SUMMARY
Obesity is associated with increased cancer incidence and progression. However, the relationship between adiposity and cancer remains poorly understood at the mechanistic level. Here, we report that adipocytes from tumor-invasive mammary fat undergo de-differentiation to fibroblast-like precursor cells during tumor progression and integrate into the tumor microenvironment. Single-cell sequencing reveals that these de-differentiated adipocytes lose their original identities and transform into multiple cell types, including myofibroblast- and macrophage-like cells, with their characteristic features involved in immune response, inflammation, and extracellular matrix remodeling. The de-differentiated cells are metabolically distinct from tumor-associated fibroblasts but exhibit comparable effects on tumor cell proliferation. Inducing de-differentiation by Xbp1s overexpression promotes tumor progression despite lower adiposity. In contrast, promoting lipid-storage capacity in adipocytes through MitoNEET overexpression curbs tumor growth despite greater adiposity. Collectively, the metabolic interplay between tumor cells and adipocytes induces adipocyte mesenchymal transition and contributes to reconfigure the stroma into a more tumor-friendly microenvironment.
In brief
Zhu et al. report that in mammary tumors, adipocytes undergo metabolic reprograming and de-differentiation. They acquire myofibroblast- and macrophage-like features, a process referred to as “adipocyte mesenchymal transition,” which modifies the tumor microenvironment via ECM remodeling and activation of the immune response and likely contributes to tumor progression.
Graphical Abstract
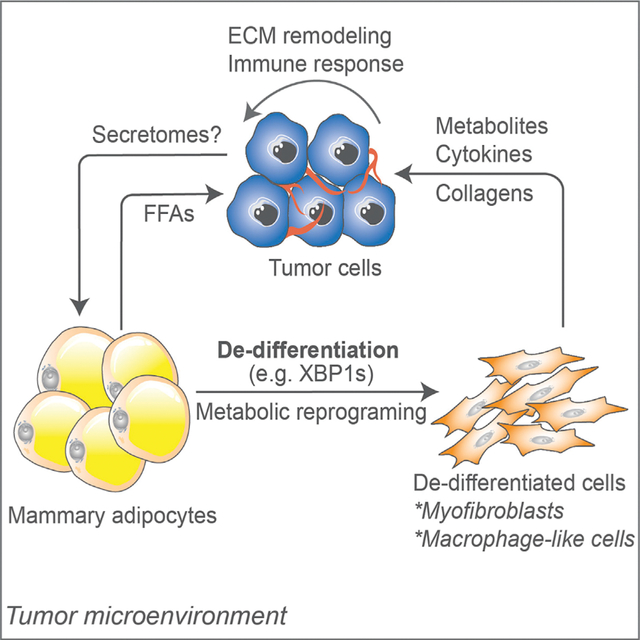
INTRODUCTION
Obesity is often considered to be a risk factor for breast cancer occurrence and worse prognosis (Khandekar et al., 2011; Park et al., 2014; Quail and Dannenberg, 2019). However, other studies have challenged this perception with findings that obesity has minimal effects on the recurrence or survival in patients with breast cancer (Elwood et al., 2018). This discrepancy raises the question as to what exactly the mechanistic involvement of adipocytes on tumor progression is. In the setting of breast cancers, tumor cells are in close contact with mammary adipocytes. These tumor-associated adipocytes exhibit enhanced lipolysis and promote tumor invasiveness (Molocea et al., 2020; Wang et al., 2017). Thus, the metabolic reprograming of adipocytes during lesion growth must be an important contributor.
Tumor stroma is an extensive heterogeneous tissue (Jackson et al., 2020) that contains many cell types beyond tumor stem cells, including fibroblasts and epithelial and immune cells; the orchestration of cellular responses and metabolic reprograming in the stroma forms a unique microenvironment favoring tumor growth (Cairns et al., 2011; Dey et al., 2021). In breast cancer progression, tumor-associated adipocytes significantly change due to a loss of lipid content. However, we do not know what the fate of these delipidated adipocytes is and what, if any, roles they play during tumor progression. As previously demonstrated, adipocytes display an unexpectedly high degree of plasticity. They can undergo trans-differentiation and de-differentiation under different metabolic conditions. The appearance of beige adipocytes is observed during breast cancer progression. These are cells with enhanced catabolism, in which “lipid-storing” white adipocytes are replaced by “lipid-burning” beige adipocytes (Kajimura et al., 2015; Singh et al., 2016; Zhu and Scherer, 2018). On the other hand, adipocyte de-differentiation is seen under several physiological and pathological conditions, including skin fibrosis (Zhang et al., 2019), lactation (Wang et al., 2018), and liposarcomas (Thway, 2019). Metabolic reprograming during adipocyte de-differentiation is an essential contributor toward the development of liposarcomas (Bi et al., 2016). Moreover, in vitro-induced de-differentiation of adipocytes via mechanical compression has a positive effect on breast tumor growth (Li et al., 2020). Although previous studies have indicated the role of adipocytes/adipocyte precursor cells (APCs) in breast tumor progression (Bochet et al., 2013; Dirat et al., 2011; Li et al., 2020; Zhang et al., 2012), no direct evidence has been provided to date in terms of adipocyte de-differentiation in the tumor context in vivo. Here, by taking advantage of a multipronged approach involving adipocyte lineage tracing and single-cell RNA sequencing, we report direct genetic evidence that the interactions between mammary tumor cells and adipocytes drive adipocyte mesenchymal transition upon de-differentiation during tumor progression. We also test this genetically by inducing adipocyte de-differentiation, thereby promoting tumor growth. In contrast, genetically enhancing adipocyte lipid-storage capacity suppresses tumor growth, regardless of adiposity. Therefore, we propose that this transition is an important mechanistic step toward generating a tumor-friendly microenvironment for tumor growth.
RESULTS
Adipocytes undergo de-differentiation during mammary tumor infiltration into the stroma
Mammary tumor growth often invades the neighboring adipose tissue both in rodent and human mammary tumors (Figures 1A and 1B). Adipocytes become smaller as a function of proximity to tumor lesions (Figures 1C and 1D; Table S1), a phenomenon that is irrespective of tumor subtypes in patients (Table S2). This goes hand in hand with a downregulation of adipocyte markers, including Adipoq, Lep, Pparg, and Plin1 (Figure 1E). These data suggest that the tumor-induced alterations in cellular composition in mammary fat may be due to the loss of adipocytes in either their number or their characteristics. In addition, the tumor-adipose interacting regions exhibit a high density of vascularization, as indicated by the endothelial cell marker CD31, suggesting a more active state of the tumor cells due to the nutritionally rich environment in these regions (Figure 1B). The synchronization of metabolic programs among different cell types in the tumor stroma establishes a growth-supporting microenvironment for tumor progression. This raises the question as to the long-term fate of the tumor proximal adipocytes during this process. We thus took advantage of our doxycycline (dox)-dependent “pulse-labeling” adipocyte-tracer system, the “AdipoChaser mouse” (Zhang et al., 2019) (Figure 1F), in which we not only permanently label all mature adipocytes with EGFP but also all cells subsequently derived from these cells. We further crossed these mice into a mammary tumor model, the MMTV-PyMT mouse (PyMTChaser), and labeled the adipocytes before the tumors arise. In the absence of dox, cells remain unlabeled, whereas upon dox treatment, mature adipocytes express EGFP, demonstrating specific and efficient labeling (Figures 1Ga and 1Gb). Interestingly, at the tumor invasive edge and some deeper areas of the tumor, cells labeled with EGFP appear smaller and exhibit a fibroblast-like morphology, indicating the transition of mature adipocytes to APCs (Figure 1Gc). Moreover, we implant EO771 cells in these mice to induce an ectopic mammary tumor (AlloChaser). Similarly, allograft tumor lesions also induce adipocyte de-lipidation and an APC-like transformation, a process occurring in the tumor invasive regions but not in areas of the mammary fat more distal from the tumor (Figures 1Gd–1Gf). In addition, a similar pattern of tumor induced-APC-like transformation of adipocytes is also observed in a dox-independent tracing system (Figure S1). Thus, both ectopic tumor implants and genetically induced tumors arising from the endogenous ductal epithelium induce an adipocyte de-differentiation phenotype, and these de-differentiated adipocytes fully integrate into the tumor stroma rather than undergoing apoptosis, suggesting that a process we refer to as “adipocyte mesenchymal transition” (AMT) occurs in the tumor.
Figure 1. Adipocytes undergo de-differentiation during mammary tumor infiltration into the stroma.
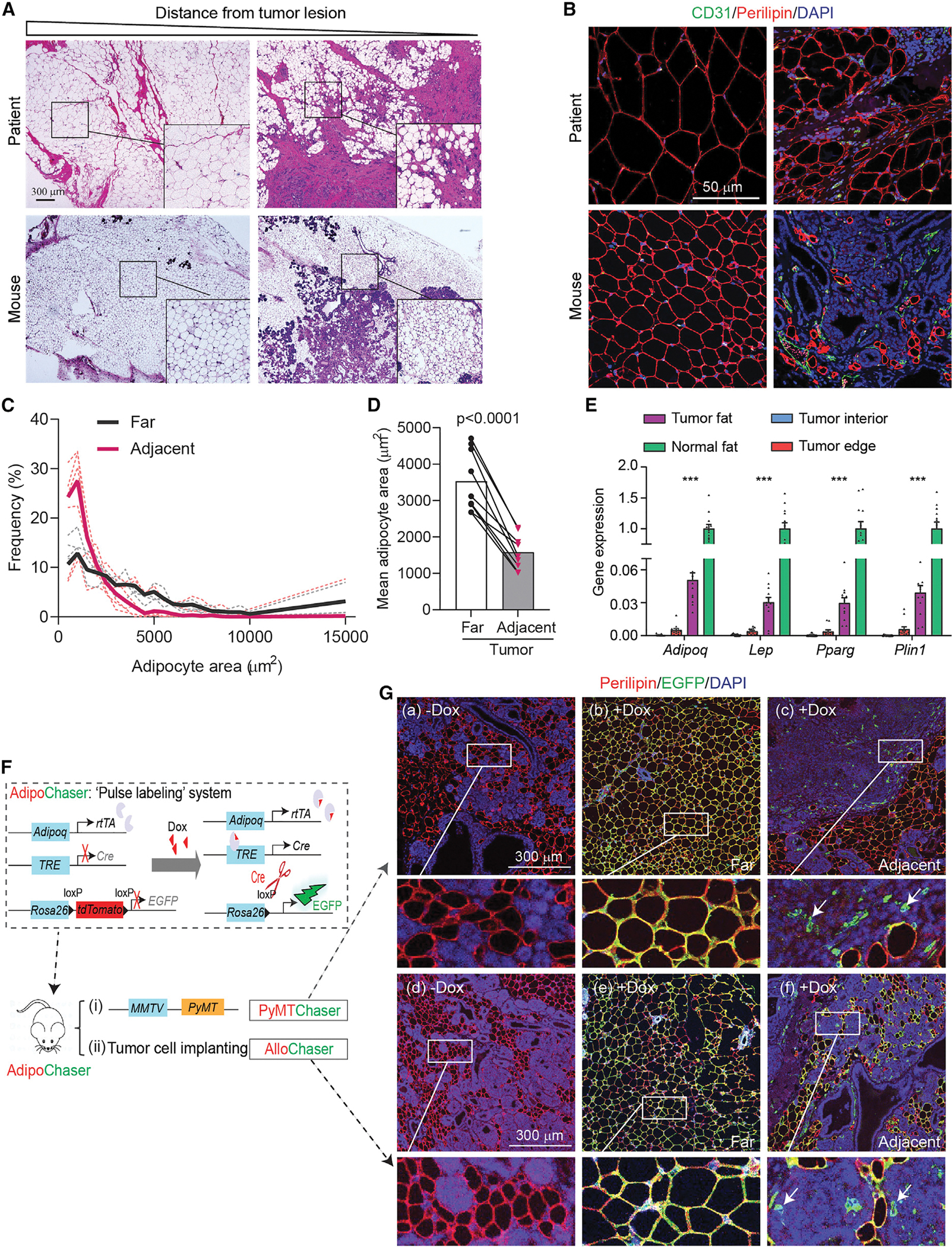
(A) H&E staining reveals smaller adipocytes upon mammary tumor invasion in patients and PyMT mice. Scale bar, 300 μm.
(B) Immunofluorescent staining of adipocyte marker Perilipin (red) and endothelial cell marker CD31 (green) for patient and mouse tumor samples (n = 3). Scale bar, 50 μm.
(C and D) Frequency and average adipocyte size in regions distant (>500 μm) and adjacent (<500 μm) to the tumor lesion in patient samples (n = 9).
(E) Down-regulation of adipocyte marker genes upon tumor invasion in mammary adipose tissues from PyMT-mice (n = 12). “Normal fat” indicates corresponsive fat from mice without tumor, “tumor fat” is distal fat from PyMT-tumor, “tumor edge” is fat connected with tumor, and “tumor interior” is tissues from tumor core.
(F) Scheme of PyMTChaser and AlloChaser mice.
(G) Both PyMTChaser (a-c) and AlloChaser (d-f) reveal mammary adipocyte de-differentiation upon tumor infiltration (n = 3). (a, d) No dox control; (b, e) distal fat from the tumor lesion; (c, f) tumor adjacent region. Arrows indicate de-differentiated EGFP+/Perilipin− cells with fibroblast-like morphology. Scale bar, 300 μm. Each dot represents an individual sample. Data are presented as mean ± SEM. Unpaired Student’s t test was used in (D), and one-way ANOVA with Holm-Sidak’s multiple comparisons test was used in (E). ***p < 0.001.
Single-cell RNA sequencing reveals AMT during mammary tumor progression
It is important to understand the exact nature of these newly dedifferentiated adipocytes. To that end, we performed single-cell RNA sequencing with these de-differentiated adipocytes (EGFP+, tdTomato−) from PyMTChaser or AlloChaser mice (PyMT-EGFP+ or Allo-EGFP+ cells), respectively. A total of 576 (PyMT) and 2,135 (allografts) EGFP+/tdTomato− cells are analyzed, and a high degree of heterogeneity, with distinct gene-expression signatures, is seen in both cell populations (Figures 2A and 2B). In both cases, mature adipocyte markers Adipoq and Lep are undetectable, and Plin1 and Fabp4 are only expressed in a few of these EGFP+ cells (Figure S2A). In contrast, in PyMT-EGFP+ cells, the adipogenic master regulators Pparg and Cebpa and other adipocyte early-differentiation markers, such as Lpl, are mainly present in groups 2 or 3 (Figure 2C, left). A similar pattern is observed in Allo-EGFP+ cells, in which Pparg, Cebpa, and Lpl are mainly present in groups 1, 3, 4, or 6 (Figure 2C, right). These results reflect that these cells have lost their adipocyte identity and have transitioned to new cell types. Moreover, prominent populations throughout the groups from both cases exhibit mesenchymal stem cell or adipocyte progenitor gene signatures, as judged by the expression of Cd44, Ly6a, Cd24a, Cd9, Icam1, or Cd38 (Carrière et al., 2017; Cawthorn et al., 2012; Hepler et al., 2018; Merrick et al., 2019) (Figure 2D). Thus, adipocytes undergo a process of de-differentiation driven by tumor lesions, leading to the AMT.
Figure 2. Single cell-RNA sequencing reveals adipocyte mesenchymal transition during mammary tumor progression.
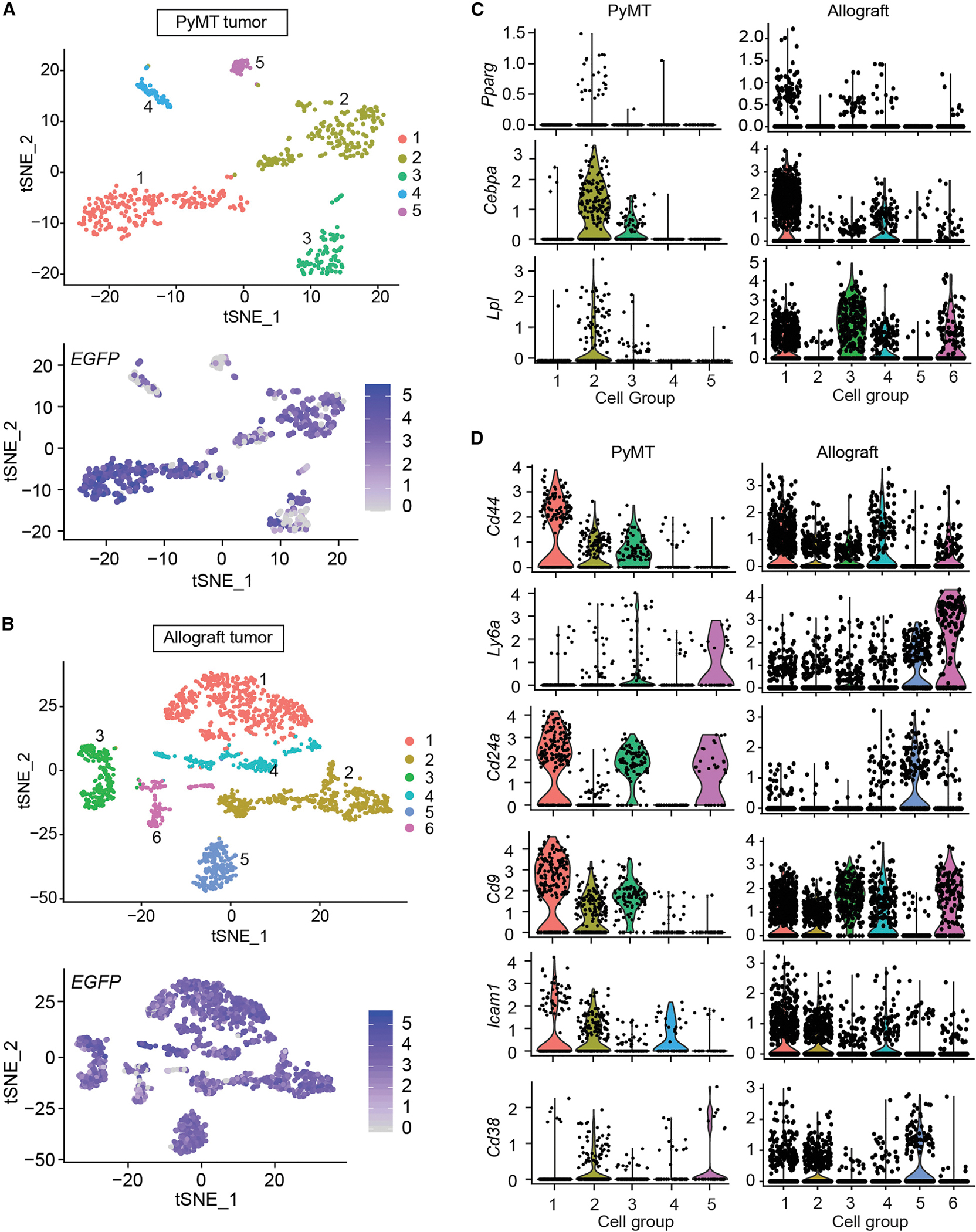
EGFP-labeled de-differentiated adipocytes from PyMTChaser and AlloChaser mice were isolated by FACS and subjected to scRNA-seq.
(A and B) T-distributed stochastic neighbor embedding (t-SNE) plots. Clustering of PyMT-EGFP + or allograft (Allo)-EGFP + cells, with mean number of genes per cell equals 1, 163 or 2, 173, respectively.
(C) Representative adipocyte early differentiation marker genes.
(D) Representative mesenchymal stem cell/adipocyte precursor marker genes. The y axis is the log-scale normalized read count.
Mammary tumor-induced de-differentiated adipocytes contribute to the tumor microenvironment via inflammation and extracellular matrix (ECM) remodeling
What, if any, role do these tumor-induced de-differentiated adipocytes play? We analyzed the prominent groups that commonly express Lpl from Allo-EGFP+ cells. Although these groups display distinct gene-expression patterns (Figure 3A), Gene Ontology (GO) enrichment analyses suggest that these de-differentiated cells (groups 3 and 6 from Allo-EGFP+ cells) are engaged in multiple pathways related to the biological process of ECM remodeling, including a response to hypoxia, collagen fibril organization, extracellular fibril organization, and cell adhesion (Figure 3B). Moreover, cells in groups 1 and 4 exhibit a gene-expression profile invoking pathways involved in the immune response and inflammation, including chemotaxis, inflammatory responses, and an immune system response (Figure 3B). In addition, PyMT-EGFP+ cells exhibit a similar pattern in the pathway analyses (Figure S3A). These data suggest that these de-differentiated cells might be at different stages of transitioning during the harvest.
Figure 3. Mammary tumor-induced de-differentiated adipocytes contribute to tumor microenvironment concerning inflammation and ECM remodeling.
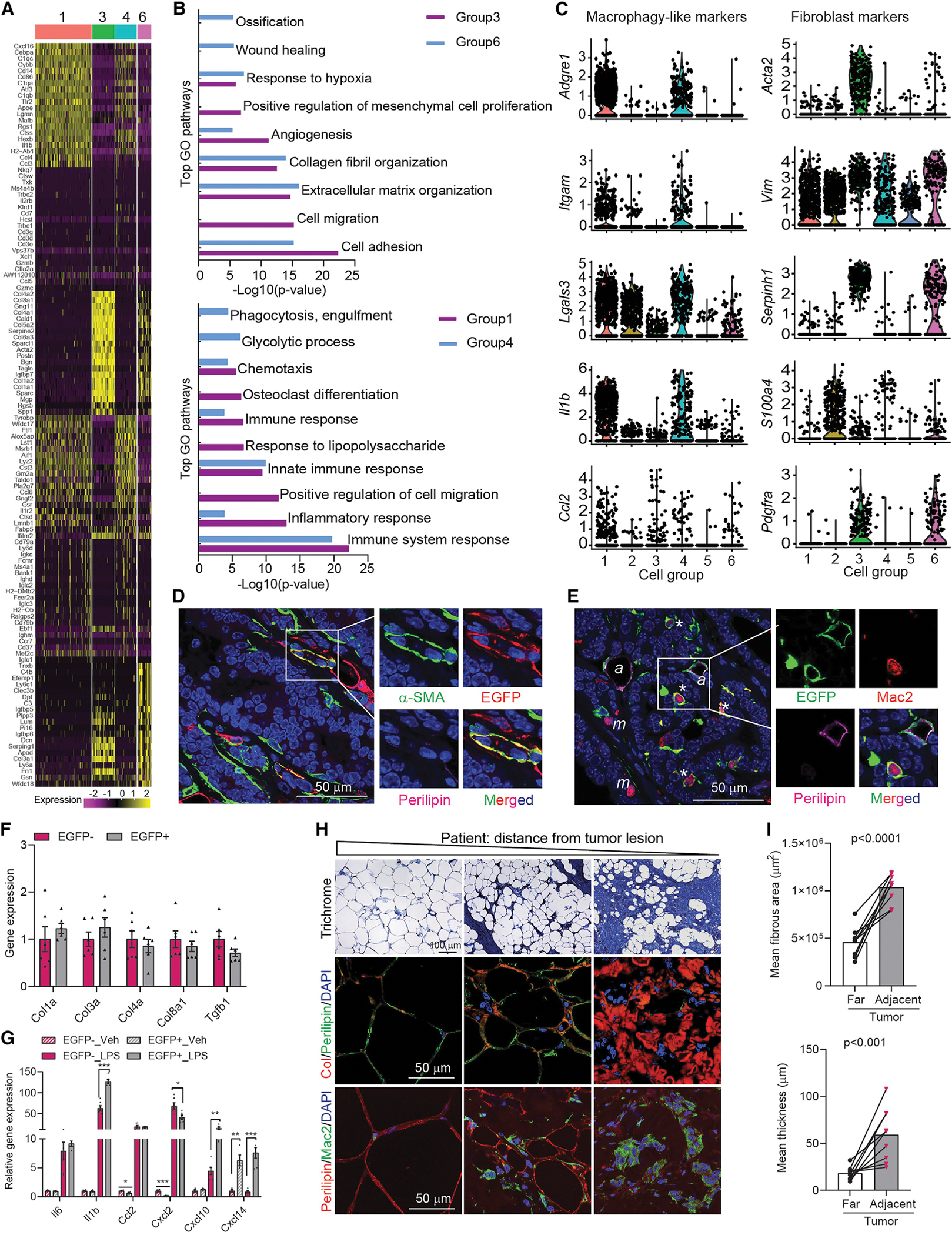
(A) Heat map of top 20 genes across different cell groups from allograft (Allo)-EGFP + cells.
(B) Gene ontology (GO) pathway enrichment analysis.
(C) Representative macrophage and fibroblast markers genes. The y-axis is the log-scale normalized read count.
(D) Co-staining of α-SMA (green), EGFP (red), and Perilipin (magenta) of PyMTChaser-tumor. Scale bar, 50 μm.
(E) Co-staining of EGFP (green), Mac2 (red), and Perilipin (magenta) of PyMTChaser-tumor. ‘*’ indicates macrophage-like cells (EGFP+/Perilipin−/Mac2+); ‘a’ indicates adipocytes (EGFP+/Perilipin+/Mac2−); ‘m’ indicates macrophages (EGFP−/Perilipin−/Mac2+). Scale bar, 50 μm.
(F) mRNA levels of genes related to ECM remodeling in cultures of de-differentiated adipocytes (EGFP+, tdTomato−) and tumor-associated fibroblasts (EGFP−, tdTomato+) isolated from AlloChaser-tumor (n = 6).
(G) mRNA levels of cytokines/chemokines in cultures of EGFP+ and EGFP− cells treated with vehicle (PBS) or LPS (100 ng/mL) for 3 h (n = 6).
(H) Trichrome, collagen (Col), and Mac2 staining indicates enhanced fibrosis, collagen production and inflammation in the mammary fat at a distance from the tumor lesion in patients (n = 9). Scale bar, 100 μm for trichrome staining and 50 μm for Col and Mac2 staining.
(I) Average fibrous area and thickness in distant and adjacent regions from the tumor lesion in patients (n = 9). Each dot represents an individual sample. Data are presented as mean ± SEM. Unpaired Student’s t test was used in (F), (G), and (I). *p < 0.05; **p < 0.01; ***p < 0.001.
Interestingly, in addition to their mesenchymal stem cell or adipocyte progenitor gene signatures, these de-differentiated cells, mainly in groups 1 and 2 from PyMT-EGFP+ cells and groups 1 and 4 from Allo-EGFP+ cells, also display macrophage markers, including Adgre1, Itgam, and Lgals3, suggesting that these populations gain “macrophage-like” features (Figures 3C and S3B). In contrast, cells mainly in group 3 from PyMT-EGFP+ cells and groups 3 and 6 from Allo-EGFP+ cells express fibroblast markers, such as Acta2, Vim, or Serpinh1 (Figures 3C and S3B). In addition, there are small cell populations (groups 4 and 5 from PyMT-EGFP+ cells and groups 2 and 5 form Allo-EGFP+ cells) that express Cd3e (T cell marker) or Cd19 (B cell marker), in addition to the adipocyte precursor markers, indicating a mixed phenotype of these cells (Figure S2B). We further found some of the de-differentiated EGFP+/Perilipin− cells are also α-smooth muscle actin (α-SMA) positive (Figure 3D), suggesting a transition from adipocytes to myofibroblasts. Moreover, we also observed that a number of de-differentiated EGFP+/Perilipin− cells are Mac2 positive (Figure 3E), further substantiating the macrophage-like nature of these cells. In addition, to test the broad differentiation potential of the de-differentiated cells, we compared the adipogenic and myogenic processes with the de-differentiated cells (EGFP+, tdTomato−) and tumor-associated fibroblasts (EGFP−, tdTomato+) that do not derive from adipocytes in vitro. Isolated EGFP+ cells display a fibroblast morphology, producing collagen and expressing the fibroblast marker FSP1 (Figure S4A). Interestingly, compared with the EGFP− cells, these cells re-differentiate with greater ease into adipocytes (Figures S4B and S4C), reflecting an intrinsic propensity to resume their adipocyte identity under adipogenic conditions. In addition, the de-differentiated adipocytes also gain muscle cell-like characteristics with desmin (a muscle-specific marker) expression upon a myogenic stimulus, a phenomenon not observed in EGFP− cells (Figure S4D). Thus, the tumor-induced de-differentiated adipocytes have distinct differentiation potential compared with tumor-associated fibroblasts in vitro. These results indicate that de-differentiated adipocytes acquire features of progenitors and can transition into multiple cell types in the context of an infiltrating tumor.
We further analyzed the de-differentiated cells and tumor-associated fibroblasts in cultures with regards to ECM remodeling and the inflammatory response. Interestingly, the EGFP+ cells express comparable levels of genes related to ECM remodeling as the EGFP− cells (Figure 3F). However, the EGFP+ cells are more responsive to lipopolysaccharide (LPS) exposure (Figure 3G). Thus, these de-differentiated cells likely mediate a tumor-friendly microenvironment in terms of immunological responses and ECM remodeling. In line with these responses, enhanced collagen production and inflammation are observed in the mammary fat even at a distance from the infiltrating lesions in patients (Figure 3H). Quantitative analyses further suggest a higher degree of fibrosis in adipose tissues that is adjacent to tumor lesions, compared with regions that are further away from infiltrating tumor cells (Figure 3I), reflecting a high degree of ECM remodeling occurring upon tumor invasion.
Distinct metabolic characteristics of mammary tumor-induced de-differentiated adipocytes
What are the unique metabolic features of these de-differentiated adipocytes? Metabolite profiling in the culture medium suggests that de-differentiated cells and tumor-associated fibroblasts behave differently in their metabolic responses (Figures 4A–4D). Under fed conditions, EGFP+ cells display differential pathways, involving aminoacyl-tRNA biosynthesis and pyrimidine metabolism (Figure 4C), whereas under fasted conditions, these cells embrace pathways related to alanine, aspartate and glutamate metabolism, and glycolysis/gluconeogenesis (Figure 4D). Glycolytic stress assays indicate that the EGFP+ cells display enhanced glycolysis compared with EGFP− cells (Figures 4E and 4F). The EGFP+ cells exhibit similar basal and maximal oxygen consumption rates (OCRs) but display higher coupling efficiency (Figures 4G–4I). Thus, the de-differentiated cells are metabolically distinct from the tumor fibroblasts. Finally, we examined their effects on tumor cell proliferation. Interestingly, the conditioned media from EGFP+ and EGFP− cells exert similar effects on the proliferation rates of various human (Figure 4J), as well as mouse (Figure 4K), breast cancer cells. Thus, it seems likely that the de-differentiated cells coordinate with tumor-associated fibroblasts to contribute to a tumor-supportive microenvironment.
Figure 4. Distinct metabolic characteristics of tumor-induced de-differentiated adipocytes.
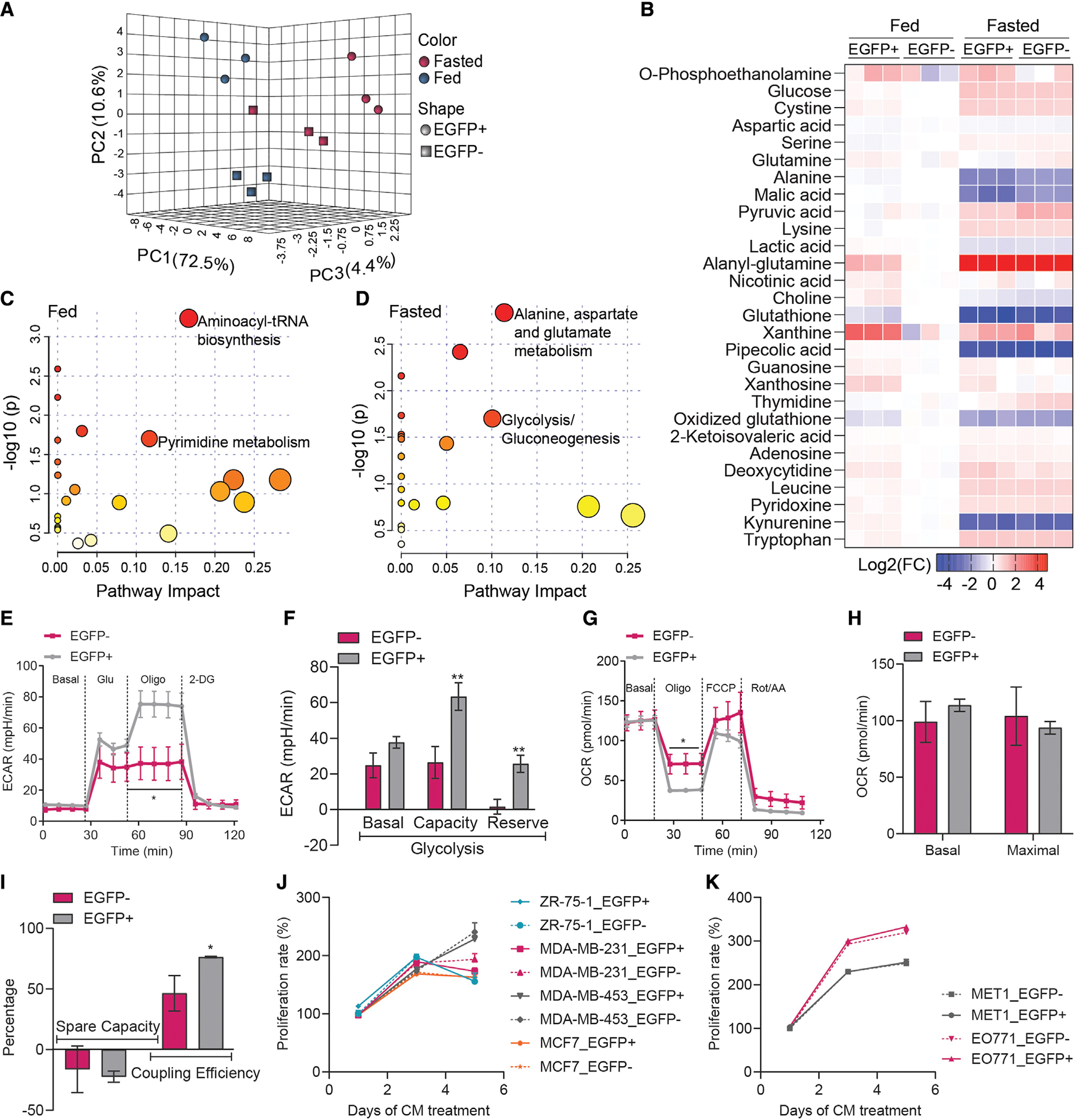
(A and B) Interactive PCA analysis and heatmap of metabolite profile in the culture medium of de-differentiated adipocytes (EGFP+, tdTomato−) and tumor-associated fibroblasts (EGFP−, tdTomato+) isolated from AlloChaser-tumor (n = 3). At fasted condition, medium was analyzed in the absence of serum for 6 hr.
(C and D) The metabolome map of matched metabolic pathways in fed and fasted condition according to the p values from the enrichment analysis and impact values from the pathway topology analysis. Colors varying from yellow to red indicate the metabolites are in enrichment analysis with different levels of significance.
(E and F) Enhance glycolysis in EGFP + cells (n = 6–7).
(G–I) EGFP + cells exhibit similar basal and maximal oxygen consumption rate (OCR) but have higher coupling efficiency (n = 6–7). Conditioned medium (CM) from EGFP + cells and EGFP− cells shows similar effects on the proliferation of various human (J) and mouse (K) breast cancer cells (n = 4). Data are presented as mean ± SEM. Unpaired Student’s t test was used in (E)-(I); two-way ANOVA with Holm-Sidak’s multiple comparisons test was used in (J) and (K). *p < 0.05; **p < 0.01.
Adipocyte de-differentiation promotes mammary tumor growth
As described above, close-knit interactions between adipocytes and tumor cells result in AMT, over the course of which adipocytes display reduced lipid-storing capacity. We thus wondered whether directly modulating AMT could affect tumor growth. To achieve this, we took advantage of mouse models with enhanced or impaired lipid-storage capacity in adipocytes. As a model of reduced lipid-storage capacity, we used a model with constitutive Xbp1s expression in adipocytes (hereby referred to as Ad-Xbp1s mice). Ad-Xbp1s mice display reduced adiposity with notably smaller fat pads (Figures 5A and 5B), a phenotype similar to that observed in mice with inducible induction of Xbp1s in adipocytes (Deng et al., 2018). Due to the fat loss, Ad-Xbp1s mice display lower adiponectin and leptin levels and hyperglycemia but without hyperinsulinemia. In addition, fed serum non-esterified fatty acids are elevated, whereas fasting triglycerides are reduced, in Ad-Xbp1s mice (Table S3). X-box binding protein 1 (XBP1) is a key transcription factor involved in the unfolded protein response pathway, and spliced XBP1s is the active form, which is highly upregulated upon various lipolytic stimuli in adipose tissues (Deng et al., 2018). Interestingly, both Xbp1t (unspliced) and Xbp1s expression are dramatically upregulated in tumor fat and within the tumor lesion (Figure 5C). Similar to what we observed in the wild-type (WT) tumor-infiltrated fat, Xbp1s-trangenic fat also expresses remarkably reduced expression of adipocyte markers (Figure 5D), reflecting a de-differentiation phenotype. We thus generate the Xbp1sChaser mice (Figure 5E). Indeed, as early as 2 weeks of Xbp1s induction in adipocytes causes dramatic de-lipidation and de-differentiation (Figure 5F). As a result, Ad-Xbp1s-PyMT double-transgenic mice display a greater tumor burden despite lower adiposity in a C57BL/6 background at 23 weeks (a model with reduced tumor aggressiveness) (Figure 5G), as well as in the FVB background at 14 weeks (a model with enhanced aggressiveness) (Figure 5H). Moreover, the lesion-associated adipose tissue almost completely disappears in Ad-Xbp1s-PyMT mice (Figure 5I). Thus, Xbp1s induces adipocyte de-lipidation and de-differentiation and promotes tumor growth.
Figure 5. Adipocyte de-differentiation promotes mammary tumor growth.
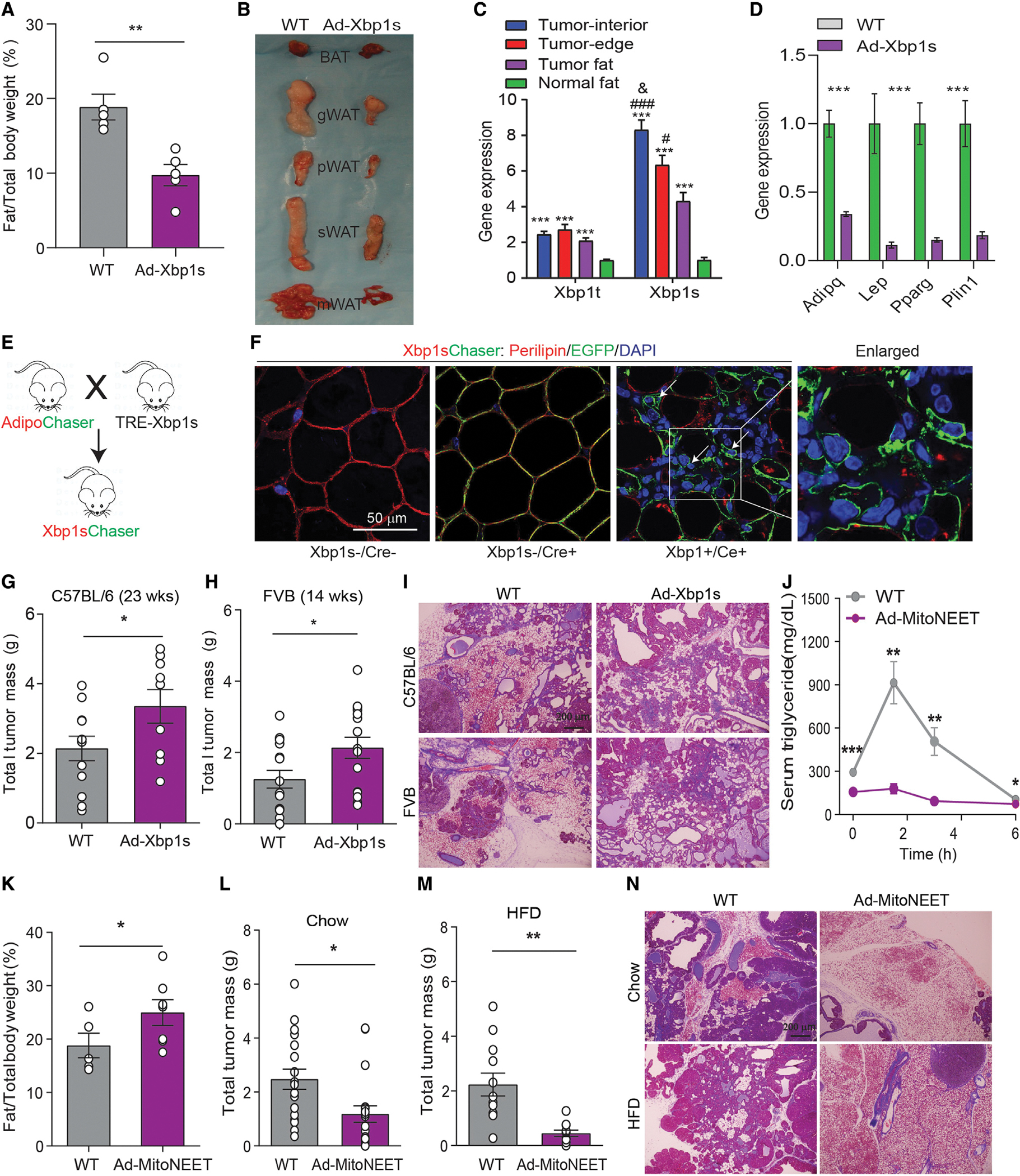
(A and B) Ad-Xbp1s mice display lower adiposity and smaller size of different fat pads (n = 5).
(C) Up-regulation of total Xbp1 (Xbp1t) and spliced Xbp1 (Xbp1s) upon tumor invasion (n = 6).
(D) Downregulation of adipocyte marker genes in mammary fat from Ad-Xbp1s mice (n = 8–9).
(E) Scheme of the Xbp1sChaser mice.
(F) Two weeks of Xbp1s induction causes adipocyte de-differentiation. Scale bar, 50 μm.
(G and H) Increased total tumor mass in Ad-Xbp1s-PyMT mice in C57B/L6 (n = 9–12) or FVB background (n = 15–16) at 23 or 14 weeks old, respectively.
(I) H&E staining indicates mammary adipose tissue is largely engulfed by tumor in Ad-Xbp1-PyMT mice (n = 3). Scale bar, 200 μm.
(J) Ad-MitoNEET-PyMT mice promotes triglycerides tolerance (n = 5–7).
(K) Increased fat mass percentage in Ad-MitoNEET-PyMT mice (n = 5–7).
(L and M) Decrease in total tumor mass at 23-week-old Ad-MitoNEET-PyMT mice fed with chow (n = 18–19) or HFD (n = 11–12), respectively.
(N) H&E staining indicates mammary adipose tissue is largely preserved in Ad-Xbp1-PyMT mice (n = 3). Scale bar, 200 μm.
Each dot represents an individual mouse. Data are presented as mean ± SEM. Unpaired Student’s t test was used in (A), (D), (G), (H), and (K)–(M); one-way ANOVA or two-way ANOVA with Holm-Sidak’s multiple comparisons test was used in (C) or (J), respectively. *, #, or &p < 0.05; **p < 0.01; ***or ###p < 0.001.
In contrast, adipocyte MitoNEET-transgenic mice display higher adiposity due to healthy expansion of adipose tissues (a model we have previously characterized in depth [Kusminski et al., 2012]). In line with our previous results, Ad-MitoNEET-PyMT double-transgenic mice also exhibit elevated lipid re-esterification and higher adiposity (Figures 5J and 5K), highlighting that these mice dramatically favor lipid storage over lipid utilization. However, in this adipose tissue-enriched microenvironment, tumor growth is curbed with a dramatically reduced tumor burden under chow-fed conditions (Figure 5L). These results became even more significant under high-fat diet (HFD)-fed conditions (Figure 5M). Histological analysis indicates that the surrounding adipocytes are largely “engulfed” by the tumor mass in WT mice, in sharp contrast to the morphology in Ad-MitoNEET-PyMT mice (Figure 5N). Thus, these results strongly suggest that adipocytes with enhanced lipid-storage capacity counteract tumor progression by limiting substrate availability for the tumor cells while at the same time limiting the potential of adipocytes to undergo AMT. This suggests that effective de-lipidation is integral to the AMT process.
DISCUSSION
Here, we show that the interactions between tumor cells and adipocytes lead to AMT upon de-differentiation. Adipocyte de-differentiation may be a unique process to adapt to physiological and pathophysiological stress settings. Our findings suggest that tumor-induced de-differentiated adipocytes have distinct metabolic features from tumor-associated fibroblasts. This metabolic reprograming may be an incentive for adipocyte dedifferentiation during tumor progression. In addition, these dedifferentiated cells have similar effects as the tumor-associated fibroblasts on tumor cell proliferation. Thus, these cells are unique components of the tumor stroma, where they further collude with tumor-associated fibroblasts, conferring to a tumor-friendly microenvironment.
We show that de-differentiated adipocytes are multipotent and acquire the ability to convert into multiple cell types. In vitro studies reveal that these cells are able to re-differentiate into multiple cell types, including adipocytes, skeletal myocytes, muscle cells, and cardiomyocytes upon appropriate exogenous stimuli (Wei et al., 2013). Our in vitro results suggest that the tumor-induced de-differentiated adipocytes can indeed be redifferentiated into adipocytes and muscle-like cells, in line with the multipotent potential of the de-differentiated adipocytes previously reported (Wei et al., 2013). However, in the context of the tumor microenvironment in vivo, these de-differentiated cells exhibit mesenchymal cell signatures, including an AMT. The myofibroblast transition can be observed for the de-differentiated adipocytes, a phenomenon also seen in de-differentiated dermal adipocytes (Zhang et al., 2019). A previous study revealed that stromal-vascular cells derived from white adipose tissue or the 3T3-L1 preadipocyte cell line can convert into a functional macrophage-like cell type when injected into the peritoneal cavity in mice (Charrière et al., 2003). Here, we show that the newly formed macrophage-like cells in the tumor microenvironment stem from adipocyte-derived mesenchymal cells during AMT. These cells may in turn exert a positive effect on tumor growth by mediating immunological changes and changes in ECM composition. As the lesions progress, tumor cells alter their dependence on the immune response and the ECM remodeling (Winkler et al., 2020). Notably, inflammatory cells are indispensable contributors to the tumor microenvironment (Coussens and Werb, 2002; Lin and Pollard, 2004). As such, an elevated degree of inflammation is commonly observed in the tumor microenvironment. Prior studies proposed that adipocyte de-differentiation leads to an elevated inflammatory response and contributes to liposarcoma development (Bi et al., 2016), though direct evidence could not be provided. As a result of the high cellular heterogeneity in the tumor stroma, various types of cells are involved in shaping the tumor microenvironment. It seems likely that the tumor microenvironment morphs the de-differentiated cells towards inflammatory and ECM-modifying cells.
The process of AMT also provides a new venue to interfere with tumor progression. Our previous studies suggested that suppressing fibrosis and inflammation, as achieved by endotrophin neutralizing antibodies, could curb tumor growth, although this intervention was not selective for adipocyte-derived precursors (Bu et al., 2019; Sun et al., 2014). Moreover, a number of studies have shown anti-inflammatory and anti-tumor proliferative activities of PPARγ ligands (Nakles et al., 2013; Vella et al., 2017). In liposarcomas, the PPARγ agonist rosiglitazone reverses adipocyte de-differentiation and slows tumor development (Bi et al., 2016). Beyond that, therapies combining rosiglitazone with MEK inhibitors have been suggested to incite partial mammary tumor cell differentiation into adipocytes, and these agents thus suppress tumor invasion and metastasis (Ishay-Ronen et al., 2019). Therefore, approaches that block adipocyte de-differentiation and curb AMT are likely to be beneficial to suppress tumor progression.
Clinically, the “obesity paradox” has been highlighted in a number of studies (Lennon et al., 2016). Obesity can also provide protective effects under some circumstances and act as a positive factor for cancer survival rather than being a driver for a worse outcome. There may be several mechanisms involved in this phenomenon, such as aging, altered tumor characteristics, and treatment responses. However, it is clear that the cellular physiology of the adipocyte is an important factor to be reckoned with. Healthy adipocytes display appropriate metabolic flexibility in response to extracellular stress via lipolysis, lipid esterification, or lipid synthesis (Zhu et al., 2022). However, this flexibility is dramatically reduced upon tumor invasion as AMT occurs, and it thus results in poor lipid-storage capacity in adipose tissues. In our study, XBP1s induces adipocyte de-differentiation and thus suppresses lipid accumulation in adipose tissues. These “altruistic” behaviors of adipocytes promote tumor growth despite lower adiposity. Conversely, MitoNEET enhances lipid uptake and favors re-esterification and storage in adipocytes. Due to these “selfish” properties, Ad-MitoNEET-PyMT mice show slower tumor progression despite greater adiposity. Thus, the metabolic features of the adipocytes, rather than adiposity per se, determine tumor progression. This highlights the potential for inhibitors of adipocyte lipolysis as potential agents to impair tumor invasiveness and progression.
Limitations of the study
Overall, our work highlights the mammary tumor-induced adipocyte de-differentiation and its mesenchymal transition during breast tumor progression. Unfortunately, we are not yet able to define the factors that drive this transition. A component of the tumor-derived “secretome” is a likely initiator. It could be alternate signals, such as hypoxia driven by the metabolic stress in the tumor microenvironment. Future efforts are warranted to determine these key drivers for the process.
STAR★METHODS
RESOURCE AVAILABILITY
Lead contact
Further information and resources and reagents should be directed to and will be fulfilled by the lead contact, Philipp Scherer (Philipp.Scherer@UTSouthwestern.edu).
Materials availability
Mouse lines generated in this study are available from the lead contact upon request.
Data and code availability
Single-cell RNA-seq data have been deposited at GEO and are publicly available as of the date of publication. Accession number is provided in the key resources table. Microscopy data reported in this paper will be shared by the lead contact upon request.
This paper did not report original code.
Any additional information required to reanalyze the data reported in this study is available from the lead contact upon request.
KEY RESOURCES TABLE.
| REAGENT or RESOURCE | SOURCE | IDENTIFIER |
|---|---|---|
|
| ||
| Antibodies | ||
| Guinea Pig anti-Perilipin antibody | Fitzgerald | Cat#20R-PP004; RRID: AB_1288416 |
| Chicken anti-GFP antibody | Abcam | Cat# ab13970; RRID: AB_300798 |
| Mouse anti- α-SMA antibody | eBioscience™ | Cat# 14-9760-80; RRID:AB_2572995 |
| Rabbit anti-CD31 antibody | Abcam | Cat# ab28364; RRID:AB_726362 |
| Rat anti-Mac2 antibody | CEDARLANE | Cat# CL8942AP; RRID:AB_10060357 |
| Rabbit anti-FSP1 antibody | Abcam | Cat# ab41532; RRID:AB_945346 |
| Rabbit anti- Desmin antibody | Cell Signaling | Cat# 5332; RRID:AB_1903947 |
|
| ||
| Biological samples | ||
|
| ||
| Human breast cancer sample slices | National Disease Research Interchange | https://ndriresource.org/ |
|
| ||
| Chemicals, peptides, and recombinant proteins | ||
|
| ||
| Collagen hybridizing peptide | Advanced BioMatrix | Cat# 5276 |
| Collagenase D | Roche | Cat# 11088882001 |
| Collagenase B | Roche | Cat# 11088831001 |
| Deoxyribonuclease | Sigma-Aldrich | Cat# DN25 |
| Intralipid | Sigma-Aldrich | Cat# I141 |
| BSA | Sigma-Aldrich | Cat# A8806 |
|
| ||
| Critical commercial assays | ||
|
| ||
| Seahorse XFe24 FluxPak | Agilent | Cat# 102340-100 |
| Mouse Insulin ELISA Jumbo Pack | ALPCO | Cat# 80-INSMS-E10 |
| Mouse Leptin ELISA Kit | Crystal Chem | Cat# 90030 |
| Mouse Adiponectin ELISA | EMD Millipore | Cat# EZMADP-60K |
| LabAssay ™ NEFA | Wako | Cat# 633-52001 |
| LabAssay ™ Triglyceride | Wako | Cat# 632-50991 |
| LabAssay ™ Cholesterol | Wako | Cat# 635-50981 |
| Oil red O stain kit | Abcam | Cat# ab150678 |
| iScript cDNA Synthesis Kit | BIO-RAD | Cat# 170-8891 |
| Sybr Green Master Mix | Applied biosystems | Cat# A25778 |
|
| ||
| Deposited data | ||
|
| ||
| Single cell RNA-seq | This paper | GSE148646 |
|
| ||
| Experimental models: Cell lines | ||
|
| ||
| Mouse: EO771 cells | Gift from Rolf Brekken Lab, University of Texas Southwestern Medical Center | N/A |
| Mouse: Met1 cells | Gift from Rolf Brekken Lab, University of Texas Southwestern Medical Center | N/A |
| Human: ZR-75-1 cells | Hamon Cancer Center Collection, University of Texas Southwestern Medical Center | N/A |
| Human: MDA-MB-231 cells | Hamon Cancer Center Collection, University of Texas Southwestern Medical Center | N/A |
| Human: MDA-MB-453 cells | Hamon Cancer Center Collection, University of Texas Southwestern Medical Center | N/A |
| Human: MCF7 cells | Hamon Cancer Center Collection, University of Texas Southwestern Medical Center | N/A |
|
| ||
| Experimental models: Organisms/strains | ||
|
| ||
| Mouse: Adipoq-rtTA | Deng et al. (2018) | N/A |
| Mouse: aP2-MitoNEET | Kusminski et al. (2012) | N/A |
| Mouse: TRE-Xbp1s | Deng et al. (2018) | N/A |
| Mouse: Adipoq-Xbp1s | Generated in-house | N/A |
| Mouse: MMTV-PyMT | Gift from Jeffrey Pollard Lab, Albert Einstein College of Medicine | N/A |
| Mouse: Adipoq-Cre | The Jackson Laboratory | JAX: 028020; RRID: IMSR_JAX:028020 |
| Mouse: TRE-Cre | The Jackson Laboratory | JAX: 006234; RRID: IMSR_JAX:006234 |
| Mouse: Rosa26-mT/mG | The Jackson Laboratory | JAX: 007676; RRID: IMSR_JAX:007676 |
|
| ||
| Oligonucleotides | ||
|
| ||
| Primers for qPCR | See Table S4 | N/A |
|
| ||
| Software and algorithms | ||
|
| ||
| Image J | NIH | https://imagej.nih.gov/ij/ |
| Prism | GraphPad | https://www.graphpad.com/ |
| R studio | RStudio | https://www.rstudio.com/ |
| Cell Ranger | 10X GENOMICS | https://www.10xgenomics.com/ |
| Seurat package 3.0 | Satija Lab and Collaborators | https://satijalab.org/seurat/ |
| MetaboAnalyst 5.0 | MetaboAnalyst | https://www.metaboanalyst.ca/ |
| DAVID 6.8 | DAVID Bioinformatics Resources | https://david.ncifcrf.gov/home.jsp |
|
| ||
| Other | ||
|
| ||
| 60% HFD paste | BioServ | S1850 |
| Normal chow diet | Lab Diet | 5058 |
| Doxycycline chow diet (600 mg/kg diet) | BioServ | S4107 |
EXPERIMENTAL MODEL AND SUBJECT DETAILS
Patient samples
Slices from female patients were randomly obtained from the National Disease Research Interchange. Patients received total mastectomy or simple radical mastectomy. No known presurgical therapy was applied. The study was approved by University of Texas Southwestern Medical Center Institutional Review Board protocol # STU 012017–043 (Endotrophin Upregulation in Tumor Tissues). The tumor characteristics of the samples are presented in Table S2.
Mice
Mice were housed at 22°C with 12-h light-dark cycles and free access to water and food (chow #5058, Lab Diet; doxycycline chow (dox, 600 mg/kg), BioServ). Female mice at 6–8 weeks of age were used to initiate the studies. All mouse protocols were approved by the Institute Animal Care and Use Committees of University of Texas Southwestern Medical Center (APN: 2015–101207G). Ad-Xbp1s-PyMT mice were either on a C57BL/6 or FVB background as indicated. All other mice were on a C57BL/6 background. Adipoq-promoter driven Adipoq-rtTA, aP2-promoter driven aP2-MitoNEET, and TRE-Xbp1s mouse lines were generated as previous described (Deng et al., 2018; Kusminski et al., 2012). Ad-Xbp1s transgenic mouse line was generated using the cDNA of the spliced version of mouse Xbp1 (Xbp1s), in which a constitutive expression of Xbp1s is achieved under the control of Adipoq-promoter. Adipoq-Cre, TRE-Cre and Rosa26-mT/mG mice were obtained from Jackson Laboratories. MMTV-PyMT mice were a kind gift of Dr. Jeffrey Pollard. In the dox-dependent ‘pulse-labeling’ adipocyte-tracer system, PyMTChaser or AlloChaser mice were fed with dox-chow diet for two weeks at age of three months and then switch to normal chow diet until sacrifice. Mice were dissected and tissues were collected for histology and cell isolation when the tumors reached approximately one centimeter in diameter. For the tumor grafting, E0771 breast cancer cells (0.25 × 106 cells) were implanted with a 21-gauge needle into mammary fat pad for five weeks.
Cell lines and primary cells
Human breast cancer cell lines, including ZR-75–1, MDA-MB-231, MDA-MB-453 and MCF7, were obtained from the Hamon Cancer Center Collection, University of Texas Southwestern Medical Center. Mouse breast cancer cell lines including Met1 and EO771 were kind gifts from Rolf Brekken Lab at University of Texas Southwestern Medical Center.
Tumor induced-dedifferentiated adipocytes (EGFP+, tdTomato−) and tumor-associated fibroblasts (EGFP−, tdTomato+) were isolated from EO771-allografts or PyMT-tumor tissues from AdipoChaser mice by flow cytometry as described below.
METHOD DETAILS
Body composition analyses
Fat, lean, and fluid mass of chow-fed and high fat-fed mice were measured using a Bruker Minispec mq10 system (Bruker).
Metabolic analyses
Blood from fed and overnight fasted mice were collected. Blood glucose levels were determined with a glucose meter. Serum insulin, leptin, and adiponectin were determined by commercially available ELISA kits (for insulin, cat# 80-INSMS-E10, ALPCO; for leptin, cat# 90030, Crystal Chem; for adiponectin, cat# EZMADP-60K, EMD Millipore) following the manufacturers’ instructions, respectively. Serum non-esterified fatty acids (NEFA), triglycerides and cholesterol were determined by using enzymatic colorimetric methods with reagents from FUJIFILM Wako Chemicals.
For triglyceride tolerance tests, mice were fast overnight, and received a 20% Intralipid emulsions (I141, Sigma) via oral gavage. Blood was collected at the indicated time points. Serum triglycerides were determined as described above.
Histological analyses and immunofluorescent staining
Patient slices were obtained from the National Disease Research Interchange. All patients were diagnosed with invasive lobular/ductal carcinoma breast, Grade 2. Patients received total mastectomy or simple radical mastectomy with no known presurgical therapy. Samples were analyzed under the approved University of Texas Southwestern Medical Center Institutional Review Board protocol # STU 012017-043 (Endotrophin Upregulation in Tumor Tissues). Mouse tissues were dissected and fixed in 10% formalin overnight. Paraffin processing, embedding, sectioning and hematoxylin/eosin staining and trichrome staining were performed by John Sheldon at UTSW Medical Center. Immunostaining was performed following standard protocols, with the following antibodies: Perilipin (1:1000, #20R-PP004, Fitzgerald), EGFP (1:500, ab13970, Abcam), α-SMA (1:500, eBioscience™, #14-9760-80), CD31 (1:200, Abcam, #ab28364), Mac2 (1:750, CL8942AP, CEDARLANE) and collagen hybridizing peptide (#5276, Advanced BioMatrix). Images were acquired with LSM510 confocal microscope (Zeiss) and analyzed by Image J software.
Adipocyte area quantification
Histological images were processed using ImageJ. Digital images were thresholded and all foreground pixels were assigned to the area occupied by the extracellular space (equivalent to fibrous area). Background pixels were used to calculate adipocyte-specific statistics. Thresholding also enabled a technique for fibrotic thickness measurement described below. Individual adipocytes were captured using the “Analyze Particles” command, with minimum and maximum particle sizes set to 150 and 30,000 square microns respectively, and with a minimum circularity of 0.4. The fibrotic thickness associated with each adipocyte was calculated as the radial distance from an adipocyte’s perimeter to an adjacent adipocyte (or other feature characterized by the image’s background). Eight such projections are measured, and each cell is assigned values equal to the mean, median, and highest-observed values of these fibrotic thicknesses. The means of these values are then recorded as statistics characterizing for each image. The statistics collected from each image are outlined in the Table S1.
Cell culture and treatments
Fibroblasts from tumor were isolated following standard protocol as previous described (Sharon et al., 2013). Briefly, mammary tumor was carefully dissected and the necrotic regions were removed. Tissues were then minced thoroughly and digested with an enzyme cocktail containing collagenase B (0.25%, w/v), collagenase D (0.25%, w/v), deoxyribonuclease (0.05%, w/v) and BSA (0.5%, w/v). Fluorescence-Activated Cell Sorting (FACS) was used to isolate de-differentiated adipocytes (EGFP+, tdTomato−) and tumor-associated fibroblasts (EGFP−, tdTomato+) cells, which was conducted by UT Southwestern Medical Center Flow Cytometry Core Facility. It ruled out any possible contamination of adipocytes from the isolation due to cell size-limitation in the flow cytometry. Cells were cultured in DMEM/F12 containing 10% FBS and penicillin/streptomycin. For LPS treatment, cells were incubated either with vehicle (PBS) or LPS (100 ng/mL) for 3 h. For adipogenic assay, cells were treated with differentiation cocktail containing 5 μg/mL insulin, 2 μg/mL dexamethasone, 0.5 mM 3-isobutyl-1-methylxanthine and 0.5 μM rosiglitazone. Two days later, cells were refreshed with maintenance medium containing 5 μg/mL insulin. Oil Red O staining was performed as previously described (Zhu et al., 2017) at day 6 of differentiation. Myogenesis was accessed by switching to differentiation medium containing 2% horse serum. Immunofluorescent staining was performed with antibodies against FSP1 (1: 200, ab41532, Abcam) and Desmin (1:100, #5332, Cell Signaling).
Metabolite profiling
Equal number of de-differentiated adipocytes (EGFP+, tdTomato−) and tumor-associated fibroblasts (EGFP−, tdTomato+) cells were seeded in 12-well-plate and the supernatant were collected for overnight (fed condition) or 6 h in the absence of serum (fasted condition). Small molecules of water-soluble metabolites in the supernatant were analyzed by LC-MS/MS (Shimadzu Scientific Instruments, Columbia, MD). Samples were processed and analyzed on a CLAM 2030 fully antomated sample preparation module for LC-MS coupled to a Nexera X2 UHPLC system and an LC-MS-8060 triple quadrupole mass spectrometer using the instrument parameters and chromatographic conditions described in the Shimadzu LC-MS/MS Method Package for Cell Culture Profiling. The sample preparation sequence was programmed as follows: addition of 80 μL of MeOH onto CLAM 2030 filtration vial; addition of 20 μL of sample; addition of 10 μL of internal standard cocktail; vortexing for 60 seconds at 1,900 rpm; addition of 100 μL of MeOH; vortexing for 60 seconds at 1,900 rpm, addition of 100 μL of water, vortexing for 90 seconds at 1,900 rpm, filtration for 70 seconds. Collected sample into the collection vial is then automatically injected for LC-MS/MS analysis (1 μL injection). Interactive principal component analysis (iPCA) and pathway analyses were conducted with MetaboAnalyst 5.0 (https://www.metaboanalyst.ca/). Pathways with p < 0.05 and impact value >0.1 are marked and considered as the most relevant pathways.
Seahorse metabolic flux assay
Cells were seeded in XF 24-well-plate (5×104 cells/well), and glycolysis and mitochondrial stress assays were performed following standard procedures, respectively. For glycolysis assay, final 10 mM glucose, 1 μM oligomycin and 50 mM 2-Deoxy-D-glucose (2-DG) were used. For mitochondrial stress assay, final 1 μM oligomycin, 4 μM FCCP, 3 μM rotenone and 2 μM antimycin A were used.
Cell proliferation
Cancer cells were seeded in 96-well-plate (7000 cells/well). After attached, cells were treated with conditioned medium from dedifferentiated adipocytes (EGFP+, tdTomato−) and tumor-associated fibroblasts (EGFP−, tdTomato+) cell cultures at a ratio of 1:1. At the indicated time points, cells were washed with PBS and stained with 0.5% crystal violet solution. OD570 values were measured by a microplate reader, and used to calculate the cell proliferation rate.
Single cell RNA sequencing
Isolated de-differentiated adipocytes (EGFP+, tdTomato−) cells from PyMT- or allograft-tumor were applied for single-cell sequencing. Standard procedures provided from 10X Genomics were used for sample preparation and library construction as described previously (Hepler et al., 2018). Sequencing was conducted on an IIlumina NextSeq 500 High Output (400M) by the UTSW McDermott Center Next Generation Sequencing Core. Paired-end reads were obtained using one flow cell with the following length input: 26 bp Read 1, 58 bp Read 2, and 8 bp Index 1. Sample demultiplexing, alignment, filtering was performed with Cell Ranger software (v2.1.0) as previously described (Hepler et al., 2018). Clustering and gene expression were visualized with the Seurat package (version 3.0) (Satija Lab, Seurat; https://satijalab.org/seurat/) in RStudio (Version 1.2.5019). Violin plots and tSNE plots were generated by using the Seurat toolkits in RStudio. Gene ontology (GO) enrichment analyses were conducted with DAVID 6.8 (https://david.ncifcrf.gov/).
RNA isolation and quantitative RT-PCR
Total RNA was extracted by using the traditional Trizol method. Quantitative gene expression was performed by two-step quantitative RT-PCR using iScript cDNA Synthesis Kits (#170–8891, Bio-Rad) and SYBR Green PCR Master Mix (Applied Biosystems). mRNA expression levels were determined using the ΔΔCT method and normalized to the housekeeping genes Rps16 and Rps18. Primers were listed in Table S4.
QUANTIFICATION AND STATISTICAL ANALYSIS
All data were expressed as mean ± SEM. Statistical significance was determined according to the number of biological replicates as indicated in the figure legends (***p ≤ 0.001, **p ≤ 0.01, *p ≤ 0.05). For two independent data sets, two tailed Student’s t-test was used. For multiple comparisons, one-way or two-way ANOVA were used with Holm-Sidak’s multiple comparisons test. The statistical analyses were performed with GraphPad Prism 8.0 (GraphPad Software, Inc. La Jolla, CA, USA).
Supplementary Material
Highlights.
Mammary adipocytes undergo de-differentiation upon breast tumor invasion
De-differentiated adipocytes display myofibroblast- and macrophage-like features
Xbp1s causes adipocyte de-differentiation and promotes tumor growth
MitoNEET enhances lipid-storage capacity in adipocytes and curbs tumor growth
ACKNOWLEDGMENTS
We thank all members of Scherer and Gupta laboratories for their support of this study. We would also like to thank the UTSW Metabolic Phenotyping Core Facility, the McDermott Center Next Generation Sequencing Core and Bioinformatics Lab, the UTSW Flow Cytometry Facility, the Histo-Pathology Core, the Live Cell Imaging Core, UTSW ARC, and Charlotte Lee for help with histology. We thank Shimadzu Scientific Instruments for the collaborative efforts in mass spectrometry technology resources. This study was supported by US National Institutes of Health grants R01-DK104789 to R.K.G.; RC2-DK118620 to P.E.S. and R.K.G.; and R01-DK55758, R01-DK099110, R01-DK118620, R01-DK127274, and R01-131537 to P.E.S. Y.Z. was supported by a NIH K99 grant (K99-DK114498). S.Z. was supported by a NIH grant (K99-AG068239). Q. Zhu was supported by American Heart Association Career Development Award 855170. O.V. and J.J.R. are supported by NIH grants P51-OD01192 for operation of the Oregon National Primate Research Center and 1S10-OD025002-01.
Footnotes
DECLARATION OF INTERESTS
The authors declare no competing interests.
SUPPLEMENTAL INFORMATION
Supplemental information can be found online at https://doi.org/10.1016/j.celrep.2022.111362.
REFERENCES
- Bi P, Yue F, Karki A, Castro B, Wirbisky SE, Wang C, Durkes A, Elzey BD, Andrisani OM, Bidwell CA, et al. (2016). Notch activation drives adipocyte dedifferentiation and tumorigenic transformation in mice. J. Exp. Med. 213, 2019–2037. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bochet L, Lehuédé C, Dauvillier S, Wang YY, Dirat B, Laurent V, Dray C, Guiet R, Maridonneau-Parini I, Le Gonidec S, et al. (2013). Adipocyte-derived fibroblasts promote tumor progression and contribute to the desmoplastic reaction in breast cancer. Cancer Res. 73, 5657–5668. [DOI] [PubMed] [Google Scholar]
- Bu D, Crewe C, Kusminski CM, Gordillo R, Ghaben AL, Kim M, Park J, Deng H, Xiong W, Liu XZ, et al. (2019). Human endotrophin as a driver of malignant tumor growth. JCI Insight 5, 125094. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cairns RA, Harris IS, and Mak TW (2011). Regulation of cancer cell metabolism. Nat. Rev. Cancer 11, 85–95. [DOI] [PubMed] [Google Scholar]
- Carrière A, Jeanson Y, Côté JA, Dromard C, Galinier A, Menzel S, Barreau C, Dupuis-Coronas S, Arnaud E, Girousse A, et al. (2017). Identification of the ectoenzyme CD38 as a marker of committed preadipocytes. Int. J. Obes. 41, 1539–1546. [DOI] [PubMed] [Google Scholar]
- Cawthorn WP, Scheller EL, and MacDougald OA (2012). Adipose tissue stem cells meet preadipocyte commitment: going back to the future. J. Lipid Res. 53, 227–246. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Charrière G, Cousin B, Arnaud E, André M, Bacou F, Pénicaud L, and Casteilla L (2003). Preadipocyte conversion to macrophage: evidence of plasticity. J. Biol. Chem. 278, 9850–9855. [DOI] [PubMed] [Google Scholar]
- Coussens LM, and Werb Z (2002). Inflammation and cancer. Nature 420, 860–867. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Deng Y, Wang ZV, Gordillo R, Zhu Y, Ali A, Zhang C, Wang X, Shao M, Zhang Z, Iyengar P, et al. (2018). Adipocyte Xbp1s overexpression drives uridine production and reduces obesity. Mol. Metab. 11, 1–17. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dey P, Kimmelman AC, and DePinho RA (2021). Metabolic codependencies in the tumor microenvironment. Cancer Discov. 11, 1067–1081. candisc.1211. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dirat B, Bochet L, Dabek M, Daviaud D, Dauvillier S, Majed B, Wang YY, Meulle A, Salles B, Le Gonidec S, et al. (2011). Cancer-associated adipocytes exhibit an activated phenotype and contribute to breast cancer invasion. Cancer Res. 71, 2455–2465. [DOI] [PubMed] [Google Scholar]
- Elwood JM, Tin Tin S, Kuper-hommel M, Lawrenson R, and Campbell I (2018). Obesity and breast cancer outcomes in chemotherapy patients in New Zealand – a population-based cohort study. BMC Cancer 18, 1–13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hepler C, Shan B, Zhang Q, Henry GH, Shao M, Vishvanath L, Ghaben AL, Mobley AB, Strand D, Hon GC, and Gupta RK (2018). Identification of functionally distinct fibro-inflammatory and adipogenic stromal subpopulations in visceral adipose tissue of adult mice. Elife 7. e39636–36. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ishay-Ronen D, Diepenbruck M, Kalathur RKR, Sugiyama N, Tiede S, Ivanek R, Bantug G, Morini MF, Wang J, Hess C, and Christofori G (2019). Gain fat—lose metastasis: converting invasive breast cancer cells into adipocytes inhibits cancer metastasis. Cancer Cell 35, 17–32.e6. [DOI] [PubMed] [Google Scholar]
- Jackson HW, Fischer JR, Zanotelli VRT, Ali HR, Mechera R, Soysal SD, Moch H, Muenst S, Varga Z, Weber WP, and Bodenmiller B (2020). The single-cell pathology landscape of breast cancer. Nature 578, 615–620. [DOI] [PubMed] [Google Scholar]
- Kajimura S, Spiegelman BM, and Seale P (2015). Brown and beige fat: physiological roles beyond Heat generation. Cell Metab. 22, 546–559. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Khandekar MJ, Cohen P, and Spiegelman BM (2011). Molecular mechanisms of cancer development in obesity. Nat. Rev. Cancer 11, 886–895. [DOI] [PubMed] [Google Scholar]
- Kusminski CM, Holland WL, Sun K, Park J, Spurgin SB, Lin Y, Askew GR, Simcox JA, McClain DA, Li C, and Scherer PE (2012). MitoNEET-driven alterations in adipocyte mitochondrial activity reveal a crucial adaptive process that preserves insulin sensitivity in obesity. Nat. Med. 18, 1539–1549. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lennon H, Sperrin M, Badrick E, and Renehan AG (2016). The obesity paradox in cancer: a review. Curr. Oncol. Rep. 18, 56. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li Y, Mao AS, Seo BR, Zhao X, Gupta SK, Chen M, Han YL, Shih TY, Mooney DJ, and Guo M (2020). Compression-induced dedifferentiation of adipocytes promotes tumor progression. Sci. Adv. 6. eaax5611–14. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lin EY, and Pollard JW (2004). Role of infiltrated leucocytes in tumour growth and spread. Br. J. Cancer 90, 2053–2058. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Merrick D, Sakers A, Irgebay Z, Okada C, Calvert C, Morley MP, Percec I, and Seale P (2019). Identification of a mesenchymal progenitor cell hierarchy in adipose tissue. Science 364, aav2501. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Molocea CE, Tsokanos FF, and Herzig S (2020). Exploiting common aspects of obesity and cancer cachexia for future therapeutic strategies. Curr. Opin. Pharmacol. 53, 101–116. [DOI] [PubMed] [Google Scholar]
- Nakles RE, Kallakury BVS, and Furth PA (2013). The PPARγ agonist efatutazone increases the spectrum of well-differentiated mammary cancer subtypes initiated by loss of full-length BRCA1 in association with TP53 haploinsufficiency. Am. J. Pathol. 182, 1976–1985. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Park J, Morley TS, Kim M, Clegg DJ, and Scherer PE (2014). Obesity and cancer - mechanisms underlying tumour progression and recurrence. Nat. Rev. Endocrinol. 10, 455–465. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Quail DF, and Dannenberg AJ (2019). The obese adipose tissue microenvironment in cancer development and progression. Nat. Rev. Endocrinol. 15, 139–154. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sharon Y, Alon L, Glanz S, Servais C, and Erez N (2013). Isolation of normal and cancer-associated fibroblasts from fresh tissues by fluorescence activated cell sorting (FACS). J. Vis. Exp. 1–6, e4425. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Singh R, Parveen M, Basgen JM, Fazel S, Meshesha MF, Thames EC, Moore B, Martinez L, Howard CB, Vergnes L, et al. (2016). Increased expression of beige/brown adipose markers from host and breast cancer cells influence xenograft formation in mice. Mol. Cancer Res. 14, 78–92. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sun K, Park J, Gupta OT, Holland WL, Auerbach P, Zhang N, Goncalves Marangoni R, Nicoloro SM, Czech MP, Varga J, et al. (2014). Endotrophin triggers adipose tissue fibrosis and metabolic dysfunction. Nat. Commun. 5, 3485. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Thway K (2019). Well-differentiated liposarcoma and dedifferentiated liposarcoma: an updated review. Semin. Diagn. Pathol. 36, 112–121. [DOI] [PubMed] [Google Scholar]
- Vella V, Nicolosi ML, Giuliano S, Bellomo M, Belfiore A, and Malaguarnera R (2017). PPAR-γ agonists as antineoplastic agents in cancers with dysregulated IGF axis. Front. Endocrinol. 8, 31. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang QA, Song A, Chen W, Schwalie PC, Zhang F, Vishvanath L, Jiang L, Ye R, Shao M, Tao C, et al. (2018). Reversible de-differentiation of mature white adipocytes into preadipocyte-like precursors during lactation. Cell Metab. 28, 282–288.e3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang YY, Attané C, Milhas D, Dirat B, Dauvillier S, Guerard A, Gilhodes J, Lazar I, Alet N, Laurent V, et al. (2017). Mammary adipocytes stimulate breast cancer invasion through metabolic remodeling of tumor cells. JCI Insight 2. e87489–20. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wei S, Zan L, Hausman GJ, Rasmussen TP, Bergen WG, and Dodson MV (2013). Dedifferentiated adipocyte-derived progeny cells (DFAT cells). Adipocyte 2, 122–127. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Winkler J, Abisoye-Ogunniyan A, Metcalf KJ, and Werb Z (2020). Concepts of extracellular matrix remodelling in tumour progression and metastasis. Nat. Commun. 11, 5120. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang Y, Daquinag AC, Amaya-Manzanares F, Sirin O, Tseng C, and Kolonin MG (2012). Stromal progenitor cells from endogenous adipose tissue contribute to pericytes and adipocytes that populate the tumor microenvironment. Cancer Res. 72, 5198–5208. [DOI] [PubMed] [Google Scholar]
- Zhang Z, Shao M, Hepler C, Zi Z, Zhao S, An YA, Zhu Y, Ghaben AL, Wang M-Y, Li N, et al. (2019). Dermal adipose tissue has high plasticity and undergoes reversible dedifferentiation in mice. J. Clin. Invest. 129, 5327–5342. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhu Q, An YA, and Scherer PE (2022). Mitochondrial regulation and white adipose tissue homeostasis. Trends Cell Biol. 32, 351–364. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhu Q, and Scherer PE (2018). Immunologic and endocrine functions of adipose tissue: implications for kidney disease. Nat. Rev. Nephrol. 14, 105–120. [DOI] [PubMed] [Google Scholar]
- Zhu Q, Ghoshal S, Tyagi R, and Chakraborty A (2017). Global IP6K1 deletion enhances temperature modulated energy expenditure which reduces carbohydrate and fat induced weight gain. Mol. Metab. 6, 73–85. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Data Availability Statement
Single-cell RNA-seq data have been deposited at GEO and are publicly available as of the date of publication. Accession number is provided in the key resources table. Microscopy data reported in this paper will be shared by the lead contact upon request.
This paper did not report original code.
Any additional information required to reanalyze the data reported in this study is available from the lead contact upon request.
KEY RESOURCES TABLE.
| REAGENT or RESOURCE | SOURCE | IDENTIFIER |
|---|---|---|
|
| ||
| Antibodies | ||
| Guinea Pig anti-Perilipin antibody | Fitzgerald | Cat#20R-PP004; RRID: AB_1288416 |
| Chicken anti-GFP antibody | Abcam | Cat# ab13970; RRID: AB_300798 |
| Mouse anti- α-SMA antibody | eBioscience™ | Cat# 14-9760-80; RRID:AB_2572995 |
| Rabbit anti-CD31 antibody | Abcam | Cat# ab28364; RRID:AB_726362 |
| Rat anti-Mac2 antibody | CEDARLANE | Cat# CL8942AP; RRID:AB_10060357 |
| Rabbit anti-FSP1 antibody | Abcam | Cat# ab41532; RRID:AB_945346 |
| Rabbit anti- Desmin antibody | Cell Signaling | Cat# 5332; RRID:AB_1903947 |
|
| ||
| Biological samples | ||
|
| ||
| Human breast cancer sample slices | National Disease Research Interchange | https://ndriresource.org/ |
|
| ||
| Chemicals, peptides, and recombinant proteins | ||
|
| ||
| Collagen hybridizing peptide | Advanced BioMatrix | Cat# 5276 |
| Collagenase D | Roche | Cat# 11088882001 |
| Collagenase B | Roche | Cat# 11088831001 |
| Deoxyribonuclease | Sigma-Aldrich | Cat# DN25 |
| Intralipid | Sigma-Aldrich | Cat# I141 |
| BSA | Sigma-Aldrich | Cat# A8806 |
|
| ||
| Critical commercial assays | ||
|
| ||
| Seahorse XFe24 FluxPak | Agilent | Cat# 102340-100 |
| Mouse Insulin ELISA Jumbo Pack | ALPCO | Cat# 80-INSMS-E10 |
| Mouse Leptin ELISA Kit | Crystal Chem | Cat# 90030 |
| Mouse Adiponectin ELISA | EMD Millipore | Cat# EZMADP-60K |
| LabAssay ™ NEFA | Wako | Cat# 633-52001 |
| LabAssay ™ Triglyceride | Wako | Cat# 632-50991 |
| LabAssay ™ Cholesterol | Wako | Cat# 635-50981 |
| Oil red O stain kit | Abcam | Cat# ab150678 |
| iScript cDNA Synthesis Kit | BIO-RAD | Cat# 170-8891 |
| Sybr Green Master Mix | Applied biosystems | Cat# A25778 |
|
| ||
| Deposited data | ||
|
| ||
| Single cell RNA-seq | This paper | GSE148646 |
|
| ||
| Experimental models: Cell lines | ||
|
| ||
| Mouse: EO771 cells | Gift from Rolf Brekken Lab, University of Texas Southwestern Medical Center | N/A |
| Mouse: Met1 cells | Gift from Rolf Brekken Lab, University of Texas Southwestern Medical Center | N/A |
| Human: ZR-75-1 cells | Hamon Cancer Center Collection, University of Texas Southwestern Medical Center | N/A |
| Human: MDA-MB-231 cells | Hamon Cancer Center Collection, University of Texas Southwestern Medical Center | N/A |
| Human: MDA-MB-453 cells | Hamon Cancer Center Collection, University of Texas Southwestern Medical Center | N/A |
| Human: MCF7 cells | Hamon Cancer Center Collection, University of Texas Southwestern Medical Center | N/A |
|
| ||
| Experimental models: Organisms/strains | ||
|
| ||
| Mouse: Adipoq-rtTA | Deng et al. (2018) | N/A |
| Mouse: aP2-MitoNEET | Kusminski et al. (2012) | N/A |
| Mouse: TRE-Xbp1s | Deng et al. (2018) | N/A |
| Mouse: Adipoq-Xbp1s | Generated in-house | N/A |
| Mouse: MMTV-PyMT | Gift from Jeffrey Pollard Lab, Albert Einstein College of Medicine | N/A |
| Mouse: Adipoq-Cre | The Jackson Laboratory | JAX: 028020; RRID: IMSR_JAX:028020 |
| Mouse: TRE-Cre | The Jackson Laboratory | JAX: 006234; RRID: IMSR_JAX:006234 |
| Mouse: Rosa26-mT/mG | The Jackson Laboratory | JAX: 007676; RRID: IMSR_JAX:007676 |
|
| ||
| Oligonucleotides | ||
|
| ||
| Primers for qPCR | See Table S4 | N/A |
|
| ||
| Software and algorithms | ||
|
| ||
| Image J | NIH | https://imagej.nih.gov/ij/ |
| Prism | GraphPad | https://www.graphpad.com/ |
| R studio | RStudio | https://www.rstudio.com/ |
| Cell Ranger | 10X GENOMICS | https://www.10xgenomics.com/ |
| Seurat package 3.0 | Satija Lab and Collaborators | https://satijalab.org/seurat/ |
| MetaboAnalyst 5.0 | MetaboAnalyst | https://www.metaboanalyst.ca/ |
| DAVID 6.8 | DAVID Bioinformatics Resources | https://david.ncifcrf.gov/home.jsp |
|
| ||
| Other | ||
|
| ||
| 60% HFD paste | BioServ | S1850 |
| Normal chow diet | Lab Diet | 5058 |
| Doxycycline chow diet (600 mg/kg diet) | BioServ | S4107 |


