Highlights
-
•
The ternary complex of SPI, β-glucan and myricetin was prepared, and the interaction mechanism was studied.
-
•
QCM-D was used to explore the binding affinity of 7S and 11S to β-glucan and myricetin.
-
•
Molecular docking analysis indicated that 11S protein has a stronger binding capacity compared with 7S.
Keywords: Soybean protein isolate, β-Glucan, Myricetin, Ternary complex, Interaction mechanism
Abstract
The complexation of soybean protein isolate (SPI) with β-glucan (DG) and myricetin (MC) was focused in this study. UV-Vis, circular dichroism and 3D fluorescence analysis jointly proved that interaction with DG and MC altered the structures of SPI, whose β-sheet decreased to 29 % and random coil increased to 35 %, respectively. Moreover, the microenvironment of tryptophan and tyrosine from protein were changed. The ternary complex performed a different molecular weight distribution, showing a larger molecular weight of 1.17×106 g/mol compared with SPI verified by gel permeation chromatography (GPC). And it was further evidenced by Quartz Crystal Microbalance with Dissipation (QCM-D) and molecular docking that glycinin (11S) possessed a better affinity toward DG and MC compared with β-conglycinin (7S), which indicated stronger binding ability through hydrogen bonds. The successful preparation of SPI-DG-MC complex will advance the application of soybean resource as a functional food ingredient.
Introduction
Protein, polysaccharide and polyphenol, important nutrients and functional substances in foods, often interact with each other when coexist (Li et al., 2022). Normally, the interactions between these compounds can be divided into non-covalent and covalent, in which non-covalent interaction dominates. The complex formed due to non-covalent interaction has the characteristics of specificity and reversibility, depending on hydrogen bond, electrostatic interaction (ion interaction) and hydrophobic interaction (Buitimea-Cantua, Gutierrez-Uribe, & Serna-Saldivar, 2018). For example, the polyphenols can be linked to arabinan backbone by hydrogen bond, which is influenced by temperature, pH, and ion strength (Fernandes, Le Bourvellec, Renard, Wessel, Cardoso, & Coimbra, 2020). Nevertheless, the enzyme-induced and non-enzyme-induced covalent interactions, including Maillard reaction, free radical graft and carbon diimide induction, affect the sensory, functional and nutritional properties of foods(de Oliveira, Coimbra, de Oliveira, Zuniga, & Rojas, 2016), Therefore, the interaction and complexation of proteins, polysaccharides and polyphenols are crucial that attracts great concern of scientists in recent years.
Soybean protein isolate (SPI) is a kind of plant protein rich in essential amino acids and has high nutritional value. SPI possesses important functional properties such as emulsification, solubility, foaming, water holding and gelation, but which is restrictedly applied with difficulties to meet the needs of different food systems. Hence, upgrading the overall performance of SPI has been regarded as a topical issue. A large number of studies have proved that complexation with typical polysaccharide and/or polyphenol can greatly improve the functional properties of protein. For instance, the composite product of SPI and maltodextrin has a more compact microstructure with enhanced water holding capacity and gel strength (C. Zhao, Yin, Yan, Niu, Qi, & Liu, 2021). The covalent complex formed by SPI and anthocyanin as emulsion stabilizer can make the emulsion system show excellent stability and significantly improve antioxidant activity (Ju et al., 2020). In addition, the formation of complex between SPI and tea polyphenols enhanced the free radical scavenging ability of polyphenols (Djuardi, Yuliana, Ogawa, Akazawa, & Suhartono, 2020). However, the one-step and synchronous complexation of SPI with polysaccharide and polyphenol are less reported, along with their impact on the structure and functional properties of protein.
β-glucan (DG) is a bioactive polysaccharide composed of glucose monomers linked by 1–3, 1–4 or 1–6 glycosidic bonds (Bai et al., 2019), which has gathered much attention due to its good biological activities such as regulation of immune (Chethan et al., 2017), lowering blood glucose and serum cholesterol levels (Ekström, Henningsson Bok, Sjöö, & Östman, 2017). Myricetin (MC, 3, 5, 7, 3′, 4′, 5′-OH), a class of hexahydroxy polyphenol, possesses excellent antioxidant activities (Song et al., 2021), as well as anti-tumor (Jiang, Zhu, Wang, & Yu, 2019) and cardiovascular-protective effects (L. Wang, Wu, Yang, & Dong, 2019). Previously, our lab has evidenced the formation of DG-MC complex with 35 % adsorption capacity of polyphenol. To be interest, it was found that the foaming and emulsifying properties of SPI were significantly improved by this binary complex (data not shown). Hence, in this work, the addition of DG and MC into SPI was further studied to verify the interaction and its possible mechanism of protein, polysaccharide and polyphenol.
Particularly, 7S and 11S globulins account for more than 70 % of the protein content (J. Li, Li, & Guo, 2014). 7S globulin (molecular weight 180–210 kDa) has three subunits, which are α' (≈71KDa), α (≈67 kDa) and β (≈50 kDa), each consisting of an acidic N-terminal α-chain and an alkaline C-terminal β-chain (C. Wu, Hua, Chen, Kong, & Zhang, 2016). 11S globulin consists of six non-covalently-linked subunit pairs, each consisting of an acidic subunit with a molecular weight of about 32 kDa (an N-terminal acid α chain) and a basic subunit with a molecular weight of about 20 kDa (a C-terminal alkaline β-chain) (Singh, Meena, Kumar, Dubey, & Hassan, 2015). However, current studies on the complex of SPI mainly focus on mixed proteins, there are few publications on the interaction of 7S and 11S globulins with other nutrients.
Therefore, Ultraviolet–visible (UV–vis) spectrum, circular dichroism (CD), three-dimensional (3D) fluorescence spectrum and field emission scanning electron microscope (FESEM) analysis of SPI-DG-MC complex were carried out. The adsorption differences of 7S and 11S (the main protein components in SPI) with DG and MC were observed by Quartz Crystal Microbalance with Dissipation (QCM-D). According to the data of adsorption thickness and the maximum binding energy, the most optimal binding globulin was obtained as well as the possible interaction mechanism. This result will lay a theoretical foundation for the further development and utilization of functional food ingredients from SPI and their complexes.
Materials and methods
Materials and chemicals
Soy protein isolate (SPI, ≥96.0 %, CAS:9010–10-0, Lot:20201016) was purchased from Henan Zhong Tai Food Chemical Co., ltd. β-glucan (DG) was derived from barley (≥95.0 %, CAS:9041-22-9, Lot:20210306) and myricetin (MC, ≥98.0 %, HPLC, CAS:529–44-2, Lot:200619) were purchased from Shandong West Asia Chemical Co., ltd. All other chemical reagents are analytical pure and obtained from Sinopharm Chemical Reagents Co., ltd.
Preparation of 7S and 11S proteins
The preparation of 7S and 11S globulin was referring to the previously reported method with slight modifications. SPI was dispersed in the deionized water (1:15, W/V), and the pH was adjusted to 8.0 with 2 M NaOH, after which the solution was stirred with magnetic force at 45 ℃ for 1 h. The insoluble residue was discarded after centrifugating at 9,000 rpm for 30 min, and the pH value of the supernatant was adjusted to 6.4. The left solution was placed at 4 ℃ overnight for cold precipitation. Afterwards, the precipitation (11S globulin) was obtained by centrifuging at 4,000 rpm for 20 min. The pH value of supernatant was adjusted to 4.8 with 2 M HCl. Consequently, after centrifuged at 9,000 rpm for 30 min at 4 ℃, the precipitate was isolated and collected as 7S globulin. All the acid precipitated globulins were finally neutralized with NaOH solution and freeze-dried.
Preparation of SPI binary and ternary complexes
Appropriate amount of SPI was added to deionized water (10 mg/mL) and stirred overnight with magnetic force until complete hydration. The protein mother liquor was centrifuged to collect the supernatant, which was tested based on the Bradford method. The preparation of SPI ternary complex was operated according to the method of Wu et al. (Z. Wu, Li, Ming, & Zhao, 2011) and made appropriate modifications. The solution of DG (2 mg/mL) was stirred magnetically at 80 ℃ for 2 h until completely dissolved. The mixture solution of SPI (10 mg/mL), MC (2.5 mg/mL) and DG (VSPI: VDG: VMC = 4:2:1, Preliminary experimental results) was stirred for 2 h at room temperature and then transferred into the pre-processed dialysis bag (1000 Da). The dialysis bag was placed in a 3 L beaker for 48 h at 40 ℃ until the adsorption equilibrium was achieved, free polyphenols were removed from the dialysis bag. The solution inside the dialysis bag was identified as SPI-DG-MC ternary complex, which was followingly lyophilized. SPI-β-glucan (SPI-DG), SPI-myricetin (SPI-MC) and β-glucan-myricetin (DG-MC) complexes were prepared in the same way.
Multiple spectroscopic analysis
Ultraviolet–visible (UV–vis)
The mass proportion of SPI, DG and MC in the ternary complex was previously determined and calculated to prepare the physical mixture of these three components. Then, the physical mixture and complex of SPI, DG and MC were dissolved to 1 mg/mL, using deionized water as reference. The UV absorption spectrum was recorded by scanning in the wavelength range of 200–500 nm on a UV–vis spectrophotometer (Evolution 220, Thermo Fisher, American).
Circular dichroism (CD)
The mixture and complex of SPI, DG and MC were configured into a 1 mg/mL of solution, using SPI as reference. The secondary structures of protein, complex and their mixture were analyzed by far ultraviolet circular dichroism (J-1500, JASCO, Japan). The scanning temperature was set at 25 ℃, the scanning rate was set at 100 nm/min, and multiple scanning was performed in the wavelength range of 190–260 nm.
Three-dimensional (3D) fluorescence spectrum
The mixture and complex of SPI, DG and MC were dissolved to 0.5 mg/mL, using SPI as reference. Three-Dimensional (3D) fluorescence spectrum (Cary Eclipse, Varian, American) determination of the samples was set at following conditions: the excitation wavelength was from 200 to 320 nm, the emission wavelength of continuous scanning recording was from 270 to 500 nm, the slit was 2 nm, and the scanning rate was 12,000 nm/min.
Field emission scanning electron microscopy (FESEM)
Firstly, the conductive adhesive was pasted on the sample table, after which the dried SPI, SPI-DG-MC complex and their mixture were fixed, and the unfixed powder was removed prior to gold spraying. Microscopic images of the samples were collected under a field emission scanning electron microscope (GeminiSEM 300, ZEISS, Germany) at 5 kV with magnifications of × 100, ×200 and × 500, respectively.
Sodium dodecyl sulphate polyacrylamide gel electrophoresis (SDS-PAGE)
The SDS-PAGE experiment was used to analysis the molecular weight of the samples. The thickness of vertical slab gel was 1.5 mm. The glue was consisted of 4 % acrylamide stacking gel and 10 % acrylamide resolving gel. The sample concentration was 2 mg/mL, 10 μL sample loading buffer was added to 30 μL protein sample, then heated in 100 ℃ water bath for 5–10 min. The loading volume of samples were 10 μL. The initial voltage was 80 V until into resolving gel, then increased to 120 V. After electrophoresis, gel bands were stained using Coomassie brilliant blue R-250.
High performance gel permeation chromatography
The molecular weight distribution of SPI, SPI-DG, SPI-MC and SPI-DG-MC complexes, was studied by high performance gel permeation chromatography (GPC). Samples (2 mg/mL, dissolved by ultrapure water) were determined using P230 GPC-gel permeation chromatography system (Elite, Dalian). The type of column applied was sEC-300 (7.8*300 mm, 5 μm). Conditions were as follows: flow rate at 1.0 mL/min, column temperature at 25 ℃, and the mobile phase was 0.2 M PBS.
Quartz crystal Microbalance with Dissipation (QCM-D)
Preparation of gold sensor
The adsorption process of different compounds was monitored using a commercial QCM-D device (Q-Sense AB, Sweden), which was equipped with Q-Sense software (Q-Soft, Q-Sense) and a temperature-controlled measuring chamber along with a peristaltic pump. Before measurement, the gold sensor was placed in a UV/ozone chamber for 10 min and immersed in a solution which consists of ammonia (NH4OH, 25 %), hydrogen peroxide (H2O2, 30 %) and ultrapure water mixed at a ratio of 1:1:5 at 75 ℃ for 10 min. Then, the dried gold sensors were placed in the UV/ozone chamber again for another 10 min.
QCM-D experiments
SPI, 7S, 11S globulin, DG, MC, DG-MC (GM) complex was prepared as a series of 2 mg/mL solution, and all samples were ultrasonicated for 20 min to remove bubbles. The cleaned gold chip was installed in the QCM-D module and excited at a fundamental frequency. The experiments were started by pumping Milli-Q water into the QCM-D chamber until a stable baseline was obtained. Subsequently, the protein samples were pumped into the chamber until equilibrium was reached (△F3 < 1 Hz within 5 min). After the layer of protein was rinsed, DG, MC and GM complex were introduced. This was then followed by rinsing gold sensor with Milli-Q water to remove the loosely adsorbed molecules. The Voigt viscoelastic model in Q-Tools software was used to fit and calculate the changes in adsorption mass and thickness. All experiments were performed at 25 ℃, a pump speed of 0.1 mL/min, which were repeated for three times.
Molecular modeling
The docking study was employed to investigate the molecular interaction of SPI, DG and MC. Previously, the crystal structures of soybean 7S protein (PDB:1UIK) and soybean 11S protein (PDB:1FXZ) were downloaded from Protein Data Bank (https://www.rcsb.org/). Then, the 3D structure of small molecule was downloaded from PubChem database according to the CAS number. Pymol2.3.0 was used to remove the protein crystal water and the original ligand, and the optimized structures of protein, polysaccharide and polyphenol were imported into AutoDockTools-1.5.6 for hydrogenation, charge calculation, charge distribution, atomic type designation and storage in “PDBQT” format.
AutoDockVina1.1.2 was used to perform the molecular docking simulations. The relevant parameters of 11S target were set as center_x = 7.019, Center_y = 47.268, center_z = 81.158. 7S target parameters were set as center_x = 103.2, center_y = 11.744, center_z = 72.578. The size of grid boxes was set to 126 × 126 × 126 (the spacing of each grid point is 0.375 Å), and other parameters were set to the default values. Substrate binding affinity was estimated using the docking score.
Data analysis
All experiments were carried out three times and the results were presented as mean ± standard deviation (SD). Origin 9.0 program was used for figure preparation, data processing and statistical analysis.
Results and discussion
Secondary and tertiary structure analysis of SPI and complexes
UV–vis spectrum
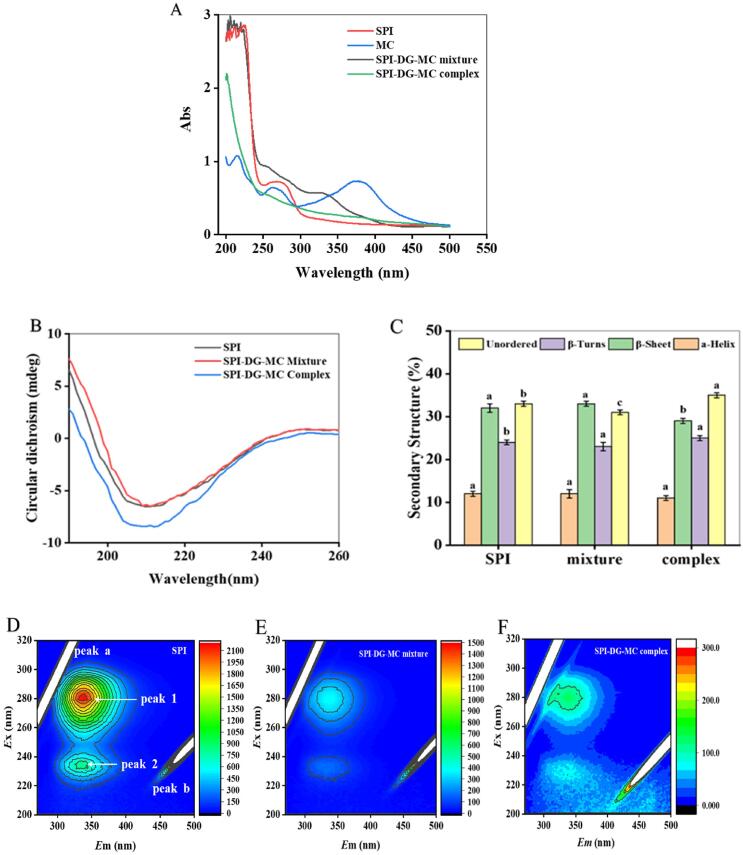
UV–vis analysis is a common technique to examine the aromatic amino acids of protein, which further reflects the interaction between protein and other substances with high sensitivity. Generally, the C—O bond from protein skeleton structure and the amino acid residues of tryptophan (Trp) and tyrosine (Tyr) have typical absorption during 200–215 nm and 260–280 nm, respectively (Chen et al., 2019, Guo et al., 2022). Thus, it can be seen from Fig. 1 A that SPI had a typical absorption around 280 nm, which was mostly coming from the aromatic residues. Moreover, due to the ring B of MC moieties, the absorption peaks at 260 nm and 380 nm were detected in MC and SPI-DG-MC mixture. However, after addition of DG and MC, all of these characteristic absorption of complexes in the UV–vis spectrum disappeared, indicating the successful grafting of polysaccharide and/or polyphenol. It has been proved that the complexation of polyphenol and protein could modify the ultraviolet properties of phenolic compounds, and the microenvironment of aromatic amino acids was changed (Fuguo Liu, Ma, McClements, & Gao, 2017). In addition, the significant absorption of gallic acid at 280 nm also disappeared in the complex conjugated with soluble dietary fiber from lotus root (S. Li, Li, Zhu, Cheng, He, & Lamikanra, 2020). However, Wang et al. (2012) found that the UV absorption value of bovine serum albumin increased gradually with the addition of chitosan, mainly due to the interaction between chitosan and bovine serum albumin (Z. Wang, Zeng, Tu, & Zhao, 2012).
Fig. 1.
UV–vis scanning results of SPI, MC, SPI-DG-MC mixture and complex (A). Far-UV CD spectra of SPI, SPI-DG-MC mixture and complex (B). Secondary structure of SPI, SPI-DG-MC mixture and complex (C). Different letters above each column represent that data are significantly different (p < 0.05). Three-dimensional (3D) fluorescence spectra of SPI (D), SPI-DG-MC mixture (E) SPI-DG-MC complex (F).
Secondary structure analysis
Circular dichroism (CD) is often used to analyze the secondary structure of protein, especially the relative ratio of α-helix, β-sheet and β-turn subunit in the components. In this section, the influence of DG and MC on the secondary structure of SPI was measured, and the quantitative analysis of α-helix, β-sheet, β-turn and random coil were fitted by Yang’s equation. As shown in Fig. 1B, the wide negative peak at 210 nm indicated the presence of α-helix in the secondary structure of SPI, while the ellipticity of this negative peak decreased on account of DG and MC. The random coil and β-sheet were also observed in the structure of SPI, which accounted for 65 % in total (Fig. 1C). However, obvious decrease in β-sheet, significant increase in β-turn and random coil were found because of the complicated combination. It could be speculated that complexation with DG and MC promoted the β-sheet in SPI transform into β-turn and random coil (Cong Ren, Xiong, Li, & Li, 2019). Moreover, the increase of random coil implied that DG and MC made the protein structure of SPI become much more loser. This is consistent with the previous report that the change in protein secondary structure was always followed by covalent and non-covalent interactions between anthocyanins and soy proteins (Ren et al., 2018). Therefore, our results further evidenced the alteration of SPI structure during interaction and suggested the possibility in formation of binary and ternary SPI complexes simultaneously (F. Liu, Wang, Sun, McClements, & Gao, 2016).
Three-dimensional (3D) fluorescence spectroscopy
Three-dimensional fluorescence spectroscopy can acquire the changes in fluorescence intensity information. The conformational changes of SPI after interacted with DG and MC were determined by 3D fluorescence spectroscopy and the results were presented in Fig. 1 D-F. As usual, peak a (λ ex = λ em) and b (λ em = 2λex) are considered as the Rayleigh scattering peak and the second-order scattering peak, respectively (Sui, Sun, Qi, Zhang, Li, & Jiang, 2018). Peak 1 (λ ex = 280 nm, λ em = 340 nm) mainly represents the fluorescence behaviors of tryptophan (Trp) and tyrosine (Tyr) residues of the protein, while peak 2 (λ ex = 230 nm, λ em = 350 nm) represents the characteristic absorption peak of polypeptide chain skeleton structure production (Li et al., 2019). Referring to the figure, the fluorescence intensity of protein decreased after adding DG and MC, as the color of the characteristic peak became lighter with thinner contour lines. Nevertheless, the fluorescence intensity of the SPI-DG-MC complex weakened more obviously compared to the mixture, which revealed a strong interaction among SPI, DG and MC. The decrease in fluorescence intensity of peak 1 may be caused by the burying of hydrophobic groups from Trp and Tyr residues, or the change in the tertiary structure, thus reducing the fluorescence value of protein. The quenching in fluorescence intensity of peak 2 possibly resulted from the extending skeleton of polypeptide chain induced by complexion with DG and MC. This is in accordance with the conclusion of Yan et al (2021) that epigallocatechin gallate (EGCG) decreased the fluorescence intensity of SPI in the presence of chitosan, and the unfolding of polypeptide chain skeleton led to the change of tertiary structure of protein (Yan, Xie, Zhang, Jiang, Qi, & Li, 2021).
FESEM analysis of SPI and complexes
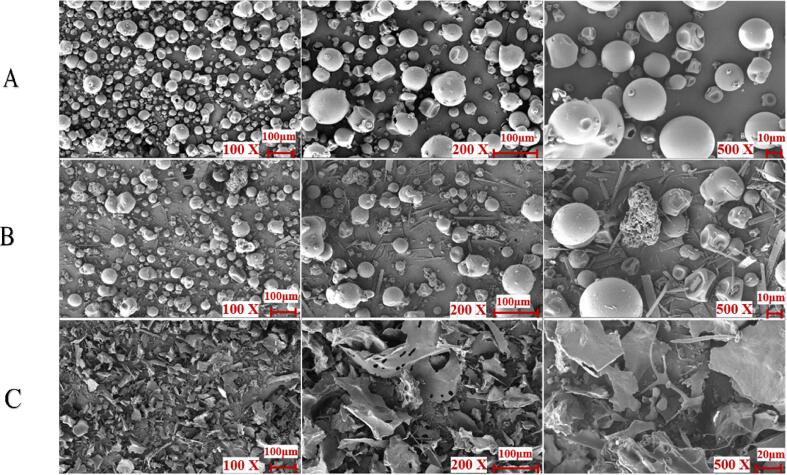
It has been found that anionic polysaccharides could make globular structure of SPI convert to distinct flaky particles by interactions, which formed tighter and smoother structure (Zhao et al., 2020). Herein, field emission scanning electron microscopy (FESEM) was applied to observe the microstructure of SPI with or without participation of DG and MC. As shown in Fig. 2, the microstructures of SPI, DG, and MC mixture were significantly different compared to the complex. According to Fig. 2 A, SPI has a saclike structure with concave holes on its surface and irregular spherical shape (Huang, Ding, Dai, & Ma, 2017). When it comes to the mixture, three varieties of morphologic structures can be detected, in which the irregular spherical shape could be SPI, the block could be DG, and the strip was MC (Fig. 2B). However, the ordered structure of proteins was found to be broken after conjugated and the ternary complexes formed a dense and irregular fragmentary sheet-like structure with uneven surface (Fig. 2C). It has been previously claimed that anionic polysaccharides could make distinct flaky particles of SPI convert to tighter and smooth structure by noncovalent interactions (Zhao et al., 2020). Furthermore, when the display multiple of the complex was enlarged to 600×, loose and porous protein structure can be observed (Wang et al., 2020). That is to say, the complexation with polysaccharide and polyphenol does alter the microstructure of protein, which further certified the hydrogen bond and hydrophobic interaction among SPI, DG and MC (G. Chen, Wang, Feng, Jiang, & Miao, 2019).
Fig. 2.
FESEM pictures of SPI (A), SPI-DG-MC mixture (B), and SPI-DG-MC complex (C).
Molecular weight distribution of different SPI complexes
SDS-PAGE analysis
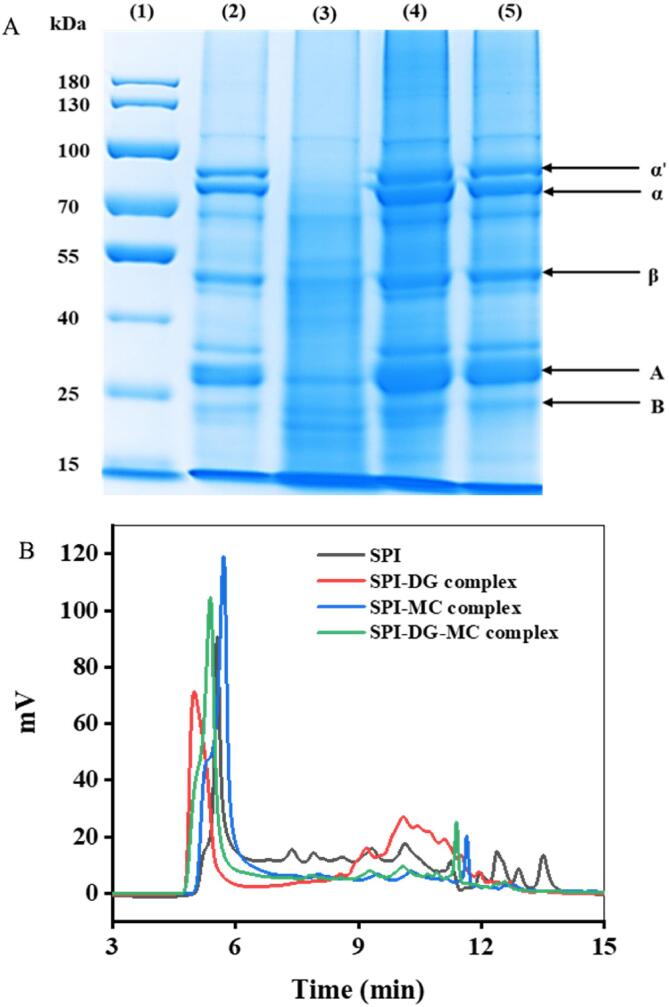
In order to explore the combination among SPI, DG and MC, the molecular weight range of SPI, SPI-DG, SPI-MC and SPI-DG-MC complexes were compared by SDS-PAGE analysis. As shown in Fig. 3A, lane (1) was the marker, while lane (2) represented SPI containing two main protein components, β-conglycinin (7S) and glycinin (11S). 7S (molecular weight 180 ∼ 210 kDa) possesses α (≈71 kDa), α (≈67 kDa) and β (≈50 kDa) subunits, and 11S has an acidic subunit A (≈32 kDa) and basic subunit B (≈20 kDa). For SPI-DG complex (lane (3)), it was found that most of the bands were similar compared to lane (2), indicating that SPI and DG may be bonded by non-covalent, however the bands became blurred, possibly due to the outer layer of SPI molecule wrapped by DG, resulting in the polymerization of a small number of SPI molecules with SDS. It was proved that polysaccharides were covered in the layer of protein molecules, reducing the binding of SPI with SDS, as illustrated by the shallow stains appearing in Lane 3 and 4 (Y. Zhao, et al., 2020). The bands of SPI-MC complex (lane (4)) and SPI-DG-MC complex (lane (5)) had no change compared with lane (2), but it was noteworthy that the bands of molecular weight in lane (4) became wider and darker, which may come from the non-covalent binding of MC and SPI. In addition, lane (5) was lighter in color compared to lane (4), suggesting that DG may be encapsulated in the outer layer of SPI-MC complex through non-covalent interactions.
Fig. 3.
Molecular weight distribution of different SPI complexes. SDS-PAGE profiles of SPI and SPI-DG-MC conjugates (A): Lane (1) molecular weight standard; lane (2) SPI, lane (3) SPI-DG complex, lane (4) SPI- MC complex, and lane (5) SPI-DG-MC complex; GPC of different SPI complexes (B).
GPC profiles
GPC is often used to separate and determine the molecular weights and distribution of polymers (Gaborieau & Castignolles, 2011). In this part, the molecular weight distribution of SPI, SPI-DG, SPI-MC and SPI-DG-MC complexes were further analyzed. As exhibited in Fig. 3B, there was a main fraction with wide range of distribution from 5 to 6 min for SPI, some polymers were eluted around 5.56 min with larger molecular weight. However, the main peak in SPI was postponed in SPI-DG and SPI-MC complexes, manifesting the modification of protein. The content of the second main peak was decreased significantly around 10 min and the first eluted peak was increased, compared to the original SPI. The larger molecular weight of other compounds existed around 5.3 min, suggesting the formation of various complexes with different binding rates. Moreover, the Mw of SPI, SPI-DG, SPI-MC and SPI-DG-MC complex were 6.68 × 105, 1.03 × 106, 9.78 × 105 and 1.17 × 106 g/mol, respectively, further proving the conjugation of polysaccharide and polyphenol with protein. The main fraction of SPI-DG-MC was eluted earlier than SPI-DG and SPI-MC, which indicated that the simultaneous presence of binary and ternary complex during interaction. Similar study was carried out that inulin and green tea polyphenols successful grafting formed complex the Mw was significantly larger and molecular weight distribution changed (Li et al., 2021).
Quartz crystal microbalance with dissipation (QCM-D) analysis
QCM-D technology has been widely used to study the adsorption of DNA and protein macromolecules, as well as the interaction behaviors among biomolecules, polymers and phospholipid membranes. With the real-time monitoring capability, QCM-D was utilized to investigate the interaction of SPI, 7S, 11S with DG, MC and their binary complex (GM) in this study. Fundamentally, the absolute change in frequency is proportional to the change in mass and the dissipation factor is related to material structure. The increase of ΔD means that the energy on the quartz crystal is dissipated rapidly and formed softer or looser of sample film, while the reduction of ΔF illustrates that the sample is adsorbed on the chip, making the sample film on the chip thicker (Q. Chen, Xu, Liu, Masliyah, & Xu, 2016). These changes mostly occur within the first few minutes, indicating that the adsorption of sample is a fast process.
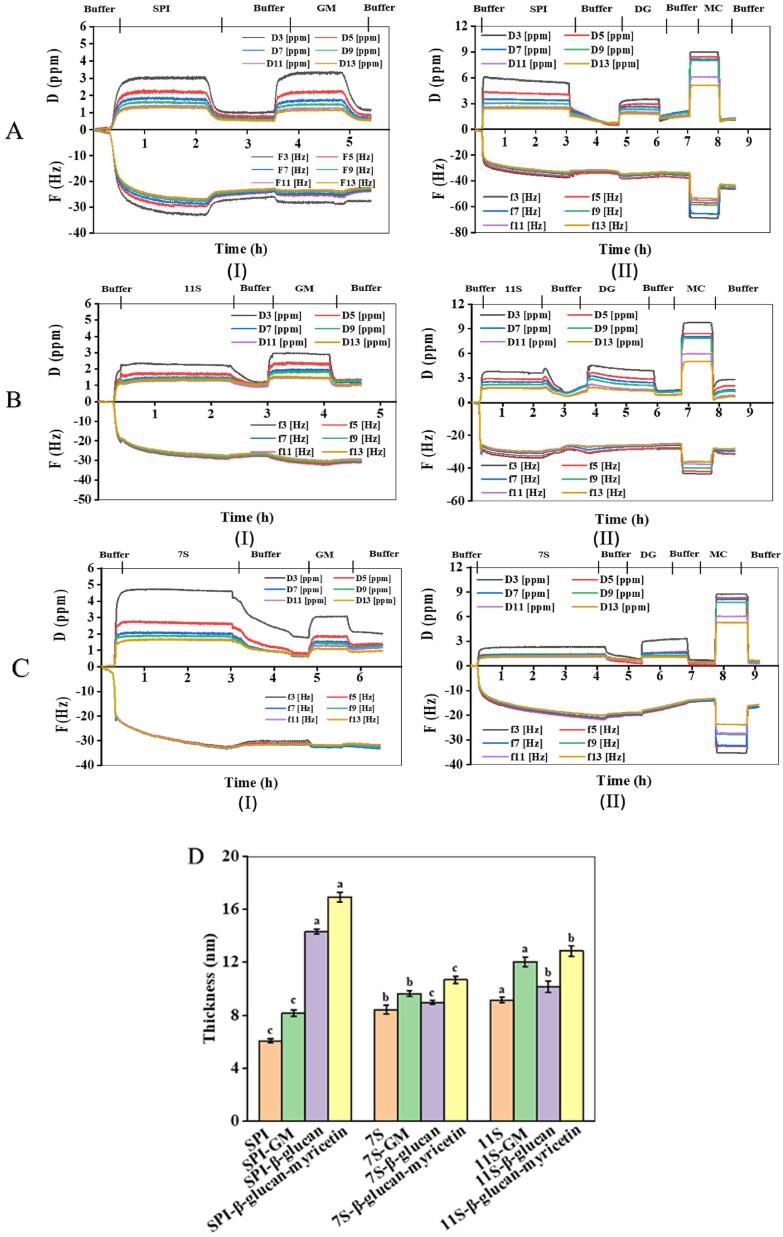
As shown in Fig. 4, the experiment was excited at a series of fundamental frequency (3, 5, 7, 9, 11 times). The measurement was started at approximate 25 min once a stable baseline was achieved. The individual SPI, 7S and 11S solution was introduced until a steady state was obtained, which was followed by rinsing the sensor with a buffer at the same pH value. Afterwards, the GM solution was introduced and a second-steady-state was acquired (Fig. 4 (Ⅰ)). Meanwhile, the DG and MC solutions were successively introduced in Fig. 4 (Ⅱ). The testing was finalized by rinsing the sensor with the original buffer to remove the loosely attached molecules. According to the ΔF value, the adsorption order of GM on proteins was 11S˃SPI˃7S, and the adsorption order of separate DG and MC on proteins was SPI˃11S˃7S, all indicating that 11S was more easily to be conjugated when interacted with other protein components, which was also stated by Cong et al. (2019) that 11S had higher affinity on cyanidin-3-O-glucoside than 7S, on the basis of binding sites and constants (Cong Ren, Xiong, Li, & Li, 2019). Moreover, the total adsorption of DG and MC was much greater than GM binary complex, which was likely that some active sites were occupied in DG when complexed with MC, and the left spatial position was limited to interact with the adsorbed proteins.
Fig. 4.
QCM-D monitoring for testing the selectivity of SPI (A), 11S (B), 7S (C) against on (Ⅰ) GM and (Ⅱ) DG and MC. The thickness of GM, DG and MC adsorbed on SPI, 11S, 7S (D). Different letters above each column represent that data are significantly different (p < 0.05).
According to the examining curves, the surface load was calculated using the Voight mode, and the thickness of SPI, 7S and 11S before and after adsorption of GM, DG and MC was presented in Fig. 4D. It was indicated that the maximum adsorption thickness of GM, separate DG and MC compounds was all found on 11S, suggesting its stronger binding capacity. The one possible reason was that the charge density of 7S was significantly greater than 11S. On the other hand, 11S has a higher surface hydrophobicity than 7S due to higher proportion of hydrophobic and uncharged amino acids which could strengthen the hydrophobic interaction with polysaccharide and polyphenol (Tang, Luo, Liu, & Chen, 2013). To be mentioned, the adsorption of DG and MC on SPI may also depend on their sequence of addition. We assume that the smaller molecules will occupy the free sites more quickly and easily. Hence, the molecular mechanism of interaction during the formation of ternary complex should be further studied by other models and techniques.
Molecular modeling
Up to date, the molecular docking technique has been usually used to simulate the interactions between ligands and biomacromolecules, predicting the binding patterns and affinity (Anighoro & Bajorath, 2016). The above data has confirmed the complicated interactions between SPI, DG and MC. 7S and 11S, the main components of SPI, can both conjugate with these two compounds but revealing different combining abilities. In order to verify this viewpoint, DG and MC were successively docked with SPI (7S and 11S). The optimal binding site and interaction mode were selected according to the maximum binding energy (Dai et al., 2020). The binding energies corresponding to ten binding sites were shown in the Table 1, in which MC was firstly docked with 7S and 11S respectively. Consequently, the SPI-MC complexes with the best docking score (7S-MC: −8.2 kcal/mol, 11S-MC: −8.5 kcal/mol) were chosen for secondary docking with DG, and the best binding site was further selected based on the highest binding energy (Zou, Xu, Zhao, Wang, & Liao, 2019). Finally, 7S-DG-MC (-6.6 kcal/mol) and 11S-DG-MC (-7.9 kcal/mol) were screened as the representatives to analyze the details of the high-affinity binding. Herein, the comparison of binding energies further implied that the interaction force among MC, DG and 11S was stronger than those with 7S protein (Dumitrascu, Stanciuc, Grigore-Gurgu, & Aprodu, 2020).
Table 1.
The binding energy of DG and MC with 11S and 7S protein.
| Type of complex | Binding energy (kcal/mol) | |||||||||
|---|---|---|---|---|---|---|---|---|---|---|
| 11S-MC | −8.5 | −8.1 | −7.8 | −7.5 | −7.3 | −7.0 | −7.0 | −6.9 | −6.8 | −6.8 |
| 11S-MC-DG | −7.9 | −7.4 | −7.3 | −7.2 | −6.6 | −6.3 | −6.3 | −6.3 | −6.2 | −6.1 |
| 7S-MC | −8.2 | −8.1 | −7.7 | −7.5 | −7.5 | −7.3 | −7.3 | −7.1 | −7.1 | −6.8 |
| 7S-MC-DG | −6.6 | −6.5 | −6.5 | −6.4 | −6.2 | −6.2 | −6.1 | −5.9 | −5.9 | −5.9 |
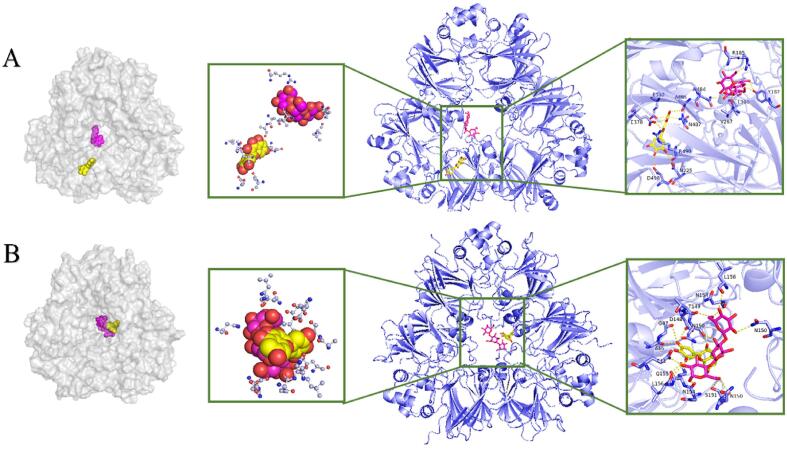
As indicated in Fig. 5A, MC and DG were wrapped in the cavity of SPI (7S, 11S) and conjugated with the surrounding amino acid residues mainly through hydrogen bonds. Concretely, MC interacted with Asp498, Asn225, Arg490, Glu378, Glu382, Ala485 and Asn487 of 7S protein, but bound to Asn150, Asp148, Gly47, Ala46, and Gln155 of 11S protein. The active amino acids involving 7S-DG conjugate were Val267, Leu390, Tyr187, Arg185, Asn484, while Asn150, Thr149, Glu11, Leu156, Asn154, Ser151 were the major contributors that participated to form 11S-DG complex. Particularly, three hydrogen bonds were found between DG and MC, also proving the presence of ternary complex. In other words, SPI, DG and MC were mostly bonded with each other by non-covalent combination, which was consistent with previous studies. It has been demonstrated by Yang et al. (2020) that EGCG changed the microenvironment of tryptophan and tyrosine within 11S and 7S, the interaction forces between them all involved electrostatic interaction, hydrogen bond interaction and hydrophobic interaction, among which hydrogen bond dominated (Yang, Wang, Lei, Li, Zhao, Zhang, et al., 2020).
Fig. 5.
The docking structures of DG and MC with 7S(A) and 11S(B) protein.
Conclusion
Due to complexation with DG and MC, the typical absorption peak of SPI in UV–vis disappeared, the 3D fluorescence intensity decreased with sparse contour lines, and the secondary structure of protein changed, increasing in β-sheet and random coil. Furthermore, the molecular weight of SPI increased from 6.68 × 105 g/mol to 1.17 × 106 g/mol after interaction, showing the successfully preparation of the ternary complex. The 11S component had a higher surface hydrophobicity and binding affinity than 7S, which could absorb DG and MC more easily with higher binding energy, mainly through hydrogen bonds based on QCM-D analysis. And this conclusion was consistent with the molecular simulation results. In a word, these findings will provide guidance to understand the interaction mechanism of protein, polysaccharide and polyphenol in foods, which would promote the application of soybean resource as a functional food ingredient.
CRediT authorship contribution statement
Dan Lei: Conceptualization, Investigation, Software, Writing – original draft. Junsheng Li: Software, Data curation. Chao Zhang: Methodology, Validation. Shuyi Li: Conceptualization, Writing – review & editing. Zhenzhou Zhu: Conceptualization, Funding acquisition, Supervision. Feifei Wang: Data curation. Qianchun Deng: Formal analysis. Nabil Grimi: Supervision.
Declaration of Competing Interest
The authors declare that they have no known competing financial interests or personal relationships that could have appeared to influence the work reported in this paper.
Acknowledgement
This work was financially supported by The National youth talent program in food industry of China and Outstanding young and middle-aged science and technology innovation team in Hubei Province (T2020012).
Contributor Information
Shuyi Li, Email: lishuyisz@sina.com.
Zhenzhou Zhu, Email: zhenzhouzhu@126.com.
References
- Anighoro A., Bajorath J. Three-dimensional similarity in molecular docking: prioritizing ligand poses on the basis of experimental binding modes. Journal of Chemical Information and Modeling. 2016;56(3):580–587. doi: 10.1021/acs.jcim.5b00745. [DOI] [PubMed] [Google Scholar]
- Bai J., Ren Y., Li Y., Fan M., Qian H., Wang L.…Rao Z. Physiological functionalities and mechanisms of β-glucans. Trends in Food Science & Technology. 2019;88:57–66. [Google Scholar]
- Buitimea-Cantua N.E., Gutierrez-Uribe J.A., Serna-Saldivar S.O. Phenolic-protein interactions: Effects on food properties and health benefits. Journal of Medicinal Food. 2018;21(2):188–198. doi: 10.1089/jmf.2017.0057. [DOI] [PubMed] [Google Scholar]
- Chen G., Wang S., Feng B., Jiang B., Miao M. Interaction between soybean protein and tea polyphenols under high pressure. Food Chemistry. 2019;277:632–638. doi: 10.1016/j.foodchem.2018.11.024. [DOI] [PubMed] [Google Scholar]
- Chen Q., Xu S., Liu Q., Masliyah J., Xu Z. QCM-D study of nanoparticle interactions. Advances in Colloid and Interface Science. 2016;233:94–114. doi: 10.1016/j.cis.2015.10.004. [DOI] [PubMed] [Google Scholar]
- Chen Z., Wang C., Gao X., Chen Y., Kumar Santhanam R., Wang C.…Chen H. Interaction characterization of preheated soy protein isolate with cyanidin-3-O-glucoside and their effects on the stability of black soybean seed coat anthocyanins extracts. Food Chemistry. 2019;271:266–273. doi: 10.1016/j.foodchem.2018.07.170. [DOI] [PubMed] [Google Scholar]
- Chethan G.E., Garkhal J., Sircar S., Malik Y.P.S., Mukherjee R., Sahoo N.R.…De U.K. Immunomodulatory potential of beta-glucan as supportive treatment in porcine rotavirus enteritis. Veterinary Immunology and Immunopathology. 2017;191:36–43. doi: 10.1016/j.vetimm.2017.07.012. [DOI] [PubMed] [Google Scholar]
- Dai T., Li T., Li R., Zhou H., Liu C., Chen J., McClements D.J. Utilization of plant-based protein-polyphenol complexes to form and stabilize emulsions: Pea proteins and grape seed proanthocyanidins. Food Chemistry. 2020;329 doi: 10.1016/j.foodchem.2020.127219. [DOI] [PubMed] [Google Scholar]
- de Oliveira F.C., Coimbra J.S., de Oliveira E.B., Zuniga A.D., Rojas E.E. Food Protein-polysaccharide Conjugates Obtained via the Maillard Reaction: A Review. Critical Reviews in Food Science and Nutrition. 2016;56(7):1108–1125. doi: 10.1080/10408398.2012.755669. [DOI] [PubMed] [Google Scholar]
- Djuardi A.U.P., Yuliana N.D., Ogawa M., Akazawa T., Suhartono M.T. Emulsifying properties and antioxidant activity of soy protein isolate conjugated with tea polyphenol extracts. Journal of Food Science and Technology. 2020;57(10):3591–3600. doi: 10.1007/s13197-020-04391-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dumitrascu L., Stanciuc N., Grigore-Gurgu L., Aprodu I. Soluble dietary fiber and polyphenol complex in lotus root: Preparation, interaction and identification. Spectrochimica Acta, Part A: Molecular and Biomolecular Spectroscopy. 2020;231 doi: 10.1016/j.saa.2020.118114. [DOI] [PubMed] [Google Scholar]
- Ekström L.M.N.K., Henningsson Bok E.A.E., Sjöö M.E., Östman E.M. Oat β-glucan containing bread increases the glycaemic profile. Journal of Functional Foods. 2017;32:106–111. [Google Scholar]
- Fernandes P.A.R., Le Bourvellec C., Renard C., Wessel D.F., Cardoso S.M., Coimbra M.A. Interactions of arabinan-rich pectic polysaccharides with polyphenols. Carbohydrate Polymers. 2020;230 doi: 10.1016/j.carbpol.2019.115644. [DOI] [PubMed] [Google Scholar]
- Gaborieau M., Castignolles P. Size-exclusion chromatography (SEC) of branched polymers and polysaccharides. Analytical and Bioanalytical Chemistry. 2011;399(4):1413–1423. doi: 10.1007/s00216-010-4221-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Guo Z., Huang Y., Huang J., Li S., Zhu Z., Deng Q., Cheng S. Formation of protein-anthocyanin complex induced by grape skin extracts interacting with wheat gliadins: Multi-spectroscopy and molecular docking analysis. Food Chemistry. 2022;385 doi: 10.1016/j.foodchem.2022.132702. [DOI] [PubMed] [Google Scholar]
- Huang L., Ding X., Dai C., Ma H. Changes in the structure and dissociation of soybean protein isolate induced by ultrasound-assisted acid pretreatment. Food Chemistry. 2017;232:727–732. doi: 10.1016/j.foodchem.2017.04.077. [DOI] [PubMed] [Google Scholar]
- Jiang M., Zhu M., Wang L., Yu S. Anti-tumor effects and associated molecular mechanisms of myricetin. Biomedicine & Pharmacotherapy. 2019;120 doi: 10.1016/j.biopha.2019.109506. [DOI] [PubMed] [Google Scholar]
- Ju M., Zhu G., Huang G., Shen X., Zhang Y., Jiang L., Sui X. A novel pickering emulsion produced using soy protein-anthocyanin complex nanoparticles. Food Hydrocolloids. 2020;99 [Google Scholar]
- Li J., Li Y., Guo S. The binding mechanism of lecithin to soybean 11S and 7S globulins using fluorescence spectroscopy. Food Science and Biotechnology. 2014;23(6):1785–1791. [Google Scholar]
- Li S., Lei D., Zhu Z., Cai J., Manzoli M., Jicsinszky L.…Cravotto G. Complexation of maltodextrin-based inulin and green tea polyphenols via different ultrasonic pretreatment. Ultrasonics Sonochemistry. 2021;74 doi: 10.1016/j.ultsonch.2021.105568. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li S., Li J., Zhu Z., Cheng S., He J., Lamikanra O. Soluble dietary fiber and polyphenol complex in lotus root: Preparation, interaction and identification. Food Chemistry. 2020;314 doi: 10.1016/j.foodchem.2020.126219. [DOI] [PubMed] [Google Scholar]
- Li S., Xu H., Sui Y., Mei X., Shi J., Cai S.…Barba F.J. Comparing the LC-MS Phenolic Acids Profiles of Seven Different Varieties of Brown Rice (Oryza sativa L.) Foods. 2022;11(11) doi: 10.3390/foods11111552. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li Y., Liu B., Jiang L., Regenstein J.M., Jiang N., Poias V.…Wang Z. Interaction of soybean protein isolate and phosphatidylcholine in nanoemulsions: A fluorescence analysis. Food Hydrocolloids. 2019;87:814–829. [Google Scholar]
- Liu F., Ma C., McClements D.J., Gao Y. A comparative study of covalent and non-covalent interactions between zein and polyphenols in ethanol-water solution. Food Hydrocolloids. 2017;63:625–634. [Google Scholar]
- Liu F., Wang D., Sun C., McClements D.J., Gao Y. Utilization of interfacial engineering to improve physicochemical stability of beta-carotene emulsions: Multilayer coatings formed using protein and protein-polyphenol conjugates. Food Chemistry. 2016;205:129–139. doi: 10.1016/j.foodchem.2016.02.155. [DOI] [PubMed] [Google Scholar]
- Ren C., Xiong W., Li J., Li B. Comparison of binding interactions of cyanidin-3-O-glucoside to β-conglycinin and glycinin using multi-spectroscopic and thermodynamic methods. Food Hydrocolloids. 2019;92:155–162. [Google Scholar]
- Ren C., Xiong W., Peng D., He Y., Zhou P., Li J., Li B. Effects of thermal sterilization on soy protein isolate/polyphenol complexes: Aspects of structure, in vitro digestibility and antioxidant activity. Food Research International. 2018;112:284–290. doi: 10.1016/j.foodres.2018.06.034. [DOI] [PubMed] [Google Scholar]
- Singh A., Meena M., Kumar D., Dubey A.K., Hassan M.I. Structural and functional analysis of various globulin proteins from soy seed. Critical Reviews in Food Science and Nutrition. 2015;55(11):1491–1502. doi: 10.1080/10408398.2012.700340. [DOI] [PubMed] [Google Scholar]
- Song X., Tan L., Wang M., Ren C., Guo C., Yang B.…Pei J. Myricetin: A review of the most recent research. Biomedicine & Pharmacotherapy. 2021;134 doi: 10.1016/j.biopha.2020.111017. [DOI] [PubMed] [Google Scholar]
- Sui X., Sun H., Qi B., Zhang M., Li Y., Jiang L. Functional and conformational changes to soy proteins accompanying anthocyanins: Focus on covalent and non-covalent interactions. Food Chemistry. 2018;245:871–878. doi: 10.1016/j.foodchem.2017.11.090. [DOI] [PubMed] [Google Scholar]
- Tang C.-H., Luo L.-J., Liu F., Chen Z. Transglutaminase-set soy globulin-stabilized emulsion gels: Influence of soy β-conglycinin/glycinin ratio on properties, microstructure and gelling mechanism. Food Research International. 2013;51(2):804–812. [Google Scholar]
- Wang L., Wu H., Yang F., Dong W. The protective effects of myricetin against cardiovascular disease. Journal of Nutritional Science and Vitaminology. 2019 doi: 10.3177/jnsv.65.470. [DOI] [PubMed] [Google Scholar]
- Wang S., Yang J., Shao G., Liu J., Wang J., Yang L.…Jiang L. pH-induced conformational changes and interfacial dilatational rheology of soy protein isolated/soy hull polysaccharide complex and its effects on emulsion stabilization. Food Hydrocolloids. 2020;109 [Google Scholar]
- Wang Z., Zeng R., Tu M., Zhao J. A novel biomimetic chitosan-based nanocarrier with suppression of the protein-nanocarrier interactions. Materials Letters. 2012;77:38–40. [Google Scholar]
- Wu C., Hua Y., Chen Y., Kong X., Zhang C. Effect of 7S/11S ratio on the network structure of heat-induced soy protein gels: A study of probe release. RSC Advances. 2016;6(104):101981–101987. [Google Scholar]
- Wu Z., Li H., Ming J., Zhao G. Optimization of adsorption of tea polyphenols into oat beta-glucan using response surface methodology. Journal of Agriculture and Food Chemistry. 2011;59(1):378–385. doi: 10.1021/jf103003q. [DOI] [PubMed] [Google Scholar]
- Yan, S., Xie, F., Zhang, S., Jiang, L., Qi, B., & Li, Y. (2021). Effects of soybean protein isolate − polyphenol conjugate formation on the protein structure and emulsifying properties: Protein − polyphenol emulsification performance in the presence of chitosan. Colloids and Surfaces A: Physicochemical and Engineering Aspects, 609.
- Yang Y., Wang Q., Lei L., Li F., Zhao J., Zhang Y.…Ming J. Molecular interaction of soybean glycinin and β-conglycinin with (−)-epigallocatechin gallate induced by pH changes. Food Hydrocolloids. 2020;108 [Google Scholar]
- Zhao C., Yin H., Yan J., Niu X., Qi B., Liu J. Structure and acid-induced gelation properties of soy protein isolate–maltodextrin glycation conjugates with ultrasonic pretreatment. Food Hydrocolloids. 2021;112 [Google Scholar]
- Zhao Y., Wang X., Li D., Tang H., Yu D., Wang L., Jiang L. Effect of anionic polysaccharides on conformational changes and antioxidant properties of protein-polyphenol binary covalently-linked complexes. Process Biochemistry. 2020;89:89–97. [Google Scholar]
- Zou H., Xu Z., Zhao L., Wang Y., Liao X. Effects of high pressure processing on the interaction of alpha-lactalbumin and pelargonidin-3-glucoside. Food Chemistry. 2019;285:22–30. doi: 10.1016/j.foodchem.2019.01.129. [DOI] [PubMed] [Google Scholar]