Abstract
Minimizing bias is critical to adoption and implementation of machine learning (ML) in clinical practice. Systematic mathematical biases produce consistent and reproducible differences between the observed and expected performance of ML systems, resulting in suboptimal performance. Such biases can be traced back to various phases of ML development: data handling, model development, and performance evaluation. This report presents 12 suboptimal practices during data handling of an ML study, explains how those practices can lead to biases, and describes what may be done to mitigate them. Authors employ an arbitrary and simplified framework that splits ML data handling into four steps: data collection, data investigation, data splitting, and feature engineering. Examples from the available research literature are provided. A Google Colaboratory Jupyter notebook includes code examples to demonstrate the suboptimal practices and steps to prevent them.
Keywords: Data Handling, Bias, Machine Learning, Deep Learning, Convolutional Neural Network (CNN), Computer-aided Diagnosis (CAD)
© RSNA, 2022
Keywords: Data Handling, Bias, Machine Learning, Deep Learning, Convolutional Neural Network (CNN), Computer-aided Diagnosis (CAD)
Summary
This report describes 12 major practices during data handling that predispose machine learning systems to mathematical bias.
Key Points
■ Systematic biases in machine learning systems produce consistent and reproducible differences between the observed and expected performance from those systems, which limit the application of these systems to real-world scenarios.
■ Although proper data handling is gaining more attention, guidelines to address the correct management of big data are scarce.
■ Strategies are introduced to prevent different mathematical biases resulting from poor data collection, data investigation, data splitting, and feature engineering of machine learning models.
Introduction
Machine learning (ML) applications in radiology have resulted in more than 8000 publications worldwide from 2000 to 2018 (1). Nonetheless, mitigation of possible mathematical bias (hereafter called bias) remains a critical concern for adopters (2,3). Although bias in medical research may be random, such as sampling variability or measurement precision, systematic bias produces consistent and reproducible differences between observed and expected performance (4). Unrecognized bias may contribute to suboptimal results.
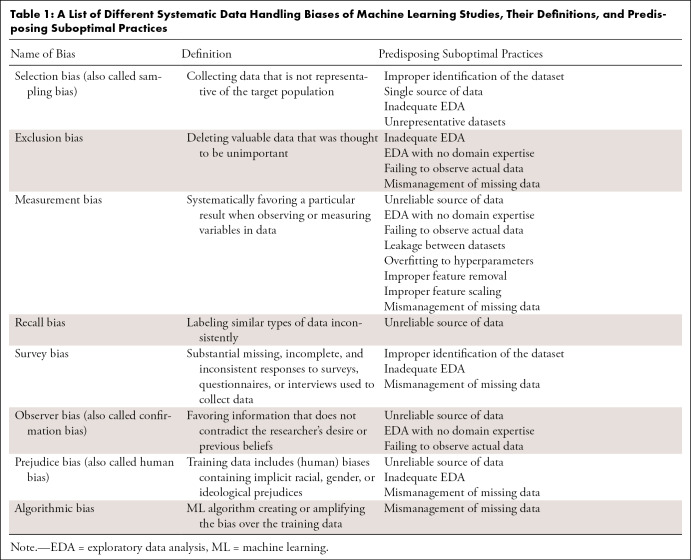
To mitigate bias, researchers should carefully design and implement a pipeline of data handling, model development, and performance evaluation (5). Each of these steps may introduce systematic or random bias. Systematic biases can reduce the fairness of ML systems; such biases must be recognized and, ideally, eliminated. Table 1 describes several systematic biases that can arise in the data handling phase of ML system development.
Table 1:
A List of Different Systematic Data Handling Biases of Machine Learning Studies, Their Definitions, and Predisposing Suboptimal Practices
Suboptimal quality of clinical data often limits the performance of ML algorithms (6,7). Therefore, developers must handle data accurately when performing data sampling, de-identification, annotation, labeling, or managing missing values. Although several guidelines exist for the proper development of ML systems, discussion of proper data handling is often insufficient (8,9). This report highlights common suboptimal data handling practices that may lead to systematic biases and briefly introduces common techniques to address them.
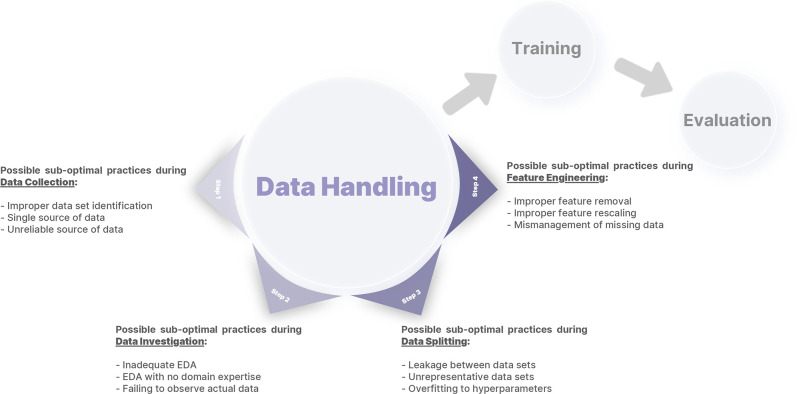
In line with Kocak et al (5), we define data handling as all data-related processes following the initial planning for an ML study up to model development and training. Although there are many ways to define data handling (10), we define four steps: data collection, data investigation, data splitting, and feature engineering (Fig 1). These steps focus on the aspects of data handling that ML researchers can control. We avoid the terms data preprocessing and data wrangling as these terms may encompass more than one step. This report, intended for radiologists interested in ML who are not expert data scientists, focuses on computer vision examples; however, most of the principles are applicable to other ML applications such as natural language processing or tabular data analysis. A Google Colaboratory Jupyter notebook is provided with code examples for each step (https://colab.research.google.com/drive/1c4G2b_ynikPsf2J5ExNS2D0or4uHX1dy?usp=sharing).
Figure 1:
An arbitrary framework for defining data handling, consisting of four different steps: data collection, data investigation, data splitting, and feature engineering. Different errors introduced in this report for each step are also summarized. EDA = exploratory data analysis.
Step 1: Data Collection
Data collection typically follows study design and usually requires researchers to access private or public data pools, query desired data types (eg, imaging, text, tabular), and transfer data to research storage. Depending on data properties, institutional policies, and study design, data de-identification may be done in this step or during feature engineering. Researchers may also access pre-existing human-labeled data, such as annotations for segmentation or classification models, or algorithm-generated data, such as synthetic images or text.
Improper Identification of the Dataset
Careful selection of data is critical for ML system development. Collecting all available data (instead of a chosen subset) is not always feasible, and feeding redundant data may hinder training by adding unnecessary complexity (11). Conversely, failing to collect all available data predisposes trained models to additional biases. An example of improper dataset identification occurs when retrieving Digital Imaging and Communications in Medicine (DICOM) files. Researchers may lose access to nonimaging data attributes available in DICOM metadata (eg, patient age and sex) if these files are converted to other file formats upon retrieval. Such attributes could be used to check study inclusion and exclusion criteria and to assure appropriate demographic representation (10).
To identify and collect the appropriate datasets, it is vital to estimate the types, attributes, and size of the data required. First, an in-depth review of available clinical and technical literature, accompanied by insights from medical experts, helps to determine the data types or attributes that are critical (12). Second, statistical power estimation techniques and knowledge of similarly developed ML systems can help determine the minimum dataset size needed to show an effect and also ensure the generalizability of the trained model (10,13). Finally, researchers may have access to more than one data type (eg, imaging from different modalities, different clinical datasets, or even human-labeled annotations for other ML purposes). Training a model on multiple data types may improve the performance of ML systems (14).
Single Source of Data
Models may not generalize well if training data are collected from a single source (15). A model may perform well with data from the pool it was trained on, but there is a substantial risk of failing to perform well on data from other sources. Data from a single source may not sample the full data distribution in the real world. For example, chest radiograph classifiers trained on publicly available datasets may have multiple racial, sex, and socioeconomic biases that reflect the patient distribution in their training data (16), resulting in poor generalizability.
Another less recognized “single source” issue is collection of data from a single point in time. Hardware and software scanner upgrades may alter image appearance. Disease incidence and appearance change with time, and new treatments may affect the appearance. Although the input data to ML systems may remain homogeneous over time, potential data drift should be anticipated (17).
Although it can be difficult to estimate how much expansion or diversification of training data will ensure generalization, there are multiple strategies to improve the heterogeneity of data sources. One strategy is to collect data from multiple institutions with different patient compositions (7). Although this strategy is not always feasible due to technical or patient privacy issues, the emergence of data de-identification tools, federated learning, and cloud data storage may help address this challenge (10,18–20). A second strategy is to collect data from different vendors (eg, imaging devices or electronic health records) within a single institution. Different makes and models of devices, including older devices, will help, although this option is likely inferior to collecting multi-institutional data (21). A third strategy is to use public datasets. Several nonmedical and medical datasets have been released publicly (22). These datasets may include human- or machine-generated labels, but even a dataset without labels may still help train self-supervised models. Such models are trained on tasks that do not need ground truth labels but still learn meaningful features from their training data, which can facilitate the training of other supervised models through a process called transfer learning. Any external data from other institutes or public sources should be checked in advance to avoid implicit biases (see below) (23). Finally, estimating the exact training data qualities needed to minimize distribution shift issues in real clinical practice is not always feasible. However, the training data should at least be sampled in a way that is representative of the population data.
Unreliable Source of Data
Data should be collected from reliable sources: Users should understand unambiguously how the data have been retrieved, processed, and transferred. For example, developers may be unable to determine if protected health information embedded in DICOM files was removed appropriately in the absence of de-identification protocols. Human-labeled annotations (eg, segmentation masks or bounding boxes) require a clear protocol and measurement of intra- and interannotator reliability to reduce recall, observer, or measurement biases (24).
It is easier to remove any ambiguity in data before collection. Access to live clinical data from picture archiving and communication system and electronic health records is usually restricted to staff physicians, technologists, and data managers. Therefore, developers may need to collaborate with clinical stakeholders to communicate needs and clarify any ambiguities (10). Public datasets may be prone to biases and may not represent or report the age, race, or sex of the original population (25). It is also critical to make sure that there is no overlap of specific patients and images in public datasets, the so-called Frankenstein problem, where one public set is used to train a model and another is used to validate, but the presence of the same case in both sets means the real-world performance is poorer than measured (26).
Step 2: Data Investigation
Developers should investigate collected data from multiple perspectives to discover properties that may assist in detecting potential data issues, a process known as exploratory data analysis (EDA) (27). The main goals of EDA are to (a) organize and summarize the raw data, (b) discover important features and patterns in the data and flag any deviations, and (c) interpret findings in the context of the problem. The analytic operations of an EDA (eg, filtering, aggregation, and visualization) are done sequentially, where the result of one operation often denotes the next operation (28).
Inadequate EDA
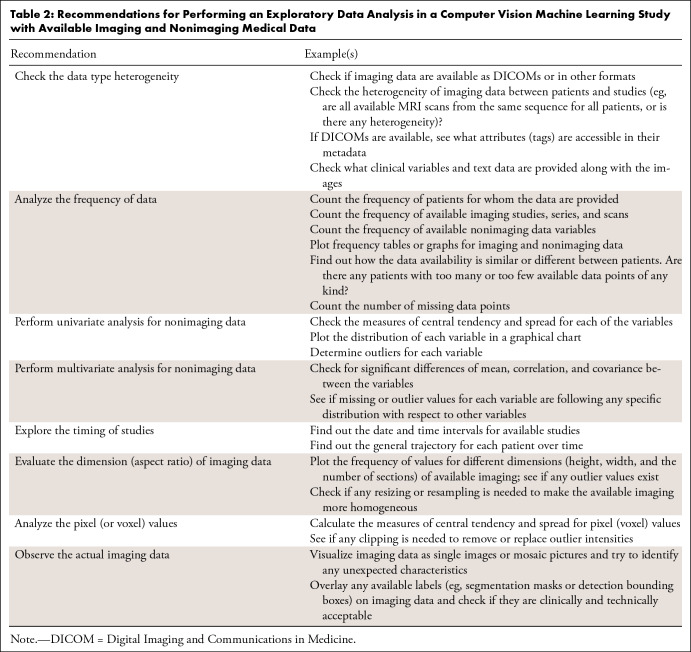
An inadequate EDA may be challenging to detect, but there may be no EDA at all if the collected data appear trivial. Many statistical aspects of the data should be investigated during an EDA, including measures of frequency, central tendency and spread, the shape of data distribution, missing data, and outlier data (27). One should investigate the interdependency within the data, such as if there is more than one imaging examination for some patients. Table 2 provides recommendations for EDA in a medical ML study.
Table 2:
Recommendations for Performing an Exploratory Data Analysis in a Computer Vision Machine Learning Study with Available Imaging and Nonimaging Medical Data
EDA tools allow data visualization in many formats, including graphs, plots, or tables. Most ML applications are coded in Python, and many packages, such as Pandas and Matplotlib, provide basic EDA tooling (29–31). DataPrep and AutoAIViz perform EDA automatically for different data types (28,32,33). Although helpful, none of these tools will replace the need for in-depth and targeted analysis of data by clinical and data science experts.
EDA with No Domain Expertise
Incorporation of domain expertise can enrich EDA (34). In fact, the quality of an EDA depends on the ability of the developers to ask the right questions and detect patterns within the data based on their cognitive ability, domain expertise, and experience in data interpretation (35). To detect bias, EDA should be conducted as a joint effort of ML statisticians, data scientists, and clinicians or other domain experts. Knowledge sharing, which can facilitate collaboration between developers and domain experts, involves identifying related domain experts, obtaining information from them using formal or informal tools (eg, meetings or interviews), and processing and storing the collected information in repositories, such as question banks or manuals, for further use. Knowledge sharing is facilitated by many communication standards (36,37).
Failing to Observe Actual Data
Although it seems trivial, personally reviewing the collected data is of utmost importance for ML developers and clinicians. While we already described several statistical measures to evaluate input data, observing the data itself (and not its statistical properties) often provides new insights. For example, in a study for developing an ML model to segment brain tumors from brain MRI studies, initial observation of a random sample from the input data may reveal varying sequences and quality of MRI studies. Such findings indicate a need to check the frequency of available sequences, as well as vendor and scan protocols. If feasible, all data points in datasets should be reviewed, and if not, researchers should draw a representative sample and inspect that subset. For example, imaging data could be visualized by programmatically building a mosaic photograph of individual imaging instances (38,39). Despite the lower resolution of each of these images (tile), they may be helpful for comparison and anomaly detection purposes (Fig 2). Similarly, conventional DICOM viewers may be used to inspect images and any human-labeled annotations. Tools like ITK-SNAP can overlay segmentation masks on underlying images to help check the quality of annotations (40). It is also necessary to review data after the feature engineering step (see below) to detect unwanted or undesirable modifications to the data.
Figure 2:
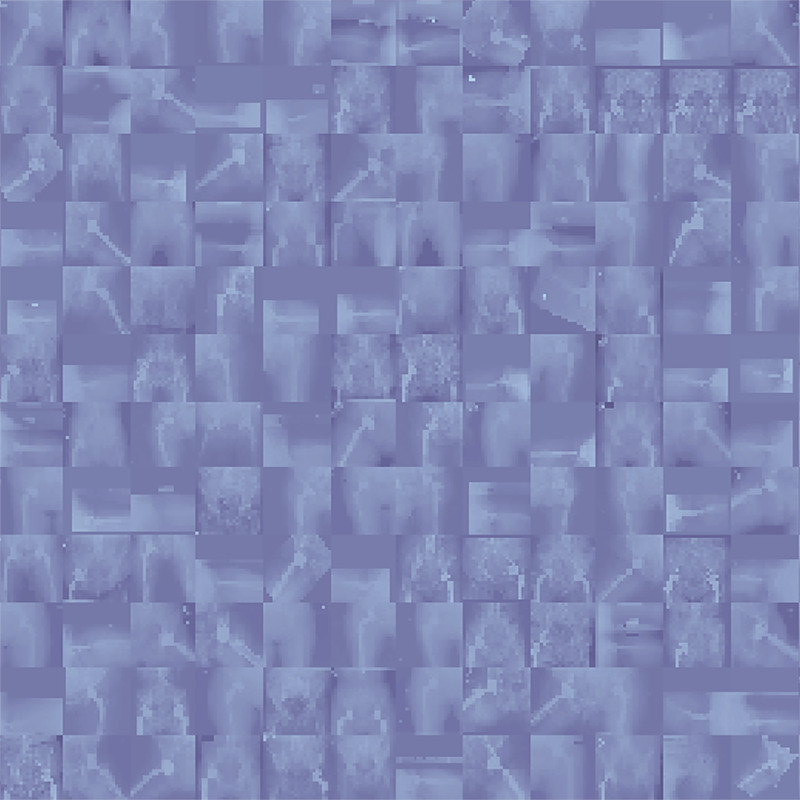
A mosaic photograph of random radiographs was collected from our institutional dataset of patients who underwent total hip arthroplasty. Despite the reduced resolution of individual images, a quick look at this photograph reveals valuable insights for developers who desire training models on this dataset; for example, radiographs have different views, not all radiographs have prostheses, radiographs are from different sexes (the anatomy of pelvis is different between male and female patients), different prosthesis brands are available in the data, some radiographs have outlier intensities (presenting darker or brighter than expected), and so forth.
Step 3: Data Splitting
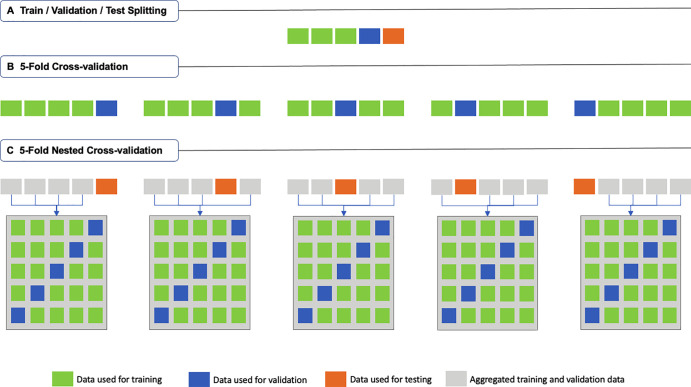
The data are partitioned into training, validation, and optionally testing sets. The training set is used to fit an ML model during training. The validation set is used to track the learning process during training but is not used directly to train the model. The test set is used to evaluate the performance and generalizability of the ML model after the training is complete (Fig 3A). Critical data handling errors may occur, particularly when working with medical data.
Figure 3:
Schematic description of (A) traditional train-validation-test splitting, (B) fivefold cross-validation (k = 5; where k is the number of folds), and (C) fivefold nested cross-validation (k = m = 5; where k is the number of folds in the first-level cross-validation, and m is the number of folds in the second-level cross-validation).
Leakage between Datasets
There should be no leakage between the training and test sets. In other words, to assure that models can distinguish meaningful signals from noise, the ML model should not “see” any of the test data during training (41). Data may “leak,” even if developers ensure that no data are repeated in both training and test sets because medical data are usually clustered at different levels. For example, a patient with a liver tumor may have four different liver MRI studies, each with more than one series, and each series with several images. In such a scenario, a random train-test split at the image level will result in biased training. Individual images are not necessarily independent, and images from the same series could go to both training and testing sets. Although the ML model will not see the same section in both the training and test sets, it will likely find very similar features from adjacent sections. This will help the model learn their inherent similarities. In such a case, both training and validation loss would decrease, but the model would not have learned general features and will subsequently perform poorly if applied to other patients. Even splitting based on series or study levels is not sufficient to prevent data leakage. Different scans from the same patient will have similarities, and the model’s performance may still be overestimated when applied to validation or test sets.
The standard way to prevent data leakage is to split medical data at the patient level. When training and test sets consist of data from different patients, developers can be more assured that their models will not have a chance to find similar noise between the sets. Splitting at the patient level is not always easy, as patients may have a different number of data points available, and multiple variables must be balanced between the sets. Python packages such as scikit-learn provide algorithmic tools that can speed up data splitting while grouping by a patient-level variable, such as the PatientID attribute of DICOM files, and stratifying the data based on other variables (see the notebook, https://colab.research.google.com/drive/1c4G2b_ynikPsf2J5ExNS2D0or4uHX1dy?usp=sharing) (42). Finally, there are instances where data split at levels higher than the patient level may be beneficial. For example, if there are consistent differences in how a hospital handles patient scans, it may be valuable to separate data at the institution level.
Imbalanced Datasets
The training and test sets should both represent real-world data. This condition is not easily met in medical domains for two important reasons: (a) the size of medical datasets is substantially smaller than most nonmedical datasets (eg, ImageNet), and (b) even the available data may have imbalanced distribution based on several variables. The causes of this imbalance go beyond ML and may pertain to population-level frequencies or patient recruitment protocols. Suppose developers want to train an ML model on hip radiographs from 10 000 patients who have undergone total hip arthroplasty to predict the risk of a complication. If the incidence of the complication in their institution is 2%, they would have 9800 patients with no complications versus 200 with complications, yielding a highly imbalanced dataset. As the conventional train-test split often holds out 10%–20% of data for testing, the test set will contain the data for only 20–40 patients with complications. In addition to the many training challenges that such a dataset will cause, there is a substantial risk that the few patients with complications from the test set are not representative of the population. Therefore, developers cannot rely on the reported model performance applied to their test set, as it may not generalize to the real-world data.
Another data splitting technique, k-fold cross-validation (Fig 3B), splits the data into k nonoverlapping folds (usually five or 10) (43). Each fold is used once as the test set, while all other folds are used as a training dataset. A total of k models are trained (one for each test set), and the mean performance is reported. Using k-fold cross-validation in our example, developers can ensure that the data from each of the 200 patients with complications have been used once to evaluate the model’s performance. Also, as the k models have been trained on different data, it is more likely that their mean performance is generalizable to real-world data, particularly if the variance is low. Using k-fold cross-validation is typically better than using the conventional train-test data splitting, specifically when dealing with small and imbalanced datasets. If used, we strongly recommend reporting not only the mean but also the range and the CIs of model performance to assess reliability.
Overfitting to Hyperparameters
Overfitting is a known issue of training ML models when they do not perform as well on the test set as on the training data, denoting the lack of generalizability of the model. Although overfitting is generally a training issue, a particular case of it may be traced to data splitting: overfitting to hyperparameters or the test set. While training ML models, developers use various approaches like grid searching to choose the best hyperparameters. To compare each set of hyperparameters, one might evaluate their effect on model performance on the test set. Therefore, there is a risk that developers may gradually find the hyperparameters that work best on their test set but are not necessarily generalizable to other unseen data (44). To avoid this issue in the conventional train-test split, a distinct subset of the training set (aka, validation set) would be used for hyperparameter tuning (and no more for training), and the test set will be used in neither of these tasks.
We noted above that cross-validation can be useful for imbalanced and small datasets. Nested cross-validation is a variant of the k-fold cross-validation technique that tries to address the issue of overfitting to the hyperparameters (45). In this technique (Fig 3C), the kth fold will be held out, and data from the k − 1 other folds, which were previously regarded as the training set, will collectively be split into m distinct folds using a second (inner-layer) m-fold cross-validation. The resulting m folds will then be used to train m models and find the best hyperparameter settings. The best model will finally be applied to the holdout kth fold, and its performance will be reported. In summary, m × k models will be trained in a nested cross-validation approach, while no hyperparameter tuning happens on the holdout sets. It should be noted that both the k-fold cross-validation and nested cross-validation approaches are prone to methodologic biases and may not be always feasible; therefore, specific guidelines must be followed to obtain their most reliable performance (46).
Step 4: Feature Engineering
Feature engineering transforms or removes features from the input data before fitting an ML model (46). The idea is to change the input data so that ML models see more meaningful and less redundant features. Many techniques could be incorporated into feature engineering, including techniques that aim to reduce noise in the data (eg, denoising, kernel-based transformation, organ windowing followed by normalization, and cropping followed by zero padding of imaging) before feeding to ML models (47). Because of the complicated nature of feature engineering, suboptimal practices can occur during feature engineering and make the ML models prone to underfitting and bias.
Improper Feature Removal
One routine question in feature engineering is “What features can be removed from the input data to make the learning easier for ML models?” The underlying assumption for this question is that there is noise in the data that, if removed, enables easier fitting of an ML model. Whether this assumption is valid depends on the input data, the algorithm, and the task at hand. Suppose some developers would like to train a deep learning classifier for differentiating COVID-19 pneumonia from other viral pneumonia using chest radiographs. Model performance may not necessarily improve if they use another model to first segment the lungs from the original chest radiograph and then train the classifier only on cropped lung regions. While noise may be reduced, the classifier could look at nonlung regions of the chest radiograph (eg, the heart) and find additional helpful signals. Another problem with that assumption is that ML models do not necessarily learn as humans do. What is considered noise to developers may be a source of valuable signal data for models.
Evaluating all scenarios in separate experiments is the best way to understand if removing features will cause the model to fit better or worse (thus reducing or increasing bias). Although such a solution could increase training costs, developers should consider it whenever possible while also relying on common sense or implicit clinical knowledge and experience. Figure 4 demonstrates an example of how improper feature removal may lead to subsequent bias in a model’s performance.
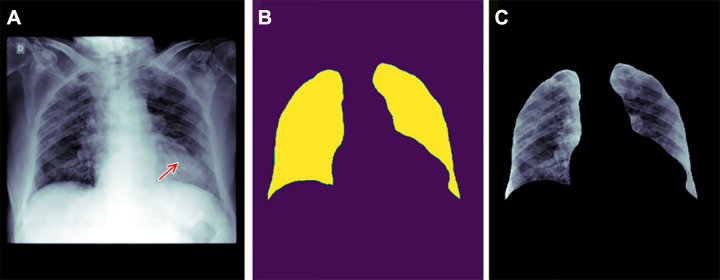
Figure 4:
Example of how improper feature removal from imaging data may lead to bias. (A) Chest radiograph in a male patient with pneumonia. (B) Segmentation mask for the lung, generated using a deep learning model. (C) Chest radiograph is cropped based on the segmentation mask. If the cropped chest radiograph is fed to a subsequent classifier for detecting consolidations, the consolidation that is located behind the heart will be missed (arrow, A). This occurs because primary feature removal using the segmentation model was not valid and unnecessarily removed the portion of the lung located behind the heart.
Improper Feature Scaling
Feature scaling means applying arithmetic operations on feature values so that most (if not all) of the features from the input data are brought to a similar scale. This way, features will not have dramatically different magnitudes, and the learning process (eg, the back propagation in gradient descent algorithms) is more efficient. Many techniques are available for feature scaling, but two common ones are (a) normalization, which changes the feature values to a common scale (eg, 0–1) and (b) standardization (or z score normalization), which scales feature values to have zero mean and unit SD.
Although these techniques are applied to improve model fit and generalizability, they can result in poorer fit, and even bias, if misapplied. Seeking consultation from expert statisticians may help choose the appropriate scaling techniques for the data and algorithms. For example, standardization is more effective if the feature values have an almost Gaussian distribution, and the ML algorithm assumes the distribution of data to be normal. Normalization, on the other hand, does not have this limitation. Therefore, while normalization is more appropriate for algorithms like k-nearest neighbors and artificial neural networks, standardization works better for regression algorithms. As with other examples, normalization may amplify the noise in features with almost constant values, and standardization can significantly distort data values in the presence of outliers (48).
There are several tips to ensure feature scaling results in more benefit than harm for the input data. First, feature scaling must be deemed necessary. For example, ML algorithms such as tree-based models do not consider the scale of the data and often do not need feature scaling. One should also check the current scale and distribution of data and apply the appropriate feature scaling technique. For example, it is needed to remove outlier values in data before applying standardization. To do so, voxel values in medical imaging data like radiographs or MRI can be clipped to a specific value range (eg, between 5% and 95% percentile of intensity distribution or between the lower and upper narrow tales of a histogram obtained from probability distribution function of the image intensity) (47).
Mismanagement of Missing Data
All real-world data are likely to have missing data. In fact, some ML repositories may have more than 40% of data missing (49). While discussing why missing data exists in ML is outside the scope of this report, how missing data are handled does affect bias and fairness. Previous research has shown that missing data may not be evenly distributed, leading to unwanted effects on the fairness of the data and the resulting models (50). For example, underrepresented groups may be reluctant to provide sensitive information on medical or demographic questionnaires and are more likely to miss follow-up medical visits (51). They may even receive more or fewer diagnostic and treatment options than necessary (52). Therefore, the medical data for such underrepresented groups may be less available and ignored (or interpreted mistakenly) by ML systems that have not addressed missing values properly during training, resulting in prejudice and selection biases.
The simplest (but not recommended) way to deal with missing data during ML development is to remove all patients for whom data points are missing. This approach can result in bias and fairness issues, as (a) removal of cases may exacerbate the problem of small or imbalanced datasets, and (b) the missing data are not random and more commonly affects a specific population of patients; thus, the final model’s performance may be significantly worse on underrepresented populations (53,54). Moreover, various studies have shown how the performance of models may be degraded when missing data are discarded. A better strategy is to substitute missing data with synthetic values (so-called imputation). Imputation may be as simple as replacing the missing values with the mean or median of the available data; however, it could also be done using more sophisticated techniques and even by using ML to predict the missing values (55). While some imputation techniques (especially those that use predictive ML) may amplify algorithmic bias in the data, they generally improve the model’s performance while maintaining its fairness (50). Exhaustive discussions of the pros and cons of different imputation methods are beyond the scope of this report but are available elsewhere (53–55).
Conclusion
ML has been widely deployed, from image reconstruction and hypothesis testing to improving diagnostic, prognostic, and monitoring tools (56). This report discussed how ML tools could be susceptible to biases in their data handling phase and how developers might prevent those biases. Our report covered items 7–13 of the Checklist for Artificial Intelligence in Medical Imaging and items 1–5 of the Guiding Principles for Good Machine Learning Practice for Medical Device Development, recently introduced by the U.S. Food and Drug Administration, Health Canada, and the United Kingdom’s Medicines and Healthcare products Regulatory Agency (24,57).
The recommendations here should be interpreted and applied with caution. First, the discussion is based on an arbitrary data handling framework of four successive steps: data collection, investigation, data splitting, and feature engineering. This framework is oversimplified, and we acknowledge that ML systems are not developed as a linear process. Second, this report has considered only the mathematical forms of bias. Social and ethical forms of bias can harm underrepresented groups or benefit those with more resources. Addressing such biases often needs political decisions and time-consuming changes in stakeholders’ behaviors. Third, ML bias is an active field of research, and this report is not an exhaustive list of problems or solutions. Additionally, bias mitigation strategies are problem specific, and certain strategies described above may be unnecessary or even inappropriate in specific situations. Finally, not all bias-prone practices in ML can be easily replaced or improved. Limitations do exist in ML studies, and ML systems may be biased in different ways. Depending on the data and task at hand, such biases could be tolerated or even considered helpful (58). However, developers must identify erroneous practices during ML development, mitigate as many of them as possible, and explicitly report the remaining limitations of their ML systems. In further reports, we will focus on biases that exist in two other phases of ML studies, namely model development and performance evaluation.
Disclosures of conflicts of interest: P.R. No relevant relationships. B.K. No relevant relationships. S.F. No relevant relationships. M.M. No relevant relationships. D.V.V.G. No relevant relationships. Y.S. No relevant relationships. K.Z. No relevant relationships. G.M.C. Member of the Radiology: Artificial Intelligence trainee editorial board. B.J.E. Grant from NCI; stock/stock options in FlowSIGMA, VoiceIT, and Yunu; consultant to the editor for Radiology: Artificial Intelligence.
Abbreviations:
- DICOM
- Digital Imaging and Communications in Medicine
- EDA
- exploratory data analysis
- ML
- machine learning
References
- 1. West E , Mutasa S , Zhu Z , Ha R . Global Trend in Artificial Intelligence-Based Publications in Radiology From 2000 to 2018 . AJR Am J Roentgenol 2019. ; 213 ( 6 ): 1204 – 1206 . [DOI] [PubMed] [Google Scholar]
- 2. Tariq A , Purkayastha S , Padmanaban GP , et al . Current Clinical Applications of Artificial Intelligence in Radiology and Their Best Supporting Evidence . J Am Coll Radiol 2020. ; 17 ( 11 ): 1371 – 1381 . [DOI] [PubMed] [Google Scholar]
- 3. Liew C . The future of radiology augmented with Artificial Intelligence: A strategy for success . Eur J Radiol 2018. ; 102 : 152 – 156 . [DOI] [PubMed] [Google Scholar]
- 4. Krishna R , Maithreyi R , Surapaneni KM . Research bias: a review for medical students . J Clin Diagn Res 2010. ; 4 ( 2 ): 2320 – 2324 . https://www.jcdr.net/article_abstract.aspx?issn=0973-709x&year=2010&volume=4&issue=2&page=2320%20-%202324&issn=0973-709x&id=677 . [Google Scholar]
- 5. Kocak B , Kus EA , Kilickesmez O . How to read and review papers on machine learning and artificial intelligence in radiology: a survival guide to key methodological concepts . Eur Radiol 2021. ; 31 ( 4 ): 1819 – 1830 . [DOI] [PubMed] [Google Scholar]
- 6. Wang F , Casalino LP , Khullar D . Deep Learning in Medicine-Promise, Progress, and Challenges . JAMA Intern Med 2019. ; 179 ( 3 ): 293 – 294 . [DOI] [PubMed] [Google Scholar]
- 7. Beam AL , Manrai AK , Ghassemi M . Challenges to the Reproducibility of Machine Learning Models in Health Care . JAMA 2020. ; 323 ( 4 ): 305 – 306 . [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8. Kumar A , Boehm M , Yang J . Data Management in Machine Learning: Challenges, Techniques, and Systems . In: Proceedings of the 2017 ACM International Conference on Management of Data . New York, NY: : Association for Computing Machinery; , 2017. ; 1717 – 1722 . [Google Scholar]
- 9. Polyzotis N , Roy S , Whang SE , Zinkevich M . Data Management Challenges in Production Machine Learning . In: Proceedings of the 2017 ACM International Conference on Management of Data . New York, NY: : Association for Computing Machinery; , 2017. ; 1723 – 1726 . [Google Scholar]
- 10. Willemink MJ , Koszek WA , Hardell C , et al . Preparing Medical Imaging Data for Machine Learning . Radiology 2020. ; 295 ( 1 ): 4 – 15 . [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11. Ohno-Machado L , Fraser HS , Ohrn A . Improving machine learning performance by removing redundant cases in medical data sets . Proc AMIA Symp 1998. ; 523 – 527 . [PMC free article] [PubMed] [Google Scholar]
- 12. Rahman P . Amplifying domain expertise in medical data pipelines . https://search.proquest.com/openview/74c5d6f5beae52e809a135fcaf95341b/1?pq-origsite=gscholar&cbl=18750&diss=y. Published 2020. Accessed October 2021 . [DOI] [PMC free article] [PubMed]
- 13. Eng J . Sample size estimation: how many individuals should be studied? Radiology 2003. ; 227 ( 2 ): 309 – 313 . [DOI] [PubMed] [Google Scholar]
- 14. Huang S-C , Pareek A , Seyyedi S , Banerjee I , Lungren MP . Fusion of medical imaging and electronic health records using deep learning: a systematic review and implementation guidelines . NPJ Digit Med 2020. ; 3 ( 1 ): 136 . [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15. Doshi-Velez F , Kim B . Considerations for Evaluation and Generalization in Interpretable Machine Learning . In: Escalante HJ , Escalera S , Guyon I , et al. , eds. Explainable and Interpretable Models in Computer Vision and Machine Learning. The Springer Series on Challenges in Machine Learning . Cham, Switzerland: : Springer; , 2018. ; 3 – 17 . [Google Scholar]
- 16. Seyyed-Kalantari L , Liu G , McDermott M , Chen IY , Ghassemi M . CheXclusion: Fairness gaps in deep chest X-ray classifiers . Pac Symp Biocomput 2021. ; 26 : 232 – 243 . [PubMed] [Google Scholar]
- 17. Guo X , Gichoya JW , Trivedi H , Purkayastha S , Banerjee I . MedShift: identifying shift data for medical dataset curation . arXiv 2112.13885 [preprint] http://arxiv.org/abs/2112.13885. Posted December 27, 2021. Accessed October 2021 . [DOI] [PMC free article] [PubMed]
- 18. Yang X , Lyu T , Li Q , et al . A study of deep learning methods for de-identification of clinical notes in cross-institute settings . BMC Med Inform Decis Mak 2019. ; 19 ( Suppl 5 ): 232 . [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19. Sheller MJ , Edwards B , Reina GA , et al . Federated learning in medicine: facilitating multi-institutional collaborations without sharing patient data . Sci Rep 2020. ; 10 ( 1 ): 12598 . [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20. Kagadis GC , Kloukinas C , Moore K , et al . Cloud computing in medical imaging . Med Phys 2013. ; 40 ( 7 ): 070901 . [DOI] [PubMed] [Google Scholar]
- 21. Biondetti GP , Gauriau R , Bridge CP , Lu C , Andriole KP . “Name that manufacturer” . Relating image acquisition bias with task complexity when training deep learning models: experiments on head CT arXiv 2008.08525 [preprint] http://arxiv.org/abs/2008.08525. Posted August 19, 2020. Accessed October 2021 .
- 22. Li J , Zhu G , Hua C , et al . A Systematic Collection of Medical Image Datasets for Deep Learning . arXiv 2106.12864 [preprint] http://arxiv.org/abs/2106.12864. Posted June 24, 2021. Accessed October 2021 .
- 23. Cheplygina V , de Bruijne M , Pluim JPW . Not-so-supervised: A survey of semi-supervised, multi-instance, and transfer learning in medical image analysis . Med Image Anal 2019. ; 54 : 280 – 296 . [DOI] [PubMed] [Google Scholar]
- 24. Mongan J , Moy L , Kahn CE Jr . Checklist for Artificial Intelligence in Medical Imaging (CLAIM): A Guide for Authors and Reviewers . Radiol Artif Intell 2020. ; 2 ( 2 ): e200029 . [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25. Garcia Santa Cruz B , Bossa MN , Sölter J , Husch AD . Public Covid-19 X-ray datasets and their impact on model bias - A systematic review of a significant problem . Med Image Anal 2021. ; 74 : 102225 . [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26. Roberts M , Driggs D , Thorpe M , et al . Common pitfalls and recommendations for using machine learning to detect and prognosticate for COVID-19 using chest radiographs and CT scans . Nat Mach Intell 2021. ; 3 ( 3 ): 199 – 217 . [Google Scholar]
- 27. Morgenthaler S . Exploratory data analysis . WIREs Comput Stat 2009. ; 1 ( 1 ): 33 – 44 . [Google Scholar]
- 28. Milo T , Somech A . Automating Exploratory Data Analysis via Machine Learning: An Overview . In: Proceedings of the 2020 ACM SIGMOD International Conference on Management of Data . New York, NY: : Association for Computing Machinery; ; 2020. ; 2617 – 2622 . [Google Scholar]
- 29. Sahoo K , Samal AK , Pramanik J , Pani SK . Exploratory data analysis using Python . Int J Innov Technol Explor Eng 2019. ; 8 ( 12 ): 4727 – 4735 . [Google Scholar]
- 30. Reback J , McKinney W , et al . pandas-dev/pandas: Pandas 1.0.5. Zenodo . Published June 17, 2020. Accessed October 2021 . [Google Scholar]
- 31. Hunter JD . Matplotlib: A 2D Graphics Environment . Comput Sci Eng 2007. ; 9 ( 3 ): 90 – 95 . [Google Scholar]
- 32. Weidele DKI , Weisz JD , Oduor E , et al . AutoAIViz . In: Proceedings of the 25th International Conference on Intelligent User Interfaces , 2020. ; 308 – 312 . [Google Scholar]
- 33. Peng J , Wu W , Lockhart B , et al . DataPrep.EDA: Task-Centric Exploratory Data Analysis for Statistical Modeling in Python . In: Proceedings of the 2021 International Conference on Management of Data . New York, NY: : Association for Computing Machinery; ; 2021. ; 2271 – 2280 . [Google Scholar]
- 34. Langer T , Meisen T . Towards Utilizing Domain Expertise for Exploratory Data Analysis . In: Proceedings of the 12th International Symposium on Visual Information Communication and Interaction . New York, NY: : Association for Computing Machinery; ; 2019. ; 1 – 5 . [Google Scholar]
- 35. Grolemund G , Wickham H . A Cognitive Interpretation of Data Analysis . Int Stat Rev 2014. ; 82 ( 2 ): 184 – 204 . [Google Scholar]
- 36. Park S , Wang AY , Kawas B , Liao QV , Piorkowski D , Danilevsky M . Facilitating Knowledge Sharing from Domain Experts to Data Scientists for Building NLP Models . In: 26th International Conference on Intelligent User Interfaces . New York, NY: : Association for Computing Machinery; ; 2021. ; 585 – 596 . [Google Scholar]
- 37. Ackerman MS , Dachtera J , Pipek V , Wulf V . Sharing Knowledge and Expertise: The CSCW View of Knowledge Management . Comput Support Coop Work 2013. ; 22 ( 4 ): 531 – 573 . [Google Scholar]
- 38. Himite B . How to Create a Photo Mosaic in Python: A look under the hood of photo mosaics in Python . towardsdatascience . https://towardsdatascience.com/how-to-create-a-photo-mosaic-in-python-45c94f6e8308 . Published 2021. Accessed October 2021.
- 39. Sadri AR , Janowczyk A , Zhou R , et al . Technical Note: MRQy - An open-source tool for quality control of MR imaging data . Med Phys 2020. ; 47 ( 12 ): 6029 – 6038 . [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40. Yushkevich PA , Piven J , Hazlett HC , et al . User-guided 3D active contour segmentation of anatomical structures: significantly improved efficiency and reliability . Neuroimage 2006. ; 31 ( 3 ): 1116 – 1128 . [DOI] [PubMed] [Google Scholar]
- 41. Saravanan N , Sathish G , Balajee JM . Data Wrangling and Data Leakage in Machine Learning for Healthcare . https://papers.ssrn.com/abstract=3708142. Published 2018. Accessed June 22, 2022 . [Google Scholar]
- 42. Pedregosa F , Varoquaux G , Gramfort A , et al . Scikit-learn: Machine Learning in Python . J Mach Learn Res 2011. ; 12 : 2825 – 2830 . https://www.jmlr.org/papers/volume12/pedregosa11a/pedregosa11a.pdf . [Google Scholar]
- 43. Stone M . Cross-Validatory Choice and Assessment of Statistical Predictions (With Discussion) . J R Stat Soc Ser B Methodol 1976. ; 38 ( 1 ): 102 . [Google Scholar]
- 44. Cawley GC , Talbot NLC . On Over-Fitting in Model Selection and Subsequent Selection Bias in Performance Evaluation . J Mach Learn Res 2010. ; 11 ( 70 ): 2079 – 2107 . https://jmlr.org/papers/v11/cawley10a.html . [Google Scholar]
- 45. Varma S , Simon R . Bias in error estimation when using cross-validation for model selection . BMC Bioinformatics 2006. ; 7 ( 1 ): 91 . [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46. Zheng A , Casari A . Feature Engineering for Machine Learning: Principles and Techniques for Data Scientists . Sebastopol, Calif: : O’Reilly Media; , 2018. . [Google Scholar]
- 47. Masoudi S , Harmon SA , Mehralivand S , et al . Quick guide on radiology image pre-processing for deep learning applications in prostate cancer research . J Med Imaging (Bellingham) 2021. ; 8 ( 1 ): 010901 . [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48. Cendrero SM . Warning About Normalizing Data . business.blogthinkbig.com. https://business.blogthinkbig.com/warning-about-normalizing-data/. Published 2018. Accessed October 2021 .
- 49. Dua D , Graff C . UCI Machine Learning Repository . http://archive.ics.uci.edu/ml. Published 2017. Accessed October 2021.
- 50. Martínez-Plumed F , Ferri C , Nieves D , Hernández-Orallo J . Missing the missing values: The ugly duckling of fairness in machine learning . Int J Intell Syst 2021. ; 36 ( 7 ): 3217 – 3258 . [Google Scholar]
- 51. Gilbert DT , Fiske ST , Lindzey G . The Handbook of Social Psychology . Boston, Mass: : McGraw-Hill; , 1998. . [Google Scholar]
- 52. Pierson E , Cutler DM , Leskovec J , Mullainathan S , Obermeyer Z . An algorithmic approach to reducing unexplained pain disparities in underserved populations . Nat Med 2021. ; 27 ( 1 ): 136 – 140 . [DOI] [PubMed] [Google Scholar]
- 53. Schafer JL , Graham JW . Missing data: our view of the state of the art . Psychol Methods 2002. ; 7 ( 2 ): 147 – 177 . [PubMed] [Google Scholar]
- 54. Johnson DR , Young R . Toward best practices in analyzing datasets with missing data: Comparisons and recommendations . J Marriage Fam 2011. ; 73 ( 5 ): 926 – 945 . [Google Scholar]
- 55. Thomas T , Rajabi E . A systematic review of machine learning-based missing value imputation techniques . Data Technol Appl 2021. ; 55 ( 4 ): 558 – 585 . [Google Scholar]
- 56. May M . Eight ways machine learning is assisting medicine . Nat Med 2021. ; 27 ( 1 ): 2 – 3 . [DOI] [PubMed] [Google Scholar]
- 57. U.S. Food and Drug Administration (FDA) . Good Machine Learning Practice for Medical Device Development: Guiding Principles . https://www.fda.gov/medical-devices/software-medical-device-samd/good-machine-learning-practice-medical-device-development-guiding-principles. Published 2021. Accessed October 2021 .
- 58. Pot M , Kieusseyan N , Prainsack B . Not all biases are bad: equitable and inequitable biases in machine learning and radiology . Insights Imaging 2021. ; 12 ( 1 ): 13 . [DOI] [PMC free article] [PubMed] [Google Scholar]