Abstract
During spore formation in Bacillus subtilis, the SpoIVB protein is a critical component of the ςK regulatory checkpoint. SpoIVB has been shown to be a serine peptidase that is synthesized in the spore chamber and which self-cleaves, releasing active forms. These forms can signal proteolytic processing of the transcription factor ςK in the outer mother cell chamber of the sporulating cell. This forms the basis of the ςK checkpoint and ensures accurate ςK-controlled gene expression. SpoIVB has also been shown to activate a second distinct process, termed the second function, which is essential for the formation of heat-resistant spores. In addition to the serine peptidase domain, SpoIVB contains a PDZ domain. We have altered a number of conserved residues in the PDZ domain by site-directed mutagenesis and assayed the sporulation phenotype and signaling properties of mutant SpoIVB proteins. Our work has revealed that the SpoIVB PDZ domain could be used for up to four distinct processes, (i) targeting of itself for trans proteolysis, (ii) binding to the protease inhibitor BofC, (iii) signaling of pro-ςK processing, and (iv) signaling of the second function of SpoIVB.
PDZ domains are relatively small (≈100 amino acids) domains involved in protein-protein interactions (21, 23). Many of these interactions occur at the interface of the plasma membrane, enabling the recruitment and formation of larger complexes (24). PDZ domains have been shown to allow high selectivity in the targeting of proteins and can bind to short COOH-terminal peptide motifs. There are two main classes of binding site, h-X-V-COO− (where h is a hydrophobic amino acid) and S/T-X-V-COO−, based on the sequences of these motifs (1, 23, 24, 32, 34, 35). In addition, PDZ domains have been shown to be able to bind to internal motifs, as well as to other PDZ domains (14). Some PDZ proteins contain more than one domain; for example, the Drosophila InaD scaffolding protein carries five discrete PDZ domains (36). These multivalent PDZ domain proteins enable a series of distinct protein-protein interactions which can be used to build a protein complex in steps. PDZ domains can be carried as discrete modules within a multidomain protein, and pertinent examples of these modular PDZ proteins for this work are two families of bacterial serine peptidases, the Prc (also called Tsp) family (15) and the HtrA (also called DegP) family (22). In these proteases, the PDZ domain enables substrate recognition, which is thought to occur at the C terminus of the target. The crystal structures of four PDZ domains, in complex with their cognate peptide ligands, have provided invaluable insight into how these domains interact with their targets (7, 8, 14). The PDZ domains consist of a compact arrangement of six β strands and two α helices (Fig. 1C). Peptides bind in a groove between βB and α2 in an antiparallel manner to βB that extends the β-sheet structure. The peptide bound in this orientation places the carboxyl group of the C-terminal residue in a position to interact with a loop between βA and βB. This loop has the consensus sequence h-G-h (where h is a hydrophobic residue) and forms a “carboxylate-binding pocket.” Remarkably, recognition of such a short, degenerate motif, coupled with the presence of a free carboxyl group, is sufficient to confer high selectivity of binding, and artificial PDZ constructs have been demonstrated to bind new targets and efficiently transport them to a defined subcellular location (32).
FIG. 1.
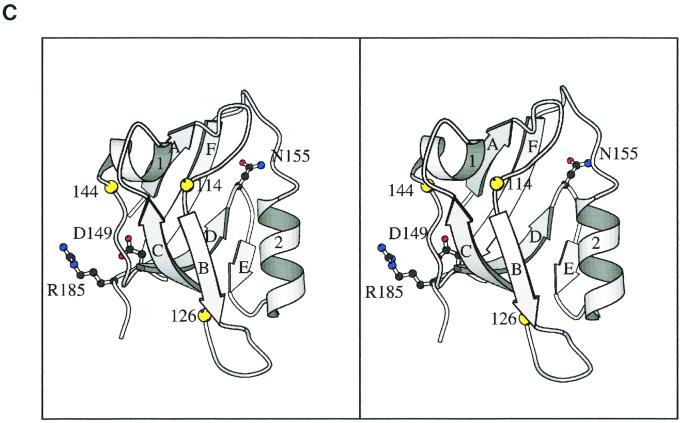
The SpoIVB PDZ domain. (A) Schematic diagram showing the position of the PDZ domain of SpoIVB (residues 102 to 187). The arrow indicates the site of the first self-cleavage reaction in the SpoIVB polypeptide. Also shown are the propeptide sequence, the region containing the serine peptidase domain, and a region thought to be involved in SpoIVB's putative second function (shaded box). (B) Structure-based sequence alignment of the putative PDZ domain in B. subtilis SpoIVB (accession no. P17896; residues 102 to 187; Sp4B, center) with four PDZ domains of known structure (above the Sp4B sequence) and four SpoIVB homologues (below the Sp4B sequence). Amino acid identity and similarity between the groups are indicated (h, hydrophobic; s, small; −, negative charge; +, positive charge). A consensus secondary structure (α1 and α2, α helices; βA to -F, β strands) is given above the PDZ sequences. Note that all gaps and insertions required for maximal primary sequence alignment are placed outside of these structural elements. Residues known to contact ligands based on the structures of PDZ-peptide complexes are underlined. The positions of mutations in B. subtilis SpoIVB are indicated by arrows. These correspond to three positions that are conserved between the two groups (G114, D149, and N155) and three which are conserved within the SpoIVB group (G126, G144, and R185). Of the 286 PDZ domains identified by the Simple Modular Architecture Research Tool (33), Gly114 is conserved in 266 sequences, Gly126 is conserved in 114, Gly144 is conserved in 156, Asp149 is conserved in 269, Asn155 is conserved in 209, and Arg185 is conserved in 37. Preliminary sequence data were obtained from The Institute for Genomic Research (www.tigr.org), the B. stearothermophilus Genome Sequencing Project at the University of Oklahoma (www.genome.ou.edu), and Genome Therapeutics Corp. (www.cric.com). Pdz1, brain postsynaptic density protein 95 (residues 312 to 397); Pdz2, rabbit α-syntrophin (residues 80 to 164); Pdz3, neuronal nitric oxide synthase (residues 15 to 101); Pdz4, hCASK (residues 482 to 574). Bant, B. anthracis; Bste, B. stearothermophilus; Bhal, B. halodurans (BAB06494); Cace, Clostridium acetobutylicum. (C) Stereoscopic representation of the SpoIVB PDZ domain structure. A homology model of the B. subtilis SpoIVB PDZ domain was constructed based on the crystal structures of PDZ 1 to 4 (Protein Data Bank codes 1be9, 1qav, 1qau, and 1kwa; [7, 8, 14]) by using the program Modeller (29). The peptide-binding groove is between βB and α2, and the carboxylate-binding loop is between βA and βB. The positions of glycine residues 114, 144, and 126 are shown as yellow balls. The side chains of residues Asp149, Asn155, and Arg185 are drawn as ball-and-stick images and colored with carbon atoms in grey, nitrogen atoms in blue, and oxygen atoms in red. The image was drawn with Molscript (16).
A PDZ domain has been identified in the Bacillus subtilis regulatory protein SpoIVB (Fig. 1) (21). SpoIVB is a multifunctional protein which plays a crucial role in the ςK checkpoint by providing the signal that activates proteolytic processing of pro-ςK (2, 3). SpoIVB has been shown to be a serine peptidase that is synthesized in the forespore chamber and is secreted across the inner forespore membrane (IFM), where it somehow activates the proteolysis of pro-ςK (37). Pro-ςK is an inactive transcription factor synthesized in the outer mother cell chamber of the sporulating cell and is cleaved by a proposed complex of three proteins, SpoIVFA, SpoIVFB, and BofA, which are embedded in the outer forespore membrane (5, 26, 27, 40). The SpoIVFB protein has been identified as the zinc metalloprotease which cleaves pro-ςK to its active form, ςK (17, 28). When activated by proteolytic cleavage, ςK directs the final program of gene expression in the mother cell chamber of the sporulating cell. The important feature of this regulatory checkpoint is that ςK-directed gene expression must wait until the appropriate signal is received from the forespore. Accurate signaling is essential to maintaining the fidelity of spore formation, since premature signaling leads to a marked decrease in spore-forming efficiency (3). How this is achieved is revealed by the extraordinary number of regulatory elements in the ςK checkpoint which inhibit premature signaling. Initially, ςF-directed transcription of the spoIVB gene is repressed at stage II (10, 12); it has been shown that should any inadvertent expression occur then, the BofC protein would inhibit SpoIVB autoproteolysis, most probably by direct protein-protein interaction (11, 38). Premature signaling is also prevented by two inhibitors, SpoIVFA and BofA, which are thought to maintain the SpoIVFB protease in an inactive state (5, 25, 27). The C termini of both of these inhibitors protrude into the space between the IFM and outer forespore membrane and could interact with SpoIVB (13).
Potentially, the PDZ domain of SpoIVB could be used for protein-protein interactions to control these events, by activating and targeting its peptidase function or providing surfaces to direct inhibition by binding to protein partners. In this work, we have used site-directed mutagenesis to analyze the function of the SpoIVB PDZ domain. The results suggest that the PDZ domain is involved in multiple roles, including autoproteolysis, interaction with BofC, and signaling of pro-ςK processing.
MATERIALS AND METHODS
Bacterial strains.
The strains used in this work are listed in Table 1 and were all congenic with prototrophic spo+ strain PY79. To construct lysogens of SPβ::gerE-lacZ, a phage lysate was prepared from strain SC433 and used for transduction of the appropriate recipient strain. For integration of DNA at the amyE locus, cells were transformed with linearized DNA. Strain constructions using DNA-mediated transformation are outlined briefly in Table 1. SC2373 was constructed by transformation of competent cells of SC2221 (spoIVBΔ::spc bofB8) with linearized pNH1134 (see pDG364-spoIVBDN149 below) plasmid DNA, followed by selection for chloramphenicol resistance (Cmr; encoded by the pDG364 plasmid).
TABLE 1.
B. subtilis strains used in this study
| Strain | Genotype | Construction or reference |
|---|---|---|
| NH573 | spoIVBΔ::spc amyE::spoIVBND155 | pNH487 into SC1836 |
| NH577 | spoIVBΔ::spc amyE::pDG364 | pDG364 into SC1836 |
| NH578 | spoIVBΔ::spc amyE::spoIVB+ | pNH470 into SC1836 |
| NH723 | spoIIIGΔ1 spoIVBΔ::spc | SC1836 into SC500 |
| NH587 | spoIVBΔ::spc amyE::spoIVBGA144 | pNH534 into SC1836 |
| NH685 | spoIVBΔ::spc amyE::spoIVBGA114 | pNH674 into SC1836 |
| NH687 | spoIVBΔ::spc amyE::spoIVBGA144/ND155 | pNH676 into SC1836 |
| NH987 | spoIVBΔ::spc amyE::spoIVBGA126 | pNH973 into SC1836 |
| NH990 | spoIVBΔ::spc amyE::spoIVBRK185 | pNH970 into SC1836 |
| NH991 | spoIVBΔ::spc amyE::spoIVBRH185 | pNH985 into SC1836 |
| NH1001 | spoIVBΔ::spc amyE::spoIVBGQ114 | pNH977 into SC1836 |
| NH1003 | spoIVBΔ::spc amyE::spoIVBGQ126 | pNH979 into SC1836 |
| NH1005 | spoIVBΔ::spc amyE::spoIVBGA144 | pNH981 into SC1836 |
| NH1007 | spoIVBΔ::spc amyE::spoIVBNY155 | pNH983 into SC1836 |
| NH1042 | spoIIIGΔ1 bofCΔ::neo spoIVBΔ::spc | SC1836 into PW71 |
| NH1097 | spoIIIGΔ1 spoIVBΔ::spc amyE::spoIVB+ | pNH470 into NH723 |
| NH1099 | spoIIIGΔ1 spoIVBΔ::spc amyE::spoIVBGQ114 | pNH977 into NH723 |
| NH1101 | spoIIIGΔ1 spoIVBΔ::spc amyE::spoIVBGA144 | pNH534 into NH723 |
| NH1103 | spoIIIGΔ1 spoIVBΔ::spc amyE::spoIVBND155 | pNH487 into NH723 |
| NH1105 | spoIIIGΔ1 spoIVBΔ::spc amyE::spoIVBGA144/ND155 | pNH676 into NH723 |
| NH1107 | spoIIIGΔ1 spoIVBΔ::spc amyE::spoIVBRK185 | pNH970 into NH723 |
| NH1135 | spoIVBΔ::spc amyE::spoIVBDN149 | pNH1134 into SC1836 |
| NH1140 | spoIIIGΔ1 bofCΔ::neo spoIVBΔ::spc amyE::spoIVB+ | pNH470 into NH1042 |
| NH1151 | spoIIIGΔ1 spoIVBΔ::spc amyE::spoIVBDN149 | pNH1134 into NH723 |
| NH1210 | spoIIIGΔ1 spoIVBΔ::spc amyE::spoIVBGA126 | pNH973 into NH723 |
| NH1212 | spoIIIGΔ1 spoIVBΔ::spc amyE::spoIVBGQ126 | pNH979 into NH723 |
| NH1214 | spoIIIGΔ1 spoIVBΔ::spc amyE::spoIVBGQ144 | pNH534 into NH723 |
| NH1216 | spoIIIGΔ1 spoIVBΔ::spc amyE::spoIVBNY155 | pNH983 into NH723 |
| NH1218 | spoIIIGΔ1 spoIVBΔ::spc amyE::spoIVBRH185 | pNH985 into NH723 |
| NH1248 | spoIIIGΔ1 bofCΔ::neo spoIVBΔ::spc amyE::pDG364 | pDG364 into NH1042 |
| NH1250 | spoIIIGΔ1 bofCΔ::neo spoIVBΔ::spc amyE::spoIVBGA114 | pNH674 into NH1042 |
| NH1252 | spoIIIGΔ1 bofCΔ::neo spoIVBΔ::spc amyE::spoIVBGA126 | pNH973 into NH1042 |
| NH1254 | spoIIIGΔ1 bofCΔ::neo spoIVBΔ::spc amyE::spoIVBGA144 | pNH534 into NH1042 |
| NH1256 | spoIIIGΔ1 bofCΔ::neo spoIVBΔ::spc amyE::spoIVBND155 | pNH487 into NH1042 |
| NH1258 | spoIIIGΔ1 bofCΔ::neo spoIVBΔ::spc amyE::spoIVBGA144/ND155 | pNH676 into NH1042 |
| NH1260 | spoIIIGΔ1 bofCΔ::neo spoIVBΔ::spc amyE::spoIVBRK185 | pNH970 into NH1042 |
| NH1262 | spoIIIGΔ1 bofCΔ::neo spoIVBΔ::spc amyE::spoIVBGQ114 | pNH977 into NH1042 |
| NH1264 | spoIIIGΔ1 bofCΔ::neo spoIVBΔ::spc amyE::spoIVBGQ126 | pNH979 into NH1042 |
| NH1266 | spoIIIGΔ1 bofCΔ::neo spoIVBΔ::spc amyE::spoIVBGQ144 | pNH981 into NH1042 |
| NH1268 | spoIIIGΔ1 bofCΔ::neo spoIVBΔ::spc amyE::spoIVBDN149 | pNH1134 into NH1042 |
| NH1270 | spoIIIGΔ1 bofCΔ::neo spoIVBΔ::spc amyE::spoIVBNY155 | pNH983 into NH1042 |
| NH1272 | spoIIIGΔ1 bofCΔ::neo spoIVBΔ::spc amyE::spoIVBRH185 | pNH985 into NH1042 |
| NH1278 | spoIIIGΔ1 spoIVBΔ::spc amyE::spoIVBGA114 | pNH674 into NH723 |
| PW71 | spoIIIGΔ1 bofCΔ::neo | 38 |
| PY79 | spo+ | 39 |
| SC433 | SPβ::gerE-lacZ | 4 |
| SC500 | spoIIIGΔ1 | 3 |
| SC1836 | spoIVBΔ::spc | 20 |
| SC2373 | spoIVBΔ::spc bofB8 amyE::spoIVBDN149 | This work |
General methods.
The general Bacillus methods used (transduction, transformation, antibiotic selection, etc.) were those described by Cutting and Vander-Horn (6). Sporulation was induced by the resuspension method (19). Determination of heat and lysozyme resistance and measurements of gerE-directed β-galactosidase synthesis were done as described previously (19).
Site-specific mutagenesis.
Two oligonucleotide primers were used to amplify a 1,429-bp spoIVB product by PCR using chromosomal DNA from B. subtilis strain PY79 as a template. The primers used were P1 (5′-TTATGGATCCCGTGCACATCCATTCGTTC-3′), which annealed to nucleotides −146 to −127 from the spoIVB start codon, and P2 (5′-AACAAGCTTAGTCAGCTTGCTTTTTCTTTTCC-3′), which annealed to the spoIVB stop codon (in bold) and a further 18 bases upstream. The PCR product carried either a BamHI (P1) or a HindIII (P2) restriction site (underlined), enabling direct cloning into pBluescript II KS(+). The resultant clone, pNH252, was sequenced completely to verify the presence of an unmodified spoIVB cistron. Next, mutations were created with mismatch oligonucleotides by using the method of Kunkel as described by Sambrook et al. (30). In each pBluescript clone, the presence of a single amino acid change was verified by DNA sequencing. Finally, the spoIVB genes were subcloned as 1.4-kb HindIII-BamHI fragments into pDG364 (6). pDG364 clones were pNH674 (spoIVBGA114), pNH973 (spoIVBGA126), pNH534 (spoIVBGA144), pNH1134 (spoIVBDN149), pNH487 (spoIVBND155), pNH676 (spoIVBGA144/ND155), NH970 (spoIVBRK185), NH977 (spoIVBGQ114), NH979 (spoIVBGQ126), NH981 (spoIVBA144), NH983 (spoIVBNY155), and pNH985 (spoIVBRH185).
pDG364 enables insertion of cloned DNA, in trans, at the amyE locus by double-crossover marker replacement. In each case, we linearized the pDG364 subclones by digestion with XhoI and introduced them into SC1836 (spoIVBΔ::spc) cells by DNA-mediated transformation, followed by selection for Cmr (encoded by pDG364). Insertion at the amyE locus was confirmed by testing for an Amy− phenotype (failure to digest starch) as described elsewhere (6). Mutant strains had the genotype spoIVBΔ::spc amyE::spoIVB. To make the spoIVBGA144/ND155 double mutant, we first constructed the spoIVBGA144 allele and then used the resultant mutant plasmid as a template to create a second spoIVBND155 mutation.
We also constructed two isogenic control strains, NH578 (spoIVBΔ::spc amyE::spoIVB+) and NH577 (spoIVBΔ::spc amyE::pDG364). NH578 was created by integrating a pDG364 subclone, pNH470, carrying the full-length, 1,429-bp, wild-type spoIVB gene into the amyE locus, and NH577 was created by integrating the unmodified pDG364 plasmid into the chromosome by a double-crossover recombinational event at amyE.
Preparation of extracts for Western blotting.
Samples (1 ml) were taken from sporulating cultures, and cells were harvested by centrifugation and frozen in liquid N2. To break the cells, pellets were suspended in 50 μl of TS buffer (25 mM Tris-HCl, pH 7.4; 0.1 M NaCl) containing lysozyme (0.2 μg/ml) and incubated for 10 min on ice. A 50-μl volume of 2× sodium dodecyl sulfate (SDS)-polyacrylamide gel electrophoresis (PAGE) loading dye was then added, and the samples were sonicated for 10 s before gel loading (approximately 20 μl of sample per well).
Western analysis.
Immunoblotting of sporulating extracts with polyclonal antiserum to pro-ςK or SpoIVB was done as described previously (13, 37).
RESULTS
Site-specific mutagenesis of the PDZ domain.
We used the alignment of bacterial PDZ domains, including SpoIVB, created by Pallen and Ponting (21) to identify key residues within the SpoIVB PDZ domain for site-directed mutagenesis (Fig. 1B). These residues span the entire PDZ domain and represent three (Gly114, Asp149, and Asn155) which are conserved in almost all PDZ domains and Gly126, Gly144, and Arg185, which are conserved within SpoIVB homologues. Two types of amino acid alteration were made as shown in Table 2, i.e., semiconservative (GA114, GA126, GA144, DN149, ND155, and RK185) and nonconservative (GQ114, GQ126, GQ144, NY155, and RH185) changes. In addition, we constructed a double mutant carrying two changes, GA144 and ND155, that we refer to here as spoIVBGA144/ND155.
TABLE 2.
Mutations in the SpoIVB PDZ domain
| Strain | Relevant allele | Heatra | Lysra | Gerb | Signalingc | Pro-ςK processingd | BofCe | Self-cleavagef |
|---|---|---|---|---|---|---|---|---|
| NH578 | spoIVB+ | 70.3 | 92.2 | + | + | + | + | |
| NH577 | spoIVBΔ::spc | 0.0009 | 0.235 | ND | − | − | ||
| NH685 | spoIVBGA114 | 53.2 | 97 | + | + | + | − | + |
| NH987 | spoIVBGA126 | 67.9 | 80 | + | + | + | − | + |
| NH587 | spoIVBGA144 | 59.7 | 79.6 | + | Delayed | + | − | + |
| NH1135 | spoIVBDN149 | <0.001g | 0.261 | ND | Delayed | (−) | − | Impaired |
| NH573 | spoIVBND155 | 66.2 | 81.5 | + | Delayed | + | − | + |
| NH990 | spoIVBRK185 | 97.9 | 100 | + | Delayed | + | − | + |
| NH687 | spoIVBGA144/ND155 | 41.2 | 89 | + | Delayed | + | − | + |
| NH1001 | spoIVBGQ114 | 87.7 | 79.9 | + | Delayed | Delayed | − | Impaired |
| NH1003 | spoIVBGQ126 | 97.3 | 78 | + | + | + | − | + |
| NH1005 | spoIVBGQ144 | 91.9 | 100 | + | + | + | + | |
| NH1007 | spoIVBNY155 | 76.2 | 63.8 | + | + | + | − | + |
| NH991 | spoIVBRH185 | 69.1 | 69.6 | + | + | + | − | + |
Heat resistance (Heatr) or lysozyme resistance (Lysr) of cultures 24 h after the initiation of sporulation in DS medium. Values are expressed as the percentage of CFU per milliliter in the untreated culture. All values are averages of at least two independent experiments.
Germination proficiency (Ger) was examined by using 7-day-old colonies grown on agar plates as described by Cutting and Vander-Horn (6). ND, not determined.
Signaling was defined on the basis of two criteria: (i) formation of pigmented colonies (Pig+) associated with the production of the ςK-expressed CotA protein (31) and (ii) expression of the ςK-controlled reporter gene gerE-lacZ in cells containing an SPβ::gerE-lacZ lysogen as described previously (3).
Pro-ςK processing (Fig. 3). In the case of spoIVBDN149, although processing could not be detected by immunoblotting, gerE-lacZ expression was detectable, indicating that at least some processing must occur, but at extremely low levels.
Involved in interaction with the BofC protein (Fig. 5).
Measurable defect in or impairment of SpoIVB autoproteolysis (Fig. 6).
Temperature-sensitive phenotype (see Table 3).
As described in Materials and Methods, the mutated spoIVB alleles were introduced at the amyE locus in cells carrying a spoIVBΔ::spc insertion-and-deletion mutation. By using appropriate spoIVB and spoIVB+ congenic controls, we examined spore-forming efficiency during sporulation (Table 2). Only one mutation, spoIVBDN149, substantially impaired the formation of heat-resistant and lysozyme-resistant spores. For the other alleles, which had no effect on spore formation, we examined the capacity of spores to germinate correctly since impaired activation of the ςK checkpoint has been shown, in some circumstances, to lead to the production of germination-defective spores. We found that, in each case (with the exception of spoIVBDN149), spores germinated normally.
Since the spoIVBDN149 allele caused a direct impairment of signaling, we determined whether it is dominant or recessive. We integrated the spoIVBDN149 gene at the amyE locus of spo+ (strain PY79) cells. In trans at the amyE locus, we found that spoIVBDN149 is recessive, with the merodiploid producing essentially (99%) the same amount of heat-resistant spores (data not shown) as a control spo+ strain (NH578 spoIVBΔ::spc amyE::spoIVB+).
We also examined whether the spoIVBDN149 mutation is involved in both of SpoIVB's sporulation-specific functions, i.e., signaling in the ςK checkpoint and the development of heat resistance through a ςK-independent process (20). This can be established by engineering spoIVBDN149 cells to express active ςK constitutively and determine whether heat-resistant spores develop. If so, then the spoIVBDN149 mutation only affects signaling of pro-ςK processing. Accordingly, we constructed a strain (SC2373 amyE::spoIVBDN149 spoIVBΔ::spc bofB8) carrying both the bofB8 and spoIVBDN149 alleles. In these cells, the bofB8 suppressor mutation renders the pro-ςK processing enzyme, SpoIVFB, constitutively active. We found that in SC2373 cells, spore formation was blocked at stage IV-V with the production of phase grey spores. Phenotypically, then, spore formation had advanced, which is attributed to the premature synthesis and assembly of spore coat proteins onto the forespore, leading to the production of phase grey spores and referred to as the Bof phenotype, in contrast to a SpoIVB null phenotype, where stable phase grey spores are not produced (3). However, heat-resistant, phase bright spores were not formed at 37°C (Table 3). Failure to restore spore formation demonstrated that the spoIVBDN149 mutation disrupts both functions. As described below, the DN149 allele is temperature sensitive, so we also examined the phenotype of SC2373 at 30°C (Table 3). We found that at this permissive temperature, signaling in the ςK checkpoint was restored in SC2373 cells, which was confirmed by the presence of phase grey stage IV-V spores and normal gerE-lacZ expression. However, 10 times fewer spores were produced than in cells carrying only the spoIVBDN149 allele. This phenomenon has been observed before and is due to premature signaling of pro-ςK processing, which results in a 10-fold reduction in spore-forming efficiency (3).
TABLE 3.
Temperature sensitivitya of the spoIVBDN149 allele
| Strain | Relevant genotype | 37°C
|
30°C
|
||||||
|---|---|---|---|---|---|---|---|---|---|
| Viable count (CFU/ml) | Heatr (CFU/ml) | % Spores | Signalingb | Viable count (CFU/ml) | Heatr (CFU/ml) | % Spores | Signalingb | ||
| NH578 | spoIVBΔ::spc amyE::spoIVB+ | 3.6 × 108 | 2 × 108 | 55 | + | 1.22 × 108 | 9.2 × 107 | 75.4 | + |
| NH577 | spoIVBΔ::spc amyE::pDG364 | 9.50 × 107 | 1 × 101 | 1.05 × 10−5 | − | 1.03 × 108 | 1 × 101 | 9.7 × 10−6 | − |
| NH1135 | spoIVBΔ::spc amyE::spoIVBDN149 | 5.4 × 107 | 4.9 × 102 | 9.0 × 10−4 | + (delayed) | 6.2 × 107 | 2.2 × 106 | 3.5 | + |
| SC2373 | spoIVBΔ::spc amyE::spoIVBDN149 bofB8 | 3.2 × 107 | 4.7 × 102 | 2 × 10−4 | + (premature) | 2.9 × 107 | 1.2 × 105 | 0.45 | + |
Sporulation was induced by the resuspension method at 30 or 37°C. Samples were taken at 24 h (37°C) or 34 h (30°C) and measured for heat resistance (Heatr). Values are expressed as the percentage of CFU per milliliter in the untreated culture. All values are averages of at least two independent experiments.
Signaling in the ςK checkpoint, representing (i) gerE-directed β-galactosidase synthesis in liquid medium or on agar plates, (ii) pigmented colonies due to synthesis of the CotA spore coat protein, and/or (iii) immunoblotting of cell extracts with anti-pro-ςK serum.
Effects of PDZ domain mutations on signaling of pro-ςK processing.
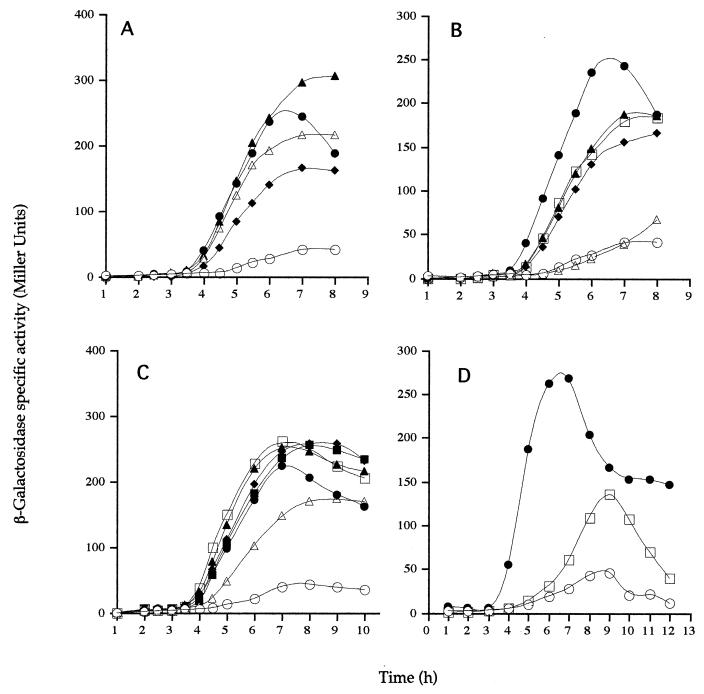
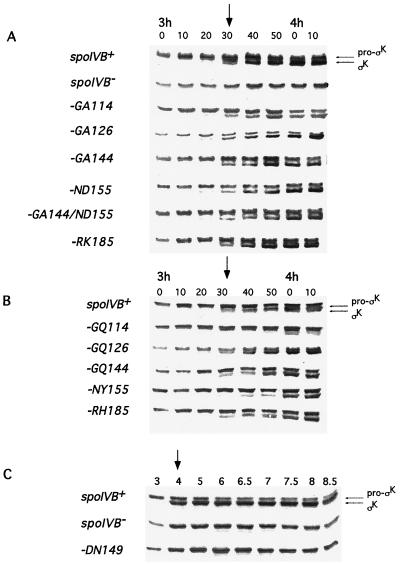
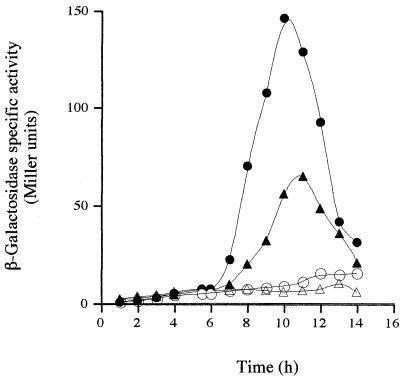
To examine the effects of PDZ mutations on signaling in the ςK checkpoint, we used two methods: first, expression of a ςK-controlled gene, gerE, and second, proteolytic processing of pro-ςK during spore formation. In the first approach, cells carrying the spoIVB PDZ allele at the amyE locus were lysogenized with the bacteriophage SPβ::gerE-lacZ. Cells carrying this reporter gene were induced to sporulate by the resuspension method, and gerE-directed β-galactosidase synthesis was measured during spore formation. Our results are summarized in Table 2, and representative profiles of gerE-lacZ expression are given in Fig. 2A to D. We found that four of the mutations in the PDZ domain, GA144 (Fig. 2A), ND155 (Fig. 2B), RK185 (Fig. 2B), and GQ114 (Fig. 2C), as well as the double mutation GA144/ND155 (Fig. 2B), produced a modest yet reproducible delay in gerE-lacZ expression. We have repeated these experiments at least three times and found a delay of 20 to 30 min, together with a partial reduction in the level of gerE expression. Finally, the spoIVBDN149 allele produced strong impairment of gerE-lacZ expression (Fig. 2B), which was consistent with the block in spore formation observed with this mutant. Careful analysis of the profile of gerE-lacZ expression suggested that reduced levels (although higher than that of the spoIVB null mutant) were being produced but gerE-lacZ expression initiated 2 h later than in wild-type cells and peaked at 8 to 9 h instead of 6 to 7 h (Fig. 2D). We also probed wild-type and mutant sporulating cultures for active ςK and inactive ςK (pro-ςK) by using a polyclonal antiserum to pro-ςK (Fig. 3A and B). Two PDZ alleles produced a noticeable effect on ςK processing. spoIVBGQ114 produced a marked delay in pro-ςK processing of approximately 20 to 30 min (Fig. 3A), and in a spoIVBDN149 mutant, we could not detect any processing of pro-ςK even at the eighth hour of spore formation (Fig. 3B). Since we have shown that gerE is expressed, albeit at a later time and at reduced levels, in the spoIVBDN149 mutant, we assume that extremely low levels of ςK are being produced by proteolysis of pro-ςK but at levels undetectable by immunoblotting. That gerE could be transcribed by very low threshold levels of ςK-RNAP has been proposed before for ςK-controlled genes (18) and is consistent with the SpoIVBDN149 phenotype.
FIG. 2.
ςK-directed gene expression in spoIVB PDZ mutants. β-Galactosidase synthesis was measured at the indicated times following the initiation of sporulation in cells carrying an SPβgerE::lacZ reporter. In each case, congenic strains were used and the relevant alleles (at the amyE locus) are given here (Table 1 contains the complete genotypes). (A) NH578, spoIVB+ (●); NH577, spoIVBΔ::spc (○); NH685, spoIVBGA114 (▵); NH987, spoIVBGA126 (▴); NH587, spoIVBGA144 (♦). (B) NH578, spoIVB+ (●); NH577, spoIVBΔ::spc (○); NH1135, spoIVBDN149 (▵); NH573, spoIVBND155 (▴); NH687, spoIVBGA144/ND155 (♦); NH990, spoIVBRK185 (□). (C) NH578, spoIVB+ (●); NH577, spoIVBΔ::spc (○); NH1001, spoIVBGQ114 (▵); NH1003, spoIVBGQ126 (▴); NH1005, spoIVBGQ144 (□); NH1007, spoIVBNY155 (▪); NH991, spoIVBRH185 (♦). (D) NH578, spoIVB+ (●); NH577, spoIVBΔ::spc (○); NH1135, spoIVBDN149 (□). Background levels of gerE-directed β-galactosidase synthesis present in cells containing no reporter have been subtracted.
FIG. 3.
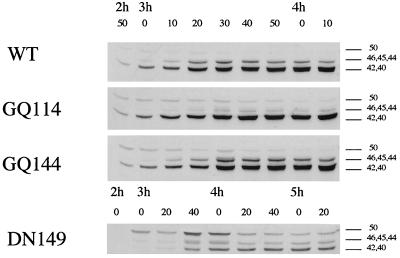
Pro-ςK processing during spore formation. Sporulation was induced in wild-type and mutant cells, and at the indicated times, samples were removed, cell extracts were prepared, and proteins were size fractionated by SDS–12% PAGE and then probed with a polyclonal antiserum to pro-ςK. Panels A and B show 10-min time points taken between 3 h and 4 h 10 min for cells carrying semiconservative (A) or nonconservative (B) changes in the spoIVB PDZ domain. The strains used were NH578 (spoIVB+), NH577 (spoIVBΔ::spc), NH685 (spoIVBGA114), NH987 (spoIVBGA126), NH587 (spoIVBGA144), NH573 (spoIVBND155), NH990 (spoIVBRK185), NH687 (spoIVBGA144/ND155), NH1001 (spoIVBGQ114), NH1003 (spoIVBGQ126), NH1005 (spoIVBGQ144), NH1007 (spoIVBNY155), and NH991 (spoIVBRH185). Panel C shows a similar immunoblot but using 30- or 60-min sampling between 3 and 8.5 h for spoIVB, spoIVBΔ::spc, and spoIVBDN149 (NH1135) cells. In each panel, the onset of pro-ςK processing is indicated by an arrow.
We constructed additional strains in which the bofCΔ::neo mutation was introduced into spoIIIGΔ1 spoIVBΔ::spc amyE::spoIVBPDZ cells. In these cells, low levels of SpoIVB would be synthesized due to ςF-controlled gene expression (12). Normally, wild-type SpoIVB cannot signal under these conditions due to the BofC inhibition. However, with BofC absent, we would expect delayed activation of pro-ςK processing and ςK-directed gene expression, as has been shown previously (11, 38).
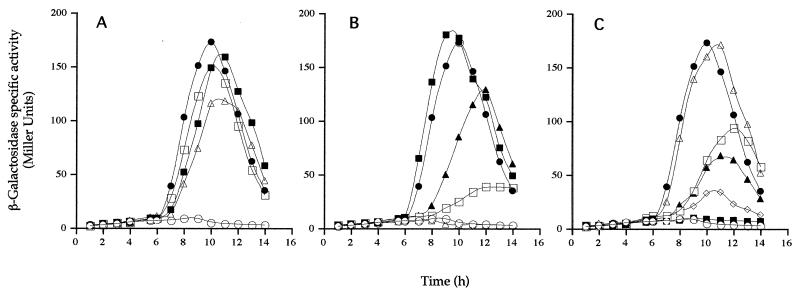
Strains were lysogenized with SPβ::gerE-lacZ, sporulation was induced, and gerE-directed β-galactosidase synthesis was determined (Fig. 4A to C). In cells carrying a wild-type spoIVB gene at the amyE locus (strain NH1140), gerE expression commenced at 6 h, reaching a maximum at 10 to 11 h, while in cells devoid of an intact spoIVB gene (strain NH1248), no gerE expression was detected. We found essentially no detectable gerE expression in cells carrying the spoIVBDN149 (Fig. 4C) and spoIVBGQ114 (Fig. 4C) alleles. In strains carrying the spoIVBGA144 (Fig. 4A), spoIVBRK185 (Fig. 4B), spoIVBGQ126 (Fig. 4C), spoIVBNY155 (Fig. 4C), and spoIVBRH185 (Fig. 4C) alleles, as well as the double mutation spoIVBGA144/ND155 (Fig. 4B), gerE-lacZ expression was clearly delayed and reduced. Both of the experiments outlined above (Fig. 3 and 4) show that the PDZ mutations were interfering with signaling of pro-ςK processing.
FIG. 4.
Effects of PDZ mutations on ςK-directed gene expression when spoIVB is expressed prematurely. β-Galactosidase synthesis was measured at the indicated times following the initiation of sporulation in cells lysogenized with SPβ::gerE-lacZ. Cells carried both spoIIIGΔ1 and bofCΔ::neo, allowing signaling in the ςK checkpoint. In addition, cells carried the following spoIVB alleles at the amyE locus. (A) NH1140, spoIVB+ (●); NH1248, spoIVB− (○); NH1250, spoIVBGA114 (▪); NH1252, spoIVBGA126 (□); NH1254, spoIVBGA144 (▵). (B) NH1140, spoIVB+ (●); NH1248, spoIVB− (○); NH1256, spoIVBND155 (▪); NH1258, spoIVBGA144/ND155 (□); NH1260, spoIVBRK185 (▴); NH1268, spoIVBDN149 (▵). (C) NH1140, spoIVB+ (●); NH1248, spoIVB− (○); NH1262, spoIVBGQ114 (▪); NH1264, spoIVBGQ126 (□); NH1266, spoIVBGQ144 (▵); NH1270, spoIVBNY155 (▴); NH1272, spoIVBRH185 (⋄). Background levels of gerE-directed β-galactosidase synthesis present in cells containing no reporter have been subtracted.
Temperature-sensitive nature of the spoIVBDN149 allele.
When grown on sporulation agar plates at 37°C, spoIVBDN149 mutant cells (NH1135) were Spo− and indistinguishable from spoIVBΔ::spc cells (NH577), producing low levels of phase grey spores which were not released from the mother cell. However, prolonged incubation of these plates at room temperature revealed a low number of phase bright spores (Spo+) which were released from the sporangial cell. This suggested that the spoIVBDN149 mutation could be temperature sensitive. To test this, we induced sporulation in spo+, spoIVBΔ::spc, and spoIVBDN149 cells at 30 and 37°C by the resuspension method. Samples were removed at 34 and 24 h from the 30 and 37°C cultures, respectively, and the numbers of heat-resistant spores (65°C, 45 min) were determined. The different assay times were chosen because spore formation at 30°C is slower at the lower temperature. As shown in Table 3, we found that at 30°C, approximately 3.5% of the culture consisted of heat-resistant spores. Moreover, this was almost 4,000-fold more than were present when sporulation was induced at 37°C. Since the number of spores was unaffected in the spoIVBΔ::spc mutant, these results show that the spoIVBDN149 allele is temperature sensitive.
Genetic evidence that the PDZ domain can interact with BofC.
If the PDZ domain is involved in interaction with the BofC inhibitor, then mutations in the PDZ domain may release SpoIVB from BofC's inhibitory action. To address this possibility, we constructed strains carrying the spoIIIGΔ1 mutation, the wild-type or mutant spoIVB gene at the amyE locus, and (to monitor ςK activity) the reporter phage SPβ::gerE-lacZ. These cells were induced to sporulate, and gerE-directed β-galactosidase synthesis was measured. We found that cells carrying the spoIVBGQ144 allele at the amyE locus allowed measurable levels of gerE-directed β-galactosidase synthesis with expression beginning at 6 h (Fig. 5). With all other mutants, we observed no effect on gerE-lacZ expression (not shown), although, as an example, expression in cells carrying spoIVBGA114 is shown in Fig. 5. As an internal control, we also measured gerE expression in spoIIIGΔ1 cells carrying the bofC deletion-and-insertion mutation bofCΔ::neo. In spoIIIGΔ1 bofCΔ::neo cells, expression of gerE commenced at 6 h, reaching maximum levels at 11 h, which was similar to the result obtained with spoIIIGΔ1 amyE::spoIVBGQ144 cells, although the maximum levels of gerE expression were higher. Delayed expression of gerE-lacZ in a bofC mutant is thought to occur because of the low levels of SpoIVB produced in this mutant, where spoIVB is transcribed under ςF control and must therefore accumulate to sufficient levels in order to signal (38). The simplest explanation for these results is that the GQ144 allele affects the interaction of SpoIVB with BofC.
FIG. 5.
Effects of PDZ mutations on ςK-directed gene expression in the absence of ςG. β-Galactosidase synthesis was measured at the indicated times following the initiation of sporulation in cells lysogenized with SPβ::gerE-lacZ. Cells carried a spoIIIGΔ1 mutation, as well as (i) a wild-type or modified spoIVB gene carried at the amyE locus or (ii) a bofCΔ::neo insertional mutation, as indicated (Table 1 contains the complete genotypes). PW71, spoIIIGΔ1 bofCΔ::neo (●); NH1097, spoIIIGΔ1 spoIVB+ (○); NH214, spoIIIGΔ1 spoIVBGQ144 (▴); NH1278, spoIIIGΔ1 spoIVBGA114 (▵). Background levels of gerE-directed β-galactosidase synthesis present in cells containing no reporter have been subtracted.
Autoproteolysis of SpoIVB in PDZ mutants.
The spoIVBDN149 and spoIVBGQ114 mutations produced substantial effects on signaling of pro-ςK processing. One possible explanation for their phenotype is that in these PDZ mutants, processing of SpoIVB was perturbed, leading to a consequential effect on signaling. Such an explanation implies that the PDZ domain is important for autoproteolysis of SpoIVB instead of, or in addition to, having a direct role in signaling by protein-protein interaction. To address this possibility, we examined the proteolysis of SpoIVB in sporulating cells. Figure 6 shows Western blots of sporulating cell samples taken from spo+ cells (NH578) and cells carrying the spoIVBGQ114 (NH1001) and spoIVBDN149 (NH1135) mutations. In spo+ cells, SpoIVB is synthesized as a 50-kDa protein which is subject to rapid autoproteolysis beginning at about 3 h. Self-cleavage yields intermediate-size products of 46, 45, and 44 kDa, but these forms are rapidly inactivated by secondary cleavage to yield 42- and 40-kDa products. The 46-, 45-, and 44-kDa species are thought to be the active forms of SpoIVB, and these are seen only intermittently during spore formation while the 42- and 40-kDa forms accumulate. Our blots showed that in spoIVBGQ114 cells there appeared less of the intermediate and presumably active forms of SpoIVB (the 46-, 45-, and 44-kDa forms). In addition, the 50-kDa, full-length form of SpoIVB appeared to be more stable, unlike in wild-type cells, where the 50-kDa form was gradually processed. While this is a small difference, it was reproducible and suggests that processing of SpoIVB was impaired or reduced compared with that in wild-type cells. In spoIVBDN149 cells, the 50-kDa form of SpoIVB persisted longer during the time course we examined, suggesting that SpoIVB was being processed less efficiently.
FIG. 6.
Autoproteolysis of SpoIVB in PDZ domain mutants. Sporulation was induced in NH578 (wild type [WT]; spo+), NH1001 (GQ114; spoIVBGQ114), NH1005 (GQ144; spoIVBGQ144), and NH1135 (DN149; spoIVBDN149) cells, and samples were taken every 10 min. Samples were fractionated by SDS–12% PAGE and examined with a polyclonal antiserum to SpoIVB (2-min enhanced-chemiluminescence exposure time). The full-length, 50-kDa, unprocessed form of SpoIVB is marked, as are 46-, 45-, and 44-kDa intermediate SpoIVB cleavage products and the 42- and 40-kDa cleavage products produced by secondary cleavage.
We have shown GQ144 to be important for PDZ interaction with BofC, so we also examined SpoIVB autoproteolysis in this mutant and found little, if any, effect on SpoIVB cleavage, although some retardation of the cleavage of the intermediate forms of SpoIVB was apparent. We have also examined all of the other PDZ alleles (data not shown) and detected no significant effect on autoproteolysis or on the levels of intermediate forms.
DISCUSSION
SpoIVB has been extensively studied primarily because it is the signal which activates pro-ςK processing at the ςK checkpoint (2, 10, 12, 20, 37). This protein has recently been shown to be a serine peptidase (37) and so resembles the Prc and HtrA serine peptidases, which carry both a PDZ and a serine peptidase domain (15, 22). In work to be published elsewhere (N. T. Hoa, J. A. Brannigan, and S. M. Cutting, unpublished data), we have shown that the catalytic triad for the SpoIVB serine peptidase is confined to the C terminus and is downstream of the PDZ domain (Fig. 1A).
SpoIVB is synthesized in the forespore and is secreted across the IFM. Since this protein lacks a normal signal sequence, it is probably not released by signal peptide cleavage from the membrane and, instead, releases itself by self-cleavage, which can occur in trans. At the time when SpoIVB is synthesized, the BofC protein is made in the forespore and is secreted across the IFM. Unlike SpoIVB, BofC carries a typical secretory signal sequence that is cleavable by a signal peptidase. BofC has been shown to inhibit autoproteolysis of SpoIVB by stabilizing SpoIVB in an inactive form (38). BofC is only important at stage II of development and provides a mechanism by which to ensure that any inadvertent transcription of the spoIVB gene does not lead to premature signaling. Most probably, BofC would inhibit self-cleavage of SpoIVB by direct interaction and it appears to act stoichiometrically, suggesting that once the level of SpoIVB molecules exceeds that of BofC, self-cleavage of SpoIVB would commence, leading to signaling (38). When proteolytically active, SpoIVB signals two distinct events. The first is processing of pro-ςK, and the second is an unidentified process, termed the second function, which leads to the formation of heat-resistant spores (20). The second function was illuminated in genetic experiments in which bypassing of the requirement of SpoIVB for processing of pro-ςK did not restore heat resistance, implying that SpoIVB must, therefore, have two distinct roles. We can assign four processes to SpoIVB, each of which could involve protein-protein interaction: (i) autoproteolysis, (ii) interaction with the BofC protein, (iii) signaling of pro-ςK processing, and (iv) signaling of the second function. Our work has shown that the PDZ domain could be used in all four of these putative interactions.
Our results have shown that most changes in the PDZ domain produced only slight phenotype changes. In part, this was expected due to the size of the PDZ domain. However, we have revealed the importance of the Asp149 and Gly114 residues in the PDZ domain. These residues correspond to the most highly conserved positions within PDZ sequences. Gly114 in the motif h-G-h is important for maintaining the conformation of the carboxylate-binding loop. Interactions with a substrate carboxylate are mediated by backbone amides from the loop, and so the amino acid residue side chains can vary significantly. Substitutions for glycine in such a structural role are acceptable; however, there are energetic penalties, as the introduced amino acid must take up unfavorable conformations to preserve the architecture. In the case of the SpoIVB PDZ, we assume that the smaller Ala side chain is more easily accommodated than a glutamine. The most drastic effects are produced by mutation of Asp149. It is likely that alterations at this position lead to significant changes in the PDZ structure, and the pleiotropic nature of DN149 mutants suggests that the PDZ domain of SpoIVB is important to all of the functions tested. In some crystal structures of PDZ domains, Asp149 forms a salt bridge between β strands A and D (8). The temperature-sensitive phenotype of a spoIVBDN149 mutant (see below) indicates that there may be a similar structural role for Asp149 in SpoIVB.
Interestingly, the spoIVBDN149 allele has been isolated previously, in a classical genetic screen for new spoIVB alleles (20). Characterization of this allele, known as spoIVB57, differed somewhat from our results described here. Specifically, the spoIVB57 strain was found to allow very low levels of ςK-directed gene expression and higher levels of heat-resistant phase bright spores (1.2%) at 37°C. In contrast, our work shown here demonstrated a much lower level of spore formation (<0.0001%) and moderate-to-low levels of gerE-lacZ expression. We have no immediate explanation for these results, although the spoIVB57 mutant was analyzed by inducing spore formation by the exhaustion method (using DS medium [19]), whereas here we used the resuspension method. Although it is an unsatisfactory explanation, we now know that the spoIVBDN149 allele is temperature sensitive and we cannot be sure that these two studies were performed under identical conditions.
Possibly, the most important discovery in this work is that proteolysis of SpoIVB was defective in the spoIVBDN149 and spoIVBGQ114 mutants. Self-cleavage of SpoIVB still occurred in these mutants but at a reduced rate. This provides strong support for the hypothesis that the PDZ domain is used for self-cleavage. That there are other families of prokaryotic serine peptidases (e.g., the Prc and HtrA serine peptidases) with PDZ domains makes it likely that the PDZ domain has been exploited by bacterial proteases for substrate recognition. We cannot predict how SpoIVB would recognize itself at this stage, although head-to-tail oligomerization has been proposed for some PDZ-PDZ interactions (14). Another important finding is that the spoIVBDN149 allele is temperature sensitive, which suggests a weaker interaction more easily disrupted at the higher temperature. Moreover, even at the restrictive temperature, signaling of pro-ςK processing occurred, although this was at levels undetectable by immunoblotting. We can conclude that since, after a pronounced delay, gerE-lacZ expression reached 50% of the wild-type level, then expression of gerE must be susceptible to a very low threshold level of active ςK, an observation which has been made before, i.e., that some ςK-controlled genes require different levels of ςK for expression (18). This is supported by the GQ114 allele, where the defect in SpoIVB autoproteolysis was less severe than in the DN149 mutant, producing a delay in pro-ςK processing of 30 min. An important question is whether the defective signaling is due to impaired PDZ-mediated interaction of SpoIVB with one or more components of the pro-ςK processing complex (SpoIVFA, BofA, or SpoIVFB) or whether a simple reduction in active SpoIVB cleavage products is required for signaling. In the first model, the PDZ domain is required specifically for targeting of SpoIVB to the pro-ςK processing complex in the outer forespore membrane, while in the second model, the PDZ domain would be required only to enable SpoIVB self-cleavage and the generation of cleavage products which signal by a different mechanism. Our work does not provide direct evidence that SpoIVB uses its PDZ domain for signaling in the ςK checkpoint or signaling of the second function, although it is attractive to propose this. The simplest way in which a PDZ domain would achieve this is to interact specifically with C-terminal motifs. However, we are unable to identify any known PDZ-binding sequences at the C termini of SpoIVFA, BofA, SpoIVFB, or, indeed, SpoIVB.
A further reason for speculating that the PDZ domain is involved in other interactions (other than self-cleavage) is our genetic evidence that the PDZ domain interacts with BofC. This was revealed by the GQ144 allele, which permitted substantial levels of gerE-lacZ expression under conditions in which the BofC protein inhibits SpoIVB-mediated signaling. This confirms our previous report, in which we suggested that SpoIVB and BofC interact (38). Again, we have failed to identify any potential PDZ recognition motif within BofC, which suggests that the SpoIVB recognition sequence represents a new class of motif. Since our work shows interaction of the SpoIVB PDZ domain with BofC, as well as with itself, it seems reasonable to predict that this domain is involved in other SpoIVB-mediated interactions, such as signaling of pro-ςK processing and the second function.
Our work has revealed that the SpoIVB PDZ domain is involved in at least two, if not more, distinct partner interactions. Unlike other multivalent PDZ-domain proteins, though, SpoIVB appears to use a single motif to interact with a number of target proteins. Some single PDZ domains mediate a number of interactions, which supports the multiple roles proposed for SpoIVB's PDZ domain. For instance, PDZ2 of the 95-kDa postsynaptic density protein can heterodimerize with neuronal nitric oxide synthase or α-syntrophin and interact with the C terminus of the Shaker-type potassium ion channel Kv1.4. Single mutations in PDZ2 can alter the specificity and affinity of one interaction without affecting the other (9). One plausible model is that the level of SpoIVB would dictate with which protein SpoIVB could interact, and indeed, we have proposed previously that the level of SpoIVB is important for escaping inhibition by the BofC protein (38).
ACKNOWLEDGMENTS
We thank Tony Wilkinson and Phil Wakeley for help and advice.
This work was supported by a grant from the Biotechnology and Biological Sciences Research Council (BBSRC) to S.M.C. J.A.B. is supported by the BBSRC-funded Structural Biology Centre at York.
REFERENCES
- 1.Beebe K D, Shin J, Peng J, Chaudhury C, Khera J, Pei D. Substrate recognition through a PDZ domain in tail-specific protease. Biochemistry. 2000;39:3149–3155. doi: 10.1021/bi992709s. [DOI] [PubMed] [Google Scholar]
- 2.Cutting S, Driks A, Schmidt R, Kunkel B, Losick R. Forespore-specific transcription of a gene in the signal transduction pathway that governs pro-ςK processing in Bacillus subtilis. Genes Dev. 1991;5:456–466. doi: 10.1101/gad.5.3.456. [DOI] [PubMed] [Google Scholar]
- 3.Cutting S, Oke V, Driks A, Losick R, Lu S, Kroos L. A forespore checkpoint for mother cell gene expression during development in B. subtilis. Cell. 1990;62:239–250. doi: 10.1016/0092-8674(90)90362-i. [DOI] [PubMed] [Google Scholar]
- 4.Cutting S, Panzer S, Losick R. Regulatory studies on the promoter for a gene governing synthesis and assembly of the spore coat in Bacillus subtilis. J Mol Biol. 1989;207:393–404. doi: 10.1016/0022-2836(89)90262-3. [DOI] [PubMed] [Google Scholar]
- 5.Cutting S, Roels S, Losick R. Sporulation operon spoIVF and the characterization of mutations that uncouple mother-cell from forespore gene expression in Bacillus subtilis. J Mol Biol. 1991;221:1237–1256. doi: 10.1016/0022-2836(91)90931-u. [DOI] [PubMed] [Google Scholar]
- 6.Cutting S M, Vander-Horn P B. Genetic analysis. In: Harwood C R, Cutting S M, editors. Molecular biological methods for Bacillus. Chichester, England: John Wiley & Sons, Ltd.; 1990. pp. 27–74. [Google Scholar]
- 7.Daniels D L, Cohen A R, Anderson J M, Brunger A T. Crystal structure of the hCASK PDZ domain reveals the structural basis of class II PDZ domain target recognition. Nat Struct Biol. 1998;5:317–325. doi: 10.1038/nsb0498-317. [DOI] [PubMed] [Google Scholar]
- 8.Doyle D A, Lee A, Lewis J, Kim E, Sheng M, MacKinnon R. Crystal structures of a complexed and peptide-free membrane protein-binding domain: molecular basis of peptide recognition by PDZ. Cell. 1996;85:1067–1076. doi: 10.1016/s0092-8674(00)81307-0. [DOI] [PubMed] [Google Scholar]
- 9.Gee S H, Quenneville S, Lombardo C R, Chabot J. Single-amino acid substitutions alter the specificity and affinity of PDZ domains for their ligands. Biochemistry. 2000;39:14638–14646. doi: 10.1021/bi001633t. [DOI] [PubMed] [Google Scholar]
- 10.Gomez M, Cutting S, Stragier P. Transcription of spoIVB is the only role of ςG that is essential for pro-ςK processing during spore formation in Bacillus subtilis. J Bacteriol. 1995;177:4825–4827. doi: 10.1128/jb.177.16.4825-4827.1995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Gomez M, Cutting S M. bofC encodes a putative forespore regulator of the Bacillus subtilis ςK checkpoint. Microbiology. 1997;143:157–170. doi: 10.1099/00221287-143-1-157. [DOI] [PubMed] [Google Scholar]
- 12.Gomez M, Cutting S M. Expression of the Bacillus subtilis spoIVB gene is under dual ςF/ςG control. Microbiology. 1996;142:3453–3457. doi: 10.1099/13500872-142-12-3453. [DOI] [PubMed] [Google Scholar]
- 13.Green D H, Cutting S M. Membrane topology of the Bacillus subtilis pro-ςK processing complex. J Bacteriol. 2000;182:278–285. doi: 10.1128/jb.182.2.278-285.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Hillier B J, Christopherson K S, Prehoda K E, Bredt D S, Lim W A. Unexpected modes of PDZ domain scaffolding revealed by structure of nNOS-syntrophin complex. Science. 1999;284:812–815. [PubMed] [Google Scholar]
- 15.Keiler K C, Waller P R, Sauer R T. Role of a peptide tagging system in degradation of proteins synthesised from damaged messenger RNA. Science. 1996;271:990–993. doi: 10.1126/science.271.5251.990. [DOI] [PubMed] [Google Scholar]
- 16.Kraulis P J. Molscript: a program to produce both detailed and schematic plots of protein structure. J Appl Crystallogr. 1991;24:946–950. [Google Scholar]
- 17.Lewis A P, Thomas P J. A novel clan of zinc-metalloproteases with possible intramembrane cleavage properties. Protein Sci. 1999;8:439–442. doi: 10.1110/ps.8.2.439. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Lu S, Kroos L. Overproducing the Bacillus subtilis mother cell sigma factor precursor, pro-ςK, uncouples ςK-dependent gene expression from dependence on intercompartmental communication. J Bacteriol. 1994;176:3936–3943. doi: 10.1128/jb.176.13.3936-3943.1994. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Nicholson W L, Setlow P. Sporulation, germination and outgrowth. In: Harwood C R, Cutting S M, editors. Molecular biological methods for Bacillus. Chichester, United Kingdom: John Wiley & Sons, Ltd.; 1990. pp. 391–450. [Google Scholar]
- 20.Oke V, Shchepetov M, Cutting S. SpoIVB has two distinct functions during spore formation in Bacillus subtilis. Mol Microbiol. 1997;23:223–230. doi: 10.1046/j.1365-2958.1997.2091573.x. [DOI] [PubMed] [Google Scholar]
- 21.Pallen M J, Ponting C P. PDZ domains in bacterial proteins. Mol Microbiol. 1997;26:411–415. doi: 10.1046/j.1365-2958.1997.5591911.x. [DOI] [PubMed] [Google Scholar]
- 22.Pallen M J, Wren B W. The HtrA family of serine proteases. Mol Microbiol. 1997;26:209–221. doi: 10.1046/j.1365-2958.1997.5601928.x. [DOI] [PubMed] [Google Scholar]
- 23.Ponting C P. Evidence for PDZ domains in bacteria, yeast, and plants. Protein Sci. 1997;6:464–468. doi: 10.1002/pro.5560060225. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Ponting C P, Philips C, Davies K E, Blake D J. PDZ domains: targeting signalling molecules to sub-membraneous sites. Bioessays. 1997;19:469–479. doi: 10.1002/bies.950190606. [DOI] [PubMed] [Google Scholar]
- 25.Resnekov O. Role of the sporulation protein BofA in regulating activation of the Bacillus subtilis developmental transcription factor ςK. J Bacteriol. 1999;181:5384–5388. doi: 10.1128/jb.181.17.5384-5388.1999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Resnekov O, Alper S, Losick R. Subcellular localization of proteins governing the proteolytic activation of a developmental transcription factor in Bacillus subtilis. Genes Cells. 1996;1:529–542. doi: 10.1046/j.1365-2443.1996.d01-262.x. [DOI] [PubMed] [Google Scholar]
- 27.Resnekov O, Losick R. Negative regulation of the proteolytic activation of a transcription factor in Bacillus subtilis. Proc Natl Acad Sci USA. 1998;95:3162–3167. doi: 10.1073/pnas.95.6.3162. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Rudner D Z, Fawcett P, Losick R. A family of membrane-embedded metalloproteases involved in regulated proteolysis of membrane-associated transcription factors. Proc Natl Acad Sci USA. 1999;96:14765–14770. doi: 10.1073/pnas.96.26.14765. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Sali A, Blundell T L. Comparative protein modelling by satisfaction of spatial restraints. J Mol Biol. 1993;234:779–815. doi: 10.1006/jmbi.1993.1626. [DOI] [PubMed] [Google Scholar]
- 30.Sambrook J, Fritsch E F, Maniatis T. Molecular cloning: a laboratory manual. 2nd ed. Cold Spring Harbor, N.Y: Cold Spring Harbor Laboratory Press; 1989. [Google Scholar]
- 31.Sandman K, Kroos L, Cutting S, Youngman P, Losick R. Identification of the promoter for a spore coat protein gene in Bacillus subtilis and studies on the regulation of its induction at a late stage of sporulation. J Mol Biol. 1988;200:461–473. doi: 10.1016/0022-2836(88)90536-0. [DOI] [PubMed] [Google Scholar]
- 32.Schneider S, Buchert M, Georgiev O, Catimel B, Halford M, Stacker S A, Baechi T, Moelling K, Hovens C M. Mutagenesis and selection of PDZ domains that bind new protein targets. Nat Biotechnol. 1999;17:170–175. doi: 10.1038/6172. [DOI] [PubMed] [Google Scholar]
- 33.Schultz J, Copley R R, Doerks T, Ponting C P, Bork P. SMART: a web-based tool for the study of genetically mobile domains. Nucleic Acids Res. 2000;28:231–234. doi: 10.1093/nar/28.1.231. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Songyang Z, Fanning A S, Fu C, Xu J, Marfatia S M, Chishti A H, Crompton A, Chan A C, Anderson J M, Cantley L C. Recognition of unique carboxyl-terminal motifs by distinct PDZ domains. Science. 1997;275:73–77. doi: 10.1126/science.275.5296.73. [DOI] [PubMed] [Google Scholar]
- 35.Stricker N L, Christopherson K S, Byungdoo A Y, Schatz P J, Raab R W, Dawes G, Bassett D E, Bredt D S, Li M. PDZ domain of neuronal nitric oxide synthase recognises novel C-terminal peptide sequences. Nat Biotechnol. 1997;15:336–342. doi: 10.1038/nbt0497-336. [DOI] [PubMed] [Google Scholar]
- 36.Tsunoda S, Sierralta J, Sun Y, Suzuki E, Becker A, Socolich M, Zuker C S. A multivalent PDZ-domain protein assembles signalling complexes in a G-protein-coupled cascade. Nature. 1997;388:243–249. doi: 10.1038/40805. [DOI] [PubMed] [Google Scholar]
- 37.Wakeley P, Dorazi R, Hoa N T, Bowyer J R, Cutting S M. Proteolysis of SpoIVB is a critical determinant in signalling of pro-ςK processing in Bacillus subtilis. Mol Microbiol. 2000;36:1336–1348. doi: 10.1046/j.1365-2958.2000.01946.x. [DOI] [PubMed] [Google Scholar]
- 38.Wakeley P, Hoa N T, Cutting S M. BofC negatively regulates SpoIVB-mediated signalling in the B. subtilis ςK-checkpoint. Mol Microbiol. 2000;36:1415–1424. doi: 10.1046/j.1365-2958.2000.01962.x. [DOI] [PubMed] [Google Scholar]
- 39.Youngman P, Perkins J, Losick R. Construction of a cloning site near one end of Tn917 into which foreign DNA may be inserted without affecting transposition in Bacillus subtilis or expression of the transposon-borne erm gene. Plasmid. 1984;12:1–9. doi: 10.1016/0147-619x(84)90061-1. [DOI] [PubMed] [Google Scholar]
- 40.Zhang B, Hofmeister A, Kroos L. The prosequence of pro-ςK promotes membrane association and inhibits RNA polymerase core binding J. Bacteriol. 1998;180:2434–2441. doi: 10.1128/jb.180.9.2434-2441.1998. [DOI] [PMC free article] [PubMed] [Google Scholar]