Abstract
Brain metastasis is a common characteristic of late-stage lung cancers. High doses of targeted radiation can control tumor growth in the brain but can also result in radiation-induced necrosis. Current methods are limited for distinguishing whether new parenchymal lesions following radiotherapy are recurrent tumors or radiation-induced necrosis, but the clinical management of these two classes of lesions differs significantly. Here we developed, validated, and evaluated a new MRI technique termed selective size imaging using filters via diffusion times (SSIFT) to differentiate brain tumors from radiation necrosis in the brain. This approach generates a signal filter that leverages diffusion time dependence to establish a cell-size-weighted map. Computer simulations in silico, cultured cancer cells in vitro, and animals with brain tumors in vivo were used to comprehensively validate the specificity of SSIFT for detecting typical large cancer cells and the ability to differentiate brain tumors from radiation necrosis. SSIFT was also implemented in patients with metastatic brain cancer and radiation necrosis. SSIFT showed high correlation with mean cell sizes in the relevant range of less than 20 μm. The specificity of SSIFT for brain tumors and reduced contrast in other brain etiologies allowed SSIFT to differentiate brain tumors from peri-tumoral edema and radiation necrosis. In conclusion, this new, cell size-based MRI method provides a unique contrast to differentiate brain tumors from other pathologies in the brain.
Keywords: cell size, brain cancer, radiation, necrosis, MRI
Introduction
Approximately 1.7 million new cancer cases are projected to occur in the United States each year, and 50% of lung cancer (1) as well as 10 – 30% of all cancer patients (2,3) will develop brain metastases. With the remarkable improvements in systemic therapies, cancer patients are living longer and have higher chances of developing brain metastases. Stereotactic radiosurgery (SRS) is a common therapy using high radiation doses to provide the local control of tumors without the late neurocognitive sequelae associated with whole-brain radiotherapy (4). With SRS, the median survival for patients with brain metastases from non-small cell lung cancer (NSCLC) with EGFR mutation has increased from 4 – 9 months (5) to 46 months (6). However, 10-20% of SRS treated patients will develop new parenchymal lesions that are either radiation induced necrosis (radionecrosis) or recurrent tumors. These two types of lesions usually occur within a similar time frame but need to be treated very differently: radionecrosis can be managed conservatively to reduce patient morbidity associated with treatment; by contrast, tumor recurrence is often best managed early and more aggressively to control lesion growth. To complicate matters, both chemotherapy and the increasingly used immunotherapy with SRS can increase the occurrence of radionecrosis (7,8). Therefore, there is a need in clinical radiation oncology to develop a reliable method to differentiate radionecrosis from tumor progression.
Computer-assisted stereotactic biopsy is a standard method to address this clinical problem (9), but it is an invasive procedure that results in unwanted risks such as hemorrhage that can lead to increased patient mortality (10). Non-invasive magnetic resonance imaging with gadolinium (Gd)-based contrast agents (Gd-MRI) is the current standard of care imaging method for the detection and post-treatment management of brain metastases. Unfortunately, Gd enhancement relies on an indirect effect, i.e., the breakdown of the blood-brain barrier (BBB) and hence is incapable of reliably differentiating brain metastases from radionecrosis because the BBB is usually disrupted in both types of lesions (11). This hinders the early and accurate diagnosis necessary for timely and effective treatments.
Several imaging methods have been found useful to differentiate recurrent brain metastases from radionecrosis (12). Although amino acid and FDG-PET/CT has a sensitivity of 85% and a specificity of 88% to differentiate radionecrosis from current brain metastases (13), its intrinsic low resolution, added cost, time, radiation exposure, and high background in the brain (FDG-PET) are hindrances to the adoption of PET/CT in routine clinical imaging (14). Dynamic susceptibility contrast (DSC) MRI with relative cerebral blood volume (rCBV) has been widely used for imaging brain cancer (15) and recurrent brain metastases (16) but significant heterogeneity in different DSC-MRI studies has been reported (17). Additionally, there is increasing interest in developing contrast-agent-free MRI methods to reduce the extra burden and safety concerns associated with MR contrast agents. Proton MR spectroscopy (1H MRS) is a molecular imaging technique that probes brain metabolites such as N-acetylaspartate (NAA), total choline (tCho), and total creatine (tCr). Extensive studies (14,18,19) show that 1H MRS, particularly with ratios of tCho:tCr and tCho:NAA, is a promising tool to differentiate brain metastases from radionecrosis. However, MRS has a low spatial resolution and the possible partial volume effects are disadvantageous, especially for small brain metastases. Amide proton transfer weighted (APTw) imaging based on chemical exchange saturation transfer (CEST) probes mobile proteins and peptides (20) and has shown promise to distinguish recurrent brain tumor from radionecrosis in animals (21). However, varying results have been reported recently in patients (22). Diffusion tensor imaging (DTI) (23) allows estimation of the mean diffusivity (MD), or apparent diffusion coefficient (ADC), and fractional anisotropy (FA) of tissues, which can be indicators of cellularity and coherence of axonal fiber directions, respectively. DTI has been investigated to differentiate tumor from radionecrosis (24) but may suffer from lack of specificity (25). This suggests the complex, highly heterogeneous microenvironments in tumors and radionecrosis are a remarkable hurdle to their differentiation. It is still desirable to develop improved imaging methods that can capture more intrinsic molecular or pathological differences between tumors and radionecrosis.
Cell size is one of the intrinsic differences between cancerous cells and normal brain cells. Although brain tumors can be remarkably heterogeneous, most primary and metastatic brain cancers have relatively high volume fractions of cells 10 – 20 μm in diameter. By contrast, normal brain cells are much smaller. For example, the majority of brain cells are glia (26) with a reported average size of 5.26 μm (27), while the diameter of most brain neuron axons is < 3 μm (28). Cortical neurons have been measured to have larger sizes (11.7-15.0 μm) but make up a small volume fraction (27). Although radiation induces complex etiologies such as vasogenic edema, cystic fluid, and hemorrhage, no accompanying significant changes in cell size have been reported. Therefore, information on cell size may provide new insights into the differentiation of brain tumors and radionecrosis.
Diffusion MRI (dMRI) has been used to estimate cell sizes in vivo using multiple diffusion times (tdiff) and b-values. Model-based dMRI approaches have quantified cell size in human prostate (VERDICT (29) and IMPULSED (30)) and breast (IMPULSED (31)) tumors. However, extending these methods to the brain is non-trivial. Each method assumes isotropic diffusion, which might be true for select extracranial tumors but is not appropriate in the brain, where the microstructure is generally more complex. Previous attempts to apply these models to the brain have required isolating the fitting to tumor ROIs (32,33), which may hinder the ability to differentiate between tumors and other neuropathologies. While acquisitions with multiple tdiff, b-values, and gradient directions combined with complex biophysical models are a possible approach, they lead to longer scan times that are not desirable in clinical practice. In the present study, we therefore propose a new fast, model-free approach to address this problem. In lieu of more complex modeling to quantify cell size in brain tissue, our approach is to generate a signal filter that leverages diffusion time dependence to generate a cell-size-weighted map. This filter selectively enhances sensitivity to cells within a range of sizes similar to the relatively large cells present in brain tumors, while simultaneously suppressing signals from relatively smaller normal brain cells and CSF. In contrast to ADC dependence on the density of cells of all sizes, this filter is only sensitive to cells within the selective size range. This results in high specificity to cancerous tissues and hence provides a unique means to distinguish brain tumors from other pathologies such as radionecrosis. Experiments using computer simulations, cultured cells in vitro, and animals in vivo were performed to comprehensively validate this technique. Moreover, preliminary results of this approach performed in patients with metastatic brain cancer and radionecrosis to demonstrate the feasibility of translating this technique to routine clinical practice.
Materials and Methods
Selective size imaging using filters via diffusion times (SSIFT)
The detection sensitivity of diffusion MRI for detecting different hinderances to free diffusion depends on the root-mean-square displacement , where tdiff is the diffusion time, and D is the water diffusivity. Therefore, adjustments of tdiff can be used to tune the detection sensitivity of dMRI to different length scales (34,35). Conventional dMRI uses pulsed gradient spin echo (PGSE) sequences and can probe only long tdiff > 30 ms on regular MRI scanners due to hardware limitations (35). To alleviate this restriction, oscillating gradient spin echo (OGSE) sequences may be used to extend the sensitivity to much shorter tdiff that are not accessible by conventional PGSE (36,37). Figure 1a shows the calculated dependence of dMRI signals on three representative restriction sizes (5, 15, and 50 μm) for a clinically achievable range of tdiff using both PGSE and OGSE. The incremental area under the curve (iAUC; Figure 1b) is a metric of signal change with respect to tdiff and shows a strong dependence on cell size. iAUC is calculated as:
| [1] |
where M(tdiff,m) indicates the directionally averaged measured dMRI signal with the tdiff of the mth measurement and Ntdiff is the number of unique tdiff values. iAUC peaks at ~20 μm covering typical brain cancer cell sizes (10 – 20 μm) and approaches zero when cell sizes are small (< 5 μm) or large (> 50 μm) as shown in Figure 1b. Therefore, iAUC with an appropriate tdiff range can serve as a filter to selectively detect signals arising from relatively large cancer cells with simultaneous suppression of signals from normal brain cells, vasogenic edema, and cystic fluid.
Figure 1.
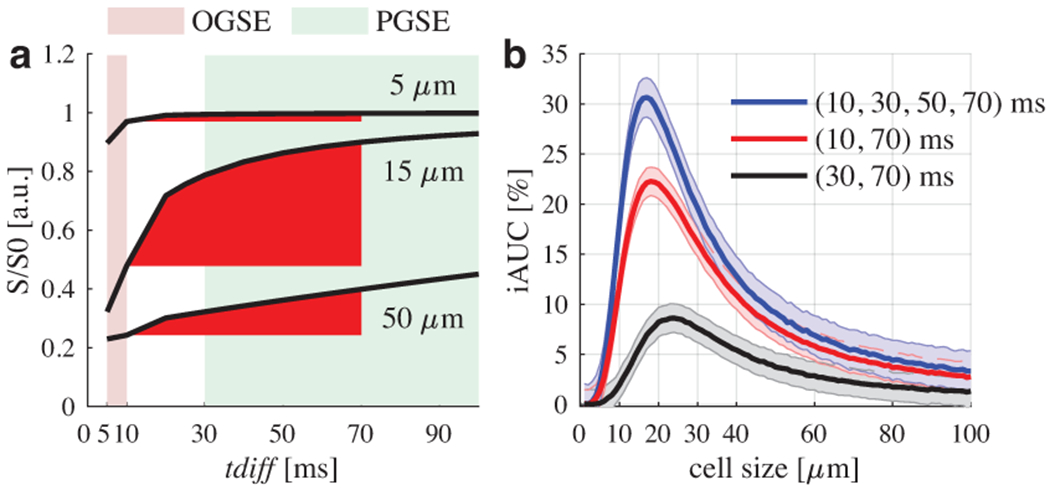
(a) Simulated dependence of intracellular dMRI signals on cell size and diffusion time tdiff. The red incremental area under curve (iAUC) shows a strong dependence on cell size. Colored backgrounds represent typical measurable tdiff ranges in clinics. (b) Normalized iAUC of intracellular signals with three different combinations of tdiff values may serve as a filter to selectively enhance sensitivity to typical cancer cell sizes (i.e., 10 – 20 μm) in brain tumors. Solid lines and shaded areas indicate means and standard deviations of 500 runs with a signal-to-noise ratio of 20.
Although Figure 1 demonstrates the biophysical basis of selective cell size imaging using tdiff, it is not trivial to implement SSIFT in the brain due to complex anisotropic microstructures and multiple tissue compositions. Each measurement depends on signal fractions and local diffusion tensor parameters from varying compartments in the tissue microstructure (Supplementary Figure S1). Directional averaging removes the dependence on diffusion gradient directions (38) and fiber organization such as undulation, crossing, and dispersion (39). Previous studies have shown that tdiff dependence in healthy tissue is small in the PGSE (40) and OGSE (41,42) regimes used in this study (10 – 70 ms). The tdiff dependence analysis in Figure 1 then suggests that the directional averaged iAUC is dominated by diffusion compartments with large cell sizes. This in turn provides a more specific imaging method to detect brain tumors based on sensitivity to the density of cells within a size filter with minimal influences from axons, small normal brain cells, and unrestricted fluid. We term this method as selective size imaging using filters via diffusion times (SSIFT).
SSIFT protocol
Although more tdiff values may further enhance the selective sensitivity to large cancer cells as shown in Figure 1, this would increase the total scan time which is not desirable in clinical practice. Therefore, a minimum of two tdiff values, 10 ms and 70 ms obtained using OGSE and PGSE, respectively, were used throughout the studies in this work. All dMRI acquisitions used b = 0 and 1000 mm2/s with 32 gradient directions. For PGSE, gradient duration δ = 12 ms and gradient time interval Δ = 74 ms. For OGSE, δ = 40 ms, Δ = 52.4 ms, number of oscillating cycles = 1, Details of the implementation of OGSE sequences on both animal and human scanners have been reported previously (31). Additional dMRI measurements using PGSE acquisitions with Δ = 54 and 34 ms were also performed in the animal studies and one human subject.
Computer simulations in silico
Finite difference simulations (43) were performed to investigate the dependence of SSIFT on cell size and density. Tissues were modeled as regularly packed spherical cells with both intra- and extracellular spaces following previous studies (31,34,44,45). Cell sizes ranged from 2 to 32 μm in increments of 2 μm and intracellular volume fractions were 10.0%, 15.1%, 19.6%, 24.2%, 30.4% 35.7%, 38.9%, 46.4%, 50.9%, 56.0%, and 61.8%. The intracellular diffusivity of both cell types was fixed to a previously measured value Din = 1.58 μm2/ms (46) and extracellular diffusivity was 2.0 μm2/ms. Relaxation times were assumed to be equal in both compartments and cell boundaries were considered impermeable. Signal was simulated with diffusion times of 10 and 70 ms with b-value 1,000 s/mm2.
Cell lines in vitro
Cells
Six different cancer cell lines were used to investigate the influences of various cancer cell types and sizes:
C6 (rodent glioma), 9L (rodent gliosarcoma) and GL261 (murine glioma) to mimic primary brain cancer; and
A549 (human lung carcinoma), B16 (mouse melanoma), and MDA-MB-231 (human breast adenocarcinoma) to mimic the three most common sources of metastatic brain cancers, i.e., lung, skin (melanoma), and breast cancers (47).
Moreover, human lymphocyte and Jurkat cell data from a previous study (31) were used as a reference for small cells, and a doped water sample with 0.1 mM MnCl2 was used to mimic CSF. All tumor cell lines were purchased from ATCC and cultured in RPMI Medium 1640 supplemented with 10% FBS, 50 Units/ml penicillin, 50 μg/mL streptomycin (and 5 μg/mL recombinant insulin for breast cell lines) (Invitrogen, CA) in a humidified incubator maintained with 5% CO2 at 37°C. Cell lines were periodically tested for mycoplasma contamination by a PCR kit (Universal Mycoplasma Detection Kit ATCC® 30-1012K™) and were not authenticated following purchase. Cells were cultured in 150 mm dishes to full confluence, then harvested by trypsinization, washed and resuspended with PBS. Lymphocytes were extracted from human peripheral blood by using the Ficoll method (48), briefly, blood was diluted with an equal volume of PBS, and carefully added to the top of an equal volume of Ficoll-Paque in a centrifuge tube. After centrifugation at 800g for 20 minutes, the cells in the interface layers were collected, the residual red cells were removed by hypotonic lysis and washing, and the final lymphocytes were pelleted and re-suspended with PBS.
For MR experiments, cells were washed with PBS after fixation with 4% paraformaldehyde for over one hour, about 3 x 107 cultured cells (or 1x109 lymphocytes) were centrifuged at 2000g x 2 minutes in a 0.65 mL of Eppendorf tube to obtain a tight cell pellet. All the liquid on the top was carefully removed, and the tube with cell pellet was used for MRI measurements.
MRI experiments of cells
MRI measurements of cells in vitro were performed on a Varian/Agilent 4.7T MRI scanner (Agilent Technologies, Santa Clara, CA). A 2-mm-thick slice through the center of each cell pellet was imaged with a field of view (FOV) 16 × 16 mm2 and a matrix size 32 × 32, yielding a spatial resolution of 500 μm.
Light microscopy
A small aliquot of cells from each sample pool used in MRI experiments was spotted on a glass slide and covered by coverslip. Digital images of cells were recorded at both 20x and 40x, amplification. A stage micrometer was used with the same microscope and settings for size calibration. Average cell sizes from over 200 cells were calculated with the help of NIH ImageJ software (49). Supplementary Table S1 summarizes the calculated cell sizes.
Animals in vivo
Animals
Animal studies of rats and mice were approved by the local IACUCs and performed at Vanderbilt University Medical Center and Washington University in St. Louis, respectively. The use of different species, tumor types, and methods of inducing radionecrosis at two different institutions provided an opportunity to interrogate the ability of SSIFT to differentiate brain tumors from radionecrosis under various conditions.
Rat study at Vanderbilt University Medical Center
Brain tumor in rats. To generate 9L gliosarcoma xenografts, male Fischer rats (Envigo, Indianapolis, IN, USA) were immobilized in a stereotactic head holder under anesthesia (via a 5:95% isoflurane/oxygen mixture) and were inoculated with 1×105 9L cells in 5 μL by using a 10-μl gastight syringe (Hamilton, Reno, NV, USA). The site of the inoculation was 1 mm anterior and 3 mm lateral to the bregma on the right side of the head, at a depth of 4 mm relative to the dural surface. Tumor growth and size were estimated by MRI after 2 weeks. 8 subjects grew large tumors and were imaged using SSIFT and DTI protocols.
Radionecrosis in rats. 6 rats were treated replicating clinical high-dose stereotactic radiosurgery (SRS) via a clinical Novalis TX™ unit by Varian Medical Systems (Palo Alto, California). The animals were stabilized on the couch with a bite bar and head bar. Cone-beam CT (CBCT) was used to localize the target and register the dose plan. 140 Gy was delivered to isocenter via 10 noncoplanar arcs using a 4 mm diameter cone. All 6 rats developed sufficiently large radionecrotic lesions by 8 weeks.
Histology. Following imaging, rat brain tissues containing 9L gliosarcoma or radionecrosis were fixed by cardiac perfusion using 10% formalin, immersed in formalin for two days, and immersed in 70% ethanol. 8 μm-thick slices were stained with hematoxylin and eosin (H&E) and assessed digitally using QuPath version 0.3.1 (50).
Mice study at Washington University
Brain tumor in mice. Tumor cells were implanted in mice, as described previously (51). Briefly, mice were anesthetized with isoflurane and secured in a stereotactic head holder. Murine DBT glioblastoma cells were implanted (~10,000 cells suspended in 10 μL per mouse) over three minutes at a site 2-mm posterior and 3-mm to the left of bregma, 2-mm below the cortical surface.
Radionecrosis in mice. Six- to eight-week-old Balb/c mice were irradiated focally using a clinical Leksell Gamma Knife (GK) Perfexion™ (Elekta AB; Stockholm, Sweden) with a single 50 Gy (50% isodose) radiation dose in the left hemisphere. Mice were scanned by MR 13 weeks post irradiation, at which time radionecrosis occupied a significant fraction of the irradiated hemisphere (52).
MRI experiments of animals
Animal MRI scans of rats and mice were acquired separately at Vanderbilt University Medical Center and Washington University, but both used the same protocol on 4.7T Varian/Agilent horizontal small animal scanners. A single-shot echo-planar imaging (EPI) diffusion sequence was used in both studies with fat suppression and saturation bands to minimize confounding signals from outside the brain. Rats were scanned using a Litz38 volume coil for both transmission and reception. Axial slice thickness = 1 mm, FOV = 16 × 16 mm and matrix size 64 × 64, resulting in a 250 μm in-plane resolution. Mice were scanned using an actively decoupled coil pair: a 9-cm inner diameter volume coil (transmit) and a 1.5-cm outer diameter surface coil (receive). Axial slices with 1-mm thickness were acquired with FOV = 12 × 9 mm and matrix size 48 × 32, yielding an in-plane resolution of 250 × 285 μm. TR/TE = 4000/98 ms.
Patients
Human studies were approved by the local IRB at Vanderbilt University Medical Center and written informed consent was obtained from all subjects. Two patients with brain metastases, one with non-small cell lung cancer (NSCLC) and one with melanoma, were scanned pre-treatment with the SSIFT protocol. One patient with extensive radiation necrosis was scanned following treatment of renal cell carcinoma metastatic to the brain. Scans were performed using a Philips 3T dStream Ingenia scanner with a 32-channel head coil. Acquisition sequence parameters were: TR/TE = 15 s / 118 ms; FOV = 224 × 224 mm; half scan factor 0.64; slice thickness = 5 mm; an isotropic resolution of 2 mm; and fat suppression with spectral adiabatic inversion recovery. Total scan time of SSIFT was ≈ 16.5 minutes. In addition, DTI measurements using PGSE acquisitions with Δ = 54 and 34 ms were performed in one subject with NSCLC to further investigate ADC dependence on diffusion times, shown in Supplementary Figure S4. For comparison, anatomical T1w gradient echo images with TR/TE = 8 ms / 3.76 ms were acquired with image size 256 x 256 and 1 mm3 isotropic voxel size, and T2w FLAIR images with TR/TE = 11 s / 125 ms, image size 512 x 512, and 0.5 x 0.5 x 3 mm3 voxel size. The patient with NSCLC received clinical standard contrast-enhancement while the melanoma and radionecrosis patients were unable to receive gadolinium due to concerns of kidney dysfunction.
Data analysis
All diffusion images were processed using MRtrix3 (53) for noise (54) and Gibbs ringing (55) correction, FSL (56–58) for eddy current correction (59) (human data only), ANTS for N4 field bias correction (60) (human data only), and purpose-written Matlab (The MathWorks Inc., Natick, MA, USA) scripts. The average of signals over all diffusion directions of the same b value was used to remove the effects of diffusion anisotropy. Voxelwise normalization by b=0 intensity was avoided, as compartments with free water or small restrictions would not be filtered out of the b=0 signal and could reduce cell-size-weighted contrast due to a difference in T2-weighted contrast (61). Instead, the directionally averaged diffusion signals were normalized using median b=0 signals in white matter for each scan as determined with an FA mask (as done for intersubject comparison in (62)); however, rodent data were normalized by the median contralateral gray matter signal because white matter was difficult to isolate. The incremental area under curve (iAUC, see Fig. 1) was calculated using the normalized signal and values are reported as the iAUC as a percentage of normalization signal. DTI metrics (i.e., FA and ADC) were obtained using the PGSE acquisitions for comparison. ROIs were manually drawn on pre- and post-contrast T2w EPI images: tumors and radionecrosis were contrast-enhancing regions (including hyperintensities in radionecrosis), peritumor edema was the hyperintense but non-enhancing region next to the tumor, and contralateral regions were the subcortical area contralateral to the lesions excluding the hippocampus and CSF. The contrast-to-noise ratio (CNR) of an MRI metric P between two types of tissues I and II was calculated as .
Data availability statement
The data generated in this study are available upon request from the corresponding author.
Results
Simulations
The simulated influences of intracellular volume fraction vin and cell size are shown in Figure 2. Note that ΔADC = ADC(tdiff=10ms) – ADC(tdiff=70ms), which has shown tdiff dependent sensitivity to subtle microstructural changes in the brain (63). Figure 2a and 2b show ADC values acquired with PGSE and OGSE acquisitions. Depending on vin, conventional ADC of brain tissues with tdiff = 70 ms could be higher or lower than that of cancerous tissue, suggesting that conventional ADC is not a reliable biomarker to distinguish cancer from brain tissues. In contrast, ΔADC (Figure 2c) and iAUC (Figure 2d) have high sensitivity to regions with cancer-sized cells and large volume fractions. Both ΔADC and iAUC are able to distinguish tumors from brain tissues, but iAUC has a tighter peak in the 10-20 μm region and higher contrast against low volume fractions (such as edema).
Figure 2.
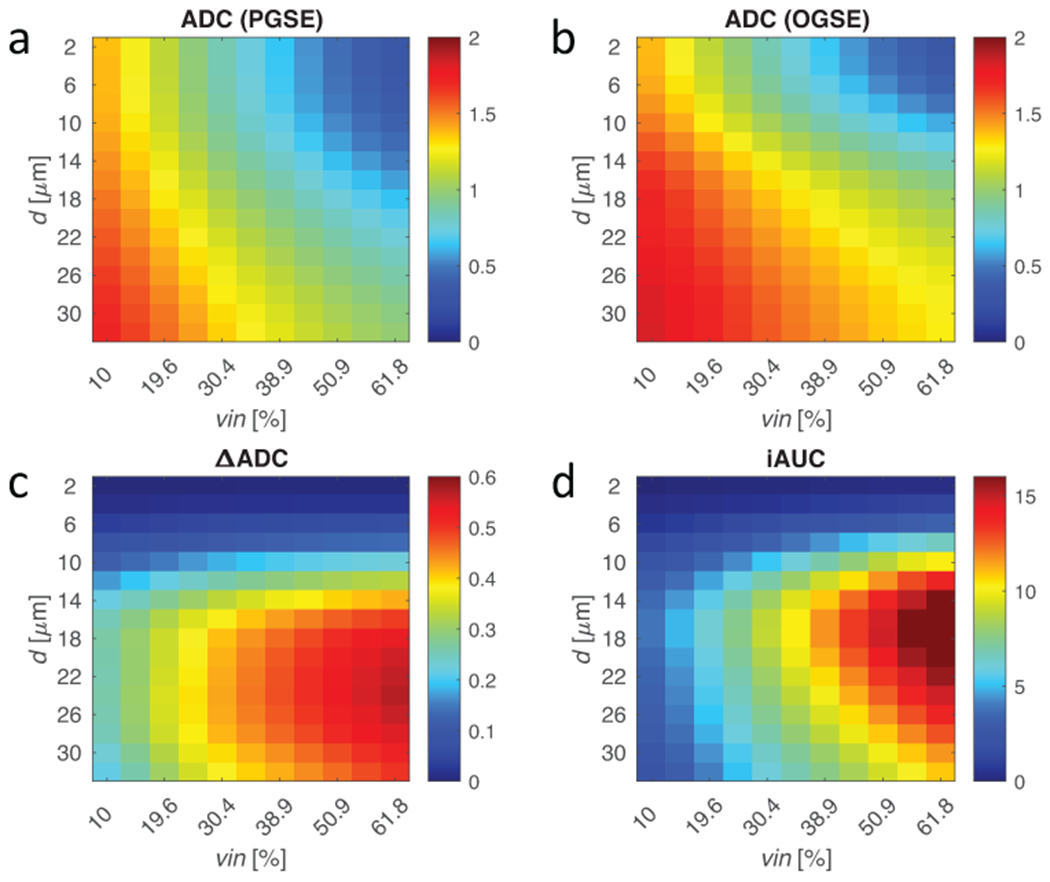
Finite difference simulations of regularly packed cells with varying cell diameter (d) and volume fraction (vin) using 70 ms (PGSE) and 10 ms (OGSE) diffusion times. (a-b) ADC is fitted from signals with both diffusion times. The diffusion time dependence using (c) the difference in ADC and (d) the iAUC change the sensitivity landscape. Where ADC is mostly sensitive to volume fraction at small restriction sizes, such as axons (1-5 μm) or glia (5-10 μm), ΔADC and iAUC filter out these components to give contrast driven by size-selective cellular density. iAUC provides more selective sensitivity to cancer cells ~10-20 μm than ΔADC.
Cells
Cell experimental results are summarized in Figure 3, showing a correlation between iAUC and mean cell size obtained using light microscopy with r = 0.92 and p < 0.01 obtained using the Spearman rank correlation. There are significant discrepancies in iAUC between all cancer cells (12.6 – 15.6 μm) and the lymphocytes and jurkat cells with a smaller cell size (10.3 – 11.9 μm). This is consistent with the simulation results shown above that SSIFT iAUC selectively enhances sensitivity to large cancer cells with simultaneous suppression of signals from water and small cells. It is notable, however, that the smaller cells are suppressed more than would be predicted from simulation.
Figure 3.
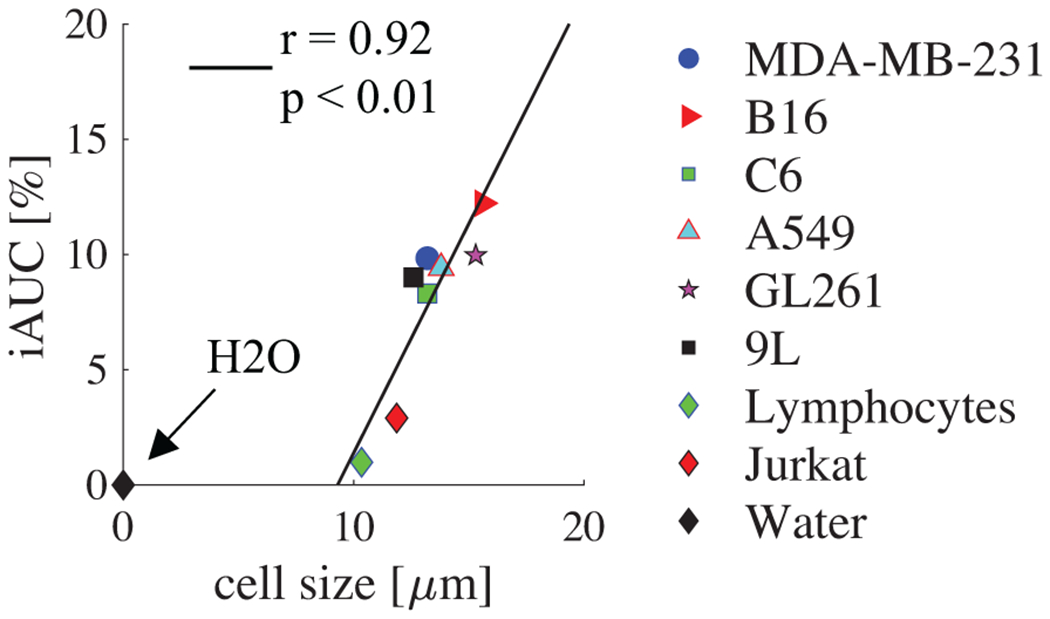
The correlation between iAUC and mean cell size obtained using light microscopy. Doped water iAUC is also provided as a reference. Correlation coefficient and p-value were determined by Spearman rank correlation.
Animals
Figure 4 shows the multi-parametric images of both rats and mice with brain tumors and radionecrosis. ADC alone provides ambiguous results due to the variations of the CSF volume fraction. This provides challenges to distinguishing viable tumors (highlighted on T2w with contrast agents) from peri-tumoral edema. SSIFT iAUC, however, provides a clear contrast that emphasizes the tumor. SSIFT iAUC shows diminished contrast in radionecrosis and contralateral regions, which is expected because there are no significant changes in cell size in radionecrosis.
Figure 4.
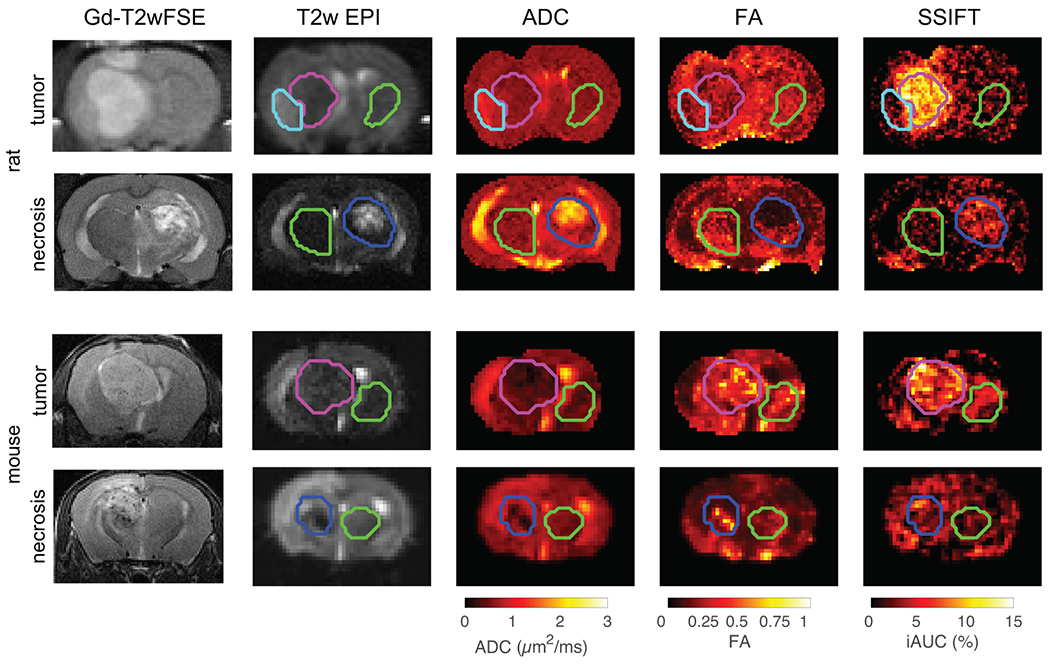
Multi-parametric images of both rats and mice with brain tumors and radionecrosis. ROIs of tumor (pink), radionecrosis (blue), and contralateral regions (green) were manually drawn on T2w EPI images. Peritumor edema (cyan) was delineated in rats by hyperintense T2w EPI signal that is unaffected by Gd contrast and absent in the contralateral anatomy. ROIs were overlayed ADC, FA, and SSIFT maps registered to the T2w EPI. SSIFT shows more selective contrast in the tumor ROI than ADC or FA.
Figure 5 summarizes all iAUC (Figure 5a and 5c) and ADC (Figure 5b and 5d) values of all ROIs. Using a two-sided Wilcoxon rank sum test, in both mice (N=4 tumor, N=4 radionecrosis) and rats (N=8 tumor, N=6 radionecrosis) ROI-averaged SSIFT iAUC values are significantly different in tumor vs radionecrosis (p < 0.05 mice, p < 0.001 rats) and tumor vs contralateral tissue (p < 0.01 mice, p < 0.001 rats). Similarly, ADC shows significantly different distributions in tumor vs radionecrosis (p < 0.05 mice, p < 0.01 rats). Contrast between peritumor edema and radionecrosis was not significant (ADCrat,edema = 0.92 ± 0.10 μm2/ms, ADCrat,RN = 0.99 ± 0.14 μm2/ms, p = 0.35). FA decrease in radionecrosis relative to all other tissues (p = 0.11 mice, p < 0.01 rats in radionecrosis vs tumor) and better separates necrosis from peritumoral edema (FArat,RN = 0.16 ± 0.03, FArat,edema = 0.24 ± 0.04, p < 0.01) than ADC, but is insensitive to tumor.
Figure 5.
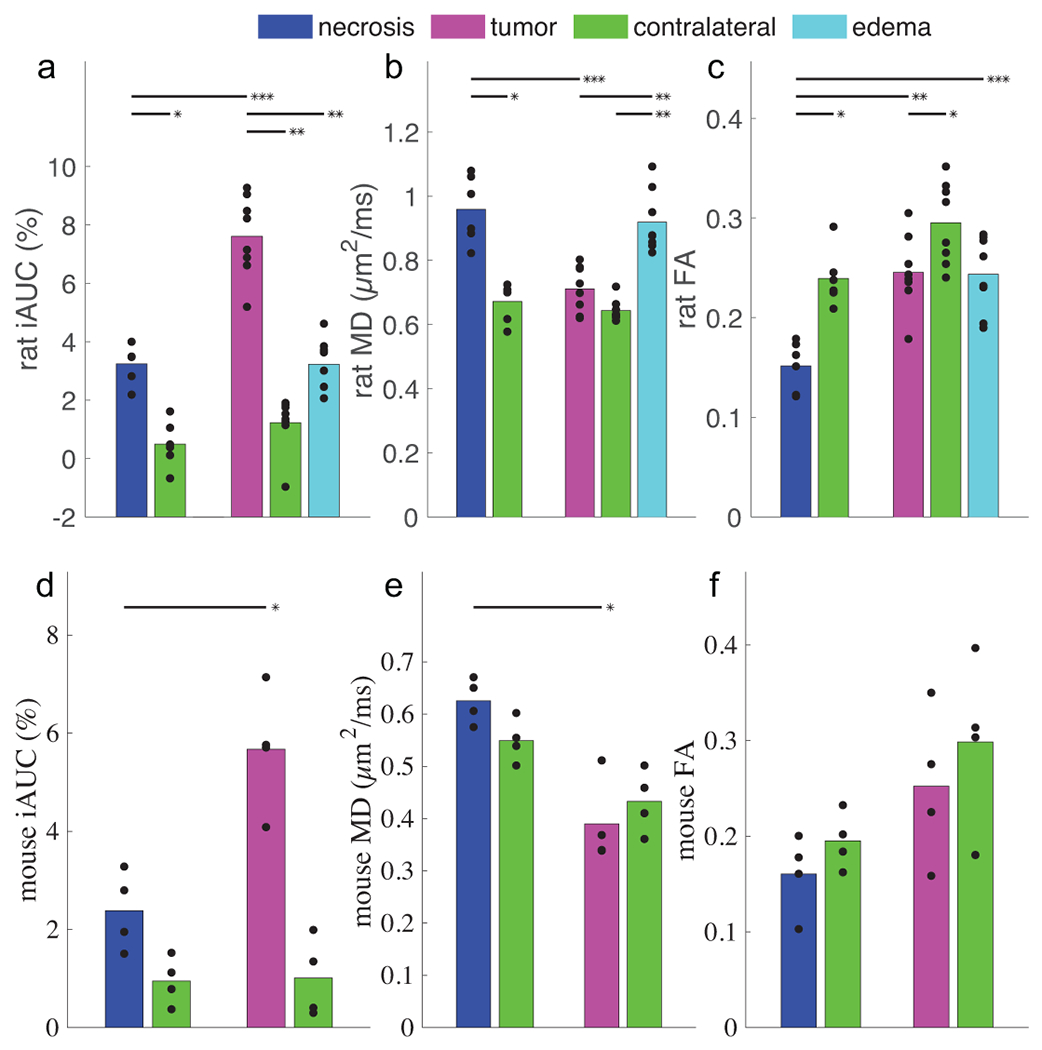
Summarized results of SSIFT iAUC (a,d), ADC (b,e) and FA (c,f) in different tissue types in rat (a-c) and mouse (d-f) subjects. Bar colors match the ROIs in Figure 4. Scatter points represent the ROI-averaged metric in individual subjects. Significance was tested with an unsigned and signed Wilcoxon rank sum test for inter- and intra-cohort comparisons, respectively. *** p < 0.001, ** p < 0.01, * p < 0.05.
Representative slices of H&E staining from rat tumor and radionecrosis are shown in Supplementary Figure S3. The cross sections from rat brains following radiation therapy demonstrate heterogeneous features including vascular telangiectasias, intravascular thrombosis, fibrinoid necrosis, cavitation, tissue necrosis, inflammatory cell infiltrates including neutrophils and lymphocytes, macrophages and microglia, and gliosis in addition to remnant neural tissue with neurons and normal glial cells. The cells demonstrate a broad range in size from approximately 27-562 μm2, with the largest cells representing neurons, foamy macrophages, or reactive astrocytes and the smallest cells representing inflammatory cells. The cell density is relatively low, averaging approximately 2600 cells/mm2. Sections from rats with gliosarcoma demonstrate a relatively well-circumscribed neoplasm comprising enlarged neoplastic cells with ovoid-to-pleomorphic, hyperchromatic nuclei and eosinophilic cytoplasm. Cell density within the tumor measured around 6000 cells/mm2. The nonneoplastic and nonreactive brain tissue in similar regions of the brain (e.g., hippocampus and thalamus) averages approximately 1900 cells/mm2 with cell size ranging from approximately 22-502 μm 2.
Patients with Brain Metastases and Radionecrosis
Figure 6 shows representative anatomical MRI, FA, and ADC (ADC) maps from conventional DTI, and SSIFT iAUC maps of two brain cancer patients. The non-small cell lung cancer patient received Gd contrast while the melanoma patient was unable to receive Gd due to kidney dysfunction. T2w FLAIR shows regions of viable tumors and peri-tumor edema, which are challenging to distinguish on FA or ADC maps. By contrast, SSIFT iAUC maps show distinct contrast of brain metastases alone, providing a similar contrast as the post-contrast T1w image. The radionecrosis lesion shows slight elevation in ROI-averaged iAUC (1.9 ± 2.0 %), while the NSCLC tumor shows much higher values (11.9 ± 3.4 %). Contralateral values in both are 0.5 ± 2.5 % and −0.3 ± 1.0 %, respectively. Supplementary Figure S4 shows ADC values in the NSCLC tumor calculated from four diffusion times to demonstrate the importance of including OGSE ADC at short diffusion times.
Figure 6.
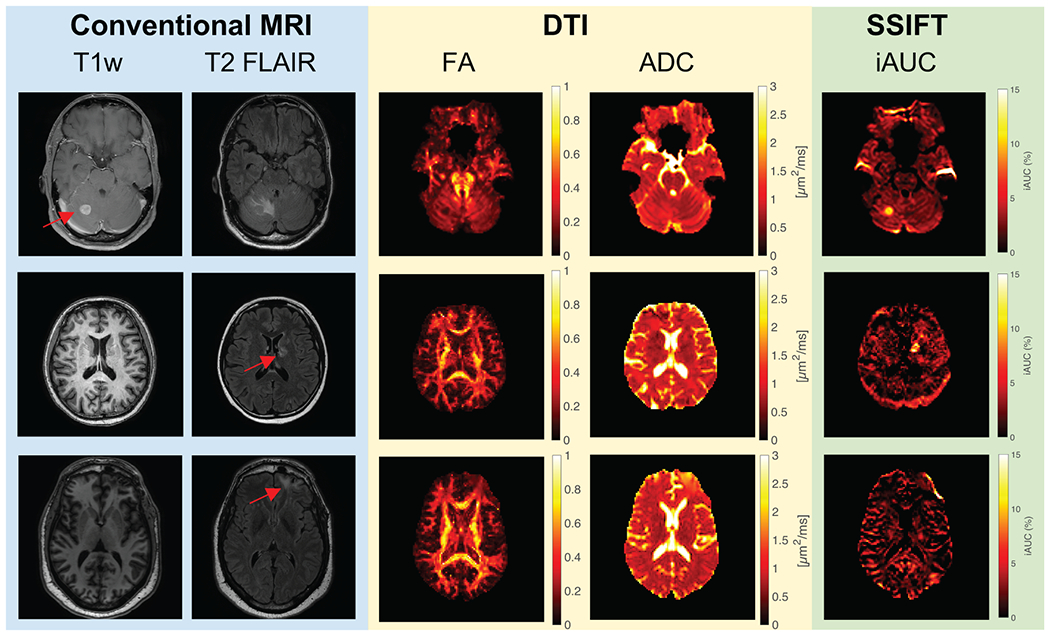
Representative images of patients with non-small cell lung cancer metastatic to the right cerebellum (top row) and melanoma adjacent to the left ventricle (middle row). The T1w image on the top is Gd-enhanced while the T1w images in the middle and bottom rows were collected without Gd contrast due to kidney dysfunction. Red arrows indicate lesions identified on the Gd-T1w image when available or T2 FLAIR otherwise. The cerebellar lesion demonstrates considerable associated peritumor edema (evident in T2 FLAIR) that is not apparent on the SSIFT map, while the tumor itself shows high signal contrast on SSIFT imaging relative to the surrounding tissue. The melanoma lesion apparent on T2 FLAIR has a high iAUC relative to the contralateral tissue. iAUC in the right frontal brain in radionecrosis (bottom row) is slightly elevated, but is substantially lower than in the tumors.
Discussion
Diffusion MRI has been implemented to differentiate recurrent tumors from radionecrosis. However, a meta-analysis shows that dMRI has only moderate diagnostic performance (64). One explanation is that dMRI is influenced by multiple tissue microstructural features simultaneously including tissue orientation and composition. All types of tissues in an image voxel contribute to mixed dMRI signals. Therefore, other concurrent brain processes, such as edema and radionecrosis, may sometimes appear indistinguishable from brain metastases (65). The SSIFT method focuses on a key intrinsic difference between brain tumors and other brain etiologies, i.e., the fraction of cells distinguished by cell size, and uses this distinct pathological feature of brain tumor and radionecrosis to distinguish these two types of lesions. This was validated in simulations, cells in vitro, and animals in vivo in the current work. The application of this technique in patients with neoplasms metastatic to the brain further confirms this and demonstrates the feasibility of the SSIFT method in human imaging. By targeting a physiological feature different from other dMRI methods, SSIFT may improve the diagnostic accuracy in differentiating radionecrosis from tumor. SSIFT additionally shows higher contrast between tumor and all other tissues in each study than is observed in conventional imaging, suggesting it may be useful for characterizing highly heterogeneous lesions.
The SSIFT method is based on the premise that cancer cells usually have larger cell sizes than those of most brain cells, and the contrast is dependent on both the differences in cell size, i.e., larger cancer cells resulting in larger SSIFT iAUC values (Figure 3), and the signal fraction of cells (i.e., cell density) within the SSIFT filter (Figure 2). As opposed to ADC which is dependent on all the density of cells of all sizes , SSIFT iAUC provides higher specificity to select cancer cell sizes and their corresponding size-selective cell density, which in turn may provide new insights into brain cancer in vivo. The in vivo animal validations in this study comprised 9L gliosarcoma and DBT glioblastoma cell lines, which showed high contrast in SSIFT relative to normal brain tissue and other malignancies. Cell counts from H&E histology indicate the tumors in rats have a high density of 9L cells that are in the size range detectable by SSIFT, as measured in Figure 3. Future studies are of interest to determine the cause of this diffusion time-dependence in necrosis, as well as validating SSIFT using robust 3D histology to better understand its source of contrast and to determine whether the measured contrast purportedly driven by size-selective cellularity differs from that measured by ADC. The cell study (Figure 3) consisted of human cell lines from the most common sites of origin of brain metastases: lung, breast, and melanoma. However, brain cancers can be very heterogenous and SSIFT may have different performance in different types of brain neoplasms with different cancer cell sizes and cellularities. More investigation is necessary to evaluate SSIFT in specific types of human brain cancers. Further investigation into SSIFT in patients with radionecrosis will also be needed, as the animal subjects in this study showed slightly elevated SSIFT iAUC values relative to normal tissue. This is likely due to the increased cellularity in radionecrosis lesions observed in rats, which could diminish contrast relative to some cancer cell lines if present in humans. Nevertheless, SSIFT is a promising imaging method particularly for NSCLC which has large mean cancer cell sizes of 17.3 – 20.6 μm (66). Because the most common source of brain metastases is lung cancer (67.2%) (47) and 80 – 85% of all cases of lung cancer are NSCLC (67) SSIFT has the potential to play a role in imaging NSCLC in the clinical setting.
Further development of the method may improve human implementation. IVIM (intravoxel incoherent motion) effects were not explicitly considered in the current work but it may adversely influence SSIFT contrast. Because OGSE sequences are flow-compensated and less influenced by blood perfusion (68), IVIM shows a tdiff dependence, i.e., pseudo-diffusion increases with larger tdiff (69). If the pseudo-diffusion (perfusion) component is sufficiently large that it may conceal the cell size restriction induced tdiff dependence, which is the biophysical basis of SSIFT. This effect has been observed in the liver with a high blood volume (70). Our results in animals and patients in vivo did not show this effect, presumably because of the relatively much lower cerebral blood volume. However, if the IVIM effect becomes a concern, additional acquisitions with lower b values (e.g., 250 s/mm2) can be used to remove the IVIM effects (70), although this could increase the scan time. Additionally, the propagation of imaging artifacts to the iAUC has not been fully characterized. For instance, PGSE and OGSE diffusion-encoding gradients may induce different eddy currents which, if not fully corrected, could appear as diffusion time dependence and lead to aberrant values in SSIFT. It is also unknown how much variance in iAUC is due to noise or physiology. The CNR does not change significantly as the number of direction averages is decreased (Supplementary Figure S2), indicating the variance may not be due to random noise. Whether due to noise or diffusion time dependence in normal tissue, the contralateral variations are small compared to the iAUC values in tumor (Figure 6) such that sufficient contrast can still be generated.
The total scan time of the SSIFT method in humans was 16.5 mins because of the long TR (15 sec), a limitation imposed by diffusion gradient duty cycle in the OGSE sequence used in our Philips Ingenia 3T scanner. The inclusion of multi-band techniques can significantly reduce TR and the scan time. For example, our recent OGSE brain scan with a whole-brain coverage can achieve TR = 5.2 sec with a multi-band factor of 3 on a clinical Philips Ingenia 3T scanner. This cuts the total scan time of SSIFT with 32 directions to ~ 6 mins. Moreover, the number of gradient directions can be decreased while maintaining similar imaging contrast. Supplementary Figure S2 shows that fewer gradient directions, e.g., 6 directions, can still provide a significant contrast to differentiate tumors from peri-tumor edema, with which the total scan time of SSIFT could be further reduced to ~ 1.5 mins. While this shows contrast between tumor and normal-appearing tissues is sufficient with very few directions, this should be further optimized by comparing human tumor and radionecrosis data in future studies. Although further optimization of SSIFT is needed, the scan time is not a limiting factor for the translation of SSIFT to clinics, and may be an advantage over alternative quantitative models. Additionally, since PGSE images with multiple diffusion encoding directions are acquired, information from DTI can also be obtained. Combining iAUC with information from ADC or FA could add to SSIFT’s diagnostic ability to classify lesions at no cost to scan time.
Although contrast-enhanced T1w MRI is a standard method for imaging brain metastases, its usage can be limited in the setting of kidney dysfunction or severe contrast allergy. Moreover, some brain tumors, such as low-grade glioma, do not show contrast enhancement, which results in challenges for treatment planning in radiation oncology. Other non-contrast-enhanced MRI methods such as T2 FLAIR and DTI suffer from complex tissue compositions and cannot reliably differentiate tumors from peri-tumor edema. By contrast, Figure 6 shows SSIFT provides reasonably high conspicuity and delineation of brain metastases, suggesting SSIFT may be an alternative method for imaging brain metastases when contrast-enhanced MRI is not an option. However, SSIFT is a dMRI based method that suffers from drawbacks such as distortions and relatively low resolutions (e.g, 2 mm). Corrections for eddy-current and susceptibility induced distortions can alleviate such confounding effects. SSIFT will also benefit from recent progress in super-high resolution of DTI (71), smaller than the typical 2 mm resolution used in standard clinical practice.
Supplementary Material
Statement of significance.
This work introduces and provides preclinical validation of a new diffusion MRI method that exploits intrinsic differences in cell sizes to distinguish brain tumors and radiation necrosis.
Acknowledgments
The authors thank MR technologists Marisa Bush, Clair Jones, Joshua Hageman, and Christopher Thompson for assistance in data acquisitions, and Ms. Zou Yue for assistance in tumor induction in animals.
Grant Support:
This work was funded by NIH grants K25CA168936, R01CA109106, R01CA173593, UL1TR002243, S10OD021771, F32CA216942, American Cancer Society grant IRG#58-009-56, and Vanderbilt Radiology/VUIIS Catalyst Award Program. Studies were carried out, in part, using the Small Animal MR Facility of the Mallinckrodt Institute of Radiology, with support from the Small-Animal Cancer Imaging Shared Resource of the Siteman Cancer Center, Washington University in St. Louis.
Footnotes
Conflict of interest disclosure statement: No potential conflicts of interest were disclosed by the authors.
References
- 1.Owonikoko TK, Arbiser J, Zelnak A, Shu HKG, Shim H, Robin AM, et al. Current approaches to the treatment of metastatic brain tumours. Nat Rev Clin Oncol. 2014; 11:203–22. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Barnholtz-Sloan JS, Sloan AE, Davis FG, Vigneau FD, Lai P, Sawaya RE. Incidence proportions of brain metastases in patients diagnosed (1973 to 2001) in the Metropolitan Detroit Cancer Surveillance System. J Clin Oncol. 2004; 22:2865–72. [DOI] [PubMed] [Google Scholar]
- 3.Siegel RL, Miller KD, Jemal A. Cancer statistics, 2016. CA Cancer J Clin. 2016; 66:7–30. [DOI] [PubMed] [Google Scholar]
- 4.Chang EL, Wefel JS, Hess KR, Allen PK, Lang FF, Kornguth DG, et al. Neurocognition in patients with brain metastases treated with radiosurgery or radiosurgery plus whole-brain irradiation: a randomised controlled trial. Lancet Oncol. 2009; 10:1037–44. [DOI] [PubMed] [Google Scholar]
- 5.Lagerwaard FJ, Levendag PC, Nowak PJCM, Eijkenboom WMH, Hanssens PEJ, Schmitz PIM. Identification of prognostic factors in patients with brain metastases: A review of 1292 patients. Int J Radiat Oncol Biol Phys. 1999; 43:795–803. [DOI] [PubMed] [Google Scholar]
- 6.Magnuson WJ, Lester-Coll NH, Wu AJ, Yang TJ, Lockney NA, Gerber NK, et al. Management of brain metastases in tyrosine kinase inhibitor-Naïve epidermal growth factor receptor-mutant non-small-cell lung cancer: A retrospective multi-institutional analysis. J Clin Oncol. 2017; 35:1070–7. [DOI] [PubMed] [Google Scholar]
- 7.Ruben JD, Dally M, Bailey M, Smith R, McLean CA, Fedele P. Cerebral radiation necrosis: Incidence, outcomes, and risk factors with emphasis on radiation parameters and chemotherapy. Int J Radiat Oncol Biol Phys. 2006; 65:499–508. [DOI] [PubMed] [Google Scholar]
- 8.Colaco RJ, Martin P, Kluger HM, Yu JB, Chiang VL. Does immunotherapy increase the rate of radiation necrosis after radiosurgical treatment of brain metastases? J Neurosurg. 2016; 125:17–23. [DOI] [PubMed] [Google Scholar]
- 9.Forsyth PA, Kelly PJ, Cascino TL, Scheithauer BW, Shaw EG, Dinapoli RP, et al. Radiation necrosis or glioma recurrence: Is computer-assisted stereotactic biopsy useful? J Neurosurg. 1995; 82:436–44. [DOI] [PubMed] [Google Scholar]
- 10.Malone H, Yang J, Hershman DL, Wright JD, Bruce JN, Neugut AI. Complications Following Stereotactic Needle Biopsy of Intracranial Tumors. World Neurosurg. 2015; 84:1084–9. [DOI] [PubMed] [Google Scholar]
- 11.Valk PE, Dillon WP. Radiation injury of the brain. AJNR Am J Neuroradiol. 1991; 12:45–62. [PMC free article] [PubMed] [Google Scholar]
- 12.Pope WB. Brain metastases: neuroimaging. Handb Clin Neurol. 2018; 149:89–112. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Li H, Deng L, Bai HX, Sun J, Cao Y, Tao Y, et al. Diagnostic Accuracy of Amino Acid and FDG-PET in Differentiating Brain Metastasis Recurrence from Radionecrosis after Radiotherapy: A Systematic Review and Meta-Analysis. Am J Neuroradiol. 2018; 39:280–8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Chiang GC, Kovanlikaya I, Choi C, Ramakrishna R, Magge R, Shungu DC. Magnetic resonance spectroscopy, positron emission tomography and radiogenomics-Relevance to glioma. Front Neurol. 2018; 9:33. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Welker K, Boxerman J, Kalnin A, Kaufmann T, Shiroishi M, Wintermark M. ASFNR recommendations for clinical performance of MR dynamic susceptibility contrast perfusion imaging of the brain. Am J Neuroradiol. 2015; 36:E41–51. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Mitsuya K, Nakasu Y, Horiguchi S, Harada H, Nishimura T, Bando E, et al. Perfusion weighted magnetic resonance imaging to distinguish the recurrence of metastatic brain tumors from radiation necrosis after stereotactic radiosurgery. J Neurooncol. 2010; 99:81–8. [DOI] [PubMed] [Google Scholar]
- 17.Kwee RM, Kwee TC. Dynamic susceptibility MR perfusion in diagnosing recurrent brain metastases after radiotherapy: A systematic review and meta-analysis. J Magn Reson Imaging. 2020; 51:524–34. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Fink K, Fink J. Imaging of brain metastases. Surg Neurol Int. 2013; 4:209. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Vellayappan B, Tan CL, Yong C, Khor LK, Koh WY, Yeo TT, et al. Diagnosis and Management of Radiation Necrosis in Patients With Brain Metastases. Front. Oncol 2018. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Zhou J, Lal B, Wilson DA, Laterra J, Van Zijl PCM. Amide Proton Transfer (APT) Contrast for Imaging of Brain Tumors. Magn Reson Med. 2003; 50:1120–6. [DOI] [PubMed] [Google Scholar]
- 21.Zhou J, Tryggestad E, Wen Z, Lal B, Zhou T, Grossman R, et al. Differentiation between glioma and radiation necrosis using molecular magnetic resonance imaging of endogenous proteins and peptides. Nat Med. 2011; 17:130–4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Mehrabian H, Desmond KL, Soliman H, Sahgal A, Stanisz GJ. Differentiation between radiation necrosis and tumor progression using chemical exchange saturation transfer. Clin Cancer Res. 2017; 23:3667–75. [DOI] [PubMed] [Google Scholar]
- 23.Basser PJ, Mattiello J, LeBihan D. MR diffusion tensor spectroscopy and imaging. Biophys J. 1994; 66:259–67. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Xu JL, Li YL, Lian JM, Dou SW, Yan FS, Wu H, et al. Distinction between postoperative recurrent glioma and radiation injury using MR diffusion tensor imaging. Neuroradiology. 2010; 52:1193–9. [DOI] [PubMed] [Google Scholar]
- 25.Alexiou GA, Zikou A, Tsiouris S, Goussia A, Kosta P, Papadopoulos A, et al. Comparison of diffusion tensor, dynamic susceptibility contrast MRI and 99mTc-Tetrofosmin brain SPECT for the detection of recurrent high-grade glioma. Magn Reson Imaging. 2014; 32:854–9. [DOI] [PubMed] [Google Scholar]
- 26.Herculano-Houzel S The glia/neuron ratio: How it varies uniformly across brain structures and species and what that means for brain physiology and evolution. Glia. 2014; 62:1377–91. [DOI] [PubMed] [Google Scholar]
- 27.Rajkowska G, Selemon LD, Goldman-Rakic PS. Neuronal and glial somal size in the prefrontal cortex: A postmortem morphometric study of schizophrenia and huntington disease. Arch Gen Psychiatry. 1998; 55:215–24. [DOI] [PubMed] [Google Scholar]
- 28.Aboitiz F, Scheibel AB, Fisher RS, Zaidel E. Fiber composition of the human corpus callosum. Brain Res. 1992; 598:143–53. [DOI] [PubMed] [Google Scholar]
- 29.Panagiotaki E, Chan RW, Dikaios N, Ahmed HU, O’Callaghan J, Freeman A, et al. Microstructural characterization of normal and malignant human prostate tissue with vascular, extracellular, and restricted diffusion for cytometry in tumours magnetic resonance imaging. Invest Radiol. 2015; 50:218–27. [DOI] [PubMed] [Google Scholar]
- 30.Wu D, Jiang K, Li H, Zhang Z, Ba R, Zhang Y, et al. Time-Dependent Diffusion MRI for Quantitative Microstructural Mapping of Prostate Cancer. Radiology. 2022; :211180. [DOI] [PubMed] [Google Scholar]
- 31.Xu J, Jiang X, Li H, Arlinghaus LR, McKinley ETET, Devan SPSP, et al. Magnetic resonance imaging of mean cell size in human breast tumors. Magn Reson Med. 2020; 83:2002–14. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Zaccagna F, Riemer F, Priest AN, McLean MA, Allinson K, Grist JT, et al. Non-invasive assessment of glioma microstructure using VERDICT MRI: correlation with histology. Eur Radiol. 2019/03/19. 2019; 29:5559–66. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Roberts TA, Hyare H, Agliardi G, Hipwell B, d’Esposito A, Ianus A, et al. Noninvasive diffusion magnetic resonance imaging of brain tumour cell size for the early detection of therapeutic response. Sci Rep. 2020; 10:9223. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Xu J, Does MD, Gore JC. Sensitivity of MR diffusion measurements to variations in intracellular structure: Effects of nuclear size. Magn Reson Med. 2009; 61:828–33. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Gore JC, Xu J, Colvin DC, Yankeelov TE, Parsons EC, Does MD. Characterization of tissue structure at varying length scales using temporal diffusion spectroscopy. NMR Biomed. 2010; 23:745–56. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Does MD, Parsons EC, Gore JC. Oscillating gradient measurements of water diffusion in normal and globally ischemic rat brain. Magn Reson Med. 2003; 49:206–15. [DOI] [PubMed] [Google Scholar]
- 37.Jiang X, Li H, Devan SP, Gore JC, Xu J. MR cell size imaging with temporal diffusion spectroscopy. Magn Reson Imaging. 2021; 77:109–23. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Kaden E, Kruggel F, Alexander DC. Quantitative mapping of the per-axon diffusion coefficients in brain white matter. Magn Reson Med. 2016; 75:1752–63. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Devan S, Jiang X, Bagnato F, Xu J. Optimization and numerical evaluation of multi-compartment diffusion MRI using the spherical mean technique for practical multiple sclerosis imaging. Magn Reson Imaging. 2020; 74:56–63. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Clark CA, Hedehus M, Moseley ME. Diffusion time dependence of the apparent diffusion tensor in healthy human brain and white matter disease. Magn Reson Med. 2001; 45:1126–9. [DOI] [PubMed] [Google Scholar]
- 41.Xu J, Li H, Harkins KD, Jiang X, Xie J, Kang H, et al. Mapping mean axon diameter and axonal volume fraction by MRI using temporal diffusion spectroscopy. Neuroimage. 2014; 103:10–9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Baron C, Beaulieu C. Oscillating gradient spin-echo (OGSE) diffusion tensor imaging of the human brain. Magn Reson Med. 2014; 72:726–36. [DOI] [PubMed] [Google Scholar]
- 43.Xu J, Does MD, Gore JC. Numerical study of water diffusion in biological tissues using an improved finite difference method. Phys Med Biol. 2007; 52:N111–26. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Xu J, Does MD, Gore JC. Dependence of temporal diffusion spectra on microstructural properties of biological tissues. Magn Reson Imaging. 2011; 29:380–90. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Jiang X, Li H, Xie J, Zhao P, Gore JC, Xu J. Quantification of cell size using temporal diffusion spectroscopy. Magn Reson Med. 2016; 75:1076–85. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Jiang X, Li H, Xie J, McKinley ET, Zhao P, Gore JC, et al. In vivo imaging of cancer cell size and cellularity using temporal diffusion spectroscopy. Magn Reson Med. 2017; 78:156–64. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Schouten LJ, Rutten J, Huveneers HAM, Twijnstra A. Incidence of brain metastases in a cohort of patients with carcinoma of the breast, colon, kidney, and lung and melanoma. Cancer. 2002; 94:2698–705. [DOI] [PubMed] [Google Scholar]
- 48.Boyle W, Chow A. Isolation of Human Lymphocytes by a Ficoll Barrier Method. Transfusion. 1969; 9:151–5. [DOI] [PubMed] [Google Scholar]
- 49.Schneider CA, Rasband WS, Eliceiri KW. NIH Image to ImageJ: 25 years of image analysis. Nat Methods. 2012; 9:671–5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Bankhead P, Loughrey MB, Fernández JA, Dombrowski Y, McArt DG, Dunne PD, et al. QuPath: Open source software for digital pathology image analysis. Sci Reports 2017 71. 2017; 7:1–7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Jost SC, Hope A, Kiehl E, Perry A, Travers S, Garbow JR. A Novel Murine Model for Localized Radiation Necrosis and its Characterization using Advanced Magnetic Resonance Imaging. Int J Radiat Oncol Biol Phys. 2009; 75:527–33. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Jiang X, Engelbach JA, Yuan L, Cates J, Gao F, Drzymala RE, et al. Anti-VEGF antibodies mitigate the development of radiation necrosis in mouse brain. 2014; 20:2695–702. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Tournier JD, Smith R, Raffelt D, Tabbara R, Dhollander T, Pietsch M, et al. MRtrix3: A fast, flexible and open software framework for medical image processing and visualisation. Neuroimage. 2019; 202:116137. [DOI] [PubMed] [Google Scholar]
- 54.Veraart J, Novikov DS, Christiaens D, Ades-aron B, Sijbers J, Fieremans E. Denoising of diffusion MRI using random matrix theory. Neuroimage. 2016; 142:394. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Kellner E, Dhital B, Kiselev VG, Reisert M. Gibbs-ringing artifact removal based on local subvoxel-shifts. Magn Reson Med. 2016; 76:1574–81. [DOI] [PubMed] [Google Scholar]
- 56.Jenkinson M, Beckmann CF, Behrens TEJ, Woolrich MW, Smith SM. FSL. Neuroimage. 2012; 62:782–90. [DOI] [PubMed] [Google Scholar]
- 57.Woolrich MW, Jbabdi S, Patenaude B, Chappell M, Makni S, Behrens T, et al. Bayesian analysis of neuroimaging data in FSL. Neuroimage. 2009; 45. [DOI] [PubMed] [Google Scholar]
- 58.Smith SM, Jenkinson M, Woolrich MW, Beckmann CF, Behrens TEJ, Johansen-Berg H, et al. Advances in functional and structural MR image analysis and implementation as FSL. Neuroimage. 2004. [DOI] [PubMed] [Google Scholar]
- 59.Andersson JLR, Sotiropoulos SN. An integrated approach to correction for off-resonance effects and subject movement in diffusion MR imaging. Neuroimage. 2016; 125:1063. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Tustison NJ, Avants BB, Cook PA, Zheng Y, Egan A, Yushkevich PA, et al. N4ITK: Improved N3 bias correction. IEEE Trans Med Imaging. 2010; 29:1310–20. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Oh J, Cha S, Aiken AH, Han ET, Crane JC, Stainsby JA, et al. Quantitative apparent diffusion coefficients and T2 relaxation times in characterizing contrast enhancing brain tumors and regions of peritumoral edema. J Magn Reson Imaging. 2005; 21:701–8. [DOI] [PubMed] [Google Scholar]
- 62.Raffelt D, Tournier JD, Rose S, Ridgway GR, Henderson R, Crozier S, et al. Apparent Fibre Density: A novel measure for the analysis of diffusion-weighted magnetic resonance images. Neuroimage. 2012; 59:3976–94. [DOI] [PubMed] [Google Scholar]
- 63.Gao F, Shen X, Zhang H, Ba R, Ma X, Lai C, et al. Feasibility of oscillating and pulsed gradient diffusion MRI to assess neonatal hypoxia-ischemia on clinical systems. J Cereb Blood Flow Metab. 2021; 41:1240–50. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Daducci A, Canales-Rodríguez EJ, Zhang H, Dyrby TB, Alexander DC, Thiran JP. Accelerated Microstructure Imaging via Convex Optimization (AMICO) from diffusion MRI data. Neuroimage. 2015; 105:32–44. [DOI] [PubMed] [Google Scholar]
- 65.Pronin IN, Kornienko VN, Fadeeva LM, Rodionov PV, Golanov AV. [Diffusion-weighted image in the study of brain tumors and peritumoral edema] . Zh Vopr Neirokhir Im N N Burdenko. 2000; :14–6. [PubMed] [Google Scholar]
- 66.Hosokawa M, Kenmotsu H, Koh Y, Yoshino T, Yoshikawa T, Naito T, et al. Size-based isolation of circulating tumor cells in lung cancer patients using a microcavity array system. PLoS One. 2013; 8:e67466–e67466. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.Molina JR, Yang P, Cassivi SD, Schild SE, Adjei AA. Non-small cell lung cancer: Epidemiology, risk factors, treatment, and survivorship. Mayo Clin Proc. 2008; 83:584–94. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68.Van AT, Holdsworth SJ, Bammer R. In vivo investigation of restricted diffusion in the human brain with optimized oscillating diffusion gradient encoding. Magn Reson Med. 2014; 71:83–94. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69.Wu D, Zhang J. The Effect of Microcirculatory Flow on Oscillating Gradient Diffusion MRI and Diffusion Encoding with Dual-Frequency Orthogonal Gradients (DEFOG). Magn Reson Med. 2017; 77:1583–92. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70.Jiang X, Xu J, Gore JC. Mapping hepatocyte size in vivo using temporal diffusion spectroscopy MRI. Magn Reson Med. 2020/04/25. 2020; 84:2671–83. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 71.Holdsworth SJ, O’Halloran R, Setsompop K. The quest for high spatial resolution diffusion-weighted imaging of the human brain in vivo. NMR Biomed. 2019; 32:e4056. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Data Availability Statement
The data generated in this study are available upon request from the corresponding author.


