Highlights
-
•
A systematic strategy for detection and annotation of carboxyl compound was developed.
-
•
197 carboxyl compounds were detected in Chinese Baijiu for the first time.
-
•
Annotation was based on MS1, tR, in-silico MS/MS, and characteristic fragments.
-
•
Three of carboxyl compounds were newly identified in Chinese Baijiu.
-
•
Distribution of carboxyl compounds in Baijiu with different flavors was revealed.
Keywords: High-coverage analysis, Carboxyl compounds, Chinese Baijiu, Chemical derivatization, High-resolution mass spectrometry
Abstract
Carboxyl compounds have a significant influence on the flavor of Chinese Baijiu. However, because of the structural diversity and low concentration, the deep profiling of carboxyl compounds in Chinese Baijiu is still challenging. In this work, a systematic method for comprehensive analysis of carboxyl compounds in Chinese Baijiu was established. After derivatized under optimized conditions, 197 p-dimethylaminophenacyl bromide-derived carboxylic compounds were annotated by multidimensional information including accurate mass, predicted tR, in-silico MS/MS, and diagnostic ions for the first time. In addition, 48 of the 197 carboxyl compounds were positively identified, and three of them were newly identified in Chinese Baijiu. Moreover, we found the number and the concentration of carboxyl compounds in sauce-flavor Baijiu were more abundant than in strong-flavor Baijiu. This work provides a novel method for the analysis of carboxyl compounds in Baijiu and other complex samples.
1. Introduction
Chinese Baijiu has a long history and a good market in many countries for its pleasing flavors. According to the differences in raw materials, production processes, and storage conditions, there are twelve flavors of Chinese Baijiu (Duan et al., 2022, Son et al., 2018), among them sauce-flavor and strong-flavor are the most famous. Carboxyl compounds are widely found in Chinese Baijiu and have a great influence on the flavor of Chinese Baijiu (Yang, Fan, & Xu, 2017). Therefore, high-coverage detection and annotation of carboxyl compounds in Chinese Baijiu would be helpful to reveal their flavors and improve the quality.
Gas chromatography-mass spectrometry (GC–MS) and Liquid chromatography-mass spectrometry (LC-MS) have been used to detect carboxyl compounds in Chinese Baijiu (He et al., 2021, Liao & Li, 2013, Liao & Zhou, 2019). However, GC–MS is suitable to detect non-polar and semi-polar compounds (Baroudi, Al-Alam, Chimjarn, Delhomme, Fajloun, & Millet, 2020), GC–MS with derivatization and LC-MS are specifically targeted at polar and semi-polar compounds (Proestos et al., 2006, Sharma et al., 2018), meanwhile, carboxyl compounds in Chinese Baijiu are polarity diversity and exist in a low concentration, leading to a poor-coverage detection of carboxyl compounds in Chinese Baijiu (Jia et al., 2020, Son et al., 2018). Chemical derivatization is capable of improving the overall analytical performance of LC-MS (Zhao & Li, 2020). Therefore, various chemical derivatization methods have been developed to enhance the analytical performance of carboxyl compounds in LC-MS analysis. For example, p-dimethylaminophenacyl bromide (DmPABr) was used to profile 2286 potential carboxyl compounds in yeast (Luo, Zhao, Huan, Sun, Friis, Schultz, et al., 2016). With the assistance of 2-Dimethylaminoethyl-amine, 269 carboxyl compounds in plasma were detected (He et al., 2019). After being labeled by N-methylpheny-lethylamine, 403 potential carboxyl compounds in HepG2 cells were detected (Zheng, Gong, Zheng, Zhang, & Feng, 2020). Among all of the derivatization reagents, DmPABr has been proven to reveal good performance in tuning the retention on reversed-phase liquid chromatography, improving detection sensitivity, and facilitating the identification of carboxyl compounds (Guo & Li, 2010). Therefore, with the help of DmPABr, high-coverage detection of carboxyl compounds in Chinese Baijiu is feasible.
Currently, there are still some difficulties in conducting chemical derivatization for comprehensive annotation of carboxyl compounds in complex samples. On one hand, for the annotation of carboxyl compounds, most derivatization methods applied the chemical isotope labeling strategy, in which labeled compounds were extracted by peak pairs obtained from light and heavy isotope derivatization reagents (An et al., 2021, Li et al., 2019, Xiong et al., 2021). However, heavy isotope derivatization reagents are expensive and even commercially unavailable, which limits the application of the chemical isotope labeling strategy. On the other hand, the retention times (tR) and MS/MS spectra of derivatives are different from those of unlabeled compounds, which increases the difficulty of annotating carboxyl compounds. Some efforts have been made to solve the above problems. For example, to achieve high-coverage identification of carboxyl compounds without heavy isotope-labeled derivatization reagents, three characteristic fragment ions from the derivatization group were hunted, and 1054 carboxylic acids were defined after derivatized by 5-(diisopropylamino) amylamine (Bian et al., 2020). The predicted tR was also used for the identification of carboxyl-containing compounds in plasma and yeast (Zhao, Li, Han, Chan, & Li, 2019). In addition, in-silico predicted MS/MS spectra were applied to the identification of carboxyl exposure biomarkers in urine samples (Jia et al., 2019). Moreover, characteristic fragments were put forward to testify the candidates, such as the neutral loss of NH3 obtained from amino acids, the neutral loss of H2O obtained from hydroxyl, as well as a serious difference of 14 Da from fatty acids (Bian et al., 2017, Wei et al., 2020, Zheng et al., 2020). Although the methods mentioned above were useful for the annotation of carboxyl compounds, we believed that the integrated application of these methods is more efficient.
In the present study, chemical derivatization and ultrahigh-performance liquid chromatography coupled to high-resolution mass spectrometry (UHPLC−HRMS) were used for the high-coverage analysis of carboxyl compounds in different flavors of Chinese Baijiu. The Chinese Baijiu samples were labeled by DmPABr under optimized conditions. Characteristic fragments related to derivatization were utilized to define compounds with the carboxyl acid group. Subsequently, the carboxyl compounds were annotated with the assistance of the accurate mass of precursor ion, predicted tR, in-silico MS/MS, as well as multiple fragments from the characteristic structure. Finally, the difference of carboxyl compounds in 6 commercial Chinese Baijiu with different flavors was explored.
2. Experiment section
2.1. Chemicals and reagents
HPLC-grade ethanol, methanol, and acetonitrile (ACN) were purchased from Merck (Darmstadt, Germany), formic acid of analytical grade was purchased from J&K Scientific ltd. (Beijing, China). Ultra-pure water was obtained from a Milli-Q system (Millipore, Billerica, MA, USA). DmPABr and triethylamine (TEA) were purchased from Aladdin (Shanghai, China). Sodium chloride, hydrogen chloride, and ethyl acetate were purchased from Macklin (Shanghai, China). All standards were purchased from Sigma-Aldrich (St Louis, MO, USA).
2.2. Sample preparation
The carboxyl standards (Table S1) were dissolved in methanol and ultra-pure water with appropriate ratios, then stored at −20 °C. Chinese Baijiu (S1, S2, S3, S4, S5, and S6) were purchased from the local market, the detailed information of samples was shown in Table S2. The blank sample was ethanol–water (53:47, v/v). Pooled Baijiu sample was prepared by mixing equal volumes of Baijiu samples. The 50 mL of Chinese Baijiu or blank sample were evaporated by a vacuum distillation system (70 rpm/min, 45 °C, and −0.1 MPa) till the volume was concentrated to 2 mL, then the concentrated samples were stored at −20 °C before derivatization.
The chemical derivatization reaction was performed as previously described with some modifications (Zhao, Li, Han, Chan, & Li, 2019). Briefly, 30 μL of samples or blank samples were mixed with 2 μL of mixed internal standards (IS, myristic acid-d27, fumaric acid-d2, succinic acid-d4, cholic acid-d4, chenodeoxycholic acid-d4, palmitic acid-d31, and 2-ketoglutaric acid-d6). Subsequently, 7.5 μL of hydrogen chloride (6 M), 15 μL of saturated sodium chloride, and 150 μL of ethyl acetate were added and shaken for 5 min, after centrifugation, the organic phase was taken out, 30 μL of TEA (180 mg/mL in ACN) was added, then the solution was freeze-dried. After that, 64 µL of TEA solution (20 mg/mL in ACN) was used to re-dissolve the residues, and 40 µL of newly prepared DmPABr (10 mg/mL in ACN) was added for 100 min derivatization at 76 °C. Finally, the reaction solution was centrifuged and 80 μL of the supernatant was transferred into a vial for UHPLC−HRMS analysis.
2.3. UHPLC−HRMS analysis
The chromatographic separation was performed on a UHPLC system (Waters, Milford, MA, USA), with a BEH C8 column (2.1 mm × 100 mm, 1.7 μm, Waters, Milford, USA) at 50 °C. Mobile phase A and mobile phase B consisted of 0.1% (v/v) formic acid in water and 0.1% (v/v) formic acid in ACN, respectively. The flow rate was 0.35 mL/min. A gradient started with 5% B and held for 1 min, then increased to 100% B at 23 min and maintained for 4 min. Lastly, the gradient was back to 5% B in 0.1 min and held for 1.9 min. The volume for injection was 1 µL and the sample manager temperature was 4 °C.
The MS data acquisition was carried out on a Q Exactive HF (Thermo Fisher Scientific, Rockford, IL, USA) system. All spectra were obtained in positive ion mode, the resolutions were 120,000 (full width at half-maximum, FWHM) in full scan mode and 30,000 in data-dependent MS/MS (ddMS2) mode. The automatic gain control target of the full scan was 3 × 10 6 and the maximum injection time of the full scan was set as 100 ms. For ddMS2, the automatic gain control target and maximum injection time were 1 × 105 and 50 ms, respectively. Normalized collision energy (NCE) was set as 15%, 30%, and 45%. The top N was 10, spray voltage was 3.5 kV. The aux gas heater temperature and capillary temperatures were 350 °C and 300 °C, respectively. Aux gas flow rate was 10 arbitrary units and sheath gas was 45 arbitrary units. The S-lens RF level was 50 arbitrary units.
2.4. Establishment of quantitative structure−retention relationship (QSRR) model
To establish a model for predicting tR, we measured tR of 100 authentic carboxyl standards with various structures. Meanwhile, simplified molecular input line entry specification (SMILES) of 100 carboxyl standards were generated (Table S1). Then, 75 tR and SMILES were imported to QSRR_Automator software, and the software transformed SMILES into molecular descriptors (Naylor et al., 2020), the best machine learning model and valid molecular descriptors were chosen automatically for establishing the QSRR model. Subsequently, to evaluate the performance of the established model, SMILES of the remaining 25 authentic standards were imported, and the established model was used to predict their tR. Finally, the predicted tR and actual tR were fitted with a linear model.
2.5. Response surface methodology for optimizing derivatization conditions
To detect as many carboxyl compounds as possible, a 3-factorial central composite design was carried out with the pooled Baijiu samples. A total of 20 experiments (Table S3) were designed in a randomized order. The range of each factor was set based on previous studies and operation limits (Luo et al., 2016, Peng and Li, 2013, Zhao et al., 2019). In detail, the reaction time was in the range of 42 to 100 min, the reaction temperature was in the range of 40 to 85 °C, and the volume of DmPABr was in the range of 10 to 33 µL.α = 1.6818. The number of potential carboxyl compounds was used to evaluate the efficiency of derivatization. The process order was a quadratic model, and the optimal reaction condition was predicted by Design Expert 12 software (Stat-Ease Inc, Minneapolis, USA).
2.6. Qualitative analysis
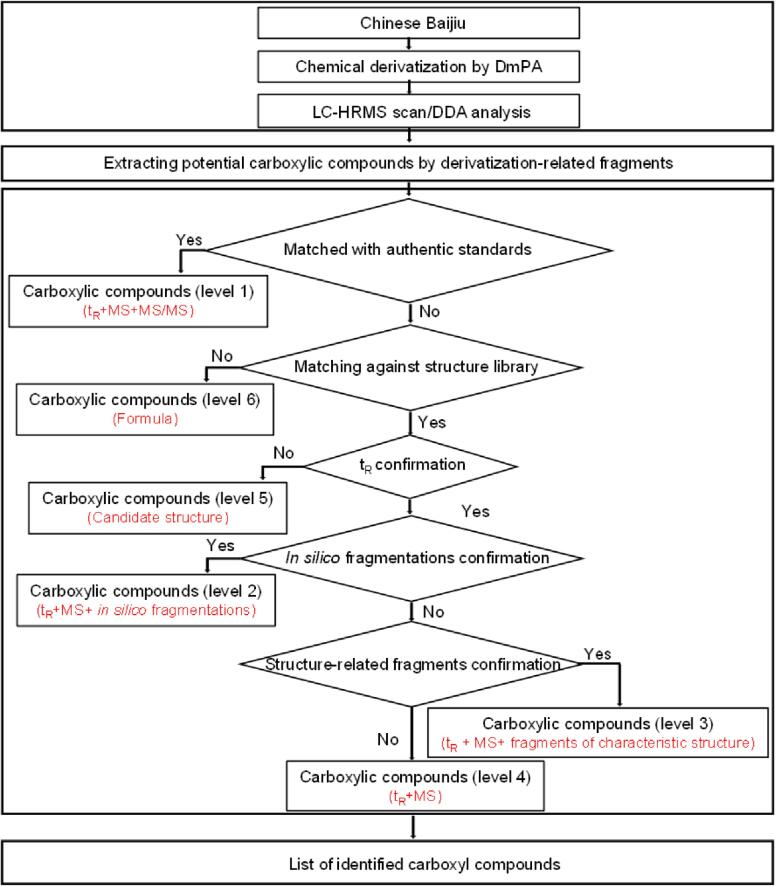
A workflow for the annotation of carboxyl compounds in Chinese Baijiu is shown in Fig. 1. Firstly, UHPLC-full scan MS/ddMS2 acquisition was used to analyze labeled Chinese Baijiu samples. Following that, in-house python programming (https://github.com/Xxy0903/annotation-of-carboxyl-compounds) was used to pick off the potential carboxyl compounds by searching characteristic fragment ions of m/z 180.1020, 134.0962, and [M+H-179.0946] from MS/MS spectra. Then, the potential carboxyl compounds were unambiguously identified by matching the tR, precursor ions and MS/MS with authentic standards (level 1). After that, with the assistance of in-house python programming (https://github.com/Xxy0903/annotation-of-carboxyl-compounds), accurate m/z of precursor ions of potential carboxyl compounds were used to search against structure library with a mass tolerance of 10 ppm. For precursor ions without candidates in the library, their formulas were calculated and set as level 6. On the contrary, for the precursor ions with candidates, a QSRR mode was used to predict the tR of candidate structures, if the difference of tR between the candidate structures and corresponding carboxyl compounds was greater than 50 s, they were classified as level 5. Otherwise, CFM-ID was used to predict the MS/MS spectra of candidate structures (Wang, Liigand, Tian, Arndt, Greiner, & Wishart, 2021). Then, the in-silico MS/MS spectra were matched against actual MS/MS spectra, and a reverse dot product algorithm was used to evaluate the similarity between in-silico and actual MS/MS spectra. If the score was greater than 0.5 (Li et al., 2021), they were set as level 2. In the condition that the score was less than 0.5, characteristic fragments from characteristic structures were used to verify the candidate structures. If the characteristic fragments matched with candidate structures, they were defined as level 3 otherwise set as level 4.
Fig. 1.
Strategy for the annotation of carboxyl compounds in Chinese Baijiu.
2.7. Data processing and statistical analysis
Thermo Xcalibur software (version 2.2, Thermo Fisher Scientific, Rockford, IL, USA) was used to collect raw data and calculate the formula of precursor ions (mass tolerance of 5 ppm). The peak list including average tR, average m/z, peak area, and MS/MS spectra was generated by MS-DIAL software (version 4.24), the detailed parameters are listed in Table S4. An in-house Python programming was applied to pick out the characteristic fragments related to derivatization (mass tolerance of 5 ppm) and calculate the relative intensity of characteristic fragments. An In-house python programming was used to search various databases including compounds obtained from published literature (In-house database), the Yeast Metabolome Database (YMDB), and Collective Molecular Activities of Useful Plants (CMAUP), and fatty acids from Lipids Maps (LM-FA). To calculate the formula of precursor ions, the fundamental elements were set as C, H, O, N, and S. The formula of carboxyl compounds was obtained by removing the DmPABr residue (C10H11NO).
Partial least squares-discriminant analysis (PLS-DA) was performed with SIMCA-P 13 software (Umetrics, Umea, Sweden). The student’s t-test was conducted with IBM SPSS 25.0 software (International Business Machines Corporation, New York).
3. Results and discussion
3.1. Fragmentations of DmPABr-Labeled carboxyl compounds
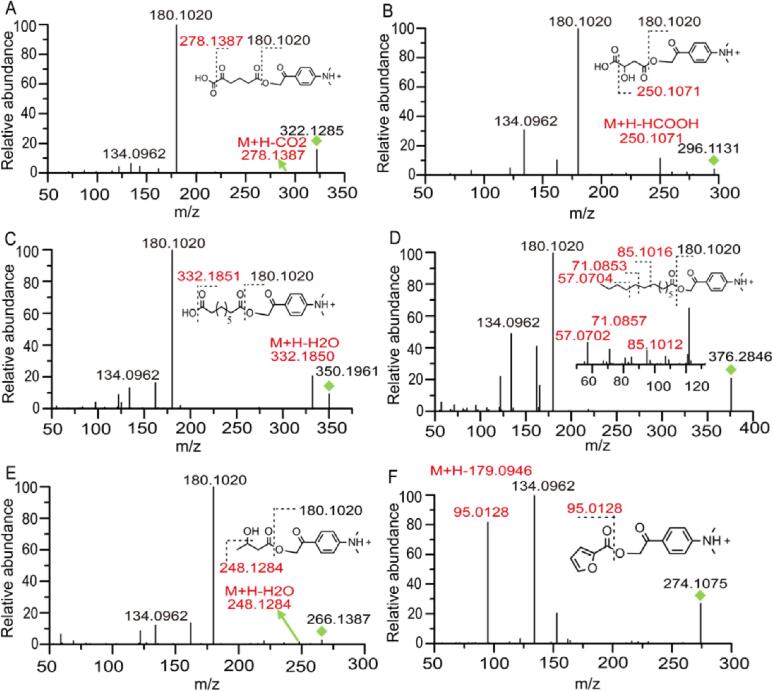
Firstly, the MS/MS spectra of 100 authentic standards after derivatization (Table S1) were analyzed. We found that fragments of m/z 180.1020 and 134.0962 appeared in those carboxyl compounds frequently (Fig. 2). However, among the 100 compounds, not all of them produced the above two fragments at the same time. A previous study indicated that the identification based on only one fragmentation will lead to serious false-positive results (Bian et al., 2020). Therefore, the fragments after the neutral loss of 179.0946 Da were added for defining carboxyl compounds. According to that, we suggested the precursor ions generating two of the three fragments mentioned above were potential carboxyl compounds.
Fig. 2.
Characteristic MS/MS fragmentation of DmPABr–labeled carboxylic acids (A) DmPABr–labeled 2-oxohexanedioic acid, (B) DmPABr–labeled 2-hydroxybutanedioic acid, (C) DmPABr–labeled azelaic acid, (D) DmPABr–labeled tridecanoic acid, (E) DmPABr–labeled 3-Hydroxybutanoic acid. (F) DmPABr–labeled furan-2-carboxylic acid. Red shows characteristic fragmentations from the characteristic structure. (For interpretation of the references to colour in this figure legend, the reader is referred to the web version of this article.)
As shown in Fig. 2A–C, after derivatized, multi-carboxylic acids including 2-oxohexanedioic acid, 2-hydroxybutanedioic acid, and azelaic acid produced the character fragment ions of m/z 278.1387, 250.1071, and 332.1850, which are assigned to the fragments after the neutral loss of 43.9898 Da (CO2), 46.0054 Da (HCOOH), and 18.0106 Da (H2O), respectively. Similarly, tridecanoic acid is representative of fatty acid. According to MS/MS spectra (Fig. 2D), a series of ions with a difference of 14.0156 Da were produced from the loss of methylene. In addition, 3-hydroxybutanoic acid is one of the compounds with carboxyl and hydroxyl, the fragment ion peak at m/z 248.1284 represented the neutral loss of H2O (Fig. 2E). Interestingly, we found that the carboxyl compounds containing carbon–carbon double bond at the alpha position of their carboxyl groups tended to generate a neutral loss of 179.0946 Da and showed a more abundant response than other carboxyl compounds (Fig. 2F, Fig. S1). This can be explained as, after the neutral loss of 179.0946 Da, the formed carbocations will conjugate with the carbon–carbon double bond, making the carbocations more stable (Shirakawa, 2001). It can be seen from Fig. S1 that when the relative abundance of the fragments with neutral loss of 179.0946 Da reached 50%, the corresponding structures of the precursor ions should have a carbon–carbon double bond at the alpha position of the carboxyl group.
3.2. QSRR model for tR prediction
After 75 SMILES of carboxyl compounds were imported into QSRR_Automator software, a series of automatic optimization was performed. Finally, 11 features and linear regression were chosen to form a QSRR model. To validate the performance of this model, external validation was performed. As shown in Fig. S2, the value of R2 was 0.97 and the average absolute error (AAE) between the predicted tR and the actual tR was 42 s, suggesting the good performance of the model in predicting tR.
3.3. Optimization of derivatization conditions for the pooled Baijiu sample
The main factors of the derivatization conditions including reaction time, reaction temperature, and the volume of DmPABr were optimized. Table S3 presented the experimental conditions and the number of potential carboxyl compounds. As shown in Table S3, the numbers of potential carboxyl compounds ranged from 152 to 438, which indicated that the optimized experimental conditions had a significant influence on derivatization reaction. Fisher’s F-test was used to evaluate the model, the detailed information of the result is shown in Table S5. The coefficient of variation (CV) was lower than 10% and the “Adeq Precision” was greater than 4, which suggested the good reproducibility and precision of the model (Ghaedi et al., 2019, Xie et al., 2020). In addition, the difference between the predicted R2 (0.8925) and adjusted R2 (0.9604) was less than 0.2 indicating the good accuracy of the model (Su, Vera, & Nerín, 2020). Lastly, the p-value was less than 0.0001 and the lack of fit was not significant (p > 0.05), demonstrating that the model was suitable to clarify the relationship between independent variables and dependent variable (Pinto et al., 2021).
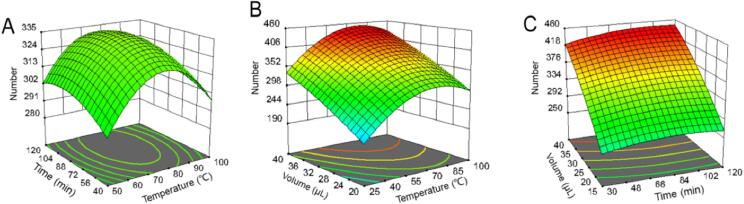
To present the relationship between experimental conditions and the number of potential carboxyl compounds, 3D surface plots were generated. It can be observed from Fig. 3 that the critical factors were the reaction temperature and volume of DmPABr, while the reaction time had a slight influence on the reaction efficiency. In addition, the interaction of reaction temperature and reaction time had significant effects on reaction efficiency, while the interaction of the volume of DmPABr and reaction time or the interaction of the volume of DmPABr and reaction temperature had insignificant effects on reaction efficiency. As shown in Fig. 3A, in the initial stage the increase of the temperature and time led to an increase in the responses, until the temperature was around 76 °C and time around 100 min, respectively, after that, the responses decreased. Finally, based on the signature model and 3D contour, the optimized experimental conditions were defined as follows: reaction time: 100 min, reaction temperature: 76 °C, volume of DmPABr: 40 μL.
Fig. 3.
Response surface 3D plots for the interaction effects of independent variables on the dependent variable. (A) The interaction effects of time and temperature (Temp) on the number of potential carboxyl compounds, (B) the interaction effects of temperature and volume on the number of potential carboxyl compounds, (C) the interaction effects of time and volume on the number of potential carboxyl compounds.
3.4. Chemical profiling of carboxyl compounds in pooled Baijiu sample
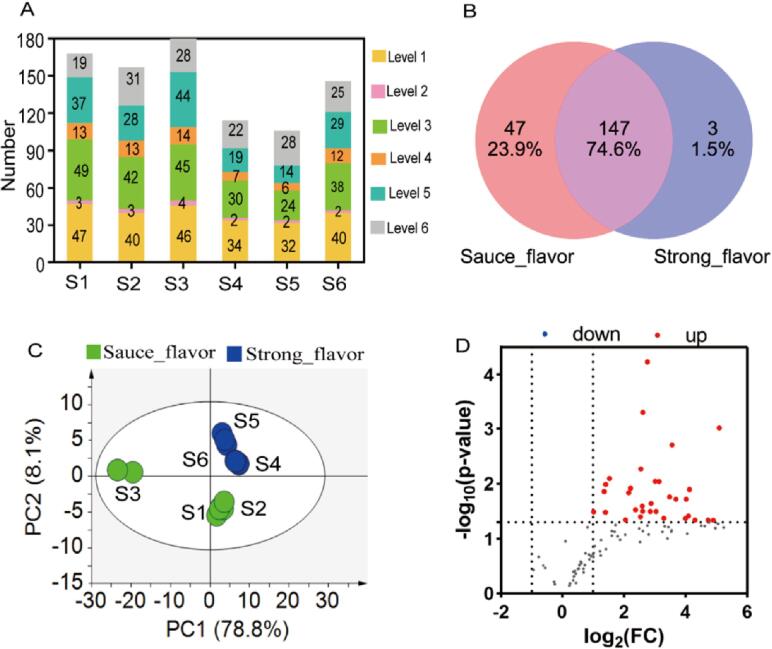
We analyzed carboxyl compounds in pooled Baijiu sample. As shown in Table S6, a total of 197 carboxyl compounds were detected in 6 Baijiu samples, more than double the carboxyl compounds in Chinese Baijiu detected by the most comprehensive method (Naylor et al., 2020) to date. With the annotation strategy, 48 (Nos. 1–48), 5 (Nos. 49–53), 51 (Nos. 54–104), 14 (Nos. 105–118), 44 (Nos. 119–162), and 35 (Nos. 163–197) were at level 1, level 2, level 3, level 4, level 5, and level 6 respectively. In addition, of the 197 carboxyl compounds, 162 (82.2%) carboxyl compounds were positively identified or putatively identified, and higher annotation coverage was achieved than using other existing methods ( Zheng, Gong, Zheng, Zhang, & Feng, 2020). Besides that, by searching the databases (PubChem and The Good Scents Company Information System), 31 of the 48 compounds in level 1 were found to be flavor compounds or related to flavor (Table S6). Moreover, 3-hydroxyoctanoic acid, 2-phenylbutanoic acid, and 5-hydroxyisovanillic acid were identified in Baijiu for the first time.
It is not surprising that some peaks were matched to several candidates in the annotation. As mentioned in Section 2.6, the candidates were obtained from searching against structure libraries by precursor ions, then predicted tR, in-silico MS/MS and diagnostic ions were used to narrow down the number of possible candidate structures. However, even with one of the top tools (CFM-ID 4.0) for predicting MS/MS, 100 % accuracy is impossible (Wang, Liigand, Tian, Arndt, Greiner, & Wishart, 2021), and a similar situation existed in predicting tR (Naylor et al., 2020). In addition, not all carboxyl compounds possess diagnostic ions. Therefore, it is possible for a peak to match several candidates using this annotation strategy. Although several candidates occurred for some peaks, it still provides an opportunity to get the right structure.
3.5. Comparison of carboxyl compounds in different flavors of Chinese Baijiu
To explore the differences of carboxyl compounds in different flavors of commercial Chinese Baijiu, we analyzed carboxyl compounds in three sauce-flavor Chinese Baijiu (S1, S2, and S3) and three strong-flavor Chinese Baijiu (S4, S5, and S6), respectively. It can be seen from Fig. 4A, the numbers of carboxyl compounds detected were 168, 157, 181, 114, 106, and 146 in samples S1, S2, S3, S4, S5, and S6, respectively. Different samples showed different numbers of carboxyl compounds. As shown in Fig. 4B, the number of carboxyl compounds in sauce-flavor samples was more than in strong-flavor samples, with 47 carboxyl compounds being unique in sauce-flavor Baijiu. In addition, as shown in Fig. 4C, sauce-flavor and strong-flavor Chinese Baijiu were well separated with 86.9% of the total variance, this result suggested that there were great differences in the carboxylic acids between sauce-flavor and strong-flavor Chinese Baijiu. As shown in Fig. 4D, the relative contents of 32 carboxyl compounds were significantly changed (p < 0.05 and fold change >2.0), and all of them were higher in sauce-flavor Chinese Baijiu than in strong-flavor Chinese Baijiu, which was in line with the result that the total concentrations of the non-volatile organic acids in sauce and sesame flavor Baijiu were more than in other types (Wang et al., 2022). The more number and higher contents of carboxyl compounds in sauce-flavor Chinese Baijiu may be related to the higher temperature of fermentation in sauce-flavor Chinese Baijiu than in strong-flavor Chinese Baijiu (Kim et al., 2009, Zheng et al., 2011).
Fig. 4.
Carboxyl compounds in Chinese Baijiu from different flavors. (A) The number of identified carboxyl compounds in different Chinese Baijiu, (B) Venn diagram of carboxyl compounds numbers, (C) PLS-DA score plot based on all detected carboxyl compounds, (D) Volcano plot of carboxyl compounds in sauce-flavor Chinese Baijiu and strong-flavor Chinese Baijiu.
4. Conclusions
In this study, we developed a comprehensive method for the detection and annotation of carboxyl compounds in Chinese Baijiu. A total of 197 carboxyl compounds were detected in 6 Chinese Baijiu, and 48 of them were positively identified. 3-hydroxyoctanoic acid, 2-phenylbutanoic acid, and 5-hydroxyisovanillic acid were identified in Baijiu for the first time. In addition, we found the number and content of carboxyl compounds in sauce-flavor Chinese Baijiu were more abundant than in strong-flavor Chinese Baijiu. The developed method is suitable for the high-coverage annotation of carboxyl compounds in various Chinese Baijiu.
CRediT authorship contribution statement
Xiaoyu Xie: Conceptualization, Methodology, Validation, Formal analysis, Investigation, Writing – original draft, Visualization. Xin Lu: Conceptualization, Writing – review & editing. Xiuqiong Zhang: Methodology, Writing – review & editing. Fujian Zheng: Software, Visualization. Di Yu: Visualization. Chao Li: Software. Sijia Zheng: Investigation. Bo Chen: Writing – review & editing. Xinyu Liu: Writing – review & editing. Ming Ma: Writing – review & editing, Supervision. Guowang Xu: Conceptualization, Methodology, Validation, Formal analysis, Resources, Writing – review & editing, Supervision, Funding acquisition.
Declaration of Competing Interest
The authors declare that they have no known competing financial interests or personal relationships that could have appeared to influence the work reported in this paper.
Acknowledgements
This work was supported by the Key Foundation (21934006) from the National Natural Science Foundation of China, the foundation from the Youth Innovation Promotion Association CAS (2021186), the Innovation program (DICP&QIBEBT UN201806, DICP I201918) of Science and Research from the DICP.
Footnotes
Supplementary data to this article can be found online at https://doi.org/10.1016/j.fochx.2022.100440.
Contributor Information
Ming Ma, Email: mingma@hunnu.edu.cn.
Guowang Xu, Email: xugw@dicp.ac.cn.
Appendix A. Supplementary data
The following are the Supplementary data to this article:
References
- An N., Zhu Q.-F., Wang Y.-Z., Xiong C.-F., Hu Y.-N., Feng Y.-Q. Integration of chemical derivatization and in-source fragmentation mass spectrometry for high-coverage profiling of submetabolomes. Analytical Chemistry. 2021;93(32):11321–11328. doi: 10.1021/acs.analchem.1c02673. [DOI] [PubMed] [Google Scholar]
- Baroudi F., Al-Alam J., Chimjarn S., Delhomme O., Fajloun Z., Millet M. Conifers as environmental biomonitors: A multi-residue method for the concomitant quantification of pesticides, polycyclic aromatic hydrocarbons and polychlorinated biphenyls by LC-MS/MS and GC-MS/MS. Microchemical Journal. 2020;154 doi: 10.1016/j.microc.2019.104593. [DOI] [Google Scholar]
- Bian X., Qian Y., Tan B., Li K., Hong X., Wong C.C., et al. In-depth mapping carboxylic acid metabolome reveals the potential biomarkers in colorectal cancer through characteristic fragment ions and metabolic flux. Analytica Chimica Acta. 2020;1128:62–71. doi: 10.1016/j.aca.2020.06.064. [DOI] [PubMed] [Google Scholar]
- Bian X., Sun B., Zheng P., Li N., Wu J.-L. Derivatization enhanced separation and sensitivity of long chain-free fatty acids: Application to asthma using targeted and non-targeted liquid chromatography-mass spectrometry approach. Analytica Chimica Acta. 2017;989:59–70. doi: 10.1016/j.aca.2017.08.009. [DOI] [PubMed] [Google Scholar]
- Duan J., Yang S., Li H., Qin D., Shen Y., Li H., et al. Why the key aroma compound of soy sauce aroma type baijiu has not been revealed yet? LWT. 2022;154 doi: 10.1016/j.lwt.2021.112735. [DOI] [Google Scholar]
- Ghaedi A.M., Karamipour S., Vafaei A., Baneshi M.M., Kiarostami V. Optimization and modeling of simultaneous ultrasound-assisted adsorption of ternary dyes using copper oxide nanoparticles immobilized on activated carbon using response surface methodology and artificial neural network. Ultrasonics Sonochemistry. 2019;51:264–280. doi: 10.1016/j.ultsonch.2018.10.007. [DOI] [PubMed] [Google Scholar]
- Guo K., Li L. High-performance isotope labeling for profiling carboxylic acid-containing metabolites in biofluids by mass spectrometry. Analytical Chemistry. 2010;82(21):8789–8793. doi: 10.1021/ac102146g. [DOI] [PubMed] [Google Scholar]
- He F., Duan J., Zhao J., Li H., Sun J., Huang M., et al. Different distillation stages Baijiu classification by temperature-programmed headspace-gas chromatography-ion mobility spectrometry and gas chromatography-olfactometry-mass spectrometry combined with chemometric strategies. Food Chemistry. 2021;365 doi: 10.1016/j.foodchem.2021.130430. [DOI] [PubMed] [Google Scholar]
- He Y., Luo Y., Chen H., Chen J., Fu Y., Hou H., et al. Profiling of carboxyl-containing metabolites in smokers and non-smokers by stable isotope labeling combined with LC-MS/MS. Analytical Biochemistry. 2019;569:1–9. doi: 10.1016/j.ab.2018.12.006. [DOI] [PubMed] [Google Scholar]
- Jia S., Xu T., Huan T., Chong M., Liu M., Fang W., et al. Chemical isotope labeling exposome (CIL-EXPOSOME): One high-throughput platform for human urinary global exposome characterization. Environmental Science Technology. 2019;53(9):5445–5453. doi: 10.1021/acs.est.9b00285. [DOI] [PubMed] [Google Scholar]
- Jia W., Fan Z., Du A., Li Y., Zhang R., Shi Q., et al. Recent advances in Baijiu analysis by chromatography based technology–A review. Food Chemistry. 2020;324 doi: 10.1016/j.foodchem.2020.126899. [DOI] [PubMed] [Google Scholar]
- Kim J.-S., Kam S.F., Chung H.Y. Comparison of the volatile components in two Chinese wines, Moutai and Wuliangye. Journal of the Korean Society for Applied Biological Chemistry. 2009;52(3):275–282. doi: 10.3839/jksabc.2009.049. [DOI] [Google Scholar]
- Li S., Gao D., Song C., Tan C., Jiang Y. Isotope labeling strategies for acylcarnitines profile in biological samples by liquid chromatography-mass spectrometry. Analytical Chemistry. 2019;91(3):1701–1705. doi: 10.1021/acs.analchem.8b05120. [DOI] [PubMed] [Google Scholar]
- Li T., Yin Y., Zhou Z., Qiu J., Liu W., Zhang X., et al. Ion mobility-based sterolomics reveals spatially and temporally distinctive sterol lipids in the mouse brain. Nature Communications. 2021;12(1):1–13. doi: 10.1038/s41467-021-24672-x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liao Q, Li Y, et al. Determination of higher fatty acids in liquor by LC-QTOF. Liquor-Making Science & Technology. 2013;10:97–99. doi: 10.13746/j.njkj.2013.10.031. [DOI] [Google Scholar]
- Liao Q, Zhou H, et al. Identification of Carboxylic Acid Compounds in Baijiu by LC-QTOF. Liquor-Making Science & Technology. 2019;12:77–81. doi: 10.13746/j.njkj.2019154. [DOI] [Google Scholar]
- Luo X., Zhao S., Huan T., Sun D., Friis R.M.N., Schultz M.C., et al. High-performance chemical isotope labeling liquid chromatography–mass spectrometry for profiling the metabolomic reprogramming elicited by ammonium limitation in yeast. Journal of Proteome Research. 2016;15(5):1602–1612. doi: 10.1021/acs.jproteome.6b00070. [DOI] [PubMed] [Google Scholar]
- Naylor B.C., Catrow J.L., Maschek J.A., Cox J.E. QSRR automator: A tool for automating retention time prediction in lipidomics and metabolomics. Metabolites. 2020;10(6):237. doi: 10.3390/metabo10060237. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Peng J., Li L. Liquid–liquid extraction combined with differential isotope dimethylaminophenacyl labeling for improved metabolomic profiling of organic acids. Analytica Chimica Acta. 2013;803:97–105. doi: 10.1016/j.aca.2013.07.045. [DOI] [PubMed] [Google Scholar]
- Pinto D., Vieira E.F., Peixoto A.F., Freire C., Freitas V., Costa P., et al. Optimizing the extraction of phenolic antioxidants from chestnut shells by subcritical water extraction using response surface methodology. Food Chemistry. 2021;334 doi: 10.1016/j.foodchem.2020.127521. [DOI] [PubMed] [Google Scholar]
- Proestos C., Sereli D., Komaitis M. Determination of phenolic compounds in aromatic plants by RP-HPLC and GC-MS. Food Chemistry. 2006;95(1):44–52. doi: 10.1016/j.foodchem.2004.12.016. [DOI] [Google Scholar]
- Sharma A., Rai P.K., Prasad S. GC-MS detection and determination of major volatile compounds in Brassica juncea L. leaves and seeds. Microchemical Journal. 2018;138:488–493. doi: 10.1016/j.microc.2018.01.015. [DOI] [Google Scholar]
- Shirakawa H. The discovery of polyacetylene film: The dawning of an era of conducting polymers (Nobel lecture) Angewandte Chemie International Edition. 2001;40:2574–2580. doi: 10.1002/1521-3773(20010716)40:14<2574::AID-ANIE2574>3.0.CO;2-N. [DOI] [PubMed] [Google Scholar]
- Son E.Y., Lee S.M., Kim M., Seo J.-A., Kim Y.-S. Comparison of volatile and non-volatile metabolites in rice wine fermented by Koji inoculated with Saccharomycopsis fibuligera and Aspergillus oryzae. Food Research International. 2018;109:596–605. doi: 10.1016/j.foodres.2018.05.008. [DOI] [PubMed] [Google Scholar]
- Su Q.-Z., Vera P., Nerín C. Direct immersion–solid-phase microextraction coupled to gas chromatography-mass spectrometry and response surface methodology for nontarget screening of (Semi-) volatile migrants from food contact materials. Analytical Chemistry. 2020;92(7):5577–5584. doi: 10.1021/acs.analchem.0c00532. [DOI] [PubMed] [Google Scholar]
- Wang F., Liigand J., Tian S., Arndt D., Greiner R., Wishart D.S. CFM-ID 4.0: More accurate ESI-MS/MS spectral prediction and compound identification. Analytical Chemistry. 2021;93(34):11692–11700. doi: 10.1021/acs.analchem.1c01465. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang G., Song X., Zhu L., Li Q., Zheng F., Geng X., et al. A flavoromics strategy for the differentiation of different types of Baijiu according to the non-volatile organic acids. Food Chemistry. 2022;374 doi: 10.1016/j.foodchem.2021.131641. [DOI] [PubMed] [Google Scholar]
- Wei J., Xiang L., Li X., Song Y., Yang C., Ji F., et al. Derivatization strategy combined with parallel reaction monitoring for the characterization of short-chain fatty acids and their hydroxylated derivatives in mouse. Analytica Chimica Acta. 2020;1100:66–74. doi: 10.1016/j.aca.2019.11.009. [DOI] [PubMed] [Google Scholar]
- Xie Y., Wu B., Wu Z., Tu X., Xu S., Lv X., et al. Ultrasound-assisted one-phase solvent extraction coupled with liquid chromatography-quadrupole time-of-flight mass spectrometry for efficient profiling of egg yolk lipids. Food Chemistry. 2020;319 doi: 10.1016/j.foodchem.2020.126547. [DOI] [PubMed] [Google Scholar]
- Xiong C.-F., Zhu Q.-F., Chen Y.-Y., He D.-X., Feng Y.-Q. Screening and identification of epoxy/dihydroxy-oxylipins by chemical labeling-assisted ultrahigh-performance liquid chromatography coupled with high-resolution mass spectrometry. Analytical Chemistry. 2021;93(28):9904–9911. doi: 10.1021/acs.analchem.1c02058. [DOI] [PubMed] [Google Scholar]
- Yang H., Fan W., Xu Y. Characterization of non-volatile organic acids in baijius (Chinese liquors) based on BSTFA derivatization coupled with GC-MS. Food and Fermentation Industries. 2017;43(5):192–197. doi: 10.13995/j.cnki.11-1802/ts.201705031. [DOI] [Google Scholar]
- Zhao S., Li H., Han W., Chan W., Li L. Metabolomic coverage of chemical-group-submetabolome analysis: Group classification and four-channel chemical isotope labeling LC-MS. Analytical Chemistry. 2019;91(18):12108–12115. doi: 10.1021/acs.analchem.9b03431. [DOI] [PubMed] [Google Scholar]
- Zhao S., Li L. Chemical derivatization in LC-MS-based metabolomics study. Analytical Chemistry. 2020;131 doi: 10.1016/j.trac.2020.115988. [DOI] [Google Scholar]
- Zheng J., Gong G.-G., Zheng S.-J., Zhang Y., Feng Y.-Q. High coverage profiling of carboxylated metabolites in HepG2 cells using secondary amine-assisted ultrahigh-performance liquid chromatography coupled to high-resolution mass spectrometry. Analytical Chemistry. 2020;93(3):1604–1611. doi: 10.1021/acs.analchem.0c04048. [DOI] [PubMed] [Google Scholar]
- Zheng X.W., Tabrizi M.R., Nout M.R., Han B.Z. Daqu—a traditional Chinese liquor fermentation starter. Journal of the Institute of Brewing. 2011;117(1):82–90. doi: 10.1002/j.2050-0416.2011.tb00447.x\. [DOI] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.