Background:
Marine recruits training at Parris Island experienced an unexpectedly high rate of severe acute respiratory syndrome coronavirus 2 (SARS-CoV-2) infection, despite preventive measures including a supervised, 2-week, pre-entry quarantine. We characterize SARS-CoV-2 transmission in this cohort.
Methods:
Between May and November 2020, we monitored 2,469 unvaccinated, mostly male, Marine recruits prospectively during basic training. If participants tested negative for SARS-CoV-2 by quantitative polymerase chain reaction (qPCR) at the end of quarantine, they were transferred to the training site in segregated companies and underwent biweekly testing for 6 weeks. We assessed the effects of coronavirus disease 2019 (COVID-19) prevention measures on other respiratory infections with passive surveillance data, performed phylogenetic analysis, and modeled transmission dynamics and testing regimens.
Results:
Preventive measures were associated with drastically lower rates of other respiratory illnesses. However, among the trainees, 1,107 (44.8%) tested SARS-CoV-2-positive, with either mild or no symptoms. Phylogenetic analysis of viral genomes from 580 participants revealed that all cases but one were linked to five independent introductions, each characterized by accumulation of mutations across and within companies, and similar viral isolates in individuals from the same company. Variation in company transmission rates (mean reproduction number R0; 5.5 [95% confidence interval [CI], 5.0, 6.1]) could be accounted for by multiple initial cases within a company and superspreader events. Simulations indicate that frequent rapid-report testing with case isolation may minimize outbreaks.
Conclusions:
Transmission of wild-type SARS-CoV-2 among Marine recruits was approximately twice that seen in the community. Insights from SARS-CoV-2 outbreak dynamics and mutations spread in a remote, congregate setting may inform effective mitigation strategies.
Keywords: Congregate setting, Mathematical modeling, Phylogenetic analysis, Severe acute respiratory syndrome coronavirus 2, Superspreader events, Transmission dynamics, U.S. Military
Initially reported in December 2019,1 coronavirus disease 2019 (COVID-19) has rapidly emerged as a worldwide pandemic associated with substantial morbidity and mortality. While asymptomatic COVID-19 infections have been more highly represented among young people (for review, see2), transmission of the virus by this group to the rest of the population is an important factor in community spread.3,4 Severe acute respiratory syndrome coronavirus 2 (SARS-CoV-2) infections have been associated with outbreaks in many types of congregate settings, including assisted living communities,5,6 churches,7 cruise ships,8 prisons,9,10 and the military.11–18 Limiting viral spread in such settings represents a crucial public health challenge, and the implementation of effective nonpharmaceutical interventions can contribute to reducing SARS-CoV-2 transmission.19,20 Marine recruits going through basic training at Parris Island constitute a homogeneous population of young and healthy individuals living together in a geographically isolated location and operating under highly controlled public health practices. This congregate setting provides a unique case study for investigating factors affecting virus transmission and the effectiveness of mitigation measures.20
Between May and November 2020, Letizia and his colleagues21 conducted the prospective, longitudinal COVID-19 Health Action Response for Marines (CHARM) study, which followed new recruits upon arrival at a supervised 2-week quarantine location, through the subsequent first 6 weeks of their basic training at Marine Corps Recruit Depot Parris Island (MCRDPI). Epidemiologic and phylogenetic study during the supervised quarantine away from MCRDPI found that 0.9% of recruits arrived with a positive SARS-CoV-2 quantitative polymerase chain reaction (qPCR) test and 1.9% later tested positive during quarantine.14 At the end of quarantine, recruits who tested negative for SARS-CoV-2 were transferred to MCRDPI for basic training. During the 6 weeks at MCRDPI, a high rate of new SARS-CoV-2 infections was detected.21 This finding was astounding when considering this unique, tightly controlled study setting, characterized by: (i) a strict 2-week isolation of subjects before training, (ii) highly structured activities, (iii) an absence of physical interaction between companies during training, and (iv) the additional implementation of nonpharmaceutical interventions at MCRDPI to limit the spread of COVID-19. To explain the high infection rate, we conducted a careful investigation of SARS-CoV-2 infections among Marine recruits through biweekly qPCR testing and phylogenetic analysis of viral genomes from infected individuals. Similar to other reports,22–24 non-SARS-CoV-2 respiratory infections declined significantly during the study, thus strongly suggesting that the Marine recruits complied with preventive measures. Additionally, these measures were given as orders and therefore were mandatory with compliance strictly enforced. We reconstructed viral introduction events and transmission clusters, traced mutations that occurred during local transmission as the virus spread within and between companies, and modeled transmission dynamics to guide improved control measures.
METHODS
Study Design and Participants
Participants enrolled in CHARM 2 weeks before basic training, within 48 hours of arrival at an isolated supervised quarantine location where they were tested by qPCR for SARS-CoV-2 on enrollment and on day 7 (see eAppendix; http://links.lww.com/EDE/B957). All recruits received a Marine Corps-mandated SARS-CoV-2 test at the end of quarantine. Recruits testing negative were transferred to basic training at MCRDPI.
Upon arrival at MCRDPI, recruits were assigned to a company of 400–500 (either male or female) individuals. Multiple companies overlapped in time but did not interact. Study participants were retested by qPCR at the end of weeks 2, 4, and 6.
This study was approved by the Naval Medical Research Center (NMRC) institutional review board (IRB), in compliance with all applicable U.S. federal regulations governing the protection of human subjects. All participants provided written informed consent.
Procedures
The rates of other respiratory diseases (obtained through passive surveillance specifically at MCRDPI) were acquired from Military Public Health Disease and Injury Surveillance records25 from 1 January 2017, to 31 December 2020 (eAppendix; http://links.lww.com/EDE/B957).
Mid-turbinate nasal swab specimens were tested by qPCR for SARS-CoV-2 within 48 hours of collection by Lab24 (Boca Raton, FL) and the Naval Infectious Diseases Diagnostic Laboratory (Naval Medical Research Center, Silver Spring, MD), as detailed in the eAppendix; http://links.lww.com/EDE/B957. For SARS-CoV-2 genome recovery, RNA was isolated from viral transport media samples, reverse transcribed, qPCR amplified, sequenced, and analyzed to identify consensus genomes from each sample. Median joining haplotype networks were built using PopART version 1.7 (http://popart.otago.ac.nz)26,27 (eAppendix; http://links.lww.com/EDE/B957).
We modeled point prevalence data collected at 2-week intervals during the 6-week basic training period that followed the supervised 2-week quarantine by extending the Susceptible-Exposed-Infectious-Recovered (SEIR) model, including an additional post-infection (P) compartment before recovery, during which a person has detectable viral load but can no longer transmit. A similar model was previously used to infer the spread of SARS-CoV-2 from point prevalence data.28 Model parameters, including the basic reproduction number R0 and the mean duration of each infection stage, were estimated by fitting the deterministic model to prevalence time series for each company using Hamiltonian Monte Carlo in Stan.29 Further details on the modeling are in the eAppendix; http://links.lww.com/EDE/B957.
Statistical Analysis
We analyzed the rates of infection of other non-SARS-CoV-2 infections or illnesses before and after institution of preventive measures in 2020, and during these periods in previous years, by two-sample t tests, and compared the change in 2020 to that in the previous years combined using analysis of variance (ANOVA). For the comparison of symptoms in the presence and absence of the viral mutation S3883A, we used median and interquartile range (IQR) to summarize the data.
RESULTS
Cohort Characteristics and SARS-CoV-2 Infection During Basic Training
A total of 3,249 Marine recruits volunteered for enrollment in CHARM at the beginning of a supervised 2-week quarantine in either Charleston, SC (11 May 2020 to 15 July 2020) or Atlanta, GA (10 August 2020, to 7 September 2020).21 Of these, 28 tested qPCR positive for SARS-CoV-2 at the time of enrollment and another 45 during quarantine (Figure 1). A total of 707 were lost to follow-up. From 25 May 2020, to 5 November 2020, the remaining 2,469 participants who had been qPCR negative throughout the quarantine proceeded to MCRDPI and were tested for SARS-CoV-2 after 2, 4, and 6 weeks of basic training.
FIGURE 1.
CHARM study flow diagram. Marine Corps recruits arrived for a supervised quarantine following a 2-week home quarantine. Those who enrolled in the CHARM study were tested by qPCR for SARS-CoV-2 upon arrival and then weekly during a 2-week supervised quarantine. Participants who tested positive were excluded from the study. Participants who had three negative qPCR tests (at quarantine days 0, 7, and 14) proceeded to basic training at MCRDPI where qPCR follow-up testing was performed every 2 weeks for the first 6 weeks of their basic training. Nearly complete viral genomes were recovered from 597 samples from 580 recruits who tested positive for SARS-CoV-2.
Participants were primarily young adults (2,108 [85.4%] were 18–20 years old) and predominantly male (2,250 [91.1%]; eTable 1; http://links.lww.com/EDE/B957). Altogether, 1,107 (44.8%) participants tested SARS-CoV-2 qPCR positive, for an overall incidence rate of 15.5 per 1,000 person–days. There was no difference in the incidence rate (rate ratio = 0.95; 95% confidence interval [CI] = 0.80, 1.17) of SARS-CoV-2 between males and females (15.5 vs. 16.0 per 1,000 person–days, respectively). Most infections were asymptomatic, with only 250 (22.6%) reporting symptoms 2 weeks preceding the initial qPCR positive test, and an additional 174 (15.7%) reporting symptoms the following 2 weeks. We assessed symptoms at each follow-up encounter by questionnaire in a group setting by company. No participants sought healthcare at that time.
Effects of Public Health Measures on Non-SARS-CoV-2 Respiratory Infections
In response to the COVID-19 pandemic, before the beginning of CHARM, military public health officials instituted nonpharmaceutical preventive measures at MCRDPI. These measures were in addition to the supervised quarantine and negative qPCR test before transfer to MCRDPI, and included masking of recruits and staff except during long runs, increased spacing during formations, head-to-toe sleeping arrangements, increased hand hygiene and surface cleaning, controlled movement of recruits, and reduced company size. Additionally, travel to and from the base was limited, visitors were no longer allowed on base, and base amenities were closed or had limited access.
We evaluated the impact of the public health measures on non-SARS-CoV-2 acute respiratory infections (ARIs) and pneumonia among all Marine recruits at MCRDPI using long-term passive surveillance data. New ARI cases decreased from 47.0 to 3.6 per 1,000 recruits per week following implementation of SARS-CoV-2 mitigation measures, whereas they ranged from 18 to 27 per 1,000 recruits per week in the previous 3 years (Figure 2). Likewise, new pneumonia cases were less than 2 per 1,000 recruits per week during the May–November CHARM period versus 6.9 to 9.7 in the previous 3 years. Conversely, new COVID-19 cases, detected by active surveillance qPCR testing of CHARM participants, rose to 205.8 per 1,000 study participants per week by mid-July 2020, despite the implementation of mitigation measures in the preceding months. Comparison of the incidence of ARI (change of difference, –28.9 [–35.3 to –22.5; P < 0.001]) and pneumonia (change of difference, –9.5 [–11.8 to –7.1; P < 0.001]; Figure 2; eTable 2; http://links.lww.com/EDE/B957) in the months after introduction of preventive measures in 2020 versus the same periods in the 3 previous years demonstrated a substantial decline.
FIGURE 2.
Impact of COVID-19 preventive measures on incidence of other respiratory illnesses and SARS-CoV-2. Historical incidences for acute respiratory infections (Top) and pneumonia (Middle) from 2017 to 2019 compared with 2020 following the implementation of the public health infection control measures in 2020 as described (arrows). Shown are 3-week moving averages of new cases per 1,000 persons per week. (Bottom) New SARS-CoV-2 cases per 1,000 study participants per week detected during CHARM. Gray box denotes the 2020 CHARM study period. (i) Implementation of Centers for Disease Control and Prevention (CDC) guidelines on increased hand hygiene and social distancing; established isolation barracks for confirmed cases; no travel for official duty and personal travel limited to local area; quarantine for those returning from high-risk areas; no visitors to base. (ii) Mandatory wearing of cloth masks at all times except on long-distance runs; changes in select group training to limit contact among recruits; base amenities including fitness center and pool closed; limited access to grocery store and commercial venues. (iii) Implementation of mandatory Marine-supervised 2-week quarantine for all new recruits before entry into basic training. (iv) Resumed base access for retirees and family members, but not visitors; resumed group training including long-distance runs. (v) Resumed official and personal travel outside of local area.
Genetic Characteristics and Transmission of SARS-CoV-2 During Training
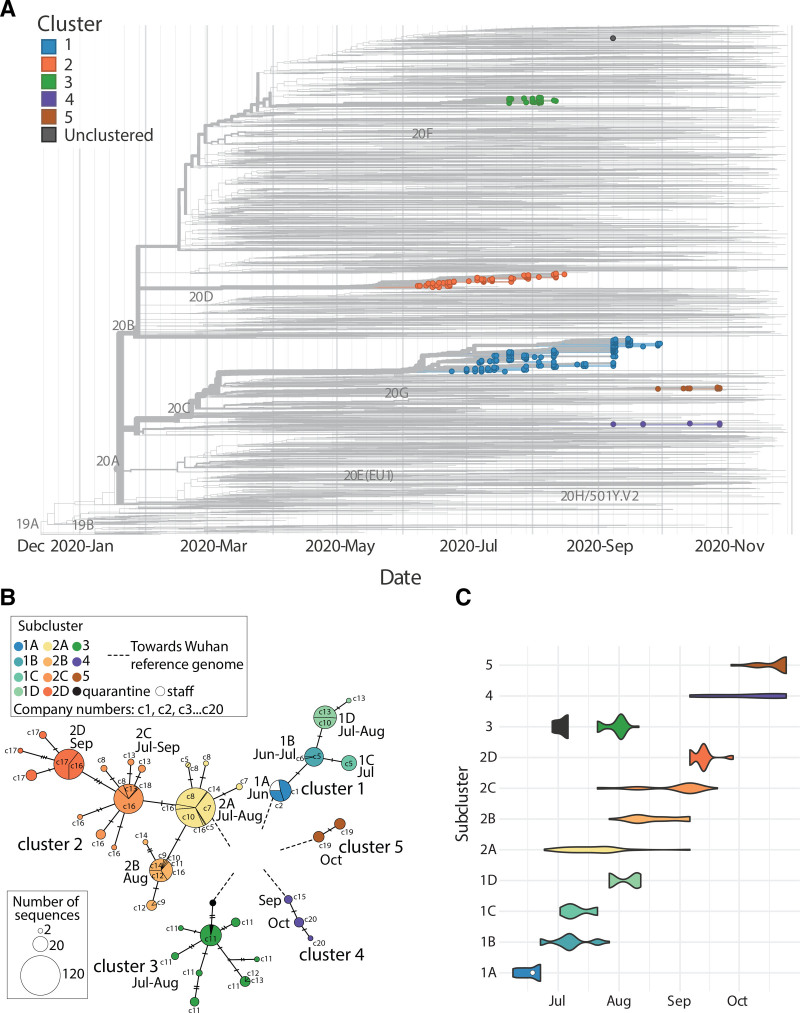
To characterize the transmission of SARS-CoV-2 among Marine recruits during basic training, we sequenced the viral genomes from 597 specimens collected from 580 participants who had tested positive by qPCR, had high viral load, and were representative of multiple companies at different time points. The selection of those 580 individuals out of the 1,107 who tested SARS-CoV-2-positive was skewed towards low Ct values (higher virus levels), making recovery of full length virus sequence feasible. Phylogenetic analysis using a global background with a focus on South Carolina revealed that 596 of these isolates belonged to one of five independent monophyletic transmission clusters, numbered 1 through 5 in order of their first detection (Figure 3A). For each participant with repeated sampling, the sequences of the different timepoints matched the first timepoint, and thus were collapsed and represented by the earliest isolate in the phylogenetic inferences. Cluster 1 was first detected on 26 May and shared ancestry with contemporaneous US isolates. Cluster 2 shared ancestry with 4 other sequences originating from South Carolina, suggesting possible local community introduction. Sequences reported from South Carolina were positioned basal to cluster 2, with collection dates between 4 June 2020, and 7 July 2020. One of these sequences was identical to the early cases in cluster 2 and was collected one day before the earliest detected case in MCRDPI on 24 June. Cluster 3 matched the viral genome from participants who had tested positive during the supervised quarantine,14 suggesting that the virus had spread from the quarantine site to MCRDPI. A single isolate that did not cluster with any of the outbreak clusters fell within other contemporaneous US isolates. In all clusters, mutations accumulated following each introduction, as the virus was transmitted from company to company over time, leading to subclusters with shared mutational profiles (Figure 3B and C; eTable 3; http://links.lww.com/EDE/B957). We also analyzed nine SARS-CoV-2 sequences (available in GenBank) obtained via public health surveillance from staff members who tested positive at MCRDPI on June 18. All nine staff infections overlapped in time with the first cluster outbreak at MCRDPI and belonged to subcluster 1A.
FIGURE 3.
Phylogenetic analysis and identification of transmission clusters. A, Time-calibrated maximum likelihood phylogenetic tree of the SARS-CoV-2 isolates from this study placed on a South Carolina-focused global background of sequenced isolates from GISAID. The five monophyletic clusters are indicated by color and numbered in chronological order of the first isolate of each. Each marker indicates the viral isolate from one participant B, Median joining network of SARS-CoV-2 isolates. Each node corresponds to isolates with a shared genetic profile, with the size of the node indicating the number of isolates with that profile. Edges indicate mutations separating isolates, with the number of separating mutations specified by the number of tick marks on the corresponding edge. The approximate month of subcluster identification in each company is indicated. Subclusters are color-coded, as indicated on the key. The companies in which the clusters were found are designated as c1 through c20. C, Violin plots showing the emergence of each transmission cluster and subcluster over time. Sequences from staff cases are included in cluster 1 as the white circle, and the cases from cluster 1 that occurred in the preceding quarantine are indicated in black.
No substitutions in the five transmission clusters involved loci mutated in known SARS-CoV-2 variants of concern. During CHARM, six subcluster-defining nonsynonymous mutations occurred after cluster introduction to MCRDPI, only one of which, S3883A, was present in more than a single subcluster (eTable 3; http://links.lww.com/EDE/B957). This mutation, which was found in the 1B subcluster and persisted in the later emerging 1C and 1D subclusters (eFigure 1A; http://links.lww.com/EDE/B957), is in the region encoding one of the accessory subunits (nsp7 S24A) of the RNA-dependent RNA polymerase (RdRp). S3883 is conserved among closely related sarbecoviruses, although alanine substitutions were found among a broader alignment of evolutionarily diverse orthocoronaviruses (eFigures 2 and 3; http://links.lww.com/EDE/B957). Although study participants were either asymptomatic or had mild disease, the presence of the S3883A mutation was associated with fewer symptoms. Because relatively few participants were women, we compared symptoms in the presence and absence of this mutation for all participants (eFigure 4; http://links.lww.com/EDE/B957) and for males only (eFigure 1B; http://links.lww.com/EDE/B957), with similar results (see eTable 4; http://links.lww.com/EDE/B957 for the summary data). Querying the GISAID database (https://www.gisaid.org/) on 8 July 2021, indicated that S3883A was reported in only 34 out of the 2,270,310 deposited SARS-CoV-2 sequences. Isolates with this mutation have been identified in Europe and North America (the regions with the majority of available sequences), with only seven reported in 2020. They are scattered among different genetic lineages, suggesting independent emergence of this mutation in different genetic backgrounds. No differences in transmission rates among the different isolates from the MCRDPI clusters were observed. Given the localization of S3883A in an RdRp subunit, we investigated whether this mutation affected viral replication. We investigated the replication of three viral isolates from subclusters 1B and 1C (A3883 variant) and two isolates from subcluster 1A (S3883) in cultured cells using qPCR quantification of SARS-CoV-2 E gene expression. All five viral isolates showed similar expression of the E gene, both in the commonly used Vero E6 cell line and in primary normal human epithelial (NHBE) cells (eFigure 5; http://links.lww.com/EDE/B957). These results suggest that the S3883A mutation does not alter viral replication efficiency.
Mathematical Modeling of SARS-CoV-2 Transmission Dynamics Within Recruit Companies
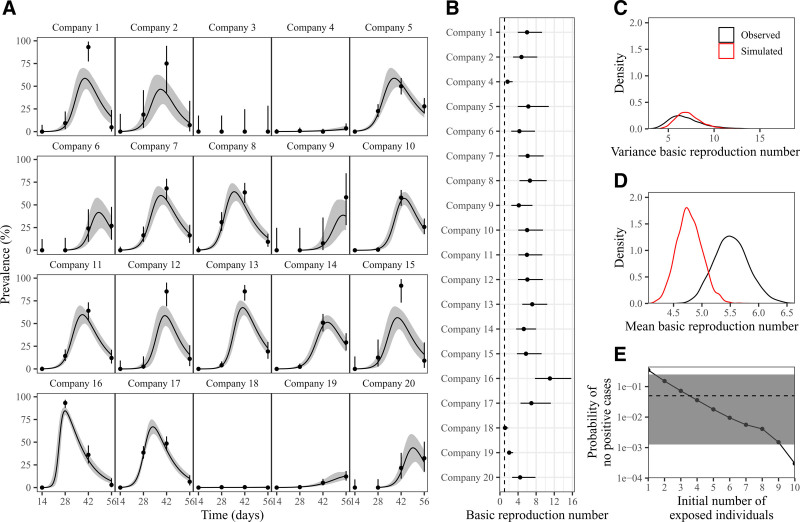
Next, we sought to better understand SARS-CoV-2 spread in the recruit setting and guide the implementation of more effective mitigation strategies. We explored transmission dynamics within companies using a deterministic model accounting for the temporal dynamics of viral infection (Figure 4A, eAppendix; http://links.lww.com/EDE/B957). The model underestimation of peak prevalence in some cases (e.g., companies 1, 12, 13, 15) may be due to stochasticity in real outbreaks; namely, if a large proportion of the company became infected at the same time (due to superspreading events), a deterministic model that assumes continuous changes in infection rates will poorly describe a large number of simultaneous infections. We were unable to fit our model to company 3 because no cases were detected.
FIGURE 4.
Modeling SARS-CoV-2 outbreaks in 20 companies. A, Model fits to qPCR prevalence data of SARS-CoV-2 in each company. Points and error bars represent the observed qPCR positivity and associated 95% confidence intervals. Solid lines and shaded regions represent the posterior median and 95% credible intervals of the predicted prevalence. B, Posterior median (points) and 95% credible intervals (error bars) of the estimated basic reproduction number. C, The distribution of variance of estimated basic reproduction numbers for real data (black) and simulated data (red). D, The distribution of mean of estimated basic reproduction numbers for real data (black) and simulated data (red). E, Quantification of the probability of not observing any positive cases across four biweekly periods when increasing the initial number of infections. Gray horizontal line and shaded areas represent the observed probability and the associated 95% binomial confidence interval.
We found considerable variation in basic reproduction number R0 estimates (from 1.1 (95% CI = 0.6, 1.8) in company 18 to 11.0 (95% CI = 7.7, 15.7) in company 16; Figure 4B), reflecting the variation in outbreak dynamics (Figure 4A). Since our model neglects the role of stochasticity, high R0 estimates might emerge by chance as a result of superspreading events. Nonetheless, compared with the median R0 of 2.79 reported for wild-type SARS-CoV-2,30 we estimate a high mean R0 (5.5; 95% CI = 5.0–6.1), indicating rapid SARS-CoV-2 spread within companies. Other transmission dynamics parameters are provided in eFigures 6–8; http://links.lww.com/EDE/B957.
To test the hypothesis that superspreading events alone could account for the variation in R0 estimates, we simulated 20 outbreaks using a stochastic model with R0 = 5.5 and low overdispersion parameter (k = 0.1), and fitted the aforementioned deterministic model. Using this model fitting, we matched the observed variance of R0 estimates (Figure 4C), suggesting that variation in R0 estimates is likely driven by deterministic approximation of a stochastic process rather than by real differences in R0. Similarly, model fits to simulated outbreaks matched the dynamics observed in actual outbreaks (eFigure 9; http://links.lww.com/EDE/B957).
Despite consistent variance estimates, fits to simulated outbreaks gave lower mean R0 than actual outbreaks (Figure 4D). This may be due to frequent fade-out events from stochasticity. We examined whether introducing a greater number of infections at the beginning of an epidemic may result in a better match. When quantifying the probability of no positive cases across three biweekly periods (Figure 4E), we found that initially having a single infected (exposed) individual results in substantially more frequent fade-out events than observed. This supports the hypothesis that company-to-company spread occurred via staff, i.e., by initial infection of multiple company members simultaneously.
The stochastic model matched the distribution of cumulative proportion of infections among companies, further supporting our hypothesis about the role of stochasticity in real outbreaks (eFigure 10; http://links.lww.com/EDE/B957). We found that R0 = 4.5 and k = 0.75 best described the data, despite an uncertainty in k estimates. While these estimates suggest a slightly lower R0 than found by fitting deterministic models and a higher k than previously reported,31 we find that a wide range of overdispersion parameter k is consistent with the data. While it is difficult to assess whether transmission patterns in this cohort were substantially less dispersed than previously reported, we are still able to rule out high values of k (>2). Our analysis supports the notion that superspreading events were likely drivers of the outbreak dynamics within companies.
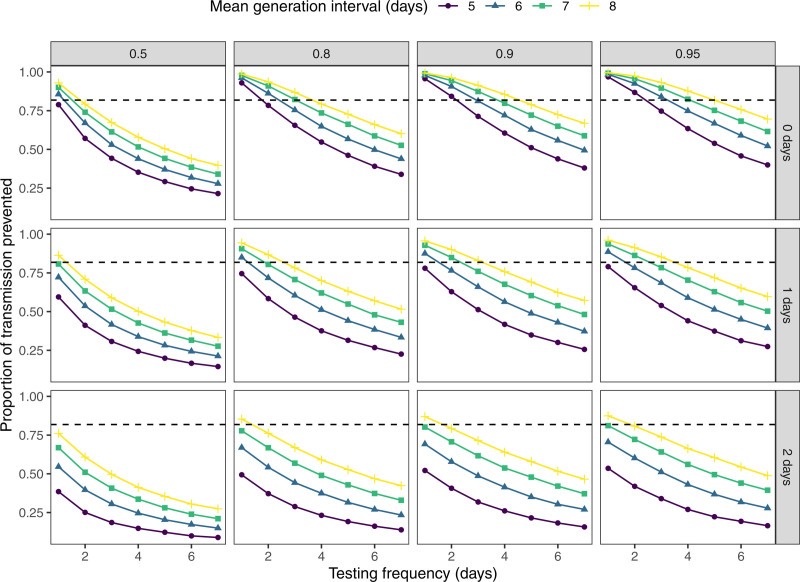
Finally, we used stochastic simulations to evaluate the effects of additional control measures in this high transmission setting, namely varying test sensitivity, test rapidity, and mean generation interval (i.e., time between infection and transmission).32 In particular, we simulate 50,000 infector–infectee pairs and vary the timing of transmission and the duration of latent period for each pair. We then ask what proportion of transmission can be prevented with frequent testing—for example, pairs with shorter latent periods and faster transmission are less likely to be caught by testing (see eAppendix; http://links.lww.com/EDE/B957). We found that frequent testing and rapid isolation alone can sufficiently reduce transmission to bring Reff below 1 even when R0 is as high as 5.5 (Figure 5). Generation interval plays a critical role in determining effectiveness of the testing strategy because faster transmission implies that fewer tests will be administered before transmission. Nonetheless, even when test sensitivity is as low as 0.5, we estimate that 75% of transmissions could be prevented by daily testing and immediate isolation of positive cases.
FIGURE 5.
Model-based prediction of the impact of testing frequency and generation intervals on viral transmission. Each point represents proportion of transmission prevented from periodic testing, calculated by simulating 50,000 infector–infectee pairs for a given value of the mean generation interval (i.e., time between infection and transmission; 5, 6, 7, or 8 days), testing sensitivity (0.5, 0.8, 0.9, or 0.95), and delay from testing to positive result and isolation (0, 1, or 2 days). Dashed, horizontal lines represent the amount of transmission that needs to be prevented to reduce Reff below 1 when R0 = 5.5.
DISCUSSION
The longitudinal CHARM study, in which serial testing was performed prospectively before infection, provides a unique window into epidemiology and cluster evolution in an isolated population that was almost entirely free of active or previous infection. Over 90% of participants tested negative for SARS-CoV-2 antibodies upon arrival at the supervised quarantine.21 The decreased incidence of non-SARS-CoV-2 ARI following implementation of COVID-19 mitigation strategies is consonant with the reported reduction in respiratory illness in military settings following similar control measures.33,34 Still, nearly half of participants became SARS-CoV-2-positive within 6 weeks, providing insight into the virus outbreak dynamics.
Our R0 estimate is about double that observed in community settings for wild-type infection.30,35 SARS-CoV-2 variants at MCRDPI predate and lack the mutations seen in recent highly transmissible variants of concern. Thus, the increased transmissibility cannot be attributed to the virus strains’ characteristics and is presumably related to the environmental setting. Close-quartered living conditions for military trainees increase the risk of respiratory infections.36 The high transmission rate of SARS-CoV-2 despite masking and social distancing indicates a disproportionate capacity to transmit during recruit training. High reproductive rates have been reported in other congregate settings, with high rates of asymptomatic infection.8,10,13,37 One factor likely contributing to the high R0 is the prevalence of asymptomatic SARS-CoV-2 infections in young people, which confounds identification and isolation.3
No staff was shared between the quarantine site and MCRDPI. We speculate that cluster 3, which was first identified during quarantine, may have been introduced by a recruit who, despite being infected, tested qPCR negative just before transferring to basic training. This illustrates the challenge of complete containment through quarantine, and the importance of implementing quarantine procedures including isolation of infected individuals and administration of multiple tests over a quarantine period. Otherwise, low-level transmission during quarantine may lead to an early infection at the time of qPCR testing, increasing the likelihood of a false negative.38 Phylogenetics and modeling suggest that other clusters may have been introduced by contact with staff. Besides the infection of nine staff members by subcluster 1A, additional observations implicating staff in the virus spread include: limited direct contact between members of different companies; simultaneous rapid spread of subclusters into multiple companies; mathematical modeling suggesting that multiple company members were often infected at the onset of detection within a company. Although we sequenced samples from 52% of participants, specimen Ct levels and limitations of our sequencing throughput precluded sequencing a greater percentage participant samples. However, the emergence of a mutation associated with decreased symptom reporting during the study illustrates the rapidity with which SARS-CoV-2 can evolve. Our work provides a high-resolution map of transmission and the spread of mutations among young, healthy participants, which could contribute to understanding viral transmission in other densely populated settings.
False-negative qPCR results or missed infections between biweekly sampling times might have biased our estimates of transmission, and loss-to-follow-up (21% of participants). Factors that may have contributed to loss-to-follow-up include participants dropping out to focus on training, leaving the Marines, or being removed from the base for medical or administrative reasons. Despite these limitations, the magnitude of R0, superspreading scale, and pattern of mutation spread are likely to be robust estimates. Given the data, there was little insight into how disease severity might modulate transmission dynamics. Separating the independent impact of specific nonpharmaceutical interventions was difficult, as they were implemented cumulatively, and ascertaining their potential effect on curbing SARS-CoV-2 transmission. After rapidly spreading through individual companies, the first three cluster strains were no longer detected within a few months. Thus, besides the value of company isolation, additional measures such as reducing contact among staff and monitoring their infection status might further suppress company-to-company transmission.
Our analysis of the effect of serial testing on preventing transmission relies on several simplifying assumptions. While our generation-interval framework implicitly accounts for variation in the timing of infectiousness, we do not account for the possibility that the duration of infectiousness may be correlated with the test sensitivity. For example, individuals with shorter infectious periods may have be less likely to test positive due to low viral load and faster viral decay. In addition, our analysis assumes that all individuals participate in the testing plan at a fixed interval—in practice, some individuals may be less compliant with the testing schedule than others. Therefore, while our analysis is likely to overestimate the effectiveness of frequent testing, it can still help prevent transmission, especially when they are coupled with other intervention measures.
Detection bias also limits our ability to draw an inference regarding causation from the role of nonpharmaceutical interventions on the decrease in other respiratory illnesses due to: (i) changes in health seeking behavior and provider testing patterns in 2020 and (ii) the likelihood of a partial immunity against other respiratory illnesses and their lower transmission rate compared with SARS-CoV-2. However, such a striking reduction in rates from 2020 as compared with the three previous years is likely related to the implementation of those mitigation efforts and is largely supported by other studies.22–24
Understanding the drivers of efficient propagation and increased transmissibility of new SARS-CoV-2 variants is increasingly important for defining ways to control transmission in general and in congregate settings. Our results support the hypothesis that, besides the mitigation measures implemented, rapid and frequent testing followed by quickly isolating positive cases can effectively control SARS-CoV-2 spread despite the high transmission rate in this setting. Thus, despite the concerns raised about the lower sensitivity of some rapid SARS-CoV-2 tests,39 low-cost rapid readout tests that compromise sensitivity could play an important role in augmenting public health measures to suppress SARS-CoV-2 transmission.
ACKNOWLEDGMENTS
We gratefully acknowledge the authors, originating and submitting laboratories of the genetic sequence and metadata made available through GISAID that were used as background to place our samples in the context of the local and global diversity. We thank the Navy and Marine Corps Public Health Center for historical passive surveillance data on acute respiratory infection (ARI) and pneumonia; Mary Anne Amper, Nitish Seenarine, Mital Vasoya, Sagie Mofsowitz, and Natalia Mendelev for technical assistance; the many U.S. Navy corpsmen who assisted in the logistics and sample acquisition; and the Marine Corps recruits who volunteered for this study. We thank Lisa Miorin and Teresa Aydillo for their help in establishing the in vitro assays of severe acute respiratory syndrome coronavirus 2 (SARS-CoV-2) infections in the BSL3 laboratory. We thank Stacie Dodgson from Life Science Editors for editorial comments on the article.
Supplementary Material
Footnotes
R.A.L., R.S.G.S., S.W.P., G.R.S., C.K.P., A.S.G.-R., Y.G., C.M.M., A.G.L., and S.C.S. contributed equally to this work.
This work was supported by a grant (9700130) from the Defense Health Agency through the Naval Medical Research Center and by the Defense Advanced Research Projects Agency (contract number N6600119C4022). Computational resources and staff expertise were provided by Scientific Computing at the Icahn School of Medicine at Mount Sinai (award numbers S10OD018522 and S10OD026880). Sequencing and bioinformatics analyses were supported by Work Unit Number (WUN) A1417, COVID-19 Health Action Response for Marines (CHARM), and Global Emerging Infections Surveillance (GEIS) P0013_20_AH_01.01 to KAB-L.
R.A.L., C.K.P., C.W.G., M.S.T., S.E.L., R.Z.C., C.E.A., J.J.M., E.N., E.S.A., M.A.S., D.R.S., P.S., D.L.W., K.A.B.L., and A.G.L. are military service members or GS employees. The other authors have no conflicts to report. This work was prepared as part of their official duties. Title 17, U.S.C., §105 provides that copyright protection under this title is not available for any work of the U.S. Government. Title 17, U.S.C., §101 defines a U.S. Government work as a work prepared by a military service member or employee of the U.S. Government as part of that person’s official duties.
The views expressed in the article are those of the authors and do not necessarily express the official policy and position of the US Navy, the Department of Defense, the US government or the institutions affiliated with the authors.
All sequencing data and code used in the analysis were deposited in the Global Initiative on Sharing Avian Influenza Data (GISAID) database (Accession Numbers EPI_ISL_2894633 to 2894868 and EPI_ISL_5441341 to EPI_ISL_5441705) and in a dedicated GitHub Repository (https://github.com/parksw3/navy). Sequences obtained from staff members are available in GenBank as Accession Numbers MW940533 through MW940541.
Supplemental digital content is available through direct URL citations in the HTML and PDF versions of this article (www.epidem.com).
REFERENCES
- 1.Wang D, Hu B, Hu C, et al. Clinical characteristics of 138 hospitalized patients with 2019 novel coronavirus-infected pneumonia in Wuhan, China. JAMA. 2020;323:1061–1069. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Han D, Li R, Han Y, Zhang R, Li J. COVID-19: insight into the asymptomatic SARS-COV-2 infection and transmission. Int J Biol Sci. 2020;16:2803–2811. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Johansson MA, Quandelacy TM, Kada S, et al. SARS-CoV-2 transmission from people without COVID-19 symptoms. JAMA Netw Open. 2021;4:e2035057. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Sah P, Fitzpatrick MC, Zimmer CF, et al. Asymptomatic SARS-CoV-2 infection: a systematic review and meta-analysis. Proc Natl Acad Sci U S A. 2021;118:e2109229118. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.McMichael TM, Clark S, Pogosjans S, et al. ; Public Health – Seattle & King County, EvergreenHealth, and CDC COVID-19 Investigation Team. COVID-19 in a long-term care facility - King County, Washington, February 27-March 9, 2020. MMWR Morb Mortal Wkly Rep. 2020;69:339–342. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Roxby AC, Greninger AL, Hatfield KM, et al. Outbreak investigation of COVID-19 among residents and staff of an independent and assisted living community for older adults in Seattle, Washington. JAMA Intern Med. 2020;180:1101–1105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.James A, Eagle L, Phillips C, et al. High COVID-19 attack rate among attendees at events at a Church - Arkansas, March 2020. MMWR Morb Mortal Wkly Rep. 2020;69:632–635. [DOI] [PubMed] [Google Scholar]
- 8.Moriarty LF, Plucinski MM, Marston BJ, et al. ; CDC Cruise Ship Response Team; California Department of Public Health COVID-19 Team; Solano County COVID-19 Team. Public health responses to COVID-19 outbreaks on cruise ships - worldwide, February-March 2020. MMWR Morb Mortal Wkly Rep. 2020;69:347–352. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Hagan LM, Williams SP, Spaulding AC, et al. Mass testing for SARS-CoV-2 in 16 prisons and jails - six jurisdictions, United States, April-May 2020. MMWR Morb Mortal Wkly Rep. 2020;69:1139–1143. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Njuguna H, Wallace M, Simonson S, et al. Serial laboratory testing for SARS-CoV-2 infection among incarcerated and detained persons in a correctional and detention facility - Louisiana, April-May 2020. MMWR Morb Mortal Wkly Rep. 2020;69:836–840. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Baettig SJ, Parini A, Cardona I, Morand GB. Case series of coronavirus (SARS-CoV-2) in a military recruit school: clinical, sanitary and logistical implications. BMJ Mil Health. 2021;167:251–254. [DOI] [PubMed] [Google Scholar]
- 12.Crameri GAG, Bielecki M, Züst R, Buehrer TW, Stanga Z, Deuel JW. Reduced maximal aerobic capacity after COVID-19 in young adult recruits, Switzerland, May 2020. Euro Surveill. 2020;25:2001542. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Kasper MR, Geibe JR, Sears CL, et al. An outbreak of Covid-19 on an aircraft carrier. N Engl J Med. 2020;383:2417–2426. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Letizia AG, Ramos I, Obla A, et al. SARS-CoV-2 transmission among marine recruits during quarantine. N Engl J Med. 2020;383:2407–2416. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Michael NL. SARS-CoV-2 in the U.S. military - lessons for civil society. N Engl J Med. 2020;383:2472–2473. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Pirnay JP, Selhorst P, Cochez C, et al. Study of a SARS-CoV-2 outbreak in a Belgian military education and training center in Maradi, Niger. Viruses. 2020;12:949. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Talmy T, Tsur A, Shabtay O. Duration of SARS-CoV-2 detection in Israel Defense Forces soldiers with mild COVID-19. J Med Virol. 2021;93:608–610. [DOI] [PubMed] [Google Scholar]
- 18.Ramos I, Goforth C, Soares-Schanoski A, et al. Antibody responses to SARS-CoV-2 following an outbreak among marine recruits with asymptomatic or mild infection. Front Immunol. 2021;12:681586. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Liu Y, Morgenstern C, Kelly J, Lowe R, Jit M; CMMID COVID-19 Working Group. The impact of non-pharmaceutical interventions on SARS-CoV-2 transmission across 130 countries and territories. BMC Med. 2021;19:40. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Marcus JE, Frankel DN, Pawlak MT, et al. COVID-19 monitoring and response among U.S. Air Force Basic Military Trainees - Texas, March-April 2020. MMWR Morb Mortal Wkly Rep. 2020;69:685–688. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Letizia AG, Ge Y, Vangeti S, et al. SARS-CoV-2 seropositivity and subsequent infection risk in healthy young adults: a prospective cohort study. Lancet Respir Med. 2021;9:712–720. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.McNeil JC, Flores AR, Kaplan SL, Hulten KG. The indirect impact of the SARS-CoV-2 pandemic on invasive group A Streptococcus, Streptococcus pneumoniae and Staphylococcus aureus infections in Houston area children. Pediatr Infect Dis J. 2021;40:e313–e316. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Nagakubo Y, Hirotsu Y, Maejima M, et al. Non-pharmaceutical interventions during the COVID-19 epidemic changed detection rates of other circulating respiratory pathogens in Japan. PLoS One. 2022;17:e0262874. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Tanislav C, Kostev K. Fewer non-COVID-19 respiratory tract infections and gastrointestinal infections during the COVID-19 pandemic. J Med Virol. 2022;94:298–302. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Stewart JN. DoD Instruction 6490.03, “Deployment Health.” 2019. Available at: https://www.esd.whs.mil/Portals/54/Documents/DD/issuances/dodi/649003p.pdf. Accessed June 19, 2019.
- 26.Bandelt HJ, Forster P, Röhl A. Median-joining networks for inferring intraspecific phylogenies. Mol Biol Evol. 1999;16:37–48. [DOI] [PubMed] [Google Scholar]
- 27.Leigh JW, Bryant D. POPART: full-feature software for haplotype network construction. Methods Ecol Evol. 2015;6:1110–1116. [Google Scholar]
- 28.Lavezzo E, Franchin E, Ciavarella C, et al. ; Imperial College COVID-19 Response Team; Imperial College COVID-19 Response Team. Suppression of a SARS-CoV-2 outbreak in the Italian municipality of Vo’. Nature. 2020;584:425–429. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Carpenter B, Gelman A, Hoffman MD, et al. Stan: a probabilistic programming language. J Stat Softw. 2017;76:32. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Liu Y, Rocklov J. The reproductive number of the Delta variant of SARS-CoV-2 is far higher compared to the ancestral SARS-CoV-2 virus. J Travel Med. 2021;28:taab124. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Endo A, Abbott S, Kucharski AJ, et al. ; Centre for the Mathematical Modelling of Infectious Diseases. Estimating the overdispersion in COVID-19 transmission using outbreak sizes outside China. Wellcome Open Res. 2020;5:67. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Svensson A. A note on generation times in epidemic models. Math Biosci. 2007;208:300–311. [DOI] [PubMed] [Google Scholar]
- 33.Kak V. Infections in confined spaces: cruise ships, military barracks, and college dormitories. Infect Dis Clin North Am. 2007;21:773–784, ix–x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Lee T, Jordan NN, Sanchez JL, Gaydos JC. Selected nonvaccine interventions to prevent infectious acute respiratory disease. Am J Prev Med. 2005;28:305–316. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Petersen E, Koopmans M, Go U, et al. Comparing SARS-CoV-2 with SARS-CoV and influenza pandemics. Lancet Infect Dis. 2020;20:e238–e244. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Gray GC, Callahan JD, Hawksworth AW, Fisher CA, Gaydos JC. Respiratory diseases among U.S. military personnel: countering emerging threats. Emerg Infect Dis. 1999;5:379–385. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Pettirosso E, Giles M, Cole S, Rees M. COVID-19 and pregnancy: a review of clinical characteristics, obstetric outcomes and vertical transmission. Aust N Z J Obstet Gynaecol. 2020;60:640–659. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Kucirka LM, Lauer SA, Laeyendecker O, Boon D, Lessler J. Variation in false-negative rate of reverse transcriptase polymerase chain reaction-based SARS-CoV-2 tests by time since exposure. Ann Intern Med. 2020;173:262–267. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Mina MJ, Peto TE, García-Fiñana M, Semple MG, Buchan IE. Clarifying the evidence on SARS-CoV-2 antigen rapid tests in public health responses to COVID-19. Lancet. 2021;397:1425–1427. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.