Abstract
Recently emerged SARS-CoV-2 Omicron subvariant, BA.2.75, displayed a growth advantage over circulating BA.2.38, BA.2.76, and BA.5 in India. However, the underlying mechanisms for enhanced infectivity, especially compared with BA.5, remain unclear. Here, we show that BA.2.75 exhibits substantially higher affinity for host receptor angiotensin-converting enzyme 2 (ACE2) than BA.5 and other variants. Structural analyses of BA.2.75 spike shows its decreased thermostability and increased frequency of the receptor binding domain (RBD) in the “up” conformation under acidic conditions, suggesting enhanced low-pH-endosomal cell entry. Relative to BA.4/BA.5, BA.2.75 exhibits reduced evasion of humoral immunity from BA.1/BA.2 breakthrough-infection convalescent plasma but greater evasion of Delta breakthrough-infection convalescent plasma. BA.5 breakthrough-infection plasma also exhibits weaker neutralization against BA.2.75 than BA.5, mainly due to BA.2.75’s distinct neutralizing antibody (NAb) escape pattern. Antibody therapeutics Evusheld and Bebtelovimab remain effective against BA.2.75. These results suggest BA.2.75 may prevail after BA.4/BA.5, and its increased receptor-binding capability could support further immune-evasive mutations.
Keywords: SARS-CoV-2, Omicron, BA.2.75, neutralizing antibody, spike glycoprotein, cryo-EM structure, COVID-19, immune evasion
Graphical abstract
SARS-CoV-2 BA.2.75 is growing rapidly and globally. Cao et al. solved the structure of BA.2.75 spike and show it has stronger binding to human ACE2 than previous variants. BA.2.75 also exhibited distinct antigenicity compared with BA.5, escaping neutralizing antibodies targeting various epitopes and evading convalescent plasma from BA.5 breakthrough infections.
Introduction
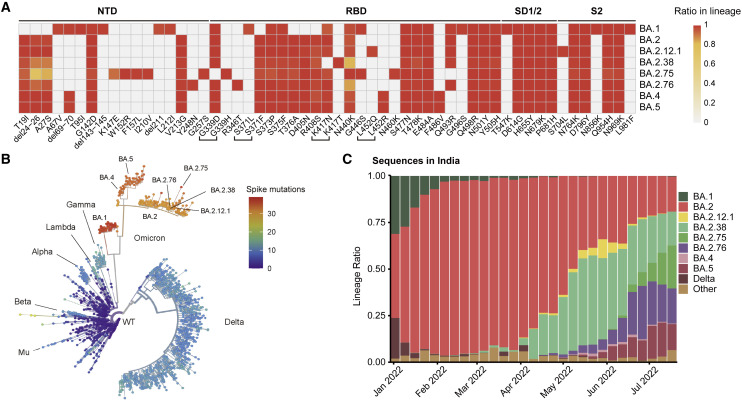
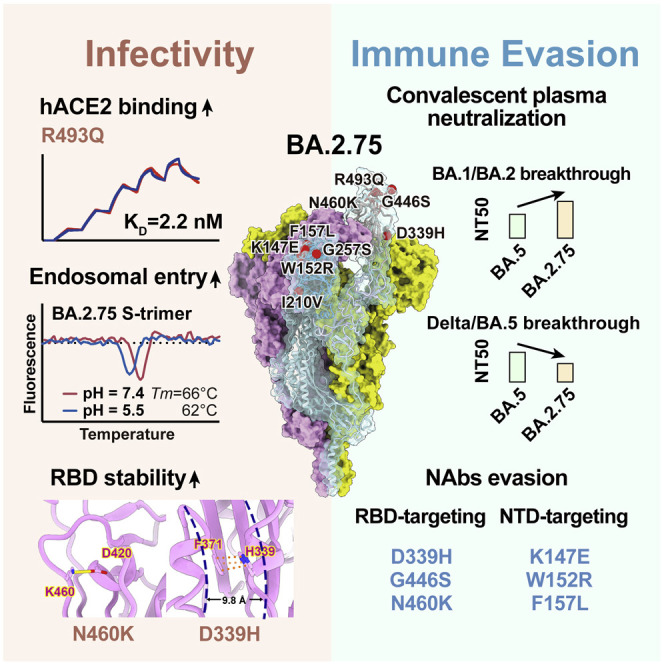
Severe acute respiratory coronavirus (SARS-CoV-2) Omicron variants have been continuously evolving and dominating the pandemic (Chen et al., 2021; Tegally et al., 2022). Globally, BA.1 was rapidly replaced by the antigenically distinct descendant BA.2, whereas BA.4, BA.5, and BA.2.12.1, derived from BA.2, exhibited further increased humoral immunity evasion and have outcompeted BA.2 (Cao et al., 2022b; Tuekprakhon et al., 2022; Wang et al., 2022b). More recently, a novel BA.2 subvariant designated as BA.2.75, a variant of concern (VOC) lineage under monitoring by the World Health Organization (WHO), is spreading rapidly in India and around the globe, contributing to over 20% of recently reported sequences in India and is continuously increasing (Chen et al., 2021; WHO, 2022). Compared with the BA.2 spike (S-trimer), BA.2.75 carries nine additional mutations, among which five are on the N-terminal domain (NTD), including K147E, W152R, F157L, I210V, and G257S, and four on the receptor binding domain (RBD), namely D339H, G446S, N460K, and R493Q (Figures 1A and 1B). Among them, G446S appeared in BA.1, and R493Q reversion was observed in BA.4/BA.5. N460K and D339H mutations have not been observed on prevailing variants, and their functions remain unclear.
Figure 1.
BA.2.75 and BA.2.76 displayed growth advantage in India
(A) Main mutations on the spike glycoprotein appearing in SARS-CoV-2 Omicron sublineages.
(B) Phylogenetic tree of existing SARS-CoV-2 variants. Color scales indicate number of mutations on the spike.
(C) Lineage distribution of recent sequences from India by time. BA.2.75 and BA.2.76 is growing rapidly in India, showing advantage compared with other lineages.
Importantly, BA.2.75 displayed a local growth advantage in India compared with BA.2.38 (BA.2+N417T), BA.2.76 (BA.2+R346T+Y248N), and BA.5 (Figure 1C). The enhanced transmissibility over BA.5 questions whether BA.2.75 would prevail after the global BA.4/BA.5 wave. To address this, the receptor-binding affinity and humoral immune evasion capability of BA.2.75, especially under the immune background after BA.4/BA.5 infection, need immediate evaluation.
Results
BA.2.75 displays higher ACE-binding affinity than BA.4/BA.5
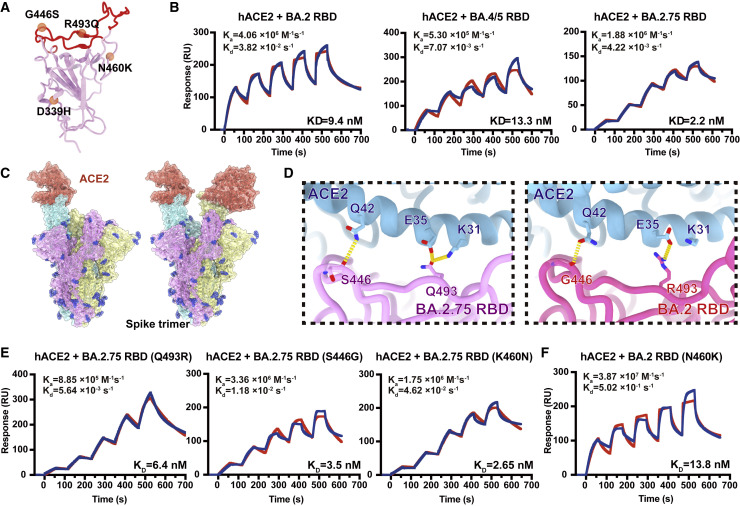
Recent studies have revealed that Omicron subvariants have further improved binding affinities for human ACE2 (hACE2) (Cao et al., 2022b; Li et al., 2022; Tuekprakhon et al., 2022; Wang et al., 2022b) and stability in vitro, which, to some extent, correlates with their increased viral transmission and infection. The difference in amino acid composition of the RBD between BA.2 and BA.2.75 is four substitutions: D339H, G446S, N460K, and R493Q, among which G446S and R493Q lie in the RBD/hACE2 interface and N460K and D339H locate close to and far away from the interface, respectively (Figure 2 A). To explore the impact of these mutations on the hACE2 binding of BA.2.75 RBD, we assessed the binding affinities between hACE2 and RBDs from the six Omicron subvariants (BA.1, BA.2, BA.3, BA.4/5, BA.2.12.1, and BA.2.75), together with the other earlier four VOCs (Alpha, Beta, Gamma, and Delta) by surface plasmon resonance (SPR) (Figures 2B and S1A). Surprisingly, BA.2.75 displayed a 4- to 6-fold increased binding affinity to hACE2 compared with other Omicron variants, substantially higher than the other four VOCs as well, reaching the highest binding activity measured in circulating SARS-CoV-2 strains so far.
Figure 2.
BA.2.75 exhibited enhanced human ACE2 binding
(A) Position distribution of important amino acids in BA.2.75 RBD. The mutated residues relative to BA.2 RBD are marked as orange globules. RBM is colored in red.
(B) Binding affinities of RBDs of BA.2, BA.4/5, and BA.2.75 subvariants to hACE2 measured by SPR.
(C) Overall structure of the BA.2.75 S-trimer in complex with hACE2. Three copies of S monomer are colored in yellow, cyan, and magenta, respectively. The hACE2 molecules bound to RBD are colored in orange.
(D) Changes at the interfaces between BA.2.75 RBD (left) and BA.2 RBD (PDB: 7ZF7, right) with hACE2. Key mutated residues are shown as sticks and hydrogen bonds are shown as yellow dash lines.
(E) Binding affinity of hACE2 with BA.2.75 RBD with single substitution Q493R, S446G, and K460N measured by SPR.
(F) Binding affinity of hACE2 with BA.2+N460K RBD measured by SPR.
See also Figure S1.
To further unveil the molecular details of the enhanced hACE2 binding, we determined the near-atomic structure of the BA.2.75 S-trimer in complex with hACE2 (Figures 2C and S1B; Table S1). Like most complex structures, one or two copies of hACE2 are bound to the RBDs in the up conformation (Figure 2C). Structural comparisons revealed that the substitution of Q493R in BA.2.75 established one extra hydrogen bond with K31 on ACE2, increasing its binding capability (Figure 2D) (Cao et al., 2022b; Nutalai et al., 2022). By contrast, the hydrophilic interaction between the main-chain carbonyl group of either G446 or S446 with Q42 from hACE2 is retained (Figure 2D), and mutations of N460K and D339H cause no notable alterations in the binding interface.
To further deconvolute the contribution on hACE2 binding of each mutation on BA.2.75 RBD, we evaluated the individual reverse mutation of H339D, S446G, K460N, or Q493R in BA.2.75 RBD on the hACE2 affinity. SPR assays revealed that only Q493R site mutation induced an approximately 3-fold affinity decrease, whereas the other three reversions did not substantially alter the binding affinity (Figures 2E and S1C). Notably, previous deep mutational scanning (DMS) results suggested that N460K would slightly enhance the ACE2-binding affinity in BA.2 background (Starr et al., 2022). To further confirm whether N460K could impact hACE2 binding, we measured the hACE2-binding affinity of BA.2+N460K RBD. Results demonstrated that N460K did not significantly affect hACE2-binding in both BA.2 and BA.2.75 background (Figure 2F).
Structural analyses of BA.2.75 spike suggest enhanced endosomal pathway usage
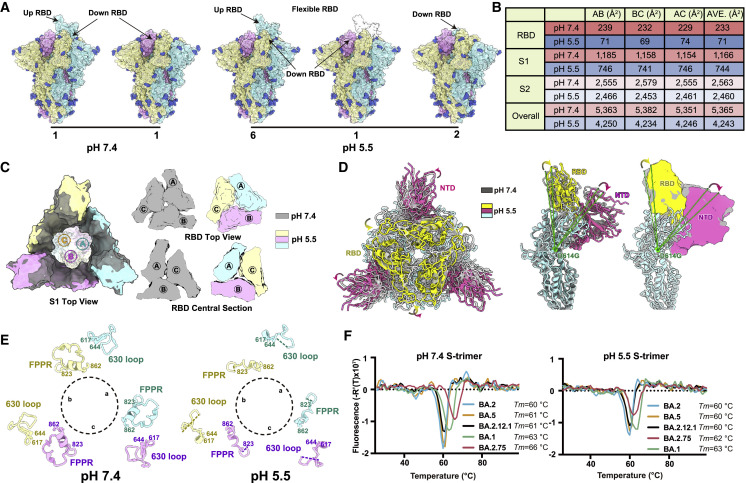
Omicron can enter into host cells endosomally as well as through TMPRSS2 but prefers endosomal fusion. TMPRSS2-dependent cell-surface fusion involves engaging at neutral pH, whereas the endosomal pathway proceeds with acidic pH (Cui et al., 2022; Meng et al., 2022; Zhou et al., 2020). To investigate the putative alterations in cell-entry properties of BA.2.75, we determined the asymmetric cryo-EM reconstructions of the BA.2.75 spike (S-trimer) at an overall resolution of 2.8–3.5 Å at serological (pH 7.4) and endosomal pH levels (pH 5.5) (Figures 3 A, S2A, and S2B; Table S1). Like BA.2 and BA.2.12.1, the BA.2.75 S-trimer exhibit two distinct conformational states (mol ratio ≈ 1:1) corresponding to a closed-form with all three RBDs “down” and an open form with one RBD “up” at neutral pH (Figure 3A). Interestingly, single-RBD-up conformations dramatically dominated at pH 5.5 with mol ratio of 3:1, which is line with a pH-dependent conformational switch observed in D614G, but contrast to BA.1 in one sole configuration with undistinguishable structure either at pH 7.5 or pH 5.5 (Cui et al., 2022). In addition, multiple orientations of RBD in the S-trimer were observed at pH 5.5, revealing structural heterogeneity in BA.2.75, akin to structural observations in D614G and Delta (Zhou et al., 2020), suggesting putative enhanced viral fusion efficiency (Figure 3A). In spite of adopting a similar inter-subunit organization in S2, the neutral BA.2.75 S-trimer exhibits a more compact architecture in the regions formed by three copies of S1 and RBD with a 1.6- and 3.3-fold increased inter-subunit interactions, when compared with its counterparts at pH 5.5 (Figures 3B, 3C, and S2C). Accompanied with alterations in inter-subunit arrangement, a pivotal and correlated NTD-RBD rotation pivoting around D614 within one subunit was observed between the pH 7.4 and pH 5.5 structures (Figure 3D). Apart from these, the fusion peptide proximal region (FPPR) segment (residues 828–853) and 630 loop (residues 620–640) were well resolved in the pH 7.4 structure, whereas they were largely disordered in the pH 5.5 structure, indicating that they might be involved in modulating the spike stability and structural rearrangements (Figure 3E). Remarkably, these pH-mediated structural differences observed in BA.2.75 are much greater than those in other variants (Figures S2D–S2F), suggesting a specific role played by low pH in BA.2.75. In vitro thermal-stability evaluation indicated that the BA.2.75 S-trimer was the most stable among Omicron variants with a melting temperature of 66°C at neutral pH, 3°C improved compared with BA.1 at neutral pH (Figure 3F). Unlike the other four subvariants, BA.2.75 exhibited a distinct shift in stability, less stable than BA.1 at endosomal pH. Together, results suggest BA.2.75 may have evolved to further utilize the low-pH-endosomal cell-entry pathway.
Figure 3.
Structural characteristics of BA.2.75 spike glycoprotein
(A) Surface representation for structures of S-trimer of BA.2.75 at neutral (pH = 7.4) and acidic conditions (pH = 5.5); three protomers were colored in yellow, light blue, and pink, respectively, and N-glycans were highlighted in deep blue.
(B) Buried surface areas between two neighboring protomers, S2-subunits, S1-subunits, or RBD domains.
(C) Structural organization of three S1-subunits and RBDs from the neutral (gray) and acidic BA.2.75 S-trimer (yellow, light blue, and pink). Top view of S2 subunit (left), top view of the RBD (upper right), and central section of RBD (bottom right) show the inter-subunit contacts of the BA.2.75 S-trimers in different pH.
(D) Superimposition of the neutral BA.2.75 S-trimer structure (gray) onto the structure of the acidic BA.2.75 S-trimer (RBD, yellow; NTD, hot pink); Structural rotations and shifts between these two structures were marked by green lines and arrows.
(E) Structural alterations in the 630 loop (residues 617–644, light blue) and FPPR (residues 823–862, yellow) of the three protomers (a, b, and c) from these two S-trimers were shown. Dashed lines indicate gaps in the chain trace (disordered loops).
(F) Thermal stability analysis of BA.1 (green), BA.2 (blue), BA.2.12.1 (black), BA.2.75 (red), and BA.5 (yellow) S-trimers at neutral and acidic pHs.
See also Figure S2.
Structural features underpinning the stability
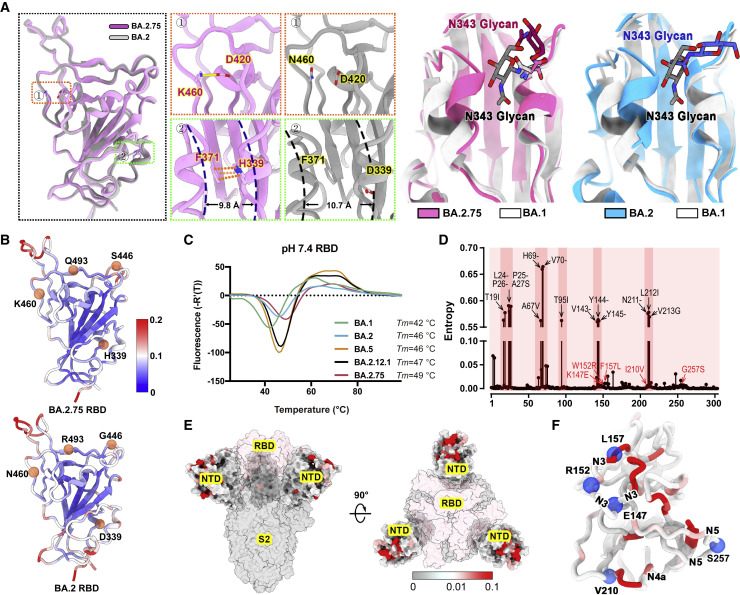
Compared with other Omicron subvariants, increased structural heterogeneity in BA.2.75 is mainly contributed by the reduced hydrophilic and hydrophobic interactions between two contacted RBDs due to the reversion of R493Q and local conformational alterations in the hairpin loop (residues 373–380), respectively (Figure S2G). Surprisingly, the BA.2.75 RBD displays one more rigid and compact configuration than other subvariants, presumably representing improved stability and immunogenicity (Figure 4 A). Substitution of N460K established one new salt bridge with D420 and mutation of D339H with altered rotamer formed π-π interactions with F371, pulling α1 and α2 helices closer compared with BA.2 and BA.1, respectively (Figures 4A and S3). Perhaps correlated with this, molecular simulation analysis also revealed overall improved stability for the RBD and the distal RBM in BA.2.75, which might facilitate the protein folding and increase protein expression (Figure 4B). These structural analyses are in line with experimental observations revealed by thermal stability and DMS results of N460K/D339H (Figure 4C; Starr et al., 2020, 2022; Zahradník et al., 2021).
Figure 4.
Structural features of BA.2.75 spike RBD and NTD
(A) Structural comparisons of RBDs of BA.2.75, BA.2, and BA.1. The newly established interaction of BA.2.75 (pink) with respect to BA.2 (gray) on the RBD is shown on the left. Salt bridges formed between D420 and K460 and π-π stack formed between H339 and F371 in BA.2.75 RBD are highlighted. The distances between α1 and α2 helices on RBD are also marked. A diagram presentation of N343 glycan conformational differences among BA.1 (gray), BA.2 (blue), and BA.2.75 (pink) is shown on the right.
(B) The stability landscapes of BA.2.75 and BA.2 RBD. The cartoons of BA.2.75 and BA.2 RBD are colored by root mean square fluctuation (RMSF) calculated from the last 2 ns of the MD stimulations. Residues 339, 446, 460, and 493 are shown as red spheres.
(C) Thermal stability measurements of the RBD from BA.1 (green), BA.2 (blue), BA.5 (orange), BA.2.12.1 (black), and BA.2.75 (red) at pH 7.4.
(D) Entropy of SARS-CoV-2 NTD variants among circulating isolates. The residues with higher entropy are highlighted by dark red background. The dominant mutations on SARS-CoV-2 NTD and the mutations on BA.2.75 NTD are labeled in black and red, respectively.
(E) The heatmap for circulating variants with mutations on the NTD. Mutation frequency for each residue is calculated based on the datasets from Global Initiative on Sharing All Influenza Data (GISAID).
(F) Cartoon representation of NTD colored by mutation frequency as same as (E). Five mutations in the BA.2.75 NTD are displayed as blue balls and labeled. The secondary structure these mutations locate are also labeled.
See also Figure S3.
The NTD shows the most diversity with a larger number of prevalent mutations and deletions compared with other regions of the S-trimer (Figure 4D). Interestingly, the majority of these substitutions are highly enriched in the peripheral region of the S-trimer, adjacent to the NTD super-site (Figure 4E), presumably escaping neutralizing antibodies and modulating entry efficiency (Cerutti et al., 2021; McCallum et al., 2021). Among five mutations in the BA.2.75 NTD, K147E, W152R, and F157L locate at the super-site, whereas the other two partially overlap with epitopes targeted by other neutralizing antibody classes (Figure 4F).
BA.2.75 significantly evades plasma from Delta and BA.4/BA.5 convalescents
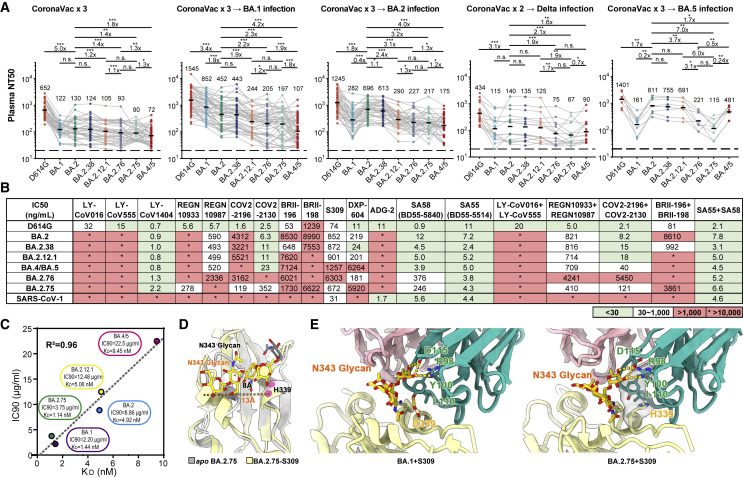
Next, we evaluated the effect of BA.2.75 on the neutralizing activities of vaccinees/convalescents plasma. We found all of the samples exhibited highest 50% neutralization titers (NT50s) against D614G, in comparison with other variants, including the corresponding infected variant. This could be explained by the immune imprinting or “original antigenic sin” of Omicron breakthrough infection reported recently (Cao et al., 2022b; Quandt et al., 2022; Reynolds et al., 2022). BA.2.38 is neutralized exactly similarly to BA.2, which indicates N417T mutations does not significantly cause more evasion of NAbs. BA.2.75 exhibits significantly stronger humoral immune evasion than BA.2 in plasma samples from individuals who had received 3 doses of CoronaVac before or after BA.1/BA.2 breakthrough infection, resulting in a 1.4- to 3.2-fold reduction in NT50 (Figure 5 A; Table S2). BA.2.75 is also slightly more neutralization-evasive than BA.2.12.1 in post-vaccination BA.2 convalescents, but less than BA.4/BA.5 (Figure 5A). However, BA.2.75 is more humoral immune evasive than BA.4/BA.5 in plasma from Delta breakthrough-infection convalescents, which may explain BA.2.75’s substantial growth advantage in India (Figure 5A). This phenomenon could be contributed by the R452 stimulation of Delta, which could result in potent antibodies that are highly effective against BA.4/BA.5. In plasma samples from vaccinees or convalescents from BA.1/BA.2 breakthrough infection, BA.2.12.1 exhibited strong immune evasion and significant lower NT50 than that against BA.2, as reported previously (Cao et al., 2022b; Kimura et al., 2022; Wang et al., 2022b). However, comparable neutralization against BA.2 and BA.2.12.1 was observed in plasma from Delta or BA.5 convalescents, indicating a proportion of NAbs induced by infection with Delta or BA.5, which harbors R452 are cross-reactive to RBD with L452 or Q452. Most importantly, BA.2.75 also displays strong humoral immune evasion in the convalescent plasma from BA.5 breakthrough-infection convalescents (Figure 5A). Compared with BA.5, the NT50 against BA.2.75 by BA.5 convalescent plasma showed a nearly 4-fold reduction. Considering these samples also showed significantly decreased neutralization against BA.1, G446S, which also appeared in BA.1, should contribute greatly. In addition, since NTD mutations in BA.1 and BA.2 are significantly different and BA.2.75 also harbors additional mutations on NTD, NTD mutations could contribute a lot as well.
Figure 5.
BA.2.38, BA.2.75, and BA.2.76 showed distinct antibody evasion
(A) Half neutralization titers (NT50s) against SARS-CoV-2 D614G and Omicron variants pseudoviruses by plasma samples from individuals who received 3 doses CoronaVac (n = 40), 3 doses CoronaVac followed by BA.1 infection (n = 50), 3 doses CoronaVac followed by BA.2 infection (n = 39), 2 doses CoronaVac followed by Delta infection (n = 16), or 3 doses CoronaVac followed by BA.5 (n = 8) infection. Geometric mean titers (GMTs) are annotated above each group. ∗p < 0.05; ∗∗p < 0.01; ∗∗∗p < 0.001. p values are calculated using two-tailed Wilcoxon signed-rank test of paired samples. Pair-wise fold changes were calculated using the NT50 against the strain on the left divided by that against the strain on the right.
(B) Neutralizing activities against SARS-CoV-2 D614G, Omicron variants, and SARS-CoV-1 pseudovirus of therapeutic neutralizing antibodies. Background colors indicate neutralization levels. Green, IC50 < 30 ng/mL; white, 30 ng/mL < IC50 < 1,000 ng/mL; red, IC50 > 1,000 ng/mL. ∗IC50 > 10,000 ng/mL.
(C) Correlation plots of binding affinities and neutralizing activities (IC90) of S309 against BA.1, BA,2, BA.2.12.1, BA.4/5, and BA.2.75.
(D) Local conformational alterations in the BA.2.75 RBD upon S309 binding. The N343 glycan of the apo BA.2.75 RBD (gray) and S309-bounded BA.2.75 RBD (yellow) are shown as sticks and the distances between α1 and α2 helices from two configurations are also labeled.
(E) Interaction details of the BA.1 RBD (left) and BA.2.75 RBD (right) in complex with S309. Hydrogen bonds and hydrophobic patches are presented as yellow dashed lines and gray surfaces, respectively. The light chain and heavy chain of S309 are colored in pink and cyan, respectively.
See also Figures S4 and S5.
Efficacy of NAb drugs against BA.2.75
As for antibody therapeutics, we tested the pseudovirus neutralizing activity of 14 NAb drugs in clinical development against SARS-CoV-2 variants, including BA.2.38, BA.2.75, and BA.2.76 (Du et al., 2020; Hansen et al., 2020; Jones et al., 2021; Pinto et al., 2020; Rappazzo et al., 2021; Shi et al., 2020; Wang et al., 2021; Westendorf et al., 2022; Zost et al., 2020). REGN10933 and COV2-2196 partially recovered their activities against BA.2.75 due to R493Q reversion (Figure 5B). However, REGN10987 and COV2-2130 were also affected by G446S, resulting in only a mild change in the neutralizing activity of the corresponding cocktails against BA.2.75 (Figure 5B; Cao et al., 2022a; Hansen et al., 2020; Zost et al., 2020). LY-CoV1404 (bebtelovimab) remains highly potent against BA.2.75 and BA.2.76 (Westendorf et al., 2022).
Notably, multiple studies have reported the efficacy of S309 (Sotrovimab; Pinto et al., 2020) against BA.2.75; however, some suggest S309 recovered potency against BA.2.75, whereas others suggest the opposite (Sheward et al., 2022; Wang et al., 2022c; Yamasoba et al., 2022). We noticed that although D339H is a charge-reversing mutation on the S309 binding interface, the neutralizing activity of S309 was not affected and even exhibited slightly improved neutralization activity against BA.2.75 compared with BA.2 and BA.4/5 (Figure 5B). To further confirm this observation, we measured the binding affinity of S309 against BA.1, BA.2, BA.2.12.1, BA.4/5, and BA.2.75 RBD using biolayer interferometry (BLI) (Figure S4A). Interestingly, we found that S309 recovered its binding activity against BA.2.75 RBD as to that against BA.1, and the binding affinities of S309 to these five Omicron subvariants matched well with the IC90s, but not the IC50s (Figures 5C and S4B). The neutralization of these non-ACE2-competing NAbs may be associated with N343 glycosylation, RBD up/down dynamics, and inhibition of membrane fusion; thus, the measured IC50 is largely affected by the utilized pseudovirus types and cell lines and may not be as robust as the measurement of IC90 (Luo et al., 2022). Although S309 itself had been largely escaped by previous Omicron variants and could hardly be effective, it targets an epitope, which is also targeted by a special group of non-ACE2-competing Nabs, which broadly and potently neutralize SARS-CoV-2-related sarbecoviruses. Therefore, to explore the underlying mechanism and detailed impacts of D339H, we determined the cryo-EM structure of the BA.2.75 S-trimer in complex with S309 at 3.5 Å, together with structures of the apo BA.2.75 and previously reported BA.1-S309 complex, allowing us to dissect the detailed structural variations (Figures S5A and S5B). Upon S309 binding, BA.2.75 displays a pivotal conformational alteration in the hairpin loop (residues 366–377), separating α1 and α2 helices away, akin to structural arrangement in apo BA.1 and BA.1-S309 complexes (Figures 5D and S5C). The mutation D339H lost one hydrogen bond that would be established by Y100 from heavy-chain complementary-determing region (HCDR3) in BA.1-S309 complex but established more hydrophobic interactions with Y100 and L110 from S309 in BA.2.75, slightly increasing its binding affinity to BA.2.75 compared with BA.1 (Figure 5E). These suggest Sotrovimab may become active against BA.2.75, which requires authentic virus validations.
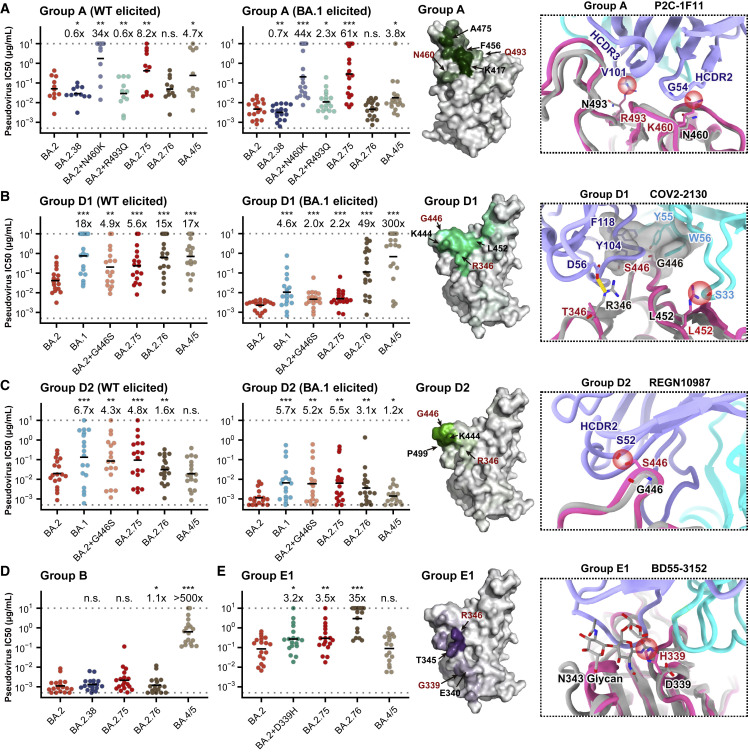
BA.2.75 displays distinct RBD antibody evasion patterns from BA.4/BA.5
To further investigate the antibody escaping mechanism of BA.2.75, we determined the neutralizing activities against BA.2.75, BA.2.76, and BA.4/BA.5 of a panel of BA.2-effective NAbs from five epitope groups, defined by unsupervised clustering based on high-throughput yeast-display-based DMS, that could be potentially affected by D339H, N460K, R493Q, and G446S (Cao et al., 2022a, 2022b; Greaney et al., 2021; Table S3). BA.2.75 can cause a global reduction in neutralizing activities of group A NAbs, represented by DXP-604 and P2C-1F11 (BRII-196) (Figure 6 A; Cao et al., 2020; Du et al., 2020; Ge et al., 2021). N460K is the major mutation that caused this antibody evasion. Analyses of the representative structure of P2C-1F11 (BRII-196) suggest K460 with a longer side chain could induce steric clashes that result in NAb evasions (Figure 6A; Ge et al., 2021). Interestingly, R493Q can cause neutralization reduction of group A NAbs isolated from individuals infected by BA.1, which carries R493, whereas the reversion can also induce neutralization recovery of group A NAbs isolated from individuals stimulated by wild-type (WT) SARS-CoV-2, which carries Q493 (Figure 6A). Importantly, although F486V of BA.4/BA.5 could induce neutralization reduction of group A NAbs, BA.2.75 causes more severe group A antibody evasion, especially for those NAbs that remain highly effective against BA.4/BA.5 (Table S3).
Figure 6.
Evasion of NAbs targeting various RBD epitopes by BA.2.38, BA.2.75, and BA.2.76
Neutralizing activities against SARS-CoV-2 Omicron variants of NAbs in (A) group A (n = 11 isolated from SARS-CoV-2WT convalescents or vaccinees; n = 18 from post-vaccination BA.1 convalescents), (B) group D1 (n = 17, 18, respectively), (C) group D2 (n = 18, 17, respectively), (D) group B (n = 18), and (E) group E1 (n = 18). Deep mutational scanning (DMS) profiles of antibodies in groups A, D1, D2, and E1 are projected onto RBD structure to show interacting hotspots of each group. Color shades indicate escape scores of RBD residues. Interface structural models of representative antibodies in groups A, D1, D2, and E1, in complex of RBD, show potential escaping mechanism of BA.2.75 and BA.2.76. Geometric mean of IC50 fold changes compared with BA.2 are annotated above the bars. ∗p < 0.05; ∗∗p < 0.01; ∗∗∗p < 0.001. p values are calculated using two-tailed Wilcoxon signed-rank test of paired samples.
Reduced activities against BA.2.75 compared with against BA.2 were also observed for NAbs in groups D1 and D2, mostly due to G446S via disruption of the hydrophobic interactions, conferring that the neutralizing activities of these NAbs against BA.2.75 are quite similar to those against BA.1 (Figures 6B and 6C). Due to the L452R mutation, BA.4/BA.5 could heavily escape from neutralization by D1 antibodies (e.g., COV2-2130, a representative antibody from group D1). Similarly, the R346T mutation carried by BA.2.76 could also decrease the neutralizing activity of group D1 NAbs. Notably, D1 NAbs isolated from BA.1 convalescents exhibit stronger resistance to BA.2.75, and overall, D1 NAbs neutralize BA.2.75 better than BA.4/BA.5; however, this property is not observed in group D2 NAbs, whose activities against BA.2.75 are lower than against BA.4/BA.5, since D2 NAbs are not sensitive to L452R, but to G446S (e.g., REGN10987, a representative antibody from group D2) (Cao et al., 2022b; Starr et al., 2021).
Furthermore, neutralizing activities of group B antibodies were not affected by mutations of BA.2.75, but most of them were completely escaped by F486V of BA.4/BA.5, as reported previously (Figure 6D). D339H, a new specific mutation observed in BA.2.75, could cause an overall neutralization reduction for BA.4/5-effective NAbs in group E1, which could be largely attributed to the altered configuration of N343 glycan (e.g., BD55-3152, a representative antibody from group E1) (Figure 6E; Cao et al., 2022b). Together, the results suggest all four additional RBD mutations of BA.2.75 compared with BA.2 are capable of escaping certain groups of NAbs, and the RBD antigenicity of BA.2.75 is distinct from that of BA.4/BA.5, which may partially explain BA.2.75’s strong humoral immune evasion observed in the convalescent plasma from BA.5 breakthrough infection.
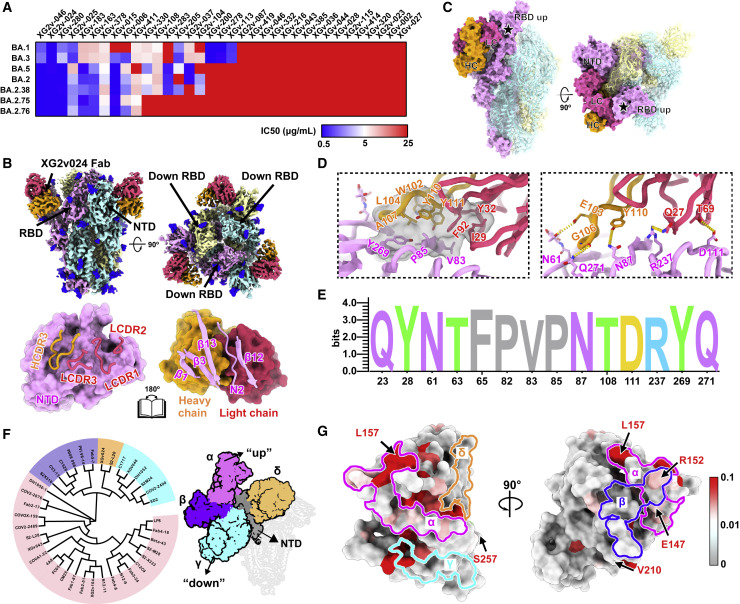
BA.2.75 evades BA.5 effective anti-NTD NAbs
Besides mutations on RBD, BA.2.75 also harbors multiple NTD mutations that may cause anti-NTD NAb evasion (Wang et al., 2022c). In previous studies, we isolated 323 monoclonal antibodies (mAbs) derived from memory B cells in participants who had received 3 doses of CoronaVac, ∼50% and ∼30% of which recognized RBD and NTD, respectively. Among 71 anti-NTD mAbs, 25 neutralized at least one of SARS-CoV-2 VOCs (Tables S3 and S6A). Interestingly, anti-NTD NAbs displayed diverse reactivity against Omicron variants (Figure 7 A). A proportion of these NAbs, such as XG2v-024, could broadly neutralize all VOCs, including BA.2.75, with good potency, whereas some NAbs could efficiently neutralize BA.2 and BA.5 but showed decreased activity against BA.2.75 and BA.2.76 (Figures 7A and S6A). This suggests that NTD-targeting NAbs also form diverse epitopes and reacts differently to Omicron subvariants due to their distinct mutations on NTD. To dissect the neutralizing breadth of XG2v-024-like NTD-targeting NAbs, we first examined the cryo-EM structure of XG2v-024 in complex with BA.2.75 spike (Figure S7). The XG2v024/spikeBA.2.75 structure reveals only one configuration: three XG2v024 Fabs bound to a completely closed S-trimer with all three RBDs in the down state (Figure 7B). This is contrary to the two conformations observed in the apo BA.2.75 S-trimer, indicative of a role of XG2v-024 in allosteric modulation on RBD “down” disposition. Interestingly, the binding of XG2v-024 created steric clashes with the adjacent “up” RBD or its bound hACE2, which reveals the neutralization mechanism via distant RBD conformation modulation and further blockade of ACE2 binding (Figure 7C). Remarkably, all XG2v-024 epitope residues are extremely conserved epitopes across nearly all circulating SARS-CoV-2 variants, enabling XG2v-024’s broad SARS-CoV-2 neutralizing ability (Figures 7D and 7E).
Figure 7.
BA.2.75 and BA.2.76 evolve mutations to escape NTD-targeting NAbs
(A) Heatmap of pseudo-typed virus neutralization by antibodies recognized NTD.
(B) Cryo-EM structures of the BA.2.75 S-trimer in complex with XG2v024 Fab (top). Structures are shown as surface. The three subunits of S protein are colored in yellow, cyan, and magenta, respectively. The heavy chain and light chain are colored in hot pink and orange, respectively. Interactions between the XG2v024 and BA.2.75 NTD (bottom). The CDRs of the XGv024 that interact with BA.2.75 NTD are displayed as thick tubes over the magenta surface of the NTD. The XG2v024 epitope is shown as a cartoon representation over the surface of the XG2v024 Fab.
(C) XG2v024 neutralizing mechanism. The BA.2.75 S-trimer and XG2v024 Fab are shown as surface. The color scheme remains unchanged from the previous panels. Steric clash is marked as black stars.
(D) Interactions details of BA.2.75 NTD in complex with XG2v024 Fab. The hydrophobic (top) and hydrophilic (bottom) interactions are shown, respectively. Hydrogen bonds and hydrophobic patches are presented as yellow dashed lines and gray surfaces, respectively.
(E) Structural landscapes of the four classes of NTD NAbs. Antigenic patches recognized by four types of NAbs are outlined in the assigned color scheme. Five mutations in the BA.2.75 NTD are labeled.
(F) Structure-based antigenic clustering of SARS-CoV-2 NTD Nabs (left). A total of 38 NTD NAbs with available structures are classified into four clusters (α, β, γ, and δ). Surface representative model of four types of NAbs bound to the NTD (right). Fab fragments of four representative antibody are shown in different colors and the NTD is colored in gray.
(G) Analysis of sequence conservation of XG2v024 epitope. The logo plot represents the conservation of XG2v024 epitope residues from 26 SARS-CoV-2 lineages.
See also Figures S6 and S7.
By further examining the available structures of 38 SARS-CoV-2 NTD-targeting NAbs, including XG2v-024 presented here, we found that the NTD NAbs can be classified into four classes and the NTD mutations of BA.2.75 affects different classes (Figures 7F and 7G). Class α antibodies, targeting the NTD super-site and facing away from the viral membrane (facing up), possess limited neutralizing breadth due to highly frequent mutations on the epitopes (Figures S6B and S6C; McCallum et al., 2021; Wang et al., 2022d). Sites β and δ (e.g., targeted by XG2v-024), as the left and right flank clusters, construct a shallow groove and locate at the back of the groove, respectively, eliciting relatively broad neutralizing antibodies, albeit with less potency (Figure 7F). Importantly, the new NTD mutations K147E, W152R, F157L, and G257S carried by BA.2.75 and the Y248N carried by BA.2.76 are located on the epitope of Class α and β antibodies, likely causing the escape of those BA.5 effective NTD NAbs. By contrast, γ antibodies bound to a patch beneath the groove have their Fab constant domains directed downward toward the virus membrane (facing down) and were proved to enhance infection efficiency in vitro but exhibit protection in vivo (Li et al., 2021; Liu et al., 2021); unfortunately, the V213G harbored by all Omicron variants could cause large-scale escapes of these NAbs (Figures 7F and S6C). Noteworthy, the classification is highly similar to the classes assigned in BLI competition experiments (Wang et al., 2022d). Together, we showed that besides the RBD mutations, the new NTD mutations evolved by BA.2.75 could also induce further neutralization escape of BA.2 and BA.5 effective anti-NTD NAbs.
Discussion
Previous studies revealed that Omicron is less dependent on TMPRSS2 cleavage but used endosomal pathway more for cell entry, which is associated with its preference to upper airway, instead of lung cells with higher TMPRSS2 expression, resulting in the relatively mild symptoms caused by Omicron compared with early variants, especially Delta (Meng et al., 2022; Peacock et al., 2022). Correlated with this, to overcome the environmental exposure, Omicron variants continuously evolve to improve their stability, and BA.2.75 exhibits further increased stability than other variants in neutral conditions. In spite of facilitating viral attachment to the host cells, viral membrane fusion may be compromised by such improved stability. To achieve an effective fusion step, BA.2.75 displays a substantial pH-dependent conformational switch to mediate RBD positioning and inter-subunit destabilization at endosomal microenvironments, thereby conferring improved fusion efficiency, indicative of enhanced endosomal pathway usage. These structural analysis is also largely supported by observed improved cell-cell fusion for BA.2.75 in comparison to BA.2, which is primarily driven by the N460K mutation (Qu et al., 2022). In spite of not altering binding affinity to hACE2, this substitution can alter positively charged distribution on the BA.2.75 RBD surface, presumably facilitating potential charge-charge contacts with host cell membrane or negatively charged sugars on membrane proteins to increase its fusion efficiency.
The rapid evolution of SARS-CoV-2 Omicron variants greatly challenged the efficacy of vaccines and therapeutics (Bruel et al., 2022; Cele et al., 2022; Dejnirattisai et al., 2022; Quandt et al., 2022; Tuekprakhon et al., 2022; Yamasoba et al., 2022). In this paper, we demonstrated that the local growth advantage of BA.2.75 compared with BA.5 could be explained by the previous widespread infections with Delta VOC in India (Yang and Shaman, 2022). As Delta and BA.4/5 both harbor L452R mutation on the RBD, and arginine generally being an immunogenic residue, convalescent plasma from Delta infection may contain 452R-targeting BA.5-effective NAbs, which could hinder the transmission of BA.4/BA.5 in India.
More importantly, we showed that the BA.5 convalescent plasma could not neutralize BA.2.75 well, suggesting that BA.2.75 and its subvariants could potentially reinfect BA.5 convalescents and become the next dominant strain after the current global BA.4/5 wave. In addition, it is worrisome that the significantly enhanced hACE2-binding affinity of BA.2.75, mainly due to R493Q reversion, could lead to its higher capability to tolerate additional immune-evasive mutations, which usually results in the reduction of ACE2-binding, such as mutations on F486 like BA.4/5 (Cao et al., 2022b; Wang et al., 2022b). Indeed, several mutations that were proved antibody evasive, such as R346T, F486S, and L452R, have been identified in sublineages of BA.2.75, which would further increase its humoral evasion capability and transmission advantage. BA.2.75.2, a representative highly antibody-escaping subvariant, has been detected in India, Singapore, Australia, United States, and many other regions. Our results urge the close monitoring of the spread of BA.2.75 and its subvariants.
Author contributions
Y.C., X.S.X., and X.W. designed the study. Y.C. and X.W. and wrote the manuscript with inputs from all authors. F.J. and J.W. (BIOPIC) performed and analyzed the yeast-display-based DMS experiments. Y.C., W.S., A.Y., Y.Y., T.X., P.W., J.W. (Changping Laboratory), S.Y., L.L., R.A., Yao Wang, N.Z., X.N., L.Y., C.Y., and F.S. performed and analyzed the pseudovirus neutralization assays. Y.Y. and Youchun Wang prepared the vesicular stomatitis virus (VSV)-based SARS-CoV-2 pseudovirus. L.W., P.L., C.Y., Q.Z., J.D., W.F., and X.W. performed the structural analyses. B.M. and R.K.G. performed infectivity and fusion analysis. Q.G. proofed the manuscript. C.Y., J.D., and X.W. performed SPR experiments. X.H. and R.J. recruited the SARS-CoV-2 vaccinees and Omicron/Delta convalescents.
STAR★Methods
Key resources table
| REAGENT or RESOURCES | SOURCE | IDENTIFIER |
|---|---|---|
| Bacterial | ||
| Trelief 5α Chemically Competent Cell | Tsingke | Cat# TSC-C01 |
| G∗ΔG-VSV | Kerafast | Cat# EH1020-PM |
| DH5α Chemically Competent Cell | Invitrogen | Cat# 12034013 |
| Cell lines | ||
| HEK293F cells | Thermo Fisher | Cat# 11625019 |
| Huh-7 cells | Japanese Collection of Research Bioresources [JCRB] | 0403 |
| 293T-hACE2 | SinoBiological | N/A |
| Chemicals and recombinant proteins | ||
| StarFect 293 Transient Transfection Medium | GenStar | Cat# C104-1L |
| 1x PBS, pH 7.4 | Thermo Fisher | Cat# 10010031 |
| 0.4% (w/v) Trypan blue stain | ThermoFisher | 15250061 |
| DMEM | Hyclone | SH30243.01 |
| D-luciferin reagent | PerkinElmer | 6066769 |
| FBS | Gibco | 10099141C |
| DMSO | Solarbio | D8371 |
| PBS | Invitrogen | C10010500BTC |
| Penicillin–Streptomycin solution | Gibco | 15140122 |
| HEPES | Gibco | 15630080 |
| 0.25% Trypsin-EDTA | Gibco | 25200072 |
| Critical commercial reagents | ||
| Superose 6 Increase 10/300 GL | Cytiva | Cat# 29091596 |
| Superdex 200 Increase 10/300 GL | Cytiva | Cat# 28990944 |
| SYPRO Orange Protein Gel Stain | Thermo Fisher | Cat# S6650 |
| Protein A Biosensors | Octet | Cat# 18-5010 |
| CM5 Biosensor | Cytiva | Lot# 10310113 |
| Recombinant DNA | ||
| SARS-CoV-2 Omicron/BA.1 S gene, residues 1-1208, 6P and A67V, Δ69-70, T95I, G142D, Δ143-145, Δ211, L212I, ins214EPE, G339D, S371L, S373P, S375F, K417N, N440K, G446S, S477N, T478K, E484A, Q493R, G496S, Q498R, N501Y, Y505H, T547K, D614G, H655Y, N679K, P681H, N764K, D796Y, N856K, Q954H, N969K, L981F, T4 fibritin trimerization motif, 6xHis, pcDNA | This manuscript | GenBank: UOT56373.1 |
| SARS-CoV-2 Omicron/BA.2 S gene, residues 1-1208, 6P and T19I, L24S, Δ25-27, G142D, V213G, G339D, S371F, S373P, S375F, T376A, D405N, R408S, K417N, N440K, S477N, T478K, E484A, Q493R, Q498R, N501Y, Y505H, D614G, H655Y, N679K, P681H, N764K, D796Y, Q954H, N969K, T4 fibritin trimerization motif,6xHis, pcDNA | This manuscript | GenBank: UMX32778.1 |
| SARS-CoV-2 Omicron/BA.2.12.1 S gene, residues 1-1208, 6P and T19I, L24S, Δ25-27, G142D, V213G, G339D, S371F, S373P, S375F, T376A, D405N, R408S, K417N, N440K, L452Q, S477N, T478K, E484A, Q493R, Q498R, N501Y, Y505H, D614G, H655Y, N679K, P681H, S704L, N764K, D796Y, Q954H, N969K, T4 fibritin trimerization motif, 6xHis, pcDNA | This manuscript | N/A |
| SARS-CoV-2 Omicron/BA.2.75 S gene, residues 1-1208, 6P and T19I, L24S, Δ25-27, G142D, K147E, W152R, F157L, I210V, V213G, G257S, G339H, S371F, S373P, S375F, T376A, D405N, R408S, K417N, N440K, G446S, N460K, S477N, T478K, E484A, Q498R, N501Y, Y505H, D614G, H655Y, N679K, P681H, N764K, D796Y, Q954H, N969K, T4 fibritin trimerization motif, 2xStrep, 6xHis, pcDNA | This manuscript | N/A |
| SARS-CoV-2 Omicron/BA.5 S gene, residues 1-1208, 6P and T19I, L24S, Δ25-27, Δ69-70, G142D, V213G, G339D, S371F, S373P, S375F, T376A, D405N, R408S, K417N, N440K, L452R, S477N, T478K, E484A, F486V, Q498R, N501Y, Y505H, D614G, H655Y, N679K, P681H, N764K, D796Y, Q954H, N969K, T4 fibritin trimerization motif, 2xStrep, 6xHis, pcDNA | This manuscript | GenBank: UTF37239.1 |
| Deposited data | ||
| pH7.4 BA.2.75 Spike Trimer (1 RBD Up) | This manuscript | PDB ID 7YQT, EMD-34034 |
| pH7.4 BA.2.75 Spike Trimer (3 RBD Down) | This manuscript | PDB ID 7YQU, EMD-34035 |
| pH5.5 BA.2.75 Spike Trimer (1 RBD Up) | This manuscript | PDB ID 7YQV, EMD-34036 |
| pH5.5 BA.2.75 Spike Trimer (3 RBD Down) | This manuscript | PDB ID 7YQW, EMD-34037 |
| BA.2.75 Spike Trimer in complex with S309 (state1) | This manuscript | PDB ID 7YQX, EMD-34038 |
| BA.2.75 Spike Trimer in complex with S309 (state2) | This manuscript | PDB ID 7YQY, EMD-34039 |
| BA.2.75 Spike Trimer in complex with S309 (state3) | This manuscript | PDB ID 7YQZ, EMD-34040 |
| BA.2.75 Spike Trimer in complex with S309 (local) | This manuscript | PDB ID 7YR0, EMD-34041 |
| BA.2.75 Spike Trimer in complex with XG2v024 | This manuscript | PDB ID 7YR1, EMD-34042 |
| BA.2.75 Spike Trimer in complex with ACE2 (state1) | This manuscript | PDB ID 7YR2, EMD-34043 |
| BA.2.75 Spike Trimer in complex with ACE2 (state2) | This manuscript | PDB ID 7YR3, EMD-34044 |
| BA.2.75 Spike Trimer in complex with ACE2 (local) | This manuscript | PDB ID 7YR4, EMD-34045 |
| Software | ||
| igraph (1.2.5) | Csardi and Nepusz, 2006 | https://cran.r-project.org/web/packages/ igraph/index.html |
| Gctf program (v1.06) | Zhang, 2016 | https://www.mrc-lmb.cam.ac.uk/kzhang/Gctf |
| RELION (v3.07) | Zivanov et al., 2018 | https://www2.mrc-lmb.cam.ac.uk/relion |
| UCSF Chimera | Pettersen et al., 2004 | https://www.cgl.ucsf.edu/chimera |
| UCSF ChimeraX | Goddard et al., 2018 | https://www.rbvi.ucsf.edu/chimerax/ |
| PHENIX | Adams et al., 2010 | https://www.phenix-online.org |
| Coot | Emsley et al., 2010 | https://www2.mrc-lmb.cam.ac.uk/ Personal/pemsley/coot |
| cryoSPARC 3.2.4 | Punjani et al., 2017 | https://cryosparc.com |
Resource availability
Lead contact
Further information and requests for resources and reagents should be directed to and will be fulfilled by the lead contact, Xiangxi Wang (xiangxi@ibp.ac.cn)
Materials availability
All reagents generated in this study are available from the lead contact with a completed Materials Transfer Agreement.
Experimental model and subject details
Cell lines
HEK293T and Huh-7 cells were cultured in Dulbecco’s Modified Eagle’s Medium (DMEM) supplemented with 10% fetal bovine serum (FBS). The cultures were maintained at 37°C in an incubator supplied with 8% CO2.
SARS-CoV-2 pseudovirus
The SARS-CoV-2 pseudovirus was constructed as previously described using VSV pseudotyped virus (G∗ΔG-VSV) (Nie et al., 2020). Pseudovirus carrying spikes of SARS-CoV-2 D614G (SARS-CoV-2 spike (GenBank: MN908947) +D614G), BA.1 (A67V, H69del, V70del, T95I, 142-144del, Y145D, N211del, L212I, ins214EPE, G339D, S371L, S373P, S375F, K417N, N440K, G446S, S477N, T478K, E484A, Q493R, G496S, Q498R, N501Y, Y505H, T547K, D614G, H655Y, N679K, P681H, N764K, D796Y, N856K, Q954H, N969K, L981F), BA.2 (GISAID: EPI_ISL_7580387, T19I, del24-26, A27S, G142D, V213G, G339D, S371F, S373P, S375F, T376A, D405N, R408S, K417N, N440K, S477N, T478K, E484A, Q493R, Q498R, N501Y, Y505H, D614G, H655Y, N679K, P681H, N764K, D796Y, Q954H, N969K), BA.2.38 (BA.2+N417T), BA.2.12.1 (BA.2+L452Q+S704L), BA.2.75 (BA.2+K147E+W152R+F157L+I210V+G257S+D339H+G446S+N460K+R493Q), BA.2.76 (BA.2+R346T+Y248N), BA.4/BA.5 (T19I, L24S, del25-27, del69-70, G142D, V213G, G339D, S371F, S373P, S375F, T376A, D405N, R408S, K417N, N440K, L452R, S477N, T478K, E484A, F486V, Q498R, N501Y, Y505H, D614G, H655Y, N679K, P681H, N764K, D796Y, Q954H, N969K) was constructed and used, as described previously (Cao et al., 2022b).
Method details
Protein expression and purification
The full-length Spike (S) proteins of Omicron (BA.1) and its variants BA.2, BA2.12.1, BA.4/5 were derived from previous constructions. BA.2 RBD and its mutation N460K, BA.2.75 spike (T19I, L24S, Δ25-27, G142D, K147E, W152R, F157L, I210V, V213G, G257S, G339H, S371F, S373P, S375F, T376A, D405N, R408S, K417N, N440K, G446S, N460K, S477N, T478K, E484A, Q498R, N501Y, Y505H, D614G, H655Y, N679K, P681H, N764K, D796Y, Q954H, N969K ) and BA.2.75 RBD and its mutations H339D, S446G, K460N, Q493R were realized by overlapping PCR with BA.2 Spike gene as template. To facilitate protein expression and stabilize the trimer conformation, all S gene constructs have proline substitutions at residues 817, 892, 899, 942, 986, 987 and alanine substitutions at residues 683, 685, and the T4 fibrin fold domain added to the C-terminal target sequence (ACRO Biosystems, cat. no. SPN-C5523). In addition, His or Strep II tags were connected to the C-terminal of spike and RBD sequences to facilitate protein purification. For the selected antibody, the light and heavy chains need to be constructed separately. All the constructed plasmids were transiently transfected into suspended HEK293 F cells and cultured in a constant temperature shaker of 8% CO2 and 37 °C. After 72 hours of culture(Antibodies require 120 hours), cell supernatants were collected and Spike and RBD were initially purified by affinity chromatography using Ni-NTA or StrepTactin resin, while antibodies required protein A purification. The proteins were repurified using Superdex 200 10/300GL (Cytiva) or Superose 6 10/300 (Cytiva) in 20 mM Tris-HCl, 200 mM NaCl, pH 8.0.
Determination of thermal stability
The thermofluor stability assay was performed to evaluate the stability of Spike and RBD of omicron and its variants BA.1, BA.2, BA.2.12.1, BA.2.75 and BA.5(ACRO Biosystems). For the Spike protein, we performed two different pH conditions (pH = 7.4 and pH = 5.5), while for the RBD, only stability assay was performed at pH = 7.4. All protein samples were set up as 25 uL reaction system, including 5 mg protein and 5000× SYPRO Orange (Invitrogen, Carlsbad, USA) as fluorescence probe. The MX3005 qPCR instrument (Agilent, Santa Clara, USA) was used to detect the fluorescent signal generated by the protein during heating from 25°C to 99°C at a rate of 1°C/min. GraphPad Prism 9.0.1 (GraphPad Software Inc.) was used to draw the temperature curve.
Bio-layer interferometry
Bio-layer interferometry (BLI) experiments were performed on an Octet Red 96 instrument (Fortebio) to measure the binding affinities of S309 NAb. S309 was immobilized onto Protein A biosensors (Fortebio). BA.1 RBD, BA.2 RBD, BA.2.12.1 RBD, BA.2.75 RBD and BA.4/5 RBD and in PBS used as analytes were diluted by threefold serial dilutions. The Octet BLI Analysis 9.1 (Fortebio) software was used to analyze the experimental data using a 1:1 fitted model.
Fab generation
Fab fragments were prepared using the Pierce Fab Preparation Kit (Thermo Scientific) as previously described (Wang et al., 2022a). Briefly, the sample first needs to be subjected to a desalting column to remove salts. Then, the effluent was collected and incubated with papain attached to the beads. The Fab fragments were cleaved from the antibody for 5 hours at 37 °C. The mixture was then transferred to a Protein A column to purify the Fab fragments (ThermoFisher, Catalog (Cat.) No.). 10010023)
MD stimulation and RMSF calculating
Initial models of BA.2 and BA.2.75 RBD were from the structure 7XIW downloading from PDB and the structure of apo S-Trimer of BA.2.75 at neutral pH determined by this study, respectively. Before final stimulation, we used CHARMM-GUI to generate the inputs for simulation packages GROMACS. After PDB checking, waterbox size specifying, water model specifying (TIP3P), ions adding, periodic boundary condition setting and force fielding specifying (OPLS-AA/M), the data generated was submitted to GROMACS-2021 to Energy Minimization, NVT Equilibration, NPT Equilibration and 10 ns MD simulation. NVT ensemb1e via the Nose-Hoover method at 300 K and NPT ensemble at 1 bar with the Parinello-Rahman algorithm were employed to make the temperature and the pressure equilibrated, respectively. The last 2 ns frames were extracted to calculated RMSF.
Surface plasmon resonance
Surface plasmon resonance (SPR) experiments were performed on the Biacore 8K (GE Healthcare). Immobilization of human ACE2 on a CM5 sensor chip and injection of purified Omicron and its variant RBDs with its corresponding single point mutation. The response units were recorded by Biacore 8K Evaluation Software (GE Healthcare) at room temperature, and the raw data curves were fitted to a 1:1 binding model using Biacore 8K Evaluation Software (GE Healthcare).
Cryo-EM sample preparation and data collection
3 μL of purified SARS-CoV-2 BA.2.75 variant Spike trimer (pH 7.4 and pH5.5) at 1.2 mg/mL and BA.2.75 S protein mixed with S309 Fab, XG2v024 Fab and human ACE2 at 1.0 mg/mL in purification buffer solution (20 mM Tris, 200 mM NaCl, pH 7.4) were dropped onto the pre-glow-discharged holey carbon-coated gold grid (C-flat, 300-mesh, 1.2/1.3, Protochips In.) Grids were blotted for 6 s in 100% relative humidity and room temperature for plunge-freezing (Vitrobot; FEI) in liquid ethane. Cryo-EM data sets were collected at a 300 kV Titan Krios microscope (Thermo Fisher) equipped with a K2 or K3 detector. Movies (32 frames, total dose 60 e− Å−2) were recorded with a defocus of from -1.5 to -2.7 μm using SerialEM, which yields a final pixel size of 1.07 Å or 1.04 Å.
Data processing and model refinement
A total of 6,459 micrographs of pH 7.4 BA.2.75 S protein, 4,886 micrographs of pH 5.5 BA.2.75 S protein, 5,761 micrographs of BA.2.75 S in complex with S309 Fab, 5,606 micrographs of BA.2.75 S in complex with XG2v024 and 7,225 micrographs of BA.2.75 S in complex with ACE2 were recorded. The CTF values were estimated by Patch CTF in cryoSPARC (v3.3.2). Particles were picked based on templates and extracted for 2D Classification. High-quality particles were selected and passed to Heterogeneous Refinement for 3D classification. Then, Homogeneous Refinement and Non-uniform Refinement were performed for high-resolution reconstruction. To obtain reliable interface density, Local Refinement was used to further improve the resolution of interfaces between BA.2.75 S and S309 Fab, XG2v024 Fab and ACE2. The atom models were generated by fitting the apo BA.2 S trimer (PDBID:7XIW, 7XIX), S309 Fab (PDBID:7TLY), ACE2 (6M0J), XG2v024 (PDBID:7CHS for heavy chain and 7N4L for light chain) into the cryo-EM density by Chimera. Then the models were adjusted manually in Coot and real-space refinement in Phenix.
Plasma isolation
Blood samples were obtained from SARS-CoV-2 vaccinee convalescent individuals who had been infected with Delta, BA.1, BA.2, and BA.5. Written informed consent was obtained from each participant in accordance with the Declaration of Helsinki. Whole blood samples were diluted 1:1 with PBS+2% FBS and then subjected to Ficoll (Cytiva, 17-1440-03) gradient centrifugation. After centrifugation, plasma was collected from the upper layer. Plasma samples were aliquoted and stored at −20 °C or less and were heat-inactivated before experiments.
Pseudovirus neutralization assay
SARS-CoV-2 variants Spike pseudovirus was prepared based on a vesicular stomatitis virus (VSV) pseudovirus packaging system. Variants’ spike plasmid is constructed into pcDNA3.1 vector. G∗ΔG-VSV virus (VSV G pseudotyped virus, Kerafast) and spike protein plasmid were transfected to 293T cells (American Type Culture Collection [ATCC], CRL-3216). After culture, the pseudovirus in the supernatant was harvested, filtered, aliquoted, and frozen at −80°C for further use.
Huh-7 cell line (Japanese Collection of Research Bioresources [JCRB], 0403) was used in pseudovirus neutralization assays. Plasma samples or antibodies were serially diluted in culture media and mixed with pseudovirus, and incubated for 1 h in a 37°C incubator with 5% CO2. Digested Huh-7 cells were seeded in the antibody-virus mixture. After one day of culture in the incubator, the supernatant was discarded. D-luciferin reagent (PerkinElmer, 6066769) was added into the plates and incubated in darkness for 2 min, and cell lysis was transferred to the detection plates. The luminescence value was detected with a microplate spectrophotometer (PerkinElmer, HH3400). IC50 was determined by a four-parameter logistic regression model.
Quantification and statistical analysis
Surface plasmon resonance assays were performed in three, and biolayer interferometry assays were performed in two biological replicates. SPR and BLI data were fitted to a 1:1 binding model by Biacore 8K Evaluation Software 3.0 and ForteBio Data Analysis Software 9.1, respectively. Neutralization assays were performed in at least two biological replicates. IC50 values were determined by a four-parameter logistic regression model. Wilcoxon signed-rank tests were used to determine the significance of differences in neutralizing activities (NT50 or IC50), and levels of significance were marked as: ∗, p < 0.05; ∗∗, p < 0.01; ∗∗∗, p < 0.001, as described in the corresponding figure legends (Figures 5 and 6).
Acknowledgments
We thank Dr. Yunjiao Zhou and Dr. Qiao Wang from Fudan University for providing XGv and XG2v antibody sequences and cloning XGv 1-50 constructs. This project is financially supported by the Ministry of Science and Technology of China and Changping Laboratory under the project number (CPL-1233 and CPL-2022A-03-03), Chinese Academy of Sciences (YSBR-010 and XDB29010000), Beijing Natural Science Foundation (Z210014) and National Key Research and Development Program (2020YFA0707500).
Declaration of interests
X.S.X. and Y.C. are inventors on the provisional patent applications of BD series antibodies, which includes BD30-604 (DXP-604), BD55-5840 (SA58), and BD55-5514 (SA55). X.S.X. and Y.C. are founders of Singlomics Biopharmaceuticals.
Published: October 4, 2022
Footnotes
Supplemental information can be found online at https://doi.org/10.1016/j.chom.2022.09.018.
Supplemental information
Data and code availability
-
•
The cryo-EM maps have been deposited at the Electron Microscopy Data Bank (www.ebi.ac.uk/emdb) and are available under accession numbers: EMDB: 34034 (BA.2.75 spike 1-up), EMDB: 34035 (BA.2.75 spike 3-down), EMDB: 34036 (BA.2.75 spike 1-up at pH 5.5), EMDB: 34037 (BA.2.75 spike 3-down at pH 5.5), EMDB: 34038 (BA.2.75-S309 complex state 1), EMDB: 34039 (BA.2.75-S309 complex state 2), EMDB: 34040 (BA.2.75-S309 complex state 3), EMDB: 34041 (BA.2.75-S309 complex interface), EMDB: 34042 (BA.2.75-XG2v024 complex), EMDB: 34043 (BA.2.75-ACE2 complex state 1), EMDB: 34044 (BA.2.75-ACE2 complex state 2), EMDB: 34045 (BA.2.75-ACE2 complex interface). Atomic models corresponding to them have been deposited in the Protein Data Bank (www.rcsb.org) and are available under accession numbers, PDB: 7YQT, 7YQU, 7YQV, 7YQW, 7YQX, 7YQY, 7YQZ, 7YR0, 7YR1, 7YR2, 7YR3, 7YR4, respectively.
-
•
Supplemental Tables are available from Mendeley Data at https://doi.org/10.17632/wyf5vvrzpy.1
-
•
This study did not generate custom computer code.
-
•
Any additional information required to reanalyze the data reported in this paper is available from the lead contact upon request.
References
- Adams P.D., Afonine P.V., Bunkóczi G., Chen V.B., Davis I.W., Echols N., Headd J.J., Hung L.W., Kapral G.J., Grosse-Kunstleve R.W., et al. Phenix: a comprehensive Python-based system for macromolecular structure solution. Acta Crystallogr. D Biol. Crystallogr. 2010;66:213–221. doi: 10.1107/S0907444909052925. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bruel T., Hadjadj J., Maes P., Planas D., Seve A., Staropoli I., Guivel-Benhassine F., Porrot F., Bolland W.H., Nguyen Y., et al. Serum neutralization of SARS-CoV-2 Omicron sublineages BA.1 and BA.2 in patients receiving monoclonal antibodies. Nat. Med. 2022;28:1297–1302. doi: 10.1038/s41591-022-01792-5. [DOI] [PubMed] [Google Scholar]
- Cao Y., Su B., Guo X., Sun W., Deng Y., Bao L., Zhu Q., Zhang X., Zheng Y., Geng C., et al. Potent neutralizing antibodies against SARS-CoV-2 identified by high-throughput single-cell sequencing of convalescent patients' B cells. Cell. 2020;182:73–84.e16. doi: 10.1016/j.cell.2020.05.025. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cao Y., Wang J., Jian F., Xiao T., Song W., Yisimayi A., Huang W., Li Q., Wang P., An R., et al. Omicron escapes the majority of existing SARS-CoV-2 neutralizing antibodies. Nature. 2022;602:657–663. doi: 10.1038/s41586-021-04385-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cao Y., Yisimayi A., Jian F., Song W., Xiao T., Wang L., Du S., Wang J., Li Q., Chen X., et al. BA.2.12.1, BA.4 and BA.5 escape antibodies elicited by Omicron infection. Nature. 2022;608:593–602. doi: 10.1038/s41586-022-04980-y. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cele S., Jackson L., Khoury D.S., Khan K., Moyo-Gwete T., Tegally H., San J.E., Cromer D., Scheepers C., Amoako D.G., et al. Omicron extensively but incompletely escapes Pfizer BNT162b2 neutralization. Nature. 2022;602:654–656. doi: 10.1038/s41586-021-04387-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cerutti G., Guo Y., Zhou T., Gorman J., Lee M., Rapp M., Reddem E.R., Yu J., Bahna F., Bimela J., et al. Potent SARS-CoV-2 neutralizing antibodies directed against spike N-terminal domain target a single Supersite. Cell Host Microbe. 2021;29:819–833.e7. doi: 10.1016/j.chom.2021.03.005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chen C., Nadeau S., Yared M., Voinov P., Xie N., Roemer C., Stadler T. CoV-spectrum: analysis of globally shared SARS-CoV-2 data to identify and characterize new variants. Bioinformatics. 2021;38:1735–1737. doi: 10.1093/bioinformatics/btab856. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Csardi G., Nepusz T. The igraph software package for complex network research. InterJournal Complex Syst. 2006;1695:1–9. [Google Scholar]
- Cui Z., Liu P., Wang N., Wang L., Fan K., Zhu Q., Wang K., Chen R., Feng R., Jia Z., et al. Structural and functional characterizations of infectivity and immune evasion of SARS-CoV-2 Omicron. Cell. 2022;185:860–871.e13. doi: 10.1016/j.cell.2022.01.019. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dejnirattisai W., Huo J., Zhou D., Zahradník J., Supasa P., Liu C., Duyvesteyn H.M.E., Ginn H.M., Mentzer A.J., Tuekprakhon A., et al. SARS-CoV-2 Omicron-B.1.1.529 leads to widespread escape from neutralizing antibody responses. Cell. 2022;185:467–484.e15. doi: 10.1016/j.cell.2021.12.046. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Du S., Cao Y., Zhu Q., Yu P., Qi F., Wang G., Du X., Bao L., Deng W., Zhu H., et al. Structurally resolved SARS-CoV-2 antibody shows high efficacy in severely infected hamsters and provides a potent cocktail pairing strategy. Cell. 2020;183:1013–1023.e13. doi: 10.1016/j.cell.2020.09.035. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Emsley P., Lohkamp B., Scott W.G., Cowtan K. Features and development of coot. Acta Crystallogr. D Biol. Crystallogr. 2010;66:486–501. doi: 10.1107/S0907444910007493. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ge J., Wang R., Ju B., Zhang Q., Sun J., Chen P., Zhang S., Tian Y., Shan S., Cheng L., et al. Antibody neutralization of SARS-CoV-2 through ACE2 receptor mimicry. Nat. Commun. 2021;12:250. doi: 10.1038/s41467-020-20501-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Goddard T.D., Huang C.C., Meng E.C., Pettersen E.F., Couch G.S., Morris J.H., Ferrin T.E. UCSF ChimeraX: meeting modern challenges in visualization and analysis. Protein Sci. 2018;27:14–25. doi: 10.1002/pro.3235. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Greaney A.J., Starr T.N., Gilchuk P., Zost S.J., Binshtein E., Loes A.N., Hilton S.K., Huddleston J., Eguia R., Crawford K.H.D., et al. Complete mapping of mutations to the SARS-CoV-2 spike receptor-binding domain that escape antibody recognition. Cell Host Microbe. 2021;29:44–57.e9. doi: 10.1016/j.chom.2020.11.007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hansen J., Baum A., Pascal K.E., Russo V., Giordano S., Wloga E., Fulton B.O., Yan Y., Koon K., Patel K., et al. Studies in humanized mice and convalescent humans yield a SARS-CoV-2 antibody cocktail. Science. 2020;369:1010–1014. doi: 10.1126/science.abd0827. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jones B.E., Brown-Augsburger P.L., Corbett K.S., Westendorf K., Davies J., Cujec T.P., Wiethoff C.M., Blackbourne J.L., Heinz B.A., Foster D., et al. The neutralizing antibody, LY-CoV555, protects against SARS-CoV-2 infection in nonhuman primates. Sci. Transl. Med. 2021;13:eabf1906. doi: 10.1126/scitranslmed.abf1906. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kimura I., Yamasoba D., Tamura T., Nao N., Suzuki T., Oda Y., Mitoma S., Ito J., Nasser H., Zahradnik J., et al. Virological characteristics of the SARS-CoV-2 Omicron BA.2 subvariants including BA.4 and BA.5. Cell. 2022 doi: 10.1016/j.cell.2022.09.018. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li D., Edwards R.J., Manne K., Martinez D.R., Schäfer A., Alam S.M., Wiehe K., Lu X., Parks R., Sutherland L.L., et al. In vitro and in vivo functions of SARS-CoV-2 infection-enhancing and neutralizing antibodies. Cell. 2021;184:4203–4219.e32. doi: 10.1016/j.cell.2021.06.021. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li L., Liao H., Meng Y., Li W., Han P., Liu K., Wang Q., Li D., Zhang Y., Wang L., et al. Structural basis of human ACE2 higher binding affinity to currently circulating Omicron SARS-CoV-2 sub-variants BA.2 and BA.1.1. Cell. 2022;185:2952–2960.e10. doi: 10.1016/j.cell.2022.06.023. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liu Y., Soh W.T., Kishikawa J.I., Hirose M., Nakayama E.E., Li S., Sasai M., Suzuki T., Tada A., Arakawa A., et al. An infectivity-enhancing site on the SARS-CoV-2 spike protein targeted by antibodies. Cell. 2021;184:3452–3466.e18. doi: 10.1016/j.cell.2021.05.032. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Luo S., Zhang J., Kreutzberger A.J.B., Eaton A., Edwards R.J., Jing C., Dai H.Q., Sempowski G.D., Cronin K., Parks R., et al. An antibody from single human VH-rearranging mouse neutralizes all SARS-CoV-2 variants Through BA.5 by Inhibiting Membrane Fusion. Sci. Immunol. 2022 doi: 10.1126/sciimmunol.add5446. eadd5446. [DOI] [PMC free article] [PubMed] [Google Scholar]
- McCallum M., De Marco A., Lempp F.A., Tortorici M.A., Pinto D., Walls A.C., Beltramello M., Chen A., Liu Z., Zatta F., et al. N-terminal domain antigenic mapping reveals a site of vulnerability for SARS-CoV-2. Cell. 2021;184:2332–2347.e16. doi: 10.1016/j.cell.2021.03.028. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Meng B., Abdullahi A., Ferreira I.A.T.M., Goonawardane N., Saito A., Kimura I., Yamasoba D., Gerber P.P., Fatihi S., Rathore S., et al. Altered TMPRSS2 usage by SARS-CoV-2 Omicron impacts infectivity and fusogenicity. Nature. 2022;603:706–714. doi: 10.1038/s41586-022-04474-x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nie J., Li Q., Wu J., Zhao C., Hao H., Liu H., Zhang L., Nie L., Qin H., Wang M., et al. Establishment and validation of a Pseudovirus neutralization assay for SARS-CoV-2. Emerg. Microbes Infect. 2020;9:680–686. doi: 10.1080/22221751.2020.1743767. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nutalai R., Zhou D., Tuekprakhon A., Ginn H.M., Supasa P., Liu C., Huo J., Mentzer A.J., Duyvesteyn H.M.E., Dijokaite-Guraliuc A., et al. Potent cross-reactive antibodies following Omicron breakthrough in vaccinees. Cell. 2022;185:2116–2131.e18. doi: 10.1016/j.cell.2022.05.014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Peacock T.P., Brown J.C., Zhou J., Thakur N., Sukhova K., Newman J., Kugathasan R., Yan A.W.C., Furnon W., De Lorenzo G., et al. The altered entry pathway and antigenic distance of the SARS-CoV-2 Omicron variant map to separate domains of spike protein. bioRxiv. 2022 doi: 10.1101/2021.12.31.474653. Preprint at. [DOI] [Google Scholar]
- Pettersen E.F., Goddard T.D., Huang C.C., Couch G.S., Greenblatt D.M., Meng E.C., Ferrin T.E. UCSF Chimera–a visualization system for exploratory research and analysis. J. Comput. Chem. 2004;25:1605–1612. doi: 10.1002/jcc.20084. [DOI] [PubMed] [Google Scholar]
- Pinto D., Park Y.J., Beltramello M., Walls A.C., Tortorici M.A., Bianchi S., Jaconi S., Culap K., Zatta F., De Marco A., et al. Cross-neutralization of SARS-CoV-2 by a human monoclonal SARS-CoV antibody. Nature. 2020;583:290–295. doi: 10.1038/s41586-020-2349-y. [DOI] [PubMed] [Google Scholar]
- Punjani A., Rubinstein J.L., Fleet D.J., Brubaker M.A. cryoSPARC: algorithms for rapid unsupervised cryo-EM structure determination. Nat. Methods. 2017;14:290–296. doi: 10.1038/nmeth.4169. [DOI] [PubMed] [Google Scholar]
- Qu P., Evans J.P., Zheng Y.M., Carlin C., Saif L.J., Oltz E.M., Xu K., Gumina R.J., Liu S.L. Evasion of neutralizing antibody response by the SARS-CoV-2 BA.2.75 Variant. bioRxiv. 2022 doi: 10.1101/2022.08.14.503921. Preprint at. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Quandt J., Muik A., Salisch N., Lui B.G., Lutz S., Krüger K., Wallisch A.K., Adams-Quack P., Bacher M., Finlayson A., et al. Omicron BA.1 breakthrough infection drives cross-variant neutralization and memory B cell formation against conserved epitopes. Sci. Immunol. 2022;7:eabq2427. doi: 10.1126/sciimmunol.abq2427. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rappazzo C.G., Tse L.V., Kaku C.I., Wrapp D., Sakharkar M., Huang D., Deveau L.M., Yockachonis T.J., Herbert A.S., Battles M.B., et al. Broad and potent activity against SARS-like viruses by an engineered human monoclonal antibody. Science. 2021;371:823–829. doi: 10.1126/science.abf4830. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Reynolds C.J., Pade C., Gibbons J.M., Otter A.D., Lin K.M., Muñoz Sandoval D., Pieper F.P., Butler D.K., Liu S., Joy G., et al. Immune boosting by B.1.1.529 (Omicron) depends on previous SARS-CoV-2 exposure. Science. 2022;377:eabq1841. doi: 10.1126/science.abq1841. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sheward D.J., Kim C., Fischbach J., Muschiol S., Ehling R.A., Björkström N.K., Karlsson Hedestam G.B., Reddy S.T., Albert J., Peacock T.P., Murrell B. Evasion of neutralizing antibodies by Omicron sublineage BA.2.75. bioRxiv. 2022 doi: 10.1101/2022.07.19.500716. Preprint at. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shi R., Shan C., Duan X., Chen Z., Liu P., Song J., Song T., Bi X., Han C., Wu L., et al. A human neutralizing antibody targets the receptor-binding site of SARS-CoV-2. Nature. 2020;584:120–124. doi: 10.1038/s41586-020-2381-y. [DOI] [PubMed] [Google Scholar]
- Starr T.N., Greaney A.J., Addetia A., Hannon W.W., Choudhary M.C., Dingens A.S., Li J.Z., Bloom J.D. Prospective mapping of viral mutations that escape antibodies used to treat COVID-19. Science. 2021;371:850–854. doi: 10.1126/science.abf9302. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Starr T.N., Greaney A.J., Hilton S.K., Ellis D., Crawford K.H.D., Dingens A.S., Navarro M.J., Bowen J.E., Tortorici M.A., Walls A.C., et al. Deep mutational scanning of SARS-CoV-2 receptor binding domain reveals constraints on folding and ACE2 binding. Cell. 2020;182:1295–1310.e20. doi: 10.1016/j.cell.2020.08.012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Starr T.N., Greaney A.J., Stewart C.M., Walls A.C., Hannon W.W., Veesler D., Bloom J.D. Deep mutational scans for ACE2 binding, RBD expression, and antibody escape in the SARS-CoV-2 Omicron BA.1 and BA.2 receptor-binding domains. bioRxiv. 2022 doi: 10.1101/2022.09.20.508745. Preprint at. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tegally H., Moir M., Everatt J., Giovanetti M., Scheepers C., Wilkinson E., Subramoney K., Makatini Z., Moyo S., Amoako D.G., et al. Emergence of SARS-CoV-2 Omicron lineages BA.4 and BA.5 in South Africa. Nat. Med. 2022;28:1785–1790. doi: 10.1038/s41591-022-01911-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tuekprakhon A., Nutalai R., Dijokaite-Guraliuc A., Zhou D., Ginn H.M., Selvaraj M., Liu C., Mentzer A.J., Supasa P., Duyvesteyn H.M.E., et al. Antibody escape of SARS-CoV-2 Omicron BA.4 and BA.5 from vaccine and BA.1 serum. Cell. 2022;185:2422–2433.e13. doi: 10.1016/j.cell.2022.06.005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang K., Jia Z., Bao L., Wang L., Cao L., Chi H., Hu Y., Li Q., Zhou Y., Jiang Y., et al. Memory B cell repertoire from triple vaccinees against diverse SARS-CoV-2 variants. Nature. 2022;603:919–925. doi: 10.1038/s41586-022-04466-x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang Q., Guo Y., Iketani S., Nair M.S., Li Z., Mohri H., Wang M., Yu J., Bowen A.D., Chang J.Y., et al. Antibody evasion by SARS-CoV-2 Omicron subvariants BA.2.12.1, BA.4 and BA.5. Nature. 2022;608:603–608. doi: 10.1038/s41586-022-05053-w. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang Q., Iketani S., Li Z., Guo Y., Yeh A.Y., Liu M., Yu J., Sheng Z., Huang Y., Liu L., Ho D.D. Antigenic characterization of the SARS-CoV-2 Omicron subvariant BA.2.75. bioRxiv. 2022 doi: 10.1101/2022.07.31.502235. Preprint at. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang R., Zhang Q., Ge J., Ren W., Zhang R., Lan J., Ju B., Su B., Yu F., Chen P., et al. Analysis of SARS-CoV-2 variant mutations reveals neutralization escape mechanisms and the ability to use ACE2 receptors from additional species. Immunity. 2021;54:1611–1621.e5. doi: 10.1016/j.immuni.2021.06.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang Z., Muecksch F., Cho A., Gaebler C., Hoffmann H.H., Ramos V., Zong S., Cipolla M., Johnson B., Schmidt F., et al. Analysis of memory B cells identifies conserved neutralizing epitopes on the N-terminal domain of variant SARS-Cov-2 spike proteins. Immunity. 2022;55:998–1012.e8. doi: 10.1016/j.immuni.2022.04.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Westendorf K., Žentelis S., Wang L., Foster D., Vaillancourt P., Wiggin M., Lovett E., van der Lee R., Hendle J., Pustilnik A., et al. LY-CoV1404 (bebtelovimab) potently neutralizes SARS-CoV-2 variants. Cell Rep. 2022;39:110812. doi: 10.1016/j.celrep.2022.110812. [DOI] [PMC free article] [PubMed] [Google Scholar]
- WHO Tracking SARS-CoV-2 variants. 2022. https://www.who.int/activities/tracking-SARS-CoV-2-variants
- Yamasoba D., Kimura I., Kosugi Y., Uriu K., Fujita S., Ito J., Sato K. Neutralization sensitivity of Omicron BA.2.75 to therapeutic monoclonal antibodies. bioRxiv. 2022 doi: 10.1101/2022.07.14.500041. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yang W., Shaman J. COVID-19 pandemic dynamics in India, the SARS-CoV-2 Delta variant and implications for vaccination. J. R. Soc. Interface. 2022;19:20210900. doi: 10.1098/rsif.2021.0900. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zahradník J., Marciano S., Shemesh M., Zoler E., Harari D., Chiaravalli J., Meyer B., Rudich Y., Li C., Marton I., et al. SARS-CoV-2 variant prediction and antiviral drug design are enabled by RBD in vitro evolution. Nat. Microbiol. 2021;6:1188–1198. doi: 10.1038/s41564-021-00954-4. [DOI] [PubMed] [Google Scholar]
- Zhang K. Gctf: real-time CTF determination and correction. J. Struct. Biol. 2016;193:1–12. doi: 10.1016/j.jsb.2015.11.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhou T., Tsybovsky Y., Gorman J., Rapp M., Cerutti G., Chuang G.Y., Katsamba P.S., Sampson J.M., Schön A., Bimela J., et al. Cryo-EM structures of SARS-CoV-2 spike without and with ACE2 reveal a pH-dependent switch to mediate endosomal positioning of receptor-binding domains. Cell Host Microbe. 2020;28:867–879.e5. doi: 10.1016/j.chom.2020.11.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zivanov J., Nakane T., Forsberg B.O., Kimanius D., Hagen W.J., Lindahl E., Scheres S.H. New tools for automated high-resolution cryo-EM structure determination in RELION-3. eLife. 2018;7:e42166. doi: 10.7554/eLife.42166. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zost S.J., Gilchuk P., Case J.B., Binshtein E., Chen R.E., Nkolola J.P., Schäfer A., Reidy J.X., Trivette A., Nargi R.S., et al. Potently neutralizing and protective human antibodies against SARS-CoV-2. Nature. 2020;584:443–449. doi: 10.1038/s41586-020-2548-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Data Availability Statement
-
•
The cryo-EM maps have been deposited at the Electron Microscopy Data Bank (www.ebi.ac.uk/emdb) and are available under accession numbers: EMDB: 34034 (BA.2.75 spike 1-up), EMDB: 34035 (BA.2.75 spike 3-down), EMDB: 34036 (BA.2.75 spike 1-up at pH 5.5), EMDB: 34037 (BA.2.75 spike 3-down at pH 5.5), EMDB: 34038 (BA.2.75-S309 complex state 1), EMDB: 34039 (BA.2.75-S309 complex state 2), EMDB: 34040 (BA.2.75-S309 complex state 3), EMDB: 34041 (BA.2.75-S309 complex interface), EMDB: 34042 (BA.2.75-XG2v024 complex), EMDB: 34043 (BA.2.75-ACE2 complex state 1), EMDB: 34044 (BA.2.75-ACE2 complex state 2), EMDB: 34045 (BA.2.75-ACE2 complex interface). Atomic models corresponding to them have been deposited in the Protein Data Bank (www.rcsb.org) and are available under accession numbers, PDB: 7YQT, 7YQU, 7YQV, 7YQW, 7YQX, 7YQY, 7YQZ, 7YR0, 7YR1, 7YR2, 7YR3, 7YR4, respectively.
-
•
Supplemental Tables are available from Mendeley Data at https://doi.org/10.17632/wyf5vvrzpy.1
-
•
This study did not generate custom computer code.
-
•
Any additional information required to reanalyze the data reported in this paper is available from the lead contact upon request.