Abstract
In contrast to recA of other bacteria, the recA gene of Streptomyces lividans has been described as indispensable for viability (G. Muth, D. Frese, A. Kleber, and W. Wohlleben, Mol. Gen. Genet. 255:420–428, 1997.). Therefore, a closer analysis of this gene was performed to detect possible unique features distinguishing the Streptomyces RecA protein from the well-characterized Escherichia coli RecA protein. The S. lividans recA gene restored UV resistance and recombination activity of an E. coli recA mutant. Also, transcriptional regulation was similar to that of E. coli recA. Gel retardation experiments showed that S. lividans recA is also under control of the Streptomyces SOS repressor LexA. The S. lividans recA gene could be replaced only by simultaneously expressing a plasmid encoded recA copy. Surprisingly, the recA expression plasmid could subsequently be eliminated using an incompatible plasmid without the loss of viability. Besides being UV sensitive and recombination deficient, all the mutants were blocked in sporulation. Genetic complementation restored UV resistance and recombination activity but did not affect the sporulation defect. This indicated that all the recA mutants had suffered from an additional mutation, which might allow toleration of a recA deficiency.
The RecA protein is the central enzyme in homologous recombination, DNA strand exchange, and recombinational DNA repair (reviewed in reference 15). In response to DNA damage, RecA becomes activated by the presence of single-stranded DNA and supports as a coprotease the autocatalytic cleavage of the SOS repressor LexA, UmuDC, and phage repressors (17). Digestion of LexA results in the induction of the SOS regulon, a set of more than 30 genes in Escherichia coli that are required for DNA repair, UV-induced mutagenesis, and inhibition of cell division (34). The E. coli recA gene has been analyzed in great detail. By three-dimensional structural, biochemical, and mutagenesis studies, protein regions have been proposed which are associated with distinct enzymatic activities of RecA (13). These regions include amino acid sequences for DNA binding, monomer-monomer interaction, filament formation, and LexA cleavage. Sequencing studies of more than 70 different procaryotic recA genes demonstrated that the deduced RecA proteins are highly conserved, with an overall similarity of between 43 and 100% (3). Only the N- and C-terminal regions, which are located on the outer surfaces of RecA filaments (32) and are involved in monomer interaction, display species-specific variety.
In Streptomyces, RecA is believed to be involved in genetic instability, manifested by the occurrence of large deletions comprising up to 1,000 kb, DNA amplifications, and DNA rearrangements (39). Treatment of Streptomyces cultures with agents inducing a SOS response enhances genetic instability (37). Although the recA genes of several Streptomyces strains have been cloned (1, 21, 26, 44), it was not possible to inactivate recA by targeted gene replacement. Only C-terminally truncated recA mutants with residual activity were isolated (1, 24). One of these mutants, FRECD3, missing the last 87 amino acid residues, was severely impaired in homologous recombination, highly UV sensitive, and defective in DNA amplification. The genetic instability of FRECD3 was about 70 times enhanced, and mutants that had lost the ends of the linear Streptomyces lividans chromosome (16) were segregated with a frequency of about 32% (38). Since a partial inactivation of recA had dramatic effects and since no completely defective recA mutants of S. lividans could be isolated, an essential role of recA for the viability of Streptomyces was suggested (24). A plausible hypothesis for a specific function of RecA in ensuring the viability of Streptomyces was proposed by Volff and Altenbuchner (38). In this model, RecA is required for the repair of single-stranded gaps which would cause the replication fork to collapse. Without the RecA-dependent reconstitution of the replication fork, a chromosomal end becomes lost (38). Recently, a Streptomyces rimosus mutant in which recA was disrupted was described (20). The mutant was UV sensitive, but its ability to perform homologous recombination was not analyzed. However, the presence of such a mutant indicated that at least in a specific strain background, recA could be inactivated without interfering with viability.
In this article, we address the question of what distinguishes the Streptomyces RecA protein from the RecA proteins of other bacteria, in our attempt to explain the different viability phenotypes of recA mutants. From gene inactivation studies in the presence of a second recA copy, we obtained evidence that recA could be inactivated only in strains that had suffered from an additional mutation, probably suppressing the lethal effects of RecA deficiency.
MATERIALS AND METHODS
Bacterial strains and media.
The E. coli strains used for subcloning and gene expression were XL1-Blue (4) and JM109 (43). The parental Streptomyces strain was S. lividans TK64 (12). E. coli cells were grown at 37°C in Luria-Bertani (LB) medium. Streptomyces strains were cultured as described previously (12). The plasmids used are listed in Table 1. Antibiotics were added supplementally, where appropriate, at the following concentrations: ampicillin, 150 μg ml−1; kanamycin, 50 μg ml−1; thiostrepton, 25 μg ml−1; gentamicin, 5 μg ml−1; chloramphenicol, 10 μg ml−1, hygromycin, 50 μg ml−1; tetracycline, 15 μg ml−1.
TABLE 1.
Plasmids used in this work
| Plasmid | Description | Reference(s) |
|---|---|---|
| 4H8 | S. coelicolor cosmid, carrying recA region; aphII | 28 |
| pUC18 | bla lacZ | 43 |
| pUC18rec | pUC18, carrying S. lividans recA on a 2,820-bp fragment; bla | Present study |
| pGM8 | Temperature-sensitive pSG5 derivative; tsr aacC1 | 25 |
| pGMhyg | pSG5 derivative carrying the hygromycin phospho-transferase gene hph | 19; Muth, unpublished |
| pSVXS | pUC18 derivative carrying the 3,271-bp XhoI-SalI (partial digest) fragment (recA) of cosmid 4H8 | 28; present study |
| pEXrecA | recA expression plasmid; tsr aacC1 cat recA | 36 |
| pKOrecA | Temperature-sensitive recA replacement vector; hph aphII | Present study |
| pRErecA | Replacement vector for the reconstitution of recA; tsr aacC1 bla | Present study |
| pTWSl1 | pSVB30 derivative carrying the 1,293-bp ApaI-BamHI fragment containing the S. lividans recA gene | Present study |
| pSVB30 | Cloning vector; bla | 2 |
| pIJ920 | SCP2 derivative; vph | 18 |
| pCK3S | pSG5 derivative carrying a 550-bp fragment of snpR; tsr | 24 |
| pJOE2702 | E. coli expression vector, rhamnose induction; bla | 33 |
| pJOE2702lexA | pJOE2702, carrying the S. lividans lexA gene | Present study |
DNA manipulations.
Standard procedures were as described by Hopwood et al. (12) and Sambrook et al. (30). Hybridization was performed with digoxigenin (DIG)-labeled dUTP and a DIG detection kit (Roche, Mannheim, Germany). Gene replacement mutants were selected as described by Wohlleben and Muth (42).
Assay for UV sensitivity.
E. coli cultures were grown till they reached an optical density at 600 nm of 0.8, harvested by centrifugation, and resuspended in 0.8% NaCl. Serial dilutions were plated onto LB agar containing 1 mM isopropyl-β-d-thiogalactopyranoside and irradiated with UV light (VL115c, 254 nm, 730 μW/cm2; Vilber Lourmat, Marne-La-Vallée, France) at a distance of 10 cm for various periods (2, 5, 10, 15, and 20 s), followed by incubation in the dark. UV resistance of S. lividans strains was determined as described by Muth et al. (24).
Assay for genetic instability.
The genetic instability of S. lividans strains was measured as the ratio of chloramphenicol-sensitive colonies, as described by Vierling et al. (36).
Assay for homologous recombination.
To assay the efficiency of homologous recombination in E. coli, matings with the Hfr donor strain KH500 (Hantke, Tübingen, Germany), which carries a tetracycline resistance marker (Tn10) near the F plasmid integration site, and a recA deletion strain, DK1 (14), harboring the plasmid pTWSl1 (Table 1), were performed. The mobilization frequency (Table 2) of the tetracycline resistance marker into DK1 was determined on isopropyl-β-d-thiogalactopyranoside-containing medium. Recombination activity in S. lividans was measured by its ability to integrate the temperature-sensitive plasmid pCK3S via homologous recombination into the chromosome as described by Vierling et al. (36).
TABLE 2.
Recombination activity of the E. coli recA mutant DK1, carrying the S. lividans recA gene
| Recipient strain (plasmid) | Titers
|
Transfer frequency (%) | Relative recombination activity | ||
|---|---|---|---|---|---|
| Recipient | Donor | Transconjugants | |||
| JM83 | 8.20 × 108 | 3.59 × 109 | 1.81 × 107 | 2.20 × 10−2 | 1 |
| DK1 (pSVB30) | 1.50 × 106 | 3.59 × 109 | 0 | <6.67 × 10−7 | <0.00003 |
| DK1 (pTWSl1) | 2.58 × 108 | 3.59 × 109 | 6.05 × 106 | 2.35 × 10−2 | 1.06 |
Gel retardation experiments.
The lexA gene of S. lividans TK64 was amplified by PCR using chromosomal DNA of S. lividans TK64 and the primers 5′-GGAATTCCATATGCACGCGATGAGCGACGC and 5′-CGGGATCCTCAGACGCGACGCAGTACGGCC, which were derived from the Streptomyces coelicolor lexA gene located on cosmid 5B8 (ftp://ftp.sanger.ac.uk/pub/S_coelicolor/sequences). PCR products were cloned under control of the rhamnose-inducible promoter in the expression plasmid pJOE2702 (33). E. coli JM109 (pJOE2702lexA) was grown at 37°C until it reached an optical density at 578 nm of 0.2 and induced with rhamnose (0.2%). Six hours after induction, the cells were harvested and disrupted with a French press. The upstream region of recA containing the putative SOS box was amplified with the primers 5′-GGAATTCCGTACGCTCGGAAGTGC and 5′-CGGGATCCTCGACATCACCCGTCA. The resulting fragment was 3′-end labeled with DIG-11-dUTP (Roche) according to the manufacturer's instructions. Thirty femtomoles of the labeled fragment was incubated at room temperature for 15 min with 11 μg of LexA-containing soluble crude extract in a total volume of 20 μl (binding buffer, 4 μl; poly[d(I-C)], 1 μl; poly-l-lysine, 1 μl [Digoxigenin Gelshift Kit; Roche]). Subsequently the reaction mixture was run on a 5% polyacrylamide gel and transferred to a nylon membrane by Southern blotting, and the DIG-labeled DNA complexes were visualized using anti-DIG-alkaline-phosphatase-conjugated antibody.
Immunoblotting.
Immunoblotting was performed as described by Engels et al. (9) using polyclonal rabbit antisera raised against purified His-tagged S. lividans RecA protein (Vierling and Muth, unpublished data).
Construction of the recA replacement plasmid.
A pUC18 subclone (pUC18rec) carrying a 2,820-bp chromosomal fragment of S. lividans TK64 that contained recA with its upstream and downstream regions was digested with BamHI and NcoI. Following Klenow treatment, the recA-containing BamHI-NcoI fragment was replaced by an aphII cassette. The NcoI site overlaps the putative start codon of recA, while the BamHI site is located 99 bp downstream of the recA stop codon in a noncoding region. The resulting plasmid was subsequently fused with pGMhyg (Muth, unpublished data), a hygromycin resistance-encoding, temperature-sensitive pSG5 derivative, yielding the recA replacement plasmid pKOrecA.
Construction of the replacement plasmid for the reconstitution of recA.
A 3,271-bp fragment of the S. coelicolor cosmid 4H8 resulting from a XhoI/partial SalI digest was subcloned into pUC18, resulting in pSVXS. In order to distinguish the reconstituted recA gene from the wild-type gene, the single BamHI site located downstream of recA was eliminated by Klenow treatment. Subsequently the resulting plasmid was fused via EcoRI with the temperature-sensitive pGM8, yielding pRErecA.
Fixation of Streptomyces colonies for scanning electron microscopy.
The S. lividans wild type, S. lividans SV64ΔrecA, and the reconstituted mutant SVRErecA were grown on R2YE agar for 5 days. Agar plugs were cut out with a cork borer and fixed for 10 min in 2.5% glutaraldehyde–100 mM cacodylate (pH 7.5). Subsequently the plugs were washed in 100 mM cacodylate (10 min) and H20 (10 min) and dehydrated (10 min) in 30, 50, 70, 90, and 100% EtOH. After critical point drying under CO2, the mycelium was coated in a vacuum evaporator with a thin layer of Au-Pd. Observations were made with a Hitachi S-2460N scanning electron microscope with a secondary electron mode operating at 10 kV.
RESULTS
S. lividans recA complements the E. coli recA deletion mutant DK1 efficiently.
In order to analyze whether the Streptomyces RecA protein has the same activities as the well-characterized E. coli RecA protein, we attempted complementation of an E. coli mutant devoid of recA. The presence of the plasmid pTWSl1, containing the recA gene under control of the lac promoter, complemented the UV sensitivity of the recA deletion mutant DK1 to wild-type levels (data not shown). This indicated the proficiency of the S. lividans RecA protein for recombinational repair and the ability to support cleavage of the E. coli LexA repressor. The ability to perform homologous recombination was studied by Hfr matings using DK1 (pTWSl1) as the recipient. The outgrowth of tetracycline-resistant DK1 (pTWSl) colonies demonstrated that the S. lividans recA gene was able to restore recombination activity in DK1 (Table 2). Therefore, the S. lividans RecA protein possesses the same basic activities as the E. coli RecA protein.
S. lividans recA is regulated by the LexA repressor.
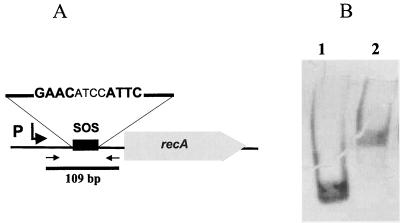
Next, we tested whether regulation of the Streptomyces recA gene differs from that for other bacteria. Previously it was shown that transcription of the recA operon in S. lividans was induced following treatment with the DNA-damaging methane methylsulfonate (36). This indicated that as in all other bacteria, recA is regulated by the SOS repressor LexA, which binds to so-called SOS boxes (Cheo box) in the promoter region of the genes of the SOS response. In the putative promoter region of the S. lividans recA gene, there is a sequence, GAACATCCATTC, which resembles (as indicated by boldface type) the Bacillus subtilis SOS box GAACNNNNGTT(C/T). To analyze transcriptional regulation of recA by LexA, we expressed the S. lividans lexA gene in E. coli as described in Materials and Methods. A 109-bp fragment containing the putative SOS box of the S. lividans recA gene was amplified by PCR and 3′ labeled with DIG. After incubation with LexA, the reaction mixture was separated on a 5% Tris-borate-polyacrylamide gel, blotted onto a nylon membrane, and visualized with anti-DIG antibody conjugate. The retardation of the SOS box-containing fragment (Fig. 1) showed that the Streptomyces LexA protein is able to bind the proposed SOS box, indicating that LexA controls recA expression. When the respective fragments were incubated with an E. coli crude extract containing GlnR, a transcriptional regulator that binds in the promoter region of the glnA gene (N. Weisschuh and A. Engels, personal communication), no retardation was observed (data not shown).
FIG. 1.
Transcriptional regulation of the S. lividans recA gene. A 109-bp fragment containing the putative SOS box of the S. lividans TK64 recA gene was amplified by PCR (A), labeled with DIG, and incubated with S. lividans LexA-containing crude extract. Following electrophoresis on a 5% polyacrylamide gel and capillary transfer to a nylon membrane, the shifted and unshifted fragments were visualized by alkaline phosphatase-conjugated anti-DIG antibody (B) (Roche). Lane 1, 109-bp fragment, incubated with LexA-free crude extracts; lane 2, 109-bp fragment with LexA-containing crude extract.
The chromosomal recA gene of S. lividans TK64 can be deleted in the presence of a plasmid-borne recA copy.
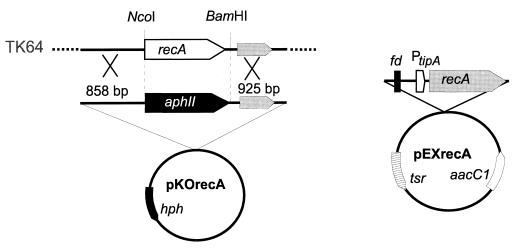
Since it was not possible to detect any significant difference between the activities conferred by the S. lividans and E. coli recA genes or their regulation, it was essential to confirm that the inability to remove recA from the genetic background was not the result of methodological complications. Therefore, we proceeded to demonstrate that the replacement plasmid was functional and that the homologous DNA fragments are sufficient in size to allow efficient recombination. To analyze whether the chromosomal recA fragment could be deleted while expressing a plasmid-borne recA copy, the recA gene of S. lividans was cloned under control of the thiostrepton-inducible tipA promoter (23). Since S. lividans did not tolerate the transformation with recA on a multicopy plasmid (unpublished results), the expression cassette was inserted into a single-copy SCP2 derivative, yielding pEXrecA (Fig. 2).
FIG. 2.
Replacement of the S. lividans TK64 recA gene by the simultaneous expression of a plasmid-borne recA copy. Schematic maps of the S. lividans chromosomal recA region, gene replacement plasmid pKOrecA and the recA expression plasmid pEXrecA, carrying the terminator region of phage fd and the thiostrepton-inducible tipA promoter (PtipA), are given. The sizes of the homologous regions and relevant restriction sites are indicated.
In the temperature-sensitive recA gene replacement plasmid pKOrecA, the complete recA coding region was replaced with the aphII gene. The aphII gene was inserted in the same orientation as recA to minimize any polar effects on the downstream recX gene, which is cotranscribed with recA after induction of the SOS response (36). pKOrecA contained fragments of 858 and 925 bp, corresponding to the upstream and downstream regions of recA for recombination with the chromosome. It carried only a 75-bp region identical to pEXrecA to minimize the risk of recombination between the two plasmids. S. lividans TK64 was cotransformed with the plasmids pEXrecA and pKOrecA. Transformants carrying both plasmids were selected on gentamicin- and kanamycin-containing agar. Subsequently, colonies that carried the kanamycin resistance gene integrated into the chromosome were selected under inducing (thiostrepton-kanamycin) conditions at 39°C. From 400 picked colonies, four were found to be kanamycin resistant and hygromycin sensitive, indicating that the chromosomal recA gene was replaced. By PCR and Southern blotting experiments, the correct replacement of the chromosomal recA gene via double crossover and the loss of vector sequences were confirmed in all of these clones (data not shown). In contrast, if the replacement plasmid pKOrecA was introduced into S. lividans TK64 without pEXrecA, replacement of the chromosomal recA gene could not be achieved. From 3,000 picked colonies that were selected at 39°C on kanamycin-containing agar, all still carried the hygromycin resistance gene of the vector, indicating that the whole plasmid had integrated into the chromosome via a single crossover. This result confirmed our previous observations (24) that recA might be indispensable in S. lividans and that it was not possible to inactivate recA without concomitant expression of a recA copy.
A mutant deficient for recA can be generated by curing the recA expression plasmid.
To study the presumed detrimental effects of recA inactivation by switching the tipA promoter on and off, first the inducibility of recA expression in pEXrecA was analyzed. The recA gene of pEXrecA was replaced by the promoterless aphII gene from the transposon Tn5. Without induction, the tipA promoter mediated resistance to kanamycin (50 μg ml−1). On 0.5-μg ml−1, 1 μg-ml−1, and 5-μg-ml−1 thiostrepton, respectively, a resistance to kanamycin at concentrations of 150, 200, and 400 μg ml−1 was observed. The highest level of resistance (at a kanamycin concentration of 600 μg ml−1) was obtained by induction with 25 μg of thiostrepton ml−1. Due to the basic activity of the tipA promoter even in the absence of thiostrepton, it was necessary to cure the recA mutant strains of plasmid pEXrecA in order to analyze whether the strains survived in the absence of RecA.
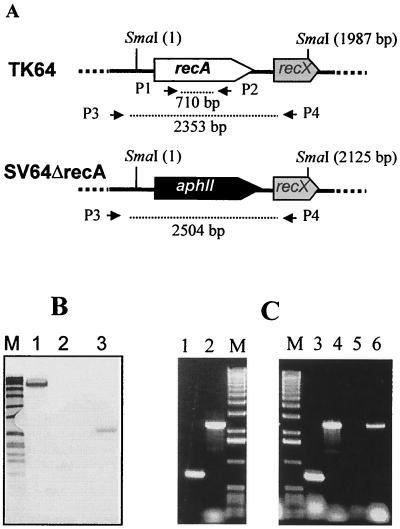
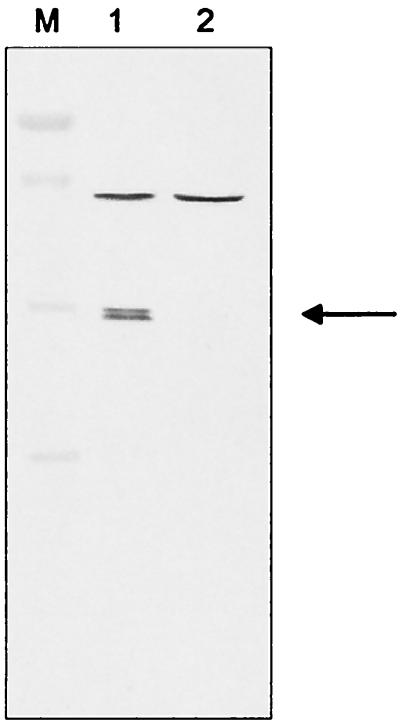
The four SV64ΔrecA strains were transformed with the plasmid pIJ920. pIJ920 is an SCP2 derivative containing the viomycin resistance gene vph (18) and is incompatible with the recA expression plasmid pEXrecA, which is also based on the SCP2 replicon. By selecting for the viomycin resistance gene of pIJ920, the pEXrecA plasmid could be displaced from the isolated recA replacement mutants. Five out of 80 tested viomycin-resistant transformants had lost the gentamicin and thiostrepton resistance of pEXrecA. Southern blotting and PCR experiments using internal recA primers confirmed the absence of recA (Fig. 3). Furthermore, immunoblots with RecA-specific antisera were negative (Fig. 4).
FIG. 3.
Replacement of the recA gene of S. lividans TK64. A schematic drawing (A), Southern blot (B), and PCR analysis (C) are shown. (A) Relevant restriction sites, primers used for PCR amplification, and the sizes of the respective fragments are indicated. (B) SmaI-digested total DNA of S. lividans TK64 and the recA mutant SV64ΔrecA was hybridized against a DIG-labeled recA PCR fragment. Lane M, Bio-VII-Marker (Roche): 8,576, 7,427, 6,106, 4,899, 3,639, 2,799, 1,953, 1,882, 1,515, 1,482, 1,164, 992, 710, 492, and 359 bp; lane 1, SV64ΔrecA (pEXrecA); lane 2, SV64ΔrecA; lane 3, S. lividans TK64. (C) Agarose gel electrophoresis of PCR fragments. Lane M, 1-kb ladder (Roche): 12,216, 11,198, 10,180, 9,162, 8,144, 7,126, 6,108, 5,090, 4,072, 3,054, 2,036, 1,636, 1,018, 517, 506, 396, 344, 298, 220, 201, 154, 134, and 75 bp; lane 1, TK64, P1/P2; lane 2, TK64, P3/P4; lane 3, SV64ΔrecA (pEXrecA), P1/P2; lane 4, SV64ΔrecA (pEXrecA), P3/P4; lane 5, SV64ΔrecA, P1/P2; lane 6, SV64ΔrecA, P3/P4.
FIG. 4.
Detection of RecA by immunoblotting. Total proteins were separated on a 12.5% sodium dodecyl sulfate-polyacrylamide gel and blotted to a nylon membrane. RecA was detected using a polyclonal antiserum raised against purified S. lividans RecA protein. The arrow indicates the RecA-specific band. Lane M, marker, Prestained Low Range Standard (Bio-Rad, Munich, Germany): 116, 80, 52.5, 34.9, 29.9, and 21.8 kDa; lane 1, S. lividans TK64; lane 2, SV64ΔrecA.
The recA mutant SV64 displayed a classical recA phenotype.
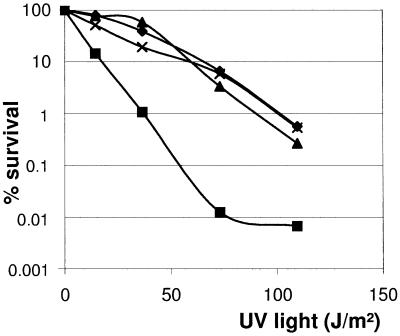
To assay for recombinational activity, the SV64ΔrecA mutant was transformed with the plasmid pCK3S (24), a temperature-sensitive pGM derivative that carries a 550-bp fragment of the TK64 snpR gene, encoding the regulator of the metalloprotease SnpA. Following a temperature shift to 39°C to eliminate autonomously replicating plasmids, the cultures were homogenized, and serial dilutions of the mycelial fragments were plated in parallel on LB agar and LB containing thiostrepton. The ratio of the titer obtained on thiostrepton plates allowing the outgrowth only of colonies with pCK3S in their chromosome to the titer on LB agar revealed the recombination frequency. While the plasmid pCK3S was integrated into the chromosome of TK64 at a frequency of about 54% (titer on LB agar, 4.5 × 106; titer on thiostrepton, 2.4 × 106), integration of pCK3S into the SV64ΔrecA chromosome did not occur (titer on LB agar, 5.0 × 106; titer on thiostrepton, 0). Furthermore, the recA deletion mutant S. lividans SV64ΔrecA was highly sensitive to UV irradiation. Although still more than 10% of the wild-type fragments survived a UV dose of 73 J/m2, about 99.99% of S. lividans SV64ΔrecA mycelial fragments were destroyed (Fig. 5). To analyze the effects of recA deficiency on genetic instability, mycelial fragments of the S. lividans wild type and the recA mutant SV64ΔrecA were plated on soja-mannitol-agar. After 7 days, the mycelium was scraped off, homogenized, and replated. After three rounds, dilutions were plated and single colonies were subsequently picked and placed on chloramphenicol-containing and chloramphenicol-free medium. The recA mutant, SV64ΔrecA, had segregated chloramphenicol-sensitive colonies with a frequency of 6.2%, about 12.5 times that of the wild type.
FIG. 5.
UV sensitivity of the S. lividans recA mutant SV64ΔrecA. Spores or aerial mycelial fragments of S. lividans TK64 (♦), the recA mutant SV64ΔrecA (▪), SV64ΔrecA carrying the recA expression plasmid pEXrecA (▴), and the reconstituted SVRErecA strain (×) were plated on LB agar and irradiated with UV light (254 nm, 730 μW/cm2) for various periods. SV64ΔrecA(pEXrecA) was grown on LB containing thiostrepton for induction of recA expression.
The recA mutant SV64 represents a whi mutant.
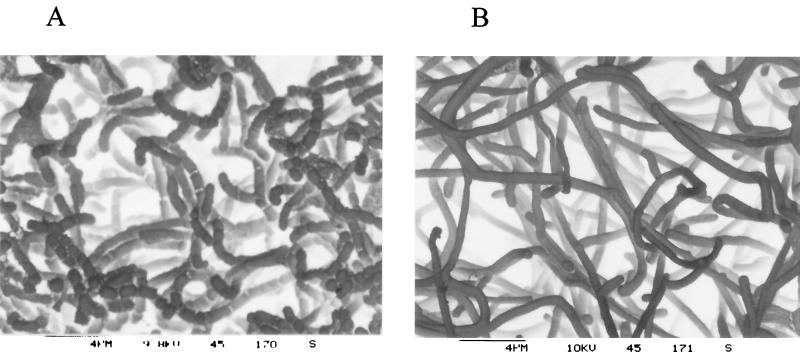
Besides the defects in homologous recombination, UV resistance, and genetic instability, all recA mutants were impaired in sporulation. On R5- or soja-mannitol-agar, a white aerial mycelium was formed that contained no spores. The aerial mycelium was further studied by scanning electron microscopy. The mutant SV64ΔrecA formed long straight unseptated hyphae with little or no curling (Fig. 6). Obviously, sporulation was blocked at an early time point in the life cycle of S. lividans. Thus, the recA mutant had a phenotype similar to that described for the S. coelicolor whi mutants (6).
FIG. 6.
Scanning electron micrographs of the surfaces of colonies of the wild-type strain S. lividans TK64 (A) and the recA mutant SV64ΔrecA (B). Colonies were grown for 5 days on soja-mannitol plates before being prepared for electron microscopy.
Reconstitution of recA does not complement the sporulation defect.
To analyze whether the sporulation defect was an effect of recA inactivation or the recA mutant had suffered from an additional mutation, we complemented the mutant by reconstituting a wild-type recA gene into the chromosome of S. lividans SV64ΔrecA: using the plasmid pRErecA, the aphII gene was replaced by the S. coelicolor recA gene, which differs from the S. lividans recA gene by two base-pair substitutions (see Discussion). The plasmid pRErecA carries a 3,172-bp chromosomal fragment of S. coelicolor A(3)2 with 1,161 bp upstream and 885 bp of the downstream region of recA. To distinguish the reconstituted recA gene from the wild-type recA gene, the BamHI site located in the intergenic region 96 bp downstream of recA was eliminated by Klenow treatment. Following a temperature shift, transformants were picked on thiostrepton- and kanamycin-containing media to screen for tsr and aphII sensitive colonies that probably had replaced the aphII gene by a double crossover event. The correct replacement event was confirmed by Southern blot analysis and PCR. By reverse transcription-PCR analysis, the inducibility of recA transcription (data not shown) in response to the DNA-damaging methane methylsulfonate was found to be indistinguishable from that of the parent S. lividans TK64 strain (36). The reconstituted mutant was fully complemented with regard to UV sensitivity (Fig. 5) and recombination activity. The integration of the recombination test plasmid occurred with a frequency of 29%, which is on the same order as in the wild type. However, the reconstitution of recA did not affect the sporulation deficiency of S. lividans SV64ΔrecA. Scanning electron microscopy also revealed no difference in the S. lividans SV64ΔrecA mutant (data not shown). This clearly demonstrates that the mutants have suffered from an additional mutation affecting morphologic differentiation and that the sporulation defect was not a consequence of inactivation of recA.
DISCUSSION
To investigate whether the Streptomyces RecA protein had functions different from those of other RecA proteins, we complemented an E. coli recA mutant with the S. lividans recA gene. Expression of the S. lividans recA gene in DK1 (14) restored UV resistance and recombination activity in Hfr matings to the wild-type level. This suggests that the S. lividans RecA protein fulfills all the enzymatic activities that have been ascribed to E. coli RecA, namely protease activity to support cleavage of the LexA repressor, proficiency for recombinational DNA repair, and the ability to perform homologous recombination (13). Furthermore, there is no evidence that the Streptomyces RecA protein might have a distinct activity, since the deduced amino acid sequence of the S. lividans RecA protein is, besides the species-specific C terminus, highly similar to that of other bacterial RecA proteins (3).
In all bacteria, it has been shown that transcription of DNA damage-inducible genes is controlled by LexA, which binds to so-called SOS boxes in promoter regions (17). By sequence comparison and site-directed mutagenesis combined with gel retardation assays and hydroxyl radical footprint protection assays, Winterling et al. proposed a new consensus sequence (CGAACRNRYGTTYC) for SOS boxes of gram-positive bacteria (40, 41). Although the SOS box of the S. lividans recA gene (CGAACATCCATTCT) differs from this consensus sequence in three positions (shown in bold) and does not form a perfect palindrome, the binding of S. lividans LexA and the inducibility by DNA damaging agents demonstrated its functionality. The same sequence, CGAACATC(C/T)ATTCT, is also found in front of all the other Streptomyces recA genes where sequence information is available (EMBL accession no. AL020958) (1). As in Mycobacterium tuberculosis and M. smegmatis (22), this SOS box overlaps with a consensus sequence of a heat shock promoter. This putative heat shock promoter of the Streptomyces recA genes has been postulated by sequence similarity (26), but in M. smegmatis, a transcriptional start site of the recA gene corresponding to this heat shock promoter was mapped by primer extension (27). The overlap of the SOS box with the −10 or −35 promoter region is a common feature and was described for various genes of the SOS response (10). A second putative SOS box (TGAACG(G/C)CA(G/A)TTCG) (conserved bases shown in bold) is present within the N-terminal coding region of Streptomyces recA (amino acid position +18) (1). However, its involvement in SOS regulation has not been investigated. Two LexA binding sites have also been described for several other LexA-regulated genes, e.g., B. subtilis dinC and dinR, recN or lexA from E. coli (10). Although the Streptomyces SOS box is not a perfect palindrome, the presence of two binding sites may indicate tight regulation by LexA. Note that the promoter region of the S. coelicolor (EMBL accession no. AL022268) and Streptomyces clavuligerus (EMBL accession no. AJ224870) lexA genes also contain putative SOS boxes. These boxes lie 148 and 76 bp upstream of the putative translational start of lexA, respectively. The lexA SOS boxes differ from the recA SOS box in six positions (shown in bold) (CGAACGTGTGTTTG) and fit perfectly the proposed consensus sequence. A gel retardation reaction performed with an 86-bp PCR fragment containing the putative SOS box of lexA showed that the Streptomyces LexA is able to bind the proposed SOS box (Vierling and Muth, unpublished results), indicating that LexA is autoregulative also in S. coelicolor.
Because the S. lividans recA gene neither conferred a function distinct from that of E. coli recA nor differed in its regulation from that of other bacteria, we tested whether it was possible to replace the chromosomal recA gene in the presence of a plasmid-borne recA copy. This turned out to be a successful approach. The chromosomal recA gene could be efficiently replaced with a frequency of about 1%, whereas it was not possible (>0.03%) without the simultaneous recA expression.
Since the tipA promoter in pEXrecA is not tightly repressed in the absence of thiostrepton, resulting in a basal level of recA expression, it was not possible to study the presumed toxicity of recA inactivation by switching the tipA promoter on and off. Therefore, we had to cure the recA expression plasmid to demonstrate the indispensability of recA. To our surprise, we observed that following the replacement of the chromosomal recA fragment, it was possible to cure the recA expression plasmid pEXrecA without a lethal effect. Displacing the resident recA expression plasmid by the incompatible plasmid pIJ920 (18) was a very efficient curing technique. In contrast to other described curing methods, such as growth at elevated temperatures or treatment with intercalating dyes (8), plasmid curing by incompatibility is not associated with any mutagenic side effects.
There are two possible explanations of why the generation of a completely defective recA mutant succeeded only by this procedure whereas it was not possible by the classical protocol.
(i) For unknown reasons, the recA-containing DNA fragment is only a poor substrate for recombination enzymes. Overexpression of the RecA protein from the thiostrepton-inducible tipA promoter could confer an enhanced recombination activity that allowed even the recombination of poor substrates. A 10-fold stimulation of homologous recombination by the overexpression of a bacterial recA gene has already been reported for plant and mammalian cells (29, 31). However, in S. lividans (pEXrecA), induction of recA overexpression did not result in an enhanced recombination rate. Neither under inducing conditions nor under noninducing conditions was the integration rate of a test plasmid with a 540-bp fragment, suitable for homologous recombination, increased (unpublished results). In contrast, the presence of the recA expression plasmid had only negative effects on the integration frequency of the test plasmid. This was probably due to the detrimental effects of recA overexpression (36) interfering with the survival of the integrants.
(ii) The recA mutant had acquired an additional mutation which suppresses the toxic effects of recA inactivation. Since up to now no suppressor mutations for recA have been described (15), the mutation must affect a function that allows the cell to survive with recA deficiency. The plasmid-borne copy of recA which is under control of the tipA promoter might be just sufficient to override the lethal effect of RecA deficiency but might not be able to complement recA with wild-type efficiency. Therefore, selection pressure could exist to select for such suppressing mutations. The reasons for the lethal effects of inactivation of recA in Streptomyces are not known, but a role of RecA in the repair of damaged replication forks was suggested (38). In an alternative model, the recombination patching model, RecA activity is required for the replication of the ends of the linear chromosome. Since the recA mutant SV64ΔrecA still contained a linear chromosome, it was possible recently to disprove this model (C.-H. Huang, H.-H. Lee, S.-H. Chou, and C. W. Chen, personal communication). Although E. coli recA mutants are viable, they are also severely affected and show slower growth, probably due to the generation of up to 50% dead cells (5). If the lethal effect of recA inactivation reflects a defect in DNA repair, a suppressing mutation could stimulate RecA-independent repair mechanisms or delay cell division to provide more time for the repair. The requirement for a second mutation would explain why neither by classical mutagenesis (11) nor by conventional gene inactivation techniques (1, 24) has it been possible to isolate recA mutants of S. coelicolor, S. lividans, or Streptomyces ambofaciens. A recA mutant was described only for S. rimosus (20). Since the genotype of this strain was not characterized in detail, there is no information available about the presence of any additional defects.
Beside the classical recA phenotype, UV sensitivity, deficiency in homologous recombination, and enhanced genetic instability, all the mutants were sporulation deficient. It should be stressed that some of the mutants were isolated in independent experiments. S. lividans SV64ΔrecA strains had the morphology of so-called whi mutants (6). This was confirmed by scanning electron microscopy of the recA mutant. Obviously, the differentiation was blocked in an early stage before the formation of septation. Thus, the mutant resembles whiA, whiB, whiG, whiH, whiI, or whiJ mutants of S. coelicolor, with which the formation of sporulation septa is essentially abolished (7). Since the recA mutant formed long straight hyphae with little to no curling, the morphology was similar to that described for whiG mutants (35).
All defects of the classical recA phenotype could be fully restored by the S. lividans recA gene when placed under control of the thiostrepton-inducible tipA promoter on a single-copy SCP2 derivative or by the reintroduction of the S. coelicolor recA gene at the original chromosomal position. The S. coelicolor recA gene differs from that of S. lividans by two base-pair substitutions (shown in bold): a CGT-CGG exchange that had no effect on the amino acid composition and a GCG to ACG substitution that changes an alanine to a threonine. This amino acid exchange is localized at position 369 in the C-terminal end, which is not conserved in bacterial RecA proteins (3). However, complementation did not affect the sporulation deficiency, demonstrating that the block in morphologic differentiation was not caused by the inactivation of recA. This might be a clear indication that S. lividans SV64ΔrecA had acquired an additional mutation that could suppress the toxic effects of recA deficiency. During vegetative growth of the Streptomyces substrate mycelium, only very few cross walls are formed in the growing hyphae. The formation of cross walls, which corresponds to the cell division of unicellular bacteria, occurs in the Streptomyces life cycle mainly during differentiation. The aerial mycelium erected from the substrate mycelium becomes fragmented into spore chains and finally is released (6). A mutation blocking the septation of the Streptomyces aerial mycelium could have an effect for Streptomyces similar to the inhibition of cell division by SulA during the SOS response in E. coli, in preventing Streptomyces from producing nonviable spores with damaged DNA.
ACKNOWLEDGMENTS
This research was supported by the Deutsche Forschungsgemeinschaft (SFB-323).
We thank D. Fink for critical reading of the manuscript, A. Radunz for preparing the antibodies, C. F. Bardele and H. Schoepmann for taking the electron micrographs, and K. Hantke for providing strain KH500.
REFERENCES
- 1.Aigle B, Holl A C, Angulo J F, Leblond P, Decaris B. Characterization of two Streptomyces ambofaciens recA mutants: identification of the RecA protein by immunoblotting. FEMS Microbiol Lett. 1997;149:181–187. doi: 10.1111/j.1574-6968.1997.tb10326.x. [DOI] [PubMed] [Google Scholar]
- 2.Arnold W, Pühler A. A family of high-copy-number plasmid vectors with single end-label sites for rapid nucleotide sequencing. Gene. 1988;70:171–179. doi: 10.1016/0378-1119(88)90115-1. [DOI] [PubMed] [Google Scholar]
- 3.Brendel V, Brocchieri L, Sandler S J, Clark A J, Karlin S. Evolutionary comparisons of RecA-like proteins across all major kingdoms of living organisms. J Mol Evol. 1997;44:528–541. doi: 10.1007/pl00006177. [DOI] [PubMed] [Google Scholar]
- 4.Bullock W O, Fernandez J M, Short J M. XL1-blue, a high efficiency plasmid transforming recA Escherichia coli strain with beta galactosidase selection. BioTechniques. 2000;5:376–378. [Google Scholar]
- 5.Capaldo F N, Ramsey G, Barbour S D. Analysis of the growth of recombination-deficient strains of E. coli K-12. J Bacteriol. 1974;118:242–249. doi: 10.1128/jb.118.1.242-249.1974. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Chater K F. Genetics of differentiation in Streptomyces. Annu Rev Microbiol. 1993;47:685–713. doi: 10.1146/annurev.mi.47.100193.003345. [DOI] [PubMed] [Google Scholar]
- 7.Chater K F. Taking a genetic scalpel to the Streptomyces colony. Microbiology (United Kingdom) 1998;144:1465–1478. doi: 10.1099/00221287-144-6-1465. [DOI] [PubMed] [Google Scholar]
- 8.Crameri R, Davies J E, Hütter R. Plasmid curing and generation of mutations induced with ethidium bromide in streptomycetes. J Gen Microbiol. 1986;132:819–824. doi: 10.1099/00221287-132-3-819. [DOI] [PubMed] [Google Scholar]
- 9.Engels A, Kahmann U, Ruppel H G, Pistorius E K. Isolation, partial characterization and localization of a dihydrolipoamide dehydrogenase from the Cyanobacterium Synechocystis PCC6803. Biochim Biophys Acta. 1997;1340:33–44. doi: 10.1016/s0167-4838(97)00025-3. [DOI] [PubMed] [Google Scholar]
- 10.Friedberg E C, Walker G C, Siede W. DNA repair and mutagenesis. Washington, D.C.: ASM Press; 1995. [Google Scholar]
- 11.Harold R J, Hopwood D A. Ultraviolet-sensitive mutants of Streptomyces coelicolor. II. Genetics. Mutat Res. 1970;10:439–448. doi: 10.1016/0027-5107(70)90004-7. [DOI] [PubMed] [Google Scholar]
- 12.Hopwood D A, Bibb M J, Chater K F, Kieser T, Bruton C J, Kieser H M, Lydiate D J, Smith C P, Ward J M, Schrempf H. Genetic manipulation of Streptomyces, a laboratory manual. Norwich, United Kingdom: The John Innes Institute; 1985. [Google Scholar]
- 13.Kowalczykowski S C, Dixon D A, Eggleston A K, Lauder S D, Rehrauer W M. Biochemistry of homologous recombination in Escherichia coli. Microbiol Rev. 1994;58:401–465. doi: 10.1128/mr.58.3.401-465.1994. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Kurnit D M. Escherichia coli recA deletion strains that are highly competent for transformation and for in vivo phage packaging. Gene. 1989;82:313–315. doi: 10.1016/0378-1119(89)90056-5. [DOI] [PubMed] [Google Scholar]
- 15.Kuzminov A. Recombinational repair of DNA damage in Escherichia coli and bacteriophage lambda. Microbiol Mol Biol Rev. 1999;63:751–813. doi: 10.1128/mmbr.63.4.751-813.1999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Lin Y-S, Kieser H M, Hopwood D A, Chen C W. The chromosomal DNA of Streptomyces lividans 66 is linear. Mol Microbiol. 1993;10:923–933. doi: 10.1111/j.1365-2958.1993.tb00964.x. [DOI] [PubMed] [Google Scholar]
- 17.Little J W. Mechanism of specific LexA cleavage: autodigestion and the role of RecA coprotease. Biochimie. 1991;73:411–421. doi: 10.1016/0300-9084(91)90108-d. [DOI] [PubMed] [Google Scholar]
- 18.Lydiate D J, Malpartida F, Hopwood D A. The Streptomyces plasmid SCP2*: its functional analysis and development into useful cloning vectors. Gene. 1985;35:223–235. doi: 10.1016/0378-1119(85)90001-0. [DOI] [PubMed] [Google Scholar]
- 19.Malpartida F, Zalacain M, Jimenez A, Davies J. Molecular cloning and expression in Streptomyces lividans of a hygromycin B phosphotransferase gene from Streptomyces hygroscopicus. Biochem Biophys Res Commun. 1983;117:6–12. doi: 10.1016/0006-291x(83)91533-4. [DOI] [PubMed] [Google Scholar]
- 20.Mikoc A, Ahel I, Gamulin V. Construction and characterization of a Streptomyces rimosus recA mutant: the RecA-deficient strain remains viable. Mol Gen Genet. 2000;264:227–232. doi: 10.1007/s004380000284. [DOI] [PubMed] [Google Scholar]
- 21.Mikoc A, Vujaklija D, Gamulin V. The recA gene from Streptomyces rimosus R6: sequence and expression in Escherichia coli. Res Microbiol. 1997;148:397–403. doi: 10.1016/S0923-2508(97)83870-3. [DOI] [PubMed] [Google Scholar]
- 22.Movahedzadeh F, Colston M J, Davis E O. Determination of DNA sequences required for regulated Mycobacterium tuberculosis RecA expression in response to DNA-damaging agents suggests that two modes of regulation exist. J Bacteriol. 1997;179:3509–3518. doi: 10.1128/jb.179.11.3509-3518.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Murakami T, Holt T G, Thompson C J. Thiostrepton-induced gene expression in Streptomyces lividans. J Bacteriol. 1989;171:1459–1466. doi: 10.1128/jb.171.3.1459-1466.1989. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Muth G, Frese D, Kleber A, Wohlleben W. Mutational analysis of the Streptomyces lividans recA gene suggests that only mutants with residual activity remain viable. Mol Gen Genet. 1997;255:420–428. doi: 10.1007/s004380050514. [DOI] [PubMed] [Google Scholar]
- 25.Muth G, Nussbaumer B, Wohlleben W, Pühler A. A vector system with temperature-sensitive replication for gene disruption and mutational cloning in streptomycetes. Mol Gen Genet. 1989;219:341–348. [Google Scholar]
- 26.Nussbaumer B, Wohlleben W. Identification, isolation and sequencing of the recA gene of Streptomyces lividans TK24. FEMS Microbiol Lett. 1994;118:57–64. doi: 10.1111/j.1574-6968.1994.tb06803.x. [DOI] [PubMed] [Google Scholar]
- 27.Papavinasasundaram K G, Movahedzadeh F, Keer J T, Stoker N G, Colston M J, Davis E O. Mycobacterial recA is cotranscribed with a potential regulatory gene called recX. Mol Microbiol. 1997;24:141–153. doi: 10.1046/j.1365-2958.1997.3441697.x. [DOI] [PubMed] [Google Scholar]
- 28.Redenbach M, Kieser H M, Denapaite D, Eichner A, Cullum J, Kinashi H, Hopwood D A. A set of ordered cosmids and a detailed genetic and physical map for the 8 Mb Streptomyces coelicolor A3(2) chromosome. Mol Microbiol. 1996;21:77–96. doi: 10.1046/j.1365-2958.1996.6191336.x. [DOI] [PubMed] [Google Scholar]
- 29.Reiss B, Klemm M, Kosak H, Schell J. RecA protein stimulates homologous recombination in plants. Proc Natl Acad Sci USA. 1996;93:3094–3098. doi: 10.1073/pnas.93.7.3094. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Sambrook J, Fritsch E F, Maniatis T. Molecular cloning: a laboratory manual. Cold Spring Harbor, N.Y: Cold Spring Harbor Laboratory Press; 1989. [Google Scholar]
- 31.Scherbakova O G, Lanzov V A, Ogawa H, Filatov M V. Overexpression of bacterial RecA protein stimulates homologous recombination in somatic mammalian cells. Mutat Res. 2000;459:65–71. doi: 10.1016/s0921-8777(99)00059-2. [DOI] [PubMed] [Google Scholar]
- 32.Story R M, Weber I T, Steit T A. The structure of the E. coli RecA protein monomer and polymer. Nature. 1992;355:318–325. doi: 10.1038/355318a0. [DOI] [PubMed] [Google Scholar]
- 33.Stumpp T, Wilms B, Altenbuchner J. Ein neues, L-Rhamnose-induzierbares Expressionssystem für Escherichia coli. BioSpektrum. 2000;6:33–36. [Google Scholar]
- 34.Sutton M D, Smith B T, Godoy V G, Walker G C. The SOS response: recent insights into umuDC-dependent mutagenesis and DNA damage tolerance. Annu Rev Genet. 2000;34:479–497. doi: 10.1146/annurev.genet.34.1.479. [DOI] [PubMed] [Google Scholar]
- 35.Tan H R, Yang H H, Tian Y Q, Wu W, Whatling C A, Chamberlin L C, Buttner M J, Nodwell J, Chater K F. The Streptomyces coelicolor sporulation-specific ςWhiG form of RNA polymerase transcribes a gene encoding a ProX-like protein that is dispensable for sporulation. Gene. 1998;212:137–146. doi: 10.1016/s0378-1119(98)00152-8. [DOI] [PubMed] [Google Scholar]
- 36.Vierling S, Weber T, Wohlleben W, Muth G. Transcriptional and mutational analyses of the Streptomyces lividans recX gene and its interference with RecA activity. J Bacteriol. 2000;182:4005–4011. doi: 10.1128/jb.182.14.4005-4011.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Volff J-N, Vandewiele D, Simonet J-M, Decaris B. Stimulation of genetic instability in Streptomyces ambofaciens ATCC 23877 by antibiotics that interact with DNA gyrase. J Gen Microbiol. 1993;139:2551–2558. doi: 10.1099/00221287-139-11-2551. [DOI] [PubMed] [Google Scholar]
- 38.Volff J N, Altenbuchner J. Influence of disruption of the recA gene on genetic instability and genome rearrangement in Streptomyces lividans. J Bacteriol. 1997;179:2440–2445. doi: 10.1128/jb.179.7.2440-2445.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Volff J N, Altenbuchner J. Genetic instability of the Streptomyces chromosome. Mol Microbiol. 1998;27:239–246. doi: 10.1046/j.1365-2958.1998.00652.x. [DOI] [PubMed] [Google Scholar]
- 40.Winterling K W, Chafin D, Hayes J J, Sun J L A S, Yasbin R E, Woodgate R. The Bacillus subtilis DinR binding site: redefinition of the consensus sequence. J Bacteriol. 1998;180:2201–2211. doi: 10.1128/jb.180.8.2201-2211.1998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Winterling K W, Levine A S, Yasbin R E, Woodgate R. Characterization of DinR, the Bacillus subtilis SOS repressor. J Bacteriol. 1997;179:1698–1703. doi: 10.1128/jb.179.5.1698-1703.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Wohlleben W, Muth G. Streptomyces plasmid vectors. In: Hardy K G, editor. Plasmids—a practical approach. 2nd ed. New York, N.Y: Oxford University Press; 1993. pp. 147–175. [Google Scholar]
- 43.Yanish-Perron C R, Viera J, Messing J. Improved M13 phage cloning vectors and host strains: nucleotide sequences of the M13mp18 and pUC19 vectors. Gene. 1985;33:103–119. doi: 10.1016/0378-1119(85)90120-9. [DOI] [PubMed] [Google Scholar]
- 44.Yao W, Vining L C. Cloning and sequence analysis of a recA-like gene from Streptomyces venezuelae ISP5230. FEMS Microbiol Lett. 1994;118:51–56. doi: 10.1111/j.1574-6968.1994.tb06802.x. [DOI] [PubMed] [Google Scholar]