Abstract
Background:
Parkinson's disease (PD) is the second most common neurodegenerative disease after Alzheimer's dementia. Mitochondrial dysfunction is involved in the pathology of PD. Coiled-coil-helix-coiled-coil-helix domain-containing 2 (CHCHD2) was identified as associated with autosomal dominant PD. However, the mechanism of CHCHD2 in PD remains unclear.
Methods:
Short hairpin RNA (ShRNA)-mediated CHCHD2 knockdown or lentivirus-mediated CHCHD2 overexpression was performed to investigate the impact of CHCHD2 on mitochondrial morphology and function in neuronal tumor cell lines represented with human neuroblastoma (SHSY5Y) and HeLa cells. Blue-native polyacrylamide gel electrophoresis (PAGE) and two-dimensional sodium dodecyl sulfate-PAGE analysis were used to illustrate the role of CHCHD2 in mitochondrial contact site and cristae organizing system (MICOS). Co-immunoprecipitation and immunoblotting were used to address the interaction between CHCHD2 and Mic10. Serotype injection of adeno-associated vector-mediated CHCHD2 and 1-methyl-4-phenyl-1,2,3,6-tetrahydropyridine (MPTP) administration were used to examine the influence of CHCHD2 in vivo.
Results:
We found that the overexpression of CHCHD2 can protect against methyl-4-phenylpyridinium (MPP+)-induced mitochondrial dysfunction and inhibit the loss of dopaminergic neurons in the MPTP-induced mouse model. Furthermore, we identified that CHCHD2 interacted with Mic10, and overexpression of CHCHD2 can protect against MPP+-induced MICOS impairment, while knockdown of CHCHD2 impaired the stability of MICOS.
Conclusion:
This study indicated that CHCHD2 could interact with Mic10 and maintain the stability of the MICOS complex, which contributes to protecting mitochondrial function in PD.
Keywords: CHCHD2, MICOS complex, Mic10, Parkinson's disease
Background
Parkinson's disease (PD) is the second most common neurodegenerative disorder in the central nervous system. About 2% of the population above the age of 60 is affected by the disease worldwide.[1,2] To date, however, there are no disease-modifying therapies that directly target the underlying disease mechanisms or halt the progression of the disease. Hence, elucidation of the molecular mechanisms underlying the pathogenesis of PD represents the most fundamental basis for the development of disease-modifying therapies.[3] Several theories have been suggested for the pathogenesis of PD, of which mitochondrial dysfunction plays a pivotal role in both sporadic and familial forms of PD.[4,5]
Coiled-coil-helix-coiled-coil-helix domain-containing 2 (CHCHD2) is a mitochondria-localized protein encoded by the CHCHD2 gene. This protein belongs to the coiled-coil-helix-coiled-coil-helix domain (CHCHD) protein family, which contains twin cysteine-x9-cysteine motifs. These motifs are characterized primarily by four cysteine residues spaced ten amino acids apart from one another (cysteine-x9-cysteine motif),[6] In 2015, Funayama et al[7] have identified that CHCHD2 mutations might be a cause of autosomal dominant PD. Previous studies have suggested that CHCHD2 influences complex I and complex IV mitochondrial biogenesis and stability, mitochondrial cristae structure, and the regulation of apoptosis.[8–10] However, the mechanisms of CHCHD2 underlying PD are still obscure.
Mitochondrial cristae are the main site of oxidative phosphorylation and their structural integrity is crucial for mitochondrial normal functions.[11] Physiologically, cris-tae biogenesis and maintenance depend on the mitochon-drial contact site and cristae organizing system (MICOS), which plays a crucial role in the formation of crista junctions (CJs) and oligomers of the F1Fo-adenosine triphosphate (ATP) synthase and the generation of cristae rims and tubules.[12] In Saccharomyces cerevisiae (S. cerevisiae), the MICOS is composed of six subunits: Mic10, Mic12, Mic19, Mic26, Mic28, and Mic60. In mammalian systems, nine subunits (Mic60/Mitofilin, Mic10/MINOS1, Mic19/CHCHD3, Mic25/CHCHD6, Mic26/Apoo, Mic27/Apool, Mic19/QIL1, Mic14/CHCHD10, and DnaJC11) have been described, and the interactions among them are much more complicated.[13–19] Mic10 and Mic60 are the two most functionally important components of MICOS both in S. cerevisiae and mammalian systems.[20,21] Deletion of any component of the MICOS complex can lead to dysfunction in the formation of CJs and a loss of cristae.[12,22]
In the present study, we aim to address the role of CHCHD2 in the regulation of mitochondrial function in an experimental model of PD. And we testified that CHCHD2 can interact with Mic10 and protect against MICOS impairment in PD model. Taken together, our findings suggest that CHCHD2 can protect mitochondrial function by maintaining the MICOS complex in PD.
Methods
Ethics approval
All experimental procedures involving animals were reviewed and approved by the Institutional Animal Ethics Committee of The First Affiliated Hospital of Guangzhou Medical University, Guangzhou, China (No. 202059).
Antibodies and reagents
Anti-Flag was purchased from Sigma Aldrich (F1804, Sigma Aldrich, Saint Louis, MO, USA). Voltage-dependent anion channel (VDAC) (D73D12) antibody was purchased from Cell Signaling Technology (Danvers, MA, USA). Antibodies against Mic60 (ab137057), Mic10 (ab84969), CHCHD3 (ab224565), translocase of the outer membrane 20 (Tom20) (ab56783), heat shock protein 60 (Hsp60) (ab46798), glyceraldehydes-3-phosphate dehydrogenase (ab181602), β-actin (ab8226), and tyrosine hydroxylase (TH) (ab112) were from Abcam (Cambridge, UK). The anti-mouse and anti-rabbit secondary antibodies (1:1000) were purchased from Cell Signaling Technology. MitoTracker Deep Red was obtained from Life Technologies (Carlsbad, CA, USA) (M22426). 1-methyl-4-phenyl-1,2,3,6-tetrahydropyri-dine (MPTP) hydrochloride and methyl-4-phenylpyridi-nium (MPP+) were purchased from Santa Cruz Biotechnology (Dallas, TX, USA).
Cell culture and animal husbandry
HeLa and neuronal tumor cell lines represented with human neuroblastoma (SHSY5Y) cells were cultured using Dulbecco's modified Eagle's medium supplemented with 10% fetal bovine serum (Gibco, Waltham, MA, USA), 1% penicillin and streptomycin (Gibco), and maintained in a 5% CO2 incubator at 37°C. A total of 24 adult male C57BL6 mice at 7 to 8 weeks of age and 21 to 23 g in weight were used. All animals were housed with food and water provided ad libitum under a 12:12 light-dark cycle (from 7:00 to 19:00 as light time) with controlled temperature (20–26°C) and humidity.
Cell line generation
A lentiviral vector containing human CHCHD2 was constructed by Shanghai Genechem (China). Briefly, complementary deoxyribonucleic acid encoding human CHCHD2 was cloned into the GV341 vector using AgeI and NheI restriction sites, and an empty vector was used as a control. For the generation of stable knockdown cell lines, we used pLV-Puro-U6 short hairpin ribonucleic acid (shRNA) constructs and scramble-shRNA as a control. For lentiviral transduction, HeLa and SHSY5Y cells were seeded in 24-well plates at a density of 1 × 105 cells per well and transduced with lentiviral particles according to the manufacturer's instructions; a multiplicity of infection of 15 was selected on the basis of the company's instructions. After approximately 48 h of infection, stable cell line selection was performed by replacing culture medium with fresh medium containing puromycin at a concentration of 5 μg/mL. After 2 weeks of puromycin selection, a cell line stably overexpressing human CHCHD2 and a knockdown human CHCHD2 cell line were obtained, and CHCHD2 expression levels were confirmed by Western blotting or reverse transcription-polymerase chain reaction (RT-PCR) [Supplementary Figure 1 A-D].
Mitochondria isolation
Mitochondria were isolated with a Mitochondria Isolation Kit (Abcam, ab110170). Briefly, cells were collected by centrifugation at 1000 × g. The cells were resuspended to 5 mg/mL in Reagent A, incubated for 10 min on ice, and disrupted with a Dounce Homogenizer. The homogenized cellular extract was then centrifuged at 1000 × g to obtain a post-nuclear supernatant. Mitochondria were pelleted by centrifugation at 12,000 × g for 10 min. The pellet was resuspended in Reagent C and stored at − 80°C for subsequent analysis.
Electron microscopy
HeLa cells of control, shRNA, and CHCHD2-shRNA were rinsed twice with 0.1 mol/L phosphate-buffered saline (PBS) and fixed with 2.5% glutaraldehyde in 0.1 mol/L PBS (pH 7.4) for 1 h at room temperature. Then, cells were subsequently stained with 1% osmium tetroxide for 30 min, rinsed twice with Milli-Q water, and dehydrated in a series of graded ethanol (50, 70, 90, and 100 vol%). Aided by centrifugation, the cells were then infiltrated with increasing concentrations of Epon 812/acetonitrile resin. The final resin-infiltrated samples were then polymerized at 65°C for 24 h. The embedded cells were sectioned to a thickness of approximately 50 to 100 nm onto a water bath using an ultramicrotome (Leica EM UC6, German) equipped with a 35° diamond knife and imaged under a Jeol transmission electron microscope (Japan). Mitochondrial crista length was assessed using Image J and the data were compared by control cells.
Protein extraction and Western blotting
For protein extraction, the cells were lysed in ice-cold radioimmunoprecipitation assay buffer (Thermo, 89901, USA) supplemented with protease and phosphatase inhibitor cocktail (Thermo, 78440, USA), and then incubated on ice for 20 min. Cell lysates were sonicated and centrifuged at 12,000 × g, 4°C for 20 min to obtain a supernatant. The protein concentrations were determined by the bicinchoninic acid method. 40 μg protein was loaded and ran in 10% to 12% sodium dodecyl sulfate-polyacrylamide gel electrophoresis (SDS-PAGE) and then transferred onto a polyvinylidene fluoride (PVDF) membrane using an electroblotting apparatus. The membrane was then blocked with 5% bovine serum albumin for 1 h. After being washed three times with tris-buffered saline with 0.1% Tween®20 detergent (TBS-T, Life Tech, 85113, USA) (200 mmol/L tris, 1.5 mol/L NaCl, 0.1% Tween-20, pH 7.5), the membranes were incubated overnight with primary antibodies (1:1000). After being washed three times with TBS-T, membranes were then incubated with the appropriate horseradish peroxidase-conjugated secondary antibodies for 1 h and washed three times. Bands were visualized by enhanced chemiluminescence (Millipore) using an ImageQuant LAS 4000 system (General Electric Healthcare).
Blue-native PAGE (BN-PAGE) and two-dimensional SDS-PAGE analysis
Mitochondria samples were prepared in 1× NativePA-GETM Sample Buffer (50 mmol/L BisTris, 6 mol/L HCl, 50 mmol/L NaCl, 10% w/v Glycerol, 0.001% Ponceau, Life Tech, BN2003) and detergents (1% Digitonin) according to the NativePAGETM Sample Prep Kit (Life Tech, BN2008) protocol. The NativePAGE™ 5% G-250 Sample Additive (2.5 μL per 10 μL sample) was added prior to electrophoresis. In total, 25 μL of sample and 10 μL of NativeMark™ Unstained Protein Standard were loaded in a NativePAGE 3% to 12% Bis-Tris Protein Gel (Life Tech, BN1001BOX). The inner chamber was filled with cold cathode buffer, and the outer/lower chamber was filled with cold anode buffer in the dry wells at 4°C. The samples in each well were overlaid with cold Cathode Buffer. After running at 100 V for 90 min at room temperature, the proteins in the gel were transferred to a PVDF membrane, and the proteins were detected using the indicated antibodies. For two-dimensional SDS-PAGE analysis, a standard 10% SDS-gel was prepared with a single large lane for the one-dimensional BN-PAGE lane and one regular lane for the molecular weight marker. The BN-PAGE gel in the plates was removed from the electrophoresis apparatus, and the lanes of the BN-PAGE gel containing the proteins of interest were cut out. The BN-PAGE gel slice was placed in a 2 × SDS sample buffer, incubated for 10 min at room temperature, and then boiled briefly (not >20 s) in a microwave. The BN-PAGE gel slice was incubated in a hot SDS sample buffer for another 15 min at room temperature and then loaded in the large well over the stacking gel of the SDS-PAGE gel, avoiding air bubbles. The slice was overlaid with SDS sample buffer, and electrophoresis was performed according to the standard protocols.[23]
Immunofluorescence and immunohistochemical staining
For immunofluorescence, cells grown on 15-mm glass coverslips or mouse brain sections were pre-incubated with PBS 0.1% Triton X-100 for 10 min and then blocked with 5% bovine serum albumin (BSA) for 1 h. The primary antibodies used for immunofluorescence were rabbit anti-Mic60 (1:500), rabbit anti-TH (1:500), and mouse anti-Flag (1:200) antibody was used to label CHCHD2. To determine the localization of CHCHD2 and Mic60 in the HeLa cells, double immunofluorescence staining was performed. Colocalization of CHCHD2-flag and Mic60 was observed by merged images and 4’,6-diamidino-2-phenylindole was used for nuclei staining, and images were captured using a confocal microscope (Olympus IX71, Tokyo, Japan).
For immunohistochemical staining, brain sections were incubated with 3% H2O2 in PBS for 15 min and rinsed with PBS three times. After blocking in PBS containing 5% BSA and 0.3% Triton X-100 for 1 h, brain sections were incubated with anti-TH antibody (1:500) overnight at 4°C, followed by incubation at room temperature with the appropriate secondary antibodies (ZSGB BIO, China). The sections were developed with 3,3’-diaminobenzidine and examined via light microscopy.
Co-immunoprecipitation (Co-IP) assays
Co-IP assays were performed in whole-cell lysates of HeLa cells. Cells were resuspended in lysis buffer (150 mmol/L NaCl, 50 mmol/L Tris-HCl, pH 7.6, 1% NP-40, 0.1% SDS) supplemented with protease and phosphatase inhibitors. Samples were pre-cleared by incubation with protein A agarose beads (Life Technologies). Beads were removed by centrifugation, and the samples were incubated overnight at 4°C with mouse anti-Flag (1:200), rabbit anti-Mic60 (1:200), rabbit anti-Mic10 (1:200), or rabbit anti-CHCHD3 (1:200) antibodies. A total of 50 μL of Protein A agarose beads were added to the samples, and the total sample volume of the protein mixture was added up to 500 μL with IP buffer (25 mmol/L Tris, 150 mmol/L NaCl; pH 7.2). The mixture was then incubated on a rotator for 2 h. The beads were washed and resuspended in a loading buffer containing dithiothreitol (Life Technologies), and finally, the proteins were released from the beads by incubation at 95°C for 5 min. After centrifugation, the samples were analyzed by immunoblotting (IB).
ATP measurement
The ATP concentration of SH-SY5Y cells was determined using a luminescent ATP detection assay (ab113849, Abcam) according to the manufacturer's protocol.
Mitochondrial morphology assessment
To observe mitochondrial morphology, cells were plated in glass-bottom confocal dishes. Cells were pre-incubated with 100 nmol/L MitoTracker Deep Red for 15 min, and images were taken from each group under confocal microscopy (Leica Biosystems, Newcastle, UK).
Mitochondrial cristae morphology assessment
HeLa cells with control shRNA or CHCHD2-shRNA were rinsed twice with 0.1 mol/L PBS and fixed with 2.5% glutaraldehyde in 0.1 mol/L PBS (pH 7.4) for 1 h at room temperature. Then, cells were subsequently stained with 1% osmium tetroxide for 30 min, rinsed twice with Milli-Q water, and dehydrated in a series of graded ethanol (50, 70, 90, and 100 vol%). Aided by centrifugation, the cells were then infiltrated with increasing concentrations of Epon 812/acetonitrile resin. The final resin-infiltrated samples were then polymerized at 65°C for 24 h. The embedded cells were sectioned to a thickness of approximately 50 to 100 nm onto a water bath using an ultramicrotome (Leica EM UC6) equipped with a 35° diamond knife and imaged under a Jeol transmission electron microscope.
Adeno-associated vector (AAV) serotype injection and MPTP administration
AAV9 was designed by Shanghai Genechem. Deeply anesthetized mice were stereotactically injected unilaterally into the right substantia nigra with a microinjector (Stoelting, Kiel, WI, USA) at a rate of 0.2 μL/min with either 1.5 μL of empty AAV9 or 1.5 μL of AAV9 expressing human CHCHD2 both at a concentration of 5.16 × 1012 genomic particles (gp)/mL. The following coordinates from Bregma were used for injection: anterioposterior –3.1 mm; mediolateral –1.4 mm; dorsoventral −4.4 mm. Twelve mice were injected with AAV9-CHCHD2, and 12 mice were injected with an empty AAV9 vector. One month after AAV injection, six mice from both groups were intraperitoneally injected four times in 1 day with MPTP (20 mg/kg) at 2 h intervals. Control mice were injected with an equivalent volume of saline. All mice were sacrificed 7 days after the last injection.
Statistical analysis
Statistical analyses were performed by using SPSS (version 20, IBM, Armonk, NY, USA) software. Data were presented as the mean ± standard deviation (SD). The one-way analysis of variance Post hoc Tukey test was performed for comparisons among multiple groups. The t test or Mann-Whitney U test was performed for comparisons between two groups. A value of P < 0.05 was considered to be statistically significant.
Results
CHCHD2 protects mitochondrial cristae stability and ATP production
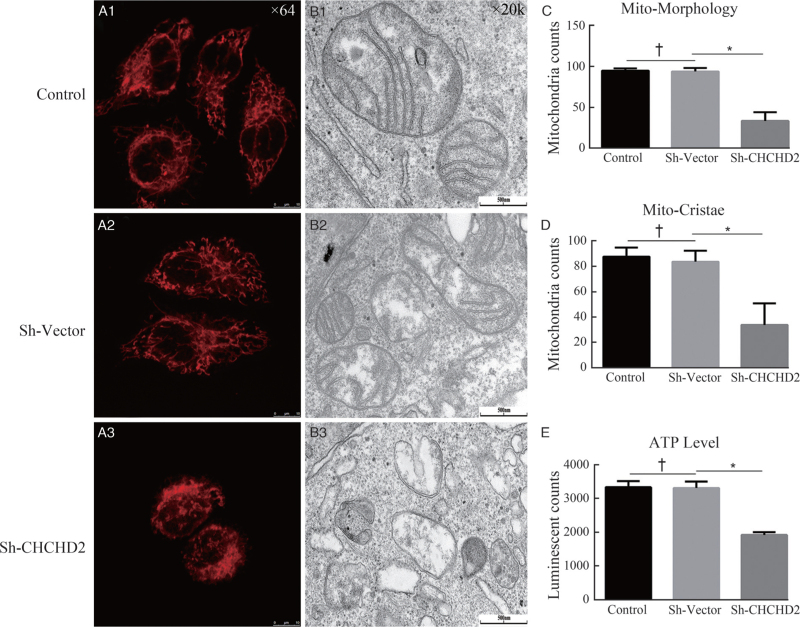
We first analyzed the effect of CHCHD2 on mitochondrial morphology changes in HeLa cells via live cell imaging. We found that CHCHD2-knockdown can destroy the morphology of mitochondria, which shows interconnected network-like, functionally united organelles shifting to more fragment fission in the Sh-CHCHD2 group with the comparison of the control group [Figure 1A1-A3, C]. Next, we examined the effect of CHCHD2 on mitochondrial cristae morphology by transmission electron microscopy. The results showed that the mitochondria in scrambled control cells had well-defined cristae structures; however, CHCHD2 depletion can lead to dramatic changes in cristae organization which lacked any visible connection to the inner boundary membrane [Figure 1B1-B3, D]. As mitochondria play a prominent role in energy metabolism, the ATP level was assessed in Hela cells by using a Luminescent ATP Detection Assay Kit. There was no difference in ATP level between Sh-vector and control group, while ATP production was significantly decreased in the Sh-CHCHD2 group (Figure 1E).
Figure 1.
Loss of CHCHD2 impairs the morphology and function of mitochondria and mitochondrial crista. (A1-A3) Representative Images taken by live cell Imaging showed alteration In mitochondrial morphology in control, Sh-Vector, and Sh-CHCHD2 groups. (B1-B3) Mitochondrial crista in control, Sh-Vector, and Sh-CHCHD2 groups were analyzed by transmission electron microscope. (C, D) Quantitation of the number of cells with normal mitochondrial morphology and mitochondrial crista in control, Sh-Vector and Sh-CHCHD2 groups (n = 100 cells; mean ± SD; ∗P < 0.05, †P > 0.05). (E) ATP production in different groups were measured in Hela cells (mean ± SD; ∗P < 0.05, †P > 0.05). ATP: Adenosine triphosphate; CHCHD2: Coiled-coil-helix-coiled-coil-helix domain-containing 2; SD: Standard deviation.
CHCHD2 interacts with Mic10 subunit of MICOS complex
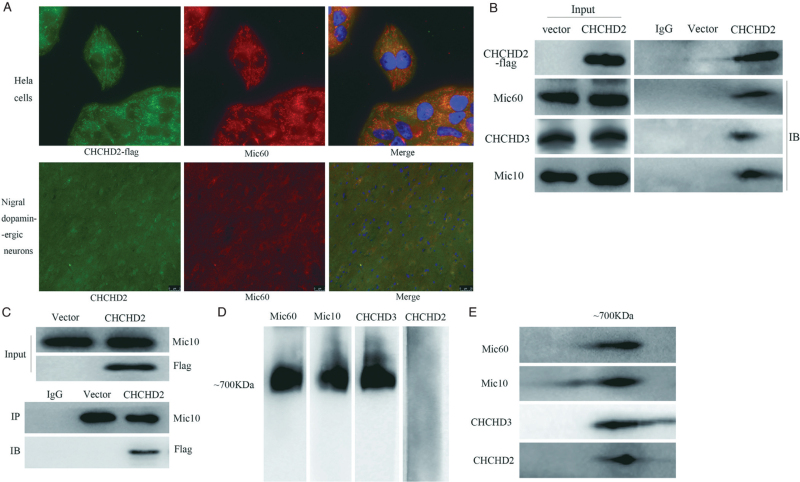
Co-localization analysis showed that Flag-tagged CHCHD2 co-localized with Mic60 of MICOS complex in HeLa cells and in nigral dopaminergic neurons by immunofluorescence microscopy [Figure 2A]. Furthermore, Co-IP assays identified that CHCHD2-flag interacted with Mic60, Mic10, and CHCHD3, respectively [Figure 2B]. However, results from reciprocal Co-IP showed that only Mic10 could coimmunoprecipitate with CHCHD2-flag [Figure 2C, Supplementary Figure 2]. To further clarify the role of CHCHD2 in MICOS, mitochondria were isolated from HeLa cells stably expressing Flag-tagged CHCHD2. Then, the mitochondria were lysed in 1% digitonin and subjected to BN-PAGE followed by IB analysis. As expected, Mic60, Mic10, and CHCHD3 were found to be localized at ∼700 kDa, which is in accordance with the molecular weight of the MICOS complex, whereas CHCHD2-flag was not detected in the corresponding band [Figure 2D]. However, Western blotting analysis of 2D gels containing 700 kDa complex showed that CHCHD2-flag, along with Mic60, Mic10, and CHCHD3 were detected in this 700 kDa complex [Figure 2E]. Taken together, these results indicate that CHCHD2 may interact with MICOS instead of contributing to one subunit of the MICOS complex.
Figure 2.
The interaction of CHCHD2 with MICOS complex. (A) Immunofluorescent staining for CHCHD2-Flag (Green) and Mic60 (Red) In HeLa cells (First line) and in the nigral dopaminergic neurons. DNA was visualized with DAPI (blue). (B) Co-IP assays to detect the interaction between CHCHD2-Flag and subunits of the MICOS. Co-IP was performed using an anti-Flag antibody, followed by IB analysis with anti-Mic60, anti-CHCHD3, and anti-Mic10. (C) The reciprocal Co-IP of Mic10 and CHCHD2-flag. (D) Mitochondria of HeLa cells stably expressing CHCHD2-flag were solubilized indigitonin buffer, separated by BN-PAGE, and analyzed by Western blotting using Mic60, CHCHD3, Flag, and Mic10 antibodies. (E) Two-dimensional analysis of the gel by SDS-PAGE and analysis by Western blotting with Mic60, CHCHD3, and Mic10 antibodies. CHCHD2 was detected with an anti-flag antibody. BN-PAGE: Blue native polyacrylamide gel electrophoresis; CHCHD2: Coiled-coil-helix-coiled-coil-helix domain-containing 2; Co-IP: Co-immunoprecipitation; DAPI: 4’,6-diamidino-2-phenylindole; IB: Immunoblot; IP: Immunoprecipitation; MICOS: Mitochondrial contact site and cristae organizing system; SDS-PAGE: Sodium dodecyl sulfate-polyacrylamide gel electrophoresis.
CHCHD2 protects mitochondrial morphology and its function in MPP+-induced cell model
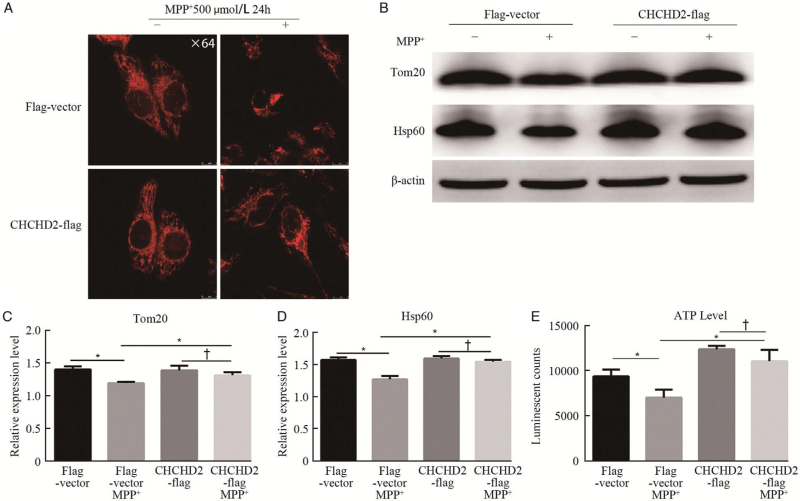
We then analyzed the effect of CHCHD2 on MPP+-induced mitochondrial morphology changes in HeLa cells via live cell imaging. The results demonstrated that there was no obvious alteration in mitochondrial morphology between the flag-vector group and the CHCHD2-flag group, while MPP+ could damage the mitochondrial network and lead to a reduced branching in the vector group but had little effect on mitochondrial morphology changes in the CHCHD2-flag group [Figure 3A]. We further evaluated changes in mitochondrial proteins, including Tom20, and mitochondrial matrix protein Hsp60, in SHSY5Y cells for elaborating mitochondrial function. As shown in Figure 3B-D, compared to Flag-vector without MPP+ group, both Tom20 and Hsp60 showed a significant decrease after MPP+ treatment (P < 0.05), indicating the mitochondrial dysfunction in the MPP+-induced cell model. However, overexpression of CHCHD2 can restore the loss of Tom20 and Hsp60 compared with the corresponding controls. In addition, the ATP level was assessed in SHSY5Y cells. There was no difference in ATP level between the flag-vector and CHCHD2-flag groups. After 24 h of MPP+ treatment for the Flag-vector group, the ATP level was significantly reduced compared to the non-MPP Flag-vector group (P < 0.05). Whereas, the overexpression of CHCHD2 can rescue MPP+-induced loss of ATP [Figure 3E]. These results indicate that CHCHD2 can prevent MPP+-induced mitochondrial dysfunction.
Figure 3.
Overexpression of CHCHD2 attenuates MPP+-induced mitochondrial dysfunction and mitochondrial protein loss. (A) Representative images taken by live cell imaging showed obvious alteration in mitochondrial morphology in Flag-Vector and CHCHD2-flag group with or without MPP+ treatment (500 μmol/L, 24 h). (B) The expression of outer mitochondrial membrane protein (Tom20) and mitochondrial matrix protein (Hsp 60) was measured by Western blotting. (C, D) Densitometry analysis of Tom20 and Hsp60 on IBs (mean ± SD; P < 0.05, †P > 0.05). (E) ATP production in different groups was measured in SHSY5Y cells (mean ± SD; ∗P < 0.05, †P > 0.05). ATP: Adenosine triphosphate; CHCHD2: Coiled-coil-helix-coiled-coil-helix domain-containing 2; Hsp60: Heat shock protein 60; IBs: Immunoblots; MPP: Methyl-4-phenylpyridinium; SD: Standard deviation; Tom20: Translocase of the outer membrane 20.
CHCHD2 maintained MICOS stability and protected against MPP+-induced MICOS impairment
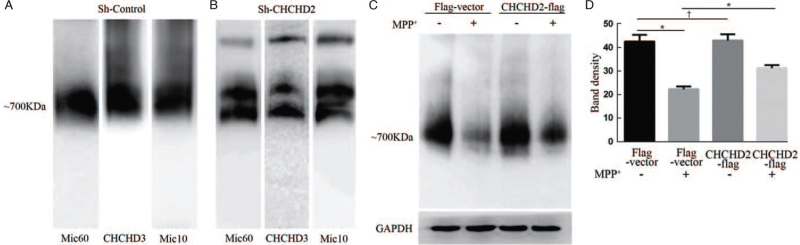
For further exploring the role of CHCHD2 in the stability of the MICOS complex, we studied the Mic60, Mic10, and CHCHD3 as MICOS markers for IB by using blue native electrophoresis in CHCHD2 knockdown cells and corresponding controls. In control cells, the MICOS complex migrated at a molecular weight of approximately 700 kDa, which indicated that the MICOS complex was complete [Figure 4A]. However, in the absence of CHCHD2, the MICOS complex separated into two parts with the molecular weights approximately 550 and 850 kDa [Figure 4B], which suggests that loss of CHCHD2 results in the destruction of the MICOS complex. Next, we evaluated the role of CHCHD2 on the MICOS complex in the MPP+-induced PD model in SH-SY5Y cells. CHCHD2 overexpression or the control group was treated with 500 μmol/L MPP+ for 24 h, and the mitochondria were isolated for BN-PAGE analysis. As shown in Figure 4C and 4D, MPP+ resulted in a loss of the MICOS complex, while CHCHD2 overexpression could partially reverse the loss of the MICOS. These results indicate that CHCHD2 is essential for the stability of the MICOS complex and can protect against MPP+-induced MICOS impairment.
Figure 4.
CHCHD2 maintains MICOS stability and protects against MPP+-induced MICOS decrease. (A, B) Blue-native gel analysis of mitochondria of CHCHD2 knockdown (Sh-CHCHD2) or scrambled vector (Sh-Control) cells. Blots were probed with anti-MIC60, anti-CHCHD3, and anti-Mic10 to examine the MICOS complex. (C) Blue-native gel analysis of the mitochondrial MICOS of SHSY5Y cells in which CHCHD2 was overexpressed (CHCHD2-flag) or control vector (Flag-Vector) with (+) or without (−) M PP+ treatment (500 μM, 24 h). GAPDH was assayed as protein-loading control. (D) Densitometric analysis was carried out by using National Institutes of Health (NIH) ImageJ. (mean ± SD; ∗P < 0.05, †P > 0.05). CHCHD2: Coiled-coil-helix-coiled-coil-helix domain-containing 2; GAPDH: Glyceraldehydes-3-phosphate dehydrogenase; MICOS: Mitochondrial contact site and cristae organizing system; MPP: Methyl-4-phenylpyridinium; SD: Standard deviation.
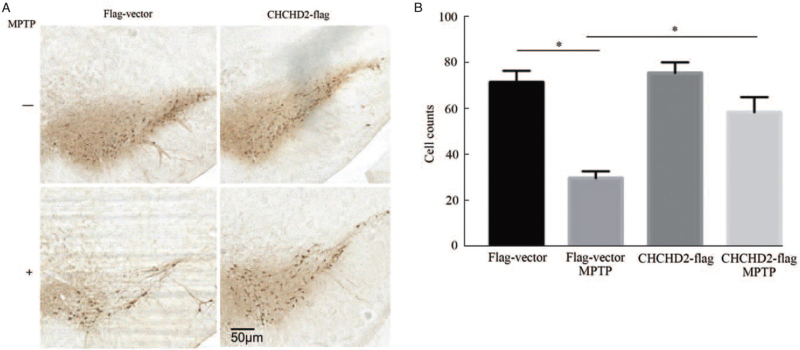
CHCHD2 protects dopaminergic neurons in MPTP-induced mouse model
To explore the effects of CHCHD2 on dopaminergic neurodegeneration in MPTP-induced mouse model, adeno-associated viral vectors (GV411/AAV9) expressing Flag-tagged human CHCHD2 and vector were delivered to the substantia nigra pars compacta (SNpc) of mice by unilateral stereotactic injection [Supplementary Figure 1E]. Exposure to MPTP resulted in a significant loss of TH-positive neurons in the SNpc [P < 0.05; Figure 5A and 5B; Supplementary Figure 3]. However, mice in CHCHD2 overexpressing group significantly reverse the loss of TH when compared to the Flag-Vector plus MPTP group [P < 0.05; Figure 5A and 5B; Supplementary Figure 3]. These results demonstrate that CHCHD2 can protect against MPTP-induced dopaminergic neuronal degeneration.
Figure 5.
Effect of CHCHD2 on MPTP-induced dopaminergic neuronal loss in the substantia nigra. (A) Mice with CHCHD2 overexpression were more resistant than control mice to MPTP-induced dopaminergic cell loss. Representative images of TH-positive neurons in the SNpc of CHCHD2-Flag mice and Flag-vector mice at 7 days after saline or MPTP (20 mg/kg, i.p.) administration. (B) Quantification of the number of TH-positive neurons in substantia nigra pars compacta (SNPc) in different groups. (n = 6, mean ± SD; ∗P < 0.05). CHCHD2: Coiled-coil-helix-coiled-coil-helix domain-containing 2; MPTP: 1-methyl-4-phenyl-1,2,3,6-tetrahydropyridine; SD: Standard deviation; SNpc: Substantia nigra pars compacta; TH: Tyrosine hydroxylase.
Discussion
In the present study, we identified that CHCHD2 could interact with the MICOS complex. Depletion of CHCHD2 results in the destruction of the MICOS and mitochondrial morphology, function, and crista structure. We also showed the evidence that CHCHD2 can protect against the MICOS damage and mitochondrial dysfunction in a PD cell model and prevent dopaminergic neuron loss in an MPTP-induced animal model.
CHCHD2 belongs to the CHCH domain family, which is known to be primarily in the intermembrane space (IMS) of mitochondria.[24] CHCHD2 is encoded by CHCHD2 and was imported into the IMS via Mia40-mediated pathway.[25] Silencing CHCHD2 resulted in the destabili-zation of OxPhos complexes and a marked loss of OxPhos enzymatic activity.[8,26] CHCHD2 was also reported to inhibit apoptosis by interacting with Bcl-xL to regulate Bax activation.[9] A recent study showed that CHCHD2 can maintain mitochondrial crista integrity and bind to cytochrome C along with mitochondrial morphology and cristae structure 1 (MICS1) to modulate cell death signaling.[10] These findings provide clues to explore the mechanisms of CHCHD2 in mitochondria. However, the location and regulatory mechanisms of CHCHD2 in mitochondria are still obscure.
The MICOS complex is embedded in the inner membrane with domains facing the IMS that mediates the formation of heterologous structures localized to the inner boundary membrane.[27,28] To date, six conserved MICOS subunits are found in yeast (Mic60, Mic10, Mic19, Mic27, Mic26, Mic12), and the MICOS in mammals consisting of many more subunits seems more complicated.[12,29] Disrupted in schizophrenia 1 and CHCHD10 were recently added to the growing list of subunits of the MICOS.[30,31] Among the subunits of MICOS, Mic10 and Mic60 are the two core components.[21,32] The MICOS complex makes contact sites between the mitochondrial inner and outer membranes to anchor cristae and bends membranes to form CJs. The MICOS is also important for the proper import of proteins into the mitochondria and for lipid transfer to and from mitochondrial membranes.[27,33]
To address the relationship between CHCHD2 and the MICOS, we firstly explored the interactions of CHCHD2 with subunits of MICOS (Mic60, Mic10, CHCHD3). We found that CHCHD2-flag can interact with Mic60, Mic10, and CHCHD3. However, reciprocal Co-IP showed that only Mic10 can coimmunoprecipitate with CHCHD2-flag. In BN-PAGE analysis, MIC60, MIC10, and CHCHD3 were found to be localized at ∼700 kDa, which was in accordance with the molecular weight of MICOS. However, CHCHD2-Flag was not detected in the corresponding band. Interestingly, Western blotting analyses of 2D gels containing 700 kDa complex showed that CHCHD2-Flag, along with Mic60, Mic10, and CHCHD3 can be detected in the 700 kDa complex. These results indicate that CHCHD2 can interact with the MICOS and the binding site may locate at the Mic10 subcomplex of the MICOS. Next, we explored the role of CHCHD2 in the stability of MICOS. Our results showed that loss of CHCHD2 impaired the stability of M+ICOS. CHCHD2 overexpression can protect against MPP+-induced MICOS impairment. Our findings highlighted the role of CHCHD2 in MICOS. Meng et al[10] found that loss of CHCHD2 in Drosophila affected mitochondrial crista structure and the introduction of human CHCHD2 rescued the PD-associated phenotypes. These findings suggested the protective role of CHCHD2 in PD, which was in accordance with our results. Recently, Zhou et al[34] showed that CHCHD2 interacted with CHCHD10 and localized at the rim of the mitochondria in the proximity with MICOS complex by using super-resolution microscopy, and CHCHD2 knockdown can disrupt the MICOS. These findings indicate the role of CHCHD2 in the stability of the MICOS. However, Huang et al[35] showed that CHCHD2 and CHCHD10 localized at the mitochondrial cristae and are dispensable for MICOS complex assembly and cristae structure. As there are different opinions on the role of CHCHD2 in MICOS, our results showed evidence that CHCHD2 can interact with Mic10, the core subunit of the MICOS, and is essential for MICOS stability. Further studies are still required to fully understand the regulation of CHCHD2 in the MICOS.
In conclusion, this work indicated that CHCHD2 could interact with Mic10 and maintain the stability of the MICOS complex, which contributes to protecting mito-chondrial function in the MPP+-induced PD cell model and MPTP-induced PD mouse model.
Funding
This work was supported by research grants from the National Natural Science Foundation of China (Nos. 81901282, 81870992, 82071444), the Nature Science Foundation of Guangdong Province (Nos. 2019A1515011189, 2020A1515010985), and the Technology Project of Guangzhou (Nos. 202102020029, 2019ZD09).
Conflicts of interest
None.
Supplementary Material
Footnotes
How to cite this article: Lu L, Mao H, Zhou M, YL, Dai W, Qiu J, Xiao Y, Mo M, Zhu X, Wu Z, Pei Z, Guo W, Xu P, Chen X. CHCHD2 maintains mitochondrial contact site and cristae organizing system stability and protects against mitochondrial dysfunction in an experimental model of Parkinson's disease. Chin Med J 2022;135:1588–1596. doi: 10.1097/CM9.0000000000002053
Supplemental digital content is available for this article.
References
- 1.Poewe W, Seppi K, Tanner CM, Halliday GM, Brundin P, Volkmann J, et al. Parkinson disease. Nat Rev Dis Primers 2017; 3:17013.doi: 10.1038/nrdp.2017.13. [DOI] [PubMed] [Google Scholar]
- 2.Lee A, Gilbert RM. Epidemiology of Parkinson disease. Neurol Clin 2016; 34:955–965. doi: 10.1016/j.ncl.2016.06.012. [DOI] [PubMed] [Google Scholar]
- 3.Franco-Iborra S, Vila M, Perier C. The Parkinson disease mitochondrial hypothesis: where are we at? Neuroscientist 2016; 22:266–277. doi: 10.1177/1073858415574600. [DOI] [PubMed] [Google Scholar]
- 4.Larsen SB, Hanss Z, Kruger R. The genetic architecture of mitochondrial dysfunction in Parkinson's disease. Cell Tissue Res 2018; 373:21–37. doi: 10.1007/s00441-017-2768-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Bose A, Beal MF. Mitochondrial dysfunction in Parkinson's disease. J Neurochem 2016; 139: (Suppl 1): 216–231.doi: 10.1111/jnc.13731. [DOI] [PubMed] [Google Scholar]
- 6.Modjtahedi N, Tokatlidis K, Dessen P, Kroemer G. Mitochondrial proteins containing Coiled-Coil-Helix-Coiled-Coil-Helix (CHCH) domains in health and disease. Trends Biochem Sci 2016; 41:245–260. doi: 10.1016/j.tibs.2015.12.004. [DOI] [PubMed] [Google Scholar]
- 7.Funayama M, Ohe K, Amo T, Furuya N, Yamaguchi J, Saiki S, et al. CHCHD2 mutations in autosomal dominant late-onset Parkinson's disease: a genome-wide linkage and sequencing study. Lancet Neurol 2015; 14:274–282. doi: 10.1016/s1474-4422(14)70266-2. [DOI] [PubMed] [Google Scholar]
- 8.Baughman JM, Nilsson R, Gohil VM, Arlow DH, Gauhar Z, Mootha VK. A computational screen for regulators of oxidative phosphor-ylation implicates SLIRP in mitochondrial RNA homeostasis. PLoS Genet 2009; 5:e1000590.doi: 10.1371/journal.pgen.1000590. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Liu Y, Clegg HV, Leslie PL, Di J, Tollini LA, He Y, et al. CHCHD2 inhibits apoptosis by interacting with Bcl-x L to regulate Bax activation. Cell Death Differ 2015; 22:1035–1046. doi: 10.1038/cdd.2014.194. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Meng H, Yamashita C, Shiba-Fukushima K, Inoshita T, Funayama M, Sato S, et al. Loss of Parkinson's disease-associated protein CHCHD2 affects mitochondrial crista structure and destabilizes cytochrome c. Nat Commun 2017; 8:15500.doi: 10.1038/ncomms15500. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Kuhlbrandt W. Structure and function of mitochondrial membrane protein complexes. BMC Biol 2015; 13:89.doi: 10.1186/s12915-015-0201-x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Kozjak-Pavlovic V. The MICOS complex of human mitochondria. Cell Tissue Res 2017; 367:83–93. doi: 10.1007/s00441-016-2433-7. [DOI] [PubMed] [Google Scholar]
- 13.Munoz-Gomez SA, Slamovits CH, Dacks JB, Wideman JG. The evolution of MICOS: ancestral and derived functions and interactions. Commun Integr Biol 2015; 8:e1094593.doi: 10.1080/19420889.2015.1094593. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Barbot M, Jans DC, Schulz C, Denkert N, Kroppen B, Hoppert M, et al. Mic10 oligomerizes to bend mitochondrial inner membranes at cristae junctions. Cell Metab 2015; 21:756–763. doi: 10.1016/j.cmet.2015.04.006. [DOI] [PubMed] [Google Scholar]
- 15.Guarani V, McNeill EM, Paulo JA, Huttlin EL, Frohlich F, Gygi SP, et al. QIL1 is a novel mitochondrial protein required for MICOS complex stability and cristae morphology. Elife 2015; 4:e06265.doi: 10.7554/eLife.06265. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Van Laar VS, Berman SB, Hastings TG. Mic60/mitofilin overexpression alters mitochondrial dynamics and attenuates vulnerability of dopaminergic cells to dopamine and rotenone. Neurobiol Dis 2016; 91:247–261. doi: 10.1016/j.nbd.2016.03.015. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Darshi M, Mendiola VL, Mackey MR, Murphy AN, Koller A, Perkins GA, et al. ChChd3, an inner mitochondrial membrane protein, is essential for maintaining crista integrity and mitochondrial function. J Biol Chem 2011; 286:2918–2932. doi: 10.1074/jbc.M110.171975. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.An J, Shi J, He Q, Lui K, Liu Y, Huang Y, et al. CHCM1/CHCHD6, novel mitochondrial protein linked to regulation of mitofilin and mitochondrial cristae morphology. J Biol Chem 2012; 287:7411–7426. doi: 10.1074/jbc.M111.277103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Ott C, Dorsch E, Fraunholz M, Straub S, Kozjak-Pavlovic V. Detailed analysis of the human mitochondrial contact site complex indicate a hierarchy of subunits. PLoS One 2015; 10:e0120213.doi: 10.1371/journal.pone.0120213. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Milenkovic D, Larsson NG. Mic10 oligomerization pinches off mitochondrial cristae. Cell Metab 2015; 21:660–661. doi: 10.1016/j.cmet.2015.04.020. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Li H, Ruan Y, Zhang K, Jian F, Hu C, Miao L, et al. Mic60/Mitofilin determines MICOS assembly essential for mitochondrial dynamics and mtDNA nucleoid organization. Cell Death Differ 2016; 23:380–392. doi: 10.1038/cdd.2015.102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Huynen MA, Muhlmeister M, Gotthardt K, Guerrero-Castillo S, Brandt U. Evolution and structural organization of the mitochondrial contact site (MICOS) complex and the mitochondrial intermembrane space bridging (MIB) complex. Biochim Biophys Acta 2016; 1863:91–101. doi: 10.1016/j.bbamcr.2015.10.009. [DOI] [PubMed] [Google Scholar]
- 23.Fiala GJ, Schamel WW, Blumenthal B. Blue native polyacrylamide gel electrophoresis (BN-PAGE) for analysis of multiprotein complexes from cellular lysates. J Vis Exp 2011; 48:2164.doi: 10.3791/2164. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Grossman LI, Purandare N, Arshad R, Gladyck S, Somayajulu M, Huttemann M, et al. MNRR1, a biorganellar regulator of mitochondria. Oxid Med Cell Longev 2017; 2017:6739236.doi:10.1155/2017/6739236. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Aras S, Bai M, Lee I, Springett R, Huttemann M, Grossman LI. MNRR1 (formerly CHCHD2) is a bi-organellar regulator of mitochondrial metabolism. Mitochondrion 2015; 20:43–51. doi:10.1016/j.mito.2014.10.003. [DOI] [PubMed] [Google Scholar]
- 26.Aras S, Pak O, Sommer N, Finley R, Jr, Huttemann M, Weissmann N, et al. Oxygen-dependent expression of cytochrome c oxidase subunit 4-2 gene expression is mediated by transcription factors RBPJ, CXXC5 and CHCHD2. Nucleic Acids Res 2013; 41:2255–2266. doi:10.1093/nar/gks1454. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Wideman JG, Munoz-Gomez SA. Cell biology: functional conservation, structural divergence, and surprising convergence in the MICOS complex of trypanosomes. Curr Biol 2018; 28:R1245–R1248. doi:10.1016/j.cub.2018.09.057. [DOI] [PubMed] [Google Scholar]
- 28.Schorr S, van der Laan M. Integrative functions of the mitochondrial contact site and cristae organizing system. Semin Cell Dev Biol 2018; 76:191–200. doi: 10.1016/j.semcdb.2017.09.021. [DOI] [PubMed] [Google Scholar]
- 29.van der Laan M, Horvath SE, Pfanner N. Mitochondrial contact site and cristae organizing system. Curr Opin Cell Biol 2016; 41:33–42. doi: 10.1016/j.ceb.2016.03.013. [DOI] [PubMed] [Google Scholar]
- 30.Genin EC, Plutino M, Bannwarth S, Villa E, Cisneros-Barroso E, Roy M, et al. CHCHD10 mutations promote loss of mitochondrial cristae junctions with impaired mitochondrial genome maintenance and inhibition of apoptosis. EMBO Mol Med 2016; 8:58–72. doi: 10.15252/emmm.201505496. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Pinero-Martos E, Ortega-Vila B, Pol-Fuster J, Cisneros-Barroso E, Ruiz-Guerra L, Medina-Dols A, et al. Disrupted in schizophrenia 1 (DISC1) is a constituent of the mammalian mitochondrial contact site and cristae organizing system (MICOS) complex, and is essential for oxidative phosphorylation. Hum Mol Genet 2016; 25:4157–4169. doi: 10.1093/hmg/ddw250. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Rampelt H, Bohnert M, Zerbes RM, Horvath SE, Warscheid B, Pfanner N, et al. Mic10, a core subunit of the mitochondrial contact site and cristae organizing system, interacts with the dimeric F1Fo-ATP synthase. J Mol Biol 2017; 429:1162–1170. doi: 10.1016/j.jmb.2017.03.006. [DOI] [PubMed] [Google Scholar]
- 33.Harner ME, Unger AK, Geerts WJ, Mari M, Izawa T, Stenger M, et al. An evidence based hypothesis on the existence of two pathways of mitochondrial crista formation. Elife 2016; 5:e18853.doi:10.7554/eLife.18853. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Zhou W, Ma D, Sun AX, Tran HD, Ma DL, Singh BK, et al. PD-linked CHCHD2 mutations impair CHCHD10 and MICOS complex leading to mitochondria dysfunction. Hum Mol Genet 2019; 28:1100–1116. doi: 10.1093/hmg/ddy413. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Huang X, Wu BP, Nguyen D, Liu YT, Marani M, Hench J, et al. CHCHD2 accumulates in distressed mitochondria and facilitates oligomerization of CHCHD10. Hum Mol Genet 2018; 27:3881–3900. doi: 10.1093/hmg/ddy270. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.