Highlights
-
•
The 2019 coronavirus (COVID-19) caused by severe acute respiratory syndrome coronavirus 2 (SARS-CoV-2) has become a global epidemic threatening the lives and health of people worldwide. Currently, there are no effective therapies or available vaccines for COVID-19.
-
•
In SARS-CoV-2 infection, lncRNAs are demonstrated to be closely related to viral infection, interferon and cytokine storm in COVID-19. LncRNA NEAT1, DANCR, MALAT1, C058791.1, TTTY15 and TPTEP1 played a role in the infection of SARS-CoV-2. lncRNAs also can modulate the expression of interferon. Inversely, interferon can stimulate the expression of lncRNAs. Besides, lncRNAs can regulate the cytokine storm by downregulating the expression of cytokines.
-
•
Thus, we can speculate that developing drugs targeting some specific sites in the pathway or relation network of lncRNAs may be promising strategy to treat SARS-CoV-2 infection.
Keywords: LncRNAs, COVID-19, SARS-CoV-2, Viral infection, Interferon, Cytokine storms
Abstract
Coronavirus disease 2019 (COVID-19) caused by severe acute respiratory syndrome coronavirus 2 (SARS-CoV-2) has become a global pandemic threatening the lives and health of people worldwide. Currently, there are no effective therapies or available vaccines for COVID-19. The molecular mechanism causing acute immunopathological diseases in severe COVID-19 is being investigated. Long noncoding RNAs (lncRNAs) have been proven to be involved in many viral infections, such as hepatitis, influenza and acquired immune deficiency syndrome. Many lncRNAs present differential expression between normal tissue and virus-infected tissue. However, the role of lncRNAs in SARS-CoV-2 infection has not been fully elucidated. This study aimed to review the relationship between lncRNAs and viral infection, interferon and cytokine storms in COVID-19, hoping to provide novel insights into promising targets for COVID-19 treatment.
1. Introduction
Coronavirus disease 2019 (COVID-19) is caused by severe acute respiratory syndrome coronavirus 2 (SARS-CoV-2). COVID-19 spread at an alarming rate, threatening the lives and health of the entire human race. SARS-CoV-2, a single-stranded RNA virus, is a member of the Coronaviridae family (Pontecorvi et al., 2020). Patients with COVID-19 present with fever, dry cough and respiratory failure, combined with gastrointestinal and neurological symptoms (Wiersinga et al., 2020). It is projected that asymptomatic dissemination is responsible for nearly half of SARS-CoV-2 transmission (Al-Tawfiq, 2020). Respiratory failure accounts for the highest mortality of patients with COVID-19 (Ruan et al., 2020). SARS-CoV-2 can lead to acute respiratory distress syndrome (ARDS) mediated by proinflammatory cytokines (Ziegler et al., 2020). Proinflammatory cytokines can cause severe damage to vital organs and lead to multiple organ dysfunction syndrome (MODS) (Xiong et al., 2020). The main therapeutic measures include convalescent plasma, remdesivir and cytokine blocker drugs (Chen et al., 2020). Specific vaccination represents a key strategy against SARS-CoV-2 infection. According to the World Health Organization (WHO), more than 360 vaccines have been developed, and 45 vaccines have been evaluated in phase III clinical trials to date (Fiolet et al., 2022). These COVID-19 vaccines seem to be well tolerated and display a preventive effect on the original strain and variants of SARS-CoV-2 (Fiolet et al., 2022). However, the regulatory mechanism of host genetic expression in response to infection remains unclear (Casagrande et al., 2021). It is necessary to explore viral pathogenesis and biological reactions against SARS-CoV-2 in the host.
Long noncoding RNAs (lncRNAs), a subclass of noncoding RNAs, lack an open reading frame and have a length of more than 200 nucleotides (Yang et al., 2014).
By competing with endogenous RNA (ceRNA), lncRNA serves as a miRNA sponge, regulating the expression of target genes. LncRNA, miRNA, and mRNA were used to construct a lncRNA‒miRNA-mRNA competing endogenous (ceRNA) network (Liu et al., 2018). LncRNAs are involved in a broad spectrum of biological processes, especially immunity and inflammatory reactions (Atianand et al., 2017; Stojic et al., 2020). The lncRNA XIST plays a role in inflammation via the NF-κB/NLRP3 pathway (Ma et al., 2019). Chen et al. found that the lncRNA MALAT1 functions as a negative factor in inflammation in sepsis (Chen et al., 2018). It has been revealed that lncRNA SNHG14 enhances the inflammatory response by increasing the level of ROCK1 and downregulating the expression of miR-136–5p (Zhong et al., 2019). Several lncRNAs have been identified in multiple viral infections, such as hepatitis and AIDS (Imam et al., 2015; Sur et al., 2018). Notably, lncRNAs were also identified in the process of SARS-CoV-2 infection. LncRNA H19 regulates spike transcription of SARS-CoV-2 by binding to the genome of SARS-CoV-2, thereby affecting SARS-CoV-2 infection (Natarelli et al., 2021). However, the relationship of lncRNAs with viral infection and the antiviral inflammatory response in COVID-19 needs to be clarified.
2. LncRNAs in SARS-CoV-2 viral infection
Some lncRNAs, such as NEAT1 and EGOT, increase in influenza A virus and HIV infection (Laha et al., 2021). In addition, NEAT1 suppresses the replication and growth of HIV and Hantaan virus (HTNV) (Pan et al., 2019; Maarouf et al., 2019; Zhang et al., 2013; Ma et al., 2017). Similarly, in SARS-CoV-2 viral infection, lncRNA NEAT1 and DANCR change the level of inflammatory transcripts by regulating the expression of immune-related genes (Meydan et al., 2020). Approximately 500 lncRNAs were found to be differentially expressed during SARS-CoV-2 infection (Peng et al., 2010). HOTAIRM1, PVT1 and AL392172.1 can bind to the SARS-CoV-2 genome with high affinity, suggesting the major regulatory role of lncRNAs in COVID-19 (Moazzam-Jazi et al., 2021). Transcriptome analysis of bronchial epithelial cells in patients with SARS-CoV-2 infection showed that the lncRNA MALAT1 had significantly different expression levels. The lncRNA MALAT1 may be a potential biomarker of SARS-CoV-2 infection (Vishnubalaji et al., 2020). An in silico study detected the expression levels of lncRNAs both in cell lines and lung tissue. In vitro, there were a total of 20 overexpressed lncRNAs and 4 downregulated lncRNAs during SARS-CoV-2 infection. Moreover, NEAT1 was upregulated in the lung tissue of patients with SARS-CoV-2 infection (Laha et al., 2021). Another study found that a total of 898 lncRNAs (414 overexpressed lncRNAs; 484 downregulated lncRNAs) were differentially expressed between healthy individuals and patients with SARS-CoV-2 infection. In addition, these differentially expressed lncRNAs were likely to be associated with exosomes and regulate the inflammatory process (Wu et al., 2021). Transcriptome analysis of SARS-CoV-2-infected lung tissues showed that three lncRNAs (C058791.1, TTTY15 and TPTEP1) were enriched with maximum target genes. These three lncRNAs interact with genes via the lncRNA‒miRNA-mRNA pathway and release the mRNA for translation. The activity of AC058791.1 has not been identified; TTTY15 is involved in proteolysis and ubiquitin-dependent catabolism (Lei et al., 2019), and TPTEP1 inhibits STAT3 phosphorylation (Ding et al., 2019).
Furthermore, lncRNA H19 can bind to the 5′UTR of the SARS-CoV-2 genome and modulate the spike transcript in viral infection (Natarelli et al., 2021). LncRNA-based oligosequences can be candidates to combat SARS-CoV-2. Of note, these results need to be experimentally validated at the patient level. Clinically, a comprehensive profile of COVID-19-related lncRNAs in peripheral blood mononuclear cells of patients with SARS-CoV-2 infection and healthy individuals was built. More specifically, 1072 lncRNAs were differentially expressed between the two populations. The top three increased lncRNAs are ENSG00000231412 (AC005392.2), followed by ENSG00000274173 (AL035661.1) and ENSG00000231535 (LINC00278). The top decreased lncRNAs are ENSG00000229807 (XIST) and ENSG00000273160 (AL359962.2) (Cheng et al., 2021). These differentially expressed lncRNAs may be potential biomarkers for the diagnosis and prognosis of patients with SARS-CoV-2 viral infection. Collectively, these COVID-19-related lncRNAs may be possible innovative candidates against SARS-CoV-2 infection. Further studies are needed to fully identify and understand the functions of lncRNAs in COVID-19.
3. LncRNAs and interferon in COVID-19
LncRNAs were shown to modulate interferon-stimulated genes (ISGs) in the inflammatory process. LncRNA#32/LUARIS can combat EMCV, HBV and HCV infection by regulating the transcription factor ATF2 to alter the expression of ISG (Nishitsuji et al., 2016). Interestingly, angiotensin I converting enzyme 2 (ACE2), the receptor for COVID-19, is an ISG. SARS-CoV-2 could exploit species-specific interferon-driven upregulation of ACE2 to aggravate the host immune response and enhance infection (Ziegler et al., 2020). Additionally, lncRNAs can be stimulated by interferon (IFN). The expression of NRIR/lncCMPK2 can be activated by IFN and then modulated by signal transducer and activator of transcription 2 (STAT2). There was a reliable association between the expression of IFN and SARS-CoV-2 infection both in laboratory and clinical studies (Vishnubalaji et al., 2020). It was observed that lncRNAs can modify ISG levels in different viral infections (Nishitsuji et al., 2016; Kambara et al., 2014; Xie et al., 2018; Carpenter et al., 2013). In contrast, Laha et al. found that the expression of lncRNAs had no correlation with interferons. Then, they investigated the interaction between lncRNAs and heterogeneous nuclear ribonucleoproteins (hnRNPUs), a group of RNA-binding proteins. They found that several lncRNAs could be regulated by interferon regulatory factors (IRFs) and STAT in response to SARS-CoV-2 infection (Laha et al., 2021). LncRNAs might be involved in the antiviral response by regulating IFN. However, several host lncRNAs can repress the viral immune response by downregulating type I interferons (IFN-1). In the early stage of SARS-CoV-2 viral infection in asymptomatic patients, the production of IFN, especially IFN-b, was delayed. More importantly, SARS-CoV-2 N protein can inhibit IFN production by disturbing the retinoic acid-induced gene I (RIG-I) pathway (Yang et al., 2021). Clinically, IFN-α is a common interferon for antiviral therapy in patients with COVID-19. These lncRNAs mediate IFN regulation in COVID-19 and should be validated to explore effective antiviral strategies in future studies.
4. LncRNAs regulate cytokine storms in COVID-19
The organ damage caused by the inflammatory reaction accounts for the high mortality and morbidity in COVID-19 (Tay et al., 2020; Perlman and Dandekar, 2005). Studies in human and animal models implied that immunopathological events might lead to the death of patients with SARS-CoV-2 viral infection (Channappanavar et al., 2016; Rockx et al., 2009). In addition, the lung tissue of patients with SARS-CoV-2 infection displayed pathological infiltration of immune cells such as macrophages and monocytes (Yao et al., 2020). It has been validated that an inflammatory cytokine storm, an overwhelming inflammatory immune response with hyperproduction of mainly proinflammatory cytokines, such as IL-1, IL-6, IL-12, IFN-γ, and TNF-α, contributes to the development of SARS-CoV-2 infection and is the cause of severe COVID-19 (Zumla et al., 2020; Zhang et al., 2020; Costela-Ruiz et al., 2020). LncRNAs can target cytokines in inflammatory cytokine storms in COVID-19. LncRNAs with the potential to regulate the inflammatory response showed differential expression in patients with SARS-CoV-2 infection compared with healthy individuals (Wu et al., 2021). LncRNAs MALAT1 and NEAT1 may contribute to the development of inflammation in SARS-CoV-2-infected cells (Moazzam-Jazi et al., 2021). In contrast, 22 lncRNAs bound to 10 important cytokines, and 8 of 22 lncRNAs targeted multiple cytokines. RAD51-AS1 and lnrCXCR4 each can target 3 cytokines. Notably, the lncRNA NORAD, which is activated by DNA damage, can bind with 5 cytokines, including interleukin (IL)−6, IL-10, CSF3, tumor necrosis factor (TNF)-α and CXCL10 (Morenikeji et al., 2020). LncRNAs may target and bind important cytokine nucleotide sequences and possibly decrease the expression of cytokines, thus reducing the emergence of cytokine storms in the infection. Given the interaction between lncRNAs and inflammatory cytokines, some methods, such as viral gene therapy, RNAi knockdown, viral vectors and antisense oligonucleotides, have been used in clinical practice (Fatemi et al., 2014; Roberts et al., 2020). Agents targeting lncRNAs show promise in enhancing the anti-SARS-CoV-2 response by inhibiting the cytokine storm.
5. LncRNAs function as potential targets for COVID-19
Several functional lncRNAs involved in viral infection and cytokine storms in COVID-19 are summarized in Table 1. In a coexpression network analysis of human lung epithelial cell lines and bronchoalveolar lavage fluid from patients with SARS-CoV-2 infection, four lncRNAs (WAKMAR2, EGOT, EPB41L4A-AS1, and ENSG00000271646) were upregulated. These four lncRNAs were associated with multiple cytokine pathways and overactivated inflammatory responses (Mukherjee et al., 2021). LncRNAs can regulate the expression of IL-6 and NLRP3 through epigenetic, transcriptional, and post-transcriptional mechanisms. Agents targeting these signaling pathways have been developed. Tocilizumab, an IL-6 receptor antagonist, effectively inhibits the IL-6 receptor (IL-6R) and reduces the serum levels of C-reactive protein (CRP) and serum amyloid A (SAA) (Nishimoto et al., 2003; Nishida et al., 2009). Similarly, BML-111, an IL-6 blocker, can increase the level of lncRNA MALAT1 and then downregulate the expression of inflammatory factors, such as monocyte chemotactic protein-1 (MCP-1) and IL-6 (Li et al., 2018; Gong et al., 2012). A drug perturbation analysis found that digoxin and proscillaridin can regulate gene expression levels by increasing or decreasing the expression levels of some lncRNAs. By conducting molecular docking and drug perturbations on gene expression, we found that digoxin and proscillaridin can be used to treat severe COVID-19 infections (Aishwarya et al., 2020). Moreover, a bioinformatic analysis implied that the TGF-beta signaling pathway is interactive and involved in the network of lncRNAs, human proteins, and miRNAs. The TGF-beta signaling pathway may be a promising target for COVID-19 treatment (Yousefi et al., 2020). These studies suggested that lncRNAs can interact with several inflammatory factors, including inflammatory genetic processes and cytokine release in COVID-19. LncRNAs may serve as potential therapeutic enhancers in combatting SARS-CoV-2.
Table 1.
A summary of functional lncRNAs involved in viral infection and cytokine storm in COVID-19.
| LncRNA | Function |
|---|---|
| LncRNAs in SARS-CoV-2 viral infection | |
| NEAT1 | Upregulated in the lung tissue of patients with SARS-CoV-2 infection (Laha et al., 2021) |
| MALAT1 | May be a potential biomarker of SARS-CoV-2 infection (Vishnubalaji et al., 2020) |
| HOTAIRM1/PVT1 /AL392172.1 | Bind to the SARS-CoV-2 genome with the high affinity (Moazzam-Jazi et al., 2021) |
| TTTY15 | Regulates proteolysis, ubiquitin dependent catabolism (Lei et al., 2019) |
| TPTEP1 | Inhibits STAT3 phosphorylation (Ding et al., 2019) |
| LncRNA H19 | Binds to the 5′UTR of SARSCoV-2 genome and modulates the Spike transcript (Natarelli et al., 2021) |
| LncRNAs regulate cytokine storm in COVID-19 | |
| MALAT1 | Induces inflammatory responses and release of IL-6 and TNF-α (Li et al., 2018) |
| DANCR | Regulates ncRNA-mRNA network in inflammation (Li et al., 2018) |
| HOTAIR | Regulates activation of NF-κB and IL-6 and iNOS expression (Obaid et al., 2018) |
| NEAT1 | Promotes activation of inflammasomes in macrophages (Zhang et al., 2019) |
| NORAD | Binds with IL-6, IL-10, CSF3, TNF-a and CXCL10 (Morenikeji et al., 2020) |
| RAD51-AS1 | Leads to expression of pro-inflammatory cytokines (Morenikeji et al., 2020) |
6. Summary
In SARS-CoV-2 infection, lncRNAs have been demonstrated to be related to viral infection, interferon and cytokine storms in COVID-19 (Fig. 1). These lncRNAs interacted with genes of SARS-CoV-2 via the lncRNA‒miRNA-mRNA pathway. LncRNAs can also modulate the expression of interferon. Conversely, interferon can stimulate the expression of lncRNAs. In addition, lncRNAs bind with important or multiple cytokine storm cytokines. LncRNAs identified in the COVID-19 cytokine storm have the potential to serve as disease markers or drug targets. Thus, we can speculate that developing drugs targeting some specific sites in the pathway or network of lncRNAs may be a promising strategy to treat SARS-CoV-2 infection. More experimental studies are needed to further confirm the regulatory mechanism of lncRNAs in COVID-19.
Fig. 1.
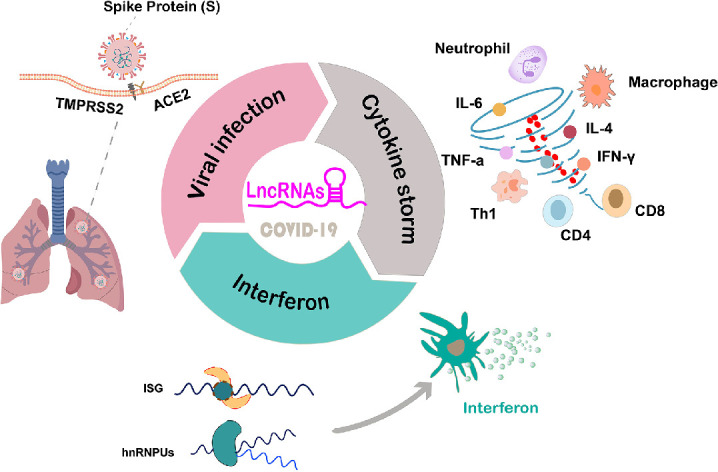
LncRNAs are related to viral infection, interferon and cytokine storm in COVID-19.
Funding
This work was financially supported by National Natural Science Foundation of China (82,002,580).
CRediT authorship contribution statement
Jing Ding: Data curation, Formal analysis, Funding acquisition, Investigation, Methodology, Writing – original draft, Validation. Jing Chen: Writing – review & editing, Validation. Xude Yin: Resources, Supervision. Jin zhou: Conceptualization, Validation.
Declaration of Competing Interest
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
Acknowledgments
None.
Contributor Information
Jing Chen, Email: chenjingmaomao111@163.com.
Jin zhou, Email: zhoujt521@163.com, dingjing511@163.com.
Data availability
Data will be made available on request.
References
- Aishwarya S., Gunasekaran K., Margret A.A. Computational gene expression profiling in the exploration of biomarkers, non-coding functional RNAs and drug perturbagens for COVID-19. J. Biomol. Struct. Dyn. 2020:1–16. doi: 10.1080/07391102.2020.1850360. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Al-Tawfiq J.A. Asymptomatic coronavirus infection MERS-CoV and SARS-CoV-2 (COVID-19) Travel Med. Infect. Dis. 2020;35 doi: 10.1016/j.tmaid.2020.101608. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Atianand M.K., Caffrey D.R., Fitzgerald K.A. Immunobiology of Long Noncoding RNAs. Annu. Rev. Immunol. 2017;35:177–198. doi: 10.1146/annurev-immunol-041015-055459. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Carpenter S., Aiello D., Atianand M.K., Ricci E.P., Gandhi P., Hall L.L., et al. A long noncoding RNA mediates both activation and repression of immune response genes. Science. 2013;341(6147):789–792. doi: 10.1126/science.1240925. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Casagrande M., Fitzek A., Spitzer M., Püschel K., Glatzel M., Krasemann S., et al. Detection of SARS-CoV-2 genomic and subgenomic RNA in retina and optic nerve of patients with COVID-19. Br. J. Ophthalmol. 2021 doi: 10.1136/bjophthalmol-2020-318618. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Channappanavar R., Fehr A.R., Vijay R., Mack M., Zhao J., Meyerholz D.K., et al. Dysregulated type I interferon and inflammatory monocyte-macrophage responses cause lethal pneumonia in SARS-CoV-infected mice. Cell Host Microbe. 2016;19(2):181–193. doi: 10.1016/j.chom.2016.01.007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chen H., Wang X., Yan X., Cheng X., He X., Zheng W. LncRNA MALAT1 regulates sepsis-induced cardiac inflammation and dysfunction via interaction with miR-125b and p38 MAPK/NFκB. Int. Immunopharmacol. 2018;55:69–76. doi: 10.1016/j.intimp.2017.11.038. [DOI] [PubMed] [Google Scholar]
- Chen L., Xiong J., Bao L., Shi Y. Convalescent plasma as a potential therapy for COVID-19. Lancet Infect. Dis. 2020;20(4):398–400. doi: 10.1016/s1473-3099(20)30141-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cheng J., Zhou X., Feng W., Jia M., Zhang X., An T., et al. Risk stratification by long non-coding RNAs profiling in COVID-19 patients. J. Cell. Mol. Med. 2021;25(10):4753–4764. doi: 10.1111/jcmm.16444. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Costela-Ruiz V.J., Illescas-Montes R., Puerta-Puerta J.M., Ruiz C., Melguizo-Rodríguez L. SARS-CoV-2 infection the role of cytokines in COVID-19 disease. Cytokine Growth Factor Rev. 2020;54:62–75. doi: 10.1016/j.cytogfr.2020.06.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ding H., Liu J., Zou R., Cheng P., Su Y. Long non-coding RNA TPTEP1 inhibits hepatocellular carcinoma progression by suppressing STAT3 phosphorylation. J. Exp. Clin. Cancer Res. 2019;38(1):189. doi: 10.1186/s13046-019-1193-0. [DOI] [PMC free article] [PubMed] [Google Scholar] [Retracted]
- Fatemi R.P., Velmeshev D., Faghihi M.A. De-repressing LncRNA-targeted genes to upregulate gene expression focus on small molecule therapeutics. Mol. Ther. Nucleic Acids. 2014;3(11):e196. doi: 10.1038/mtna.2014.45. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fiolet T., Kherabi Y., MacDonald C.J., Ghosn J., Peiffer-Smadja N. Comparing COVID-19 vaccines for their characteristics, efficacy and effectiveness against SARS-CoV-2 and variants of concern a narrative review. Clin. Microbiol. Infect. 2022;28(2):202–221. doi: 10.1016/j.cmi.2021.10.005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gong J., Guo S., Li H.B., Yuan S.Y., Shang Y., Yao S.L. BML-111, a lipoxin receptor agonist, protects haemorrhagic shock-induced acute lung injury in rats. Resuscitation. 2012;83(7):907–912. doi: 10.1016/j.resuscitation.2011.12.035. [DOI] [PubMed] [Google Scholar]
- Imam H., Bano A.S., Patel P., Holla P., Jameel S. The lncRNA NRON modulates HIV-1 replication in a NFAT-dependent manner and is differentially regulated by early and late viral proteins. Sci. Rep. 2015;5:8639. doi: 10.1038/srep08639. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kambara H., Niazi F., Kostadinova L., Moonka D.K., Siegel C.T., Post A.B., et al. Negative regulation of the interferon response by an interferon-induced long non-coding RNA. Nucleic. Acids. Res. 2014;42(16):10668–10680. doi: 10.1093/nar/gku713. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Laha S., Saha C., Dutta S., Basu M., Chatterjee R., Ghosh S., et al. In silico analysis of altered expression of long non-coding RNA in SARS-CoV-2 infected cells and their possible regulation by STAT1, STAT3 and interferon regulatory factors. Heliyon. 2021;7(3):e06395. doi: 10.1016/j.heliyon.2021.e06395. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lei F., Zhang H., Xie X. Comprehensive analysis of an lncRNA-miRNA-mRNA competing endogenous RNA network in pulpitis. PeerJ. 2019;(7):e7135. doi: 10.7717/peerj.7135. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li H., Shi H., Ma N., Zi P., Liu Q., Sun R. BML-111 alleviates acute lung injury through regulating the expression of lncRNA MALAT1. Arch. Biochem. Biophys. 2018;649:15–21. doi: 10.1016/j.abb.2018.04.016. [DOI] [PubMed] [Google Scholar]
- Liu H., Zhang Z., Wu N., Guo H., Zhang H., Fan D., et al. Integrative analysis of dysregulated lncRNA-associated ceRNA network reveals functional lncRNAs in gastric cancer. Genes (Basel) 2018;9(6) doi: 10.3390/genes9060303. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ma H., Han P., Ye W., Chen H., Zheng X., Cheng L., et al. The long noncoding RNA NEAT1 exerts antihantaviral effects by acting as positive feedback for RIG-I signaling. J. Virol. 2017;91(9) doi: 10.1128/jvi.02250-16. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ma M., Pei Y., Wang X., Feng J., Zhang Y., Gao M.Q. LncRNA XIST mediates bovine mammary epithelial cell inflammatory response via NF-κB/NLRP3 inflammasome pathway. Cell Prolif. 2019;52(1):e12525. doi: 10.1111/cpr.12525. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Maarouf M., Chen B., Chen Y., Wang X., Rai K.R., Zhao Z., et al. Identification of lncRNA-155 encoded by MIR155HG as a novel regulator of innate immunity against influenza A virus infection. Cell. Microbiol. 2019;21(8):e13036. doi: 10.1111/cmi.13036. [DOI] [PubMed] [Google Scholar]
- Meydan C., Madrer N., Soreq H. The neat dance of COVID-19 NEAT1, DANCR, and Co-modulated cholinergic RNAs link to inflammation. Front. Immunol. 2020;11 doi: 10.3389/fimmu.2020.590870. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Moazzam-Jazi M., Lanjanian H., Maleknia S., Hedayati M., Daneshpour M.S. Interplay between SARS-CoV-2 and human long non-coding RNAs. J. Cell. Mol. Med. 2021;25(12):5823–5827. doi: 10.1111/jcmm.16596. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Morenikeji O.B., Bernard K., Strutton E., Wallace M., Thomas B.N. Evolutionarily conserved long non-coding rna regulates gene expression in cytokine storm during COVID-19. Front. Bioeng. Biotechnol. 2020;8 doi: 10.3389/fbioe.2020.582953. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mukherjee S., Banerjee B., Karasik D., Frenkel-Morgenstern M. mRNA-lncRNA Co-expression network analysis reveals the role of lncRNAs in immune dysfunction during severe SARS-CoV-2 infection. Viruses. 2021;13(3) doi: 10.3390/v13030402. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Natarelli L., Parca L., Mazza T., Weber C., Virgili F., Fratantonio D. MicroRNAs and long non-coding RNAs as potential candidates to target specific motifs of SARS-CoV-2. Noncoding RNA. 2021;7(1) doi: 10.3390/ncrna7010014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nishida S., Hagihara K., Shima Y., Kawai M., Kuwahara Y., Arimitsu J., et al. Rapid improvement of AA amyloidosis with humanised anti-interleukin 6 receptor antibody treatment. Ann. Rheum. Dis. 2009;68(7):1235–1236. doi: 10.1136/ard.2008.099267. [DOI] [PubMed] [Google Scholar]
- Nishimoto N., Yoshizaki K., Maeda K., Kuritani T., Deguchi H., Sato B., et al. Toxicity, pharmacokinetics, and dose-finding study of repetitive treatment with the humanized anti-interleukin 6 receptor antibody MRA in rheumatoid arthritis. Phase I/II clinical study. J. Rheumatol. 2003;30(7):1426–1435. [PubMed] [Google Scholar]
- Nishitsuji H., Ujino S., Yoshio S., Sugiyama M., Mizokami M., Kanto T., et al. Long noncoding RNA #32 contributes to antiviral responses by controlling interferon-stimulated gene expression. Proc. Natl. Acad. Sci. USA. 2016;113(37):10388–10393. doi: 10.1073/pnas.1525022113. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Obaid M., Udden S.M.N., Deb P., Shihabeddin N., Zaki M.H., Mandal S.S. LncRNA HOTAIR regulates lipopolysaccharide-induced cytokine expression and inflammatory response in macrophages. Sci. Rep. 2018;8(1):15670. doi: 10.1038/s41598-018-33722-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pan Q., Zhao Z., Liao Y., Chiu S.H., Wang S., Chen B., et al. Identification of an interferon-stimulated long noncoding RNA (LncRNA ISR) involved in regulation of influenza a virus replication. Int. J. Mol. Sci. 2019;20(20) doi: 10.3390/ijms20205118. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Peng X., Gralinski L., Armour C.D., Ferris M.T., Thomas M.J., Proll S., et al. Unique signatures of long noncoding RNA expression in response to virus infection and altered innate immune signaling. MBio. 2010;1(5) doi: 10.1128/mBio.00206-10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Perlman S., Dandekar A.A. Immunopathogenesis of coronavirus infections implications for SARS. Nat. Rev. Immunol. 2005;5(12):917–927. doi: 10.1038/nri1732. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pontecorvi G., Bellenghi M., Ortona E., Carè A. microRNAs as new possible actors in gender disparities of Covid-19 pandemic. Acta Physiol. (Oxf) 2020;230(1):e13538. doi: 10.1111/apha.13538. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Roberts T.C., Langer R., Wood M.J.A. Advances in oligonucleotide drug delivery. Nat. Rev. Drug Discov. 2020;19(10):673–694. doi: 10.1038/s41573-020-0075-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rockx B., Baas T., Zornetzer G.A., Haagmans B., Sheahan T., Frieman M., et al. Early upregulation of acute respiratory distress syndrome-associated cytokines promotes lethal disease in an aged-mouse model of severe acute respiratory syndrome coronavirus infection. J. Virol. 2009;83(14):7062–7074. doi: 10.1128/jvi.00127-09. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ruan Q., Yang K., Wang W., Jiang L., Song J. Clinical predictors of mortality due to COVID-19 based on an analysis of data of 150 patients from Wuhan, China. Intensive Care Med. 2020;46(5):846–848. doi: 10.1007/s00134-020-05991-x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stojic L., Lun A.T.L., Mascalchi P., Ernst C., Redmond A.M., Mangei J., et al. A high-content RNAi screen reveals multiple roles for long noncoding RNAs in cell division. Nat. Commun. 2020;11(1):1851. doi: 10.1038/s41467-020-14978-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sur S., Sasaki R., Devhare P., Steele R., Ray R., Ray R.B. Association between MicroRNA-373 and long noncoding RNA NORAD in Hepatitis C Virus-infected hepatocytes impairs wee1 expression for growth promotion. J. Virol. 2018;92(20) doi: 10.1128/jvi.01215-18. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tay M.Z., Poh C.M., Rénia L., MacAry P.A., Ng L.F.P. The trinity of COVID-19 immunity, inflammation and intervention. Nat. Rev. Immunol. 2020;20(6):363–374. doi: 10.1038/s41577-020-0311-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vishnubalaji R., Shaath H., Alajez N.M. Protein coding and long noncoding RNA (lncRNA) Transcriptional landscape in SARS-Cov-2 infected bronchial epithelial cells highlight a role for interferon and inflammatory response. Genes (Basel) 2020;11(7) doi: 10.3390/genes11070760. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wiersinga W.J., Rhodes A., Cheng A.C., Peacock S.J., Prescott H.C. Pathophysiology, transmission, diagnosis, and treatment of coronavirus disease 2019 (COVID-19) a review. JAMA. 2020;324(8):782–793. doi: 10.1001/jama.2020.12839. [DOI] [PubMed] [Google Scholar]
- Wu Y., Zhao T., Deng R., Xia X., Li B., Wang X. A study of differential circRNA and lncRNA expressions in COVID-19-infected peripheral blood. Sci. Rep. 2021;11(1):7991. doi: 10.1038/s41598-021-86134-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Xie Q., Chen S., Tian R., Huang X., Deng R., Xue B., et al. Long noncoding RNA ITPRIP-1 positively regulates the innate immune response through promotion of oligomerization and activation of MDA5. J. Virol. 2018;92(17) doi: 10.1128/jvi.00507-18. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Xiong Y., Liu Y., Cao L., Wang D., Guo M., Jiang A., et al. Transcriptomic characteristics of bronchoalveolar lavage fluid and peripheral blood mononuclear cells in COVID-19 patients. Emerg. Microbes Infect. 2020;9(1):761–770. doi: 10.1080/22221751.2020.1747363. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yang G., Lu X., Yuan L. LncRNA a link between RNA and cancer. Biochim. Biophys. Acta. 2014;1839(11):1097–1109. doi: 10.1016/j.bbagrm.2014.08.012. [DOI] [PubMed] [Google Scholar]
- Yang Q., Lin F., Wang Y., Zeng M., Luo M. Long noncoding RNAs as emerging regulators of COVID-19. Front. Immunol. 2021;12 doi: 10.3389/fimmu.2021.700184. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yao X.H., Li T.Y., He Z.C., Ping Y.F., Liu H.W., Yu S.C., et al. A pathological report of three COVID-19 cases by minimal invasive autopsies. Zhonghua Bing Li Xue Za Zhi. 2020;49(5):411–417. doi: 10.3760/cma.j.cn112151-20200312-00193. [DOI] [PubMed] [Google Scholar]
- Yousefi H., Poursheikhani A., Bahmanpour Z., Vatanmakanian M., Taheri M., Mashouri L., et al. SARS-CoV infection crosstalk with human host cell noncoding-RNA machinery an in-silico approach. Biomed. Pharmacother. 2020;130 doi: 10.1016/j.biopha.2020.110548. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang Q., Chen C.Y., Yedavalli V.S., Jeang K.T. NEAT1 long noncoding RNA and paraspeckle bodies modulate HIV-1 posttranscriptional expression. MBio. 2013;4(1) doi: 10.1128/mBio.00596-12. e00596-12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang P., Cao L., Zhou R., Yang X., Wu M. The lncRNA Neat1 promotes activation of inflammasomes in macrophages. Nat. Commun. 2019;10(1):1495. doi: 10.1038/s41467-019-09482-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang C., Wu Z., Li J.W., Zhao H., Wang G.Q. Cytokine release syndrome in severe COVID-19 interleukin-6 receptor antagonist tocilizumab may be the key to reduce mortality. Int. J. Antimicrob. Agents. 2020;55(5) doi: 10.1016/j.ijantimicag.2020.105954. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhong Y., Yu C., Qin W. LncRNA SNHG14 promotes inflammatory response induced by cerebral ischemia/reperfusion injury through regulating miR-136-5p /ROCK1. Cancer Gene Ther. 2019;26(7–8):234–247. doi: 10.1038/s41417-018-0067-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ziegler C.G.K., Allon S.J., Nyquist S.K., Mbano I.M., Miao V.N., Tzouanas C.N., et al. SARS-CoV-2 receptor ACE2 is an interferon-stimulated gene in human airway epithelial cells and is detected in specific cell subsets across tissues. Cell. 2020;181(5):1016–1035. doi: 10.1016/j.cell.2020.04.035. e19. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zumla A., Hui D.S., Azhar E.I., Memish Z.A., Maeurer M. Reducing mortality from 2019-nCoV host-directed therapies should be an option. Lancet. 2020;395(10224):e35–e36. doi: 10.1016/s0140-6736(20)30305-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Data Availability Statement
Data will be made available on request.


