Abstract
Desulfovibrio gigas neelaredoxin is an iron-containing protein of 15 kDa, having a single iron site with a His4Cys coordination. Neelaredoxins and homologous proteins are widespread in anaerobic prokaryotes and have superoxide-scavenging activity. To further understand its role in anaerobes, its genomic organization and expression in D. gigas were studied and its ability to complement Escherichia coli superoxide dismutase deletion mutant was assessed. In D. gigas, neelaredoxin is transcribed as a monocistronic mRNA of 500 bases as revealed by Northern analysis. Putative promoter elements resembling ς70 recognition sequences were identified. Neelaredoxin is abundantly and constitutively expressed, and its expression is not further induced during treatment with O2 or H2O2. The neelaredoxin gene was cloned by PCR and expressed in E. coli, and the protein was purified to homogeneity. The recombinant neelaredoxin has spectroscopic properties identical to those observed for the native one. Mutations of Cys-115, one of the iron ligands, show that this ligand is essential for the activity of neelaredoxin. In an attempt to elucidate the function of neelaredoxin within the cell, it was expressed in an E. coli mutant deficient in cytoplasmic superoxide dismutases (sodA sodB). Neelaredoxin suppresses the deleterious effects produced by superoxide, indicating that it is involved in oxygen detoxification in the anaerobe D. gigas.
Sulfate-reducing bacteria (SRB) were shown to occur in oxic surface layers of aquatic habitats and to be active in microbial mats at oxygen tensions near saturation (9, 21). Moreover, they can survive prolonged exposure to oxygen (14, 49). These observations suggest that oxygen may play a physiological role in these so-called strict anaerobes, although it has not been clearly demonstrated that SRB can grow in the presence of oxygen as the sole electron acceptor (30). Indeed, in the presence of oxygen the sulfate reducer Desulfovibrio gigas is capable of generating ATP from internal reserves of polyglucose (16, 48). Analysis of oxygen utilization by this bacterium revealed that the full reduction of oxygen directly to water is linked to NADH oxidation through a three-component soluble electron transfer chain (11, 20, 25).
The fact that SRB can tolerate transient contact with oxygen indicates that these microorganisms have protective mechanisms that will eliminate toxic oxygen products, which causes serious damage at the protein and DNA levels (18, 28, 45, 58). Indeed, catalase and superoxide dismutase (SOD) were found in some Desulfovibrio species (15, 27). Moreover, SRB contain several other nonheme and mononuclear iron proteins, such as rubredoxin, desulfoferrodoxin, neelaredoxin, and rubrerythrin, whose functions are possibly related to defense mechanisms against oxidative stress (41, 46, 47, 50, 59). Furthermore, besides the presence of oxygen-utilizing or -detoxifying enzymes, some SRB may also have chemosensory pathways that trigger physiological responses to the presence of oxygen (22). It was reported that D. gigas can avoid the deleterious effect of reactive oxygen species (ROS), either through scavenging oxygen by reducing it to water (11) or through reactions involving canonical ROS-detoxifying enzymes such as FeSOD and catalase (15). Another protein that is likely to be involved in defense mechanisms against ROS in D. gigas cells is neelaredoxin (Nlr) (50).
Nlr is a small (∼15-kDa) iron-containing protein, first discovered in D. gigas (12). Its name comes from the fact that it was the first blue iron protein (Neela), due to the presence of a so far unique iron center, penta-coordinated by four histidine residues in the basal plane and a cysteine residue in the axial position (13, 50, 60). This metal site is identical to center II of another mononuclear iron protein, desulfoferrodoxin (Dfx), also first found in SRB (13, 43, 55). Dfx contains another metal site, at the N-terminal domain, similar to that of D. gigas desulforedoxin (Dx), a small rubredoxin-like protein with no attributed function and with one iron atom bound in a distorted tetrahedral configuration to four cysteines (2, 13, 42). It is now clear that Nlr and Dfx proteins are widespread in anaerobic prokaryotes; in particular, they are found in all genomes of anaerobes so far sequenced, which shows their general biological relevance.
These proteins have superoxide-scavenging activity, either as SODs or as superoxide reductases (1, 29, 31, 39, 40, 47, 50). Their activity is due to the common Neela center, the His4Cys iron site, since it was shown that the overproduction of Dfx from Desulfoarculus barsii complements the SOD− phenotype of an Escherichia coli mutant strain deficient in cytoplasmic SODs, whereas overproduction of D. gigas Dx, corresponding to the Dfx first domain, does not (46).
In spite of a wealth of structural and spectroscopic data on this protein family and of the numerous reports on the in vitro activity of its members as superoxide scavengers, very little is known at the physiological level, i.e., the level of genomic organization, expression, and function.
The present report shows that the D. gigas nlr gene is contained in a monocistronic unit and that it is constitutively expressed. The mRNA levels are not increased if cells are subjected to oxidative stress. Data obtained with recombinant wild-type Nlr proteins and those mutated at the cysteine iron ligand show that this residue is essential for the catalytic activity. Furthermore, it is shown that nlr overexpression suppresses the deleterious effects produced by the lack of SOD activity in the double-mutant E. coli sodA sodB strain.
MATERIALS AND METHODS
Bacterial strains and growth conditions.
D. gigas ATCC 19364 was grown as previously described (12). The E. coli strains XL2-Blue (Stratagene, La Jolla, Calif.), BL21(DE3) (5), and QC1799 [F− Δ(argF-lac)U169 rpsL ΔsodA3 φ(sodB-kan)Δ2] (57) were grown aerobically in Luria-Bertani (LB) broth (containing, per liter, tryptone [Difco], 10 g; yeast extract [Difco], 5 g; and sodium chloride, 5 g) or in minimal medium. Minimal medium was M9 (4) supplemented with 0.4% glucose, gluconate, or succinate and with thiamine and biotin (1 μg/ml each). This medium was also supplemented with 0.5 mM amino acids when required. Growth of E. coli strains carrying the pT7-7/pT7-7-derived plasmids and pCYTEXP/pCYTEXP-derived plasmids was measured in media supplemented with ampicillin (100 μg/ml). Strains with the pZErO-1 (Invitrogen, San Diego, Calif.) and pZErO-1-derived vectors were grown in LB broth supplemented with 20 μg of zeocin per ml or with 20 and 100 μg of zeocin and isopropyl-β-d-thiogalactopyranoside (IPTG) per ml, respectively. Plasmid DNA was prepared using the standard protocols or the plasmid purification kit from Gibco BRL (Life Technologies). E. coli XL2-Blue and E. coli QC1799 competent cells were prepared as specified by Invitrogen. E. coli BL21(DE3) competent cells were prepared by an electroporation transformation procedure (Bio-Rad).
Biochemical reagents and materials.
Restriction enzymes, T4 polynucleotide kinase, T4 DNA ligase, and Pfu DNA polymerase were from either Boehringer Mannheim or Stratagene (San Diego) and were used as specified by the manufacturer. Ampicillin and IPTG were from Promega (Madison, Wis.). Diethylpyrocarbonate, paraquat (methyl viologen), nitroblue tetrazolium (NBT), and riboflavin were from Sigma. Potassium cyanide (KCN) was from Merck. The Thermo Sequenase cycle-sequencing kit used for sequencing was purchased from Amersham Life Science. [α-35S]dATP and [γ-32P]dATP were from ICN. The oligonucleotides used were synthesized by Gibco BRL. Q-Sepharose and Superdex/75 columns were from Pharmacia Biotech Inc. All solutions used were prepared in MilliQ (Millipore Corp.) ultrapure water or in diethylpyrocarbonate treated ultrapure water.
RNA isolation, Northern blotting and primer extension analyses.
Total RNA was isolated from exponential and stationary D. gigas cells grown anaerobically and from E. coli cells producing wild-type and mutated Nlr recombinant proteins grown aerobically as described by Silva et al. (51). The amount of RNA loaded was calculated by reading the absorbance at 260 nm, and the RNA was then separated on a 1.5 to 2% agarose in 1× morpholinepropanesulfonic acid (MOPS) buffer (4)–6% (vol/vol) formaldehyde gel using a 0.24- to 9.5-kb RNA ladder (Gibco BRL, Life Technologies) as a size marker. Northern blotting was carried out essentially as described previously (4). Filters were prehybridized and hybridized as described previously (51) at 53 to 58°C. 32P-labeled oligonucleotides homologous to the nlr coding unit (BP5R and BP3′ [see below]), to ORF1 (5′-GGAACTTGCGATGAGCCGAGTGC-3′), and to ORF2 (5′-GGCGCCGATGGCCGCCACG-3′), together with a 32P-labeled 23S rRNA primer, were used as probes. The 23S rRNA oligonucleotide (5′-CTTTTCRCCTTTCCCTCACGGTAC-3′) was used as an internal control of loading and hybridization conditions. It was designed based on a search for gene sequence homology between the 23S rRNA gene sequences from different microorganisms, using the Sequence Retrieval System (SRS) and ClustalW (56). After hybridization, the filters were washed in 2× SSC (1× SSC is 0.15 M NaCl plus 0.015 M sodium citrate)–0.1% sodium dodecyl sulfate (SDS) and 0.2× SSC–0.1% SDS under homologous hybridization temperatures. For removal of probe, a boiling solution of 0.1% SDS was poured onto the membrane. Quantification analysis of Northern blots was performed using ImageQuant version 3.1 (Molecular Dynamics). Primer extension was carried out as described previously (51) using two 32P-labeled primers complementary to the nlr sequence from +51 to +72, primer BP5R (5′-GTCGCACTCGATGGCGGGCACG-3′), and from +100 to +120, primer BP5ext (5′-GCCCAGGCTCACGGTGACGGG-3′).
Overexpression and purification of recombinant neelaredoxin.
The nlr gene was cloned as described previously (50). The neelaredoxin coding unit was amplified from the recombinant plasmid pZ3NI by PCR using oligonucleotides BP5′ (5′-CGGGATCCATATGAAAATGTGCGACATGTTTCAAAC-3′) and BP3′ (5′-TTAAGCTTACTTGAGGGCCACGGCCTTG-3′), with NdeI and HindIII restriction sites (underlined), respectively. The cycling conditions were as follows: 1 cycle of 96°C for 1 min followed by 40 cycles of 94°C for 30 s, 60°C for 30 s, and 72°C for 1 min, with a final cycle of 72°C for 10 min. The amplified fragments were gel purified and subcloned into the EcoRV site of pZErO-1, and the resulting plasmid, pZBP, was transformed in competent E. coli XL2-Blue cells. Positive clones were identified by restriction analysis, and nlr gene integrity was subsequently verified by sequencing analysis using the plasmid reverse and universal primers. The nlr gene was excised from pZBP with NdeI and HindIII and subcloned into the NdeI-HindIII sites of the expression vector pT7-7 to give the recombinant vector pT7Nlr. After transformation of competent E. coli BL21(DE3), we obtained BL21(DE3)[pT7Nlr]. Overexpression of the nlr gene was performed by growing BL21(DE3)[pT7Nlr] aerobically at 37°C overnight in LB broth containing ampicillin (100 μg/ml). After overnight growth, the culture was diluted 1:400 and left to grow until it reached an optical density at 600 nm (OD600) of 0.5 to 0.6. At this point, nlr expression was induced by adding 1 mM IPTG, and the cell cultures were then grown for an additional 6 h. The cells were then chilled, harvested by centrifugation at 3,000 × g for 15 min, washed in 10 mM Tris-HCl buffer (pH 7.6), and suspended in the same buffer at about 0.4 g (wet cell weight)/ml. A cell extract (total-protein soluble extract) was obtained after cell disruption using a French pressure cell at 10,000 lb/in2 and ultracentrifugation at 42,000 × g for 2 h. The presence of Nlr overproduction was checked by SDS-polyacrylamide gel electrophoresis (PAGE) and its functionality was detected by using the NBT reduction technique described elsewhere (5). Neelaredoxin expression was also induced with 0.6 and 0.8 mM IPTG, and the cell cultures were grown for an additional 10 and 8 h, respectively. Neelaredoxin expression was also induced using different iron concentrations, by adding 1 and 4 mM FeSO4 to the medium.
All purification procedures were performed at pH 7.6 and at 4°C, using anion-exchange and gel filtration chromatography of soluble cell extracts. Total-protein soluble extracts from Nlr overexpressed in the absence and in the presence of 1 and 4 mM FeSO4 were concentrated to 5 ml using a Diaflo apparatus equipped with a YM3 membrane (Amicon, Inc., Beverly, Mass.). This blue-green soluble extract was loaded onto a Q-Sepharose ion-exchange column (1.8 by 18 cm) previously equilibrated with 10 mM Tris-HCl (pH 7.6) (buffer A) and eluted with a linear gradient of 0 to 1.0 M NaCl in 10 mM Tris-HCl at a flow rate of 1 ml/min. The presence of recombinant protein in blue fractions, eluted mostly at 0.190 to 0.270 mM NaCl, was assayed by UV-visible spectroscopy. Protein fractions with similar purity levels were combined, analyzed by SDS-PAGE, concentrated as described above, separately applied to a Superdex/75 gel filtration column (2.6 by 100 cm), equilibrated with 50 mM Tris-HCl (pH 7.6)–100 mM NaCl, and eluted with the same buffer at a flow rate of 0.5ml/min. After this step, the protein was considered pure as established by SDS-PAGE.
Analysis of Nlr superoxide-scavenging activity.
The superoxide-scavenging activity of the recombinant Nlr was evaluated by the NBT reduction technique (5) in nondenaturating 10% polyacrylamide gels using riboflavin in both resolving and concentrating gels as described previously (26).
Analytical determinations.
SDS-PAGE was carried out in a 15% polyacrylamide gel in the presence of glycine (36). SDS was omitted from all buffers when PAGE was performed under nondenaturating conditions in 10% polyacrylamide gels. Gels were calibrated with protein high-molecular-weight Rainbow standards from Amersham Life Science and stained with Coomassie brilliant blue R-250. The protein concentration was determined using either the Bio-Rad protein assay reagent (7) with bovine serum albumin as a standard or the bicinchoninic acid protein assay kit from Pierce (53). The total iron content was determined by the 2,4,6-tripyridyl-s-triazine (TPZT) method (17) and by atomic absorption spectroscopy. UV-visible spectra and SOD activity measurements were recorded on a Shimadzu UV-3100 spectrophotometer with 1-cm quartz cells. Electron paramagnetic resonance spectra were recorded using a Bruker EPS 380 spectrometer equipped with an ESR 900 continuous-flow helium cryostat from Oxford Instruments.
Site-directed mutagenesis.
Nlr mutant proteins in unique amino acid residues were obtained by site-directed mutagenesis, using complementary mutagenic primers with the desired mutation and pCYTNlr as the DNA template, as specified in the QuikChange site-directed mutagenesis kit instructions. pCYTNlr was obtained by subcloning the nlr gene excised from pT7Nlr with NdeI and EcoRI into the NdeI-EcoRI sites of the expression vector pCYTEXP (pCYT) (6). Primer NlrC4S (5′-ATGAAAATGTCCGACATGTTTCAAACTGC-3′) and its complementary primer were used to replace the TGC Cys-4 codon (boldface) by a TCC Ser (S) codon. Primer NlrC23S (5′-CCCGCCATCGAGTCCGACGATGCCG-3′) and its complementary primer were used for the replacement of the TGC Cys-23 codon by the TCC Ser (S) codon. Primers NlrC115A (5′-GCTTCGTCTTTCGCCAATATTCACGGG-3′), NlrC115D (5′-GCTTCGTCTTTCGACAATATTCACGGG-3′), and NlrC115S (5′-GCTTCGTCTTTCTCCAATATTCACGGG-3′) and their complementary primers were used to replace the TGC Cys-115 codon with GCC Ala (A), GAC Asp (D), and TCC Ser (S) codons, respectively. The PCR conditions used were as follows: 1 cycle at 96°C for 15 s followed by 16 cycles of denaturation at 94°C for 30 s, annealing at 55°C for 1 min, and extension at 68°C for 12 min. After the reaction was stopped, DpnI was added to the reaction mixture at 37°C and the mixture was incubated for 30 min to destroy the original, nonmutagenized methylated target plasmid. The obtained mutated plasmids, pCYTNlrC4S, pCYTNlrC23S, pCYTNlrC115A, pCYTNlrC115D, and pCYTNlrC115S, were analyzed by gel agarose electrophoresis and introduced into competent cells of E. coli QC1799, which were then plated in LB agar with ampicillin. Transformants were screened by digestion of the corresponding plasmid DNA and sequencing analysis to confirm the desired mutations and to detect unexpected mutations, using the pCYT primers 5′-GATACGAAACGAAGCATTG-3′ and 5′-GTTGTTCCTTTCTATTCTCACTCCG-3′. Plasmids with mutated nlr genes obtained by PCR amplifications can have errors introduced during the PCR procedure. Thus, mutated nlr genes from these plasmids were then excised, recloned into a new pCYTEXP vector with compatible ends, and transformed into E. coli QC1799 competent cells. Sequencing analysis was once again performed with pCYTEXP primers and the primer BP5R (see above), to confirm nlr mutated genes and pCYTEXP promoter regions. Wild-type and mutated nlr genes were expressed by growing E. coli QC1799 strains carrying recombinant pCYTEXP vectors harboring wild-type and mutated nlr genes aerobically at 28°C overnight in LB broth containing ampicillin (100 μg/ml). These cultures were then diluted 1:400 in the same medium, and the cells were grown to an OD595 of ≈0.5, at which stage the cultures were transferred to a 42°C incubator and grown for an additional 3 h. Production of wild-type and mutated nlr genes was verified by SDS-PAGE analysis as described above. SOD activity of wild-type and mutated Nlr recombinant proteins was tested on NBT-stained gels as described elsewhere (5).
Effect of Nlr on the E. coli sodA sodB mutant strain.
The studies concerning the effect of Nlr on the sensitivity of the E. coli sodA sodB mutant strain to paraquat and to H2O2, and on the growth of this strain in minimal medium with glucose, gluconate, or succinate as the carbon source were done essentially as described previously (10, 46).
RESULTS
Characterization of the neelaredoxin operon of D. gigas.
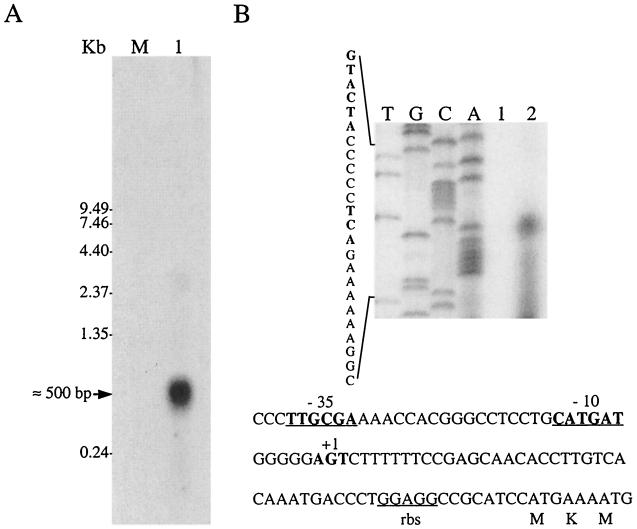
Previous sequencing analysis suggested that Nlr was encoded by a polycistronic unit, which contains two additional open reading frames (ORF1 and ORF2) coding for chemotaxis-like proteins (50). To investigate whether these genes are located in the same operon, total RNA extracted from mid-log-phase (OD550 ≈0.3 to 0.4) cells of D. gigas was probed with ORF1, ORF2, and nlr probes. Unexpectedly, Northern blot hybridization analysis revealed that under the growth conditions used, no mRNA was detected with either the ORF1 or ORF2 probe. Moreover, a transcript of approximately 500 bases hybridizing with the nlr probe was observed in large amounts (Fig. 1A), indicating that the nlr gene is highly transcribed as a single coding unit. The transcription start site was identified by primer extension analysis and appears to be located 50 to 52 bp upstream of the ATG start codon (Fig. 1B). The transcript size indicates that the nlr mRNA terminates in the inverted repeat located downstream of the stop codon, as previously proposed (50). At −10 and −35 upstream of the transcription start site, promoter elements resembling those recognized by E. coli ς70 were detected. They match four of the six bases (boldface) with either the −35 (TTGACA) or the −10 (TATAAT) consensus sequence (Fig. 1B). This observation indicates that E. coli ς70 -RNA polymerase might recognize the nlr promoter from D. gigas. Indeed, using plasmid constructs in which the nlr promoter elements were present or absent, it was observed by Northern analysis that E. coli RNA polymerase is able to transcribe nlr mRNA from its own promoter. Moreover, the transcription start site of nlr mRNA expressed in E. coli cells was shown to be located in the same region as the one determined using total RNA from D. gigas (data not shown).
FIG. 1.
Transcriptional analysis of the D. gigas nlr operon. (A) Northern blot analysis of D. gigas total RNA using an nlr homologous probe. M represents molecular mass standards, with corresponding sizes marked on the left; lane 1 contains 10 μg of total RNA from D. gigas. The arrow indicates the size of the nlr mRNA. (B) Determination of the transcription start site of nlr mRNA. Lanes 1, primer extension control (no RNA); 2, primer extension product. T, G, C, and A show the DNA sequence obtained with the same primer. The −10, and −35 promoter regions are in boldface. The nucleotides which might correspond to the transcriptional start site of nlr mRNA are also highlighted in boldface. rbs, ribosome binding site.
Expression of neelaredoxin gene under oxidative stress conditions and in stationary-phase cells.
D. gigas cells were grown anaerobically to an OD550 of ≈0.3, after which they were saturated with O2 (1.27 mM) or exposed to H2O2 (100 μM) and left to grow for an additional 1 h at 37°C. Aliquots were taken for total-RNA extraction at 0, 10, 30, and 60 min after O2 or H2O2 exposure. Northern blot hybridization and densitometric analyses of total RNA from these cultures, revealed that nlr mRNA is transcribed at the same level in cultures saturated with O2 and in cultures treated with H2O2 for different times (data not shown). Northern blot analysis of anaerobically grown D. gigas cells (OD550 ≈0.8) showed that nlr expression decreases upon entry into the stationary phase. Studies concerning the expression of ORF1 and ORF2 genes in D. gigas cells saturated with oxygen were also performed; however, no expression of these genes was observed by Northern blot hybridization analysis.
Taken together, these results strongly indicate that nlr gene is transcribed from its own promoter and that it is constitutively and abundantly expressed and is not induced under oxidative stress conditions.
Overexpression and characterization of recombinant neelaredoxin.
To better characterize Nlr, the gene for D. gigas Nlr was cloned using PCR with primers homologous to both ends of the nlr gene and subcloned into the expression vector pT7-7. A subclone of E. coli BL21(DE3) containing the recombinant expression vector, BL21(DE3)[pT7Nlr], was shown to overproduce the recombinant Nlr after induction with 0.6, 0.8, or 1 mM IPTG, as evidenced by the appearance of a blue soluble protein extract and by the gene product identified as a 15-kDa protein on SDS-PAGE (12). The highest expression of the recombinant protein was observed with 1 mM IPTG. Recombinant Nlr was also obtained by growing BL21(DE3)[pT7Nlr] in the presence of 1 or 4 mM iron with 1 mM IPTG. The respective UV-visible and electron paramagnetic resonance spectra did not show any significant differences between them and were identical to those of the native Nlr, as revealed by the existence of its characteristic absorption bands at 666 and at 325 nm (data not shown). The analysis for the presence of iron and zinc in both recombinant Nlr samples revealed that zinc was either absent or present only in trace amounts whereas values of 0.7 mol of Fe/mol of monomer were found for all recombinant Nlr proteins, as determined both by the TPTZ method and by atomic absorption spectroscopy. These data indicate that Nlr contains only one iron atom per monomer, as expected based on its amino acid sequence and on the comparison with the respective orthologues (1, 50, 60). SOD activity of recombinant Nlr proteins was monitored by the NBT test, and in all cases recombinant Nlr clearly exhibited superoxide-scavenging activity.
Expression of neelaredoxin gene in the E. coli sodA sodB mutant strain.
Wild-type E. coli contains three superoxide dismutases: FeSOD, MnSOD, and CuZnSOD. Thus, an E. coli sodA sodB mutant, deficient in cytoplasmic SODs-E. coli QC1799 (57)-was used to express the nlr gene, using the expression vector pCYTEXP. Although this strain has only two sod genes mutated (sodA and sodB), the superoxide-scavenging activity in total soluble protein extract of E. coli QC1799 was detected only when it expressed the D. gigas nlr gene (QC1799[pCYTNlr]), as shown by the NBT test (Fig. 2B, compare lane 1 with lane 0).
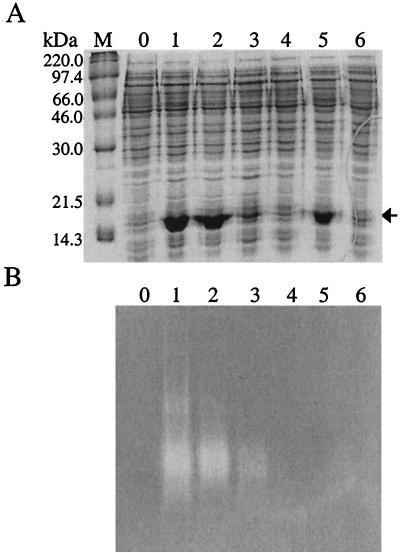
FIG. 2.
Analysis of mutated recombinant Nlr proteins. (A) SDS-PAGE of 10 μg of total soluble protein extracts from E. coli QC1799 cells expressing the wild-type and mutated Nlr proteins. (B) native PAGE gel stained for detection of superoxide-scavenging activity of mutated Nlr proteins by the NBT test. M, molecular mass standards with corresponding molecular masses on the left; lanes: 0, cell extract from E. coli QC1799 harboring pCYTEXP (not expressing nlr); 1 to 6, contain cell extracts from E. coli QC1799 strains expressing wild-type Nlr and C4S, C23S, C115A, C115D, and C115S mutated proteins, respectively. Recombinant wild-type and mutated Nlr proteins are indicated by an arrow in panel A.
Expression and characterization of mutated Nlr proteins.
The observation that not only the ligands to Dfx center II but also the surrounding amino acid residues are conserved in Nlr and Nlr orthologues led us to propose that these amino acid residues could be involved in iron binding in D. gigas Nlr (50), which was proven to be the case for the homologous protein from Pyroccocus furiosus (60). Besides these conserved amino acid residues and in contrast to the other homologous proteins, D. gigas Nlr contains extra cysteines (50). Therefore, to assess the possible function of some of them, Cys-4 and Cys-23 were mutated to serine. The cysteine bound to the iron center (Cys-115) was also mutated to serine, alanine, or aspartate, allowing us to assess its relevance for the catalytic function (it should be noted that FeSODs, although also having a penta-coordinated iron site, do not have any cysteine ligand). The mutated proteins were respectively designated C4S, C23S, C115S, C115A, and C115D.
SDS-PAGE analysis of cell extracts from strains producing wild-type and mutated Nlr proteins revealed that both the C4S and C115D mutants were produced at almost the same level as that of the recombinant wild type (Fig. 2A, compare lanes 2 and 5 with lane 1) whereas C23S was produced less abundantly (lane 3). For C115A and C115S, it is not clear whether these mutated proteins were produced at low levels (lanes 4 and 6), in spite of the fact that both coding and promoter regions were correct as confirmed by sequencing analysis. Both C4S and C23S mutants had superoxide-scavenging activity (Fig. 2B, lanes 2 and 3) whereas C115D did not (lane 5), although it was produced in large amounts (Fig. 2A, lane 5).
Overexpression of nlr gene suppresses superoxide damage in E. coli sodA sodB double mutant.
Bacterial mutants devoid of both FeSOD and MnSOD are hypersensitive to oxidizing agents (10, 33). Furthermore, although they can survive aerobically in rich medium, growth occurs slowly in minimal medium and is induced only on addition of the 20 amino acids, suggesting a drastic effect of ROS on amino acid biosynthesis (10, 33, 35). The growth rate of these mutants in minimal medium containing gluconate or succinate as the carbon source is also lower, due to 6-phosphogluconate and aconitase inactivation (23, 24). Thus, overexpression of proteins with SOD activity in these mutants is expected to suppress the deleterious effect caused by superoxide anion.
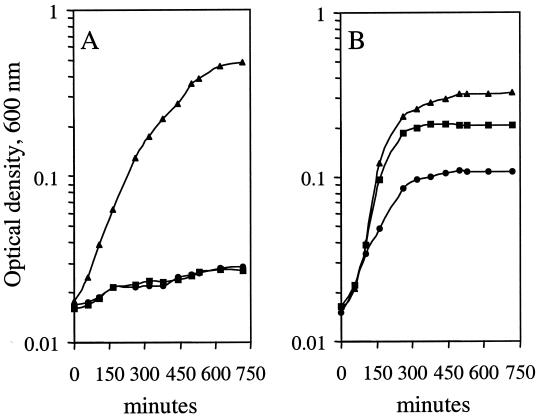
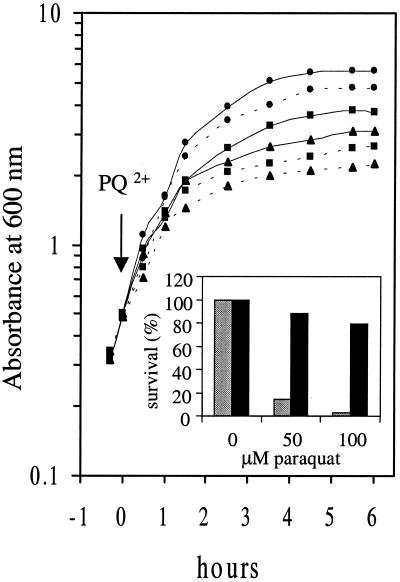
The observation that Nlr has superoxide-scavenging activity prompted us to test if it could contribute to minimizing the damage arising from the lack of SOD activity in E. coli SOD mutants. Indeed, as shown in Fig. 3B, an E. coli sodA sodB mutant strain expressing the D. gigas nlr gene, QC1799[pCYTNlr], has reestablished growth in enriched minimal medium in the absence of branched amino acids, in contrast to the E. coli sodA sodB mutant strain in the absence of nlr expression, QC1799[pCYT] (Fig. 3A). Moreover, the growth of QC1799[pCYTNlr] was also higher in the absence of all amino acids. The growth of E. coli sodA sodB mutant strains on gluconate or succinate as the carbon source was also measured, and again the growth rate of QC1799[pCYTNlr] was higher than that of QC1799[pCYT] (data not shown). Similarly, Nlr enhanced the resistance of sodA sodB mutant strain both to paraquat (Fig. 4) and to 2.5 mM H2O2 (data not shown). The effect of Nlr on the sensitivity of the sodA sodB mutant to paraquat was much more evident when the survival after a 6-h treatment with paraquat was measured, as shown in the inset of Fig. 4. Furthermore, although all experiments performed with either QC1799 or QC1799[pCYT] showed a slow growth in rich medium and their colonies were very different in size, suggesting serious stress to the cells, this was not observed with QC1799[pCYTNlr]. These findings clearly show that nlr gene expression, similarly to Dfx expression, suppresses the deleterious effects produced by superoxide in the E. coli sodA sodB mutant strain.
FIG. 3.
Effect of Nlr on growth of the E. coli sodA sodB strain in minimal medium. sodA sodB strains were grown overnight in enriched M9 medium diluted to an OD600 of ≈0.015 in preheated M9 glucose (circles), enriched M9 without branched amino acids (squares), or enriched M9 (triangles) at 42°C. (A) sodA sodB strain carrying the expression vector pCYTEXP, QC1799[pCYT]. (B) sodA sodB strain carrying the recombinant expression vector with the nlr coding unit, QC1799[pCYTNlr]. Growth of the sodA sodB strain without the expression vector, QC1799, was similar to that of QC1799[pCYT]. When growth of sodA sodB strains was at 28°C, a temperature at which nlr is not expressed, the growth rate of QC1799[pCYTNlr] was similar to that observed with QC1799[pCYT].
FIG. 4.
Effect of Nlr on sensitivity of E. coli sodA sodB strains to paraquat. sodA sodB strains carrying the expression vector, QC1799[pCYT] (dotted lines), and the expression vector harboring the nlr coding unit, QC1799[pCYTNlr] (full lines), were grown in the presence of paraquat. Overnight cultures grown in LB broth at 42°C were diluted to an OD600 of ≈0.02 in the same medium and grown to an OD600 of ≈0.5, at which point paraquat was added at 0 μM (circles), 50 μM (squares), or 100 μM (triangles) (final concentrations). The results are the average values of four different experiments. The inset represents the percent survival of QC1799[pCYT] and QC1799[pCYTNlr] after paraquat treatment. Following a 6-h treatment with 0, 50, or 100 μM paraquat, cells were diluted and plated on LB plates at 37°C to monitor cell viability. The percent survival was calculated as the ratio of the number of colonies grown in the presence of paraquat to those in the absence of the drug. Values are the average of two identical experiments. Grey and black bars represent QC1799[pCYT] and QC1799[pCYTNlr], respectively.
DISCUSSION
The results presented in this report clearly show that the D. gigas nlr gene is transcribed monocistronically from an upstream promoter, with sequence elements resembling those of an E. coli ς70 promoter. Furthermore, the nlr gene is constitutively and abundantly expressed, and its expression is not affected by oxidative stress. A decrease in gene expression is, however, observed in stationary-phase cells, a fact which is a general feature of microorganisms under these conditions.
Replacement of either Cys-4 or Cys-23 led to the formation of Nlr mutants with wild-type-like characteristics, indicating that these residues are not functionally important. On the other hand, replacement of the iron ligand Cys-115 by other amino acid residues resulted either in the overproduction of catalytically inactive Nlr or in the absence of Nlr production, as detected by SDS-PAGE (Fig. 2A). These data clearly demonstrate that Cys-115 is essential for Nlr catalytic activity, in contrast to the histidine-carboxylate binding in FeSODs (for an example, see reference 37). Furthermore, the low C23S production and the absence of C115A and C115S production suggest that these mutated Nlr proteins might be more sensitive to proteases.
In spite of being an anaerobe, D. gigas was shown not only to have an oxygen-utilizing pathway (11, 48) but also to contain classical enzymes involved in O2 detoxification such as catalase and FeSOD (15). Given that Nlr has superoxide-scavenging activity, it appears reasonable that in D. gigas cells neelaredoxin would be involved in cell protection against oxidative stress, together with FeSOD and catalase. Because deletion of genes in this bacterium is still difficult, an E. coli mutant deficient in cytoplasmic SODs (sodA sodB) was used to probe in vivo the physiological function of Nlr. Our results obtained with nlr overexpression in an E. coli sodA sodB mutant clearly show that Nlr efficiently suppresses the deleterious effects produced by the lack of SODs (Fig. 3 and 4). The ability of E. coli sodA sodB mutants to grow aerobically in minimal medium is restored when this strain overexpresses the D. gigas nlr gene (Fig. 3). Furthermore, neelaredoxin expression also increases the resistance of this double-mutant strain toward oxidative stress agents such as paraquat (Fig. 4) and H2O2 (data not shown). These data indicate that Nlr reduces the steady-state level of the superoxide anion within the cell, protecting it from the deleterious effect of superoxide.
In contrast to our own results, Lumppio et al. (41) have recently stated that D. gigas Nlr is not able to complement the SOD deficiency of an E. coli mutant. It is possible that this discrepancy occurs because D. gigas Nlr might not be functionally produced in the E. coli sodA sodB mutant. Indeed, It is already known that the active center of this protein family may incorporate zinc instead of iron, resulting in inactive proteins (3). Our data are corroborated by those obtained with Treponema pallidum Nlr, which, like D. gigas Nlr, although not containing Dfx center I, was shown to fully complement the SOD deficiency of an E. coli mutant strain (40), indicating that Nlr and Dfx proteins are involved in protecting cells against superoxide anion (40, 46).
The data obtained by Voordouw and Voordouw (59), showing that deletion of the dfx gene increased the oxygen sensitivity of D. vulgaris when exposed to transient aerobic conditions, strongly indicate that both Dfx and Nlr proteins are involved in defense mechanisms against ROS in the anaerobic SRB. This protein family has been proposed to function either as SOD or superoxide reductase proteins (1, 29, 31, 38–41). The “bifunctionality” of these kind of enzymes could provide an advantage for these anaerobic microorganisms, since it would give them the versatility to be protected from the toxic effect caused by ROS using either the SOD cycle or a superoxide reduction reaction, depending on the cell redox status. However, given the abundant and constitutive expression of the nlr gene, an additional role for this protein during anaerobiosis cannot be ruled out, a fact that makes it interesting for future studies.
The observations that Nlr, Nlr orthologues, and Dfx proteins are found in all the available genome sequences from anaerobes (8, 19, 32, 34, 44, 52) but not from aerobes, as well as the terminal oxygen reductase (ROO) discovered in D. gigas (11, 20), raise the question whether these proteins are the ancestors of enzymes involved in protective mechanisms against oxidative stress in anaerobes.
ACKNOWLEDGMENTS
This work was supported by PRAXIS XXI 32/96 and 11074/98 to C.R.-P. and PRAXIS XXI/BD/9016 to G.S. from Fundação para a Ciência e Tecnologia and by NIH grant GM 56000 to J.L.G.
We are indebted to Danièle Touati for generously providing E. coli QC1799. We thank Paula Fareleira, Maria Manuel Sampaio, and João Carita (ITQB) for their collaboration in the growth of D. gigas cells. We also thank Isabel Pacheco and Célia Romão (ITQB) for valuable assistance in protein purification techniques and in analysis of the superoxide-scavenging activity.
REFERENCES
- 1.Abreu I A, Saraiva L M, Carita J, Huber H, Stetter K O, Cabelli D, Teixeira M. Oxygen detoxification in the strict anaerobic archaeon Archaeoglobus fulgidus: superoxide scavenging by neelaredoxin. Mol Microbiol. 2000;38:322–334. doi: 10.1046/j.1365-2958.2000.02121.x. [DOI] [PubMed] [Google Scholar]
- 2.Archer M, Huber R, Tavares P, Moura I, Moura J J G, Carrondo M A, Sieker L C, LeGall J, Romão M J. Crystal structure of desulforedoxin from Desulfovibrio gigas determined at 1.8 Å resolution: a novel non-heme iron protein structure. J Mol Biol. 1995;251:690–702. doi: 10.1006/jmbi.1995.0465. [DOI] [PubMed] [Google Scholar]
- 3.Ascenso C, Rusnak F, Cabrito I, Lima M J, Naylor S, Moura I, Moura J J G. Desulfoferrodoxin: a modular protein. J Biol Inorg Chem. 2001;5:720–729. doi: 10.1007/s007750000161. [DOI] [PubMed] [Google Scholar]
- 4.Ausubel F M, Brent R, Kingston R E, Moore D D, Seidman J G, Smith J A, Struhl K. Current protocols in molecular biology. New York, N.Y: Greene Publishing Associates and Wiley-Interscience; 1995. [Google Scholar]
- 5.Beauchamp C, Fridovich I. Superoxide dismutase: improved assays and an assay applicable to acrylamide gels. Anal Biochem. 1971;44:276–287. doi: 10.1016/0003-2697(71)90370-8. [DOI] [PubMed] [Google Scholar]
- 6.Belev T N, Singh M, McCarthy J E G. A fully modular vector system for the optimization of gene expression in Escherichia coli. Plasmid. 1991;26:147–150. doi: 10.1016/0147-619x(91)90056-3. [DOI] [PubMed] [Google Scholar]
- 7.Bradford M M. A rapid and sensitive method for the quantification of microgram quantities of protein utilizing the principle of protein-dye binding. Anal Biochem. 1976;72:248–254. doi: 10.1016/0003-2697(76)90527-3. [DOI] [PubMed] [Google Scholar]
- 8.Bult C J, White O, Olsen G J, Zhou L, Fleischmann R D, et al. Complete genome sequence of the methanogenic archaeon, Methanococcus jannaschii. Science. 1996;273:1058–1073. doi: 10.1126/science.273.5278.1058. [DOI] [PubMed] [Google Scholar]
- 9.Canfield D E, Des Marais D J. Aerobic sulfate reduction in microbial mats. Science. 1991;251:1471–1473. doi: 10.1126/science.11538266. [DOI] [PubMed] [Google Scholar]
- 10.Carlioz A, Touati D. Isolation of superoxide dismutase mutants in Escherichia coli: is superoxide dismutase necessary for aerobic life? EMBO J. 1986;5:623–630. doi: 10.1002/j.1460-2075.1986.tb04256.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Chen L, Liu M-Y, LeGall J, Fareleira P, Santos H, Xavier A V. Purification and characterization of an NADH-rubredoxin oxidoreductase involved in the utilization of oxygen by Desulfovibrio gigas. Eur J Biochem. 1993;216:443–448. doi: 10.1111/j.1432-1033.1993.tb18162.x. [DOI] [PubMed] [Google Scholar]
- 12.Chen L, Sharma P, LeGall J, Mariano A M, Teixeira M, Xavier A V. A blue non-heme iron protein from Desulfovibrio gigas. Eur J Biochem. 1994;226:613–618. doi: 10.1111/j.1432-1033.1994.tb20087.x. [DOI] [PubMed] [Google Scholar]
- 13.Coelho A V, Matias P, Fülöp V, Thompson A, Gonzalez A, Carrondo M A. Desulfoferrodoxin structure determined by MAD phasing and refinement to 1.9-Å resolution reveals a unique combination of a tetrahedral FeS4 centre with a square pyramidal FeSN4 centre. J Biol Inorg Chem. 1997;2:680–689. [Google Scholar]
- 14.Dilling W, Cypionka H. Aerobic respiration in sulfate-reducing bacteria. FEMS Microbiol Lett. 1990;71:123–128. [Google Scholar]
- 15.Dos Santos W G, Pacheco I, Liu M-Y, Teixeira M, Xavier A V, LeGall J. Purification and characterization of an iron superoxide dismutase and a catalase from the sulfate-reducing bacterium Desulfovibrio gigas. J Bacteriol. 2000;182:796–804. doi: 10.1128/jb.182.3.796-804.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Fareleira P, LeGall J, Xavier A V, Santos H. Pathways for utilization of carbon reserves in Desulfovibrio gigas under fermentative and respiratory conditions. J Bacteriol. 1997;179:3972–3980. doi: 10.1128/jb.179.12.3972-3980.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Fisher D S, Price D C. Iron determination with TPTZ (tripyridyl-s-triazine: acid labile iron) in protein. Clin Chem. 1964;10:21–31. [PubMed] [Google Scholar]
- 18.Flint D H, Tuminello J F, Emptage M H. The inactivation of Fe-S cluster containing hydro-lyases by superoxide. J Biol Chem. 1993;268:22369–22376. [PubMed] [Google Scholar]
- 19.Fraser C M, Norris S J, Weinstock G M, White O, Sutton G G, et al. Complete genome sequence of Treponema pallidum, the syphilis spirochete. Science. 1998;281:375–388. doi: 10.1126/science.281.5375.375. [DOI] [PubMed] [Google Scholar]
- 20.Frazão C, Silva G, Gomes C M, Matias P, Coelho R, Sieker L, Macedo S, Liu M-Y, Oliveira S, Teixeira M, Xavier A V, Rodrigues-Pousada C, Carrondo M A, LeGall J. Structure of a dioxygen reduction enzyme from Desulfovibrio gigas. Nat Struct Biol. 2000;7:1041–1045. doi: 10.1038/80961. [DOI] [PubMed] [Google Scholar]
- 21.Fründ C, Cohen Y. Diurnal cycles of sulfate reduction under oxic conditions in cyanobacterial mats. Appl Environ Microbiol. 1992;58:70–77. doi: 10.1128/aem.58.1.70-77.1992. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Fu R, Wall J D, Voordouw G. DcrA, a c-type heme-containing methyl-accepting protein from Desulfovibrio vulgaris Hildenborough, senses the oxygen concentration or redox potential of the environment. J Bacteriol. 1994;176:344–350. doi: 10.1128/jb.176.2.344-350.1994. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Gardner P R, Fridovich I. Superoxide sensitivity of Escherichia coli 6-phosphogluconate dehydratase. J Biol Chem. 1991;266:1478–1483. [PubMed] [Google Scholar]
- 24.Gardner P R, Fridovich I. Superoxide sensitivity of Escherichia coli aconitase. J Biol Chem. 1991;266:19328–19333. [PubMed] [Google Scholar]
- 25.Gomes C M, Silva G, Oliveira S, LeGall J, Liu M-Y, Xavier A V, Rodrigues-Pousada C, Teixeira M. Studies on the redox centers of the terminal oxidase from Desulfovibrio gigas and evidence for its interaction with rubredoxin. J Biol Chem. 1997;272:22502–22508. doi: 10.1074/jbc.272.36.22502. [DOI] [PubMed] [Google Scholar]
- 26.Hames B D. One-dimensional polyacrylamide gel electrophoresis. In: Hames B D, Rickwood D, editors. Gel electrophoresis of proteins: a practical approach. Oxford, United Kingdom: IRL Press; 1990. pp. 30–50. [Google Scholar]
- 27.Hatchikian E C, LeGall J, Bell G R. Significance of superoxide dismutase and catalase activities in the strict anaerobes, sulfate-reducing bacteria. In: Michelson A M, McCord J M, Fridovich I, editors. Superoxide and superoxide dismutase. London, United Kingdom: Academic Press; 1977. pp. 159–172. [Google Scholar]
- 28.Imlay J A, Linn S. DNA damage and oxygen radical toxicity. Science. 1988;240:1302–1309. doi: 10.1126/science.3287616. [DOI] [PubMed] [Google Scholar]
- 29.Jenney F E, Jr, Verhagen M F J M, Cui X, Adams M W W. Anaerobic microbes: oxygen detoxification without superoxide dismutase. Science. 1999;286:306–309. doi: 10.1126/science.286.5438.306. [DOI] [PubMed] [Google Scholar]
- 30.Johnson M S, Zhulin I B, Gapuzan M-E R, Taylor B L. Oxygen-dependent growth of the obligate anaerobe Desulfovibrio vulgaris Hildenborough. J Bacteriol. 1997;179:5598–5601. doi: 10.1128/jb.179.17.5598-5601.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Jovanović T, Ascenso C, Hazlett K R O, Sikkink R, Krebs G, Litwiller R, Benson L M, Moura I, Moura J J G, Radolf J D, Huynh B H, Naylor S, Rusnak F. Neelaredoxin, an iron-binding protein from syphilis spirochete, Treponema pallidum, is a superoxide reductase. J Biol Chem. 2000;275:28439–28448. doi: 10.1074/jbc.M003314200. [DOI] [PubMed] [Google Scholar]
- 32.Kawarabayasi Y, Sawada M, Horikawa H, Haikawa Y, Hino Y, et al. Complete sequence and gene organization of the genome of a hyper-thermophilic archaebacterium, Pyrococcus horikoshii OT3. DNA Res. 1998;5:55–76. doi: 10.1093/dnares/5.2.55. [DOI] [PubMed] [Google Scholar]
- 33.Kim Y C, Miller C D, Anderson A J. Superoxide dismutase activity in Pseudomonas putida affects utilization of sugars and growth on root surfaces. Appl Environ Microbiol. 2000;66:1460–1467. doi: 10.1128/aem.66.4.1460-1467.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Klenk H-P, Clayton R A, Tomb J-F, White O, Nelson K E, et al. The complete genome sequence of the hyperthermophilic, sulphate-reducing archeon Archaeoglobus fulgidus. Nature. 1997;390:364–370. doi: 10.1038/37052. [DOI] [PubMed] [Google Scholar]
- 35.Kuo C F, Mashino T, Fridovich I. α,β-dihydroxyisovalerate dehydratase, a superoxide-sensitive enzyme. J Biol Chem. 1987;262:4724–4727. [PubMed] [Google Scholar]
- 36.Laemmli U K. Clevage of structural proteins during the assembly of the head of bacteriophage T4. Nature. 1970;227:680–685. doi: 10.1038/227680a0. [DOI] [PubMed] [Google Scholar]
- 37.Lah M S, Dixon M M, Pattridge K A, Stallings W C, Fee J A, Ludwig M L. Structure-function in Escherichia coli iron superoxide dismutase: comparisons with the manganese enzyme from Thermus thermophilus. Biochemistry. 1995;34:1646–1660. doi: 10.1021/bi00005a021. [DOI] [PubMed] [Google Scholar]
- 38.Liochev S I, Fridovich I. A mechanism for complementation of the sodA sodB defect in Escherichia coli by overproduction of the rbo gene product (desulfoferrodoxin) from Desulfoarculus baarsii. J Biol Chem. 1997;272:25573–25575. doi: 10.1074/jbc.272.41.25573. [DOI] [PubMed] [Google Scholar]
- 39.Lombard M, Fontecave M, Touati D, Nivière V. Reaction of the desulfoferrodoxin from Desulfoarculus baarsii with superoxide anion. J Biol Chem. 2000;275:115–121. doi: 10.1074/jbc.275.1.115. [DOI] [PubMed] [Google Scholar]
- 40.Lombard M, Touati D, Fontecave M, Nivière V. Superoxide reductase as a unique defense system against superoxide stress in the microarophile Treponema pallidum. J Biol Chem. 2000;275:27021–27026. doi: 10.1074/jbc.M004201200. [DOI] [PubMed] [Google Scholar]
- 41.Lumppio L H, Shenvi N V, Summers A O, Voordouw G, Kurtz D M., Jr Rubrerythrin and rubredoxin oxidoreductase in Desulfovibrio vulgaris: a novel oxidative stress protection system. J Bacteriol. 2001;183:101–108. doi: 10.1128/JB.183.1.101-108.2001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Moura I, Bruschi M, Le Gall J, Moura J J G, Xavier A V. Isolation and characterization of desulforedoxin, a new type of non-heme iron protein from Desulfovibrio gigas. Biochem Biophys Res Comm. 1977;75:1037–1044. doi: 10.1016/0006-291x(77)91486-3. [DOI] [PubMed] [Google Scholar]
- 43.Moura I, Tavares P, Moura J J G, Ravi N, Huynh B H, Liu M-Y, LeGall J. Purification and characterization of desulfoferrodoxin—a novel protein from Desulfovibrio desulfuricans (ATCC 27774) and from Desulfovibrio vulgaris (strain Hildenborough) that contains a distorted rubredoxin center and a mononuclear ferrous center. J Biol Chem. 1990;265:21596–21602. [PubMed] [Google Scholar]
- 44.Nelson K E, Clayton R A, Gill S R, Gwinn L, Dodson R J, et al. Evidence for lateral gene transfer between Archaea and Bacteria from genome sequence of Thermotoga maritima. Nature. 1999;399:323–329. doi: 10.1038/20601. [DOI] [PubMed] [Google Scholar]
- 45.Nunoshiba T, Obata F, Boss A C, Oikawa S, Mori T, Kawanishi S, Yamamoto K. Role of iron and superoxide for generation of hydroxyl radical, oxidative DNA lesions and mutagenesis in Escherichia coli. J Biol Chem. 1999;274:34832–34837. doi: 10.1074/jbc.274.49.34832. [DOI] [PubMed] [Google Scholar]
- 46.Pianzzola M J, Soubes M, Touati D. Overproduction of the rbo gene product from Desulfovibrio species suppresses all deleterious effects of lack of superoxide dismutase in Escherichia coli. J Bacteriol. 1996;178:6736–6742. doi: 10.1128/jb.178.23.6736-6742.1996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Romão C V, Liu M-Y, LeGall J, Gomes C M, Braga V, Pacheco I, Xavier A V, Teixeira M. The superoxide dismutase activity of desulfoferrodoxin from Desulfovibrio desulfuricans (ATCC 27774) Eur J Biochem. 1999;261:438–443. doi: 10.1046/j.1432-1327.1999.00278.x. [DOI] [PubMed] [Google Scholar]
- 48.Santos H, Fareleira P, Xavier A V, Chen L, Liu M-Y, LeGall J. Aerobic metabolism of carbon reserves by the “obligate anaerobe” Desulfovibrio gigas. Biochem Biophys Res Commun. 1993;195:551–557. doi: 10.1006/bbrc.1993.2081. [DOI] [PubMed] [Google Scholar]
- 49.Sass H, Berchtold M, Branke J, König H, Cypionka H, Babenzien H-D. Psychrotolerant sulfate-reducing bacteria from an oxic freshwater sediment, description of Desulfovibrio cuneatus sp. nov. and Desulfovibrio litoralis sp. nov. Syst Appl Microbiol. 1998;21:212–219. doi: 10.1016/S0723-2020(98)80025-8. [DOI] [PubMed] [Google Scholar]
- 50.Silva G, Oliveira S, Gomes C M, Pacheco I, Liu M-Y, Xavier A V, Teixeira M, LeGall J, Rodrigues-Pousada C. Desulfovibrio gigas neelaredoxin—a novel superoxide dismutase integrated in a putative oxygen sensory operon of an anaerobe. Eur J Biochem. 1999;259:235–243. doi: 10.1046/j.1432-1327.1999.00025.x. [DOI] [PubMed] [Google Scholar]
- 51.Silva G, Oliveira S, LeGall J, Xavier A V, Rodrigues-Pousada C. Analysis of the Desulfovibrio gigas transcriptional unit containing rubredoxin (rd) and rubredoxin-oxygen oxidoreductase (roo) genes and upstream ORFs. Biochem Biophys Res Commun. 2001;280:491–502. doi: 10.1006/bbrc.2000.4147. [DOI] [PubMed] [Google Scholar]
- 52.Smith D R, Doucette-Stamm L A, Deloughery C, Lee H-M, Dubois J, Aldredge T, et al. Complete genome sequence of Methanobacterium thermoautotrophicum delta H: functional analysis and comparative genomics. J Bacteriol. 1997;179:7135–7155. doi: 10.1128/jb.179.22.7135-7155.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Smith P K, Krohn R I, Hermanson G T, Mallia A K, Gartner F H, Provenzano M D, Fujimoto E K, Goeke N M, Olson B J, Klenk D C. Measurement of protein using bicinchoninic acid. Anal Biochem. 1985;150:76–85. doi: 10.1016/0003-2697(85)90442-7. [DOI] [PubMed] [Google Scholar]
- 54.Tabor S. Expression using the T7 RNA polymerase/promoter system. In: Ausubel F M, Brent R, Kingston R E, Moore D D, Seidman J G, Smith J A, Struhl K, editors. Current protocols in molecular biology. New York, N.Y: Greene Publishing Associates and Wiley Interscience; 1990. pp. 16.2.1–16.2.11. [Google Scholar]
- 55.Tavares P, Ravi N, Moura J J G, LeGall J, Huang Y H, Crouse B R, Johnson M K, Huynh B H, Moura I. Spectroscopic properties of desulfoferrodoxin from Desulfovibrio desulfuricans (ATCC 27774) J Biol Chem. 1994;269:10504–10510. [PubMed] [Google Scholar]
- 56.Thompson J D, Higgins D G, Gibson T J. CLUSTAL W: improving the sensitivity of progressive multiple sequence alignment trough sequence weighting, position-specific gap penalties and weight matrix choice. Nucleic Acids Res. 1994;22:4673–4680. doi: 10.1093/nar/22.22.4673. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Touati D, Jacques M, Tardat B, Bouchard L, Despied S. Lethal oxidative damage and mutagenesis are generated by iron in Δfur mutants of Escherichia coli: protective role of superoxide dismutase. J Bacteriol. 1995;177:2305–2314. doi: 10.1128/jb.177.9.2305-2314.1995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Valentine J S, Wertz D L, Lyons T J, Liou L-L, Goto J J, Gralla E B. The dark side of dioxygen biochemistry. Curr Opin Chem Biol. 1998;2:253–262. doi: 10.1016/s1367-5931(98)80067-7. [DOI] [PubMed] [Google Scholar]
- 59.Voordouw J K, Voordouw G. Deletion of the rbo gene increases the oxygen sensitivity of the sulfate-reducing bacterium Desulfovibrio vulgaris Hildenborough. Appl Environ Microbiol. 1998;64:2882–2887. doi: 10.1128/aem.64.8.2882-2887.1998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Yeh A P, Hu Y, Jenney F E, Jr, Adams M W W, Rees D C. Structures of the superoxide reductase from Pyrococcus furiosus in the oxidized and reduced states. Biochemistry. 2000;39:2499–2508. doi: 10.1021/bi992428k. [DOI] [PubMed] [Google Scholar]