helix packs against the rigid β subunit, which restricts its movement compared to that of M1 in the αδ subunit; this restraint appears to be sufficient to keep αγ M4 tethered to the TMD bundle and coupling region (Figs. 2c and 4b, c). Notably, the M4 helix has been proposed as a lipid sensing domain for pentameric ligand gated ion channels45-49. Its peripheral location, conformational sensitivity in α subunits, and functional relationship to desensitization further support how changes in lipidic environment could tune gating through altering M4 stability.
d-Tubocurarine stabilizes a desensitized channel conformation
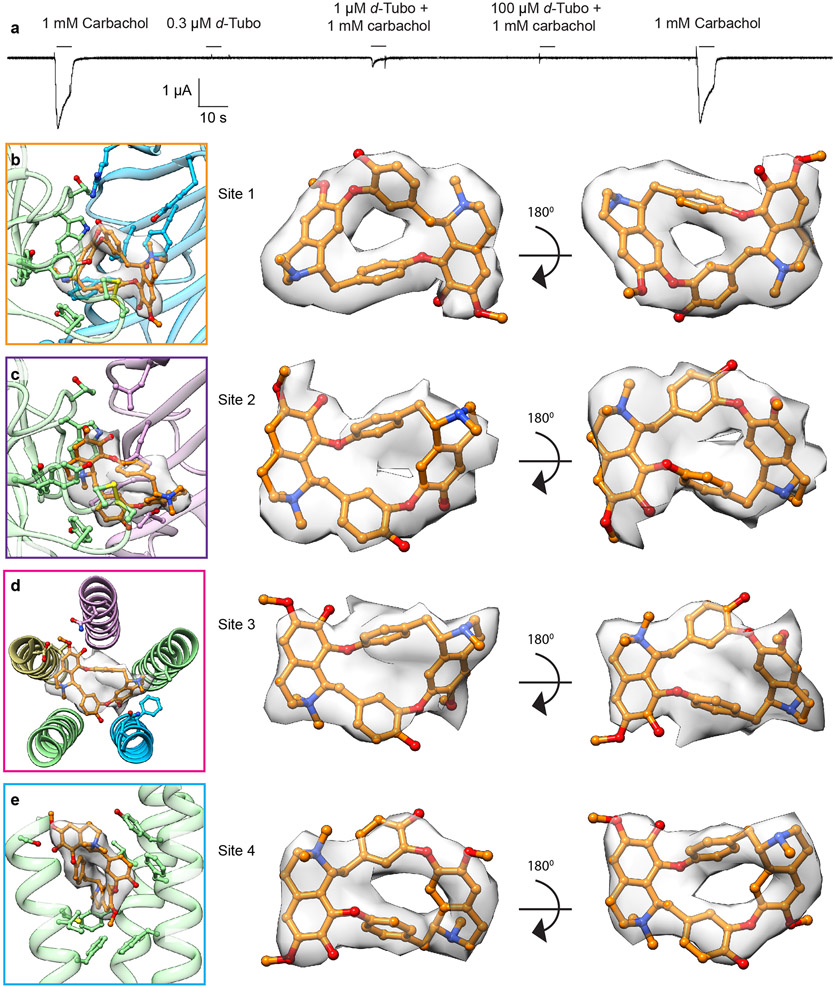
d-Tubo is a plant alkaloid and competitive antagonist of the muscle-type nicotinic receptor. This toxin was initially thought to simply compete with acetylcholine binding and stabilize a resting-like state50. Affinity labelling and functional studies suggested that d-tubo, surprisingly, desensitizes the receptor, and enhances agonist-induced desensitization51,52. Further, d-tubo is a low efficacy agonist at the fetal muscle nicotinic receptor but does not have agonist activity on the adult muscle receptor53,54, like Torpedo. We determined Torpedo receptor structures with d-tubo bound, and separately with a combination of d-tubo and carbachol bound, to interrogate structural mechanisms of inhibition by this enigmatic toxin (Figs. 5a, b and Extended Data Fig. 6a). The structure with only the antagonist bound was determined in the presence of 500 μM d-tubo, a concentration 50-fold higher than the necessary to occupy both agonist sites. Both d-tubo bound structures adopt similar desensitized-like conformations (Fig. 5c). In the presence of d-tubo alone, clear toxin density is present in both orthosteric agonist sites and in two unexpected allosteric sites, one in the extracellular mouth of the pore (site 3) and another wedged among the extracellular ends of the M1, M3 and M4 helices of the αγ subunit (site 4) (Fig. 5a).
Fig. 5: Structural pharmacology of d-tubocurarine.
a, b, Side views of pure d-tubo bound, and d-tubo plus carbachol bound structures, respectively. α- green, β- khaki, γ- blue, δ- violet; carbachol is shown as red spheres and d-tubo as orange spheres. c, Pore diameter comparison. d, d-Tubo at α/γ interface (site 1). e, d-Tubo at α/δ interface (site 2). f, d-Tubo at pore (site 3). g, d-Tubo at M1, M3 and M4 helices of αγ subunit (site 4). Ligands and interacting residues are shown as sticks and corresponding densities are shown as semitransparent surfaces. Electrostatic interactions are indicated by dashed lines.
In the d-tubo plus carbachol bound structure, d-tubo occupies the α/γ site and carbachol the α/δ site (Fig. 5b). d-Tubo and carbachol are known to have higher affinities for opposite orthosteric sites. d-Tubo has 400-fold higher affinity for the α/γ site while carbachol has a 10-fold higher affinity for the α/δ site51,55,56. Comparison between the desensitized and pure d-tubo bound structures reveals that carbachol binds similarly in its two orthosteric sites (Figs. 3b, c), while the d-tubo orientations between its two orthosteric sites are strikingly different, where opposite faces of its ring orient toward the back of the pocket. (Figs. 5d, e, and Extended Data Fig. 6b, c, and Movies S3-4). As the two α-subunits are identical, the affinity differences between the two orthosteric sites arise from interactions with the complementary subunits (γ and δ)57. The resting structure reveals that both sites are accessible to solvent, which would facilitate neurotransmitter binding. However, the α/γ site is more electronegative compared to the α/δ site (Extended Data Figs. 7a, b). Superposition of the two neurotransmitter binding pockets highlights two key residue differences: Y111/γ to R113/δ and Y117/γ to T119/δ (Extended Data Fig. 7c). These two tyrosines are positioned near the quaternary ammonium group of d-tubo in the higher affinity site (α/γ). Among the two tyrosines, Y117/γ is positioned to make a cation-π interaction with the quaternary ammonium group and an electrostatic interaction between its hydroxyl oxygen and a d-tubo ether (Fig. 5d). Mutagenesis supports that Y111 and Y117 in γ are two of the key determinants of high d-tubo affinity58,59. Furthermore, d-tubo, which carries two positive charges at physiological pH, may bind more favorably to the more electronegative α/γ interface site, while the carbonyl group of carbachol lends itself to tighter binding at the more electropositive α/δ site (Extended Data Figs. 7a, b).
In addition to the predicted competitive orthosteric binding interactions, when added to the EM sample at a higher concentration, d-tubo binds in two well-resolved allosteric sites (Fig. 5a). The d-tubo in the pore (site 3) wedges between 16′ and 20′ (Fig. 5f and Extended Data Fig. 6d, and Movie S5). Pore blocking is a well-documented mechanism for non-competitive antagonists and has been suggested for d-tubo53,60,61. d-Tubo at site 4 interacts with the extracellular end of the M1, M3 and M4 helices of αγ (Fig. 5g and Extended Data Fig. 6e, and Movie S6), stabilizing a detached M4 helix conformation, with the flipped F414 side chain, as in the carbachol-bound αδ subunit (Fig. 4, and Extended Data Fig. 7d, e). Using functional analysis, we concluded that this uncoupling of M4 relates to the slower recovery from desensitization (Fig. 4). This fourth binding site is to our knowledge previously undescribed, and a potential negative allosteric modulator site for the muscle nicotinic receptor. Together, the d-tubo structures suggest that its physiological inhibition is dominated by binding to the higher affinity orthosteric site, consistent with functional measurements62, that d-tubo and carbachol can bind simultaneously to desensitize the channel, and that sites 3 and 4 may contribute to functional inhibition of the muscle-type receptor by some drugs.
Conclusion
The muscle type nicotinic receptor has served as the archetype for ligand gated ion channels, yet high resolution structural information has lagged behind other pentameric receptors. The apo Torpedo receptor structure defines the first bona fide resting state for a heteromeric nicotinic receptor. The desensitized state structure reveals the agonist recognition mechanism and a desensitization gate at the 2′ position, which is distinct from the activation gate encompassing the 16′ and 9′ positions. The transition from resting to desensitized is asymmetric, wherein the α-subunits undergo large conformational changes relative to all other subunits. We find that M4 uncoupling is linked to the recovery from desensitization. The asymmetric transitions observed here make the muscle-type receptor distinct from the neuronal nicotinic receptors structurally characterized to date, and are in line with functional experiments on the related GABAA receptor showing subunit-dependent contributions to desensitization63. d-Tubo structurally defines a new mode of channel antagonism, which is distinct from α-bungarotoxin and stabilizes a desensitized-like state. d-Tubo further reveals a new type of allosteric site near the M4 helix of the α-subunit, and the first structural example of pore block in cation-selective Cys-loop receptors, which include the nicotinic and 5-HT3 families. This structural information provides a foundation for developing novel therapeutics selective for muscle receptors that are safer neuromuscular blocking drugs and anesthetics.
METHODS
Receptor purification
The native nicotinic receptor was purified from Tetronarce californica (formerly, Torpedo californica). The protocol for membrane fraction preparation is the same as previously described9,10. We received liquid nitrogen frozen electroplaque tissue from EastCoast Bio and stored in −80 °C. For each preparation, 100 g frozen tissue were thawed in a 500 mL beaker with 300 mL of buffer A (400 mM NaCl, 20 mM NaH2PO4, pH 7.4) and 150 mg of NEM (N-ethylmaleimide, Sigma) at room temperature (RT). Subsequent steps were carried out at 4 °C or on ice. The tissue with buffer A was homogenized in a tissue homogenizer (Kinematica Polytron Homogenizer, GmbH) and centrifuged at 3,220g for 15 min. The supernatant was collected after filtering through cheesecloth and gently stirred for 5 min with a protease inhibitor tablet (cOmplete mini, Sigma). This material was centrifuged at 105,000g for 30 min and the membrane pellet was collected. Pellet was incubated for 30 minutes after re-suspending in buffer B (80 mM NaCl, 20 mM Tris pH 11.0, 1 mM EDTA, 20% sucrose). The sample was centrifuged again at 105,000g for 30 min and the membrane pellet was collected. Finally, the membrane pellet was washed with buffer C (80 mM NaCl, 20 mM NaH2PO4, pH 7.4) and stored at −80 °C for further use. 2 g of membrane pellets were taken out of the freezer and thawed on ice. The pellets were suspended with 50 mL of cold buffer C (80 mM NaCl, 20 mM NaH2PO4, pH 7.4) using a homogenizer. Phenylmethylsulfonyl fluoride (PMSF, final concentration 1 mM) and Triton X-100 (final concentration, 1.5% v/v) were added to the sample and gently stirred for 1 hr. The supernatant was collected after centrifugation at 105,000g for 30 min at 4 °C and was diluted with 100mL of cold buffer C. The affinity reagent ATM (2-[(4-aminobutanoyl)amino]-N,N,N-trimethylethanaminium) was synthesized and incorporated into NHS Sepharose resin (Cytiva) as previously described9,10. 5 mL packed ATM resin was equilibrated with buffer C. Resin was mixed with the supernatant and nutated for 1 hour. The resin was washed with 50 mL of buffer D (80 mM NaCl, 20 mM Tris pH 7.4, 1 mM EDTA, 1 mM n-dodecyl-β-D-maltoside, DDM, Anatrace). For the preparation of apo and pure d-tubo bound forms, the receptors were eluted with buffer E (80 mM NaCl, 20 mM Tris 7.4, 1 mM EDTA, 1 mM DDM, 50 mM βME, and 1.2 M choline). To prepare the carbachol and d-tubo plus carbachol bound receptor samples, we used elution Buffer E containing 50 mM carbachol instead of 1.2 M choline. To obtain the apo sample, we used a high concentration of choline instead of carbachol in the elution buffer, after which weak-binding choline was straightforward to remove from the receptor by SEC. The eluted samples were analyzed by fluorescence size-exclusion chromatography monitoring intrinsic tryptophan fluorescence. The sample was concentrated to an A280 of ~7-8 with 100 kDa cutoff Amicon Ultra concentrators (Millipore).
Reconstitution in nanodiscs
For nanodisc reconstitution, the molar ratio of receptor: saposin: soy polar lipid (Avanti Polar Lipids) was 1:25:150. For the apo plus cholesterol sample, cholesterol was added in a 1:4 molar ratio to soy polar lipids. The Saposin A expression plasmid was provided by Salipro Biotech64. The receptor was mixed with lipids and incubated for 20 minutes at RT. Saposin was then added and incubated at RT for another 2 minutes. Approximately 120 mg of Bio-Beads (SM2, Bio-Rad) that were washed with methanol once, 3 times with Milli-Q water and one time with TBS (80 mM NaCl, 20 mM Tris pH 7.4). 500-600 μL of reaction sample was mixed with the washed Bio-Beads and rotated overnight at 4 °C. Fresh washed Bio-Beads were exchanged in the morning and rotated for an additional hour. The saposin-reconstituted receptors were separated from the Bio-Beads using a 1 mL syringe with needle and one round of centrifugation at 98,600g for 40 min to remove any precipitation or Bio-Beads. The samples were then run through a size-exclusion chromatography (SEC) column (Superose 6 10/300 GL Increase) equilibrated in TBS to remove empty nanodiscs and βME from the sample.
Cryo-EM sample preparation
To obtain the carbachol-bound structure, the final sample was supplemented with 5 mM carbachol. The addition of 100 μM d-tubo in the final sample that was affinity purified with carbachol resulted in a structure with both d-tubo and carbachol bound. To solve the pure d-tubo bound structure, the apo receptor was supplemented with 500 μM d-tubo. Samples were then concentrated to A280 of ~7. Freshly prepared fluorinated Fos-Choline-8 (Anatrace) was added to the samples just before freezing grids, to a final concentration of 1 mM. Grids (Quantifoil R 1.2/1.3, 200 mesh, Cu/Au) were plunge frozen into liquid ethane using a Vitrobot Mark IV (FEI).
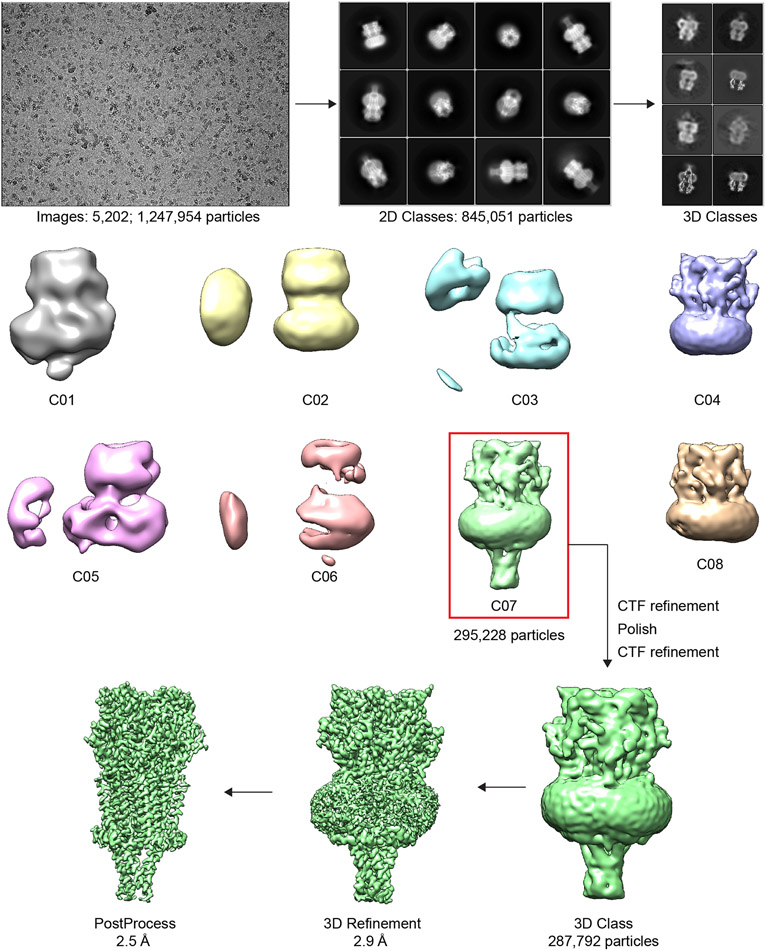
Cryo-EM data collection and processing
Cryo-EM samples were screened on the Talos Arctica at UT Southwestern Medical Center and the final datasets were collected on a 300 kV Titan Krios microscope (FEI) at the PNCC (Table S1). The cryo-EM data were processed using RELION 3.165. The potential problem of pseudo-symmetry in particle alignment turned out to not present a challenge, likely due to the large and well-ordered C-termini and F-loops visible at low resolution in the γ and δ subunits9. Dose-fractionated images were gain normalized, 2x Fourier binned, aligned, dose weighted, and summed using MotionCor266. GCTF was used to estimate defocus values. 67. Particle picking was done using crYOLO68. Bad particles were removed by 2D classification. Particles 2D classes with high resolution features were used to generate a de novo initial model for 3D classification. The 3D class with visible intracellular domain (ICD) density was low pass filtered and used as an initial model for 3D refinement followed by CTF refinement. Particles were then polished, followed by another round of CTF refinement. Polished and CTF refined particles were then used for another round of 3D classification. Refinement was done using good particles from final 3D classes. Software packages for structural biology were compiled by SBGrid69.
Model building, refinement and validation
The α-Bungarotoxin complex structure of the Torpedo receptor (PDB: 6UWZ) was used as a template for modeling all new structures. The chemical structure of carbachol was adopted from PDB ID: 1UV621 and d-tubo from PDB ID: 2XYT70. Iterative cycles of manual building in Coot71 with global real space refinement in Phenix72 resulted in good stereochemistry assessed by Molprobity73. Model-map correlation was assessed using Phenix validation tools. The pore diameter was calculated by HOLE74 and ligand binding sites were analyzed by LigPlot+75. The map and structural figures were generated using UCSF Chimera76, ChimeraX77 and Pymol (Schrodinger, LLC). The lipids in the apo cholesterol model were built in the unsharpened map and occupancy refinement was done with Phenix version dev-4227. In other structures, we built lipids based on the density and the apo cholesterol structure as a reference; occupancies were not refined in these other structures. In the d-tubo bound structure, the density of d-tubo at the lower affinity α/δ orthosteric site suggested two different orientations of binding as was observed in a d-tubo bound AChBP structure70 and further consistent with a modeling study78. We built the best fitted d-tubo orientation in this site. The d-tubo density in the extracellular side of the pore was poorly resolved, suggesting that d-tubo may bind in two or more orientations at that site. The unsharpened map was used to position d-tubo in the pore site and also for figure making related to this ligand. The C-terminus of the αγ subunit likely adopts multiple conformations and the model built corresponds to the best agreement with the density map.
Lipid Assays
Total phosphorus in samples was determined as previously described79. Replicate 125 μg protein samples (as determined by BCA protein assay) or soy polar lipids (Avanti, from stock solution), were air dried in glass vials at 80°C for 15 minutes. Briefly, 225 μL 8.9 M sulfuric acid were added to dried samples and standards and heated for 25 min at 200–215 °C. Linear dilutions of 0.5 mg/mL phosphatidylcholine (PC14) in 1% CHAPS were used as standards. Samples were cooled down to room temperature and 75 μL H2O2 was added followed by heating for an additional 30 min; 1.95 mL deionized H2O, 250 μL 2.5% ammonium molybdate tetrahydrate and 250 μL 10% ascorbic acid were added into samples previously cooled to room temperature. The samples were incubated at 100 °C for 7 min and after cooling to room temperature the absorbance was measured at 820 nm using a Molecular Devices Spectramax M5.
Total cholesterol was determined using the enzymatic cholesterol oxidase assay from Cayman Chemical (catalog 10007640, lot 0459016). Briefly, the cholesterol detector (ADHP) was reconstituted in DMSO, aliquoted, frozen and stored at −20°C prior to use. The assay cocktail was comprised of 949 μL Assay Buffer, 30 μL cholesterol detector solution (15 μL Cholesterol Detector in DMSO +15 μL Water), 10 μL HRP, 10 μL Cholesterol Oxidase and 1 μl Cholesterol Esterase. Assays were performed by adding 50 μL of sample (or 50 μL of cholesterol standards) to 50 μL of freshly prepared assay cocktail and incubated at 37°C for 30 min in the dark. Fluorescence was measured in top read mode using a Molecular Devices Spectramax M5 using 530 nm excitation 590 nm emission. A standard curve using pure cholesterol (Avanti) was used to determine absolute cholesterol relative to protein as determined by BCA. Torpedo native membrane ratios for both cholesterol and phospholipid were corrected for the 40% observed concentration of receptor to total protein in isolated membranes16.
Electrophysiology
Determination of EC50:
The open reading frames of Torpedo α, β, γ and δ constructs, generously provided by Dr. Steven Sine at the Mayo Clinic, were subcloned into the pGH19 vector. Mutations in the α subunit were introduced into pGH19-α by site-directed mutagenesis. cRNAs of α, β, γ, δ and α mutants were synthesized in vitro using the MEGAscript T7 Transcription Kit (Thermo Fisher). Xenopus laevis ovary lobes, purchased from the National Xenopus Resource, were treated with 1-2 mg/mL of collagenase I (Gibco) in Barth’s solution (in mM: 88 NaCl, 1 KCl, 2.4 NaHCO3, 0.82 MgSO4, 0.33 Ca(NO3)2, 0.68 CaCl2, 10 Hepes, pH 7.4) for 1 hour, and hypertonic shock for 10 min in 130 mM KH2PO4 pH 6.5, then washed with ND96 solution (in mM: 96 NaCl, 2 KCl, 1 MgCl2. 1.8 CaCl2, and 5 Hepes pH 7.4). Stages V–VI oocytes were collected and kept at 16°C in ND96 solution with 100 μg/ml gentamycin. Oocytes were injected with 0.1-5 ng of cRNA mixture in the ratio 2:1:1:1 of α:β:γ:δ. 24-72 hours after injection of cRNA, receptor activity was measured by TEVC recording in ND96 solution, at a holding potential of −60 mV, using an Axoclamp 900A amplifier (Molecular Devices), and the signal was digitized with a Digidata 1550B (Molecular Devices). Data were analyzed using pClamp 10.7. Pipettes were filled with 1 M KCl and showed resistances of 0.2 - 1.2 MΩ. Ligands were dissolved in ND96.
Rate of recovery from desensitization:
Whole cell voltage-clamp recordings were made from adherent HEK293S GnTI− cells transiently transfected with mouse α, β, γ and δ constructs. Each well of cells in a 12-well dish was transfected with 0.1 μg mixture of α, β, γ and δ plasmids in a ratio of 2:1:1:1, using Lipofectamine 2000 Reagent (ThermoFisher). The transfected cells were incubated at 30°C. 24 h post-transfection, cells were re-plated on 35 mm dishes and allowed to settle for at least 3 hours. Recodings were made 24-96 hours after transfection. Bath solution contains (in mM): 140 NaCl, 2.4 KCl, 4 MgCl2, 4 CaCl2, 5 HEPES and 10 glucose pH 7.3. Borosilicate pipettes were pulled and polished to an initial resistance of 2 - 4 MΩ, filled with the pipette solution that contained (in mM): 100 CsCl, 30 CsF, 10 NaCl, 10 EGTA, and 20 HEPES pH 7.3. The recordings were made with an Axopatch 200B amplifier, sampled at 5 kHz, and low-pass filtered at 2 kHz using a Digidata 1440A (Molecular Devices) and analyzed with pClamp 10 software (Molecular Devices). Cells were held at −75 mV. Carbachol solutions were prepared in bath solution from 1 M of concentrated stock. Solution exchange was achieved using a gravity driven RSC-200 rapid solution changer (Bio-Logic). Recovery from desensitization was recorded in two-pulse protocols with increasing interval wash time. The apparent parameters of recovery were analyzed by fitting with two-phase exponential association equation (GraphPad Prism 9.2.0.). Error bars represent the mean ± S.D.
Sequence alignment
The following sequences from the UniProt data base were aligned with Clustal Omega80 using default parameters; P02718 (ACHD_TETCF), P09628 (ACHD_XENLA), P02716 (ACHD_MOUSE), P18916 (ACHG_RAT), P13536 (ACHG_BOVIN), P04760 (ACHG_MOUSE), P25110 (ACHD_RAT), P07510 (ACHG_HUMAN), Q07001(ACHD_HUMAN), P04759(ACHD_BOVIN), P02713(ACHG_CHICK), P02717(ACHD_CHICK), P05376(ACHG_XENLA), P02714(ACHG_TETCF), P04758(ACHB_BOVIN), P49580(ACHE_XENLA), P11230(ACHB_HUMAN), P09690(ACHB_MOUSE), P25109(ACHB_RAT), P09660(ACHE_RAT), P20782(ACHE_MOUSE), Q04844(ACHE_HUMAN), P02712(ACHB_TETCF), P02715(ACHE_BOVIN), P49579(ACHB_XENLA), P02708(ACHA_HUMAN), P02711(ACHA_TORMA), P02710(ACHA_TETCF), P05377(ACHAB_XENLA), P22456(ACHAA_XENLA), P04756(ACHA_MOUSE), P25108(ACHA_RAT), P02709(ACHA_BOVIN), P09479(ACHA_CHICK), Q98880(ACHA_DANRE), P14144(ACHA_NATTE), P16005(ACHB_CHICK), P14143(ACHA_NAJNA), P54250(ACHA_FELCA), P54251(ACHA_HERIC), P54248(ACHA_CRORS), P54249(ACHA_ERICO). The alignment LOGO in Extended Data Fig. 3b was created with WebLogo381.
Extended Data
Extended Data Fig. 1. Relion 3.1 workflow for data processing.
Representative processing approach for apo dataset.
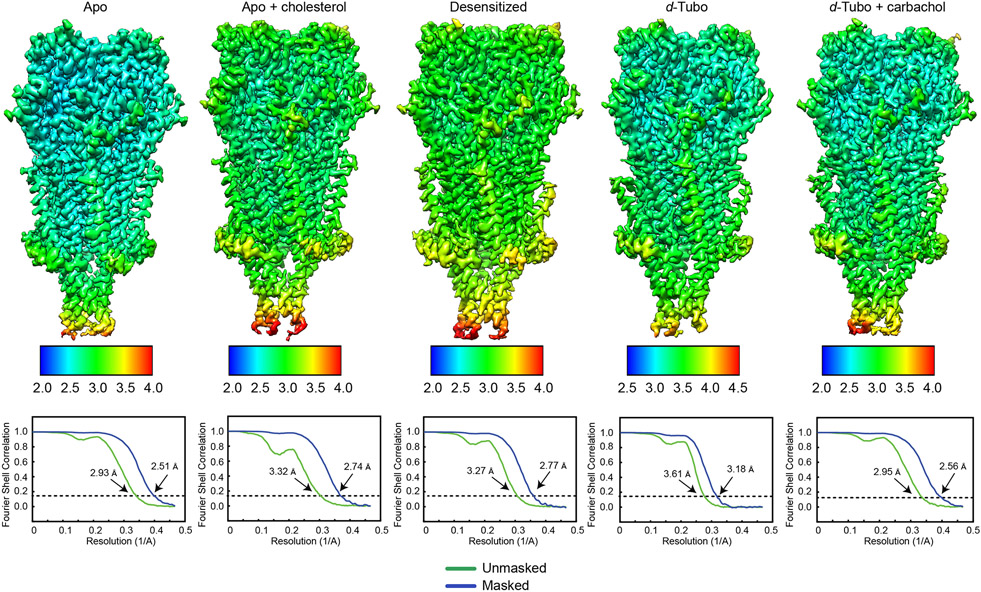
Extended Data Fig. 2. Local and global resolution estimates for cryo-EM maps.
Local resolution is illustrated by variation in map surface color and was estimated using RELION. Global resolution was estimated at FSC = 0.143 (dotted line) from half maps.
Extended Data Fig. 3. Lipid-receptor interactions.
a, Cholesterol (red) and phospholipid (yellow) binding sites in the receptor. α subunit - green, β subunit - khaki, γ subunit - blue, δ subunit – violet. High-affinity cholesterol binding sites are near the MX helices and low-affinity binding sites are near outer membrane leaflet. Among the five subunit interfaces, the high affinity cholesterols occupy the same positions in three interfaces: α/γ, α/δ, and β/α, but not γ/αδ and δ/β. The environment of that site is more polar in γ and δ subunits, where an asparagine replaces I291/α or L297/β (in M3). b, Conservation of the residues in the cholesterol high affinity binding site, residues numbers are according to T. californica α-subunit. Accession numbers for sequences used to determine conservation are given in the Methods.
Extended Data Fig. 4. Transition between resting and desensitized states.
a, Two electrode voltage clamp (TEVC) recording show activation and desensitization of the Torpedo nicotinic receptor by carbachol; 5 mM carbachol was supplemented into the cryo-EM sample for structure determination of the desensitized state. b, Asymmetry in the TMD conformational change between resting and desensitized states; residues at the 16′ position are shown as sticks. c, Conformational changes in loop C of an α-subunit and Loop F of a complementary subunit after agonist and antagonist binding compared to resting state. Carbachol is shown as green spheres. d, Conformational differences in a representative coupling region; representative α and γ subunits.
Extended Data Fig. 5. Permeation pathway and pore profile.
a, Pore diameter comparison of Torpedo in resting and desensitized states vs. other Cys-loop receptors: resting, α7 nAChR (PDB ID: 7EKI), 5-HT3AR (PDB ID: 6BE1), GlyR (PDB ID: 6UBS); and desensitized, α7 nAChR (PDB ID: 7KOQ), α4β2 nAChR (PDB ID: 6CNJ), α3β4 nAChR (PDB ID: 6PV7), 5-HT3AR (PDB ID: 6HIQ), GlyR (PDB ID: 6UBT). b, Permeation pathway cutaway colored by electrostatic potential. c, Water molecules at the 2′ gate of the desensitized state structure. d, −1′ residue orientation in the Torpedo receptor in resting and desensitized states. Transparent surface is experimental density map. e, Two α-subunits superposition of resting state structure. f, Dose response parameters measured by TEVC of WT and mutants. Nonlinear regression was carried out using GraphPad Prism 8. Replicate measurements are from independent oocytes.
Extended Data Fig. 6. Receptor activation and antagonism by carbachol and d-tubo; d-tubo binding sites with corresponding density maps.
a, Two-electrode voltage clamp (TEVC) recording illustrates receptor antagonism by d-tubo. b, d-Tubo at α/γ interface (site 1). c, d-Tubo at α/δ interface (site 2). d, d-Tubo in the pore (site 3). e, d-Tubo at junction of M1, M3 and M4 helices of the αγ subunit (site 4). Corresponding d-tubo densities are shown as semitransparent surfaces. The d-tubo density was very clear at sites 1 and 4. Site 2 density suggested two different orientations of d-tubo and we modeled the best fitted one. The density at site 3 is poorly resolved suggesting multiple orientations of d-tubo in the pore; we used the unsharpened map to model d-tubo there. Density map was contoured at a threshold of 0.02 for site 1, 2 & 4 and 0.013 for site 3 in UCSF chimera.
Extended Data Fig. 7. Orthosteric binding-sites details and carbachol vs. d-tubo complex superposition.
a, b, Electrostatic potential of the two orthosteric ligand-binding sites in the apo form; binding pockets are indicated by asterisks. c, Residue differences between two orthosteric ligand-binding sites; αγ/γ as colored (α- green, γ- blue) and αδ/δ as gray. d, Superposition of desensitized and d-tubo bound structures; d-Tubo model is colored (α, green; β, khaki; γ, blue; δ, violet) and desensitized structure is in gray. (E) Conformational difference in M4 of two α-subunits in resting, desensitized and pure d-tubo bound structures.
Supplementary Material
Source Data Fig. 1 Cholesterol and phospholipid assay
Source Data Fig. 2 Dose-response curve and desensitization recovery
Supplementary Video 1 Structural transition between resting and desensitized states. Morphing movie illustrates changes from resting state to desensitized state to resting state in top view. α subunit - green, β subunit - khaki, γ subunit - blue, δ subunit - violet.
Supplementary Video 2 Structural transition between resting and desensitized states. Morphing movie illustrates changes from resting state to desensitized state to resting state in side view. α subunit - green, β subunit - khaki, γ subunit - blue, δ subunit - violet.
Supplementary Video 4 d-Tubo interactions at α/δ interface (site 2). d-Tubo is showing as stick (orange) and corresponding densities are shown as semitransparent surfaces. Interacting residues are also shown as sticks and colored by subunits; α subunit - green, δ subunit - violet.
Supplementary Video 3 d-Tubo interactions at α/γ interface (site 1). d-Tubo is showing as stick (orange) and corresponding densities are shown as semitransparent surfaces. Interacting residues are also shown as sticks and colored by subunits; α subunit - green, γ subunit – blue.
Supplementary Video 5 d-Tubo interactions at pore (site 3). d-Tubo is showing as stick (orange) and corresponding densities are shown as semitransparent surfaces. Interacting residues are also shown as sticks and colored by subunits; α subunit - green, β subunit - khaki, γ subunit - blue, δ subunit - violet.
Supplementary Video 6 d-Tubo interactions at M1, M3 and M4 helices of αγ subunit (site 4). d-Tubo is showing as stick (orange) and corresponding densities are shown as semitransparent surfaces. Interacting residues are also shown as sticks and colored by subunit; α subunit - green.
Acknowledgements:
We thank S. Sine for Torpedo receptor cDNAs, and C. Noviello and S. Zhu for assistance in cryo-EM sample screening. We are grateful to S. Burke, J.J. Kim, C. Noviello, S. Sine, and M. Klymkowsky for critical feedback on the manuscript, D. Borek for model building discussion, and A. Sobolevsky for helpful discussion related to measuring recovery from desensitization. Single-particle cryo-EM grids were screened at the University of Texas Southwestern Medical Center Cryo-Electron Microscopy Facility, which is supported by the CPRIT Core Facility Support Award RP170644. We thank Harry Scott for cryo-EM data collection at the PNCC under user proposals 50839 and 51574. A portion of this research was supported by NIH grant U24GM129547 and performed at the PNCC at OHSU and accessed through EMSL (grid.436923.9), a DOE Office of Science User Facility sponsored by the Office of Biological and Environmental Research. M.M.R. acknowledges a postdoctoral fellowship from the American Heart Association (827474). This work was supported by grants from the NIH (DA042072 to R.E.H., AG061829 to M.H.B.S and NS120496 to R.E.H and M.H.B.S) and the MCDB Neurodegenerative Disease Fund to M.H.B.S.
Footnotes
Competing interests:
The authors declare no competing interests.
Editor recognition statement:
Florian Ullrich was the primary editor on this article and managed its editorial process and peer review in collaboration with the rest of the editorial team.
Data and code availability
Cryo-EM maps and atomic model coordinates have been deposited in the EMDB and RCSB respectively; apo (EMD-25202 and PDB 7SMM), apo plus cholesterol (EMD-25205 and PDB 7SMQ), carbachol-bound desensitized state: (EMD-25202 and PDB 7SMM), d-tubocurarine bound: (EMD-25207 and PDB 7SMS) and d-tubocurarine plus carbachol bound: (EMD-25208 and PDB 7SMT). Source data are provided with this paper.
References
- 1.Gharpure A, Noviello CM & Hibbs RE Progress in nicotinic receptor structural biology. Neuropharmacology 171, 108086, doi: 10.1016/j.neuropharm.2020.108086 (2020). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Albuquerque EX, Pereira EF, Alkondon M & Rogers SW Mammalian nicotinic acetylcholine receptors: from structure to function. Physiological reviews 89, 73–120, doi: 10.1152/physrev.00015.2008 (2009). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Karlin A Emerging structure of the nicotinic acetylcholine receptors. Nat Rev Neurosci 3, 102–114, doi: 10.1038/nrn731 (2002). [DOI] [PubMed] [Google Scholar]
- 4.Katz B & Thesleff S A study of the desensitization produced by acetylcholine at the motor end-plate. J Physiol 138, 63–80, doi: 10.1113/jphysiol.1957.sp005838 (1957). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Raghavendra T Neuromuscular blocking drugs: discovery and development. J R Soc Med 95, 363–367, doi: 10.1258/jrsm.95.7.363 (2002). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Bennett MR The concept of transmitter receptors: 100 years on. Neuropharmacology 39, 523–546, doi: 10.1016/s0028-3908(99)00137-9 (2000). [DOI] [PubMed] [Google Scholar]
- 7.King H Curare. Nature 135, 469–470, doi: 10.1038/135469b0 (1935). [DOI] [Google Scholar]
- 8.King H 330. Curare alkaloids. Part I. Tubocurarine. Journal of the Chemical Society (Resumed), 1381–1389, doi: 10.1039/JR9350001381 (1935). [DOI] [Google Scholar]
- 9.Rahman MM et al. Structure of the Native Muscle-type Nicotinic Receptor and Inhibition by Snake Venom Toxins. Neuron 106, 952–962 e955, doi: 10.1016/j.neuron.2020.03.012 (2020). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Rahman MM, Worrell BT, Stowell MHB & Hibbs RE in Methods in enzymology Vol. 653 (eds Minor Daniel L. & Colecraft Henry M.) 189–206 (Academic Press, 2021). [DOI] [PubMed] [Google Scholar]
- 11.Raftery MA, Hunkapiller MW, Strader CD & Hood LE Acetylcholine receptor: complex of homologous subunits. Science 208, 1454–1456 (1980). [DOI] [PubMed] [Google Scholar]
- 12.Noda M et al. Structural homology of Torpedo californica acetylcholine receptor subunits. Nature 302, 528–532, doi: 10.1038/302528a0 (1983). [DOI] [PubMed] [Google Scholar]
- 13.Miyazawa A, Fujiyoshi Y, Stowell M & Unwin N Nicotinic acetylcholine receptor at 4.6 A resolution: transverse tunnels in the channel wall. J Mol Biol 288, 765–786, doi: 10.1006/jmbi.1999.2721 (1999). [DOI] [PubMed] [Google Scholar]
- 14.Changeux JP The nicotinic acetylcholine receptor: a typical 'allosteric machine'. Philos Trans R Soc Lond B Biol Sci 373, doi: 10.1098/rstb.2017.0174 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Brannigan G, Henin J, Law R, Eckenhoff R & Klein ML Embedded cholesterol in the nicotinic acetylcholine receptor. Proc Natl Acad Sci U S A 105, 14418–14423, doi: 10.1073/pnas.0803029105 (2008). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Baenziger JE, Domville JA & Therien JPD The Role of Cholesterol in the Activation of Nicotinic Acetylcholine Receptors. Curr Top Membr 80, 95–137, doi: 10.1016/bs.ctm.2017.05.002 (2017). [DOI] [PubMed] [Google Scholar]
- 17.Hamouda AK, Sanghvi M, Sauls D, Machu TK & Blanton MP Assessing the lipid requirements of the Torpedo californica nicotinic acetylcholine receptor. Biochemistry 45, 4327–4337, doi: 10.1021/bi052281z (2006). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Unwin N Protein-lipid architecture of a cholinergic postsynaptic membrane. IUCrJ 7, 852–859, doi: 10.1107/S2052252520009446 (2020). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Sridhar A et al. Regulation of a pentameric ligand-gated ion channel by a semi-conserved cationic-lipid binding site. J Biol Chem, 100899, doi: 10.1016/j.jbc.2021.100899 (2021). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Zhong W et al. From ab initio quantum mechanics to molecular neurobiology: a cation-pi binding site in the nicotinic receptor. Proc Natl Acad Sci U S A 95, 12088–12093, doi: 10.1073/pnas.95.21.12088 (1998). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Celie PH et al. Nicotine and carbamylcholine binding to nicotinic acetylcholine receptors as studied in AChBP crystal structures. Neuron 41, 907–914, doi: 10.1016/s0896-6273(04)00115-1 (2004). [DOI] [PubMed] [Google Scholar]
- 22.Howard RJ Elephants in the Dark: Insights and Incongruities in Pentameric Ligand-gated Ion Channel Models. J Mol Biol, 167128, doi: 10.1016/j.jmb.2021.167128 (2021). [DOI] [PubMed] [Google Scholar]
- 23.Nemecz A, Prevost MS, Menny A & Corringer PJ Emerging Molecular Mechanisms of Signal Transduction in Pentameric Ligand-Gated Ion Channels. Neuron 90, 452–470, doi: 10.1016/j.neuron.2016.03.032 (2016). [DOI] [PubMed] [Google Scholar]
- 24.Mitra A, Cymes GD & Auerbach A Dynamics of the acetylcholine receptor pore at the gating transition state. Proc Natl Acad Sci U S A 102, 15069–15074, doi: 10.1073/pnas.0505090102 (2005). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Grandl J, Danelon C, Hovius R & Vogel H Functional asymmetry of transmembrane segments in nicotinic acetylcholine receptors. Eur Biophys J 35, 685–693, doi: 10.1007/s00249-006-0078-2 (2006). [DOI] [PubMed] [Google Scholar]
- 26.Grosman C & Auerbach A Asymmetric and independent contribution of the second transmembrane segment 12' residues to diliganded gating of acetylcholine receptor channels: a single-channel study with choline as the agonist. The Journal of general physiology 115, 637–651, doi: 10.1085/jgp.115.5.637 (2000). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Unwin N, Miyazawa A, Li J & Fujiyoshi Y Activation of the nicotinic acetylcholine receptor involves a switch in conformation of the alpha subunits. J Mol Biol 319, 1165–1176, doi: 10.1016/S0022-2836(02)00381-9 (2002). [DOI] [PubMed] [Google Scholar]
- 28.Unwin N & Fujiyoshi Y Gating movement of acetylcholine receptor caught by plunge-freezing. J Mol Biol 422, 617–634, doi: 10.1016/j.jmb.2012.07.010 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Noviello CM et al. Structure and gating mechanism of the alpha7 nicotinic acetylcholine receptor. Cell 184, 2121–2134 e2113, doi: 10.1016/j.cell.2021.02.049 (2021). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Dwyer TM, Adams DJ & Hille B The permeability of the endplate channel to organic cations in frog muscle. J Gen Physiol 75, 469–492, doi: 10.1085/jgp.75.5.469 (1980). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Revah F et al. Mutations in the channel domain alter desensitization of a neuronal nicotinic receptor. Nature 353, 846–849, doi: 10.1038/353846a0 (1991). [DOI] [PubMed] [Google Scholar]
- 32.Yakel JL, Lagrutta A, Adelman JP & North RA Single amino acid substitution affects desensitization of the 5-hydroxytryptamine type 3 receptor expressed in Xenopus oocytes. Proc Natl Acad Sci U S A 90, 5030–5033, doi: 10.1073/pnas.90.11.5030 (1993). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Basak S et al. Cryo-EM structure of 5-HT3A receptor in its resting conformation. Nat Commun 9, 514, doi: 10.1038/s41467-018-02997-4 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Wilson G & Karlin A Acetylcholine receptor channel structure in the resting, open, and desensitized states probed with the substituted-cysteine-accessibility method. Proc Natl Acad Sci U S A 98, 1241–1248, doi: 10.1073/pnas.031567798 (2001). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Zhao Y et al. Structural basis of human alpha7 nicotinic acetylcholine receptor activation. Cell Res 31, 713–716, doi: 10.1038/s41422-021-00509-6 (2021). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Kumar A et al. Mechanisms of activation and desensitization of full-length glycine receptor in lipid nanodiscs. Nat Commun 11, 3752, doi: 10.1038/s41467-020-17364-5 (2020). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Auerbach A & Akk G Desensitization of mouse nicotinic acetylcholine receptor channels. A two-gate mechanism. The Journal of general physiology 112, 181–197, doi: 10.1085/jgp.112.2.181 (1998). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Gharpure A et al. Agonist Selectivity and Ion Permeation in the alpha3beta4 Ganglionic Nicotinic Receptor. Neuron, doi: 10.1016/j.neuron.2019.07.030 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Morales-Perez CL, Noviello CM & Hibbs RE X-ray structure of the human alpha4beta2 nicotinic receptor. Nature 538, 411–415, doi: 10.1038/nature19785 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Walsh RM Jr., et al. Structural principles of distinct assemblies of the human alpha4beta2 nicotinic receptor. Nature 557, 261–265, doi: 10.1038/s41586-018-0081-7 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Polovinkin L et al. Conformational transitions of the serotonin 5-HT3 receptor. Nature 563, 275–279, doi: 10.1038/s41586-018-0672-3 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Basak S, Gicheru Y, Rao S, Sansom MSP & Chakrapani S Cryo-EM reveals two distinct serotonin-bound conformations of full-length 5-HT3A receptor. Nature 563, 270–274, doi: 10.1038/s41586-018-0660-7 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Kim JJ et al. Shared structural mechanisms of general anaesthetics and benzodiazepines. Nature 585, 303–308, doi: 10.1038/s41586-020-2654-5 (2020). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Masiulis S et al. GABAA receptor signalling mechanisms revealed by structural pharmacology. Nature 565, 454–459, doi: 10.1038/s41586-018-0832-5 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Thompson MJ, Domville JA & Baenziger JE The functional role of the alphaM4 transmembrane helix in the muscle nicotinic acetylcholine receptor probed through mutagenesis and coevolutionary analyses. J Biol Chem 295, 11056–11067, doi: 10.1074/jbc.RA120.013751 (2020). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Carswell CL et al. Role of the Fourth Transmembrane alpha Helix in the Allosteric Modulation of Pentameric Ligand-Gated Ion Channels. Structure 23, 1655–1664, doi: 10.1016/j.str.2015.06.020 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Butler AS et al. Importance of the C-terminus of the human 5-HT3A receptor subunit. Neuropharmacology 56, 292–302, doi: 10.1016/j.neuropharm.2008.08.017 (2009). [DOI] [PubMed] [Google Scholar]
- 48.Cory-Wright J et al. Aromatic Residues in the Fourth Transmembrane-Spanning Helix M4 Are Important for GABArho Receptor Function. ACS Chem Neurosci 9, 284–290, doi: 10.1021/acschemneuro.7b00315 (2018). [DOI] [PubMed] [Google Scholar]
- 49.Pons S et al. Critical role of the C-terminal segment in the maturation and export to the cell surface of the homopentameric alpha 7-5HT3A receptor. Eur J Neurosci 20, 2022–2030, doi: 10.1111/j.1460-9568.2004.03673.x (2004). [DOI] [PubMed] [Google Scholar]
- 50.Jenkinson DH The antagonism between tubocurarine and substances which depolarize the motor end-plate. J Physiol 152, 309–324, doi: 10.1113/jphysiol.1960.sp006489 (1960). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Pedersen SE & Papineni RV Interaction of d-tubocurarine analogs with the Torpedo nicotinic acetylcholine receptor. Methylation and stereoisomerization affect site-selective competitive binding and binding to the noncompetitive site. J Biol Chem 270, 31141–31150, doi: 10.1074/jbc.270.52.31141 (1995). [DOI] [PubMed] [Google Scholar]
- 52.Moore MA & McCarthy MP Snake venom toxins, unlike smaller antagonists, appear to stabilize a resting state conformation of the nicotinic acetylcholine receptor. Biochim Biophys Acta 1235, 336–342, doi: 10.1016/0005-2736(95)80022-8 (1995). [DOI] [PubMed] [Google Scholar]
- 53.Sine SM & Steinbach JH Acetylcholine receptor activation by a site-selective ligand: nature of brief open and closed states in BC3H-1 cells. J Physiol 370, 357–379, doi: 10.1113/jphysiol.1986.sp015939 (1986). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Steinbach JH & Chen Q Antagonist and partial agonist actions of d-tubocurarine at mammalian muscle acetylcholine receptors. J Neurosci 15, 230–240 (1995). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Neubig RR & Cohen JB Equilibrium binding of [3H]tubocurarine and [3H]acetylcholine by Torpedo postsynaptic membranes: stoichiometry and ligand interactions. Biochemistry 18, 5464–5475, doi: 10.1021/bi00591a032 (1979). [DOI] [PubMed] [Google Scholar]
- 56.Chiara DC & Cohen JB Identification of amino acids contributing to high and low affinity d-tubocurarine sites in the Torpedo nicotinic acetylcholine receptor. J Biol Chem 272, 32940–32950, doi: 10.1074/jbc.272.52.32940 (1997). [DOI] [PubMed] [Google Scholar]
- 57.Sine SM & Claudio T Gamma- and delta-subunits regulate the affinity and the cooperativity of ligand binding to the acetylcholine receptor. J Biol Chem 266, 19369–19377 (1991). [PubMed] [Google Scholar]
- 58.Chiara DC, Xie Y & Cohen JB Structure of the agonist-binding sites of the Torpedo nicotinic acetylcholine receptor: affinity-labeling and mutational analyses identify gamma Tyr-111/delta Arg-113 as antagonist affinity determinants. Biochemistry 38, 6689–6698, doi: 10.1021/bi9901735 (1999). [DOI] [PubMed] [Google Scholar]
- 59.Sine SM Molecular dissection of subunit interfaces in the acetylcholine receptor: identification of residues that determine curare selectivity. Proc Natl Acad Sci U S A 90, 9436–9440, doi: 10.1073/pnas.90.20.9436 (1993). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Strecker GJ & Jackson MB Curare binding and the curare-induced subconductance state of the acetylcholine receptor channel. Biophys J 56, 795–806, doi: 10.1016/S0006-3495(89)82726-2 (1989). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Colquhoun D, Dreyer F & Sheridan RE The actions of tubocurarine at the frog neuromuscular junction. J Physiol 293, 247–284, doi: 10.1113/jphysiol.1979.sp012888 (1979). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Sine SM & Taylor P Relationship between reversible antagonist occupancy and the functional capacity of the acetylcholine receptor. J Biol Chem 256, 6692–6699 (1981). [PubMed] [Google Scholar]
- 63.Gielen M, Barilone N & Corringer PJ The desensitization pathway of GABAA receptors, one subunit at a time. Nat Commun 11, 5369, doi: 10.1038/s41467-020-19218-6 (2020). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Zivanov J et al. New tools for automated high-resolution cryo-EM structure determination in RELION-3. Elife 7, doi: 10.7554/eLife.42166 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Frauenfeld J et al. A saposin-lipoprotein nanoparticle system for membrane proteins. Nature methods 13, 345–351, doi: 10.1038/nmeth.3801 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Zheng SQ et al. MotionCor2: anisotropic correction of beam-induced motion for improved cryo-electron microscopy. Nature methods 14, 331–332, doi: 10.1038/nmeth.4193 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.Zhang K Gctf: Real-time CTF determination and correction. J Struct Biol 193, 1–12, doi: 10.1016/j.jsb.2015.11.003 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68.Wagner T et al. SPHIRE-crYOLO is a fast and accurate fully automated particle picker for cryo-EM. Commun Biol 2, 218, doi: 10.1038/s42003-019-0437-z (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69.Morin A et al. Collaboration gets the most out of software. Elife 2, e01456, doi: 10.7554/eLife.01456 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70.Brams M et al. A structural and mutagenic blueprint for molecular recognition of strychnine and d-tubocurarine by different cys-loop receptors. PLoS Biol 9, e1001034, doi: 10.1371/journal.pbio.1001034 (2011). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 71.Emsley P, Lohkamp B, Scott WG & Cowtan K Features and development of Coot. Acta Crystallogr D Biol Crystallogr 66, 486–501, doi: 10.1107/S0907444910007493 (2010). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 72.Afonine PV et al. Real-space refinement in PHENIX for cryo-EM and crystallography. Acta Crystallogr D Struct Biol 74, 531–544, doi: 10.1107/S2059798318006551 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 73.Williams CJ et al. MolProbity: More and better reference data for improved all-atom structure validation. Protein Sci 27, 293–315, doi: 10.1002/pro.3330 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 74.Smart OS, Neduvelil JG, Wang X, Wallace BA & Sansom MS HOLE: a program for the analysis of the pore dimensions of ion channel structural models. J Mol Graph 14, 354–360, 376 (1996). [DOI] [PubMed] [Google Scholar]
- 75.Wallace AC, Laskowski RA & Thornton JM LIGPLOT: a program to generate schematic diagrams of protein-ligand interactions. Protein Eng 8, 127–134, doi: 10.1093/protein/8.2.127 (1995). [DOI] [PubMed] [Google Scholar]
- 76.Pettersen EF et al. UCSF Chimera--a visualization system for exploratory research and analysis. J Comput Chem 25, 1605–1612, doi: 10.1002/jcc.20084 (2004). [DOI] [PubMed] [Google Scholar]
- 77.Pettersen EF et al. UCSF ChimeraX: Structure visualization for researchers, educators, and developers. Protein Sci 30, 70–82, doi: 10.1002/pro.3943 (2021). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 78.Gao F et al. Curariform antagonists bind in different orientations to acetylcholine-binding protein. J Biol Chem 278, 23020–23026, doi: 10.1074/jbc.M301151200 (2003). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 79.Basta T et al. Self-assembled lipid and membrane protein polyhedral nanoparticles. Proc Natl Acad Sci U S A 111, 670–674, doi: 10.1073/pnas.1321936111 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 80.McWilliam H et al. Analysis Tool Web Services from the EMBL-EBI. Nucleic Acids Res 41, W597–600, doi: 10.1093/nar/gkt376 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 81.Crooks GE, Hon G, Chandonia JM & Brenner SE WebLogo: a sequence logo generator. Genome Res 14, 1188–1190, doi: 10.1101/gr.849004 (2004). [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Source Data Fig. 1 Cholesterol and phospholipid assay
Source Data Fig. 2 Dose-response curve and desensitization recovery
Supplementary Video 1 Structural transition between resting and desensitized states. Morphing movie illustrates changes from resting state to desensitized state to resting state in top view. α subunit - green, β subunit - khaki, γ subunit - blue, δ subunit - violet.
Supplementary Video 2 Structural transition between resting and desensitized states. Morphing movie illustrates changes from resting state to desensitized state to resting state in side view. α subunit - green, β subunit - khaki, γ subunit - blue, δ subunit - violet.
Supplementary Video 4 d-Tubo interactions at α/δ interface (site 2). d-Tubo is showing as stick (orange) and corresponding densities are shown as semitransparent surfaces. Interacting residues are also shown as sticks and colored by subunits; α subunit - green, δ subunit - violet.
Supplementary Video 3 d-Tubo interactions at α/γ interface (site 1). d-Tubo is showing as stick (orange) and corresponding densities are shown as semitransparent surfaces. Interacting residues are also shown as sticks and colored by subunits; α subunit - green, γ subunit – blue.
Supplementary Video 5 d-Tubo interactions at pore (site 3). d-Tubo is showing as stick (orange) and corresponding densities are shown as semitransparent surfaces. Interacting residues are also shown as sticks and colored by subunits; α subunit - green, β subunit - khaki, γ subunit - blue, δ subunit - violet.
Supplementary Video 6 d-Tubo interactions at M1, M3 and M4 helices of αγ subunit (site 4). d-Tubo is showing as stick (orange) and corresponding densities are shown as semitransparent surfaces. Interacting residues are also shown as sticks and colored by subunit; α subunit - green.