Abstract
Spontaneous, erythromycin-resistant mutants of Thermus thermophilus IB-21 were isolated and found to carry the mutation A2058G in one of two 23S rRNA operons. The heterozygosity of these mutants indicates that A2058G confers a dominant or codominant phenotype in this organism. This mutation provides a valuable tool for the genetic manipulation of the 23S rRNA genes of Thermus.
Genetic analysis of rRNAs and ribosomal proteins has been an essential tool in elucidating the mechanism of protein synthesis. While numerous ribosomal protein and rRNA mutants have been isolated, none has been examined structurally in atomic detail. This is primarily due to the inadequacy of ribosomes from genetically tractable organisms such as Escherichia coli for forming crystals that diffract X-rays at sufficient resolution. Ribosomes suitable for X-ray crystallographic analysis have thus far been obtained exclusively from thermophiles (4, 23, 33) or halophiles (1). Regrettably, only limited effort has been devoted to studying the ribosomes of these organisms using genetic approaches (see, however, reference 26). Determining the structures of mutant ribosomes of such an organism (an organism producing ribosomes suitable for X-ray crystallographic analysis) should define conformational perturbations that result in specific functional consequences. We have, therefore, initiated a genetic dissection of ribosomes of the extreme thermophile Thermus thermophilus. Despite the promise of Thermus as a genetic system (16, 21), the lack of selectable genetic markers such as transposons or antibiotic resistance mutations makes the mapping of mutations to ribosomal protein and RNA genes problematic. Here we report the isolation of a 23S rRNA mutation conferring resistance to the antibiotics erythromycin and lincomycin. This selectable genetic marker will facilitate the identification, by transformation, of 23S rRNA mutations affecting 50S subunit structure and function.
Thermus spp. have an unusual rRNA gene composition of two 16S rRNA genes unlinked from two 23S rRNA-5S rRNA-tRNAGly operons (3, 12, 13). Thus, rRNA mutations are potentially obtainable by direct selection if they exhibit a dominant or codominant phenotype or if gene conversion occurs at a sufficiently high frequency. For these experiments we chose the halotolerant strain T. thermophilus IB-21 (ATCC 43615). Although the strain was originally described as Thermus sp. strain IB-21 (17), the designation T. thermophilus is indicated by DNA-DNA hybridization experiments (32) and our own DNA sequence analysis (S. Gregory, J. H. D. Cate, and A. E. Dahlberg, unpublished results).
Isolation of spontaneous erythromycin-resistant mutants.
We conducted a search for erythromycin resistance mutations in 23S rRNA which have been isolated for a number of organisms with multiple rRNA operons (reviewed in reference 30). In several instances, including in the cases of Streptomyces ambofaciens (22), Helicobacter pylori (29), and Streptococcus pneumoniae (27), heterozygous mutants indicate that this mutation is dominant or codominant, as has been previously demonstrated using multicopy plasmid-encoded rrn operons in E. coli (9, 24, 31). Spontaneous, erythromycin-resistant mutants were selected by plating approximately 109 cells from an overnight culture grown at 72.5°C in medium 162 (7) (supplemented with 0.25% tryptone and 0.25% yeast extract) onto plates of the same medium solidified with 2.8% agar (Difco) and containing 100 μg of erythromycin per ml. After 3 days of incubation at 72.5°C, mutants appeared at a frequency of approximately 10−7 and were purified by restreaking once onto medium 162-erythromycin plates and a second time onto antibiotic-free plates. Six resistant isolates, designated HG48, HG49, HG50, HG51, HG52, and HG53, respectively, were kept for analysis.
Identification of a 23S rRNA mutation.
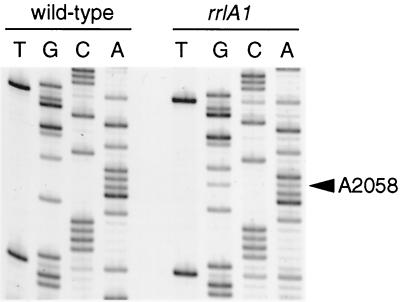
The most frequently occurring macrolide resistance mutations occur at A2058 of 23S rRNA (30), which is footprinted by macrolide, lincosamide, and streptogramin B antibiotics in chemical probing experiments (10, 20). We therefore amplified a segment of the T. thermophilus 23S rRNA genes extending from positions 1660 to 2293 (according to E. coli 23S rRNA numbering) by PCR using Taq DNA polymerase (Promega) and oligonucleotide primers Tth23SD (5′-GGACCTTTGGGCGCCTCCGTTACC-3′) and Tth23SE (5′-CGCCAAGGAACTCTGCAAGTTGGC-3′) (both primers from Operon Technologies) based on the T. thermophilus HB8 23S rRNA sequence (14). Direct sequencing of PCR products revealed that three of the six isolates (HG48, HG50, and HG53) carry the mutation A2058G (numbered according to E. coli 23S rRNA numbering). Moreover, concurrent terminations in the A and G lanes at the position corresponding to position 2058 indicated that all three isolates are heterozygous (Fig. 1).
FIG. 1.
Sequencing of PCR-amplified 23S rRNA operons in the region of A2058. Sequences from wild-type T. thermophilus IB-21 and from the rrlA1 mutant HG48. The presence of mixed termination products at position A2058 indicates that this mutant is heterozygous for the rrlA1 mutation.
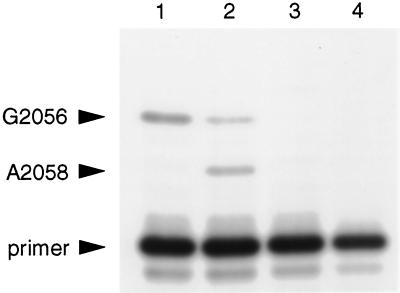
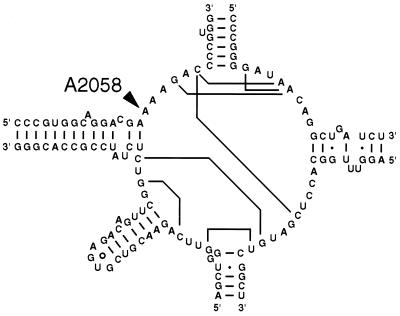
Strong-stop primer extension analysis (25) of total RNA from the A2058G mutants using the oligonucleotide primer Tth2058RT (5′-CAGTAAAGCTCCACGGGGTC-3′), complementary to positions G2061 to G2080, confirmed the presence of both mutant and wild-type 23S rRNA (Fig. 2), with mutant RNA comprising between 50 and 60% of the total 23S rRNA in the three mutants. We have designated the 23S rRNA gene bearing the A2058G mutation from one of the isolates (HG48) rrlA and the wild-type 23S rRNA gene rrlB (these designations, though conforming to standard nomenclature [2, 8], were assigned arbitrarily). The A2058G allele has been designated rrlA1. The site of this mutation in the T. thermophilus 23S rRNA secondary structure (11) is shown in Fig. 3.
FIG. 2.
Primer extension analysis of total RNA from wild-type T. thermophilus IB-21 and from the rrlA1 mutant HG48 (25). Lane 1, extension from wild-type RNA using dATP, dGTP, dTTP, and ddCTP; lane 2, extension from RNA from HG48 using dATP, dGTP, dTTP, and ddCTP; lane 3, extension from wild-type RNA using four deoxynucleoside triphosphates (dNTPs) and no ddNTPs; lane 4, extension without RNA using four dNTPs. The positions of G2056 and A2058 and of an unextended primer are indicated. The presence of A2058G mutant 23S rRNA is apparent in lane 2.
FIG. 3.
Secondary structure of the central loop of domain V of T. thermophilus HB8 23S rRNA (11) and the site of the A2058G erythromycin resistance mutation.
The doubling times of both wild-type IB-21 and the rrlA1 mutant HG48 were determined to be approximately 48 min, whereas the doubling time of HG48 in the presence of 100 μg of erythromycin per ml was about 81 min. This increase in doubling time is consistent with roughly 60% of the ribosomes carrying the resistance mutation. Some excess of mutant ribosomes in cells grown in erythromycin is not unexpected given the drug's property of inhibiting assembly of sensitive 50S subunits (6).
Macrolide-resistant mutants of H. pylori have been shown to contain A2058G in one or both 23S rRNA-5S rRNA operons (29), indicating that homozygosity can result from either gene conversion or an independent mutational event at A2058 in the second operon. Attempts to obtain second-step homozygous rrlA rrlB double mutants of T. thermophilus by a second round of selection on higher levels of erythromycin (400, 600, and 800 μg/ml) were unsuccessful. This suggests either that gene conversion to homozygosity is rare or that such events do not generate significantly higher levels of resistance. We prefer the latter explanation, since Thermus spp. are proficient at homologous recombination (16) and because rrlB mutations are expected to appear spontaneously (at a frequency similar to that at which rrlA mutations appear) in a second round of selection.
Cross-resistance to lincomycin.
The A2058G mutation has been shown to confer resistance to clindamycin (a lincosamide) in vitro to reconstituted 50S subunits of Thermus aquaticus (15) and has been demonstrated to confer resistance to macrolide, lincosamide, and streptogramin B antibiotics in vivo to several organisms (30). Not unexpectedly, we found that it confers high-level cross-resistance to lincomycin in vivo to T. thermophilus. Levels of drug resistance were determined by streaking overnight cultures for single colonies on medium 162 plates containing closely spaced concentrations of either erythromycin or lincomycin. Wild-type T. thermophilus IB-21 is not highly sensitive to erythromycin and was inhibited at an antibiotic concentration of only 50 μg/ml. The rrlA1 mutants are only weakly resistant to erythromycin and were inhibited by an antibiotic concentration of 400 μg/ml. In contrast, these mutants exhibited high-level resistance to lincomycin. The MIC for the wild-type strain was found to be only 2 μg/ml, whereas rrlA1 mutants were resistant to lincomycin at concentrations higher than 800 μg/ml.
The three remaining mutants (HG49, HG51, and HG52), with an unidentified mutation of a gene tentatively designated eryA, behaved uniformly with regard to antibiotic resistance phenotypes. They are weakly resistant to erythromycin (completely inhibited by an antibiotic concentration of 400 μg/ml) and only very weakly resistant to lincomycin, showing marginal inhibition at an antibiotic concentration of 10 μg/ml and complete inhibition at a antibiotic concentration of 40 μg/ml. They are thus phenotypically distinct from the rrlA1 mutants. Sequencing of several candidate sites within rrlA(B) and within rplD and rplV (encoding ribosomal proteins L4 and L22, respectively [see reference 5]) did not reveal the nature of the mutation. While a ribosomal mutation has not been excluded, it is also quite possible that eryA affects antibiotic uptake.
Utility of the A2058G mutation.
We have demonstrated that it is possible to obtain 23S rRNA mutants of T. thermophilus exhibiting a dominant or codominant antibiotic-resistance phenotype, despite the presence of two 23S rRNA gene copies. The principle value of this mutation as a selectable genetic marker is as a tool for the identification of other 23S rRNA mutations of interest, such as ram mutations, or other mutations producing a functional defect for which there is a genetic selection. Such mutations could be mapped genetically to rrlA by transformation to erythromycin resistance (using DNA from the erythromycin-resistant mutant) and screening for loss of the original phenotype. Alternatively, mutations in cis to A2058G could be enriched by transforming a wild-type recipient with DNA from pooled mutants selected in the rrlA1 background, simultaneously selecting for both phenotypes. Finally, site-directed mutations may be introduced into the chromosomal rRNA operons by homologous recombination using the rrlA1 mutation as a selectable marker. Such an approach has been used with Halobacterium halobium (19). To facilitate such experiments as well as to obtain strains producing uniform populations of mutant ribosomes for structural studies, a strain of T. thermophilus bearing a deletion of one of the 23S rRNA-5S rRNA-tRNAGly operons can be constructed using the same methodology employed to delete the Thermus genes leuB (28), slpA (18), and rpsQ encoding ribosomal protein S17 (26).
Acknowledgments
This work was supported by a grant (GM19756) from the National Institutes of Health to A.E.D. and a grant from the Whitehead Institute and a Searle Scholar Award to J.H.D.C.
The authors are grateful to Michael O'Connor, Jill Thompson, and Mark Bayfield for critical reading of the manuscript, to Sun-Thorn Pond-Tor for technical assistance, and to George Q. Pennabble for insight. S.T.G. gives special thanks to Elaine Fredrick and Hanako Gregory.
REFERENCES
- 1.Ban N, Nissen P, Hansen J, Moore P B, Steitz T A. The complete atomic structure of the large ribosomal subunit at 2.4 A resolution. Science. 2000;289:905–920. doi: 10.1126/science.289.5481.905. [DOI] [PubMed] [Google Scholar]
- 2.Berlyn M K. Linkage map of Escherichia coli K-12, edition 10: the traditional map. Microbiol Mol Biol Rev. 1998;62:814–984. doi: 10.1128/mmbr.62.3.814-984.1998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Borges K M, Berquist P L. Genomic restriction map of the extreme thermophilic bacterium Thermus thermophilus HB8. J Bacteriol. 1993;175:103–110. doi: 10.1128/jb.175.1.103-110.1993. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Cate J H, Yusopov M M, Yusopova G Z, Earnest T N, Noller H F. X-ray crystal structures of 70S ribosome functional complexes. Science. 1999;285:2095–2104. doi: 10.1126/science.285.5436.2095. [DOI] [PubMed] [Google Scholar]
- 5.Chittum H S, Champney W S. Ribosomal protein gene sequence changes in erythromycin-resistant mutants of Escherichia coli. J Bacteriol. 1994;176:6192–6198. doi: 10.1128/jb.176.20.6192-6198.1994. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Chittum H S, Champney W S. Erythromycin inhibits the assembly of the large ribosomal subunit in growing Escherichia coli cells. Curr Microbiol. 1995;30:273–279. doi: 10.1007/BF00295501. [DOI] [PubMed] [Google Scholar]
- 7.Degryse E, Glansdorff N, Pierard A. A comparative analysis of extreme thermophilic bacteria belonging to the genus Thermus. Arch Microbiol. 1978;117:189–196. doi: 10.1007/BF00402307. [DOI] [PubMed] [Google Scholar]
- 8.Demerec M, Adelberg E A, Clark A J, Hartman P E. A proposal for a uniform nomenclature in bacterial genetics. Genetics. 1966;54:61–76. doi: 10.1093/genetics/54.1.61. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Douthwaite S. Functional interactions within 23S rRNA involving the peptidyl transferase center. J Bacteriol. 1992;174:1333–1338. doi: 10.1128/jb.174.4.1333-1338.1992. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Douthwaite S. Interaction of the antibiotics clindamycin and lincomycin with Escherichia coli 23S ribosomal RNA. Nucleic Acids Res. 1992;20:4717–4720. doi: 10.1093/nar/20.18.4717. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Gutell R R, Gray M W, Schnare M N. A compilation of large subunit (23S and 23S-like) ribosomal RNA structures: 1993. Nucleic Acids Res. 1993;21:3055–3074. doi: 10.1093/nar/21.13.3055. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Hartmann R K, Ulbrich N, Erdmann V A. An unusual rRNA operon constellation: in Thermus thermophilus HB8 the 23S/5S rRNA operon is a separate entity from the 16S rRNA operon. Biochimie. 1987;69:1097–1104. doi: 10.1016/0300-9084(87)90009-5. [DOI] [PubMed] [Google Scholar]
- 13.Hartmann R K, Erdman V A. Thermus thermophilus 16S rRNA is transcribed from an isolated transcription unit. J Bacteriol. 1989;171:2933–2941. doi: 10.1128/jb.171.6.2933-2941.1989. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Hopfl P, Ulrich N, Hartmann R K, Ludwig W, Schleifer K H. Complete nucleotide sequence of a 23S ribosomal RNA gene from Thermus thermophilus HB8. Nucleic Acids Res. 1988;16:9043. doi: 10.1093/nar/16.18.9043. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Khaitovich P, Tenson T, Kloss P, Mankin A S. Reconstitution of functionally active Thermus aquaticus large ribosomal subunits with in vitro-transcribed rRNA. Biochemistry. 1999;38:1780–1788. doi: 10.1021/bi9822473. [DOI] [PubMed] [Google Scholar]
- 16.Koyama Y, Hoshino T, Tomizuka N, Furukawa K. Genetic transformation of the extreme thermophile Thermus thermophilus and of other Thermus spp. J Bacteriol. 1986;166:338–340. doi: 10.1128/jb.166.1.338-340.1986. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Kristjansson J K, Hreggvidsson G O, Alfredsson G A. Isolation of halotolerant Thermus spp. from submarine hot springs in Iceland. Appl Environ Microbiol. 1986;52:1313–1316. doi: 10.1128/aem.52.6.1313-1316.1986. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Lasa I, Caston J R, Fernandez-Herrero L A, de Pedro M A, Berenguer J. Insertional mutagenesis in the extreme thermophilic eubacteria Thermus thermophilus HB8. Mol Microbiol. 1992;6:1555–1564. doi: 10.1111/j.1365-2958.1992.tb00877.x. [DOI] [PubMed] [Google Scholar]
- 19.Mankin A S, Zyrianova I M, Kagramanova V K, Garrett R A. Introducing mutations into the single-copy chromosomal 23S rRNA gene of the archaeon Halobacterium halobium by using an rRNA operon-based transformation system. Proc Natl Acad Sci USA. 1992;89:6535–6539. doi: 10.1073/pnas.89.14.6535. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Moazed D, Noller H F. Chloramphenicol, erythromycin, carbomycin and vernamycin B protect overlapping sites in the peptidyl transferase region of 23S ribosomal RNA. Biochimie. 1987;69:879–884. doi: 10.1016/0300-9084(87)90215-x. [DOI] [PubMed] [Google Scholar]
- 21.Oshima T. Genes and genetic manipulation in Thermus thermophilus. In: Sharp R, Williams R, editors. Thermus species. New York, N.Y: Plenum Press; 1995. pp. 185–205. [Google Scholar]
- 22.Pernodet J, Boccard F, Alegre M, Blondelet-Rouault M, Guerineau M. Resistance to macrolides, lincosamides and streptogramin B antibiotics due to a mutation in an rRNA operon of Streptomyces ambofaciens. EMBO J. 1988;7:277–282. doi: 10.1002/j.1460-2075.1988.tb02810.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Schluenzen F, Tocilj A, Zarivach R, Harms J, Gluehmann M, Janell D, Bashan A, Bartels H, Agmon I, Franceschi F, Yonath A. Structure of functionally activated small ribosomal subunit at 3.3 angstroms resolution. Cell. 2000;102:615–623. doi: 10.1016/s0092-8674(00)00084-2. [DOI] [PubMed] [Google Scholar]
- 24.Sigmund C D, Ettayebi M, Morgan E A. Antibiotic resistance mutations in 16S and 23S ribosomal RNA genes of Escherichia coli. Nucleic Acids Res. 1984;12:4653–4663. doi: 10.1093/nar/12.11.4653. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Sigmund C D, Ettayebi M, Borden A, Morgan E A. Antibiotic resistance mutations in ribosomal RNA genes of Escherichia coli. Methods Enzymol. 1988;164:673–690. doi: 10.1016/s0076-6879(88)64077-8. [DOI] [PubMed] [Google Scholar]
- 26.Simitsopoulou M, Avila H, Franceschi F. Ribosomal gene disruption in the extreme thermophile Thermus thermophilus HB8. Eur J Biochem. 1999;266:524–532. doi: 10.1046/j.1432-1327.1999.00887.x. [DOI] [PubMed] [Google Scholar]
- 27.Tait-Kamradt A, Davies T, Cronan M, Jacobs M R, Appelbaum P C, Sutcliffe J. Mutations in 23S rRNA and ribosomal protein L4 account for resistance in pneumococcal strains selected in vitro by macrolide passage. Antimicrob Agents Chemother. 2000;44:2118–2125. doi: 10.1128/aac.44.8.2118-2125.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Tamakoshi M, Yaoi T, Oshima T, Yamagishi A. An efficient gene replacement and deletion system for an extreme thermophile, Thermus thermophilus. FEMS Microbiol Lett. 1999;173:431–437. doi: 10.1111/j.1574-6968.1999.tb13535.x. [DOI] [PubMed] [Google Scholar]
- 29.Versalovic J, Shortridge D, Kibler K, Griffy M V, Beyer J, Flamm R K, Tanaka S K, Graham D Y, Go M F. Mutations in 23S rRNA are associated with clarithromycin resistance in Helicobacter pylori. Antimicrob Agents Chemother. 1996;40:477–480. doi: 10.1128/aac.40.2.477. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Vester B, Douthwaite S. Macrolide resistance conferred by base substitutions in 23S rRNA. Antimicrob Agents Chemother. 2001;45:1–12. doi: 10.1128/AAC.45.1.1-12.2001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Vester B, Garrett R A. A plasmid-encoded and site-directed mutation in Escherichia coli 23S RNA that confers resistance to erythromycin: implications for the mechanism of action of erythromycin. Biochimie. 1987;69:891–900. doi: 10.1016/0300-9084(87)90217-3. [DOI] [PubMed] [Google Scholar]
- 32.Williams R A, Smith K E, Welch S G, Micallef J, Sharp R J. DNA relatedness of Thermus strains, description of Thermus brockianus sp. nov., and proposal to reestablish Thermus thermophilus (Oshima and Imahori) Int J Syst Bacteriol. 1995;45:495–499. doi: 10.1099/00207713-45-3-495. [DOI] [PubMed] [Google Scholar]
- 33.Wimberly B T, Brodersen D E, Clemons W M, Morgan-Warren R J, Carter A P, Vonrhein C, Hartsch T, Ramakrishnan V. The structure of the 30S ribosomal subunit. Nature. 2000;407:327–339. doi: 10.1038/35030006. [DOI] [PubMed] [Google Scholar]