Abstract
Expression of ExoU by Pseudomonas aeruginosa is correlated with acute cytotoxicity in a number of epithelial and macrophage cell lines. In vivo, ExoU is responsible for epithelial injury. The absence of a known motif or significant homology with other proteins suggests that ExoU may possess a new mechanism of toxicity. To study the intracellular effects of ExoU, we developed a transient-transfection system in Chinese hamster ovary cells. Transfection with full-length but not truncated forms of ExoU inhibited reporter gene expression. Inhibition of reporter activity after cotransfection with ExoU-encoding constructs was correlated with cellular permeability and death. The toxicity of truncated versions of ExoU could be restored by coexpression of the remainder of the molecule from separate plasmids in trans. This strategy was used to map N- and C-terminal regions of ExoU that are necessary but not sufficient for toxicity. Disruption of a middle region of the protein reduces toxicity. This portion of the molecule is postulated to allow the N- and C-terminal regions to functionally complement one another. In contrast to ExoS and ExoT, native and recombinant ExoU molecules do not oligomerize or form aggregates. The complex domain structure of ExoU suggests that, like other P. aeruginosa-encoded type III effectors (ExoS and ExoT), ExoU toxicity may result from a molecule that possesses more than one activity.
Bacterial pneumonia due to Pseudomonas aeruginosa is a frequent occurrence in critically ill patients, particularly those under mechanical ventilation. Among nosocomial pneumonias, infection due to P. aeruginosa has a relatively high mortality rate, with death resulting from septic shock and multiple organ failure (25). The bacterial proteins that are linked to the ability of P. aeruginosa to cause fatal infections in animals include a type III secretion-intoxication system and the effector proteins delivered by this system (2, 12, 20, 32). Currently, four known effectors, ExoS, ExoT, ExoY, and ExoU, are translocated directly from the bacterial cell into the eukaryotic host cell cytoplasm by the P. aeruginosa type III secretion mechanism (8, 19, 34, 40, 41). Expression of the effectors and distribution of the genes encoding the effectors appear to vary among P. aeruginosa strains, an observation which may explain differences in clinical outcome (8, 10, 22). ExoS and ExoT exert their toxic effect by disrupting host signal transduction through their ADP-ribosyltransferase activity (15, 16, 26, 28, 36) and by promoting actin skeleton modifications (29). ExoY is an adenylate cyclase, which induces elevation of intracellular cyclic AMP in vitro (41). The translocation of ExoU induces either a cytotoxic phenotype in tissue culture models (8, 19, 22, 34) or a fatal outcome in an acute lung infection model (1, 2, 8, 11, 31, 32). The acute cytotoxicity mediated by ExoU appears to play a major role in septic shock (25) and may also aid in evasion of host responses by decreasing macrophage viability and phagocytic function (4, 31, 32).
Although the expression and delivery of ExoU by the P. aeruginosa type III system result in a clear cytotoxic response, as measured by the release of intracellular markers and the permeability of cells to certain dyes, the mechanism of action of ExoU is unclear. ExoU is a large protein of 687 amino acids with a molecular mass of approximately 74 kDa (8). The molecule is predicted to be hydrophilic and slightly acidic, with a pI of approximately 5.9 (8). The amino acid sequence possesses no significant homology, motifs, or predicted secondary structure that would aid in defining possible enzymatic or functional aspects of the molecule. Cell death mediated by ExoU occurs within 3 to 4 h of infection and is characteristic of necrosis rather than apoptosis or oncosis (2, 6, 18). In preliminary studies, purified recombinant ExoU possesses no ADP-ribosyltransferase, kinase, phosphatase, hemolytic, or cytotoxic activity (V. Finck-Barbançon and D. Frank, unpublished observations). Less than 15% of the molecule inserts into artificial liposomes, indicating that the cytotoxic activity of ExoU is likely not related to the direct formation of pores or channels in cellular membranes (J. Feix, V. Finck-Barbançon and D. Frank, unpublished results).
We developed a transient-transfection system to begin to define functional regions of ExoU and to identify a minimal domain required for cytotoxic activity. An important advantage of this type of assay is that only the protein of interest is expressed intracellularly, eliminating the potential confounding factors or toxicity associated with type III-mediated intoxication and bacterial infection. Deletion mapping and cotransfection experiments with ExoU-encoding constructs identified three domains that are important for biological activity. Our structural analysis suggests that toxicity can be mediated by N- and C-terminal domains in a trans configuration as long as one or both domains are physically linked to an internal domain. These studies indicate that ExoU is a complex molecule and that its mode of action may involve novel functional activities.
MATERIALS AND METHODS
Materials.
Reagents were purchased from Sigma unless otherwise indicated. Primers for PCR were purchased from Operon (Alameda, Calif.). pCMV-MCS, pCMV-luciferase, and pCMV-β galactosidase were gifts from Kent Wilcox, Medical College of Wisconsin. pEGFP-N1 was purchased from Clontech (Palo Alto, Calif.).
Bacterial strains, cell lines, and growth conditions.
Escherichia coli host strain DH5α was grown on Luria-Bertani (LB) agar or LB broth supplemented with either ampicillin (100 μg ml−1) or kanamycin (50 μg ml−1) when containing plasmids. CHO-K1 cells were grown in Ham's F12 medium supplemented with 10% newborn calf serum as previously described (35).
Plasmid construction.
In vivo expression vectors were constructed in plasmid pEGFP-N1 (Clontech), which contains the immediate-early (IE) promoter of cytomegalovirus (CMV) upstream of the multiple cloning site (MCS) and the green fluorescent protein (GFP) coding sequence downstream of the MCS. Translational fusions of either full-length or truncated exoU were constructed by PCR cloning procedures as previously described (9), using DeepVent polymerase (New England Biolabs, Beverly, Mass.), and inserted in the pEGFP-N1 vector. Primers were designed to allow in-frame expression of an enhanced version of GFP. Alternatively, pCMV-MCS, a eukaryotic expression vector (37) containing the IE promoter of CMV, was used in transient-transfection experiments.
To construct 1-687U-GFP (WT2), DNA encoding full-length exoU was amplified with the thermal profile described previously (9) using the primers described in Table 1. A HindIII site 5′ to the NsiI (ATGCAT) start site of exoU was introduced, and a BamHI site replaced the exoU stop codon in order to allow positional cloning in frame into the MCS of the pEGFP-N1 vector digested with the corresponding enzymes. To construct pCMV-His-1-687U (WT3), an XbaI-BamHI fragment from pETexoU (9) containing histidine-tagged exoU was ligated to the pCMV-MCS vector digested with Xba and BglII. This full-length construct harbored exoU with its own stop codon and an amino-terminal 10-histidine tag (WT3, Fig. 1). To construct a plasmid corresponding to the full-length form of exoU, not fused to the GFP coding region or a histidine tag, the coding region was amplified using primers which inserted HindIII sites 5′ to the ATG start site of exoU and 3′ to the exoU stop codon. Amplified DNA was ligated to pCMV-MCS (WT1) digested with the corresponding enzyme. Proper reading frame orientation was confirmed by restriction digest analysis. With the primers listed in Table 1 and the same strategy, DNA corresponding to N- and C-terminal deletions of exoU was amplified and inserted into pEGFP-N1 as HindIII/-BamHI inserts. Initial effector plasmids used for transient transfection of CHO-K1 cells are represented in Fig. 1. Insert DNA was sequenced on an ABI Prism 377 DNA sequencer using the BigDye terminator cycle sequencing kit (Applied Biosystems, Foster City, Calif.). Expressed products were confirmed by Western blot analysis using rabbit anti-ExoU serum and/or monoclonal anti-GFP antibodies or fluorescence-activated cell sorting (FACS) analysis.
TABLE 1.
Primers used
| Clone | Primer sequencea |
|---|---|
| pCMV-exoU (WT1) | 5′-G CTG AAG CTT ATG CAT ATC CAA TCG TTG GG |
| 5′-CCG AAG CTT GGA TCC TCA TGT GAA CTC CTT | |
| p1-687U-GFP (WT2) | 5′-G CTG AAG CTT ATG CAT ATC CAA TCG TTG GG |
| 5′-CGG TGG ATC CCA TGT GAA CTC CTT ATT CC | |
| p1-633U-GFP (N1) | 5′-G CTG AAG CTT ATG CAT ATC CAA TCG TTG GG |
| 5′-CGG TAA GCT TGG ATC CCG ACG TAA CAG AG | |
| p1-351U-GFP (N2) | 5′-G CTG AAG CTT ATG CAT ATC CAA TCG TTG GG |
| 5′-TC CGG ATC CAC GTT AAT CAT CAC C | |
| p1-123U-GFP (N3) | 5′-G CTG AAG CTT ATG CAT ATC CAA TCG TTG GG |
| 5′-GT AGG ATC CAT TGC TCC CGG GTA TGC C | |
| p22-351U-GFP (N4) | 5′-CTG AAG CTT ATG CAT TCG CAG GCA GCG CA |
| 5′-TC CGG ATC CAC GTT AAT CAT CAC C | |
| p52-351U-GFP (N5) | 5′-CTG AAG CTT ATG CAT TCC GGG AAG TTG CCG |
| 5′-TC CGG ATC CAC GTT AAT CAT CAC C | |
| p100-351U-GFP (N6) | 5′-GCG AAG CTT ATG CAT AGT CGG CCA CCA TTG |
| 5′-TC CGG ATC CAC GTT AAT CAT CAC C | |
| p155-351U-GFP (N7) | 5′-CGC AAG CTT ATG AGC CCG GCG GCG T |
| 5′-TC CGG ATC CAC GTT AAT CAT CAC C | |
| p52-123U-GFP (N9) | 5′-CTG AAG CTT ATG CAT TCC GGG AAG TTG CCG |
| 5′-GT AGG ATC CAT TGC TCC CGG GTA TGC C | |
| p52-155U-GFP (N10) | 5′-CTG AAG CTT ATG CAT TCC GGG AAG TTG CCG |
| 5′-TG CGG ATC CAT ACC TGA GGC CAA AAG G | |
| p52-201U-GFP (N11) | 5′-CTG AAG CTT ATG CAT TCC GGG AAG TTG CCG |
| 5′-TTA CGG ATC CTT GTT GCC CAA GCC CTT T | |
| p52-249U-GFP (N12) | 5′-CTG AAG CTT ATG CAT TCC GGG AAG TTG CCG |
| 5′-AGT AGG ATC CGT CGG CTG GCG TGC | |
| p52-300U-GFP (N13) | 5′-CTG AAG CTT ATG CAT TCC GGG AAG TTG CCG |
| 5′-CGC AGG ATC CGC ATT GAA CAC CAC TAA | |
| p52-578-GFP (N14) | 5′-CTG AAG CTT ATG CAT TCC GGG AAG TTG CCG |
| 5′-ATC TGG ATC CGC ATC GAG TAA CTG CTG | |
| p124-687U-GFP (C1) | 5′-G CTG AAG CTT ATG CAT CTG GCG CTA GAA GAG |
| 5′-CGG TGG ATC CCA TGT GAA CTC CTT ATT CC | |
| p343-687U-GFP (C2) | 5′-TTG AAG CTT ATG CAT CAG GAT GGC GGG GTG |
| 5′-CGG TGG ATC CCA TGT GAA CTC CTT ATT CC | |
| p409-687U-GFP (C3) | 5′-C GTC AAG CTT CTT GAG TTC GAG GGC |
| 5′-CGG TGG ATC CCA TGT GAA CTC CTT ATT CC | |
| p437-687U-GFP (C4) | 5′-GAT AAG CTT GAG CGC GGT GAT TTC AG |
| 5′-CGG TGG ATC CCA TGT GAA CTC CTT ATT CC | |
| p580-687U-GFP (C6) | 5′-ATC AAG CTT ATG CAT ATG CGC GGG CAG ACG GT |
| 5′-CGG TGG ATC CCA TGT GAA CTC CTT ATT CC | |
| p352-687U-GFP (C7) | 5′-CTG AAG CTT ATG CCG GTC CCT GAG ATG |
| 5′-CGG TGG ATC CCA TGT GAA CTC CTT ATT CC | |
| p203-687-GFP (C8) | 5′-TTGA AAG CTT ATG GGC GGC TTC TCT GAG |
| 5′-CGG TGG ATC CCA TGT GAA CTC CTT ATT CC |
Letters in bold type correspond to either HindIII or BamHI restriction endonuclease cleavage sites.
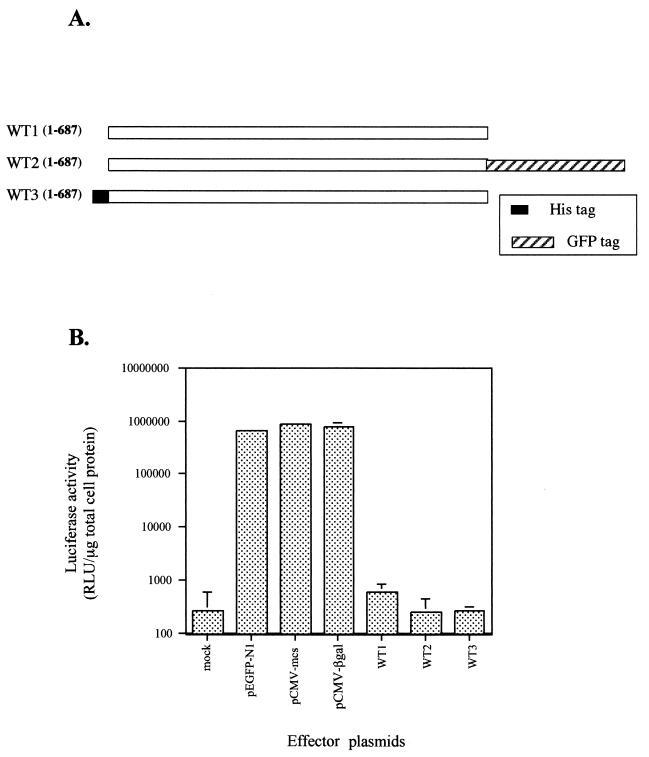
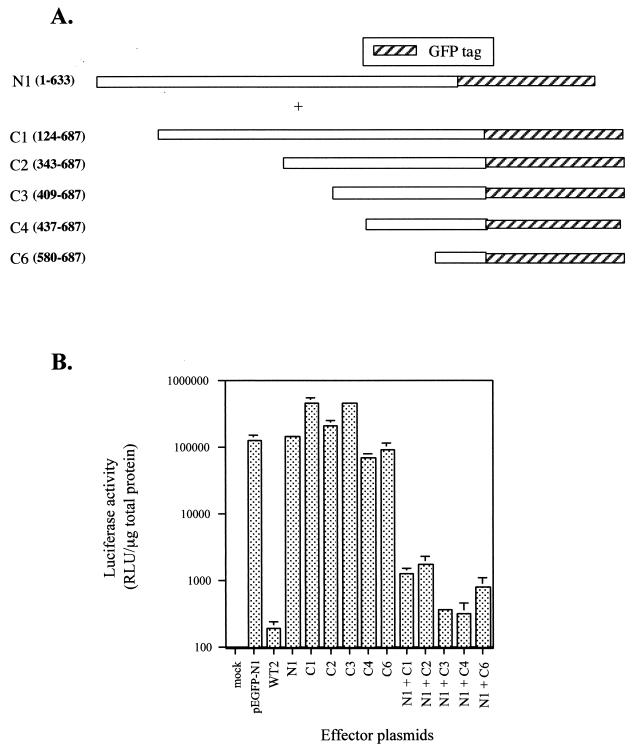
FIG. 1.
(A) Initial constructs used in transfection experiments to determine the effect of untagged and tagged versions of ExoU on reporter gene expression. The solid rectangle illustrates an amino-terminal, in-frame fusion encoding a 10-amino-acid histidine tag. The hatched box illustrates an in-frame fusion with an enhanced GFP-encoding gene. All constructs were transcribed from the CMV early promoter. ExoU amino acids are shown in open bars. (B) Analysis of luciferase expression in transfected CHO cells. Cells were cotransfected with either a plasmid control (pEGFP-N1, pCMV-mcs, or pCMV-βgal) or a plasmid encoding an effector protein (WT1, WT2, or WT3) and a reporter plasmid encoding the luciferase gene. The total amount of DNA transfected was normalized with CT DNA. Cells were harvested 24 h posttransfection and assayed for luciferase activity as described in the text. The data (relative light units [RLU]) were averaged from experiments performed in triplicate.
Transient transfection and luciferase assays.
Transient transfections were performed at 37°C in a 5% CO2 incubator as described (28) with the following modifications. CHO-K1 cells were seeded at 3 × 105 cells/well in 12-well plates the day prior to transfection. Transfections were performed in triplicate with Lipofectamine Plus (Life Technologies, Grand Island, N.Y.) in 500 μl of Opti-MEM (Life Technologies) according to the manufacturer's recommendations unless otherwise indicated. Medium was then replaced with complete Ham's F-12 medium, and the cells were harvested 24 h after the start of transfection unless a time course experiment was performed. DNA concentrations (total, 400 ng) were equalized when appropriate with the addition of sheared calf thymus (CT) DNA. Control experiments showed that pCMV-MCS, pCMV-β-galactosidase plasmid vectors, and CT DNA could be used interchangeably to normalize DNA concentrations without affecting reporter plasmid expression as measured by luciferase enzymatic activity (Fig. 1). Transfection frequency varied from 15 to 40%. In each data set, mock vector, pEGFP-N1 vector, and WT-2 (containing full-length exoU) were transfected, and luciferase activities measured to control for day-to-day variations in transfection frequency.
After transfection, cells were washed twice with 2 ml of cold Dulbecco's phosphate-buffered saline (D-PBS) (Life Technologies) and harvested for luciferase determinations. Cells were scraped in 200 μl of 1× reporter lysis buffer and treated according to the manufacturer's instructions (Promega, Madison, Wis.). Light emission was measured in a luminometer for 10 s at room temperature. Control reactions showed that luciferase activity measured in the lysates was within the linear range of the luminometer (data not shown). Determination of the total amount of protein in cell lysates was performed using the bicinchoninic acid (BCA) protein assay (Pierce Chemical Company, Rockford, Ill.). Luciferase activity was normalized to the total protein concentration.
Gel filtration chromatography.
Purified recombinant ExoU (rExoU), an E. coli BL21(DE3) lysate expressing rExoU from pETexoU (9), or ammonium sulfate-precipitated extracellular proteins from P. aeruginosa PA103 were subjected to Sephacryl S-200-HR gel filtration chromatography. After ultracentrifugation (100,000 × g for 45 min at 4°C), 380 μl of the soluble fraction was loaded onto a 38-ml column equilibrated in 10 mM Tris (pH 8.4)–100 mM NaCl–1 mM EDTA. Column fractions (500 μl) were monitored for their absorbance at 280 nm and analyzed by sodium dodecyl sulfate-polyacrylamide gel electrophoresis (SDS-PAGE) and Western blot analysis with polyclonal rabbit anti-ExoU and anti-ExoS/T immunoglobulin G IgG. Low- and high-molecular-weight gel filtration calibration kits (Amersham/Pharmacia, Piscataway, N.J.) were used to obtain a calibration curve and determine the molecular weights of proteins. Peak fractions were determined by spot densitometry of stained gels or immunoblots using AlphaEase software (Alpha Innotech Corporation, San Leandro, Calif.).
Glycerol density gradient centrifugation.
Extracellular proteins from P. aeruginosa PA103 were prepared as described previously (12). Dialyzed ammonium sulfate-precipitated proteins were centrifuged for 45 min at 100,000 × g, and 120-μl aliquots were layered on top of 5 ml of 10 to 35% glycerol gradients prepared in 60 mM Tris (pH 7.6)–100 mM NaCl (5). Fractions were collected from the top after 13.5 h of centrifugation at 40,000 rpm in an SW55 rotor (Beckman Instruments, Palo Alto, Calif.) at 4°C. Standards (albumin [4.6 S] and catalase [11.2S]) were fractionated either in separate but parallel gradients or in the same tube with our test samples. Fractions were analyzed by immunoblotting using a combination of rabbit anti-ExoU and anti-ExoS/T IgG preparations and peroxidase-labeled goat anti-rabbit Ig secondary antibodies (Boehringer Mannheim, Indianapolis, Ind.). The substrate to visualize secondary antibody binding was an enhanced chemiluminescence (ECL) reagent (SuperSignal substrate; Pierce Chemical Company).
FACS analysis of expressed ExoU fusions to GFP.
A fluorescence-activated cell sorter was used to measure the signal intensity from cells transfected with constructs encoding ExoU-GFP fusions as a means to ensure that each construct was translated. CHO-K1 cells were seeded the day prior to transfection in 24-well plates. Cell density and the amount of effector DNA were scaled down in proportion to the reduction in plate surface area but otherwise performed as described above for luciferase assays using Lipofectamine Plus (Life Technologies). After transfection (18 h), cells were washed with 0.5 ml of D-PBS, trypsinized (100 μl), and fixed with 0.5 ml of 1.0% paraformaldehyde in PBS. Quantitative measurements were determined using a FACSCAN instrument (Becton Dickinson, San Jose, Calif.) and CellQuest software. Fluorescence data for each sample were collected from 10,000 events. The geometric mean of fluorescence intensity for each construct is reported as an average of duplicate transfections (see Fig. 9A).
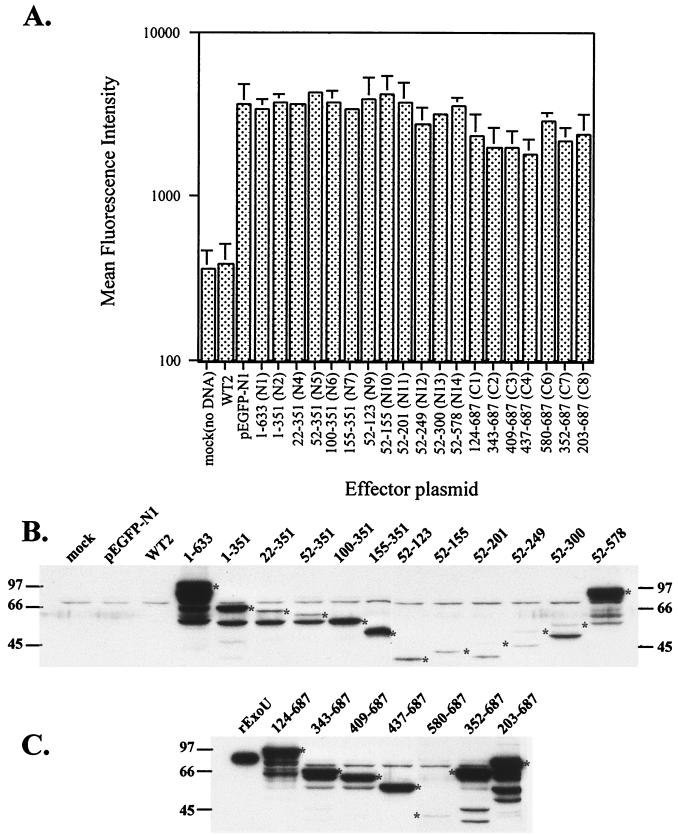
FIG. 9.
Expression levels of N-and C-terminal deletions of ExoU in CHO cells. (A) FACS analysis of cells transfected with the indicated ExoU clones fused to GFP. CHO cells were transfected in 24-well plates with 75 ng of effector plasmid (exoU fusions), 50 ng of reporter plasmid (pLuciferase), and CT DNA in duplicate. The mean fluorescence intensity of 10,000 events was measured for each well and is shown as a histogram profile of fluorescence intensity. (B) Western blot of cellular lysates made from CHO cells transfected with various N-terminal ExoU-GFP expression constructs. CHO cells (12-well plates) were transfected as described in Materials and Methods for luciferase assays. After 18 h, cells were washed, recovered, and lysed. Each lane was loaded with 20 μl of cell lysate, which was resolved by SDS-PAGE and analyzed by Western blot with anti-ExoU antibodies. The size of the expected ExoU derivative fused to GFP is indicated with an asterisk. The lanes contained (from left to right) mock-transfected cells or cells transfected with the pEGFP-N1 vector without insert, WT2, N1 (1 to 633), N2 (1 to 351), N4 (22 to 351), N5 (52 to 351), N6 (100 to 351), N7 (155 to 351), N9 (52 to 123), N10 (52 to 155), N11 (52 to 201), N12 (52 to 249), N13 (52 to 300), or N14 (52 to 578). (C) Western blot demonstrating the expression of C-terminal expression constructs of ExoU fused to GFP. The lanes contained (from left to right) purified recombinant ExoU or lysates from cells transfected with C1 (124 to 687), C2 (343 to 687), C3 (409 to 687), C4 (437 to 687), C6 (580 to 687), C7 (352 to 687), or C8 (203 to 687). The expected size of the translational fusion is indicated with an asterisk.
SDS-PAGE and immunoblot analysis of fusion proteins.
To determine the size and immunoreactivity of translational fusions from transfected CHO cells, cells were seeded into 12-well plates, transfected, and harvested as described except that SDS-PAGE buffer was used instead of 1× reporter lysis buffer. Chromosomal DNA was sheared by repetitive passage of the harvested material through a 26 3/8-gauge needle. Cellular lysates were normalized by loading the same volume per lane and subjected to SDS–10% PAGE. The separated proteins were transferred to nitrocellulose filters. Filters were probed with rabbit anti-ExoU serum (1:20,000 dilution) and a secondary horseradish peroxidase-conjugated anti-rabbit Ig antibody (1:7,000; Boehringer-Mannheim). Immunoreactive bands were detected using ECL Western blot reagents (Pierce).
For detection of combinations of translational fusions from transfected CHO cells, two 100-mm culture dishes were seeded at 5 × 106 cells/dish 24 h prior to transfection. Cells were transfected at 70 to 80% confluency with Lipofectamine Plus (Life Technologies) according to the manufacturer's recommendations and harvested 24 h after the start of transfection. Transfection efficiency for these experiments was determined to be 30% by counting three random fields. Cell lysates were prepared as previously described (38) with a few modifications. Briefly, cells were washed twice with 5 ml of ice-cold PBS and scraped off the dishes in 1 ml of HES (20 mM HEPES [pH 7.4], 1 mM EDTA, 255 mM sucrose) buffer in the presence of the protease inhibitors aprotinin (20 μg/ml), leupeptin (10 μg/ml), pepstatin (1 μg/ml), benzamidine (10 μg/ml), and phosphoramidon (3.4 μM). Cells were pelleted at 4,000 × g at 4°C for 3 min, suspended in 250 μl of HES buffer plus inhibitors, and broken by passage through a 251/2-gauge needle (15 times). Nuclei and unbroken cells were removed by a low-speed (6,000 × g) centrifugation at 4°C for 5 min. Supernatants were collected and assayed for protein content using the BCA assay. Equal amounts of protein were analyzed by SDS-PAGE. Western blots were performed using a monoclonal anti-rGFP antibody (Clontech, Palo Alto, Calif.; dilution, 1:1,000) and a secondary peroxidase-conjugated goat anti-mouse Ig antibody (Sigma Chemical Company, St. Louis, Mo.; dilution, 1:20,000). Antibody binding was detected by chemiluminescence.
Ethidium bromide vital staining.
After the start of transfection (8 or 24 h), ethidium bromide was added to the culture medium to a final concentration of 1 μM. Ethidium bromide is a nonfluorescent compound that will only enter cells that have become permeable. Upon binding nuclear DNA of dead cells, ethidium bromide will produce a bright red fluorescent signal, which was monitored using a Diaphot-200 fluorescence microscope (Nikon Corporation, Tokyo, Japan) and recorded with a Spot camera (Diagnostic Instruments, Sterling Heights, Mich.).
RESULTS
Intracellular expression of full-length ExoU inhibits reporter gene expression.
To investigate the intracellular effect of ExoU expression without interference from other effectors or secreted proteins delivered by the type III system of P. aeruginosa, we constructed various eukaryotic expression vectors encoding exoU. Transient-cotransfection experiments were performed in the presence of a luciferase reporter plasmid. Three plasmid constructs encoding full-length ExoU (687 amino acids) were designed to be expressed under the control of the CMV promoter (Fig. 1A). One plasmid (WT2) encoded a fusion to GFP which yielded a hybrid protein with a 27-kDa GFP addition at the C terminus of ExoU. Another fusion was engineered to express ExoU with a short amino-terminal tag of 10 histidine residues (WT3). To eliminate potential effects of either the GFP protein or the histidine tag on ExoU biological activity, ExoU was also expressed without any tag sequences (WT1). Each plasmid expressing full-length ExoU was cotransfected into Chinese hamster ovary (CHO) cells with a reporter plasmid encoding luciferase, and luciferase activity in cellular lysates was quantified and normalized to the amount of total cell protein. The total amount of DNA in the transfection reaction was kept constant.
Cotransfection of CHO cells with any of the full-length ExoU-encoding constructs resulted in reduced luciferase activity compared to control plasmids (Fig. 1B). The addition of a histidine peptide to the amino terminus or GFP to the carboxy terminus of ExoU did not affect the inhibition of luciferase activity. Cells transfected with the pEGFP-N1 vector alone exhibited green fluorescence; however, fluorescence was undetectable in a transfection using p1-687U-GFP (WT2).
The inhibitory effect of ExoU on reporter gene expression was followed over time by comparison of the luciferase activity from cells expressing either GFP or WT2 and the reporter plasmid (Fig. 2A). A significant (>10-fold) inhibitory effect was detectable as early as 3 h after the start of transfection with WT2. The inhibitory effect persisted throughout the experiment. In contrast, luciferase activity accumulated over time in cells cotransfected with the pEGFP-N1 vector control.
FIG. 2.
(A) Time course of luciferase expression from cells cotransfected with the indicated plasmid (pEGFP-N1 or WT2), or no plasmid (mock) and the luciferase reporter plasmid. Cells were harvested at the indicated time posttransfection and assayed for luciferase activity. The data in RLU were averaged from experiments performed in triplicate. (B) DNA dose-response for the inhibition of reporter gene expression by ExoU. CHO cells were cotransfected with the luciferase reporter plasmid and 2, 4, 8, 16, or 32 ng of effector plasmid (WT2) versus a control vector (pEGFP-N1). After 24 h, cells were harvested and assayed for luciferase activity. The data were averaged from experiments performed in triplicate.
To determine whether the inhibition of luciferase activity by ExoU was specific, dose-response experiments were performed by cotransfection of a constant amount of luciferase reporter plasmid and various amounts of the WT2 effector plasmid. Transfection of increasing amounts (2 to 32 ng) of WT2 correlated with increasing inhibition of luciferase activity, whereas no inhibitory effect on luciferase activity was observed for the pEGFP-N1 control plasmid (Fig. 2B). Overall, the dose-response and time course patterns of luciferase inhibition suggest that ExoU is a potent inhibitor of luciferase transcription-translation-activity or is generally toxic when expressed in the host cytosol. The apparent lack of GFP expression indicated that the amounts of fusion protein necessary to generate a biological effect were below the threshold of detection for GFP.
Intracellular expression of 1-687U-GFP results in cell permeability.
To determine whether ExoU was cytotoxic when expressed in CHO cells, we performed vital staining of transfected CHO cells with ethidium bromide (Fig. 3). Only cells that are permeable will allow ethidium bromide to bind nucleic acids and produce a signal detectable by fluorescence microscopy. Less than 5% of the mock-transfected cells take up the dye when stained 24 h after the start of transfection (Fig. 3B). In contrast, cells that have been transfected with WT2 are bright red after 24 h (Fig. 3C). The number of dead cells appears similar to the transfection efficiency, as determined by a control well of cells that express GFP only (Fig. 3A). Ethidium bromide uptake in a GFP control well was similar to that of the mock-transfected cells (data not shown).
FIG. 3.
Vital staining of transfected CHO cells. CHO cells from a 12-well plate were transfected either with the pEGFP-N1 vector (A), no plasmid (B), or the WT2 plasmid encoding full-length ExoU in-frame with the GFP protein (C). After 3 h the low-serum medium (OptiMem) was replaced with complete medium, and the cells were kept at 37°C for 24 h. Ethidium bromide (1 μM, final) was added to the medium in panels B and C. Cells were subjected to fluorescence microscopic analysis using a fluorescein filter (HGF712; Nikon) to detect GFP-transfected cells (A) and a tetramethyl rhodamine isocyanate filter (G-1A; Nikon) to detect ethidium bromide fluorescence (B and C).
A different experiment was performed to compare the number of dead cells induced by intracellular expression of the full-length toxin versus a nontoxic truncation such as N1 (1 to 633) and control plasmids (pEGFP-N1) after a short transfection time (8 h). As shown in Table 2, cells transfected with either the N1 effector plasmid (1 to 633) or the vector alone have less than 5% of cells taking up the dye when stained 8 h after the start of transfection. In contrast, cells that have been transfected with the WT2 (1 to 687) plasmid show 14.7 ± 1.8% dead cells after 8 h. The number of dead cells detected is in the same range as the transfection efficiency of that experiment, as determined from control wells expressing the GFP protein only or the nontoxic truncated form of ExoU (1 to 633). We concluded that the inhibition of luciferase activity was due not to a specific inhibition of luciferase transcription, translation, or enzymatic activity but to the cytotoxic effects of intracellular ExoU expression. Either cell death inhibits luciferase synthesis, or luciferase is spontaneously released when the plasma membrane is compromised.
TABLE 2.
Transfected ExoU is cytotoxic
| Transfected expression construct | % of cells expressing GFPa | % Dead cellsb |
|---|---|---|
| pEGFP-N1 | 20.4 ± 2.8 | 2.6 ± 1.0 |
| N1 (1-633GFP) | 12.9 ± 0.82 | 1.9 ± 1.7 |
| WT-2 (1-687GFP) | ND | 14.7 ± 1.8 |
Microscopic examination of cells was done 8 h posttransfection. The mean percentage of cells expressing a green fluorescent signal (± standard deviation) was determined by microscopy and counting three independent fields. ND, not detectable.
Mean percentage of cells permeable to ethidium bromide (± standard deviation).
Multiple domains are required for ExoU toxicity.
Transposon insertions in either the 5′ or 3′ end of exoU in the P. aeruginosa chromosome result in a nontoxic strain in both in vitro and in vivo assays (8, 19). The lack of cytotoxicity associated with an insertion in the 5′ region is explained by the absence of ExoU expression (8, 9). The insertion at the 3′ end of the gene, however, resulted in a truncated form of ExoU that was secretion competent, indicating that this molecule could potentially be delivered into the cellular cytoplasm by the type III secretory mechanism (19). These data suggest that sequences within the carboxy-terminal region are important for either translocation or cytotoxicity. Our results in transient-transfection experiments with the full-length forms of ExoU indicated that we could use this assay to map a putative minimal cytotoxic domain by measuring the effects on luciferase activity expressed from a cotransfected reporter plasmid. Low luciferase activity would correlate to toxicity, while the accumulation of luciferase activity would indicate that the ExoU derivative was defective for cytotoxicity. Plasmids were constructed to express amino- or carboxy-terminal regions of ExoU fused in-frame to GFP and cotransfected with the reporter plasmid. Luciferase activity was not significantly different from control values when either amino- or carboxy-terminal portions of ExoU were expressed (Fig. 4B, N1, N2, N3, C1, and C2). As a control in all experiments, transient transfection with full-length ExoU (WT2) significantly reduced luciferase activity. These data suggested that small deletions of ExoU resulted in a loss of biological activity and that the entire molecule may be required for a toxic response to occur. Alternatively, multiple domains within the molecule may encode ExoU-mediated toxicity.
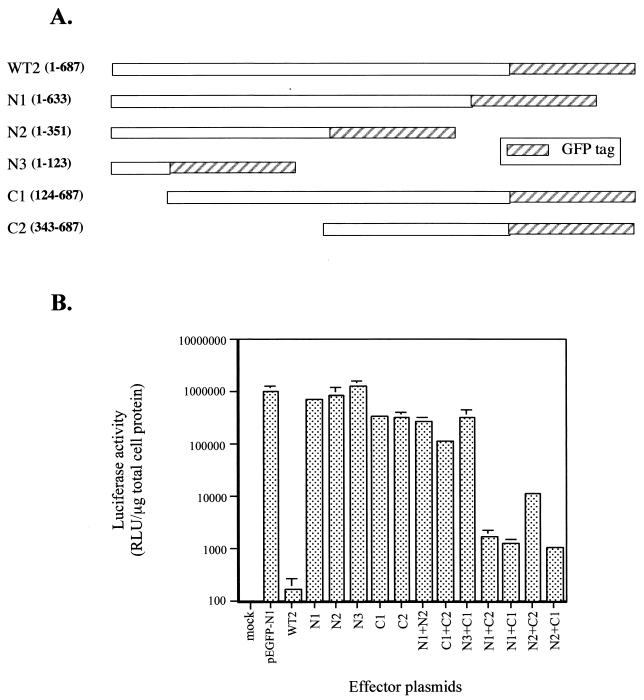
FIG. 4.
Amino- and carboxy-terminal sequences of ExoU complement in trans to affect luciferase activity. CHO cells were cotransfected with the luciferase reporter plasmid and single plasmids expressing ExoU fragments or triply transfected with reporter and two separate ExoU expression constructs as described in the text. After transfection (24 h) with 150 ng each of effector plasmid (or CT DNA) and 100 ng of reporter plasmid, cells were lysed and assayed for luciferase activity. (A) Map of each effector plasmid showing the amino acids of ExoU encoded within each clone. (B) Luciferase activity (in RLU) measured 24 h posttransfection. The data were averaged from experiments performed in triplicate.
To test the hypothesis that ExoU may require multiple domains to exert a toxic response, we performed triple transfections to introduce purified plasmids encoding separate regions of ExoU and the reporter plasmid. Effector plasmids expressing amino-terminal or carboxy-terminal fragments of ExoU (Fig. 4A) were transfected at the same molar ratio simultaneously with a luciferase reporter plasmid. Analysis of the transfected cells revealed that coexpression of N1 and either the C1 or C2 C-terminal truncation caused a significant reduction in luciferase activity (Fig. 4B). A less pronounced but significant decrease in luciferase activity was observed when N2 was coexpressed with C2 (compared to the cotransfection of N2 and C1). These results suggested that separate regions of ExoU functionally complement one another in trans when expressed intracellularly. One functional region appears to be located between amino acids 1 and 351. The second region maps to a region encompassed by amino acids 343 to 687. Both regions were required for ExoU-mediated reduction of reporter activity. Coexpression of either two N-terminal domains (N1 and N2) or two carboxy-terminal regions of ExoU (C1 and C2) resulted in a nontoxic phenotype, consistent with the hypothesis that expression of the complementary domain is needed to induce this response (Fig. 4B). The inhibition observed when the ExoU domains were coexpressed was not as potent as the one induced by the entire molecule (WT2). A transfection beyond 24 h or higher doses of effector DNA may be required to fully restore the biological effect of the full-length molecule.
Establishing the domain boundaries of N- and C-terminal regions of ExoU.
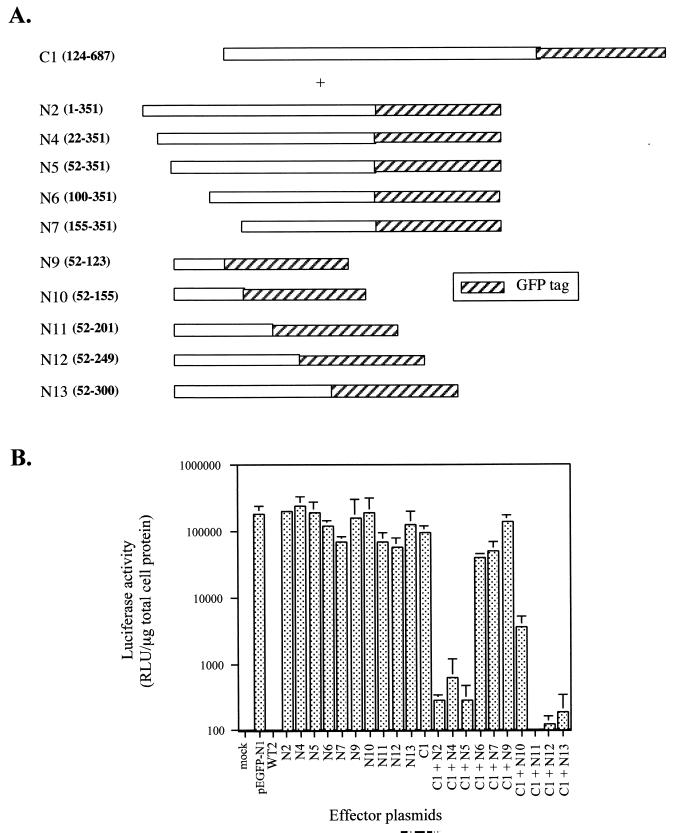
Our initial transfection results suggested that an amino- and a carboxy-terminal region are both required for the intracellular activity of ExoU, but the reduced potency of the N2-C2 combination indicated that a central domain may contribute to the effects on reporter activity (Fig. 4A and B). To exclusively map the N- and C-terminal domains of ExoU, large complementation clones consisting of intact N or C termini linked to the middle region of the molecule were used in triple transfection analyses. The boundaries of the N terminus were defined using the C1 construct encoding amino acids 124 to 687. Cotransfection of C1 with successive deletions from the first amino acid of ExoU to position 155 (Fig. 5A) indicated that the first 52 amino acids are not required for toxicity (Fig. 5B). These results are consistent with the hypothesis that the secretion and/or chaperone binding domain of ExoU (previously mapped as residing between amino acids 1 and 124 [9]) is a separate functional region and likely not required for intracellular activity (7, 33, 39). To test whether the entire amino-terminal domain from amino acids 52 to 351 is required for toxicity, plasmids encoding successive deletions from amino acid 351 to 123 (Fig. 5A) were cotransfected with C1 (124 to 687), and the luciferase activity of the transfected cell lysates was measured. Based on the reduction in reporter activity, the N-terminal boundary of the amino-terminal domain resides between amino acid 52 and 100. The C-terminal boundary of the amino-terminal domain is between positions 155 and 201 of ExoU (Fig. 5B).
FIG. 5.
Mapping the minimal functional amino-terminal domain of ExoU. (A) Map of various deletions of ExoU used in cotransfection experiments with the C1 (124 to 687) plasmid in the presence of the luciferase reporter gene. (B) After transfection (24 h), cells were lysed and assayed for luciferase activity. The data (in RLU) were averaged from experiments performed in triplicate.
Cotransfections of the N1 (1 to 633) plasmid with plasmids encoding successive truncations from the amino terminus of ExoU to amino acid 580 were performed to map the minimal carboxy-terminal domain (Fig. 6A). The luciferase activity of all the mutants tested in the presence of N1 was significantly reduced (Fig. 6B). In contrast, transfection of the mutants in the absence of the N1 plasmid in trans induced a luciferase activity similar to that of the control. We conclude that amino acid residues 580 to 687 encode a functional C-terminal domain.
FIG. 6.
Mapping the minimal functional carboxyl-terminal domain of ExoU. (A) Map of the various deletions of ExoU used in cotransfection experiments with the N1 (1 to 633) plasmid construct in the presence of the luciferase reporter gene. (B) After transfection (24 h), cells were lysed and assayed for luciferase activity. The data (in RLU) were averaged from experiments performed in triplicate.
Our strategy for mapping N- and C-terminal functional domains always included a complementary clone expressing a large and overlapping proportion of ExoU. While this strategy made the contribution of a potential middle domain constant in each experiment, it did not eliminate the possibility that the underlying mechanism of toxicity involved DNA rearrangements between transfection clones, resulting in a complete coding region for toxic activity. Although this would be considered an exceedingly rare event considering the supercoiled nature of the template DNA and the number of independent recombination sites involved, nonoverlapping N- and C-terminal domains (C8 [203 to 687] and N14 [52 to 578]) were constructed to test this possibility. As shown in Fig. 7A and B, transfection with individual clones did not affect luciferase expression. Cotransfection of a combination of expression clones encoding the mapped N- and C-terminal regions (N11 + C8 or N14 + C6) inhibited the accumulation of luciferase activity. Expression combinations representing only the mapped N- and C-terminal regions without a middle domain (e.g., N11 + C6) were, however, found to be nontoxic in terms of reporter activity (data not shown).
FIG. 7.
Intracellular expression of ExoU as N- and C-terminal nonoverlapping fragments and middle-domain deletions or disruptions. (A) Map of the deletions of ExoU cloned in the pEGFP-N1 vector as described in the text and used in cotransfection experiments in the presence of the luciferase reporter gene. (B) After transfection (24 h), cells were lysed and assayed for luciferase activity. The data (in RLU) were averaged from experiments performed in triplicate.
To confirm that ExoU required a functional middle domain, transfection experiments were repeated with additional expression clone combinations (N13 + C2 and N5 + C7) that possessed intact N- and C-terminal regions but disrupted the middle part of the molecule. Luciferase reporter activity is not significantly different from vector controls when the boundary of expression clones interrupts a region between amino acids 300 and 352. The results shown in Fig. 7 confirmed the localization of N- and C-terminal domains and the fact that these domains can function in trans. Our analysis also indicates that disruption of a region between the mapped N- and C-terminal domains eliminates the toxicity of ExoU, suggesting that a third functional domain is located in the middle of the protein.
Oligomeric state of ExoU.
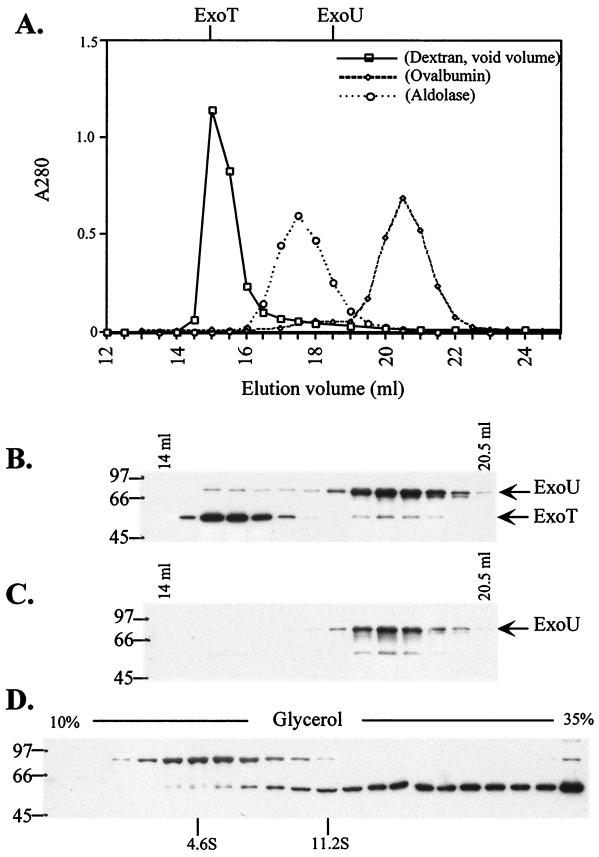
Mapping studies and transfection analysis indicate that three regions of ExoU are required for intracellular toxicity. Our data further indicate that N- and C-terminal domains can function in trans as long as a central region is included in one or both expression clones. The pattern of intramolecular complementation suggests that one or more of the ExoU domains may mediate the oligomerization of the molecule into a functional complex. To determine whether ExoU exists as a dimer or higher oligomer, recombinant histidine-tagged ExoU and native ExoU secreted by P. aeruginosa were subjected to sizing chromatography. Native ExoU (73.9 kDa) from a culture supernatant of P. aeruginosa PA103 eluted in peak fractions that corresponded to an 80- to 90-kDa molecule (Fig. 8A to C). ExoT served as an internal control for S-200 column chromatography. ExoT and ExoU are both present in the culture supernatants of P. aeruginosa strain PA103 when the bacteria are induced for type III protein expression. In contrast to ExoU, ExoT eluted as a soluble high-molecular-mass aggregate in the void volume of the S-200 gel filtration column (fractionation range of the resin, 5 to 250 kDa for globular proteins) (Fig. 8B). To confirm the sizing chromatography results, we analyzed the sedimentation of ExoT and ExoU in a glycerol gradient (5). Native ExoU sedimented as a monomeric protein predominantly in fractions corresponding to ∼4.6S, while ExoT appeared to sediment beyond catalase (11.2S) and across the gradient (Fig. 8D) as a soluble aggregate. High-molecular-mass aggregates of ExoT were detected in the bottom pellet fraction. ExoU also behaved as a monomeric protein when purified protein fractions or bacterial lysates containing histidine-tagged ExoU were subjected to S-200 sizing chromatography (data not shown). We conclude that native ExoU secreted by the P. aeruginosa type III system, like the rExoU expressed as a cytoplasmic protein by E. coli, is monomeric. ExoT secreted from P. aeruginosa forms soluble aggregates similar to the highly homologous type III-secreted toxin ExoS (21, 23, 24). Our analysis indicates that ExoT and ExoU do not form complexes (Fig. 8B and C). By these criteria, the functional domains of ExoU that have been mapped most likely do not mediate an oligomerization of the molecule.
FIG. 8.
Velocity sedimentation and gel filtration data suggest that ExoU is a monomer. (A) An ammonium sulfate concentrate of extracellular proteins from a culture of P. aeruginosa PA103 grown in the presence of 10 mM nitrilotriacetic acid to induce the type III secretion-translocation system was subjected to gel filtration chromatography. (A) Elution profiles on Sephacryl S-200-HR of dextran blue (void), aldolase (158 kDa), and ovalbumin (43 kDa) as determined by their A280. The position of the peak of immunoreactivity of ExoT and ExoU is represented above the graph. (B) Type III-secreted proteins from P. aeruginosa PA103 were fractionated on a calibrated gel filtration column (Sephacryl S200-HR, 38 ml). Column fractions (0.5 ml) were analyzed by Western blot using a combination of anti-ExoU and -ExoT rabbit IgG. Elution peaks indicate that ExoT (15 ml) is present in the void volume, while ExoU elutes as a monomer (18.5 ml). (C) Western blot of a duplicate gel using only anti-ExoU IgG indicates that ExoT and ExoU do not associate after secretion to the extracellular medium. (D) Type III-secreted proteins from PA103 were centrifuged through 10 to 35% glycerol gradients as described in the text. Gradients (5 ml) were fractionated from the top, and aliquots were resolved on SDS–10% polyacrylamide gels. ExoU and ExoT were detected by immunoblotting with anti-ExoU and -ExoS/T IgG and ECL reagents. The positions of standards sedimented in parallel or in conjunction and analyzed by densitometric analysis of Coomassie blue-stained gels are shown (albumin [4.6S] and catalase [11.2S]). The peak of ExoU immunoreactivity (upper band) corresponds to ∼4.6S. The peak of ExoT immunoreactivity (lower band) was detected in fractions beyond catalase, and high-molecular-mass aggregates of ExoT were detected in the bottom pellet fraction. Sizes are shown at the left (in kilodaltons).
Representative patterns of ExoU-GFP fusion protein expression in CHO-K1 cells.
As a control for translational efficiency of each clone, the mean fluorescence intensity was determined for the ExoU-GFP expression constructs (Fig. 9A) by FACS analysis. A fluorescent signal was not detected in mock-transfected cells or cells transfected with a full-length ExoU expression construct. Expression of the GFP signal was relatively constant when truncated versions of ExoU were fused to GFP, indicating that translation was proceeding through the C-terminal GFP tag. These results are consistent with microscopic determinations of GFP expression in our transfection experiments.
To determine the molecular weight of the expressed products, Western blot analysis of cells transfected with each ExoU construct was performed (Fig. 9B and C). Antisera to ExoU (Fig. 9B and C) and GFP (Fig. 10) were used to probe for the expression of recombinant products. When probed with antisera specific for ExoU, a background band appears (above the 66-kDa molecular size marker) in all lanes, including the mock and vector control lanes (Fig. 9B). When full-length ExoU-GFP is transfected an immunoreactive band is not seen with either the anti-ExoU (Fig. 9B, WT-2) or anti-GFP antiserum (data not shown). The products of ExoU truncations fused to GFP, however, are detectable at various intensities. Extracts from some transfections also show cross-reactive breakdown products or alternative sites of translation initiation. The immunoreactivity of the products to anti-ExoU and the predicted molecular weights are consistent with intracellular ExoU expression. Combined, our microscopic, FACS, and Western blot analyses confirm that each clone is expressing a product corresponding to the cloned DNA.
FIG. 10.
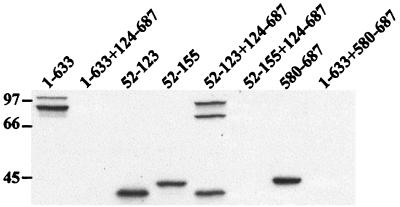
Expression of translational protein fusions in transfected cells. CHO cells (5 × 106 per 100 mm dish) were cotransfected with pairs of effector plasmids harboring various domains of ExoU (4 μg of each) or with single effector plasmids (4 μg) and CT DNA (4 μg). Twenty-four hours after transfection, cells were washed, harvested, and lysed in HES buffer in the presence of protease inhibitors as described in the text. Equivalent amounts (25 μg) of lysate were analyzed by Western blot using a monoclonal anti-rGFP and ECL reagents. The lanes contained (from left to right) N1 (1 to 633), N1 (1 to 633) and C1 (124 to 687), N9 (52 to 123), N10 (52 to 155), N9 (52 to 123) and C1 (124 to 687), N10 (52 to 155) and C1 (124 to 687), C6 (580 to 687), or N1 (1 to 633) and C6 (580 to 687) plasmids.
To detect intracellular expression of two cotransfected ExoU derivatives, postnuclear supernatants were prepared from transfected CHO cells. Cell lysates (25 μg) were subjected to Western blot analyses using a monoclonal antibody recognizing the GFP (Clontech) portion of the hybrid protein (Fig. 10). Immunoreactive products were identified when extracts from cells transfected with a single expression construct encoding a truncated form of ExoU were tested (Fig. 10, lanes 1–633, 52–123, 52–155, and 580–687). Protein expression was detectable from a combination of constructs that did not affect luciferase activity (Fig. 10, lane 52–123 + 124–687). In contrast, immunoreactive products were undetectable when CHO cells were transfected with plasmid combinations that caused a decrease in luciferase activity (Fig. 10, lanes 1–633 + 124–687, 52–155 + 124–687, and 1–633 + 580–687). In summary, when combinations of constructs that resulted in the diminution of luciferase activity were cotransfected, immunoreactive proteins were not present in cellular extracts and GFP fluorescence was not visible by microscopy.
DISCUSSION
A transient-transfection assay was developed to begin structure-function studies of ExoU, a type III-secreted toxin of P. aeruginosa. This strategy circumvents the use of bacterial infection assays, which restrict the types of truncations that can be constructed and can potentially introduce other variables that affect cell viability or protein expression. Transfection assays have been used to examine expression of specific domains of ExoS (28, 29), to demonstrate that YopJ production is sufficient for the downregulation of mitogen-activated protein kinases in COS-1 cells (27), and to identify the minimal functional domain of VacA, a toxin secreted from Helicobacter pylori (42). To map the minimal cytotoxic domain of ExoU, we constructed three full-length derivatives (no tag, an amino-terminal histidine tag, and a carboxy-terminal GFP tag) in eukaryotic expression vectors and transiently transfected CHO-K1 cells. The biological effect of intracellular ExoU expression was quantitated by measuring luciferase activity from a cotransfected reporter plasmid. All full-length forms of ExoU expression constructs inhibited reporter gene expression compared to a control effector plasmid, regardless of the presence or absence or localization of the encoded tags. Cells transfected with full-length forms of ExoU demonstrate the same permeable phenotype as cells that are intoxicated with ExoU by infection with P. aeruginosa. Reporter expression was inhibited in a DNA dose-dependent fashion when different concentrations of the effector plasmid encoding ExoU were subjected to transfection. From these data we conclude that ExoU expression from a transfected plasmid is sufficient to mediate a cytotoxic response in eukaryotic cells and that this cytotoxicity severely reduces reporter gene expression from cotransfected constructs.
Based on the relatively low doses of effector DNA and the early time points at which reporter gene expression was diminished, ExoU appears to be a potent cytotoxin. This conclusion is supported by the use of GFP-tagged molecules as secondary reporters to ensure translation of each ExoU fusion construct. Using fluorescence microscopy, the detection threshold of GFP is approximately 10,000 molecules. While transfection of ExoU truncations (C-terminal GFP fusions) and nontoxic plasmid combinations resulted in detection of GFP, expression of full-length forms of the molecule or toxic plasmid combinations appeared to prevent the accumulation of sufficient amounts of GFP for a detectable fluorescent signal microscopically and by FACS analysis. In a time course analysis, toxic effects were detectable 3 h after transfection. The early biological effects and low dose of ExoU argue against general effects on cellular protein, RNA, or DNA synthesis but may be explained by a low abundance of a critical intracellular target. Although our structural studies suggest a complex mode of action, the mechanism of ExoU-mediated toxicity remains to be defined.
Using transfection analysis, expression of multiple regions of ExoU is required to elicit a toxic response. An N-terminal domain is encoded by amino acids 52 to 100 to 155 to 202. The boundaries of the carboxy-terminal domain of ExoU appear to encompass a region located between residues 580 and 687. The absence of interference of the GFP tags with the functional complementation of the truncations suggests that the two domains of ExoU essential for biological effect are not contiguous. We noted that reporter gene activity was enhanced when the boundaries of cotransfection constructs impinged in an area that roughly mapped from amino acids 300 to 400. Moreover, transfection experiments using clones encoding only the N- and C-terminal domains resulted in high reporter gene activities, suggesting a nontoxic phenotype. To map the boundaries of the N- and C-terminal domains, overlapping constructs were used to normalize the potential contribution of this middle region. To ensure that the mapped boundaries were correct and that the toxic phenotype was not due to recombination between expression plasmids at the DNA level, we constructed nonoverlapping clones. Transfection of nonoverlapping N- or C-terminal clones with the appropriate complementation construct recapitulated the toxic phenotype, confirming the N- and C-terminal domain mapping results obtained with overlapping constructs. To demonstrate that a middle region of the molecule was required for the toxic response, we generated additional clones with deletions or boundaries located between amino acids 300 and 400. Reporter gene expression was uninhibited when the middle region of the molecule was disrupted. From these data we conclude that ExoU consists of at least three functional regions.
The requirement for multiple domains may imply that a possible role for one or more regions of the molecule is to maintain a functionally active conformation. This conformation could include oligomerization of ExoU into a protein complex. To test this hypothesis we subjected ExoU to sizing chromatography and glycerol gradient centrifugation. These experiments were performed with purified protein, crude bacterial cytoplasmic extracts, and culture supernatants from P. aeruginosa that contained ExoT as well as ExoU. Results from all experiments indicate that ExoU is expressed as a monomer irrespective of being histidine tagged at the amino terminus and expressed in the cytoplasm of E. coli or secreted to the extracellular medium from its native host, P. aeruginosa. In similar experiments, ExoT formed higher-molecular-mass aggregates, which is consistent with the predicted properties of the protein based on its similarity to ExoS. The ExoT aggregates did not contain ExoU, indicating that these proteins do not associate after secretion from P. aeruginosa. Thus, even in the absence of its cognate chaperone, SpcU, ExoU was monomeric. If ExoU-mediated cytotoxicity requires that the molecule form higher-order oligomers, a eukaryotic cofactor may be necessary to catalyze this reaction.
Several type III secreted proteins have multiple domains with different functions. YopH, a protein tyrosine phosphatase from Yersinia species (3), has an amino-terminal substrate-binding domain as well as a carboxy-terminal catalytic domain. Each domain contributes to substrate recognition. ExoS, a type III effector protein from P. aeruginosa, has been shown to be a bifunctional cytotoxin. The ADP-ribosylating domain is located within the C-terminal part of ExoS (15, 21, 26, 28, 36), and the N terminus is a GTPase activating protein for rho GTPases (17, 29). StpP, a tyrosine phosphatase of Salmonella enterica serovar Typhimurium, possesses two independent effector domains (14) that appear to affect similar cell functions. VacA, an exotoxin from Helicobacter pylori, is cleaved into two moieties (P33 and P70) that are proposed to function as an A-B-type toxin (30, 42). Functional data, however, indicate that both moieties expressed intracellularly from separate plasmids resulted in vacuolation (42). One can speculate that each domain of ExoU may have a distinct function whose additive effects are required for toxicity. Alternatively, ExoU may target separate intracellular molecules, and the sequential modification of these targets is required for cytotoxicity. Our current structure-function analysis indicates that the middle domain has to be linked to either the N- or C-terminal domain. However, we have not eliminated the possible independent functions of all three domains in transfection analysis. The boundaries of the middle domain will have to be more clearly established before this experiment can be designed. Intermolecular interactions of ExoU do not seem to occur when the protein is isolated from a prokaryotic environment; however, this property may be different after eukaryotic cell contact and toxin translocation. The possible requirement for a eukaryotic factor would be consistent with the observation that three other type III effectors (ExoS, -T, and -Y) of P. aeruginosa require a cellular cofactor for enzymatic activity (13, 41).
The toxicity of ExoU and the complementation pattern of N- and C-terminal regions allowed the use of the transfection assay to identify the amino acid boundaries of each domain. In all cases of a toxic combination, luciferase activities were diminished by 10- to 1,000-fold. Controls for translational efficiency and protein expression included the assessment of GFP by FACS analysis and microscopy and the expression of proteins in Western blot analysis with antibodies to either the GFP tag or ExoU itself. While all reported constructs expressed immunoreactive proteins, two limitations to the mapping analysis include the intracellular stability of the expressed protein and the ability of eukaryotes to initiate translation at several sites within coding sequences. Thus, in the transfection-mapping system, there is some expression variability that could impact the interpretation of domain boundaries. The toxicity of even low DNA doses encoding ExoU, however, appeared to enhance the sensitivity of the assay, since a biological effect on luciferase activity was clearly detectable even when the expressed full-length ExoU or toxic domain combinations could not be detected. Moreover, expression constructs that gave a relatively low Western blot signal were clearly toxic when paired with the appropriate complementation clone. Overall, these data indicate that ExoU has a potent effect on eukaryotic cells.
Although we have used the inhibition of reporter gene expression in transient-transfection assays as a sensitive readout for cytotoxicity, we have yet to resolve whether increased cellular permeability is an early or late event in ExoU-mediated intoxication of cells. Mapping the functional domains of ExoU is the first step in the investigation of the mechanism of ExoU cytotoxicity and promises to contribute to the identification of intracellular targets and/or potential eukaryotic cofactors.
ACKNOWLEDGMENTS
We gratefully acknowledge the technical assistance of Amy J. Vallis and Monika S. Casey for tissue culture expertise. We thank the Medical College of Wisconsin Protein and Nucleic Acid Core Facility for performing nucleotide sequence analysis on our fusion derivatives. We are grateful to R. L. Truitt and the Flow Cytometry Core Facility at the Medical College of Wisconsin for performing the FACS analyses of our fusion derivatives. Anne Delcour of the University of Houston Department of Biochemistry and Jim Feix, Medical College of Wisconsin Department of Biophysics, contributed preliminary information on the interaction of rExoU with liposomes.
This work was supported by grants HL59239 (D.W.F.) and RG009L (American Lung Association of Wisconsin) (V.F-B.).
REFERENCES
- 1.Allewelt M, Coleman F T, Grout M, Priebe G P, Pier G B. Acquisition of expression of the Pseudomonas aeruginosa ExoU cytotoxin leads to increased bacterial virulence in a murine model of acute pneumonia and systemic spread. Infect Immun. 2000;68:3998–4004. doi: 10.1128/iai.68.7.3998-4004.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Apodaca G, Bomsel M, Lindstedt R, Engel J, Frank D W, Mostov K E, Wiener-Kronish J P. Characterization of Pseudomonas aeruginosa-induced MDCK cell injury: glycosylation defective host cells are resistant to bacterial killing. Infect Immun. 1995;63:1541–1551. doi: 10.1128/iai.63.4.1541-1551.1995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Black D S, Montagna L G, Zitsman S, Bliska J B. Identification of an amino terminal substrate-binding domain in the Yersinia tyrosine phosphatase that is required for efficient recognition of focal adhesion targets. Mol Microbiol. 1998;29:1263–1274. doi: 10.1046/j.1365-2958.1998.01014.x. [DOI] [PubMed] [Google Scholar]
- 4.Coburn J, Frank D W. Macrophages and epithelial cells respond differently to the Pseudomonas aeruginosa type III-secretion system. Infect Immun. 1999;67:3151–3154. doi: 10.1128/iai.67.6.3151-3154.1999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Cover T L, Hanson P I, Heuser J E. Acid-induced dissociation of VacA, the Helicobacter pylori vacuolating cytotoxin, reveals its pattern of assembly. J Cell Biol. 1997;138:759–769. doi: 10.1083/jcb.138.4.759. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Dacheux D, Toussaint B, Richard M, Brochier G, Croize J, Attree I. Pseudomonas aeruginosa cystic fibrosis isolates induce rapid, type III secretion-dependent, but ExoU-independent, oncosis of macrophages and polymorphonuclear neutrophils. Infect Immun. 2000;68:2916–2924. doi: 10.1128/iai.68.5.2916-2924.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Day J B, Plano G V. A complex composed of SycN and YscB functions as a specific chaperone for YopN in Yersinia pestis. Mol Microbiol. 1998;30:777–788. doi: 10.1046/j.1365-2958.1998.01110.x. [DOI] [PubMed] [Google Scholar]
- 8.Finck-Barbançon V, Goranson J, Zhu L, Sawa T, Wiener-Kronish J P, Fleiszig S M J, Wu C, Mende-Mueller L, Frank D W. ExoU expression by Pseudomonas aeruginosa correlates with acute cytotoxicity and epithelial injury. Mol Microbiol. 1997;25:547–557. doi: 10.1046/j.1365-2958.1997.4891851.x. [DOI] [PubMed] [Google Scholar]
- 9.Finck-Barbançon V, Yahr T L, Frank D W. Identification and characterization of SpcU, a chaperone required for efficient secretion of the ExoU cytotoxin. J Bacteriol. 1998;180:6224–6231. doi: 10.1128/jb.180.23.6224-6231.1998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Fleiszig S M J, Wiener-Kronish J P, Miyazaki H, Vallas V, Mostov K E, Kanada D, Sawa T, Yen T S B, Frank D W. Pseudomonas aeruginosa-mediated cytotoxicity and invasion correlate with distinct genotypes at the loci encoding exoenzyme S. Infect Immun. 1997;65:579–586. doi: 10.1128/iai.65.2.579-586.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Frank D W. The exoenzyme S regulon of Pseudomonas aeruginosa. Mol Microbiol. 1997;26:621–629. doi: 10.1046/j.1365-2958.1997.6251991.x. [DOI] [PubMed] [Google Scholar]
- 12.Frank D W, Nair G, Schweizer H P. Construction and characterization of chromosomal insertions of the Pseudomonas aeruginosa trans-regulatory locus. Infect Immun. 1994;62:554–563. doi: 10.1128/iai.62.2.554-563.1994. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Fu H, Coburn J, Collier R J. The eukaryotic host factor that activates exoenzyme S of Pseudomonas aeruginosa is a member of the 14-3-3 protein family. Proc Natl Acad Sci USA. 1993;90:2320–2324. doi: 10.1073/pnas.90.6.2320. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Fu Y, Galan J E. The Salmonella typhimurium tyrosine phosphatase SptP is translocated into host cells and disrupts the actin cytoskeleton. Mol Microbiol. 1998;27:359–368. doi: 10.1046/j.1365-2958.1998.00684.x. [DOI] [PubMed] [Google Scholar]
- 15.Ganesan A K, Frank D W, Misra R P, Schmidt G, Barbieri J T. Pseudomonas aeruginosa exoenzyme S ADP-ribosylates Ras at multiple sites. J Biol Chem. 1998;273:7332–7337. doi: 10.1074/jbc.273.13.7332. [DOI] [PubMed] [Google Scholar]
- 16.Ganesan A K, Mende-Mueller L, Selzer J, Barbieri J T. Pseudomonas aeruginosa exoenzyme S, a double ADP-ribosyl transferase, resembles vertebrate mono-ADP-ribosyl transferases. J Biol Chem. 1999;274:9503–9508. doi: 10.1074/jbc.274.14.9503. [DOI] [PubMed] [Google Scholar]
- 17.Goehring U M, Schmidt G, Pederson K, Aktories K, Barbieri J T. The N-terminal domain of Pseudomonas aeruginosa is a GTPase-activating protein for Rho GTPases. J Biol Chem. 1999;274:36369–36372. doi: 10.1074/jbc.274.51.36369. [DOI] [PubMed] [Google Scholar]
- 18.Hauser A R, Engel J N. Pseudomonas aeruginosa induces type III-secretion-mediated apoptosis of macrophages and epithelial cells. Infect Immun. 1999;67:5530–5537. doi: 10.1128/iai.67.10.5530-5537.1999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Hauser A R, Kang P J, Engel J N. PepA, a secreted protein of Pseudomonas aeruginosa, is necessary for cytotoxicity and virulence. Mol Microbiol. 1998;27:807–818. doi: 10.1046/j.1365-2958.1998.00727.x. [DOI] [PubMed] [Google Scholar]
- 20.Kang P J, Hauser A R, Apodaca G, Fleiszig S M J, Wiener-Kronish J P, Mostov K, Engel J N. Identification of Pseudomonas aeruginosa genes required for epithelial cell injury. Mol Microbiol. 1997;24:1249–1262. doi: 10.1046/j.1365-2958.1997.4311793.x. [DOI] [PubMed] [Google Scholar]
- 21.Knight D A, Finck-Barbançon V, Kulich S M, Barbieri J T. Functional domains of Pseudomonas aeruginosa exoenzyme S. Infect Immun. 1995;63:3182–3186. doi: 10.1128/iai.63.8.3182-3186.1995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Kudoh I, Wiener-Kronish J P, Hashimoto S, Pittet J F, Frank D W. Exoproduct secretions of Pseudomonas aeruginosa strains influence severity of alveolar epithelial injury. Am J Physiol. 1994;267:L551–L556. doi: 10.1152/ajplung.1994.267.5.L551. [DOI] [PubMed] [Google Scholar]
- 23.Kulich S M, Frank D W, Barbieri J T. Purification and characterization of exoenzyme S from Pseudomonas aeruginosa 388. Infect Immun. 1993;61:307–313. doi: 10.1128/iai.61.1.307-313.1993. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Kulich S M, Frank D W, Barbieri J T. Expression of recombinant exoenzyme S of Pseudomonas aeruginosa. Infect Immun. 1995;63:1–8. doi: 10.1128/iai.63.1.1-8.1995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Kurahashi K, Kajikawa O, Sawa T, Ohara M, Gropper M A, Frank D W, Martin T R, Wiener-Kronish J P. Pathogenesis of septic shock in Pseudomonas aeruginosa pneumonia. J Clin Investig. 1999;104:743–750. doi: 10.1172/JCI7124. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.McGuffie E M, Frank D W, Vincent T S, Olson J C. Modification of Ras in eukaryotic cells by Pseudomonas aeruginosa exoenzyme S. Infect Immun. 1998;66:2607–2613. doi: 10.1128/iai.66.6.2607-2613.1998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Palmer L E, Pancetti A R, Greenberg S, Bliska J B. YopJ of Yersinia spp. is sufficient to cause downregulation of multiple mitogen-activated protein kinases in eukaryotic cells. Infect Immun. 1999;67:708–716. doi: 10.1128/iai.67.2.708-716.1999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Pederson K J, Barbieri J T. Intracellular expression of the ADP-ribosyltransferase domain of Pseudomonas aeruginosa exoenzyme S is cytotoxic to eukaryotic cells. Mol Microbiol. 1998;30:751–759. doi: 10.1046/j.1365-2958.1998.01106.x. [DOI] [PubMed] [Google Scholar]
- 29.Pederson K J, Vallis A J, Aktories K, Frank D W, Barbieri J T. The amino-terminal domain of Pseudomonas aeruginosa exoenzyme S disrupts actin filaments via small-molecular-weight GTP-binding proteins. Mol Microbiol. 1999;32:393–401. doi: 10.1046/j.1365-2958.1999.01359.x. [DOI] [PubMed] [Google Scholar]
- 30.Reyrat J-M, Pelicic V, Papini E, Montecucco C, Rappuoli R, Telford J. Towards deciphering the Helicobacter pylori cytotoxin. Mol Microbiol. 1999;34:197–204. doi: 10.1046/j.1365-2958.1999.01592.x. [DOI] [PubMed] [Google Scholar]
- 31.Sawa T, Ohara M, Kurahashi K, Twining S S, Frank D W, Doroques D B, Long T, Gropper M A, Wiener-Kronish J P. In vitro cellular toxicity predicts Pseudomonas aeruginosa virulence in lung infections. Infect Immun. 1998;66:3242–3249. doi: 10.1128/iai.66.7.3242-3249.1998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Sawa T, Yahr T L, Ohara M, Kurahashi K, Gropper M A, Wiener-Kronish J P, Frank D W. Active and passive immunization with the Pseudomonas V antigen protects against type III intoxication and lung injury. Nat Med. 1999;5:392–398. doi: 10.1038/7391. [DOI] [PubMed] [Google Scholar]
- 33.Sory M P, Boland A, Lambermont I, Cornelis G R. Identification of the YopE and YopH domains required for secretion and internalization into the cytosol of macrophages, using the cyaA gene fusion approach. Proc Natl Acad Sci USA. 1995;92:11998–1002. doi: 10.1073/pnas.92.26.11998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Vallis A J, Finck-Barbançon V, Yahr T L, Frank D W. Biological effects of Pseudomonas aeruginosa type III-secreted proteins on CHO cells. Infect Immun. 1999;67:2040–2044. doi: 10.1128/iai.67.4.2040-2044.1999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Vallis A J, Yahr T L, Barbieri J T, Frank D W. Regulation of ExoS production and secretion by Pseudomonas aeruginosa in response to tissue culture conditions. Infect Immun. 1999;67:914–920. doi: 10.1128/iai.67.2.914-920.1999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Vincent T S, Fraylick J E, McGuffie E M, Olson J C. ADP-ribosylation of oncogenic Ras proteins by Pseudomonas aeruginosa exoenzyme S in vivo. Mol Microbiol. 1999;32:1054–1064. doi: 10.1046/j.1365-2958.1999.01420.x. [DOI] [PubMed] [Google Scholar]
- 37.Xiao W, Pizer L I, Wilcox K W. Identification of a promoter-specific transactivation domain in the herpes simplex virus regulatory protein ICP4. J Virol. 1997;71:1757–1765. doi: 10.1128/jvi.71.3.1757-1765.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Xu Y, Barbieri J T. Pertussis toxin-mediated ADP-ribosylation of target proteins in Chinese hamster ovary cells involves a vesicle trafficking mechanism. Infect Immun. 1995;63:825–832. doi: 10.1128/iai.63.3.825-832.1995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Yahr T L, Goranson J, Frank D W. Exoenzyme S of Pseudomonas aeruginosa is secreted by a type III pathway. Mol Microbiol. 1996;22:991–1003. doi: 10.1046/j.1365-2958.1996.01554.x. [DOI] [PubMed] [Google Scholar]
- 40.Yahr T L, Mende-Mueller L, Friese M B, Frank D W. Identification of type III secreted products of the Pseudomonas aeruginosa exoenzyme S regulon. J Bacteriol. 1997;179:7165–7168. doi: 10.1128/jb.179.22.7165-7168.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Yahr T L, Vallis A J, Hancock M K, Barbieri J T, Frank D W. ExoY, an adenylate cyclase secreted by the Pseudomonas aeruginosa type III system. Proc Natl Acad Sci USA. 1998;95:13899–13904. doi: 10.1073/pnas.95.23.13899. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Ye D, Willhite D C, Blanke S R. Identification of the minimal intracellular vacuolating domain of the Helicobacter pylori vacuolating toxin. J Biol Chem. 1999;274:9277–9282. doi: 10.1074/jbc.274.14.9277. [DOI] [PubMed] [Google Scholar]