Schwann cells undergo c-Jun–mediated reprogramming in pancreatic cancer, analogous to their trans-differentiation following trauma, and collectively organize into dynamic tracks that promote cancer cell invasion.
Abstract
Nerves are a component of the tumor microenvironment contributing to cancer progression, but the role of cells from nerves in facilitating cancer invasion remains poorly understood. Here we show that Schwann cells (SC) activated by cancer cells collectively function as tumor-activated Schwann cell tracks (TAST) that promote cancer cell migration and invasion. Nonmyelinating SCs form TASTs and have cell gene expression signatures that correlate with diminished survival in patients with pancreatic ductal adenocarcinoma. In TASTs, dynamic SCs form tracks that serve as cancer pathways and apply forces on cancer cells to enhance cancer motility. These SCs are activated by c-Jun, analogous to their reprogramming during nerve repair. This study reveals a mechanism of cancer cell invasion that co-opts a wound repair process and exploits the ability of SCs to collectively organize into tracks. These findings establish a novel paradigm of how cancer cells spread and reveal therapeutic opportunities.
Significance:
How the tumor microenvironment participates in pancreatic cancer progression is not fully understood. Here, we show that SCs are activated by cancer cells and collectively organize into tracks that dynamically enable cancer invasion in a c-Jun–dependent manner.
See related commentary by Amit and Maitra, p. 2240.
This article is highlighted in the In This Issue feature, p. 2221
INTRODUCTION
The migration of cancer cells and their invasion away from primary sites lead to metastasis, the principal cause of death in patients with cancer. The mechanisms of cancer cell migration and invasion are complex (1, 2) and dependent on contributions from the tumor microenvironment (TME). The TME provides promigratory factors (2) and a matrix with distinct mechanical properties that modulate cancer cell motility (3–5). The TME cells known to affect cancer invasion through these mechanisms are immune cells and fibroblasts (4, 5). Nerves are also an important component of the TME that stimulate cancer progression. Chemical and surgical denervation experiments in animals show inhibition of tumor growth in models of pancreatic, prostate, breast, skin, and head and neck cancers (6–11). Some cancer cells migrate, proliferate, and invade around nerves in a process called perineural invasion (PNI). Through PNI, cancer cells may spread outside the organ of the tumor's origin, representing an unusual form of metastasis. PNI is associated with pain, paralysis, and worse patient survival (12–14). Paracrine signaling has been studied in nerve cells, and a variety of secreted factors stimulate cancer cell migration (15). However, there are currently no treatments designed to inhibit PNI, cancer invasion induced by nerves, or cancer innervation (16).
Pancreatic ductal adenocarcinoma (PDAC) is an aggressive cancer with a 5-year survival rate of just 8% (17) and an incidence of PNI up to 100% (12). We demonstrated that Schwann cells (SC), the most abundant cell type in nerves, are a mediator of PNI and facilitate pancreatic cancer cell dispersion (18). We and others have reported an increase of an SC subtype expressing elevated levels of glial fibrillary acidic protein (GFAP) when in close proximity to pancreatic cancer cells in patient specimens (18, 19). A similar increased expression of GFAP is also found in SCs responding to nerve injury. These nonmyelinating repair SCs play a dynamic role in nerve regeneration and originate from GFAP-negative SCs, myelinating SCs, and nonmyelinating (Remak) SCs expressing moderate GFAP levels that ensheathe axons in uninjured nerves (20, 21).
SCs are highly plastic cells that undergo cellular reprogramming in response to nerve injury (22–25) or infection (26). SC reprogramming during nerve repair results in the altered expression of approximately 4,000 genes and is driven by c-Jun, Notch, Sox2, and MAPK signaling in addition to other factors (27, 28). Quiescent myelinating and Remak SCs are reprogrammed into nonmyelinating nerve repair SCs that exhibit dynamic motility, release neurotrophic factors and chemokines, recruit immune cells, clear myelin by autophagy and phagocytosis, and reorganize into cellular tracks called Büngner bands. These bands are composed of elongated and aligned SCs that form a path to guide the regeneration of damaged axons (29).
Here, we found that diminished survival in patients with PDAC correlates with nonmyelinating SC gene expression signatures. Cancer cells activate SCs, inducing a c-Jun–dependent reprogramming that is related to that triggered by nerve injury and shifts SC differentiation state toward that of nonmyelinating/repair cells. Linear tracks of such SCs, which we have named tumor-activated Schwann cell tracks (TAST), function as active pathways for pancreatic cancer cell migration and invasion. This study presents a novel mechanism of cancer cell invasion that exploits dynamic cellular tracks formed by reprogrammed SCs that drive cancer migration and propagation.
RESULTS
Nonmyelinating SC Signature Scores Correlate with Diminished Survival in Patients with Pancreatic Adenocarcinoma and with Pathways Related to Cancer Invasion
We first assessed the relationship of SCs with outcomes for patients with pancreatic cancer. We obtained several signatures of human SCs, including myelinating SC and nonmyelinating SC signatures from the Tabula Sapiens portal using OnClass (ref. 30; Fig. 1A; Supplementary Data S1). SC precursors differentiate into immature SCs that diverge to form myelinating SCs or nonmyelinating (Remak) SCs. Other SCs that do not form myelin include terminal SCs and repair SCs that form from myelin and Remak cells after injury (22). We computationally derived an inferred pathway activation/suppression (IPAS) SC signature score for each available signature (31–33) for 178 patients with pancreatic adenocarcinoma (PAAD) from The Cancer Genome Atlas (TCGA) using their gene expression levels (Supplementary Data S2). We tested for correlations between the SC signature scores and clinical outcomes [survival and progression-free survival (PFS); Fig. 1B–F; Supplementary Fig. S1A]. Results revealed significant correlations between clinical outcomes and nonmyelinating, precursor, terminal, and global SC signature scores, with higher scores corresponding to worse outcomes (Fig. 1B and C; Supplementary Fig. S1A). There was a similar trend, but no significant clinical correlation was identified for the myelinating SC signatures (Fig. 1E and F). The reported correlations were for a stratification of patients that yielded the best significance. This stratification often corresponded to a 10:90 ratio, with the highest proportion of patients being in the high SC signature group. When stratified by mean or median, significance was not reached (Supplementary Data S3). We show that the differences in outcomes between the two sets of patients at this stratification are truly significant and are highly unlikely to be reproduced by random (Supplementary Data S3, Prand).
Figure 1.
Nonmyelinating SC signature scores correlate with diminished survival in patients with pancreatic adenocarcinoma and with pathways related to cancer invasion. A, Hierarchical organization of SCs from Tabula Sapiens (full lines), with dashed arrows indicating transitions in the SC lineage (22). B, Forest plot with hazard ratio and 95% confidence interval. ADEX, aberrantly differentiated endocrine exocrine. C–F, Kaplan–Meier curves of overall survival (C, E) and progression-free survival (PFS; D, F) with high or low scores for signatures of nonmyelinating SCs (C, D) and myelinating SCs (E, F) in 178 TCGA pancreatic adenocarcinoma (PAAD) patients. NS, not signficant. G, Nonmyelinating and myelinating SC signature scores in PDAC subtypes (A, ADEX; I, immunogenic; P, progenitor; S, squamous). *, P < 0.05; **, P < 0.01; ***, P < 0.001; ****, P < 0.0001. H, Heat map of gene sets correlating with high and low scores for nonmyelinating SC signature in TCGA PAAD patients. I, Heat map of gene sets correlating with high and low scores for myelinating SC signature in TCGA PAAD patients. Columns represent TCGA PAAD samples that have been rank ordered by the top row signature. ECM, extracellular matrix; HMDB, Human Metabolic Database; KEGG, Kyoto Encyclopedia of Genes and Genomes; SMPDB, Small Molecule Pathway Database. J, Top seven enriched pathways in HEI-286 cocultured with MiaPaCa-2 as compared with HEI-286 SCs alone (EnrichR, human KEGG 2019 data set). K, Kaplan–Meier curve of overall survival with high or low scores for the cancer-exposed HEI-286 SC (HEI-mix) signature in 178 TCGA PAAD patients. L, Heat map of gene sets correlating with high and low scores for cancer-exposed HEI-286 SC (HEI-mix) signature in TCGA PAAD patients. Columns represent TCGA PAAD samples that have been rank ordered by the top row signature. M, Kaplan–Meier of overall survival of PDAC patients with high or low GFAP expression in SCs determined histologically.
Furthermore, using Cox regression modeling, we found that the SC scoring is associated with survival in both univariate and multivariate analyses (Supplementary Data S4). Age, cellularity, and stage showed some association with survival as well: Patients of older age and higher stage have a poorer outcome as compared with younger patients and patients of lower stages. Tumor cellularity is significantly inversely correlated with SC scores, consistent with the presence of nerves and SCs in stroma. Neither of the confounding factors overrode the statistical significance of the SC scoring.
We also stratified TCGA patients into four PDAC subtypes previously described (ref. 34; Supplementary Fig. S1B): squamous, pancreatic progenitor, immunogenic, and aberrantly differentiated endocrine exocrine (ADEX) subtypes. SC signatures are associated with increased risk at a similar level to the squamous subtype (Fig. 1B). Cox analysis does not show an association between SC signature and subtype when competing with other variables. However, SC signature scores did vary with PDAC subtype (Fig. 1G; Supplementary Fig. S1C; Supplementary Data S5). The lowest SC scores were in the progenitor group, and the highest scores were in the immunogenic group. The overall order of the SC scores is: immunogenic > ADEX = squamous > progenitor. These data suggest that the PDAC subtypes are associated with varying degrees of SC expression and activity.
The correlations of SC signatures with survival were also found in a Memorial Sloan Kettering (MSK) cohort of short-survivor patients, which has a similar ratio of PDAC subtypes (Supplementary Fig. S1D; Supplementary Data S6). In contrast, a cohort of rare MSK long-survivor patients (>4 years) shows that elevated SC signatures were associated with improved survival. This cohort has fewer squamous PDAC subtypes (Supplementary Data S6; Supplementary Fig. S1D).
Further analysis indicated that the gene expression signature of nonmyelinating SCs positively correlated with multiple pathways related to cancer invasion in the data set from TCGA, including epithelial–mesenchymal transition (EMT), MAPK signaling, PI3K–Akt signaling, extracellular matrix organization, and others. The nonmyelinating SC signature also correlated with axonal guidance gene sets (Fig. 1H). The myelinating SC signature positively correlated with gene sets related to immune cells and immune function (Fig. 1I; Supplementary Fig. S1E and S1F). Similarly, the nonmyelinating SC signature is associated with EMT/invasive pathways and myelinating SC signature with immune-related pathways in the MSK patient cohort of short and long survivors (Supplementary Fig. S1G). The negatively correlated gene sets were similar for both nonmyelinating and myelinating signatures and involved pathways with lipoic acid metabolism, mitochondrial activity, and tRNA processing (Fig. 1H and I).
We created a cancer cell–exposed SC signature (HEI-mix) experimentally using the nonneoplastic human SC line HEI-286 that was cocultured either with or without MiaPaCa-2 pancreatic cancer cells. Gene Ontology (GO) pathway analysis of the transcriptome of SCs sorted by FACS after cancer cell coculture revealed a significant enrichment of genes related to axon guidance and MAPK signaling pathways (Fig. 1J; Supplementary Data S7). We then tested correlations between our cancer cell–exposed SC signature score (HEI-mix) with clinical outcomes using the data set from TCGA. HEI-mix scores were significantly correlated with patient survival, with higher scores corresponding to worse patient survival (Fig. 1K). Furthermore, the gene sets and pathways that correlated the most with this signature were nearly identical to those that correlated with the nonmyelinating SC signature and are linked to cancer invasion (Fig. 1L; Supplementary Data S8). Among all SC signatures tested, the nonmyelinating SC signature was the cell signature that most closely correlated with the HEI-mix signature (Supplementary Fig. S1H). Altogether, these findings link both nonmyelinating SCs and SCs whose transcriptome profile has been altered by the presence of cancer cells with a worse clinical severity of PAAD and transcriptomic profiles relating to cancer cell invasion.
SCs Wrap and Align Cancer Cells In Vivo and In Vitro
We previously reported the presence of nonmyelinating GFAP+ SCs in PDAC (18). We found that the high expression of GFAP+ SCs on histologic sections of patients with PDAC correlates with poor survival (Fig. 1M), consistent with the signature-based survival data. To explore the role of these cells in cancer invasion, we examined their distribution in human specimens of PDAC. GFAP+ SCs closely associated with cancer cell clusters in the nerves with PNI (Fig. 2A–H). Nerves invaded by cancer cells exhibit SC heterogenicity in GFAP expression, with strongly GFAP+ SCs located near the cancer cells (Fig. 2B–D; Supplementary Fig. S2A). The GFAP+ SCs appeared to align cancer cells within the nerves (Fig. 2E–H). Orthogonal views (xy and xz) of confocal images revealed that GFAP+ SCs enveloped the cancer cells (Fig. 2H). In PDAC stroma rich with nerve fibers, we also found that GFAP+ SCs formed close associations with cancer cells. Sometimes a GFAP+ SC encircled a single cancer cell or a small cluster of cancer cells (Fig. 2I–K). Other times, columns of cancer cells were found intertwined with SCs insinuating around them (four of six patients with PNI; Fig. 2L and M; Supplementary Fig. S2B). We examined SC and cancer cell interactions using a PNI model in which Panc02 cancer cells injected into sciatic nerves of mice migrate toward the spinal cord (35, 36). As in PNI human samples, longitudinal sections of the mouse sciatic nerves revealed that GFAP+ SCs surrounded the clusters of cancer cells (Fig. 2N and O).
Figure 2.
SCs wrap and align cancer cells. A, Hematoxylin and eosin (H&E) section of a human PDAC specimen with PNI. Scale bar, 200 μm. N, nerve; S, stroma; T, tumor. B, GFAP (green) and cytokeratin (CK; magenta) staining in a section adjacent to A showing cancer cells surrounded by GFAP+ SCs in a nerve. The yellow and orange dotted regions of the nerve are adjacent (A) or distal (D) to the cancer cells. Scale bar, 200 μm. C, Quantification of nerves with an uneven GFAP distribution comparing when cancer is visibly present or absent. D, Quantification of GFAP mean intensity in adjacent and distal regions of nerves (paired t test, P < 0.0001). E, Enlargement of rectangle in A. Scale bar, 50 μm. F, GFAP and CK staining of an adjacent section of E. Scale bar, 50 μm. G, Confocal images corresponding to rectangle 1 in F. Scale bar, 10 μm. H, Confocal images corresponding to rectangle 2 in F. Top image is an xy maximum projection, and the bottom image is an xz image corresponding to the dotted line. Scale bars, 10 μm. I, H&E section of a human PDAC specimen with a nerve (N) and tumor cells (T) in the neighboring stroma (S). Scale bar, 200 μm. J, Enlargement of I. Arrows indicate two cancer cells. Scale bar, 20 μm. K, GFAP and CK staining of a section adjacent to J showing GFAP+ SCs wrapping two cancer cells (arrows). Scale bar, 20 μm. L and M, H&E staining and adjacent GFAP and CK staining of a human PDAC specimen showing GFAP+ SCs around aligned cancer cells. Scale bars, 20 μm. N and O, H&E staining and adjacent GFAP staining of a longitudinal section of a murine sciatic nerve injected with cancer cells. Scale bars, 500 μm. P, Schematic representation of HEI-286 SCs and MiaPaCa-2 cancer cells in a 3D Matrigel invasion assay. The MiaPaCa-2-RFP cancer cells are placed on the surface of a Matrigel chamber in which HEI-286 SCs have grown. Cancer cells invade the gel as a chain of cancer cells surrounded by HEI-286 SC tracks. Q, Maximum projection of confocal images showing a chain of cancer cells lined up within a tubular structure of HEI-286 SCs in Matrigel. Scale bar, 20 μm. R, Single focal planes of confocal images showing MiaPaCa-2 cancer cells and HEI-286 SCs organized into columns. Longitudinal (left) and transverse (right) images correspond to the indicated positions. Scale bars, 20 μm.
We next examined SC and cancer cell interactions with an in vitro 3D model of cancer invasion to assess if SCs wrap cancer cells. HEI-286 SCs expressing GFP facilitate the downward invasion of red fluorescent human pancreatic cancer cells MiaPaCa-2 and Panc01 seeded on top of Matrigel chambers (18). Three days after the addition of MiaPaCa-2 or Panc01, HEI-286 SCs were found closely associated with chains of cancer cells (Fig. 2P–R; Supplementary Fig. S2C). HEI-286 SCs wrapped around and aligned with chains of cancer cells (Fig. 2Q and R), creating invasive columns of cells. In contrast, cancer cells seeded on Matrigel chambers lacking HEI-286 SCs, or in the presence of NIH3T3 fibroblasts, failed to invade and did not organize into chains (Supplementary Fig. S2D and S2E). Collectively, the in vivo and in vitro images revealed a consistent pattern of organization in which SCs wrapped cancer cells and created columns of SCs and cancer cells. These observations suggest that SCs reorganize cancer cells into chains and create tracks for migration, which we have named TASTs.
SCs Organize into Dynamic Tracks that Allow Cancer Cell Migration
We used microfabricated channels to investigate TASTs. They allow the alignment of HEI-286 SCs in 3D as in TASTs and the visualization of cell motility and cell behavior. To determine if cancer cells migrate along TASTs, we assessed dynamic interactions between HEI-286 SCs and MiaPaCa-2 cells. HEI-286 SCs and MiaPaCa-2 cells, seeded together in wells, migrated into the channels and cells organized into columns of SCs and cancer cells (Fig. 3A and B). Confocal microscopy showed that HEI-286 and MiaPaCa-2 cells occupied the full width of the 10- to 20-μm-wide channels (Supplementary Fig. S3A, xz 2 and xz 4). HEI-286 SCs wrapped around MiaPaCa-2 cells at 80% of contact sites, either partially (Supplementary Fig. S3A, xz 3) or completely (Fig. 3B, xz). This wrapping behavior around centrally positioned cancer cells mirrored observations from our 3D Matrigel invasion assay (Fig. 2P and R) and PDAC sample (Fig. 2H and K). We examined cancer cell dynamics in the 3D microchannels seeded first with HEI-286 SCs (Supplementary Fig. S3B). Cancer cells migrated within the microchannels lined by HEI-286 SCs (Fig. 3C; Supplementary Video S1). HEI-286 wrapped themselves around MiaPaCa-2, lined themselves along the microchannel walls, and permitted cancer cell migration through the microchannel (Fig. 3C and D; Supplementary Video S2).
Figure 3.
SCs form dynamic tracks for cancer cells. A, Schematic of microchannels with HEI-286 SCs (green) and MiaPaCa-2 cells (magenta). Both cell types are seeded in the adjacent well (left, 2D) and enter microchannels (3D) where they make contact with each other. PDMS, polydimethylsiloxane. B, Confocal images of HEI-286 SCs and MiaPaCa-2 within microchannels in longitudinal (xy) and transverse (xz) sections. Scale bars, 50 μm. C, Confocal images of time-lapse movies showing a cancer cell moving in a microchannel lined by HEI-286 SCs. Time is h:min. Scale bars, 15 μm. D, Confocal images of time-lapse movies showing an HEI-286 SC wrapping around a cancer cell. Scale bar, 50 μm. E, Fluorescent images of time-lapse movie showing an HEI-286 SC pushing a cancer cell. Arrows indicate cancer cell displacement. Circles indicate intracellular movement within the SC (* and ** are time points shown in E). F, Fluorescent images of an HEI-286 SC and cancer cell from E at two time points (* and **) overlaid with vectors obtained by PIV analysis indicating vector direction. G, Quantification of the mean instantaneous velocity of the HEI-286 SC and cancer cell in E, F, and G. Corresponding time points are indicated by * and **. H, Fluorescent images of time-lapse movie showing HEI-286 SCs pulling a cancer cell. Arrows indicate cancer cell displacement. I, Fluorescent images of the HEI-286 SCs and cancer cells from H at one time point (*) overlaid with vectors obtained by PIV analysis. J, Quantification of the mean instantaneous velocity. *, Corresponding time point in I. Dotted circles indicate periods with synchronized increases in velocity for both cancer cells and SCs.
SCs Exert Forces on Cancer Cells that Propel Migration
To assess if SCs influence the movement of cancer cells within TASTs, we analyzed time-lapse microscopy and particle image velocimetry (PIV). A single HEI-286 SC was able to propel a cancer cell through a pushing effect (Fig. 3E; Supplementary Video S3). Circles show a propulsive wave within an HEI-286 SC that propagated from the center of the cell toward its contact point with a static cancer cell. The wave then created an SC protrusion at the site of contact, which propelled the cancer cell away from the HEI-286 SC. PIV analysis of both cells revealed intracellular movement corresponding to an intracellular propulsive wave within the SC prior to the cancer cell displacement (Fig. 3F and G), with the movement of both cells in the same direction (Fig. 3G). We also observed squeezing motions by HEI-286 SCs surrounding a MiaPaCa-2 cell. The combined effect of wrapping and squeezing of the MiaPaCa-2 cells led to propulsive forces moving the cancer cell (Supplementary Fig. S3C; Supplementary Video S4). HEI-286 SCs also exerted pulling forces, making contact with MiaPaCa-2 cells and then retracting the cancer cells toward themselves (Fig. 3H; Supplementary Video S5). PIV analysis revealed that HEI-286 SCs and cancer cells displayed coordinated directional vectors (Fig. 3I) and instantaneous velocity intensities (Fig. 3J). Increases in SC velocities correlated temporally with increases in cancer cell velocities (circles in Fig. 3J). The HEI-286 SCs that formed the TASTs permitted MiaPaCa-2 cells to pass through them centrally and directly applied forces on the cancer cells. HEI-286 SCs were able to push, squeeze, or pull cancer cells, contributing to cancer cell motility.
Cancer Cells Activate c-Jun in SCs
Following nerve trauma, myelinating and Remak SCs transdifferentiate into SCs that actively engage in nerve repair. This transition is controlled in part by the transcription factor c-Jun (27), which reprograms myelinating and Remak SCs into SCs that align into Büngner bands to enable axonal guidance and nerve regeneration. This SC organization during nerve repair shows similarities to our observations of SC alignment into tracks enabling cancer cell migration in vitro. In the PAAD TCGA data set, we identified a significant correlation between JUN expression level and the nonmyelinating SC signature (P = 0.0004 r = 0.2608), but not with the myelinating signature (P = 0.8334, r = 0.01588). Furthermore, higher JUN expression correlated with worse patient survival (Fig. 4A; P = 0.035).
Figure 4.
Cancer cells induce SC c-Jun activation and reprogramming. A, Kaplan–Meier curve of overall survival with high or low JUN expression in TCGA PAAD patients. B, P-c-Jun staining in S100-labeled nerves from PDAC specimens that are close to tumor as compared with nerves from adjacent (Adj.) normal tissue. Scale bars, 20 μm. C and D, Assessment of nerves from within pancreatic specimens of non-PDAC pathology, PDAC, and the normal tissues adjacent to PDAC. C, Quantification of the number of nerves per slide with no (-), low (+)-, or high (++)-intensity P-c-Jun staining. Each x-axis number represents a patient. D, Percentage of P-c-Jun–positive nerves (scored + or ++) in the pancreatic specimens of non-PDAC pathology, PDAC, and the normal tissues adjacent to PDAC (non-PDAC, n = 4; PDAC and adjacent normal, n = 6 each; mean ± SEM). E and F, Immunofluorescence of P-c-Jun (white) in a Panc02-injected murine sciatic nerve expressing GFP+ SCs. P-c-Jun is expressed in the green SCs but not in the Panc02 cancer cells (CC). Scale bars, 50 μm. G, Quantification of P-c-Jun fluorescence intensity in SCs of uninjected nerves and in SCs close to or far from injected Panc-02 tumor cells (near tumor: n = 6; far from tumor: n = 4; PBS-injected: n = 7; mean ± SEM, representative of three independent experiments). Fluorescence intensity is the mean intensity measured per area of nerve covered by green SCs. H, P-c-Jun staining in a sciatic nerve injected with Panc-02. (1) Region adjacent to the tumor. (2) Region far from the tumor. Scale bars, 1,000 and 100 μm. I, P-c-Jun staining in a normal nerve without cancer cells. Scale bar, 100 μm. J, Immunofluorescence of P-c-Jun in HEI-286 GFP SCs grown alone or grown mixed with MiaPaCa-2-RFP. Top images show P-c-Jun staining alone in HEI-286 SCs alone (left) and cocultured HEI-286 mixed with MiaPaCa-2 (right). Bottom overlay images allow identification of HEI-286 SCs (green) and MiaPaCa-2 (magenta). Scale bar, 50 μm. K, Quantification of P-c-Jun fluorescence intensity in HEI-286 SCs grown alone or mixed with MiaPaCa-2. Mean fluorescence intensity was measured per HEI-286 SC (n > 15 cells/group, mean ± SEM, representative of three independent experiments). L and M, GSEA assessing axon guidance genes in cancer cocultured HEI-286 SCs compared with HEI-286 SCs alone (L and in cancer cocultured HEI-286 compared with cancer cocultured c-Jun KO HEI-286 SCs (M). NES, normalized enrichment score. N, IPAS scores for the axon guidance genes (KEGG 2019).
To explore whether c-Jun–dependent reprogramming occurs during SC and cancer cell interactions, we assessed c-Jun phosphorylation (P-c-Jun) in the nuclei of SCs in proximity to cancer cells in human surgical specimens. We identified strong P-c-Jun expression in nerves within PDAC, reduced P-c-Jun expression in nerves in normal tissue adjacent to PDAC, and minimal P-c-Jun expression in nerves from non-PDAC pancreatic specimens (Fig. 4B–D; Supplementary Fig. S4A). This pattern suggests that cancer cells trigger SC c-Jun activity.
We also assessed SC P-c-Jun expression in the murine model of nerve invasion (Fig. 2N and O) using transgenic P0-CRE mT/mG mice with GFP-expressing SC after injecting Panc02 cancer cells into the sciatic nerves. P-c-Jun localized in GFP-expressing SCs but not in cancer cells (Fig. 4E and F). We assessed P-c-Jun expression in SCs that were (i) close to the cancer cells, (ii) far from the cancer cells, or (iii) in non–cancer-bearing mice. Immunofluorescence staining revealed a significant increase of SC P-c-Jun staining only when in close proximity to the cancer cells (Fig. 4G–I).
We tested if SC c-Jun activation could be mediated directly by cancer cells in the absence of the TME using HEI-286 SCs expressing GFP cocultured with MiaPaCa-2 cells expressing RFP. P-c-Jun expression was significantly increased in the HEI-286 cells mixed with MiaPaCa-2-RFP as compared with HEI-286 cells alone (Fig. 4J and K). These results demonstrate that cancer cells directly induce SC c-Jun activation.
To assess the transcriptional effects of cancer cell–mediated SC c-Jun activation, we generated c-Jun knockout (KO) HEI-286 SCs (Supplementary Fig. S4B–S4D). We noted that NCAM1 and GDNF molecules, which are expressed by SCs during PNI (18, 35) and in a c-Jun–dependent manner during nerve repair (27), showed reduced expression in c-Jun KO HEI-286 (Supplementary Fig. S4E). We examined the gene expression profiles of c-Jun KO HEI-286 SCs cocultured with MiaPaCa-2 cancer cells as compared with c-Jun KO HEI-286 SCs grown alone, and derived the cancer-exposed c-Jun KO SC signature (KO-HEI-mix). The correlation of this signature score with patient survival was less significant (P = 0.0480) than the correlation of the control HEI-mix signature score, with patient survival reported previously in Fig. 1I (P = 0.0182). In HEI-286 cells, GO pathway analysis and gene set enrichment analysis (GSEA) revealed enrichment of axon guidance genes in cocultured HEI-286 SCs as compared with the HEI-286 SCs grown alone (Fig. 1H; Fig. 4L) and of other c-Jun–related pathways such as the MAPK signaling pathway (Fig. 1H). To assess the relevance of c-Jun in SC axon guidance activation by cancer, we examined the gene expression profile of HEI-286 SCs as compared with c-Jun KO HEI-286 SCs cocultured with MiaPaCa-2 cells. GSEA revealed an enrichment in axon guidance genes for HEI-286 SCs with intact c-Jun [normalized enrichment score (NES) = 1.56, FDR q-value (FDRq) = 0.0015; Fig. 4M; Supplementary Fig. S4F]. The IPAS scores of axon guidance genes [Kyoto Encycopledia of Genes and Genomes (KEGG) 2019] for the different conditions revealed the comparative enrichment to be HEI_mix>KO_mix>HEI>KO (Fig. 4N). Both of these analyses implicate a role for SC c-Jun in the regulation of axon guidance gene expression following cancer exposure.
SC gene expression changes during nerve repair that are dependent on c-Jun have been identified (27). We tested enrichment of the gene set upregulated in injured sciatic nerves of wild-type (WT) as compared with c-Jun KO mice (27) in our cocultured SCs. GSEA revealed an enrichment of c-Jun nerve repair genes in cocultured HEI-286 SCs (FDRq <0.25) as compared with HEI-286 SCs grown alone (NES = 1.191, FDRq = 0.188; Supplementary Fig. S4G) despite species differences. Similarly, IPAS revealed a higher enrichment of c-Jun nerve repair genes in the cocultured HEI-286 SCs as compared with HEI-286 SCs grown alone (Supplementary Fig. S4H). We then tested whether our identified genes upregulated in cocultured HEI-286 SCs as compared with cocultured c-Jun KO HEI-286 SCs (452 genes with FC >2, P < 0.05) were enriched in the injured (cut) or intact (uncut) murine sciatic nerve from the same study (27). GSEA revealed an enrichment of these human genes in the murine-injured sciatic nerves (NES = 1.318, FDRq = 0.015; Supplementary Fig. S4I). These findings indicate that transcriptomic changes in HEI-286 SCs following cancer exposure show parallels with SC reprogramming following nerve injury. Both axon guidance– and c-Jun–related nerve repair gene sets in SCs are induced by cancer.
SC c-Jun Enhances Cancer Migration along TASTs in Microchannels
We assessed how SC c-Jun affects cancer cell migration within microchannels. The c-Jun KO HEI-286 SCs did not migrate as fast as control HEI-286 SCs (Supplementary Fig. S5A–S5C), made shorter columns than control HEI-286 SCs when combined with MiaPaCa-2 (Supplementary Fig. S5D), and were less able to wrap cancer cells. Although 80% of control HEI-286 SCs wrapped cancer cells at the site of contact as described (Fig. 3A and B), only about 50% of the c-Jun KO HEI-286 SCs wrapped cancer cells (Fig. 5A and B). The c-Jun KO HEI-286 SCs sometimes clustered in the central part of the microchannel, surrounded by cancer cells that lined the periphery (Fig. 5A).
Figure 5.
SC c-Jun facilitates cancer migration along SC tracks in microchannels. A, Confocal images of HEI-286 with MiaPaCa-2 (top) and c-Jun KO HEI-286 with MiaPaCa-2 (bottom) within microchannels showing longitudinal (xy) and transverse (xz) sections. Scale bars, 10 μm. B, Quantification of percentage of HEI-286 SCs wrapping around MiaPaCa-2 cells for control or c-Jun KO HEI-286 SCs (n = 5–7 recordings per group with a total of 23 to 27 cells/group, mean ± SEM). C, Quantification of distance migrated by cancer cells in microchannels of different widths and occupied by HEI-286 SCs (n = 9–20 cells per channel size). D, Tracks of cancer cells in microchannels occupied by control and c-Jun KO HEI-286 SCs. Average representation of 21 MiaPaCa-2 cells in each group (F test with F = 930.1; P < 0.0001). E, Quantification of cancer cell speed (absolute values) in microchannels occupied by control versus c-Jun KO HEI-286 SCs (n = 21 cells in each group). F, Quantification of MiaPaCa-2 cell directional persistence (absolute values) in microchannels occupied by control versus c-Jun KO HEI-286 SCs (n = 21 cells in each group). G, Fluorescent images of time-lapse movies showing the behavior of a MiaPaCa-2 after HEI-286 SC contact. A cancer cell passes by a control HEI-286 SC, whereas another is blocked by a c-Jun KO HEI-286 SC. Scale bars, 15 μm. H, Quantification of G showing the percentage of cancer cells (CC) passing by an HEI-286 SC (n = 3 experiments per group with at least 11 cells/group in each experiment, mean ± SEM). I, Images showing stiffness maps of cocultured HEI-286 SCs versus c-Jun KO HEI-286 SCs. Scale bar, 15 μm. J, Quantification of stiffness of HEI-286 SCs versus c-Jun KO HEI-286 SCs, measured by AFM (n = 12–13 cells/group, mean ± SEM, representative of two independent experiments). K, Images showing actin staining in cocultured HEI-286 SCs versus c-Jun KO HEI-286 SCs. Scale bar, 20 μm.
When HEI-286 SCs were seeded first in 10- to 16-μm-wide microchannels, MiaPaCa-2 migrated significantly longer distances combined with control HEI-286 SCs as compared with c-Jun KO HEI-286 SCs (Fig. 5C). Interestingly, this difference was not seen in channels of wider diameter (Fig. 5C), showing that HEI-286-SC c-Jun–dependent enhancement of MiaPaCa-2 migration occurs only under confinement conditions. Similar results were obtained using Panc01 cancer cells (Supplementary Fig. S5E).
We tracked individual MiaPaCa-2 cell movement within microchannels of 10- to 16-μm width occupied by HEI-286 SCs to assess migration dynamics over 20 hours. Cancer cells displayed persistent migration and reached an average speed of 7.80 ± 1.04 μm/hour in the presence of control HEI-286 SCs. In contrast, the average cancer cell speed was significantly reduced at 2.04 ± 0.34 μm/hour in the presence of c-Jun KO HEI-286 SCs (Fig. 5D and E; Supplementary Fig. S5F). The average directional persistence of cancer cell migration was significantly greater in the microchannels seeded with control HEI-286 SCs as compared with c-Jun KO HEI-286 SCs (Fig. 5F). MiaPaCa-2 often either failed to move or moved backward in the presence of c-Jun KO HEI-286 SCs (Supplementary Fig. S5E).
We next evaluated the ability of a migrating cancer cell to either pass or be blocked by HEI-286 SCs in a microchannel after making contact. Our experiments demonstrated that control HEI-286 SCs enabled the passage of cancer cells in a confined microchannel (about 80% of the time after contact), whereas c-Jun KO HEI-286 SCs were prone to blocking cancer cell passage (about 40% of the time; Fig. 5G and H; Supplementary Video S6).
To test whether differences in the physical properties of control as compared with c-Jun KO HEI-286 SCs contribute to cancer cell transit, we measured the physical stiffness of HEI-286 SCs using atomic force microscopy (AFM). We reasoned that softer and more pliable HEI-286 SCs might facilitate the passage of MiaPaCa-2 cells by being more dynamic and better able to mold their shape when contacting cancer cells. AFM analysis revealed that c-Jun KO HEI-286 SCs were indeed stiffer than control HEI-286 SCs (Fig. 5I and J). The difference in physical properties might originate from a rearrangement of actin. We looked for actin organization in cocultured HEI-286 SCs. Phalloidin staining revealed a difference in actin organization between control and c-Jun KO HEI-286 SCs. Actin was organized in stress fibers in control cells and as cortical actin in c-Jun KO cells (Fig. 5K). These findings suggest that SC c-Jun regulates actin organization and modifies cellular physical properties. In addition, c-Jun enables SCs to wrap cancer cells, to conform to the microchannel, and to create tracks supporting the fast directional migration of cancer cells.
c-Jun Coordinates SC Collective Organization into TASTs that Allow Cancer Invasion
We characterized the role of c-Jun in the 3D organization of SCs in Matrigel. The control HEI-286 SCs adopted an elongated shape and self-organized into linear, branched structures in the Matrigel, forming frequent end-to-end cellular contacts (Fig. 6A, left). In contrast, the c-Jun KO HEI-286 SCs adopted a more rounded shape (Fig. 6A, right), comparable with that of c-Jun KO murine SCs in vivo and in vitro (27), and formed disorganized clusters, demonstrating a fundamental role of c-Jun in the 3D self-organization of SCs into linear structures. No difference was observed in 2D between control and c-Jun KO SCs (Fig. 6A, insets), showing the importance of the dimensional environment in SC organization. P-c-Jun was expressed in the 3D SC structures (Supplementary Fig. S6A). SCs treated with SP600125, a c-Jun N-terminal Kinase (JNK) inhibitor that blocks c-Jun phosphorylation (Supplementary Fig. S6B), also exhibited an inability to collectively organize into linear structures in 3D (Supplementary Fig. S6C). We next followed the organization of HEI-286 SCs using time-lapse microscopy and measured the maximum length of the SC structures formed from a single starting cell in 3D. After 80 hours and about three to four mitoses, the daughter cells of a control SC connected to create a branched, linear structure (Supplementary Video S7, left), which was longer than the structure created by c-Jun KO HEI-286 SCs (Fig. 6B; Supplementary Fig. S6D) or SP600125-treated cells (Fig. 6C; Supplementary Fig. S6E). Tracking analysis of time-lapse movies indicated that the control daughter cells separated from one another and initially migrated in opposite directions before stretching out and oscillating back and forth to eventually connect with one another (Supplementary Fig. S6F and S6G). This sequence was a consistent initial step for replicating HEI-286 SCs to link together into 3D branched, linear structures. Single c-Jun KO HEI-286 SCs or SP600125-treated control SCs also underwent mitosis at least three to four times but remained bunched together (Fig. 6C–F; Supplementary Videos S7 and S8). They exhibited defects in separation and migration away from each other after mitosis (Supplementary Fig. S6E and S6G; Supplementary Videos S7 and S8). To quantify daughter cell separation following mitosis, we performed time-lapse microscopy in microchannels. As in Matrigel, after cell division, daughter control HEI-286 SCs consistently separated and migrated away from each other (Supplementary Fig. S6H; Supplementary Video S9, top). They remained separated until the cells became more confluent in the microchannel. In contrast, daughter c-Jun KO HEI-286 SCs failed to migrate away from one another and reestablished contact after mitosis (Supplementary Fig. S6H; Supplementary Video S9, bottom). This was quantified by measuring the maximum distance of separation between two daughter HEI-286 SCs following mitosis (Supplementary Fig. S6I) and the percentage of daughter cells that separated from each other following mitosis (Supplementary Fig. S6J). These data provide insights into how cell separation is an integral component of HEI-286 SC self-organization into 3D branched, linear structures. This process is driven by c-Jun and forms the basis for HEI-286 SC alignment into tracks.
Figure 6.
c-Jun coordinates SC collective organization and enhances cancer cell invasion in 3D Matrigel through track formation. A, Confocal images of control and c-Jun KO HEI-GFP SCs in Matrigel showing a lack of SC organization in c-Jun KO HEI-286 SCs. Scale bars, 30 μm. B and C, Quantification of the length of the SC structures created in Matrigel from one cell after 80 hours for control and c-Jun KO cells (control n = 10, c-Jun KO n = 6, mean ± SEM; B) and JNK inhibitor–treated cells (JNK inh; control n = 14, JNK inh n = 30, mean ± SEM; C). D, Quantification of cancer cell invasion into a 3D Matrigel chamber in the presence of control versus c-Jun KO HEI-286 SCs grown in the Matrigel (n = 8–12 measurements/condition, mean ± SEM). E, Maximum projection view of confocal images of MiaPaCa-2 (magenta) invasion in the presence of HEI-286-SCs (green) growing in Matrigel. Scale bar, 50 μm. F, Confocal image enlarged from E and schematic showing a chain of cancer cells (CC) aligned within a tubular, linear structure of control SCs in Matrigel. Scale bar, 50 μm. G, Confocal image enlarged from E and schematic showing branched structures created by aligned SCs in Matrigel. Scale bar, 50 μm. H, Maximum projection view of confocal images of MiaPaCa-2 invasion in the presence of c-Jun KO HEI-286 SCs growing in Matrigel. Scale bar, 50 μm. I, Confocal image enlarged from F and schematic showing a disorganized cluster of c-Jun KO HEI-286 SCs lacking organization. Scale bar, 50 μm.
We assessed the role of SC c-Jun in the 3D model of cancer cell invasion in which we showed HEI-286 SCs organizing around cancer cells (Fig. 2P–R). Analysis of confocal images revealed that c-Jun KO HEI-286 SCs were less able to support cancer invasion of MiaPaCa-2, Panc01, and KPC cells than control HEI-286 SCs (Fig. 6D). As previously described, cancer cells were associated with the branched, linear structures of control HEI-286 SCs and aligned along these tracks extending into the Matrigel (Fig. 6E–G). In contrast, the c-Jun KO HEI-286 SCs failed to form linear structures but largely remained in disorganized clusters (Fig. 6H and I). MiaPaCa-2 did not show affinity for the unstructured c-Jun KO HEI-286 SCs and failed to align as chains of cells, although they were occasionally associated with individual c-Jun KO HEI-286 SCs (Fig. 6H and I). These data demonstrate a fundamental role of c-Jun in the 3D self-organization of SCs into linear structures that serve as pathways for cancer cells.
SC c-Jun Promotes Cancer Cell Neural Invasion In Vivo
To investigate the role of SC c-Jun in PNI in vivo, we injected Panc02 or KPC pancreatic cancer cells into the sciatic nerves of c-Jun KO and WT mice to assess the resulting length of PNI proximally toward the spinal cord from the site of injection (36). Histologic analysis of hematoxylin and eosin (H&E) staining of nerve longitudinal sections after 7 days showed a significant reduction of PNI length along the sciatic nerve in the c-Jun KO mice as compared with WT mice (Fig. 7A and B). Quantifying sciatic nerve function (37, 38) indicated that WT mice developed more severe nerve paralysis and hind limb dysfunction 6, 10, and 14 days after injection as compared with c-Jun KO SC mice (Fig. 7C–E).
Figure 7.
c-Jun–reprogrammed SCs promote cancer invasion in vivo.A, Histologic analysis of injected murine sciatic nerves in P0-CRE− (WT) and P0-CRE+ (c-Jun KO) c-Junfl/fl mice. Representative samples of cancer invasion detected by H&E staining. Boxes show the invasion of areas away from the site of injection. Scale bars, 2,000 μm. Box scale bars, 500 μm. B, Quantification of distance of nerve invasion by Panc02 cells (WT n = 7, c-Jun KO n = 11, mean ± SEM) and KPC cells (WT n = 13, c-Jun KO n = 11, mean ± SEM). C, Representative images of murine hind limbs after Panc02 cancer cell injections show less paralysis in c-Jun KO SC mice. D, Quantification of the maximum width of hind limb paw in P0-CRE− (WT) versus P0-CRE+ (c-Jun KO) c-Junfl/fl mice 10 days after cancer injection (n = 8 mice/condition, mean ± SEM). E, Quantification of sciatic nerve function in P0-CRE− (WT) versus P0-CRE+ (c-Jun KO) c-Junfl/fl mice (n = 8 mice/condition, mean ± SEM). F, Effect of SP600125 on sciatic nerve invasion. Quantification of the length of nerve invasion in mice injected with Panc02 and SP600125 [JNK inhibitor (JNKi)] or Panc02 and DMSO (n = 11 mice/treatment, mean ± SEM). G, Representative H&E staining images of sciatic nerves injected with Panc02 and SP600125 versus Panc02 and DMSO. Top images are from proximal nerve regions at the spinal cord. No cancer is seen within the SP600125-injected nerve. Bottom images are from the site of injection, distal to the spinal cord. CC, cancer cells. Scale bars, 200 μm. H, Effect of SP600125 on flank tumor growth. Quantification of tumor volume in mice coinjected with Panc02 and SP600125 and with Panc02 and DMSO (n = 5 mice/treatment, mean ± SEM). NS, not significant. I, Survival analysis of WT and c-Jun KO mice injected with Panc02 in the sciatic nerve (WT n = 9, c-Jun KO = 11, P = 0.0003).
To corroborate the role of SC c-Jun in PNI and to address the therapeutic utility of targeting JNK, we applied the JNK inhibitor SP600125 to the same experiment described above using the murine sciatic nerve model. The addition of SP600125 significantly decreased the length of PNI caused by Panc02 cancer cells as compared with controls (Fig. 7F and G). SP600125 did not affect cancer growth or tumor volumes when Panc02 cells were injected into the murine flank (Fig. 7H), demonstrating that inhibitory effects of SP600125 were not due to a direct effect on Panc02 proliferation. In addition, a survival study on this animal model shows that c-Jun KO mice survived longer than WT mice (Fig. 7I). Together, these data suggest that c-Jun activity in SCs can be effectively targeted by pharmacologically blocking JNK and that this modulation of the nerve microenvironment can have therapeutic effects in inhibiting cancer progression along nerves.
DISCUSSION
Although the concept that cells from the TME contribute to cancer invasion is established, most reported mechanisms have focused on well-identified cells such as fibroblasts and immune cells. Our bioinformatics approach revealed the importance of nonmyelinating SCs, which are more difficult to detect than other cells from the TME using conventional tools. Our experiments demonstrate how these cells unconventionally lead to the invasive phenotype of the cancer cells. Our study describes a mechanism of invasion in which nonmyelinating, activated SCs create TASTs, 3D structures that serve as a living scaffold to promote cancer invasion. However, this scaffold is not simply a passive structure. These SC tracks are composed of highly dynamic SCs that directly apply forces onto cancer cells to enhance their migration. These SCs exhibit a variety of complex behaviors that induce pushing, squeezing, and pulling forces on cancer cells. They are also highly pliable and allow cancer cells to pass alongside them in a confined space.
Our computational analysis of TCGA gene expression data suggests the clinical importance of SCs in PDAC for a variety of different SC types that each carry different functions. The two main SC types are myelinating and nonmyelinating SCs. PDAC has recently been categorized into subtypes based on their transcript analysis (34). The SC signature scores vary within these different subtypes, suggesting that the nature of cancer might influence the presence of SCs. The progenitor subtype of PDAC, characterized by the expression of genes involved in early pancreatic development (FOXA2/3, PDX1, and MNX1), has the lowest SC scores and is known to be less invasive than the squamous type, which is enriched in p53 and KDM6A mutations and is associated with a poor prognosis. Higher SC scores are seen in the immunogenic subtype, which is known to be rich in immune cells and stroma. This is consistent with the presence of nerves in the stroma and our data showing higher SC scores in lower cellularity samples. Higher SC scores in the immunogenic subtype might also be related to the known role of SCs in immune regulation (39). The SC signatures do not associate with specific PDAC subtypes. However, all SC signatures do follow the same pattern of preference, with the highest being for the immunogenic subtype and the lowest for the progenitor subtype. In contrast, nonmyelinating and myelinating SCs show strong differences in their associated pathways, which likely reflect their different functions. Nonmyelinating SCs associate with gene sets that correspond to cancer invasion pathways (EMT, extracellular matrix modifications), whereas myelinating SCs associate with immune-related gene sets. In this study, we expand on the nonmyelinating SCs, in which we report activation of SC c-Jun in cancer tissue and how these SCs organize in TASTs.
Activated SCs have been shown to be present in a variety of cancers (18, 40–43). We demonstrate that cancer cells activate SCs in a manner related to that seen after nerve injury and activate the transcription factor c-Jun. This c-Jun activity is consistent with the previously reported stimulatory roles of SC NCAM1 (18) and GDNF (35) in PNI, since their expression is regulated by c-Jun in the HEI-286 SC line, and in SCs involved in nerve repair (27). SCs organized in linear structures function to promote cancer invasion, recapitulating to some extent SC behavior following nerve injury. Following nerve transection, repair SCs migrate to traverse the gap, creating a linear bridge that links the cut ends together (29). SCs along the denervated distal nerve stump adopt an elongated and often branched morphology, enabling them to form regeneration tracks, or Büngner bands, that guide regrowing axons (44). We show here that SCs adopt a similar phenotype in 3D gel, creating branching patterns, with elongated cells forming long tracks. In the cancer setting, these SC tracks enable opportunistic cancer invasion, allowing cancer cells to align and invade along TASTs extending into the 3D matrix. These findings suggest that cancer cells hijack the SC nerve repair program and are guided by SC tracks in a manner that is analogous to how regenerating axons are guided during nerve repair.
It remains unclear as to the identity of the exact stimulus or paracrine signaling axis that leads to c-Jun activation in SCs. One candidate factor is the proinflammatory cytokine IL6, which plays a role in SC activation under hypoxic conditions in pancreatic cancer (19). Blockade of IL6 signaling suppresses SC activation around pancreatic cancer precursor lesions (19). IL6 is produced during nerve repair (45), but IL6 KO mice do not lose their ability to undergo nerve repair (46). In addition, IL6 has been shown to be regulated by JNK (46) rather than stimulating JNK. c-Jun does appear to serve as a general alert signal in SCs activated by a disturbance in homeostasis.
The identification of c-Jun as a key regulator of this cancer-associated SC phenotype may be harnessed for the treatment of SC-induced cancer invasion. We demonstrate that SC self-organization into TASTs, SC-mediated cancer cell migration and invasion, SC association with cancer cells in vivo, and SC-mediated increase in PNI in vivo are all attenuated with the loss of c-Jun activity. Importantly, the use of a JNK inhibitor was able to impede PNI in a mouse model, demonstrating that this mechanism can be therapeutically targeted.
METHODS
Contact for Reagents and Resource Sharing
Further information and requests for resources and reagents should be directed to and will be fulfilled by the lead contact Richard J. Wong (wongr@mskcc.org).
Experimental Model and Subject Details
Mice.
All mouse procedures were performed in accordance with an institutional animal care and use committee (IACUC)–approved protocol at Memorial Sloan Kettering Cancer Center (MSKCC) and in compliance with all relevant ethical regulations. C57BL/6J mice were obtained from The Jackson Laboratory (Stock No. 000664). The protein myelin zero promoter-specific Cre recombinase mice P0-CRE and P0-CRE c-Junfl/fl were previously described (27). P0-CRE mT/mG were generated by crossing P0-CRE mice with mT/mG mice obtained from The Jackson Laboratory (Stock No. 007676).
Cell Lines and Cell Culture.
The human nonneoplastic HEI-286-GFP stable cell lines and the pancreatic cancer cells MiaPaCa-2-RFP and Panc01-RFP were previously described (18). The murine pancreatic cancer cell line Panc02 was from Dr. Min Li (45) and KPC was from Dr. Robert Vonderheide (46). KPC-Tomato was generated using pEF1a-Tdtomato vector (Clontech). c-Jun KO HEI-GFP cell lines were generated using CRISPR–Cas9 technology. Two constructs were made at the MSK Gene Editing and Screening Core Facility using lentiCRISPRv2 vector (Addgene 52961) and the following JUN-targeting oligonucleotide sequences:
| JUN-1F | CACCGCCGTCCGAGAGCGGACCTTA |
| JUN-1R | AAACTAAGGTCCGCTCTCGGACGGC |
| JUN-2F | CACCGTCGGCGGCGCAGCCGGTCAA |
| JUN-2R | AAACTTGACCGGCTGCGCCGCCGAC |
DNA constructs were nucleofected in HEI-286-GFP using Amaxa (program T-020). A control cell line was generated using an empty vector. Stable cell lines were generated after selection with puromycin (3 μg/mL). We generated both polyclonal cell lines and a monoclonal cell line for sequence number 2. Depletion of c-Jun in c-Jun K0-HEI was verified by Western blotting and immunofluorescence assays using standard protocols.
All cells were cultured in 5% CO2 at 37°C in Dulbecco's modified Eagle's medium (DMEM, Cellgro) containing 10% FBS (Gemini) and 50 U/mL penicillin/streptomycin (Cellgro). The culture medium for c-Jun KO HEI-286-GFP cells was supplemented with puromycin (3 μg/mL). The culture medium for cells expressing GFP and RFP and Far red-PM670 was supplemented with G418 (50 μg/mL). Cell lines were routinely screened to avoid Mycoplasma contamination and maintained in a humidified chamber with 5% CO2 at 37°C.
Patient Materials.
Written informed consent was obtained from each patient. The studies were conducted in accordance with recognized ethical guidelines [e.g., Declaration of Helsinki, Council for International Organizations of Medical Sciences (CIOMS), Belmont Report, U.S. Common Rule] and approved by the MSK Institutional Review Board. The non-PDAC pancreatic specimens included one patient with a benign pancreatic cyst (with a history of rhabdomyosarcoma; 1), two patients with well-differentiated pancreatic neuroendocrine tumors (2 and 3), and one patient with a solid pseudopapillary neoplasm (4).
Method Details
Murine Sciatic Nerve Injection.
Sciatic nerve injection procedures were carried out as previously described (35, 36). Briefly, mice (P0-CRE c-Junfl/fl, P0-CRE mT/mG, and C57BL/6J mice) were anesthetized using isoflurane (1%–3%), and their sciatic nerve was exposed. Sterile PBS or Panc02 murine pancreatic cancer cells (50,000) were injected into the sciatic nerve under loop magnification using a 10 μL Hamilton syringe. For the assessment of SP600125 effect on PNI, Panc02 were injected into the nerves with SP600125 (Tocris, 3 mg/kg) or DMSO (control). After treatment with meloxicam for analgesia, the mouse was closed up with surgical sutures. Mice were followed for recovery every day for 72 hours and monitored for nerve function. For the survival analysis, animals were monitored until death or criteria mandating sacrifice were reached including complete limb paralysis.
Murine Sciatic Nerve Function.
Sciatic nerve function was measured in control and c-Jun KO mice 6, 10, and 14 days after injection of Panc02 cells using the nerve function score (37, 38). It was graded from 4 (normal) to 1 (total paw paralysis), according to hind limb paw response to the manual extension of the body. The maximum hind paw width was measured on day 10 after injection using a digital caliper.
Preparation of Murine Sciatic Nerve Sections.
Seven days after injection, murine sciatic nerves were dissected up to the spinal cord and embedded in Tissue-Tek OCT (Electron Microscopy Sciences). The specimen-frozen blocks were serially sectioned longitudinally at respectively 5 μm thickness using Cryostat microtome (Leica CM1950). Sections were used for H&E and immunofluorescence staining. Morphologic assessment and quantification of neural invasion by cancer cells in sciatic nerves were performed using Panoramic Viewer (3DHISTECH) on the H&E sections.
Immunofluorescence of Murine Sciatic Nerve Sections and Human Specimen Sections.
Murine frozen sections were fixed using 4% paraformaldehyde. Sections from human pancreatic specimens were obtained from paraffin blocks that were prepared using a standard protocol. Sections were permeabilized and blocked in 3% horse serum and 0.1% Triton X-100/PBS for 1 hour. Primary antibodies (anti–P-c-Jun 1:200, anti-GFAP 1:1,000, anti-cytokeratin 1:200) diluted in 0.1% horse serum and 0.1% Triton X-100/PBS were incubated overnight at 4°C. Sections were washed with PBS, and detection was performed using an appropriate fluorescent secondary antibody (Alexa Fluor 488, 568, or 647; Invitrogen). Samples were mounted in DAPI containing antifade mounting medium. Slides were scanned using Flash Scanner (PerkinElmer). Immunofluorescent sections were analyzed using Panoramic Viewer (3DHISTECH).
Immunofluorescence and Actin Staining of HEI-286 SCs Cocultured with MiaPaCa-2.
GFP HEI-286 and RFP MiaPaCa-2 (30,000 cells total, 1/1) were seeded on a 35-mm, glass-bottom dish precoated with 20% Matrigel for 3 days. Cells were fixed in 4% paraformaldehyde/PBS for 15 minutes and washed 3 times with PBS. For immunofluorescence, cells were blocked in 3% horse serum and 0.1% Triton X-100/PBS for 1 hour and incubated with antibody anti–P-c-Jun diluted in 0.1% horse serum and 0.1% Triton X-100/PBS overnight at 4°C. After washing, cells were incubated with Alexa Fluor 647 secondary antibody diluted in 0.1% horse serum and 0.1% Triton X-100/PBS containing DAPI. For actin staining, cells were incubated with Alexa Fluor Plus 647 Phalloidin (1:100; #A2287, Invitrogen) for 30 minutes at room temperature and stained with DAPI for 30 minutes.
Images were taken using an inverted Axiovert 200M microscope (Zeiss) equipped with an EC Plan-Neofluar 40 × 0.3NA Ph1 lens and analyzed using Axiovision software and Fiji.
Invasion Assay.
Invasion assays were performed as previously described (18). Briefly, HEI-286-GFP or HEI-286 c-Jun KO were seeded in 40 μL Matrigel matrix at 70 cells/μL in a two-chamber insert (Ibidi GmbH) placed in glass-bottom, 35-mm MatTek dishes and grown for 6 days in DMEM 10% FBS. Cancer cells (MiaPaCa-2-RFP, Panc01-RFP, and KPC-Tomato; 50,000 cells) were then added on top of the Matrigel. After 3 days, samples were fixed in 4% paraformaldehyde for 30 minutes for imaging. The coculture was imaged using a Leica SP5- or SP8-inverted confocal microscope with a 10 × 0.4 NA lenses. [The z-stacks obtained with the 10× lens were used for quantification of invasion and were 1 mm thick with 5-μm steps. Images were recorded at 12-bit resolution. Stacks were reconstructed in three dimensions (3D) using Imaris software (Bitplane), and red fluorescent structures corresponding to cancer cells were quantified using the Imaris software. The software determined the number of red cancer cells in an area of interest (size x = 66, y = 296, z = 552 μm), which was about 300 μm below the surface of the Matrigel. The invasion was about 60 cells/area of interest and was considered 100%.]
Samples were turned upside down for imaging the interaction of HEI-286-GFP (control and c-Jun KO) and cancer cells at the top of Matrigel. The lens used was 20×, and the stack thickness was set for optimal resolution. At the top of the Matrigel, we counted 5.5 ± 1.73 chains of cancer cells surrounded by HEI-286-GFP per mm squared surface area with control HEI-286-GFP and about four disorganized KO HEI-286-GFP clusters per mm squared surface area.
Imaging SCs in 3D Matrigel.
HEI-GFP (control and c-Jun KO) were mixed at 4°C with 100% Matrigel at a concentration of 70 cells/μL. Drops of 10 μL of the Matrigel containing SCs were placed in glass-bottom dishes. The samples were then incubated at 37°C for 30 minutes to allow polymerization. A culture medium was added, and the samples were incubated at 37°C 5% CO2 until imaging. In some experiments, 10 μmol/L SP600125 or DMSO was added to the medium at the time of plating. Images of cell clusters were made 5 days after seeding using Leica confocal microscope with a 20 × 0.7 NA lenses. Time-lapse imaging was performed 24 hours after seeding at the two-cell stage using a motorized stage inverted Axiovert 200M microscope (Zeiss) equipped with an EC Plan-Neofluar 10 × 0.3NA Ph1 lens and temperature and CO2 controllers. Images were recorded every 10 minutes for 72 hours.
Imaging Microchannels.
Microchannels (10–20 μm width, 10 μm height; 4Dcell) were coated with fibronectin (10 μg/mL; Sigma-Aldrich F1141) for 1 hour at RT and washed 3 times with PBS before incubating with the cell culture medium for 1 hour at 37°C and 5% CO2. A total of 10,000 cells were placed in the wells. When SCs were combined with cancer cells, 5,000 of each cell type were placed in the wells that connect the channels. Cells were recorded 24 hours or at the indicated time after loading. Migrating cells were recorded overnight with an inverted microscope (Zeiss) at 37°C with 5% CO2 atmosphere and a 10× objective (0.3NA) at one image every 10 minutes for 24 hours. For some experiments, still confocal images and time-lapse images were taken using an inverted confocal microscope (Leica, SP5). Time-lapse images were taken for 12 hours every 10 minutes.
AFM.
Glass-bottom petri dishes (FluoroDish FD5040, World Precision Instruments) containing HEI-286 SCs with MiaPaCa-2 cancer cells were used for the acquisition of stiffness maps. Experiments were performed at 37°C with an MFP-3D-BIO AFM microscope (Oxford Instruments). Fluorescent images of GFP-expressing HEI-286 SCs and RFP-expressing MiaPaCa-2 cancer cells were acquired together with the stiffness maps using the inverted optical microscope (Zeiss AxioObserver Z1) integrated with the AFM microscope. Cantilevers with colloidal borosilicate probes were used for experiments, having a diameter of 5 μm and nominal spring constant k = 0.1 N/m (Novascan). Before each experiment, the exact cantilever spring constant was determined with the thermal noise method, and the optical sensitivity was determined using a glass-bottom petri dish filled with PBS as an infinitely stiff substrate. Stiffness maps of 80 × 80 μm2 (20 × 20 points) were collected in areas containing the cells and the substrate, used as a reference, at the rate of 1.5 Hz for a complete single approach/withdraw cycle. A trigger point of 1 nN was used to ensure maximum sample penetration of less than 1 μm. Force curves in each map were fitted according to the Hertz model using the routine implemented in the MFP3D AFM (Igor Pro, WaveMetrics). Data fitting was performed in the range from 0% to 60% of the maximum applied force, by setting tip Poisson νtip = 0.19, tip Young's modulus Etip = 68 GPa, and sample Poisson νsample = 0.45 (47). To account for possible substrate effects, a threshold of 500 nm was chosen for the selection of the stiffness data, from the corresponding topographical maps of each cell collected in situ with the stiffness maps, to account for possible substrate effects. Extraction of the stiffness values from the raw Igor Binary Wave (.ibw) data within the mask region was obtained by means of a home-built routine implemented in Igor (Igor Pro, WaveMetrics) and a custom script in MATLAB (MathWorks). Data visualization was performed using OriginPro (OriginLab) and GraphPad Prism. Stiffness analysis was performed only on SCs physically connected to cancer cells, as inferred from the respective fluorescence signal.
Cell Sorting, RNA Sequencing Library Preparation, and Analysis.
RNA sequencing (RNA-seq) data were from three independent biological replicates. Four million GFP-expressing HEI-286 cells were grown alone or mixed with RFP-expressing MiaPaCa-2 for 4 days (1:1). Cells were trypsinized, pelleted, resuspended in PBS, and passed through a 35-μm cell strainer to yield single cells. Live (DAPI-negative) GFP cells were sorted on a BD SORP FACSAria IIu equipped with a 488-nm laser and a 525/50-nm bandpass filter to excite and detect GFP, a 561-nm laser and a 610/20-nm bandpass filter to excite and detect mRFP, and a 405-nm laser and a 450/40-nm bandpass filter to excite and detect DAPI.
RNA was extracted from cells by using the RNeasy mini kit (Qiagen, #74104) as per the manufacturer's instructions. RNA-seq was performed by the MSK Integrated Genomics Operation. After RiboGreen quantification and quality control by Agilent BioAnalyzer, 500 ng of total RNA underwent polyA selection and TruSeq library preparation according to instructions provided by Illumina (TruSeq Stranded mRNA LT Kit, #RS-122-2102), with 8 cycles of PCR. Samples were barcoded and run on a HiSeq 2500 in a 50 bp/50 bp paired-end run using the HiSeq SBS Kit v4 (Illumina). An average of 58 million paired reads was generated per sample. At the most, the ribosomal reads represented 5% of the total reads generated and the percentage of mRNA bases averaged 79%. Sequence data processing was performed by the Bioinformatics Core at MSK. Output data (FASTQ files) were mapped to the human genome using the rnaStar aligner that mapped reads genomically and resolved reads across splice junctions. Output SAM files were post-processed using PICARD tools and converted into BAM format. The expression count matrix from the mapped reads was computed using HTSeq (http://htseq.readthedocs.io/), and the raw count matrix was processed using the R/Bioconductor package DEseq (https://www.bioconductor.org/packages//2.10/bioc/html/DESeq.html) to normalize the full data set and analyze differential expression between sample groups. One thousand and ninety-two of 20,245 genes were found differentially regulated in the cocultured HEI-286 SCs as compared with HEI-286 SCs alone (fold change >2, P < 0.05). Of those, 878 were upregulated. Because SCs are phagocytic and internalized material from cancer cells, consistent with its phagocytic role in Wallerian degeneration, genes found highly expressed in cancer cells were filtered out from the list of upregulated genes in SCs grown in the presence of cancer cells using microarray data of HEI-286 and MiaPaCa-2 cells. The RNA of these HEI-286 and MiaPaCa-2 cells was extracted, and cDNA was generated and applied to the Illumina HumanHT12 v4. Six hundred and ninety-seven genes remained upregulated and were used for GO pathway analysis using the KEGG 2019 human database. Genes found highly expressed in cocultured HEI-286 compared with HEI-286 grown alone, and cocultured c-Jun KO HEI-286 compared with c-Jun KO HEI-286 grown alone, were submitted for GO enrichment analysis using a Web-based tool (https://amp.pharm.mssm.edu/Enrichr/) for pathway analysis using the KEGG 2019 data set. Heat maps were generated using Morpheus.
Quantification and Statistical Analysis
Correlations between SC Gene Signature Scores or JUN Expression and Clinical Outcomes in TCGA and MSK Patients.
Signatures for SCs were obtained from Tabula Sapiens using OnClass (ref. 30; http://tabula-sapiens-onclass.ds.czbiohub.org/), PanglaoDB (ref. 48; https://panglaodb.se/markers.html), and a single-cell analysis study of the pancreas (49). The activities of SCs and other pathways were compared across tumor samples using newly developed IPAS scores (ref. 31; http://pathwayassessor.com). TCGA clinical data, including survival and PFS, were downloaded from the data portal of the Broad Institute (https://gdac.broadinstitute.org). Cox proportional hazards regression analysis was performed to examine the correlation between SC signature IPAS scores and patient outcomes (survival and PFS). The Survival package of R.3.6.0 was utilized to calculate log-rank P values. The optimal patient separation by IPAS score that yielded the lowest P value in clinical outcome was selected to separate the patients into high- versus low-score groups as an exploratory approach. Log-rank tests were used to detect differences in survival and PFS. The survival differences were visualized by generating Kaplan–Meier survival plots using Prism 9.
Clustering of TCGA Patients in Subtypes.
To attribute PDAC subtypes to TCGA patients, we used the list of feature genes (GP) determined in Bailey and colleagues (34). GP scores were computed as the average log-transformed expression of marker genes (50). GP9, GP6, GP2, and GP1 were used to predict the ADEX, immunogenic, squamous, and progenitor subtypes, respectively.
Cellularity.
Tumor cellularity data were generated by a pathologic review of TCGA H&E slides on the NCI data portal (https://portal.gdc.cancer.gov/). Some data were obtained from Dr. Shivan Sivakumar (51).
Quantification of GFAP in 15 MSK PDAC Patients.
GFAP levels were assessed on histologic sections immunofluorescently stained for GFAP and S100. Nerves with S100 and strong GFAP staining were considered positive. Patients with five or more GFAP+ nerves per section were considered as high GFAP and patients with less than five GFAP+ nerves per section were low GFAP. Kaplan–Meier survival plots were generating using Prism 9.
Quantification of Cancer Cell Invasion in a 3D Assay.
The z-stacks obtained with a 10× lens were used for quantification of invasion and were 1-mm thick with 5-μm steps. Images were recorded at 12-bit resolution. Stacks were reconstructed in 3D using Imaris software (Bitplane), and red fluorescent structures corresponding to cancer cells were quantified using the Imaris software. The software determined the number of red cancer cells in an area of interest (size x = 66, y = 296, z = 552 μm), which was about 300 μm below the surface of the Matrigel. The invasion was about 60 cells/area of interest and was considered 100%.
Quantification of Length of SC Structures in Matrigel, Distance after Division, SC Separation after Division, and Cancer Cell Passing an SC.
Axiovision software (Zeiss) was used to measure: (i) the length of the structures created in Matrigel from two SCs after 72 hours, (ii) the distance between two SCs in microchannels after division, (iii) cell separation after division in microchannels, and (iv) the number of times a cancer cell passed an SC in microchannels. To measure the distance between two SCs after division in microchannels, we selected dividing cells that had sufficient space to migrate away from each other. Distance measurement was done within 400 minutes after the division and included the space between the cells and the size of the cells. To quantify SC separation after division, dividing cells that had sufficient space to migrate away from each other were selected and counted as positive if the cells were separated 400 minutes after division. To quantify the number of events, cancer cells passing a SC in microchannels, single moving cancer cells that encountered an SC were selected. They were counted as positive if the cancer cell went through the SC. Analyses were performed visually.
Quantification of SC Wrapping.
Confocal xz projection images of SCs and cancer cells in contact within microchannels were analyzed to quantify SC wrapping around cancer cells. SCs were considered as wrapping cancer cells if they covered more than 60% of the cancer cell.
Quantification of Instantaneous Velocity.
PIV analysis was performed by PIVlab version 2.36 (Time-Resolved Digital Particle Image Velocimetry Tool for MATLAB, developed by W. Thielicke and E.J. Stamhuis; http://pivlab.blogspot.com). Images from time-lapse movies were first doubled in size and saved as bmp files in Fiji. Using PIV, the cells were selected for every individual frame. Images were calibrated and analyzed.
Statistical Analysis.
The multivariate Cox regression modeling of survival data was conducted using R package “survival.” One-way ANOVA was used to compare SC signature cores in PDAC subtypes. Pairwise comparisons were conducted using an unpaired or paired (for Fig. 2D) two-tailed Student t test. Statistical significance was defined at P values less than 0.05. Statistical analyses were performed using Prism 7 or 9 (GraphPad Software, Inc.).
Randomized control in survival analysis was performed using the original software (52) developed for the assessment of statistical significance of the difference between potential disease subtypes represented by two sets of gene expression profiles. In this test, 104 random groupings of molecular profiles of sizes equal to sizes of tested subtypes (for example, nonmyelinating SC 17:161) were generated. The P value of survival difference was computed for each of 104 random groupings and compared with PSC value of the SC-specific patient grouping; a total number of cases when P values were lower than or equal to PSC was counted and then divided by 104, the total number of tests (Table 1 from Supplementary Data S3, Prand).
Supplementary Material
Acknowledgments
We acknowledge the technical services provided by MSK core facilities: the Molecular Cytology and Flow Cytometry facilities, the Integrated Genomics Operation Core, the Bioinformatics Core, and the Animal Imaging Core. We thank Ning Fan for assistance with sectioning specimens. We thank Dr. Shivan Sivakumar (University of Oxford, United Kingdom) for sharing cellularity data in patients from the TCGA. We thank Luc G.T. Morris and Nicolas Lecomte for critical reading of the manuscript. The Integrated Genomics Operation Core was funded by Cycle for Survival and the Marie-Josée and Henry R. Kravis Center for Molecular Oncology. MSK Core facilities were supported by NIH P30CA008748. This work was supported by NIH R01CA219534 (R.J. Wong).
The costs of publication of this article were defrayed in part by the payment of page charges. This article must therefore be hereby marked advertisement in accordance with 18 U.S.C. Section 1734 solely to indicate this fact.
Footnotes
Note: Supplementary data for this article are available at Cancer Discovery Online (http://cancerdiscovery.aacrjournals.org/).
Data and Software Availability
The accession numbers for the cell RNA-seq and microarray data reported in this paper are GSE180710 and GSE180971, respectively. The accession number corresponding to the patients from MSKCC is GSE184585.
Authors’ Disclosures
No disclosures were reported.
Authors’ Contributions
S. Deborde: Conceptualization, resources, data curation, formal analysis, supervision, validation, investigation, visualization, methodology, writing–original draft, writing–review and editing. L. Gusain: Resources, data curation, validation, investigation, methodology. A. Powers: Data curation, validation, investigation, methodology. A. Marcadis: Data curation, validation, investigation, methodology. Y. Yu: Data curation, validation, investigation, methodology. C.-H. Chen: Data curation, validation, investigation, methodology. A. Frants: Data curation, validation, investigation, methodology. E. Kao: Data curation, validation, investigation, methodology. L.H. Tang: Resources, investigation, visualization, methodology. E. Vakiani: Resources, investigation, visualization, methodology. M. Amisaki: Resources, data curation. V.P. Balachandran: Resources, data curation. A. Calo: Resources, data curation, formal analysis, methodology. T. Omelchenko: Formal analysis, investigation, methodology, writing–review and editing. K.R. Jessen: Resources, formal analysis, writing–review and editing. B. Reva: Resources, data curation, software, formal analysis. R.J. Wong: Conceptualization, resources, data curation, software, formal analysis, supervision, funding acquisition, writing–original draft, project administration, writing–review and editing.
References
- 1. Chambers AF, Groom AC, MacDonald IC. Dissemination and growth of cancer cells in metastatic sites. Nat Rev Cancer 2002;2:563–72. [DOI] [PubMed] [Google Scholar]
- 2. Friedl P, Wolf K. Tumour-cell invasion and migration: diversity and escape mechanisms. Nat Rev Cancer 2003;3:362–74. [DOI] [PubMed] [Google Scholar]
- 3. Kai F, Drain AP, Weaver VM. The extracellular matrix modulates the metastatic journey. Dev Cell 2019;49:332–46. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4. Winkler J, Abisoye-Ogunniyan A, Metcalf KJ, Werb Z. Concepts of extracellular matrix remodelling in tumour progression and metastasis. Nat Commun 2020;11:5120. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5. van Helvert S, Storm C, Friedl P. Mechanoreciprocity in cell migration. Nat Cell Biol 2018;20:8–20. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6. Magnon C, Hall SJ, Lin J, Xue X, Gerber L, Freedland SJ, et al. Autonomic nerve development contributes to prostate cancer progression. Science 2013;341:1236361. [DOI] [PubMed] [Google Scholar]
- 7. Peterson SC, Eberl M, Vagnozzi AN, Belkadi A, Veniaminova NA, Verhaegen ME, et al. Basal cell carcinoma preferentially arises from stem cells within hair follicle and mechanosensory niches. Cell Stem Cell 2015;16:400–12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8. Raju B, Haug SR, Ibrahim SO, Heyeraas KJ. Sympathectomy decreases size and invasiveness of tongue cancer in rats. Neuroscience 2007;149:715–25. [DOI] [PubMed] [Google Scholar]
- 9. Saloman JL, Albers KM, Li D, Hartman DJ, Crawford HC, Muha EA, et al. Ablation of sensory neurons in a genetic model of pancreatic ductal adenocarcinoma slows initiation and progression of cancer. Proc Natl Acad Sci U S A 2016;113:3078–83. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10. Zhao C-M, Hayakawa Y, Kodama Y, Muthupalani S, Westphalen CB, Andersen GT, et al. Denervation suppresses gastric tumorigenesis. Sci Transl Med 2014;6:250ra115. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11. Zahalka AH, Frenette PS. Nerves in cancer. Nat Rev Cancer 2020;144:646–15. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12. Bapat AA, Hostetter G, Hoff DDV, Han H. Perineural invasion and associated pain in pancreatic cancer. Nat Rev Cancer 2011;11:695–707. [DOI] [PubMed] [Google Scholar]
- 13. Deborde S, Wong RJ. How Schwann cells facilitate cancer progression in nerves. Cell Mol Life Sci 2017;341:1236361–16. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14. Liebig C, Ayala G, Wilks JA, Berger DH, Albo D. Perineural invasion in cancer. Cancer 2009;115:3379–91. [DOI] [PubMed] [Google Scholar]
- 15. Amit M, Na'ara S, Gil Z. Mechanisms of cancer dissemination along nerves. Nat Rev Cancer 2016;6:399–408. [DOI] [PubMed] [Google Scholar]
- 16. Demir IE, Reyes CM, Alrawashdeh W, Ceyhan GO, Deborde S, Friess H, et al. Clinically actionable strategies for studying neural influences in cancer. Cancer Cell 2020;38:11–4. [DOI] [PubMed] [Google Scholar]
- 17. Siegel RL, Miller KD, Jemal A. Cancer statistics, 2018. CA Cancer J Clin 2018;68:7–30. [DOI] [PubMed] [Google Scholar]
- 18. Deborde S, Omelchenko T, Lyubchik A, Zhou Y, He S, McNamara WF, et al. Schwann cells induce cancer cell dispersion and invasion. J Clin Invest 2016;126:1538–54. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19. Demir IE, Tieftrunk E, Schorn S, Saricaoglu ÖC, Pfitzinger PL, Teller S, et al. Activated Schwann cells in pancreatic cancer are linked to analgesia via suppression of spinal astroglia and microglia. Gut 2016;6:1001–14. [DOI] [PubMed] [Google Scholar]
- 20. Boerboom A, Dion V, Chariot A, Franzen R. Molecular mechanisms involved in Schwann cell plasticity. Front Mol Neurosci 2017;10:38. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21. Jessen KR, Mirsky R. The repair Schwann cell and its function in regenerating nerves. J Physiol 2016;13:3521–31. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22. Jessen KR, Mirsky R. The origin and development of glial cells in peripheral nerves. Nat Rev Neurosci 2005;6:671–82. [DOI] [PubMed] [Google Scholar]
- 23. Jessen KR, Mirsky R, Arthur-Farraj P. The role of cell plasticity in tissue repair: adaptive cellular reprogramming. Dev Cell 2015;34:613–20. [DOI] [PubMed] [Google Scholar]
- 24. Jessen KR, Mirsky R, Lloyd AC. Schwann cells: development and role in nerve repair. Cold Spring Harb Perspect Biol 2015;7:a020487. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25. Jopling C, Boue S, Belmonte JCI. Dedifferentiation, transdifferentiation and reprogramming: three routes to regeneration. Nat Rev Mol Cell Biol 2011;12:79–89. [DOI] [PubMed] [Google Scholar]
- 26. Masaki T, Qu J, Cholewa-Waclaw J, Burr K, Raaum R, Rambukkana A. Reprogramming adult Schwann cells to stem cell-like cells by leprosy bacilli promotes dissemination of infection. Cell 2013;152:51–67. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27. Arthur-Farraj PJ, Latouche M, Wilton DK, Quintes S, Chabrol E, Banerjee A, et al. c-Jun reprograms Schwann cells of injured nerves to generate a repair cell essential for regeneration. Neuron 2012;75:633–47. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28. Jessen KR, Arthur-Farraj P. Repair Schwann cell update: adaptive reprogramming, EMT, and stemness in regenerating nerves. Glia 2019;67:421–37. [DOI] [PubMed] [Google Scholar]
- 29. Jessen KR, Mirsky R. The success and failure of the Schwann cell response to nerve injury. Front Cell Neurosci 2019;13:33. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30. Wang S, Pisco AO, McGeever A, Brbic M, Zitnik M, Darmanis S, et al. Leveraging the cell ontology to classify unseen cell types. Nat Commun 2021;12:5556. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31. Reva B, Omelchenko T, Calinawan A, Nair S, Schadt E, Tewari A. Prioritization of prostate cancer to immune checkpoint therapy by ranking tumors along IFN-γ axis and identification of immune resistance mechanisms. BioRxiv 2020.10.19.345629 [Preprint]. 2021. Available from: 10.1101/2020.10.19.345629. [DOI] [Google Scholar]
- 32. Reva B, Omelchenko T, Nair SS, Tewari AK. Immune escape in prostate cancer known and predicted mechanisms and targets. Urol Clin North Am 2020;47:e9–16. [DOI] [PubMed] [Google Scholar]
- 33. Satpathy S, Krug K, Beltran PMJ, Savage SR, Petralia F, Kumar-Sinha C, et al. A proteogenomic portrait of lung squamous cell carcinoma. Cell 2021;184:4348–71. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34. Bailey P, Chang DK, Nones K, Johns AL, Patch AM, Gingras MC, et al. Genomic analyses identify molecular subtypes of pancreatic cancer. Nature 2016;531:47–52. [DOI] [PubMed] [Google Scholar]
- 35. Gil Z, Cavel O, Kelly K, Brader P, Rein A, Gao SP, et al. Paracrine regulation of pancreatic cancer cell invasion by peripheral nerves. J Natl Cancer Inst 2010;102:107–18. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36. Deborde S, Yu Y, Marcadis A, Chen CH, Fan N, Bakst RL, et al. An in vivo murine sciatic nerve model of perineural invasion. J Vis Exp 2018;e56857. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37. Bakst RL, Lee N, He S, Chernichenko N, Chen CH, Linkov G, et al. Radiation impairs perineural invasion by modulating the nerve microenvironment. PLoS One 2012;7:e39925. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38. Bakst RL, Xiong H, Chen CH, Deborde S, Lyubchik A, Zhou Y, et al. Inflammatory monocytes promote perineural invasion via CCL2-mediated recruitment and cathepsin B expression. Cancer Res 2017;77:6400–14. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39. Deborde S, Wong RJ. The role of Schwann cells in cancer. Adv Biol (Weinh) 2022Jun 4 [Epub ahead of print]. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40. Demir IE, Boldis A, Pfitzinger PL, Teller S, Brunner E, Klose N, et al. Investigation of Schwann cells at neoplastic cell sites before the onset of cancer invasion. J Natl Cancer Inst 2014;106:dju184. [DOI] [PubMed] [Google Scholar]
- 41. Silva VM, Gomes JA, Tenório LPG, de Omena Neta GC, da Costa Paixão K, Duarte AKF, et al. Schwann cell reprogramming and lung cancer progression: a meta-analysis of transcriptome data. Oncotarget 2019;10:7288–307. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42. Shurin GV, Kruglov O, Ding F, Lin Y, Hao X, Keskinov AA, et al. Melanoma-induced reprogramming of Schwann cell signaling aids tumor growth. Cancer Res 2019;79: 2736–47. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43. Weiss T, Taschner-Mandl S, Janker L, Bileck A, Rifatbegovic F, Kromp F, et al. Schwann cell plasticity regulates neuroblastic tumor cell differentiation via epidermal growth factor-like protein 8. Nat Commun 2021;12:1624–19. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44. Gomez-Sanchez JA, Pilch KS, van der Lans M, Fazal SV, Benito C, Wagstaff LJ, et al. After nerve injury, lineage tracing shows that myelin and remak Schwann cells elongate extensively and branch to form repair Schwann cells, which shorten radically on remyelination. J Neurosci 2017;37:9086–99. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45. Wang Y, Zhang Y, Yang J, Ni X, Liu S, Li Z, et al. Genomic sequencing of key genes in mouse pancreatic cancer cells. Curr Mol Med 2012;12:331–41. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46. Winograd R, Byrne KT, Evans RA, Odorizzi PM, Meyer ARL, Bajor DL, et al. Induction of T-cell immunity overcomes complete resistance to PD-1 and CTLA-4 blockade and improves survival in pancreatic carcinoma. Cancer Immunol Res 2015;3:399–411. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47. Calò A, Romin Y, Srouji R, Zambirinis CP, Fan N, Santella A, et al. Spatial mapping of the collagen distribution in human and mouse tissues by force volume atomic force microscopy. Sci Rep 2020;10:15664–12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48. Franzén O, Gan L-M, Björkegren JLM. PanglaoDB: a web server for exploration of mouse and human single-cell RNA sequencing data. Database 2019;2019:baz046. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49. Tosti L, Hang Y, Debnath O, Tiesmeyer S, Trefzer T, Steiger K, et al. Single-nucleus and in situ RNA–sequencing reveal cell topographies in the human pancreas. Gastroenterology 2021;160:1330–44. [DOI] [PubMed] [Google Scholar]
- 50. Danaher P, Warren S, Dennis L, D'Amico L, White A, Disis ML, et al. Gene expression markers of tumor infiltrating leukocytes. J Immunother Cancer 2017;5:18. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51. SI deY C, Heij L, Middleton MR, Markowetz F, Grabsch HI, et al. Immunophenotypes of pancreatic ductal adenocarcinoma: meta-analysis of transcriptional subtypes. Int J Cancer 2019;145:1125–37. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52. Rykunov D, Beckmann ND, Li H, Uzilov A, Schadt EE, Reva B. A new molecular signature method for prediction of driver cancer pathways from transcriptional data. Nucleic Acids Res 2016;44:e110. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Data Availability Statement
The accession numbers for the cell RNA-seq and microarray data reported in this paper are GSE180710 and GSE180971, respectively. The accession number corresponding to the patients from MSKCC is GSE184585.



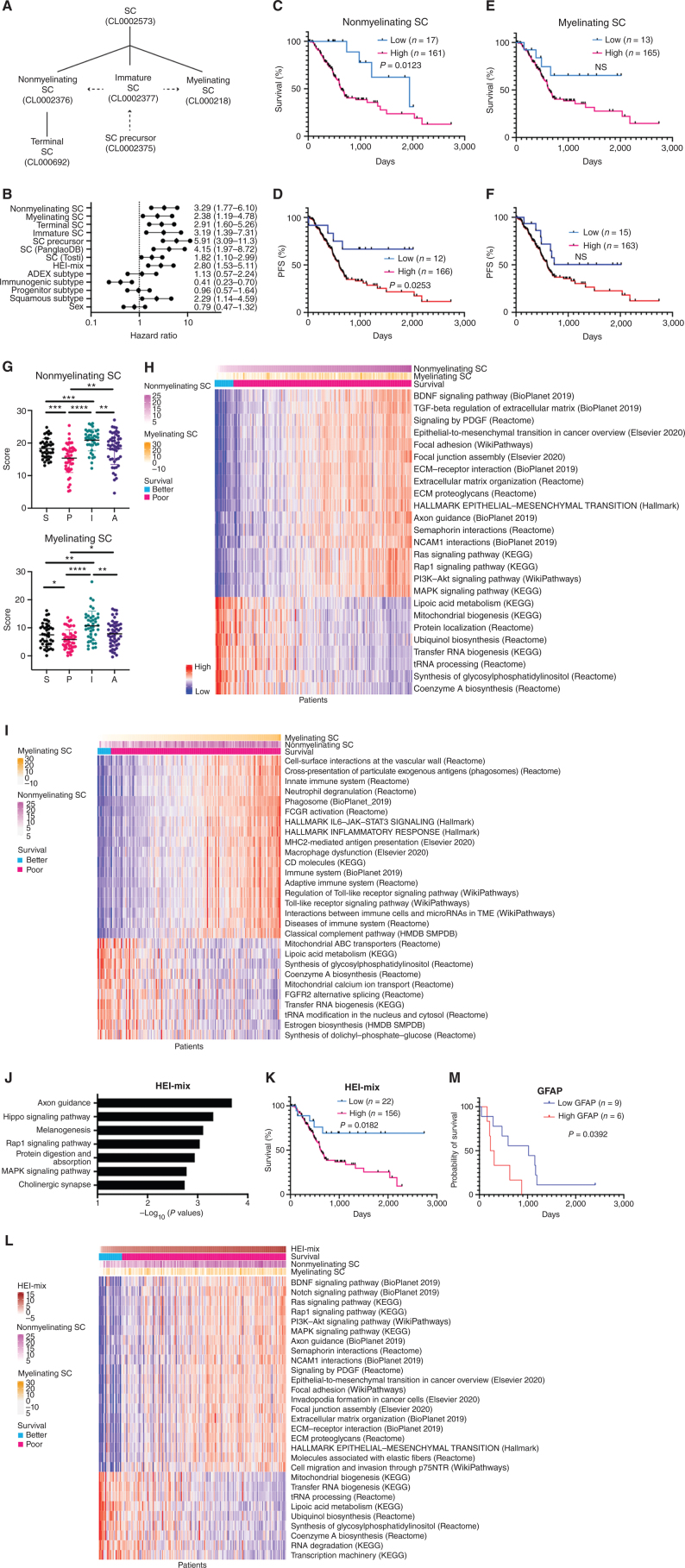
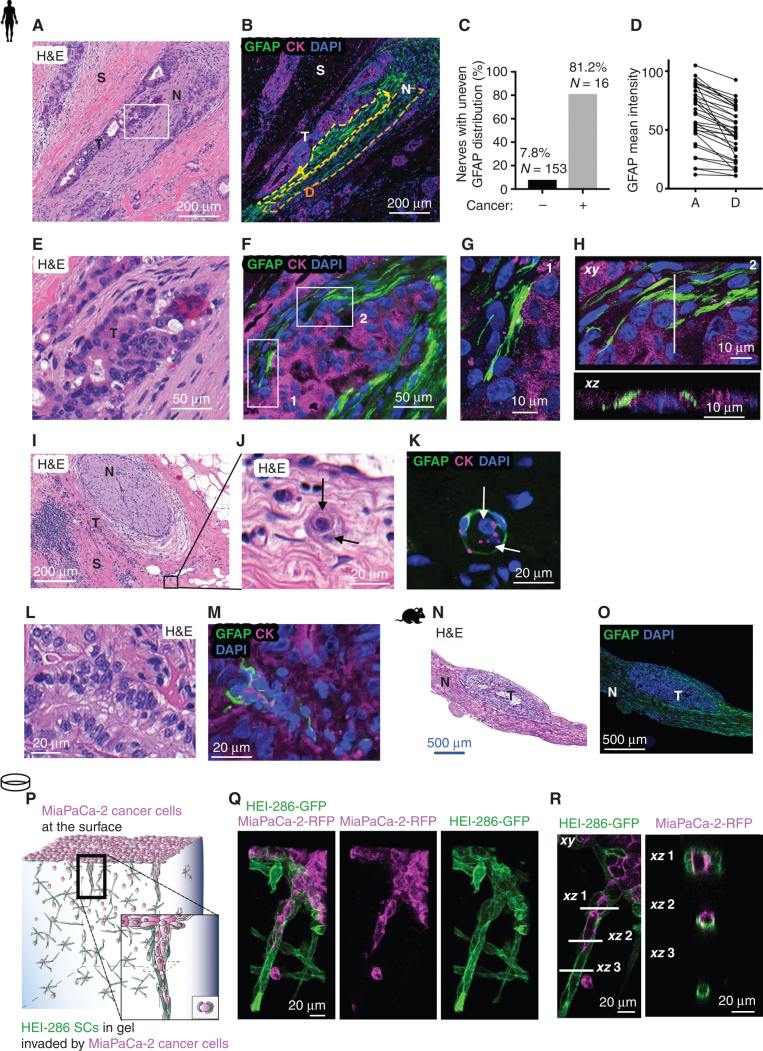
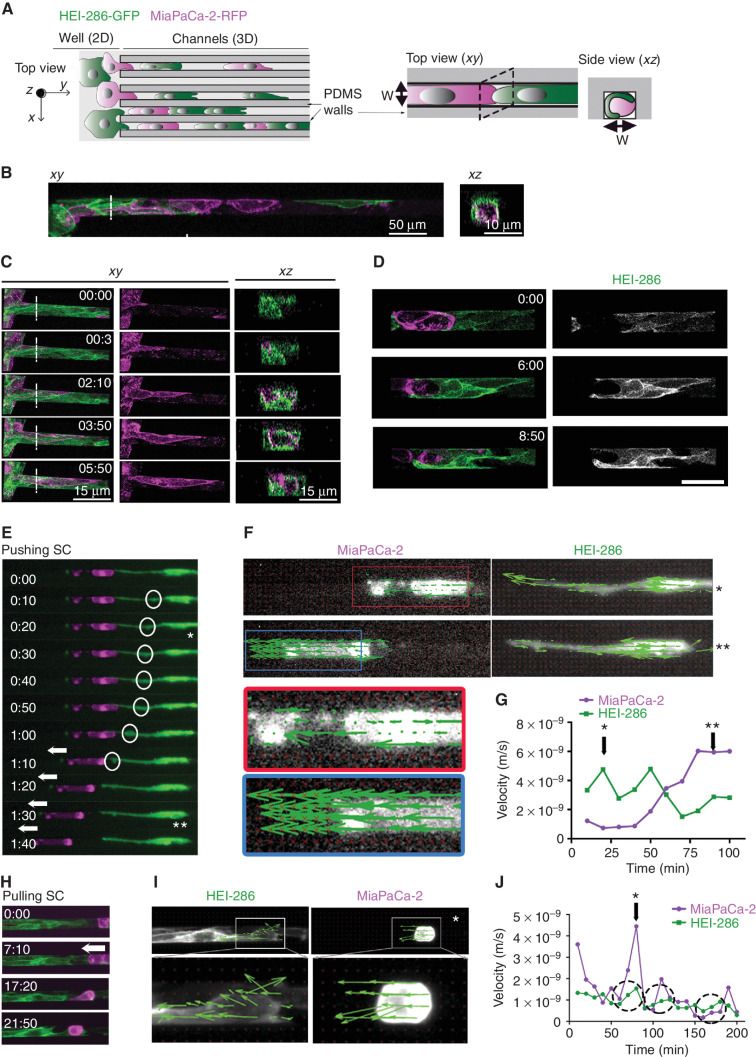
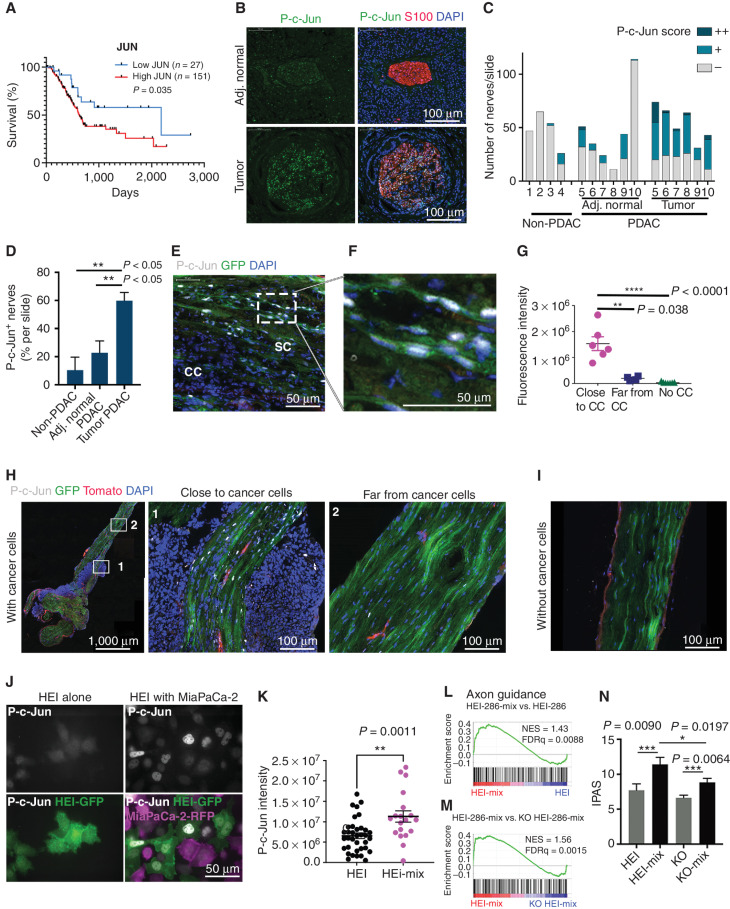
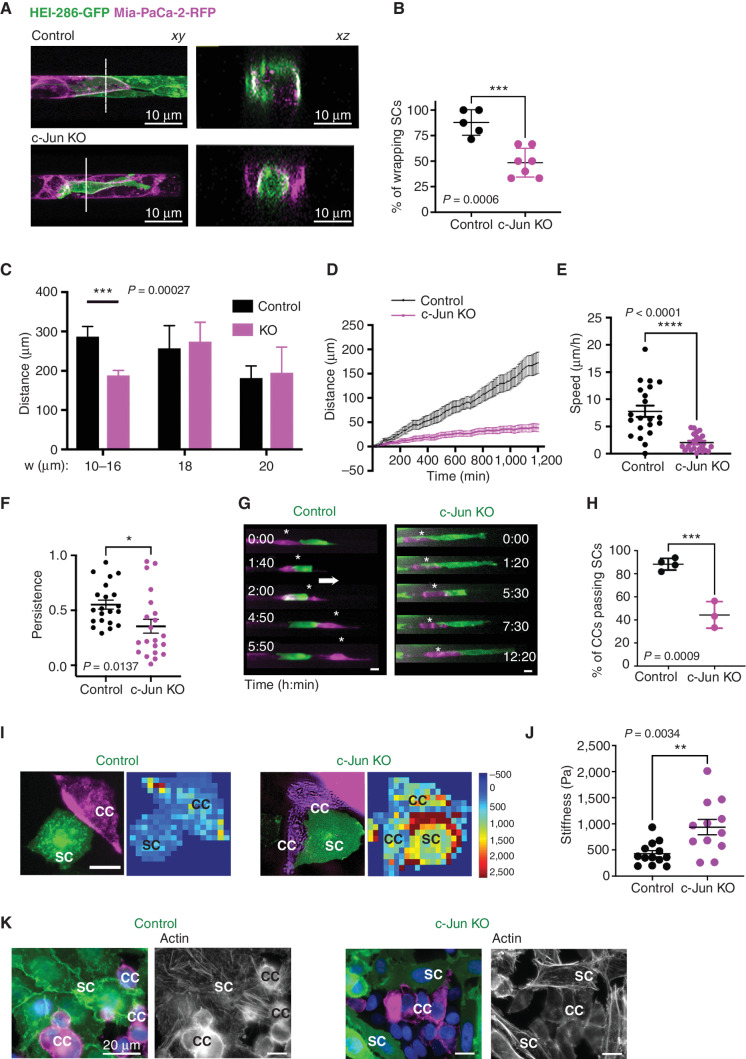
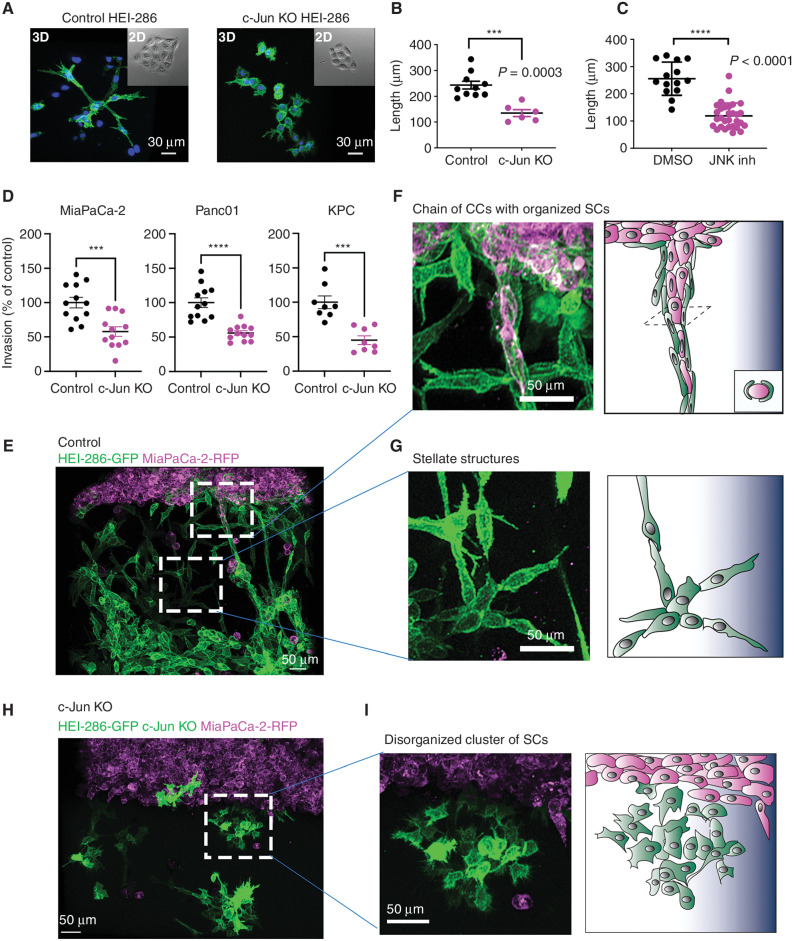
![Figure 7. c-Jun–reprogrammed SCs promote cancer invasion in vivo. A, Histologic analysis of injected murine sciatic nerves in P0-CRE− (WT) and P0-CRE+ (c-Jun KO) c-Junfl/fl mice. Representative samples of cancer invasion detected by H&E staining. Boxes show the invasion of areas away from the site of injection. Scale bars, 2,000 μm. Box scale bars, 500 μm. B, Quantification of distance of nerve invasion by Panc02 cells (WT n = 7, c-Jun KO n = 11, mean ± SEM) and KPC cells (WT n = 13, c-Jun KO n = 11, mean ± SEM). C, Representative images of murine hind limbs after Panc02 cancer cell injections show less paralysis in c-Jun KO SC mice. D, Quantification of the maximum width of hind limb paw in P0-CRE− (WT) versus P0-CRE+ (c-Jun KO) c-Junfl/fl mice 10 days after cancer injection (n = 8 mice/condition, mean ± SEM). E, Quantification of sciatic nerve function in P0-CRE− (WT) versus P0-CRE+ (c-Jun KO) c-Junfl/fl mice (n = 8 mice/condition, mean ± SEM). F, Effect of SP600125 on sciatic nerve invasion. Quantification of the length of nerve invasion in mice injected with Panc02 and SP600125 [JNK inhibitor (JNKi)] or Panc02 and DMSO (n = 11 mice/treatment, mean ± SEM). G, Representative H&E staining images of sciatic nerves injected with Panc02 and SP600125 versus Panc02 and DMSO. Top images are from proximal nerve regions at the spinal cord. No cancer is seen within the SP600125-injected nerve. Bottom images are from the site of injection, distal to the spinal cord. CC, cancer cells. Scale bars, 200 μm. H, Effect of SP600125 on flank tumor growth. Quantification of tumor volume in mice coinjected with Panc02 and SP600125 and with Panc02 and DMSO (n = 5 mice/treatment, mean ± SEM). NS, not significant. I, Survival analysis of WT and c-Jun KO mice injected with Panc02 in the sciatic nerve (WT n = 9, c-Jun KO = 11, P = 0.0003).](https://cdn.ncbi.nlm.nih.gov/pmc/blobs/e26a/9533012/0a0f31fcb2e1/2454fig7.jpg)