Highlights
-
•
The oxidation of soybean protein has the regular temporal evolution mechanism.
-
•
The oxidation with proper time can enhance functionality properties of soy protein.
-
•
Excessive oxidation leads to the decline of functionality properties of soy protein.
Keywords: Soy protein, Oxidation, Aggregation, Structure, Emulsifying activity
Abstract
The emulsifying activity of soy protein would decrease after long-term storage, which caused huge economic losses to food processing plants. This study explored the temporal evolution mechanism of oxidation on the structure and function of soy protein aggregates, which would improve the application of soy protein in food industry. Decreased α-helix and increased random coil were observed at the initial oxidation stage (0–4 h), which induced increases in hydrophobicity and disulfide bond content. In addition, emulsibility increased significantly. However, when the oxidation time extended to 6–12 h, the soluble aggregates transformed into insoluble aggregates with large particle size, low solubility, and molecular flexibility. Surface hydrophobicity and emulsifying activity were reduced, resulting in bridging flocculation of emulsion droplets. Mutual transformation between components is affected by factors that include spatial conformation and intermolecular forces, which eventually lead to functional changes in the protein molecules.
Introduction
Oxidation is an inevitable phenomenon that occurs during protein storage and processing. Oxidation can change the structure of proteins, leading to the formation of soluble or insoluble protein aggregates (Chen et al., 2013, Chen et al., 2013), which affects the properties of proteins (Sante-Lhoutellier, Aubry, & Gatellier, 2007). Previous studies have suggested that when free radicals modify amino acid side chains and attack the protein backbone, the polypeptide chain breaks, protein unfolds, tertiary structure opens, and spatial conformation changes (Davies, 2005). The sulfhydryl group of cysteine can be oxidized to disulfide bonds (Davies, Delsignore, & Lin, 1987), which can drive intramolecular and intermolecular cross-linking aggregation between proteins. Therefore, moderate oxidative modification can promote partial unfolding of proteins, generate soluble protein aggregates with high molecular flexibility, and enhance the water holding capacity and emulsifying properties of proteins (Wu, Li, & Wu, 2020). These observations about moderate oxidative modification have been made for soybean protein (Cui, Xiong, Kong, Zhao, & Liu, 2012), whey protein (Tan, Wang, Chen, Niu, & Yu, 2016), and myofibrillar protein (Xia et al., 2022). Liu, Lu, Han, Chen, and Kong (2015) demonstrated that mild oxidation markedly improved the functional properties of soybean protein, which is related to the formation of soluble aggregates and the level of aggregation. Excessive oxidation can promote the formation of disulfide bonds and the aggregation of proteins. These changes can induce the formation of insoluble aggregates, resulting in the loss of flexibility and functionality (Chen et al., 2013, Chen et al., 2013). Therefore, the oxidation degree of protein will greatly change the structure and affect the functional characteristics of protein.
Soy protein isolate (SPI) is an excellent nutrient with good processing properties, such as solubility and emulsifying properties. These attributes have made SPI the most widely used plant protein globally (Liu et al., 2015). Many studies have addressed the effects of oxidation on soybean protein structure and function. Soy protein modification by oxidation was related to loss of α-helix and increase of β-sheet structure, moreover, the emulsifying characteristics of soy protein increased and antioxidant activities of the digests from oxidized SPI decreased because of the oxidation modification. Most of this research has focused on the effects of different degrees of oxidative attack on soybean protein (Chen et al., 2013, Chen et al., 2013, Cui et al., 2012, Liu et al., 2015, Wu et al., 2010). The increased demand for more soybean protein products has resulted in huge pressure related to the storage and transportation of raw soybean meal and soybean protein. Research shows that during the storage of proteins, various reactive oxygen species are generated, including free radicals, lipid hydroperoxides, and reactive aldehydes, which are key factors for protein oxidation (Chen et al., 2013, Chen et al., 2013). During the production of soy protein, nearly 1% of lipids and highly reactive lipoxygenases (LOX) were left behind, resulting in a constant oxidative environment for soy protein, which made soybean protein susceptible to oxidation during storage (Harel & Kanner, 1985). After excessive oxidative denaturation, the solubility and interfacial activity of soybean protein would decrease significantly, resulting in the reduction of its functionality, which greatly limited the application of SPI in the field of food and produced severe economic losses for the food processing plants (Hinderink, Schrder, Sagis, Schron, & Berton-Carabin, 2021). Most of this research has focused on the effects of different degrees of oxidative attack on soybean protein at present. The mechanism of evolution of oxidative aggregates of soybean protein with respect to their behavior, structure, and function with oxidation time has not been thoroughly studied. By constructing the soybean protein oxidative aggregate model with different oxidation time, the change mechanism of the behavior, structure, and function of soybean protein oxidative aggregate with oxidation time can be clarified, which can simulate the whole process of oxidative denaturation of soybean protein during storage, help to improve the storage conditions of soybean protein and improve the utilization rate of protein.
2,2-Azobis (2-amidinopropane) dihydrochloride (AAPH) can be degraded due to heat to produce peroxyl radicals ROO· at a stable temperature of 37 °C. The amount of peroxyl radicals produced due to the thermal decomposition of AAPH is proportional to its concentration. In addition, AAPH is a free radical initiator with good controllability, stability, repeatability, and applicability, which is often used as inducer of protein oxidation model (Gieseg, Duggan, & Gebicki, 2000). Hence, we selected peroxy radicals produced by thermal decomposition of AAPH under aerobic conditions to represent lipid radicals in the process of lipid oxidation, and construct the oxidative aggregation system of soybean protein, assessing the effects of different oxidation times on the degree of aggregation of SPI in terms of particle size and turbidity, structural characteristics (secondary structure, tertiary structure, surface hydrophobicity, and disulfide bond), and emulsifying properties (solubility, emulsifying activity, emulsifying stability, particle size of emulsion, zeta (ζ)-potential, and confocal laser scanning microscopy [CLSM] appearance). The results of this study provide guidance for modification, transportation, and storage of SPI at industrial level.
Materials and methods
Materials
The Yuwang Group (Shangdong, China) supplied soy flakes without fat (52.27% crude protein concentration). Sigma-Aldrich (St. Louis, MO, USA) provided bovine serum albumin and AAPH. Analytical grade reagents and chemicals were used for the experiments.
Soy protein production
A previously method described by Jiang, Chen, and Xiong (2009) was used to produce the soy protein. 10% (w/v) defatted soy flake solution was adjusted to pH 8.0 and stirred for 2 h to extract protein and then centrifuged (13500g, 30 min) at 4 °C. The supernatant was adjusted to pH 4.5 with 2 mol/L HCl and centrifuged (3300g, 20 min). The pellet was washed twice with distilled water and neutralized to pH 7.0 with 2 mol/L NaOH. The sample was freeze-dried and stored at 2 °C. Protein concentrations were determined at 95% (w/w) by the biuret assay using bovine serum albumin as a standard (Gornall, 1949).
Oxidized soy protein
A previously method described by Wu, Zhang, Kong, and Hua (2009) was used for the oxidation of SPI. An appropriate amount of soy protein was dissolved in phosphate buffer (0.01 mol/L, pH 7.2, containing 0.5 mg/mL NaN3) to prepare sample solution (10 mg/mL). AAPH (0.5 mmol/L final concentration) was then added. A mixture solution was incubated at 37 °C in the dark for 12 h. Taking out part of solution every 2 h and reactions were stopped by immediately chilling them to 4 °C in an ice bath. Residual AAPH and salt were removed by centrifugation at 10,000g for 20 min and dialyzed (14,000 Da) for 72 h. And then samples were dried using a freeze-dryer (Marin Christ, Germany) and stored at 4 °C.
Identification of carbonyl groups
The carbonyl groups content of samples were analyzed using TU-1901 ultraviolet (UV) spectrophotometry (Persee, Beijing) (Huang, Hua, & Qiu, 2005). Samples was dissolved in deionized water to prepared 5 mg/mL protein solution. The sample solution was mixed with 0.01 mol/L 2,4-dinitrophenylhydrazine (DNPH) in 2 mol/L HCl with the volume ratio of 1:3. The sample was then dissolved in 0.1 mol/L sodium phosphate 6 mol/L guanidine hydrochloride buffer without DNPH as the blank control group. For control group and experimental group, 20% trichloroacetic acid was added and mingled. The mixed solution was centrifuged (10,000g, 10 min) at 4 °C, after standing for 20 min to discard the supernatant, and the ethanol/ethyl acetate solution (1:1, v/v) was used to wash pellet three times. The blank control group was used for numerical correction at 367 nm. Soluble protein concentration in guanidine hydrochloride solution were determined by the biuret method (Gornall, 1949), with bovine serum albumin (Sigma) as the standard. The results were expressed as nmole of carbonyl groups per milligram with molar extinction coefficient of 22,000 M−1 cm−1.
Particle size distribution
Dynamic light scattering (DLS) technique was used to determine the particle size distribution in the samples through Malvern ZetasizerNano ZS (Malvern Instruments Ltd., Malvern, UK). Briefly, the suspension (0.2 mg/mL) was centrifuged (10,000g, 15 min) and the supernatant was filtered through cellulose acetate membranes with a pore size of 0.45 μm to remove any other insoluble particles. Dispersion Technology Software version 4.20 was used to estimate the SPI particle size distribution. Results are expressed as volume percentage (%) versus particle size diameter (nm).
Turbidity
A previously method described by Huang et al. (2005) was used to analyze the turbidity of the samples. Briefly, the sample was dissolved in sodium phosphate buffer (0.01 mol/L, pH 7.0) by stirring on a magnetic stirrer (MYP19-2, CHIJIU, Shanghai, China) at 25 °C for 1 h to prepare 1 mg/mL sample solution. The measurement of dispersion absorbance was done with F7000 fluorescence spectrophotometer (Hitachi Co., Tokyo, Japan) at 600 nm to express turbidity.
Fourier transform infrared spectroscopy (FTIR)
The secondary structure of the samples was detected with an FTIR spectral analysis (Thermo Nexus 470, Nicolet Co. Ltd., USA). The sample was further dried in a desiccator with P2O5, mixed with potassium bromide, and then pressed into small tablet to determine FTIR. A total of 64 scans were evaluated at 0.5 cm−1 resolution within the range 400–4000 cm−1. Peakfit 4.2.0 software was used to calculate the relative content of each secondary structure by using the integral area. Of these, the band from 1646 to 1662 cm−1 could be assigned to α-helix. The bands from 1608 to 1622 cm−1 could be assigned to anti-parallel intermolecular β-sheet (β1), and the bands from 1622 to 1637 cm−1 as well as bands from 1682 to 1700 cm−1 could be assigned to intramolecular β-sheet and parallel intermolecular β-sheet (β2) of the protein respectively. The band from 1637 to 1645 cm−1 could be assigned to the random coil structure, while the bands from 1662 to 1681 cm−1 could be assigned to the β-turn.
Intrinsic fluorescence spectrum
A previously procedure described by Jiang et al. (2014) was used to measure the fluorescence spectrum. 0.2 mg/mL protein solution was prepared by phosphate buffer (0.01 mol/L, pH 7.0). A fluorescence spectrophotometer (F7000; Hitachi Co., Japan) was used to record the fluorescence spectrum of samples. The specific parameters were excitation wavelength at 290 nm and emission wavelength from 300 nm to 400 nm.
Surface hydrophobicity (H0)
A previously procedure described by Huang et al. (2005) was used to measure the surface hydrophobicity. The samples were prepared through phosphate buffer to obtain concentrations of 0.05–0.4 mg/mL. Then, 40 μL ANS (8 mmol/L) and 4 mL protein solution was mixed by incubating at room temperature for 30 min. The fluorescence intensity was measured through Hitachi F-7000 fluorescence spectrometer (Hitachi, Tokyo, Japan). Excitation and emission wavelength were 390 nm and 468 nm respectively, the scanning slit was 10 nm, and the scanning speed was 10 nm/s. Linear regression was used to fit the plot of fluorescence intensity vs protein concentration. The slope of the curve was used to define the H0 of samples.
Sulfhydryl and disulfide content
A previously procedure described by Beveridge, Toma, and Nakai (1974) was used to measure the Sulfhydryl and disulfide content. 5 mg/mL SPI samples were prepared by Trs-Gly buffer (containing 8 mol/L urea, pH 8.0) at 25 °C for 0.5 h. The suspension was centrifuged (10,000g, 15 min) at 4 °C. Then 3 mL supernatant was added to 0.02 mL of Ellman's reagent (4 mg/mL DTNB dissolved in Tris-Gly buffer) at 25 °C for 1 h, the absorbance was measured at 412 nm using TU-1901 UV spectrophotometer (Persee, Beijing). When the free sulfhydryl content was determined, buffer without urea was used.
where D is the dilution factor of the samples and C is the initial protein content (mg/mL).
where SHtotal was total sulfhydryl content (μmol/g) and SHfree was free sulfhydryl content (μmol/g).
Solubility
A previously procedure described by Lowry, Rosebrough, Farr, and Randall (1951) was used to measure protein solubility of different samples. Briefly, samples were dispersed in distilled water (pH 7.0) and continuously stirred for 30 min to prepare 20 mg/mL solution. After centrifugation (4000 r/min, 15 min), the micro-Kjeldahl process was used to determine the concentration of the proteins present in the supernatant (Technical, 2009).
Emulsion preparation
A previously procedure described by Liu, Damodaran, and Heinonen (2018) was used to prepare emulsion. The SPI was dissolve in sodium phosphate buffer (0.01 mol/L, pH 7.0) to prepare protein dispersion (20 mg/mL). Then the protein solution with soybean oil in a volume ratio of 2:1 was mixed and homogenized (Ultra-Turrax T18, ANGNI Co. Ltd., Shanghai, China) at 20,000 rpm for 5 min to obtain coarse emulsion. Then, a high-pressure (60 MPa) homogenizer (FB-110Q, LiTu Mechanical Equipment Co., Ltd, Shanghai, China) was used for 10 min to form stable oil-in-water emulsions.
Emulsifying activity index (EAI) and emulsion stability index (ESI)
A previously procedure described by Kevin et al. (1978) with some modifications was used to measure EAI and ESI. Briefly, the fresh emulsion prepared by 2.12 method was absorbed 50 μL from the bottom at 0 min and 10 min respectively, and then mixed with 0.1% (w/v) SDS solution at 1:100 (v/v). The absorbance of the sample was measured at 500 nm through a TU-1800 spectrophotometer (Beijing Purkinje General Instrument Co., Ltd. Beijing, China). The following formulas were used to estimate the EAI and ESI.
where A0 represents the absorbance at 500 nm, dilution factor is 100, C is the protein concentration (g/mL) before emulsification, and the oil volume fraction (v/v) of the emulsion (φ) is 0.20. The emulsion stability index was calculated as:
where A10 and A0 represent the absorbance at 500 nm after 10 and 0 min, respectively.
Emulsion ζ-potential
The ζ-potential of the prepared samples was measured using a Zetasizer Nano-ZS instrument (Malvern Panalytical, Malvern, UK). The freshly prepared emulsion was diluted 100 times with sodium phosphate buffer (0.01 mol/L, pH 7.0), and then measured the ζ-potential after equilibration at room temperature for 120 s. Each sample was measured in parallel for 3 times.
Emulsion droplet size distribution
A previously procedure described by Chen, Zhao, Sun, Ren et al. (2013), Chen, Zhao, and Sun (2013) was used to measure the emulsion droplet size distribution. Briefly, the emulsion was suspended directly in distilled water and estimated the droplet size distribution through Zetasizer Nano-ZS instrument (Malvern Panalytical, Malvern, UK).
Confocal laser scanning microscopy (CLSM)
A previously procedure described by Li, Zheng, Ge, Zhao, and Sun (2019) was used to visualize the microstructure of the emulsion samples. Briefly, emulsion samples were mixed with 40 μL mixed dye (0.02% Nile red dye and 0.1% Nile blue) and placed on a slide, covered with a coverslip, and the slides were placed on the focal stage of the microscope. Under the conditions of 488 nm Ar ion laser light and 633 nm He/Ne ion laser light, CLSM images of emulsion samples were obtained through the TCS SP2 microscope (Leica Microsystems, Wetzlar, Germany). The resulting fluorescence images were produced at a density of 1024 × 1024 pixels.
Statistical analysis
All generated data from this study were analyzed using the SPSS 26.0 statistical package (SPSS Inc., Chicago, IL, USA), followed by a one-way analysis of variance (ANOVA). Mean comparisons were performed using the least significant difference technique. All analyses were performed in triplicate. A P-value < 0.05 denoted a significant difference.
Results and discussion
Protein carbonyl contents
Amino acids with NH or NH2 groups on their side chains are highly reactive to OH•, which induced the transformation to carbonyl groups during protein oxidation (Sante-Lhoutellier et al., 2007). The protein carbonyl content in the SPI samples increased with increasing oxidation time, and the carbonyl content first increases slowly, then increases rapidly, and finally increases slowly again (P < 0.05; Fig. 1A). The modified soy protein content increased 11-fold in the carbonyl groups compared to that of the control after a 12-h post-incubation period. Soy protein reacts with the NH or NH2 groups of cysteine, histidine, and lysine during AAPH oxidation to form Schiff bases and carbonyl compounds (Davies, 2005). However, when the oxidation time was 8–12 h, the increase rate in the content of carbonyl groups decreased. The increase in protein carbonyl content may be due to the loose structure, high flexibility, and easy oxidation of SPI during the early stages of oxidation. However, when the duration of oxidization too long, SPI is cross-linked to a certain extent, and nucleophilic groups are buried. The overall structure is more compact and not easy to oxidize, resulting in the decrease in the rate of carbonyl formation.
Fig. 1.
The carbonyl content, PSD, turbidity, and structure of unoxidized and oxidized treated soybean protein aggregates at a different time (2, 4, 6, 8, 10, and 12 min).
Particle size distribution analysis, PDI, and turbidity
DLS and turbidity analyses can reveal the degree of soy protein aggregation and size change after oxidative attack, thereby characterizing the process of protein aggregate formation. Oxidation treatment substantially increased the average particle size of the control SPI (from 181.59) by 22.09% and 2012% after 2 and 12 h, respectively (Fig. 1B, Table 1). The particle size distribution of oxidized soy protein gradually changed from a trimodal to bimodal distribution as the oxidation time increased, and further changed to a unimodal distribution when the oxidation time reached 12 h. In the early stages of oxidation, protein unfolding is triggered, which lowers the surface electrostatic charge density, resulting in the formation of large-sized oxidized protein aggregates.
Table 1.
Average particle size, PDI, sulfhydryl content, emulsion capacity and stability of unoxidized, oxidized treated soybean protein aggregates at a different time (2, 4, 6, 8, 10, and 12 min).
| Sample | Average particle size (nm) | PDI | total sulfhydryl (nmol/mg) | free sulfhydryl (nmol/mg) | disulfide bond (nmol/mg) | EAI/(m2·g−1) | ESI/min |
|---|---|---|---|---|---|---|---|
| 0 | 181.59 ± 2.77 g | 0.76 ± 0.11a | 11.72 ± 0.15a | 15.64 ± 0.32a | 1.96 ± 0.11f | 91.61 ± 1.17c | 186.19 ± 3.89d |
| 2 | 221.71 ± 4.16f | 0.36 ± 0.04c | 11.18 ± 0.24b | 15.44 ± 0.28ab | 2.13 ± 0.16ef | 112.40 ± 2.06a | 226.66 ± 2.87b |
| 4 | 264.85 ± 5.03e | 0.21 ± 0.01e | 10.41 ± 0.17c | 15.03 ± 0.13b | 2.31 ± 0.18e | 108.90 ± 1.86b | 244.50 ± 3.14a |
| 6 | 683.49 ± 7.72d | 0.24 ± 0.03de | 9.50 ± 0.19d | 14.50 ± 0.19c | 2.50 ± 0.11d | 85.66 ± 1.97d | 203.85 ± 2.69c |
| 8 | 1884.42 ± 29.15c | 0.27 ± 0.01d | 7.83 ± 0.22e | 13.45 ± 0.20d | 2.81 ± 0.12c | 71.16 ± 1.73e | 188.81 ± 2.73e |
| 10 | 2511.70 ± 41.85b | 0.47 ± 0.05b | 6.10 ± 0.19f | 12.26 ± 0.12e | 3.08 ± 0.14b | 59.80 ± 2.24f | 157.20 ± 2.81f |
| 12 | 3836.18 ± 66.82a | 0.19 ± 0.03e | 3.09 ± 0.23 g | 9.83 ± 0.14f | 3.37 ± 0.13a | 46.97 ± 2.24 g | 137.20 ± 3.51 g |
Note: Comparisons were carried out between values of the same column; values with a different letter(s) indicate a significant difference at p ≤ 0.05.
Variations in the mass and size of the aggregates in the solution influence turbidity levels and indicate noncovalent protein–protein associations (Cromwell, Hilario, & Jacobson, 2006). We observed that turbidity increased with oxidation time (Fig. 1C), which could be because of the following two reasons: 1) increase in protein particle size caused by oxidation, and 2) the mass and size increase as insoluble aggregates are generated from soluble aggregates, resulting in elevated turbidity levels.
Fourier transform-infrared spectroscopy (FT-IR)
FT-IR is an optical method for analyzing protein secondary structure. The IR spectrum has three notable levels: amide I, II, and III bands. The amide I region is characterized by variations in the protein α-helix, β-sheet, β-turn, and random coil conformations (Feng et al., 2015). With increase in the oxidation time, the proportions of reverse parallel folding structure (β1) and random coil of oxidized SPI gradually increased, the proportion of α-helix gradually decreased, and the proportion of β-turn first decreased and then increased, reaching the lowest value at 4 h of oxidation. The proportions of parallel folding structure (β2) and intermolecular β-sheet showed no obvious change (Table 2).
Table 2.
Secondary structure content of unoxidized, oxidized treated soybean protein aggregates at a different time (2, 4, 6, 8, 10, and 12 min).
| Content/% | Anti-parallel intermolecular β-sheet (β1) | parallel intermolecular β-sheet (β2) | Intramolecular β-sheet | α-Helix | β-Turn | γ-Random coil |
|---|---|---|---|---|---|---|
| 0 | 9.27 ± 0.28f | 15.95 ± 0.23a | 18.41 ± 0.18a | 26.35 ± 0.18a | 21.33 ± 0.17e | 8.69 ± 0.15f |
| 2 | 10.18 ± 0.14e | 11.24 ± 0.21c | 16.12 ± 0.19d | 25.62 ± 0.25b | 21.61 ± 0.19e | 15.23 ± 0.19e |
| 4 | 10.67 ± 0.26d | 9.64 ± 0.19e | 17.22 ± 0.21b | 25.42 ± 0.22b | 20.06 ± 0.18f | 16.99 ± 0.21bc |
| 6 | 11.16 ± 0.19c | 10.24 ± 0.22d | 16.40 ± 0.20 cd | 23.21 ± 0.21c | 22.95 ± 0.22d | 16.04 ± 0.18d |
| 8 | 11.99 ± 0.24b | 12.02 ± 0.18b | 16.64 ± 0.19c | 19.40 ± 0.23d | 23.39 ± 0.18c | 16.56 ± 0.13c |
| 10 | 12.02 ± 0.16b | 10.70 ± 0.24c | 17.09 ± 0.24b | 18.23 ± 0.24e | 24.24 ± 0.24b | 17.72 ± 0.15a |
| 12 | 13.77 ± 0.19a | 7.16 ± 0.18f | 17.18 ± 0.13b | 17.82 ± 0.19e | 26.66 ± 0.20a | 17.41 ± 0.20b |
Note: Comparisons were carried out between values of the same column; values with a different letter(s) indicate a significant difference at p ≤ 0.05.
Aggregated protein molecules have β1 structures that play critical roles in their overall protein conformation. After oxidative induction, the soy protein unfolds and the interaction between the partial amino acid sequence and specific protein molecules is destroyed. As a consequence, the α-helix content is decreased (Cao et al., 2021), and the content of random coils is increased. In the present study, the proportion of β1 increased gradually and the proportion of β2 showed an irregular trend. These findings indicated that the peptide bonds C O and C—N between peptide units in the peptide chain of the oxidized aggregates of soy protein were arranged in the trans configuration. This may be because the structure of the trans construct of oxidized protein aggregates has a lower free energy than that of the cis arrangement. Similarly, Sun, Zhou, Sun, and Zhao (2013) confirmed that oxidation can significantly reduce the α-helix content of myofibrillar protein. Wu et al. (2010) also showed that oxidation treatment results in decreased content of α-helices and β-turns in soy protein.
Intrinsic fluorescence spectroscopy
The current study also investigated tertiary structural changes in SPI in the presence of oxidative treatment. The redshift of λmax indicates that the protein expands and the local microenvironment of tryptophan residues becomes more polar. A blue shift of λmax indicates aggregation of proteins in the embedding of tryptophan residues and the weakening of the polarity of protein molecules. Tryptophan oxidation levels and changes in the SPI microenvironment are key indices of intrinsic fluorescence. The intrinsic fluorescence spectra of intact and oxidized SPI are shown in Fig. 1E. As the oxidation time increased, the intrinsic fluorescence intensity of SPI gradually decreased, and the maximum fluorescence emission wavelength (λmax) showed an increasing–decreasing pattern and reached a maximum at 4 h. The findings indicate that in the initial stage of oxidation (0–4 h) the structure of SPI gradually unfolded due to oxidation, and internal tryptophan and other aromatic amino acid residues were gradually exposed to the external polar environment. As the oxidation further intensified, free radicals induced intermolecular cross-linking and aggregation of SPI. The exposed tryptophan residues were re-wrapped inside the protein and the microenvironment polarity of the fluorescence-emitting group was reduced. However, the fluorescence intensity of SPI exhibited a significant downward trend with increasing oxidation time. This is because tryptophan residue is vulnerable to peroxyl radicals, and loss of intrinsic tryptophan fluorescence is an early phenomenon of peroxyl radical-mediated protein oxidation (Chao, Ma, & Stadtman, 1997). A previous study demonstrated that peroxyl radicals can lower tryptophan levels, which could explain the reduced levels of intrinsic fluorescence intensity (Wu et al., 2009).
Surface hydrophobicity
Oxidative treatment induces the unfolding of hydrophobic protein clusters. Therefore, a change in H0 reflected a change in SPI structure. With increasing oxidation time, the hydrophobicity of SPI gradually increased from 0 to 4 h (Fig. 2A). The results showed that oxidation contributed to the increase of hydrophobic interaction. Combined with the results of intrinsic fluorescence spectroscopy, when free radicals attack the protein in the external oxidizing environment, oxidation can destroy the non-covalent interaction between proteins, such as hydrogen bond, resulting in the unfolding of protein structure. More hydrophobic amino acids are exposed, and the hydrophobic interaction between proteins improve, which result in significant increase in H0. This indicates that appropriate oxidation time can lead to the gradual formation of large protein oxidation aggregates by changing the structure and hydrophobic interaction of SPI (Jiang et al., 2009). However, when the oxidation time exceeds 4 h, the H0 of SPI decreases significantly, indicating that too long oxidation time will reduce the hydrophobic interaction between SPI. This may be due to the mutual polymerization of protein molecules because of long-term oxidative attack, and the exposed hydrophobic groups will be buried again, resulting in the reduction of surface hydrophobicity (Xu et al., 2017). This is consistent with the results of fluorescence spectrum and particle size distribution.
Fig. 2.
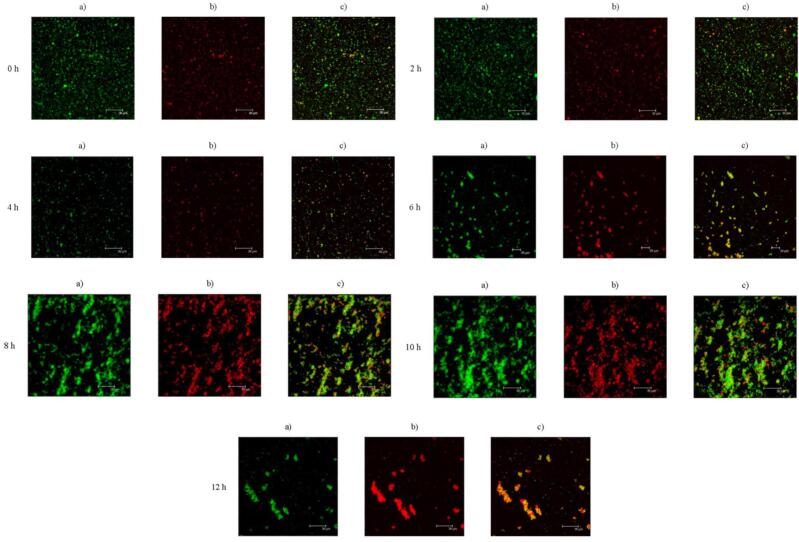
The surface hydrophobicity, solubility, and emulsion properties of unoxidized and oxidized treated soybean protein aggregates at a different time (2, 4, 6, 8, 10, and 12 min). Note: Protein and oil were stained by Nile Blue (a) and Nile Red (b), respectively. (c) was combined image of a and b.
Free sulfhydryl and disulfide bonds
Cysteine has been reported to be susceptible to oxidation. This feature is often masked by its carbonyl protein content (Cao et al., 2021). An alternative estimation is the sulfhydryl group and disulfide bond in cysteine, as they are essential components of protein functionality. Generally, the higher the disulfide bond content, the more compact is the spatial structure of the protein (Platt & Gieseg, 2003). As shown in Table 1, an increase in oxidation time significantly decreased the content of the free SH groups of the oxidized SPI, which is believed to induce the formation of disulfide bonds. The finding shows that oxidation treatment increases the tightness of the soy protein molecular space structure by forming a disulfide bond. In an oxidizing environment, the sulfhydryl group of the protein undergoes reversible and non-reversible oxidation reactions. The reversible oxidation reaction can generate disulfide bonds and sulfinic acid, and the irreversible oxidation reaction can generate non-disulfide bond sulfur compounds. As the oxidation time increased, the total sulfhydryl group content gradually decreased, and the rate of decrease of the free sulfhydryl group content was significantly higher than that of the disulfide bond formation. These observations indicate that the oxidation reaction of soy protein gradually changed from reversible oxidation to irreversible oxidation, and the change promoted the conversion of free sulfhydryl groups into non-disulfide bond sulfur compounds.
In the early stage of oxidation (0–4 h), the generation rate of disulfide bonds is slow because the expansion of the protein exposes the hydrophobic groups embedded initially in the protein. Hydrophobic interaction promotes protein aggregation and affects the SH/SS exchange in protein molecules (Hoffmann & Mil, 1997). When the oxidation time exceeds 4 h, the surface hydrophobicity of the proteins began to decrease. The formation rate of disulfide bonds increases, and the content of sulfhydryl groups decreases more rapidly, and this pattern is supported by the changes in the particle size distribution observed in this study. In contrast, all these changes can promote protein aggregation in radical-mediated oxidation, which increases the protein particle size.
Protein solubility
Protein insolubility can be an indicator of protein aggregation. In the present study, after 4 h of oxidation, an inverse relationship between oxidized SPI solubility levels and AAPH concentration was observed (P < 0.05; Fig. 2B). In the initial oxidation stage (0–4 h), the solubility of oxidized SPI was not significantly different from that of intact SPI. This may be because ROO· can oxidize the main peptide chain and side-chain groups of SPI during a short oxidation period, making the peptide chain structure loose and enhancing the ability of the protein to bind to water molecules (Arzeni et al., 2012). With increased oxidation time, the oxidized protein aggregates further aggregate through covalent cross-linking, resulting in the transformation of soluble aggregates into insoluble aggregates and decreased solubility. In the present study, AAPH treatment induced the production of insoluble protein aggregates.
EAI and ESI
The water-attracting and-repelling groups in protein molecules can serve as emulsifiers depending on their interactions with water and oil phases, respectively (Pearce & Kinsella, 1978). In this study, short-term oxidation treatment enhanced the EAI and ESI of the intact SPI samples (Table 1). The EAI and ESI reached a maximum at 2 h and 4 h of oxidation, respectively. This is because an appropriate oxidation treatment opens the tertiary structure of SPI, exposing hydrophobic groups, improving molecular flexibility, and changing the hydrophilic/lipophilic properties of the protein molecular surface. The changes promote the binding of more protein molecules to the oil–water interface layer. This reduces the interfacial tension of the oil–water interface, enhances the adsorption capacity of the protein molecules at the oil/water interface, and increases the viscosity of the emulsion (Keerati-U-Rai & Corredig, 2009). Ultimately, the EAI and ESI of the emulsion are increased. Earlier research suggested that increased protein surface hydrophobicity could promote emulsification. However, after reaching a peak, both indices showed a significant downward trend in the present study (P < 0.05). This is because as the degree of oxidation of SPI increases, a highly ordered β1 structure is formed between the molecules, and flexibility reduces and rearrangement at the oil–water interface becomes more difficult. At the same time, SPI aggregates, causing the hydrophobic groups to remain embedded, which reduces H0 and solubility. These changes ultimately lead to a significant decline in the EAI and ESI of oxidized SPI.
ζ-Potential of emulsion
ζ-Potential influences the physicochemical properties of a given emulsion. In the current study, all emulsion samples had negative ζ-potential values (˃20 mV). When the oxidation time was 4 h, the absolute value of ζ-potential was the highest, but it had an inverse relationship with the oxidation time. An increase in surface net charge improves the electrostatic repulsive force of the emulsion droplets, which is often stronger than the attractive forces (Kim, Decker, & Mcclements, 2002). At the same time, because the anionic interfacial layer can adsorb cations, the strong negative charge on the interface also affects the stability of the emulsion (Mcclements & Decker, 2000), thereby forming a more stable oil–water interface and enhancing the emulsification performance of oxidized SPI. Changes in protein surface charge could be caused by an imbalance in the acid/basic amino acid ratio and oxidation of selected amino acid groups. In addition, the emulsifying indices of moderately oxidized SPI can be increased by increasing its negative surface charges (Liu et al., 2015).
Particle size of emulsion
The larger the particle size of the emulsion, the more unstable is the emulsion. Therefore, the particle size of the protein emulsion can help in the characterization of the emulsification performance. As shown in Fig. 2D, as the oxidation time increased, the particle size distribution of the oxidized SPI emulsion exhibited diverse characteristics, including unimodal, bimodal, and trimodal distributions. The particle size distribution of the intact SPI emulsion was unimodal. However, after 2 h of oxidation, the particle size peak of the emulsion shifted to the left; after 4 h of oxidation, the particle size peak of the emulsion exhibited a trimodal distribution; after oxidation for 6 h, the particle size of the emulsion returned to a unimodal distribution; and after 8 h of oxidation, the particle size peak of the emulsion exhibited a bimodal distribution. With an increase in the oxidation time, the particle size of the SPI emulsion returned to the unimodal distribution, and the peak gradually shifted to the right. Compared to that of the intact SPI, the particle size of the SPI emulsion after oxidation treatment for 2 h and 4 h was reduced, which is in agreement with results from previous studies (Sun et al., 2013). Combined with the results of protein structure, EAI and ESI, moderate oxidation (0–4 h) causes the protein structure to unfold and the increase of surface hydrophobicity and molecular flexibility. These hydrophobic groups are easier to adsorb on the oil–water interface, which helps to improve the emulsification performance of SPI through strengthening the attraction between protein molecules and oil droplets and reducing the surface tension of the water–oil interface (Liu & Tang, 2016). Therefore, appropriate oxidation time can enhance the emulsification performance of SPI and form emulsion droplets with small particle size. Long time oxidation will lead to the aggregation of soybean protein to form large particle size aggregates and decrease the emulsification performance, which do not easily bind to the oil–water interface during homogenization and cannot form a stable oil–water interface film, leading to increase particle size of emulsion (Chen et al., 2013, Chen et al., 2013).
CLSM
To further observe the effect of different oxidation times on the oil–water interface of the emulsion, the microstructures of the emulsions were visualized using CLSM. Fig. 3 shows representative CLSM images of the emulsions formed using SPI at different oxidation times. The emulsion droplets formed by SPI after short-term oxidation treatment (2–4 h) were smaller than those formed by intact SPI. SPI oxidized for 4 h formed a relatively uniform emulsion and smaller emulsified droplets, consistent with the particle size results. Droplet flocculation and merging of droplets were observed after SPI was further oxidized (>4h). These findings indicate that moderate oxidation treatment helps more protein molecules to adsorb at the oil–water interface to form a more stable interface film, thereby reducing the particle size distribution of the emulsion droplets and improving particle size uniformity. In general, the oxidative aggregation of SPI affects the hydrophobicity/hydrophilicity of its surface, culminating in changes in emulsion stability. These changes are influenced by creaming rate and size of the oil droplet. Reduction in the size of oil droplets is triggered by moderate oxidation activities, which results in lowered chance of emulsion instability (Li et al., 2019). A previous study demonstrated that SPI aggregates could increase the rheological properties of the sample, thus serving as a good stabilizing agent (Tang, 2017). Excessive oxidation leads to an increase in the degree of aggregation of protein molecules and a decrease in solubility. Because the diffusion rate of insoluble proteins at the interface is much slower than that of soluble proteins, it is impossible to form a stable interface film through interface rearrangement and relaxation during the emulsification process, which causes rapid polymerization-bridging flocculation of emulsion droplets (Dickinson, 2010). This results in a large collection area of red oil droplets on the image of Fig. 3-12h.
Fig. 3.
The CLSM of unoxidized and oxidized treated soybean protein aggregates at a different time (2, 4, 6, 8, 10, and 12 min).
Conclusion
The results indicate that with increased oxidation time from 0 to 4 h, free radicals attack soy protein, resulting in the expansion of structure, change in secondary structure, exposure of internal hydrophobic groups, formation of disulfide bonds between protein molecules, and formation of oxidation aggregates with high molecular flexibility and increased particle size. However, with a further increase in the oxidation treatment time (6 h to 12 h), the oxidation attack changes the spatial structure of soy protein, affecting the intermolecular interaction and cross-linking reaction, and promoting the mutual transformation from free sulfhydryl to disulfide bonds and formation of the β1 structure. These changes induce the formation of oxidized aggregates with large particle size, and low solubility and molecular flexibility. Thus, controlling oxidation time could lead to the directional regulation of conformational factors that play a decisive role in the activity of the protein interface, thereby improving the emulsifying activity of soy protein. Our future work will focus on the investigation of the evolutionary mechanisms of the components involved in the oxidative aggregation of proteins and the competitive interfacial adsorption mechanism between the components.
CRediT authorship contribution statement
Yanan Guo: Conceptualization, Software, Writing – original draft. Zhongjiang Wang: Data curation. Zhaodong Hu: Investigation. Zongrui Yang: Visualization. Jun Liu: Methodology. Bin Tan: Investigation. Zengwang Guo: . Bailiang Li: Funding acquisition, Project administration. He Li: Investigation.
Declaration of Competing Interest
The authors declare that they have no known competing financial interests or personal relationships that could have appeared to influence the work reported in this paper.
Acknowledgments
The authors would like to thank the Heilongjiang Province “Hundred, Thousand, Ten Thousand” Engineering Science and Technology Major Project (2021ZX12B02); Shandong key R&D plan (major scientific and technological innovation project) (2022CXGC010603); Youth talent promotion project of China Association for Science and Technology (2019QNRC001); Key R&D projects in Heilongjiang Province (GY2021ZB0204); National key R&D plan in the 14th five year plan (2021YFD2100400); Transformation project of major scientific and technological achievements in Heilongjiang Province (CG19A002) and Major industrial key projects for the transformation of new and old kinetic energy in Shandong Province for the support.
Contributor Information
Yanan Guo, Email: gynname@163.com.
Zhongjiang Wang, Email: wzjname@126.com.
Zhaodong Hu, Email: nodanger2021@163.com.
Zongrui Yang, Email: yzrname@163.com.
Jun Liu, Email: liujun@yuwangcn.com.
Bin Tan, Email: 1079826176@qq.com.
Zengwang Guo, Email: gzwname@163.com.
Bailiang Li, Email: 15846092362@163.com.
He Li, Email: 326790339@qq.com.
References
- Arzeni C., Martinez K., Zema P., Arias A., Perez O.E., Pilosof A. Comparative study of high intensity ultrasound effects on food proteins functionality. Journal of Food Engineering. 2012;108(3):463–472. [Google Scholar]
- Beveridge T., Toma S.J., Nakai S. Determination of SH- and SS- groups in some food protein using Ellman's reagrent. Journal of Food Science. 1974;39(1):49–51. [Google Scholar]
- Cao H., Sun R., Shi J., Li M., Guan X., Liu J.…Zhang Y. Effect of ultrasonic on the structure and quality characteristics of quinoa protein oxidation aggregates. Ultrasonics Sonochemistry. 2021;77 doi: 10.1016/j.ultsonch.2021.105685. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chao C.C., Ma Y.S., Stadtman E.R. Modification of protein surface hydrophobicity and methionine oxidation by oxidativesystems. Proceedings of the National Academy of Sciences of the United States of America. 1997;94(7):2969–2974. doi: 10.1073/pnas.94.7.2969. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chen N., Zhao M., Sun W. Effect of protein oxidation on the in vitro digestibility of soy protein isolate. Food Chemistry. 2013;141(3):3224–3229. doi: 10.1016/j.foodchem.2013.05.113. [DOI] [PubMed] [Google Scholar]
- Chen N., Zhao M., Sun W., Ren J., Cui C. Effect of oxidation on the emulsifying properties of soy protein isolate. Food Research International. 2013;52(1):26–32. [Google Scholar]
- Cromwell M., Hilario E., Jacobson F. Protein aggregation and bioprocessing. Aaps Journal. 2006;8(3):572–579. doi: 10.1208/aapsj080366. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cui X., Xiong Y.L., Kong B., Zhao X., Liu N. Hydroxyl Radical-Stressed Whey Protein Isolate: Chemical and Structural Properties. Food & Bioprocess Technology. 2012;5(6):2454–2461. [Google Scholar]
- Davies M.J. The oxidative environment and protein damage. Biochimica et Biophysica Acta. 2005;1703(2):93–109. doi: 10.1016/j.bbapap.2004.08.007. [DOI] [PubMed] [Google Scholar]
- Davies K., Delsignore M.E., Lin S. Protein damage and degradation by oxygen radicals. II. Modification of amino acids. Journal of Biological Chemistry. 1987;262(20):9902–9907. [PubMed] [Google Scholar]
- Dickinson E. Food emulsions and foams: Stabilization by particles. Current Opinion in Colloid & Interface Science. 2010;15(1):40–49. [Google Scholar]
- Feng X., Li C., Ullah N., Cao J., Lan Y., Ge W.…Chen L. Susceptibility of whey protein isolate to oxidation and changes in physicochemical, structural, and digestibility characteristics. Journal of Dairy Science. 2015;98(11):7602–7613. doi: 10.3168/jds.2015-9814. [DOI] [PubMed] [Google Scholar]
- Gieseg S., Duggan S., Gebicki J.M. Peroxidation of proteins before lipids in U937 cells exposed to peroxyl radicals. Biochemical Journal. 2000;350(1):215–218. [PMC free article] [PubMed] [Google Scholar]
- Gornall A.G. Determination of serum proteins by means of the biuret reaction. Journal of Biological Chemistry. 1949;177(2):751–766. [PubMed] [Google Scholar]
- Harel S., Kanner J. Muscle membranal lipid peroxidation initiated by hydrogen peroxide-activated metmyoglobin. Journal of Agricultural & Food Chemistry. 1985;33(6):1188–1192. [Google Scholar]
- Hinderink E., Schrder A., Sagis L., Schron K., Berton-Carabin C.C. Physical and oxidative stability of food emulsions prepared with pea protein fractions. LWT- Food Science and Technology. 2021;146(5) [Google Scholar]
- Hoffmann M., Mil P. Heat induced aggregation of -lactoglobulin: Role of the free thiol group and disulphide bonds. Journal of Agricultural & Food Chemistry. 1997;45(8):2942–2948. [Google Scholar]
- Huang Y., Hua Y., Qiu A. Soybean protein aggregation induced by lipoxygenase catalyzed linoleic acid oxidation. Food Research International. 2005;39(2):240–249. [Google Scholar]
- Jiang J., Chen J., Xiong Y.L. Structural and emulsifying properties of soy protein isolate subjected to acid and alkaline pH-shifting processes. Journal of Agricultural & Food Chemistry. 2009;57(16):7576–7583. doi: 10.1021/jf901585n. [DOI] [PubMed] [Google Scholar]
- Jiang L., Wang J., Li Y., Wang Z., Liang J., Wang R.…Zhang M. Effects of ultrasound on the structure and physical properties of black bean protein isolates. Food Research International. 2014;62:595–601. [Google Scholar]
- Keerati-U-Rai M., Corredig M. Heat-induced changes in oil-in-water emulsions stabilized with soy protein isolate. Food Hydrocolloids. 2009;23(8):2141–2148. [Google Scholar]
- Kim H.J., Decker E.A., Mcclements D.J. Role of Postadsorption Conformation Changes of β-Lactoglobulin on Its Ability To Stabilize Oil Droplets against Flocculation during Heating at Neutral pH. Langmuir. 2002;18(20):7577–7583. [Google Scholar]
- Li Q., Zheng J., Ge G., Zhao M., Sun W. Impact of heating treatments on physical stability and lipid-protein co-oxidation in oil-in-water emulsion prepared with soy protein isolates. Food Hydrocolloids. 2019;100 [Google Scholar]
- Liu C., Damodaran S., Heinonen M. Effects of microbial transglutaminase treatment on physiochemical properties and emulsifying functionality of faba bean protein isolate. LWT- Food Science and Technology. 2018;99(3):396–403. [Google Scholar]
- Liu Q., Lu Y., Han J., Chen Q., Kong B. Structure-modification by moderate oxidation in hydroxyl radical-generating systems promote the emulsifying properties of soy protein isolate. Food Structure. 2015;6:21–28. [Google Scholar]
- Liu F., Tang C. Soy glycinin as food-grade Pickering stabilizers: Part. I. Structural characteristics, emulsifying properties and adsorption/arrangement at interface. Food Hydrocolloids. 2016;60:606–619. [Google Scholar]
- Lowry O.H., Rosebrough N.J., Farr A.L., Randall R.J. Protein measurement with the Folin phenol reagent. Journal of Biological Chemistry. 1951;193:265–275. [PubMed] [Google Scholar]
- Mcclements D.J., Decker E.A. Lipid oxidation in oil-in-water emulsions: Impact of molecular environment on chemical reactions in heterogeneous food systems. Blackwell Publishing Ltd. 2000;65(8):1270–1282. [Google Scholar]
- Pearce K.N., Kinsella J.E. Emulsifying properties of proteins: Evaluation of a turbidimetric technique. Journal of Agricultural & Food Chemistry. 1978;26(3):716–723. [Google Scholar]
- Platt A.A., Gieseg S.P. Inhibition of protein hydroperoxide formation by protein thiols. Redox Report. 2003;8(2):81–86. doi: 10.1179/135100003125001288. [DOI] [PubMed] [Google Scholar]
- Sante-Lhoutellier V., Aubry L., Gatellier P. Effect of Oxidation on In Vitro Digestibility of Skeletal Muscle Myofibrillar Proteins. Journal of Agricultural & Food Chemistry. 2007;55(13):5343–5348. doi: 10.1021/jf070252k. [DOI] [PubMed] [Google Scholar]
- Sun W., Zhou F., Sun D., Zhao M. Effect of Oxidation on the Emulsifying Properties of Myofibrillar Proteins. Food & Bioprocess Technology. 2013;6(7):1703–1712. [Google Scholar]
- Tan Y., Wang J., Chen F., Niu S., Yu J. Effect of protein oxidation on kinetics of droplets stability probed by microrheology in O/W and W/O emulsions of whey protein concentrate. Food Research International. 2016;85:259–265. doi: 10.1016/j.foodres.2016.05.005. [DOI] [PubMed] [Google Scholar]
- Tang C. Emulsifying properties of soy proteins: A critical review with emphasis on the role of conformational flexibility. Critical Reviews in Food Technology. 2017;57(12):2636–2679. doi: 10.1080/10408398.2015.1067594. [DOI] [PubMed] [Google Scholar]
- Technical, A. (2009). Crude Protein--Micro-Kjeldahl Method: AACC International Approved Methods.
- Wu X., Li F., Wu W. Effects of rice bran rancidity on the oxidation and structural characteristics of rice bran protein. LWT- Food Science and Technology. 2020;120 [Google Scholar]
- Wu W., Wu X., Hua Y. Structural modification of soy protein by the lipid peroxidation product acrolein. LWT - Food Science and Technology. 2010;43:133–140. [Google Scholar]
- Wu W., Zhang C., Kong X., Hua Y. Oxidative modification of soy protein by peroxyl radicals. Food Chemistry. 2009;116(1):295–301. [Google Scholar]
- Xia T., Xu Y., Zhang Y., Xu L., Kong Y., Song S.…Xu X. Effect of oxidation on the process of thermal gelation of chicken breast myofibrillar protein. Food Chemistry. 2022;132368 doi: 10.1016/j.foodchem.2022.132368. [DOI] [PubMed] [Google Scholar]
- Xu J., Chen Z., Han D., Li Y., Sun X., Wang Z., Jin H. Structural and functional properties changes of β-conglycinin exposed to hydroxyl radical-generating systems. Molecules. 2017;22(11):1893. doi: 10.3390/molecules22111893. [DOI] [PMC free article] [PubMed] [Google Scholar]