Loss-of-function alterations in LZTR1, a Cullin-3 RING E3 ubiquitin ligase adapter, and impaired ubiquitin-mediated degradation of RAS proteins via mutations in RIT1 drive clonal hematopoietic transformation.
Abstract
Recently, screens for mediators of resistance to FLT3 and ABL kinase inhibitors in leukemia resulted in the discovery of LZTR1 as an adapter of a Cullin-3 RING E3 ubiquitin ligase complex responsible for the degradation of RAS GTPases. In parallel, dysregulated LZTR1 expression via aberrant splicing and mutations was identified in clonal hematopoietic conditions. Here we identify that loss of LZTR1, or leukemia-associated mutants in the LZTR1 substrate and RAS GTPase RIT1 that escape degradation, drives hematopoietic stem cell (HSC) expansion and leukemia in vivo. Although RIT1 stabilization was sufficient to drive hematopoietic transformation, transformation mediated by LZTR1 loss required MRAS. Proteolysis targeting chimeras (PROTAC) against RAS or reduction of GTP-loaded RAS overcomes LZTR1 loss-mediated resistance to FLT3 inhibitors. These data reveal proteolysis of noncanonical RAS proteins as novel regulators of HSC self-renewal, define the function of RIT1 and LZTR1 mutations in leukemia, and identify means to overcome drug resistance due to LZTR1 downregulation.
Significance:
Here we identify that impairing proteolysis of the noncanonical RAS GTPases RIT1 and MRAS via LZTR1 downregulation or leukemia-associated mutations stabilizing RIT1 enhances MAP kinase activation and drives leukemogenesis. Reducing the abundance of GTP-bound KRAS and NRAS overcomes the resistance to FLT3 kinase inhibitors associated with LZTR1 downregulation in leukemia.
This article is highlighted in the In This Issue feature, p. 2221
INTRODUCTION
Oncogenic mutations in canonical RAS GTPases (NRAS, HRAS, and KRAS) are among the most common genetic events in cancer, and there are intense efforts to develop small-molecule inhibitors of individual oncogenic RAS alleles. In addition to somatic mutations, germline variants in these genes can be found in a class of neurodevelopmental disorders, termed RASopathies, characterized by dysmorphic features, cardiovascular and lymphatic abnormalities, and predisposition to neoplasia. Recently, forward screens of genes mediating resistance to ABL kinase and FLT3 inhibitors identified leucine zipper-like transcriptional regulator 1 (LZTR1) as a critical mediator of sensitivity to inhibitors of a number of signaling pathways in leukemia (1, 2). This finding, along with several other recent studies (3, 4), resulted in the discovery that LZTR1 serves to regulate the levels of different canonical and noncanonical RAS GTPase proteins. LZTR1 acts as a substrate adapter for the RING E3 ubiquitin ligase Cullin-3 (CRL3) and has been proposed to mediate ubiquitination and proteasomal degradation of a variety of RAS proteins, including NRAS, KRAS, HRAS, MRAS, and RIT1 (1, 3, 4).
Consistent with the newly discovered mechanistic role of LZTR1, loss-of-function germline variants in LZTR1 have been identified in individuals with Noonan syndrome, the most common RASopathy (5). In addition, LZTR1 somatic mutations have been found in clonal hematopoiesis (6), and recent work has identified that LZTR1 is misspliced and consequently downregulated in myeloid neoplasms bearing mutations in the RNA splicing factor ZRSR2 (7). In addition, mutations in RIT1, which encodes one of LZTR1's noncanonical RAS GTPase substrates, were identified in myeloid neoplasms (8) and in Noonan syndrome (9). Distinct from cancer-associated mutations in KRAS or NRAS, which affect GTPase cycling, leukemia-associated mutations in RIT1 mostly allow the RIT1 oncoprotein to escape LZTR1-mediated ubiquitination, accumulate in cells, and drive MAPK activation (3).
Despite the above discoveries, the role of LZTR1 and RIT1 mutations in normal and malignant hematopoiesis remains unknown. In addition, the precise substrates of LZTR1 and consequences of LZTR1-mediated ubiquitination differ across studies and may differ across tissues (1, 3, 4). Although some publications posit that LZTR1 regulates the abundance of the canonical RAS GTPases KRAS, HRAS, and NRAS (1, 10), we have demonstrated that endogenous LZTR1 only binds and regulates the abundance of the noncanonical RAS GTPases MRAS and RIT1 (3, 11). Finally, genetically engineered mouse models demonstrating oncogenic roles of LZTR1 or RIT1 mutations have yet to be published and knowledge of the functional impact of RIT1 mutations is much less well developed than our understanding of the effects of mutations in KRAS or NRAS.
Here, we identify that loss of LZTR1 or expression of leukemia-associated mutations in RIT1 that abrogate LZTR1-mediated RIT1 proteolysis strongly promotes hematopoietic stem cell (HSC) self-renewal, MAP kinase activation, and development of leukemias in vivo. Hematopoietic transformation downstream of LZTR1 required MRAS and SHOC2, the two of which function in a complex to activate RAF kinases (12). Given that activation of the MAPK pathway by the MRAS/SHOC2 complex still requires the function of canonical RAS proteins, targeted degradation of KRAS and NRAS, as well as attenuation of GTP loading via SOS1 inhibition, could overcome the resistance to FLT3 inhibition seen with LZTR1 loss. These data thereby identify an important role for the function of LZTR1 in malignant hematopoiesis and underscore oncogenic RIT1 and LZTR1 mutations as novel bona fide drivers of leukemogenesis.
RESULTS
Loss of Lztr1 Enhances HSC Self-renewal and Drives Leukemia Development
Given recent data suggesting that loss of LZTR1 drives clonal hematopoiesis and myeloid neoplasms, we first functionally evaluated the impact of LZTR1 somatic mutations identified in subjects with clonal hematopoiesis (Fig. 1A; ref. 7). We developed a reporter for RIT1 degradation using a vector recently used to evaluate IKZF3 degradation (13). In this assay, RIT1 cDNA is linked to eGFP and when coexpressed with wild-type (WT) LZTR1 cDNA, RIT1-eGFP fluorescence is downregulated. In contrast, four distinct clonal hematopoiesis–associated LZTR1 somatic variants located in the Kelch repeats required for substrate recognition (T7fsX33, H71Q, S144F, and Q286R) failed to elicit eGFP downregulation to the extent seen with WT LZTR1 (Fig. 1B and C). These data suggest that clonal hematopoiesis associated with LZTR1 variants impair degradation of a well-described LZTR1 substrate.
Figure 1.
Loss of Lztr1 enhances HSC self-renewal and drives leukemia development. A, Lollipop plot of LZTR1 mutations identified in the blood of subjects with clonal hematopoiesis (7). B, Representative histograms of GFP in 293T cells encoding RIT1 fused to eGFP and empty vector (EV), WT LZTR1, or any of four CH-associated LZTR1 mutations. The percentage of eGFP+ cells is indicated. Red dotted line indicates the cutoff for GFP+. C, Quantification of data from B. Mean ± SD. *, P < 0.05; **, P < 0.01; ***, P < 0.001. D, Schema of the allele for Lztr1 constitutive or conditional gene disruption. E, Western blots of Lztr1 and RAS GTPases in E14.5 fetal liver cells from Lztr1 WT or knockout embryos. F, Immunofluorescence images of Lztr1 WT or null embryos for cleaved caspase-3 (green), Ter119 (red), and DAPI (blue) in whole mount (top; WT bar: 100 μm; KO bar: 50 μm) or focused on fetal livers (bottom; bar, 20 μm). G, Schema of experiments evaluating the effects of Lztr1 deletion on fetal hematopoietic cells in vitro and in vivo. H, Colony number in methylcellulose replating assays using 20 × 103 fetal liver hematopoietic cells from Lztr1+/+, Lztr1+/−, and Lztr1−/− fetus. Mean ± SD. n = 3. ***, P < 0.001; ****, P < 0.0001. I, The percentage of CD45.2+ cells in the peripheral blood (PB) of CD45.1+ recipient mice following primary (1°) and secondary (2°) competitive transplantation. n = 5–10. **, P < 0.01; ****, P < 0.0001. J, Box-and-whisker plots of the percentage of total CD45.2+ cells in the bone marrow (BM). n = 5. **, P < 0.01. K, The percentage of CD45.2+ hematopoietic stem and progenitors following 16 weeks of competitive transplantation (as shown in the schema in Fig. 1G). n = 5. *, P < 0.05; **, P < 0.01; ****, P < 0.0001. CMP, common myeloid progenitor; GMP, granulocyte–macrophage progenitor; LT-HSC, long-term HSC; LSK, lineage-negative Sca-1+ c-Kit+; MEP, megakaryocyte–erythroid progenitor; MPP, multipotent progenitor; ST, short-term HSC. L, Kaplan–Meier curve of primary and secondary transplant recipient mice. Pie chart indicates the number and proportion of analyzed mice developing lethal hematopoietic malignancies across both primary and secondary transplantation. Lztr1+/+ primary (1o) recipient mice n = 7, Lztr1−/− primary (1o) recipient mice n = 7, Lztr1−/− secondary (2o) recipient mice n = 30. *, P < 0.05; ****, P < 0.0001. M, Peripheral blood counts of CD45.1+ recipient mice transplanted with CD45.2+Lztr1+/+ or Lztr1−/− fetal liver cells. n = 6–10. **, P < 0.01; ****, P < 0.0001. HGB, hemoglobin; WBC, white blood cell. N, Flow-cytometric analysis of live CD45.2+Lztr1−/− cells from secondary transplant recipient mice developing myeloid neoplasms or B-ALL.
We next evaluated the impact of Lztr1 loss on hematopoiesis in vivo using an Lztr1 allele that allows for constitutive global as well as conditional tissue-specific deletion of Lztr1 (Fig. 1D). Complete loss of Lztr1 was embryonic lethal between embryonic day 17.5 (E17.5) and birth (Supplementary Fig. S1A and S1B; ref. 11). E14.5 Lztr1-null fetal liver cells displayed clear loss of Lztr1 protein, as well as increased expression of the noncanonical RAS GTPases Rit1 and Mras, as well as canonical RAS GTPases Kras and Nras (Fig. 1E). Although the basis for embryonic lethality of Lztr1 knockout (KO) has recently been attributed to cardiovascular defects (11), E16.5 Lztr1-null embryos were characterized by pallor and massive apoptosis in the fetal liver compared with littermate Lztr1 WT counterparts (Fig. 1F; Supplementary Fig. S1C–S1E). Annexin V/DAPI flow cytometry of specific cell types in Lztr1+/+ and Lztr1−/− fetal liver cells at E15.5 revealed clear increases in apoptosis in both the erythroid and myeloid lineage at this stage, with the majority of apoptotic cells being erythroid (Ter119+) cells (Supplementary Fig. S1F and S1G). Moreover, Lztr1 KO was associated with clear dyserythropoiesis with a highly evident increase in circulating nucleated erythroid cells (Supplementary Fig. S1D).
In order to characterize the functional effects of Lztr1 loss on hematopoiesis in a WT Lztr1 microenvironment, we next evaluated the in vitro and in vivo clonogenic capacity of nucleated cells from E14.5 fetal liver from Lztr1 KO and littermate WT control mice (Fig. 1G). Interestingly, this revealed a clear enhanced clonogenic capacity of Lztr1-null hematopoietic cells in vitro as well as a reduction in granulocyte–macrophage progenitor and erythroid colonies derived from Lztr1 KO cells (Fig. 1H; Supplementary Fig. S1H). CD45.2+Lztr1-null cells outcompeted CD45.1+ WT adult bone marrow hematopoietic cells from 6-week-old donors following transplantation into WT 6-week-old CD45.1+ congenic recipient mice (Fig. 1I), an effect seen at the level of mature cells in peripheral blood as well as at the level of HSCs in bone marrow (Fig. 1J nd K). The competitive advantage of Lztr1-null hematopoietic cells persisted in primary and secondary transplantation and exceeded the reconstitution capacity of littermate Lztr1 WT fetal liver cells (Fig. 1I). Hematopoietic cells from Lztr1 KO donor fetal liver cells exhibited increased myeloid chimerism (Supplementary Fig. S2A).
Importantly, recipient animals reconstituted with Lztr1 KO fetal liver cells developed fatal hematologic malignancies characterized by anemia, thrombocytopenia, and increased mature myeloid cells [consistent with a myeloproliferative neoplasm (MPN)] or B-lymphoid cells (Fig. 1L–N; Supplementary Fig. S2B). Animals developing lethal B-cell malignancy had a massively increased frequency of CD19+B220+CD43+ immature B cells, which were monomorphic in appearance, with a near-complete absence of mature (IgD+) B cells (Supplementary Fig. S2C and S2D). Upon serial transplantation into lethally irradiated recipients, recipients developed a lethal hematopoietic malignancy consistent with a mature MPN, whereas the remainder were consistent with an immature B-cell malignancy (Fig. 1L–N; Supplementary Fig. S3A and S3B). Of the mice analyzed, 47% (n = 7) developed an MPN, whereas 53% (n = 8) developed B-cell acute lymphoblastic leukemia (B-ALL) across primary and secondary transplants. Targeted genomic analysis of all protein-coding exons and select introns of 611 cancer-related genes in bone marrow cells from three recipient mice developing myeloid neoplasms following transplantation of Lztr1−/− fetal liver cells revealed rare genomic alterations coexisting with Lztr1 loss in two out of three cases. This included genomic alterations in Braf (mouse Braf K544N corresponding to human K547N, which is a variant of unknown significance) as well as alterations in Zfhx3 and KMT2D (Supplementary Fig. S3C). Overall, these data clearly indicate a role for Lztr1 in regulating normal HSC self-renewal.
Characteristics of RIT1 Mutations in Patients with Hematopoietic Malignancies
Given the impact of Lztr1 loss on Rit1 protein levels in vivo and one prior report describing RIT1 mutations in myeloid leukemia (8), we next evaluated the frequency and characteristics of RIT1 mutations across a cohort of 4,113 clinically annotated and uniformly sequenced patients with myeloid malignancies [acute myeloid leukemia (AML), myelodysplastic syndromes (MDS), MPNs, and MDS/MPN overlap syndromes]. This revealed 40 patients with somatic RIT1 mutations, which were most common in patients with MDS/MPN overlap syndromes (13 of 3,838; 0.34%) and AML (17 of 1,818; 0.94%; Fig. 2A). Similar to lung cancer and Noonan syndrome, RIT1 mutations clustered in a region near the switch II domain of RIT1 (Fig. 2B). The most common co-occurring mutations were in ASXL1 (70%), SRSF2 (40%), SETBP1 (25%), and ZRSR2 (15%), in keeping with mutations identified in MDS/MPN overlap syndromes (Supplementary Fig. S4A and S4B). Similar to other RAS pathway mutations in leukemia (14, 15), RIT1 mutations tended to be subclonal (Fig. 2C). Serial sequencing of RIT1 mutations demonstrated that RIT1 mutations were acquired at the transformation of MDS to AML, and RIT1 subclones were both lost and newly identified at disease relapse (Fig. 2D).
Figure 2.
Characterization of RIT1 mutations in patients with hematologic malignancies. A, Histogram of the number of patients with RIT1 mutations based on myeloid malignancy diagnosis. MF, myelofibrosis; No Dx, no diagnosis. B, Diagram of the location of RIT1 mutations identified. C, Variant allele frequency (VAF) of mutations in RAS GTPases or regulators of RAS-GTP abundance relative to mutations in transcriptional modifiers in patients with myeloid leukemia. D, Fishtail representation plots of VAFs of mutations across a serial genomic analysis of three patients with RIT1 mutations. E, Western blot of LZTR1 and RIT1 following cycloheximide (CHX) treatment of TF-1 cells with or without LZTR1 deletion. sg, single guide. F, Representative histograms of GFP in cells encoding WT or mutant RIT1 fused to eGFP along with empty vector (EV), WT LZTR1, or mutant LZTR1. The percentage of eGFP+ cells is indicated. The red dotted line indicates the cutoff for GFP+. G, Levels of phosphorylated and total MEK1/2 and ERK1/2 as well as RIT1 in TF-1 cells with LZTR1 deletion or expression of EV RIT1 WT or mutant cDNAs. H, Western blot of p-ERK and total ERK levels in 293T cells transfected with increasing amounts of FLAG-RIT1 WT and mutant cDNAs.
RIT1 hotspot mutations have previously been shown in other cellular contexts to result in incomplete degradation of RIT1 (3). Consistent with these data, loss of LZTR1 increased stability and accumulation of RIT1 protein in TF-1 human myeloid leukemia cells (Fig. 2E). Moreover, although LZTR1 could downregulate WT RIT1, LZTR1 failed to degrade the cancer-associated RIT1 M90I mutant (Fig. 2F; Supplementary Fig. S4C). Similarly, either KO of LZTR1 or expression of any of the three most common hotspot mutations in RIT1 (E81Q, F82C, and M90I) increased RIT1 protein and phosphorylation of MEK1/2 and ERK1/2 (Fig. 2G). Transfection of increasing amounts of WT RIT1 cDNA and the RIT1 M90I mutant revealed that the effects of RIT1 mutations on ERK1/2 phosphorylation are dose dependent, and the RIT1 M90I mutation results in the accumulation of RIT1 protein levels and increased ERK1/2 activation relative to WT RIT1 (Fig. 2H). Of note, these experiments also included the RIT1 S35N dominant-negative mutant, demonstrating that RIT1 activity is required for increased ERK1/2 activation (RIT1 S35N is equivalent to a dominant-negative mutation in K/N/HRAS at S17N, which is unable to bind GTP; ref. 16).
TF-1 cells are normally dependent on GM-CSF for cell growth, and KO of LZTR1 or expression of leukemia-associated mutations in RIT1 conferred cytokine-independent growth (Fig. 3A). Moreover, deletion of LZTR1 or expression of RIT1 oncogenic mutations impaired erythroid differentiation of TF-1 cells in the setting of erythropoietin (EPO) exposure (Fig. 3B and C). These data demonstrate that RIT1 mutations are recurrent in myeloid neoplasms and that RIT1 hotspot mutations enhance RIT1 protein abundance, alter hematopoietic differentiation, and stimulate MAPK signaling in a manner analogous to that seen with LZTR1 loss.
Figure 3.
Convergent effects of Lztr1 deletion and leukemia-associated mutations in the Lztr1 substrate Rit1 in normal hematopoiesis. A, Growth of cells from G following cytokine depletion. Mean ± SD. n = 3. **, P < 0.01; ***, P < 0.001. sg, single guide. B, Left, representative FACS histograms of glycophorin A levels in TF-1 cells ± LZTR1 deletion in the absence or presence of erythropoietin (EPO; 2 IU/mL × 4 days). Right, enumeration of the median fluorescence intensity (MFI) of glycophorin A. n = 3. ***, P < 0.001. C, As in B but in TF-1 cells with RIT1 mutants. n = 3. ****, P < 0.0001. D, Schema for competitive transplantation of hematopoietic cells with postnatal deletion of Lztr1 or expression of Rit1M90I/WT. pIpC, polyinosinic:polycytidylic acid. E, Peripheral blood (PB) chimerism of CD45.1 recipient mice from D. n = 6–9. ns, not significant; *, P < 0.05; **, P < 0.01; ****, P < 0.0001. F, percentage of CD45.2+ hematopoietic stem and progenitor cells in the bone marrow (BM) of CD45.1 recipient mice from D at 16 weeks after transplantation. n = 6–9. *, P < 0.05. For box-and-whisker plots, bar indicates median; box edges, first and third quartile values; and whisker edges, minimum and maximum values. CMP, common myeloid progenitor; GMP, granulocyte–macrophage progenitor; LT-HSC, long-term HSC; LSK, lineage-negative Sca-1+ c-Kit+; MEP, megakaryocyte–erythroid progenitor; MPP, multipotent progenitor; ST, short-term HSC. G, Uniform manifold approximation and projection (UMAP) dimensionality reduction of 20,536 bone marrow lineage-negative cells from Cre-negative control mice, Mx1-cre Lztr1fl/fl mice, and Mx1-cre Rit1M90I/WT mice. Baso, basophil progenitor; B cell, B-cell progenitor; CD3d+ T cell, CD3d+ T-cell progenitor; CLP, common lymphoid progenitor; Ery, erythroid progenitor; HSPC, hematopoietic stem/progenitor cell; IMP, immature myeloid progenitor; MkP, megakaryocyte progenitor; Mono, monocyte progenitor; Neu, neutrophil/granulocyte progenitor; Neu/Ery, neutrophil/erythroid progenitor; T prog, T-cell progenitor. H, Alluvial plots of key clusters from A, including all differentially expanded or reduced populations in mutant mice compared with controls. Shaded sections represent populations differentially represented in mutant animals compared with WT. I, Violin plots of log-transformed normalized gene expression for genes differentially expressed in the Neu/Ery–1 cluster in mutant animals compared with controls. Superimposed box-and-whisker plots represent median values within the interquartile range (IQR; boxes) and 1.5 × IQR (whiskers).
Convergent Effects of Lztr1 Loss and Leukemia-Associated Mutations in the Lztr1 Substrate Rit1
To evaluate the impact of expressing RIT1 mutations from the endogenous Rit1 allele in vivo and to directly compare the impact of LZTR1 loss to RIT1 accumulation, we evaluated the impact of expressing heterozygous Rit1M90I/WT mutation on adult hematopoiesis using Mx1-cre Rit1M90I/WT conditional knockin mice (Supplementary Fig. S4D and S4E). In parallel, we generated Mx1-cre Lztr1fl/fl mice for accurate comparison with Rit1M90I mutant mice and to evaluate the impact of Lztr1 deletion on postnatal adult hematopoiesis (Supplementary Fig. S5A and S5B).
One common characteristic of expressing oncogenic NRAS and KRAS alleles in hematopoietic cells is that these mutations confer GM-CSF hypersensitivity to HSCs (17, 18). Similarly, expression of Rit1M90I/WT or deletion of Lztr1 enhanced GM-CSF colony formation compared with littermate Mx1-cre control cells (Supplementary Fig. S5C and S5D). Extensive prior studies of the impact of oncogenic mutations in RAS alleles in mice have identified a varying impact of RAS activation on HSC frequency and self-renewal (17–19). We therefore sought to evaluate the impact of Lztr1 loss or Rit1M90I/WT mutation on HSC frequency and self-renewal by in vivo competition assays (Fig. 3D). This revealed that either loss of Lztr1 or expression of Rit1M90I/WT resulted in a clear increase in peripheral blood chimerism, splenomegaly, as well as increased frequencies of hematopoietic stem/progenitor cells (HSPC) in the bone marrow, with a greater frequency of proliferating HSPCs (Fig. 3E and F; Supplementary Fig. S5E–S5G).
To further characterize the impact of Lztr1 deletion versus Rit1M90I mutation on adult hematopoiesis, we performed single-cell RNA sequencing (RNA-seq) on lineage-negative bone marrow cells from 12-week-old Mx1-cre Lztr1fl/fl, Mx1-cre Rit1M90I/+, and Cre-negative control mice 1 month after Cre-mediated recombination. Dimensionality reduction and clustering identified a total of 28 transcriptional distinct clusters (Fig. 3G; Supplementary Fig. S5H), consistent with prior reports (20). Lztr1 deletion at this time point resulted in an increase in monocyte and neutrophil/erythroid precursor cells as well as a reduction in T-cell precursors (Fig. 3H). Interestingly, a similar expansion in neutrophil/erythroid precursor cells was seen in the Rit1 mutant mice. Moreover, neutrophil/erythroid precursor cells across both mutant genotypes were characterized by reduced expression of a number of myeloid tumor suppressor genes including Gata2, Ybx1, Hmgb1, ApoE, Mif, Cdc42, Ikzf1, and Ezh2 (Fig. 3I). Overall, these data identify convergent effects of Lztr1 loss and Rit1 accumulation via the M90I mutation on myeloid skewing and gene expression in myeloid precursor cells.
Rit1 Mutants that Escape Proteolysis Drive Development of Myeloid Neoplasms In Vivo
To date, genetically engineered primary animal models demonstrating a transforming capacity of RIT1 mutations have not been presented. We found that activation of the Rit1M90I/WT mutation at 6 weeks of age in primary mice resulted in the development of a spectrum of myeloid neoplasms (Fig. 4A). These phenotypes were consistent with MPNs in nearly 30% of mice and mixed MDS/MPN disorders in the remaining ∼70% of mice (Fig. 4B–D). Animals died of a fatal anemia, thrombocytopenia, and leukocytosis, with increased monocytes, neutrophils, and extramedullary hematopoiesis in the lung, liver, and spleen (Fig. 4E; Supplementary Fig. S6A–S6C). The myeloid neoplasm was serially transplantable in sublethally irradiated recipient mice in which Rit1M90I/WT leukemic cells expanded in peripheral blood and bone marrow over time and resulted in a fatality in recipients at a median of 21 weeks with splenomegaly and anemia (Fig. 4F and G; Supplementary Fig. S7A and S7B). The malignant cells in the blood of secondary recipients were nearly entirely myeloid, and there was an expansion of granulocyte–macrophage progenitors in the bone marrow of recipient mice at the time of death (Supplementary Fig. S7C–S7G). Importantly, deletion of Lztr1 in HSPCs also resulted in the development of a lethal hematopoietic malignancy in recipient mice with age (Supplementary Fig. S8A–S8C). Eighty percent of these animals died of a lethal myeloproliferative disorder, whereas the rest died of acute B-cell leukemia (phenotypes similar to those seen with Lztr1 constitutive KO mice; Supplementary Fig. S8A).
Figure 4.
Rit1 M90I/WT mutation drives the development of myeloid neoplasms in vivo. A, Kaplan–Meier curve of primary Mx1-cre Rit1M90I/WT mice following polyinosinic:polycytidylic acid (pIpC) treatment. Venn diagram indicates diagnoses of Rit1M90/+ mice at the time of death and proportion developing MPN and myelodysplasia/MPN (MDS/MPN). n = 12. ***, P < 0.001. Wright–Giemsa stain of peripheral blood (B) and bone marrow (C) cytospins of Mx1-cre control and Mx1-cre Rit1M90I/WT mice at the time of disease onset of the Rit1M90I/WT mice. Red arrows indicate dysplastic erythroid precursors. Green arrows indicate dysplastic neutrophils. Yellow arrow indicates dysplastic myeloid precursors. Two representative animals are shown for the Rit1M90I/WT mice. D, Hematoxylin–eosin stain of bone marrow (yellow arrows indicate dysplastic megakaryocytes; scale bars = 100 μm). Two representative animals are shown for the Rit1M90I/WT mice. E, Box-and-whisker plots of peripheral blood counts of primary Mx1-cre Rit1M90I/WT mice and their age-matched Mx1-cre control mice from A. For box-and-whisker plots, bar indicates median; box edges, first and third quartile values; and whisker edges, minimum and maximum values. n = 12. *, P < 0.05; **, P < 0.01; ***, P < 0.001. WBC, white blood cell. F, The percentage of CD45.2+ cells in peripheral blood of sublethally irradiated CD45.1+ recipient mice following transplantation of Mx1-cre Rit1M90/WT bone marrow cells from the time of disease onset from D. n = 5–10. Mean ± SD. ****, P < 0.001. G, Kaplan–Meier curve of CD45.1 recipient mice from F. n = 5–10. ***, P < 0.001.
Given the age-dependent phenotype of Rit1M90I/WT mice, we evaluated for potential coexisting genomic alterations in the bone marrow of >1-year-old leukemic mutant and littermate WT mice using both whole-exome sequencing of bone marrow cells and skin. This revealed a consistent presence of the heterozygous Rit1M90I/WT mutation and several coexisting genomic alterations in mutant mice but no clear recurrent individual mutation (although some genes were recurrently mutated in the mutant mice, including Kmt2d; Supplementary Fig. S8D and S8E).
Overall, these data identify that leukemia-associated mutations in RIT1, which escape LZTR1-mediated proteolysis, are bona fide drivers of leukemogenesis and confer a clonal advantage to HSCs.
Lztr1-Null Leukemic Cells Depend on MRAS for Transformation
Given the convergent effects of Lztr1 loss and Rit1 accumulation on HSCs, we next sought to determine whether RIT1 was required for the transforming effects of LZTR1 loss on hematopoietic cells. KO of LZTR1 in TF-1 cells, a cell line where LZTR1 loss is transforming (Fig. 3A), resulted in an accumulation of RIT1, MRAS, and KRAS (Supplementary Fig. S9A). To evaluate if RIT1 is required for LZTR1-mediated transformation, we utilized single-guide RNAs (sgRNA) to delete RIT1 alone as well as RIT1 and LZTR1 simultaneously in TF-1 cells (Supplementary Fig. S9B). Although RIT1 deletion reduced the proliferation of LZTR1-null TF-1 cells in cytokine-free media, RIT1 deletion alone was not sufficient to abolish the LZTR1-mediated transformation of TF-1 cells (Supplementary Fig. S9C).
To further evaluate the requirement of RIT1 on the effect of LZTR1 deletion on hematopoiesis, we next developed mice with combined germline deletion of Rit1 and Lztr1 (along with single KO controls). Whole-cell extracts of the E14.5 fetal liver from these mice revealed clear increases in Rit1, Mras, and Kras in Lztr1-null embryos and elimination of Rit1 in Lztr1/Rit1 double-KO embryos (Fig. 5A). Given the consistent effect of Lztr1 deletion on increasing the clonogenic capacity of hematopoietic cells, we evaluated the requirement of Rit1 on the in vivo self-renewal potential of Lztr1 KO hematopoietic cells (Fig. 5B). Although deletion of Rit1 rescued the profound cell death seen in fetal livers with germline Lztr1 KO, Rit1 deletion was only able to partially rescue the enhanced self-renewal of Lztr1-null HSCs (Fig. 5C; Supplementary Fig. S9D–S9F). These data, combined with those from human TF-1 cells in vitro, suggest that the transforming cellular effects of LZTR1 loss on hematopoietic cells are not entirely reliant on RIT1.
Figure 5.
Lztr1-null cells depend on multiple RAS GTPases. A, Western blot of E14.5 fetal liver cells from mice with germline deletion of Lztr1, Rit1, or both Lztr1 and Rit1. B, Schema of experiments evaluating effects of Lztr1 or Rit1 deletion, alone or together, on fetal hematopoietic cells in vivo. C, Peripheral blood chimerism of the experiment in B. n = 10/group. *, P < 0.05; **, P < 0.01; ***, P < 0.001; ****, P < 0.001. For box and whiskers plots, bar indicates median; box edges, first and third quartile values; and whisker edges, minimum and maximum values. D, Schema of positive enrichment custom CRISPR–Cas9 pooled lentiviral screen to identify genes required for cytokine-independent growth following LZTR1 deletion or expression of RIT1 mutations in TF-1 cells. E, Heat map of sgRNAs depleted in RIT1 F82C, RIT1 M90I, or LZTR1 KO TF-1 cells following cytokine depletion. Log2 fold change (FC) is shown. F, Schema of growth competition assay to evaluate effects of RIT1, MRAS, or SHOC2 suppression on LZTR1 WT or KO cells. G, Relative ratio of shRNA-expressing (dsRED+) LZTR1 KO TF-1 cells following culture in doxycycline and removal of GM-CSF. Doxycycline induces expression of dsRED simultaneously with expression of shRNAs targeting Renilla (“shRen,” a negative control), RIT1, MRAS, or SHOC2. H, Schema of the effects of LZTR1 loss on signaling in hematopoietic cells. At baseline, LZTR1 restrains the abundance of multiple RAS GTPases, including RIT1, KRAS, and MRAS (left). Upon receptor tyrosine kinase (RTK) stimulation, RIT1, KRAS, and MRAS exchange GDP for GTP and enable RAF activation of MEK and ERK (middle box). LZTR1 depletion results in the accumulation of RIT1, KRAS, and MRAS, and the resultant cytokine hypersensitivity and transformation of LZTR1-null cells requires MRAS–SHOC2–PP1 activity.
To more systematically dissect genes required downstream of LZTR1 loss, we performed a genome-wide CRISPR screen in Cas9-expressing variants of TF-1 cells with LZTR1 KO or expression of leukemia-associated RIT1 F82C and M90I mutations (Fig. 5D). We used the published Brunello library (21), and cells were infected with a multiplicity of infection of 0.5 while growing in cytokine-free conditions. We then evaluated the sgRNAs that are depleted upon cytokine withdrawal in LZTR1 KO versus RIT1 mutant conditions to understand the determinants that are required for either genetic state. sgRNAs targeting multiple spindle assembly checkpoint-related genes (BUB1 and BUB3) as well as CDK7 were required in the transformation of TF-1 cells following either LZTR1 KO or expression of RIT1 mutants (Fig. 5E; Supplementary Fig. S10A–S10C). These data are consistent with recent data identifying that pathogenic levels of RIT1 silence the spindle assembly checkpoint (22, 23).
In addition to the above, the CRISPR screen also identified that SHOC2 was preferentially required in LZTR1 KO cells over RIT1 mutants (Supplementary Fig. S10B and S10C). This is relevant, as MRAS, a direct interactor of SHOC2, was consistently upregulated in LZTR1 KO normal and malignant hematopoietic cells. Although MRAS shares most regulatory and effector interactions with canonical RAS oncoproteins, unique among RAS proteins, MRAS is also part of a phosphatase holoenzyme when in complex with the scaffolding protein SHOC2 and the phosphatase PP1 (12, 24). This MRAS–SHOC–PP1 complex dephosphorylates an inhibitory site in RAF kinases (S259 in CRAF, S365 in BRAF, and S214 in ARAF), which allows RAF proteins to dimerize and activate downstream signaling. Small hairpin RNA (shRNA) competition assays evaluating whether LZTR1 KO TF-1 cells could survive in cytokine-free conditions in the absence of RIT1, MRAS, or SHOC2 revealed that MRAS and SHOC2 were absolutely required for the transformation of LZTR1 KO cells (Fig. 5F and G; Supplementary Fig. S10D). In these experiments, LZTR1 KO TF-1 cells expressing a construct that simultaneously expresses a shRNA and the fluorescent protein dsRED only upon doxycycline treatment were mixed in an equal ratio with LZTR1 KO cells lacking the shRNA. Then, the ratio of fluorescent cells was evaluated following doxycycline induction and GM-CSF withdrawal (a condition in which LZTR1 KO TF-1 cells normally survive). In this manner, we were able to identify that MRAS and SHOC2 were absolutely required for the growth of LZTR1-null cells in cytokine-free conditions, whereas RIT1 was not. At the same time, deletion of either RIT1 or MRAS partially rescued the impaired erythroid differentiation of TF-1 cells upon LZTR1 loss (Supplementary Fig. S10E and S10F). Finally, although LZTR1 deletion increased ERK activation in TF-1 cells, these cells are still dependent on MEK/ERK signaling as indicated by their sensitivity to pharmacologic MEK1/2 inhibition with cobimetinib (Supplementary Fig. S11A and S11B). Overall, these data identify that although the noncanonical RAS GTPases RIT1 and MRAS are upregulated by LZTR1 loss in hematopoietic cells, MRAS (and its PP1 phosphatase complex members) is specifically required for LZTR1-mediated transformation (Fig. 5H).
Degradation of KRAS and NRAS or SOS1 Inhibition Overcomes LZTR1-Mediated Kinase Inhibitor Resistance
Prior work has shown that deletion of LZTR1 in AML cell lines results in reduced in vitro sensitivity to the FLT3 inhibitor quizartinib (1). More recently, an inverse correlation was identified between LZTR1 expression and sensitivity of FLT3 mutant AML to the multikinase inhibitor sorafenib (2). In fact, based on ex vivo drug experiments using AML patient samples from the BeatAML study (25), LZTR1 expression levels are inversely correlated with sensitivity to the broad class of FDA-approved (gilteritinib) and investigational FLT3 inhibitors in the late stages of clinical development (crenolanib and quizartinib; Fig. 6A). KO of LZTR1 consistently reduced the sensitivity of FLT3 mutant AML cell lines to each of these FLT3 inhibitors in vitro (Supplementary Fig. S11C–S11E) as well as in cell line xenografts in vivo (Fig. 6B and C; Supplementary Fig. S12A and S12B). These latter studies were conducted by treating NOD scid gamma (NSG) mice engrafted with luciferase-labeled LZTR1 WT or KO MOLM-13 cells with vehicle or gilteritinib (at 30 mg/kg/day every 5 days) for 3 weeks.
Figure 6.
LZTR1-mediated drug resistance can be overcome by modulating RAS-GTP abundance. A, Scatter plots of RNA expression levels [shown as normalized reads per kilobase of transcript per million reads mapped (RPKM)] plotted against drug-sensitivity measured ex vivo as the area under the curve (AUC) from AML patient samples harboring FLT3-ITD mutation. n ≥ 111. *, P < 0.05. Pearson correlation coefficients are shown as (r) values. Statistical significance is indicated. B, Schema of the MOLM-13 cell line xenograft experiment. Luciferase-expressing MOLM-13 cells treated with anti-LZTR1 or control sgRNAs were systemically engrafted with 1 × 105 cells/animal via a tail-vein injection after sublethal (225 cGy) irradiation. Seven days later, animals were treated with either vehicle or 30 mg/kg/day of gilteritinib via oral gavage. Animals underwent bioluminescence imaging weekly. C, Kaplan–Meier curve of experiment in B. n = 7–10. **, P < 0.01; ***, P < 0.001. D, Schema of protein-based degrader of RAS proteins. A chimeric protein consisting of a high-affinity target-binding domain for RAS proteins (DARPin K27) is fused to an engineered E3 ligase adapter (SPOP) to confer ubiquitin-mediated degradation of RAS proteins. E, Western blot demonstrating levels of LZTR1 as well as total and GTP-bound KRAS/NRAS/MRAS and RIT1 in MOLM-13 cells ± LZTR1 KO upon doxycycline-mediated induction of the RAS biodegrader (performed in biological triplicate). EV, empty vector. F, Seventy-two-hour cell proliferation assay on parental and LZTR1 KO MOLM-13 cells ± induction of RAS degradation. Cell viability was measured in triplicate using CellTiter-Glo. G, 2D synergy plots using the Zero Interaction Potency (ZIP) model of control sgRNA (“WT”) or anti-LZTR1 sgRNA (“LZTR1 KO”) MOLM-13 cells treated for 72 hours with BI-3406 and/or gilteritinib at various concentrations. Western blot demonstrating p-MEK, p-ERK, and total MEK, ERK, KRAS/NRAS/MRAS, and RIT1 levels (H) as well as RAS-GTP levels (I) in MOLM-13 cells ± LZTR1 KO treated with increasing concentration of DMSO or BI-3406 alone 4 hours after drug treatment.
Given that treatment-emergent activating mutations in NRAS and KRAS have been observed in patients with clinical resistance to FLT3 inhibition, we also evaluated the impact of LZTR1 loss in the context of NRAS mutations. Combining NRAS G12D mutation with LZTR1 deletion revealed that the combination increases ERK activation more than either alteration alone and renders MOLM-13 cells less susceptible to cell killing by gilteritinib or quizartinib in vitro (Supplementary Fig. S12C and S12D). Reduced LZTR1 expression was also associated with resistance to ponatinib (an ABL and multikinase inhibitor with activity against T315I mutant BCR–ABL) and numerous JAK family kinase inhibitors in patient samples (Supplementary Fig. S13A and S13B).
In hopes of identifying means to overcome drug-resistant effects of LZTR1 downregulation, we tested whether concomitant KO of either RIT1 or MRAS could sensitize LZTR1 KO cells to FLT3 inhibition. Although RIT1 loss did not rescue drug resistance in LZTR1-null cells, MRAS deletion partially restored FLT3 inhibitor sensitivity to LZTR1-null cells (Supplementary Fig. S13C–S13G). We next evaluated if degradation of the canonical RAS proteins KRAS and NRAS, which are absolutely required for MAPK activation (26), could alter the sensitivity of LZTR1-null cells to FLT3 inhibition. To accomplish this, we utilized a recently published protein-based degrader (so-called “biodegrader”) of canonical RAS proteins (27). This RAS-targeting biodegrader fuses a high-affinity target-binding domain using designed ankyrin repeat protein (DARPin) K27, specific for canonical RAS proteins, to the E3 ligase CRL3SPOP (Fig. 6D). Intracellular expression of a construct encoding this chimeric protein thereby results in the degradation of total and GTP-bound KRAS and NRAS (Fig. 6E). Interestingly, KRAS/NRAS degradation was able to overcome FLT3 inhibitor resistance resulting from LZTR1 KO (Fig. 6F).
Although drugs to degrade WT canonical RAS proteins are being developed, we also sought to use existing pharmacologic means to overcome LZTR1-mediated FLT3 inhibitor resistance. We therefore also tested the impact of SOS1 inhibition with BI-3406 (28) on response to FLT3 inhibition in these same LZTR1 WT or KO FLT3 mutant AML cells. SOS1 is a guanine exchange factor for RAS that binds and activates GDP-bound RAS family proteins and thereby promotes the exchange of GDP for GTP. SOS1 inhibition promisingly reduced GTP-loaded KRAS and NRAS as well as MAPK activation downstream of LZTR1 loss and improved response of LZTR1-null AML cells to gilteritinib (Fig. 6G–I). In fact, BI-3406 exhibited a synergistic interaction with gilteritinib in MOLM-13 cells upon LZTR1 KO, a finding not seen in the LZTR1 WT setting (Fig. 6G). These data thereby nominate several means to manipulate canonical RAS protein abundance and/or activation to overcome drug-resistant effects of LZTR1 loss.
DISCUSSION
Mutations activating the canonical RAS proteins KRAS and NRAS by altering the abundance of their GTP-bound forms are associated with resistance to FLT3 and IDH1/2 inhibitors and with progression to leukemia in the setting of MDS. Although KRAS and NRAS mutations have been heavily studied in leukemias, the role of LZTR1 or its noncanonical RAS substrates RIT1 and MRAS has been less investigated in this setting. Here we identify that the loss of LZTR1 leads to upregulation of RIT1 and MRAS in hematopoietic cells, drives HSC self-renewal, and promotes malignant transformation of hematopoietic cells in vitro and in vivo. Consistently, leukemia-associated mutations in RIT1 that are resistant to LZTR1-mediated degradation were also sufficient to drive clonal expansion and leukemia development in vivo. Unexpectedly, we find that during hematopoietic transformation, MRAS acts as the critical substrate of LZTR1. Following LZTR1 deletion, the MRAS–SHOC2–PP1 phosphatase complex was required for the transformation and activation of the RAF/MEK/ERK pathway.
The fact that LZTR1 deletion resulted in the increased HSC self-renewal and development of lethal myeloid neoplasms, as well as acute B-cell leukemias, is interesting given that mouse models of oncogenic mutations in canonical RAS proteins do not result in similar phenotypes (17, 29). The differences in hematopoietic phenotypes between LZTR1 loss and oncogenic mutations in canonical RAS proteins are likely related to the fact that the biochemical impact of LZTR1 loss is not equivalent to oncogenic mutations in canonical RAS proteins. Mutations in canonical RAS GTPases (such as KRAS/NRAS/HRAS) increase the proportion of GTP bound to an individual RAS paralog, whereas LZTR1 loss simultaneously upregulates the protein abundance of the noncanonical RAS proteins RIT1 and MRAS. Moreover, downstream effectors of RIT1 are distinct from those of canonical RAS proteins. For example, RIT1 mutations have recently been shown to weaken mitotic fidelity in a manner not recapitulated by oncogenic mutations in canonical RAS proteins (23, 30).
Both Lztr1 deletion and oncogenic Rit1 mutations result in strong enhancement in HSC self-renewal. Overall, these data indicate that Rit1 protein stabilization, due to oncogenic mutations that abrogate its ubiquitin-mediated degradation, is sufficient to induce myeloid transformation. In contrast, Lztr1 deletion causes both myeloid and B-lymphoid neoplasms in mice. The phenotypic differences between Lztr1 loss and Rit1 stabilization strongly suggest that in the hematopoietic lineage, LZTR1 relies on substrates beyond Rit1. Thus, although there are some convergent phenotypic effects of Lztr1 deletion and Rit1 mutation during hematopoiesis, Rit1 mutations alone cannot fully recapitulate the full spectrum of disease phenotypes seen with Lztr1 deletion.
By focusing our studies on a single tissue and cell type and using models of physiologic expression of LZTR1 and RIT1 alterations, the present data address discrepancies in the literature on the substrates of endogenous LZTR1 and the importance of distinct RAS GTPase members on transformation following LZTR1 loss. For example, our data from hematopoietic cells from Lztr1 constitutive KO mice, conditional KO mice, and multiple human leukemia cell lines identify upregulation of the noncanonical RAS proteins RIT1 and MRAS as well as the canonical RAS proteins KRAS and NRAS following LZTR1 deletion. However, although oncogenic mutations stabilizing RIT1 protein abundance have convergent effects with LZTR1 deletion, we also identify a critical requirement for MRAS during transformation. Hence, it is tempting to speculate that the requirements of LZTR1 substrates are likely to depend on different tissue lineages and cellular processes. For instance, the cardiac phenotypes resulting from Lztr1 deletion in mice can be fully rescued by codeleting Rit1 (11), but Lztr1/Rit1 double-KO HSCs still have aberrant self-renewal capacity.
Our study also provides a number of unique genetically engineered mouse models and cell lines of leukemia driven by mutant LZTR1 or RIT1. Given the paucity of cancer models with these alterations available to date, the models developed here will be an important resource for future therapeutic and mechanistic studies of LZTR1 and RIT1 mutations.
It is important to note that LZTR1, RIT1, MRAS, and SHOC2 are mutated not only in myeloid neoplasms but also in RASopathies with germline variants in the same genes. Although RIT1 somatic mutations are relatively rare, demonstrating experimentally that LZTR1 and RIT1 alterations are drivers of leukemogenesis is clinically important given the association of RAS pathway mutations with resistance to a number of targeted therapies used in the treatment of patients with AML. These data suggest that clinical evaluation of RIT1 mutations and LZTR1 alterations or downregulation may have clinical implications in myeloid malignancies.
From a therapeutic perspective, our data reveal that degradation of KRAS and NRAS or attenuating the abundance of GTP-bound KRAS and NRAS may serve as a potential strategy to overcome resistance associated with LZTR1 downregulation. We propose that this therapeutic approach is efficacious given the role of canonical RAS proteins in activating the downstream RAF/MEK/ERK pathway. Selectively degrading canonical RAS proteins, or reducing their GTP loading, attenuates the convergent activation of RAF mediated by RIT1 and MRAS. Currently, the SOS1 inhibitor BI-1701963 is being investigated in phase I clinical trials in solid tumor patients with KRAS mutations in combination with either MEK or KRASG12C inhibitors. However, our data here suggest an additional therapeutic potential for SOS1 inhibition in combination with FLT3 inhibitors. Moreover, our data support continued efforts to develop pharmacologic means to degrade canonical and noncanonical RAS proteins given the strong connection between RAS protein abundance and malignant transformation of hematopoietic cells.
METHODS
Patient Mutational Analyses
Gene mutations in blood or bone marrow specimens were determined at the time of diagnosis using the clinical targeted next-generation sequencing of genes recurrently mutated in AML performed as part of routine clinical care at Dana-Farber Cancer Institute (DFCI; ref. 31). Variants were interpreted for pathogenicity as previously described (15, 32). The DFCI Institutional Review Board approved retrospective deidentified analyses of clinical and genetic data in this cohort.
Lztr1 KO, Rit1M90I Conditional Knockin, and Rit1 KO Mice
All animals were housed at the Memorial Sloan Kettering Cancer Center (MSKCC). All animal procedures were completed in accordance with the Guidelines for the Care and Use of Laboratory Animals and were approved by the Institutional Animal Care and Use Committees at MSKCC. All mouse experiments were performed in accordance with a protocol approved by the MSKCC Institutional Animal Care and Use Committee (11-12-029). Eight-week-old, female CD45.1+ C57BL/6 mice were purchased from The Jackson Laboratory (Stock No: 002014).
Lztr1 KO mice (Lztr1fl/fl) were generated by breeding constitutive Lztr1+/− mice with C57BL/6 mice containing Flp recombinase (The Jackson Laboratory, Stock No: 016226) to remove the lacZ-neo cassette. Lztr1fl/fl and Rit1M90I conditional knockin mice were bred to Mx1-cre mice (The Jackson Laboratory, Stock No: 003556) to generate Mx1-cre Lztr1fl/fl and Mx1-cre Rit1M90I/WT mice. Eight-week-old mice were treated with three doses of polyinosinic:polycytidylic acid (pIpC; 12 mg/kg; GE Healthcare) every other day via intraperitoneal injection to achieve recombination in hematopoietic tissue. Genotyping methods are listed in the Supplementary Methods and primers are in Supplementary Table S1.
RIT1 Degradation Reporter Assay
RIT1 and RIT1 M90I mutant cDNAs were cloned into an enhanced green fluorescent protein (eGFP) degradation reporter (Addgene #74459). LZTR1 WT and mutant cDNAs were cloned into pLenti-EF1a vector (EF1a-IRES-Hygromycin-T2A-mBFP). RIT1-eGFP degradation reporter plasmid and LZTR1 WT or mutant-expressing plasmid were cotransfected into 293T cells using polyethylenimine hydrochloride. Culture media were refreshed 8 hours after transfection. Cells were harvested 48 hours after transfection for flow cytometry analysis.
Virus Packaging and Transduction
RIT1 WT and mutant cDNAs were cloned into the pMIG-II retroviral vector (MSCV-IRES-GFP). sgRNAs against nontargeting control, LZTR1, and RIT1 were cloned into lentiCRISPRV2-GFP or tet-pLKO-sgRNA-puro. shRNAs against RIT1, MRAS, KRAS, and SHOC2 were cloned into the LT3REPIR lentiviral vector (T3G-dsRED-mirE-PGK-Puro-IRES-rtTA3). RAS biodegrader Flag-K27-SPOP cDNAs were subcloned into pCW-cas9-puro plasmids to replace cas9. The pLenti-Cas9-RFP plasmid was used to express Cas9 in TF-1 cells expressing RIT1 mutants. The MSCV-Red2-IRES construct expressing NrasG12D cDNA was a gift from Dr. Scott Lowe's laboratory (MSKCC). sgRNA and shRNA sequences are given in Supplementary Table S2.
Retroviral supernatants were produced by transfecting GP-Ampho cells with retroviral cDNA-expressing constructs and the packaging plasmids pGagpol and pAmpho using ProFection Mammalian Transfection System (Promega). Lentiviral supernatants were produced by transfecting HEK293T cells with lentiviral constructs and the packaging plasmids pVSVG and psPAX2 using PEI. Virus supernatants were used for transduction in the presence of polybrene (8 μg/mL). Cells were FACS-sorted or puromycin-selected (2 μg/mL) for 7 days to obtain transduced cells. To induce shRNA expression or RAS biodegrader K27-SPOP expression, doxycycline (1 μg/mL, Sigma-Aldrich) was added to the culture medium.
Bone Marrow Transplantation
Freshly dissected femora and tibiae were isolated from Mx1-cre, Mx1-cre Lztr1fl/fl, and Mx1-cre Rit1M90I/WT CD45.2+ mice. Bone marrow was flushed with a 3-cm3 insulin syringe into cold PBS supplemented with 0.5% fetal calf serum (FCS; heat-inactivated). The bone marrow was spun by centrifugation at 1,500 rpm for 5 minutes, and red blood cells were lysed in ammonium chloride–potassium bicarbonate lysis (ACK) buffer for 3 minutes on ice. Cells were then resuspended in PBS + 0.5% FBS, passed through a 40-μm cell strainer, and counted. For competitive transplantation experiments, 0.1 × 106 fetal liver cells from CD45.2+Lztr1+/+, Lztr1−/−, Lztr1+/+Rit1−/−, and Lztr1−/−Rit1−/− E14.5 embryos were mixed with 0.3 × 106 WT CD45.1+ competitor bone marrow, or 0.5 × 106 total bone marrow cells from Mx1-cre, Mx1-cre Lztr1fl/fl, and Mx1-cre Rit1M90I/WT CD45.2+ mice were mixed with 0.5 × 106 WT CD45.1+ competitor bone marrow and transplanted via tail-vein injection into lethally irradiated (900 cGy) CD45.1+ recipient mice. To activate the conditional alleles, mice were treated with 3 doses of pIpC every other day via intraperitoneal injection. Serial transplantation was performed by transplanting 1 × 106 total bone marrow cells from recipient mice into lethally irradiated (900 cGy) or sublethally irradiated (450 cGy) CD45.1+ recipient mice. Peripheral blood chimerism was assessed every 4 weeks by flow cytometry. Whole blood cell counts were measured by Procyte Dx Hematology Analyzer (IDEXX Veterinary Diagnostics).
In Vitro Colony-Forming Assays
fetal liver cells (20 × 103) from Lztr1+/+ and Lztr1−/− E14.5 embryos were seeded into cytokine-supplemented methylcellulose medium (Methocult M3434; STEMCELL Technologies). Colonies propagated in culture were scored on day 7. The remaining cells were resuspended and counted, and a portion was taken for replating every week.
To test GM-CSF hypersensitivity, live (DAPI−) LSK cells were FACS-sorted from the bone marrow of Mx1-cre, Mx1-cre Lztr1fl/fl, and Mx1-cre Rit1M90I/WT mice 4 weeks after pIpC administration and seeded at a density of 800 cells/replicate into the methylcellulose medium (Methocult M3231; STEMCELL Technologies) with murine GM-CSF (PeproTech) at doses of 0, 0.01, 0.1, 1, and 10 ng/mL. Colonies were scored on day 7.
Histologic Analysis
Mice were sacrificed and autopsied, and dissected tissues were fixed in 4% paraformaldehyde, dehydrated, and embedded in paraffin. Paraffin blocks were sectioned at 4 μm thickness and stained with hematoxylin and eosin (H&E). Differential blood counts were realized on blood smears stained using Wright–Giemsa staining. Images were acquired using an Axio Observer A1 microscope (Carl Zeiss) or a BX-61 microscope (Olympus), or they were scanned using a MIRAX Scanner (Zeiss). All histologic, IHC, and immunofluorescence tissue analyses underwent a blinded review by both a board-certified veterinary pathologist (S. Monette) and a board-certified hematopathologist (B.H. Durham).
IHC for cleaved caspase-3 (CC3) was performed on a Leica Bond RX automated stainer as follows: After heat-induced epitope retrieval in a pH 6.0 citrate buffer, the primary antibody was applied (rabbit polyclonal anti-CC3 antibody, Cell Signaling Technology 9661, 1:250 dilution), followed by a polymer detection kit used as instructed by the vendor (DS9800, Novocastra Bond Polymer Refine Detection, Leica Biosystems). The chromogen was 3,3 diaminobenzidine tetrachloride (DAB), and sections were counterstained with hematoxylin. H&E- and CC3-stained slides of embryos were examined in all major organs, and CC3-positive cell quantification on whole liver sections was performed in Qupath 0.3.0 (33).
Immunofluorescent Staining
Immunofluorescence for CC3 and TER-119 was performed manually on paraffin sections of embryos. After heat-induced epitope retrieval in a pH 6.0 citrate buffer, the primary antibodies were applied (rabbit polyclonal anti-CC3 antibody, Cell Signaling Technology, 9661, rat monoclonal anti–TER-119 antibody, BD Pharmingen, 550565, both at 1:200 dilution), followed by the application of secondary antibodies (anti-rabbit IgG conjugated with Alexa Fluor 488, Thermo Fisher, A-21206, anti-rat IgG conjugated with Alexa Fluor 594, Thermo Fisher, A-21209, both at 1:500 dilution), DAPI, and an autofluorescence quenching kit (TrueVIEW, Vector Laboratories, SP-8400-15). Slides were imaged on a BX-61 microscope, and CC3-positive cell quantification on whole liver sections was performed in Qupath 0.3.0.
Cell Lines and Tissue Culture
All cell lines were purchased from either ATCC or DSMZ. HEK293T cells were grown in DMEM + 10% FCS (heat-inactivated). TF-1 (human erythroleukemia cell line) cells were grown in RPMI + 10% FCS with 2 ng/mL recombinant human GM-CSF (R&D Systems; 215-GM) unless noted otherwise. RPMI + 10% FCS with 2 IU/mL recombinant human EPO (PeproTech; 100-64) was used to induce erythroid differentiation in TF-1 cells. MOLM-13 cells were grown in RPMI + 10% FCS. All cell lines were cultured at 37°C and 5% CO2 in the presence of penicillin (100 U/mL) and streptomycin (100 μg/mL). Human cell lines have been authorized using ATCC fingerprinting or by short tandem repeat (STR) profiling assay at the MSKCC Integrated Genomics Operation Core. All cell lines in this study were Mycoplasma-free and routinely tested by the Antibody and Bioresource Core at MSKCC (MycoAlert Mycoplasma Detection Kit, Lonza, LT07-701; MycoAlert Assay Control Set, Lonza, LT07-518).
Antibodies, Flow Cytometry, and Western Blot Analysis
The following antibodies were used for flow cytometry: anti–B220-APC-Cy7 (clone RA3-6B2, BioLegend, 103224, 1:200 dilution), anti–B220-PerCP-Cy5.5 (RA3-6B2, eBioscience, 45-0452-82, 1:200), anti–B220-BV605 (RA3-6B2, BioLegend, 103244, 1:200), anti–CD3-APC-Cy7 (17A2, BioLegend, 100222, 1:200), anti–CD3-PE-Cy7 (17A2, BioLegend, 100220, 1:200), anti–CD3-APC-Cy7 (17A2, BioLegend, 100222, 1:200), anti–Gr-1-APC (RB6-8C5, eBioscience, 25-5931-82, 1:500), anti–CD11b-FITC (M1/70, BioLegend, 101206, 1:200), anti–CD11b-APC-Cy7 (M1/70, BioLegend, 101226, 1:200), anti–Ter119-APC-Cy7 (Ter119, BioLegend, 116223, 1:200), anti–c-Kit-PerCP-Cy5.5 (2B8, BioLegend, 105824, 1:100), anti–c-Kit-BV605 (ACK2, BioLegend, 135120, 1:100), anti–Sca-1-PE-Cy7 (D7, BioLegend, 108102, 1:100), anti–CD48-PerCP-Cy5.5 (HM48-1, BioLegend, 103422, 1:100), anti–CD150-PE (9D1, eBioscience, 12-1501-82, 1:100), anti–CD16/CD32 (FcγRII/III)-Alexa 700 (93, eBioscience, 56-0161-82, 1:100), anti–CD34-FITC (RAM34, BD Biosciences, 553731, 1:50), anti–CD45.1-FITC (A20, BioLegend, 110706, 1:200), anti–CD45.1-BV711 (A20, BioLegend, 110739, 1:200), anti–CD45.2-PE (104, eBioscience, 12-0454-82, 1:200), anti–CD19-PE-cy7 (eBio1D3, eBioscience, 25-0193-82, 1:200), anti–CD43-FITC (eBioR2/60, eBioscience, 11-0431-85, 1:200), anti-IgM-PE (II/41, eBioscience, 12-5790-82, 1:200), anti–IgD-APC (11-26c.2a, BioLegend, 405714, 1:200), and anti-human CD235a (glycophorin A)-APC (HI264, BioLegend, 349114, 1:100). Annexin V/DAPI staining was performed using the FITC Annexin V Apoptosis Detection Kit (BD Biosciences, 556457) following the manufacturer's protocol. Flow cytometry data acquisition or cell sorting was performed by BD LSRFortessa or BD FACSAria II. Flow cytometry data were analyzed by FlowJo 10 software.
The above antibodies were used to define cell-surface immunophenotypes for hematopoietic stem and progenitor cells as follows: LSK, long-term HSCs (lineage-negative Sca1+ c-KIT+ CD150+ CD48−), short-term HSCs (lineage-negative Sca1+ c-KIT+ CD150+ CD48+), multipotent progenitors (lineage-negative Sca1+ c-KIT+ CD150− CD48+), granulocyte–monocyte progenitors (lineage-negative Sca1− c-KIT+ CD16/32+ CD34+), common myeloid progenitors (lineage-negative Sca1− c-KIT+ CD16/32− CD34+), and megakaryocytic–erythroid progenitors (lineage-negative Sca1− c-KIT+ CD16/32− CD34−).
For western blot analysis, cells were lysed by IP lysis buffer (Pierce) containing the Protease/Phosphatase Inhibitor Cocktail (Sigma-Aldrich). Protein concentration was measured using the BCA Protein Assay Kit (Pierce). Equivalent amounts of each sample were loaded on 4% to 12% Bis-Tris gels (Invitrogen), transferred to 0.45 μm PVGF membrane, and blotted with 5% nonfat milk in 0.1% TBS-T buffer. The following antibodies were used for western blot analysis: LZTR1 (Santa Cruz Biotechnology, sc-390166X), RIT1 (Abcam, ab53720), MRAS (Abcam, ab176570), KRAS (Sigma-Aldrich, WH0003845M1), NRAS (Invitrogen, 703435), HRAS (Thermo Fisher, 18295-1-AP), SHOC2 (Cell Signaling Technology, 53600S), phospho-ERK (Cell Signaling Technology, 9101L), ERK1/2 (Cell Signaling Technology, 4695S), phospho-MEK1/2 (Cell Signaling Technology, 9154S), MEK1/2 (Cell Signaling Technology, 9126S), SPOP (Invitrogen, PA5-28522), HSP90 (Cell Signaling Technology, 4877S), FLAG (Sigma-Aldrich, F1804), and b-Actin (Sigma-Aldrich, A-5441). All primary antibodies for western blotting were diluted to a final concentration of 1:1,000 in either 5% BSA or 5% nonfat milk in 0.1% TBS-T.
For the detection of GTP-bound RAS proteins, cells were lysed and processed using the Active RAS Detection Kit (Cell Signaling Technology, 8821) following the manufacturer's instructions. After pulldown, levels of GTP-loaded RAS isoforms were determined by western blotting.
In Vitro Competition Assay
For in vitro competition assay to evaluate the cellular effect of knockdown of RIT1, MRAS, or SHOC2, TF-1 cell lines were transduced with a doxycycline-inducible LT3REPIR lentiviral vector (T3G-dsRED-mirE-PGK-Puro-IRES-rtTA3) expressing shRNAs against RIT1, MRAS, or SHOC2 or a nontargeting shRNA against Renilla (sequences provided in Supplementary Table S2). Transduced cells were selected in puromycin (2 μg/mL) containing medium for 7 days. Transduced cells were mixed with nontransduced cells at a 1:1 ratio. shRNAs were then induced with the addition of doxycycline (0.6 μg/mL, Sigma-Aldrich). The percentage of dsRED-expressing cells was measured over time after doxycycline induction using BD LSRFortessa. Normalized dsRED-positive rates in living cells at each point were compared with that of day 5 after doxycycline induction.
CellTiter-Glo Proliferation Assay
ATP luminescence readings were taken every 24 hours after seeding using CellTiter-Glo (Promega) according to the manufacturer's instructions.
In Vivo Xenograft Experiment
MOLM-13 luciferase-expressing cells were transduced with the lentiCRISPRV2-GFP construct with either nontargeting control or LZTR1 sgRNAs. GFP+ cells were sorted by FACS 2 days after transduction. One hundred thousand GFP+ cells with either sgNTC or sgLZTR1 were intravenously injected into sublethally irradiated (225 cGy) 8-week-old NSG mice. All whole-body bioluminescent imaging was performed by the intraperitoneal injection of Luciferin (Goldbio) at a 50 mg/kg concentration, and imaging was performed after 10 minutes using an IVIS imager. Engraftment was confirmed, and treatment began at day 10 after inoculation. Bioluminescent signals (radiance) were quantified using Living Image software with standard regions of interest rectangles.
For FLT3 inhibitor treatment, gilteritinib (Selleck Chemicals, S7754) was dissolved in 0.5% CMC-NA (Selleck Chemicals, S6703) to obtain a final concentration of 4 mg/mL. Upon disease onset as measured by bioluminescent imaging, mice were orally administrated with either 30 mg/kg gilteritinib or vehicle (0.5% CMC-NA) once daily for 5 days/week for 3 weeks.
In Silico and In Vitro Drug Sensitivity Assays
Drug sensitivity and LZTR1 expression level in AML patient samples were collected from http://vizome.org/aml/inhibitor/, and Prism software (GraphPad) was used to produce nonlinear fit and Pearson r correlation value.
Small-molecule inhibitors, gilteritinib, quizartinib, and ponatinib purchased from MedChemExpress and BI-3406 purchased from Selleck Chemicals were reconstituted in DMSO. MOLM-13 cells were seeded in white flat-well, 96-well plates (Costar) at a density of 2,000 cells per well with different concentrations of drugs. ATP luminescence readings were taken 72 hours after drug treatment using CellTiter-Glo (Promega) according to the manufacturer's instructions. Relative cell viability was calculated by normalizing to untreated control wells. Prism software (GraphPad) was used to produce nonlinear fit and IC50.
The synergyfinder (v2.4.16) software package (34) was used to calculate Zero Interaction Potency (ZIP) synergy scores across a dose–response matrix of drug x and drug y. Figures were generated in Rstudio (v1.3.1073).
Whole-Genome CRISPR–Cas9 Screening
Lentivirus carrying human CRISPR Brunello lentiviral pooled sgRNA library was produced in 293T cells. Virus titer was determined by measuring the percentage of puromycin-resistant cells after transduction. A titer resulting in 30% of transduced cells (puromycin resistant) was selected for the following experiments. TF-1 RIT1 mutant cells expressing Cas9-RFP or TF-1 LZTR1 KO cells were transduced with lentivirus expressing human CRISPR Brunello sgRNA library, and puromycin selection (2 μg/mL) was performed in human GM-CSF containing media for 7 days. Then, GM-CSF (day 0) was washed out, and surviving cells were harvested at 14 days after cytokine depletion (day 14). Cell pellets were lysed, and genomic DNA was extracted (NucleoSpin Blood XL, Takara) and quantified by Qubit (Thermo Scientific). A quantity of gDNA covering 1,000× representation of gRNAs was PCR-amplified using TaKaRa Ex Taq DNA Polymerase (Takara) to add Illumina adapters and multiplexing barcodes. Amplicons were quantified by Qubit and Bioanalyzer (Agilent) and sequenced on an Illumina HiSeq 2500. Sequencing reads were aligned to the screened library, and counts were obtained for each gRNA. We used standard methods from the R/Bioconductor package, and the specific package was edgeR. For the probe level analysis, we used the standard workflow with the glmLRT option for the model fitting/statistical test.
Single-Cell RNA-seq
Eight-week-old Mx1Cre control, Rit1M90I/+Mx1Cre+, and Lztr1fl/fl; Mx1Cre+ mice were treated with 3 doses of pIpC (12 mg/kg) every other day. Bone marrow cells were harvested from the mice 4 weeks after pIpC treatment and stained with antibodies, including anti–CD3-APCcy7, anti–CD11b-APCcy7, anti–Gr1-APCcy7, anti–B220-APCcy7, and anti–Ter119-APCcy7. Live lineage-negative (APCcy7− and DAPI−) cells were sorted and suspended in PBS + 0.04% BSA. Sorted cells were stained with Trypan blue, and the Countess II Automated Cell Counter (Thermo Fisher) was used to assess both cell number and viability (90%–91%). Following quality control, the single-cell suspension was loaded onto Chromium Chip B (10X Genomics; PN 2000060) and GEM generation, cDNA synthesis, cDNA amplification, and library preparation of 3 to 6,700 cells proceeded using the Chromium Single-Cell 3′ Reagent Kit v3 (10X Genomics; PN 1000075) according to the manufacturer's protocol. cDNA amplification included 11 cycles, and 240 to 370 ng of the material was used to prepare sequencing libraries with 10 cycles of PCR. Indexed libraries were pooled in an equimolar ratio and sequenced on a Illumina NovaSeq 6000 in a PE28/91 paired-end run using the NovaSeq 6000 S1 Reagent Kit (100 cycles; Illumina). An average of 30,000 reads was generated per cell. Demultiplexing of each sample was performed using the “cellranger mkfastq” command from 10X Genomics’ Cell Ranger software (version 6.1.1). Preprocessing was performed with the “cellranger count” command for the alignment of sequencing reads to the mouse Mm10 reference genome (“mm10-2020-A”). The next steps of data processing, analysis, and visualization were all performed in Rstudio (v1.3.1073) using Seurat (v4.0.5). Initial quality control was performed on each of the samples to remove (i) genes detected in at least four cells and (ii) cells containing at least 200 genes. We also ensured that the percentage of mitochondrial-derived reads was less than 15%, and that the total number of molecules within each cell was filtered by 500 < nCount_RNA < 20,000. Next, cell-cycle scores were calculated using the following command: CellCycleScoring(Object, s.features = m.s.genes, g2m.features = m.g2m.genes,set.ident = TRUE) using S-phase and G2–M phase genes preloaded into Seurat. Cells were then scaled and normalized using the SCTransform function of the Seurat package using the following command: SCTransform(Object, method = “glmGamPoi,” vars.to.regress = c(“percent.mt,” “nFeature_RNA,” “nCount_RNA,” “percent.ribo,” “S.Score,” “G2M.Score”), new.assay.name = “SCT”). Following filtering, the Cre-negative sample retained 7,528 cells, Mx1-cre Lztr1fl/fl retained 10,610 cells, and Mx1-cre Rit1M90I/+ retained 2,398 cells. Each sample was subsequently merged together into a single object using the Seurat “merge” function and the “merge.data = TRUE” setting to aggregate the normalized values. To determine the intersample anchors for integration, the “FindIntegrationAnchors” function was used with the top 3,000 highly variable genes, which were themselves identified using the “SelectIntegrationFeatures” command. Next, sample integration was performed using “PrepSCTIntegration” and “IntegrateData” functions on SCTransform normalized data. Dimensionality reduction was performed through principal component analysis using the “runPCA” function, followed by the uniform manifold approximation and projection (UMAP) method for visualization of unsupervised clustering using the “runUMAP” function, in which the first 20 dimensions (“dims = 1:20”) are used for downstream unbiased clustering with a resolution of 0.8. Cell clustering was performed using the “FindNeighbors” and “FindCluster” functions in Seurat. Annotation of cluster identities was performed manually by examining differentially expressed and cluster-specific genes identified using the “FindAllMarkers” function in Seurat. To explore differentially expressed genes within a given cluster between genotypes, we used the “FindMarkers” function in Seurat. Plots were produced in Rstudio using the “VlnPlot,” “geom_alluvium,” and “DimPlot” functions.
Statistics and Reproducibility
Statistical significance was determined by an unpaired two-sided Student t test after testing for normal distribution as well as one-way or two-way ANOVA followed by the Tukey, Sidak, or Dunnett multiple comparisons test. Data were plotted using GraphPad Prism 9 software as mean values, with error bars representing the standard error of the mean. Representative western blot and PCR results are shown from at least three biologically independent experiments. Representative flow cytometry profiles are shown from biological replicates (n ≥ 3). *, P < 0.05; **, P < 0.01; ***, P < 0.001; ****, P < 0.0001.
Data Availability
The mouse whole-exome sequencing, CRISPR screen, and single-cell RNA-seq data have been deposited in the Gene Expression Omnibus under accession ID GSE190857.
Supplementary Material
Acknowledgments
We acknowledge the use of the Integrated Genomics Operation Core, funded by the NCI Cancer Center Support Grant (CCSG, P30 CA08748), the Bioinformatics Core, Cycle for Survival, and the Marie-Josée and Henry R. Kravis Center for Molecular Oncology. This work was supported by the Lady Tata Memorial Trust (S. Chen), the American Society of Hematology (ASH; S. Chen and D. Inoue), the Leukemia & Lymphoma Society (S. Chen, D. Inoue, and O. Abdel-Wahab), MSK-ICTTP T32-CA009207 (R.F. Stanley), ASH Research Training Award for Fellows (R.F. Stanley), NIH R01 CA251138 (O. Abdel-Wahab), NIH/NCI 1P50 254838-01 (O. Abdel-Wahab), NIH K08 CA204734 (R.C. Lindsley), the Geoffrey Beene Cancer Research Center of Memorial Sloan Kettering Cancer Center (O. Abdel-Wahab), the Edward P. Evans MDS Foundation (O. Abdel-Wahab and R.S. Vedula), NIH/NCI F31CA265066 (A. Cuevas-Navarro), R35CA197709 (F. McCormick), R00CA245122 (P. Castel), DOD CDMRP Neurofibromatosis Research Program (W81XWH-20-1-0391; P. Castel), and NIH P30 CA008748 (O. Abdel-Wahab). D. Inoue is supported by AMED (JP20cm0106165), the Takeda Science Foundation, the Yasuda Medical Foundation, the Kanae Foundation for the Promotion of Medical Science, the MSD Life Science Foundation, the Inoue Foundation for Science, and JSPS KAKENHI (JP20H00537).
The costs of publication of this article were defrayed in part by the payment of page charges. This article must therefore be hereby marked advertisement in accordance with 18 U.S.C. Section 1734 solely to indicate this fact.
Footnotes
Note: Supplementary data for this article are available at Cancer Discovery Online (http://cancerdiscovery.aacrjournals.org/).
Authors’ Disclosures
A. Cuevas-Navarro reports grants from the National Science Foundation and the NCI during the conduct of the study. N. Rosen reports personal fees and other support from Beigene, Zai Lab, MAPCure, Ribon, and Chugai, other support from Kura and Effector, grants from Revolution Medicines, Boehringer Ingelheim, and Pfizer-Array, and personal fees from Ikena and AstraZeneca outside the submitted work. R.C. Lindsley reports personal fees from Takeda Pharmaceuticals, Bluebird Bio, Qiagen, Nuprobe, and Thermo Fisher outside the submitted work. P. Castel reports personal fees and other support from Venthera, Inc. outside the submitted work. O. Abdel-Wahab reports grants from Nurix Therapeutics, Loxo Oncology, and H3B Biomedicine Inc., personal fees from Pfizer (Boulder), and other support from Envisagenics Inc., AIChemy Inc., and Harmonic Discovery Inc. outside the submitted work. No disclosures were reported by the other authors.
Authors’ Contributions
S. Chen: Conceptualization, data curation, formal analysis, methodology, writing–original draft, writing–review and editing. R.S. Vedula: Data curation, formal analysis, visualization, writing–review and editing. A. Cuevas-Navarro: Conceptualization, data curation, formal analysis, investigation. B. Lu: Investigation. S.J. Hogg: Investigation, visualization, methodology, writing–original draft. E. Wang: Investigation. S. Benbarche: Investigation. K. Knorr: Investigation. W.J. Kim: Investigation. R.F. Stanley: Investigation, writing–review and editing. H. Cho: Investigation. C. Erickson: Investigation. M. Singer: Investigation. D. Cui: Formal analysis, investigation. S. Tittley: Investigation. B.H. Durham: Investigation, visualization. T.S. Pavletich: Investigation. E. Fiala: Investigation. M.F. Walsh: Investigation. D. Inoue: Investigation. S. Monette: Formal analysis, investigation, visualization, methodology. J. Taylor: Investigation, writing–review and editing. N. Rosen: Formal analysis, supervision, writing–review and editing. F. McCormick: Conceptualization, resources, writing–review and editing. R.C. Lindsley: Data curation, formal analysis, methodology, writing–original draft. P. Castel: Conceptualization, resources, supervision, investigation, visualization, writing–original draft, writing–review and editing. O. Abdel-Wahab: Conceptualization, resources, formal analysis, supervision, funding acquisition, investigation, visualization, writing–original draft, project administration, writing–review and editing.
References
- 1. Bigenzahn JW, Collu GM, Kartnig F, Pieraks M, Vladimer GI, Heinz LX, et al. LZTR1 is a regulator of RAS ubiquitination and signaling. Science 2018;362:1171–7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2. Damnernsawad A, Bottomly D, Kurtz SE, Eide CA, McWeeney SK, Tyner JW, et al. Genome-wide CRISPR screen identifies regulators of MAPK and MTOR pathways mediating sorafenib resistance in acute myeloid leukemia. Haematologica 2022;107:77–85. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3. Castel P, Cheng A, Cuevas-Navarro A, Everman DB, Papageorge AG, Simanshu DK, et al. RIT1 oncoproteins escape LZTR1-mediated proteolysis. Science 2019;363:1226–30. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4. Steklov M, Pandolfi S, Baietti MF, Batiuk A, Carai P, Najm P, et al. Mutations in LZTR1 drive human disease by dysregulating RAS ubiquitination. Science 2018;362:1177–82. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5. Chen PC, Yin J, Yu HW, Yuan T, Fernandez M, Yung CK, et al. Next-generation sequencing identifies rare variants associated with Noonan syndrome. Proc Natl Acad Sci U S A 2014;111:11473–8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6. Pich O, Reyes-Salazar I, Gonzalez-Perez A, Lopez-Bigas N. Discovering the drivers of clonal hematopoiesis. Nat Commun 2022;13:4267. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7. Inoue D, Polaski JT, Taylor J, Castel P, Chen S, Kobayashi S, et al. Minor intron retention drives clonal hematopoietic disorders and diverse cancer predisposition. Nat Genet 2021;53:707–18. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8. Gomez-Segui I, Makishima H, Jerez A, Yoshida K, Przychodzen B, Miyano S, et al. Novel recurrent mutations in the RAS-like GTP-binding gene RIT1 in myeloid malignancies. Leukemia 2013;27:1943–6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9. Aoki Y, Niihori T, Banjo T, Okamoto N, Mizuno S, Kurosawa K, et al. Gain-of-function mutations in RIT1 cause Noonan syndrome, a RAS/MAPK pathway syndrome. Am J Hum Genet 2013;93:173–80. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10. Abe T, Umeki I, Kanno SI, Inoue SI, Niihori T, Aoki Y. LZTR1 facilitates polyubiquitination and degradation of RAS-GTPases. Cell Death Differ 2020;27:1023–35. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11. Cuevas-Navarro A, Rodriguez-Munoz L, Grego-Bessa J, Cheng A, Rauen KA, Urisman A, et al. Cross-species analysis of LZTR1 loss-of-function mutants demonstrates dependency to RIT1 orthologs. Elife 2022;11:e76495. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12. Young LC, Hartig N, Del Rio IB, Sari S, Ringham-Terry B, Wainwright JR, et al. SHOC2-MRAS-PP1 complex positively regulates RAF activity and contributes to Noonan syndrome pathogenesis. Proc Natl Acad Sci U S A 2018;115:E10576–E85. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13. Sievers QL, Gasser JA, Cowley GS, Fischer ES, Ebert BL. Genome-wide screen identifies cullin-RING ligase machinery required for lenalidomide-dependent CRL4(CRBN) activity. Blood 2018;132:1293–303. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14. Papaemmanuil E, Gerstung M, Malcovati L, Tauro S, Gundem G, Van Loo P, et al. Clinical and biological implications of driver mutations in myelodysplastic syndromes. Blood 2013;122:3616–27; quiz 3699. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15. Papaemmanuil E, Gerstung M, Bullinger L, Gaidzik VI, Paschka P, Roberts ND, et al. Genomic classification and prognosis in acute myeloid leukemia. N Engl J Med 2016;374:2209–21. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16. Shao H, Kadono-Okuda K, Finlin BS, Andres DA. Biochemical characterization of the Ras-related GTPases Rit and Rin. Arch Biochem Biophys 1999;371:207–19. [DOI] [PubMed] [Google Scholar]
- 17. Li Q, Haigis KM, McDaniel A, Harding-Theobald E, Kogan SC, Akagi K, et al. Hematopoiesis and leukemogenesis in mice expressing oncogenic NrasG12D from the endogenous locus. Blood 2011;117:2022–32. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18. Van Meter ME, Diaz-Flores E, Archard JA, Passegue E, Irish JM, Kotecha N, et al. K-RasG12D expression induces hyperproliferation and aberrant signaling in primary hematopoietic stem/progenitor cells. Blood 2007;109:3945–52. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19. Wong JC, Perez-Mancera PA, Huang TQ, Kim J, Grego-Bessa J, Del Pilar Alzamora M, et al. KrasP34R and KrasT58I mutations induce distinct RASopathy phenotypes in mice. JCI Insight 2020;5:e140495. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20. Izzo F, Lee SC, Poran A, Chaligne R, Gaiti F, Gross B, et al. DNA methylation disruption reshapes the hematopoietic differentiation landscape. Nat Genet 2020;52:378–87. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21. Doench JG, Fusi N, Sullender M, Hegde M, Vaimberg EW, Donovan KF, et al. Optimized sgRNA design to maximize activity and minimize off-target effects of CRISPR-Cas9. Nat Biotechnol 2016;34:184–91. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22. Vichas A, Riley AK, Nkinsi NT, Kamlapurkar S, Parrish PCR, Lo A, et al. Integrative oncogene-dependency mapping identifies RIT1 vulnerabilities and synergies in lung cancer. Nat Commun 2021;12:4789. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23. Cuevas-Navarro A, Van R, Cheng A, Urisman A, Castel P, McCormick F. The RAS GTPase RIT1 compromises mitotic fidelity through spindle assembly checkpoint suppression. Curr Biol 2021;31:3915–24. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24. Rodriguez-Viciana P, Oses-Prieto J, Burlingame A, Fried M, McCormick F. A phosphatase holoenzyme comprised of Shoc2/Sur8 and the catalytic subunit of PP1 functions as an M-Ras effector to modulate Raf activity. Mol Cell 2006;22:217–30. [DOI] [PubMed] [Google Scholar]
- 25. Tyner JW, Tognon CE, Bottomly D, Wilmot B, Kurtz SE, Savage SL, et al. Functional genomic landscape of acute myeloid leukaemia. Nature 2018;562:526–31. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26. Drosten M, Dhawahir A, Sum EY, Urosevic J, Lechuga CG, Esteban LM, et al. Genetic analysis of Ras signalling pathways in cell proliferation, migration and survival. EMBO J 2010;29:1091–104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27. Lim S, Khoo R, Juang YC, Gopal P, Zhang H, Yeo C, et al. Exquisitely specific anti-KRAS biodegraders inform on the cellular prevalence of nucleotide-loaded states. ACS Cent Sci 2021;7:274–91. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28. Hofmann MH, Gmachl M, Ramharter J, Savarese F, Gerlach D, Marszalek JR, et al. BI-3406, a potent and selective SOS1-KRAS interaction inhibitor, is effective in KRAS-driven cancers through combined MEK inhibition. Cancer Discov 2021;11:142–57. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29. Li Q, Bohin N, Wen T, Ng V, Magee J, Chen SC, et al. Oncogenic Nras has bimodal effects on stem cells that sustainably increase competitiveness. Nature 2013;504:143–7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30. Lo A, Holmes K, Kamlapurkar S, Mundt F, Moorthi S, Fung I, et al. Multiomic characterization of oncogenic signaling mediated by wild-type and mutant RIT1. Sci Signal 2021;14:eabc4520. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31. Kluk MJ, Lindsley RC, Aster JC, Lindeman NI, Szeto D, Hall D, et al. Validation and implementation of a custom Next-generation sequencing clinical assay for hematologic malignancies. J Mol Diagn 2016;18:507–15. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32. Lindsley RC, Mar BG, Mazzola E, Grauman PV, Shareef S, Allen SL, et al. Acute myeloid leukemia ontogeny is defined by distinct somatic mutations. Blood 2015;125:1367–76. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33. Bankhead P, Loughrey MB, Fernandez JA, Dombrowski Y, McArt DG, Dunne PD, et al. QuPath: open source software for digital pathology image analysis. Sci Rep 2017;7:16878. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34. Ianevski A, Giri AK, Aittokallio T. SynergyFinder 2.0: visual analytics of multi-drug combination synergies. Nucleic Acids Res 2020;48:W488–W93. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Data Availability Statement
The mouse whole-exome sequencing, CRISPR screen, and single-cell RNA-seq data have been deposited in the Gene Expression Omnibus under accession ID GSE190857.



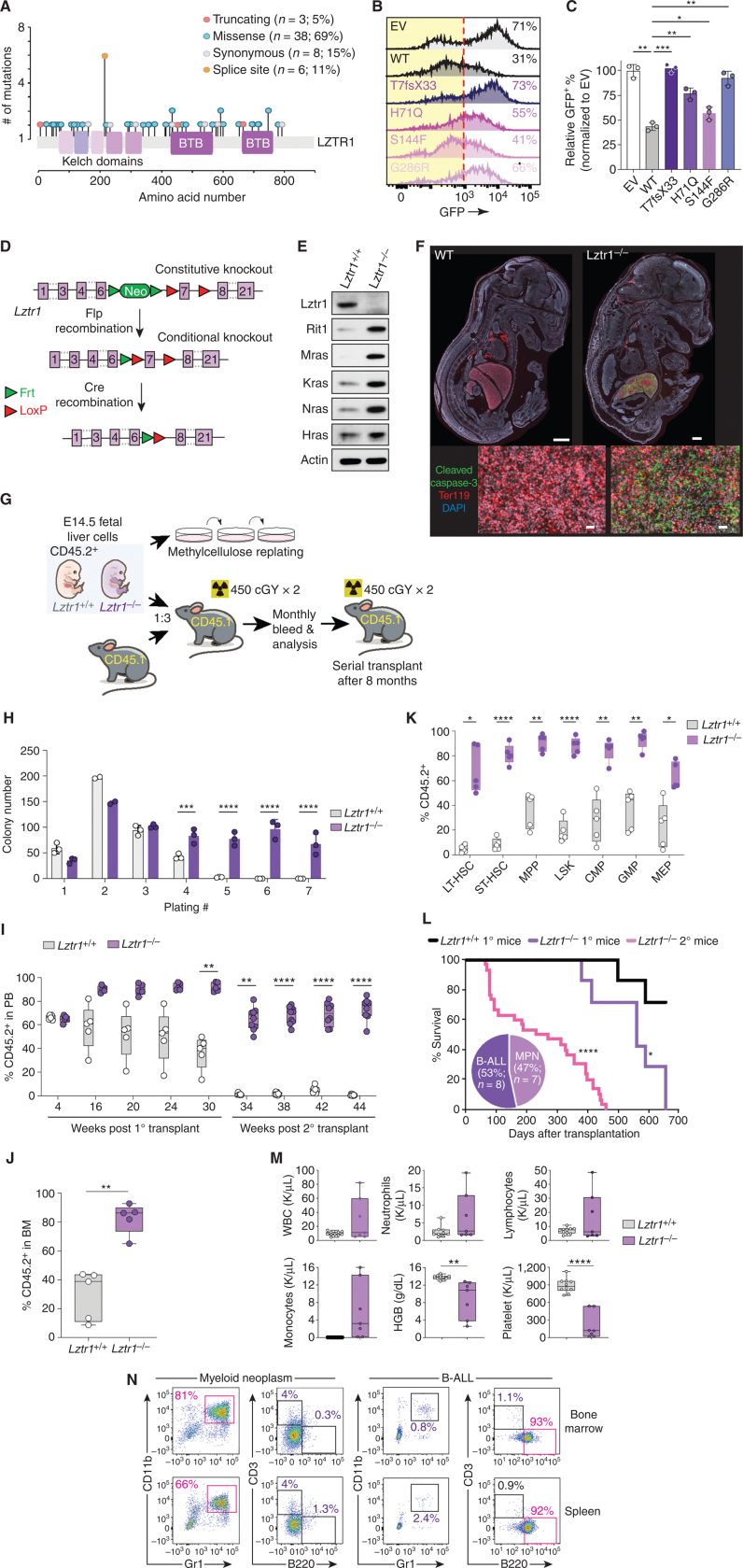
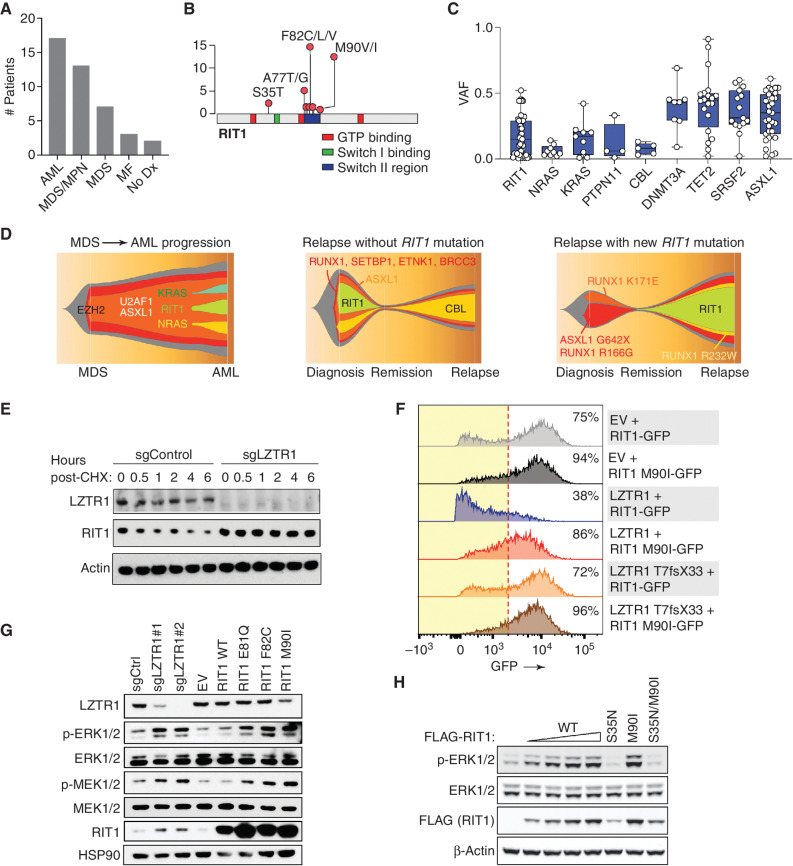
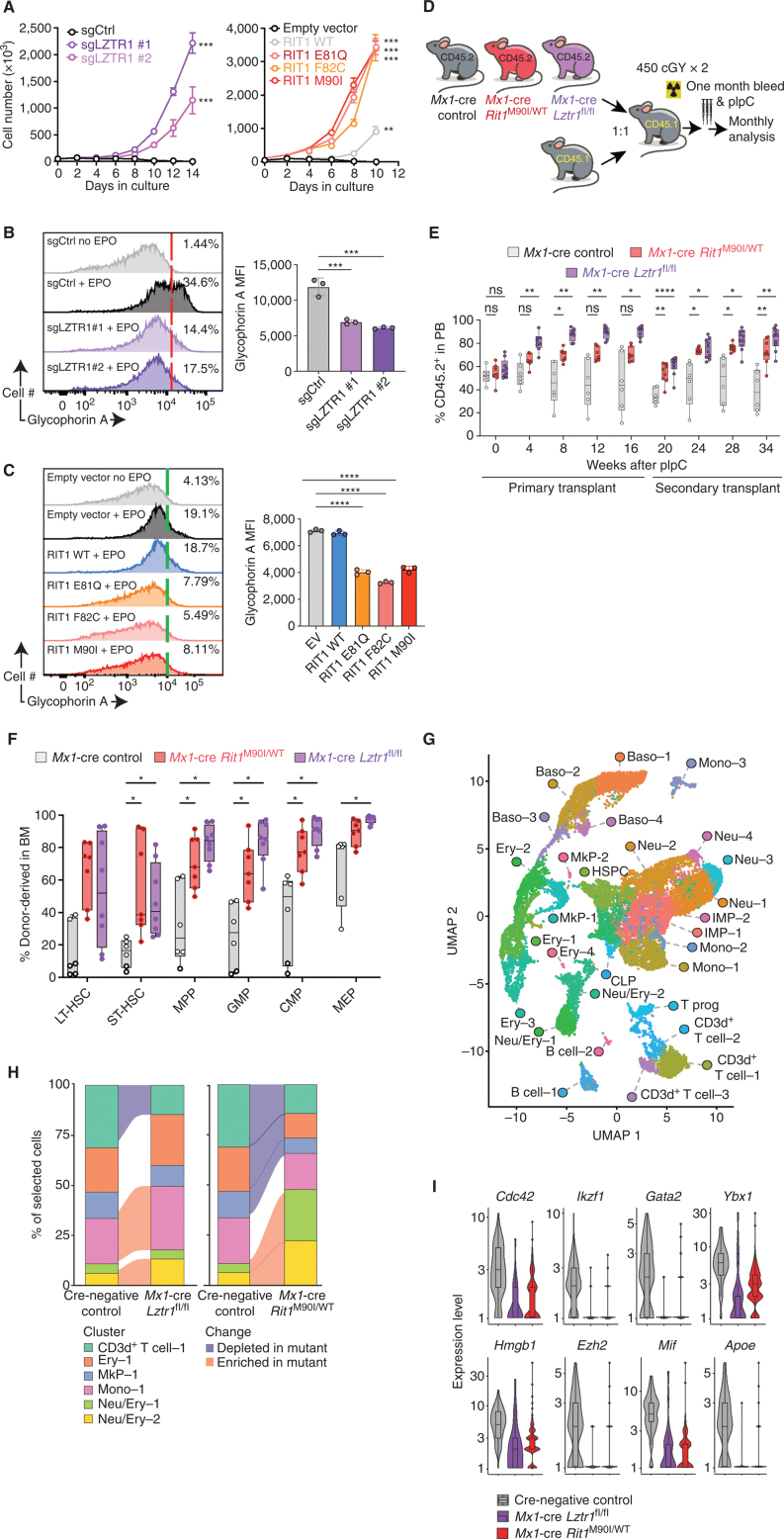
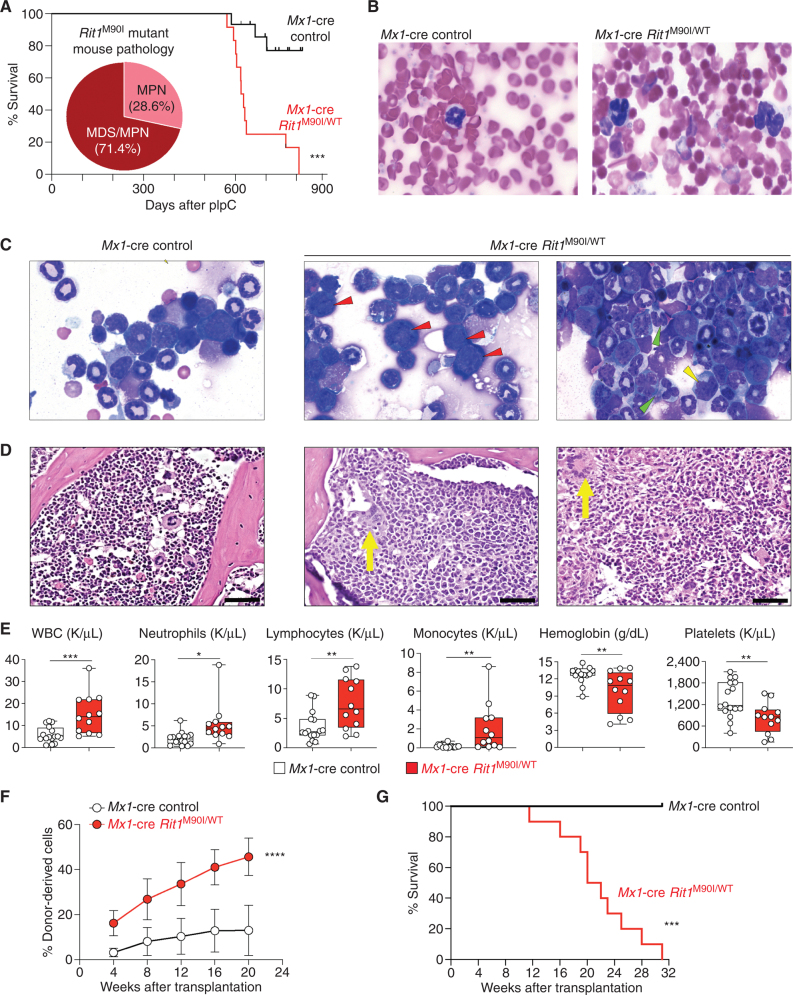
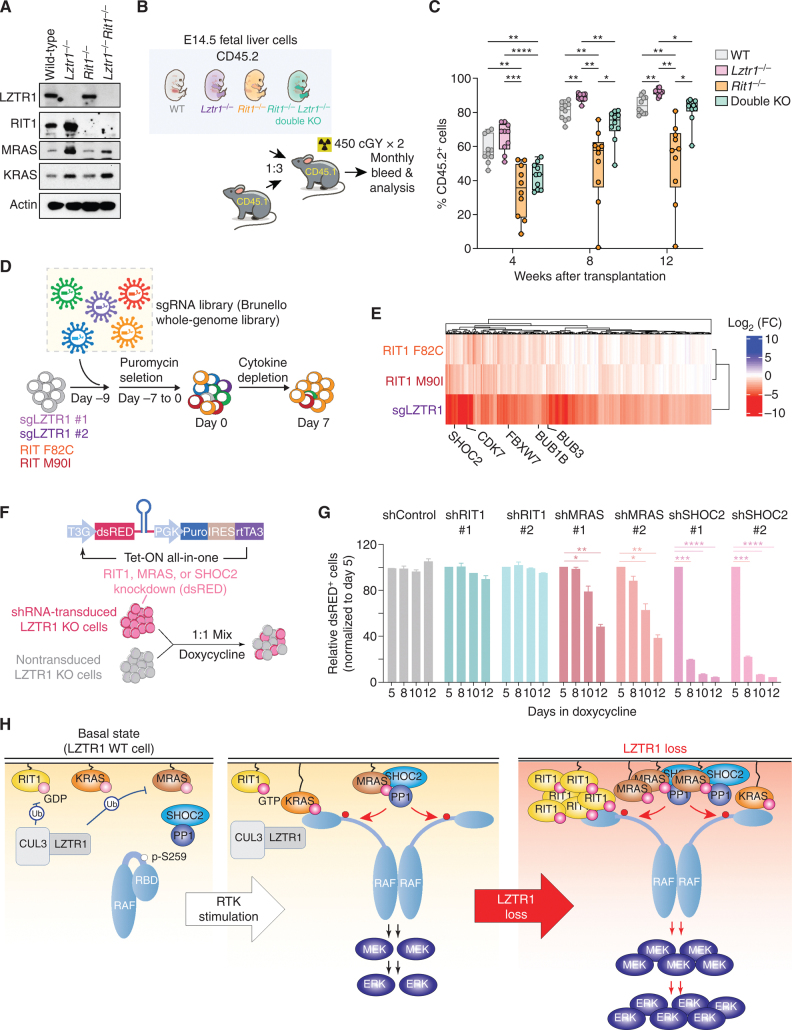
![Figure 6. LZTR1-mediated drug resistance can be overcome by modulating RAS-GTP abundance. A, Scatter plots of RNA expression levels [shown as normalized reads per kilobase of transcript per million reads mapped (RPKM)] plotted against drug-sensitivity measured ex vivo as the area under the curve (AUC) from AML patient samples harboring FLT3-ITD mutation. n ≥ 111. *, P < 0.05. Pearson correlation coefficients are shown as (r) values. Statistical significance is indicated. B, Schema of the MOLM-13 cell line xenograft experiment. Luciferase-expressing MOLM-13 cells treated with anti-LZTR1 or control sgRNAs were systemically engrafted with 1 × 105 cells/animal via a tail-vein injection after sublethal (225 cGy) irradiation. Seven days later, animals were treated with either vehicle or 30 mg/kg/day of gilteritinib via oral gavage. Animals underwent bioluminescence imaging weekly. C, Kaplan–Meier curve of experiment in B. n = 7–10. **, P < 0.01; ***, P < 0.001. D, Schema of protein-based degrader of RAS proteins. A chimeric protein consisting of a high-affinity target-binding domain for RAS proteins (DARPin K27) is fused to an engineered E3 ligase adapter (SPOP) to confer ubiquitin-mediated degradation of RAS proteins. E, Western blot demonstrating levels of LZTR1 as well as total and GTP-bound KRAS/NRAS/MRAS and RIT1 in MOLM-13 cells ± LZTR1 KO upon doxycycline-mediated induction of the RAS biodegrader (performed in biological triplicate). EV, empty vector. F, Seventy-two-hour cell proliferation assay on parental and LZTR1 KO MOLM-13 cells ± induction of RAS degradation. Cell viability was measured in triplicate using CellTiter-Glo. G, 2D synergy plots using the Zero Interaction Potency (ZIP) model of control sgRNA (“WT”) or anti-LZTR1 sgRNA (“LZTR1 KO”) MOLM-13 cells treated for 72 hours with BI-3406 and/or gilteritinib at various concentrations. Western blot demonstrating p-MEK, p-ERK, and total MEK, ERK, KRAS/NRAS/MRAS, and RIT1 levels (H) as well as RAS-GTP levels (I) in MOLM-13 cells ± LZTR1 KO treated with increasing concentration of DMSO or BI-3406 alone 4 hours after drug treatment.](https://cdn.ncbi.nlm.nih.gov/pmc/blobs/a403/9533010/5b444dfc9322/2434fig6.jpg)