Abstract
The GerAA, -AB, and -AC proteins of the Bacillus subtilis spore are required for the germination response to l-alanine as the sole germinant. They are likely to encode the components of the germination apparatus that respond directly to this germinant, mediating the spore's response; multiple homologues of the gerA genes are found in every spore former so far examined. The gerA operon is expressed in the forespore, and the level of expression of the operon appears to be low. The GerA proteins are predicted to be membrane associated. In an attempt to localize GerA proteins, spores of B. subtilis were broken and fractionated to give integument, membrane, and soluble fractions. Using antibodies that detect Ger proteins specifically, as confirmed by the analysis of strains lacking GerA and the related GerB proteins, the GerAA protein and the GerAC+GerBC protein homologues were localized to the membrane fraction of fragmented spores. The spore-specific penicillin-binding protein PBP5∗, a marker for the outer forespore membrane, was absent from this fraction. Extraction of spores to remove coat layers did not release the GerAC or AA protein from the spores. Both experimental approaches suggest that GerAA and GerAC proteins are located in the inner spore membrane, which forms a boundary around the cellular compartment of the spore. The results provide support for a model of germination in which, in order to initiate germination, germinant has to permeate the coat and cortex of the spore and bind to a germination receptor located in the inner membrane.
A spore germination response requires the interaction of the germinant with a specific receptor on the spore (17). The gerA operon of Bacillus subtilis, required for germination in l-alanine (19, 33, 34), was the first characterized of a family of operons that is present in all spore formers so far examined; multiple family members are present in each genome. B. subtilis also contains the gerB (6) and gerK (10) operons required for germination in the alternative germinative combination of an amino acid, such as l-alanine or l-asparagine, in combination with sugars. Both gerA and gerB operons have been shown to be expressed in the developing forespore under the control of sigma G-associated RNA polymerase (5, 7). The gerA operon encodes three proteins: GerAA, which is predicted to comprise both hydrophobic and hydrophilic domains; GerAB, which is predicted to be an integral membrane protein and is a member of the single-component amino acid/polyamine/organocation transporter superfamily (11); and GerAC, a predicted lipoprotein. Spores of triple mutants in the gerA, gerB, and gerK operons will not germinate on nutrient media (22). Two other homologous operons in the B. subtilis genome (12) presumably encode components that respond to as-yet-unidentified germinants. In addition to the five operons in B. subtilis, there are homologues of demonstrated importance to inosine germination in B. cereus (4) and to germination in macrophages, encoded in the virulence cluster of plasmid pXO1 of B. anthracis (8). The levels of expression from gerA-lacZ fusions are very low and are only detectable by using fluorogenic substrates for β-galactosidase (7). If the Ger proteins are expressed only at a low level during sporulation, it is not likely to be practicable to detect the location of these proteins in spores by immunogold labeling. Fractionation of spores provides an alternative approach to defining their position(s) in the spore. The definition of the location of these proteins in the spore is of crucial importance to our formulation of models for the mechanism of spore germination.
MATERIALS AND METHODS
Bacterial strains and plasmids.
The strains of B. subtilis used in this study are listed in Table 1. The gerA601 deletion mutant has a chromosomal deletion from the SstI site in gerAA to the next SstI site downstream of the operon (32, 34). The gerB null mutation was achieved by insertional inactivation of the first gene of the operon, gerBA. Bases 567 to 923 of the gerB operon sequence, as based on the published numbering (6), were cloned as a Sau3A-BamHI fragment into integrational vector pMTL20EC (3) to yield p13S1, and the resulting plasmid was introduced into the chromosome of B. subtilis strain 1604 by transformation.
TABLE 1.
Bacterial strains
| B. subtilis strain | Genotype | Source or reference |
|---|---|---|
| 1604 | trpC2 | 19 |
| AM047 | gerE36 leu-2 | 16 |
| WB600 | nprΔ aprAΔ eprΔ bpfΔ mpr nprB | 30 |
| AM418 | gerAΔ601 trpC2 | 32 |
| AM1422 | gerAΔ601 trpC2 gerB::p13S1 | This work |
Antibody production.
Peptide TGKKKTRSLTEPTTEKV (amino acids 115 to 131 of GerAA) was coupled, via an added N-terminal cysteine residue, to ovalbumin, and 200 μg of peptide-carrier protein conjugate, emulsified in Freund complete adjuvant, was injected subcutaneously into a rabbit. Plasmid pQEAC was constructed by cloning the open reading frame of GerAC, without the signal sequence, as a 1.7-kb BspEI fragment from pAAM6 (18), into the XmaI site of vector pQE32 (Qiagen, Inc.). This created a gene encoding a protein with an N-terminal histidine tag and linker region fused upstream of amino acid residue 17 of GerAC. This modified GerAC protein, with a His6 tag, was overexpressed from the host Escherichia coli M15(pREP4), purified on a nickel-nitrilotriacetic acid-resin column and used to raise polyclonal antibodies in rabbits. For both types of antigen, rabbits were boosted at 4 and 8 weeks (and at 12 and 16 weeks for the coupled peptide) with 200 μg of protein, emulsified in Freund incomplete adjuvant. The serum was checked at intervals for the presence of antibodies against GerAA or GerAC by Western blotting. Sera were cleaned of anti-E. coli antibodies by treating aliquots with immobilized E. coli cell lysates (Perbio) according to the manufacturer's instructions.
Preparation of spores.
Flasks with 700 ml of CCY broth (27) were inoculated with 7 ml of mid-logarithmic-phase B. subtilis cells in Oxoid nutrient broth and incubated at 37°C until the culture contained free spores (ca. 3 days). The spores were harvested and washed at 4°C by repeated centrifugation in sterile distilled water. The washed spore preparations contained >98% phase bright spores and were free of vegetative cell debris, as judged by phase-contrast microscopy. Spores were stored in distilled water at −20°C.
Fractionation of spores.
Spore breakage was accomplished using a FastPrep cell disintegrator (Bio 101). Spores (170 to 200 mg, dry weight) were suspended in 3 ml of breakage buffer (50 mM Tris HCl, 0.5 mM EDTA, 1 mM phenylmethylsulfonyl fluoride [PMSF]). The spore suspension was then divided equally among 10 FastRNA tubes, supplied preloaded with breakage beads (Grade Blue [Bio 101 product 6020601]). Tubes were subjected to two 30-s bursts of agitation, with 1 min of cooling in an ice bath between bursts. A total of >98% breakage was confirmed by microscopy, and the volume in each tube was raised to 1 ml with breakage buffer. The tubes were mixed by inversion, and the beads allowed to settle for up to 30 s. The crude extract was pipetted off the settled beads and stored on ice for fractionation. Crude spore extract (7.5 ml) from the above procedure was centrifuged at 4°C for 10 min at 16,000 rpm in a Beckman JA20 rotor. The pellet, containing integuments, was washed three times in 20 ml of ice-cold breakage buffer, followed by a final spin for 2 min at 12,000 rpm (15,600 × g) at 4°C in a Jouan MR22i benchtop centrifuge. The pellet was then resuspended in 1 ml of breakage buffer, frozen in liquid nitrogen, and stored at −70°C. The supernatant from the initial centrifugation step, containing membrane and soluble fraction, was centrifuged again for 10 min at 16,000 rpm. The supernatant from this step was then centrifuged at high speed (35,000 rpm, 70 min, at 4°C in a Beckman 50.2 Ti rotor) to sediment the membrane fraction, and the pellet at this stage was checked for the absence of integument material by phase-contrast microscopy. This membrane pellet was resuspended in 250 μl of membrane buffer (100 mM Tris HCl, pH 7.5; 10 mM MgCl2) by six 30-s bursts of sonication in a sonic bath, interspersed with a 30-s cooling period on ice, and then the volume was increased to 18 ml and the membrane pelleting procedure was repeated. The final pellet was resuspended by sonication into 300 μl of membrane buffer, frozen in liquid nitrogen, and stored at −70°C. The soluble fraction from the initial centrifugation was concentrated to 600 μl in an Amicon ultrafiltration cell with a 10-kDa molecular mass cutoff filter. The sample was kept at 4°C during filtration, and the concentrated material was stored at −70°C.
Fractionation of germinated spores.
Spores were germinated at 37°C for 50 min in 10 mM Tris HCl buffer (pH 7.5) with 10 mM l-alanine and 20 mM KCl. At the end of this period, germination was effectively complete: 95% of the spores were phase dark, and the optical density of the suspension was 52% of the original. Germinated spores were harvested and then broken and fractionated as described above for dormant spores.
Electron microscopy.
Electron microscopy was done as described in detail by Leatherbarrow et al. (15). Essentially, after formaldehyde-glutaraldehyde fixation, the material was postfixed in 2% osmium tetroxide, dehydrated, and embedded in Araldite. Sections were stained in uranyl acetate and then in lead citrate.
Preparation of coat-extracted spores for Western blotting.
Coat-extracted spores were prepared by an adaptation of the method of Vary (29). Dormant spores (at 10 mg [dry weight] per ml) were incubated in extraction buffer (0.1 M NaCl, 0.1 M dithiothreitol [DTT], 0.5% sodium dodecyl sulfate [SDS]; adjusted to pH 10.0 with NaOH) at 37°C for 2.5 h and then harvested by centrifugation (5,000 × g, 10 min) at 4°C. The extracted spores were then washed six times in 1 M Tris-HCl (pH 8.0) at 4°C. The spores were confirmed as >90% phase bright, resuspended at 70 mg per ml in breakage buffer with PMSF, and then broken by the FastPrep procedure described above. SDS-polyacrylamide gel electrophoresis (PAGE) sample buffer was added to the tube, the extracts were boiled for 4 min and then centrifuged, and the supernatants were stored at −20°C. The material that was solubilized in the alkaline SDS-DTT extraction procedure was dialyzed against distilled water, the precipitate was removed by centrifugation, and the supernatant was concentrated ca. 15-fold in an Amicon ultrafiltration cell with a 10-kDa molecular mass cutoff filter; the precipitate removed was then resuspended in the concentrated extract, and the resulting suspension was solubilized by boiling for 4 min in SDS-PAGE sample buffer for protein separation by SDS-PAGE.
Isolation of control spore membranes for detection of penicillin-binding proteins.
Isolation of total (inner and outer) membranes was carried out using gerE36 spores by the method of Buchanan and Neyman (2), which involves protoplast preparation from these spores by lysozyme treatment in osmotically supportive medium, harvesting of the protoplasts, and then vigorous vortexing of the mixture in membrane buffer. Isolation of spore inner membranes (from permeabilized spores of wild-type strain 1604) and isolation of vegetative cell membranes was done according to the methods of Buchanan and Neyman (2). Penicillin-binding protein profiles were generated by incubation of 6 μl of membrane protein (4 mg/ml) for 10 min at 37°C with 2 μl of 3H-penicillin (18 μCi/mmol; Amersham). The reaction was terminated by the addition of unlabeled penicillin (to 10 mg/ml). An equal volume of sample buffer was added, samples were boiled for 4 min, and proteins were separated by SDS-PAGE. After staining was done for evaluation of total protein, gels were impregnated in Amplify fluorographic reagent (Amersham Pharmacia) and autoradiographed.
Gel electrophoresis and Western blotting.
SDS-PAGE (14) and Western blotting were done by standard techniques. Proteins were blotted on polyvinylidene difluoride (PVDF) membrane and detected using the Amersham enhanced chemiluminescence (ECL) procedures, according to the manufacturer's instructions. Secondary antibody was horseradish peroxidase (HRP)-linked anti-rabbit immunoglobulin G. Biotinylated markers were detected by using streptavidin-HRP conjugate (Amersham).
RESULTS
Antibodies against Ger proteins.
Polyclonal antibodies were raised against residues 115 to 131 of GerAA, coupled to ovalbumin, and against an overexpressed histidine-tagged GerAC protein lacking the N-terminal prelipoprotein signal sequence cloned in and purified from E. coli cells. Antibodies were preadsorbed with immobilized E. coli proteins and used in Western blots to probe B. subtilis spore proteins separated by SDS-PAGE. In control experiments, these antibodies were able to detect overexpressed and purified cognate proteins in Western blots at high antibody dilutions. For the anti-GerAA antibody, this was a GerA-LacZ fusion protein (this fusion protein had been too unstable to use as an immunogen directly). For the anti-GerAC antibody, the histidine-tagged overexpressed GerAC protein was used. The specificity and any cross-reactivity of the antibodies with other Ger proteins were tested using spores of null mutants in the gerA and gerB operons, as discussed below.
Detection of GerAC and GerBC proteins in spore fractions.
Dormant spores were broken with glass beads, and proteins from crude extracts and from fractions were detected by Western blotting. The strains used were 1604, our laboratory wild type, and WB600, a strain lacking six extracellular proteases, from which spore extracts were more stable (30). Identical results were obtained for both strains (9). Upon probing with anti-GerAC antibody, a band of the expected size for GerAC was detected in extracts of the broken spores of wild type (Fig. 1). This band was visible but normally fainter in equivalent extracts of a gerA-null mutant and was absent in extracts of spores from a gerA gerB double null mutant (Fig. 1). This suggests that, at the concentrations used, the polyclonal antibody preparation detects both GerAC and GerBC proteins but none of the other B. subtilis homologues. Some cross-reaction is not entirely surprising, since GerAC and GerBC are the most closely related in sequence (36% amino acid identity). The GerAC protein had already been confirmed as running at this position in SDS-PAGE analyses by expression of DNA carrying the gerA region under the control of lambda pL in an in vitro-coupled transcription-translation assay (A. Moir, J. McCarvil, and I. M. Feavers, unpublished results). Radiolabeled proteins could be detected in this system, but we have never been able to overexpress either GerAA or GerAB in vivo from this or other expression vectors. The apparent molecular size of GerAC and/or GerBC compares well with predicted sizes of 40,454 and 40,360 for GerAC and GerBC, after cleavage of their prelipoprotein signals, respectively. In the Western blots, depending on the efficiency of the preadsorption with the immobilized E. coli lysates, additional bands were sometimes seen; in Fig. 1, for example, a low-molecular-weight band can be seen that is still present in the gerA gerB double null mutant spores.
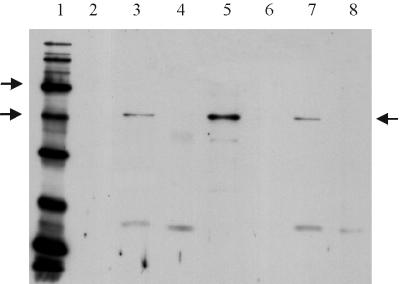
FIG. 1.
Proteins in extracts from broken and fractionated spores were separated on an SDS–12% polyacrylamide gel and transferred to PVDF membrane for Western blotting by the Amersham ECL chemiluminescence detection system. Each sample track contains ca. 11 μg of protein. Detection of GerAC was achieved using anti-GerAC antibody, diluted at 1/500. The secondary antibody was used at 1/15,000 dilution. Lane 1 contained biotinylated protein standards (Sigma SDS-6B: sizes of 205, 116, 97, 58, 40, 29, 20, 14.3, and 6.5 kDa). Arrows by the marker lane indicate the 58 and 40-kDa markers, and the arrow on the right of the gel indicates the GerAC band. Lane 2 contained prestained protein markers, lane 3 contained total extract from broken spores of strain 1604, and lanes 4 to 6 contained integument, membrane, and soluble fractions, respectively, of strain 1604. Lanes 7 and 8 contained total spore extracts of strains AM418 (gerAΔ) and AM1422 (gerAΔ gerB null), respectively.
In spore fractions (Fig. 1), the GerAC and GerBC proteins are detectable in the “membrane” fraction, but not in the integument or soluble fractions. The signal in the membrane fraction is more intense than in the total spore extract, since equivalent amounts of protein were loaded in each track, and the GerAC and GerBC proteins are relatively enriched in the membrane fraction. In fractionated germinated spores, the GerAC and GerBC proteins remained in the same membrane fraction and retained the same apparent molecular weight, as in dormant spores (9). Since only wild-type spores were fractionated, there is a formal possibility that only one of the two homologous proteins, GerAC or GerBC, is present in this fraction, but since no signal is detected in the other fractions, the other homologue would have to have been destroyed completely and specifically in the fractionation procedure; this is extremely unlikely.
Detection of GerAA protein in spore fractions.
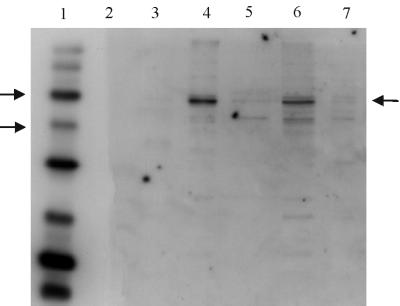
The detection of GerAA was less sensitive, since the anti-peptide antibody had a lower effective titer, but a band of the expected size was detected in broken spore extracts (Fig. 2). The observed position of the band, at ca. 56 kDa, is consistent with the predicted molecular weight of 53,789 for GerAA, and our unpublished data from in vitro transcription-translation experiments. No band was detected at this position in spore extracts from a gerA gerB double null mutant (Fig. 2) or from a gerA mutant (shown below in Fig. 6) using this concentration of antibody. The smaller band (ca. 42 kDa) detected in extracts in Fig. 2 was not gerA related, since it was still present in the double null mutant. These data confirm that the protein detected at 56 kDa is GerAA; like GerAC, it is found in the “membrane” fraction.
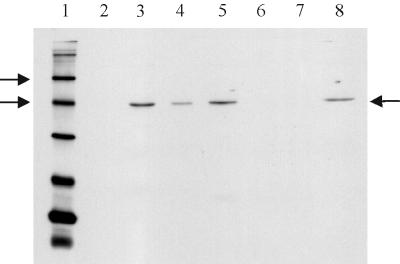
FIG. 2.
Western blot of spore extracts probed with anti-GerAA anti-peptide antibody. Conditions used were as for Fig. 1, except that the secondary antibody was used at a 1 in 10,000 dilution. Lanes 1 and 2 contained biotinylated and prestained protein markers, respectively. Arrows by the marker lane indicate the 58 and 40-kDa markers, and the arrow on the right of the gel indicates the GerAA band. Lanes 3 to 5 contained integument, membrane, and soluble fractions, respectively, of dormant spores of strain WB600. Lane 6 contains total extract from WB600 spores, and lane 7 contained an extract of spores of strain AM1422 (gerAΔ gerB null).
FIG. 6.
Detection of GerAA protein in Western blots of coat-stripped spores. Lanes 3 to 9 contain 11 μg of protein. Conditions were as described for Fig. 2. Lanes 1 and 2 are markers, as in the other Western blots. Arrows by the marker lane indicate the 58- and 40-kDa biotinylated protein standards in Sigma SDS-6B, and the arrow on the right indicates the position of the GerAA band. Lane 3 and 4 contained total spore extracts of WB600 and AM047, respectively. Lanes 5 and 6 contained extracts from the gerA and gerAB null mutant spores, respectively. Lane 7 contains concentrated extracted coat and outer membrane material from spores of strain AM047, and lane 8 contains protein from the broken, decoated, AM047 spores.
Composition of the fractions.
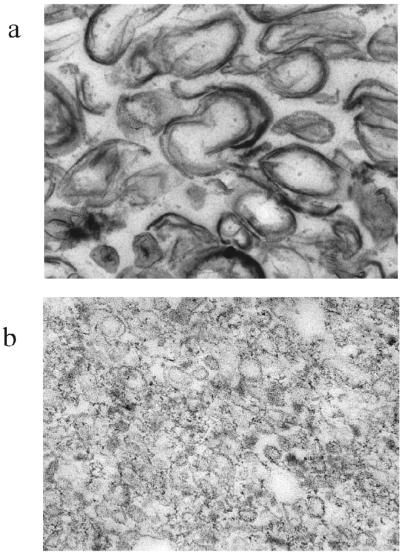
The spore breakage procedure used generated very large integument fragments, as visualized by light microscopy. Transmission electron microscopy of sections taken from samples of those pellets from differential centrifugation that were to be resuspended to form the integument and membrane fractions confirmed the expected distribution of the surface layers. In the integument fraction, material from inner and outer coats can be observed, as well as less clearly staining fibrillar cortex material (Fig. 3a). The membrane fraction (Fig. 3b) was primarily vesicular and appeared to be essentially free of coat or cortex.
FIG. 3.
Electron micrographs of material in integument (a) and membrane (b) fractions. Magnification: ×21,000 and ×78,000, respectively.
Absence of PBP5∗ in the spore membrane fraction.
The degree of integrity of the spore outer membrane is not certain, but at least remnants of it, and proteins associated with it, persist in the spore. For example, Buchanan and Neyman (2) reported that penicillin-binding protein PBP5∗, which is involved in synthesis of the spore cortex peptidoglycan, is located exclusively in the mother cell membrane that becomes the outer membrane of the mature spore.
It might be expected that the outer spore membrane, between coats and cortex, would be retained in the integument fraction, but it was important to establish directly whether the outer membrane was absent from our “membrane” fraction from broken spores. We repeated the experiments of Buchanan and Neyman (2) to generate controls that would demonstrate the spore penicillin-binding protein profile (Fig. 4). These included a total (inner plus outer) spore membrane preparation from spores of a coat-defective gerE mutant, from which it is easy to extract both membranes. The expected penicillin-binding protein profile, including PBP5∗, was seen in this total extract. Membranes extracted from wild-type spores after removal of the coat layers, which would also remove the outer membrane, resulted in a preparation lacking PBP5∗, as previously described. These controls were compared with the penicillin-binding protein profile of the spore membrane fractions (prepared from dormant and germinated spores) used in our Western blot analysis. The PBP5 band was present at normal levels in the membrane fraction, but PBP5∗ was absent, even when the autoradiograph was overexposed to highlight minor bands (Fig. 4). This provides strong evidence that the “membrane fraction” of our GerA protein localization experiments does not contain a significant amount of spore outer membrane, therefore confirming that the membrane material in this fraction, and the GerA proteins detected, are derived from the inner membrane only.
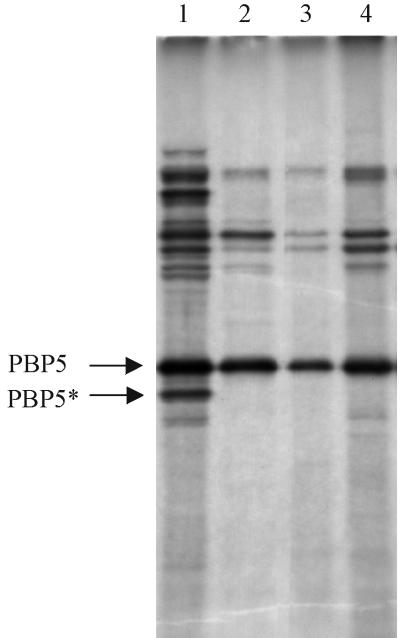
FIG. 4.
Penicillin-binding protein profiles. Penicillin-binding proteins were labeled with [3H]penicillin and separated on an SDS–10% polyacrylamide gel. Bands corresponding to PBP5 and PBP5∗ are arrowed. Lane 1, total spore membranes from AM047; lane 2, the membrane fraction from dormant spores of strain 1604; lane 3, the membrane fraction from germinated spores of 1604, as used for Western blotting; lane 4, membrane from coat-stripped spores of 1604. The autoradiograph has been overdeveloped to expose faint bands to establish that there is no evidence of PBP5∗ in the membrane fractions in lanes 2 and 3.
GerAC and GerAA remain associated with the spore after spore coat extraction.
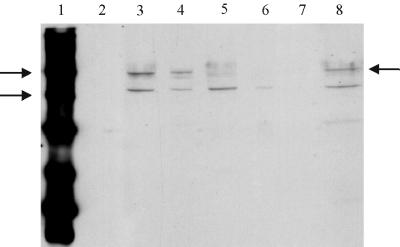
Spore coat extraction was used to provide a second independent line of evidence that the GerAA and GerAC proteins are not located in the outer layers of the spore. A gerE36 mutant was used for this experiment, since the coat layers are incomplete in this mutant and can be extracted efficiently. Although coat defective, this mutant shows a normal early germination response to alanine (16). The coat extraction treatment would be expected to remove both coats and the consequently accessible spore outer membrane material. The decoated spore preparation was examined by electron microscopy: ca. 90% of the spores had lost their coats and associated layers and were composed of cores surrounded by spore cortex (9). The decoated spores were then broken. Proteins from the total broken spore preparation were extracted into gel loading buffer and separated by SDS-PAGE, and the Ger proteins were detected immunologically. Figure 5 shows the results for GerAC. A single band corresponding to GerAC plus GerBC was seen in extracts from whole spores of WB600 and AM047 (gerE36). It was present in a gerAΔ strain and absent in a gerAΔ gerB null double mutant, confirming the specificity of the antibody for GerAC and GerBC proteins. These proteins were retained in AM047 (gerE36) spores after coat extraction, and none was detected in the extracted material. The same conclusion was reached for GerAA (Fig. 6), although additional cross-reacting bands that were not GerAA related are also present on the blot.
FIG. 5.
Western blot of GerAC and GerBC proteins from coat-stripped spores. Each sample track contains ca. 11 μg of protein. Conditions for electrophoresis and Western blotting were as for Fig. 1. Arrows by the marker lane indicate the 58- and 40-kDa markers, and the arrow on the right of the gel indicates the GerAC band. Lane 1, biotinylated markers; lane 2, prestained protein markers; lane 3, total spore extract of strain WB600; lane 4, total spore extract of strain AM047 (gerE36). Lanes 5 and 6 contained total spore extracts of the gerAΔ and gerAΔ gerBΔ strains respectively. Lane 7 contains concentrated extracted coat and outer membrane material from spores of strain AM047, and lane 8 contains protein from the broken, decoated, AM047 spores.
DISCUSSION
The GerA proteins represent the paradigm of a group of proteins found in spores that are responsible for germinant recognition and transduction of the germination signal. The evidence presented above suggests very strongly that these are located in a membrane fraction that contains spore inner, but not outer, membrane. The appearance of GerAA in a membrane fraction is not surprising, given its extensive hydrophobic domain of ca. 200 amino acids. Since it is expressed in the forespore, the most likely final target of the protein would be the forespore membrane rather than the mother cell-derived outer spore membrane. Similarly, the GerAC protein is predicted to be a lipoprotein and would be synthesized in the forespore and then exported across the forespore membrane and associated with it via a lipid anchor. All “membrane” fractions will also contain particulate material that sediments at similar g forces but, given the predicted membrane association of GerA proteins, it is reasonable to suggest that an inner membrane location is more likely. The GerAB protein, as an integral membrane protein synthesised in the forespore, is also likely to be located in the same membrane, although this has not been tested experimentally. In our experience from in vitro expression experiments, the GerAB protein was not reproducibly detectable by SDS-PAGE under our conditions, making a similar Western blot analysis impractical.
Spore formers contain multiple copies of ger operons involved in the germination response to different nutrient germinants. The conservation of sequence similarities and hydrophobicity profiles suggests that all retain the same general features. Although there is only direct evidence for the compartmentalization of gene expression for the gerA and gerB operons (5, 7), and evidence here for the inner membrane location of GerAA, GerAC, and GerBC proteins, it is expected that the proteins from all of the homologues would be similarly located.
Since GerAC and GerBC are predicted lipoproteins, it is likely that these are transferred across the forespore membrane and anchored to its surface. Not all forespore-expressed lipoproteins need be retained in such a position, however; the GerD protein, also synthesized in the forespore compartment and a prelipoprotein, is localized in the integument fraction of mature spores and can be released from it by extraction in gel loading buffer or by lysozyme digestion of the peptidoglycan in the integument fraction (24).
We suggest that the evidence strongly supports an inner membrane location for a spore germination receptor. The coevolution and noninterchangeability of the homologues in different ger operons suggests that the GerAA, -AB, and -AC proteins are likely to form a complex, and genetic evidence for interaction between GerBA and GerBB has been obtained (21). There is strong genetic evidence for the role of the GerA and GerB proteins as receptors in the spore responding to l-alanine. Mutations in gerBA and gerBB have been isolated that alter the germinant specificity of the gerB system to recognize d-alanine (21). The gerAB38 and gerAB44 mutations alter the concentration of germinant required for the spore's germination response (26, 33) and affect residues in the likely membrane-spanning regions of this protein (B. M. Corfe and A. Moir, unpublished results). The germination phenotypes of gerE mutant spores (16) and coat-stripped spores (1, 13, 29) have demonstrated that the initial response of the spores to germinant is unaltered, confirming the expectation that the signal recognition apparatus is not located in the outer layers of the spore.
The proposed location for the GerA proteins based on the data presented here is in disagreement with the suggestions (from immunogold labeling experiments) of Sakae et al. (25), who have argued that the GerAB protein, and therefore the germination receptor apparatus, is located at the outer edge of the cortex, and those of Yasuda et al. (31), who reported that antibodies to all three proteins bound to the surface of cortex-less coat fragments. These experiments were carried out with anti-peptide antibodies raised against GerA peptide sequences, but no evidence was provided to demonstrate that these antibodies would detect exclusively GerA proteins. In our work, two separate approaches—spore fractionation and stripping of the coat layers—both suggest the same membrane location for the GerAA and for the GerAC+GerBC proteins. In an earlier review (20) we suggested from preliminary evidence that the GerAA protein would be located in a membrane fraction; Paidhungat and Setlow (22a) also have found an inner membrane location for the GerBA protein, which was not examined in our work.
The current model of germination requires that the germinant penetrates to the inner membrane of the spore, where it initiates an as-yet-undefined signal transduction process that leads to the loss of heat resistance. It interacts with at least the GerAA and GerAB proteins. Since the GerAB family of proteins are evolutionarily related to a superfamily of membrane transport proteins, these may have evolved from such to act as signal transducers, binding germinant within the membrane. The gerN gene product, an Na/H antiporter homologue, is required for the earliest stages of inosine germination in B. cereus (28), suggesting a role for ion movement in the germination process. For all these reasons, the initial signal transduction process is likely to be membrane associated. Hydrolysis of the cortex is necessary for later stages of germination, though not for the initial loss of heat resistance in response to germinant (23). The precise events of germination remain to be clarified, but the data presented here suggest strongly that these events occur at the inner spore membrane.
ACKNOWLEDGMENTS
This work was funded by BBSRC project grant 50/F08208 and a BBSRC CASE studentship with Unilever Research (K.D.H.).
We thank John Proctor for help with electron microscopy and Penny Thackray for reading the manuscript.
REFERENCES
- 1.Behravan J, Chirakkal H, Masson A, Moir A. Mutations in the gerP locus of Bacillus subtilis and Bacillus cereus affect the access of germinants to their target in the spore. J Bacteriol. 2000;182:1987–1994. doi: 10.1128/jb.182.7.1987-1994.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Buchanan C E, Neyman S L. Correlation of penicillin binding protein composition with different functions of two membranes in Bacillus subtilis forespores. J Bacteriol. 1986;165:498–503. doi: 10.1128/jb.165.2.498-503.1986. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Chambers S P, Prior S E, Barstow D A, Minton N. The pMTL nic-cloning vectors. I. Improved pUC polylinker regions to facilitate the use of sonicated DNA for nucleotide sequencing. Gene. 1988;68:139–149. doi: 10.1016/0378-1119(88)90606-3. [DOI] [PubMed] [Google Scholar]
- 4.Clements M O, Moir A. Role of the gerI operon of Bacillus cereus 569 in the response of spores to germinants. J Bacteriol. 1998;180:6729–6735. doi: 10.1128/jb.180.24.6729-6735.1998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Corfe B M, Moir A, Popham D, Setlow P. Analysis of the expression and regulation of the gerB spore germination operon of Bacillus subtilis 168. Microbiology. 1994;140:3079–3083. doi: 10.1099/13500872-140-11-3079. [DOI] [PubMed] [Google Scholar]
- 6.Corfe B M, Sammons R L, Smith D A, Mauel C. The gerB region of the Bacillus subtilis 168 chromosome encodes a homologue of the gerA spore germination operon. Microbiology. 1994;140:471–478. doi: 10.1099/00221287-140-3-471. [DOI] [PubMed] [Google Scholar]
- 7.Feavers I M, Foulkes J, Setlow B, Sun D, Nicholson W, Setlow P, Moir A. The regulation of transcription of the gerA spore germination operon of Bacillus subtilis. Mol Microbiol. 1990;4:275–282. doi: 10.1111/j.1365-2958.1990.tb00594.x. [DOI] [PubMed] [Google Scholar]
- 8.Guidi-Rontani C, Pereira Y, Ruffie S, Sirard J-C, Weber-Levy M, Mock M. Identification and characterization of a germination operon on the virulence plasmid pXO1 of Bacillus anthracis. Mol Microbiol. 1999;33:407–414. doi: 10.1046/j.1365-2958.1999.01485.x. [DOI] [PubMed] [Google Scholar]
- 9.Hudson K D. The localisation of germination proteins in spores of Bacillus subtilis. Ph.D. thesis. Sheffield, United Kingdom: University of Sheffield; 1999. [Google Scholar]
- 10.Irie R, Fujita Y, Kobayashi M. Nucleotide sequence and gene organisation of the gerK spore germination locus of Bacillus subtilis 168. J Gen Appl Microbiol. 1996;42:141–153. [Google Scholar]
- 11.Jack D L, Paulsen I T, Saier M H. The amino acid/polyamine/organocation APC superfamily of transporters specific for amino acids, polyamines and organocations. Microbiology. 2000;146:1797–1814. doi: 10.1099/00221287-146-8-1797. [DOI] [PubMed] [Google Scholar]
- 12.Kunst F, Ogasawara N, Moszer I, Albertini A M, Alloni G, Azevedo V, et al. The complete genome sequence of the gram-positive bacterium Bacillus subtilis. Nature. 1997;390:249–256. doi: 10.1038/36786. [DOI] [PubMed] [Google Scholar]
- 13.Kutima P M, Foegeding P M. Involvement of the spore coat in germination of Bacillus cereus T spores. Appl Environ Microbiol. 1987;53:47–52. doi: 10.1128/aem.53.1.47-52.1987. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Laemmli U K. Cleavage of structural proteins during assembly of the head of bacteriophage T4. Nature. 1970;227:680–689. doi: 10.1038/227680a0. [DOI] [PubMed] [Google Scholar]
- 15.Leatherbarrow A J H, Yazdi M A, Curson J P, Moir A. The gerC locus of Bacillus subtilis, required for menaquinone biosynthesis, is concerned only indirectly with spore germination. Microbiology. 1998;144:2125–2130. doi: 10.1099/00221287-144-8-2125. [DOI] [PubMed] [Google Scholar]
- 16.Moir A. Germination properties of a spore coat-defective mutant of Bacillus subtilis 168. J Bacteriol. 1981;146:1106–1116. doi: 10.1128/jb.146.3.1106-1116.1981. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Moir A, Smith D A. The genetics of bacterial spore germination. Annu Rev Microbiol. 1990;44:531–553. doi: 10.1146/annurev.mi.44.100190.002531. [DOI] [PubMed] [Google Scholar]
- 18.Moir A, Feavers I M, Guest J R. Characterization of the fumarase gene of Bacillus subtilis 168 cloned and expressed in Escherichia coli K12. J Gen Microbiol. 1984;130:3009–3017. doi: 10.1099/00221287-130-11-3009. [DOI] [PubMed] [Google Scholar]
- 19.Moir, Lafferty A E, Smith D A. Genetic analysis of spore germination mutants of Bacillus subtilis 168: the correlation of phenotype with map location. J Gen Microbiol. 1979;111:165–180. doi: 10.1099/00221287-111-1-165. [DOI] [PubMed] [Google Scholar]
- 20.Moir A, Kemp E H, Robinson C, Corfe B M. The genetic analysis of bacterial spore germination. J Appl Bacteriol. 1994;76:9S–16S. [PubMed] [Google Scholar]
- 21.Paidhungat M, Setlow P. Isolation and characterization of mutations in Bacillus subtilis that allow spore germination in the novel germinant d-alanine. J Bacteriol. 1999;181:3341–3350. doi: 10.1128/jb.181.11.3341-3350.1999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Paidhungat M, Setlow P. Role of Ger proteins in nutrient and non-nutrient triggering of spore germination in Bacillus subtilis. J Bacteriol. 2000;182:2513–2519. doi: 10.1128/jb.182.9.2513-2519.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22a.Paidhungat M, Setlow P. Localization of a germinant receptor protein (GerBA) to the inner membrane of Bacillus subtilis spores. J Bacteriol. 2001;183:3982–3990. doi: 10.1128/JB.183.13.3982-3990.2001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Popham D L, Helin J, Costello C E, Setlow P. Muramic lactam in peptidoglycan of Bacillus subtilis spores is required for spore outgrowth but not for spore dehydration or heat resistance. Proc Natl Acad Sci USA. 1996;93:15405–15410. doi: 10.1073/pnas.93.26.15405. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Robinson C. A molecular analysis of the gerD and gerF spore germination genes of Bacillus subtilis 168. Ph.D. thesis. Sheffield, United Kingdom: University of Sheffield; 1996. [Google Scholar]
- 25.Sakae Y, Yasuda Y, Tochikubo K. Immunoelectron microscopic localization of one of the spore germination proteins, GerAB, in Bacillus subtilis spores. J Bacteriol. 1995;177:6294–6296. doi: 10.1128/jb.177.21.6294-6296.1995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Sammons R L, Moir A, Smith D A. Isolation and properties of spore germination mutants of Bacillus subtilis 168 defective in the initiation of germination. J Gen Microbiol. 1981;124:229–241. [Google Scholar]
- 27.Stewart G S A B, Johnstone K, Hagelberg E, Ellar D J. Commitment of bacterial spores to germinate: a measure of the trigger reaction. Biochem J. 1981;198:101–106. doi: 10.1042/bj1980101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Thackray P D, Behravan J, Southworth T W, Moir A. GerN, an antiporter homologue important in the germination of Bacillus cereus endospores. J Bacteriol. 2001;183:476–482. doi: 10.1128/JB.183.2.476-482.2001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Vary J C. Germination of Bacillus megaterium spores after various extraction procedures. J Bacteriol. 1973;116:797–802. doi: 10.1128/jb.116.2.797-802.1973. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Wu X C, Lee W, Tran L, Wong S L. Engineering a Bacillus subtilis expression-secretion system with a strain deficient in 6 extracellular proteases. J Bacteriol. 1991;173:4952–4958. doi: 10.1128/jb.173.16.4952-4958.1991. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Yasuda Y, Sakae Y, Tochikubo K. Immunological detection of the GerA spore germination proteins in the spore integuments of Bacillus subtilis using scanning electron microscopy. FEMS Lett. 1996;139:235–238. doi: 10.1111/j.1574-6968.1996.tb08208.x. [DOI] [PubMed] [Google Scholar]
- 32.Zuberi M A. A molecular analysis of the gerA spore germination locus of Bacillus subtilis 168. Ph.D. thesis. Sheffield, United Kingdom: University of Sheffield; 1985. [Google Scholar]
- 33.Zuberi M A, Feavers I M, Moir A. Identification of three complementation units in the spore germination gerA locus of Bacillus subtilis 168. J Bacteriol. 1985;162:756–762. doi: 10.1128/jb.162.2.756-762.1985. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Zuberi M A, Moir A, Feavers I M. The nucleotide sequence and gene organisation of the gerA spore germination operon of Bacillus subtilis 168. Gene. 1987;51:1–11. doi: 10.1016/0378-1119(87)90468-9. [DOI] [PubMed] [Google Scholar]