Abstract
Vascular endothelial cells form the inner layer of blood vessels where they have a key role in the development and maintenance of the functional circulatory system and provide paracrine support to surrounding non-vascular cells. Technical advances in the past 5 years in single-cell genomics and in in vivo genetic labelling have facilitated greater insights into endothelial cell development, plasticity and heterogeneity. These advances have also contributed to a new understanding of the timing of endothelial cell subtype differentiation and its relationship to the cell cycle. Identification of novel tissue-specific gene expression patterns in endothelial cells has led to the discovery of crucial signalling pathways and new interactions with other cell types that have key roles in both tissue maintenance and disease pathology. In this Review, we describe the latest findings in vascular endothelial cell development and diversity, which are often supported by large-scale, single-cell studies, and discuss the implications of these findings for vascular medicine. In addition, we highlight how techniques such as single-cell multimodal omics, which have become increasingly sophisticated over the past 2 years, are being utilized to study normal vascular physiology as well as functional perturbations in disease.
Subject terms: Cardiovascular biology, Developmental biology, Angiogenesis, Organogenesis
Advances in single-cell RNA sequencing technologies in the past 5 years have led to a greater understanding of endothelial cell development and heterogeneity. In this Review, Red-Horse and Trimm discuss the most up-to-date research on vascular endothelial cell development and diversity, and highlight the latest findings on organ-specific endothelial cells in the heart, brain, lungs, kidneys and liver.
Key points
Artery differentiation is coupled to cell cycle arrest, and new arteries expand during development by recruiting endothelial cells from capillaries and veins.
Single-cell RNA sequencing (scRNA-seq) data and multimodal omics atlases provide insights into the heterogeneity of endothelial cells across tissues, as well as conserved transcription codes during development.
Novel methods of endothelial cell lineage tracing combined with scRNA-seq has facilitated the identification of functionally distinct capillary populations in the lung, liver and kidney, as well as their transcriptional response to disease.
Capillary zonation, or the gradual phenotypic continuum of endothelial cells along an axis, has been identified in multiple organs including the brain, heart and liver, and communication between endothelial cells and mural cells has a key role in maintaining capillary zonation and important structures such as the blood–brain barrier.
Comparison of scRNA-seq atlases from humans and mice reveals that endothelial cell populations are largely conserved between species across multiple organs and that although unique species-specific gene expression patterns exist for endothelial subtypes, the high degree of similarity between species demonstrates the utility of mice as a model organism for vascular biology.
Advances in bioengineering have led to the creation of organoids with increasingly functional vasculature; key insights from single-cell studies are expected to facilitate improved differentiation protocols for human pluripotent stem cells as well as the in vitro development of organ-specific vasculature.
Introduction
The development of functional vasculature is crucial for the health of every cell in the body. Endothelial cells, which line the inside of all blood and lymphatic vessels, have key roles in delivering oxygen and nutrients, regulating blood flow, modulating immune cell trafficking and maintaining tissue homeostasis. Vascular endothelial cell dysfunction is central to the progression of most chronic conditions, as well as ischaemic heart disease and stroke, the top two global causes of mortality1. However, many questions remain about the fundamental biology of endothelial cells; for example, how do endothelial cell progenitors differentiate into distinct subtypes; how do they achieve organotypic heterogeneity; and how is the function of specialized endothelial cells perturbed in disease? In the past 5 years, the advent of single-cell RNA sequencing (scRNA-seq) technologies, combined with increasingly sophisticated strategies for genetic lineage tracing, have allowed the field to address these questions2,3. In this Review, we highlight new discoveries that can be generalized to blood vessel development across all tissues, such as the role of cell cycling in the differentiation of endothelial cell subtypes as well as the phenomenon of ‘pre-artery’ specification. In addition, we discuss new findings for organ-specific endothelial cells in several key organs such as the heart, brain, lungs, kidneys and liver. Lymphatic endothelial cell development and heterogeneity have been reviewed elsewhere4 and are beyond the scope of this Review. While this Review is not intended to comprehensively cover all aspects of organ-specific vasculature, we aim to demonstrate the wealth of new insights into endothelial cell biology that can be gained from scRNA-seq and integrated multimodal omic atlases. Particularly groundbreaking discoveries include the identification of functionally distinct capillary subpopulations in the lung, kidney and liver, as well as capillary zonation in the brain and heart. The identification of novel endothelial cell subtypes and molecular markers, as well as cell–cell interactions, has implications for tissue engineering and the development of novel therapeutic strategies for cardiovascular disease.
Arterial and venous differentiation
During embryogenesis, blood vessels must first arise de novo in a process known as vasculogenesis. The differentiation of angioblasts (endothelial cell progenitors) and their transition to endothelial cells are tightly regulated by transcription factors and signalling pathways that have been extensively studied5–9. The first blood vessels arise from the rapid proliferation of endothelial cells, which subsequently form vascular tubes to create separate arteries and veins. Much research over the past two decades has described the crucial role of vascular endothelial growth factor (VEGF) and Notch signalling during arteriovenous differentiation. In the broader context, increased VEGF and Notch signalling supports arterial specification, whereas reduced activation of VEGF and COUP transcription factor 2 (COUP-TFII) (also known as NR2F2) is required for venous specification10–15. Although angiogenesis (the formation of new vessels from existing vessels) is mechanistically distinct from vasculogenesis, angiogenesis is also regulated by VEGF and Notch signalling. VEGF activates angiogenesis, whereas Notch restricts it13,16–27. Despite the decades-old knowledge that the VEGF and Notch signalling pathways are required for arterial differentiation, evidence indicating that these pathways directly regulate the expression of genes associated with arterial specification is scarce. However, studies from the past 5 years have begun to elucidate the underlying mechanisms involved in how these pathways alter the fate of endothelial cells.
Cell cycling and endothelial cell fate
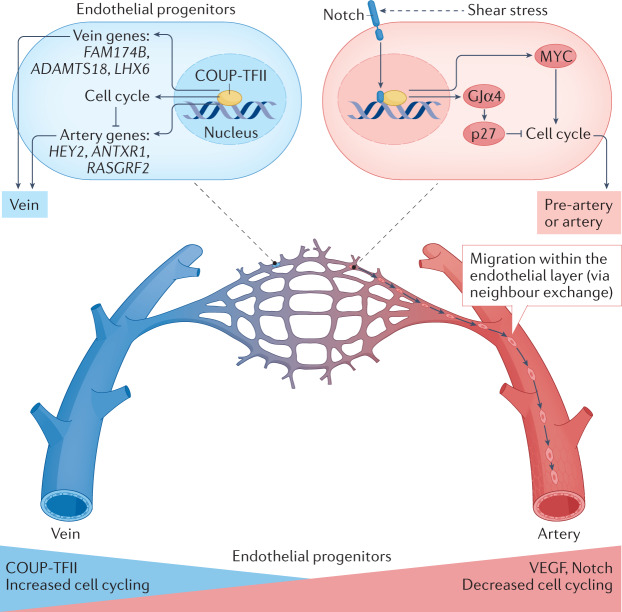
Insights from scRNA-seq and mosaic analyses have spurred the advances in our understanding of how Notch and COUP-TFII signalling support and restrict arterial differentiation, respectively28–31. Surprisingly, the mechanism seems to involve the effects of Notch and COUP-TFII signalling on cell cycling and metabolism, rather than the direct regulation of arterial and venous cell fate determinants. The first study describing the relationship between cell cycling and arterial differentiation used the retina as a model system28. During blood vessel formation, initiation of blood flow resulted in shear stress-induced Notch signalling and subsequent upregulation of gap junction α4 protein (also known as connexin 37). Gap junction α4 protein signalling led to flow-induced endothelial cell cycle arrest via p27 expression, enabling the expression of arterial specification genes28. A similar relationship between arterial differentiation and cell cycling was subsequently described in the developing heart, although in the heart, arterial differentiation was initiated in single endothelial cells within the immature vascular plexus before the initiation of blood flow29. These individual pre-artery endothelial cells were lineage-traced to mature coronary arteries, establishing these endothelial cells as pre-specified progenitors. Pre-arterial differentiation was augmented by Notch activation, but blocked by COUP-TFII overexpression, owing to the transcriptional activation of cell cycle genes29,30.
Numerous epigenetic studies support the link between cell cycle genes and the inhibitory effects of COUP-TFII on arterial differentiation. In a genome-wide analysis comparing COUP-TFII DNA binding with artery-specific and vein-specific transcriptional enhancers, COUP-TFII was over-represented at vein-specific enhancers, the majority of which were near actively expressed cell cycle genes32. The association between proliferating endothelial cells and capillary clusters with enriched expression of genes characteristic of venous endothelial cells had previously been identified in the brain as well as in regenerating aortas, adding further evidence supporting the idea that actively cycling cells are skewed towards a venous fate33,34. Investigators are also beginning to identify the molecular mechanisms underlying the role of the cell cycle in arterial endothelial cell differentiation31. Genomic analyses of mice expressing the fluorescent ubiquitination cell cycle indicator reporter specifically in endothelial cells showed that endothelial cells in the late G1 state are more sensitive to transforming growth factor-β1 (TGFβ1)-stimulated arterial specification-associated gene transcription than those in the early G1 state31. These newer studies correlating arterial endothelial differentiation with cell cycle exit contribute to our mechanistic understanding of arterial differentiation during development and form an important embryological parallel to older observations that mature arterial endothelial cells rarely proliferate in adults20,26.
Pre-specification of arterial endothelial cells has also been described in other vascular beds such as the zebrafish trunk and the mouse developing retina in cell populations with low proliferative rates and high Notch activity35–39. Live imaging in zebrafish revealed activation of Notch in tip cells, which upregulated cxcr4a (encoding the chemokine C-X-C motif receptor 4a) expression in endothelial cells, enabling them to connect with and join arterial vessels36. In the mouse retina, a subset of cells within the tip cell subpopulation similarly receives higher Notch signalling, which facilitates arterial differentiation (as indicated by the expression of Cxcr4) and migration of cells to form arteries35,40. Pre-specification at the tip cell location is supported by data from scRNA-seq analyses showing that tip cells and arterial endothelial cells are closely related at the transcriptional level33. In addition to Notch, increased VEGF–ERK signalling in both the retinal tip cells and coronary vessel plexus is another upstream trigger of cell cycle exit and arterial differentiation35,41,42. VEGF is normally expressed where arterial pre-specification occurs, notably the retinal tip cell region and the intramyocardium41,42. A mosaic analysis that involved genetic alteration of individual cells to express high levels of either the VEGF receptor 2 or Notch showed that both stimuli caused endothelial cells to exit the cell cycle and increased their propensity to colonize arteries30,43. To further investigate the downstream effects of Notch signalling on endothelial cell differentiation, an RNA-seq analysis was performed in mice with normal, diminished or increased Notch signalling30. Notch-induced arterial endothelial cell differentiation was not dependent on upregulation of genes that determine arterial fate, but instead was dependent on the suppression of MYC-induced transcription of cell cycle and/or metabolism genes.
Arterial expansion via cell recruitment
The findings discussed above support a model in which arterial differentiation is coupled to cell cycle arrest (Fig. 1), which presents an intriguing question: how do arteries expand in size during development if their endothelial cell population does not proliferate? Numerous lineage-tracing studies from the past 5 years have shown that arteries grow by recruiting cells from veins and capillaries, rather than by expansion via endothelial cell division26,34,39,44,45. In the mouse heart, timed labelling of Apj-expressing capillary plexus cells resulted in progressive pre-arterial specification and accumulation of endothelial cells in arteries during permissive developmental and postnatal time windows29,46. Three studies have assessed lineage-labelled arteries (BMXCreER), capillaries (Mfsd2aCreER) and veins (Gm5127(BAC)CreER) during postnatal retinal vasculature development in mice and showed that endothelial cells migrate in a vein-to-capillary-to-artery direction as the retinal vascular network expands and matures, but not in the opposite direction39,44,47. Interestingly, these migration patterns are restricted to development and not seen in the mature eye. Furthermore, another study combined scRNA-seq analysis with endothelial cell lineage tracing and showed that endothelial cell migration from the vein to the artery is a common theme during development45. The trajectory analysis performed on scRNA-seq data from mice at embryonic days 8–11 and matched human data predicted differentiation from vein to artery at multiple sites in the embryo in this early period45. Computational predictions were validated by overlaying scRNA-seq data from lineage-labelled mouse vein endothelial cells (Nr2f2CreER)45. This propensity of endothelial cells to migrate into arteries might also explain why endocardium-derived vessels preferentially populate arterial segments in the outer wall of postnatal hearts48.
Fig. 1. Artery and vein endothelial cell differentiation in a developing vascular bed.
Artery differentiation is coupled to cell cycle arrest. Shear stress activation of Notch signalling upregulates the levels of gap junction α4 protein (GJα4), which subsequently inhibits the expression of cell cycle genes via p27 expression. Notch signalling also inhibits MYC-induced transcription of genes related to cell cycle and metabolism. Together, Notch signalling indirectly supports arterial cell fate via cell cycle inhibition. Pre-artery endothelial cells then migrate against the direction of blood flow in the vein-to-artery direction. Adjacent endothelial cells exchange positions in a process known as ‘neighbour exchange’ to expand arteries. By contrast, COUP transcription factor 2 (COUP-TFII) (also known as NR2F2) activates the expression of cell cycle genes, inhibits the expression of certain genes associated with arterial specification and induces vein-specific gene expression. VEGF, vascular endothelial growth factor.
Endothelial cells migrate against blood flow
The physical forces of blood flow drive endothelial cell migration towards arteries because endothelial cells are programmed to move against the direction of flow26,49. Many questions remain regarding how endothelial cells are guided to perform this task, but several mechanisms have been proposed, including involvement of the bone morphogenetic protein (BMP) signalling pathway. Inhibiting the BMP receptor signalling components activin receptor type 1 (ALK1), endoglin or SMAD4 blocks blood flow-guided migration of endothelial cells in many settings50–53. Loss of these components and subsequent loss of proper endothelial cell migration leads to arteriovenous malformations that form direct shunts between arteries and veins, bypassing capillaries39,44,54,55. Zebrafish intersomitic vessel development is balanced by vein endothelial cell proliferation and polarization or migration against flow into arterial segments38,56–58, a process that requires the actin cytoskeletal regulator Wasb (the homologue of human Wiskott–Aldrich syndrome protein)58 and flow-activated Notch38,57.
Overactivation of endothelial cell migration against blood flow and into arteries has been achieved in mice by overexpressing Eng (encoding endoglin) or Dach1 (encoding dachshund homologue 1)51,59,60, the latter of which results in increased numbers of artery branches. Interestingly, loss of endothelial cell-derived non-canonical WNT5A and WNT11 caused endothelial cells to become over-responsive to flow, increasing their polarization and migration against flow61. In the plexus, this augmented polarization and migration against flow increased pruning (the regression of a subset of microvessels within a growing vasculature), although the effect on arterial branching was not investigated62. Further investigation into the mechanisms that drive endothelial cell migration towards arteries for arterial expansion might provide the foundation for therapeutic applications in ischaemic diseases60.
Compared with arterial endothelial cells, less is known about the development and maintenance of endothelial cells in veins and capillaries. Veins have transcriptional profiles that are distinct from those of arteries and the signals involved in inducing the venous cell state have mostly been studied in the early embryo6–8,11. The establishment of veins requires cell autonomous endothelial cell phosphoinositide 3-kinase signalling and expression of COUP-TFII6. Venous gene expression has been shown to be induced directly by BMP–BMP receptor type 1A (BMPR1A)–SMAD signalling in both mice and fish, ultimately leading to the formation of the first veins of the embryo63. This discovery was made through the identification of a vein-specific enhancer in Ephb4 (encoding ephrin type B receptor 4) and SMAD1 and SMAD5 binding sites as regulators of venous gene expression. Further experiments showed that Bmp4 is expressed preferentially in cells around early veins that express Bmpr1a64. Smad4 or Bmpr1a deletion in mouse endothelial cells abolished the expression of Ephb4 and Nr2f2 (which encodes COUP-TFII), repressing the development of the veins64. Less is known about the induction of capillary fates; however, emerging evidence indicates that capillary endothelial cells are less of an established cell state than artery and vein endothelial cells, and might instead exist along a continuum, known as zonation29,45,65–69. This zonation has been observed in capillaries in the brain and heart of both mice and humans.
scRNA-seq and multimodal omics
Endothelial cells have heterogeneous phenotypes that match the diverse structures and metabolic needs of each tissue and organ. For example, capillary endothelial cells of the brain form the blood–brain barrier (BBB) with tight intercellular junctions to restrict transport, whereas glomerular capillary endothelial cells in the kidney are highly fenestrated to support filtration of blood to Bowman’s space70. The anatomical heterogeneity of blood vessels has been well characterized and extensively reviewed70. Technical innovations in genomics in the past decade have provided insights into the mechanisms underlying this molecular heterogeneity.
Despite substantial functional differences across organ systems, all endothelial cells share the same genome. Heterogeneity must, therefore, stem from how and when transcriptional networks are activated, as well as from epigenetic modifications. Transcriptional profiling of endothelial cells, especially at the single-cell level through scRNA-seq, has greatly informed our understanding of endothelial cell heterogeneity. The pace of technical innovations for single-cell level genomic measurements is staggering, such that it is now possible to profile both gene expression and chromatin accessibility in each cell with the use of ‘multiome’ platforms71,72. In this section, we discuss studies that have incorporated these techniques to create atlases for individual tissues or entire organisms29,33,65–69,73–101 (Table 1).
Table 1.
Multi-organ and organ-specific endothelial cell scRNA-seq and multiome atlases
| Organ system | Species | Developmental stage | Dataset contents | Single-cell capture and scRNA-seq method | Refs. |
|---|---|---|---|---|---|
| Multi-organ | Mouse | Adult | scRNA-seq | Smart-seq2 and 10× Genomics 3′ | 95 |
| scRNA-seq | Microwell-seq | 73 | |||
| scRNA-seq | Smart-seq2 | 74 | |||
| scRNA-seq, RNA-seq, ATAC-seq, MethylC-seq | 10× Genomics GemCode | 33 | |||
| sci-ATAC-seq | sci-ATAC-seq | 75 | |||
| Human | Embryonic (E72–E129) | sci-RNA-seq, sci-ATAC-seq | sci-RNA-seq3 | 76,92 | |
| Adult (22–74 years) | scRNA-seq | Smart-seq2 and 10× Genomics 3′ | 93 | ||
| Heart | Mouse | Embryonic (E12.5–E14.5) | scRNA-seq | 10× Genomics 3′ | 29 |
| Mouse, human | Embryonic mouse (E12, E17.5), embryonic human (11 weeks, 14 weeks, 22 weeks), adult mouse | scRNA-seq | 10× Genomics 3′ | 66 | |
| Human | Embryonic (5–25 weeks) | scRNA-seq | STRT-seq | 77 | |
| Adult (40–75 years) | scRNA-seq | 10× Genomics 3′ | 78 | ||
| Lung | Mouse | Embryonic (E12.5–E17.5), postnatal (P3–P42) | scRNA-seq, scATAC-seq | 10× Genomics 3′ | 79 |
| Postnatal (P1) | scRNA-seq | Drop-seq | 80 | ||
| Postnatal (P3, P7, P14) | scRNA-seq | MULTI-seq | 81 | ||
| Human | Adult | scRNA-seq | Smart-seq2 and 10× Genomics 3′ | 82 | |
| Adult | scRNA-seq | 10× Genomics 3′ | 83 | ||
| Brain | Mouse | Embryonic (E14.5) | RNA-seq (TRAP-seq), scRNA-seq | TruSeq v2 | 84 |
| Adult | scRNA-seq | Smart-seq2 | 65 | ||
| Mouse, human | Adult mouse, paediatric and adult human | snRNA-seq | 10× Genomics 3′ | 67 | |
| Human | Fetal, adult | scRNA-seq | 10× Genomics 3′ | 69 | |
| Adult | scRNA-seq, RNA-seq | 10× Genomics 3′ | 68 | ||
| Kidney | Mouse | Embryonic | scRNA-seq | 10× Genomics 3′ | 85 |
| Embryonic E14.5 | scRNA-seq | 10× Genomics 3′ | 87 | ||
| Embryonic E14.5 | scRNA-seq | Drop-seq, Chromium 10× Genomics and Fluidigm C1 | 86 | ||
| Embryonic, adult | snRNA-seq, sn-ATAC-seq | Smart-seq2 and 10× Genomics 3′ | 88 | ||
| Human | Embryonic, paediatric, adult | scRNA-seq | 10× Genomics 3′ | 89 | |
| Embryonic (12–18 weeks) | scRNA-seq | Drop-seq | 90 | ||
| Embryonic (9–19 weeks) | scRNA-seq | 10× Genomics 3′ | 91 | ||
| Adult | snRNA-seq, sn-ATAC-seq | 10× Genomics 3′ v2 | 96 | ||
| Liver | Mouse | Embryonic (E12–E18), postnatal (P2–P30) | scRNA-seq, bulk RNA-seq | 10× Genomics 3′ | 97 |
| Adult | scRNA-seq | MARS-seq | 98 | ||
| scRNA-seq | 10× Genomics 3′ | 99 | |||
| Human | Adult | scRNA-seq | CEL-seq2 | 100 | |
| scRNA-seq | 10× Genomics 3′ | 101 |
ATAC-seq, assay for transposase-accessible chromatin using sequencing; CEL-seq2, cell expression by linear amplification and sequencing; Drop-seq, droplet sequencing; E, embryonic day; MARS-seq, massively parallel RNA single-cell sequencing; MULTI-seq, multiplexing using lipid-tagged indices for single-cell and single-nucleus RNA sequencing; P, postnatal day; scRNA-seq, single-cell RNA sequencing; snRNA-seq, single-nucleus RNA sequencing; sci-ATAC-seq, single-cell combinatorial indexing assay for transposase-accessible chromatin using sequencing; sci-RNA-seq, single-cell combinatorial indexing RNA sequencing; Smart-seq2, switching mechanism at the 5′ end of the RNA template sequencing; sn-ATAC-seq, single-nucleus combinatorial indexing assay for transposase-accessible chromatin using sequencing; STRT-seq, single-cell tagged reverse transcription sequencing; TRAP-seq, targeted purification of polysomal mRNA.
Global insights from multi-organ studies
Bulk RNA-seq provided the first transcriptomic insights into the molecular heterogeneity of endothelial cells among different tissues, facilitating the identification of several tissue-specific endothelial markers for the heart, muscle, brain, liver and lung33,102. ScRNA-seq corroborates the finding that endothelial cells from different tissues have specialized transcription programmes, which can be observed when endothelial cells in large multi-tissue datasets typically computationally separate (or cluster) into organ-specific subtypes2,103,104. A detailed summary of scRNA-seq techniques that are commonly used in cardiovascular research is available elsewhere105. Interestingly, organotypic endothelial cell specialization in mammals might be more exaggerated in blood endothelial cells because lymphatic endothelial cells cluster together regardless of the tissue of origin2. Single-cell analyses also facilitate the characterization of heterogeneity according to endothelial cell subtype (that is, artery, arteriole, capillary, venule and vein) (Tables 2,3). Endothelial cell heterogeneity across tissues is largely attributable to differential transcription in capillary endothelial cells; the endothelial cells in arteries and veins from different organs are more similar at the transcriptional level104,106. Accordingly, ageing in the brain also affects capillary endothelial cells to a greater extent than arterial or venous endothelial cells107,108. The level of resolution afforded by single-cell analyses also establishes that organs with highly specialized vasculature, such as the brain, liver, lungs and kidneys, have more distinct transcriptomic profiles, whereas the gene expression of heart, skeletal muscle, diaphragm, mammary gland and adipose tissue tends to overlap93,95. In addition, whereas endothelial cells from different organs share several major developmental pathways, including those relating to WNT signalling, cytokine regulation and metabolism, the expression patterns of the majority of genes associated with these pathways are unique to each organ2,104.
Table 2.
Representative selection of marker genes for mouse organ-specific vascular subtypes
| Organ system | Developmental stage | Vascular EC subtype | Marker genes | Refs. |
|---|---|---|---|---|
| Multi-organ | Embryonic or adult | Pan-endothelial | Pecam1, Kdr, Cldn5, Emcn, Cdh5, Tie1, Egfl7 | 2,80,85,131 |
| Vein | Apj, Nr2f2, Vwf | 29,65,66 | ||
| Artery | Gja4, Mecom | 29,66 | ||
| Heart | Embryonic | Artery | Gja4, Gja5 | 66 |
| Pre-artery or arteriole | Gja4, Gja5, Unc5b, Hey1, Slc45a4 | 29,66 | ||
| Vein | Nr2f2, Apj, Nr2f2 | 29,66 | ||
| Capillary | Aplnr, Car4 | 66 | ||
| Adult | Tissue-specific EC | Wt1, Slc28a2, Eepd1, Kcna5, Car8, Fbln1, Meox2, Rftn1, Lamb1, Myadm | 2 | |
| Artery | Apln, Dll4, Notch1, Mecom, Igfbp3, Cxcr4, Cx40 | 29 | ||
| Lung | Adult | Artery | Mgp, Cdh13, Htra1, Bmx, Fbln2, Sulf1 | 131 |
| Vein | Slc6a2, Bst1, Car8, Amigo2, Mutn1, Csrp2 | 131 | ||
| Tissue-specific EC | Grtp1, Adrb1, Scn7a, Tmem100, Hpgd, Foxf1a, Nckap5, Rasgef1a, Fendrr, Prx | 2 | ||
| Capillary (aCap) | Car4, Ednrb, Fibin, Tbx2, Rprml, Chst1, Apln | 81,131 | ||
| Capillary (gCap) | Aplnr, Gpihbp1, Plvap, Cd93, Ptprb, Cemip2, Tek, Cxcl12 | 81,131 | ||
| Brain | Embryonic | Tissue-specific EC | Foxf2, Foxl2, Foxq1, Lef1, Ppard, Zfp551, Zic3 | 84 |
| Adult | Tissue-specific EC | Slco1c1, Slco1a4, Slc22a8, Mfsd2a, Slc38a3, Spock2, Foxf2, Edn3, Stra6, Slc38ak | 2 | |
| Artery | Gkn3, Sema3g, Efnb2 | 65 | ||
| Venule | Slc38a5 | 65 | ||
| Capillary or vein | Slc16a1 | 65 | ||
| Kidney | Adult | Tissue-specific EC | Egfl7, Dram1, Dkk2, Esm1, Igfbp5, Pbx1, Boc, Igfbp3, Irx3, Tnfaip2, Ptpru | 87,88 |
| Glomerular EC | Pi16, Plat, Ehd3, Cyb4b1, Tspan7, Lpl | 143 | ||
| Cortical EC | Igfbp3, Plvap, Npr3 | 143 | ||
| Medullary EC | Igf1, Cryab, Igfbp7, Cd36, Aqp1, Ifi27l2a | 143 | ||
| Liver | Embryonic or adult | Portal vein | Ly6a, Cd34, Cd9, Ephb2, Gja5, Sox17 | 97 |
| LSEC | Mrc1, Fcgr2b, Stab1, Stab2 | 97 | ||
| Adult | Tissue-specific EC | c-Maf, Clec4g, Fcgr2b, Stab2, Oit3, Bmp2, Aass, Mrc1, Plxnc1, Wnt2 | 2,136 | |
| LSEC zone 1 | Dll4, Efnb2, Ltbp4, Msr1, Ntn4 | 98,99 | ||
| LSEC zone 2 or 3 | Rspo3, Wnt2, Wnt9b | 98,99 |
aCap, aerocyte capillary; EC, endothelial cell; gCap, general capillary; LSEC, liver sinusoidal endothelial cell.
Table 3.
Representative selection of marker genes for human organ-specific vascular subtypes
| Organ system | Developmental stage | Vascular EC subtype | Marker genes | Refs. |
|---|---|---|---|---|
| Multi-organ | Embryonic or adult | Pan-endothelial | PECAM1, CDH5, VWF, KDR, FLT1, TEK, CLDN5 | 68,78,82,91,93 |
| Artery | GJA4, GJA5, HEY1, GATA2, CXCR4, SOX17, MECOM | 66,67,93 | ||
| Vein | ACKR1, NR2F2, PLVAP | 68,69,78 | ||
| Heart | Embryonic | Artery | GABBR2, GRIA2, SSUH2, JAG1 | 66 |
| Vein | ACKR1, LHX6, SELE | 66 | ||
| Capillary | PRDM1, INMT, APLNR, CA4 | 66 | ||
| Adult | Tissue-specific EC | SLC14A1 | 93 | |
| Artery | SEMA3G, EFNB2, DLL4, | 78 | ||
| Vein | NR2F231,ACKR1 | 78 | ||
| Capillary | RGCC, CA4 | 78 | ||
| Lung | Adult | Tissue-specific EC | VIPR1 | 93 |
| Artery | DKK2, SERPINE2 | 131 | ||
| Vein | CPE, PTGDS, C7, PLA1A | 131 | ||
| Bronchial EC | VWA1, HSPG2, PLVAP, MYC, HBEGF | 82 | ||
| Capillary (aCap) | ADIRF, S100A4, EMCN, HPGD, EDNRB, SOSTDC1, IL1RL1, APLN | 131 | ||
| Capillary (gCap) | FCN3, EDN1, SLC6A4, GPIHBP1, CD36, IL7R, VWF, PTPRB, PLVAP | 131 | ||
| Brain | Adult | Large artery | LTBP4 | 69 |
| Artery | INTS6, HSPA1A, JUNB, MECOM, ARL15, TXNIP, MGP, ADAMTS1 | 68,69 | ||
| Arteriole | VEGFC, ARL15, BMX, EFNB2, WISR, AIF1L, CD320 | 67 | ||
| Venule | TSHZ2, ADGRG6, SLC38A5, LRRC1, BNC2, ETV6, TMEM132C, ATP10A, JAM2, PRCP, PRSS23, RAMP3 | 67–69 | ||
| Vein | ACKR1, IL1R1, TSHZ2, PTGDS, POSTN, DNASE1 | 68,69 | ||
| Large vein | CCL2 | 69 | ||
| Capillary | MFSD2A, SLC7A5, TFRC, SLC38A5, SRARP, RGCC, SLC3A2, BSG, SLC16A1, SLCO1A2 | 67–69 | ||
| Kidney | Embryonic | Tissue-specific EC | NOTCH4 | 90 |
| Paediatric or adult | Glomerular EC | SEMA3G, CLDN5 | 89 | |
| Liver | Adult | Tissue-specific EC | OIT3 | 93 |
| Portal vein | AQP1, CD9, IFITM1, RAMP3, INMT, DNASE1L3, LIFR | 97,101 | ||
| LSEC | CLEC4G, STAB1, STAB2, CD14 | 97 | ||
| LSEC zone 1 | MGP, SPARCL1, TM4SF1, CLEC14A, BTNL9, ANPEP | 100,101 | ||
| LSEC zone 2 or 3 | CCL14, CLEC1B, FCN2, S100A13, LYVE1, FCN3 | 100,101 | ||
| Central vein | SELP, SELE, VWF | 97 |
aCap, aerocyte capillary; EC, endothelial cell; gCap, general capillary; LSEC, liver sinusoidal endothelial cell.
Numerous studies have incorporated chromatin architecture information to gain further insights into the tissue-specific transcriptional regulation in endothelial cells, first by identifying DNA binding motifs in accessible chromatin regions and then comparing these motifs with the expression of genes encoding transcription factors to computationally predict endothelial transcription codes109–111. Some consistent observations have emerged from these studies. Transcription factor motifs from the ETS transcription factor family, including transcriptional regulator ERG and Friend leukaemia integration 1 transcription factor, are universally enriched in endothelial cells33,109. This finding is consistent with data showing that heterogeneous endothelial cell gene expression requires the combined function of both the endothelial cell subtype-specific transcription factors and ETS factors at enhancers110. The source of heterogeneity in gene expression between tissues might stem from differences in distal, rather than proximal, cis-regulatory elements. In a study comparing assay for transposase-accessible chromatin using sequencing (ATAC-seq) peaks in endothelial cells from the liver, lung, brain and kidney, the majority of ATAC-seq peaks at <2 kb from transcription start sites (proximal peaks) were shared between tissues, whereas the majority of ATAC-seq peaks at >2 kb from transcription start sites (distal peaks) were unique to individual tissues33. Predicted tissue-specific endothelial cell transcriptional networks include lymphoid enhancer-binding factor or T cell factor family members in the brain, which are well-known to be downstream effectors of canonical WNT signalling. GATA transcription factors seem to regulate heart-specific and liver-specific endothelial cell transcription, whereas homeobox transcription factor family members are enriched in the lungs and kidneys33,109.
Multi-organ datasets have provided support for earlier observations that surrounding cells induce tissue-specific behaviour in endothelial cells, whereas endothelial cells in turn affect the transcriptomic profiles of their neighbours. For example, subpopulations of cardiac endothelial cells express cardiac muscle-specific genes and sinusoidal endothelial cells have similar gene expression profiles to those of adjacent hepatocytes2,98,111. Numerous studies have begun to investigate the functional implications of these interactions across a number of organ systems. A study combining scRNA-seq analysis with bone marrow transplantation experiments characterized the crucial role of arterial endothelial cells in establishing haematopoietic stem cell colonization of the bone marrow112,113. Another study similarly used scRNA-seq to probe the complex response of endothelial cells to pericyte loss in the brain, a phenomenon seen in a wide range of neurological diseases114.
Finally, sexual dimorphisms are known to contribute to differences in organ function, susceptibility to disease and ageing, processes that involve blood vessels that permeate almost every tissue115. Transcriptomic and proteomic analyses have begun to establish that these differences, at least in the heart, are attributable to both hormonal control and sex chromosome composition115. Many organism-wide scRNA-seq analyses control for the possibility of sex-dependent differences in gene expression by utilizing tissues from a single sex, but only a few studies have assessed the effects of sex on organ-specific transcriptomic profiles2,106. Using the Tabula Muris dataset, sex was found to be an important factor underlying endothelial cell transcriptome heterogeneity, but only in some tissues, including the aorta, brain and lungs2,95. Future cell atlas studies should incorporate sex as a parameter to better understand its influence on vascular physiology.
Organ-specific findings at high resolution
Cardiac vasculature
The majority of coronary blood vessels develop via convergent differentiation of endothelial cells from two distinct progenitor sources: the inlet vein known as the sinus venosus and the endocardium that lines the heart chamber42,116–120. Cells from these two populations ultimately localize to complementary yet overlapping regions of the heart. The sinus venosus mostly forms vessels in the outer ventricular walls and the endocardium mostly forms vessels in the septum and inner ventricular wall. Despite this spatial divergence, endothelial cells from both progenitor sources begin to sprout between embryonic days 11 and 12.5 in mice116. The sinus venosus and endocardium initially sprout in response to distinct molecular signals, specifically epicardial VEGFC–apelin receptor early endogenous ligand signalling and SMAD signalling for sinus venosus-derived cells, and intramyocardial VEGFA for endocardium-derived cells64,117,118,120. After sprouting onto the developing heart, previously fully differentiated sinus venosus vein endothelial cells or endocardium will undergo extensive cell state changes within the coronary plexus and remodel into coronary arteries, capillaries and veins118.
After birth, neonatal cardiac ventricles undergo rapid growth and remodelling. The remaining trabecular myocardium, which lacks intramyocardial vessels during embryogenesis, undergoes compaction and acquires rich coronary vascularization121. Endocardium-derived vessels have been shown to expand into this region in the weeks after birth, and new vessels have been suggested to arise from postnatal lineage conversion of endocardial cells to coronary vessel endothelial cells42. Stage-specific lineage-tracing experiments have further refined this model by showing that late fetal and neonatal endocardial cells contribute minimally to the formation of postnatal coronary vessels, even with injury48,122,123. Differentiation of endocardial cells into coronary endothelial cells occurs mostly during early heart development (before embryonic day 13.5 in mice) and endocardium-derived vascular tunnels do not adopt a mature artery fate until postnatal day 7 in mice9,48. Instead of endocardial conversion, postnatal endothelial cells from coronary vessels in the inner myocardium express high levels of VEGFR3 and undergo angiogenic expansion in a delta-like protein 4–NOTCH 1-dependent manner to vascularize the newly compacted myocardium and extend the endocardium-derived vascular tunnels into mature arteries48,122. Several studies have found that this endocardium-directed coronary growth can be enhanced under certain conditions such as with VEGFB transduction or overexpression of Kdr (encoding VEGFR2) in adult mice124,125. Comparing scRNA-seq data of all cardiac endothelial cells from early (embryonic day 12.5) and late (embryonic day 17.5) stages revealed that Bmp2 expression is specific to the embryonic time period when angiogenesis occurs123. BMP2 expression could also reactivate endocardial angiogenesis in neonatal mouse models of myocardial infarction123.
Direct and deep comparisons of capillary endothelial cell transcriptional states from lineage-traced hearts indicated that adult coronary endothelial cells originating from distinct sources do not retain the molecular signatures of their progenitors66. This phenomenon of ‘convergent differentiation’ has also been seen in olfactory projection neurons126, but not in certain populations of haematopoietic cells or tissue-resident macrophages, which retain markers of primed fate potential127,128. Furthermore, no functional differences have been observed in endothelial cells from either lineage, such that they have largely identical proliferative capacity in response to injury66.
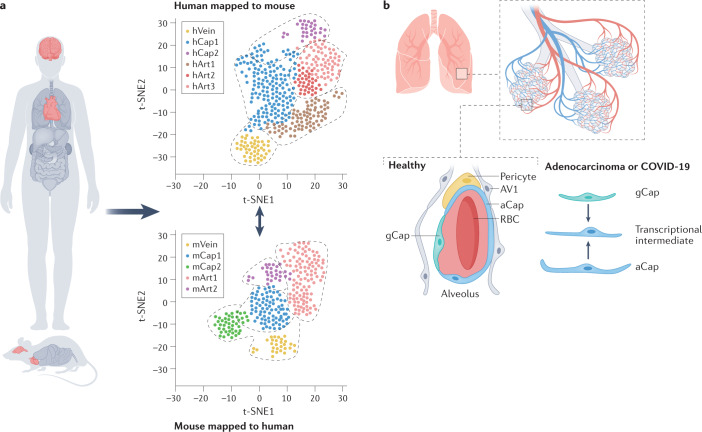
Techniques such as reference mapping, as well as comparisons of mouse and human scRNA-seq datasets through robust integration, provide information about the transcriptional similarities and differences between species. These analyses of developing mouse and human hearts revealed that the developmental environment and endothelial cell subtype transcriptional states are similar in both species66,129, validating the use of mice as a model for many aspects of human coronary development (Fig. 2a). Follow-up functional experiments used these data in an induced pluripotent stem cell culture model to determine that the histone–lysine N-methyltransferase MECOM is crucial for human artery endothelial cell differentiation129. Furthermore, trajectory analyses of human data predicted a transitory cell population from the endocardium129, which had a gene expression pattern validated using lineage tracing that overlapped with that of the transitory cell population in mice123. Therefore, despite the dearth of lineage-tracing techniques for human studies, overlapping human and mouse datasets provide some evidence that coronary vessels from humans also arise from the endocardium.
Fig. 2. scRNA-seq reveals novel endothelial cell types.
a | Comparison of human and mouse single-cell RNA sequencing (scRNA-seq) datasets for heart and brain vasculature reveals similar endothelial cell states between species. Although expression of unique genes exists for endothelial cell subtypes within each species, these analyses support the use of mouse models for studying human development and pathology. Key findings from these datasets reveal that the heart has conserved endothelial cell populations and similar capillary endothelial cell states in mice and humans and unique human artery specification genes. Furthermore, the brain also shows conserved endothelial cell populations and zonation between mice and humans and a greater diversity of human perivascular cells. b | Two transcriptionally distinct populations of capillary endothelial cells exist in the lung: general capillaries (gCap) and aerocyte capillaries (aCap). These two capillary populations have different roles in lung maturation and mature lung function. In disease states such as adenocarcinoma, a transcriptional intermediate between gCap and aCap arises in both humans and mice. This transcriptional intermediate has also been seen in the lungs of patients with coronavirus disease 2019 (COVID-19). Overlapping features of inflammation and cellular stress can also be seen in both capillary subtypes in advanced chronic obstructive pulmonary disease. AV1, alveolar type 1 cell; RBC, red blood cell; t-SNE, t-distributed stochastic neighbour embedding.
Lung vasculature
Alveoli, the terminal airspaces in the mammalian lung and a major site of gas exchange, are surrounded by a dense network of capillaries. Single-cell studies have identified heterogeneities in alveolar vasculature that are crucial for structural integrity. Alveolar development beginning during late embryogenesis is accompanied by the emergence of a specialized capillary subtype called CAR4+ aerocyte capillaries (aCaps), which intermingle with the ‘general capillaries’ (gCaps)130–132. CAR4 encodes carbonic anhydrase 4, which catalyses the reversible hydration of carbon dioxide into bicarbonate and protons. Although gCaps have similar gene markers to capillary endothelial cells from other organs, aCaps express markers such as ICAM1 (encoding intercellular adhesion molecule 1) and EDNRB (encoding endothelin receptor type B), and are unique to the lung. aCaps are also atypically large with a ‘Swiss cheese’-like morphology, spreading over the thin alveolar type 1 epithelium where gas exchange occurs130,131. This location and their gene expression patterns indicate that aCaps have specialized roles in gas exchange and leukocyte trafficking.
Trajectory analysis of scRNA-seq and lineage-tracing data revealed that gCaps are progenitors for aCaps during alveolar development79,130–132 and after injury in adults131,133. Development and maintenance of aCaps specifically require VEGF from airway epithelium, given that Vegf deletion results in depleted aCap numbers but does not affect gCap numbers. aCaps also seem to be required for lung maturation because lack of aCaps in mice results in enlarged alveoli despite the presence of myofibroblasts130.
Together, this deeper understanding of capillary heterogeneity has informed human pathology. As in the heart, intraspecies comparisons have revealed that all endothelial cell subtypes found in mice are present in humans, and that CAR4+ aCaps also emerge during late embryogenesis79,83,131. Analysis of a human lung adenocarcinoma sample revealed a loss of aCaps and gain of a transcriptionally intermediate population that co-expressed markers of aCaps and gCaps131. This same phenomenon was observed in single-cell atlases of lungs from patients who died of coronavirus disease 2019 (COVID-19)134 (Fig. 2b). Interestingly, endothelial cells were the second-most abundant site of viral RNA load in these patients, despite low-to-absent expression of the entry receptor for SARS-CoV-2 (ref.135), consistent with the knowledge that endothelial cells are a major site of pathology in patients with COVID-19 (ref.136). A comparison of hyperoxia-induced bronchopulmonary dysplasia in mice with matched human samples showed that aCaps mediate the increased expression of INHBA, which encodes inhibin-β A chain81, a member of the TGFβ superfamily that has been suggested to contribute to bronchopulmonary dysplasia. Furthermore, the study showed that capillary endothelial cell subtypes, rather than artery and vein endothelial cells, contain the most transcriptional changes that are potentially pathological in nature. Capillary endothelial cell subtypes also show the greatest change in inflammatory signalling and stress response in patients with advanced chronic obstructive pulmonary disease137. These and other similar findings demonstrate the power of single-cell level atlases to provide a more granular understanding of pulmonary disease.
Brain vasculature
Given the prominent role of the central nervous system blood vessels in human disease and ageing138, these vessels have been the subject of many single-cell transcriptomic studies in the past 5 years. Indeed, single-cell analyses of brain-specific capillaries led to the identification of a previously unappreciated feature of many capillary beds. Capillaries comprise the vast majority of blood vessels and are the main site of oxygen exchange, yet multi-organ studies have thus far failed to identify a unifying capillary molecular marker2. This lack of a unifying capillary marker might be attributable to the observation that instead of being a homogeneous entity, cells within a single capillary bed exist along an arterial–venous transcriptional continuum, referred to as zonation, which depicts gradual cellular phenotypic changes along an anatomical axis (Fig. 1). This feature of capillaries was first confirmed in the brain (via an approach that assessed and visualized data using distance matrices) and later in the heart (using artery or vein gene scores)29,65. Interestingly, zonation is not a phenotype shared by endothelial cell-adjacent mural cells in the central nervous system vasculature. Arteriole smooth muscle cells abruptly transition to become pericytes at the arteriole–capillary boundary in both mice and humans65,139.
Other studies have also identified capillary zonation in human brains, early mouse and human embryos and developing human hearts45,66–69 (Fig. 2a). Loss of communication between pericytes — a cell type reported to regulate the formation of the BBB140,141 — and endothelial cells was also found to skew capillary zonation towards a venous identity rather than causing widespread loss of BBB function and ectopic tip cells114.
Finally, single-cell analyses can also provide insights into the source of disease that results from genetic variants. Findings from the cross-referencing of cell subtype gene expression in mice with gene variants associated with Alzheimer disease or genes with altered expression in brain tissue from patients with Alzheimer disease suggest that capillary endothelial cells are a potential source of pathology, possibly through changes in BBB-related genes108. The same analysis using human data pinpointed endothelial cells and mural cells as potential sites of disease139.
Kidney vasculature
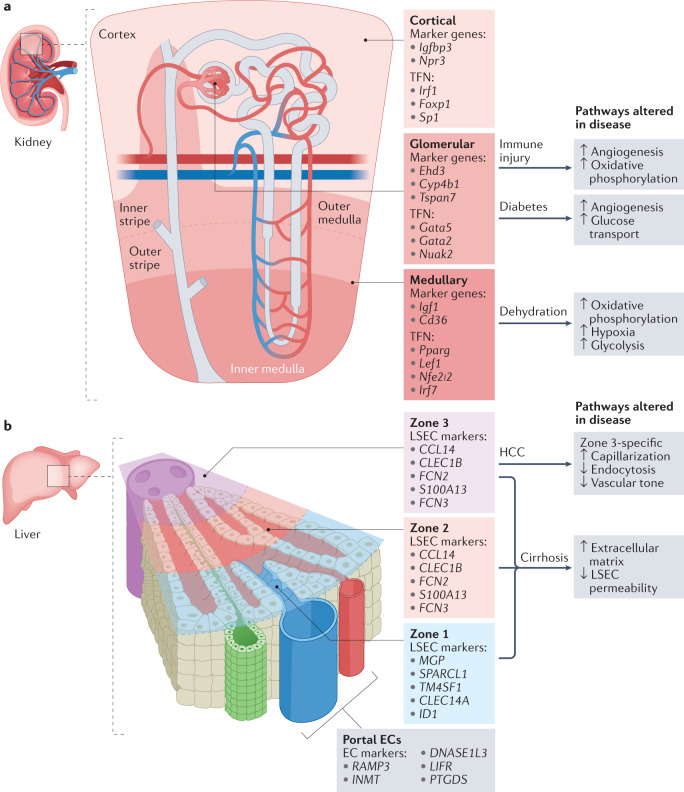
Given that the kidney has diverse and distinctive endothelial cell populations, the renal vasculature is a good candidate for single-cell analysis96,142. The renal microvasculature regulates blood flow, facilitates filtration, modulates inflammation and maintains physiological blood pressure. These functions are compartmentalized to specific regions: glomerular endothelial cells make up the filtration barrier, whereas tubular and peritubular capillary endothelial cells absorb and secrete fluids and other substances96,142. Single-cell analyses of mouse models of kidney disease have provided insights into the functional divisions between cortical, medullary and glomerular endothelial cells and identified cell type-specific responses to pathology. Cortical endothelial cells express high levels of Igfbp3 (encoding insulin-like growth factor binding protein 3) and Npr3 (encoding natriuretic peptide receptor 3) among other genes, whereas medullary endothelial cells express Igf1 (encoding insulin-like growth factor 1) and Cd36 (encoding platelet glycoprotein 4). Glomerular endothelial cell-enriched genes include Ehd3 (encoding EH domain-containing protein 3), Cyp4b1 (encoding cytochrome P450 4B1) and Tspan7 (encoding tetraspanin 7)143–146 (Fig. 3a). In response to acute water deprivation, medullary endothelial cells show the greatest transcriptional changes, upregulating the expression of genes related to oxidative phosphorylation in an effort to promote survival and concentrate urine143. A mouse model of nephritis (via nephrotoxic serum administration) showed that the disease involves an endothelial cell trajectory towards an activated cell state, whereas endothelial cell states in a mouse model of diabetes mellitus (ob/ob) overlapped completely with that of wild-type mice147,148.
Fig. 3. scRNA-seq reveals inter-organ heterogeneity and heterogeneous endothelial cell responses to disease.
a | Single-cell RNA sequencing (scRNA-seq) has revealed transcriptomic heterogeneity between endothelial cells in each compartment of the kidney. Cortical endothelial cells, glomerular endothelial cells and medullary endothelial cells have differential responses to injury. Marker genes and enriched transcriptional factor networks (TFN) for each subtype are indicated. Mouse models have been developed to assess kidney injury during disease states including immune-mediated injury (nephritis and systemic lupus erythematosus), diabetic kidney injury and dehydration. Key pathways that are altered in endothelial cells from the different renal compartments of mice are highlighted. b | Endothelial cell zonation in the liver has been characterized in both humans and mice. Broadly, liver sinusoid endothelial cells (LSECs) can be grouped into three zones based on their localization with respect to the portal vein and central vein. The top five differentially expressed genes for human LSECs are indicated as marker genes for each zone. Studies in mice have revealed that LSECs in zone 3 are the most susceptible to damage associated with cirrhosis. EC, endothelial cell; HCC, hepatocellular carcinoma.
Liver vasculature
Liver endothelial cells have long been known to have crucial roles in liver function, including supporting its dual blood supply from the hepatic artery and portal vein, scavenging macromolecules and regulating the host immune response to pathogens149. ScRNA-seq has provided insights into the transcriptome that support these diverse functions in both mice and humans, and new studies have demonstrated how liver sinusoidal endothelial cells become dysregulated in the context of cirrhosis and hepatocarcinoma99,100.
In a landmark study, investigators developed a strategy for spatially resolved scRNA-seq in the liver by mapping the positions of attached hepatocyte–endothelial cell pairs back to their expression-inferred tissue coordinates150. Molecular signatures for subpopulations of pericentral liver endothelial cells were identified and the zonation of endothelial cells across lobules was characterized98 (Fig. 3b). An analysis of the spatially resolved proteome revealed a high level of concordance with the transcriptomic data. Interestingly, although zonation in the brain corresponds to a continuum of arterial–venous transcriptional states, several studies have found that zonation in the liver relates to functional differences across the lobule axis98,101. Endothelial cell zonation was found to drive much of this functional zonation in hepatocytes via a TIE–WNT signalling axis150. The initial establishment of zonation in embryonic endothelial cell progenitors depends on MAF, a transcription factor crucial for sinusoid (or liver capillary) identity97.
As with capillaries in the kidney, liver sinusoid endothelial cells have zone-specific susceptibility to injury inflicted during disease, such as sinusoid endothelial cells in lobule zone 3 that are particularly affected in cirrhotic liver of mice99. Just as aCaps and gCaps in the lung lose their specialized transcriptional states in disease states, cirrhosis causes zone 3 sinusoid endothelial cells to lose their specialized fenestrae through a process known as capillarization99.
Advances in organoid vasculature
Recent developments in single-cell genomics have revolutionized our understanding of the heterogeneity of vascular cells and their exquisitely regulated development within tissue microenvironments. As well as facilitating the identification of rare and novel endothelial cell types, such as specialized capillary subtypes in the lungs, liver, kidneys and retina, scRNA-seq has offered a higher resolution window into the transcriptional changes that occur during disease47,98,131,143. Furthermore, scRNA-seq data have provided new insights into the interactions between endothelial cells and adjacent cell types, such as smooth muscle cells, pericytes and immune cells, and the roles that these interactions have in both the development and the maintenance of organ-specific vasculature. These findings, combined with results from sophisticated lineage-tracing studies, raise important considerations for tissue engineering. A lack of physiologically accurate perfusable vasculature is one of the most important challenges limiting the expansion of multicellular ‘mini-organs’ known as organoids151. Advances in differentiation protocols and culture techniques for human pluripotent stem cells (hPSCs)152 have resulted in the development of 3D self-organizing human blood vessel organoids, which are capable of forming complex vascular networks that recapitulate many of the mural cell–endothelial cell interactions known to be crucial for supporting structures such as the BBB153. These organoids, as well as several hPSC-derived liver154,155, kidney156 and brain157 organoids, have shown promising, though often partial, levels of host vascularization in vivo after transplantation into immunodeficient mice.
Although in vivo transplants are an exciting demonstration of the therapeutic potential of organoids, the generation of endogenous organ-specific vasculature remains crucial. By using scRNA-seq data, one group demonstrated that hPSC-derived intestinal organoids are capable of inducing the differentiation of endothelial cells with an intestinal-specific transcriptional signature158. Other efforts have focused on increasing the complexity and functionality of organ-specific vasculature by modulating key signalling pathways and exposing in vitro systems to physiologically relevant physical forces159. Through a combination of these methods, several groups have developed increasingly sophisticated cardiac organoids complete with internal lumens and vascular networks160–162. Other groups have shown that modulating morphogen gradients and fluid shear stress conditions using microfluidic systems improves the expansion of endothelial progenitors in the kidney163 and brain164, although further analysis is required to determine whether faithful endothelial cell subtype differentiation has been achieved with these methods.
Finally, 3D bioprinting offers an alternative approach to embedding perfusable vascular channels for larger regions of tissue. In a newly developed biomanufacturing method, multiple hPSC-derived organoids, embryoid bodies or spheroids can be combined into a granular tissue matrix. Sacrificial ink is injected and then removed to create channels within the tissue that can be seeded with endothelial cells165. This approach has resulted in the successful vascularization of a contractile cardiac organoid matrix165 and might be capable of producing more structurally accurate organ-specific vasculature than other methods.
Conclusions
In this Review, we discuss the current molecular understanding of the differentiation of endothelial progenitors into arteries and veins. New insights are used to form a model in which artery differentiation is coupled to cell cycle arrest, whereas vein differentiation is associated with cell proliferation and direct induction of the BMP–BMPR1A–SMAD pathway. Arteries then expand by recruiting cells from capillaries and veins, which migrate against the direction of blood flow to expand the arterial network. Numerous multi-organ scRNA-seq and single-cell multiomic atlases developed in the past 5 years have led to the discovery of novel endothelial cell-specific findings for several critical cardiovascular organs. Capillary endothelial cells are the primary source of vascular heterogeneity across organs, and findings from the past 5 years have shown that they are also the primary source of transcriptional changes in multiple disease states. A general trend observed in the liver, lungs and brain is that disease promotes a loss of specialized capillary function and a reversal to a more general transcriptional state. These findings have implications for the development of targeted therapies and for the differentiation of improved organ-specific vasculature for tissue-engineering applications.
Acknowledgements
E.T. is supported by the Fannie and John Hertz Foundation.
Author contributions
The authors contributed substantially to all aspects of the article.
Peer review
Peer review information
Nature Reviews Cardiology thanks Bin Zhou and the other, anonymous, reviewer(s) for their contribution to the peer review of this work.
Competing interests
The authors declare no competing interests.
Footnotes
Publisher’s note
Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Related links
Tabula Muris: https://tabula-muris.ds.czbiohub.org/
References
- 1.World Health Organization. The Top 10 Causes of Death (WHO, 2020).
- 2.Paik DT, et al. Single-cell RNA sequencing unveils unique transcriptomic signatures of organ-specific endothelial cells. Circulation. 2020;142:1848–1862. doi: 10.1161/CIRCULATIONAHA.119.041433. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Elmentaite R, Domínguez Conde C, Yang L, Teichmann SA. Single-cell atlases: shared and tissue-specific cell types across human organs. Nat. Rev. Genet. 2022 doi: 10.1038/s41576-022-00449-w. [DOI] [PubMed] [Google Scholar]
- 4.den Braanker H, van Stigt AC, Kok MR, Lubberts E, Bisoendial RJ. Single-cell RNA sequencing reveals heterogeneity and functional diversity of lymphatic endothelial cells. Int. J. Mol. Sci. 2021 doi: 10.3390/ijms222111976. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Corada M, Morini MF, Dejana E. Signaling pathways in the specification of arteries and veins. Arterioscler. Thromb. Vasc. Biol. 2014;34:2372–2377. doi: 10.1161/ATVBAHA.114.303218. [DOI] [PubMed] [Google Scholar]
- 6.Fish JE, Wythe JD. The molecular regulation of arteriovenous specification and maintenance. Dev. Dyn. 2015;244:391–409. doi: 10.1002/dvdy.24252. [DOI] [PubMed] [Google Scholar]
- 7.Dejana E, Hirschi KK, Simons M. The molecular basis of endothelial cell plasticity. Nat. Commun. 2017;8:14361. doi: 10.1038/ncomms14361. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Qiu J, Hirschi KK. Endothelial cell development and its application to regenerative medicine. Circ. Res. 2019;125:489–501. doi: 10.1161/CIRCRESAHA.119.311405. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Apte RS, Chen DS, Ferrara N. VEGF in signaling and disease: beyond discovery and development. Cell. 2019;176:1248–1264. doi: 10.1016/j.cell.2019.01.021. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Phng L-K, Gerhardt H. Angiogenesis: a team effort coordinated by notch. Dev. Cell. 2009;16:196–208. doi: 10.1016/j.devcel.2009.01.015. [DOI] [PubMed] [Google Scholar]
- 11.You L-R, et al. Suppression of Notch signalling by the COUP-TFII transcription factor regulates vein identity. Nature. 2005;435:98–104. doi: 10.1038/nature03511. [DOI] [PubMed] [Google Scholar]
- 12.Lawson ND, Vogel AM, Weinstein BM. sonic hedgehog and vascular endothelial growth factor act upstream of the Notch pathway during arterial endothelial differentiation. Dev. Cell. 2002;3:127–136. doi: 10.1016/S1534-5807(02)00198-3. [DOI] [PubMed] [Google Scholar]
- 13.Benedito R, Hellström M. Notch as a hub for signaling in angiogenesis. Exp. Cell Res. 2013;319:1281–1288. doi: 10.1016/j.yexcr.2013.01.010. [DOI] [PubMed] [Google Scholar]
- 14.Kohli V, et al. Arterial and venous progenitors of the major axial vessels originate at distinct locations. Dev. Cell. 2013;25:196–206. doi: 10.1016/j.devcel.2013.03.017. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Casie Chetty S, et al. Vegf signaling promotes vascular endothelial differentiation by modulating etv2 expression. Dev. Biol. 2017;424:147–161. doi: 10.1016/j.ydbio.2017.03.005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Zygmunt T, et al. In parallel interconnectivity of the dorsal longitudinal anastomotic vessels requires both VEGF signaling and circulatory flow. J. Cell Sci. 2012;125:5159–5167. doi: 10.1242/jcs.108555. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Bahary N, et al. Duplicate VegfA genes and orthologues of the KDR receptor tyrosine kinase family mediate vascular development in the zebrafish. Blood. 2007;110:3627–3636. doi: 10.1182/blood-2006-04-016378. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Covassin LD, Villefranc JA, Kacergis MC, Weinstein BM, Lawson ND. Distinct genetic interactions between multiple Vegf receptors are required for development of different blood vessel types in zebrafish. Proc. Natl Acad. Sci. USA. 2006;103:6554–6559. doi: 10.1073/pnas.0506886103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Bussmann J, Wolfe SA, Siekmann AF. Arterial-venous network formation during brain vascularization involves hemodynamic regulation of chemokine signaling. Development. 2011;138:1717–1726. doi: 10.1242/dev.059881. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Ehling M, Adams S, Benedito R, Adams RH. Notch controls retinal blood vessel maturation and quiescence. Development. 2013;140:3051–3061. doi: 10.1242/dev.093351. [DOI] [PubMed] [Google Scholar]
- 21.Hellström M, et al. Dll4 signalling through Notch1 regulates formation of tip cells during angiogenesis. Nature. 2007;445:776–780. doi: 10.1038/nature05571. [DOI] [PubMed] [Google Scholar]
- 22.Leslie JD, et al. Endothelial signalling by the Notch ligand delta-like 4 restricts angiogenesis. Development. 2007;134:839–844. doi: 10.1242/dev.003244. [DOI] [PubMed] [Google Scholar]
- 23.Lobov IB, et al. Delta-like ligand 4 (Dll4) is induced by VEGF as a negative regulator of angiogenic sprouting. Proc. Natl Acad. Sci. USA. 2007;104:3219–3224. doi: 10.1073/pnas.0611206104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Noguera-Troise I, et al. Blockade of Dll4 inhibits tumour growth by promoting non-productive angiogenesis. Nature. 2006;444:1032–1037. doi: 10.1038/nature05355. [DOI] [PubMed] [Google Scholar]
- 25.Suchting S, et al. The Notch ligand delta-like 4 negatively regulates endothelial tip cell formation and vessel branching. Proc. Natl Acad. Sci. USA. 2007;104:3225–3230. doi: 10.1073/pnas.0611177104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Red-Horse K, Siekmann AF. Veins and arteries build hierarchical branching patterns differently: bottom-up versus top-down. BioEssays. 2019;41:1800198. doi: 10.1002/bies.201800198. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Siekmann AF, Covassin L, Lawson ND. Modulation of VEGF signalling output by the Notch pathway. BioEssays. 2008;30:303–313. doi: 10.1002/bies.20736. [DOI] [PubMed] [Google Scholar]
- 28.Fang JS, et al. Shear-induced Notch-Cx37-p27 axis arrests endothelial cell cycle to enable arterial specification. Nat. Commun. 2017 doi: 10.1038/s41467-017-01742-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Su T, et al. Single-cell analysis of early progenitor cells that build coronary arteries. Nature. 2018;559:356–362. doi: 10.1038/s41586-018-0288-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Luo W, et al. Arterialization requires the timely suppression of cell growth. Nature. 2021;589:437–441. doi: 10.1038/s41586-020-3018-x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Chavkin, N. W. et al. Endothelial Cell Cycle State Determines Propensity for Arterial-Venous Fate (Cold Spring Harbor Laboratory, 2020). [DOI] [PMC free article] [PubMed]
- 32.Sissaoui S, et al. Genomic characterization of endothelial enhancers reveals a multifunctional role for NR2F2 in regulation of arteriovenous gene expression. Circ. Res. 2020;126:875–888. doi: 10.1161/CIRCRESAHA.119.316075. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Sabbagh MF, et al. Transcriptional and epigenomic landscapes of CNS and non-CNS vascular endothelial cells. eLife. 2018 doi: 10.7554/elife.36187. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.McDonald AI, et al. Endothelial regeneration of large vessels is a biphasic process driven by local cells with distinct proliferative capacities. Cell Stem Cell. 2018;23:210–225.e6. doi: 10.1016/j.stem.2018.07.011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Pitulescu ME, et al. Dll4 and Notch signalling couples sprouting angiogenesis and artery formation. Nat. Cell Biol. 2017;19:915–927. doi: 10.1038/ncb3555. [DOI] [PubMed] [Google Scholar]
- 36.Hasan SS, et al. Endothelial Notch signalling limits angiogenesis via control of artery formation. Nat. Cell Biol. 2017;19:928–940. doi: 10.1038/ncb3574. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Quillien A, et al. Distinct Notch signaling outputs pattern the developing arterial system. Development. 2014;141:1544–1552. doi: 10.1242/dev.099986. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Geudens I, et al. Artery-vein specification in the zebrafish trunk is pre-patterned by heterogeneous Notch activity and balanced by flow-mediated fine tuning. Development. 2019;146:dev181024. doi: 10.1242/dev.181024. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Lee H-W, et al. Role of venous endothelial cells in developmental and pathologic angiogenesis. Circulation. 2021;144:1308–1322. doi: 10.1161/CIRCULATIONAHA.121.054071. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Xu C, et al. Arteries are formed by vein-derived endothelial tip cells. Nat. Commun. 2014;5:5758. doi: 10.1038/ncomms6758. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Blanco R, Gerhardt H. VEGF and Notch in tip and stalk cell selection. Cold Spring Harb. Perspect. Med. 2013;3:a006569. doi: 10.1101/cshperspect.a006569. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Tian X, et al. De novo formation of a distinct coronary vascular population in neonatal heart. Science. 2014;345:90–94. doi: 10.1126/science.1251487. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Pontes-Quero S, et al. High mitogenic stimulation arrests angiogenesis. Nat. Commun. 2019 doi: 10.1038/s41467-019-09875-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Park H, et al. Defective flow-migration coupling causes arteriovenous malformations in hereditary hemorrhagic telangiectasia. Circulation. 2021;144:805–822. doi: 10.1161/CIRCULATIONAHA.120.053047. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Hou S, et al. Heterogeneity in endothelial cells and widespread venous arterialization during early vascular development in mammals. Cell Res. 2022 doi: 10.1038/s41422-022-00615-z. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Das S, et al. A unique collateral artery development program promotes neonatal heart regeneration. Cell. 2019;176:1128–1142.e18. doi: 10.1016/j.cell.2018.12.023. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Zarkada G, et al. Specialized endothelial tip cells guide neuroretina vascularization and blood-retina-barrier formation. Dev. Cell. 2021;56:2237–2251.e6. doi: 10.1016/j.devcel.2021.06.021. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Tang J, et al. Extension of endocardium-derived vessels generate coronary arteries in neonates. Circ. Res. 2022;130:352–365. doi: 10.1161/CIRCRESAHA.121.320335. [DOI] [PubMed] [Google Scholar]
- 49.Roman BL, Hinck AP. ALK1 signaling in development and disease: new paradigms. Cell. Mol. Life Sci. 2017;74:4539–4560. doi: 10.1007/s00018-017-2636-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Rochon ER, Menon PG, Roman BL. Alk1 controls arterial endothelial cell migration in lumenized vessels. Development. 2016;143:2593–2602. doi: 10.1242/dev.135392. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Jin Y, et al. Endoglin prevents vascular malformation by regulating flow-induced cell migration and specification through VEGFR2 signalling. Nat. Cell Biol. 2017;19:639–652. doi: 10.1038/ncb3534. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Poduri A, et al. Endothelial cells respond to the direction of mechanical stimuli through SMAD signaling to regulate coronary artery size. Development. 2017;144:3241–3252. doi: 10.1242/dev.150904. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Capasso TL, et al. BMP10-mediated ALK1 signaling is continuously required for vascular development and maintenance. Angiogenesis. 2020;23:203–220. doi: 10.1007/s10456-019-09701-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Sugden WW, Siekmann AF. Endothelial cell biology of Endoglin in hereditary hemorrhagic telangiectasia. Curr. Opin. Hematol. 2018;25:237–244. doi: 10.1097/MOH.0000000000000419. [DOI] [PubMed] [Google Scholar]
- 55.Deng H, et al. Activation of Smad2/3 signaling by low fluid shear stress mediates artery inward remodeling. Proc. Natl Acad. Sci. USA. 2021 doi: 10.1073/pnas.2105339118. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Kwon H-B, et al. In vivo modulation of endothelial polarization by Apelin receptor signalling. Nat. Commun. 2016;7:11805. doi: 10.1038/ncomms11805. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Weijts B, et al. Blood flow-induced Notch activation and endothelial migration enable vascular remodeling in zebrafish embryos. Nat. Commun. 2018;9:5314. doi: 10.1038/s41467-018-07732-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Rosa A, et al. WASp controls oriented migration of endothelial cells to achieve functional vascular patterning. Development. 2022;149:dev200195. doi: 10.1242/dev.200195. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Chang AH, et al. DACH1 stimulates shear stress-guided endothelial cell migration and coronary artery growth through the CXCL12-CXCR4 signaling axis. Genes Dev. 2017;31:1308–1324. doi: 10.1101/gad.301549.117. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Raftrey B, et al. Dach1 extends artery networks and protects against cardiac injury. Circ. Res. 2021 doi: 10.1161/circresaha.120.318271. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Franco CA, et al. Non-canonical Wnt signalling modulates the endothelial shear stress flow sensor in vascular remodelling. eLife. 2016;5:e07727. doi: 10.7554/eLife.07727. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Franco CA, et al. Dynamic endothelial cell rearrangements drive developmental vessel regression. PLoS Biol. 2015;13:e1002125. doi: 10.1371/journal.pbio.1002125. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Neal A, et al. Venous identity requires BMP signalling through ALK3. Nat. Commun. 2019;10:453. doi: 10.1038/s41467-019-08315-w. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Payne S, et al. Regulatory pathways governing murine coronary vessel formation are dysregulated in the injured adult heart. Nat. Commun. 2019 doi: 10.1038/s41467-019-10710-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Vanlandewijck M, et al. A molecular atlas of cell types and zonation in the brain vasculature. Nature. 2018;554:475–480. doi: 10.1038/nature25739. [DOI] [PubMed] [Google Scholar]
- 66.Phansalkar R, et al. Coronary blood vessels from distinct origins converge to equivalent states during mouse and human development. eLife. 2021 doi: 10.7554/elife.70246. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.Garcia FJ, et al. Single-cell dissection of the human brain vasculature. Nature. 2022 doi: 10.1038/s41586-022-04521-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68.Winkler EA, et al. A single-cell atlas of the normal and malformed human brain vasculature. Science. 2022;375:eabi7377. doi: 10.1126/science.abi7377. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69.Wälchli, T. et al. Molecular Atlas of the Human Brain Vasculature at the Single-Cell Level (Cold Spring Harbor Laboratory, 2021).
- 70.Augustin HG, Koh GY. Organotypic vasculature: from descriptive heterogeneity to functional pathophysiology. Science. 2017;357:eaal2379. doi: 10.1126/science.aal2379. [DOI] [PubMed] [Google Scholar]
- 71.Stoeckius M, et al. Simultaneous epitope and transcriptome measurement in single cells. Nat. Methods. 2017;14:865–868. doi: 10.1038/nmeth.4380. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 72.Zhu C, Preissl S, Ren B. Single-cell multimodal omics: the power of many. Nat. Methods. 2020;17:11–14. doi: 10.1038/s41592-019-0691-5. [DOI] [PubMed] [Google Scholar]
- 73.Han X, et al. Mapping the mouse cell atlas by Microwell-Seq. Cell. 2018;172:1091–1107.e17. doi: 10.1016/j.cell.2018.02.001. [DOI] [PubMed] [Google Scholar]
- 74.He L, et al. Single-cell RNA sequencing of mouse brain and lung vascular and vessel-associated cell types. Sci. Data. 2018;5:180160. doi: 10.1038/sdata.2018.160. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 75.Cusanovich DA, et al. A single-cell atlas of in vivo mammalian chromatin accessibility. Cell. 2018;174:1309–1324.e18. doi: 10.1016/j.cell.2018.06.052. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 76.Domcke S, et al. A human cell atlas of fetal chromatin accessibility. Science. 2020 doi: 10.1126/science.aba7612. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 77.Cui Y, et al. Single-cell transcriptome analysis maps the developmental track of the human heart. Cell Rep. 2019;26:1934–1950.e5. doi: 10.1016/j.celrep.2019.01.079. [DOI] [PubMed] [Google Scholar]
- 78.Litviňuková M, et al. Cells of the adult human heart. Nature. 2020;588:466–472. doi: 10.1038/s41586-020-2797-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 79.Zepp JA, et al. Genomic, epigenomic, and biophysical cues controlling the emergence of the lung alveolus. Science. 2021;371:eabc3172. doi: 10.1126/science.abc3172. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 80.Guo M, et al. Single cell RNA analysis identifies cellular heterogeneity and adaptive responses of the lung at birth. Nat. Commun. 2019 doi: 10.1038/s41467-018-07770-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 81.Hurskainen M, et al. Single cell transcriptomic analysis of murine lung development on hyperoxia-induced damage. Nat. Commun. 2021 doi: 10.1038/s41467-021-21865-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 82.Travaglini KJ, et al. A molecular cell atlas of the human lung from single-cell RNA sequencing. Nature. 2020;587:619–625. doi: 10.1038/s41586-020-2922-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 83.Schupp JC, et al. Integrated single-cell atlas of endothelial cells of the human lung. Circulation. 2021;144:286–302. doi: 10.1161/CIRCULATIONAHA.120.052318. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 84.Hupe M, et al. Gene expression profiles of brain endothelial cells during embryonic development at bulk and single-cell levels. Sci. Signal. 2017;10:eaag2476. doi: 10.1126/scisignal.aag2476. [DOI] [PubMed] [Google Scholar]
- 85.Park J, et al. Single-cell transcriptomics of the mouse kidney reveals potential cellular targets of kidney disease. Science. 2018;360:758–763. doi: 10.1126/science.aar2131. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 86.Magella B, et al. Cross-platform single cell analysis of kidney development shows stromal cells express Gdnf. Dev. Biol. 2018;434:36–47. doi: 10.1016/j.ydbio.2017.11.006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 87.Bais AS, et al. Single-cell RNA sequencing reveals differential cell cycle activity in key cell populations during nephrogenesis. Sci. Rep. 2021;11:22434. doi: 10.1038/s41598-021-01790-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 88.Miao Z, et al. Single cell regulatory landscape of the mouse kidney highlights cellular differentiation programs and disease targets. Nat. Commun. 2021;12:2277. doi: 10.1038/s41467-021-22266-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 89.Young MD, et al. Single-cell transcriptomes from human kidneys reveal the cellular identity of renal tumors. Science. 2018;361:594–599. doi: 10.1126/science.aat1699. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 90.Menon R, et al. Single-cell analysis of progenitor cell dynamics and lineage specification in the human fetal kidney. Development. 2018 doi: 10.1242/dev.164038. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 91.Hochane M, et al. Single-cell transcriptomics reveals gene expression dynamics of human fetal kidney development. PLoS Biol. 2019;17:e3000152. doi: 10.1371/journal.pbio.3000152. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 92.Cao J, et al. A human cell atlas of fetal gene expression. Science. 2020 doi: 10.1126/science.aba7721. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 93.The Tabula Sapiens Consortium The Tabula Sapiens: a multiple-organ, single-cell transcriptomic atlas of humans. Science. 2022;376:eabl4896. doi: 10.1126/science.abl4896. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 94.Calvanese V, et al. Mapping human haematopoietic stem cells from haemogenic endothelium to birth. Nature. 2022;604:534–540. doi: 10.1038/s41586-022-04571-x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 95.Tabula Muris Consortium et al. Single-cell transcriptomics of 20 mouse organs creates a Tabula Muris. Nature. 2018;562:367–372. doi: 10.1038/s41586-018-0590-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 96.Muto Y, et al. Single cell transcriptional and chromatin accessibility profiling redefine cellular heterogeneity in the adult human kidney. Nat. Commun. 2021;12:2190. doi: 10.1038/s41467-021-22368-w. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 97.Gómez-Salinero JM, et al. Specification of fetal liver endothelial progenitors to functional zonated adult sinusoids requires c-Maf induction. Cell Stem Cell. 2022;29:593–609.e7. doi: 10.1016/j.stem.2022.03.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 98.Halpern KB, et al. Paired-cell sequencing enables spatial gene expression mapping of liver endothelial cells. Nat. Biotechnol. 2018;36:962–970. doi: 10.1038/nbt.4231. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 99.Su T, et al. Single-cell transcriptomics reveals zone-specific alterations of liver sinusoidal endothelial cells in cirrhosis. Cell. Mol. Gastroenterol. Hepatol. 2021;11:1139–1161. doi: 10.1016/j.jcmgh.2020.12.007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 100.Aizarani N, et al. A human liver cell atlas reveals heterogeneity and epithelial progenitors. Nature. 2019;572:199–204. doi: 10.1038/s41586-019-1373-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 101.MacParland SA, et al. Single cell RNA sequencing of human liver reveals distinct intrahepatic macrophage populations. Nat. Commun. 2018 doi: 10.1038/s41467-018-06318-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 102.Nolan DJ, et al. Molecular signatures of tissue-specific microvascular endothelial cell heterogeneity in organ maintenance and regeneration. Dev. Cell. 2013;26:204–219. doi: 10.1016/j.devcel.2013.06.017. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 103.Feng W, Chen L, Nguyen PK, Wu SM, Li G. Single cell analysis of endothelial cells identified organ-specific molecular signatures and heart-specific cell populations and molecular features. Front. Cardiovasc. Med. 2019 doi: 10.3389/fcvm.2019.00165. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 104.Kalucka J, et al. Single-cell transcriptome atlas of murine endothelial cells. Cell. 2020;180:764–779.e20. doi: 10.1016/j.cell.2020.01.015. [DOI] [PubMed] [Google Scholar]
- 105.Paik DT, Cho S, Tian L, Chang HY, Wu JC. Single-cell RNA sequencing in cardiovascular development, disease and medicine. Nat. Rev. Cardiol. 2020;17:457–473. doi: 10.1038/s41569-020-0359-y. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 106.Brulois K, et al. A molecular map of murine lymph node blood vascular endothelium at single cell resolution. Nat. Commun. 2020;11:3798. doi: 10.1038/s41467-020-17291-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 107.Chen MB, et al. Brain endothelial cells are exquisite sensors of age-related circulatory cues. Cell Rep. 2020;30:4418–4432.e4. doi: 10.1016/j.celrep.2020.03.012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 108.Zhao L, et al. Pharmacologically reversible zonation-dependent endothelial cell transcriptomic changes with neurodegenerative disease associations in the aged brain. Nat. Commun. 2020 doi: 10.1038/s41467-020-18249-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 109.Gutierrez MEC, Hill MC, Largoza G, Martin JF, Wythe JD. Defining the transcriptional and epigenetic basis of organotypic endothelial diversity in the developing and adult mouse. bioRxiv. 2021 doi: 10.1101/2021.11.15.468651. [DOI] [Google Scholar]
- 110.Neal A, et al. ETS factors are required but not sufficient for specific patterns of enhancer activity in different endothelial subtypes. Dev. Biol. 2021;473:1–14. doi: 10.1016/j.ydbio.2021.01.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 111.Yucel N, et al. Cardiac endothelial cells maintain open chromatin and expression of cardiomyocyte myofibrillar genes. Elife. 2020 doi: 10.7554/eLife.55730. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 112.Liu Y, et al. A specialized bone marrow microenvironment for fetal haematopoiesis. Nat. Commun. 2022;13:1327. doi: 10.1038/s41467-022-28775-x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 113.Tikhonova AN, et al. The bone marrow microenvironment at single-cell resolution. Nature. 2019;569:222–228. doi: 10.1038/s41586-019-1104-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 114.Mäe MA, et al. Single-cell analysis of blood-brain barrier response to pericyte loss. Circ. Res. 2021 doi: 10.1161/circresaha.120.317473. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 115.Shi W, et al. Cardiac proteomics reveals sex chromosome-dependent differences between males and females that arise prior to gonad formation. Dev. Cell. 2021;56:3019–3034.e3017. doi: 10.1016/j.devcel.2021.09.022. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 116.Red-Horse K, Ueno H, Weissman IL, Krasnow MA. Coronary arteries form by developmental reprogramming of venous cells. Nature. 2010;464:549–553. doi: 10.1038/nature08873. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 117.Wu B, et al. Endocardial cells form the coronary arteries by angiogenesis through myocardial-endocardial VEGF signaling. Cell. 2012;151:1083–1096. doi: 10.1016/j.cell.2012.10.023. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 118.Chen HI, et al. The sinus venosus contributes to coronary vasculature through VEGFC-stimulated angiogenesis. Development. 2014;141:4500–4512. doi: 10.1242/dev.113639. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 119.Zhang H, et al. Endocardium minimally contributes to coronary endothelium in the embryonic ventricular free walls. Circ. Res. 2016;118:1880–1893. doi: 10.1161/CIRCRESAHA.116.308749. [DOI] [PubMed] [Google Scholar]
- 120.Sharma B, et al. Alternative progenitor cells compensate to rebuild the coronary vasculature in Elabela- and Apj-deficient hearts. Dev. Cell. 2017;42:655–666.e3. doi: 10.1016/j.devcel.2017.08.008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 121.Sedmera D, Pexieder T, Vuillemin M, Thompson RP, Anderson RH. Developmental patterning of the myocardium. Anat. Rec. 2000;258:319–337. doi: 10.1002/(SICI)1097-0185(20000401)258:4<319::AID-AR1>3.0.CO;2-O. [DOI] [PubMed] [Google Scholar]
- 122.Lu P, et al. Perinatal angiogenesis from pre-existing coronary vessels via DLL4–NOTCH1 signalling. Nat. Cell Biol. 2021;23:967–977. doi: 10.1038/s41556-021-00747-1. [DOI] [PubMed] [Google Scholar]
- 123.D’Amato, G. et al. Endocardium-to-coronary Artery Differentiation During Heart Development and Regeneration Involves Sequential Roles of Bmp2 and Cxcl12/Cxcr4 (Cold Spring Harbor Laboratory, 2021). [DOI] [PMC free article] [PubMed]
- 124.Jiang Z, et al. Overexpression of Kdr in adult endocardium induces endocardial neovascularization and improves heart function after myocardial infarction. Cell Res. 2021;31:485–487. doi: 10.1038/s41422-020-00436-y. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 125.Räsänen M, et al. VEGF-B promotes endocardium-derived coronary vessel development and cardiac regeneration. Circulation. 2021;143:65–77. doi: 10.1161/CIRCULATIONAHA.120.050635. [DOI] [PubMed] [Google Scholar]
- 126.Li H, et al. Classifying Drosophila olfactory projection neuron subtypes by single-cell RNA sequencing. Cell. 2017;171:1206–1220.e22. doi: 10.1016/j.cell.2017.10.019. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 127.Dick SA, et al. Self-renewing resident cardiac macrophages limit adverse remodeling following myocardial infarction. Nat. Immunol. 2019;20:29–39. doi: 10.1038/s41590-018-0272-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 128.Weinreb C, Rodriguez-Fraticelli A, Camargo FD, Klein AM. Lineage tracing on transcriptional landscapes links state to fate during differentiation. Science. 2020;367:eaaw3381. doi: 10.1126/science.aaw3381. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 129.McCracken IR, et al. Mapping the developing human cardiac endothelium at single-cell resolution identifies MECOM as a regulator of arteriovenous gene expression. Cardiovasc. Res. 2022 doi: 10.1093/cvr/cvac023. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 130.Vila Ellis L, et al. Epithelial Vegfa specifies a distinct endothelial population in the mouse lung. Dev. Cell. 2020;52:617–630.e6. doi: 10.1016/j.devcel.2020.01.009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 131.Gillich A, et al. Capillary cell-type specialization in the alveolus. Nature. 2020;586:785–789. doi: 10.1038/s41586-020-2822-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 132.Negretti NM, et al. A single-cell atlas of mouse lung development. Development. 2021 doi: 10.1242/dev.199512. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 133.Niethamer TK, et al. Defining the role of pulmonary endothelial cell heterogeneity in the response to acute lung injury. eLife. 2020 doi: 10.7554/elife.53072. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 134.Wang S, et al. A single-cell transcriptomic landscape of the lungs of patients with COVID-19. Nat. Cell Biol. 2021;23:1314–1328. doi: 10.1038/s41556-021-00796-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 135.McCracken IR, et al. Lack of evidence of angiotensin-converting enzyme 2 expression and replicative infection by SARS-CoV-2 in human endothelial cells. Circulation. 2021;143:865–868. doi: 10.1161/CIRCULATIONAHA.120.052824. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 136.Delorey TM, et al. COVID-19 tissue atlases reveal SARS-CoV-2 pathology and cellular targets. Nature. 2021;595:107–113. doi: 10.1038/s41586-021-03570-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 137.Sauler M, et al. Characterization of the COPD alveolar niche using single-cell RNA sequencing. Nat. Commun. 2022 doi: 10.1038/s41467-022-28062-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 138.Watanabe C, et al. Aging of the vascular system and neural diseases. Front. Aging Neurosci. 2020 doi: 10.3389/fnagi.2020.557384. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 139.Yang AC, et al. A human brain vascular atlas reveals diverse mediators of Alzheimer’s risk. Nature. 2022 doi: 10.1038/s41586-021-04369-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 140.Daneman R, Zhou L, Kebede AA, Barres BA. Pericytes are required for blood–brain barrier integrity during embryogenesis. Nature. 2010;468:562–566. doi: 10.1038/nature09513. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 141.Armulik A, et al. Pericytes regulate the blood–brain barrier. Nature. 2010;468:557–561. doi: 10.1038/nature09522. [DOI] [PubMed] [Google Scholar]
- 142.Jourde-Chiche N, et al. Endothelium structure and function in kidney health and disease. Nat. Rev. Nephrol. 2019;15:87–108. doi: 10.1038/s41581-018-0098-z. [DOI] [PubMed] [Google Scholar]
- 143.Dumas SJ, et al. Single-cell RNA sequencing reveals renal endothelium heterogeneity and metabolic adaptation to water deprivation. J. Am. Soc. Nephrol. 2020;31:118–138. doi: 10.1681/ASN.2019080832. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 144.Brunskill EW, Potter SS. Gene expression programs of mouse endothelial cells in kidney development and disease. PLoS ONE. 2010;5:e12034. doi: 10.1371/journal.pone.0012034. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 145.Karaiskos N, et al. A single-cell transcriptome atlas of the mouse glomerulus. J. Am. Soc. Nephrol. 2018;29:2060–2068. doi: 10.1681/ASN.2018030238. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 146.Takemoto M, et al. Large-scale identification of genes implicated in kidney glomerulus development and function. EMBO J. 2006;25:1160–1174. doi: 10.1038/sj.emboj.7601014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 147.Fu J, et al. Single-cell RNA profiling of glomerular cells shows dynamic changes in experimental diabetic kidney disease. J. Am. Soc. Nephrol. 2019;30:533–545. doi: 10.1681/ASN.2018090896. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 148.Chung J-J, et al. Single-cell transcriptome profiling of the kidney glomerulus identifies key cell types and reactions to injury. J. Am. Soc. Nephrol. 2020;31:2341–2354. doi: 10.1681/ASN.2020020220. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 149.Aird WC. Phenotypic heterogeneity of the endothelium. Circ. Res. 2007;100:174–190. doi: 10.1161/01.RES.0000255690.03436.ae. [DOI] [PubMed] [Google Scholar]
- 150.Inverso D, et al. A spatial vascular transcriptomic, proteomic, and phosphoproteomic atlas unveils an angiocrine Tie–Wnt signaling axis in the liver. Dev. Cell. 2021;56:1677–1693.e10. doi: 10.1016/j.devcel.2021.05.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 151.Shirure VS, Hughes CCW, George SC. Engineering vascularized organoid-on-a-chip models. Annu. Rev. Biomed. Eng. 2021;23:141–167. doi: 10.1146/annurev-bioeng-090120-094330. [DOI] [PubMed] [Google Scholar]
- 152.Ang LT, et al. Generating human artery and vein cells from pluripotent stem cells highlights the arterial tropism of Nipah and Hendra viruses. Cell. 2022;185:2523–2541.e30. doi: 10.1016/j.cell.2022.05.024. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 153.Wimmer RA, et al. Human blood vessel organoids as a model of diabetic vasculopathy. Nature. 2019;565:505–510. doi: 10.1038/s41586-018-0858-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 154.Takebe T, et al. Vascularized and functional human liver from an iPSC-derived organ bud transplant. Nature. 2013;499:481–484. doi: 10.1038/nature12271. [DOI] [PubMed] [Google Scholar]
- 155.Takebe T, et al. Massive and reproducible production of liver buds entirely from human pluripotent stem cells. Cell Rep. 2017;21:2661–2670. doi: 10.1016/j.celrep.2017.11.005. [DOI] [PubMed] [Google Scholar]
- 156.Van Den Berg CW, et al. Renal subcapsular transplantation of PSC-derived kidney organoids induces neo-vasculogenesis and significant glomerular and tubular maturation in vivo. Stem Cell Rep. 2018;10:751–765. doi: 10.1016/j.stemcr.2018.01.041. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 157.Mansour AA, et al. An in vivo model of functional and vascularized human brain organoids. Nat. Biotechnol. 2018;36:432–441. doi: 10.1038/nbt.4127. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 158.Holloway EM, et al. Differentiation of human intestinal organoids with endogenous vascular endothelial cells. Dev. Cell. 2020;54:516–528.e7. doi: 10.1016/j.devcel.2020.07.023. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 159.Garreta E, et al. Rethinking organoid technology through bioengineering. Nat. Mater. 2021;20:145–155. doi: 10.1038/s41563-020-00804-4. [DOI] [PubMed] [Google Scholar]
- 160.Lewis-Israeli YR, et al. Self-assembling human heart organoids for the modeling of cardiac development and congenital heart disease. Nat. Commun. 2021;12:5142. doi: 10.1038/s41467-021-25329-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 161.Hofbauer P, et al. Cardioids reveal self-organizing principles of human cardiogenesis. Cell. 2021;184:3299–3317.e22. doi: 10.1016/j.cell.2021.04.034. [DOI] [PubMed] [Google Scholar]
- 162.Drakhlis L, et al. Human heart-forming organoids recapitulate early heart and foregut development. Nat. Biotechnol. 2021;39:737–746. doi: 10.1038/s41587-021-00815-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 163.Homan KA, et al. Flow-enhanced vascularization and maturation of kidney organoids in vitro. Nat. Methods. 2019;16:255–262. doi: 10.1038/s41592-019-0325-y. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 164.Campisi M, et al. 3D self-organized microvascular model of the human blood-brain barrier with endothelial cells, pericytes and astrocytes. Biomaterials. 2018;180:117–129. doi: 10.1016/j.biomaterials.2018.07.014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 165.Skylar-Scott MA, et al. Biomanufacturing of organ-specific tissues with high cellular density and embedded vascular channels. Sci. Adv. 2019;5:eaaw2459. doi: 10.1126/sciadv.aaw2459. [DOI] [PMC free article] [PubMed] [Google Scholar]