Abstract
Bordetella pertussis and Bordetella bronchiseptica are capable of obtaining iron from hemin and hemoglobin. Genes encoding a putative bacterial heme iron acquisition system (bhu, for Bordetella heme utilization) were identified in a B. pertussis genomic sequence database, and the corresponding DNA was isolated from a virulent strain of B. pertussis. A B. pertussis bhuR mutant, predicted to lack the heme outer membrane receptor, was generated by allelic exchange. In contrast to the wild-type strain, bhuR mutant PM5 was incapable of acquiring iron from hemin and hemoglobin; genetic complementation of PM5 with the cloned bhuRSTUV genes restored heme utilization to wild-type levels. In parallel studies, B. bronchiseptica bhu sequences were also identified and a B. bronchiseptica bhuR mutant was constructed and confirmed to be defective in heme iron acquisition. The wild-type B. bronchiseptica parent strain grown under low-iron conditions produced the presumptive BhuR protein, which was absent in the bhuR mutant. Furthermore, production of BhuR by iron-starved B. bronchiseptica was markedly enhanced by culture in hemin-supplemented medium, suggesting that these organisms sense and respond to heme in the environment. Analysis of the genetic region upstream of the bhu cluster identified open reading frames predicted to encode homologs of the Escherichia coli ferric citrate uptake regulators FecI and FecR. These putative Bordetella regulators may mediate heme-responsive positive transcriptional control of the bhu genes.
Pathogenic microorganisms encounter severe iron limitation in mammalian hosts, where the concentration of free iron is several orders of magnitude less than that required to support microbial growth (18). Iron in serum and on mucosal surfaces is sequestered by the host iron binding proteins transferrin and lactoferrin, respectively, while the majority of host iron is found intracellularly in the form of heme and hemoproteins (58). To overcome iron sequestration by the host, pathogenic bacteria have evolved two general types of high-affinity iron acquisition systems that enable them to scavenge iron. In siderophore-dependent microbial iron acquisition systems, high-affinity iron-chelating siderophores are excreted and utilized to obtain nutritional iron (45, 54), while siderophore-independent systems employ cell surface proteins that mediate the direct binding and utilization of host-derived iron compounds (31, 50, 65, 76).
Many gram-negative pathogens use siderophore-independent systems to acquire iron from heme and hemoglobin (31, 76), and expression of the systems studied to date is negatively regulated at the transcriptional level by the ferric uptake regulator (Fur) protein, with ferrous iron as the corepressor (35, 57, 74). One type of heme utilization system described for Serratia marcescens (46) and Pseudomonas spp. (39, 47) relies on the secretion of small hemophore proteins, which bind heme and deliver it to heme-hemophore-specific outer membrane receptors. A second general mechanism of heme iron acquisition is exemplified by that of certain Neisseria spp. which express a bipartite hemoglobin receptor consisting of a TonB-dependent outer membrane receptor component and an accessory outer membrane lipoprotein (20, 48). Yet a third class of heme iron utilization system identified in organisms including Pseudomonas aeruginosa (57), Yersinia spp. (72, 74), and Shigella dysenteriae (51) utilizes a single-component TonB-dependent outer membrane receptor specific for heme, hemoglobin, or other heme compounds.
Bordetella pertussis and Bordetella bronchiseptica are gram-negative respiratory pathogens of mammals. In response to iron starvation, they produce the macrocyclic dihydroxamate siderophore alcaligin (15, 53) and also use a variety of heterologous siderophores, including enterobactin (7), ferrichrome, and desferrioxamine B (6), for iron retrieval. These organisms can also obtain iron from host sources transferrin (60, 61), lactoferrin (61), heme (1, 55), and hemoglobin (55).
In this study, we identified a cluster of B. pertussis genes (designated bhu, for Bordetella heme utilization) predicted to encode proteins highly similar to those of bacterial heme iron acquisition systems with single-component TonB-dependent outer membrane receptors. Mutational and phenotypic analyses confirmed that these Bordetella genes were required for acquisition of iron from hemin and hemoglobin in B. pertussis as well as in the closely related species B. bronchiseptica. Nucleotide sequence analysis of the region immediately upstream of the heme utilization gene cluster identified two open reading frames predicted to encode homologs of the Escherichia coli ferric citrate uptake system positive regulators FecI and FecR (11), suggesting that a similar positive regulatory mechanism may exist for the Bordetella heme system.
MATERIALS AND METHODS
Bacterial strains and culture conditions.
E. coli DH5α (Gibco-BRL, Gaithersburg, Md.) was used as the host strain for general cloning procedures and as the donor strain in triparental matings. DH5α harboring plasmid pRK2013 (30) provided mobilization functions in triparental matings. E. coli reporter strain H1717 (fhuF-lacZ aroB), used for Fur repressor titration assays, has been described previously (71). B. bronchiseptica B013N (4) and a spontaneous streptomycin-resistant derivative of wild-type B. pertussis UT25 (29), UT25Sm1 (14), have also been described previously.
E. coli strains were grown in Luria-Bertani (LB) broth or on LB agar plates. B. pertussis and B. bronchiseptica strains were cultured on Bordet-Gengou agar (9) and LB agar, respectively. Stainer-Scholte (SS) broth (68), modified as described previously (64), was used for growth of Bordetella strains in defined liquid medium. For iron-depleted cultures, SS basal medium was deferrated by treatment with Chelex100 (Bio-Rad, Richmond, Calif.) as described previously (4); iron-replete SS medium contained 36 μM FeSO4, and iron-depleted SS medium contained no iron supplements. Growth of Bordetella liquid cultures was monitored using a Klett-Summerson colorimeter equipped with a no. 54 filter (Klett Manufacturing Co., Long Island City, N.Y.). The medium used to culture B. pertussis for growth stimulation bioassays was modified LB agar (pertussis LB [PLB] agar), which was LB broth supplemented with 0.12% Molecusol MB cyclodextrin (Pharmatec, Inc., Alachua, Fla.) and 0.15% bovine serum albumin (BSA) (Sigma, St. Louis, Mo.) and solidified with Noble agar (Difco Laboratories, Detroit, Mich.). Antibiotics were used at the following concentrations: ampicillin, 100 μg/ml; gentamicin, 10 μg/ml; kanamycin, 50 μg/ml; streptomycin, 50 μg/ml; tetracycline, 15 (for B. bronchiseptica and E. coli) or 10 μg/ml (for B. pertussis).
Plasmids and genetic methods.
Plasmid cloning vectors pGEM3Z (Promega, Madison, Wis.) and pRK415 (42) were used in the construction of recombinant plasmids. Plasmid pBSL86 (2) was the source of the kanamycin resistance cassette used in the construction of B. pertussis ΔbhuR::kan mutant strain PM5. The cosmid-based B. pertussis UT25 genomic DNA library has been described previously (17). Suicide vector pSS1129 (69) was used for allelic exchange in the construction of B. pertussis mutant PM5. Conjugal transfer of plasmids from E. coli donors to Bordetella recipients was accomplished as described previously (14).
Nucleotide sequence data were accessed from the incomplete and unannotated B. pertussis Tohama I genome sequence. These sequence data were produced by the Bordetella pertussis Sequencing Group at the Sanger Centre and can be obtained from http://www.sanger.ac.uk/Projects/B_pertussis. The incomplete genomic sequence of B. bronchiseptica strain RB50 was also accessed from the Sanger Centre (http://www.sanger.ac.uk/Projects/B_bronchiseptica). Nucleotide sequences determined in this laboratory were derived from cloned DNA of B. pertussis strain UT25 by primer walking on both DNA strands. Nucleotide sequencing services were provided by the Advanced Genetic Analysis Center at the University of Minnesota. Oligonucleotide primers used in sequencing were synthesized by Gibco-BRL. Management and analysis of nucleotide and protein sequence data were performed with the Lasergene sequence analysis software package for the Macintosh PowerPC computer (DNASTAR, Inc., Madison, Wis.). Database searches were accomplished using the BLAST (3) servers provided by the Sanger Centre and the National Center for Biotechnology Information (NCBI) at the National Library of Medicine. The deduced BhuR amino acid sequence was analyzed for the presence of conserved patterns using the Conserved Domain Database and Search Service analysis (reverse position-specific BLAST) algorithm at the NCBI. Putative B. pertussis Fur-binding sequences were identified by using the MegAlign module of the Lasergene program to locate DNA regions of at least 50% identity over a 30-nucleotide (nt) region with the proposed consensus E. coli Fur-binding site 5′-GATAATGATAATCATTATC-3′ (19, 24). Amino acid sequence alignments were performed by the CLUSTAL (38) or Jotun-Hein (34) method using the MegAlign software module. The putative BhuR signal sequence cleavage site was predicted using the SignalP server at the Center for Biological Sequence Analysis (http://www.cbs.dtu.dk/services/SignalP/index.html).
The Fur repressor titration assay (71) was used to test the DNA regions upstream of hurI and bhuR for functional Fur-binding sites. E. coli indicator strain H1717 (fhuF-lacZ aroB) carrying the hurI or bhuR upstream DNA regions subcloned to pGEM3Z was plated on lactose MacConkey agar supplemented with 40 μM ferrous ammonium sulfate and appropriate antibiotics. Strain H1717(pGEM3Z) was the negative control, and the positive control was H1717 carrying p3ZFBS (T. J. Brickman, unpublished data), which contains a cloned copy of the E. coli consensus Fur-binding DNA sequence (19, 24).
Southern and in situ DNA hybridizations were performed at high stringency as described previously (62). Oligonucleotide probes for in situ DNA hybridizations (Hem1, 5′-GCAAGGACGAAAACACCGGCC-3′; Hem2, 5′-CTGGTAGGTCAACGATACGCG-3′) were synthesized by Gibco-BRL and end-labeled with [γ32-P]ATP (ICN Radiochemicals, Costa Mesa, Calif.) using T4 polynucleotide kinase as described previously (62). bhuR-specific DNA hybridization probes and the 1.2-kb HincII kanamycin resistance cassette were radiolabeled with [α32-P]dCTP (ICN) by the random-priming method (28) using the Random Primers DNA-labeling system (Gibco-BRL).
Construction of Bordetella bhuR mutants.
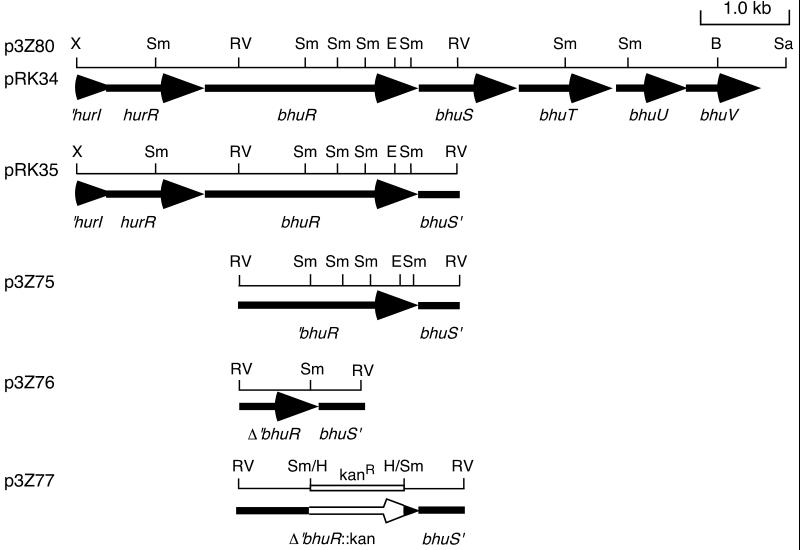
A 2.4-kb EcoRV DNA fragment encompassing the 3′ region of bhuR and 5′ region of bhuS was subcloned from B. pertussis UT25 recombinant cosmid pCPbhu1 carrying bhu sequences to produce plasmid p3Z75 (Fig. 1). This plasmid was digested with SmaI and religated, resulting in the deletion of a 1.2-kb SmaI DNA region internal to the putative bhuR coding region. This deletion derivative, p3Z76, was linearized with SmaI and ligated with a 1.2-kb HincII DNA fragment containing the kanamycin resistance cassette from pBSL86 (2), resulting in ΔbhuR::kan plasmid p3Z77 (Fig. 1). A 2.5- kb fragment of p3Z77 encompassing the mutated bhuR region was excised using plasmid vector EcoRI and BamHI sites and ligated to suicide vector pSS1129. The resulting plasmid, pSS8, was mated to B. pertussis strain UT25Sm1, and the mutation was transferred to the chromosome by homologous recombination as described by Stibitz (69) to produce ΔbhuR::kan mutant PM5. Allelic exchange in B. pertussis mutant PM5 was verified by Southern hybridization using bhuR- and kanamycin resistance cassette-specific DNA probes.
FIG. 1.
Spatial organization of the B. pertussis bhu genetic regions used to construct recombinant plasmids. The construction of the cosmid subclones and B. pertussis ΔbhuR::kan mutant PM5 is described in Materials and Methods. Solid arrows, open reading frames (arrowheads denote the direction of transcription); open arrow (DNA insert of plasmid p3Z77), kanamycin resistance gene. The open reading frames upstream of bhuR (initially termed orfI and orfR) are designated hurIR, for heme uptake regulators. Abbreviations: B, BamHI; E, EcoRI; RV, EcoRV; H, HincII; Sa, SalI; Sm, SmaI; X, XhoI.
A B. bronchiseptica bhuR mutant was constructed by delivery of ΔbhuR::kan plasmid p3Z77 (Fig. 1), which cannot replicate in Bordetella strains, to B. bronchiseptica strain B013N by electroporation. Transformants with the plasmid integrated into the chromosome were selected on agar medium containing kanamycin and ampicillin. Southern hybridization using DNA probes specific for bhuR and the kanamycin resistance gene was used to confirm the insertion of the plasmid into the B. bronchiseptica bhuR locus in mutant strain BRM21.
Hemin and hemoglobin growth stimulation bioassays.
Aqueous stock solutions of bovine hemin chloride (Sigma) at concentrations of 1 or 10 mM were made in 0.02 N NaOH. Bovine (Becton-Dickinson, Cockeysville, Md.) and human, pig, turkey, and rabbit (Sigma) hemoglobin stock solutions were prepared in 10 mM HEPES buffer, pH 7.4. Concentrations of hemoglobin stock solutions were confirmed using a plasma hemoglobin diagnostic kit (Sigma) and were adjusted to 3 or 1 mg/ml. Complexes of hemin-BSA (1:1 molar ratio) at 100 μM and of human hemoglobin-haptoglobin (hemoglobin concentration, 100 μM) were prepared by methods described previously (10). Alcaligin was purified as the deferrisiderophore as described by Brickman and coworkers (15) and used as a positive control at an aqueous concentration of 50 μM. Distilled-water diluent was used as the negative control in the bioassays.
For growth stimulation bioassays, B. pertussis strains were cultured on PLB agar plates for 3 days. PLB agar plate growth was suspended in deferrated SS basal medium to an optical density (600 nm) of 2.0; 200 μl of this suspension was seeded into 25 ml of molten PLB agar (at 50°C) containing 50 μg of ethylenediamine-di-[(o-hydroxyphenyl)acetic acid] (EDDA)/ml and poured into a 90-mm-diameter petri dish. Wells were punched in the seeded solidified agar, and 50-μl volumes of the test solutions were added. The diameters of the growth stimulation zones around wells containing the specified iron source were measured after 60 h of incubation at 37°C. B. bronchiseptica bioassays on iron-restricted medium were performed as described previously using LB agar supplemented with EDDA (15). Growth stimulation is reported as the mean diameter from three replicate bioassays and is representative of four experimental trials.
Analysis of hemin-responsive protein expression.
B. bronchiseptica strain B013N and bhuR mutant BRM21 were grown on LB agar at 37°C for 24 h and used to inoculate iron-replete SS broth cultures to an initial density of 15 Klett units. For each strain, the SS culture was grown with shaking at 37°C for 24 h, at which time cells were harvested, washed with SS basal medium, and used to inoculate one iron-replete and two iron-depleted SS cultures. After 15 h of growth, the iron starvation status of the iron-depleted cultures was confirmed by measuring the production of alcaligin using the chrome-azurol S universal siderophore detection assay (66). One of the iron-depleted cultures was supplemented with hemin to a final concentration of 5 μM, and the cultures were allowed to continue growing. All cultures were sampled at 1, 4, and 8 h after the addition of hemin. Cell samples were disrupted using a French press (American Instrument Company, Silver Spring, Md.), and the insoluble total-membrane fractions were prepared as described previously (41). Proteins were treated in solubilization buffer at 100°C for 6 min and resolved by sodium dodecyl sulfate-polyacrylamide gel electrophoresis (SDS-PAGE) on 10% polyacrylamide gels containing 0.5 M urea as described previously (64); approximately 30 μg of protein was applied to each gel lane, and proteins were visualized by Coomassie blue staining.
Nucleotide sequence accession number.
The GenBank accession number assigned to the 871-nt B. pertussis UT25 hurR-bhuR region is AY032627.
RESULTS
Identification of putative heme utilization genes.
To identify B. pertussis DNA sequences potentially encoding a heme acquisition system, the incomplete Sanger Centre B. pertussis genomic sequence assembly was subjected to a TBLASTN search using the amino acid sequence of the P. aeruginosa PhuR heme receptor as the query. PhuR was selected as a representative TonB-dependent heme outer membrane receptor, in part because a BLASTP database search revealed that it was highly similar to heme receptors of several gram-negative bacterial species (57). Although the Sanger Centre B. pertussis genomic sequence is presently at the “finishing/gap closure” stage and is thus considered preliminary, the search identified a contig with a 5′-truncated open reading frame (bhuR) predicted to encode a protein with significant amino acid sequence similarity to PhuR (Table 1). Downstream of bhuR were four closely spaced open reading frames which, based on similarity to components of the Pseudomonas Phu system (57), are predicted to encode a hemin-degrading factor (BhuS), a hemin-specific periplasmic binding protein (BhuT), a cytoplasmic membrane permease (BhuU), and an ATP-binding protein (BhuV) (Table 1). The bhuRSTUV region of the B. pertussis contig was used to search the translated-nucleotide sequence database at the NCBI. The predicted BhuRSTUV proteins were highly similar to proteins of heme utilization systems of several gram-negative organisms, including Yersinia pestis (74), Yersinia enterocolitica (72), and S. dysenteriae (51), as well as P. aeruginosa (57) (Table 1). In addition, the organization of the B. pertussis bhu open reading frames was very similar to the organization of heme utilization genes in Yersinia spp. and P. aeruginosa. A search of the Sanger Centre's incomplete genomic sequence database for taxonomically related strain B. bronchiseptica RB50 identified homologous DNA sequences having substantial identity with the B. pertussis bhu sequences.
TABLE 1.
Characteristics of B. pertussis proteins predicted to be involved in heme utilization
| B. pertussis protein | Deduced molecular mass (kDa) | Predicted function | Homologous proteins (% similarity)a |
|---|---|---|---|
| HurI | 19 | Transcriptional activator | FecI (44), PupI (46) |
| HurR | 35 | Sensor/regulator | FecR (27), PupR (31) |
| BhuR | 92 | Outer membrane heme receptor | PhuR (28), PfhR (28), HutA (26), ChuA (26), ShuA (26), HmbR (26) |
| BhuS | 38 | Heme-degrading protein | PhuS (46), HemS (41), HmuS (41), ShuS (38) |
| BhuT | 30 | Periplasmic binding protein | PhuT (25), HemT (36), HmuT (36), ShuT (35) |
| BhuU | 26 | Membrane permease | PhuU (60), HemU (52), HmuU (53), ShuU (50) |
| BhuV | 28 | ATP-binding protein | PhuV (50), HemV (40), HmuV (39), ShuV (38) |
Percent similarities were determined by the Jotun-Hein method with the PAM250 residue weight table using the MegAlign application of the Lasergene software package. Heme utilization proteins are from the following organisms (GenBank accession numbers are in parentheses): Phu, P. aeruginosa (AF055999); PfhR, P. fluorescens (AF127222); HutA, V. cholerae (L27149); ChuA, E. coli (U67920); Shu, S. dysenteriae (U64516); HmbR, N. meningitidis (AF105339); Hem, Y. enterocolitica (X68147 and X77867); Hmu, Y. pestis (U60647). FecI and FecR, ferric citrate uptake regulators of E. coli (M63115); PupI and PupR, regulators of the pupB receptor gene for transport of ferric pseudobactins BN7 and BN8 in P. putida WCS358 (X77918).
Oligonucleotides Hem1 and Hem2, corresponding to B. pertussis internal bhuR DNA sequences, were used to identify five recombinant cosmids in a B. pertussis UT25 genomic library by colony hybridization. Cosmid pCPbhu1 was chosen for limited nucleotide sequencing using the Hem1 oligonucleotide primer to establish the presence of the predicted bhu sequences. The results confirmed that the pCPbhu1 DNA sequence was identical to the contig sequence from the Sanger Centre database over the bhuR region sequenced (data not shown). Further, restriction enzyme mapping of pCPbhu1 concurred with maps deduced from the contig nucleotide sequence, confirming that the cloned B. pertussis UT25 DNA encompassed the desired bhu region.
Construction and phenotypic analysis of B. pertussis bhuR mutant PM5.
To determine if the bhu genes encoded a functional B. pertussis heme iron acquisition system, bhuR heme receptor mutant PM5 was constructed (Fig. 1). The ability of isogenic wild-type and ΔbhuR::kan mutant strains of B. pertussis to use hemin and hemoglobin as iron sources was assessed in growth stimulation bioassays. Wild-type parental strain UT25Sm1 was capable of utilizing hemin and human hemoglobin as sources of iron in these bioassays, exhibiting dose-responsive growth stimulation in response to increasing concentrations of these iron compounds (Table 2). In contrast, ΔbhuR::kan mutant PM5 was unable to use either iron source at any concentration tested. Both wild-type and bhuR mutant strains formed similar growth haloes around wells containing positive control alcaligin, indicating that alcaligin siderophore-mediated iron utilization was unaffected by the bhuR mutation.
TABLE 2.
Growth stimulation of B. pertussis strains by hemin and human hemoglobin
| Straina | Diam of
growth zoneb (mm) surrounding wells containing
the indicated concn (μM) of iron source:
|
|||||||
|---|---|---|---|---|---|---|---|---|
| Hemin
|
Hemoglobin
|
Alcaligin, 50 | ||||||
| 25 | 50 | 100 | 0.8 | 1.6 | 4 | 8 | ||
| UT25Sm1 | 15 | 17 | 20 | 19 | 21 | 22 | 25 | 35 |
| PM5 | — | — | — | — | — | — | — | 36 |
| PM5(pRK34) | 15 | 18 | 23 | 19 | 22 | 23 | 25 | 34 |
| PM5(pRK415) | — | — | — | — | — | — | — | 36 |
UT25Sm1, B. pertussis bhu+ strain; PM5, UT25Sm1 ΔbhuR::kan; PM5(pRK34), PM5 carrying plasmid-borne bhuRSTUV; PM5(pRK415), PM5 carrying plasmid vector control.
Diameters of growth zones include the diameter of the well (6 mm). No growth occurred around wells containing the distilled-water diluent. —, no detectable growth stimulation.
In genetic complementation experiments, plasmid pRK34, which carries the B. pertussis UT25 8-kb bhuRSTUV DNA insert (Fig. 1), restored the ability of mutant PM5 to obtain iron from both hemin and hemoglobin to wild-type levels (Table 2). A smaller subclone containing bhuR (pRK35; Fig. 1) did not restore heme iron utilization to PM5 (data not shown), suggesting that the ΔbhuR::kan chromosomal mutation may exert polar effects on the downstream bhuSTUV genes.
Growth stimulation bioassays were also used to determine whether B. pertussis could obtain iron from hemoglobins of various animal species. Wild-type strain UT25Sm1 showed similar levels of dose-dependent growth stimulation in response to bovine, porcine, rabbit, turkey, and human hemoglobins (data not shown). As predicted, ΔbhuR::kan mutant PM5 was incapable of utilizing any of the hemoglobins. Other bioassay experiments demonstrated that wild-type B. pertussis used both hemoglobin-haptoglobin and hemin-BSA complexes as iron sources, while the bhuR mutant strain was unable to utilize those complexes (data not shown). These data demonstrate that the bhu gene cluster is involved in the acquisition of iron from hemin and hemoglobin, as well as from haptoglobin and heme complexed with BSA. Furthermore, the data are consistent with the hypothesis that outer membrane receptor BhuR is capable of recognizing a range of heme compounds and of mediating transport of heme iron at levels sufficient to stimulate growth in an iron-limited medium.
Characterization of DNA sequences upstream of bhuRSTUV
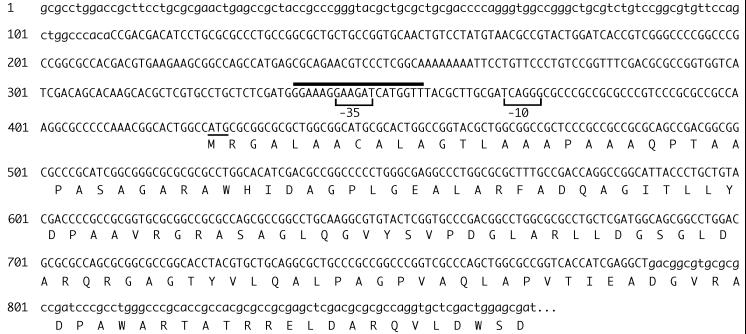
The B. pertussis bhu contig identified in the Sanger Centre database did not include the genetic region encoding the putative BhuR start codon. To identify the 5′ limit of the bhu genetic system and to analyze potential upstream regulatory sequences, the nucleotide sequence of the bhu region absent from the database was determined using the cloned B. pertussis UT25 bhu DNA (Fig. 2). Analysis of this 871-bp B. pertussis UT25 nucleotide sequence and that of the overlapping bhu contig from the database predicted that the complete bhuR open reading frame encoded a precursor protein with a molecular mass of 92 kDa; upon cleavage of a predicted 21-amino-acid signal peptide, the mature BhuR protein would have a molecular mass of 90 kDa. An RPS-BLAST search for conserved protein domains predicted a TonB box C sequence (Pfam protein family [5] database domain: pfam 00593) in the carboxy-terminal region of BhuR. The FRAP and NPNL amino acid sequence motifs, which are highly conserved among hemin/hemoglobin receptors (67, 76), were also present in the corresponding regions of the deduced BhuR proteins of both B. pertussis and B. bronchiseptica, except that a tyrosine residue is substituted for phenylalanine in the FRAP motif and a serine replaces the second asparagine in the NPNL motif. A potential Fur-binding sequence identified 68 bases upstream of the BhuR gene start codon (Fig. 2) exhibited 58% identity with the 19-nt E. coli consensus Fur-binding sequence (19, 24). However, there was a G in place of the highly conserved T at nucleotide position 16 of the consensus Fur-binding sequence (71) and there was poor conservation of the characteristic inverted repeat, suggesting that this sequence may have limited Fur-binding activity.
FIG. 2.
B. pertussis nucleotide sequence encompassing the 5′ region of bhuR. An 871-bp region of the chromosome of strain UT25 was sequenced on both strands. The BhuR putative start codon is underlined, and the N-terminal 149-amino-acid region of BhuR is shown. The line above nucleotides 338 through 356 indicates the position of a potential Fur-binding site. Bracketed nucleotides represent the proposed ECF ς factor −10 and −35 promoter elements. Lowercase letters indicate nucleotides of the 871-bp region that overlap B. pertussis nucleotide sequence contigs found in the Sanger Centre database.
Two contigs overlapping the 871-bp sequence (Fig. 2) were identified in the Sanger Centre B. pertussis genomic database. The original bhu contig exhibited 100% identity with the UT25 sequence over an 84-nt region, while a newly identified contig was 100% identical to the corresponding 110-nt end of the 871-bp sequence.
Identification of open reading frames encoding FecI and FecR homologs.
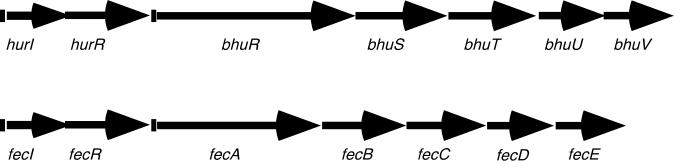
Analysis of the two B. pertussis Sanger Centre contigs and the overlapping sequences obtained in this laboratory allowed examination of the bhu upstream region. A 5-kb DNA sequence was used in a BLASTX search at the NCBI which revealed two potential open reading frames (orfI and orfR) located 210 bp upstream of bhuR (Fig. 3). The OrfI and OrfR proteins showed the highest-scoring alignments with the FecI and FecR positive regulatory proteins, which control the ferric citrate uptake genes in E. coli (11, 75), and with Pseudomonas putida WCS358 PupI and PupR, which positively regulate the pupB ferric pseudobactin receptor gene (44) (Table 1). FecI and PupI function as alternative ς factors, and FecR and PupR are cytoplasmic membrane-bound regulatory proteins. An RPS-BLAST search using the predicted OrfI sequence revealed a domain highly characteristic of ς factors of the extracytoplasmic function (ECF) family (pfam domain: pfam 00776) (52).
FIG. 3.
Genetic organization of the B. pertussis heme iron uptake system and comparison with the E. coli ferric citrate uptake system. (Top) B. pertussis DNA region containing the bhu genes and the upstream hurIR open reading frames. Vertical bars upstream of hurI and bhuR show the positions of Fur-binding sequences which were predicted by nucleotide sequence analysis and which were assessed in vivo with the Fur repressor titration assay. (Bottom) Organization of the fec genetic system of E. coli (11), with putative Fur-binding sites denoted by vertical bars upstream of fecI and fecA. fecI and fecR encode positive regulatory proteins; fecA codes for the outer membrane ferric citrate receptor; fecB, fecC, fecD, and fecE encode periplasmic binding protein and cytoplasmic membrane transport components.
No open reading frames that would be obvious targets of OrfIR regulation were identified upstream of orfIR, nor were any genes with apparent relevance to iron acquisition or bhu gene regulation found downstream of the bhu cluster. In the DNA sequence immediately upstream of orfI, two overlapping putative Fur-binding sequences exhibiting 64 and 74% identity to the consensus Fur-binding sequence were identified. A search for presumptive promoter region sequences revealed ς70-like −10 and −35 regions (33) near the putative Fur boxes upstream of orfI, while putative ECF ς factor −10 and −35 regions (26, 52) were identified upstream of bhuR (Fig. 2). Due to the high degree of similarity among FecIR, PupIR, and the deduced Bordetella OrfIR proteins and the potential for OrfIR involvement in bhu gene regulation, we have tentatively designated these B. pertussis genes hurIR, for heme uptake regulators I and R.
Fur repressor titration analyses.
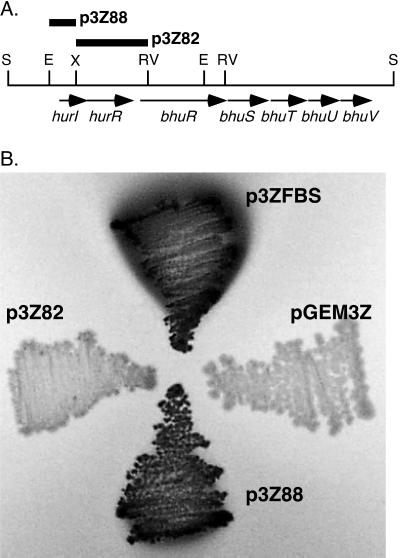
The 2.4-kb XhoI-EcoRV DNA region 5′ to bhuR and the 0.8-kb EcoRI-XhoI DNA region upstream of hurI were each cloned to high-copy-number plasmid pGEM3Z and tested for functional Fur-binding activity by the Fur repressor titration assay employing E. coli indicator strain H1717 (71) (Fig. 4). In this in vivo assay, introduction of a functional Fur-binding site in multicopy relieves the repressive influence of Fur on the expression of the chromosomal fhuF-lacZ fusion under normally repressing high-iron growth conditions. These experiments demonstrated that the bhuR upstream region (p3Z82) had no apparent in vivo Fur-binding activity. This result is consistent with the low level of similarity of the bhuR upstream sequences to the consensus Fur-binding DNA sequence. In contrast, the hurI upstream region (p3Z88) exhibited a strong Fur-binding function, as evidenced by a high level of expression of fhuF-lacZ that was qualitatively equal to that conferred by positive control plasmid p3ZFBS, containing the consensus E. coli Fur-binding site (19, 24).
FIG. 4.
Functional analysis of potential Fur-binding sites in the hur-bhuR DNA region. (A) Partial restriction map of the hur-bhu DNA region. Bars labeled p3Z88 and p3Z82, 0.8-kb EcoRI-XhoI fragment containing the hurI upstream region and 2.4-kb XhoI-EcoRV fragment containing the bhuR upstream region, respectively, which were cloned to high-copy-number vector pGEM3Z and tested for in vivo Fur binding. Abbreviations: E, EcoRI; RV, EcoRV; S, SalI; X, XhoI. (B) Fur repressor titration assays were carried out as described in Materials and Methods using E. coli host strain H1717 plated on lactose MacConkey agar containing 40 μM iron. Dark areas of bacterial growth demonstrate β-galactosidase activity, which is indicative of functional Fur binding by the cloned DNA regions. Plasmids p3Z88 and p3Z82 are described in the legend for panel A; p3ZFBS contains the consensus E. coli Fur-binding sequence; pGEM3Z is the plasmid vector control.
Analysis of heme-responsive protein expression.
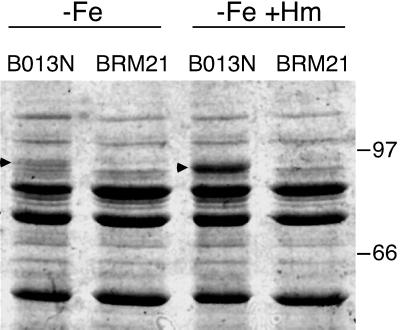
Multiple attempts to visualize the BhuR protein expressed in B. pertussis cells cultured under high- or low-iron conditions either in the presence or absence of hemin or hemoglobin were unsuccessful. In SDS-PAGE analyses, differences in stained or intrinsically radiolabeled proteins between wild-type UT25Sm1 and ΔbhuR::kan mutant PM5 could not be discerned. However, in parallel studies, B. bronchiseptica bhuR mutant BRM21 was constructed and was demonstrated in growth stimulation bioassays to be defective in heme iron acquisition (data not shown). Plasmid pRK34 carrying the wild-type B. pertussis bhu genes (Fig. 1) fully restored hemin and hemoglobin utilization to BRM21 (data not shown), indicating that the bhu system of B. pertussis can functionally complement the bhu mutation in B. bronchiseptica. Wild-type B. bronchiseptica strain B013N and bhuR mutant derivative BRM21 were cultured in SS medium under low-iron conditions with or without added hemin, and the total membranes were isolated and analyzed by SDS-PAGE. Wild-type cells grown in low-iron medium lacking hemin produced a ca. 90-kDa membrane protein, which was absent in the bhuR mutant membrane fraction (Fig. 5). This protein had an apparent molecular mass corresponding to the mass deduced for BhuR, and it was not produced by cells grown under iron-replete conditions (data not shown). Notably, production of the BhuR protein was elevated significantly in wild-type cells after 4 h of growth in the presence of hemin (data not shown) and was even more abundant after culture for 8 h in the presence of hemin (Fig. 5). The 90-kDa membrane protein was consistently absent in the bhuR mutant strain cultured under all growth conditions. This pattern of protein production by wild-type B. bronchiseptica cells suggests that iron-repressible expression of bhuR is responsive to the presence of hemin in the environment.
FIG. 5.
Hemin-responsive BhuR expression in B. bronchiseptica. The growth of iron-starved B. bronchiseptica wild-type (B013N) and isogenic bhuR mutant (BRM21) cultures with or without added hemin is described in Materials and Methods. Total-membrane fractions were prepared from cells harvested 8 h after the addition of hemin and analyzed by SDS-PAGE. −Fe, cells grown under iron-depleted conditions; −Fe +Hm, cells grown under iron-depleted conditions and supplemented with 5 μM hemin. Arrowheads, positions of the putative BhuR outer membrane receptor protein. Migration positions of the protein standards and their molecular masses in kilodaltons are designated.
DISCUSSION
When B. pertussis is starved for iron, it produces and utilizes its native siderophore alcaligin for iron acquisition (15, 53) and can also retrieve iron complexed with heterologous siderophores (6, 7), host-derived lactoferrin (61), transferrin (60, 61), hemoglobin (55), and hemin (1, 55). To date, the only Bordetella iron retrieval systems that have been characterized are those for ferric enterobactin transport (7) and for the biosynthesis and transport of alcaligin (8, 13, 32, 40, 41). In this study, we have identified a siderophore-independent iron acquisition system that is required by Bordetella spp. for the utilization of iron from heme compounds.
In the course of B. pertussis infection, virulence factors such as pertussis toxin (73), adenylate cyclase/hemolysin (21, 37), dermonecrotic toxin (49), and tracheal cytotoxin (22) are presumably elaborated, resulting in host cell dysfunction and damage to the mucosal epithelium (36). Extravasation of serum components, immune cells, and erythrocytes may ensue (59), and intracellular heme molecules may be liberated and subsequently serve as iron sources for B. pertussis. In this study, we identified the B. pertussis bhu genes, which encode functions required for utilization of iron from hemin and hemoglobin as well as other hemoproteins. Virtually identical bhu sequences were identified in the B. bronchiseptica genomic sequence database at the Sanger Centre. The Bordetella bhu genes are predicted to encode homologs of known prokaryotic heme utilization systems and are genetically organized in a cluster similar to those of other bacterial heme uptake systems. Based on amino acid sequence similarities with components of other heme utilization systems, BhuR is predicted to be the outer membrane receptor for hemin and hemoglobin. Other transport activities are hypothesized to be provided by the BhuT hemin-specific periplasmic binding protein, the BhuU cytoplasmic membrane permease protein, and BhuV, predicted to function as the ATPase required for heme transport across the cytoplasmic membrane. BhuS is similar to so-called hemin-degrading factors from P. aeruginosa (57), Y. enterocolitica (72), Y. pestis (74), and S. dysenteriae (51). Although a hemin-degrading activity for these proteins has not been demonstrated, Stojiljkovic and Hantke found that hemS was an essential gene in Y. enterocolitica and presented evidence that HemS expression prevented lethality in E. coli cells expressing the Y. enterocolitica HemR outer membrane receptor (72).
In our studies, wild-type B. pertussis was capable of acquiring iron from hemin, from hemoglobin from human, porcine, bovine, rabbit, and turkey sources, and from hemoglobin-haptoglobin and hemin-BSA complexes, while bhuR mutant PM5 was incapable of utilizing any of these compounds. These data indicate that the bhu genes are required for heme iron utilization in B. pertussis and that the BhuR outer membrane receptor is capable of recognizing a broad range of heme compounds. Y. enterocolitica receptor HemR also recognizes a variety of heme compounds including hemin, hemoglobin, myoglobin, hemopexin, and catalase and BSA- and human serum albumin-heme and haptoglobin-hemoglobin complexes (10). Though the mechanism of hemin and hemoglobin recognition by the outer membrane receptor and subsequent heme internalization remains unknown, Bracken and coworkers reported that a conserved histidine residue located between the FRAP and NPNL amino acid domains of HemR was important to the ability of Y. enterocolitica to effectively utilize hemin and heme-protein complexes (10). However, the B. pertussis and B. bronchiseptica BhuR proteins deduced from the available sequence data, as well as heme receptors PfhR (Pseudomonas fluorescens) and PhuR (P. aeruginosa) are predicted to lack the histidine residue in this region (67), suggesting that the mechanism of heme internalization by these receptors may be somewhat different from that of HemR.
In the present study, we were unable to visualize the BhuR protein by SDS-PAGE analysis of iron-starved or iron-replete wild-type B. pertussis grown in the presence or absence of hemin or hemoglobin. Because multiple proteins in the range of 80 to 95 kDa are expressed under iron starvation conditions (12), it is possible that B. pertussis BhuR comigrates in electrophoretic gels with one or more other proteins. However, comparative analysis of wild-type B. bronchiseptica and its bhuR mutant derivative revealed a membrane protein corresponding to BhuR in iron-starved wild-type cells that was absent in the bhuR mutant grown under the same low-iron conditions. Remarkably, iron-starved wild-type B. bronchiseptica cultures supplemented with hemin demonstrated dramatically enhanced production of BhuR, suggesting that bhuR expression is responsive to the presence of heme.
Expression of the Vibrio cholerae (56), Y. pestis (74), Y. enterocolitica (72), and P. aeruginosa (57) heme utilization genes is negatively regulated by iron through the Fur repressor. A potential B. pertussis Fur-binding site identified upstream of bhuR exhibited essentially no in vivo Fur-binding activity, suggesting that regulation of the bhu system differs from that of other known microbial heme systems. Transcription of bhuR may be iron repressed indirectly, perhaps through Fur repression of putative positive regulatory gene hurI, or may be unresponsive to iron concentration. However, because B. bronchiseptica produces BhuR only under iron-restricted growth conditions, the latter possibility seems unlikely. In addition to repression by Fur, positive transcriptional regulation has been demonstrated for some siderophore system genes (23), including the Bordetella alcaligin genes (8, 16) and those for ferric citrate uptake in E. coli (11). This positive regulation occurs only after Fur derepression and in the presence of the cognate iron compound. To date, no gram-negative bacterial heme iron acquisition system has been reported to require positive transcriptional regulation. In gram-positive pathogen Corynebacterium diphtheriae, the genes encoding the heme transport apparatus are expressed constitutively (25) whereas transcription of hmuO, encoding a heme oxygenase, is activated by a two-component regulatory system which responds to the presence of heme or hemoglobin in the environment (63). In our study, the hurIR open reading frames identified upstream of the B. pertussis bhu gene cluster are predicted to encode homologs of the E. coli FecI ECF ς factor/FecR family of regulatory proteins, suggesting that the bhu system may be positively regulated in a manner similar to that for the fec system. Stiefel and coworkers recently identified FecR homologs from a variety of bacterial species by genomic database BLAST searches (70). In that study, the authors identified the DNA sequence contigs in the Sanger Centre genomic database predicted to encode the HurR proteins of both B. pertussis and B. bronchiseptica. The HurR proteins of both species share the three conserved tryptophan residues characteristic for the FecR class of transmembrane regulatory proteins (70).
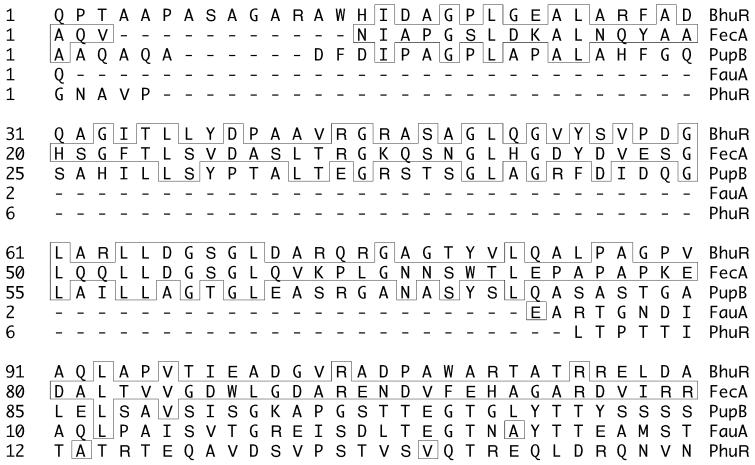
Several lines of evidence suggest that hurIR may be involved in positive transcriptional regulation of the B. pertussis bhu genes. First, no other open reading frames apparently relevant to iron acquisition or iron-regulated gene expression were identified near hurIR or the bhu genes. Second, the E. coli fecIR genes are directly adjacent to the fec genes encoding the ferric citrate transport machinery in a pattern strikingly similar to that of the hurIRbhuRSTUV genes (Fig. 3). Third, upstream putative bhuR promoter sequences are predicted to require an ECF ς factor for transcription. The hurI gene encodes a putative ECF ς factor, and upstream of hurI is a DNA sequence that exhibited strong Fur-binding activity, consistent with a role for HurI in iron metabolism. Similarly, fecI is iron regulated via Fur and the fecA ferric citrate receptor gene has an ECF ς factor-dependent promoter. The HurR protein is highly similar to the FecR and PupR cytoplasmic membrane proteins and contains one predicted membrane-spanning region. The conserved C-terminal regions of FecR and PupR are also highly conserved in HurR; the C-terminal one-third of HurR was 41 and 43% similar to the same regions of PupR and FecR, respectively. This C-terminal region of FecR interacts with the periplasmic N-terminal extension of the FecA outer membrane ferric citrate receptor to effect positive regulation of fec genes (27, 43). Most importantly, analysis of the BhuR outer membrane receptor amino acid sequence revealed the presence of this highly conserved extended N-terminal region (Fig. 6). Thus, BhuR appears to be a member of this family of outer membrane iron receptors that function in systems positively controlled by a FecIR-type transcriptional regulatory system. Amino acid sequence comparisons with the P. aeruginosa PhuR heme receptor and the B. pertussis FauA ferric alcaligin receptor revealed that these proteins lack the N-terminal extension characteristic of the FecA, PupB, and BhuR receptors (Fig. 6). The presence of this highly conserved N-terminal extension in BhuR is consistent with the notion that expression of the B. pertussis bhu genes may be positively regulated by an alternative ς factor-dependent system encoded by hurIR. Finally, the observation that iron-stressed B. bronchiseptica cells dramatically enhance the production of BhuR in response to hemin further suggests the involvement of a positive regulatory system controlling receptor gene expression. Experiments aimed at defining the potential role of hurIR in bhu gene regulation are in progress.
FIG. 6.
Amino acid sequence alignments of BhuR with selected bacterial heme and ferric siderophore receptors. The primary amino acid sequences of the deduced mature proteins were aligned using the CLUSTAL program. Shown is the alignment of the N-terminal regions of the proteins; significant similarity between the extended N-terminal domains of BhuR, FecA and PupB can be seen. Amino acid residues in boxes are those that match the FecA amino acid sequence. Proteins: BhuR, B. pertussis heme receptor; FecA (GenBank accession no. AAC77247), E. coli ferric citrate receptor; PupB (P38047), P. putida WCS358 ferric pseudobactin receptor; FauA (AAD26430), B. pertussis ferric alcaligin receptor; PhuR (AF055999), P. aeruginosa heme receptor.
The specific iron sources upon which B. pertussis relies for in vivo growth are unknown. It is clear, however, that this organism possesses genes encoding multiple iron acquisition systems. During the course of infection, from the inhalation of bacteria in microaerosols to colonization and host tissue injury, it may be expected that the genes encoding all of these iron acquisition systems are first expressed after Fur derepression. These genetic systems may then be individually positively controlled primarily by the availability of the cognate iron source by a priority regulation mechanism typified by the transcriptional control of the native siderophore system by the AlcR regulator with alcaligin as the inducer (8, 16). In the latter stages of the infectious process, when there is considerable host cell damage, intracellular heme compounds may be released, potentially providing a priority activation signal for enhanced transcription of the Bordetella bhu genes.
ACKNOWLEDGMENTS
We are grateful to Timothy Brickman for helpful discussions and critical reading of the manuscript, and we acknowledge Jessica Boeldt and Andrew Norgan for technical assistance.
This work was supported by Public Health Service grant AI-31088 from the National Institute of Allergy and Infectious Diseases.
REFERENCES
- 1.Agiato L A, Dyer D W. Siderophore production and membrane alterations by Bordetella pertussisin response to iron starvation. Infect Immun. 1992;60:117–123. doi: 10.1128/iai.60.1.117-123.1992. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Alexeyev M F. Three kanamycin resistance gene cassettes with different polylinkers. BioTechniques. 1995;18:52–56. [PubMed] [Google Scholar]
- 3.Altschul S F, Gish W, Miller W, Myers E W, Lipman D J. Basic local alignment search tool. J Mol Biol. 1990;215:403–410. doi: 10.1016/S0022-2836(05)80360-2. [DOI] [PubMed] [Google Scholar]
- 4.Armstrong S K, Clements M O. Isolation and characterization of Bordetella bronchisepticamutants deficient in siderophore activity. J Bacteriol. 1993;175:1144–1152. doi: 10.1128/jb.175.4.1144-1152.1993. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Bateman A, Birney E, Durbin R, Eddy S R, Howe K L, Sonnhammer E L L. The Pfam contribution to the annual NAR database issue. Nucleic Acids Res. 2000;28:263–266. doi: 10.1093/nar/28.1.263. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Beall B. Two iron-regulated putative ferric siderophore receptor genes in Bordetella bronchiseptica and Bordetella pertussis. Res Microbiol. 1998;149:189–201. doi: 10.1016/s0923-2508(98)80079-x. [DOI] [PubMed] [Google Scholar]
- 7.Beall B, Sanden G N. A Bordetella pertussis fepAhomologue required for utilization of exogenous ferric enterobactin. Microbiology. 1995;141:3193–3205. doi: 10.1099/13500872-141-12-3193. [DOI] [PubMed] [Google Scholar]
- 8.Beaumont F C, Kang H Y, Brickman T J, Armstrong S K. Identification and characterization of alcR, a gene encoding an AraC-like regulator of alcaligin siderophore biosynthesis and transport in Bordetella pertussis and Bordetella bronchiseptica. J Bacteriol. 1998;180:862–870. doi: 10.1128/jb.180.4.862-870.1998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Bordet J, Gengou O. Le microbe de la coqueluche. Ann Inst Pasteur (Paris) 1906;20:731–741. [Google Scholar]
- 10.Bracken C S, Baer M T, Abdur-Rashid A, Helms W, Stojiljkovic I. Use of heme-protein complexes by the Yersinia enterocoliticaHemR receptor: histidine residues are essential for receptor function. J Bacteriol. 1999;181:6063–6072. doi: 10.1128/jb.181.19.6063-6072.1999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Braun V. Surface signaling: novel transcription initiation mechanism starting from the cell surface. Arch Microbiol. 1997;167:325–331. doi: 10.1007/s002030050451. [DOI] [PubMed] [Google Scholar]
- 12.Brickman T J, Armstrong S K. Bordetella pertussis fur gene restores iron repressibility of siderophore and protein expression to deregulated Bordetella bronchisepticamutants. J Bacteriol. 1995;177:268–270. doi: 10.1128/jb.177.1.268-270.1995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Brickman T J, Armstrong S K. Essential role of the iron-regulated outer membrane receptor FauA in alcaligin siderophore-mediated iron uptake in Bordetellaspecies. J Bacteriol. 1999;181:5958–5966. doi: 10.1128/jb.181.19.5958-5966.1999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Brickman T J, Armstrong S K. The ornithine decarboxylase gene odc is required for alcaligin siderophore biosynthesis in Bordetellaspp.: putrescine is a precursor of alcaligin. J Bacteriol. 1996;178:54–60. doi: 10.1128/jb.178.1.54-60.1996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Brickman T J, Hansel J G, Miller M J, Armstrong S K. Purification, spectroscopic analysis and biological activity of the macrocyclic dihydroxamate siderophore alcaligin produced by Bordetella pertussis and Bordetella bronchiseptica. Biometals. 1996;9:191–203. doi: 10.1007/BF00144625. [DOI] [PubMed] [Google Scholar]
- 16.Brickman T J, Kang H Y, Armstrong S K. Transcriptional activation of Bordetellaalcaligin siderophore genes requires the AlcR regulator with alcaligin as inducer. J Bacteriol. 2001;183:483–489. doi: 10.1128/JB.183.2.483-489.2001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Brown D R, Parker C D. Cloning of the filamentous hemagglutinin of Bordetella pertussis and its expression in Escherichia coli. Infect Immun. 1987;55:154–161. doi: 10.1128/iai.55.1.154-161.1987. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Bullen J J. The significance of iron in infection. Rev Infect Dis. 1981;3:1127–1138. doi: 10.1093/clinids/3.6.1127. [DOI] [PubMed] [Google Scholar]
- 19.Calderwood S B, Mekalanos J J. Iron regulation of Shiga-like toxin expression in Escherichia coli is mediated by the furlocus. J Bacteriol. 1987;169:4759–4764. doi: 10.1128/jb.169.10.4759-4764.1987. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Chen C J, Elkins C, Sparling P F. Phase variation of hemoglobin utilization in Neisseria gonorrhoeae. Infect Immun. 1998;66:987–993. doi: 10.1128/iai.66.3.987-993.1998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Confer D L, Eaton J W. Phagocyte impotence caused by an invasive bacterial adenylate cyclase. Science. 1982;217:948–950. doi: 10.1126/science.6287574. [DOI] [PubMed] [Google Scholar]
- 22.Cookson B T, Tyler A N, Goldman W E. Primary structure of the peptidoglycan-derived tracheal cytotoxin of Bordetella pertussis. Biochemistry. 1989;28:1744–1749. doi: 10.1021/bi00430a048. [DOI] [PubMed] [Google Scholar]
- 23.Crosa J H. Signal transduction and transcriptional and posttranscriptional control of iron-regulated genes in bacteria. Microbiol Mol Biol Rev. 1997;61:319–336. doi: 10.1128/mmbr.61.3.319-336.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.de Lorenzo V, Wee S, Herrero M, Neilands J B. Operator sequences of the aerobactin operon of plasmid ColV-K30 binding the ferric uptake regulation (Fur) repressor. J Bacteriol. 1987;169:2624–2630. doi: 10.1128/jb.169.6.2624-2630.1987. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Drazek E S, Hammack C A, Schmitt M P. Corynebacterium diphtheriaegenes required for acquisition of iron from haemin and haemoglobin are homologous to ABC haemin transporters. Mol Microbiol. 2000;36:68–84. doi: 10.1046/j.1365-2958.2000.01818.x. [DOI] [PubMed] [Google Scholar]
- 26.Enz S, Braun V, Crosa J H. Transcription of the region encoding the ferric dicitrate-transport system in Escherichia coli: similarity between promoters for fecAand for extracytoplasmic function sigma factors. Gene. 1995;163:13–18. doi: 10.1016/0378-1119(95)00380-o. [DOI] [PubMed] [Google Scholar]
- 27.Enz S, Mahren S, Stroeher U H, Braun V. Surface signaling in ferric citrate transport gene induction: interaction of the FecA, FecR, and FecI regulatory proteins. J Bacteriol. 2000;182:637–646. doi: 10.1128/jb.182.3.637-646.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Feinberg A P, Vogelstein B. A technique for radiolabeling DNA restriction endonuclease fragments to high specific activity. Anal Biochem. 1983;132:6–13. doi: 10.1016/0003-2697(83)90418-9. [DOI] [PubMed] [Google Scholar]
- 29.Field L H, Parker C D. Differences observed between fresh isolates of Bordetella pertussis and their laboratory passaged derivatives. In: Manclark C R, Hill J C, editors. International symposium on pertussis. U.S. Washington, D.C.: Department of Health, Education, and Welfare; 1978. pp. 124–132. [Google Scholar]
- 30.Figurski D H, Helinski D R. Replication of an origin-containing derivative of plasmid RK2 dependent on a plasmid function provided in trans. Proc Natl Acad Sci USA. 1979;76:1648–1652. doi: 10.1073/pnas.76.4.1648. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Genco C A, White-Dixon D. Emerging strategies in microbial haem capture. Mol Microbiol. 2001;39:1–11. doi: 10.1046/j.1365-2958.2001.02231.x. [DOI] [PubMed] [Google Scholar]
- 32.Giardina P C, Foster L A, Toth S I, Roe B A, Dyer D W. Identification of alcA, a Bordetella bronchisepticagene necessary for alcaligin production. Gene. 1995;167:133–136. doi: 10.1016/0378-1119(95)00659-1. [DOI] [PubMed] [Google Scholar]
- 33.Hawley D K, McClure W R. Compilation and analysis of Escherichia colipromoter DNA sequences. Nucleic Acids Res. 1983;11:2237–2255. doi: 10.1093/nar/11.8.2237. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Hein J. Unified approach to alignment and phylogenies. Methods Enzymol. 1990;183:626–645. doi: 10.1016/0076-6879(90)83041-7. [DOI] [PubMed] [Google Scholar]
- 35.Henderson D P, Payne S M. Characterization of the Vibrio choleraeouter membrane heme transport protein HutA: sequence of the gene, regulation of expression, and homology to the family of TonB-dependent proteins. J Bacteriol. 1994;176:3269–3277. doi: 10.1128/jb.176.11.3269-3277.1994. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Hewlett E L. Pertussis: current concepts of pathogenesis and prevention. Pediatr Infect Dis J. 1997;16(Suppl.):S78–S84. doi: 10.1097/00006454-199704001-00002. [DOI] [PubMed] [Google Scholar]
- 37.Hewlett E L, Gordon V M. Adenylate cyclase toxin of Bordetella pertussis. In: Wardlaw A C, Parton R, editors. Pathogenesis and immunity in pertussis. New York, N.Y: Wiley; 1988. pp. 193–210. [Google Scholar]
- 38.Higgins D G, Thompson J D, Gibson T J. Using CLUSTAL for multiple sequence alignments. Methods Enzymol. 1996;266:383–402. doi: 10.1016/s0076-6879(96)66024-8. [DOI] [PubMed] [Google Scholar]
- 39.Idei A, Kawai E, Akatsuka H, Omori K. Cloning and characterization of the Pseudomonas fluorescensATP-binding cassette exporter, HasDEF, for the heme acquisition protein HasA. J Bacteriol. 1999;181:7545–7551. doi: 10.1128/jb.181.24.7545-7551.1999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Kang H Y, Armstrong S K. Transcriptional analysis of the Bordetellaalcaligin siderophore biosynthesis operon. J Bacteriol. 1998;180:855–861. doi: 10.1128/jb.180.4.855-861.1998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Kang H Y, Brickman T J, Beaumont F C, Armstrong S K. Identification and characterization of iron-regulated Bordetella pertussisalcaligin siderophore biosynthesis genes. J Bacteriol. 1996;178:4877–4884. doi: 10.1128/jb.178.16.4877-4884.1996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Keen N T, Tamaki S, Kobayashi D, Trollinger D. Improved broad-host-range plasmids for DNA cloning in gram-negative bacteria. Gene. 1988;70:191–197. doi: 10.1016/0378-1119(88)90117-5. [DOI] [PubMed] [Google Scholar]
- 43.Kim I, Stiefel A, Plantor S, Angerer A, Braun V. Transcription induction of the ferric citrate transport genes via the N-terminus of the FecA outer membrane protein, the Ton system and the electrochemical potential of the cytoplasmic membrane. Mol Microbiol. 1997;23:333–344. doi: 10.1046/j.1365-2958.1997.2401593.x. [DOI] [PubMed] [Google Scholar]
- 44.Koster M, van Klompenburg W, Bitter W, Leong J, Weisbeek P. Role for the outer membrane ferric siderophore receptor PupB in signal transduction across the bacterial cell envelope. EMBO J. 1994;13:2805–2813. doi: 10.1002/j.1460-2075.1994.tb06574.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Lankford C E. Bacterial assimilation of iron. Crit Rev Microbiol. 1973;2:273–331. [Google Scholar]
- 46.Letoffe S, Ghigo J M, Wandersman C. Iron acquisition from heme and hemoglobin by a Serratia marcescensextracellular protein. Proc Natl Acad Sci USA. 1994;91:9876–9880. doi: 10.1073/pnas.91.21.9876. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Letoffe S, Redeker V, Wandersman C. Isolation and characterization of an extracellular haem-binding protein from Pseudomonas aeruginosa that shares function and sequence similarities with the Serratia marcescensHasA haemophore. Mol Microbiol. 1998;28:1223–1234. doi: 10.1046/j.1365-2958.1998.00885.x. [DOI] [PubMed] [Google Scholar]
- 48.Lewis L A, Gray E, Wang Y P, Roe B A, Dyer D W. Molecular characterization of hpuAB, the haemoglobin-haptoglobin-utilization operon of Neisseria meningitidis. Mol Microbiol. 1997;23:737–749. doi: 10.1046/j.1365-2958.1997.2501619.x. [DOI] [PubMed] [Google Scholar]
- 49.Livey I, Wardlaw A C. Production and properties of Bordetella pertussisheat-labile toxin. J Med Microbiol. 1984;17:91–103. doi: 10.1099/00222615-17-1-91. [DOI] [PubMed] [Google Scholar]
- 50.Mietzner T A, Morse S A. The role of iron-binding proteins in the survival of pathogenic bacteria. Annu Rev Nutr. 1994;14:471–493. doi: 10.1146/annurev.nu.14.070194.002351. [DOI] [PubMed] [Google Scholar]
- 51.Mills M, Payne S M. Genetics and regulation of heme iron transport in Shigella dysenteriae and detection of an analogous system in Escherichia coliO157:H7. J Bacteriol. 1995;177:3004–3009. doi: 10.1128/jb.177.11.3004-3009.1995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Missiakas D, Raina S. The extracytoplasmic function sigma factors: role and regulation. Mol Microbiol. 1998;28:1059–1066. doi: 10.1046/j.1365-2958.1998.00865.x. [DOI] [PubMed] [Google Scholar]
- 53.Moore C H, Foster L A, Gerbig D G, Dyer D W, Gibson B W. Identification of alcaligin as the siderophore produced by Bordetella pertussis and B. bronchiseptica. J Bacteriol. 1995;177:1116–1118. doi: 10.1128/jb.177.4.1116-1118.1995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Neilands J B. Siderophores: structure and function of microbial iron transport compounds. J Biol Chem. 1995;270:26723–26726. doi: 10.1074/jbc.270.45.26723. [DOI] [PubMed] [Google Scholar]
- 55.Nicholson M L, Beall B. Disruption of tonB in Bordetella bronchiseptica and Bordetella pertussisprevents utilization of ferric siderophores, haemin and haemoglobin as iron sources. Microbiology. 1999;145:2453–2461. doi: 10.1099/00221287-145-9-2453. [DOI] [PubMed] [Google Scholar]
- 56.Occhino D A, Wyckoff E E, Henderson D P, Wrona T J, Payne S M. Vibrio cholerae iron transport: haem transport genes are linked to one of two sets of tonB, exbB, exbDgenes. Mol Microbiol. 1998;29:1493–1507. doi: 10.1046/j.1365-2958.1998.01034.x. [DOI] [PubMed] [Google Scholar]
- 57.Ochsner U A, Johnson Z, Vasil M L. Genetics and regulation of two distinct haem-uptake systems, phu and has, in Pseudomonas aeruginosa. Microbiology. 2000;146:185–198. doi: 10.1099/00221287-146-1-185. [DOI] [PubMed] [Google Scholar]
- 58.Panter S S. Release of iron from hemoglobin. Methods Enzymol. 1994;231:502–514. doi: 10.1016/0076-6879(94)31034-3. [DOI] [PubMed] [Google Scholar]
- 59.Persson C G, Erjefalt J S, Greiff L, Erjefalt I, Korsgren M, Linden M, Sundler F, Andersson M, Svensson C. Contribution of plasma-derived molecules to mucosal immune defence, disease and repair in the airways. Scand J Immunol. 1998;47:302–313. doi: 10.1046/j.1365-3083.1998.00317.x. [DOI] [PubMed] [Google Scholar]
- 60.Redhead K, Hill T. Acquisition of iron from transferrin by Bordetella pertussis. FEMS Microbiol Lett. 1991;61:303–307. doi: 10.1016/0378-1097(91)90570-z. [DOI] [PubMed] [Google Scholar]
- 61.Redhead K, Hill T, Chart H. Interaction of lactoferrin and transferrins with the outer membrane of Bordetella pertussis. J Gen Microbiol. 1987;133:891–898. doi: 10.1099/00221287-133-4-891. [DOI] [PubMed] [Google Scholar]
- 62.Sambrook J, Fritsch E F, Maniatis T. Molecular cloning: a laboratory manual. 2nd ed. Cold Spring Harbor, N.Y: Cold Spring Harbor Laboratory; 1989. [Google Scholar]
- 63.Schmitt M P. Identification of a two-component signal transduction system from Corynebacterium diphtheriaethat activates gene expression in response to the presence of heme and hemoglobin. J Bacteriol. 1999;181:5330–5340. doi: 10.1128/jb.181.17.5330-5340.1999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Schneider D R, Parker C D. Effect of pyridines on phenotypic properties of Bordetella pertussis. Infect Immun. 1982;38:548–553. doi: 10.1128/iai.38.2.548-553.1982. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Schryvers A B, Stojiljkovic I. Iron acquisition systems in the pathogenic Neisseria. Mol Microbiol. 1999;32:1117–1123. doi: 10.1046/j.1365-2958.1999.01411.x. [DOI] [PubMed] [Google Scholar]
- 66.Schwyn B, Neilands J B. Universal chemical assay for the detection and determination of siderophores. Anal Biochem. 1987;160:47–56. doi: 10.1016/0003-2697(87)90612-9. [DOI] [PubMed] [Google Scholar]
- 67.Simpson W, Olczak T, Genco C A. Characterization and expression of HmuR, a TonB-dependent hemoglobin receptor of Porphyromonas gingivalis. J Bacteriol. 2000;182:5737–5748. doi: 10.1128/jb.182.20.5737-5748.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68.Stainer D W, Scholte M J. A simple chemically defined medium for the production of phase I Bordetella pertussis. J Gen Microbiol. 1970;63:211–220. doi: 10.1099/00221287-63-2-211. [DOI] [PubMed] [Google Scholar]
- 69.Stibitz S. Use of conditionally counterselectable suicide vectors for allelic exchange. Methods Enzymol. 1994;235:458–465. doi: 10.1016/0076-6879(94)35161-9. [DOI] [PubMed] [Google Scholar]
- 70.Stiefel A, Mahren S, Ochs M, Schindler P T, Enz S, Braun V. Control of the ferric citrate transport system of Escherichia coli: mutations in region 2.1 of the FecI extracytoplasmic-function sigma factor suppress mutations in the FecR transmembrane regulatory protein. J Bacteriol. 2001;183:162–170. doi: 10.1128/JB.183.1.162-170.2001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 71.Stojiljkovic I, Baumler A J, Hantke K. Fur regulon in gram-negative bacteria. Identification and characterization of new iron-regulated Escherichia coligenes by a Fur titration assay. J Mol Biol. 1994;236:531–545. doi: 10.1006/jmbi.1994.1163. [DOI] [PubMed] [Google Scholar]
- 72.Stojiljkovic I, Hantke K. Transport of haemin across the cytoplasmic membrane through a haemin-specific periplasmic binding-protein-dependent transport system in Yersinia enterocolitica. Mol Microbiol. 1994;13:719–732. doi: 10.1111/j.1365-2958.1994.tb00465.x. [DOI] [PubMed] [Google Scholar]
- 73.Tamura M, Nogimori K, Murai S, Yajima M, Ito K, Katada T, Ui M, Ishii S. Subunit structure of islet-activating protein, pertussis toxin, in conformity with the A-B model. Biochemistry. 1982;21:5516–5522. doi: 10.1021/bi00265a021. [DOI] [PubMed] [Google Scholar]
- 74.Thompson J M, Jones H A, Perry R D. Molecular characterization of the hemin uptake locus (hmu) from Yersinia pestis and analysis of hmumutants for hemin and hemoprotein utilization. Infect Immun. 1999;67:3879–3892. doi: 10.1128/iai.67.8.3879-3892.1999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 75.van Hove B, Staudenmaier H, Braun V. Novel two-component transmembrane transcription control: regulation of iron dicitrate transport in Escherichia coliK-12. J Bacteriol. 1990;172:6749–6758. doi: 10.1128/jb.172.12.6749-6758.1990. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 76.Wandersman C, Stojiljkovic I. Bacterial heme sources: the role of heme, hemoprotein receptors and hemophores. Curr Opin Microbiol. 2000;3:215–220. doi: 10.1016/s1369-5274(00)00078-3. [DOI] [PubMed] [Google Scholar]