Abstract
Analysis of the sequence immediate upstream of ohr revealed an open reading frame, designated ohrR, with the potential to encode a 17-kDa peptide with moderate amino acid sequence homology to the MarR family of negative regulators of gene expression. ohrR was transcribed as bicistronic mRNA with ohr, while ohr mRNA was found to be 95% monocistronic and 5% bicistronic with ohrR. Expression of both genes was induced by tert-butyl hydroperoxide (tBOOH) treatment. High-level expression of ohrR negatively regulated ohr expression. This repression could be overcome by tBOOH treatment. In vivo promoter analysis showed that the ohrR promoter (P1) has organic peroxide-inducible, strong activity, while the ohr promoter (P2) has constitutive, weak activity. Only P1 is autoregulated by OhrR. ohr primer extension results revealed three major primer extension products corresponding to the 5′ ends of ohr mRNA, and their levels were strongly induced by tBOOH treatment. Sequence analysis of regions upstream of these sites showed no typical Xanthomonas promoter. Instead, the regions can form a stem-loop secondary structure with the 5′ ends of ohr mRNA located in the loop section. The secondary structure resembles the structure recognized and processed by RNase III enzyme. These findings suggest that the P1 promoter is responsible for tBOOH-induced expression of the ohrR-ohr operon. The bicistronic mRNA is then processed by RNase III-like enzymes to give high levels of ohr mRNA, while ohrR mRNA is rapidly degraded.
During bacterial interactions with hosts, bacteria are exposed to host defense responses, including increased concentrations of reactive oxygen species (ROS), such as H2O2, organic peroxide, and superoxide anion (5, 14). In addition, normal aerobic respiration produces significant levels of ROS (10, 11). ROS are toxic to biological systems and must be removed rapidly. Among different ROS, organic peroxides are highly toxic, partly due to the abilities of these compounds to participate in free radical reactions which generate reactive organic radicals by reacting with membranes and other macromolecules (11).
Bacteria have evolved complex systems for sensing, protection, and regulation against organic peroxide toxicity. Alkyl hydroperoxide reductase is the best-characterized enzyme system involved in metabolizing toxic organic peroxides to the less toxic organic alcohols (7, 24, 25). In Escherichia coli, the gene for the catalytic subunit, ahpC, has an interesting pattern of expression. Its expression is regulated by OxyR, a global peroxide sensor and transcriptional regulator (30, 32), and is highly inducible by various oxidants (16, 19, 27). In Xanthomonas campestris pv. phaseoli, ahpC is differentially regulated by OxyR. Reduced OxyR represses while oxidized OxyR activates ahpC expression (15, 16, 19). The mechanism for protection against organic peroxides in X. campestris pv. phaseoli is complex. In addition to the ahpC and catalase peroxidase systems, an organic hydroperoxide resistance (ohr) gene also provides protection against organic peroxide toxicity (20). Inactivation of ohr in Xanthomonas and several other bacteria results in increased susceptibility to organic peroxide toxicity (4, 9, 20, 22, 26).
ohr has unique patterns of oxidative stress-induced expression, unlike other genes involved in protection against oxidative stress. In several bacteria, ohr expression is highly induced by treatment with low concentrations of organic peroxides (4, 9, 20, 22). In contrast, exposure to other oxidants or stresses does not induce ohr expression (2, 9, 20, 22). The regulator of ohr expression has not been identified, but atypical patterns of gene expression suggest that a novel regulator may be involved in the process. Since ohr is widely distributed among diverse groups of gram-positive and gram-negative bacteria (4), understanding the regulatory mechanisms is important. Analyses of primary structures of Ohr homologues, alterations in the physiological properties of their mutants, and patterns of expression of the genes together suggest that Ohr probably belongs to a novel family of proteins involved in organic peroxide protection (4). At present, the biochemical mechanism of Ohr-mediated protection is not known.
In this communication, we identify a negative regulator of X. campestris pv. phaseoli ohr, OhrR. ohrR is located upstream of and forms an operon with ohr. The gene product, OhrR, functions as a negative regulator of ohr expression. Transcriptional analysis of both genes suggests that expression of ohr is regulated from a distant ohrR promoter and also involves an RNA processing step.
MATERIALS AND METHODS
Culture conditions and oxidant treatments.
Xanthomonas strains were grown aerobically in Silva-Buddenhagen medium (0.5% sucrose, 0.5% yeast extract, 0.5% peptone, 0.1% glutamic acid [pH 7.0]) at 28°C. tert-Butyl hydroperoxide (tBOOH)-induced ohr expression was achieved by the addition of 200 μM tBOOH to Xanthomonas log-phase cultures (19). The induction times for Western and primer extension experiments were 30 and 15 min, respectively.
Phylogenetic analysis.
A phylogenetic tree was constructed by the neighbor-joining method using the Tree program from the phylogenetic analysis page at http://igs-server.cnrss-mrs.fr/anrs/phylogenetics. The results were drawn using the program PHYLODENDRON (version 0.8d 1999; Department of Biology, University of Indiana [http://iubio.bio.indiana.edu]).
Northern analysis of ohr, ohrR, and ahpC.
Total RNAs from uninduced and tBOOH-induced cultures of X. campestris pv. phaseoli were purified using the hot phenol method (16, 17). Ten micrograms of purified RNA was loaded into each lane of formaldehyde agarose gels, and RNA samples were electrophoretically separated. Separated RNA samples were transferred to nylon membranes. The membranes were exposed to various probes using prehybridization, hybridization, and high-stringency washing conditions as previously described (16, 19). ohrR-specific probes were prepared from plasmid pohrR digested with SacI and KpnI. The 250-bp fragment was purified from an agarose gel. ohr- and ahpC-specific probes were prepared from plasmids pohr and pahpC, respectively, as previously described (19, 20). The gene-specific DNA fragments were radioactively labeled using a random primer kit and [α-32P]dCTP.
RT-PCR of ohrR-ohr mRNA.
Reverse transcription (RT) of ohrR-ohr mRNA was performed to confirm the bicistronic transcriptional organization of these genes. Briefly, RNA was isolated from tBOOH-induced X. campestris pv. phaseoli cultures using the hot acid-phenol method (19). Purified RNA was treated with 10 U of RNase-free DNase for 30 min to remove contaminating DNA. Primer ohr5′ (5′GCATCGGCCTCTTCGTTGGAC3′) was mixed with 10 μg of RNA, and 200 U of cloned Moloney murine leukemia virus reverse transcriptase was added. The mixture was incubated at 42°C for 60 min. Then, 5 μl of the reaction mixture was added to a PCR containing primers ohr5′ and ohrR3′ (5′GTCGAGCGCCTTGTCCGAGGA3′). PCR was performed using previously described conditions for 35 cycles, and PCR products were analyzed in an agarose gel (19).
Western analysis of Ohr and Cat.
Cell lysates were prepared from X. campestris pv. phaseoli cultures as previously described (20). Twenty micrograms of protein was loaded into each lane and separated by sodium dodecyl sulfate-polyacrylamide gel electrophoresis. Separated protein samples were transferred to nitrocellulose membranes by electroblotting (20). Immunological reactions with an anti-Ohr or an anti-Cat antibody were done as described by Mongkolsuk et al. (19, 20). The antibody reactions were detected using an alkaline phosphatase-conjugated goat anti-rabbit antibody. Subsequent detection of alkaline phosphatase activity was done using a kit from Promega in accordance with the instructions of the manufacturer.
Construction of pBBRohrR and pBBRohrRBs.
pohr (20) was digested with SfiI. The ends of the fragment were filled in with DNA polymerase I, and the fragment was redigested with SacI. The 550-bp fragment containing ohrR was purified from an agarose gel and ligated into pBBR1MCS-5 (13) digested with SmaI and SacI to give pBBRohrR. A frameshift mutation in ohrR was created by digesting pBBRohrR with BstEII located in the coding region, the ends of the fragment were filled in with DNA polymerase I, and the ends were religated. This procedure gave pBBRohrRBs.
Construction of X. campestris pv. phaseoli ohrR1, ohrR2, and ohrR3 mutants.
The 65-bp PstI-SacII and 211-bp PstI fragments from restriction enzyme-digested pohr were electrophoretically separated from other DNA fragments using a 1.5% agarose gel. The purified DNA fragments were recovered from the gel and cloned into similarly digested vector pUC18tet. This procedure gave pUCohrR1 and pUCohrR2. pohr was digested with PstI-HincII, and the 615-bp fragment was purified by electrophoresis and recovered from an agarose gel. The purified fragment was cloned into similarly digested pUC18Km to give pUCohrR3 (see Fig. 3). pUCohrR1, pUCohrR2, and pUCohrR3 were electroporated into X. campestris pv. phaseoli using previously described conditions (21), resulting in XpohrR1, XpohrR2, and XpohrR3, respectively. Transformants with pUCohrR1 and pUCohrR2 were selected for Tetr (15 μg/ml), while transformants with pUCohrR3 were selected for Kmr (15 μg/ml). Genomic DNA was isolated from these transformants and digested with appropriate restriction enzymes. After electrophoretic separation, the DNA fragments were hybridized with ohrR and pUC18 as probes (data not shown) to confirm proper integration of the plasmid into the X. campestris pv. phaseoli chromosome.
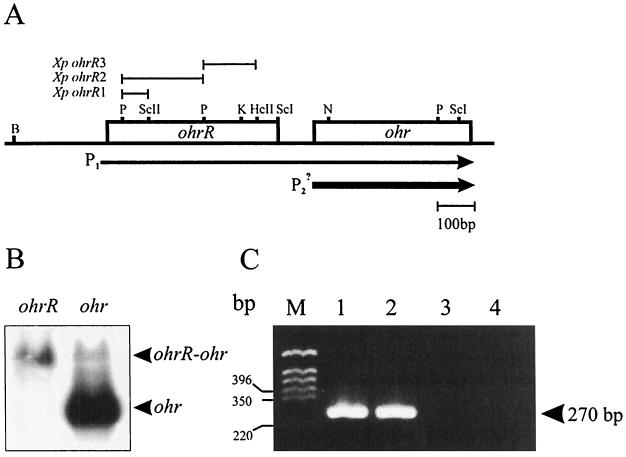
FIG. 3.
Gene order and transcriptional organization of ohrR-ohr. (A) The bars above the map of ohrR-ohr indicate the locations and sizes of the fragments used in the construction of ohrR mutants (Xp designations). The sizes and directions of the arrows represent the amounts and directions of transcription, respectively. Hc, HincII; K, KpnI; N, NotI; P, PstI; ScI, SacI; ScII, SacII. (B) Northern analysis of ohrR and ohr. Ten micrograms of RNA samples from tBOOH-induced cultures were separated in formaldehyde-agarose gels, and the RNA was transferred to nylon membranes. The membranes were hybridized separately to radioactively labeled ohrR or ohr probes. The arrowheads indicate monocistronic ohr mRNA and bicistronic ohrR-ohr mRNA. (C) RT-PCR of RNA samples from tBOOH-induced X. campestris pv. phaseoli cultures. RNA extraction and DNase I treatment were done as described in Materials and Methods. The conditions for PCR and the primers used are described in Materials and Methods. Lane M, molecular weight markers; lane 1, PCR of a positive control DNA sample; lane 2, RT-PCR of an RNA sample from tBOOH-induced X. campestris pv. phaseoli cultures; lane 3, the same RNA sample and PCR conditions as in lane 2 except that the RT step was omitted; lane 4, PCR of reagents (negative control).
Construction of pP1 and pP2.
pohr (20) was digested with Acc65I, the ends of the fragment were filled in with DNA polymerase I, and the blunt-ended DNA was redigested with BamHI. The 615-bp fragment containing P1 was separated by electrophoresis, purified from the agarose gel, and cloned into BglII-SmaI-digested promoter probe vector pUFR027cat-km (28). This procedure generated pP1 and placed the ohrR promoter in front of a promoterless cat gene. pP2 was constructed by digesting pohr (20) with NotI, filling in the ends of the fragment with DNA polymerase I, and redigesting the blunt-ended DNA with SacI. The 145-bp fragment containing P2 was recovered from the agarose gel after electrophoretic separation and cloned into SacI-SmaI-digested pUFR027catKm to give pP2.
ohr primer extension.
RNA was extracted as described above for Northern analysis (16, 17). In addition, purified RNA samples were treated with 10 U of RNase-free DNase for 30 min. Primer ohrP1 (5′GTCGAGCGCCTTGTCCGAGGA3′), located 70 bp from the translation initiation codon of ohr, was radioactively labeled using T4 polynucleotide kinase and [32P]ATP. Briefly, 10 μg of DNase I-treated RNA was added to a reverse transcriptase reaction mixture. The reaction was started by the addition of 200 U of Moloney murine leukemia virus reverse transcriptase. Products of the reaction were analyzed on sequencing gels. The sequence ladders were made using an fmol sequencing kit, ohrR1-labeled primer, and pohr (20) as the template.
Nucleotide sequence accession number.
The nucleotide sequence of ohrR has been deposited in GenBank under accession number AF036166.
RESULTS
ohr is not regulated by OxyR.
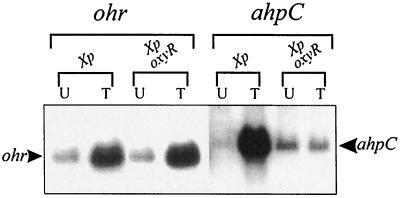
In Xanthomonas spp., Pseudomonas aeruginosa, Deinococcus radiodurans, and Bacillus subtilis, ohr expression is strongly induced by exposure to organic peroxides (tBOOH and cumene hydroperoxide [CuOOH]) but not by exposure to other oxidants and stresses (4, 9, 20, 22). This pattern of induced expression appears to be conserved in various bacteria and is unique to members of the ohr family (4). Understanding the regulatory mechanisms of ohr is likely to be generally important due to the wide distribution of ohr homologues among gram-negative and gram-positive bacteria (4, 9, 20, 22, 26). OxyR, a peroxide sensor and transcriptional regulator, is a potential regulator for organic peroxide-inducible expression of ohr. For Xanthomonas spp., it has been shown that OxyR-regulated genes are highly induced by tBOOH, suggesting that it may also be involved in sensing organic peroxides (16, 19). First, we tested whether OxyR is involved in the regulation of ohr. Total RNAs isolated from uninduced and tBOOH-induced cultures of X. campestris pv. phaseoli and an oxyR mutant (21) were probed with radioactively labeled ohr or ahpC gene-specific probes. ahpC expression was used as a positive control for an OxyR-regulated gene (19). The results of Northern analysis showed that ohr expression was highly induced by tBOOH to similar levels in both the oxyR mutant and the parent strain (Fig. 1). As expected, ahpC expression was highly induced by tBOOH only in the parent strain (Fig. 1). The data prove that OxyR is not the regulator of ohr.
FIG. 1.
OxyR-independent tBOOH induction of ohr. Northern analysis of ohr and ahpC expression in uninduced (U) or tBOOH-induced (T) cultures of X. campestris pv. phaseoli (Xp) and an oxyR mutant (Xp oxyR) is shown. The arrowheads to the left and right indicate the positions of ohr and ahpC mRNAs, respectively.
Identification of a putative ohr regulator, ohrR.
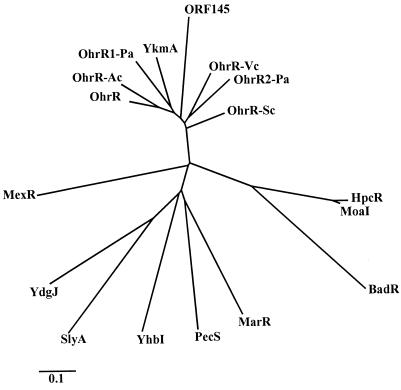
A search for a tBOOH-responsive regulator of ohr was initiated. During the analysis of ohr homologues in bacteria for which genome sequences have been completed, such as B. subtilis, P. aeruginosa, and D. radiodurans, we noticed that adjacent to the ohr homologues, there were open reading frames (ORFs) encoding proteins with moderate amino acid sequence identities to members of a family of negative regulators of gene expression including E. coli MarR (1, 2). These ORFs were candidates for regulators of ohr. Thus, additional sequencing upstream and downstream of X. campestris pv. phaseoli ohr was undertaken. Analysis of the nucleotide sequence immediately upstream of X. campestris pv. phaseoli ohr revealed an ORF encoding a 17-kDa peptide with 18% identity to E. coli MarR. We designated this ORF ohrR. The amino acid sequence of OhrR was used to search databases. These searches revealed two groups of related proteins. One group contains closely related proteins of unknown functions with amino acid identities ranging from 32 to 54% in both gram-positive and gram-negative bacteria. The genes for most members of this group are located adjacent to ohr homologues. We have designated these unknown proteins OhrR homologues. The second group has less identity to OhrR (18 to 22%). Members of this group include E. coli MarR and other known negative regulators of gene expression (1, 2, 8).
The amino acid sequences of both groups of homologues were used to construct a phylogenetic tree (Fig. 2). Analysis of the tree supported the idea that OhrR homologues belong to a larger and more diverse MarR family of transcriptional repressors. The highly conserved MarR amino acid sequence motif D-X-R-X5-L/I-T-X2-G, where X represents any amino acid (2), was found in all OhrR homologues. In addition, it was possible to extend the highly conserved MarR motif to L/M-X3-G-X3-R-X5-D-X-R-X5-L-T-X2-G by comparing members of the OhrR and MarR families. At present, the function of the conserved motif has not been clearly established.
FIG. 2.
Phylogenetic tree for OhrR and other members of the MarR family. Analysis and construction of the tree were performed as described in Materials and Methods. Proteins, GenBank accession numbers (in parentheses), and organisms are as follows: BadR (U75363), Rhodopseudomonas palustris; HpcR (S56952), E. coli; MarR (P27245), E. coli; MexR (U23763), P. aeruginosa; MoaI (D63524), Klebsiella aerogenes; OhrR (AF036166), X. campestris pv. phaseoli (this study); OhrR-Ac (Y09102), Acinetobacter sp.; OhrR1-Pa (D83290) and OhrR2-Pa (G83292), P. aeruginosa; ORF145 (Y13052), Staphylococcus sciuri; OhrR-Vc (B82389), V. cholerae; OhrR-Sc (AL163672), S. coelicolor; PecS (P42195), Erwinia chrysanthemi; SlyA (P40676), Salmonella enterica serovar Typhimurium; YdgJ (D69783), YhbI (Z99108), and YkmA (E69857), B. subtilis. The bar indicates genetic distance.
The ohrR-ohr gene order in various Xanthomonas strains was determined by PCRs using a primer set located in the 3′ region of ohrR and the 5′ region of ohr and genomic DNAs from various Xanthomonas strains. Analysis of DNA fragments generated by the PCRs showed that the ohrR-ohr gene organization was conserved among all the Xanthomonas strains tested (data not shown). The availability of bacterial genome and gene sequences in various databases allowed us to determine whether the ohrR-ohr gene organization was also conserved in other bacteria. The analysis revealed that in Acinetobacter calcoaceticus, D. radiodurans, P. aeruginosa, Vibrio cholerae, Streptomyces coelicolor, and X. campestris pv. phaseoli, ohrR is located immediately upstream of ohr (Fig. 3A). The organization in B. subtilis is slightly different, in that ohrR (ykmA) is located between two ohr homologues, yklA and ykzA (9, 31). P. aeruginosa is an exception; it has two different copies of ohrR, one copy (ohrR1-Pa) located upstream of ohr and another copy (ohrR2-Pa) located downstream of a glutathione peroxidase gene (gpx).
Transcriptional organization of ohrR-ohr.
Next, we examined the transcriptional organization of X. campestris pv. phaseoli ohrR and ohr. Northern analysis showed that ohr is transcribed as a 0.5-kb monocistronic mRNA (Fig. 1). ohrR was used to probe RNA isolated from tBOOH-induced cultures. The results showed that the ohrR probe hybridized to a 1.0-kb mRNA (Fig. 3B). This mRNA is much longer than the coding region of ohrR but is similar in size to the expected bicistronic ohrR-ohr mRNA. However, this explanation contradicted the results of the ohr Northern analysis (Fig. 1). To clarify the issue, Northern experiments using the ohr probe were repeated. Longer exposure for the ohr Northern hybridization revealed an additional positive reaction of ohr mRNA with 1.0-kb as well as 0.5-kb mRNA species (Fig. 3B). The former corresponded to the length of the expected bicistronic ohrR-ohr mRNA. More than 90% of ohr mRNA was monocistronic, while the remainder corresponded to the ohrR-ohr bicistronic form (20).
To confirm the identity of the putative operonic ohrR-ohr mRNA, it was analyzed by RT-PCR. A PCR primer set located 3′ of the ohrR and 5′ of the ohr coding regions was added to cDNA obtained by RT of total RNA from a tBOOH-induced culture. Analysis of DNA fragments from the PCRs showed the expected 270-bp fragment when the cDNA and a control Xanthomonas genomic DNA were used as templates (Fig. 3C). The 270-bp fragment was not detected in PCRs with the same RNA sample but with the RT step omitted (Fig. 3C).
Effect of OhrR on ohr expression.
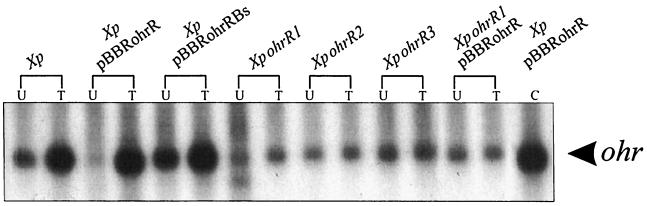
OhrR belongs to a family of negative regulators of gene expression (Fig. 2); thus, we investigated its effect on ohr expression. pBBRohrR was electroporated into X. campestris pv. phaseoli, and the levels of ohr expression in the transformants were determined. Northern analysis clearly showed that high-level expression of ohrR resulted in more than a 10-fold reduction in uninduced ohr mRNA levels (Fig. 4), while ohr expression was fully induced by exposure to tBOOH or CuOOH in both nontransformed X. campestris pv. phaseoli and the strain harboring pBBRohrR (Fig. 4). In contrast, Northern analysis of X. campestris pv. phaseoli carrying pBBRohrRBs, an ohrR frameshift mutant of pBBR1MCS-5, showed no repression of ohr expression (Fig. 4). The data provide strong evidence for the role of OhrR as a negative regulator of ohr expression.
FIG. 4.
Northern analysis of the effects of ohrR on ohr expression. Northern blotting of various X. campestris pv. phaseoli cells (Xp designations) was performed as described in Materials and Methods. The Northern blot was probed with radioactively labeled ohr. U, uninduced; T, tBOOH induced; C, CuOOH induced.
Expression from ohrR and ohr promoter fusions.
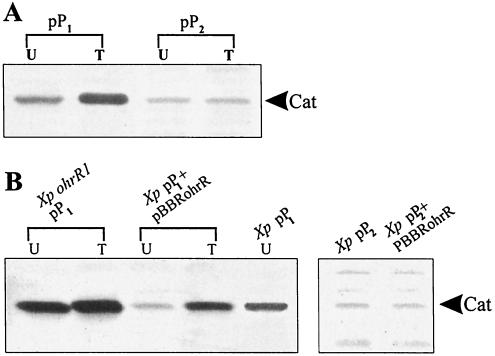
Northern analysis of ohrR and ohr expression suggested that ohrR and ohr should have weak and strong inducible promoters, respectively. Thus, the promoter activities of both genes were examined in vivo. Plasmids pP1 and pP2, containing the ohrR and ohr promoters, respectively, in front of the reporter gene, cat, in a promoter probe vector were transformed into X. campestris pv. phaseoli. Western analysis of Cat levels in the pP1- and pP2-containing strains gave unexpected results: pP1 directed tBOOH-inducible high Cat levels, whereas pP2 directed constitutive low Cat levels (Fig. 5A). These results suggest that P1 is responsible for the tBOOH-inducible expression of both ohrR and ohr, whereas ohr has a weak promoter, conclusions that contradict those drawn from the Northern analysis.
FIG. 5.
In vivo ohrR and ohr promoter analysis. Cat levels were determined by Western immunoblotting performed as described in Materials and Methods. Forty micrograms of total protein was loaded in each lane. U and I, lysates prepared from uninduced and tBOOH-induced cultures, respectively. (A) Analysis of in vivo promoter activities of ohrR (pP1) and ohr (pP2). (B) Effects of OhrR on pP1 and pP2. Western analysis of Cat levels in various strains (X. campestris pv. phaseoli [Xp] and ohrR mutant [Xp ohrR1] harboring pP1 or pP2 and with or without pBBRohrR) is shown.
OhrR is involved in autoregulation and tBOOH-induced expression from the ohrR promoter.
The effects of OhrR on ohr expression (Fig. 4) and the results of in vivo analysis of ohrR and ohr promoter activities (Fig. 5A) raised several questions regarding ohr repression and derepression mechanisms. Accordingly, experiments were undertaken to determine the consequence of high-level expression of ohrR on the P1 and P2 promoters. X. campestris pv. phaseoli harboring pP1 or pP2 was transformed with pBBRohrR, and Cat levels in the transformants were determined. The results showed that uninduced Cat levels in cells harboring pP1 and pBBRohrR were severalfold lower than those in cells harboring pP1 alone (Fig. 5B). In contrast, the repression of cat expression by pBBRohrR was relieved by tBOOH treatment; similar Cat levels were detected in tBOOH-induced cultures of X. campestris pv. phaseoli cells harboring pP1 and pBBRohrR or the vector alone (Fig. 5A and B). As expected from these results, the frameshift mutation in ohrR (pBBRohrRBs) eliminated the repression of P1 (data not shown). pBBRohrR had no effect on pP2 (data not shown).
We extended these observations by examining the promoter activities specified by pP1 and pP2 in an ohrR mutant. Densitometer analysis of Cat levels specified by pP1 in an uninduced ohrR mutant were similar to tBOOH-induced levels in the parent strain harboring the plasmid. The Cat levels in the mutant were at least fourfold higher than the levels in the uninduced parent strain. Moreover, the expression of cat from the promoter was not inducible by tBOOH in the ohrR mutant (Fig. 5B). These findings were the first indication that ohrR was required for organic peroxide-induced expression of the ohrR-ohr system. P2 promoter activity was not affected by mutations in ohrR (data not shown).
Inducible expression of ohr might involve RNA processing of ohrR-ohr transcripts.
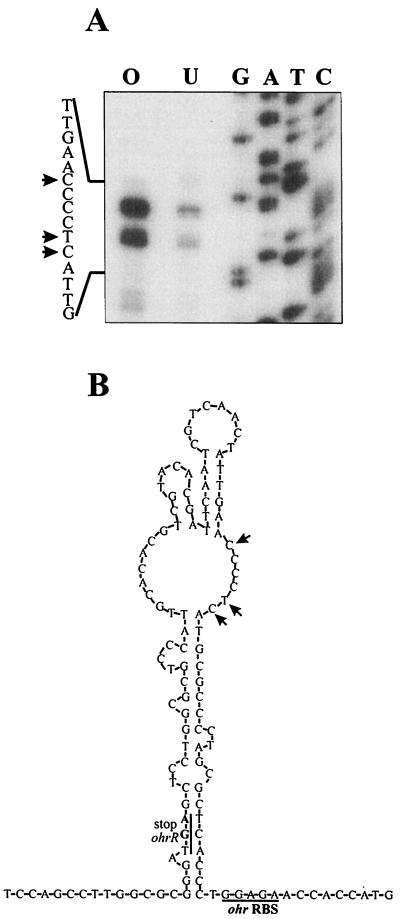
Northern analysis identified a stable 0.5-kb ohr transcript that is presumably processed from a longer, 1-kb bicistronic ohrR-ohr transcript. Primer extension experiments were done to locate the 5′ end of abundant ohr mRNA and also to determine if tBOOH exposure influenced the amounts of primer extension products. Three primer extension products were recovered (Fig. 6). The amounts of these products increased 10-fold when RNA from tBOOH-induced cultures were used (Fig. 6). The locations of primer extension products are shown in Fig. 6. Analysis of nucleotide sequences in the region showed that a stem-loop secondary structure could form upstream of ohr with the 5′ ends of ohr mRNA located in the loop section. This structure could be involved in the processing of bicistronic ohrR-ohr mRNA (Fig. 6).
FIG. 6.
Primer extension analysis of ohr mRNA and proposed processing sites of bicistronic ohrR-ohr mRNA.(A) Primer extension was performed with 10 μg of RNA isolated from uninduced (U) or tBOOH-induced (O) X. campestris pv. phaseoli cultures. G, A, T, and C are the sequence ladder. (B) Stem-loop secondary structure of the region around the putative RNA processing sites of the ohrR-ohr bicistronic mRNA. RBS, ribosome binding site. The arrows mark the locations of primer extension products in panel A and the locations of putative RNA processing sites in panel B.
Mutations in ohrR are polar.
In initial studies to determine the physiological roles of ohrR in organic peroxide resistance, insertional inactivation of the gene was generated using pUCohrR1 (Fig. 3A). In theory, inactivation of a putative negative regulator of ohr should result in a higher level of expression of ohr and thus a higher level of resistance to tBOOH. Accordingly, using the zone-of-growth-inhibition technique, the tBOOH and CuOOH resistance levels in XpohrR1 mutant strain and the parent strain were determined and found to be 25 and 18 mm for tBOOH and 27 and 19 mm for CuOOH, respectively. The results indicate that the mutant was more susceptible to tBOOH and CuOOH. Two additional disruptants of ohrR, generated with pUCohrR2 and pUCohrR3, gave similar results (data not shown). Moreover, pBBRohrR was unable to complement the phenotype. No alterations in the levels of resistance to H2O2, a superoxide generator (menadione), were observed in ohrR mutants (data not shown). The tBOOH-sensitive phenotype and the inability to complement the ohrR mutation suggest that the integration of pUCohrR1, pUCohrR2, and pUCohrR3 is polar on ohr. Northern experiments were done to determine the effect of ohrR mutations on ohr expression. The results showed lower constitutive expression of ohr in all three mutants (Fig. 4), consistent with the idea that transcription of the gene normally initiated from P1 terminates in some parts of the integrated plasmids. The low constitutive ohr mRNA levels in the ohrR mutant strains (Fig. 5) may have been due to transcription initiation from a plasmid promoter. Low ohr expression levels in ohrR mutants accounted for the reduced tBOOH-resistant phenotype of the mutants.
DISCUSSION
Organic peroxide-inducible expression of ohr expression in an oxyR mutant demonstrated that the process is independent of the global peroxide sensor and regulator OxyR (Fig. 1). This finding was the first clear indication of the existence of an additional regulatory system(s) that responds to organic peroxides. Identification of ohrR upstream of ohr suggested that it might encode a putative negative transcriptional regulator. The phylogenetic analysis (Fig. 2) revealed that OhrR homologues comprise a group of highly conserved and widely distributed proteins found in both gram-positive and gram-negative bacteria. The gene order ohrR-ohr also shows a high degree of conservation. The analyses of transcriptional organization of X. campestris pv. phaseoli ohrR-ohr by Northern blotting and RT-PCR show that these genes are coregulated and have an atypical transcriptional organization. ohrR mRNA was found as a bicistronic message with ohr, while ohr mRNA was found in both bicistronic and monocistronic forms. The monocistronic form of ohr mRNA has been observed in diverse bacteria, such as B. subtilis (9, 31), D. radiodurans, and P. aeruginosa (4, 22). Determination of ohrR and ohr transcriptional organization is seen as crucial to an understanding of the complex regulation of the expression of both genes in Xanthomonas.
OhrR is a negative regulator of ohr expression.
Identification of OhrR as a member of the E. coli MarR family of negative regulators of gene expression suggested that OhrR probably functions in a fashion similar to that of other MarR family members. However, some members of the MarR family, such as BadR (8) and SlyA (6), have been shown to act as positive regulators of their target genes; one member of the family, MexR, can act as both a negative regulator and a positive regulator (23). Nonetheless, the majority of MarR family members are transcriptional repressors (2). The working assumption that OhrR is a negative regulator of ohr was supported by the finding that the high-level expression of ohrR resulted in the repression of ohr expression (Fig. 4). The loss of repression as a consequence of a frameshift mutation in ohrR further supported the role of OhrR as a negative regulator (Fig. 4). Similar observations have been made for other bacteria, where high-level expression of MarR family members results in the repression of their target genes (1, 2, 8, 18).
A unique feature of MarR family members is the aromatic ligands recognized by these proteins. Although these are structurally diverse, all of them contain at least an aromatic ring (1, 2). It is believed that these ligands bind to the negative regulators and inactivate them, hence allowing increased expression of the target genes (1, 2). We showed that the repression of ohr by OhrR can be relieved by exposing the cells to CuOOH and tBOOH, presumably by inactivation of OhrR by these ligands (Fig. 4). Hence, OhrR probably recognizes tBOOH, a nonaromatic compound, as a ligand. In Xanthomonas, tBOOH and CuOOH (an aromatic compound) induce ohr expression equally well (20). Alternatively, organic peroxides might directly oxidize OhrR, leading to inactivation of the protein. Experiments are in progress to purify OhrR to examine the effect of tBOOH binding on OhrR function.
ohr expression probably involves processing of a bicistronic transcript.
ohr primer extension experiments showed three major primer extension products corresponding to three 5′ ends of the mRNA. All three primer extension products showed 10-fold increases in expression when RNA samples from tBOOH-treated cultures were used as templates (Fig. 6). Accordingly, we searched the sequences upstream of the 5′ ends of ohr mRNA for a possible P2 promoter. Examination of the sequences upstream of the three major primer extension products identified the sequences TTGCAC and GATTCA, which show five of six matches to the Xanthomonas promoter consensus sequence at −35 and −10, respectively (12). However, these putative promoter sequences are separated by only 11 bp and so are unlikely to function as an efficient promoter in vivo. Analysis of ohr primer extension results failed to show a constitutive primer extension product, although analysis of the P2 promoter in vivo revealed weak constitutive activity. This could have been due to a very low expression level that even the primer extension technique was unable to detect for the transcription start site. Alternatively, the weak P2 activity could have been an artifact from the cloning of the P2 promoter fragment into the promoter probe vector.
An alternative explanation for the Northern blot and primer extension results is that ohrR-ohr is transcribed as a two-gene operon from the ohrR promoter (P1) as the bicistronic mRNA is processed. The ohrR-ohr intercistronic region (98 bp; Fig. 6B) is unusually long, suggesting that the region could be involved in the regulatory process. Examination of the sequence surrounding the 5′ ends of ohr reveals that the nucleotide sequence in this region has the potential to form a stem-loop secondary structure with the three sites defining the 5′ ends of ohr mRNA located in the loop (Fig. 6B). The potential secondary structure of the mRNA sequence at this point is similar to the RNase III processing site (3). RNase III recognizes stem-loop structures and usually cleaves the mRNA in the internal loop (3). In E. coli, RNase III processing has been shown to affect the rate of mRNA degradation and to increase or decrease the levels of gene expression (29). Thus, it is likely that the ohrR-ohr mRNA is processed by an RNase III-like enzyme(s). We propose that processing results in the production of the 0.5-kb ohr mRNA and the rapid degradation of ohrR mRNA (Fig. 3B). The inability to detect the monocistronic form of ohrR mRNA supports this idea and also suggests that the processed ohrR mRNA is less stable than ohr mRNA. This would reduce the level of translation of ohrR mRNA and hence reduce the production of OhrR. Furthermore, OhrR levels would be kept low by autoregulation of the ohrR promoter by OhrR. Thus, in uninduced cells OhrR would be maintained at low levels.
The characteristics of P1, namely, organic peroxide inducibility and strong activity, fit the observed effects of organic peroxides on ohrR and ohr expression. In addition, the lack of a strong inducible promoter in front of ohr favors the idea that P1 is responsible for the organic peroxide-inducible expression of both ohrR and ohr (Fig. 3B). This explanation can be extended to account for the polar effects of ohrR insertional mutations on ohr expression. The physical separation of the ohrR promoter from ohr by insertion of pUCohrR1, pUCohrR2, and pUCohrR3 into ohrR prevented the organic peroxide induction of ohr.
OhrR is required for tBOOH-induced expression from P1.
A question arises as to whether OhrR is required for tBOOH induction of ohrR-ohr. ohrR promoter activity (P1) was constitutively high in an ohrR mutant (Fig. 5B), indicating that OhrR is not involved in the activation of operon expression. P1 could be repressed by OhrR (Fig. 5B), implying that OhrR is required to maintain the uninduced operon at low levels. This repression could be alleviated by exposure to tBOOH. These results strongly suggest that tBOOH-induced expression of the operon is due to derepression of P1. The derepression mechanism involving the inactivation of OhrR by tBOOH is likely to be the major step in organic peroxide-induced ohr expression. However, at present we cannot conclusively rule out that another, activating transcription factor also is involved in the induction process. The possibility is being investigated.
Model for ohr and ohrR tBOOH-inducible expression.
Considering all the available data, we propose a model for ohr regulation by OhrR and induction of the genes by organic peroxides. ohrR and ohr are transcribed from the strong organic peroxide-inducible P1 promoter. Then, the bicistronic 1.0-kb ohrR-ohr mRNA is processed at sites upstream of the ribosome binding site for ohr by an RNase III-like enzyme to give a 0.5-kb ohr mRNA, while ohrR mRNA is rapidly degraded. In uninduced cells, a low level of OhrR keeps P1 repressed, resulting in low levels of both OhrR and Ohr. The expression of ohrR is autoregulatory. Upon exposure to organic peroxides, binding of the ligand (organic peroxides) to OhrR leads to inactivation of the protein and prevents it from binding to P1. This process derepresses the expression of the operon and results in high-level expression of ohrR-ohr. The bicistronic ohrR-ohr mRNA is processed to give high levels of ohr mRNA and, in turn, high levels of Ohr and increased organic peroxide resistance. Concomitantly, the higher level of OhrR also produced is neutralized by the binding of the ligand to the protein. When organic peroxides have been removed, OhrR activity is restored and expression of the operon is once again repressed.
ACKNOWLEDGMENTS
We thank P. Bennett for editing the manuscript and J. H. Helmann for critical comments and suggestions.
This research was supported by grants from Chulabhorn Research Institute to the Laboratory of Biotechnology, Thailand Research Fund grant BRG 10-2543, and career development award RCF 01-40-005 from NSTDA to S.M.
REFERENCES
- 1.Alekshun M N, Levy S B. Regulation of chromosomally mediated multiple antibiotic resistance: the mar regulon. Antimicrob Agents Chemother. 1997;41:2067–2075. doi: 10.1128/aac.41.10.2067. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Alekshun M N, Levy S B. The mar regulon: multiple resistance to antibiotics and other toxic chemicals. Trends Microbiol. 1999;7:410–413. doi: 10.1016/s0966-842x(99)01589-9. [DOI] [PubMed] [Google Scholar]
- 3.Altuvia S, Kornitzer D, Kobi S, Oppenheim A B. Functional and structural elements of the mRNA of the cIII gene of bacteriophage lambda. J Mol Biol. 1991;218:723–733. doi: 10.1016/0022-2836(91)90261-4. [DOI] [PubMed] [Google Scholar]
- 4.Atichartpongkul, S., S. Loprasert, P. Vattanaviboon, W. Whangsuk, J. D. Helmann, and S. Mongkolsuk. Bacterial Ohr and OsmC paralogs define two protein families with distinct functions and patterns of expression. Microbiology 147:1775–1782. [DOI] [PubMed]
- 5.Baker C J, Orlandi E W. Active oxygen in plant pathogenesis. Annu Rev Phytopathol. 1995;33:299–321. doi: 10.1146/annurev.py.33.090195.001503. [DOI] [PubMed] [Google Scholar]
- 6.Buchmeier N, Bossie S, Chen C Y, Fang F C, Guiney D G, Libby S J. SlyA, a transcriptional regulator of Salmonella typhimurium, is required for resistance to oxidative stress and is expressed in the intracellular environment of macrophages. Infect Immun. 1997;65:3725–3730. doi: 10.1128/iai.65.9.3725-3730.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Chae H Z, Robison K, Poole L B, Church G, Storz G, Rhee S G. Cloning and sequencing of thiol-specific antioxidant from mammalian brain: alkyl hydroperoxide reductase and thiol-specific antioxidant define a large family of antioxidant enzymes. Proc Natl Acad Sci USA. 1994;91:7017–7021. doi: 10.1073/pnas.91.15.7017. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Egland P G, Harwood C S. BadR, a new MarR family member, regulates anaerobic benzoate degradation by Rhodopseudomonas palustris in concert with AadR, an Fnr family member. J Bacteriol. 1999;181:2102–2109. doi: 10.1128/jb.181.7.2102-2109.1999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Faungthong, M., S. Atichartpongkul, S. Mongkolsuk, and J. D. Helmann. Ohr is a repressor of ohrA, a key organic hydroperoxide resistance determinant in Bacillus subtillis. J. Bacteriol., in press. [DOI] [PMC free article] [PubMed]
- 10.Gonzalez-Flecha B, Demple B. Homeostatic regulation of intracellular hydrogen peroxide concentration in aerobically growing Escherichia coli. J Bacteriol. 1997;179:382–388. doi: 10.1128/jb.179.2.382-388.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Halliwell B, Gutteridge J M. Oxygen toxicity, oxygen radicals, transition metals and disease. Biochem J. 1984;219:1–14. doi: 10.1042/bj2190001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Katzen F, Becker A, Zorreguieta A, Puhler A, Ielpi L. Promoter analysis of the Xanthomonas campestris pv. campestris gum operon directing biosynthesis of the xanthan polysaccharide. J Bacteriol. 1996;178:4313–4318. doi: 10.1128/jb.178.14.4313-4318.1996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Kovach M E, Elzer P H, Hill D S, Robertson G T, Farris M A, Roop II R M, Peterson K M. Four new derivatives of the broad-host-range cloning vector pBBR1MCS, carrying different antibiotic-resistance cassettes. Gene. 1995;166:175–176. doi: 10.1016/0378-1119(95)00584-1. [DOI] [PubMed] [Google Scholar]
- 14.Levine A, Tenhaken R, Dixon R, Lamb C. H2O2 from the oxidative burst orchestrates the plant hypersensitive disease resistance response. Cell. 1994;79:583–593. doi: 10.1016/0092-8674(94)90544-4. [DOI] [PubMed] [Google Scholar]
- 15.Loprasert S, Atichartpongkun S, Whangsuk W, Mongkolsuk S. Isolation and analysis of the Xanthomonas alkyl hydroperoxide reductase gene and the peroxide sensor regulator genes ahpC and ahpF-oxyR-orfX. J Bacteriol. 1997;179:3944–3949. doi: 10.1128/jb.179.12.3944-3949.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Loprasert S, Fuangthong M, Whangsuk W, Atichartpongkul S, Mongkolsuk S. Molecular and physiological analysis of an OxyR-regulated ahpC promoter in Xanthomonas campestris pv. phaseoli. Mol Microbiol. 2000;37:1504–1514. doi: 10.1046/j.1365-2958.2000.02107.x. [DOI] [PubMed] [Google Scholar]
- 17.Loprasert S, Sallabhan R, Atichartpongkul S, Mongkolsuk S. Characterization of a ferric uptake regulator (fur) gene from Xanthomonas campestris pv. phaseoli with unusual primary structure, genome organization, and expression patterns. Gene. 1999;239:251–258. doi: 10.1016/s0378-1119(99)00412-6. [DOI] [PubMed] [Google Scholar]
- 18.Miller P F, Sulavik M C. Overlaps and parallels in the regulation of intrinsic multiple-antibiotic resistance in Escherichia coli. Mol Microbiol. 1996;21:441–448. doi: 10.1111/j.1365-2958.1996.tb02553.x. [DOI] [PubMed] [Google Scholar]
- 19.Mongkolsuk S, Loprasert S, Whangsuk W, Fuangthong M, Atichartpongkun S. Characterization of transcription organization and analysis of unique expression patterns of an alkyl hydroperoxide reductase C gene (ahpC) and the peroxide regulator operon ahpF-oxyR-orfX from Xanthomonas campestris pv. phaseoli. J Bacteriol. 1997;179:3950–3955. doi: 10.1128/jb.179.12.3950-3955.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Mongkolsuk S, Praituan W, Loprasert S, Fuangthong M, Chamnongpol S. Identification and characterization of a new organic hydroperoxide resistance (ohr) gene with a novel pattern of oxidative stress regulation from Xanthomonas campestris pv. phaseoli. J Bacteriol. 1998;180:2636–2643. doi: 10.1128/jb.180.10.2636-2643.1998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Mongkolsuk S, Sukchawalit R, Loprasert S, Praituan W, Upaichit A. Construction and physiological analysis of a Xanthomonas mutant to examine the role of the oxyR gene in oxidant-induced protection against peroxide killing. J Bacteriol. 1998;180:3988–3991. doi: 10.1128/jb.180.15.3988-3991.1998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Ochsner U A, Hassett D J, Vasil M L. Genetic and physiological characterization of ohr, encoding a protein involved in organic hydroperoxide resistance in Pseudomonas aeruginosa. J Bacteriol. 2001;183:773–778. doi: 10.1128/JB.183.2.773-778.2001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Poole K, Tetro K, Zhao Q, Neshat S, Heinrichs D E, Bianco N. Expression of the multidrug resistance operon mexA-mexB-oprM in Pseudomonas aeruginosa: mexR encodes a regulator of operon expression. Antimicrob Agents Chemother. 1996;40:2021–2028. doi: 10.1128/aac.40.9.2021. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Poole L B. Flavin-dependent alkyl hydroperoxide reductase from Salmonella typhimurium. 2. Cystine disulfides involved in catalysis of peroxide reduction. Biochemistry. 1996;35:65–75. doi: 10.1021/bi951888k. [DOI] [PubMed] [Google Scholar]
- 25.Poole L B, Ellis H R. Flavin-dependent alkyl hydroperoxide reductase from Salmonella typhimurium. 1. Purification and enzymatic activities of overexpressed AhpF and AhpC proteins. Biochemistry. 1996;35:56–64. doi: 10.1021/bi951887s. [DOI] [PubMed] [Google Scholar]
- 26.Rince A, Giard J-C, Pichereau V, Flahaut S, Auffray Y. Identification and characterization of gsp65, an organic hydroperoxide resistance (ohr) gene encoding a general stress protein in Enterococcus faecalis. J Bacteriol. 2001;183:1482–1488. doi: 10.1128/JB.183.4.1482-1488.2001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Storz G, Jacobson F S, Tartaglia L A, Morgan R W, Silveira L A, Ames B N. An alkyl hydroperoxide reductase induced by oxidative stress in Salmonella typhimurium and Escherichia coli. Genetic characterization and cloning of ahp. J Bacteriol. 1989;171:2049–2055. doi: 10.1128/jb.171.4.2049-2055.1989. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Sukchawalit S, Mongkolsuk S. Xanthomonas oryzae pv. oryzae recA is transcribed and regulated from multiple promoters. FEMS Microbiol Lett. 2001;197:35–40. doi: 10.1111/j.1574-6968.2001.tb10579.x. [DOI] [PubMed] [Google Scholar]
- 29.Takata R, Izuhara M, Hori K. Differential degradation of the Escherichia coli polynucleotide phosphorylase mRNA. Nucleic Acids Res. 1989;17:7441–7451. doi: 10.1093/nar/17.18.7441. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Toledano M B, Kullik I, Trinh F, Baird P T, Schneider T D, Storz G. Redox-dependent shift of OxyR-DNA contacts along an extended DNA-binding site: a mechanism for differential promoter selection. Cell. 1994;78:897–909. doi: 10.1016/s0092-8674(94)90702-1. [DOI] [PubMed] [Google Scholar]
- 31.Volker U, Andersen K K, Antelmann H, Devine K M, Hecker M. One of two osmC homologs in Bacillus subtilis is part of the sigmaB-dependent general stress regulon. J Bacteriol. 1998;180:4212–4218. doi: 10.1128/jb.180.16.4212-4218.1998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Zheng M, Aslund F, Storz G. Activation of the OxyR transcription factor by reversible disulfide bond formation. Science. 1998;279:1718–1721. doi: 10.1126/science.279.5357.1718. [DOI] [PubMed] [Google Scholar]