Abstract
IS1397 and ISKpn1 are IS3 family members which are specifically inserted into the loop of palindromic units (PUs). IS1397 is shown to transpose into PUs with sequences close or identical to the Escherichia coli consensus, even in other enterobacteria (Salmonella enterica serovar Typhimurium, Klebsiella pneumoniae, and Klebsiella oxytoca). Moreover, we show that homologous intergenic regions containing PUs constitute IS1397 transpositional hot spots, despite bacterial interspersed mosaic element structures that differ among the three species. ISKpn1, described here for the first time, is specific for PUs from K. pneumoniae, in which we discovered it. A sequence comparison between the two insertion sequences allowed us to define a motif possibly accounting for their specificity.
The chromosome of Escherichia coli contains various families of small extragenic sequences repeated from 6 to more than 250 times and representing almost 2% of the bacterial DNA (4). The BIME family (bacterial interspersed mosaic elements) is one of these. The basic motif of BIMEs is the palindromic unit (PU) or repetitive extragenic palindromic sequence (19). PUs are imperfect palindromes of about 40 bp that are transcribed but not translated (4). Five hundred eighty-four PUs are scattered over the chromosome of E. coli (6) and have been divided into three classes, Y, Z1, and Z2, according to slight variations in sequence and size. BIMEs have been defined as a precise combination of PUs alternating in orientation and type and associated with seven extra-PU motifs (A, B, S, L, s, l, and r), and two major BIME families can be distinguished, called BIME-1 and BIME-2 (3, 18). For a review of PUs and BIMEs, see reference 4 and the E. coli short DNA repeats section of the Unit of Molecular Programming and Genetic Toxicologyweb site (http://www.pasteur.fr/recherche/unites/pmtg/repet/index.html). The existence of a general function for BIMEs is still unclear. Some of them can stabilize mRNAs (26, 27) or play a role in transcription termination (14), translational control (38), and genomic rearrangements (36). However, their specific interactions with integration host factor (7, 28), DNA gyrase (12, 42), and DNA polymerase (16) suggest that they may play a role in the functional organization of the bacterial nucleoid.
PUs were detected originally in E. coli and Salmonella enterica serovar Typhimurium (13, 19) and later in other enterobacteria by Southern blot hybridization and sequence analysis (17). The PU consensus for S. enterica serovar Typhimurium differs slightly from the E. coli consensus in that there is an additional G between positions 10 and 11 (15). There is another PU motif, D, specific to Salmonella and Klebsiella (4). Klebsiella PUs are closely related to S. enterica serovar Typhimurium PUs (2), but they are more GC rich than the rest of the genome. The two species exhibit BIME-like structures, but extra-PU motifs seem to be less conserved than in E. coli. There is no L motif in S. enterica serovar Typhimurium, and BIMEs containing PUs in direct tandem repeats are present in these enterobacteria.
IS1397 is a 1,432-bp insertion sequence belonging to the IS3 family (5). It has been found in several natural E. coli isolates and is always inserted into the central part of a PU described as the loop (Fig. 1). We have recently shown that IS1397 is an active insertion sequence that is able to be transposed into E. coli from a donor plasmid and that it is inserted specifically into PUs (9). In this study, we analyzed whether IS1397 has the same target specificity in other Enterobacteriaceae species which also contain PUs, i.e., S. enterica serovar Typhimurium, Klebsiella pneumoniae, and K. oxytoca. We also describe a new IS, ISKpn1, that was identified during analysis of the PUs of K. pneumoniae. This insertion sequence (IS) is closely related to IS1397 and IS150 and is also specifically inserted into PUs.
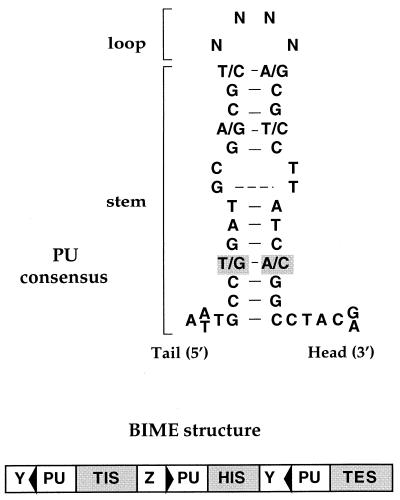
FIG. 1.
PU structure and BIME organization. (Top) Consensus sequence and hypothetical DNA structure of PUs. Two types of PUs have been defined according to two critical positions of the consensus (7 and 32, boxed in grey). In the Y PUs, these nucleotides are a G and a C, respectively, while in the Z PUs, they are a T and an A. These sequences form an imperfect palindrome, with asymmetry elements which allow orientation of the structure from the tail to the head, and confer a stem-loop structure. (Bottom) BIMEs are composed of successive PUs alternating both in type and in orientation (indicated by black triangles). They are separated by short “extra-PU” motifs located either between two heads (head internal sequences [HIS]) or two tails (tail internal sequences [TIS]). Two tail external sequences (TES), A and B, flanking the tails of the last PUs, are found in some BIMEs (18).
MATERIALS AND METHODS
Plasmids.
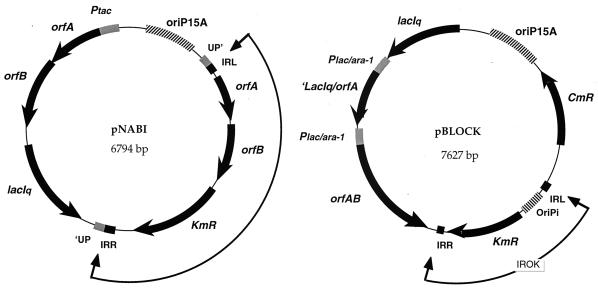
pNABI, the plasmid used to study the transposition of IS1397 into the chromosome of K. pneumoniae, has already been described (9). Its structure is shown in Fig. 2. This derivative of pACYC184 (8) carries a P15A origin of replication, an orfAB artificial gene with a disruption of the palindrome found in the putative frameshift window between orfA and orfB. However, when we sequenced this region, we found that the expected deletion of an A introduced to create an in-frame fusion between the two genes had not been achieved. As a consequence, only OrfA, and not OrfAB, could be expressed under the control of Ptac (isopropyl-β-d-thiogalactopyranoside [IPTG] inducible due to the presence of a functional lacIq gene on the plasmid). Overexpression of OrfA is toxic to the cells (J.-M. Clément and C. Wilde, unpublished observations). pNABI also contains IS1397, flanked by an interrupted PU sequence with a 4-bp duplication (as naturally found between the mtlA and mtlD genes in EPEC25 [5]) and with a Kmr-encoding gene inserted downstream of orfB.
FIG. 2.
Plasmids pNABI and pBLOCK. Both plasmids are derived from pACYC184 (p15A origin of replication, hatched box). The orientations of the different genes (thick lines) are indicated by arrows. The transposable module is located in each case, between two arrows (IROK in the case of pBLOCK). IRL and IRR are the left and right inverted repeats from IS1397, respectively.
Another plasmid, pBLOCK, was constructed to study transposition into the chromosomes of K. oxytoca and S. enterica serovar Typhimurium. It was created to allow selection and quick analysis of transposition in virtually any bacterial species. This derivative of pACYC184 carries the P15A origin of replication and lacIq. The Cmr-encoding gene is placed next to the P15 origin of replication. The artificial Plac/ara-1 promoter from pPROLar.A (Clontech) is found twice, upstream of orfA and upstream of orfAB, allowing overproduction of the two proteins upon IPTG induction. The last 93 bp of the lacIq sequence are found downstream of orfA, due to the cloning strategy. OrfA and OrfAB are both toxic to the cell. The transposable module (delineated with arrows in Fig. 2) is not flanked by the interrupted PU and is different from the one found in pNABI. The orfA and orfB sequences were replaced with the R6K origin of replication (35, 37), which allows stable maintenance of plasmids in strains expressing Π protein, such as BW19610. Most of the DNA fragments corresponding to these various parts were generated by PCR. In this case, their sequence was systematically checked after cloning into the recombinant plasmids. As for pNABI, the construction of pBLOCK involved many steps, the details of which will be supplied upon request (jclement@pasteur.fr). We checked that pNABI and pBLOCK enabled the transposition of their respective modules into the E. coli chromosome with the same target specificity (J.-M. Clément and F. Le Noanne, unpublished observations).
Media and bacterial strains.
Luria-Bertani (LB) medium was used to grow all of the strains. Kanamycin (KM) and chloramphenicol (CM) were used at concentrations of 25 and 50 μg/ml, respectively. IPTG was used at a final concentration of 10−3 M, and 5-bromo-4-chloro-3-indolyl-β-d-galactopyranoside (X-Gal) was used at 40 μg/ml.
The strains used for plasmid rescue were JM109 [recA1 endA1 gyrA96 thiA hsdR17(rK− mK+) relA1 supE44 Δ(lac-proAB) F′ traD proAB lacIqZΔM15] (43), BW19610 [DE3(lac)X74 ΔuidA::pir-116 recA1 ΔphoA532 Δ(phnC?D-P)33–30] (25), and TOP10F′ {F′ [lacIq Tn10(Tetr)] mrcA Δ(mrr-hsdRMS-mrcBC) φ80lacZΔM15 ΔlacX74 recA1 deoR araD139 Δ(ara-leu)7697 galU galK rpsL (Strr) endA1 nupG} (Invitrogen). The strains in which transposition was studied were K. pneumoniae subsp. pneumoniae ATCC 13883, K. oxytoca ATCC 13182, and S. enterica serovar Typhimurium 3261. The strains used to study ISKpn1 distribution were K. pneumoniae MGH78578 (a gift from M. McClelland, Genome Sequencing Center [GSC], Washington, University, St. Louis, Mo.), K. pneumoniae subsp. pneumoniae ATCC 13883, K. pneumoniae subsp. ozonae ATCC 11296, K. pneumoniae subsp. rhinoscleromatis ATCC 13884, K. oxytoca ATCC 13182, K. planticola ATCC 33531, K. terrigena ATCC 33257, Enterobacter aerogenes ATCC 13048, K. aerogenes W70 (20), E. coli EPEC 25 (22), E. coli C600 (1), Yersinia pestis 6.69, Y. enterocolitica 8081, and Y. pseudotuberculosis 32953. The Yersinia strains belong to the Pasteur Institute collection and were a gift from Elisabeth Carniel.
DNA techniques.
Restriction enzymes and DNA-modifying enzymes were purchased from New England Biolabs or Boehringer Mannheim and used as recommended. Plasmid DNA manipulations were carried out by using standard procedures (30). Chromosomal DNA extractions were performed with the DNA Easy Tissue kit (Qiagen). PCRs were performed by using the Amersham PCR kit as recommended with a Mastercycler gradient apparatus (Eppendorf).
Oligonucleotides.
Oligonucleotides were purchased from Genset. Two oligonucleotides were used to sequence the junctions of IS1397 chromosomal insertions. Their sequences are as follows: seqIRL, 5′CGGTTGTGGACAACAAGCCAGGG3′ (complementary to a region of the R6K origin of replication); Kmseqout, 5′CACGAGGCAGACCTCAGCGC3′ (corresponding to a region located between the end of the Kmr-encoding gene and the right inverted repeat (IRR) of the module, as found on the plasmid pBLOCK).
Two other oligonucleotides were used for ISKpn1 cloning, i.e., upKp (5′GCGAATAGCCGGCTGAAAACGTGAG3′) and downKp (5′GGTGGTCATTTCTCAAGGCGAGG3′), flanking the IS of K. pneumoniae on contig 840 (May 2000), according to the GSC web site (http://genome.wustl.edu/gsc/index.shtml).
DNA sequencing.
DNA sequencing was performed either as previously described (9) or by MWG-Biotech AG and ESGS-Cybergene.
Southern blot hybridization.
Samples (1 to 5) μg of chromosomal DNAs were loaded on a 1% agarose Tris-acetate gel. DNA transfer onto a Hybond N+ membrane (Amersham) was performed as previously described (9). Hybridization was carried out by using the DIG Nucleic Acid Labeling and Detection system (Boehringer Mannheim) at 56°C. The probes used were digoxigenin-dUTP labeled with a Promega nick translation kit.
Selection of transposition events.
Transposition events in K. pneumoniae were selected and studied as previously described for E. coli (9). We used a slightly different strategy to study transposition events in K. oxytoca and S. enterica serovar Typhimurium. Independent clones of K. oxytoca and S. enterica serovar Typhimurium transformed with pBLOCK were grown overnight at 37°C in liquid LB medium containing KM and CM. A 5-μl volume of each culture was plated on LB medium containing KM and IPTG, incubated overnight at 37°C, and replica plated onto LB medium containing IPTG and either CM or KM. After a 24-h incubation at 37°C, two colonies per plate that grew on KM but not on CM were streaked on LB medium containing KM and IPTG and grown overnight at 37°C in the same liquid medium for chromosomal DNA extraction. Chromosomal DNAs were digested with MluI (which has a single site in pBLOCK, in the lacI gene), and 1 to 5 μg was electrophoresed on a 1% agarose gel and submitted to Southern blot hybridization. The probe used was IROK (Fig. 2), which corresponds to the R6K origin-Kmr module from pBLOCK. For the cloning of chromosomal fragments containing IROK, MluI-digested chromosomal DNAs were circularized and E. coli BW19610 cells were transformed by electroporation. For long fragments, chromosomal DNAs were digested with MluI and BstEII (none of which have restriction sites in IROK) and ligated before transformation of BW19610 cells. In both cases, recombinant clones were selected on LB medium plates with KM. After DNA sequencing, chromosomal regions flanking the module were identified by using the FASTA software of Infobiogen (http://www.infobiogen.fr/) and the BLAST program of the National Center for Biotechnology Information (http://www.ncbi.nlm.nih.gov/).
K. pneumoniae MGH78578 genomic library.
Fragments of PstI-digested K. pneumoniae MGH78578 genomic DNA ranging from 2.3 to 6 kb were purified from an agarose gel (Qiaquick gel extraction kit; Qiagen) and ligated to pUC18 treated with PstI and dephosphorylated with shrimp alkaline phosphatase. TOP10F′ cells were transformed (One Shot Transformation Reaction; Invitrogen) in accordance with the manufacturer's indications with the ligation mixture. We obtained 9,000 white colonies on LB medium plates containing ampicillin, X-Gal, and IPTG. A colony blot hybridization was done with a probe obtained as follows. The IS was amplified by PCR on K. pneumoniae chromosomal DNA with the primers downKp and upKp, which are located on each side of the IS on contig 840. The PCR product was cloned into the PCRII-TOPO vector (TOPO TA Cloning; Invitrogen). The probe was the 1,067-bp AccI-HindIII fragment from ISKpn1 cloned into PCRII-TOPO. It contained the first 976 bp of the IS and 91 bp from the vector. We obtained one positive clone, which was streaked on LB medium plates with ampicillin and checked for positive hybridization with the same probe. The initial sequencing steps were done with the universal primers forward-40 and reverse, and the sequencing was completed with specific primers.
Distribution of ISKpn1.
The genomic DNAs of the strains studied were digested with HindIII and MluI, and 1 μg of each was loaded onto a 1% agarose gel for Southern blot hybridization with the probe used to screen the K. pneumoniae MGH78578 genomic library.
Determination of a global PU consensus for K. pneumoniae.
We investigated K. pneumoniae MGH78578 sequences present in the contigs released by the GSC (http://genome.wustl.edu/gsc/index.shtml) by using the E. coli PU consensus and the consensus derived from the transposition sites of IS1397 in K. pneumoniae as query sequences for the BLAST (http://www.ncbi.nlm.nih.gov/) and FASTA (http://www.infobiogen.fr/) programs, and we checked all matches by eye to see whether they were PUs. We aligned the 242 PUs detected with the clustalw software (parameters: gap opening penalty, 10; gap extension penalty, 0.20; gap separation penalty range, 8) and reformatted the result to an msf format with ftmseq in order to use Pretty (with plurality = 3), a Genetics Computer Group, Inc., program. Pretty displayed multiple-sequence alignments and determined the consensus sequence shown in Fig. 3.
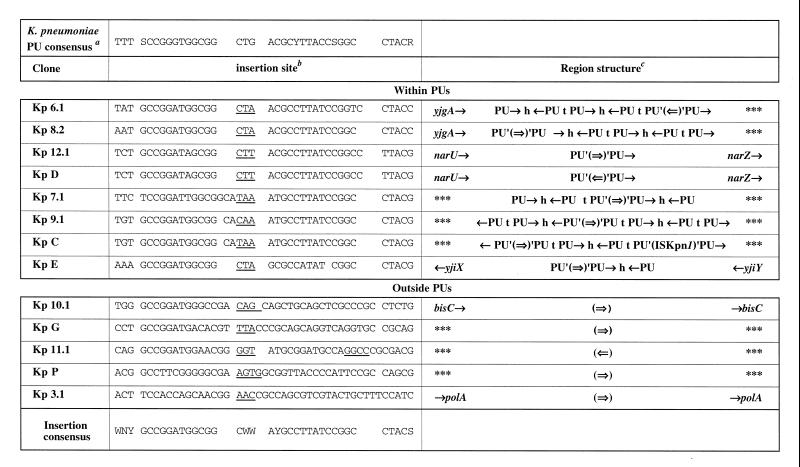
FIG. 3.
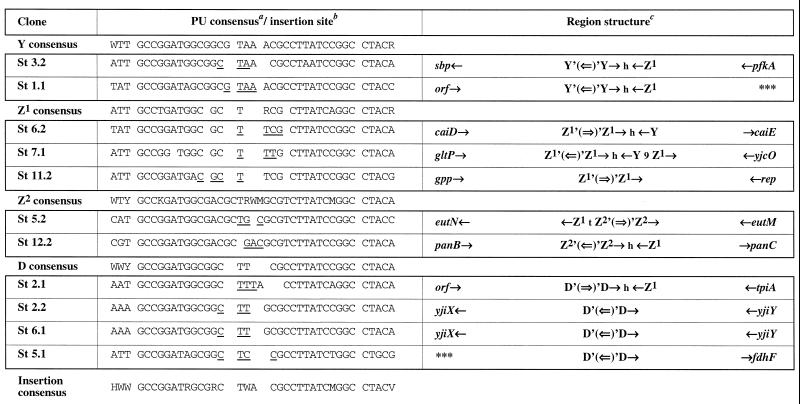
Sites of insertion of the transposable module into the chromosome of K. pneumoniae. a, The PU consensus is derived from this study. D is A, G, or T; S is G or C; W is A or T; and Y is C or T. b, Nucleotides which are duplicated after transposition are underlined. c, Single arrows indicate PU and gene orientations. The double arrow represents the oriented transposable module. The letters h and t represent head internal sequences (HIS) and tail internal sequences (TIS), respectively (Fig. 1). Gene names have been assessed by sequence homology to E. coli genes (http://genolist.pasteur.fr/Colibri/). Triple asterisks indicate a region for which no homolog has been identified.
Nucleotide sequence accession number.
The nucleotide sequence of the IS described here (see Fig. 6) was deposited in the GenBank database under accession no. AF345899.
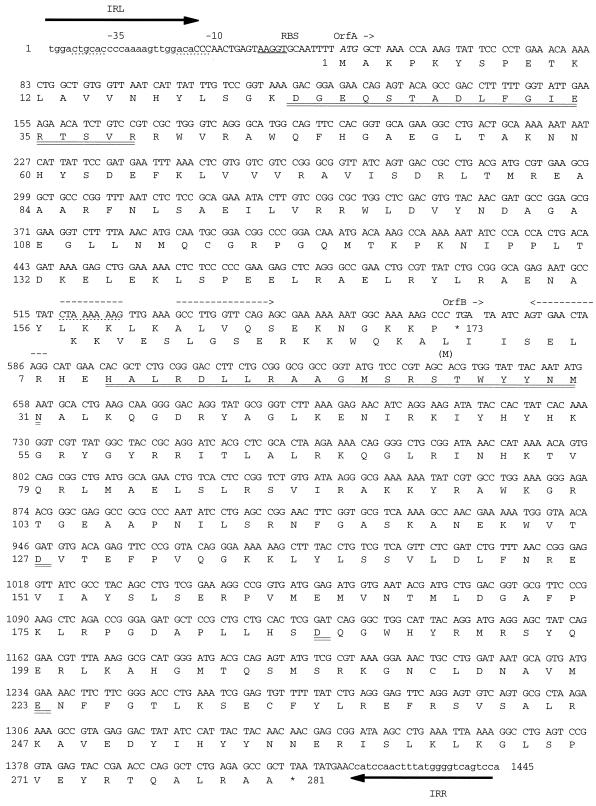
FIG. 6.
Nucleotide and amino acid sequences of the two putative ORFs of ISKpn1. The IRL and IRR are in lowercase and indicated by black arrows. The ribosome-binding site (RBS) is underlined. The initiation codons of the two ORFs are in italics, and their termination codons are indicated by stars. Nucleotide and amino acid numbers are indicated on the left. The two HTH motifs are doubly underlined. The frameshift window is overlined and followed by a palindrome indicated by dashed arrows. The DDE motif (OrfB residues 127, 187, and 223) is doubly underlined.
RESULTS
Transposition events.
We studied the transposition of IS1397 in several bacterial species by using either pNABI (9), which was previously used in E. coli, or a derivative, pBLOCK.
Selection of transposition events with pNABI relied on the Kmr IPTGr LacI− phenotype, which was the consequence of both transposition of the module into the chromosome and loss of pNABI. However, IPTGr could result from mutations in orfAB and LacI− phenotype screening of LacZ or LacI+ bacterial strains was not possible. This is why we used pBLOCK, in which the presence of both orfA and orfAB minimizes the selection of IPTGr clones due to mutations in these genes. Moreover, loss of the donor plasmid results in loss of Cmr. It should be noted that the p15A origin of replication is located between cat and orfAB (Fig. 2), so that a deletion encompassing these genes would not allow the maintenance of an autonomously replicating plasmid. Another advantage of pBLOCK is that the chromosomal insertion sites can propagate after circularization in a Pir+, strain since they are associated with the R6K origin of replication.
We grew 16 separate liquid cultures of 5 × 108 CFU of independent pNABI-containing K. pneumoniae clones, which were plated on LB medium plates containing KM and IPTG. For each culture, we obtained resistant colonies, which were tested for loss of the plasmid; they were streaked on LB medium plates with X-Gal (no IPTG added). In all but one case, we observed blue colonies. In order to check the homogeneity of the transposition events, two blue colonies originating from the same liquid culture were then examined for the presence of an IS1397-Kmr insertion in the chromosome. BglII digests of chromosomal DNA were analyzed by Southern blot hybridization using an internal IS1397 DNA fragment as a probe (data not shown). We observed a single faint band after transposition and/or several heavily labeled bands when the bacteria had kept the plasmid. In seven cases, the two clones tested displayed different profiles; in four out of these seven cases, one of the clones still had the plasmid and was discarded. In eight cases, the two clones tested exhibited the same profile; in three out of these eight cases, they had kept the plasmid and were discarded. Hence, we retained 15 clones resulting from independent transposition events for further analysis. We successfully cloned 13 different BglII chromosomal DNA fragments containing the Kmr-encoding gene in pUC18 and sequenced the junctions between the transposition module and the chromosome (Fig. 3).
Transposition events in K. oxytoca (lacZ+ lacI+) and S. enterica serovar Typhimurium (Lac−) were investigated by using pBLOCK. In the presence of IPTG, both OrfA and OrfAB are expressed, which are each lethal to the bacteria. IPTGr Kmr colonies could be isolated after overnight culture at 37°C (see Materials and Methods); such a phenotype could be due to (i) a deletion or mutation within orfA and orfAB resulting in nontoxic proteins (and the bacteria would keep the plasmid) or (ii) transposition of IROK into the chromosome with loss of pBLOCK. These two events could be discriminated by checking the sensitivity of the strains to CM, indicating loss of the plasmid.
Approximately 5 × 106 CFU from 12 liquid cultures of independent K. oxytoca and S. enterica serovar Typhimurium clones containing pBLOCK were plated on LB medium containing IPTG and KM. We obtained about 1,000 resistant colonies per plate that we replica plated on CM-containing medium. Ninety-nine percent of the S. enterica serovar Typhimurium colonies and 1% of the K. oxytoca colonies were sensitive to CM. Two Kmr IPTGr Cms clones from each plate were examined for the presence of IROK in the chromosome. MluI digests of chromosomal DNA were analyzed by Southern blot hybridization using IROK as a probe (data not shown). All S. enterica serovar Typhimurium candidates but one showed different profiles between the two clones, and one clone still contained the plasmid. No K. oxytoca candidates had kept the plasmid, and in three cases, the profiles of the two clones tested were different. We circularized the fragments and transformed E. coli BW19610, a strain allowing the replication of the circular fragments containing the R6K origin. We sequenced the junctions between the transposition module and the chromosome for 11 S. enterica serovar Typhimurium clones (Fig. 4) and 15 K. oxytoca clones (Fig. 5).
FIG. 4.
Sites of insertion of the transposable module into the chromosome of S. enterica serovar Typhimurium. a, The PU consensuses are taken from reference 4 and the Unit of Molecular Programming and Genetic Toxicology web site (http://www.pasteur.fr/recherche/unites/pmtg/repet/index.html). b, Nucleotides which are duplicated after transposition are underlined. H is A, C, or T; M is A or C; R is A or G; V is A, C, or G; and W is A or T. c, Single arrows indicate PU and gene orientations. The double arrow represents the oriented transposable module; h and t represent head internal sequences (HIS) and tail internal sequences (TIS), respectively (Fig. 2). Gene names have been assessed by sequence homology to E. coli genes (http://genolist.pasteur.fr/Colibri/). Triple asterisks indicate a region for which no homolog has been identified. orf indicates a potential coding sequence with no homolog in the databases.
FIG. 5.
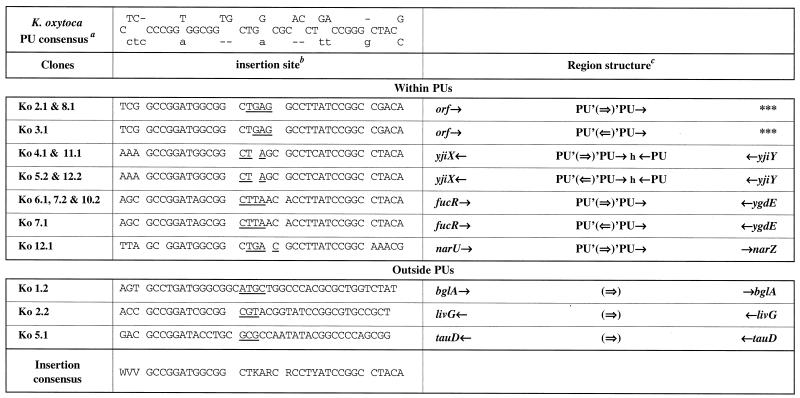
Sites of insertion of the transposable module into the chromosome of K. oxytoca. a, The PU consensus is taken from reference 2. Lowercase letters are indicative of less frequent occurrences. b, Nucleotides which are duplicated after transposition are underlined. K is G or T; R is A or G; V is A, C, or G; W is A or T; and Y is C or T. c, Single arrows indicate PU and gene orientations. The double arrow represents the orientation of the transposable module. The letters h and t represent head internal sequences (HIS) and tail internal sequences (TIS), respectively (Fig. 2). Gene names have been assessed by sequence homology to E. coli genes (http://genolist.pasteur.fr/Colibri/). Triple asterisks indicate a region for which no homolog has been identified. orf indicates a potential coding sequence without homolog in the databases.
Target specificity in enterobacteria.
Our previous results obtained with E. coli showed that IS1397 has a tight specificity of insertion into PUs (5, 9). All integrations of the transposable module into the S. enterica serovar Typhimurium, K. pneumoniae, and K. oxytoca chromosomes were transposition events with a 3- or 4-bp duplication. All of the insertions obtained in S. enterica serovar Typhimurium occurred within the loop of a PU (Fig. 4), whereas we obtained PU and non-PU integrations for the two other enterobacteria. In the case of K. pneumoniae (Fig. 3), we observed eight insertions into PUs; two insertions (Kp 10.1 and Kp G) in which the transposition module was inserted 6 nucleotides after the sequence 5′GCCGGATG3′, which is found in a PU stem; two insertions into extragenic sequences which are not PUs (Kp 11.1 and Kp P); and one intragenic transposition at the end of polA (Kp 3.1). In K. oxytoca (Fig. 5); there were 12 insertions into PUs and three into genes (Ko 1.2, Ko 2.2, and Ko 5.1), 5 to 7 nucleotides after the 5′GCC(G/T)GAT3′ sequence found in the PU stem. Interestingly, we observed hot spots of insertions in either orientation within and among the three species. There were six independent transposition events (St 2.2, St 6.1, Ko 4.1, Ko 5.2, Ko 11.1, and Ko 12.2) into a PU from the yjiX-yjiY intergenic region, four into the fucR-ygdE region (Ko 6.1, Ko 7.1, Ko 7.2, and Ko 10.2), three into a PU next to the same open reading frame (Ko 2.1, Ko 3.1, and Ko 8.1), and three (Ko 12.1, Kp 12.1, and Kp D) into a PU from the narU-narZ region.
Description of a new IS associated with K. pneumoniae PUs.
In the course of a computer analysis of the different PU types in K. pneumoniae MGH78578, we discovered a new IS that is present in five contigs (at the GSC), always inserted into the loop of a PU with a 3-bp duplication, like IS1397. We cloned this IS from a genomic library by using a PCR fragment containing the IS as a probe. The PCR was performed on genomic DNA by using unique sequences flanking the IS on contig 840 as primers and yielded a very small amount of the fragment. We established the definitive sequence of the IS from a library clone that displayed flanking regions different from what was expected, suggesting misassembly of the contigs and explaining the poor yield of the PCR (see Discussion). The IS was named ISKpn1 (Fig. 6). ISKpn1 is 1,445 bp long and belongs to the IS3 family, according to sequence homology and structural features. It is flanked with 25-bp imperfect terminal inverted repeats containing six mismatches, a left inverted repeat (IRL) and an IRR, ending with the dinucleotide 5′-CA-3′ (29). ISKpn1 contains two open reading flames on the same strand, orfA and orfB, which is in −1 frame with respect to orfA. orfA extends from ATG (position 50) to TGA (position 571) and is preceded by a ribosome-binding site located 8 bp upstream of the start codon. orfA could encode a 173-amino-acid (aa) protein, OrfA, containing a putative α-helix–turn–α-helix (or HTH) motif (aa 23 to aa 40; boxed in Fig. 6) with a probability of 71% (the standard deviation score obtained by comparison with a weight matrix was 3.98 [11]). orfB (nucleotides 568 to 1413) could begin with the rarely used CTG start codon located between the frameshift window and the HTH motif (see below) and encode a putative protein of 281 aa, OrfB, which also contains a putative HTH motif (aa 10 to 31, boxed in Fig. 6) with a probability of 71% (standard deviation score, 3.78). ISKpn1, like all members of the IS3 family, is characterized by the presence of a conserved region in OrfB called the D,D(35)E motif that is associated with several additional residues (21, 24, 29). This triad is involved in catalysis and is also present in retroviral integrases. Another characteristic of IS3 family members is the presence of an A6G sequence followed by a dyad symmetry, which enables the formation of a fusion protein containing OrfA and OrfB, called OrfAB, the putative transposase, resulting from a −1 translational frameshift. This kind of structure, called a frameshift window, has already been described for IS3 (33), IS150 (41), IS1 (31, 32), and retroviral integrases (40), for example. Screening of the SPGLOBAL database with the putative OrfA protein sequence gave the best scores with IS3 family members IS150, IS1397, and IS1223, with 38.5, 37.6, and 35.6% identity, respectively. The same screen performed with the putative OrfB protein sequence gave 63.3 and 56.6% identity with the IS150 and IS1397 OrfB proteins, respectively. Screening of the bacterial section of the GenBank database with the nucleotide sequence of ISKpn1 gave the best score with IS150 (62.3% identity). Thus, ISKpn1 is, interestingly, closer in both nucleotide and protein sequences to IS150 than to IS1397, although ISKpn1 and IS1397 share the property of insertion into PUs (see Discussion).
Distribution of ISKpn1 among several enterobacterial species.
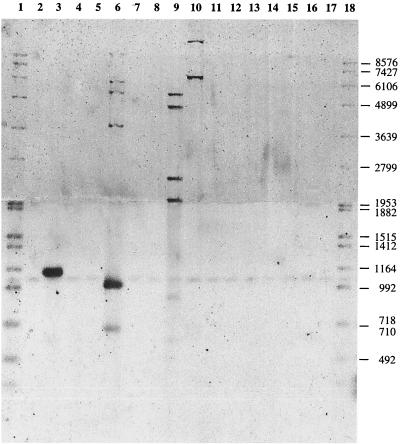
We studied the distribution of ISKpn1 among several Klebsiella and Yersinia species and two E. coli strains by Southern blot hybridization (Fig. 7). Chromosomal DNAs were digested by MluI and HindIII, and the probe (see Materials and Methods) was specific for ISKpn1 since it did not hybridize with E. coli K-12, which contains IS150, or cross-hybridize with IS1397 (the very slight signal is nonspecific, since the molar ratio of the IS1397 fragment [Fig. 7, lane 16] to the genomic DNAs ([Fig. 7, lanes 2 to 15] is 500). Figure 7 shows that ISKpn1 is present only in K. pneumoniae MGH78578, K. pneumoniae subsp. pneumoniae (used to study the transposition of IS1397), K. pneumoniae subsp. ozonae, and K. aerogenes. ISKpn1 is present in fewer than 10 copies on the chromosomes of these strains, in contrast to IS1397, which was present in high copy numbers in certain strains, such as EPEC25 (at least 25 times) and ECOR49 (about 15 times) (5). ISKpn1 is absent from K. pneumoniae subsp. rhinoscleromatis and in K. oxytoca. Since HindIII cuts into the IS (Fig. 6), the smallest fragment which can hybridize to the probe is 976 bp long and the last 469 bp of the IS do not match the probe. In K. pneumoniae subsp. ozonae and K. pneumoniae subsp. pneumoniae, we observed one fragment between 992 and 1,164 bp, indicating the presence of a HindIII or MluI restriction site close to the ISKpn1 IRL in these fragments. These bands are heavily labeled. Such a phenomenon has already been described in the case of IS1397 in several natural E. coli isolates and has been explained by a possible overrepresentation of IS-containing fragments identical or similar in size (5).
FIG. 7.
Distribution of ISKpn1. Genomic DNAs were digested by HindIII and MluI and loaded on to a 1% agarose gel for Southern blot hybridization using an ISKpn1 fragment as a probe. Lanes: 2, K. planticola; 3, K. pneumoniae subsp. ozonae; 4, K. oxytoca; 5, K. pneumoniae subsp. rhinoscleromatis; 6, K. pneumoniae subsp. pneumoniae; 7, E. aerogenes; 8, K. terrigena; 9, K. pneumoniae MGH78578; 10, K. aerogenes; 11, E. coli EPEC 25; 12, E. coli C600; 13, Y. pestis; 14, Y. pseudotuberculosis; 15, Y. enterocolitica; 1 and 18, digoxigenin-labeled DNA molecular weight marker VII (50 ng); 16, pNABI digested by EcoRV (100 ng); 17, pNABI digested by EcoRV (10 ng). The values on the right are molecular weights.
DISCUSSION
pNABI and pBLOCK harbor a transposable module containing a selectable marker (a Kmr-encoding gene) flanked by IS1397 inverted repeats. The two plasmids enabled us to select for transposition of these modules into the chromosomes of four enterobacteria: E. coli, S. enterica serovar Typhimurium, K. pneumoniae, and K. oxytoca. In all instances, a 3- or 4-bp duplication at the site of insertion was characteristic of a bona fide transposition event. The difference between pBLOCK and pNABI is that the OrfAB transposase is, in the first case, expressed from an engineered gene which is located outside of the transposable module (Fig. 2), whereas the wild-type IS1397 orfA-orfB genes are located within the module in pNABI. Both plasmids were efficient donor plamids for transposition in E. coli (not shown here). This shows that low expression of OrfAB in cis (pNABI) or its overexpression in trans (pBLOCK) can promote transposition. The same conclusion had been formulated in the case of IS903 (10). Another interesting feature is that OrfA is expressed from both pBLOCK and pNABI. The expression of OrfA has been shown to prevent transposition in the case of IS3 (34) or IS1 (23, 44). On the contrary, coexpression of OrfA and OrfAB increases transposition in the case of IS911 (39). As already mentioned (9), since we cannot accurately calculate transposition rates from our results, we do not know whether the expression of IS1397 OrfA actually reduced or increased transposition efficiency. We can only conclude that the phenomenon was not abolished. This point is currently being checked in more detail.
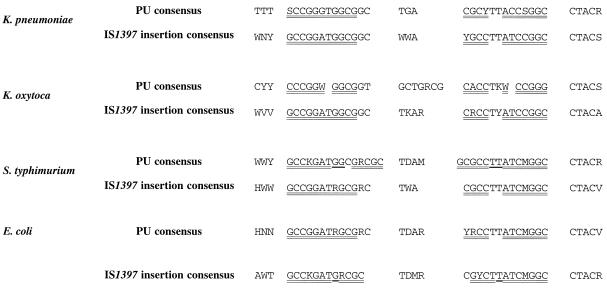
As previously described in the case of E. coli (9), we found a strong target specificity for IS1397 transposition in the three other species examined, since a large majority (80%) of the transposition sites were identified as PUs. PUs have been precisely described and classified in E. coli (3). The situation is not that clear and less well documented in the other species. Nonetheless, PU consensuses have been proposed for S. enterica serovar Typhimurium, K. pneumoniae, and K. oxytoca (2, 15). If PU sequences look very much conserved, clear differences between species can be observed and can be considered signatures of these different organisms. For instance, four PU types (Y, Z1, Z2, and D) can be distinguished in S. enterica serovar Typhimurium, whereas only three (Y, Z1, and Z2) can be found in E. coli. PUs in K. pneumoniae and K. oxytoca are more distantly related. We investigated K. pneumoniae sequences present in the contigs currently released by the GSC (http://genome.wustl.edu/gsc/index.shtml) by using the E. coli PU consensus and the consensus derived from the transposition sites of IS1397 in K. pneumoniae as query sequences for the BLAST (http://www.ncbi.nlm.nih.gov/) and FASTA (http://www.infobiogen.fr/) programs. The emerging consensus (Fig. 3 and 8) is actually rather different from E. coli. K. oxytoca PUs are less well documented due to the poor representation of sequences in currently available data banks. However, we could identify a general PU consensus for this species. This consensus is different from those of E. coli, S. enterica serovar Typhimurium, and even K. pneumoniae. A striking peculiarity of IS1397 insertion sites in the Klebsiella chromosome is that the target sequences are much closer to the consensus found in E. coli than to the general consensus found in these species. In particular, the sequence GCCGGATG is most commonly found upstream of the insertion point in eight cases) and GCCGGATA is found less often in two cases). Both sequences are found in the first part of the stem of the E. coli and Salmonella Y PU. It should also be mentioned that the equivalent region of the E. coli Z PU (GCCTGATG) has been found only once as a transposition target in K. oxytoca (Ko 1.2 in a non-PU site; Fig. 5), whereas it was the target in 9 out of 29 cases in E. coli (9). The relatively higher GC content of the Klebsiella genomes could explain this bias. However, this could not hold for S. enterica serovar Typhimurium, where we, interestingly, found five cases of insertions into Z PUs all containing GCCGGATG and never GCCTGATG, which was expected with equal probability. However, the small number of cases does not allow us to draw firm conclusions on this point. Interestingly, even in the few “aberrant” Klebsiella transpositions (i.e., not located in a PU, sometimes within an ORF; Fig. 3 and 5), GCCGGATG (exactly or with slight variations) is found upstream of the insertion point. The exact recognition site of IS1397 could thus be restricted to GCCGGATR or GCCTGATR. In Klebsiella, these sequences are not really expected to be found more frequently in PUs than in the rest of the chromosome since they are not part of the consensus. However, they are clearly preferred as transposition sites when they belong to a PU (62 to 81% of the cases), almost as frequently as in E. coli (9) or S. enterica serovar Typhimurium (86 and 100% of the cases, respectively).
FIG. 8.
Summary of PU consensuses and the transposable module insertion target consensus in Enterobacteriaceae. Data are taken from Fig. 3 to 5. The E. coli PU consensus is taken from reference 4. D is A, G, or T; H is A, C, or T; K is G or T; M is A or C; N is A, C, G, or T; S is C or G; R is A or G; V is A, C, or G; W is A or T; and Y is C or T.
From our results, we can conclude that IS1397 is specific for E. coli PUs. These PUs can be found frequently in S. enterica serovar Typhimurium and less frequently in Klebsiella and can be efficient targets for IS1397 in these cases. It will be interesting to study transposition in an enterobacterium which does not contain PUs but is phylogenetically close to E. coli. This is why we undertook the same type of work with Yersinia.
One relevant observation is that transposition hot spots were found. In K. oxytoca, we found an unidentified region three times (Ko 2.1, Ko 3.1, and Ko 8.1) and fucR-ygdE four times (Ko 6.1, Ko 7.1, Ko 7.2, and Ko 10.2). In K. pneumoniae, the region located after yjgA was found twice (Kp 6.1 and Kp 8.2). Such a phenomenon has already been observed in E. coli (9). It was more surprising that hot spots were also found between species, despite the lack of similitude between these intergenic regions. narU-narZ was found twice in K. pneumoniae (Kp 12.1 and Kp D) and once in K. oxytoca (Ko 12.1); yjiX-yjiY was found twice in S. enterica serovar Typhimurium (St 2.2 and St 6.1), four times in K. oxytoca (Ko 4.1, Ko 5.2, Ko 11.1, and Ko 12.2), and once in K. pneumoniae (Kp E). This suggests that mere sequence recognition by IS1397 transposase is not the only factor which determines transposition. Some local chromosomal features must exist, possibly shared by the three species investigated in this study, and these features are probably not related to PUs. If one considers the case of yjiX-yjiY, this intergenic region contains a solitary D PU in S. enterica serovar Typhimurium and is composed of two convergent PUs in K. oxytoca and K. pneumoniae that are similarly organized but differ in sequence. The nature of the potential for “attracting” IS1397 in these regions remains totally unknown.
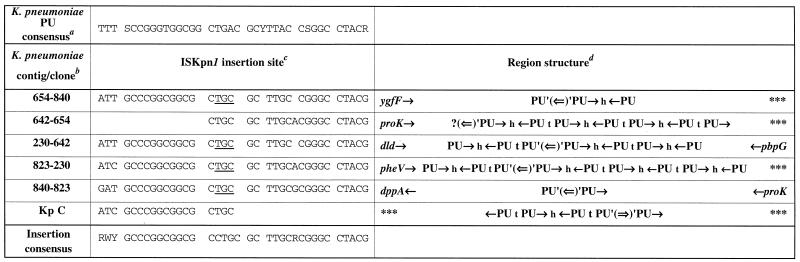
In this study, we identified a new insertion sequence in the K. pneumoniae MGH78578 genome (sequenced in St. Louis), named ISKpn1. This sequence was always found inserted into K. pneumoniae PUs (see below). We detected five copies of the IS in the contigs available in the database (May 2000). However, the flanking genes of the copy of ISKpn1 we cloned did not correspond to any of these contigs, suggesting misassemblies. We therefore checked the structure of IS flanking regions by PCR analysis. We tested each possible combination of primers and obtained fragments for only five combinations (Fig. 9). We also checked the sequences of flanking PUs and extra-PU motifs, which are extremely well conserved in each BIME. We then concluded that there were actually five copies of the IS with the same 3-bp duplication, TGC. Another example (with a partial sequence) was found in K. pneumoniae strain ATCC 13883, in which we studied IS1397 transposition. Indeed, IS1397 had been transposed into a BIME which already contained ISKpn1 (clone C; Fig. 3). Southern blot hybridization showed that ISKpn1 is only present in K. pneumoniae subsp. pneumoniae, K. pneumoniae subsp. ozonae, and K. aerogenes. This indicates that it is restricted to a small subset of species closely related to K. pneumoniae. The same observations have been made for the distribution of IS1397 in E. coli isolates (5).
FIG. 9.
Sites of insertion of ISKpn1 into the chromosome of K. pneumoniae. a, The PU consensus is derived from this study. b, The K. pneumoniae strain MGH78578 contig numbers were found at the GSC web site in May 2000 (http://genome.wustl.edu/gsc/index.shtml). The contigs have been split and reassembled by PCR analysis and BIME sequence comparison (see Discussion). In the case of 642–654, the beginning of the sequence is not available at the GSC web site. Clone Kp C is another example we sequenced after transposition of IS1397 in K. pneumoniae strain ATCC 13883 (cf. Fig. 3). c, Nucleotides which are duplicated after transposition are underlined. R is A or G, W is A or T, and Y is C or T. d, Single arrows indicate PU and gene orientations. Double arrows represent the orientation of ISKpn1; h and t represent head internal sequences (HIS) and tail internal sequences (TIS), respectively (Fig. 1). Gene names have been assessed by sequence homology to E. coli genes (http://genolist.pasteur.fr/Colibri/). Triple asterisks indicate a region for which no homolog has been identified.
Like IS1397, ISKpn1 is inserted into the central part of PUs with a 3-bp duplication. ISKpn1 is found in PUs which are specific to K. pneumoniae (Fig. 9) with a very high GC content and an additional C at position 7. On the contrary, as discussed above, IS1397 has been systematically transposed into K. pneumoniae PUs, which are closer to the E. coli consensus. The case of clone C (Fig. 3) is particularly interesting since it deals with a BIME where typical K. pneumoniae PUs alternate with PUs which are close to the E. coli consensus and which were the targets for ISKpn1 and IS1397, respectively.
ISKpn1 is a new member of the IS3 family, as shown by sequence comparisons. OrfB is known to contain the catalytic domain of the transposase, and OrfA has been demonstrated to recognize the inverted repeats of the IS specifically (see reference 24 for a review). We think that OrfA may confer the specificity of insertion. Figure 10 shows an alignment of the OrfA proteins from ISKpn1, IS150, and IS1397. ISKpn1 is closer in both its nucleotide and protein sequences to IS150 than to IS1397, although ISKpn1 and IS1397 are both inserted into PUs but not IS150. The C termini of the OrfA proteins from IS1397 and ISKpn1 share a conserved 13-aa motif, ELRYLRAENAYLK, which is not found in IS150. Thus, we can speculate that these amino acids play a role in the selection of PUs as transposition targets.
FIG. 10.
Alignment of OrfA proteins from IS150, IS1397, and ISKpn1. IS150, ISKpn1, and IS1397 were aligned by using the DNA strider alignment program (BLOCKS). Residues found to be common to all three ISs are boxed in black, and residues found to be common to of the two ISs are boxed in grey. Regions of homology among the three proteins are framed with solid lines, and regions of homology between IS150 and ISKpn1 or ISKpn1 and IS1397 are framed with dotted lines.
ACKNOWLEDGMENTS
We thank A. Cahen and F. Le Noanne for technical assistance. We thank the GSC, Washington University, St. Louis, Mo., for K. pneumoniae strain MGH78578 and for communication of DNA sequence data prior to publication. We are also thankful to P. A. D. Grimont for the other Klebsiella strains and the Salmonella strains and to E. Carniel for the Yersinia strains.
This work was supported by a grant from the Ministère Français de l'Education Nationale, de la Recherche et de la Technologie.
REFERENCES
- 1.Appleyard R K. Segregation of new lysogenic types during growth of a doubly lysogenic strain derived from E. coli K12. Genetics. 1954;39:440–452. doi: 10.1093/genetics/39.4.440. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Bachellier S, Perrin D, Hofnung M, Gilson E. Bacterial interspersed mosaic elements (BIMEs) are present in the genome of Klebsiella. Mol Microbiol. 1993;7:537–544. doi: 10.1111/j.1365-2958.1993.tb01144.x. [DOI] [PubMed] [Google Scholar]
- 3.Bachellier S, Saurin W, Perrin D, Hofnung M, Gilson E. Structural and functional diversity among bacterial interspersed mosaic elements (BIMEs) Mol Microbiol. 1994;12:61–70. doi: 10.1111/j.1365-2958.1994.tb00995.x. [DOI] [PubMed] [Google Scholar]
- 4.Bachellier S, Gilson E, Hofnung M, Hill C W. Repeated sequences. In: Neidhardt F C, Curtis III R, Ingraham J L, Lin E C C, Low K B, Magasanik B, Reznikoff W S, Riley M, Shaechter M, Umbarger H E, editors. Escherichia coli and Salmonella: cellular and molecular biology. 2nd ed. Washington, D.C.: American Society for Microbiology; 1996. p. 2012-2040. [Google Scholar]
- 5.Bachellier S, Clément J-M, Hofnung M, Gilson E. Bacterial interspersed mosaic elements (BIME) are a major source of sequence polymorphism in Escherichia coli intergenic regions including specific associations with a new insertion sequence. Genetics. 1997;145:551–562. doi: 10.1093/genetics/145.3.551. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Bachellier S, Clément J-M, Hofnung M. Short palindromic repetitive DNA elements in enterobacteria: a survey. Res Microbiol. 1999;150:627–639. doi: 10.1016/s0923-2508(99)00128-x. [DOI] [PubMed] [Google Scholar]
- 7.Boccard F, Prentki P. Specific interaction of IHF with RIBs, a class of bacterial repetitive DNA elements located at the 3′ end of transcription units. EMBO J. 1993;12:5019–5027. doi: 10.1002/j.1460-2075.1993.tb06195.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Chang A C Y, Cohen S N. Construction and characterization of amplifiable multicopy DNA cloning vehicles derived from the P15A cryptic miniplasmid. J Bacteriol. 1978;134:1141–1156. doi: 10.1128/jb.134.3.1141-1156.1978. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Clément J-M, Wilde C, Bachellier S, Lambert P, Hofnung M. IS1397 is active for transposition into the chromosome of Escherichia coli K-12 and inserts specifically into palindromic units of bacterial interspersed mosaic elements. J Bacteriol. 1999;181:6929–6936. doi: 10.1128/jb.181.22.6929-6936.1999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Derbyshire K M, Grindley N D. cis preference of the IS903 transposase is mediated by a combination of tranposase instability and inefficient translation. Mol Microbiol. 1996;21:1261–1272. doi: 10.1111/j.1365-2958.1996.tb02587.x. [DOI] [PubMed] [Google Scholar]
- 11.Dodd I B, Egan J B. Improved detection of helix-turn-helix DNA-binding motifs in protein sequences. Nucleic Acids Res. 1990;18:5019–5026. doi: 10.1093/nar/18.17.5019. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Espéli O, Boccard F. In vivo cleavage of Escherichia coli BIME-2 repeats by DNA gyrase: genetic characterization of the target and identification of the cut site. Mol Microbiol. 1997;26:767–777. doi: 10.1046/j.1365-2958.1997.6121983.x. [DOI] [PubMed] [Google Scholar]
- 13.Gilson E, Clément J-M, Brutlag D, Hofnung M. A family of dispersed repetitive extragenic palindromic DNA sequences in E. coli. EMBO J. 1984;3:1417–1421. doi: 10.1002/j.1460-2075.1984.tb01986.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Gilson E, Rousset J P, Clément J-M, Hofnung M. A subfamily of E. coli palindromic units implicated in transcription termination? Ann Inst Pasteur/Microbiol (Paris) 1986;137:259–270. doi: 10.1016/s0769-2609(86)80116-8. [DOI] [PubMed] [Google Scholar]
- 15.Gilson E, Clément J-M, Perrin D, Hofnung M. Palindromic units: a case of highly repetitive DNA sequences in bacteria. Trends Genet. 1987;3:226–230. [Google Scholar]
- 16.Gilson E, Perrin D, Hofnung M. DNA polymerase I and a protein complex bind specifically to E. coli palindromic units highly repetitive DNA: implications for bacterial chromosome organization. Nucleic Acids Res. 1990;18:3941–3952. doi: 10.1093/nar/18.13.3941. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Gilson E, Bachellier S, Perrin S, Perrin D, Grimont P A D, Grimont F, Hofnung M. Palindromic unit highly repetitive DNA sequences exhibit species specificity within Enterobacteriaceae. Res Microbiol. 1990;141:1103–1116. doi: 10.1016/0923-2508(90)90084-4. [DOI] [PubMed] [Google Scholar]
- 18.Gilson E, Saurin W, Perrin D, Bachellier S, Hofnung M. Palindromic units are part of a new bacterial interspersed mosaic element (BIME) Nucleic Acids Res. 1991;19:1375–1383. doi: 10.1093/nar/19.7.1375. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Higgins C F, Ferro-Luzzi Ames G, Barnes W M, Clément J-M, Hofnung M. A novel intercistronic regulatory element of prokaryotic operons. Nature. 1982;298:760–762. doi: 10.1038/298760a0. [DOI] [PubMed] [Google Scholar]
- 20.Katsuragi N, Takizawa N, Murooka Y. Entire nucleotide sequence of the pullulanase gene of Klebsiella aerogenes W70. J Bacteriol. 1987;169:2301–2306. doi: 10.1128/jb.169.5.2301-2306.1987. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Kulkosky F, Jones K S, Katz R A, Mack J P, Skalka A M. Residues critical for retroviral integrative recombination in a region that is highly conserved among retroviral/retrotransposon integrases and bacterial insertion sequence transposases. Mol Cell Biol. 1992;12:2331–2338. doi: 10.1128/mcb.12.5.2331. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Lim D, Gomes T A T, Maas W K. Distribution of msDNA among serotypes of enteropathogenic Escherichia coli strains. Mol Microbiol. 1990;4:1711–1714. doi: 10.1111/j.1365-2958.1990.tb00548.x. [DOI] [PubMed] [Google Scholar]
- 23.Machida C, Machida Y. Regulation of IS1 transposition by the insA gene product. J Mol Biol. 1989;208:567–574. doi: 10.1016/0022-2836(89)90148-4. [DOI] [PubMed] [Google Scholar]
- 24.Mahillon J, Chandler M. Insertion sequences. Microbiol Mol Biol Rev. 1998;62:725–774. doi: 10.1128/mmbr.62.3.725-774.1998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Metcalf W W, Jiang W, Wanner B L. Use of the rep technique for allele replacement to construct new Escherichia coli hosts for maintenance of R6K gamma origin plasmids at different copy numbers. Gene. 1994;138:1–7. doi: 10.1016/0378-1119(94)90776-5. [DOI] [PubMed] [Google Scholar]
- 26.Newbury S F, Smith N H, Higgins C F. Differential mRNA stability controls relative gene expression within a polycistronic operon. Cell. 1987;51:1131–1143. doi: 10.1016/0092-8674(87)90599-x. [DOI] [PubMed] [Google Scholar]
- 27.Newbury S F, Smith N H, Robinson E C, Hiles I D, Higgins C F. Stabilization of translationally active mRNA by prokaryotic REP sequences. Cell. 1987;48:297–310. doi: 10.1016/0092-8674(87)90433-8. [DOI] [PubMed] [Google Scholar]
- 28.Oppenheim A B, Rudd K E, Mendelson I, Teff D. Integration host factor binds to a unique class of complex repetitive extragenic DNA sequences in Escherichia coli. Mol Microbiol. 1993;10:113–122. doi: 10.1111/j.1365-2958.1993.tb00908.x. [DOI] [PubMed] [Google Scholar]
- 29.Polard P, Chandler M. Bacterial transposases and retroviral integrases. Mol Microbiol. 1995;15:13–23. doi: 10.1111/j.1365-2958.1995.tb02217.x. [DOI] [PubMed] [Google Scholar]
- 30.Sambrook J, Frisch E F, Maniatis T. Molecular cloning: a laboratory manual. 2nd ed. Cold Spring Harbor, N.Y: Cold Spring Harbor Laboratory Press; 1989. [Google Scholar]
- 31.Sekine Y, Ohtsubo E. DNA sequences required for translational frameshifting in production of the transposase encoded by IS1. Mol Gen Genet. 1992;235:325–332. doi: 10.1007/BF00279377. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Sekine Y, Nagasawa H, Ohtsubo E. Identification of the site of translational frameshifting required for production of the transposase encoded by insertion sequence IS1. Mol Gen Genet. 1992;235:317–324. doi: 10.1007/BF00279376. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Sekine Y, Eisaki N, Ohtsubo E. Translational control in production of transposase and in transposition of insertion sequence IS3. J Mol Biol. 1994;235:1406–1420. doi: 10.1006/jmbi.1994.1097. [DOI] [PubMed] [Google Scholar]
- 34.Sekine Y, Izumi K, Mizuno T, Ohtsubo E. Inhibition of transpositional recombination by OrfA and OrfB proteins encoded by insertion sequence IS3. Genes Cells. 1997;2:547–557. doi: 10.1046/j.1365-2443.1997.1440342.x. [DOI] [PubMed] [Google Scholar]
- 35.Shafferman A, Kolter R, Stalker D, Helinski D R. Plasmid R6K DNA replication. III. Regulatory properties of the pi initiation protein. J Mol Biol. 1982;161:57–76. doi: 10.1016/0022-2836(82)90278-9. [DOI] [PubMed] [Google Scholar]
- 36.Shyamala V, Schneider E, Ferro-Luzzi Ames G. Tandem chromosomal duplications: role of REP sequences in the recombination event at the joinpoint. EMBO J. 1990;9:939–946. doi: 10.1002/j.1460-2075.1990.tb08192.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Stalker D M, Kolter R, Helinski D R. Plasmid R6K DNA replication. I. Complete nucleotide sequence of an autonomously replicating segment. J Mol Biol. 1982;161:33–43. doi: 10.1016/0022-2836(82)90276-5. [DOI] [PubMed] [Google Scholar]
- 38.Stern M J, Prossnitz E, Ferro-Luzzi Ames G. Role of the intercistronic region in post-transcriptional control of gene expression in the histidine transport operon of Salmonella typhimurium: involvement of REP sequences. Mol Microbiol. 1988;2:141–152. doi: 10.1111/j.1365-2958.1988.tb00015.x. [DOI] [PubMed] [Google Scholar]
- 39.Ton-Hoang B, Polard L, Haren C, Turlan C, Chandler M. IS911 transposon circles give rise to linear forms that undergo integration in vitro. Mol Microbiol. 1999;32:617–627. doi: 10.1046/j.1365-2958.1999.01379.x. [DOI] [PubMed] [Google Scholar]
- 40.Varmus H, Brown P. Retrovirus. In: Berg D E, Howe M M, editors. Mobile DNA. Washington, D.C.: American Society for Microbiology; 1989. pp. 55–108. [Google Scholar]
- 41.Vögele K, Schwartz E, Welz C, Schiltz E, Rak B. High-level ribosomal frameshifting directs the synthesis of IS150 gene products. Nucleic Acids Res. 1991;19:4377–4385. doi: 10.1093/nar/19.16.4377. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Yang Y, Ferro-Luzzi Ames G. DNA gyrase binds to the family of prokaryotic repetitive extragenic palindromic sequences. Proc Natl Acad Sci USA. 1988;85:8850–8854. doi: 10.1073/pnas.85.23.8850. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Yanish-Perron C, Vieira J, Messing J. Improved M13 phage cloning vectors and host strains: nucleotide sequences of the M13mp18 and pUC19 vectors. Gene. 1985;33:103–119. doi: 10.1016/0378-1119(85)90120-9. [DOI] [PubMed] [Google Scholar]
- 44.Zerbid D, Polard P, Escoubas J M, Galas D, Chandler M. The regulatory role of the IS1-encoded InsA protein in transposition. Mol Microbiol. 1990;4:471–477. doi: 10.1111/j.1365-2958.1990.tb00613.x. [DOI] [PubMed] [Google Scholar]