Abstract
Bacterial antitumor therapy has great application potential given its unique characteristics, including genetic manipulation, tumor targeting specificity and immune system modulation. However, the nonnegligible side effects and limited efficacy of clinical treatment limit their biomedical applications. Engineered bacteria for therapeutic applications ideally need to avoid their accumulation in normal organs and possess potent antitumor activity. Here, we show that macrophage-mediated tumor-targeted delivery of Salmonella typhimurium VNP20009 can effectively reduce the toxicity caused by administrating VNP20009 alone in a melanoma mouse model. This benefits from tumor-induced chemotaxis for macrophages combined with their slow release of loaded strains. Inspired by changes in the tumor microenvironment, including a decrease in intratumoral dysfunctional CD8+ T cells and an increase in PDL1 on the tumor cell surface, macrophages were loaded with the engineered strain VNP-PD1nb, which can express and secrete anti-PD1 nanoantibodies after they are released from macrophages. This novel triple-combined immunotherapy significantly inhibited melanoma tumors by reactivating the tumor microenvironment by increasing immune cell infiltration, inhibiting tumor cell proliferation, remodeling TAMs to an M1-like phenotype and prominently activating CD8+ T cells. These data suggest that novel combination immunotherapy is expected to be a breakthrough relative to single immunotherapy.
Key words: Macrophage, Salmonella typhimurium VNP20009, Anti-PD1 nanobody, Tumor-targeted delivery, Immune activation
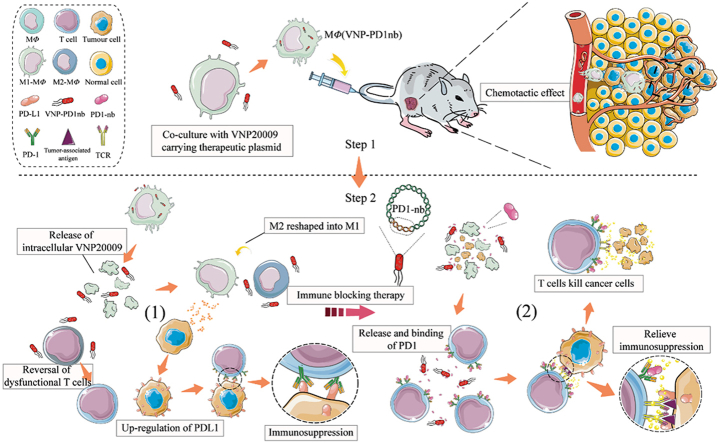
Graphical abstract
A unique "Trojan horse" macrophage-mediated tumor immunotherapy strategy was described here. This novel strategy achieved safe and efficient anti-tumor therapy by loading an engineered Salmonella strain, which can express and secrete anti-PD1 nanoantibodies, into macrophages.
1. Introduction
Since Dr. William Coley treated cancer patients with heat-inactivated Gram-positive bacteria (Streptococcus) and Gram-negative bacteria (Salmonella mucilage) in the late 19th century, an increasing number of microorganisms have been developed for tumor therapy1. These microbes, also known as oncolytic bacteria, include Salmonella typhimurium and can highly colonize tumors due to their facultative anaerobic properties and the anaerobic environment within the tumor2, 3, 4. Because of the immunosuppressive tumor environment, the amplification of these oncolytic bacteria can be implemented rapidly in tumors followed by reactivation of tumor immunity and tumor regression5, 6, 7. VNP20009 (abbreviated VNP), an attenuated strain of S. typhimurium, has received wide interest for its antitumor effects in preclinical models and relative safety8,9. However, phase I clinical trials of VNP were terminated due to its low therapeutic efficacy9. Moreover, despite significantly reducing the toxicity of VNP by the deletion of purI and msbB, the strain still induced some side effects, especially splenomegaly and hepatomegaly, in mice when administered for tumor therapy by intravenous or intraperitoneal injection10. Therefore, it is critical to improve the safety of VNP as an antitumor therapy while increasing the efficacy7,10, 11, 12.
In most cases, there is a strong innate immune response in the body, and invading microorganisms are quickly eliminated by antigen-presenting cells (APCs), such as macrophages and neutrophils13. However, Salmonella can produce a series of self-protective measures, including increasing bacterial antimicrobial peptide resistance gene expression14 and inhibiting intracellular lysosomal protein expression15, to survive after macrophage-mediated phagocytosis. Then, delayed release of strains is achieved because of the oncosis and rupture of macrophages due to the sustained intracellular stimulation of bacteria14,16,17.
In fact, macrophages in the body are often used by Salmonella as natural havens to avoid being eliminated by other strong immune cells, such as neutrophils18. Immune cells, especially macrophages, are attractive natural drug carriers due to their high chemotaxis to tumors and loading capacity19, 20, 21, 22. Therefore, a hypothesis is that macrophage-mediated tumor-targeted delivery of VNP can combine the tumor-targeting characteristics of macrophages and intracellular strains that are gradually released within tumors, which also avoids the excessive antibacterial immune response induced by direct administration of VNP. Ultimately, tumor targeting and the safety of VNP as an antitumor therapy should be improved. Our previous studies showed that this strain could activate the tumor microenvironment and promote the chemotaxis of macrophages to tumors23,24. This means that VNP released by macrophages within tumors could in turn promote the chemotaxis of macrophages loaded with the strain toward the tumor region.
Programmed cell death protein 1 (also known as CD279 and PD-1) and its ligand PD-1 ligand, PD-L1 (CD274), are critical immune checkpoints that function normally to protect against autoimmunity. Their interaction is also an important strategy that many tumors use to escape immune surveillance25. Despite the promising results of PD1/PDL1 blockade-based immunotherapy26, a majority of patients still lack a durable response to the therapy because of the low activity of effector immune cells, specifically CD8 T cells, in the tumor27,28. Therefore, increasing the response rates of cancer patients to PD1/PDL1 immune blockers is a critical challenge that needs to be overcome.
In this study, we report for the first time macrophage-mediated tumor-targeted delivery of VNP. This novel strategy combines the chemotaxis of macrophages to tumor regions, the protective and slow release of VNP by macrophages, and the colonization and tumor-killing properties of VNP. Finally, the antitumor activity of VNP was ensured, and acute organ injury caused by single VNP treatment was effectively avoided. Moreover, PDL1 levels on the surface of cancer cells were significantly upregulated after treatment with macrophages loaded with VNP, while the PD1 levels on the surface of CD8+ T cells in the tumor were significantly downregulated. This finding suggests that tumors in this state will be more sensitive to PD1/PDL1 blockade treatment26,29. Therefore, we combined this bacteria-in-cell drug with PD1/PDL1 blockade therapy for the first time, and the therapeutic efficacy in mouse melanoma was significantly improved. We anticipate the macrophage-mediated tumor-targeted delivery of engineered attenuated Salmonella combined with enhanced safety and potent treatment efficacy to be a valuable reference for the research and delivery of biologically functional bacteria, and believe that stealth bacteria loaded by macrophages represent a versatile, novel and unique tool for biomedical applications. This new drug delivery strategy also offers new ideas for combining multiple cancer therapies, such as cell therapy, bacterial therapy and immunotherapy, to achieve complementary advantages.
2. Materials and methods
2.1. Cell lines and primary cells
B16F10 mouse melanoma cells, LLC mouse lung cancer cells, MC38 mouse colon cancer cells, A20 mouse B lymphoma cells, L929 mouse fibroblasts and RAW264.7 macrophages were preserved in our laboratory. Peritoneal macrophages were extracted based on the protocol provided by Choi et al30. In brief, a 5% starch broth solution was prepared [1.8% nutritional broth (Solarbio, N8300, Beijing, China) and 5% soluble starch were dissolved in water], sterilized at 115 °C and stored at 4 °C. Eight-week-old female C57BL/6J mice were intraperitoneally administered 1 mL of starch broth solution, and peritoneal macrophages were harvested 2 or 3 days later. RAW264.7 cells were induced using 100 ng/mL LPS (Beyotime, S1732, Shanghai, China) for 12 h to obtain M1-like RAW264.7 cells.
2.2. Bacterial strains, plasmid construction and transformation
VNP20009 (abbreviated VNP), VNP-RFP (transformed with a plasmid expressing RFP with a J23100 promoter), VNP-LuxCDABE (genomic insertion of LuxCDABE with a J23100 promoter), VNP-PD1nb (transformed with a plasmid expressing PD1nb with a J23100 promoter; a flag tag was added to the N-terminus of PD1nb to facilitate subsequent detection, pJ23100-flag-PD1nb), VNP-NC (transformed with an empty plasmid) and VNP-psifB-RFP (transformed with a plasmid expressing RFP with a sifB promoter; an HA tag was added to the C-terminus of RFP to facilitate subsequent detection, psifB-RFP-HA) were preserved in our laboratory. DH5α and BL21 strains were purchased from Vazyme. All plasmids were constructed using the ClonExpress II/MultiS One Step Cloning Kit (Vazyme, C112/C113, Nanjing, China). The plasmid Pet28a-PD1nb was kindly provided by Dr. Shufeng Li (Southeast University). The construction and screening of anti-PD1 nanobodies are described in Section 2.15. The VNP strains were electrotransformed with the plasmids as described previously31.
2.3. Preparation of VNP-loaded macrophages [MΦ (VNP)]
M1-like RAW264.7 cells or peritoneal macrophages (PEMΦ cells) were harvested as previously described in Section 2.1, and then the macrophages and VNP were cocultured at a ratio of 1:10 in DMEM supplemented with 10% fetal bovine serum and no antibiotics at 37 °C for different times. The supernatant was discarded, and the cells were washed 3 or 4 times with PBS. The cells were incubated in DMEM supplemented with 10% fetal bovine serum and 50 μg/mL gentamicin at 37 °C for 60 min to kill extracellular VNP but had no significant effect on the activity of strains inside the cells32. The supernatant was discarded, and the cells were washed 2 or 3 times with PBS. RAW264.7(VNP) cells were resuspended by gentle pipetting, while PEMΦ(VNP) cells were digested with 0.2% lidocaine at 4 °C for 5–8 min and resuspended. The cell suspension was centrifuged at 300×g and 4 °C for 5 min, and MΦ(VNP) cells (including RAW264.7(VNP) and PEMΦ(VNP) cells) were harvested from the precipitate. For immunofluorescence microscopy, RAW264.7 and PEMΦ cells were cocultured with VNP-RFP for different times. The extracellular bacteria were washed and sterilized with 50 μg/mL gentamicin for 60 min. The cells were washed with PBS 3 times, fixed with 4% paraformaldehyde and permeabilized using 0.5% Triton X-100. After being washed with PBST 3 times, Actin-Tracker Green-488 (Beyotime, C2201S, Shanghai, China) was added and incubated for 1 h at 37 °C followed by DAPI (Beyotime, C1005, Shanghai, China) staining. Images were acquired using microscopy (Carl Zeiss, Axioplan 2, Oberkohen, Germany).
2.4. Intramacrophage VNP viability assays
Macrophages were cultured with VNP-RFP for different times [including RAW264.7(VNP) and PEMΦ(VNP) cells] and harvested as described in Section 2.3. RAW264.7(VNP) and PEMΦ(VNP) cells were adjusted to 1–10 × 104 cells/100 μL, and the cells were transferred to a 96-well plate at 100 μL per well. The RFP fluorescence intensity (550 nm, 585 nm) was determined by a multimode plate reader. The number of bacteria in the macrophages was determined by using a formulated standard curve, which was generated by plotting the fluorescence intensity versus the concentration of a serially diluted standard VNP-RFP solution. RAW264.7(VNP) and PEMΦ(VNP) cells were incubated in 0.5% Triton X-100 (Sigma–Aldrich, 648462, St. Louis, MO, USA) lysis buffer for 10–15 min, and intracellular VNP was released from macrophages. The lysis solution was diluted and spread on LB agar plates supplemented with kanamycin. Colonies were counted after more than 12 h of incubation, and the number of live VNP in the total VNP taken up by macrophages was calculated.
2.5. Intramacrophage bacterial release assays
B16F10 cells (20,000) were added to the lower chamber of a 3.0-μm Transwell plate (cells could not cross the pores but the bacteria could) and incubated for 6 h to adhere. Then, 40,000 RAW264.7(VNP-RFP) or PEMΦ (VNP-RFP) cells were added to the Transwell inserts, while 40,000 RAW264.7 or PEMΦ cells were added to the lower chamber [RAW264.7(VNP-RFP)-Up or PEMΦ(VNP-RFP)-Up group]. MΦ(VNP-RFP) and MΦ cells were added at opposite positions in the RAW264.7(VNP-RFP)-Low and PEMΦ(VNP-RFP)-Low groups. After 16 h of incubation, the supernatants were harvested and spread on LB agar plates with kanamycin, and the number of colonies was counted after an overnight culture at 37 °C. The adherent cells in the lower chambers were incubated in 500 μL of DMEM supplemented with 50 μg/mL gentamicin for an additional 60 min so that the extracellular bacteria could be killed. Cells were labeled with anti-CD11b antibodies and analyzed using flow cytometry (BD, Canto II, Franklin Lakes, NJ, USA).
To confirm the release behavior of macrophages, a VNP-psifB-RFP strain that could express RFP specifically inside cells was constructed. The psifB forward primer (5′-CTG CCC TAC CGC TAA ACA TCT-3′) and psifB reverse primer (5′-CCA CAA GTG ATT ATA TGA TAC-3′) were used to amplify the sifB promoter from the VNP genome. The strain obtained by amplification in LB medium was centrifuged at 12,000 rpm (Thermo, Fresco™17, Waltham, USA) for 10 min, and then the bacterial cells were adjusted to OD600 = 1.0 with PBS. Subsequently, the bacteria and RAW264.7 cells were cocultured for 60 min at a MOI of 10:1, washed with PBS 3–4 times and cultured in DMEM supplemented with 50 μg/mL gentamicin and 10% serum. After 60 min of incubation, total intracellular protein was collected for Western blot analysis. An HA-tagged antibody (CST, 3724S, Danvers, MA, USA) was used to examine the expression of RFP, and a secondary anti-rabbit IgG antibody (CST, 7074) was used. Salmonella-specific antibodies (Targetpharma, Nanjing, China) were used to track the strains in macrophages. RFP expression by intracellular activated bacteria was confirmed by fluorescence microscopy (Carl Zeiss).
2.6. Macrophage viability assays
The viability of VNP-loaded macrophages was examined using the trypan blue viability assay method. In brief, equal amounts of the cell suspension were mixed with 0.4% trypan blue solution (Invitrogen, T10282, Carlsbad, CA, USA), and 10 μL was pipetted into a Countess chamber slide (Invitrogen Countess). The slide was inserted into the automated cell counter (Invitrogen Countess), and the number of viable and nonviable cells was determined.
2.7. Bacterial growth assays
The growth curves for different VNP in LB media were obtained with the Bioscreen C (OY Growth Curves Ab Ltd., Finland). In brief, 10 μL of VNP suspension (OD600 = 1.0) was inoculated into 1 mL of LB medium, and 300 μL of solution per well was inoculated in Bioscreen C multiwell plates. The multiwell plates were incubated for 30 h at 37 °C. OD values were measured every 30 min under a brown filter with a wavelength of 600 nm.
2.8. Apoptosis assay
B16F10 cells (2.0 × 105) were plated into 12-well plates and incubated for 6–8 h until the cells adhered to the wall. Then, the collected unprocessed-VNP or released-VNP strains were cocultured with the cells at a MOI of 100 for 4 h. All cells in the plate were collected, washed and resuspended in binding buffer, stained with 1 μg of APC-conjugated annexin V protein (homemade in the laboratory) and left on ice in the dark for 30 min. All samples were added with 1 μL of PI and gently mixed before flow cytometry analysis (BD).
2.9. Macrophage chemotaxis assays
Transwell inserts (8 μm) were incubated in DMEM supplemented with 1% fetal bovine serum overnight. Approximately 3 × 104 B16F10, LLC or MC38 cells were seeded in the lower chambers, and chambers containing only DMEM served as controls. After 10 h of incubation for adherence, the medium was refreshed with 700 μL of DMEM supplemented with 10% fetal bovine serum and 50 μg/mL gentamicin. RAW264.7, RAW264.7(VNP), PEMΦ, and PEMΦ (VNP) cells were prepared, and the concentration was adjusted to 1 × 106 cells/mL using DMEM. One hundred microliters of the cell suspension was transferred to the Transwell insert and incubated at 37 °C for 16 h. Then, the Transwell inserts were removed from the plate, and the remaining cells that had not migrated from the top of the membrane were removed using a cotton-tipped applicator. After being washed 2–3 times with PBS, the Transwell inserts were placed in 4% paraformaldehyde for 20–30 min and then stained with crystal violet (KeyGen BioTech, KGA229, Nanjing, China) for 30 min at room temperature. After being washed with PBS 2–3 times, the Transwell inserts were photographed under an ortho-fluorescence microscope (Carl Zeiss).
2.10. In vitro analysis of macrophage killing of tumor cells
Approximately 60,000 B16F10, LLC or MC38 cells were plated in each well of a 0.4-μm Transwell plate (bacteria could not cross the pores, but the secreted cytokines could). After 10 h of incubation for adherence, the medium was replaced with 1200 μL of DMEM supplemented with 10% fetal bovine serum and 50 μg/mL gentamicin. RAW264.7, RAW264.7(VNP), PEMΦ, PEMΦ(VNP) and L929 cells were prepared, and the cell concentration was adjusted to 1 × 106 cells/mL. Two hundred microliters of the cell suspension was added to the Transwell inserts, and inserts containing only DMEM served as a control. The different types of cells were added to the upper chamber, and cells were not added to the lower chamber as a background control to remove interference from the upper chamber. After incubation for 12 h, the supernatant was collected. The proliferation of the tumor cells was measured using a CCK-8 assay kit (Beyotime, C0038).
2.11. qRT-PCR
qRT-PCR was performed as follows. Total RNA was extracted using TRIzol reagent (Vazyme, R401-01), and qRT-PCR was conducted on a StepOne/StepOne Plus™ Real-Time PCR System (Applied Biosystems) using a SYBR Green PCR Master mix kit (Vazyme, Q221-01) according to the manufacturer's instructions. Primers used were as follows: CD11b forward primer (5′-ATG GAC GCT GAT GGC AAT ACC-3′); CD11b reverse primer (5′-TCC CCA TTC ACG TCT CCC A-3′); TNF-α forward primer (5′-GAC GTG GAA GTG GCA GAA GAG-3′); TNF-α reverse primer (5′-TGC CAC AAG CAG GAA TGA GA-3′); iNOS forward primer (5′-CAT TGC TGA CAG GAT GCA GAA GG-3′); iNOS reverse primer (5′-TGC TGG AAG GTG GAC AGT GAG G-3′); IL-6 forward primer (5′-CTC AAT ATT AGA GTC TCA ACC CCC A-3′); IL-6 reverse primer(5′-AAG GCG CTT GTG GAG AAG G-3′); CCR5 forward primer (5′-GTC TAC TTT CTC TTC TGG ACT CC-3′); CCR5 reverse primer (5′-CCA AGA GTC TCT GTT GCC TGC A-3′); CCR2 forward primer (5′-GCT GTG TTT GCC TCT CTA CCA G-3′); CCR2 reverse primer (5′-CAA GTA GAG GCA GGA TCA GGC T-3′); CSF1R forward primer (5′-TGG ATG CCT GTG AAT GGC TCT G-3′); CSF1R reverse primer (5′-GTG GGT GTC ATT CCA AAC CTG C-3′); 18 S rRNA forward primer (5′-GTA ACC CGT TGA ACC CCA TT-3′); 18 S rRNA reverse primer (5′-CCA TCC AAT CGG TAG TAG CG-3′).
2.12. Macrophage phagocytosis assay
RAW264.7, RAW264.7(VNP), PEMΦ, and PEMΦ(VNP) cells were prepared as described in Section 2.3, mixed with red fluorescence microspheres (Sigma–Aldrich, L2778) at a ratio of 1:10 (cell: microsphere) and incubated at 37 °C for 4 h. The supernatant was discarded, and free fluorescence microspheres were washed away with PBS. The cells were resuspended carefully, and phagocytosis efficiency was determined by flow cytometry.
2.13. In vivo biodistribution
RAW264.7(VNP-RFP) and PEMΦ(VNP-RFP) cells were prepared as described in Section 2.3, and 5 × 105 VNP-RFP, 2.5 × 105 RAW264.7(VNP-RFP) and 1 × 105 PEMΦ(VNP-RFP) cells in 100 μL of PBS were injected into B16F10 tumor-bearing mice via the tail vein. The mice were sacrificed at specific times, and tumors and other organs were dissected and lysed with a tissue pulveriser. The tissue lysate was diluted and spread on LB agar plates or analyzed using flow cytometry to determine the distribution of the strain. PEMΦ(VNP-LuxCABDE) was harvested as described previously, and the cells were incubated with the near infrared (NIR) fluorescent dye DiR (Abbkine) for 45 min. PEMΦ, PEMΦ (VNP-LuxCDABE) cells (1 × 105) or VNP-LuxCDABE strains (5 × 105) in 100 μL of PBS were injected into A20 tumor-bearing mice via the tail vein. The fluorescence signals of LuxCDABE and DiR were detected using an in vivo imaging system (PerkinElmer, IVIS® Lumina III, Waltham, MA, USA).
2.14. Enzyme-linked immunosorbent assay (ELISA)
At 24 h post administration, mice were bled for sera and killed for tumor collection. The obtained tumor was added to tissue lysate (absin, abs9225, Shanghai, China) at 10 mg tumor tissue/50 μL tissue lysate and homogenized using a tissue homogenizer. The supernatant was collected by centrifugation. Both sera and tumor tissue lysates were collected for CCL2 cytokine detection by mouse CCL2 ELISA kits (absin, abs520016).
2.15. Construction and characterization of anti-PD1 nanobodies
The construction and characterization of nanobodies were performed as previously described33. In brief, a healthy dromedary camel was immunized once a week with 0.5 mg of human PD1 protein (Sino Biological Company, Beijing, China). After 7 immunizations, 100 mL of blood from the immunized dromedary was collected. Blood lymphocytes were isolated, total RNA was extracted, and cDNA was synthesized for the following nanobody library construction. Briefly, a two-step nested PCR approach was used to amplify Nbs gene fragments. The final PCR products of 500 bp were ligated into T7Select10-3b vector arms. The ligation mixtures were packaged in vitro. The library size was evaluated by gradient dilution. In addition, the insertion rate was detected with PCR amplification. PD-1 protein was immobilized onto agarose beads, and the beads were incubated with phage library and then washed unbound phage particles with PBST. The bound phages on washed beads were amplified by infecting E. coli and cultured until cell lysis occurred. The bound phage particles were recovered and used for the next round of panning. After three rounds of biopanning, the PD-1-specific phages were enriched. Then, phage ELISA was performed, and positive plaques that gave high absorbance values toward PD-1 were selected and sequenced. By cloning in the pet-32a expression vector, VHH was equipped with an HA tag and His tag at the N-terminus and utilized for Nb purification and detection. For protein expression, the recombinant plasmid was transformed into E. coli BL21 (DE3) and induced with 0.1 mmol/L IPTG at 23 °C. Nanobodies were purified by nickel Sepharose affinity chromatography and dialyzed in PBS buffer. The purity of the collected proteins was checked by SDS–PAGE. The affinity constant of nanobodies for binding to PD1 was determined by ELISA. The antibody concentration resulting in 50% of the maximum absorbance value at three antigen-coating concentrations was measured and designated [Ab]t. Three [Ab]t values were obtained and used for the calculation of three K values according to the Beatty formula. The final affinity constant is the average result of three K values.
2.16. PD1nb expression and secretion assay
VNP-PD1nb was incubated in 40 mL of LB medium with kanamycin until the OD600 was between 0.6 and 0.8. Then, the solution was centrifuged at 5000 rpm (Thermo) and 4 °C for 10 min, and the supernatant and precipitate were collected. The precipitate was resuspended in 2 mL of PBS, heated in a dry bath incubator at 110 °C for 20 min to disrupt the bacteria and release the proteins, and then centrifuged at 13,000 rpm (Thermo) for 10 min, and total proteins in the bacteria were contained in the supernatant. Proteins were collected from the supernatant obtained in the first step according to the trichloroacetic acid (TCA) protein precipitation protocol. In brief, the supernatant was transferred to a 50 mL tube (Beckman Coulter, Brea, CA, USA) and centrifuged at 15,000×g and 4 °C for 10 min using an ultracentrifuge (Beckman Coulter). Then, the supernatant was transferred to a new 50 mL centrifuge tube, mixed with 10% TCA and incubated at 4 °C for 30 min before being centrifuged at 17,000×g and 4 °C for 20 min. The precipitate was resuspended in 300 μL of PBS, transferred to a 1.5-mL EP tube containing 1.2 mL of cold acetone, and centrifuged at 17,000×g and 4 °C for 20 min. The previous step was repeated, and the total protein secreted by VNP-PD1nb was finally collected in the precipitate and resuspended in 40 μL of PBS. The concentration of total protein in both the precipitate and supernatant was determined by a BCA kit (Beyotime, P0012) and adjusted to the same concentration. The protein solution was mixed with 5 × SDS loading buffer (Yeasen, 20315ES20, Shanghai, China) and heated in a dry bath incubator at 95 °C. Twenty microliters of solution (approximately 15 μg of protein) was loaded for Western blot analysis. Flag-tagged antibodies (Sigma–Aldrich, F1804) were used to examine the expression and secretion of PD1nb, and secondary anti-mouse IgG (CST, 7076) was used.
2.17. Animal model
All procedures were conducted in compliance with all the relevant ethical regulations and were approved by the Nanjing University Institutional Animal Care and Use Committee. BALB/c (4–5 weeks, female) and C57BL/6J (6–8 weeks, female) mice were purchased from Changzhou Cavens Animals Corporation. B16F10 (2 × 105 cells per mouse) or A20 (1 × 106 cells per mouse) cells were injected subcutaneously into the right flanks of C57BL/6J mice or BALB/c mice. DTIC (dacarbazine), a recognized chemotherapeutic agent for the treatment of melanoma, was purchased from Shanghai YuanYe (S25808). The PD1nb used in this study was purified in our laboratory. When tumors grew to 80–160 mm3, DTIC (80 mg/kg) and PD1nb (5 mg/kg) were injected intraperitoneally four times every other day, while VNP (5.0 × 105 cells per mouse) and cells (1.0 × 105 cells per mouse), including PEMΦ, PEMΦ(VNP-NC) and PEMΦ(VNP-PD1nb), were injected through the caudal vein only one time. Tumors were measured by calipers 3–4 times weekly, and the volumes (V, mm3) were calculated using Eq. (1):
| V = a2b × 0.52 | (1) |
where a is the minor radius and b is the major radius.
For experimental lung metastasis, 1 × 106 B16F10 cells in 100 μL of PBS were injected into mice via the tail vein. The treatment was started on Day 6, and the mice were sacrificed on Day 18. Lung metastases were determined by ex vivo photography, and ImageJ was applied to analyze the total surface tumor area. Blood was collected by retro-orbital puncture for routine blood examination. The serum obtained from blood was cryopreserved at −80 °C until assayed for blood biochemical indexes and ELISA. Routine blood examinations, blood biochemistry analysis, H&E staining of tumor, heart, liver, spleen, lung, and kidney sections and fluorescence immunostaining of macrophages on tumor sections were prepared by Wuhan Servicebio Corporation. To plot the survival curve, tumor-bearing mice were monitored daily and sacrificed when signs of adverse effects (pain, apathy, necrotic tumor) were observed or the humane endpoint (tumor weight equal to 10% of mouse body weight) was reached.
2.18. Flow cytometry
Tumor-bearing mice were sacrificed, and the tumors were dissected and subsequently incubated in digestion medium (10 U/mL collagenase I, 400 U/mL collagenase IV, 30 U/mL DNase I, all diluted in HBSS) for 30 min. Cell clumps were removed through a 40-μm cell strainer to obtain single-cell suspensions. Blood samples were obtained by removing the eyeball after the mice were completely anesthetized, and peripheral blood lymphocytes were obtained by a peripheral blood lymphocyte isolation kit (Solarbio, P8620). The cells were stained with fixable viability dye (BD, 564407), incubated for 10–15 min at room temperature protected from light, and then stained using the following anti-mouse antibodies: CCR2-AF647 (clone SA203G11), CCR5-APC (clone HM-CCR5), CSF1R-APC (clone AFS98), CD45-PE-Cy7 (clone 30-F11), CD11b-APC (clone 561690), CD11b-PE (clone M1/70), F4/80-BV421 (clone T45-2342), CD86-PE (clone GL1), CD3e-FITC (clone 145-2C11), CD4-APC (clone RM4-5), CD25-BV421 (clone 3C7), CD8-APC (clone 53–6.7), PD1-APC (clone J43), PDL1-BV421 (clone MIH5), CD38-BV421 (clone 90/CD38), and CD127-PE (clone SB/199). Intracellular proteins, including CD206-APC (clone MR6F3), Ki67-BV421 (clone B56), IFNγ-PE (clone XMG1.2), and TNFα-PE (clone MP6-XT22), were stained after using a membrane-breaking fixative solution (BD, 565388). For analysis of peripheral blood immune cells in mice, blood was collected from the retro-orbital sinus of the mice. The blood samples were incubated with red blood cell lysis buffer (Solarbio, R1010) before staining with the viability dye and antibodies mentioned above. Blood was collected from the retro-orbital sinus of the mice, left at room temperature for 1 h and then centrifuged at 4 °C and 3000 rpm (Thermo) to obtain serum. A CBA kit (BD, 560485) was used for the analysis of cytokines in serum. Detection of bacterial load in tissues by flow cytometry: different organs were weighed, and the corresponding volume of 0.5% Triton X-100 (2 μL/mg) was added according to the weight, processed by a tissue lyser and placed at 4 °C for 30 min. After cell clump removal by a cell sieve, the population of bacteria with RFP fluorescence in the lysate was detected by flow cytometry. All analyses were performed on a BD Canto II.
2.19. Statistical analysis
Data were analyzed with GraphPad Prism software version 5 (GraphPad Software). Two groups were compared using Student's t test. Comparisons of more than two groups were analyzed by one-way analysis of variance (ANOVA) followed by Dunnett's multiple comparisons test. Data are expressed as the mean ± SEM for the in vivo mouse tumor model.
3. Results
3.1. Macrophages can function as biological carriers of VNP
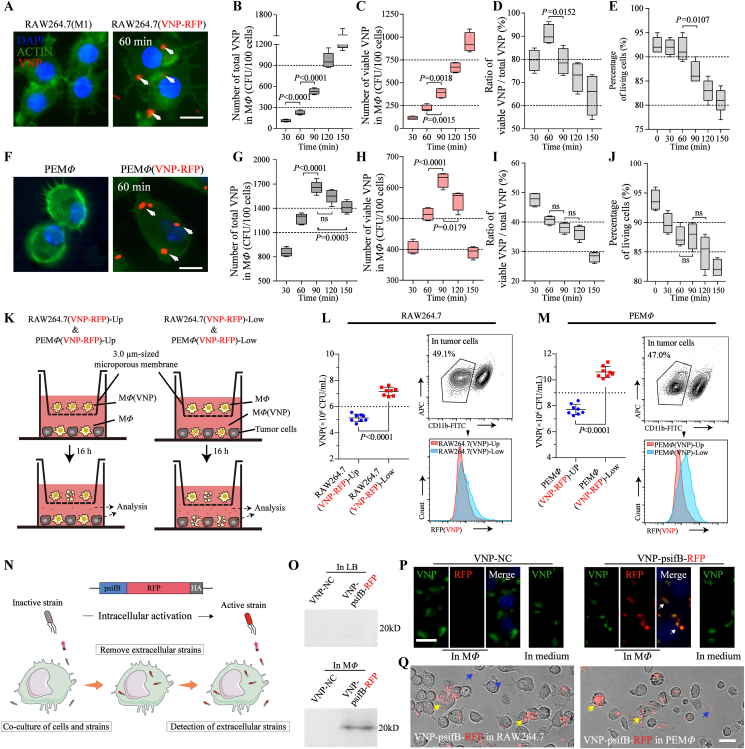
In designing a valid biological carrier, we needed to determine the appropriate preparation conditions. We first constructed VNP-RFP strains that stably express red fluorescent protein (RFP) and cocultured these bacteria with the murine macrophage line RAW264.7 after the cells were induced with lipopolysaccharide (LPS) to form an M1-like phenotype (Supporting Information Fig. S1). The cells were then treated with 50 μg/mL gentamicin for 60 min to kill extracellular bacteria (Supporting Information Fig. S2). We found that RAW264.7 cells could successfully load VNP-RFP (Fig. 1A). Fluorescence quantitative detection, which was based on the linear relationship between the fluorescence intensity and the number of bacteria (Supporting Information Fig. S3), showed that the total amount of VNP-RFP phagocytized by RAW264.7 cells positively correlated with the coculture time (Fig. 1B). Furthermore, spread plate counts were used to count the number of living VNP-RFP after macrophage phagocytosis, and the results showed that the number of live VNP-RFP loaded in RAW264.7 cells also positively correlated with the coculture time (Fig. 1C), during which the loading efficiency (live bacteria number/total bacteria number in RAW264.7) reached a peak at 60 min [220 ± 13 CFU (mean ± SEM)/100 cells] (Fig. 1D). Although the viability of RAW264.7 cells decreased with prolonged coculture with VNP-RFP, cocultured macrophages showed acceptable cell viability at 60 min (above 90%) (Fig. 1E).
Figure 1.
Preparation of MΦ(VNP) cells and the release of VNP from MΦ(VNP) cells. (A–E) RAW264.7 cells were induced with LPS (100 ng/mL, 12 h) and cocultured with VNP-RFP (1:10) for different times (30–150 min). Gentamicin (50 μg/mL) was added to kill extracellular VNP-RFP, and the quantity of the loaded bacteria in cells and cell viability were examined (n = 5). (A) Fluorescent images of the loaded cells. RAW264.7 cells were cocultured with VNP-RFP for 60 min and stained with DAPI and actin–FITC (Scale bar = 10 μm). (B) Changes in the total number of VNP-RFP strains phagocytosed by RAW264.7 cells. (C) Changes in the number of surviving VNP-RFP strains phagocytosed by RAW264.7 cells after different coculture times. (D) Changes in the loading efficiency (number of live bacteria/number of total bacteria) in macrophages. (E) The percentage of living RAW264.7 cells after different coculture times. (F–J) The results of repeating the experiments in (A–E) by replacing RAW264.7 cells with peritoneal macrophages (PEMΦ) (n = 5). (K–M) Detection of VNP-RFP released from MΦ(VNP-RFP) cells. (K) RAW264.7(VNP-RFP) or PEMΦ(VNP-RFP) (40,000 cells) cells were cultured in medium supplemented with 50 μg/mL gentamicin for 12 h, and then gentamicin was removed and cultured with B16F10 cells indirectly (RAW264.7(VNP-RFP)-Up & PEMΦ(VNP-RFP)-Up) or directly (RAW264.7(VNP-RFP)-Low & PEMΦ(VNP-RFP)-Low) by using a 3.0-μm Transwell chamber. The quantity of VNP in the supernatant or inside tumor cells was determined after 16 h. (L, M) The VNP titer in the supernatant (left) and comparison of the population of VNP inside tumor cells (right). (N) Schematic diagram of the use of VNP-psifB-RFP to examine bacterial release by macrophages. Due to the psifB promoter, VNP-sifB-RFP can only be activated and express RFP intracellularly, so extracellular red bacteria must have escaped from macrophages. (O) Western blot showing RFP expression of VNP-psifB-RFP in LB with OD600 = 0.6–0.8 or inside macrophages for 60 min. A strain carrying empty plasmid (VNP-NC) was used as a control. (P) Fluorescent images showing that VNP-psifB-RFP specifically expressed RFP within 60 min after entering into macrophages, while the strain cultured in DMEM did not express RFP. The cells were stained with Salmonella-specific antibodies (green) and DAPI (blue). (Scale bar = 10 μm). (Q) Brightfield images showing activated VNP-psifB-RFP inside RAW264.7 and PEMΦ (yellow arrows) cells and the VNP released from macrophages (blue arrows) after 3 h of culture in medium containing 50 μg/mL gentamicin (Scale bar = 20 μm). The data in (L) and (M) are reported as the mean ± SEM. Boxplot representations of the spot counts. The median, interquartile range, and minimum and maximum identifiers are shown. All data are representative of three independent experiments. Statistics were calculated using the two-tailed, unpaired Student's t test with Welch's correction.
For the translatability of the approach, we also used primary peritoneal macrophages (PEMΦ cells), which have also been used as drug carriers to target tumors30,34, to load VNP [PEMΦ(VNP) cells]. We first prepared high-purity peritoneal macrophages through starch broth stimulation followed by purification with adherent cultures, which ensured a high purity of the obtained macrophages (>98%) every time30 (Supporting Information Fig. S4). VNP-RFP strains were cocultured with peritoneal macrophages (Fig. 1F), and unlike RAW264.7 cells, effective VNP loading by PEMΦ cells decreased with time (Fig. 1I). The number of live VNP strains loaded into PEMΦ cells was sufficient at 60 min [510 ± 10 CFU (mean ± SEM)/100 cells] and reached a peak at 90 min [625 ± 12 CFU (mean ± SEM)/100 cells] (Fig. 1G and H), when both the PEMΦ(VNP) cell viability (above 85%) was acceptable (Fig. 1J). For accuracy and uniformity, a coculture time of 60 min for the two kinds of macrophages and VNP with a ratio of 1:10 was chosen to obtain VNP-loaded macrophages (RAW264.7 (VNP) and PEMΦ(VNP) cells) for all subsequent experiments.
To investigate 1) whether VNP could survive for a long time, similar to wild-type Salmonella16, after being engulfed by macrophages, and 2) the changes in the release of intracellular strains from VNP-loaded macrophages after contact with tumor cells, we cocultured RAW264.7(VNP-RFP) or PEMΦ(VNP-RFP) cells in medium supplemented with gentamicin for 12 h and then removed gentamicin and cocultured the cells with B16F10 mouse melanoma cells in vitro with or without 3.0-μm microporous membrane blocking for 16 h (Fig. 1K). When RAW264.7(VNP-RFP) or PEMΦ(VNP-RFP) cells were in indirect or direct contact with tumor cells, the VNP strains were alive and could be released from macrophages effectively (Fig. 1L and M). Moreover, when RAW264.7(VNP-RFP) and PEMΦ(VNP-RFP) cells were cocultured directly with tumor cells [RAW264.7(VNP-RFP)-Low group and PEMΦ(VNP-RFP)-Low group], more VNP strains were released into the supernatant, suggesting the acceleration of strain release by tumor cells (Fig. 1L and M left). The number of VNP strains in tumor cells, measured by flow cytometry, also showed a greater number in both the MΦ(VNP-RFP)-Low groups (Fig. 1L and M right). This contact-dependent enhancement of strain release probably results from macrophage membrane remodeling and the exocytosis of vesicles during the phagocytosis of tumor cells, which makes it easier for intracellular VNP to escape35.
To further confirm the behavior of VNP release from macrophages, we constructed an attenuated Salmonella strain that conditionally expressed RFP inside macrophages (named VNP-psifB-RFP) based on the sifB promoter6. The sifB promoter, a classic Salmonella pathogenicity island II (SPI-II) promoter, can be activated by intracellular hypoxia, a low pH and a low phosphate environment36. VNP-psifB-RFP expressed RFP only after being engulfed by macrophages (Fig. 1N). After VNP-psifB-RFP was cocultured with RAW264.7 cells for 60 min, modest protein expression by VNP-psifB-RFP was examined by Western blotting (Fig. 1O), and VNP-psifB-RFP expressing RFP was also observed inside cells (Fig. 1P). These results further indicated that the VNP-psifB-RFP strains could express RFP protein only after being activated in macrophages. Next, we prepared RAW264.7(VNP-psifB-RFP) and PEMΦ (VNP-psifB-RFP) cells and cultured them in medium supplemented with gentamicin. The appearance of VNP-psifB-RFP with red fluorescence outside the cells was observed 3 h later by fluorescence microscopy (Fig. 1Q). Since VNP-psifB-RFP activates and expresses RFP only in the intracellular environment, the extracellularly activated strain was undoubtedly released from the cells. Then, a continuous slow release of bacteria was observed for the remainder of the test period (3–15 h) (Supporting Information Fig. S5). The delayed release of the loaded VNP from cells may be due to the oncosis of macrophages and the self-escape of the strain14,16,17. Based on our results, it is possible to speculate that when the external environment is suitable, such as the immunosuppressive and relatively nutrient-sufficient environment in the tumor7, these released strains will undergo rapid proliferation.
Since the tumor microenvironment is acidic (∼pH 6.5–6.8)19, we prepared RAW264.7(VNP) and PEMΦ(VNP) cells and cultured them in medium with different pH values (7.4 and 6.7). The results showed that the acidic environment had no significant effect on the release of strains and their proliferation (Supporting Information Fig. S6). In addition, there was no significant difference between the released VNP and the unprocessed VNP in in vitro proliferation activity and ability to invade and kill tumor cells (Supporting Information Fig. S7). In conclusion, we confirmed that macrophages (RAW264.7 and PEMΦ cells) were capable of acting as natural biological carriers of VNP strains and achieved delayed slow release of intracellular bacteria without a significant effect on the proliferation activity of VNP and the invasion and killing ability of VNP on tumor cells.
3.2. Loading VNP does not weaken the phagocytosis, chemotaxis or tumoricidal activity of macrophages
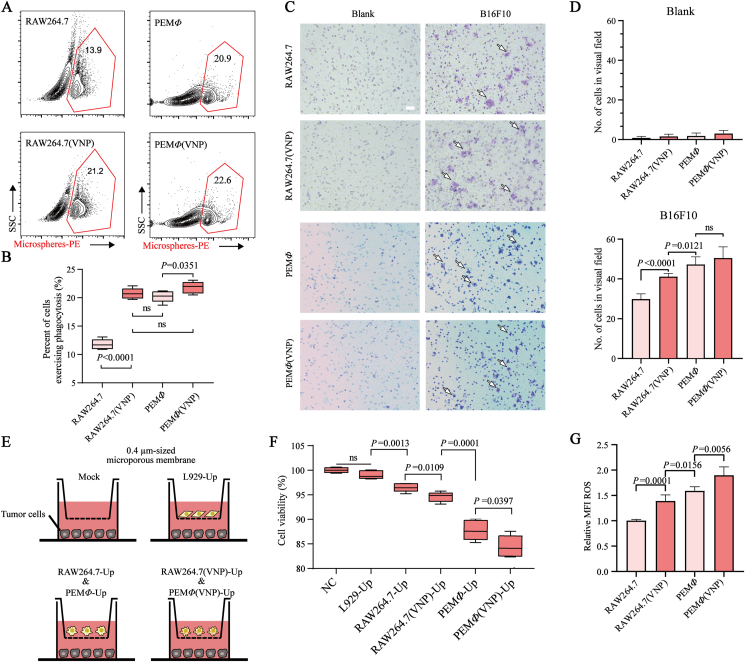
It is worth confirming whether VNP loading affects some characteristics of macrophages, including phagocytosis, chemotaxis and tumor cytotoxicity. We first examined phagocytosis and quantified it with the content of fluorescent microspheres phagocytosed in cells. The results suggested that after being loaded with VNP in RAW264.7 cells, the cells were able to phagocytose more fluorescent microspheres (Fig. 2A and B). This trend was also observed in PEMΦ loaded with VNP, although the effect was less upregulated than that in RAW264.7 cells, which could be attributed to differences in cell origins (Fig. 2A and B). The enhanced phagocytic capacity of macrophages may be due to the upregulation of CD11b (a component of integrin CR3) expression after bacterial stimulation (Supporting Information Fig. S8), which is consistent with other reports37. Considering that macrophages perform chemotactic functions through surface chemokine receptors, it is worth exploring whether phagocytosis affects the surface characterization of macrophages (i.e., changes in the membrane protein). Fluorescent microspheres replaced VNP to coincubate with macrophages, and the results of flow cytometry showed that phagocytosis by macrophages did not significantly change the content of key chemokine receptor proteins on the cell surface (Supporting Information Fig. S9).
Figure 2.
MΦ(VNP) cells achieve increased phagocytosis, chemotaxis and tumoricidal activity. (A, B) RAW264.7/RAW264.7 (VNP) and PEMΦ/PEMΦ (VNP) cells were coincubated with fluorescence microspheres at a ratio of 1:10 for 4 h. The percentage of macrophages that had engulfed microspheres was detected by flow cytometry, and the results are shown as boxplots in (B). (C, D) Transwell migration assay of RAW264.7/RAW264.7 (VNP) and PEMΦ/PEMΦ (VNP) cells toward B16F10 cells (coculture for 16 h with 50 μg/mL gentamicin). Representative images of Transwell chambers (Scale bar = 100 μm) in (C), and the number of cells crossing the pores in five random fields are visualized as boxplots in (D). (E, F) Detection of the indirect killing ability of the different groups on B16F10 tumor cells by indirect culture. (E) L929, RAW264.7, RAW264.7(VNP), PEMΦ and PEMΦ (VNP) cells were seeded in the 0.4 μm Transwell upper chamber, and the lower chamber was seeded with B16F10 cells, which were cocultured for 12 h (supplemented with 50 μg/mL gentamicin). (F) Detection of the proliferation of B16F10 tumor cells by CCK-8 assay in (E). (G) Detection of intracellular ROS in RAW264.7/RAW264.7 (VNP) and PEMΦ/PEMΦ(VNP) cells by flow cytometry. The relative mean fluorescence intensity (MFI) of ROS is shown (n = 5 in all data). Boxplot representations of the spot counts, with the median, interquartile range, and minimum and maximum identifiers, are shown. Data in (D) and (G) are reported as the mean ± SEM. All data are representative of two independent experiments. Statistics were calculated using the two-tailed, unpaired Student's t test with Welch's correction.
Transwell assays were utilized to evaluate the chemotactic properties of the cells. The results suggested that the chemotaxis of both RAW264.7(VNP) and PEMΦ(VNP) cells toward mouse melanoma tumor cells (B16F10) was not weakened, while RAW264.7(VNP) cells even showed an ∼1.6-fold improvement compared with that of RAW264.7 cells (Fig. 2C and D). The same trend was observed for other tumor cells, including LLC (mouse lung cancer cells) and MC38 (mouse colon cancer cells) cells (Supporting Information Fig. S10A). A variety of white blood cells that are used as tumor drug delivery vehicles, including macrophages, neutrophils, and T cells38, 39, 40, can cross a variety of barriers and reach tumor areas by sensing tumor-related chemokines or cytokines41. Here, we also found a significant increase in the transcript levels of key chemokine receptors, such as CCR5, CCR2 and CSF1-R42, in MΦ(VNP) cells (Supporting Information Fig. S11A), which also explained the enhanced tumor chemotaxis of MΦ(VNP) cells.
To evaluate the tumor cytotoxicity of MΦ(VNP) cells, a coculture experiment for tumor cells and MΦ(VNP) cells in vitro was performed. L929, a mouse fibroblast cell line, served as a negative control (Fig. 2E). The results revealed that both RAW264.7 and PEMΦ cells significantly inhibited tumor cell proliferation compared with L929 cells, while the suppression was more remarkable for macrophages after loading with VNP [RAW264.7(VNP) and PEMΦ(VNP-RFP) cells] (Fig. 2F). Similarly, VNP-loaded macrophages had enhanced cytotoxicity against LLC and MC38 cells (Fig. S10B). The inhibition of tumor cell proliferation in the L929 group was higher than that in the blank group, which may be due to nutritional competition between the two kinds of cells (Fig. 2F). The cytotoxic effects of macrophages without direct phagocytosis toward tumor cells could be ascribed to the significantly increased expression of proinflammatory mediators (including TNFα, iNOS and IL6)8 in RAW264.7(VNP) and PEMΦ(VNP) cells (Fig. S11B) and the release of reactive oxygen species (ROS) and NO, which could damage tumor cells (Fig. 2G and Fig. S11C)43,44. Notably, PEMΦ cells were more effective at phagocytosis, chemotaxis and tumor cell inhibition than RAW264.7 cells, which may be attributed to the strong activation of PEMΦ cells45. This was further confirmed by comparing the levels of ROS, a well-accepted marker of activated macrophages46 (Fig. 2G). Collectively, the phagocytosis, chemotaxis and tumor cytotoxicity of macrophages were not inhibited after VNP loading but were enhanced to some extent, which was beneficial for the antitumor activity of this bacteria-in-cell drug.
3.3. MΦ(VNP) cells increase tumor targeting and reduce the hepatosplenic toxicity of VNP
Typically, researchers have used intravenous or intraperitoneal injection to administer VNP, which is always accompanied by splenomegaly and hepatic inflammatory lesions. This is because some bacteria are off-target from tumors after administration8. Although intratumoral injection significantly reduces the hepatic and splenic accumulation of VNP10, this is undoubtedly not an optimal treatment for tumors. Herein, we loaded VNP into macrophages to camouflage the strain as a viable therapeutic strategy to avoid these side effects. According to the predetermined VNP-loading count by macrophages (see Section 3.1), RAW264.7(VNP) cells, hereinafter abbreviated as RAW(VNP) cells, and PEMΦ(VNP) cells were administered by tail vein at a dose of 2.5 × 105 and 1.0 × 105 cells per mouse, respectively, to ensure the appropriate and same total initial viable VNP dose for B16F10 tumor-bearing mice treatment (5 × 105 CFU per mouse of VNP47) (Fig. 3A).
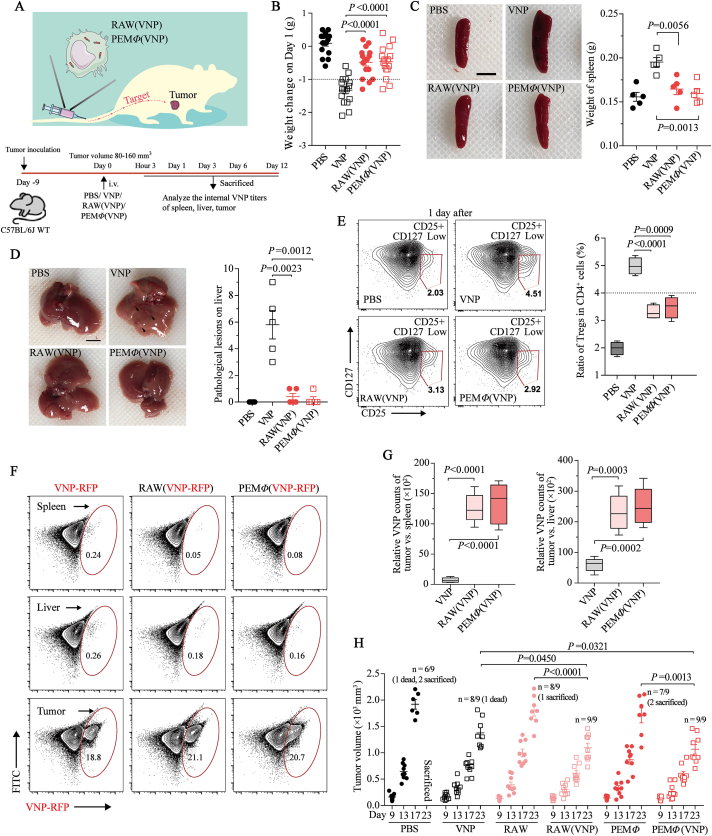
Figure 3.
MΦ(VNP) cells effectively alleviate the side effects of VNP and inhibit tumor growth. (A) B16F10 tumor-bearing C57BL/6J mice were administered VNP (5.0 × 105 CFU per mouse), RAW (VNP) (2.5 × 105 cells per mouse) and PEMΦ(VNP) (1.0 × 105 cells per mouse) cells via the tail vein. The biodistribution of VNP in different treatment groups was determined at specific time points. (B) Changes in body weight 1 day after different treatments (n = 16 mice per group). (C, D) Comparison of splenomegalia (C) and liver pathological lesions (black arrow) (D) 1 day after different treatments (n = 5 mice per group, scale bar = 5 mm). (E) Percentage of Tregs among CD4+ T cells from the peripheral blood 1 day after different treatments (n = 5 mice per group). (F) Detection of the VNP population in the spleen, liver and tumor by flow cytometry on Day 6 (n = 5 mice per group). (G) The tumor:spleen (left) and tumor:liver (right) ratios of bacterial CFU per Gram was calculated on the basis of recovered CFU from extracted organs on Day 6 (n = 5 mice per group). (H) Comparison of B16F10 tumor suppression in response to different treatments (n = 9 mice per group). “Dead” means the mice died naturally, and “sacrificed” means the mice were humanely sacrificed when the tumors reached an ethical limit. Data in (B), (C), (D) and (H) are reported as the mean ± SEM. Boxplot representations of the spot counts, with the median, interquartile range, and minimum and maximum identifiers, are shown. All data are representative of two independent experiments. Statistics were calculated using the two-tailed, unpaired Student's t test with Welch's correction.
To investigate whether MΦ(VNP) cells could lessen acute toxicity and the inflammatory response caused by VNP in mice, we first evaluated changes in body weight and the morphological characteristics of the liver and spleen at 24 h after different treatments. The results confirmed significant alleviation of weight loss, splenomegaly and hepatic inflammatory lesions induced by VNP with this strategy (Fig. 3B‒D). H&E staining and blood biochemistry analysis also revealed reductions in liver pathologic changes in the RAW (VNP) and PEMΦ(VNP) groups (Supporting Information Fig. S12). It has been reported that the content of Treg cells in the blood is related to the level of inflammation in vivo48. Thus, the CD25+ CD127low CD4+ Treg cell numbers in the blood after different treatments at 24 h were measured using flow cytometry. We found a significant decrease in Treg cell content after RAW(VNP) or PEMΦ(VNP) cell treatment compared with administration of VNP alone (Fig. 3E). All these results indicated that MΦ (VNP) cell treatment effectively avoided the side effects caused by a single administration of VNP.
The lower inflammatory response in vivo after MΦ(VNP) cell treatment is presumably because of the reduction in strain titer in normal organs. To validate this hypothesis, the colonization of VNP in vivo was determined by the plate count method. The results indicated that regardless of which kind of macrophage was used to load VNP, less VNP was observed in both the liver and spleen (Supporting Information Fig. S13B and S13C). The colonization of VNP in each tissue on Day 6 was selected as the representative. Flow cytometry showed VNP in the liver, spleen and tumor more intuitively (Fig. 3F). Within each group, we found no significant difference in the bacterial titers in the tumors (Fig. S13A). This is understandable because MΦ(VNP) cells need to gradually release the strains, causing a lower initial number of strains compared to administering VNP alone. Strains usually grow rapidly in tumors5,12. Although MΦ(VNP) cells continuously release intracellular VNP, achieving a rapid increase in the intratumoral bacterial titer later, it may have been difficult to obtain a significant difference with administering VNP alone. This made it difficult to evaluate the efficiency of strains reaching the tumor by comparing their titers. Therefore, tumor targeting, which refers to the ratio of the bacterial titer in the tumor to that in normal organs49, was used to assess the differences in the tumor-specific delivery effects of VNP between these groups. The tumor:spleen and tumor:liver ratios of bacterial CFU per Gram on Day 6 were increased by 7.8–48.8 and 2.6–12.2 times in the RAW(VNP) group, respectively, while those in the PEMΦ(VNP) group were increased by 6.9–51.7 and 2.1–12.9 times, respectively (Fig. 3G). These results highlight the increased tumor targeting of macrophage-mediated delivery of VNP. Moreover, the VNP titers in other organs, including the heart, lung and kidney, in the macrophage-loaded VNP group were also decreased compared to those in the single VNP group (Fig. S13D‒S13F), although the H&E staining of these organ sections did not reveal noticeable histological deficiency differences between these groups, suggesting the negligible treatment-induced toxicity of the cell preparations to these organs (Fig. S13G).
We hypothesized that the lower inflammatory reaction and side effects and higher tumor targeting of VNP from MΦ(VNP) cell treatment as bacterial immunogens are camouflaged by macrophages also protect the VNP from being quickly cleared by neutrophils and then load the strain into the tumor. We found that MΦ(VNP) cells exhibited great natural migration toward tumor cells in Transwell assays (Fig. 2C and D, and Fig. S10A). Furthermore, direct injection of the mixture of macrophages and VNP into mice, instead of loading, did not reduce the toxicities of VNP and failed to stabilize the body weights of mice or reduce the accumulation of VNP in the liver and spleen (Supporting Information Fig. S14). Comprehensive hematology showed that there were no significant changes in any of the indexes 1 day after MΦ(VNP) cell administration compared with PBS, although there was a tendency to lower neutrophil percentage than VNP (Supporting Information Fig. S15). These results all confirmed the reliability of this conjecture and the higher security of this delivery strategy.
The antitumor effects of different treatments were also evaluated, and we found that macrophages alone had comparatively effective antitumor abilities (Fig. 3H), which is easy to understand because we used activated M1-like macrophages that show a tumor-killing effect (Fig. 2). Compared with the PBS group, the VNP group showed significantly inhibited tumor progression, while the MΦ(VNP) group showed even better antitumor activity (Fig. 3H). These results indicated that this novel strategy not only avoided the side effects caused by the single administration of VNP but also effectively improved the therapeutic effect on tumors. Both RAW(VNP) and PEMΦ(VNP) cells showed lower toxicity and higher tumor repression than VNP, and we focused on PEMΦ(VNP) cells in subsequent studies due to their lower immunogenicity and higher translatability.
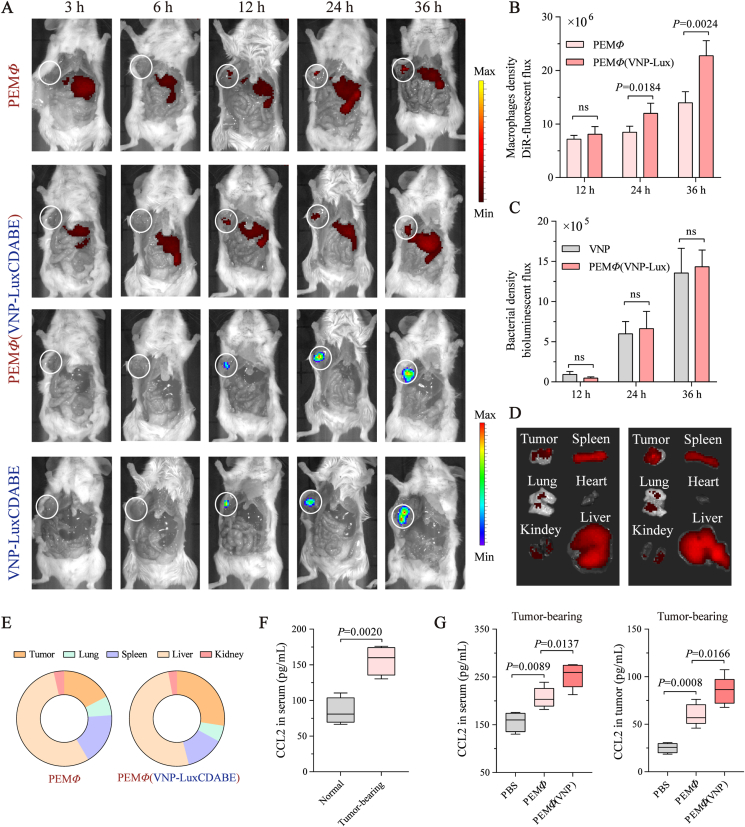
Macrophages are usually detected in the tumor area within 12 h after intravenous injection19,20. To indicate the dynamic change in tumor accumulation, the biodistribution of PEMΦ(VNP-LuxCDABE) cells at various time points was analyzed. Given the limited number of labeled cells, dissected mice bearing A20 (B lymphoma cells) subcutaneous xenograft tumors were monitored for better visibility. In vivo, both DIR-labeled PEMΦ and PEMΦ(VNP-LuxCDABE) cells were observed in A20 tumors at 12 h (Fig. 4A and B), and the released strains quickly multiplied in the tumor after 12 h, as visualized by bioluminescence imaging (Fig. 4A and C). The unprocessed VNP-LuxCDABE was used as a positive control. There was no significant difference in the bacterial bioluminescence intensity between the PEMΦ(VNP-LuxCDABE) cell group and VNP-LuxCDABE group of tumors at 24 and 36 h, which was consistent with our previous results (Fig. S13A). Chemokine secretion was analyzed by ELISA. Compared to non-tumor-bearing mice, serum CCL2, the most important chemokine for the recruitment of macrophages into tumor50, were significantly increased approximately 1.85-fold in A20 tumor-bearing mice (Fig. 4F) and 1.29-fold in B16F10 tumor-bearing mice (Supporting Information Fig. S16A). In addition, compared with PEMΦ cells, PEMΦ(VNP-LuxCDABE) cells achieved more efficient chemotaxis and enrichment in tumors (Fig. 4A, B, D and E). One hypothesis for this difference is that the released VNP strains after PEMΦ(VNP) administration promote the chemotaxis of macrophages by increasing cytokine/chemokine levels in tumors51. Compared with PEMΦ cell administration, the serum and tumor CCL2 levels of both A20 and B16F10 tumor-bearing mice were significantly increased after PEMΦ(VNP) cells administration at 36 h (Fig. 4G, Fig. S16B), further confirming our hypothesis. These results again suggest the effectiveness of macrophage-mediated tumor-targeted delivery of strains.
Figure 4.
VNP released by macrophages rapidly proliferates in tumors and feedback to accelerate the recruitment of MΦ (VNP) cells into tumors. (A) Tumor trafficking and biodistribution of DiR-labeled PEMΦ cells (1.0 × 105 cells per mouse) or PEMΦ(VNP-LuxCDABE) cells (1.0 × 105 cells per mouse) and VNP-LuxCDABE strains (5.0 × 105 CFU per mouse) in A20 tumor xenograft models were analyzed by live-animal imaging at different time points after intravenous administration. Mice were sacrificed and dissected along the abdomen for better visualization. (B, C) Changes in macrophage density DiR-fluorescent flux (B) and bacterial density bioluminescent flux (C) in tumors 12, 24 and 36 h after administration (n = 3 or 4 mice per group). (D) A20 tumor-bearing mice were killed, and organs/tumors were explanted for ex vivo imaging 36 h after a single injection of 1 × 105 PEMΦ or PEMΦ(VNP) cells (n = 4 mice per group). (E) Proportion of DiR-fluorescent flux in different organs/tumors in (D). (F) Detection of CCL2 concentration in peripheral blood of normal mice and A20 tumor-bearing mice (tumor size = 100 mm3, n = 4 mice per group). (G) Detection of CCL2 concentration in peripheral blood and tumor of A20 tumor-bearing mice (tumor size = 100 mm3) 36 h after administration (n = 4 or 5 mice per group). Data in (B) and (C) are reported as the mean ± SEM. Boxplot representations of the spot counts, with the median, interquartile range, and minimum and maximum identifiers, are shown. The experiment was performed once. Statistics were calculated using the two-tailed, unpaired Student's t test with Welch's correction.
3.4. MΦ(VNP) cells reverse CD8+ T cell dysfunction but upregulate PDL1 expression on the tumor cell surface
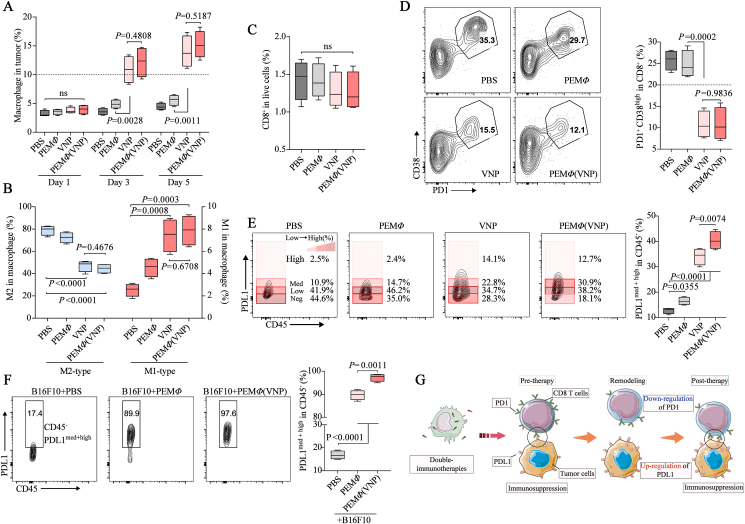
Changes in the tumor microenvironment after PEMΦ(VNP) cells treatment are worth exploring. Herein, we used macrophages as the Trojan horse of VNP for antitumor delivery and treatment, so we first examined the population of macrophages in the tumor with a flow cytometry assay. Compared with macrophages administered alone, both VNP and PEMΦ(VNP) cells recruited more macrophages after colonization in the tumor region, while there were no differences between these two groups (Fig. 5A). It has been reported that VNP can remodel macrophages from M2-like to M1-like through LPS stimulation52. Here, we also found that the number of tumor-infiltrating M2 macrophages decreased, whereas the number of M1-like macrophages increased in VNP and PEMΦ(VNP) cells on Day 3 (Fig. 5B and Supporting Information Fig. S17).
Figure 5.
MΦ(VNP) cells reverse CD8+ T cell dysfunction but upregulate PDL1 expression on the tumor cell surface. (A, B) Changes in the tumor-infiltrating macrophage population on Days 1, 3 and 5 (A) and phenotype (M1-like and M2-like) on Day 3 (B) after different treatments in the B16F10 tumor model (n = 4 mice per group). (C, D) The percentage of CD8+ T cells in tumors (C) and dysfunctional CD8+ T cells (PD1+ CD38high) among CD8+ T cells after different treatments in the B16F10 tumor model (D) (Day 3, n = 4 mice per group). (E) Proportion of PDL1 on the surface of tumor cells after different treatments on Day 3 (n = 4 mice per group). (F) Changes in PDL1 on the surface of B16F10 cells 12 h after coculture with PEMΦ and PEMΦ(VNP) cells at a ratio of 1:1 (n = 3; 50 μg/mL gentamicin was added to the medium). (G) Schematic diagram: PEMΦ (VNP) cells activate CD8+ T cells but upregulate PDL1 expression on the tumor cell surface and further immunosuppress CD8+ T cells. Boxplot representations of the spot counts, with the median, interquartile range, and minimum and maximum identifiers, are shown. All data are representative of two independent experiments. Statistics were calculated using the two-tailed, unpaired Student's t test with Welch's correction.
We next examined the population of CD8+ T cells within the tumor and found that VNP or PEMΦ(VNP) cells had no effect on the population of CD8+ T cells within the tumor compared with PBS treatment (Fig. 5C and Supporting Information Fig. S18). Remarkably, VNP or VNP-loaded macrophages decreased the proportion of PD1+ CD38High CD8+ T cells (Fig. 5D), which have been described as a population of dysfunctional cells that fail to respond to antigenic stimulation and do not exhibit effector functions53,54. This suggested that VNP could effectively reverse CD8+ T cell dysfunction. The reversal may be attributed to some inflammatory factors induced by VNP on dysfunctional CD8+ T cells10,51,54,55.
Ideally, there should be improved CD8+ T cell-mediated tumor killing after PEMΦ(VNP) cell treatment due to the reversal of dysfunctional CD8+ T cells. However, we found significantly increased expression of programmed cell death protein receptor 1 (PDL1) on the surface of tumor cells after the administration of VNP and PEMΦ(VNP) cells compared with that of PBS. The upregulation of PDL1 on the surface of tumor cells was also significant when macrophages were administered alone (Fig. 5E). It is important to note that PDL1 binds to PD1 on the surface of CD8+ T cells and inhibits the proliferation and cytotoxicity of CD8+ T cells56, ultimately impairing the antitumor efficacy of CD8+ T cells. This may help explain why VNP or VNP-loaded macrophages have a mediocre killing effect on tumors that was not much better than M1-like macrophage treatment alone (Fig. 3H). Similarly, the antitumor effect of PEMΦ(VNP) cells was not much more markedly improved than that of single VNP treatment, which could also be attributed to the greater content of PDL1 on the surface of tumor cells in the PEMΦ(VNP) group (Fig. 5E). In vitro experiments further verified that PEMΦ and PEMΦ(VNP) cell treatments upregulated PDL1 on the surface of B16F10 cells, while the upregulation was more obvious in the PEMΦ(VNP) group (Fig. 5F). This finding suggested that macrophages and VNP synergistically promote the upregulation of PDL1 on the tumor cell surface, which may be attributed to the increased inflammatory cytokines secreted by intratumoral macrophages after activation57 (Fig. S11B). Moreover, melanoma cells secrete PDL1-abundant exosomes to myeloid cells58 and then achieve stronger immunosuppression against CD8+ T cells through these myeloid cells59,60. Here, we also found an increase in PDL1 on the surface of tumor-related myeloid cells of the VNP and PEMΦ(VNP) groups (Supporting Information Fig. S19), which could further limit the effectiveness of bacteria in treating tumors.
Overall, PEMΦ(VNP) cells enhanced the fraction of M1-like macrophages and reversed dysfunctional CD8+ T cells in the tumor area, which exerted potentially stronger antitumor activity. However, the upregulation of PDL1 on the surface of tumor cells after PEMΦ(VNP) cell treatment inhibited the cytotoxicity of CD8+ T cells with PD1/PDL1 blockade. The schematic diagram in Fig. 5G shows the possible changes and interactions between CD8+ T cells and tumor cells during tumor therapy with VNP-loaded macrophages.
3.5. MΦ(VNP-PD1nb) cells potently and durably inhibit melanoma progression
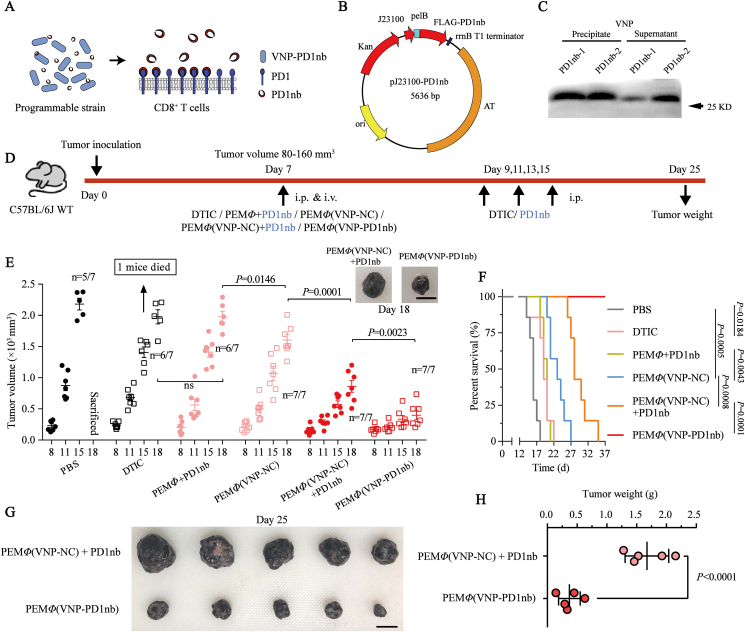
Since MΦ(VNP) cell treatments upregulate PDL1 on the surface of tumor cells, which could bind to PD1 on the surface of CD8+ T cells and inhibit their cytotoxicity, it can be hypothesized that this novel dual immunotherapy [MΦ(VNP) cell treatment] will have a significantly improved antitumor effect after combination with PD1/PDL1 blockade, especially the blockade of PD1 considering its “Buckets effect” (Fig. 6A and Supporting Information Fig. S20). To test this conjecture, we first constructed and obtained a variety of anti-PD1 nanoantibodies, including VHH1 and VHH2 (Fig. S21A‒S21F), and the affinity constants of nanobodies for binding to PD-1 were determined by ELISA. The results showed that VHH1 has better neutralization activity (Kaff = 0.82 × 107 L/mol) than VHH2 (Kaff = 1.54 × 107 L/mol) (Fig. S21G). Thus, VHH1 (subsequently called PD1nb) was used in subsequent experiments. Considering the two disadvantages of PD1 blockade therapy in clinical use, including off-target effects and the need for continuous administration, the expression of anti-PD1 antibody by programmed VNP may be a better choice. Thus, the VNP-PD1nb strain, which could stably express and secrete PD1nb, was constructed (Fig. 6B), and the VNP strain transferred with empty vector plasmids was used as a control. Western blotting verified the high expression and secretion of PD1nb by the strain in vitro (Fig. 6C).
Figure 6.
MΦ(VNP-PD1nb) cell treatment achieved significant tumor inhibition in a B16F10 tumor-bearing mouse model. (A) Engineered Salmonella typhimurium VNP20009, with macrophage-mediated tumor-targeted delivery, constitutively expresses and secretes anti-PD1 nanoantibodies that bind to PD1 on the CD8+ T cell surface in the tumor. (B) Schematic diagram of the pJ23100-PD1nb plasmid. The flag tag was inserted into the N-terminus of the protein to facilitate subsequent detection, the pelB signal peptide realized the secretion of the expressed PD1nb, and the AT element prevented plasmid loss. (C) Immunoblot analysis to check the bacterial expression and secretion of PD1nb in vitro. Samples were separated into precipitates of total strains and supernatants of culture medium fractions. (D) Experimental scheme of the antitumor effect in the six different groups. DTIC (daphnane diterpenes, 80 mg/kg) and PD1nb (5 mg/kg) were injected intraperitoneally four times every other day, and cells (1.0 × 105 per mouse), including PEMΦ, PEMΦ(VNP-NC) and PEMΦ(VNP-PD1nb) cells, were injected through the caudal vein only once. (E) Tumor growth profiles of B16F10 tumor-bearing mice after the different treatments (n = 7 mice per group, scale bar = 10 mm). (F) Survival curve of mice treated as described in (D). The survival of the mice was closely monitored several times per day, and the mice were killed when they reached a humane endpoint. (G, H) Tumors were photographed (G) and weighed (H) 25 days after B16F10 cell inoculation to compare the differences in tumor size between the PEMΦ(VNP-NC) + PD1nb and PEMΦ(VNP-PD1nb) groups (n = 5 mice per group, scale bar = 10 mm). “Dead” means the mice died naturally, and “sacrificed” means the mice were humanely sacrificed when the tumors reached an ethical limit. Data in (E) and (H) are reported as the mean ± SEM. All data are representative of two independent experiments. Statistics were calculated using the two-tailed, unpaired Student's t test with Welch's correction in (E) and (H) and the Mantel–Cox test in (F).
It is important to note that macrophages loaded more VNP-PD1nb strains than VNP-NC strains during the same time (Supporting Information Fig. S22), and the number of intracellular VNP-PD1nb strains in PEMΦ at 30 min, which reached a level similar to that of VNP-NC at 60 min (Fig. S22C), was chosen as the coculture time in the subsequent experiments. This enhanced phagocytosis of macrophages to VNP-PD1nb strains is consistent with observations that anti-PD1 antibodies effectively improve macrophage phagocytosis61. To assess whether PEMΦ(VNP-PD1nb) cells effectively release intracellular strains and whether the released VNP-PD1nb strain can still express and secrete PD1nb, we collected the culture supernatant of PEMΦ(VNP-PD1nb) cells at different time points and counted the number of strains and PD1nb content in the supernatant (Supporting Information Fig. S23A). The results revealed that the VNP-PD1nb strains were continuously released from macrophages and multiplied over time (Fig. S23B). Western blotting confirmed that the VNP-PD1nb strains, which were released from PEMΦ(VNP-PD1nb) cells, still effectively expressed and secreted PD1nb (Fig. S23C). Moreover, increasing PD1nb content secreted by the strain was detected over time in the culture supernatant because of the higher amount of strains (Fig. S23C and S23D). It is not difficult to hypothesize that the PD1nb released by the engineered VNP strains will further achieve antitumor immunotherapy through the well-known PD1/PDL1 immune blockade.
We confirmed that the macrophage-mediated dual-targeted delivery of VNP [MΦ(VNP)] to tumors significantly improves tumor targeting of the strains while alleviating the strains' side effects and ultimately enhancing the tumor treatment effect (Figure 3, Figure 4). Encouragingly, macrophage-mediated tumor-targeted delivery of VNP-PD1nb [MΦ(VNP-PD1nb)] also demonstrated fewer side effects and stronger therapeutic effects than administration of VNP-PD1nb strains (Supporting Information Fig. S24). This effectively verifies the universal applicability of this delivery strategy. To further examine the effect of PEMΦ(VNP-PD1nb) cell treatment, prepared PEMΦ(VNP-NC) and PEMΦ(VNP-PD1nb) cells (1.0 × 105 per mouse) were administered via the tail vein to B16F10 tumor-bearing mice. Daphnane diterpenes (DTIC), a recognized chemotherapeutic agent for the treatment of melanoma62, were used as a positive control (Fig. 6D). The results showed that both PEMΦ(VNP-NC) and PEMΦ(VNP-PD1nb) cells achieved superior therapeutic outcomes, while PEMΦ(VNP-PD1nb) cells achieved better antitumor effects than PEMΦ(VNP-NC) cells (Fig. 6E). Melanoma often has a high metastatic capacity because of its high degree of malignancy62. Encouragingly, both PEMΦ(VNP-NC) and PEMΦ(VNP-PD1nb) cells suppressed the lung metastasis of melanoma, while mice with PEMΦ(VNP-PD1nb) cells showed fewer lung metastases (Supporting Information Fig. S25). Since engineered bacteria could express and secrete proteins as long as they were alive, we hypothesized that PEMΦ(VNP-PD1nb) cells would inevitably achieve a more significant and persistent antitumor effect than the simple mixed treatments [PEMΦ(VNP-NC) + PD1nb]. Thus, we also investigated the therapeutic effect of treatment with PEMΦ(VNP-NC) cells mixed with exogenous PD1nb by injection (5 mg/kg, which is consistent with the typical dose of PD-1 monoclonal antibodies63). Compared with the mixed PEMΦ + PD1nb group, both the PEMΦ(VNP-NC) + PD1nb and PEMΦ(VNP-PD1nb) groups had significantly inhibited melanoma progression, which once again showed the high efficiency of VNP as a tumor immune activator (Fig. 6E). Impressively, PEMΦ(VNP-PD1nb) cell treatment exhibited a stronger and durable antitumor effect than the simple mixture of PEMΦ(VNP-NC) cells and PD1nb treatment, especially when continuous injection of PD1nb was stopped on Day 15 (Fig. 6F‒H). After treatment with PEMΦ(VNP-PD1nb) cells, there was no mortality, and all mice were alive at the end of the study period, while all of the mice treated with PEMΦ(VNP-NC) + PD1nb died within 35 days (Fig. 6F). These results suggested that the continuous production and secretion of PD1nb by engineered VNP in tumors was a more convenient and long-lasting treatment strategy. Moreover, endogenously and continuously expressed protein with strains may enable higher intratumoral PD1nb doses, which may also be one of the reasons for its better therapeutic effect than systemic injection of PD1nb.
3.6. MΦ(VNP-PD1nb) cells inhibit melanoma progression by activating the tumor microenvironment and inhibiting tumor cell proliferation
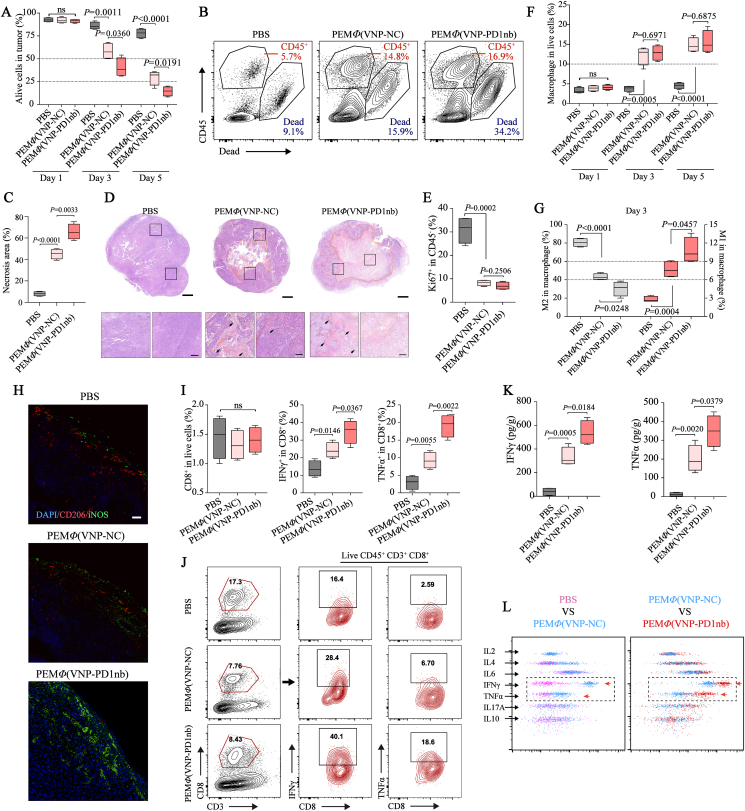
Finally, to investigate the mechanism of the improved therapeutic effect of PEMΦ(VNP-PD1nb) cell treatment, we evaluated the changes in the tumor immune microenvironment following different treatments, including PBS, PEMΦ(VNP-NC) and PEMΦ(VNP-PD1nb) cells, in melanoma. We first detected tumor necrosis. A flow cytometry assay showed that both PEMΦ(VNP-NC) and PEMΦ(VNP-PD1nb) cells caused significant intratumoral necrosis, while PEMΦ(VNP-PD1nb) cells caused more extensive necrosis than the other cells (Fig. 7A and B). H&E staining of the tumor area, as further evidence, more intuitively showed the difference in the extent of tumor necrosis after the three treatments, and the tumor necrosis areas after PEMΦ(VNP-PD1nb) cell treatment exceeded 60% within 3 days (Fig. 7C and D). Correspondingly, flow cytometry analyses revealed that both PEMΦ(VNP-PD1nb) and PEMΦ(VNP-NC) cell treatment downregulated Ki67 (a recognized cell proliferation marker) expression in tumor cells on Day 3, indicating effective inhibition of tumor cell proliferation (Fig. 7E).
Figure 7.
MΦ(VNP-PD1nb) cells accelerate tumor cell necrosis and activate TAMs and CD8+ T cells. (A, B) Detection of alive cells in the tumor on Days 1,3 and 5 by flow cytometry in (A) (n = 4 mice per group), and flow cytometry plots of representative data are shown in (B). (C, D) Comparison of the tumor necrosis proportion in different treatment groups by H&E staining on Day 3 in (C), and representative H&E images are shown in (D). Scale bars, 1 mm (complete tumor sections) and 200 μm (enlarged tumor sections). (E) The proliferation of tumor cells (CD45-) was quantified by Ki67 staining and flow cytometry. (F, G) Changes in the tumor-infiltrating macrophage population on Days 1, 3 and 5 in (F) and phenotype (M1-like and M2-like) at Day 3 in (G) after treatment with PBS, PEMΦ(VNP-NC) or PEMΦ(VNP-PD1nb) cells in the B16F10 tumor model (n = 4 mice per group). (H) Representative immunofluorescence staining of B16F10 tumor cross sections showing cell nuclei (DAPI, blue), CD206 (marker of M2-like macrophages, red) and iNOS (marker of M1-like macrophages, green) of (G) (Scale bar = 50 μm). (I, J) Profile of tumor-infiltrating CD8+ T cell and cytokine-producing CD8+ T cell subsets on Day 3 in the B16F10 tumor model (n = 4 mice per group) (I), and representative flow cytometry plots show the method of acquisition of frequency values for each T cell subset (J). (K, L) Detection of IFNγ and TNFα in peripheral blood 3 days after treatment in the B16F10 tumor model by CBA assay in (K), and flow cytometry plots of representative CBA data are shown in (L). Boxplot representations of the spot counts, with the median, interquartile range, and minimum and maximum identifiers, are shown. All data are representative of two independent experiments. Statistics were calculated using the two-tailed, unpaired Student's t test with Welch's correction.
Next, we detected changes in the population and phenotype of intratumoral macrophages. The results showed significantly more infiltration of macrophages in the tumor after PEMΦ(VNP-NC) and PEMΦ(VNP-PD1nb) cell treatment than PBS treatment (Fig. 7F), which was consistent with the increased number of infiltrating lymphocytes in the tumor (Fig. 7B), yet there were no differences between the two cell treatments. We then assessed the same tumor samples using flow-based immunophenotyping for CD206 and CD86 expression to adjudicate a macrophage phenotype on Day 3. The results suggested that both cell-treated groups significantly reduced the proportion of intratumoral M2-like macrophages and increased the proportion of M1-like macrophages, while PEMΦ(VNP-PD1nb) cell treatment showed higher efficiencies (Fig. 7G), suggesting a higher increase in the tumor-killing and antigen-presentation ability of tumor macrophages. Immunofluorescence also confirmed a significant increase in intratumoral M1-like macrophages in the PEMΦ(VNP-PD1nb) cell treatment compared with the PEMΦ(VNP-NC) cell treatment (Fig. 7H).
Cytotoxic CD8+ T cells always play a role in the promotion of oncolytic therapy, especially in the treatment of PD1/PDL1 blockade27,28,64. Thus, the cytotoxic CD8+ T cells in tumors were detected on Day 3 after different treatments as a representative time. Flow cytometry analyses revealed that there was no significant change in the total population of CD8+ T cells in the tumor among the three treatments, but markedly elevated intratumoral IFNγ+ CD8+ T cells and TNFα+ CD8+ T cells were observed after cell treatments (Fig. 7I and J, and Supporting Information Fig. S26). More activated CD8+ T cells were observed in tumors treated with PEMΦ(VNP-PD1nb) cells, which could also partially account for the stronger antitumor effect of PEMΦ(VNP-PD1nb) cells compared to PEMΦ(VNP-NC) cells.
Proinflammatory cytokines, such as IFN-γ and TNF-α, secreted by immune cells can directly kill tumor cells and induce strong tumor-specific immune responses65. Thus, the sera from B16F10 tumor-bearing mice after different treatments on Day 3 were collected to measure the blood cytokine levels using a CBA kit. The results showed that the amounts of IFN-γ and TNF-α induced by PEMΦ(VNP-NC) and PEMΦ(VNP-PD1nb) cell treatment were significantly higher than those induced by PBS treatment. More IFN-γ and TNF-α were tested in PEMΦ(VNP-PD1nb) cell treatment (Fig. 7K and L), suggesting that PEMΦ(VNP-PD1nb) cell treatment enhanced immune activation and therapeutic efficacy. Some other proinflammatory cytokines, including IL2, IL6 and IL17A, and anti-inflammatory cytokines, including IL4 and IL10, did not differ among the three groups (Fig. 7L). Collectively, treatment with PEMΦ(VNP-PD1nb) cells efficiently inhibited tumor cell proliferation, culminated in tumor necroptosis and activated the tumor microenvironment, including remodeling in TAMs and activation of CD8+ T cells, and finally prevented tumor progression.
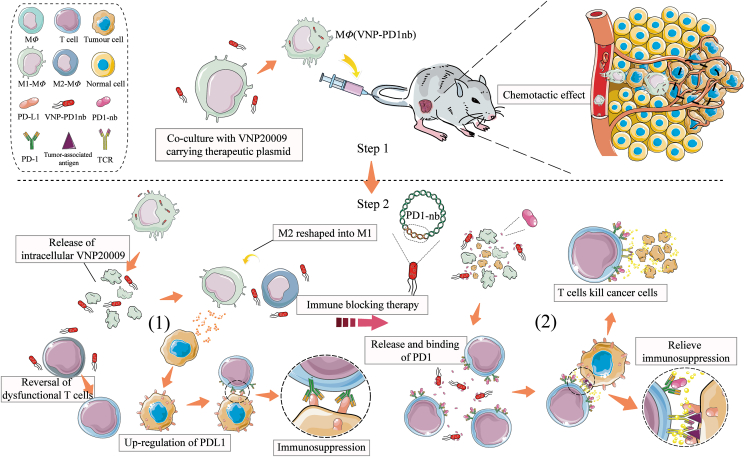
In summary, we established a combination therapy with engineered stealth bacteria camouflaged by macrophages (Fig. 8). The three treatments, including macrophages, VNP strains and PD1nb, complement each other to form a unified system. With natural tumor-targeted macrophages as Trojan horses deliver, bacteria exhibit (1) a low inflammatory response and side effects; (2) low accumulation in normal organs; and (3) unchanged antitumor bioactivities. With continuously expressed and secreted PD1nb, engineered bacterial treatment exhibits (1) high immune activation, (2) significant tumor regression, and (3) a stable and durable antitumor effect. Given these unique advantages, we anticipate promising biomedical applications of this strategy in tumoral treatment.
Figure 8.
MΦ(VNP-PD1nb) cell treatment for tumor immunotherapy. Step 1: MΦ(VNP-PD1nb) cells act as Trojan horse engineered strains and target the tumor. Step 2: (1) After being released in the tumor, VNP-PD1nb, which is loaded in MΦ(VNP-PD1nb) cells, activates intratumoral immunity and reverses dysfunctional CD8+ T cells while upregulating PDL1 in tumor cells, which results in immunosuppression. (2) PD1nb, which is released by VNP-PD1nb, can block the PD-1/PD-L1 interaction between CD8+ T cells and tumor cells, thereby inhibiting immunosuppression and enhancing the tumor killing effect.
4. Discussion
Specific tumor targeting and highly potent tumor suppression are two challenging targets using VNP and other oncolytic bacteria for tumor therapy. To attain improved tumor targeting, we first reported the use of macrophage-mediated tumor-targeted delivery of VNP in treating tumor-bearing mice. Although modification of attenuated Salmonella49 or the tumor microenvironment23 could also improve the tumor targeting of the bacteria in mice, all these improvements require the arrival of the bacteria at the tumor. A high titer of VNP was detected in the tumors of patients after intratumoral injection66, and unattenuated Salmonella could also effectively infect the tumor area67, which suggested that both Salmonella and attenuated VNP could colonize tumors and exert antitumor effects68,69. In a phase I clinical trial of intravenous treatment for sarcoma, however, VNP did not perform well in terms of effectiveness9. The investigators did not detect VNP in the tumor, and there was no evidence of colonization. Thus, the accessibility of VNP in human tumors is urgently needed to avoid excessive stimulation of the immune system by bacterial immune antigens; otherwise, the bacteria will be quickly eliminated. The use of biofilms to wrap bacteria is an effective way to avoid early exposure of the bacteria in the body12. Another effective strategy, as reported here, is to conceal VNP in macrophages to avoid premature exposure of the bacteria. This is especially valuable since the specific antigen modification of macrophages50 can further improve the tumor targeting ability of macrophages, which is worth further exploration.
One of the major challenges of cell-mediated drug delivery is the need to inject large number of cells (107‒108 cells in mice)39, which is accompanied by sophisticated operations and high costs. The novel therapeutic strategy described here only requires a dose of 105 MΦ(VNP) cells to effectively treat murine tumors, which is only 1/100–1/1000 of that required by traditional cell therapy. In fact, even if only a small amount of VNP is released into the tumor region by macrophages, a therapeutic effect can be achieved by the continuous proliferation of strains. However, the acquisition and culture of macrophages in the early stage still increases the cost of this therapy. Alternatively, low-cost allogeneic macrophages can be used to build off-the-shelf vectors that perform targeted release functions, which can then be recognized and eliminated by the host immune system. The approach proposed here of using macrophages loaded with VNP for targeted tumor therapy is a modular approach in which macrophages can also be loaded with different recombinant oncolytic bacteria. Naturally, different types of strains may have different cell-loading efficiencies; for example, during the same processing time, macrophages loaded more VNP-PD1nb strains than VNP (Fig. S22). This study demonstrates the effectiveness and universality of macrophages as VNP delivery vectors, but other kinds of cells may also be used for this type of therapy39,40,70.
For tumor suppression, we constructed an engineered VNP strain (VNP-PD1nb) that stably expressed and secreted anti-PD1 nanoantibodies and loaded the strain into macrophages for tumor treatment. This novel strategy achieved a more efficient and lasting effect than the simple combination of PD1nb and VNP-loaded macrophages for antitumor therapy. This strategy also provides a potential solution to the dilemma of a low response rate to PD1/PDL1 blockade in cancer patients27. After immunotherapy with PD1/PDL1 blockade, a small percentage of responders suffer from a series of adverse reactions due to the accumulation of drugs in normal organs or tissues71. The bispecific delivery of macrophages and VNP theoretically blocks the side effects of anti-PD1 treatment by concentrating the production of PD1nb within the tumor. The expression and secretion of PD1nb by trace levels of VNP-PD1nb in normal organs, however, may still lead to misregulated immune activation. A feasible approach would be to induce the expression of PD1nb in a controlled manner, such as hypoxia induction31 or direct regulation of bacterial survival7. The most exciting application of immunotherapy is the ability to treat tumor metastasis7 and inhibit recurrence, especially when used in combination with other therapies, including chemotherapy, photothermal therapy, photodynamic therapy, etc.72. The combined use of macrophages, attenuated Salmonella and anti-PD1 nanoantibodies in this paper also achieved significant anti-metastatic effects (Fig. S25). Moreover, due to the strong loading capacity of macrophages30 and the editable and carrier properties of VNP31, this novel immunotherapy has great room for improvement to achieve stronger effects in antitumor metastasis, suppress or even eliminate tumors, and inhibit recurrence. These possibilities warrant further investigation.
In conclusion, we programmed triple immunotherapies in one drug: macrophage-based immunotherapy, microbe-based immunotherapy and anti-PD1 immunotherapy using a Trojan horse strategy and delayed release. This study provides a promising future for exploring convergent immunotherapy approaches to address monotherapy challenges.
5. Conclusions
In this study, we designed, characterized and applied a novel cancer immunotherapy system. Briefly, this system allowed attenuated S. typhimurium VNP20009 latent in macrophages to precisely unload inside the tumor and then secrete anti-PD1 nanoantibodies. We first showed that macrophage-mediated tumor-targeted delivery of VNP20009 alleviated side effects induced by the strain and effectively suppressed melanoma in mice. Furthermore, there was a marked decrease in dysfunctional CD8+ T cells in tumors after this systemic treatment, which also suggests a synergistic response to PD1/PDL1 blockade. Thus, macrophages loaded with engineered VNP20009, which could express and secrete anti-PD1 nanoantibodies, were prepared and used to treat xenograft mouse melanoma models. The combination of the three immunotherapies (macrophages, VNP20009, PD1nb) achieved much more significant and durable tumor inhibition than simple mixed treatments by activating the tumor microenvironment, including increased immune cell infiltration, TAM remodeling to an M1-like phenotype and prominent activation of CD8+ T cells.
Acknowledgments
This study was supported in part by grants from the National Natural Sciences Foundation of China (82130106), Jiangsu Provincial Department of Science and Technology (BK20192005, China), Changzhou Bureau of Science and Technology (CJ20210024, CZ20210010, China) and Jiangsu TargetPharma Laboratories Inc., China. The PDL1nb was kindly provided by Dr. Shufeng Li (Southeast University) which was also supported by Jiangsu TargetPharma Laboratories Inc.
Footnotes
Peer review under responsibility of Chinese Pharmaceutical Association and Institute of Materia Medica, Chinese Academy of Medical Sciences.
Supporting data to this article can be found online at https://doi.org/10.1016/j.apsb.2022.05.006.
Author contributions
Zichun Hua supervised and conceived the project. Leyang Wu designed the experiments, Leyang Wu, Lin Li, Shufeng Li, Lina Liu, Wenjie Xin and Xingpeng Yin performed the experiments and analyzed the data. Leyang Wu, Lin Li and Chenyang Li wrote and Zichun Hua modified the manuscript. Xuebo Xu and Feifei Bao contributed to the discussion and provided relevant advice. All authors discussed the results and reviewed the manuscript
Conflicts of interest
The authors have applied for patents related to this study.
Appendix A. Supplementary data
The following is the Supplementary data to this article:
References
- 1.Coley W.B. The treatment of malignant tumors by repeated inoculations of erysipelas. With a report of ten original cases. Clin Orthop Relat Res. 1991;262:3–11. 1893. [PubMed] [Google Scholar]
- 2.Gurbatri C.R., Lia I., Vincent R., Coker C., Castro S., Treuting P.M., et al. Engineered probiotics for local tumor delivery of checkpoint blockade nanobodies. Sci Transl Med. 2020;12 doi: 10.1126/scitranslmed.aax0876. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Schwabe R.F., Jobin C. The microbiome and cancer. Nat Rev Cancer. 2013;13:800–812. doi: 10.1038/nrc3610. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Zhou S., Gravekamp C., Bermudes D., Liu K. Tumour-targeting bacteria engineered to fight cancer. Nat Rev Cancer. 2018;18:727–743. doi: 10.1038/s41568-018-0070-z. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Canale F.P., Basso C., Antonini G., Perotti M., Li N., Sokolovska A., et al. Metabolic modulation of tumours with engineered bacteria for immunotherapy. Nature. 2021;598:662–666. doi: 10.1038/s41586-021-04003-2. [DOI] [PubMed] [Google Scholar]
- 6.Xu X., Hegazy W.A.H., Guo L., Gao X., Courtney A.N., Kurbanov S., et al. Effective cancer vaccine platform based on attenuated Salmonella and a type III secretion system. Cancer Res. 2014;74:6260–6270. doi: 10.1158/0008-5472.CAN-14-1169. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Chowdhury S., Castro S., Coker C., Hinchliffe T.E., Arpaia N., Danino T. Programmable bacteria induce durable tumor regression and systemic antitumor immunity. Nat Med. 2019;25:1057–1063. doi: 10.1038/s41591-019-0498-z. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Clairmont C., Lee K.C., Pike J., Ittensohn M., Low K.B., Pawelek J., et al. Biodistribution and genetic stability of the novel antitumor agent VNP20009, a genetically modified strain of. Salmonella typhimurium. J Infect Dis. 2000;181:1996–2002. doi: 10.1086/315497. [DOI] [PubMed] [Google Scholar]
- 9.Toso J.F., Gill V.J., Hwu P., Marincola F.M., Restifo N.P., Schwartzentruber D.J., et al. Phase I study of the intravenous administration of attenuated Salmonella typhimurium to patients with metastatic melanoma. J Clin Oncol. 2002;20:142–152. doi: 10.1200/JCO.2002.20.1.142. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Chen G., Wei D.P., Jia L.J., Tang B., Shu L., Zhang K., et al. Oral delivery of tumor-targeting Salmonella exhibits promising therapeutic efficacy and low toxicity. Cancer Sci. 2009;100:2437–2443. doi: 10.1111/j.1349-7006.2009.01337.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Zheng J.H., Nguyen V.H., Jiang S.N., Park S.H., Tan W., Hong S.H., et al. Two-step enhanced cancer immunotherapy with engineered Salmonella typhimurium secreting heterologous flagellin. Sci Transl Med. 2017;9:eaak9537. doi: 10.1126/scitranslmed.aak9537. [DOI] [PubMed] [Google Scholar]
- 12.Cao Z., Cheng S., Wang X., Pang Y., Liu J. Camouflaging bacteria by wrapping with cell membranes. Nat Commun. 2019;10:3452. doi: 10.1038/s41467-019-11390-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Iwasaki A., Medzhitov R. Control of adaptive immunity by the innate immune system. Nat Immunol. 2015;16:343–353. doi: 10.1038/ni.3123. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Rosenberg G., Yehezkel D., Hoffman D., Mattioli C.C., Fremder M., Ben-Arosh H., et al. Host succinate is an activation signal for Salmonella virulence during intracellular infection. Science. 2021;371:400–405. doi: 10.1126/science.aba8026. [DOI] [PubMed] [Google Scholar]
- 15.Rao S., Xu T., Xia Y., Zhang H. Salmonella and S. aureus escape from the clearance of macrophages via controlling TFEB. Front Microbiol. 2020;11:573844. doi: 10.3389/fmicb.2020.573844. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Helaine S., Cheverton A.M., Watson K.G., Faure L.M., Matthews S.A., Holden D.W. Internalization of Salmonella by macrophages induces formation of nonreplicating persisters. Science. 2014;343:204–208. doi: 10.1126/science.1244705. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Sano G-i, Takada Y., Goto S., Maruyama K., Shindo Y., Oka K., et al. Flagella facilitate escape of Salmonella from oncotic macrophages. J Bacteriol. 2007;189:8224–8232. doi: 10.1128/JB.00898-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Wang M., Qazi I.H., Wang L., Zhou G., Han H. Salmonella virulence and immune escape. Microorganisms. 2020;8:407. doi: 10.3390/microorganisms8030407. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Aalipour A., Chuang H.Y., Murty S., D'Souza A.L., Park S.M., Gulati G.S., et al. Engineered immune cells as highly sensitive cancer diagnostics. Nat Biotechnol. 2019;37:531–539. doi: 10.1038/s41587-019-0064-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Li C.X., Zhang Y., Dong X., Zhang L., Liu M.D., Li B., et al. Artificially reprogrammed macrophages as tumor-tropic immunosuppression-resistant biologics to realize therapeutics production and immune activation. Adv Mater. 2019;31 doi: 10.1002/adma.201807211. [DOI] [PubMed] [Google Scholar]
- 21.Shields C.W., Evans M.A., Wang L.L.W., Baugh N., Iyer S., Wu D., et al. Cellular backpacks for macrophage immunotherapy. Sci Adv. 2020;6:eaaz6579. doi: 10.1126/sciadv.aaz6579. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Yang Q., Guo N., Zhou Y., Chen J., Wei Q., Han M. The role of tumor-associated macrophages (TAMs) in tumor progression and relevant advance in targeted therapy. Acta Pharm Sin B. 2020;10:2156–2170. doi: 10.1016/j.apsb.2020.04.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Chen J., Qiao Y., Tang B., Chen G., Liu X., Yang B., et al. Modulation of Salmonella tumor-colonization and intratumoral anti-angiogenesis by triptolide and its mechanism. Theranostics. 2017;7:2250–2260. doi: 10.7150/thno.18816. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Cheng X., Zhang X., Zhou Y., Zhang C., Hua Z.C. A Salmonella typhimurium mutant strain capable of RNAi delivery: higher tumor-targeting and lower toxicity. Cancer Biol Ther. 2014;15:1068–1076. doi: 10.4161/cbt.29185. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Ricklefs F.L., Alayo Q., Krenzlin H., Mahmoud A.B., Speranza M.C., Nakashima H., et al. Immune evasion mediated by PD-L1 on glioblastoma-derived extracellular vesicles. Sci Adv. 2018;4:eaar2766. doi: 10.1126/sciadv.aar2766. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Topalian S.L., Taube J.M., Pardoll D.M. Neoadjuvant checkpoint blockade for cancer immunotherapy. Science. 2020;367 doi: 10.1126/science.aax0182. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Ribas A., Wolchok J.D. Cancer immunotherapy using checkpoint blockade. Science. 2018;359:1350–1355. doi: 10.1126/science.aar4060. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Miller B.C., Sen D.R., Al Abosy R., Bi K., Virkud Y.V., LaFleur M.W., et al. Subsets of exhausted CD8+ T cells differentially mediate tumor control and respond to checkpoint blockade. Nat Immunol. 2019;20:326–336. doi: 10.1038/s41590-019-0312-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Herbst R.S., Giaccone G., de Marinis F., Reinmuth N., Vergnenegre A., Barrios C.H., et al. Atezolizumab for first-line treatment of PD-L1-selected patients with NSCLC. N Engl J Med. 2020;383:1328–1339. doi: 10.1056/NEJMoa1917346. [DOI] [PubMed] [Google Scholar]
- 30.Choi J., Kim H.Y., Ju E.J., Jung J., Park J., Chung H.K., et al. Use of macrophages to deliver therapeutic and imaging contrast agents to tumors. Biomaterials. 2012;33:4195–4203. doi: 10.1016/j.biomaterials.2012.02.022. [DOI] [PubMed] [Google Scholar]
- 31.Chen J., Wei D., Zhuang H., Qiao Y., Tang B., Zhang X., et al. Proteomic screening of anaerobically regulated promoters from Salmonella and its antitumor applications. Mol Cell Proteomics. 2011;10:M111. doi: 10.1074/mcp.M111.009399. 009399. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Carryn S., Van Bambeke F., Mingeot-Leclercq M.P., Tulkens P.M. Comparative intracellular (THP-1 macrophage) and extracellular activities of beta-lactams, azithromycin, gentamicin, and fluoroquinolones against Listeria monocytogenes at clinically relevant concentrations. Antimicrob Agents Chemother. 2002;46:2095–2103. doi: 10.1128/AAC.46.7.2095-2103.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Li S., Zhang W., Jiang K., Shan H., Shi M., Chen B., et al. Nanobody against the E7 oncoprotein of human papillomavirus 16. Mol Immunol. 2019;109:12–19. doi: 10.1016/j.molimm.2019.02.022. [DOI] [PubMed] [Google Scholar]
- 34.Fidler I.J. Inhibition of pulmonary metastasis by intravenous injection of specifically activated macrophages. Cancer Res. 1974;34:1074–1078. [PubMed] [Google Scholar]
- 35.Masters T.A., Pontes B., Viasnoff V., Li Y., Gauthier N.C. Plasma membrane tension orchestrates membrane trafficking, cytoskeletal remodeling, and biochemical signaling during phagocytosis. Proc Natl Acad Sci U S A. 2013;110:11875–11880. doi: 10.1073/pnas.1301766110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Garmendia J., Beuzon C.R., Ruiz-Albert J., Holden D.W. The roles of SsrA-SsrB and OmpR-EnvZ in the regulation of genes encoding the Salmonella typhimurium SPI-2 type III secretion system. Microbiology (Read) 2003;149:2385–2396. doi: 10.1099/mic.0.26397-0. [DOI] [PubMed] [Google Scholar]
- 37.Brekke O.L., Christiansen D., Fure H., Fung M., Mollnes T.E. The role of complement C3 opsonization, C5a receptor, and CD14 in E. coli-induced up-regulation of granulocyte and monocyte CD11b/CD18 (CR3), phagocytosis, and oxidative burst in human whole blood. J Leukoc Biol. 2007;81:1404–1413. doi: 10.1189/jlb.0806538. [DOI] [PubMed] [Google Scholar]
- 38.Xie Z., Su Y., Kim G.B., Selvi E., Ma C., Aragon-Sanabria V., et al. Immune cell-mediated biodegradable theranostic nanoparticles for melanoma targeting and drug delivery. Small. 2017;13:1603121. doi: 10.1002/smll.201603121. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Xue J., Zhao Z., Zhang L., Xue L., Shen S., Wen Y., et al. Neutrophil-mediated anticancer drug delivery for suppression of postoperative malignant glioma recurrence. Nat Nanotechnol. 2017;12:692–700. doi: 10.1038/nnano.2017.54. [DOI] [PubMed] [Google Scholar]
- 40.Huang B., Abraham W.D., Zheng Y., Lopez S.C.B., Luo S.S., Irvine D.J. Active targeting of chemotherapy to disseminated tumors using nanoparticle-carrying T cells. Sci Transl Med. 2015;7:291ra94. doi: 10.1126/scitranslmed.aaa5447. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Salmon H., Remark R., Gnjatic S., Merad M. Host tissue determinants of tumour immunity. Nat Rev Cancer. 2019;19:215–227. doi: 10.1038/s41568-019-0125-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.David B.A., Kubes P. Exploring the complex role of chemokines and chemoattractants in vivo on leukocyte dynamics. Immunol Rev. 2019;289:9–30. doi: 10.1111/imr.12757. [DOI] [PubMed] [Google Scholar]
- 43.Zanganeh S., Hutter G., Spitler R., Lenkov O., Mahmoudi M., Shaw A., et al. Iron oxide nanoparticles inhibit tumour growth by inducing pro-inflammatory macrophage polarization in tumour tissues. Nat Nanotechnol. 2016;11:986–994. doi: 10.1038/nnano.2016.168. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Müller E., Christopoulos P.F., Halder S., Lunde A., Beraki K., Speth M., et al. Toll-like receptor ligands and interferon-γ synergize for induction of antitumor M1 macrophages. Front Immunol. 2017;8:1383. doi: 10.3389/fimmu.2017.01383. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Pavlou S., Wang L., Xu H., Chen M. Higher phagocytic activity of thioglycollate-elicited peritoneal macrophages is related to metabolic status of the cells. J Inflamm. 2017;14:4. doi: 10.1186/s12950-017-0151-x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Xia W., Hilgenbrink A.R., Matteson E.L., Lockwood M.B., Cheng J.-X., Low P.S. A functional folate receptor is induced during macrophage activation and can be used to target drugs to activated macrophages. Blood. 2009;113:438–446. doi: 10.1182/blood-2008-04-150789. [DOI] [PubMed] [Google Scholar]
- 47.Zhang Y., Cao W., Toneri M., Zhang N., Kiyuna T., Murakami T., et al. Toxicology and efficacy of tumor-targeting Salmonella typhimurium A1-R compared to VNP 20009 in a syngeneic mouse tumor model in immunocompetent mice. Oncotarget. 2017;8:54616–54628. doi: 10.18632/oncotarget.17605. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Liberal R., Grant C.R., Yuksel M., Graham J., Kalbasi A., Ma Y., et al. Regulatory T-Cell conditioning endows activated effector T cells with suppressor function in autoimmune hepatitis/autoimmune sclerosing cholangitis. Hepatology. 2017;66:1570–1584. doi: 10.1002/hep.29307. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Massa P.E., Paniccia A., Monegal A., de Marco A., Rescigno M. Salmonella engineered to express CD20-targeting antibodies and a drug-converting enzyme can eradicate human lymphomas. Blood. 2013;122:705–714. doi: 10.1182/blood-2012-12-474098. [DOI] [PubMed] [Google Scholar]
- 50.Klichinsky M., Ruella M., Shestova O., Lu X.M., Best A., Zeeman M., et al. Human chimeric antigen receptor macrophages for cancer immunotherapy. Nat Biotechnol. 2020;38:947–953. doi: 10.1038/s41587-020-0462-y. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Chen J., Qiao Y., Chen G., Chang C., Dong H., Tang B., et al. Salmonella flagella confer anti-tumor immunological effect via activating Flagellin/TLR5 signalling within tumor microenvironment. Acta Pharm Sin B. 2021;11:3165–3177. doi: 10.1016/j.apsb.2021.04.019. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Hambleton J., Weinstein S.L., Lem L., DeFranco A.L. Activation of c-Jun N-terminal kinase in bacterial lipopolysaccharide-stimulated macrophages. Proc Natl Acad Sci U S A. 1996;93:2774–2778. doi: 10.1073/pnas.93.7.2774. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Verma V., Shrimalim R.K., Ahmadm S., Dai W., Wang H., Lu S., et al. PD-1 blockade in subprimed CD8 cells induces dysfunctional PD-1+CD38(hi) cells and anti-PD-1 resistance. Nat Immunol. 2019;20:1231–1243. doi: 10.1038/s41590-019-0441-y. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Philip M., Fairchild L., Sun L., Horste E.L., Amara S.C., Shakiba M., et al. Chromatin states define tumour-specific T cell dysfunction and reprogramming. Nature. 2017;545:452–456. doi: 10.1038/nature22367. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.McLane L.M., Abdel-Hakeem M.S., Wherry E.J. CD8 T cell exhaustion during chronic viral infection and cancer. Annu Rev Immunol. 2019;37:457–495. doi: 10.1146/annurev-immunol-041015-055318. [DOI] [PubMed] [Google Scholar]
- 56.Lin H., Wei S., Hurt E.M., Green M.D., Zhao L., Vatan L., et al. Host expression of PD-L1 determines efficacy of PD-L1 pathway blockade-mediated tumor regression. J Clin Invest. 2018;128:805–815. doi: 10.1172/JCI96113. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Cha J.H., Chan L.C., Li C.W., Hsu J.L., Hung M.C. Mechanisms controlling PD-L1 expression in cancer. Mol Cell. 2019;76:359–370. doi: 10.1016/j.molcel.2019.09.030. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Chen G., Huang A.C., Zhang W., Zhang G., Wu M., Xu W., et al. Exosomal PD-L1 contributes to immunosuppression and is associated with anti-PD-1 response. Nature. 2018;560:382–386. doi: 10.1038/s41586-018-0392-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Waldman A.D., Fritz J.M., Lenardo M.J. A guide to cancer immunotherapy: from T cell basic science to clinical practice. Nat Rev Immunol. 2020;20:651–668. doi: 10.1038/s41577-020-0306-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Oh S.A., Wu D.C., Cheung J., Navarro A., Xiong H., Cubas R., et al. PD-L1 expression by dendritic cells is a key regulator of T-cell immunity in cancer. Nat Can (Que) 2020;1:681–691. doi: 10.1038/s43018-020-0075-x. [DOI] [PubMed] [Google Scholar]
- 61.Gordon S.R., Aute R.L.M., Dulken B.W., Hutter G., George B.M., Ccracken M.N.M., et al. PD-1 expression by tumour-associated macrophages inhibits phagocytosis and tumour immunity. Nature. 2017;545:495–499. doi: 10.1038/nature22396. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Pencheva N., Buss C.G., Posada J., Merghoub T., Tavazoie S.F. Broad-spectrum therapeutic suppression of metastatic melanoma through nuclear hormone receptor activation. Cell. 2014;156:986–1001. doi: 10.1016/j.cell.2014.01.038. [DOI] [PubMed] [Google Scholar]
- 63.Zhao M., Guo W., Wu Y., Yang C., Zhong L., Deng G., et al. SHP2 inhibition triggers anti-tumor immunity and synergizes with PD-1 blockade. Acta Pharm Sin B. 2019;9:304–315. doi: 10.1016/j.apsb.2018.08.009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Siddiqui I., Schaeuble K., Chennupati V., Marraco S.A.F., Calderon-Copete S., Ferreira D.P., et al. Intratumoral Tcf1+PD-1+D8+ T cells with stem-like properties promote tumor control in response to vaccination and checkpoint blockade immunotherapy. Immunity. 2019;50:195–211. doi: 10.1016/j.immuni.2018.12.021. [DOI] [PubMed] [Google Scholar]
- 65.Kim J.E., Phan T.X., Nguyen V.H., Dinh-Vu H.V., Zheng J.H., Yun M., et al. Salmonella typhimurium suppresses tumor growth via the pro-inflammatory cytokine interleukin-1. β. Theranostics. 2015;5:1328–1342. doi: 10.7150/thno.11432. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Nemunaitis J., Cunningham C., Senzer N., Kuhn J., Cramm J., Litz C., et al. Pilot trial of genetically modified, attenuated Salmonella expressing the E-coli cytosine deaminase gene in refractory cancer patients. Cancer Gene Ther. 2003;10:737–744. doi: 10.1038/sj.cgt.7700634. [DOI] [PubMed] [Google Scholar]
- 67.Graham F.O., Coleman P.N. Infection of a secondary carcinoma by. Salmonella montevideo. Br Med J. 1952;1:1116–1117. doi: 10.1136/bmj.1.4768.1116. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68.Thamm D.H., Kurzman I.D., King I., Li Z., Sznol M., Dubielzig R.R., et al. Systemic administration of an attenuated, tumor-targeting Salmonella typhimurium to dogs with spontaneous neoplasia: phase I evaluation. Clin Cancer Res. 2005;11:4827–4834. doi: 10.1158/1078-0432.CCR-04-2510. [DOI] [PubMed] [Google Scholar]
- 69.Yi X., Zhou H., Chao Y., Xiong S., Zhong J., Chai Z., et al. Bacteria-triggered tumor-specific thrombosis to enable potent photothermal immunotherapy of cancer. Sci Adv. 2020;6 doi: 10.1126/sciadv.aba3546. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70.Huang X., Zhang F., Wang H., Niu G., Choi K.Y., Swierczewska M., et al. Mesenchymal stem cell-based cell engineering with multifunctional mesoporous silica nanoparticles for tumor delivery. Biomaterials. 2013;34:1772–1780. doi: 10.1016/j.biomaterials.2012.11.032. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 71.Moslehi J.J., Salem J.E., Sosman J.A., Lebrun-Vignes B., Johnson D.B. Increased reporting of fatal immune checkpoint inhibitor-associated myocarditis. Lancet. 2018;391:933. doi: 10.1016/S0140-6736(18)30533-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 72.Zhang W., Wang F., Hu C., Zhou Y., Gao H., Hu J. The progress and perspective of nanoparticle-enabled tumor metastasis treatment. Acta Pharm Sin B. 2020;10:2037–2053. doi: 10.1016/j.apsb.2020.07.013. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.