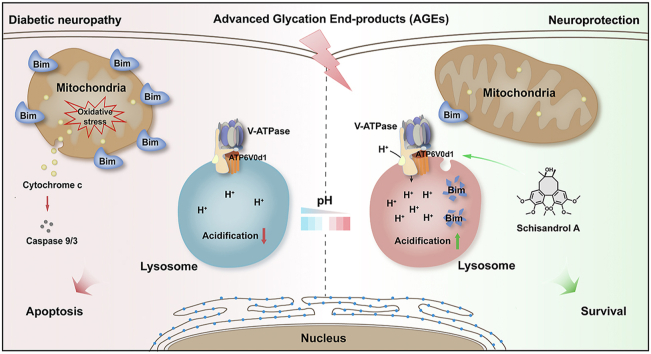
Abstract
Diabetes have been shown to cause progressive neuronal injury with pain and numbness via advanced glycation end-products (AGEs)-induced neuronal cell apoptosis; however, the valuable drug targets for diabetic neuropathy have been poorly reported so far. In this study, we discovered a natural small-molecule schisandrol A (SolA) with significant protective effect against AGEs-induced neuronal cell apoptosis. ATP6V0D1, a major subunit of vacuolar-type ATPase (V-ATPase) in lysosome was identified as a crucial cellular target of SolA. Moreover, SolA allosterically mediated ATP6V0D1 conformation via targeting a unique cysteine 335 residue to activate V-ATPase-dependent lysosomal acidification. Interestingly, SolA-induced lysosome pH downregulation resulted in a mitochondrial–lysosomal crosstalk by selectively promoting mitochondrial BH3-only protein BIM degradation, thereby preserving mitochondrial homeostasis and neuronal cells survival. Collectively, our findings reveal ATP6V0D1 is a valuable pharmacological target for diabetes-associated neuronal injury via controlling lysosomal acidification, and also provide the first small-molecule template allosterically activating V-ATPase for preventing diabetic neuropathy.
Key words: Allosteric regulation, Advanced glycation end-products, AGEs, Lysosomal acidification, Targets, V-ATPase, ATP6V0D1, Schisandrol A, Diabetic neuropathy
Graphical abstract
Schisandrol A is identified as an activator of V-ATPase by targeting ATP6V0D1 subunit, which provides a promising target against diabetic neuropathy.
1. Introduction
Diabetic neuropathy is one of the most common complications of diabetes, and most of patients with diabetes suffer from it1. Recent evidence has indicated that hyperglycemia is a crucial risk factor for the occurrence of diabetic neuropathy2. Particularly, advanced glycation end-products (AGEs) are typical harmful molecules formed when proteins and lipids combine with sugar in the bloodstream, and play key roles in hyperglycemia-associated nervous system disorders3. Recently, AGEs have been widely reported to be significantly increased in diabetic patients to induce neuronal cells injury4. However, potential therapeutic targets for diabetic neuropathy are still poorly understood.
Lysosomes are fundamental cell organelles containing a range of hydrolytic enzymes for degrading intracellular biomolecules like proteins, fats and nucleic acids5,6. Previous reports have shown that dysfunctional lysosomes cannot maintain an acidic environment to work properly in nervous system, thereby contributing to the pathogenesis of diabetic neuropathy7. Particularly, ATP-driven vacuolar-type ATPase (V-ATPase) is a highly conserved evolutionarily ancient enzyme and exerts a crucial function in lysosomal acidification8,9. Moreover, V-ATPase has been previously reported to serve as a proton pump to maintain low pH in lysosomes for hydrolase activity in neurons10, we thus purpose that selectively targeting V-ATPase may be a potential therapeutic strategy against diabetes-associated neuronal cells injury.
Current structural studies reveal that V-ATPase comprises of two major domains: the membrane-bound V0 domain and the cytosolic V1 domain11, 12, 13. V0 constitutes a channel for transporting H+ and V1 hydrolyzes ATP to produce energy for the transport process14. Previous studies have revealed a great amount of V-ATPase small-molecule inhibitors that specifically target different subunit proteins15, 16, 17. However, there have been very few reports of V-ATPase activators18. Since membrane-anchored V0 domain consists of five different subunits (a, c, c', c'', d) responsible for crucial proton translocation process in lysosomal pH control19, we speculate that V-ATPase activity may be potentially regulated via pharmacologically targeting these specific subunits.
Natural products are fundamental source of chemical diversity, which drives us to explore valuable active small molecules targeting V-ATPase. Schisandrol A (SolA), a natural small-molecule from Schisandra chinensis, has been reported to possess various biological functions including neuroprotection20, hepatoprotection21,22 and anti-tumor effect23. Particularly, SolA has been widely reported to has potential therapeutic effect for various neurodegenerative diseases. For example, SolA ameliorats SH-SY5Y cells survival by attenuating Aβ1–42-induced cellular autophagy24 and enhancing DNA methylation25 for Alzheimer's disease therapy. SolA also protects against OGD/R-induced neuronal injury by suppressing AMPK/mTOR pathways-associated autophagy26. Moreover, SolA exerts anti-inflammatory effect and ameliorates endoplasmic reticulum stress in streptozotocin-induced Alzheimer's disease rats27. Besides, SolA improves cognitive function in ovariectomized rats28 and reverses scopolamine-induced memory impairment in mice by enhancing cholinergic function29,30. Furthermore, SolA functions as sedative and hypnotic agent by modifying pentobarbital-induced sleep behaviors31. In this study, we report SolA exerts significant neuroprotective effect against AGEs-induced neuronal cells apoptosis. Moreover, we identify ATP6V0D1, a major subunit of V-ATPase in lysosome as a crucial cellular target of SolA. In particular, SolA binds to a unique cysteine 335 residue of ATP6V0D1 to induce conformational change for V-ATPase activation. Then, V-ATPase-mediated lysosomal acidification significantly promotes Bim degradation for mitochondrial homeostasis maintenance, further leading to a significant neuroprotective effect.
In this study, we report ATP6V0D1 subunit in V-ATPase as a promising druggable target for neuroprotection against AGEs-induced injury, and provide the first small-molecule template for diabetic neuropathy via allosterically activating V-ATPase to promote lysosomal acidification.
2. Materials and methods
2.1. Chemicals and reagents
Schisandrol A (SolA, C24H32O7; molecular weight 432.51) and schisandrin A (SchA, C24H32O6; molecular weight 416.507) were obtained from Baoji Herbest Bio-Tech (Baoji, China) and affirmed by 1H NMR and MS analysis. The purity was determined to be >98% by HPLC method. Bovine serum albumin (BSA) was from Equitech-Bio (Kerrville, TX, USA). High glucose Dulbecco's modified Eagle's medium (DMEM), phosphate buffer saline (PBS), trypsin, penicillin and streptomycin were purchased from Macgene (Beijing, China). Fetal bovine serum (FBS) and horse serum were from PAN-Biotech (Aidenbach, Germany). Neurobasal medium and B27 supplement were from Invitrogen (Carlsbad, CA, USA). Poly-l-lysine (PLL) and 3-(4,5-dimethyl thiazol-2-yl)-2,5-diphenyl tetrazolium bromide (MTT) were purchased from Sigma–Aldrich (St Louis, MO, USA). Isopropyl β-d-thiogalactopyranoside (IPTG) was from BBI Life Sciences (Shanghai, China). MG-132 and bafilomycin A1 (Baf-A1) were from Selleck Chemicals (Houston, TX, USA). Mito-tracker Green, Mito-tracker Red CMXRos, Lyso-tracker Red, JC-1 staining kit and DCFH-DA/ROS assay kit were purchased from Beyotime (Shanghai, China). LysoSensor™ Yellow/Blue DND-160 was purchased from Yeasen (Shanghai, China). Easy II protein quantitative kit (BCA) was purchased from TransGen (Beijing, China). LDH quantitative detection kit, V-ATPase activity colorimetric quantitative detection kit and H+ membrane transport function (V-ATPase) green fluorescence detection kit were purchased from Genmed (Shanghai, China). Hoechst 33258 staining kit, AO/EB double stain kit, E. coli DH5α and E. coli BL21 (DE3) were obtained from Solarbio (Beijing, China). Primary antibodies and secondary antibodies were from Cell Signaling Technology (Beverly, MA, USA). Chemiluminescent HRP substrate, Lipofectamine 2000, Lipofectamine RNAiMAX and opti-MEM were purchased from Thermo Fisher Scientific (Waltham, MA, USA).
2.2. Cell culture
Rat adrenal pheochromocytoma cells line PC12 were obtained from Peking Union Medical College Cell Bank (Beijing, China). The cells were cultured in high glucose DMEM supplemented with 5% fetal bovine serum, 10% horse serum, 100 U/mL penicillin and 100 μg/mL streptomycin at 37 °C in a 5% CO2 and 95% air humidified incubator.
Primary neuron cultures were prepared based on the experimental method previously reported32,33. Primary neurons were obtained from rat fetuses at gestational day. Brains were isolated and placed in HBSS. After removing meninges, cortices were isolated, shredded and dissociated in trypsin for 20 min at 37 °C in an incubator. Trypsin was neutralized with equal volume DMEM supplemented with FBS to stop the reaction. The cell suspension was filtered through a 100 μm cell strainer. Cells were suspended in DMEM and plated on culture dishes that were pre-coated with 100 mg/mL PLL at 37 °C. Neurobasal/B27 medium was added to replace DMEM after 6 h. Cells were used for follow-up experiments after 2 weeks.
2.3. MTT assay
Cell viability was determined by adding MTT (0.5 mg/mL) to each well and incubated for 4 h at 37 °C. Subsequently, formazan crystal products were dissolved in dimethylsulfoxide (DMSO) for 10 min and absorbance value was measured at 570 nm with a microplate reader (Bio-Rad Laboratories, Hercules, CA, USA).
2.4. LDH assay
LDH release was detected in both supernatant and cell lysate using the LDH assay kit (Nanjing Jiancheng Bioengineering, Nanjing, China) according to the manufacturer's instructions. Briefly, cell supernatant was collected and incubated with LDH working reagent for 30 min at 37 °C. Data were obtained by measuring absorbance at 450 nm using a microplate reader (Bio-Rad Laboratories).
2.5. Hoechst 33258 staining
Cells were fixed in 4% paraformaldehyde for 20 min. Then, cells were stained with Hoechst 33258 (1 μg/mL) for 30 min at 37 °C in darkness. Images were obtained by IX73 fluorescence microscope (Olympus, Tokyo, Japan) with excitation and emission wavelengths at 352 and 461 nm, respectively.
2.6. AO/EB staining
Cells were stained with AO/EB mixture (100 μg/mL, 1:1) for 8 min at room temperature in darkness. After washes with PBS, cells were observed under IX73 fluorescence microscope (Olympus) with excitation/emission wavelengths 490 nm/530 nm for AO staining and 520 nm/590 nm for EB staining.
2.7. Annexin V-FITC/PI assay
Cells were collected and washed with PBS. After centrifugation at 2000 rpm (5424R, Eppendorf, Hamburg, Germany) for 5 min, cells were resuspended in 500 μL binding buffer. 5 μL Annexin V-FITC and 5 μL PI were added in sequence for 10-min incubation at room temperature in darkness. Subsequently, the samples were subjected to a flowcytometer (BD Biosciences, NJ, USA). The cell population was analyzed by FlowJo v 10.6.2 software.
2.8. Mitochondrial membrane potential analysis
Mitochondrial membrane potential was detected by JC-1 staining (Beyotime, Nanjing, China). Cells were incubated with 10 μg/mL of JC-1 working solution for 20 min at 37 °C in the dark. Then, JC-1 monomer fluorescence (green) was detected at 514 nm (excitation), and JC-1 polymer fluorescence (red) was detected at 590 nm (emission). The images were captured by IX73 fluorescence microscope (Olympus).
2.9. Mitochondrial morphology analysis
Transmission electron microscopy was used to observe mitochondrial morphology. Cells were fixed in 4% glutaraldehyde at 4 °C overnight and 1% OsO4 at room temperature for 2 h, respectively. After dehydration with absolute alcohol, Eponate 812 (Ted Pella, Redding, CA, USA) was added and cells were prepared into ultrathin slices. Subsequently, mitochondrial morphology was detected using JEM-1230 transmission electron microscope (Japan Electronics, Tokyo, Japan).
2.10. Mito-tracker staining assay
Mitochondrial morphology was detected by employing Mito-tracker fluorescent dye. Briefly, Mito-tracker solution (200 nmol/L) was added to the cells and maintained for 60 min at 37 °C. Photos were taken by confocal microscopy (Leica TCS SP8 X, Leica, Wetzlar, Germany) at 490 nm (excitation) and 516 nm (emission).
2.11. ROS assay
Cells were stained with ROS sensitive dye DCFH-DA (10 μmol/L) for 30 min at 37 °C in darkness. After washes with serum-free medium, cells were observed under IX73 fluorescence microscope (Olympus) with excitation/emission wavelengths 490 nm/530 nm.
2.12. Western blotting
Cells were homogenized with ice-cold RIPA buffer for 30 min to provide whole cell lysates. Concentrations of total proteins were determined using enhanced BCA protein assay kit (TransGen Biotech, Beijing, China). Lysates were separated by 8%–12% SDS-PAGE and subsequently transferred to polyvinylidene fluoride (PVDF) membranes. Membranes were blocked in 5% non-fat milk, probed with primary antibody (1:1000 diluted) for 2 h, and further incubated with secondary antibody (1:1000 diluted) for 1 h. Then, membranes were washed and developed with enhanced chemiluminescence (ECL) substrate. Images were captured using Tanon 5200 Imaging Analysis System (Tanon, Shanghai, China). Relative protein expressions were shown by densitometry analysis using Image J software.
2.13. Pull-down assay
SolA was chemically coupled onto magnetic nanoparticles as previously reported34,35. SolA-coupled beads (SolA beads) were mixed with AGEs-induced PC12 cell lysates or recombinant ATP6V0D1, and incubated for 2 h at 4 °C. Simultaneously, un-coupled beads were used as negative control. In addition, excessive free SolA (200 μmol/L) was added to compete with SolA-coupled beads for target proteins. The beads-captured proteins were then separated using 10% SDS-PAGE and analyzed by immunoblotting or silver staining, followed by LC–MS/MS analysis using a nano-HPLC-tandem LTQ-Orbitrap Velos pro mass spectrometer (ThermoFisher Scientific).
2.14. Surface plasmon resonance (SPR) assay
Interaction between SolA and ATP6V0D1 was analyzed using Biacore T200 system (GE Healthcare, Chicago, IL, USA). Recombinant ATP6V0D1 protein (500 μg/mL) was immobilized on a carboxymethylated 5 sensor chip using a standard amine coupling method. Different concentrations of SolA (0.04–50 μmol/L) in running buffer were injected as analytes. Interaction parameters of Ka, Kd, and KD were analyzed with Biacore evaluation software (T200 Version 2.0) according to the 1:1 Langmuir model.
2.15. Microscale thermophoresis (MST) assay
Interaction between SolA and ATP6V0D1 was analyzed by MST (NanoTemper, Munich, Germany). Recombinant ATP6V0D1 protein (200 nmol/L) was labeled with His-labeling-dye (200 nmol/L) at a ratio of 1:1 in the dark for 30 min at room temperature. Different concentrations of SolA (0.03 μmol/L–1 mmol/L) were added in labeled ATP6V0D1 and centrifuged at 12,000 rpm (5424R, Eppendorf, Hamburg, Germany) for 5 min. The supernatant was transferred into the capillary for infrared scanning. The samples were monitored at LED/excitation power of 100% and MST power of 40%. Interaction parameters of KD were analyzed with MO. Affinity Analysis software.
2.16. Cellular thermal shift assay (CETSA)
For cell lysate CETSA experiment, cells were freeze-thawed five times by liquid nitrogen. Cell lysates were divided into two aliquots, with one aliquot used as control and the other being incubated with SolA (12.5 μmol/L) for 1 h at room temperature. The lysates were heated individually at indicated temperatures (44–76 °C) for 3 min, followed by cooling at room temperature. The lysates were centrifuged at 12,000 × g for 20 min at 4 °C, and then supernatants were analyzed by SDS-PAGE and immunoblotting.
For living cell CETSA experiment, cells were treated with SolA (12.5 μmol/L) for 2 h and heated at indicated temperatures (46–67 °C) for 3 min, followed by cooling at room temperature. Cell lysates were collected by 5 cycles of freeze-thawing with liquid nitrogen. The lysates were centrifuged at 12,000 × g for 20 min at 4 °C, and then supernatants were analyzed by SDS-PAGE and immunoblotting.
2.17. Drug affinity responsive target stability (DARTS) assay
Different concentrations of SolA were mixed with cell lysates for 1 h. Then, pronase (5 μg/mL) was added and incubated at room temperature for 15 min. Reactions were ceased by appending SDS-PAGE loading buffer and analyzed by immunoblotting with a specific antibody.
2.18. Lysosomal acidity analysis
Cells were incubated with preheated LysoSensor Yellow/Blue DND-160 solution (1 μmol/L) for 15 min at 37 °C in darkness. Then, cell supernatants were aspirated and replaced with fresh phenol red free medium. Images were captured using confocal microscopy (Leica TCS SP8 X, Leica) with excitation/emission wavelengths 352 nm/461 nm.
2.19. Lyso-tracker staining assay
Lysosome change was assessed with Lyso-tracker. Briefly, cells were stained with Lyso-tracker solution (50 nmol/L) for 60 min at 37 °C, washed with PBS, and examined using the confocal microscopy (Leica TCS SP8 X, Leica) at 577 nm (excitation) and 590 nm (emission).
2.20. V-ATPase activity analysis
The activity of V-ATPase was measured by V-ATPase activity colorimetric quantitative detection kit (Genmed Scientifics, Boston, MA, USA). Briefly, PC12 cells were incubated with SolA at 37 °C for 6 h. The reaction was initiated by adding cell lysates (100 μg total protein) at 37 °C. The color development of NADH/NAD+ complex indicated the progress of reactions. The absorbance at 340 nm was detected using a multifunctional microplate reader (PerkinElmer, Waltham, MA, USA) every 5 min for a total 20 min. V-ATPase activity was calculated by Eq. (1) and compared with control group for normalization.
| V-ATPase activity = [(Sample absorbance20 min–Sample absorbance0 min)–(Background absorbance20 min–Background absorbance0 min)] × System volume × Dilution factor/12.44 | (1) |
2.21. Real-time proton transport activity analysis
H+ membrane transport function (V-ATPase) green fluorescence detection kit (Genmed Scientifics) was used to detect the proton transport activity. Briefly, lysosomes were separated from PC12 cells and mixed with SolA at 37 °C for 5 min. Then the fluorescence probe 9-amino-6-chloro-2-methoxy-acridine was added into the mixture. Subsequently, the relative fluorescence unit (RFU) was detected using a multifunctional microplate reader (PerkinElmer) with excitation/emission wavelengths 490 nm/530 nm every 1 min for a total 30 min. The decreasing RFU indicated the increasing proton transport activity.
2.22. DQ Red-BSA processing assay
DQ Red-BSA (ThermoFisher Scientific) was used to visualize the lysosomal proteolytic activity. PC12 cells were treated with 10 μg/mL DQ Red-BSA at 37 °C for 2 h, followed by 4 h incubation with AGEs or SolA treatment. Then, cells were fixed using 4% paraformaldehyde and stained by 20 μg/mL DAPI for 20 min. Images were captured by confocal microscopy (Leica TCS SP8 X, Leica) (590 nm/618 nm for DQ Red-BSA and 340 nm/488 nm for DAPI).
2.23. Protein expression and purification
His-ATP6V0D1 and His-ATP6V0D1C335A plasmids were respectively transformed into E. coli BL21 (DE3) cells for the production of recombinant proteins. Cells were grown to an OD600 of 0.8 and induced with 0.5 mmol/L IPTG at 16 °C for 18 h. Cells were collected by centrifugation at 4000 rpm (5424R, Eppendorf, Hamburg, Germany) for 20 min at 4 °C and lysed in 20 mL of lysis buffer (20 mmol/L HEPES, 250 mmol/L NaCl, 10 mmol/L imidazole). Then, proteins were purified using Ni-NTA resin (GE Healthcare, Chicago, IL, USA) with HEPES buffer (20 mmol/L HEPES, 250 mmol/L NaCl, a gradient of 10–250 mmol/L imidazole). Proteins were concentrated via centrifugal filtration (Millipore, Waltham, MA, USA) and finally dissolved in PBS buffer. Purified proteins were confirmed by Coomassie brilliant blue staining before use.
2.24. Tryptophan fluorescence quenching study
Recombinant ATP6V0D1 protein was incubated with SolA (3.12–200 μmol/L) or vehicle in a quartz plate. Tryptophan fluorescence spectra of the initial protein and those upon a successive addition of SolA solutions were recorded. Fluorescence was detected by setting excitation wavelength at 280 nm and emission wavelengths from 300 to 550 nm at 1 nm increment on a fluorescence spectrophotometer (PerkinElmer). The fluorescence intensities were corrected by the buffer contribution.
2.25. Hydrogen-deuterium exchange mass spectrometry (HDXMS) analysis
HDXMS experiment was performed as previously described36. Briefly, deuterium labeling was initiated with a 20-fold dilution into D2O buffer of a pre-equilibrated (30 min, RT) aliquot of ATP6V0D1 protein and ATP6V0D1 protein with SolA stock solution. The labeling reaction was quenched with the addition of quenching buffer (37.5% [v/v] hydrochloric acid) at indicated times. Samples were then injected and online digested by a Waters ENZYMATE BEH pepsin column (2.1 mm × 30 mm, 5 μm). Subsequently, the peptides were trapped and desalted on a VanGuard Pre-Column trap (ACQUITY UPLC BEH C18, 1.7 μm) for 3 min, eluted in the trap using 15% acetonitrile at a flow rate of 100 μL/min, and separated using an ACQUITY UPLC BEH C18 column (1.7 μm, 1.0 mm × 100 mm). Relative deuterium levels of all peptides were calculated by subtracting the mass of undeuterated control sample from that of the deuterium-labeled sample.
2.26. Determination of SolA-binding site on ATP6V0D1
Recombinant ATP6V0D1 protein was incubated with SolA at 37 °C for 2 h, and the mixtures were analyzed by SDS-PAGE. Nano-LC–MS/MS experiment was performed using an EASY–nLC II system and a LTQ-Orbitrap velos pro mass spectrometer for identification of SolA-binding site on ATP6V0D1. Proteome Discoverer (1.4) workstation was applied for the elucidation of mass spectrometric data with the SEQUEST search engine (Thermo Fisher Scientific).
2.27. Transfection of siRNAs
For knockdown studies, cells were transfected with specific siRNAs designed and synthesized at GenePharma (Suzhou, Jiangsu, China). 14 μL siRNAs (10 μmol/L) and 42 μL lipofectamine RNAiMAX (Thermo) were respectively added in 700 μL Opti-MEM reduced serum medium (Thermo). Then, the siRNAs and lipofectamine RNAiMAX were premixed and applied to the cells. After 48 h, the transfected cells were used for further experiments.
2.28. Double staining of Mito-tracker and Lyso-tracker
Mitochondrial–lysosome crosstalk was detected by double staining of Mito-tracker and Lyso-tracker. Briefly, Mito-tracker solution (200 nmol/L) and Lyso-tracker solution (50 nmol/L) were added to the cells and maintained for 60 min at 37 °C. After washes with PBS, photos were taken by confocal microscopy (Leica TCS SP8 X, Leica) with excitation/emission wavelengths 490 nm/516 nm for Mito-tracker staining and 577 nm/590 nm for Lyso-tracker staining.
2.29. Co-immunoprecipitation (co-IP) analysis
PC12 cells were transfected with HA-ATP6V0D1 for 48 h and treated with or without SolA (12 μmol/L). Then the cells were lysed and incubated with anti-HA-tag antibody conjugated magnetic beads for 6 h at 4 °C. The immunoprecipitated proteins were separated by SDS-PAGE and detected by immunoblotting.
2.30. Immunofluorescence analysis
The cells were fixed in 4% paraformaldehyde for 20 min and then permeabilized with 0.5% Triton X-100 for 30 min. After washing with PBS, the cells were blocked with 5% BSA for 30 min at room temperature, washed 3 times with PBS, and probed with primary antibodies overnight at 4 °C. The cells were incubated with the secondary antibody conjugated to Dylight-594 for 2 h at room temperature. After washes for 3 times with PBS, the cells were then incubated with 20 μg/mL DAPI for 20 min. Images were captured (594 nm/618 nm for Dylight-594; 488 nm/519 nm for GFP and 340 nm/488 nm for DAPI) using a confocal microscopy (Leica TCS SP8 X, Leica).
2.31. Autophagy flux analysis
AdPlus-mCherry-GFP-LC3B was used to detect the development of autophagy flux. PC12 cells transfected with mCherry-GFP-LC3 lentivirus (Beyotime) were seeded into a 35-mm culture dish for confocal microscopy. Red and yellow puncta representing autolysosomes and autophagosomes, were detected by confocal microscopy (Leica TCS SP8 X, Leica) with excitation/emission wavelengths 594 nm/618 nm for mCherry and 488 nm/519 nm for GFP.
2.32. Zebrafish analysis
Zebrafishes were treated with 6.25 mmol/L 2,2′-azobis(2-methylpropionamidine) dihydrochloride (AAPH) alone or co-treated with AAPH and SolA (6 μmol/L). Mitochondrial function of zebrafishes was assessed at 72 h. Mito-tracker Red CMXRos (Beyotime, 200 nmol/L, 30 min) were used for the detection of mitochondrial membrane potential in situ37. The content of ROS was analyzed at 15 h. Oxidation-sensitive fluorescent probe DCFH-DA (Beyotime, 30 μmol/L, 30 min) was applied to estimate ROS generation in zebrafishes38. The stained zebrafishes were observed and photographed by a fluorescence microscope (Olympus) with excitation/emission wavelengths 490 nm/516 nm for Mito-tracker Red CMXRos and 490 nm/530 nm for ROS. The fluorescent intensity of individual zebrafish was quantified using Image Pro-Plus software.
2.33. In vivo studies
2.33.1. Animals
Male BALB/c mice (6–8 weeks old, weight 18–23 g) were obtained from Vital River Laboratories (Beijing, China) and raised at 25 ± 1 °C under 12 h dark/12 h light cycle. The mice were acclimated for two weeks with free access to feed and water. Experimental procedures were approved by the Ethical Institutional Animal Care and Use Committee of Peking University (EIACUC-PKU), Beijing, China (LA2020341).
2.33.2. Diabetic neuropathy model establishment
Mice were subjected to an intraperitoneal alloxan (ALX, 140 mg/kg, 10 mL/kg). Control mice were given an intraperitoneal injection of saline (10 mL/kg). Blood glucose testing was carried out three days later using an Accu-Check glucometer (Roche, Basel, Switzerland). Mice displayed blood glucose above 11.1 mmol/L were categorized as diabetic.
2.33.3. Animal grouping and administration
The diabetic mice were divided into two groups: diabetes group and SolA administered group. SolA (30 mg/kg) was dissolved in 0.5% sodium carboxymethylcellulose (CMC-Na) and given to mice everyday by intragastric administration. Meanwhile, the mice in control group were administered 10 mL/kg saline. The mice were anesthetized and sacrificed on the 7th day of administration. Blood samples were collected from mice and allowed to stand at 25 °C for 1 h followed by centrifuging at 3000 × g for 10 min. The supernatant was collected for the enzyme-linked immunosorbent assay (ELISA) of AGEs. The brains were quickly excised, fixed with 4% paraformaldehyde and embedded in paraffin for H&E staining and IHC analysis.
2.33.4. Enzyme-linked immunosorbent assay (ELISA)
The serum concentration of AGEs was detected using ELISA kit (Biologica, Beijing, China). Briefly, AGEs standard solutions (50, 100, 200, 400 and 800 pg/mL) were obtained by diluting the AGEs with various volumes of standard diluent. The samples and AGEs standard solutions were added into the ELISA-coated plate, which was incubated at 37 °C for 30 min. Then, the plate was washed by wash solution for five times. Enzyme labeling reagent was added to the plate, which was further incubated at 37 °C for 30 min. After washes, chromogen solution A and B were added to the plate and incubated at 37 °C for 10 min in dark. Stop solution was added to the plate and the absorbance at 450 nm was detected within 15 min. The serum concentration of AGEs was calculated according to the standard curve fitted by AGEs standard solutions and their absorbance.
2.34. Statistical analysis
All data were expressed as mean ± standard error of mean (SEM), with n representing the number of independent experiments. GraphPad Prism 6.0 was used for statistical analysis and graphs. Mean values were compared by one-way analysis of variance (ANOVA) with Dunnett's post hoc test. Student's t-test was used to analyze the statistical significance between two groups. P < 0.05 was considered as statistically significant.
3. Results
3.1. Discovery of a natural small-molecule SolA against AGEs-induced neuronal cells apoptosis
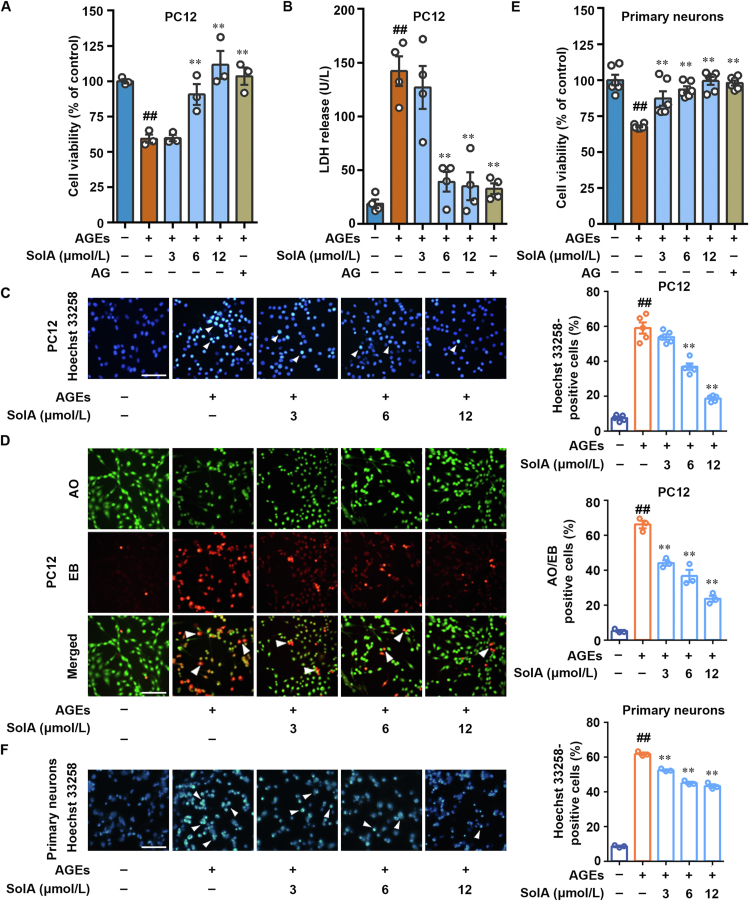
Schisandrol A (SolA) is a natural-derived small-molecule from medicinal plant S. chinensis. In this study, we found that SolA significantly increased cell viability in MTT assay against AGEs-induced PC12 pheochromocytoma cells, a convenient model for neurobiological studies (Fig. 1A). Meanwhile, SolA inhibited AGEs-induced lactate dehydrogenase (LDH) release from cytosol into medium (Fig. 1B). In particular, SolA had equivalent effect to aminoguanidine hydrochloride (AG), an agent for the inhibition of AGEs toxicity, in enhancing cell viability and reducing LDH release. Moreover, Hoechst 33258 staining revealed that SolA markedly attenuated AGEs-induced chromosome condensation, suggesting a significant anti-apoptosis effect in PC12 cells (Fig. 1C). Besides, in AO/EB staining analysis, SolA obviously downregulated the proportion of EB-positive cells, indicating that SolA inhibited PC12 cell apoptosis (Fig. 1D). Meanwhile, the result of Annexin V-FITC/PI assay by flow cytometry also revealed that SolA effectively suppressed PC12 cells apoptosis against AGEs insult in a concentration-dependent manner (Supporting Information Fig. S1A).
Figure 1.
Discovery of SolA as a neuroprotective small-molecule against AGEs-induced injury. (A) SolA increased the cell viability in AGEs-induced PC12 cells (n = 3). (B) SolA decreased LDH release in AGEs-induced PC12 cells (n = 4). (C) SolA inhibited PC12 cells apoptosis by Hoechst 33258 staining (scale bar: 100 μm, n = 5). Arrows indicate brightly stained nuclei for apoptosis. (D) SolA inhibited PC12 cells apoptosis by double staining with AO and EB (scale bar: 100 μm, n = 3). Arrows indicate orange apoptotic cells. (E) SolA increased the cell viability in AGEs-induced primary neurons (n = 6). (F) SolA inhibited primary neurons apoptosis by Hoechst 33258 staining (scale bar: 100 μm, n = 3). Arrows indicate brightly stained nuclei for apoptosis. Data are expressed as the mean ± SEM. ##P < 0.01 vs. control group, ∗∗P < 0.01 vs. AGEs group.
To further confirm the neuroprotective effect of SolA, we also established primary neurons culture from mouse cerebral cortex to test the neuroprotective effect of SolA. Our results suggest that SolA effectively suppressed cortical neurons apoptosis against AGEs insult by MTT and Hoechst 33258 assays (Fig. 1E and F). Furthermore, the immunofluorescence staining of MAP-2 showed that AGEs obviously induced neuronal cell morphological changes with shortening synapses and rounding cells, which was significantly reversed by SolA treatment (Fig. S1B). Particularly, SolA alone had no effect in the survival of PC12 cells and primary neurons (Supporting Information Fig. S1C–S1H). Collectively, these findings indicate that SolA is a valuable natural small-molecule for protecting AGEs-induced neuronal cell injury.
3.2. SolA selectively promotes mitochondrial Bim degradation
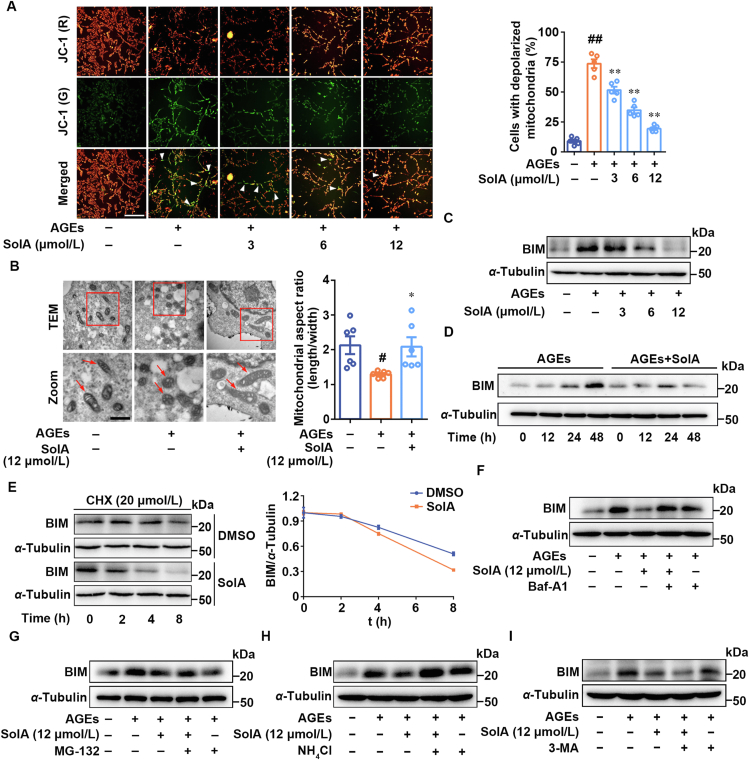
Mitochondria are highly plastic and dynamic organelles that play essential roles in neuronal cell apoptosis. Here, we performed potential-dependent JC-1 staining of mitochondria in PC12 cells to detect mitochondrial depolarization. As shown in Fig. 2A, we found that AGEs promoted a dramatic increase in the number of cells with depolarized mitochondria (green), which was significantly reversed by SolA treatment. Transmission electron microscopy analysis revealed that SolA effectively improved the damaged mitochondrial morphology from small spheres or short rod-like shape to long tubules (Fig. 2B). Moreover, we quantified mitochondrial morphology metrics by Mito-tracker staining analysis. As shown in Supporting Information Fig. S2A, we found that AGEs caused morphological shrinkage of mitochondria in cells, which was significantly improved by SolA. The mRNA expression of Tom20 and COXIV were detected by real-time PCR. As a result, we found that SolA did not significantly affect the transcription levels of Tom20 and COXIV, indicating that SolA may not regulate mitochondrial biogenesis (Fig. S2B and S2C). SolA significantly inhibited mitochondria-mediated apoptosis signal pathway (Fig. S2D and S2E). Therefore, these observations indicate an obvious protective effect of SolA on mitochondrial morphology and function.
Figure 2.
SolA maintains mitochondrial homeostasis by promoting BIM degradation. (A) SolA inhibited mitochondrial depolarization by JC-1 staining in AGEs-induced PC12 cells (scale bar: 200 μm, n = 5). Arrows indicate the cells with depolarized mitochondria. (B) SolA (12 μmol/L) inhibited mitochondrial damage by transmission electron microscope (TEM) assay in AGEs-induced PC12 cells (scale bar: 1 μm, n = 6). Arrows indicate the typical mitochondrial structure. (C) SolA suppressed AGEs-induced BIM expression in a concentration-dependent manner. (D) SolA (12 μmol/L) suppressed AGEs-induced BIM expression in a time-dependent manner. (E) SolA (12 μmol/L) promoted BIM protein degradation in a time-dependent manner. (F) Bafilomycin A1 (Baf-A1) reversed SolA (12 μmol/L)-mediated BIM degradation. (G) MG-132 failed to reverse SolA (12 μmol/L)-mediated BIM degradation. (H) NH4Cl reversed SolA (12 μmol/L)-mediated BIM degradation. (I) 3-Methyladenine (3-MA) failed to reverse SolA (12 μmol/L)-mediated BIM degradation. Data are expressed as the mean ± SEM. #P < 0.05, ##P < 0.01 vs. control group, ∗P < 0.05, ∗∗P < 0.01 vs. AGEs group.
Since BCL-2 family members precisely control mitochondrial function and bioenergetics, we then focused on the role of SolA on BCL-2 family members. Western blot was used to analyze the expression of different proteins of BCL-2 family after SolA treatment. As shown in Fig. S2F and S2G, SolA selectively inhibited BIM expression rather than other mitochondrial proteins, including BCL-2, PUMA, BCL-XL, BAK, BIK and BAX. This observation was further confirmed that SolA suppressed BIM expression in a concentration and time-dependent manner (Fig. 2C and D).
To investigate the significant role of BIM in mitochondria homeostasis and mitochondria oxidative damage, BIM was overexpressed in PC12 cells. We observed that SolA down-regulated ROS production in AGEs-induced PC12 cells, which was significantly reversed by BIM overexpression (Supporting Information Fig. S3A). Moreover, JC-1 staining showed that BIM overexpression obviously abolished SolA-induced mitochondrial protection effect (Fig. S3B). In addition, to evaluate the influence of other pro-apoptotic BCL-2 proteins on SolA efficacy, we overexpressed BAX in PC12 cells. The result showed that SolA down-regulated ROS production in AGEs-induced PC12 cells, which was partly reversed by BAX overexpression (Fig. S3C). Moreover, JC-1 staining showed that BAX overexpression modestly weakened SolA-induced mitochondrial protection effect (Fig. S3D).
Additionally, we found that AGEs treatment, in the presence or absence of SolA, did not obviously change the transcription of Bim (Supporting Information Fig. S4A). We used 20 μmol/L protein synthesis inhibitor cycloheximide to suppress newly synthesized proteins, and found that SolA significantly accelerated BIM degradation in a time-dependent fashion (Fig. 2E). Thus, we speculated that SolA may down-regulate BIM expression by promoting protein degradation rather than inhibiting its synthesis. To investigate which mechanism is involved in SolA-dependent BIM degradation, we treated the cells with bafilomycin A1 (Baf-A1, V-ATPase inhibitor) or MG-132 (proteasome inhibitor) in the presence or absence of SolA. Then, we observed that Baf-A1 rather than MG-132 effectively reversed SolA-mediated BIM degradation (Fig. 2F and G), hinting that autophagy-lysosome pathway was responsible for BIM stability regulation.
In view of the importance of autophagy-dependent protein degradation in cells, we further explored whether SolA-induced BIM degradation was regulated by autophagy processes. Interestingly, our results showed that Baf-A1 and NH4Cl, which can disrupte lysosomal function, markedly reversed SolA-induced BIM decrease (Fig. 2F and H). Meanwhile, 3-methyladenine (3-MA), which can block autophagosome formation to inhibit autophagy flux, failed to reverse SolA-induced BIM degradation (Fig. 2I). Thus, these results suggested that SolA induced BIM degradation mainly via lysosome and bypassing autophagosome formation.
Moreover, autophagy markers P62 and LC3B were detected in PC12 cells. We observed that SolA slightly reversed AGEs-dependent P62 and LC3B increase (Fig. S4B), further revealing that SolA-induced lysosomal acidity may physiologically restore lysosome function for degrading intracellular accumulated P62 and LC3B-positive cargos. Similarly, the co-staining of LC3B and lysosome marker LAMP2 showed that SolA markedly increased the co-localization of LC3B-labelled autophagosomes with LAMP2-labelled lysosomes, indicating an indirect promotion effect on autophagic flux because of the lysosomal function recovery (Fig. S4C). Furthermore, mCherry-eGFP-LC3B dual fluorescence system was performed for autophagic flux analysis. As shown in Fig. S4D, the obvious co-localization of green and red fluorescence in AGEs-treated cells suggested that the LC3B carrier flux was blocked. Meanwhile, the specific red fluorescence representing LC3B carrier flux was effectively restored upon SolA treatment, indicating that SolA reactivated autophagy flux by reactivating lysosomal acidity. Mito-Keima was used to determine the mitophagy in PC12 cells. The result revealed that both AGEs and SolA did not induce obvious red fluoresces, indicating that mitophagy may not be highly involved in SolA-dependent neuroprotection in PC12 cells (Fig. S4E). Collectively, these findings further hint that SolA may induce BIM degradation mainly by regulating lysosomal acidity, but not autophagosomes formation.
3.3. ATP6V0D1 subunit in V-ATPase is a direct cellular target of SolA
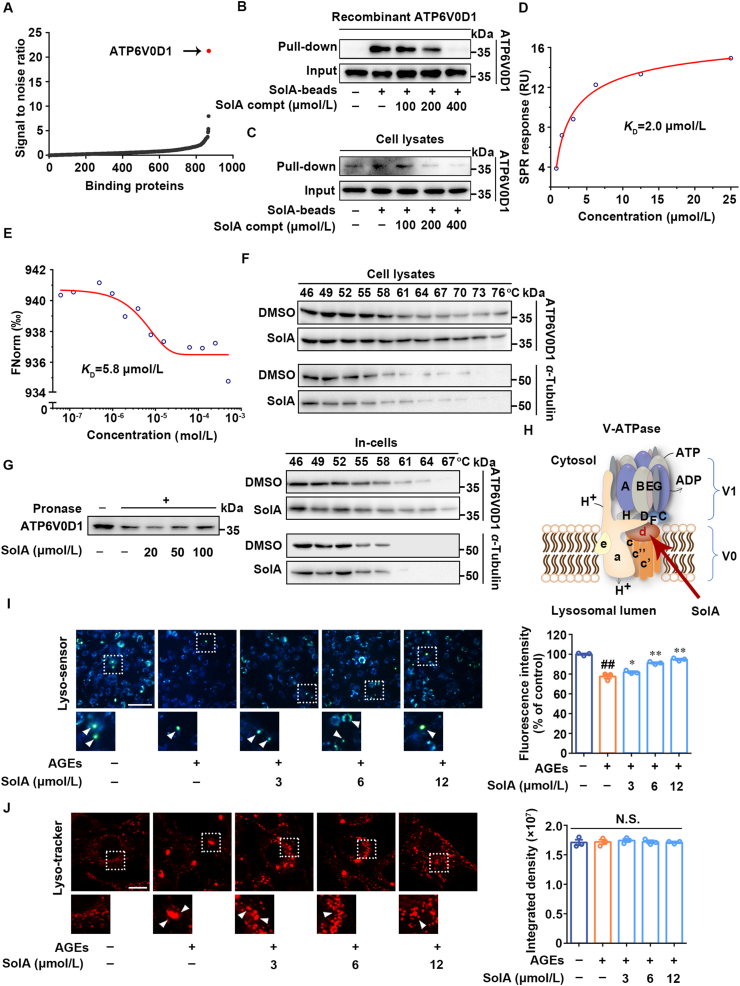
Cellular target identification of small-molecules has a key role in elucidating mechanism of action. Since Baf-A1 is a specific inhibitor of V-ATPase in lysosome and effectively abolishes SolA-dependent BIM degradation, we then speculated that V-ATPase may be a potential cellular target of SolA. To this end, proteomics-based pull-down strategy coupled with quantitative mass spectrometry was used to screen the direct cellular target of SolA. As shown in Fig. 3A, among the hotspots interacted with SolA in cell proteome, a protein showing the most obvious differential expression was identified as ATP6V0D1, which is a key component of V-ATPase. This observation was also confirmed by pull-down analysis with cell lysates and recombinant ATP6V0D1 (Fig. 3B and C). SPR analysis further verified the interaction between SolA and ATP6V0D1 with an equilibrium dissociation constant (KD) value of 2.0 μmol/L (Fig. 3D). Besides, MST analysis also showed a specific interaction between SolA and ATP6V0D1 with the KD value of 5.8 μmol/L (Fig. 3E). Moreover, CETSA showed that SolA significantly protected ATP6V0D1 from temperature-dependent denaturation, suggesting that SolA directly interacted with ATP6V0D1 (Fig. 3F and Supporting Information Fig. S5A). DARTS showed that SolA concentration-dependently inhibited ATP6V0D1 degradation induced by pronase (Fig. 3G). Meanwhile, we found that AGEs treatment with or without SolA did not cause the expression change of ATP6V0D1 protein and mRNA (Fig. S5B and S5C).
Figure 3.
Identification of V-ATPase subunit ATP6V0D1 as a cellular target of SolA. (A) Identification of cellular target of SolA in PC12 cells proteome. ATP6V0D1 showed the highest signal to noise ratio. (B) SolA selectively bound to recombinant ATP6V0D1 by pull-down analysis. (C) SolA selectively bound to endogenous ATP6V0D1 by pull-down analysis. (D) Binding affinity analysis of SolA with ATP6V0D1 determined by surface plasmon resonance (SPR). (E) Binding affinity analysis of SolA with ATP6V0D1 determined by microscale thermophoresis (MST). (F) SolA promoted ATP6V0D1 resistant to different temperature gradients by cellular thermal shift assays (CETSA) analysis. (G) SolA promoted ATP6V0D1 resistant to proteases by drug affinity responsive target stability (DARTS) analysis. (H) Structure of lysosomal proton pump V-ATPase. (I) SolA promoted lysosomal pH decrease in AGEs-induced PC12 cells (scale bar: 100 μm, n = 3). Arrows indicate highly acidic lysosomes. (J) SolA reversed aberrant lysosomal aggregation in AGEs-induced PC12 cells (scale bar: 10 μm, n = 3). Arrows indicate significantly aggregated lysosomes. Data are expressed as the mean ± SEM. ##P < 0.01 vs. control group, ∗P < 0.05, ∗∗P < 0.01 vs. AGEs group. N.S. not significant.
Since ATP6V0D1 is involved in V-ATPase-mediated lysosomal acidification (Fig. 3H), we then measured the lysosomal pH with Lyso-sensor Yellow/Blue DND160 to explore the effect of SolA on the biological function of cellular target. As shown in Fig. 3I, AGEs induced lysosome lumenal pH increase in PC12 cells with reduced fluorescence intensity, which was noticeably restored by SolA treatment. Also, DQ Red-BSA processing assay was conducted to visualize the lysosomal proteolytic activity. We observed that SolA treatment significantly reversed the loss of DQ Red-BSA fluorescence intensity induced by AGEs (Fig. S5D). Meanwhile, both AGEs and SolA treatment had no significant effects on Cathepsin B expression, suggesting that SolA rescued lysosomal activity by restoring lysosomal acidity rather than regulating the expression of cathepsins (Fig. S5E). Moreover, we found that lysosomes aggregated into large clusters after AGEs treatment, indicating the occurrence of lysosome–lysosome fusion with abnormal phagolytic function. However, SolA effectively reversed the aberrant aggregation of lysosomes (Fig. 3J).
To investigate the effect of SolA on lysosome biogenesis, the mRNA expression of Lamp1 and Lamp2 were evaluated by real-time PCR. The results showed that SolA did not significantly change the transcription levels of Lamp1 and Lamp2 in AGEs-induced PC12 cells (Fig. S5F). Additionally, as shown in Fig. S5G, SolA did not induce obvious nuclear translocation of TFEB in AGEs-induced PC12 cells. Thus, SolA showed no obvious effect on lysosomal biogenesis. Taken together, our findings supported that ATP6V0D1 was a direct cellular target of SolA for promoting lysosomal acidification.
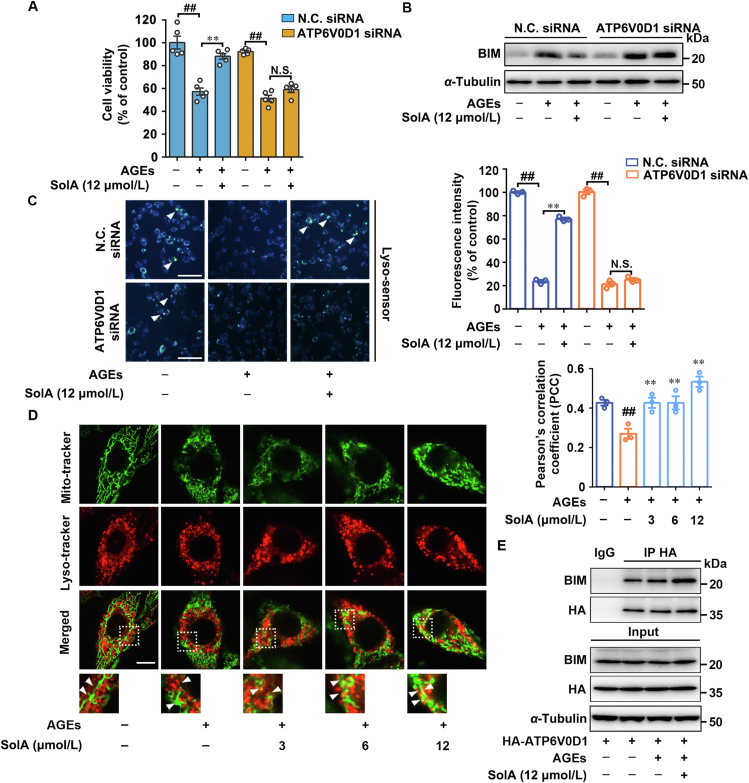
Next, biological correlation between ATP6V0D1 and BIM degradation was investigated using specific ATP6V0D1 siRNA (knockdown efficiency > 70%). Since ATP6V0D1 is essential for V-ATPase assembly and lysosomal acidification, excessive knockdown of ATP6V0D1 may cause severe damage to neuronal cells (Fig. S5H). Therefore, we controlled the relative low transfection dosage of siRNA to ensure the survival of neuronal cells to explore the neuroprotective effect of SolA. As shown in Fig. 4A, ATP6V0D1 siRNA abolished SolA-dependent neuroprotective effect under AGEs insult. Moreover, ATP6V0D1 siRNA significantly reversed SolA-induced BIM degradation (Fig. 4B), indicating that ATP6V0D1 was responsible for neuronal survival via regulating BIM stability. Furthermore, we observed that ATP6V0D1 deficiency ablated the effect of SolA-mediated lysosomal pH decrease (Fig. 4C), supporting the functional correlation between ATP6V0D1 and neuroprotection via promoting lysosomal acidification-dependent mitochondrial BIM degradation. Meanwhile, other three potential binding proteins, including multidrug resistance-associated protein 5 (ABCC5), cullin-3 (CUL-3), and scaffold attachment factor B1 (SAFB1) were investigated for their neuroprotective effects with specific siRNA knockdown. Our result shows that these proteins were not responsible for the SolA-mediated protective effect on PC12 cells, indicating that they are not the cellular targets for SolA-mediated neuroprotection (Supporting Information Fig. S6A–S6C). In addition, SolA has been reported to regulate IKKβ in non-small cell lung cancer, while our findings suggest that IKKβ cascade may be not involved in the neuroprotective activity of SolA in neuronal cells (Fig. S6D).
Figure 4.
SolA protects PC12 cells death through lysosome–mitochondria crosstalk. (A) ATP6V0D1 siRNA inhibited SolA (12 μmol/L)-dependent protection in PC12 cells by MTT assay (n = 5). (B) SolA (12 μmol/L)-induced BIM degradation was blocked by ATP6V0D1 siRNA. (C) ATP6V0D1 siRNA reversed SolA (12 μmol/L)-mediated lysosomal acidification by Lyso-sensor detection (scale bar: 100 μm, n = 3). Arrows indicate highly acidic lysosomes. (D) SolA increased co-localization of lysosomes with mitochondria (scale bar: 10 μm, n = 3). (E) SolA (12 μmol/L) induced BIM binding to ATP6V0D1 in AGEs-induced PC12 cells. Data are expressed as the mean ± SEM. ##P < 0.01 vs. control group, ∗∗P < 0.01 vs. AGEs group. N.S. not significant.
The correlation between ATP6V0D1-associated lysosomal acidification and BIM degradation suggested a potential molecular mechanism that SolA regulates mitochondrial-lysosomal crosstalk. Therefore, double fluorescence staining of lysosomes and mitochondria in PC12 cells was conducted, and result shows that SolA significantly increased the co-localization of lysosomes with mitochondria in a concentration-dependent manner (Fig. 4D). Then, we also performed co-IP assay to determine whether there was an interaction between ATP6V0D1 and BIM. Our result shows that SolA significantly promoted the association of ATP6V0D1 with BIM in AGEs-induced PC12 cells (Fig. 4E), while failed to induce BAD, BCL-XL, BAK, BAX, and BCL-2 binding to ATP6V0D1 (Fig. S6E), indicating that SolA selectively induced BIM/ATP6V0D1 interaction to promote BIM clearance in lysosome-dependent manner. Additionally, we found that BCL-2 homology domain 3 (BH3) deletion obviously abolished the association between ATP6V0D1 and BIM, indicating that BH3 is a crucial domain for mediating ATP6V0D1/BIM interaction (Fig. S6F). Subsequently, we compared the BH3 sequences in several BCL-2 family proteins, including BAD, BAX, BCL-2, BCL-XL and BIM. Analysis data showed that glutamine 148 (Gln148) and glutamic 159 (Glu159) uniquely existed in BIM, hinting that Gln148 and Glu159 might be the potential sites of BIM-ATP6V0D1 interaction. Therefore, we constructed Bim plasmids with point mutation on Gln148 (BimQ148A) or Glu159 (BimE159A), and the double mutation (BimQ148A/E159A). The co-IP analysis showed a significant decrease of the interaction between ATP6V0D1 and BIM with E159A mutant, while the interaction of ATP6V0D1 with BIMQ148A showed no obvious change, indicating that Glu159 rather than Gln148 was a crucial site mediating BIM-ATP6V0D1 interaction. Notably, the interaction of ATP6V0D1 with BIMQ148A/E159A showed the most significant decrease, suggesting that Gln148 may play a synergistic role in ATP6V0D1/BIM interaction with E159 (Fig. S6G). Taken together, these results reveal that ATP6V0D1 may serve as a direct sensor in lysosome for BIM recognition and resultant protein degradation via lysosome acidification.
3.4. Cysteine 335 is an allosteric site in ATP6V0D1 for V-ATPase activation
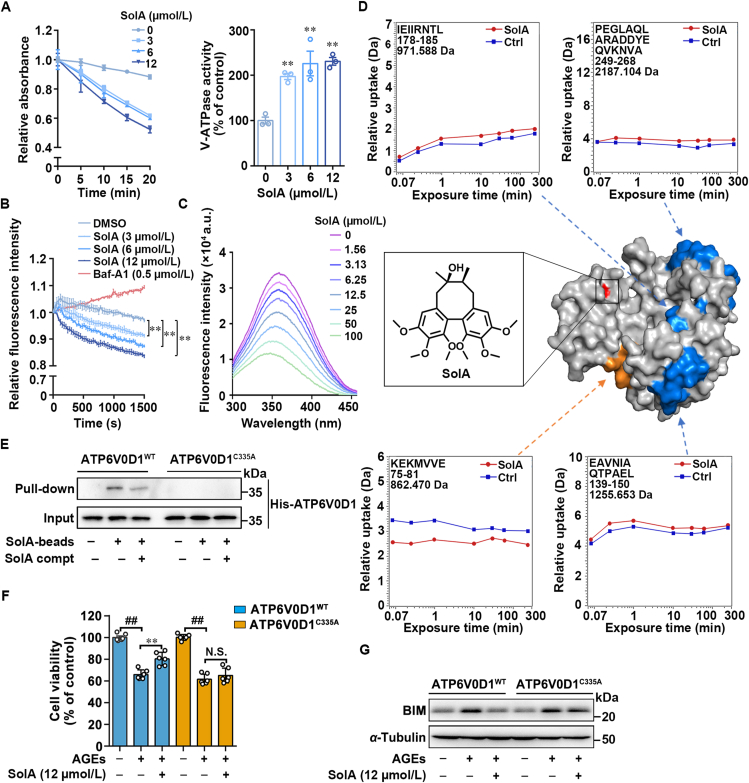
V-ATPase activity assay showed that SolA significantly increased the NAD+ production, indicating that SolA dose-dependently promoted V-ATPase activity of hydrolyzing ATP (Fig. 5A). Moreover, we performed real-time acidification assay to detect the rate of V-ATPase-dependent H+ pumping. The result showed that SolA concentration-dependently decreased the fluorescence intensity, suggesting an increase of H+ pumping rate and a promoting effect of SolA on lysosomal acidity. Meanwhile, Baf-A1 as H+ pumping inhibitor increased the fluorescence intensity (Fig. 5B).
Figure 5.
Cys335 serves as a pharmacological allosteric site of ATP6V0D1. (A) SolA promoted V-ATPase activity in a concentration-dependent manner (n = 3). (B) SolA increased the rate of H+ pumping in a concentration-dependent manner (n = 3). (C) SolA mediated ATP6V0D1 conformational change by tryptophan fluorescence quenching analysis. (D) SolA allosterically regulated ATP6V0D1 conformation by hydrogen-deuterium exchange mass spectrometry (HDXMS). SolA binding site is shown in red. Peptides with increased deuterium uptake ratio after SolA treatment are highlighted in orange, and peptides with decreased deuterium uptake ratio were highlighted in blue (PDB: 6wlw). (E) Pull-down analysis of SolA with ATP6V0D1WT or ATP6V0D1C335A. (F) ATP6V0D1 C335A mutation inhibited SolA (12 μmol/L)-mediated protection in PC12 cells by MTT assay (n = 6). (G) SolA (12 μmol/L)-induced BIM degradation was blocked by ATP6V0D1 C335A mutation. Data are expressed as the mean ± SEM. ##P < 0.01 vs. control group, ∗∗P < 0.01 vs. AGEs group. N.S. not significant.
Since allosteric effect is an important strategy for regulating protein function by small-molecules, we then tested whether SolA modulated ATP6V0D1 conformation via tryptophan fluorescence quenching assay. Here, we found the fluorescence intensity of SolA–ATP6V0D1 complex significantly decreased with SolA concentration increase, indicating that SolA induced ATP6V0D1 conformation changes (Fig. 5C). Next, we explored the binding residues of SolA on ATP6V0D1. MS analysis revealed that cysteine 335 (Cys335) in ATP6V0D1 was selectively bound by SolA (Supporting Information Fig. S7A). Then, we explored the allosteric regulation mechanism of SolA on ATP6V0D1 by hydrogen deuterium exchange (HDX) mass spectrometry (MS). As shown in Fig. 5D, SolA treatment altered the hydrogen/deuterium exchange levels of four specific peptides (peptides 75–81, 139–150, 178–185, and 249–268). Of note, the significant decrease of H/D exchange rate in peptide 75–81 during a short exchange-time suggested this sequence excreted a critical function through embedding into the hydrophobic internal region of ATP6V0D1. Moreover, peptides 139–150, 178–185, and 249–268 showed higher H/D exchange rates after SolA treatment, indicating these areas were induced to exposure in solution upon SolA binding. Meanwhile, H/D exchange rate of most regions, such as peptide 58–69 remained unchanged, revealing SolA reshaped the protein surface of ATP6V0D1 by allosteric regulation.
Since the cysteine 335 (Cys335) in ATP6V0D1 was selectively bound by SolA (Fig. S7A). Thus, we speculated that Cys335 was crucial for SolA-binding to ATP6V0D1. To this end, we constructed Atp6v0d1 plasmid with point mutation on Cys335 (Atp6v0d1C335A) and pull-down analysis showed a significant decrease of the interaction between SolA and ATP6V0D1C335A (Fig. 5E). Furthermore, C335A mutation effectively abolished the binding between SolA and ATP6V0D1 (KD > 100 μmol/L), as confirmed by SPR analysis (Fig. S7B). Meanwhile, MST analysis also showed the weakened interaction between SolA and ATP6V0D1C335A (Fig. S7C). Additionally, tryptophan fluorescence quenching assay demonstrated that C335A mutation significantly weakened the SolA-induced ATP6V0D1 conformation changes (Fig. S7D–S7E). Moreover, we found that ATP6V0D1C335A obviously antagonized SolA-induced neuronal cells survival (Fig. 5F) and Bim degradation (Fig. 5G), indicating that Cys335 of ATP6V0D1 is essential for SolA-dependent BIM degradation and neuroprotective effect. Besides, SolA down-regulated ROS production in AGEs-induced PC12 cells, which was significantly reversed by ATP6V0D1 C335A (Fig. S7F). We performed co-IP experiment by co-transfection with HA-Atp6v0d1C335A and Myc-Bim, and found that mutation of Cys335 did not affect the association between ATP6V0D1 and BIM (Fig. S7G). Collectively, these findings reveal that SolA allosterically regulated ATP6V0D1 for V-ATPase activation via selectively targeting Cys335.
The hydroxyl group connected to the octatomic ring structure in SolA is a chemically active group. To explore the importance of hydroxyl group in the interactions between SolA and Cys335, we determined the neuroprotective effects of SolA and Schisandrin A (SchA), a natural compound structurally similar to SolA but without hydroxyl group (Fig. S7H, Supporting Information Table S1). MTT assay suggested that SolA rather than SchA effectively reversed AGEs-induced PC12 cells death (Fig. S7I), revealing the essential role of hydroxyl group in SolA-dependent neuroprotective effect. Moreover, SPR result demonstrated that SchA failed to bind to ATP6V0D1 (KD N.D.), while SolA showed specific binding with ATP6V0D1 (KD = 2.0 μmol/L) (Fig. 3D, Fig. S7J). These results further support that the hydroxyl group plays a key role in mediating the interaction of SolA with ATP6V0D1 (Cys335) for neuroprotection.
3.5. SolA protects mitochondria against AGEs-associated oxidative stress via promoting lysosomal acidification in vitro and in vivo
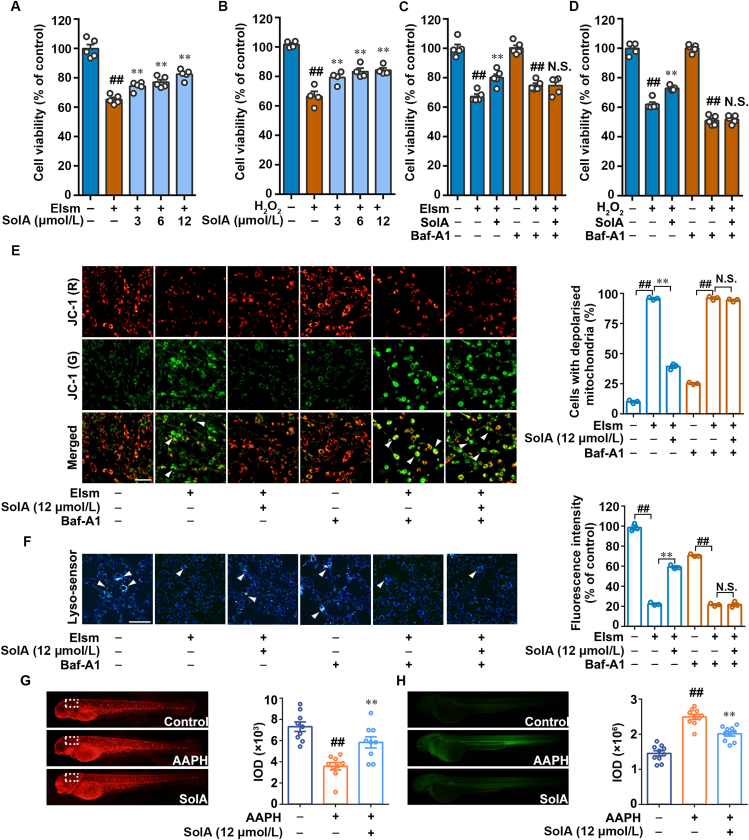
Mitochondria is one of the main sources of oxidative stress39, 40, 41, 42; thus, AGEs-induced mitochondrial dysfunction elicited ROS production to further accelerate diabetic neuropathy. For these crucial pathological characteristics, we established elesclomol and H2O2-induced oxidative damage models in PC12 cells, and tried to explore the potential neuroprotective effect of SolA. MTT assay demonstrated that SolA prevented elesclomol and H2O2-induced oxidative stress damage on PC12 cells (Fig. 6A and B), which were markedly abolished by ATPase inhibitor Baf-A1 (Fig. 6C and D). Next, we observed that SolA significantly protected elesclomol and H2O2-induced mitochondrial damage, which was reversed by ATPase inhibition with Baf-A1 (Fig. 6E, Supporting Information Fig. S8A). Moreover, Baf-A1 obviously blocked SolA-promoted lysosomal pH decrease in elesclomol and H2O2-induced PC12 cells, further confirming the important role of ATPase in SolA-mediated neuroprotection via promoting lysosomal acidification (Fig. 6F, Fig. S8B). Meanwhile, we found that SolA did not directly react with H2O2 in vitro (Fig. S8C). Besides, we analyzed the mitochondrial function and ROS content of AAPH-induced zebrafish in vivo. Mito-tracker Red staining showed that AAPH collapsed mitochondrial membrane potential in zebrafish nervous tissue, which was significantly reversed by 12 μmol/L SolA (Fig. 6G). Meanwhile, SolA dramatically decreased the content of ROS induced by AAPH in zebrafish (Fig. 6H). In addition, we found that AAPH decreased mitochondrial membrane potential in zebrafish hearts, which was not significantly improved by SolA treatment (Fig. S8D). Moreover, in the zebrafish liver, there was no significant difference in mitochondrial function with or without AAPH or SolA treatments (Fig. S8E). These results suggest that anti-mitochondrial oxidative damage may be a crucial mechanism of action involved in SolA-mediated neuroprotective effect in diabetic neuropathy.
Figure 6.
ATP6V0D1 is necessary for SolA-mediated mitochondrial protection and lysosomal acidification. (A) SolA increased the cell viability in elesclomol (Elsm)-induced PC12 cells by MTT assay (n = 5). (B) SolA increased the cell viability in H2O2-induced PC12 cells by MTT assay (n = 4). (C) Baf-A1 inhibited SolA-mediated protection in Elsm-induced PC12 cells by MTT assay (n = 5). (D) Baf-A1 inhibited SolA-mediated protection in H2O2-induced PC12 cells by MTT assay (n = 5). (E) Baf-A1 eliminated SolA-mediated mitochondrial depolarization by JC-1 staining in Elsm-induced PC12 cells (scale bar: 100 μm, n = 3). Arrows indicate the cells with depolarized mitochondria. (F) Baf-A1 reversed SolA-mediated lysosomal acidification by Lyso-sensor detection in Elsm-induced PC12 cells (scale bar: 100 μm, n = 3). Arrows indicate highly acidic lysosomes. (G) SolA (12 μmol/L) enhanced mitochondrial activity by Mito-tracker Red CMXRos staining in AAPH-induced zebrafishes (n = 9). AAPH (6.25 mmol/L) was used to induce the oxidative stress. (H) SolA (12 μmol/L) reduced mitochondrial ROS in AAPH-induced zebrafishes (n = 10). Data are expressed as the mean ± SEM. ##P < 0.01 vs. control group, ∗∗P < 0.01 vs. AGEs group. N.S. not significant.
To explore whether antioxidant enzymes and ROS-generating enzymes were regulated by SolA. We then evaluated the expression of HO-1 and iNOS proteins. The result showed that the expression of HO-1 and iNOS in the AGEs group was significantly increased. Moreover, SolA showed a slight up-regulation of HO-1 expression and a moderate down-regulation of iNOS expression, indicating that antioxidant enzymes and ROS-generating enzymes may be partly involved in the anti-oxidative stress effects mediated by SolA (Fig. S8F). Since ROS may be produced from some other organelles, such as peroxisomes; therefore, protein expression of catalase as a major peroxisome protein was detected by Western blot. The result reveals that AGEs and SolA had no obvious effect on the level of catalase (Fig. S8G). Besides, the expression of PEX5 as a redox-sensitive protein was detected. As shown in Fig. S8G, PEX5 expression showed no obvious change after AGEs and SolA treatments. Meanwhile, AGEs and SolA did not change the fluorescence intensity of catalase (Fig. S8H). Thus, we speculate that AGEs-induced ROS production may not originate from peroxisome, but damaged mitochondria in PC12 cells.
3.6. SolA exerts a neuroprotective effect in ALX-induced diabetic neuropathy (DN) mice
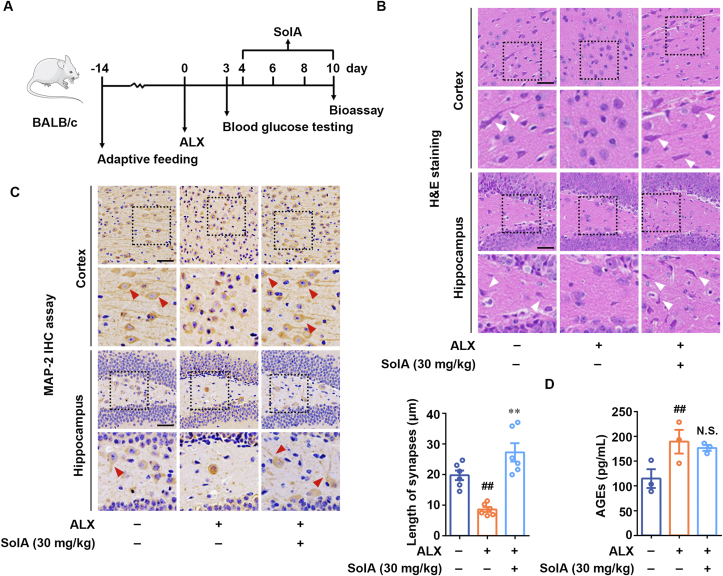
We next investigated the neuroprotective effect of SolA in a mouse DN model (Fig. 7A). H&E staining analysis showed that ALX induced significant neuronal damage in the cortex and hippocampal regions, including neuronal shrinkage and abnormal nuclei. However, SolA treatment obviously decreased the number of injured neurons in diabetic mice, indicating an obvious improvement in diabetic neuropathy (Fig. 7B). Moreover, immunohistochemistry analysis of MAP-2 was performed to demonstrate the neuronal morphological features. We observed shortening synapses and rounding neurons in the cerebral cortex and hippocampal regions of diabetic mice, which was significantly improved by SolA treatment (Fig. 7C). These results suggest that SolA markedly suppressed ALX-induced diabetic neuropathy in vivo. Thus, our results reveal that SolA exerts a significant neuroprotective effect in ALX-induced diabetic neuropathy mice. In addition, we found that the serum AGEs level of ALX-induced diabetic mice was obviously increased. Meanwhile, SolA treatment showed no significant effects on the serum AGEs level of diabetic mice (Fig. 7D).
Figure 7.
SolA exerts neuroprotection in alloxan (ALX)-induced diabetic neuropathy mouse model. (A) Scheme of neuroprotection evaluation of SolA in diabetic neuropathy model. (B) SolA decreased the number of injured neurons by H&E staining (scale bar: 40 μm). Arrows indicate the neuronal synapses. (C) SolA protected neurons from morphological damage by immunohistochemistry staining of MAP-2 (scale bar: 40 μm, n = 6). Arrows indicate the neuronal synapses. (D) SolA had no effect on the serum AGEs level of diabetic mice (n = 3). Data are expressed as the mean ± SEM. ##P < 0.01 vs. control group. N.S. not significant.
4. Discussion
The development of therapeutic options which inhibit diabetic neuropathy are critical for extending the lifespan of affected individuals. Since the formation of AGEs is a fundamental biochemical abnormality that is responsible for neuronal injury in diabetes; herein, we describe the discovery and characterization of a novel small-molecule ATPase activator SolA by targeting a unique ATP6V0D1 subunit to show neuroprotection against AGEs-induced neuronal cells apoptosis. Meanwhile, Schisandrin A (SchA), a natural compound structurally similar to SolA, showed little protective effect against AGEs-induced diabetes-associated neuronal damage. We observed that mitochondria and pro-apoptotic protein Bim are highly involved in the neuroprotective effect of SolA, while the other apoptosis regulatory proteins in mitochondria including BCL-2, BAX and BIK showed little contribution. Moreover, accumulating evidences have shown that BIM was associated with mitochondrial ROS production. For example, in neuroblastoma cells, overexpression of BIM caused ROS production and lowered mitochondrial respiration43. Moreover, BIM-mediated mitochondrial function inhibition trigger cell apoptosis by generating ROS44,45. Furthermore, BIM expression contributes to ROS production under high glucose condition46. Therefore, we speculate that SolA selectively suppress BIM expression to repair mitochondrial function.
As a major cause of failure in drug discovery is lack of effective druggable targets, our findings open a new avenue for the development of therapeutic agents for diabetic neuropathy via providing ATP6V0D1 subunit in V-ATPase as a valuable therapeutic target. To investigate the potential function of ATP6V0D1 in SolA exerting neuroprotective effect, specific ATP6V0D1 siRNA was used in our experiments. Since ATP6V0D1 is essential for V-ATPase assembly and lysosomal acidification, excessive knockdown of ATP6V0D1 may cause severe damage to neuronal cells. Therefore, the relative low transfection dosage of siRNA was used as an alternative solution to ensure the survival of neuronal cells for exploring the neuroprotective activity of SolA. Since lysosomes play an important role in controlling intracellular proteolysis, then we speculate that ATP6V0D1 may potentially interact with more functional proteins in other cell models, such as macrophages and hepatocytes to exert wide biological activities.
Although previous reports revealed V-ATPase plays essential roles on neurological function maintenance, the underlying molecular mechanism and pharmacological strategy for hyperglycemia-associated neuronal cells apoptosis is largely unknown. Here, we reported the first natural small-molecule SolA directly bound to ATP6V0D1 subunit to activate V-ATPase for neuroprotection against AGEs-induced neuronal injury. Nowadays, studies have revealed that V-ATPase contains V1 domain and V0 domain. V1 domain contains the ATP catalytic site; meanwhile, V0 domain, consisting of five different subunits: a, c, c', c'', and d, is responsible for translocation of protons across the membrane. Particularly, it has been assumed that protons are transported through a transmembrane channel formed by a and c subunits47,48. Moreover, d subunit also plays a crucial role in modulating proton transport. Particularly, as a fundamental subtype of d subunit, ATP6V0D1 is highly involved in the assembly of V0 domain, and further regulates the interaction between V0 and V1 domains49, which finally affects the protons transport. Here, we found that SolA bound to a previously undisclosed ATP6V0D1 subunit in V0 domain, which is completely different from the currently discovered compounds such as verucopeptin50, cysteine-reactive acrylamide EN618 and polymyxin B derivative17, that directly target the classical catalytic pocket in V1 domain. Therefore, our findings represent a new class of V-ATPase modulation methodology through targeting V0 domain. Moreover, existing reports indicated that Baf-A1, a potent and specific inhibitor of V-ATPase, could also directly bind to the interface of the rotating c subunits and the stationary a subunit in V0 domain48. Thus, Baf-A1 is particularly important in elucidating the physiological role of V-ATPase. Then, it was used in this study to explore the biological functions of V0 domain and the neuroprotective mechanism of SolA. Our findings highlight the importance of small-molecules targeting the unique ATP6V0D1 subunit to activate V-ATPase, which may lay a groundwork for new therapeutic strategy in diabetic neuropathy.
To explain how SolA-mediated ATP6V0D1 conformation change is involved in V-ATPase biology, we proposed two major possibilities. First, the allosteric mechanism of SolA likely accounted for its observed regulation on hydrophobic structure change to facilitate V-ATPase complex assembly, thereby resulting in proton transport and ATP hydrolysis for V-ATPase activation. The structural collapse of ATP6V0D1 residues 71–85 caused by SolA binding may promote the assembly of V0 domain via inducing the interaction of adjacent a and c subunits. Moreover, SolA-dependent allosteric regulation of ATP6V0D1 promoted the BIM–ATP6V0D1 interaction in AGEs-induced PC12 cells. We hypothesize that ATP6V0D1 may serve as a sensor in lysosome for BIM recognition and capture. After binding to lysosomal surface via interaction with ATP6V0D1, BIM may be loaded into lysosomes by classical membrane invagination-dependent manner. Particularly, further investigations about the specific molecular mechanism of lysosomal membrane invagination are needed in future. Furthermore, our results established a direct biology relationship between mitochondria and lysosome via V-ATPase/BIM/mitochondrial apoptosis signaling pathway. Thus, BIM may act as a signal hub protein for linking mitochondria and lysosome contact, and we provide a proof-of-concept strategy for pharmacologically controlling mitochondrial-lysosomal crosstalk for neuronal cell protection. Meanwhile, several reports have shown that SolA can cross the blood-brain barrier to exert pharmacological effects in mouse brain51,52, indicating an acceptable pharmacokinetic property for diabetic encephalopathy therapy in vivo.
In summary, our findings provide ATP6V0D1 subunit of V-ATPase as a promising therapeutic target against AGEs-induced neuronal injury. Moreover, the discovery of natural product SolA represents an important first step toward novel candidate drug discovery via a completely new mechanism of regulating lysosomal acidification for diabetic neuropathy.
5. Conclusions
We for the first time describe the discovery of natural small-molecule SolA as a valuable V-ATPase activator against diabetic neuropathy. The novelty of this small-molecule stems from the fact that it allosterically regulates a unique ATP6V0D1 subunit conformation via Cys355 to boost V-ATPase function, thereby promoting lysosomal acidification. Moreover, we identify mitochondrial proapoptotic protein BIM as a crucial substrate for SolA-mediated lysosomal degradation, thereby maintaining mitochondrial homeostasis for neuronal cells survival. Therefore, selective small-molecule targeting ATP6V0D1 may represent a promising proof-of-concept therapeutic strategy for diabetic neuropathy. Meanwhile, SolA may act as a crucial molecule template for interrogating new biology of V-ATPase in neuronal cells apoptosis in diabetes.
Acknowledgments
We would like to thank Yichi Zhang for her contribution to the animal experiment. This work was financially supported by National Key Research and Development Program of China (Nos. 2019YFC1708902 and 2019YFC1711000), National Natural Science Foundation of China (Nos. 81973505, 81773932 and 82104621).
Author contributions
Kewu Zeng designed the project. Xiaoqing Zhou, Shaoyang Zhao and Tingting Liu performed most of the experiments and analyzed data. Lu Yao synthesized the SolA-coupled magnetic nanoparticles. Yichi Zhang, Meimei Zhao, Xiaoming Ye, Xiaowen Zhang and Qiang Guo performed the molecular biology and cell biology experiments. Kewu Zeng, Xiaoqing Zhou and Tingting Liu wrote the manuscript. Pengfei Tu contributed to the manuscript editing. All authors contributed and approved the final version of the manuscript.
Conflicts of interest
The authors declare no conflicts of interest.
Footnotes
Peer review under the responsibility of Chinese Pharmaceutical Association and Institute of Materia Medica, Chinese Academy of Medical Sciences.
Supporting data to this article can be found online at https://doi.org/10.1016/j.apsb.2022.06.013.
Appendix A. Supplementary data
The following is the Supplementary data to this article:
References
- 1.Liu X.X., Xu Y.Y., An M.M., Zeng Q.B. The risk factors for diabetic peripheral neuropathy: a meta-analysis. PLoS One. 2019;14 doi: 10.1371/journal.pone.0212574. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Dewanjee S., Das S., Das A.K., Bhattacharjee N., Dihingia A., Dua T.K., et al. Molecular mechanism of diabetic neuropathy and its pharmacotherapeutic targets. Eur J Pharmacol. 2018;833:472–523. doi: 10.1016/j.ejphar.2018.06.034. [DOI] [PubMed] [Google Scholar]
- 3.Ahmed N. Advanced glycation end products role in pathology of diabetic complications. Diabetes Res Clin Pract. 2005;67:3–21. doi: 10.1016/j.diabres.2004.09.004. [DOI] [PubMed] [Google Scholar]
- 4.Loske C., Neumann A., Cunningham A.M., Nichol K., Schinzel R., Riederer P., et al. Cytotoxicity of advanced glycation end products is mediated by oxidative stress. J Neural Transm. 1998;105:1005–1015. doi: 10.1007/s007020050108. [DOI] [PubMed] [Google Scholar]
- 5.Alu A., Han X.J., Ma X.L., Wu M., Wei Y.Q., Wei X.W. The role of lysosome in regulated necrosis. Acta Pharm Sin B. 2020;10:1880–1903. doi: 10.1016/j.apsb.2020.07.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Ballabio A., Bonifacino J.S. Lysosomes as dynamic regulators of cell and organismal homeostasis. Nat Rev Mol Cell Biol. 2020;21:101–118. doi: 10.1038/s41580-019-0185-4. [DOI] [PubMed] [Google Scholar]
- 7.Casey J., Grinstein S., Orlowski J. Sensors and regulators of intracellular pH. Nat Rev Mol Cell Biol. 2010;11:50–61. doi: 10.1038/nrm2820. [DOI] [PubMed] [Google Scholar]
- 8.Bouch V., Espinos A.P., Leone L., Sardiello M., Ballabio A., Botas J. Drosophila Mitf regulates the V-ATPase and the lysosomal–autophagic pathway. Autophagy. 2016;12:484–498. doi: 10.1080/15548627.2015.1134081. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Mauvezin C., Neufeld T.P. Bafilomycin A1 disrupts autophagic flux by inhibiting both V-ATPase-dependent acidification and Ca-P60A/SERCA-dependent autophagosome–lysosome fusion. Autophagy. 2015;11:1437–1438. doi: 10.1080/15548627.2015.1066957. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Mauvezin C., Nagy P., Juhász G., Neufeld T.P. Autophagosome–lysosome fusion is independent of V-ATPase-mediated acidification. Nat Commun. 2015;6:7007. doi: 10.1038/ncomms8007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Toei M., Saum R., Forgac M. Regulation and isoform function of the V-ATPases. Biochemistry. 2010;49:4715–4723. doi: 10.1021/bi100397s. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Zhao J., Benlekbir S., Rubinstein J.L. Electron cryomicroscopy observation of rotational states in a eukaryotic V-ATPase. Nature. 2015;521:241–245. doi: 10.1038/nature14365. [DOI] [PubMed] [Google Scholar]
- 13.Wang L.F., Wu D., Robinson C.V., Wu H., Fu T.M. Structures of a complete human V-ATPase reveal mechanisms of its assembly. Mol Cell. 2020;80:501–511. doi: 10.1016/j.molcel.2020.09.029. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Rawson S., Phillips C., Huss M., Tiburcy F., Wieczorek H., Trinick J., et al. Structure of the vacuolar H+-ATPase rotary motor reveals new mechanistic insights. Structure. 2015;23:461–471. doi: 10.1016/j.str.2014.12.016. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Toro E.J., Ostrov D.A., Wronski T.J., Holliday L.S. Rational identification of enoxacin as a novel V-ATPase-directed osteoclast inhibitor. Curr Protein Pept Sci. 2012;13:180–191. doi: 10.2174/138920312800493151. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Chen Y.C., Backus K.M., Merkulova M., Yang C., Brown D., Cravatt B.F., et al. Covalent modulators of the vacuolar ATPase. J Am Chem Soc. 2017;139:639–642. doi: 10.1021/jacs.6b12511. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Iida M., Yamada K., Nango Y., Yamaguchi Y., Ogita A., Fujita K., et al. Vacuolar H+-ATPase subunit Vma1p functions as the molecular ligand in the vacuole-targeting fungicidal activity of polymyxin B. Microbiology. 2017;163:531–540. doi: 10.1099/mic.0.000450. [DOI] [PubMed] [Google Scholar]
- 18.Chung C.Y., Shin H.R., Berdan C.A., Ford B., Ward C.C., Olzmann J.A., et al. Covalent targeting of the vacuolar H+-ATPase activates autophagy via mTORC1 inhibition. Nat Chem Biol. 2019;15:776–785. doi: 10.1038/s41589-019-0308-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Eaton A.F., Merkulova M., Brown D. The H+-ATPase (V-ATPase): from proton pump to signaling complex in health and disease. Am J Physiol Cell Physiol. 2021;320:392–414. doi: 10.1152/ajpcell.00442.2020. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Song L., Piao Z.Y., Yao L.F., Zhang L.M., Lu Y.C. Schisandrin ameliorates cognitive deficits, endoplasmic reticulum stress and neuroinflammation in streptozotocin (STZ)-induced Alzheimer's disease rats. Exp Anim. 2020;69:363–373. doi: 10.1538/expanim.19-0146. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Jiang Y.M., Fan X.M., Wang Y., Tan H., Chen P., Zeng H., et al. Hepato-protective effects of six Schisandra lignans on acetaminophen-induced liver injury are partially associated with the inhibition of CYP-mediated bioactivation. Chem Biol Interact. 2015;231:83–89. doi: 10.1016/j.cbi.2015.02.022. [DOI] [PubMed] [Google Scholar]
- 22.Fan S.C., Liu C.H., Jiang Y.M., Gao Y., Chen Y.X., Fu K.L., et al. Lignans from Schisandra sphenanthera protect against lithocholic acid-induced cholestasis by pregnane X receptor activation in mice. J Ethnopharmacol. 2019;245 doi: 10.1016/j.jep.2019.112103. [DOI] [PubMed] [Google Scholar]
- 23.Xian H.B., Feng W.N., Zhang J.R. Schizandrin A enhances the efficacy of gefitinib by suppressing IKKβ/NF-κB signaling in non-small cell lung cancer. Eur J Pharmacol. 2019;855:10–19. doi: 10.1016/j.ejphar.2019.04.016. [DOI] [PubMed] [Google Scholar]
- 24.Song L., Yao L.F., Zhang L.M., Piao Z.Y., Lu Y.C. Schizandrol A protects against Aβ1–42-induced autophagy via activation of PI3K/AKT/mTOR pathway in SH-SY5Y cells and primary hippocampal neurons. Naunyn Schmiedeberg’s Arch Pharmacol. 2020;393:1739–1752. doi: 10.1007/s00210-019-01792-2. [DOI] [PubMed] [Google Scholar]
- 25.Zhang M., Huo D.S., Cai Z.P., Shao G., Wang H., Zhao Z.Y., et al. The effect of schizandrol A-induced DNA methylation on SH-SY5Y Aβ1–40 altered neuronal cell line: a potential use in Alzheimer's disease. J Toxicol Environ Health A. 2015;78:1321–1327. doi: 10.1080/15287394.2015.1085942. [DOI] [PubMed] [Google Scholar]
- 26.Wang G., Wang T., Zhang Y., Li F., Yu B., Kou J. Schizandrin protects against OGD/R-induced neuronal injury by suppressing autophagy: involvement of the AMPK/mTOR pathway. Molecules. 2019;24:3624–3640. doi: 10.3390/molecules24193624. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Guo L.Y., Hung T.M., Bae K.H., Shin E.M., Zhou H.Y., Hong Y.N., et al. Anti-inflammatory effects of schisandrin isolated from the fruit of Schisandra chinensis Baill. Eur J Pharmacol. 2008;591:293–299. doi: 10.1016/j.ejphar.2008.06.074. [DOI] [PubMed] [Google Scholar]
- 28.Zhao X., Liu C.M., Xu M.J., Li X.L., Bi K.S., Jia Y. Total lignans of Schisandra chinensis ameliorates Aβ1–42-induced neurodegeneration with cognitive impairment in mice and primary mouse neuronal cells. PLoS One. 2016;11 doi: 10.1371/journal.pone.0152772. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Egashira N., Kurauchi K., Iwasaki K., Mishima K., Orito K., Oishi R., et al. Schizandrin reverses memory impairment in rats. Phytother Res. 2008;22:49–52. doi: 10.1002/ptr.2258. [DOI] [PubMed] [Google Scholar]
- 30.Jiang Z.J., Wang C.Y., Xie X., Yang J.F., Huang J.N., Cao Z.P., et al. Schizandrin ameliorates ovariectomy-induced memory impairment, potentiates neurotransmission and exhibits antioxidant properties. Br J Pharmacol. 2015;172:2479–2492. doi: 10.1111/bph.13078. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Zhang C., Zhao X., Mao X., Liu A., Liu Z., Li X., et al. Pharmacological evaluation of sedative and hypnotic effects of schizandrin through the modification of pentobarbital-induced sleep behaviors in mice. Eur J Pharmacol. 2014;744:157–163. doi: 10.1016/j.ejphar.2014.09.012. [DOI] [PubMed] [Google Scholar]
- 32.Minichiello L., Casagranda F., Tatche R., Stucky C., Postigo A., Lewin G., et al. Point mutation in trkB causes loss of NT4-dependent neurons without major effects on diverse BDNF responses. Neuron. 1998;21:335–345. doi: 10.1016/s0896-6273(00)80543-7. [DOI] [PubMed] [Google Scholar]
- 33.Carla S., Liliana M. The preparation of primary cortical neuron cultures and a practical application using immunofluorescent cytochemistry. Methods Mol Biol. 2010;633:221–231. doi: 10.1007/978-1-59745-019-5_16. [DOI] [PubMed] [Google Scholar]
- 34.Liu J., Sun Z.K., Deng Y.H., Zou Y., Li C.Y., Guo X.H., et al. Highly water-dispersible biocompatible magnetite particles with low cytotoxicity stabilized by citrate groups. Angew Chem Int Ed Engl. 2009;48:5875–5879. doi: 10.1002/anie.200901566. [DOI] [PubMed] [Google Scholar]
- 35.Wang H.C., Hao S.M., Liu Y.Q., Zheng X.M., Yao L., Liu L.Y., et al. Multiple target fishing hook prepared by photochemically coupling molecules in medicines onto magnetic nanoparticles. Chin Tradit Herb Drugs. 2020;51:4142–4150. [Google Scholar]
- 36.Barclay L.A., Wales T.E., Garner T.P., Wachter F., Lee S., Guerra R.M., et al. Inhibition of pro-apoptotic bax by a noncanonical interaction mechanism. Mol Cell. 2015;57:873–886. doi: 10.1016/j.molcel.2015.01.014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Zeng H., Wang L.L., Zhang J.W., Pan T., Yu Y.H., Lu J.X., et al. Activated PKB/GSK-3β synergizes with PKC-δ signaling in attenuating myocardial ischemia/reperfusion injury via potentiation of NRF2 activity: therapeutic efficacy of dihydrotanshinone-I. Acta Pharm Sin B. 2021;11:71–88. doi: 10.1016/j.apsb.2020.09.006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Li L., Huang T., Tian C., Xiao Y.B., Kou S.M., Zhou X., et al. The defensive effect of phellodendrine against AAPH-induced oxidative stress through regulating the AKT/NF-κB pathway in zebrafish embryos. Life Sci. 2016;157:97–106. doi: 10.1016/j.lfs.2016.05.032. [DOI] [PubMed] [Google Scholar]
- 39.Zhang B.Y., Pan C.Y., Feng C., Yan C.Q., Yu Y.J., Chen Z.L., et al. Role of mitochondrial reactive oxygen species in homeostasis regulation. Redox Rep. 2022;27:45–52. doi: 10.1080/13510002.2022.2046423. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Chouchani E.T., Pell V.R., Gaude E., Aksentijević D., Sundier S.Y., Robb E.L., et al. Ischaemic accumulation of succinate controls reperfusion injury through mitochondrial ROS. Nature. 2014;515:431–435. doi: 10.1038/nature13909. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Sanz A., Pamplona R., Barja G. Is the mitochondrial free radical theory of aging intact?. Antioxidants Redox Signal. 2006;8:582–599. doi: 10.1089/ars.2006.8.582. [DOI] [PubMed] [Google Scholar]
- 42.Scialò F., Sriram A., Fernández-Ayala D., Gubina N., Lõhmus M., Nelson G., et al. Mitochondrial ROS produced via reverse electron transport extend animal lifespan. Cell Metabol. 2016;23:725–734. doi: 10.1016/j.cmet.2016.03.009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Hagenbuchner J., Kuznetsov A., Hermann M., Hausott B., Obexer P., Ausserlechner M.J. FOXO3-induced reactive oxygen species are regulated by BCL2L11 (Bim) and SESN3. J Cell Sci. 2012;125:1191–1203. doi: 10.1242/jcs.092098. [DOI] [PubMed] [Google Scholar]
- 44.Aichberger K.J., Gleixner K.V., Mirkina I., Cerny-Reiterer S., Peter B., Ferenc V., et al. Identification of proapoptotic Bim as a tumor suppressor in neoplastic mast cells: role of KIT D816V and effects of various targeted drugs. Blood. 2009;114:5342–5351. doi: 10.1182/blood-2008-08-175190. [DOI] [PubMed] [Google Scholar]
- 45.Egle A., Harris A.W., Bouillet P., Cory S. Bim is a suppressor of Myc-induced mouse B cell leukemia. Proc Natl Acad Sci U S A. 2004;101:6164–6169. doi: 10.1073/pnas.0401471101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Shin E.S., Huang Q., Gurel Z., Palenski T.L., Zaitoun I., Sorenson C.M., et al. STAT1-mediated Bim expression promotes the apoptosis of retinal pericytes under high glucose conditions. Cell Death Dis. 2014;5:e986. doi: 10.1038/cddis.2013.517. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Bowman B.J., Bowman E.J. Mutations in subunit C of the vacuolar ATPase confer resistance to bafilomycin and identify a conserved antibiotic binding site. J Biol Chem. 2002;277:3965–3972. doi: 10.1074/jbc.M109756200. [DOI] [PubMed] [Google Scholar]
- 48.Bowman E.J., Graham L.A., Stevens T.H., Bowman B.J. The bafilomycin/concanamycin binding site in subunit c of the V-ATPases from Neurospora crassa and Saccharomyces cerevisiae. J Biol Chem. 2004;279:33131–33138. doi: 10.1074/jbc.M404638200. [DOI] [PubMed] [Google Scholar]
- 49.Wang Y.Z., Zhang L., Wei Y.L., Huang W., Li L., Wu A.A., et al. Pharmacological targeting of vacuolar H+-ATPase via subunit V1G combats multidrug-resistant cancer. Cell Chem Biol. 2020;27:1359–1370. doi: 10.1016/j.chembiol.2020.06.011. [DOI] [PubMed] [Google Scholar]
- 50.Song Q., Meng B., Xu H., Mao Z. The emerging roles of vacuolar-type ATPase-dependent lysosomal acidification in neurodegenerative diseases. Transl Neurodegener. 2020;9:1–14. doi: 10.1186/s40035-020-00196-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Zhang Y.W., Lv X.Y., Qu J.M., Zhang X., Zhang M.Y., Gao H., et al. A systematic strategy for screening therapeutic constituents of Schisandra chinensis (Turcz.) Baill infiltrated blood–brain barrier oriented in lesions using ethanol and water extracts: a novel perspective for exploring chemical material basis of herb medicines. Acta Pharm Sin B. 2020;10:557–568. doi: 10.1016/j.apsb.2019.10.008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Yan T.X., Sun Y.Y., Gong G.W., Li Y., Fan K.Y., Wu B., et al. The neuroprotective effect of schisandrol A on 6-OHDA-induced PD mice may be related to PI3K/AKT and IKK/IκBα/NF-κB pathway. Exp Gerontol. 2019;128 doi: 10.1016/j.exger.2019.110743. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.