Abstract
The genes (caaD1 and caaD2) encoding the trans-3-chloroacrylic acid dehalogenase (CaaD) of the 1,3-dichloropropene-utilizing bacterium Pseudomonas pavonaceae 170 were cloned and heterologously expressed in Escherichia coli and Pseudomonas sp. strain GJ1. CaaD is a protein of 50 kDa that is composed of α-subunits of 75 amino acid residues and β-subunits of 70 residues. It catalyzes the hydrolytic cleavage of the β-vinylic carbon-chlorine bond in trans-3-chloroacrylic acid with a turnover number of 6.4 s−1. On the basis of sequence similarity, oligomeric structure, and subunit size, CaaD appears to be related to 4-oxalocrotonate tautomerase (4-OT). This tautomerase consists of six identical subunits of 62 amino acid residues and catalyzes the isomerization of 2-oxo-4-hexene-1,6-dioate, via hydroxymuconate, to yield 2-oxo-3-hexene-1,6-dioate. In view of the oligomeric architecture of 4-OT, a trimer of homodimers, CaaD is postulated to be a hexameric protein that functions as a trimer of αβ-dimers. The sequence conservation between CaaD and 4-OT and site-directed mutagenesis experiments suggested that Pro-1 of the β-subunit and Arg-11 of the α-subunit are active-site residues in CaaD. Pro-1 could act as the proton acceptor/donor, and Arg-11 is probably involved in carboxylate binding. Based on these findings, a novel dehalogenation mechanism is proposed for the CaaD-catalyzed reaction which does not involve the formation of a covalent enzyme-substrate intermediate.
Isomer-specific 3-chloroacrylic acid dehalogenases catalyze the hydrolytic cleavage of the β-vinylic carbon-chlorine bond in either cis- or trans-3-chloroacrylic acid to yield malonic acid semialdehyde and HCl. These enzymes are produced by both gram-positive and gram-negative bacteria, including Pseudomonas pavonaceae 170 (27), Pseudomonas cepacia CAA1 (11), and the coryneform bacterial strains FG41 (47) and CAA2 (11), enabling these organisms to use one or both isomers of the xenobiotic compound 3-chloroacrylic acid for growth. The dehalogenases from strain FG41 were purified to homogeneity, and trans-3-chloroacrylic acid dehalogenase (CaaD) was found to be a 50-kDa enzyme composed of different subunits of 8.7 and 7.4 kDa, whereas the cis-3-chloroacrylic acid dehalogenase was an enzyme composed of two or three identical 16-kDa subunits (47). Although large fragments of these dehalogenating enzymes were sequenced, no significant sequence similarities with other protein sequences were found when the different databases were searched in 1992 (47).
Whereas most hydrolytic dehalogenase that are active with halogenated aliphatic compounds (so-called halidohydrolases), such as haloalkane dehalogenases (26, 30, 50), haloacetate dehalogenases (15–17), and 2-haloacid dehalogenases (21, 25, 31), are only able to displace halogens bound to sp3-hybridized carbon atoms, 3-chloroacrylic acid dehalogenases are unique in that they can cleave the much more stable vinylic carbon-halogen bond, in which the halogen is bound to an sp2-hybridized carbon atom. Cleavage of the latter can also occur with 4-chlorobenzoyl-coenzyme A dehalogenases, but in that case activation of the substrate (4-chlorobenzoate) to its coenzyme A derivative is needed (2, 5, 52). 3-Chloroacrylic acid dehalogenases are, to our knowledge, the only enzymes known to dehalogenate substrates with unactivated vinylic halogens.
Nothing is known about the catalytic mechanism of 3-chloroacrylic acid dehalogenases. To obtain insight on the structure, mechanism, and ancestry of these enzymes, we sequenced the genes encoding CaaD of P. pavonaceae 170 and characterized the expressed protein. The results indicate that the dehalogenase has both structural and mechanistic similarities to 4-oxalocrotonate tautomerase (4-OT), an enzyme involved in the bacterial catabolism of catechol to metabolites in the Krebs cycle.
MATERIALS AND METHODS
Bacterial strains, plasmids, and growth conditions.
The characteristics of P. pavonaceae 170, formerly known as Pseudomonas cichorii 170, have been given elsewhere (27, 48). Escherichia coli JM101 (53) and plasmid pBluescript SK− (Stratagene) were used for subcloning experiments. E. coli HB101(pRK600) (8) was the helper strain used for mobilizing pLAFR3-derived cosmids and pDSK519-derived plasmids in triparental matings with Pseudomonas sp. strain GJ1 (13). Cosmid pLAFR3 and plasmid pDSK519 are mobilizable broad-host-range vectors (18, 35). E. coli BL21(DE3) was used in combination with the T7 expression system (pET5a system; Promega) for overexpression of the dehalogenase and the mutant enzymes (40).
Cells for general cloning and expression were cultivated at 30°C in Luria-Bertani (LB) medium (33). When required, Difco agar (15 g/liter) was added to the medium. Antibiotics were added in the following amounts: ampicillin, 100 μg/ml; tetracycline, 12.5 μg/ml; chloramphenicol, 50 μg/ml; and kanamycin, 50 μg/ml.
General methods.
Techniques for restriction enzyme digestion, ligation, transformation, and other standard molecular biology manipulations were based on methods described by Sambrook et al. (33). Triparental matings were carried out as described elsewhere (14). DNA sequencing was performed at the BioMedical Technology Centre (Groningen, The Netherlands) using a Pharmacia ALF-Express automatic sequencing machine according to the instructions provided with the Amersham Thermo Sequenase cycle-sequencing kit. The base sequence was determined by analyzing fluorescent-dye-labeled nucleotide fragments.
Nucleotide sequence data were analyzed by using the programs supplied in the DNAStar software package (DNAStar Inc., Madison, Wis.). Searches for nucleotide and amino acid sequence similarities were done by using the Blast program (1) and the DDBJ/EMBL/GenBank databases. Amino acid sequences were aligned by using ClustalW (46). Protein was analyzed by sodium dodecyl sulfate-polyacrylamide gel electrophoresis (SDS-PAGE) under denaturing conditions on gels containing 15 to 20% polyacrylamide. The gels were stained with Coomassie brilliant blue. Protein concentrations were estimated with Coomassie brilliant blue by using bovine serum albumin as the standard. N-terminal amino acid sequencing was performed by Eurosequence BV (Groningen, The Netherlands) using chemical reagents, and a sequenator (model 477A) from Applied Biosystems (Warrington, England). The native molecular mass of the purified dehalogenase was determined by Superdex 75 gel filtration using a fast protein liquid chromatography system according to the instructions provided with the Superdex 75 column and the low-molecular-weight calibration kit of Pharmacia. Circular dichroism (CD) spectra were recorded on a AVIV CD spectrometer (model no. 62A DS).
Enzyme activities.
Dehalogenase assays were performed by incubating an appropriate amount of enzyme or cell extract with 3 ml of 5 mM substrate in 50 mM Tris-sulfate buffer (pH 8.2) at 30°C. Halide liberation was monitored colorimetrically as described previously (19). All dehalogenase activities are expressed as units per milligram; 1 U was defined as the amount of enzyme that catalyzes the production of 1 μmol of halide per min. Enzyme assays were carried out twice, and the differences in specific activities were less than 10%.
Single colonies on agar plates were screened for dehalogenase activity by monitoring halide production upon incubation with cis- or trans-3-chloroacrylic acid as described previously (28).
PCR.
PCR was carried out in a Progene DNA thermal cycler (New Brunswick Scientific Benelux B.V.). The amplification reaction mixtures (100 μl) contained standard Taq amplification buffer, 250 μM each of the four deoxyribonucleoside triphosphates, 100 ng of each primer, 100 ng of template DNA, and 2 U of Taq DNA polymerase. The cycling parameters were 94°C for 5 min, followed by 30 cycles of 94°C for 60 s, 58°C for 60 s, and 72°C for 90 s, with a final elongation step of 72°C for 10 min. The reaction mixtures were subjected to electrophoresis in 1% agarose gels, and PCR products were stained with ethidium bromide.
Construction of expression vectors.
The dehalogenase expression vector pET44T2 was made using the overlap extension PCR as described by Ho et al. (12). The external PCR primers were oligonucleotides 5′-CACGGCATATGCCGATGATCTCTTGCGAC-3′ (primer A) and 5′-TTGCCCAAGCAGAGGGATCCCCTAGCT-3′ (primer D). Primer A contains an NdeI restriction site (in bold) and anneals to the 5′ end of the caaD1 gene. Primer D contains a BamHI restriction site (in bold) and anneals to the complementary sequence directly downstream of the caaD2 gene. The internal PCR primers were oligonucleotides 5′-CATGTTATCTCCTTCATTACTTGAGTT-3′ (primer B) and 5′-TCAAGTAATGAAGGAGATAACATGCCCTTC-3′ (primer C). Primer C contains the desired mutations (underlined) that result in a new ribosome-binding site (identical to the one provided by plasmid pET5a) in front of the caaD2 gene. Primer B is the complementary primer. In two separate PCRs, the AB and CD fragments were generated using cosmid pPS41, which harbors the dehalogenase genes, as the template with primers A and B in one reaction and primers C and D in a second reaction. The PCR mixtures were subjected to electrophoresis in a 1% agarose gel, and the two PCR fragments were extracted seperately using the Qiaex II gel extraction kit. Subsequently, a second PCR was carried out on a mixture of the AB and CD fragments using primers A and D. The mutated DNA fragment was isolated from a 1% agarose gel. The restriction sites NdeI and BamHI were used to clone this DNA fragment into plasmid pET5a for overexpression of the dehalogenase under control of the T7 promoter. The newly constructed plasmid, pET44T2, was sequenced in order to verify the mutations in front of the caaD2 gene.
A dehalogenase expression vector for Pseudomonas sp. strain GJ1 was constructed by cloning the SalI fragment of cosmid pPS41 into the SalI-linearized broad-host-range vector pDSK519, resulting in pDSKcaaD. Upon introduction of this vector into Pseudomonas sp. strain GJ1, high-level expression of the dehalogenase gene under control of its own promoter was obtained.
Site-directed mutagenesis.
The CaaD mutants were constructed using the coding sequence for the dehalogenase in plasmid pET44T2 as the template. The αP1A, αR11A, and αR11K mutants were generated by PCR using the primers 5′-CACGGCATATGGCGATGATCTCTTGCGAC-3′, 5′-CACGGCATATGCCGATGATCTCTTGCGACATGCGCTATGGGGCCACAGACGAACAA-3′, and 5′-CACGGCCATATGCCGATGATCTCTTGCGACATGCGCTATGGGAAAACAGACGAACAA-3′, respectively. These primers anneal to the 5′ end of the wild-type coding sequence and were used in combination with primer D. They all contain an NdeI restriction site (in bold) and the codon for the desired mutation (in italics). The αF39A, αF39Y, and βP1A mutants were generated by overlap extension PCR. Primers A and D were used as the external PCR primers. For the αF39A mutant, the internal PCR primers were oligonucleotides 5′-GAGCCCCGCGAGAACATTGCCTTTGTGATT-3′ (mutated codon in italics) and 5′-AATGTTCTCGCGGGGCTC-3′ (primer E). For the αF39Y mutant, the internal PCR primers were oligonucleotide 5′-GAGCCCCGCGAGAACATTTACTTTGTGATT-3′ (mutated codon in italics) and primer E. For the βP1A mutant, the internal PCR primers were oligonucleotides 5′-TCAAGTAATGAAGGAGATAACATGGCCTTC-3′ (mutated codon in italics) and 5′-CATGTTATCTCCTTCATTACTTGAGTT-3′.
PCRs were carried out as described above, and PCR products were purified using the QIAquick PCR purification kit or the Qiaex II gel extraction kit. The restriction sites NdeI and BamHI that were introduced during the amplification reactions were used to clone the PCR products into plasmid pET5a for overexpression of the dehalogenase mutants. The cloned dehalogenase genes were sequenced in order to verify the mutations.
Preparation of crude extracts.
CaaD and the mutant enzymes were expressed in E. coli BL21(DE3) using the pET system. Fresh BL21(DE3) transformants containing the desired plasmid were collected from a plate by resuspending them in 1 ml of LB medium and used to inoculate 100 ml of LB-ampicillin medium to a starting optical density at 600 nm of 0.1. After overnight growth at 30°C, cells were harvested by centrifugation (10 min at 10,000 × g), washed with 1 volume of 50 mM Tris-sulfate buffer (pH 8.2), and disrupted at 4°C in an appropriate amount of this buffer by sonication (10 s per ml of suspension at a 70-W output in a Vibra cell sonicator). A crude extract was obtained by centrifugation (45 min at 16,000 × g).
Purification of the dehalogenase.
For isolation of CaaD of P. pavonaceae 170, a single colony of strain 170 was used to inoculate 100 ml of LB medium. After overnight growth at 30°C, the culture was used to inoculate 1 liter of LB medium. This culture was grown at 30°C until the early stationary-growth phase. Cells were harvested by centrifugation (10 min at 10,000 × g), washed with 1 volume of TEMAG buffer (10 mM Tris-SO4, 1 mM EDTA, 1 mM β-mercaptoethanol, 0.02% sodium azide, 10% glycerol [pH 8.0]), and stored at −20°C until further use. Preparation of a crude extract and purification of the dehalogenase were done as described below for the recombinant enzyme.
High-level CaaD expression was obtained in Pseudomonas sp. strain GJ1. A single colony of strain GJ1 containing the expression vector pDSKcaaD was used to inoculate 10 ml of LB-kanamycin medium. After overnight growth at 30°C, the culture was used to inoculate 1 liter of LB-kanamycin medium. This culture was also grown overnight at 30°C. Cells were harvested by centrifugation (10 min at 10,000 × g), washed with 1 volume of TEMAG buffer, and stored at −20°C.
CaaD was purified to homogeneity by a modification of a published procedure (47). In a typical experiment, cells of a 1-liter culture were thawed and suspended in 20 ml of TEMAG buffer. The cells were disrupted at 4 to 10°C by continuous sonication, after which unbroken cells and debris were removed by centrifugation for 1 h at 50,000 rpm in a type 70 Ti rotor (Beckman). The supernatant was applied to a DEAE-cellulose column which had previously been equilibrated with TEMAG buffer. The column was washed with 1 column volume of TEMAG buffer, and the proteins were eluted with a linear gradient of 0 to 0.5 M ammonium sulfate in TEMAG. Fractions that showed the highest dehalogenase activity with trans-3-chloroacrylic acid were pooled and dialyzed overnight against PEMAG buffer (5 mM potassium phosphate, 1 mM EDTA, 1 mM β-mercaptoethanol, 0.02% sodium azide, 10% glycerol [pH 6.5]). The dialysate was loaded onto a hydroxylapatite column which had previously been equilibrated with PEMAG buffer. The column was washed with 1 column volume of PEMAG buffer, and the proteins were eluted with a linear gradient of 5 to 100 mM potassium phosphate in PEMAG. Fractions with the highest CaaD activity were analyzed by SDS-PAGE, and those that contained purified enzyme were pooled and dialyzed against TEMAG buffer. The enzyme was stored at 4 or −20°C.
Nucleotide sequence accession number.
The nucleotide sequence of the dehalogenase gene region has been deposited in the GenBank database under accession number AJ290446.
RESULTS
Cloning and characterization of the genes encoding CaaD.
The trans-specific 3-chloroacrylic acid dehalogenase of P. pavonaceae 170 was partially (∼75%) purified. After SDS-PAGE (data not shown), two dominant protein bands at about 7.5 and 8.5 kDa were observed, indicating that the dehalogenase consisted of two different subunits. Both subunits were subjected to N-terminal sequence analysis. The sequence of the 8.5-kDa subunit (designated the α-subunit) was established as P-M-I-S-C-D-M-R-Y-G-R-T-D-E-Q-K, and that of the 7.5-kDa subunit (designated the β-subunit) was P-F-I-E-C-H-I-A-T-G-L-S-V-A-R-K-Q-Q-L-I-R-D.
To isolate the genes encoding CaaD, individual clones of a previously constructed cosmid library of P. pavonaceae 170 (29) were screened for dehalogenase activity by monitoring halide production upon incubation with trans-3-chloroacrylic acid. Out of 2,500 E. coli HB101 clones tested, 1 clone expressing CaaD was found. The recombinant cosmid (pPS41) encoding the dehalogenase was isolated, and the localization of the dehalogenase genes was determined by screening subclones for dehalogenase activity.
Nucleotide sequencing of a 2.6-kb SalI subclone of pPS41 revealed several open reading frames. The two open reading frames encoding CaaD (designated caaD1 and caaD2) were identified by using the N-terminal amino acid sequences determined for both subunits of the enzyme isolated from P. pavonaceae 170. These N-terminal amino acid sequences are identical to those predicted from the DNA sequence if the initiating N-formylmethionines are removed during posttranslational processing. The caaD1 gene encodes a protein (the α-subunit) of 76 amino acids with a calculated molecular mass of 8.47 kDa. The caaD2 gene was found downstream of caaD1 and encodes a protein (the β-subunit) of 71 amino acids (7.64 kDa). The calculated masses of both subunits are in agreement with their masses estimated from SDS-PAGE.
Expression and characterization of the dehalogenase.
E. coli HB101 harboring recombinant cosmid pPS41 displayed poor expression of CaaD (Table 1). Introduction of pPS41 into Pseudomonas sp. strain GJ1 resulted in significantly better expression (Table 1). To obtain overexpression of the dehalogenase in strain GJ1, we cloned the SalI fragment that harbors the dehalogenase genes into the Pseudomonas high-copy-number vector pDSK519, resulting in pDSKcaaD. The coding sequence for the dehalogenase in pDSKcaaD is under control of its own promoter, and the dehalogenase was expressed constitutively in strain GJ1 in a soluble and active form up to a concentration equivalent to 13% of the total soluble cellular protein (Table 1). The dehalogenase was isolated from strain GJ1(pDSKcaaD) with a yield of 20 mg of pure protein, as judged by SDS-PAGE, from a 1-liter culture. The purified enzyme could be stored for several months in TEMAG buffer at 4 or −20°C without significant loss of activity.
TABLE 1.
Expression of CaaD in E. coli and Pseudomonas spp.a
| Organism | Dehalogenase sp act (mU/mg of protein) |
|---|---|
| E. coli strains | |
| HB101(pPS41) | 30 |
| BL21(DE3)(pET44T2) | 1,500 |
| Pseudomonas strains | |
| P. pavonaceae 170 | 310 |
| GJ1(pPS41) | 400 |
| GJ1(pDSKcaaD) | 2,380 |
Specific activities for trans-3-chloroacrylic acid (5 mM) were determined with crude extracts prepared from cells grown in LB broth. The specific activity measured in crude extract from strain GJ1(pDSKcaaD) corresponds to a dehalogenase concentration equivalent to 13% of the total soluble cellular protein, based on a specific activity of 18,000 mU/mg for the purified enzyme.
The native molecular mass of the dehalogenase was estimated by gel filtration chromatography to be 50 kDa. A comparison of this value to the subunit molecular masses (8.47 and 7.64 kDa) determined from the primary amino acid sequences suggests that the native dehalogenase is a hexameric protein, probably consisting of three α and three β subunits (α3β3).
The dehalogenase catalyzed the liberation of halide from trans-3-chloroacrylic acid (specific activity, 18 U/mg of protein) and trans-3-bromoacrylic acid (48 U/mg), but showed no detectable dehalogenase activity (<0.005 U/mg) toward cis-3-chloroacrylic acid, indicating that the enzyme is completely isomer selective. The saturated analog 3-chloropropionic acid was also not dehalogenated, indicating that the halogen atom is only removed when attached to an unsaturated carbon atom. Since no activity was found for 2-bromoacrylic acid and 2-chloroacrylic acid, it is essential that the halogen substituent is located at the β-position. The enzyme was also not active with trans-3-chloroallyl alcohol and trans-1,3-dichloropropene, indicating the importance of the presence of a carboxyl group in the substrate.
The dehalogenase showed a broad pH optimum around 8.5, and the temperature optimum was 40°C. By measuring the initial velocities of product formation at different trans-3-chloroacrylic acid concentrations, a Km of 0.19 mM and kcat of 6.4 s−1 were found.
Sequence similarity with 4-oxalocrotonate tautomerases/ isomerases.
Database searches identified seven related proteins as having significant sequence similarity with CaaD (Table 2). Two are well-studied enzymes involved in the bacterial catabolism of catechol to metabolites in the Krebs cycle, the 4-OT from Pseudomonas putida mt-2 (39) and the 73% identical isozyme from Pseudomonas sp. strain CF600 (41). Both are hexameric proteins that consist of identical subunits of 62 amino acid residues (4) and catalyze the isomerization of 2-oxo-4-hexene-1,6-dioate, via hydroxymuconate, to yield 2-oxo-3-hexene-1,6-dioate (51). The other five proteins that were retrieved from the similarity search have not been studied, but based on sequence similarity to 4-OT, they might be classified as putative 4-oxalocrotonate tautomerases/isomerases. Pairwise identities among the seven identified 4-OT homologues range from 35 to 92%.
TABLE 2.
4-OT sequences similar to CaaD
| Gene | Protein length (residues) | Organism | %
Identitya to:
|
Accession no. | |
|---|---|---|---|---|---|
| α-Subunit | β-Subunit | ||||
| xylH | 62 | Pseudomonas putida mt-2 | 26 | 25 | Q01468 |
| dmpI | 62 | Pseudomonas sp. strain CF600 | 26 | 22 | P49172 |
| ywhB | 61 | Bacillus subtilis 168 | 35 | 16 | CAB02512 |
| nahJ | 62 | Pseudomonas stutzeri AN10 | 26 | 24 | AAD02155 |
| xylH | 79 | Sphingomonas aromaticivorans F199 | 25 | 22 | AAD03991 |
| phnL | 76 | Pseudomonas sp. strain DJ77 | 23 | 24 | AAD03836 |
| nahJ | 62 | Pseudomonas putida G7 | 26 | 22 | AAD13221 |
Pairwise identities between the α- or β-subunit of the dehalogenase and the 4-OT sequences were calculated by using ClustalW.
Alignments optimizing identity between the amino acid sequences of the two dehalogenase subunits and the seven 4-OT homologues are shown in Fig. 1. Pairwise identities between the α-subunit (CaaD1) of the dehalogenase and the 4-OT sequences fall between 23 and 35% (Table 2), with the highest sequence similarity in the N-terminal region, particularly in the region boxed in Fig. 1A. Pairwise identities between the β-subunit (CaaD2) and the 4-OT sequences are lower and range from 16 to 25% (Table 2).
FIG. 1.
Alignments of the amino acid sequences of the α-subunit (CaaD1) and β-subunit (CaaD2) of CaaD with the seven 4-OT sequences (A and B) and with the amino-terminal sequences of the subunits of the CaaD isolated from strain FG41 (C). Residues conserved throughout all sequences are indicated by an asterisk. Dashes represent residues absent in other sequences. The catalytically important residues in 4-OT are marked with + and shown in boldface. The region of highest sequence identity among CaaD1 and the 4-OT sequences is boxed. Ppa, P. pavonaceae; Bs, B. subtilis; Psp, Pseudomonas sp.; Pp, P. putida; Ps, P. stutzeri; Sa, S. aromaticivorans; Csp, coryneform bacterial strain.
The sequence alignment in Fig. 1 suggest catalytic residues for CaaD that are in the same position in the alignment as the identified catalytic residues of 4-OT. Affinity labeling (36), kinetic analysis (37), chemical synthesis (9), nuclear magnetic resonance (38, 39), site-directed mutagenesis (6), and crystallographic studies (41) identified the amino-terminal proline as the catalytic base in the 4-OT-catalyzed tautomerization reaction. This N-terminal proline is invariant among all identified 4-OT homologues (Fig. 1). Both dehalogenase subunits also possess an N-terminal proline, indicating that one of these prolines may serve as the catalytic base in the CaaD-catalyzed dehalogenation reaction. The second catalytic residue of 4-OT, Arg-11, which is absolutely conserved among the 4-OT homologues (Fig. 1), was proposed to interact with the 6-carboxylate of the substrate (2-oxo-4-hexene-1,6-dioate) to facilitate both substrate binding and catalysis (10, 41). The sequence similarity indicates that Arg-11 is present only in the α-subunit of the dehalogenase and may perform an analogous role by interacting with the carboxylate group of trans-3-chloroacrylic acid. The third catalytic residue of 4-OT, Arg-39, which was proposed to interact with the 1-carboxylate and the 2-keto group of the substrate to promote carbonyl polarization and catalysis (10, 41), is not conserved in the dehalogenase sequence. This seems plausible since the dehalogenase substrate contains only one carboxylate group.
Characterization of dehalogenase mutants.
In the homohexameric (α6) 4-OT molecule, Pro-1 is important for tautomerase activity because it serves as the catalytic base. To test if both Pro-1 in the α-subunit and Pro-1 in the β-subunit of the heterohexameric (α3β3) CaaD molecule are important for dehalogenase activity, each proline was replaced by an alanine. The wild-type and mutant enzymes were expressed in E. coli BL21(DE3), and their specific activities with trans-3-chloroacrylic acid and trans-3-bromoacrylic acid were measured in cell extracts. Mutation of Pro-1 to alanine in the β-subunit essentially abolished catalytic activity of the dehalogenase, whereas mutation of Pro-1 to alanine in the α-subunit had no significant influence on activity (Table 3). To determine whether the loss of catalytic activity in the βP1A mutant is due to the specific alteration of the catalytic residue or to the loss of native-like protein structure, this mutant was purified to homogeneity and analyzed by gel filtration chromatography and CD. The purified βP1A mutant showed an activity of 10 mU/mg of protein with trans-3-chloroacrylic acid, which is 1,800-fold lower than the activity of the wild-type enzyme. The CD spectrum of this mutant was nearly identical to that recorded for the wild type, indicating that the mutation did not result in any gross conformational change (data not shown). It was further shown by gel filtration chromatography that the native molecular mass of mutant βP1A is similar to that of the wild type, indicating that the hexameric association was still intact. Hence, the decrease in activity found with the βP1A mutant is probably due to a direct effect of the substitution of the catalytically important proline for alanine. Taken together, the results indicate that of the two N-terminal prolines in the dehalogenase, only the N-terminal proline in the β-subunit is important for the dehalogenase-catalyzed reaction.
TABLE 3.
Activities of wild-type and mutant CaaD for trans-3-chloroacrylic acid (CAA) and trans-3-bromoacrylic acid (BAA)
| Enzyme | Dehalogenase
sp acta (mU/mg of protein)
|
|
|---|---|---|
| CAA | BAA | |
| Wild type | 1,500 | 3,250 |
| αP1A | 1,260 | 3,050 |
| βP1A | <5 | 15 |
| αR11A | <5 | <5 |
| αR11K | 110 | 240 |
| αF39A | 350 | 480 |
| αF39Y | 240 | 300 |
Activities were measured in cell extracts containing similar amounts of dehalogenase. Halide production with 5 mM substrate was determined at 30°C and pH 8.2.
Although CaaD has a low overall identity to the known 4-OTs, the region around Arg-11 in the α-subunit is highly conserved (Fig. 1A). In 4-OT, this region is related to binding of the carboxylate group of a substrate molecule that interacts with Pro-1 of another subunit (see above). The dehalogenase substrate also contains a carboxylate group, probably explaining the presence of Arg-11 in CaaD. To probe the role of α Arg-11 in the catalytic activity of CaaD, this residue was mutated to alanine or lysine. Mutation of Arg-11 to alanine resulted in an inactive enzyme, indicating that Arg-11 is essential for dehalogenase activity (Table 3). The mutant αR11K had partially restored catalytic activity compared to the catalytically inactive αR11A mutant, suggesting that a positive charge at this position is important for carboxylate binding.
In many reactions involving carbon-halogen bond cleavage, the carbon-halogen bond is weakened by functional groups that interact with the halogen substituent (26, 32, 49, 50). In the α-subunit of CaaD, Phe-39 is in the same position in the alignment as the catalytically important Arg-39 of 4-OT, suggesting that Phe-39 may be one of the residues that promote carbon-halogen bond cleavage by interacting with the chlorine atom of the dehalogenase substrate. To test if Phe-39 is catalytically important, this residue was mutated to alanine and tyrosine. The αF39A and αF39Y mutants were still able to catalyze halide release from both dehalogenase substrates, although 5- to 10-fold slower than the wild-type enzyme (Table 3), indicating that Phe-39 is not essential for dehalogenase activity.
DISCUSSION
The enzyme CaaD is produced by the soil bacterium P. pavonaceae 170 as part of a degradative pathway for the xenobiotic nematocide trans-1,3-dichloropropene (27). This hydrolytic dehalogenase, of which the properties are reported in this work, has no sequence similarity with other halidohydrolases but appears to be related to the family of 4-OTs. No other bacterial 3-chloroacrylic acid dehalogenase genes have been cloned, but the N-terminal sequences of the α and β subunits of the CaaD isolated from the gram-positive coryneform bacterial strain FG41 (47) have extensive similarity with the N-terminal parts of the α and β subunits of CaaD, respectively (Fig. 1C), suggesting that these two proteins have a common evolutionary origin and are mechanistically similar. As might be expected, the two CaaD sequences are more related to each other than to the 4-OTs.
The primary amino acid sequence of 4-OT shows no apparent similarity with those of the mammalian enzymes d-dopachrome tautomerase (DDT) (42) and macrophage migration inhibitory factor (MIF) (43), nor with that of the bacterial enzyme 5-carboxymethyl-2-hydroxymuconate isomerase (CHMI) (41), but remarkably, these four proteins have a common structural architecture (24, 42). DDT, MIF, and CHMI have an almost identical subunit topology, with two βαβ motifs related by pseudo-twofold symmetry and trimeric β-sheet packing (24, 41, 42). While CHMI, MIF, and DDT are functional as homotrimers, 4-OT is a hexamer of identical monomers. The 4-OT subunit is composed of only 62 residues and is dimerized by twofold symmetry to form a structure similar to that of the CHMI, MIF, and DDT monomer. Therefore, 4-OT is a trimer of homodimers that shows 32 symmetry; its overall hexameric structure is very similar to the trimeric structure of CHMI, MIF, and DDT (41–43). An interesting difference between the four structures is that, because of the higher symmetry of the 4-OT hexamer, there are potentially six active sites in 4-OT, yet only three are conserved in CHMI, MIF, and DDT (41, 42).
One of the characteristics of this superfamily of 4-OT-related proteins is that its members possess an amino-terminal proline that is located at the bottom of a hydrophobic pocket. CHMI and 4-OT utilize this proline as a catalytic base in their isomerization reactions (6, 9, 36–39, 41). Pro-1 of MIF is required for its d-dopachrome tautomerase and phenylpyruvate tautomerase activities (22, 34). The N-terminal proline of DDT is proposed to serve as the catalytic base in the DDT-catalyzed tautomerization reaction (42). The Pro-1 residue is conserved among all known homologues of 4-OT (Fig. 1), MIF (22, 44), and DDT (42) and is also conserved in both subunits of CaaD (Fig. 1). In the CaaD isolated from strain FG41, however, the amino-terminal proline is present only in the subunit that aligns with the β-subunit of CaaD (Fig. 1C). This suggests that Pro-1 of the β-subunit may serve as a catalytic base in both of the trans-3-chloroacrylic acid-dehalogenating enzymes. Site-directed mutagenesis experiments in which the amino-terminal prolines in CaaD were replaced by alanines indeed demonstrated that Pro-1 of the β-subunit is catalytically important, whereas Pro-1 of the α-subunit does not seem to play a role in catalysis.
On the basis of its sequence similarity to 4-OT, we conclude that CaaD also belongs to the superfamily of 4-OT-related proteins (24). The relatedness between CaaD1 and 4-OT is most apparent from the presence of a short stretch of sequence, GR(T,S)DEQK, that they have in common (Fig. 1A). This sequence motif, which includes the catalytically important Arg-11 in both 4-OT and CaaD, is not conserved in MIF and DDT but is present as GRSIEsr (lowercase letters indicate nonconserved positions) around the equivalent, catalytically important Arg-71 residue in CHMI. This functional motif is related to binding of the carboxylate group of the 4-OT, CHMI, and CaaD substrates.
In view of the oligomeric architecture of 4-OT, a trimer of homodimers (41), CaaD is postulated to function as a trimer of αβ-dimers. In contrast to the presence of potentially six active sites in the highly symmetrical 4-OT molecule (45), there are three potential active sites in CaaD. From the crystal structure of 4-OT inactivated by 2-oxo-3-pentynoate (45), the substrate should interact with Pro-1 of one subunit and Arg-11 from an adjacent subunit within the same homodimer. Consistent with this, the catalytically important Pro-1 of CaaD is located on the β-subunit, whereas Arg-11 is located on the α-subunit, showing that residues from both subunits contribute to the dehalogenase active site.
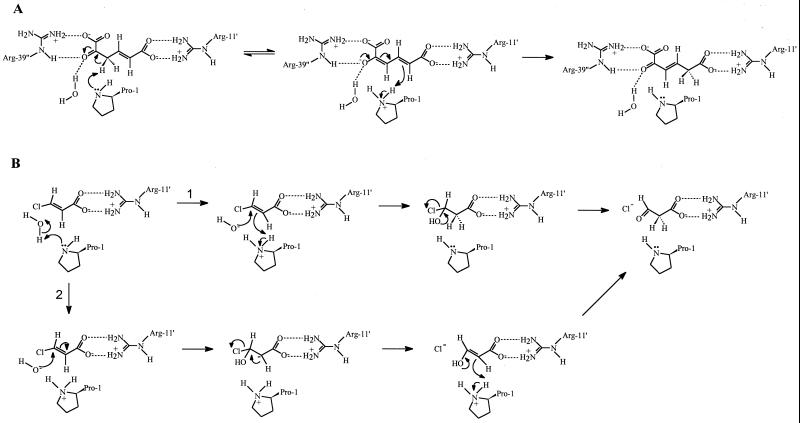
In 4-OT, Arg-11 interacts with the 6-carboxylate of the substrate (2-oxo-4-hexene-1,6-dioate) to facilitate substrate binding and catalysis, and Pro-1 transfers protons from C-3 to C-5 (Fig. 2A). We propose similar roles for βPro-1 and αArg-11 in CaaD, which leads to a minimal catalytic mechanism for the CaaD-catalyzed hydrolytic dehalogenation of trans-3-chloroacrylic acid (Fig. 2B). In principle, one can write several possible chemical pathways for the CaaD-catalyzed reaction, including a nucleophilic substitution and an addition-elimination mechanism. Although a one-step displacement of the halide by a hydroxyl ion has been suggested for the hydrolytic dehalogenation of d- and l-2-haloalkanoic acids by haloacid dehalogenase (DL-DEX 113) from Pseudomonas sp. strain 113 (25), it seems unlikely with the halide bound to an sp2 -hybridized carbon atom as in trans-3-chloroacrylic acid. Therefore, we propose that, in parallel to the hydration of monofluorofumarate by fumarase (23), CaaD catalyzes nucleophilic addition of a hydroxyl group on an sp2-hybridized carbon atom. This nucleophilic addition is favored over an electrophilic reaction because of the presence of the electron-withdrawing halogen and the carboxyl group. In this scenario, Pro-1 serves as the catalytic base that activates a water molecule to attack the β-carbon atom of the substrate. This leads to two hypothetical pathways, one involving the formation of the intermediate 3-chloro-3-hydroxypropanoic acid (route 1 in Fig. 2B) and the other involving the formation of a carbanion intermediate (route 2 in Fig. 2B). Product formation from either intermediate involves redirection of electrons, with departure of Cl− and protonation at C-2. The latter suggests the presence of an acidic residue, which may be the protonated proline or another amino acid. Loss of a proton from the hydroxyl at C-3 would produce the malonic acid semialdehyde, which may be facilitated by water or another proton acceptor. Hydration of monofluorofumarate by fumarase also yielded an unstable intermediate, α-fluorohydrin (α-fluoromalate), which subsequently decomposes to oxaloacetate and HF (23).
FIG. 2.
Comparative reaction scheme for 4-OT and CaaD. The primed residues come from other subunits. (A) Reaction catalyzed by 4-OT (adopted from references 10 and 41). (B) Two proposed reaction schemes for CaaD, one involving the formation of 3-chloro-3-hydroxypropanoic acid (route 1) and the other involving the formation of a carbanion intermediate (route 2). Pro-1 is shown as the catalytic base to activate a water molecule that attacks the substrate. Arg-11 is implicated in substrate binding.
Because CaaD catalyzes a dehalogenation reaction, it is anticipated that functional, groups involved in halogen/halide binding are required in addition to Pro-1 and Arg-11. We speculate that Phe-39 in the α-subunit of CaaD, which is in the same position in the alignment as Arg-39 in 4-OT (Fig. 1), may interact with the chlorine atom of the substrate to promote carbon-halogen bond cleavage. Indeed, aromatic ring systems are known to be partially positively charged in the plane of the ring (3). Phenylalanine residues were also proposed to contribute to halogen/halide binding in haloalkane dehalogenase (DhlA) and l-2-haloacid dehalogenase (DhlB) from Xanthobacter autotrophicus GJ10 (7, 32). However, the F39A and F39Y mutants of CaaD still had some residual activity, indicating that this residue is not essential. The presence of other functional groups interacting with the halogen atom of the substrate could explain why these mutants retained some activity. Indeed, in DhlA and DhlB, the halogen/halide-binding site is formed by more than one residue (20, 32, 49).
Screening of the cosmid library of P. pavonaceae 170 did not reveal clones that expressed the cis-3-chloroacrylic acid dehalogenase. Thus far, the only sequence information available for cis-specific 3-chloroacrylic acid dehalogenases is the N-terminal sequence of the enzyme isolated from the coryneform bacterial strain FG41 (47). This enzyme is probably a trimeric protein of 16.2-kDa subunits, and a comparison of its amino-terminal sequence with those of the CaaDs from strains FG41 and 170 revealed no overall similarity but showed that Pro-1 and Arg-11 are conserved (data not shown). Therefore, both cis- and trans-specific 3-chloroacrylic acid dehalogenases may catalyze the dehalogenation of their respective 3-chloroacrylic acid isomers through the mechanism shown in Fig. 2. This mechanism is different from that of most other hydrolytic dehalogenases in that it does not involve the formation a covalent enzyme-substrate intermediate.
ACKNOWLEDGMENTS
This study was supported by the Life Sciences Foundation (SLW), which is subsidized by the Netherlands Organization for Scientific Research (NWO), and by EC Environmental and Climate Research Program contract ENV4-CT95-0086.
We thank J. Tijmes for assistance with gel filtration chromatography and P. Terpstra (BioMedical Technology Centre, University of Groningen, Groningen, The Netherlands) for assistance with DNA sequencing.
REFERENCES
- 1.Altschul S F, Madden T L, Schäffer A A, Zhang J, Zhang Z, Miller W, Lipman D J. Gapped BLAST and PSI-BLAST: a new generation of protein database search programs. Nucleic Acids Res. 1997;25:3389–3402. doi: 10.1093/nar/25.17.3389. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Benning M M, Taylor K L, Liu R-Q, Yang G, Xiang H, Wesenberg G, Dunaway-Mariano D, Holden H. Structure of 4-chlorobenzoyl coenzyme A dehalogenase determined to 1.8 Åresolution: an enzyme catalyst generated via adaptive mutation. Biochemistry. 1996;35:8103–8109. doi: 10.1021/bi960768p. [DOI] [PubMed] [Google Scholar]
- 3.Burley S K, Petsko G A. Weakly polar interactions in proteins. Adv Protein Chem. 1988;39:125–192. doi: 10.1016/s0065-3233(08)60376-9. [DOI] [PubMed] [Google Scholar]
- 4.Chen L H, Kenyon G L, Curtin F, Harayama S, Bembenek M E, Hajipour G, Whitman C P. 4-Oxalocrotonate tautomerase, an enzyme composed of 62 amino acid residues per monomer. J Biol Chem. 1992;267:17716–17721. [PubMed] [Google Scholar]
- 5.Crooks G P, Copley S D. A surprising effect of leaving group on the nucleophilic aromatic substitution reaction catalyzed by 4-chlorobenzoyl CoA dehalogenase. J Am Chem Soc. 1993;115:6422–6423. [Google Scholar]
- 6.Czerwinski R M, Johnson W H, Jr, Whitman C P, Harris T K, Abeygunawardana C, Mildvan A S. Kinetic and structural effects of mutations of the catalytic amino-terminal proline in 4-oxalocrotonate tautomerase. Biochemistry. 1997;36:14551–14560. doi: 10.1021/bi971545h. [DOI] [PubMed] [Google Scholar]
- 7.Damborský J, Kutý M, Nemec M, Koca J. A molecular modeling study of the catalytic mechanism of haloalkane dehalogenase. I. Quantum chemical study of the first reaction step. J Chem Inf Comput Sci. 1997;37:562–568. [Google Scholar]
- 8.Finan T M, Kunkel B, De Vos G F, Signer E R. Second symbiotic megaplasmid in Rhizobium meliloticarrying exopolysaccharide and thiamine synthesis genes. J Bacteriol. 1986;167:66–72. doi: 10.1128/jb.167.1.66-72.1986. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Fitzgerald M C, Chernushevich I, Standing K G, Whitman C P, Kent S B H. Probing the oligomeric structure of an enzyme by electrospray ionization time-of-flight mass spectrometry. Proc Natl Acad Sci USA. 1996;93:6851–6856. doi: 10.1073/pnas.93.14.6851. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Harris T K, Czerwinski R M, Johnson W H, Jr, Legler P M, Abeygunawardana C, Massiah M A, Stivers J T, Whitman C P, Mildvan A S. Kinetic, stereochemical, and structural effects of mutations of the active site arginine residues in 4-oxalocrotonate tautomerase. Biochemistry. 1999;38:12343–12357. doi: 10.1021/bi991116e. [DOI] [PubMed] [Google Scholar]
- 11.Hartmans S, Jansen M W, van der Werf M J, De Bont J A M. Bacterial metabolism of 3-chloroacrylic acid. J Gen Microbiol. 1991;137:2025–2032. doi: 10.1099/00221287-137-8-2025. [DOI] [PubMed] [Google Scholar]
- 12.Ho S N, Hunt H D, Horton R M, Pullen J K, Pease L R. Site-directed mutagenesis by overlap extension using the polymerase chain reaction. Gene. 1989;77:51–59. doi: 10.1016/0378-1119(89)90358-2. [DOI] [PubMed] [Google Scholar]
- 13.Janssen D B, Scheper A, Witholt B. Biodegradation of 2-chloroethanol and 1,2-dichloroethane by pure bacterial cultures. Progr Ind Microbiol. 1984;20:169–178. [Google Scholar]
- 14.Janssen D B, Pries F, van der Ploeg J, Kazemier B, Terpstra P, Witholt B. Cloning of 1,2-dichloroethane degradation genes of Xanthobacter autotrophicus GJ10 and expression and sequencing of the dhlAgene. J Bacteriol. 1989;171:6791–6799. doi: 10.1128/jb.171.12.6791-6799.1989. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Kawasaki H, Miyoshi K, Tonomura K. Purification, crystallization and properties of haloacetate halidohydrolase from Pseudomonasspecies. Agric Biol Chem. 1981;45:543–544. [Google Scholar]
- 16.Kawasaki H, Tone N, Tonomura K. Purification and properties of haloacetate halidohydrolase specified by plasmid from Moraxellasp. strain B. Agric Biol Chem. 1981;45:35–42. [Google Scholar]
- 17.Kawasaki H, Hayashi S, Yahara H, Minami F, Tonomura K. Plasmid pUO2 determining haloacetate dehalogenase and mercury resistance in Pseudomonassp. J Ferment Technol. 1982;60:5–11. [Google Scholar]
- 18.Keen N T, Tamaki S, Kobayashi D, Trollinger D. Improved broad-host-range plasmids for DNA cloning in Gram-negative bacteria. Gene. 1988;70:191–197. doi: 10.1016/0378-1119(88)90117-5. [DOI] [PubMed] [Google Scholar]
- 19.Keuning S, Janssen D B, Witholt B. Purification and characterization of hydrolytic haloalkane dehalogenase from Xanthobacter autotrophicusGJ10. J Bacteriol. 1985;163:635–639. doi: 10.1128/jb.163.2.635-639.1985. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Krooshof G H, Ridder I S, Tepper A W J W, Vos G J, Rozeboom H J, Kalk K H, Dijkstra B W, Janssen D B. Kinetic analysis and x-ray structure of haloalkane dehalogenase with a modified halide-binding site. Biochemistry. 1998;37:15013–15023. doi: 10.1021/bi9815187. [DOI] [PubMed] [Google Scholar]
- 21.Liu J-Q, Kurihara T, Miyagi M, Esaki N, Soda K. Reaction mechanism of L-2-haloacid dehalogenase of Pseudomonas sp. YL—identification of Asp10 as the active site nucleophile by 18O incorporation experiments. J Biol Chem. 1995;270:18309–18312. [PubMed] [Google Scholar]
- 22.Lubetsky J B, Swope M, Dealwis C, Blake P, Lolis E. Pro-1 of macrophage migration inhibitory factor functions as a catalytic base in the phenylpyruvate tautomerase activity. Biochemistry. 1999;38:7346–7354. doi: 10.1021/bi990306m. [DOI] [PubMed] [Google Scholar]
- 23.Marletta M A, Cheung Y F, Walsh C. Stereochemical studies on the hydration of monofluorofumarate and 2,3-difluorofumarate by fumarase. Biochemistry. 1982;21:2637–2644. doi: 10.1021/bi00540a010. [DOI] [PubMed] [Google Scholar]
- 24.Murzin A G. Structural classification of proteins: new superfamilies. Curr Opin Struct Biol. 1996;6:386–394. doi: 10.1016/s0959-440x(96)80059-5. [DOI] [PubMed] [Google Scholar]
- 25.Nardi-Dei V, Kurihara T, Park C, Miyagi M, Tsunasawa S, Soda K, Esaki N. DL-2-haloacid dehalogenase from Pseudomonassp. 113 is a new class of dehalogenase catalyzing hydrolytic dehalogenation not involving enzyme-substrate ester intermediate. J Biol Chem. 1999;274:20977–20981. doi: 10.1074/jbc.274.30.20977. [DOI] [PubMed] [Google Scholar]
- 26.Newman J, Peat T S, Richard R, Kan L, Swanson P E, Affholter J A, Holmes I H, Schindler J F, Unkefer C J, Terwilliger T C. Haloalkane dehalogenases: structure of a Rhodococcusenzyme. Biochemistry. 1999;38:16105–16114. doi: 10.1021/bi9913855. [DOI] [PubMed] [Google Scholar]
- 27.Poelarends G J, Wilkens M, Larkin M J, van Elsas J D, Janssen D B. Degradation of 1,3-dichloropropene by Pseudomonas cichorii170. Appl Environ Microbiol. 1998;64:2931–2936. doi: 10.1128/aem.64.8.2931-2936.1998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Poelarends G J, van Hylckama Vlieg J E T, Marchesi J R, Freitas Dos Santos L M, Janssen D B. Degradation of 1,2-dibromoethane by Mycobacteriumsp. strain GP1. J Bacteriol. 1999;181:2050–2058. doi: 10.1128/jb.181.7.2050-2058.1999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Poelarends G J, Kulakov L A, Larkin M J, van Hylckama Vlieg J E T, Janssen D B. Roles of horizontal gene transfer and gene integration in evolution of 1,3-dichloropropene- and 1,2-dibromoethane-degradative pathways. J Bacteriol. 2000;182:2191–2199. doi: 10.1128/jb.182.8.2191-2199.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Pries F, Kingma J, Krooshof G H, Jeronimus-Stratingh C M, Bruins A P, Janssen D B. Histidine 289 is essential for hydrolysis of the alkyl-enzyme intermediate of haloalkane dehalogenase. J Biol Chem. 1995;270:10405–10411. doi: 10.1074/jbc.270.18.10405. [DOI] [PubMed] [Google Scholar]
- 31.Ridder I S, Rozeboom H J, Kalk K H, Janssen D B, Dijkstra B W. Three-dimensional structure of L-2-haloacid dehalogenase from Xanthobacter autotrophicusGJ10 complexed with the substrate-analogue formate. J Biol Chem. 1997;272:33015–33022. doi: 10.1074/jbc.272.52.33015. [DOI] [PubMed] [Google Scholar]
- 32.Ridder I S, Rozeboom H J, Kalk K H, Dijkstra B W. Crystal structures of intermediates in the dehalogenation of haloalkanoates by L-2-haloacid dehalogenase. J Biol Chem. 1999;274:30672–30678. doi: 10.1074/jbc.274.43.30672. [DOI] [PubMed] [Google Scholar]
- 33.Sambrook J, Fritsch E F, Maniatis T. Molecular cloning: a laboratory manual. 2nd ed. Cold Spring Harbor, N.Y: Cold Spring Harbor Laboratory; 1989. [Google Scholar]
- 34.Stamps S L, Fitzgerald M C, Whitman C P. Characterization of the role of the amino-terminal proline in the enzymatic activity catalyzed by macrophage migration inhibitory factor. Biochemistry. 1998;37:10195–10202. doi: 10.1021/bi9806955. [DOI] [PubMed] [Google Scholar]
- 35.Staskawicz B, Dahlbeck D, Keen N, Napoli C. Molecular characterization of cloned avirulence genes from race 0 and race 1 of Pseudomonas syringae pv. glycinea. J Bacteriol. 1987;169:5789–5794. doi: 10.1128/jb.169.12.5789-5794.1987. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Stivers J T, Abeygunawardana C, Mildvan A S, Hajipour G, Whitman C P, Chen L H. Catalytic role of the amino-terminal proline in 4-oxalocrotonate tautomerase: affinity labeling and heteronuclear NMR studies. Biochemistry. 1996;35:803–813. doi: 10.1021/bi951077g. [DOI] [PubMed] [Google Scholar]
- 37.Stivers J T, Abeygunawardana C, Mildvan A S, Hajipour G, Whitman C P. 4-Oxalocrotonate tautomerase: pH dependence of catalysis and pKavalues of active site residues. Biochemistry. 1996;35:814–823. doi: 10.1021/bi9510789. [DOI] [PubMed] [Google Scholar]
- 38.Stivers J T, Abeygunawardana C, Mildvan A S, Whitman C P. 15N NMR relaxation studies of free and inhibitor bound 4-oxalocrotonate tautomerase: backbone dynamics and entropy changes of an enzyme upon inhibitor binding. Biochemistry. 1996;35:16036–16047. doi: 10.1021/bi961834q. [DOI] [PubMed] [Google Scholar]
- 39.Stivers J T, Abeygunawardana C, Whitman C P, Mildvan A S. 4-Oxalocrotonate tautomerase, a 41-kDa homohexamer: backbone and side-chain resonance assignments, solution secondary structure, and location of active site residues by heteronuclear NMR spectroscopy. Protein Sci. 1996;5:729–741. doi: 10.1002/pro.5560050418. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Studier F W, Rosenberg A H, Dunn J J, Dubendorff J W. Use of T7 RNA polymerase to direct expression of cloned genes. Methods Enzymol. 1990;185:60–89. doi: 10.1016/0076-6879(90)85008-c. [DOI] [PubMed] [Google Scholar]
- 41.Subramanya H S, Roper D I, Dauter Z, Dodson E J, Davies G J, Wilson K S, Wigley D B. Enzymatic ketonization of 2-hydroxymuconate: specificity and mechanism investigated by the crystal structures of two isomerases. Biochemistry. 1996;35:792–802. doi: 10.1021/bi951732k. [DOI] [PubMed] [Google Scholar]
- 42.Sugimoto H, Taniguchi M, Nakagawa A, Tanaka I, Suzuki M, Nishihira J. Crystal structure of human d-dopachrome tautomerase, a homologue of macrophage migration inhibitory factor, at 1.54 Åresolution. Biochemistry. 1999;38:3268–3279. doi: 10.1021/bi982184o. [DOI] [PubMed] [Google Scholar]
- 43.Suzuki M, Sugimoto H, Nakagawa A, Tanaka I, Nishihira J, Sakai M. Crystal structure of the macrophage migration inhibitory factor from rat liver. Nat Struct Biol. 1996;3:259–266. doi: 10.1038/nsb0396-259. [DOI] [PubMed] [Google Scholar]
- 44.Swope M, Sun H-W, Blake P R, Lolis E. Direct link between cytokine activity and a catalytic site for macrophage migration inhibitory factor. EMBO J. 1998;17:3534–3541. doi: 10.1093/emboj/17.13.3534. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Taylor A B, Czerwinski R M, Johnson W H, Jr, Whitman C P, Hackert M L. Crystal structure of 4-oxalocrotonate tautomerase inactivated by 2-oxo-3-pentynoate at 2.4 Åresolution: analysis and implications for the mechanism of inactivation and catalysis. Biochemistry. 1998;37:14692–14700. doi: 10.1021/bi981607j. [DOI] [PubMed] [Google Scholar]
- 46.Thompson J D, Higgens D G, Gibson T J. CLUSTAL W: improving the sensitivity of progressive multiple sequence alignment through sequence weighting, position-specific gap penalties and weight matrix choice. Nucleic Acids Res. 1994;22:4673–4680. doi: 10.1093/nar/22.22.4673. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Van Hylckama Vlieg J E T, Janssen D B. Bacterial degradation of 3-chloroacrylic acid and the characterization of cis- and trans-specific dehalogenases. Biodegradation. 1992;2:139–150. doi: 10.1007/BF00124488. [DOI] [PubMed] [Google Scholar]
- 48.Verhagen C, Smit E, Janssen D B, van Elsas J D. Bacterial dichloropropene degradation in soil; screening of soils and involvement of plasmids carrying the dhlAgene. Soil Biol Biochem. 1995;27:1547–1557. [Google Scholar]
- 49.Verschueren K H G, Kingma J, Rozeboom H J, Kalk K H, Janssen D B, Dijkstra B W. Crystallographic and fluorescence studies of the interaction of haloalkane dehalogenase with halide ions—studies with halide compounds reveal a halide binding site in the active site. Biochemistry. 1993;32:9031–9037. doi: 10.1021/bi00086a008. [DOI] [PubMed] [Google Scholar]
- 50.Verschueren K H G, Seljee F, Rozeboom H J, Kalk K H, Dijkstra B W. Crystallographic analysis of the catalytic mechanism of haloalkane dehalogenase. Nature. 1993;363:693–698. doi: 10.1038/363693a0. [DOI] [PubMed] [Google Scholar]
- 51.Whitman C P, Aird B A, Gillespie W R, Stolowich N J. Chemical and enzymatic ketonization of 2-hydroxymuconate, a conjugated enol. J Am Chem Soc. 1991;113:3154–3162. [Google Scholar]
- 52.Yang G, Liang P-H, Dunaway-Mariano D. Evidence for nucleophilic catalysis in the aromatic substitution reaction catalyzed by (4-chlorobenzoyl)coenzyme A dehalogenase. Biochemistry. 1994;33:8527–8531. doi: 10.1021/bi00194a018. [DOI] [PubMed] [Google Scholar]
- 53.Yanisch-Perron C, Vieira J, Messing J. Improved M13 phage cloning vectors and host strains: nucleotide sequences of the M13mp18 and pUC19 vectors. Gene. 1985;33:103–109. doi: 10.1016/0378-1119(85)90120-9. [DOI] [PubMed] [Google Scholar]