Abstract
A new insertion sequence (IS) element, IS679 (2,704 bp in length), has been identified in plasmid pB171 of enteropathogenic Escherichia coli B171. IS679 has imperfect 25-bp terminal inverted repeats (IRs) and three open reading frames (ORFs) (here called tnpA, tnpB, and tnpC). A plasmid carrying a composite transposon (Tn679) with the kanamycin resistance gene flanked by an intact IS679 sequence and an IS679 fragment with only IRR (IR on the right) was constructed to clarify the transposition activity of IS679. A transposition assay done with a mating system showed that Tn679 could transpose at a high frequency to the F plasmid derivative used as the target. On transposition, Tn679 duplicated an 8-bp sequence at the target site. Tn679 derivatives with a deletion in each ORF of IS679 did not transpose, finding indicative that all three IS679 ORFs are essential for transposition. The tnpA and tnpC products appear to have the amino acid sequence motif characteristic of most transposases. A homology search of the databases found that a total of 25 elements homologous to IS679 are present in Agrobacterium, Escherichia, Rhizobium, Pseudomonas, and Vibrio spp., providing evidence that the elements are widespread in gram-negative bacteria. We found that these elements belong to the IS66 family, as do other elements, including nine not previously reported. Almost all of the elements have IRs similar to those in IS679 and, like IS679, most appear to have duplicated an 8-bp sequence at the target site on transposition. These elements have three ORFs corresponding to those in IS679, but many have a mutation(s) in an ORF(s). In almost all of the elements, tnpB is located in the −1 frame relative to tnpA, such that the initiation codon of tnpB overlaps the TGA termination codon of tnpA. In contrast, tnpC, separated from tnpB by a space of ca. 20 bp, is located in any one of three frames relative to tnpB. No common structural features were found around the intergenic regions, indicating that the three ORFs are expressed by translational coupling but not by translational frameshifting.
Insertion sequences (ISs) comprise a large group of bacterial transposable DNA elements. These elements vary in size from 0.7 to 3.5 kb and have imperfect terminal inverted repeat sequences (IRs) of 10 to 40 bp in length (for recent reviews, see references 16 and 20). IS elements generally encode transposase, which is required for transposition, and duplicate a sequence of several base pairs at the target site on transposition. Based on the homology of their transposase genes, IS elements are classified into a number of families (see references 16 and 20). Most IS elements have an open reading frame (ORF) which is thought to encode transposase. Some elements, such as IS1 and IS3, have two ORFs, from which the transposase is produced by translational frameshifting (9, 28, 29, 30). Unless frameshifting occurs, a protein(s) is produced that acts as a transposition inhibitor (31).
IS679, which is present in two copies in plasmid pB171 of enteropathogenic Escherichia coli (EPEC) B171, is a large IS element (2,704 bp) with imperfect 25-bp IRs (34). Unlike other IS family elements, it has three ORFs (34). IS679 is flanked by direct repeats of an 8-bp sequence at the target site (34). A homology search found that IS679 is strikingly homologous to several IS elements, including the early isolate IS66 (34). Recently, 12 IS elements related to IS66 (designated the IS66 family) have been identified in Agrobacterium and Rhizobium spp. (for a review, see reference 16). Unlike IS679, many have more than three ORFs; for example, the early isolates IS66 (15) and IS866 (2) have, respectively, four and five ORFs. No study so far, however, has addressed the transposition capability and requirement of ORFs in IS66 family elements.
We here show that IS679, an IS66 family element, can transpose and needs all three of its ORFs for transposition. Based on results of a homology search, we show that the IS66 family is composed of at least 25 elements, including nine new ones, which are widely distributed in the gram-negative bacteria belonging to the genera Agrobacterium, Rhizobium, Escherichia, Pseudomonas, and Vibrio, Structural analyses showed that many of these elements have a mutation(s) in one or more of the three ORFs that correspond to those in IS679. Based on structural features present around the intergenic regions, we discuss the involvement of a translational coupling mechanism in the production of appropriate amounts of the ORF proteins encoded by IS66 family elements.
MATERIALS AND METHODS
Bacterial strains and plasmids.
The bacterial strains used were the E. coli K-12 derivatives XL1-Blue MRF′ (Stratagene), RZ211 [Δ(lac-pro) recA56 ara rpsL srl] (13), and RZ224 [polA Δ(lac-pro) ara thi rpsL Nalr Spcr lambdar] (36).
The plasmids used were the pGEM-T Easy vector (Promega), pOX38-Gen (13), and pB171, an IS679-carrying plasmid from EPEC B171 (0111:NM) (34). pHAN plasmids (pHAN103, pHAN104, pHAN105, and pHAN106) were constructed as described below. An alkaline lysis method (24) was used to prepare plasmid DNA for cloning and nucleotide sequencing.
Media, enzymes, and oligonucleotide primers.
Culture media used were L broth and L-rich broth (37). L-agar plates contained 1.5% (wt/vol) agar (Eiken) in L broth. When necessary, antibiotics were added to the L-agar plates at the following concentrations: ampicillin, 100 μg/ml; gentamicin, 7 μg/ml; kanamycin, 30 μg/ml; nalidixic acid, 20 μg/ml; and spectinomycin, 50 μg/ml. Restriction endonucleases (SacI, SacII, and SalI [Takara] and BsaI, BspEI, BsrGI, NsiI, and RsrII [New England Biolabs]) and T4 DNA ligase (Takara) were used with the buffers recommended by the suppliers. Oligonucleotide primers (Table 1) were synthesized chemically in an OLIGO1000M DNA synthesizer (Beckman).
TABLE 1.
Primers used in this study
| Primera | Sequence (5′ to 3′) |
|---|---|
| p01 | CGTCCATGAATATCAGCAGA |
| p02 | TCTGTGGTTACCGTGCTTGT |
| p03 | aagagctcTACGTCATTGAGCATATCCA |
| p04 | aaaatgcatTCTGTGGTTACCGTGCTTGT |
| p05 | CGTGTAGATAACTACGATACG |
| p06 | actccggaGAAAATCGGTTC |
| p07 | TGACGTTAACTCCGGAGCC |
| p08 | CGTCCGCGGAAGATGAACA |
| p09 | tcacccgcggTGGAAAGGT |
| p10 | TGCTGTACATCCCCGGACT |
Five pairs of primers (p01 and p02, p03 and p04, p05 and p06, p07 and p08, and p09 and p10) were used to amplify the fragments for construction of the pHAN plasmids. The additional nucleotides with a restriction site are shown by lowercase letters. Primers p03, p04, p06, p07, p08, and p09 have the respective restriction sites SacI, NsiI, BspEI, BspEI, SacII, and SacII. Sequences that are underlined show the restriction sites.
PCR and DNA sequencing.
The PCR was carried out by the standard protocol, with the following modification: 0.1 μg of the template plasmid DNA, each pair of primers, and 2.5 U of LA-Taq DNA polymerase (Takara) were used in a 50-μl solution. The step-cycle program (total of 30 cycles) was set to denature at 96°C for 30 s, anneal at 55°C for 30 s, and extend at 72°C for 2 min and 30 s. The PCR was done in a Perkin-Elmer Cetus Thermal Cycler. PCR products were separated in a 1.0% agarose gel.
DNA was sequenced by the dideoxynucleotide chain termination method (18, 25) with dye-labeled primers (−21M13 and PR1) and an ABI PRISM Dye Primer Cycle Sequencing Ready Reaction kit (Perkin-Elmer) with the AmpliTaq DNA polymerase, FS, or with a dye-labeled terminator DyeDeoxyTerminator Cycle Sequencing kit with AmpliTaq DNA polymerase (Perkin-Elmer) and the relevant oligodeoxyribonucleotide primers. The sequencing reaction was done with Catalyst A800 (Perkin-Elmer), and the reaction products were analyzed using ABI 373S-36 DNA Sequencer.
Plasmid construction.
To construct pHAN103 carrying Tn679 (see Fig. 1B), a plasmid carrying IS679 (designated pHAN101) first was constructed by ligation of a PCR-amplified fragment bearing IS679 by use of plasmid pB171 DNA as the template; primers p01 and p02, which hybridize to the regions flanking IS679B in pB171; and the linearized pGEM-T Easy vector in a TA cloning kit (Promega). Another plasmid (designated pHAN102) was then constructed by replacement of the SacI-NsiI segment of pHAN101 with the SacI-NsiI fragment bearing the IRR region of IS679, which was obtained in a PCR with the pHAN101 template DNA and the primers p03 and p04. Finally, pHAN103 was constructed by inserting the SalI-digested kanamycin gene Genblock (Pharmacia Biotech) bearing the Kmr gene into the SalI site of pHAN102.
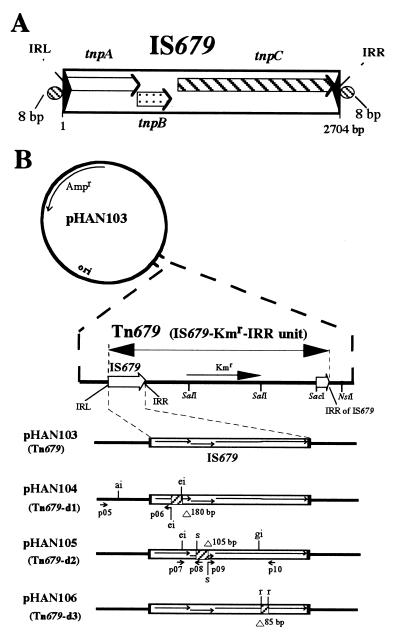
FIG. 1.
(A) Schematic representation of the IS679 structure. IS679 (2,704 bp) has imperfect 25-bp IRs. The IRs at the left and right inverted repeats (IRL and IRR) are indicated by solid triangles. Open, dotted, and cross-hatched arrows indicate, respectively, tnpA, tnpB, and tnpC. The two cross-hatched ovals flanking IS679 indicate direct repeats of an 8-bp target site sequence. (B) Schematic representations of the structures of pHAN plasmids. pHAN103 carries Tn679 with the kanamycin resistance gene (Kmr) between an intact IS679 sequence and the 3′-end region having IRR. Plasmids pHAN104, pHAN105, and pHAN106 carry a Tn679 derivative with deletions (hatched box) in tnpA, tnpB, and tnpC (thin arrows), respectively. Small solid arrows beneath the pHAN plasmid indicate primers used to construct each plasmid (see Materials and Methods). Primers with a tail indicate an additional sequence with a restriction site. s, SacII; ai, BsaI; ei, BspEI; gi, BsrGI; r, RsrII.
pHAN104 carrying Tn679-d1 was constructed by replacement of the BsaI-BspEI segment of pHAN103 with the BsaI-BspEI fragment which was amplified by PCR by using the pHAN103 template DNA and primers p05 and p06 in order to introduce a deletion in tnpA (see Fig. 1B).
pHAN105 carrying Tn679-d2 was constructed as follows (see Fig. 1B). The BspEI-SacII fragment was obtained from a PCR with the pHAN103 template DNA and primers p07 and p08. The SacII-BsrGI fragment also was obtained from a PCR with the pHAN103 template DNA and a pair of primers (p09 and p10) to introduce a deletion to tnpB (see Fig. 1B). The two fragments were mixed and treated with T4 DNA ligase. The resulting BspEI-BsrGI fragment was replaced with the BspEI-BsrGI segment of pHAN103, yielding pHAN105.
pHAN106 carrying Tn679-d3 was constructed by self-ligation of the pHAN103 DNA treated with RsrII (see Fig. 1B)
All of the ligated plasmids were introduced into E. coli XL1-Blue MRF′ by transformation. Cells harboring a plasmid were selected on L-agar plates containing ampicillin or kanamycin. The sequences of Tn679 derivatives were confirmed by DNA sequencing.
Mating assay.
The transposition of Tn679 carried by pHAN plasmids (pHAN103, pHAN104, pHAN105, and pHAN106) to the transferable plasmid pOX38-Gen was investigated with a standard mating assay that used the recA strain RZ211 harboring pOX38-Gen together with various pHAN plasmids as donors and RZ224 as the recipient. Donor cells that had been cultured overnight in 2 ml of Luria-Bertani (LB) broth containing gentamicin and kanamycin were washed and suspended in 2 ml of fresh LB broth. A 100-μl portion of this suspension was inoculated into 3 ml of fresh LB broth in a flask, and the whole was incubated without shaking at 37°C for 3 h. The recipient cells were cultured overnight in 6 ml of LB broth and then pelleted and suspended in 12 ml of fresh LB broth. This suspension was incubated with shaking at 37°C for 3 h, and 2.5 ml of this was added to the donor culture flask. Mating was done by incubating the flask at 37°C for 1 h without shaking. The mating culture was then plated on an L plate containing kanamycin, nalidixic acid, and spectinomycin, and with or without gentamicin. The transposition frequency was calculated by dividing the number of Genr Kmr Nalr Spcr transformants by the number of Kmr Nalr Spcr transformants.
Computer analysis.
The programs FASTA (21) and BLAST (1) were used for the homology search of the nucleotide sequences in the DDBJ, GenBank, and EMBL databases. Multiple sequences were aligned using the program CLUSTAL W, version 1.7 (33). Primary nucleotide sequences were analyzed with the programs HarrPlot 2.0 and GENETYX-Mac 10.1 system (Software Development Co.).
RESULTS
Transposition of the composite transposon Tn679 associated with IS679 and identification of the essential IS679 genes.
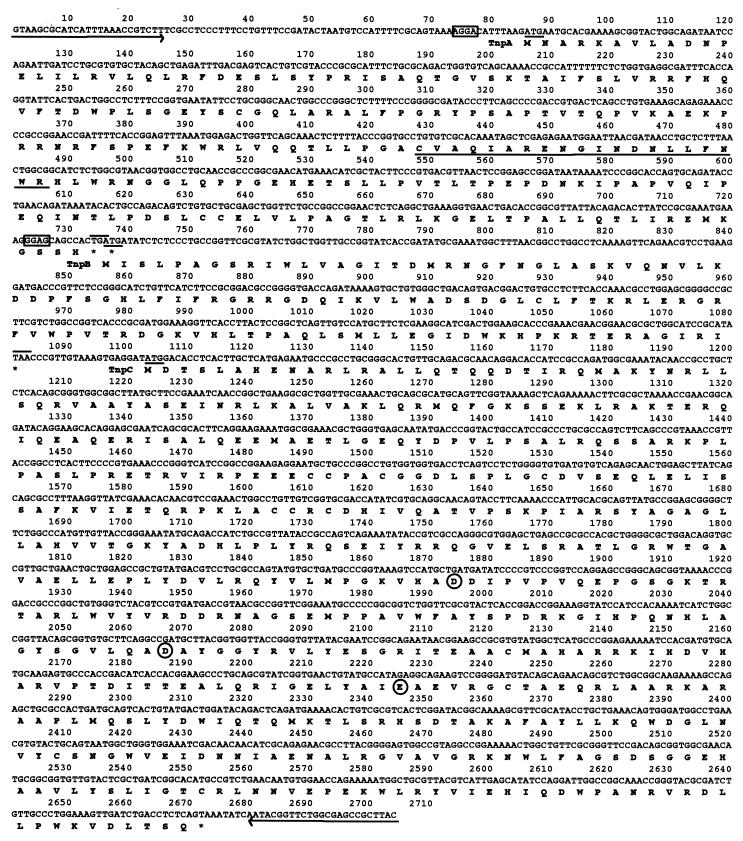
IS679, which has several structural features characteristic of an IS element, has three ORFs (here called tnpA, tnpB, and tnpC) (Fig. 1A and Fig. 2). tnpA (651 bp) and tnpB (345 bp) encode proteins of 24.2 and 13.1 kDa, respectively. tnpB is in the −1 frame with respect to tnpA, such that an ATG initiation codon of tnpB overlaps the TGA stop codon of tnpA. tnpC (1,572 bp) encodes a protein of 58.7 kDa. It is separated from tnpB by a space 20 bp in length and is located in the +1 frame with respect to tnpB.
FIG. 2.
Nucleotide sequence of IS679, showing three ORFs (tnpA, tnpB, and tnpC) and other structural features. The IRs of IS679 are shown by arrows. tnpA starts from an ATG codon at position 86 and ends with a TGA stop codon at position 736. The putative Shine-Dalgarno (SD) sequence is boxed. The potential α-helix–turn–α-helix DNA-binding motif in tnpA is underlined. tnpB starts from an ATG initiation codon at position 679 and ends with a TAA stop codon at position 1083. The putative SD sequence is boxed. tnpC starts from an ATG codon at position 1103 and ends with a TAA stop codon at position 2674. The amino acid sequences of the proteins encoded by tnpA, tnpB, and tnpC are shown below the nucleotide sequence. The potential DDE catalytic triad motif in TnpC is circled.
To examine whether IS679 with three ORFs transposes or not, we constructed the ampicillin resistance (Apr) plasmid pHAN103 with a DNA segment bearing the kanamycin resistance (Kmr) gene, which is flanked by an intact IS679 sequence and an IS679 fragment with IRR (IR on the right) (Fig. 1B). The segment (IS679-Kmr-IRR) is a composite transposon associated with IS679 and therefore was named Tn679 (Fig. 1B). Next, pHAN103 was introduced into an E. coli strain RZ211 (recA) which harbored the gentamicin resistance (Genr) plasmid pOX38-Gen, a transfer-proficient F plasmid derivative. The ability of transposition of Tn679 was investigated by a mating assay with RZ211 (recA) harboring pHAN103 and pOX38-Gen as the donor and the E. coli strain RZ224 that confers resistance to nalidixic acid (Nalr) and spectinomycin (Spcr) as the recipient. Transconjugants that had received pOX38-Gen with a Tn679 insertion were obtained as Genr Kmr Nalr Spcr colonies at the frequency of 2.0 × 10−5 (Table 2).
TABLE 2.
Frequency of the transposition of Tn679 and its derivatives
| Donor plasmid | Transposon | Frequencya |
|---|---|---|
| pHAN103 | Tn679 | 2.0 × 10−5 |
| pHAN104 | Tn679-d1 | <1.5 × 10−7 |
| pHAN105 | Tn679-d2 | <4.3 × 10−7 |
| pHAN106 | Tn679-d3 | <1.5 × 10−7 |
Transposition frequency was calculated as described in Materials and Methods.
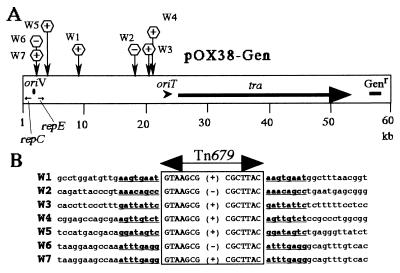
Plasmid DNAs were isolated from several Genr Kmr Nalr Spcr colonies, and their structures were examined by sequencing the junction regions between the pOX38-Gen and Tn679 sequences. Tn679 was found to be inserted into different sites on pOX38-Gen in one or the other orientation (Fig. 3A). Two plasmids (W6 and W7) had Tn679 at the same site, but their Tn679 orientations differed (Fig. 3A). An 8-bp sequence at each target site was duplicated on the transposition of Tn679 (Fig. 3B). As expected, Tn679 insertions occurred outside the replication genes of pOX38-Gen and the Genr gene used in selection of the transconjugants and the tra operon that encodes the proteins necessary for conjugation (Fig. 3B).
FIG. 3.
Target sites of Tn679 transposition. (A) Map positions and directions of the insertion of Tn679 into plasmid pOX38-Gen. Insertion products are indicated by W's plus a number. repE, oriV, and repC are the genes or sites required for pOX38-Gen replication. oriT and tra are required for plasmid transfer. Genr, gentamicin resistance gene. (B) Nucleotide sequences of pOX38-Gen around the insertion sites. Nucleotide sequences of the end regions of IS679 are boxed. The position of Tn679 is indicated at the top by a solid line with two arrowheads. Orientations of the Tn679 sequence inserted are indicated by “+” and “−,” with “+” being defined as in Fig. 1. Lowercase letters indicate the flanking sequences of Tn679, in which target site sequences duplicated on transposition are shown by underlined boldface letters.
To determine whether the three ORFs (tnpA, tnpB, and tnpC) in IS679 are essential for transposition, we constructed three mutants, each with the deletion of an IS679 ORF in Tn679 (Fig. 3B). Two of these mutants have an in-frame deletion in tnpA and tnpB which, respectively, produce proteins of 60 and 35 amino acid residues. No mutant was found to transpose (Table 2), evidence that all three ORFs in IS679 are required for transposition.
IS elements related to IS679.
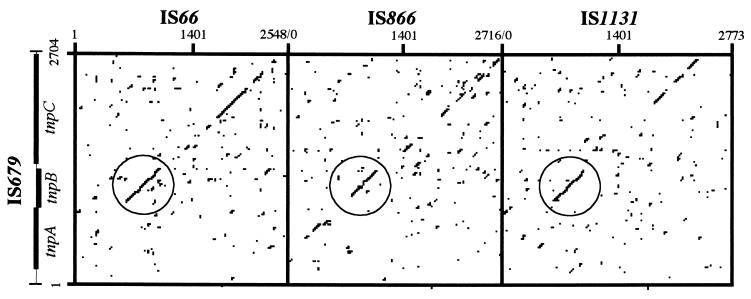
A computer-aided homology search of the databases was done with the IS679 sequence as the query. In all, 25 homologues were identified (Table 3), including 12 IS elements previously identified as IS66-related elements (16), 4 uncharacterized IS elements, and 9 new elements, here designated IS684, IS685, IS686, IS687, IS689, IS690, IS691, IS692, and IS693 (Table 3). Dot matrix analyses of IS679 with each of the three early isolates IS66, IS866, and IS1131 showed that these elements have significant homology, particularly in tnpB of IS679 (Fig. 4).
TABLE 3.
Members of the IS66 family
| IS elementa | Lengthb (bp) | IRc (bp) | TSDd (bp) | Sourcee | GenBank accession no.f | Orientationg | Positionh (bp) | Source or reference |
|---|---|---|---|---|---|---|---|---|
| IS679 | 2,704 | 25 | 8 | E. coli B171(pB171) | AB024946 | + | 41315–44018 | 34 |
| IS66 | 2,548 | 20 | 8 | A. tumefaciens(pTiA66) | M10204 | + | 31–2578 | 15 |
| IS866 | 2,716 | 27 | 8 | A. tumefaciens(pTiTm4) | M25805 | + | 1–2716 | 2 |
| IS1131 | 2,773 | 22 | 8 | A. tumefaciens PO22(pTi) | M82888 | + | 56–2828 | 35 |
| IS292 | 2,496 | 20 | 8 | Agrobacterium sp. strain X88–292 | L29283 | − | −1–2495 (+1) | 22 |
| IS71 | 2,386 | 13 | 8 | A. tumefaciens(pTi15955) | AF242881 | − | 161652–162393; 164958–166601 | 16; This study |
| IS682 | 2,533 | 24 | 8 | E. coli O157:H7 | NR | + | 1401098–1403630 | 11 |
| IS683 | (2,489) | E. coli O157:H7 | NR | + | 3854411–3856899 | 11 | ||
| IS684 | 2,040 | 30 | No | P. syringae pv. syringae B728a | AF232005 | + | 4058–6097 | This study |
| IS685 | 2,041 | 22 | No | P. putida TF4-1L carrying plasmid OCT | AJ245436 | + | 2311–4351 | This study |
| IS686 | (1,947) | Rhizobium sp. strain NGR234(pNGR234a) | AE000093 | − | 1782–3728 | This study | ||
| IS687 | (1,189) | R. leguminosarum(pRP2JI) | X84099 | − | 1–1189 | This study | ||
| IS689 | (2,584) | P. putida(pPGH1) | AF052749 | − | 1069–3652 | This study | ||
| IS690 | (1,473) | A. tumefaciens(pTi) | U96413 | + | 9209–9323 | This study | ||
| IS691 | (1,398) | V. harveyi BB7 | L26221 | + | 2915–4312 | This study | ||
| IS692 | 2,563 | 20 | 8 | A. tumefaciens(pTi15955) | AF242881 | + | 138423–140985 | This study |
| IS693 | 3,078 | 25 | 8 | Rhizobium sp. strain NGR234(pNGR234a) | AE000079 | + | 4887–7964 | This study |
| IS1313 | 2,547 | 24 | 8 | A. tumefaciens(pTiBo542) | U19149 | + | 1–2547 | 6 |
| ISEc8 | 2,442 | 22 | 8 | E. coli EDL933 | AF071034 | + | 6012–8453 | 26 |
| ISR11 | 2,495 | 20 | ? | R. leguminosarum bv. viciae 897 | L19650 | − | 1–2495 | Unpublished data |
| ISRm2 | ∼2,700 | 25 | 8 | R. meliloti 41(pRme41a and c) | M21471 | − | ND | 8 |
| ISRm14 | 2,695 | 22 | 9(?) | R. meliloti USDA1024 | AF134706 | + | 1–2695 | 26 |
| ISRm14-2 | 2,687 | 22 | 8 | R. meliloti A3 | U66830 | + | 8315–11001 | 26 |
| ISRsp1 | 3,481 | 22 | 8 | Rhizobium sp. strain NGR234(pNGR234a) | AE000077 | + | 5844–9324 | 16, 26 |
New IS elements named in this report are underlined. For their structures, see Fig. 6. ISRm2 belongs to the IS66 family, but its complete sequence is not found in the databases. IS867 in A. tumefaciens pTiTm4 is reported to have ca. 75% homology with IS866 (2), but because its complete sequence is not registered in the databases, it is not listed here. ISRm8 (1,235 bp) in R. meliloti is thought to be a part of an intact IS element (16, 27) but had only a segment with homology to tnpC and therefore is not listed.
Numbers in parentheses are the truncated element lengths.
Lengths of terminal IRs.
“No” indicates no target site duplication (TSD) in regions flanking an IS element.
A., Agrobacterium; E., Escherichia; R., Rhizobium; P., Pseudomonas; and V., Vibrio.
NR, sequence not registered in the databases.
Orientations of elements are defined as the transcriptional direction of tnpA, tnpB, and tnpC.
The position of IS292 is shown as −1 to 2495 (+1), because IS292 (2,496 bp) is registered as the 2,494-bp sequence. The position of IS71 is shown as 161652 to 162393 and 164958 to 166601 because IS71 is inserted by IS66. ND, not determined.
FIG. 4.
Dot matrix comparisons of the nucleotide sequences of IS679 with those of IS66, IS866, and IS1131. Open circles indicate the tnpB region of IS679 with significant homology to the same region in IS66, IS866, and IS1131. Dots are placed at locations where more than 25 of 50 nucleotides are identical.
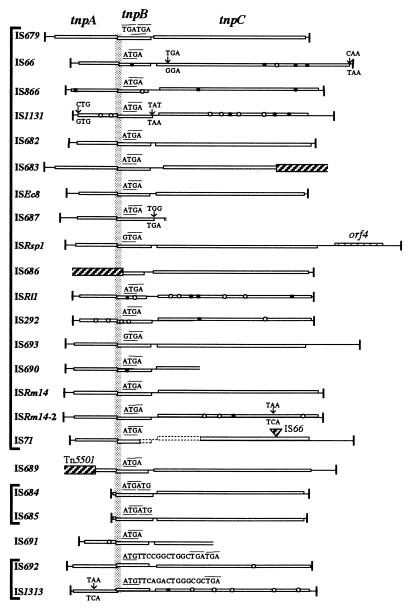
Seventeen elements were found to have imperfect IRs, 20 to 30 bp in length, whose terminal 7-bp sequences were conserved by the sequence 5′-GTAAGCG-3′ (Table 3 and Fig. 5). The other elements appeared to have a truncation at either end region, IRR or IRL, at which a non-IS sequence (such as transposon Tn5501) is present, except in elements with a partial sequence because of lack of information in the database sequences (Table 3 and Fig. 6).
FIG. 5.
Nucleotide sequences of the terminal regions of IS elements in the IS66 family. Nucleotide sequences at the left-end (IRL) and right-end (IRR) regions of each IS element are aligned from their 5′ ends. Identical nucleotides are indicated by asterisks. A consensus 7-bp terminal sequence derived from IRs of the IS66 family is shown at the top in boldface letters.
FIG. 6.
Schematic representation of the structures of IS66 family elements. Open rectangles in each element indicate three ORFs corresponding to tnpA, tnpB, and tnpC in IS679. tnpA is placed in the 0 frame on the thin solid line, which represents the length of each element; −1 and +1 frames are shown beneath and above the thin solid line, respectively. Mutations, considered to be present in an element, are suppressed at a relevant position(s) by the addition (open circles) or deletion (solid circles) of a nucleotide or by the substitution of a nucleotide in a codon (small vertical arrows) in order to deduce a coding region(s). Small thick vertical lines at the end(s) of each element indicate IRs. IS71 has an internal deletion in the middle region (dotted line) and is inserted by IS66 (2,556 bp; a cross-hatched triangle). IS689 is inserted by Tn5501 as shown by the cross-hatched large rectangles. IS683 and IS686 appear to be truncated in the 3′- and 5′-end regions due to the presence of a non-IS sequence, shown by large cross-hatched rectangles. The 5′ regions of IS687, IS686, IS689, IS690, and IS691 are not shown because of the limited information available in the databases. Vertical solid bars that cover all members indicate the intergenic regions between tnpA and tnpB in which there is a critical sequence. The termination codon of tnpA in these sequences has a line above, and the initiation codon of tnpB is underlined. An open dotted rectangle indicates an additional ORF (designated orf4) downstream of tnpC in ISRsp1.
Most elements with two IRs are flanked by direct repeat sequences of 8 bp (Table 3). Two of the new IS elements, IS684 and IS685, however, are not flanked by direct repeat sequences (Table 3). ISRm14 has been reported to be flanked by direct repeat sequences of 9 bp (26), but the existence of the sequences could not be confirmed because no such sequences are stored in the databases. A 2,687-bp IS element is present in Rhizobium meliloti A3 (26), here designated ISRm14-2 because it shows 96.5% identity to ISRm14 at the nucleotide sequence level. Noteworthy is that, like IS679, ISRm14-2 is flanked by direct repeat sequences of 8 bp (Table 3 and Fig. 5).
IS66-family elements are known to be present in a particular bacterial family, the Rhizobiaceae (genera Agrobacterium and Rhizobium) (16). The newly identified IS elements, however, are also present in the other genera, including Escherichia, Pseudomonas, and Vibrio, which belong to the gram-negative bacteria (Table 3).
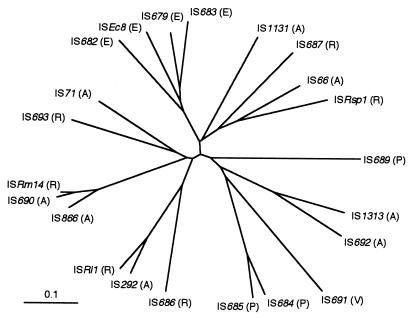
A phylogenetic analysis based on the nucleotide sequences of tnpB was done to assess the relationships of all the identified IS elements. The phylogenetic tree obtained shows that, except for E. coli, these IS elements do not fall into clusters by genera (Fig. 7).
FIG. 7.
Phylogenetic tree of IS66 family elements. The tree was constructed from tnpB nucleotide sequences of the IS66 family elements by the neighbor-joining method. The scale bar indicates a distance of 0.1.
Analysis of ORFs encoded by IS66-family elements.
Many of the IS elements identified had more than three ORFs, one or more of which appear to correspond to those in IS679 (Fig. 6). Comparison of the closely related sequences of the IS elements in the phylogenetic tree indicates that they have a substitution and/or frameshift mutation(s) within an ORF(s). In fact, sequence rearrangements produced by compensating mutations by the addition, deletion, or substitution of a nucleotide show that all of the IS elements have three ORFs that correspond to tnpA, tnpB, and tnpC in IS679 (Fig. 6).
An IS element, ISRsp1, identified in the Rhizobium sp. strain NGR234 plasmid pNGR234a, however, has an additional ORF (designated orf4) (Fig. 6) (26). The 5′-half region of orf4 is significantly homologous to the 3′-half region of the tnpR of the IS element IS1096 which belongs to a distinct IS family. IS1096 is 2,275 bp long and has tnpA and tnpR genes which are believed to encode proteins that function in transposition (3). The nucleotide sequences flanking the ISRsp1 region homologous to IS1096 are homologous to a segment in the TL-DNA of the Agrobacterium rhizogenes plasmid Ri, a root-inducing agropine-type plasmid (accession no. K03313) (32). Dot matrix analysis showed that the TL-DNA segment does not have the orf4 present in ISRsp1 (data not shown). These findings suggest that the orf4 in ISRsp1 was derived from a transposon with homology to IS1096 by insertion into an ancestor ISRsp1 element with three ORFs.
Two elements, IS684 and IS685 (2,040 and 2,041 bp, respectively), which are closely related as seen in the phylogenetic tree, are shorter than the other elements and have only two ORFs, which correspond to tnpB and tnpC in IS679 (Fig. 6). These elements, however, have a short segment that corresponds to the distal region of tnpA in IS679 (Fig. 6).
In the intergenic regions between the two ORFs that correspond to tnpA and tnpB in almost all the IS elements, the TGA termination codon of tnpA overlaps in the −1 frame with respect to the initiation codon ATG (rarely, GTG) of tnpB within the ATGA (or GTGA) sequence (Fig. 6). The two related elements, IS692 and IS1313, exceptionally have two ORFs corresponding to tnpA and tnpB, in which tnpB overlaps in a small region (11 and 14 bp, respectively) in the +1 frame with respect to tnpA between the ATG initiation codon of tnpB and the TGA termination codon of tnpA (Fig. 6).
In the three ORF products encoded by IS66 family elements, the TnpA proteins have significant homology with the OrfA protein encoded by IS2, a subfamily element of the IS3 family (see the TnpA protein from IS679 in Fig. 2), as does the protein encoded by ISRm14 (26). The homologous region between the TnpA and OrfA proteins had an α-helix–turn–α-helix DNA-binding motif (Fig. 2). The TnpC proteins encoded by IS66 family elements have a potential DDE catalytic triad motif (the motif in TnpC encoded by IS679 in Fig. 2). The TnpB proteins encoded by the IS66 family elements, however, do not have any of the motifs identified in the transposases encoded by the IS elements belonging to other IS families.
DISCUSSION
We have shown in this report that a composite transposon associated with IS679 transposes to many sites, giving rise to duplication of an 8-bp sequence at each target site. This shows that IS679 itself has the ability to transpose and duplicate an 8-bp target site sequence on its transposition, as expected from the observation that each of the two IS679 members present in plasmid pB171 of EPEC is flanked by direct repeat sequences of 8 bp (34). Moreover, in all, 25 homologues were identified, of which 13 elements with IRs having homology with those in IS679 are flanked by direct repeat sequence of 8 bp, a finding indicative that, like IS679, these elements also duplicate an 8-bp sequence at the target site on transposition. Two new IS elements (IS684 and IS685) with IRs, however, are not flanked by direct repeat sequences, suggesting that either the 5′- or 3′-end region has been removed through IS element-mediated genomic rearrangement by means of deletion or inversion, which often occurs after insertion into the initial target site.
IS66 family elements have been reported in the restricted bacterial family Rhizobiaceae (genera, Agrobacterium and Rhizobium) (16). The newly identified IS elements, however, were also present in Escherichia, Pseudomonas, and Vibrio spp. These families belong to the gram-negative bacteria, evidence that IS66 family elements are widespread in gram-negative bacteria. Phylogenetic analysis showed that, except E. coli, the IS elements do not form clusters by genera, a result indicating that IS66 family elements are transferred horizontally. Note that all the IS elements in the genus Agrobacterium are present in Ti plasmid (Table 3). The Ti plasmid has a narrow host range, being stably maintained only within Agrobacterium and Rhizobium species (12). This plasmid, however, is mobilized at a high frequency from Agrobacterium tumefaciens to E. coli and Pseudomonas fluorescens by heterologous mating, showing that the conjugal host range of the Ti plasmid extends to members of the families Enterobacteriaceae and Pseudomonadaceae (4). Interestingly, four E. coli IS elements (IS679, IS682, IS683, and ISEc8) are on one branch of the phylogenetic tree (Fig. 7). This means that horizontal transmission into E. coli occurred a long time ago.
We have shown in this report that a composite transposon associated with IS679 that has a mutation in tnpA, tnpB, or tnpC cannot transpose, providing evidence that all three ORFs are essential for IS679 transposition. We have also shown in this report that IS elements appear to have three ORFs that correspond to those of IS679, but many elements (including such early isolates as IS66 and IS866) have one or more frameshift and/or substitution mutations within an ORF(s). This suggests that, like IS679, IS66 family elements also require three ORFs for transposition, but many of them are defective ones with no transposition ability. It is notable that two related elements (IS684 and IS685) identified from different strains (P. syringae and P. putida, respectively) have two ORFs corresponding to tnpB and tnpC and a short DNA segment corresponding to the distal region of tnpA (see Fig. 6), indicating that these two IS elements have a deletion in their tnpA proximal regions. IS685 is present in an OCT plasmid, suggesting that these elements are derived from an element which has been transferred via the plasmid from one bacterium to another.
In the intergenic regions between tnpA and tnpB in IS679 and in almost all the other IS66 family elements, the initiation codon ATG (rarely GTG) of tnpB overlaps in the −1 frame with respect to the TGA termination codon of tnpA within the ATGA (or GTGA) sequence (see Fig. 6), as noted in four IS66 family elements (26). The two related elements (IS692 and IS1313), however, exceptionally had tnpB which overlapped in the +1 frame with respect to tnpA in the 11- and 14-bp regions (Fig. 6). It should be noted that the classification made by construction of a phylogenetic tree is in agreement with the structural features of the two IS elements. In contrast, in the intergenic regions between tnpB and tnpC in all of the IS elements, tnpB is separated by a 20-bp sequence and is located in the 0, −1, or +1 frame relative to tnpC (see Fig. 6).
Some IS elements have two ORFs, which are required for transposition (16, 20). The IS element IS3, for example, encodes two ORFs (orfA and orfB), in which orfB is in the −1 frame relative to orfA, and the termination codon of orfA overlaps the ATG codon of orfB in the ATGA sequence (28). The IS3 transposase is produced by a −1 translational frameshifting mechanism at the AAAAG sequence present in the overlapping region between orfA and the frame extending upward from orfB (28). Unless frameshifting occurs, both the OrfA and OrfB proteins (inhibitors of transposition) are produced by a translational coupling mechanism (31). The translational frameshifting requires a pseudoknot structure in the region downstream of the AAAAG sequence (28). No frameshifting signal sequence or pseudoknot structure was found in the intergenic regions between tnpA and tnpB or between tnpB and tnpC in IS679 and the other IS66 family elements. This suggests that the IS66 family elements may not produce a protein that is transposase by a translational frameshifting mechanism but may produce three proteins by a translational coupling mechanism, such that the ORF located distally is translated only after translation of the ORF located proximally, as in bacterial operons (7). By the translational coupling mechanism, messages from IS66 family elements may be translated to produce the amount of TnpB appropriate to that of TnpA and the amount of TnpC appropriate to that of TnpB.
Transposases encoded by many IS elements belonging to IS families other than the IS66 family have a DNA-binding domain with an α-helix–turn–α-helix DNA-binding motif and a catalytic domain with a DDE motif (16, 20). In IS66 family elements, the TnpA protein appears to have an α-helix–turn–α-helix DNA-binding motif, and the TnpC protein appears to have a potential DDE motif (Fig. 2). The tnpB proteins, however, seem to have no homology to any of the motifs identified in the transposases encoded by the IS elements of the different IS families. We assume that the TnpB protein and the TnpA and TnpC proteins are produced independently in appropriate amounts and form a complex, which acts as a transposase to promote the transposition of an IS66 family element.
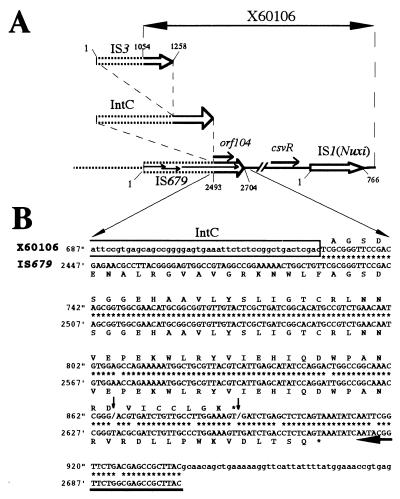
A homology search found that the distal end region of IS679 is homologous (95.8%) to a 210-bp DNA segment in the database sequence (accession no. X60106) (Fig. 8). This segment has an ORF (designated orf104 encoding a polypeptide of 53 amino acids) with significant homology to several IS66 family elements and is associated with a sequence of the group II self-splicing intron, IntC (10, 14). orf104 was found to correspond to the distal region of tnpC in IS679, and the homologue sequence is nested by IntC, and IntC is itself nested by IS3 (Fig. 8). Because multiple group II introns often are present within mobile DNA from E. coli (10), orf104 is speculated to be the signature of the presence of the group II intron IntC (14). Several kinds of group II introns were found to be inserted into various mobile genetic elements, e.g., Tn5397, the H-repeat, and several IS elements (IS629 [IS3411], IS911, and ISRm2011-2) (10, 17, 19, 23, 34), but not into IS66 family elements, a result indicating that orf104 is not necessarily a group II intron signature. As described earlier, an IS66 family element, IS689, is truncated in the 5′-end region which is the site of Tn5501 (see Fig. 6). This indicates that truncation often occurs by the insertion of a transposon, as in the case of the truncated IS679 homologue and IntC described above.
FIG. 8.
Nested structures of an IS679 member with a group II intron IntC in the enterotoxic E. coli O167:H5. (A) Proposed structure of the nested region based on the sequence (accession no. X60106) registered in the databases. Open arrows indicate IS elements [IS679, IS3, and IS1(Nuxi)] and IntC. IS679 is nested by IntC, and IntC is nested by IS3. The registered sequence (X60106) is indicated by a solid line with two arrowheads. The three ORFs in IS679 are indicated by thin arrows. Thick arrows indicate orf104, which corresponds to the distal region of tnpC of IS679, and the csvR gene, which is involved in the virulence of the enterotoxic E. coli strain (5). (B) Nucleotide sequence of a 210-bp segment showing critical structural features. The nucleotide sequence of IS679 is shown by uppercase letters. Asterisks indicate identical nucleotides in the IS679 and X60106 sequences. The nucleotide sequence of IntC, shown by lowercase letters, is boxed. Thin vertical arrows indicate possible deletion positions of nucleotides in X60106. Amino acids deduced from the IS679 and orf104 sequences are shown. A thick arrow indicates IRR of IS679.
ACKNOWLEDGMENTS
We thank W. Reznikoff for providing the E. coli strains used in this study and Y. Sekine for critical reading of the manuscript.
This research was supported by a grant-in-aid for scientific research from the Ministry of Education, Science, Sports, and Culture of Japan.
REFERENCES
- 1.Altschul S F, Gish W, Miller W, Myers E W, Lipman D J. Basic local alignment search tool. J Mol Biol. 1990;215:403–410. doi: 10.1016/S0022-2836(05)80360-2. [DOI] [PubMed] [Google Scholar]
- 2.Bonnard G, Vincent F, Otten L. Sequence and distribution of IS866, a novel T region-associated insertion sequence from Agrobacterium tumefaciens. Plasmid. 1989;22:70–81. doi: 10.1016/0147-619x(89)90037-1. [DOI] [PubMed] [Google Scholar]
- 3.Cirillo J D, Barletta R G, Bloom B R, Jacobs W R., Jr A novel transposon trap for mycobacteria: isolation and characterization of IS1096. J Bacteriol. 1991;173:7772–7780. doi: 10.1128/jb.173.24.7772-7780.1991. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Cook D M, Li P L, Ruchaud F, Padden S, Farrand S K. Ti plasmid conjugation is independent of vir: reconstitution of the tra functions from pTiC58 as a binary system. J Bacteriol. 1997;179:1291–1297. doi: 10.1128/jb.179.4.1291-1297.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.De Haan L A M, Willshaw G A, Van der Zeijst B A M, Gaastra W. The nucleotide sequence of a regulatory gene present on a plasmid in an enterotoxigenic Escherichia coli strain of serotype O167:H5. FEMS Microbiol Lett. 1991;83:341–346. doi: 10.1111/j.1574-6968.1991.tb04487.x. [DOI] [PubMed] [Google Scholar]
- 6.Deng W, Gordon M P, Nester E W. Sequence and distribution of IS1312: evidence for horizontal DNA transfer from Rhizobium meliloti to Agrobacterium tumefaciens. J Bacteriol. 1995;177:2554–2559. doi: 10.1128/jb.177.9.2554-2559.1995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Draper D E. Translational initiation. In: Neidhardt F C, et al., editors. Escherichia coli and Salmonella: cellular and molecular biology. 2nd ed. Washington, D.C.: American Society for Microbiology; 1996. pp. 902–908. [Google Scholar]
- 8.Dusha I, Kovalenko S, Banfalvi Z, Kondorosi A. Rhizobium meliloti insertion element ISRm2 and its use for identification of the fixX gene. J Bacteriol. 1987;169:1403–1409. doi: 10.1128/jb.169.4.1403-1409.1987. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Escoubas J M, Prère M F, Fayet O, Salvignol I, Galas D, Zerbib D, Chandler M. Translational control of transposition activity of the bacterial insertion sequence IS1. EMBO J. 1991;10:705–712. doi: 10.1002/j.1460-2075.1991.tb08000.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Ferat J L, Le Gouar M, Michel F. Multiple group II self-splicing introns in mobile DNA from Escherichia coli. C R Acad Sci III. 1994;317:141–148. [PubMed] [Google Scholar]
- 11.Hayashi T, Makino K, Ohnishi M, Kurokawa K, Ishii K, Yokoyama K, Han C-G, Ohtsubo E, Nakayama K, Murata T, Tanaka M, Tobe T, Iida T, Takami H, Honda T, Sasakawa C, Ogasawara N, Yasunaga T, Kuhara S, Shiba T, Hattori M, Shinagawa H. Genome sequence and comparative analysis of enterohemorrhagic E. coli O157:H7. DNA Res. 2001;8:11–22. doi: 10.1093/dnares/8.1.11. [DOI] [PubMed] [Google Scholar]
- 12.Hooykaas P J J, Klapwijk P M, Nuti M P, Schilperoort R A, Rorsch A. Transfer of Agrobacterium tumefaciens Ti plasmid to avirulent agrobacteria and Rhizobium ex planta. J Gen Microbiol. 1977;98:477–484. [Google Scholar]
- 13.Johnson R C, Reznikoff W S. Copy number control of Tn5 transposition. Genetics. 1984;107:9–18. doi: 10.1093/genetics/107.1.9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Knoop V, Brennicke A. Evidence for a group II intron in Escherichia coli inserted into a highly conserved reading frame associated with mobile DNA sequences. Nucleic Acid Res. 1994;22:1167–1171. doi: 10.1093/nar/22.7.1167. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Machida Y, Sakurai M, Kiyokawa S, Ubasawa A, Suzuki Y, Ikeda J E. Nucleotide sequence of the insertion sequence found in the T-DNA region of mutant Ti plasmid pTiA66 and distribution of its homologues in octopine Ti plasmid. Proc Natl Acad Sci USA. 1984;81:7495–7499. doi: 10.1073/pnas.81.23.7495. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Mahillon J, Chandler M. Insertion sequences. Microbiol Mol Biol Rev. 1998;62:725–774. doi: 10.1128/mmbr.62.3.725-774.1998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Martinez-Abarca F, Zekri S, Toro N. Characterization and splicing in vivo of a Sinorhizobium meliloti group II intron associated with particular insertion sequences of the IS630-Tc1/IS3 retroposon superfamily. Mol Microbiol. 1998;28:1295–1306. doi: 10.1046/j.1365-2958.1998.00894.x. [DOI] [PubMed] [Google Scholar]
- 18.Messing J. New M13 vectors for cloning. Methods Enzymol. 1983;101:20–78. doi: 10.1016/0076-6879(83)01005-8. [DOI] [PubMed] [Google Scholar]
- 19.Mullany P, Pallen M, Wilks M, Stephen J R, Tabaqchali S. A group II intron in a conjugative transposon from the gram-positive bacterium, Clostridium difficile. Gene. 1996;174:145–150. doi: 10.1016/0378-1119(96)00511-2. [DOI] [PubMed] [Google Scholar]
- 20.Ohtsubo E, Sekine Y. Bacterial insertion sequence. Curr Top Microbiol Immunol. 1996;204:1–26. doi: 10.1007/978-3-642-79795-8_1. [DOI] [PubMed] [Google Scholar]
- 21.Pearson W R, Lipman D J. Improved tools for biological sequence comparison. Proc Natl Acad Sci USA. 1988;85:2444–2448. doi: 10.1073/pnas.85.8.2444. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Ponsonnet C, Normand P, Pilate G, Nesme X. IS292: a novel insertion element from Agrobacterium. Microbiology. 1995;141:853–861. doi: 10.1099/13500872-141-4-853. [DOI] [PubMed] [Google Scholar]
- 23.Rajakumar K, Sasakawa C, Adler B. Use of a novel approach, termed island probing, identifies the Shigella flexneri she pathogenicity island which encodes a homolog of the immunoglobulin A protease-like family of proteins. Infect Immun. 1997;65:4606–4614. doi: 10.1128/iai.65.11.4606-4614.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Sambrook J, Fritsch E F, Maniatis T. Molecular cloning: a laboratory manual. 2nd ed. Cold Spring Harbor, N.Y: Cold Spring Harbor Laboratory Press; 1989. [Google Scholar]
- 25.Sanger F, Nicklen S, Coulson A R. DNA sequencing with chain-terminating inhibitors. Proc Natl Acad Sci USA. 1977;74:5463–5467. doi: 10.1073/pnas.74.12.5463. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Schneiker S, Kosier B, Puhler A, Selbitschka W. The Sinorhizobium meliloti insertion sequence (IS) element ISRm14 is related to a previously unrecognized IS element located adjacent to the Escherichia coli locus of enterocyte effacement (LEE) pathogenicity island. Curr Microbiol. 1999;39:274–281. doi: 10.1007/s002849900459. [DOI] [PubMed] [Google Scholar]
- 27.Schwedock J, Long S R. An open reading frame downstream of Rhizobium meliloti nodQ1 shows nucleotide sequence similarity to an Agrobacterium tumefaciens insertion sequence. Mol Plant-Microbe Interact. 1994;7:151–153. doi: 10.1094/mpmi-7-0151. [DOI] [PubMed] [Google Scholar]
- 28.Sekine Y, Eisaki N, Ohtsubo E. Translational control in production of transposase and in transposition of insertion sequence IS3. J Mol Biol. 1994;235:1406–1420. doi: 10.1006/jmbi.1994.1097. [DOI] [PubMed] [Google Scholar]
- 29.Sekine Y, Ohtsubo E. Frameshifting is required for production of the transposase encoded by insertion sequence 1. Proc Natl Acad Sci USA. 1989;86:4609–4613. doi: 10.1073/pnas.86.12.4609. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Sekine Y, Ohtsubo E. Translational frameshifting in IS elements and other genetic systems. In: Kimura M, Takahata N, editors. New aspects of the genetics of molecular evolution. Berlin, Germany: Springer-Verlag; 1991. pp. 243–261. [Google Scholar]
- 31.Sekine Y, Izumi K, Mizuno T, Ohtsubo E. Inhibition of transpositional recombination by OrfA and OrfB proteins encoded by insertion sequence IS3. Genes Cells. 1997;2:547–557. doi: 10.1046/j.1365-2443.1997.1440342.x. [DOI] [PubMed] [Google Scholar]
- 32.Slightom J L, Durand-Tardif M, Jouanin L, Tepfer D. Nucleotide sequence analysis of TL-DNA of Agrobacterium rhizogenes agropine type plasmid. Identification of open reading frames. J Biol Chem. 1986;261:108–121. [PubMed] [Google Scholar]
- 33.Thompson J D, Higgins D G, Gibson T J. CLUSTAL W: improving the sensitivity of progressive multiple sequence alignment through sequence weighting, position-specific gap penalties and weight matrix choice. Nucleic Acids Res. 1994;22:4673–4680. doi: 10.1093/nar/22.22.4673. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Tobe T, Hayashi T, Han C G, Schoolnik G K, Ohtsubo E, Sasakawa C. Complete DNA sequence and structural analysis of the enteropathogenic Escherichia coli adherence factor plasmid. Infect Immun. 1999;67:5455–5462. doi: 10.1128/iai.67.10.5455-5462.1999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Wabiko H. Sequence analysis of an insertion element, IS1131, isolated from the nopaline-type Ti plasmid of Agrobacterium tumefaciens. Gene. 1992;114:229–233. doi: 10.1016/0378-1119(92)90579-e. [DOI] [PubMed] [Google Scholar]
- 36.Weinreich M D, Reznikoff W S. Fis plays a role in Tn5 and IS50 transposition. J Bacteriol. 1992;174:4530–4537. doi: 10.1128/jb.174.14.4530-4537.1992. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Yoshioka Y, Ohtsubo H, Ohtsubo E. Repressor gene fin0 in plasmids R100 and F: constitutive transfer of plasmid F is caused by insertion of IS3 into fin0. J Bacteriol. 1987;169:619–623. doi: 10.1128/jb.169.2.619-623.1987. [DOI] [PMC free article] [PubMed] [Google Scholar]