Abstract
Aspartate aminotransferases have been cloned and expressed from Crithidia fasciculata, Trypanosoma brucei brucei, Giardia intestinalis, and Plasmodium falciparum and have been found to play a role in the final step of methionine regeneration from methylthioadenosine. All five enzymes contain sequence motifs consistent with membership in the Ia subfamily of aminotransferases; the crithidial and giardial enzymes and one trypanosomal enzyme were identified as cytoplasmic aspartate aminotransferases, and the second trypanosomal enzyme was identified as a mitochondrial aspartate aminotransferase. The plasmodial enzyme contained unique sequence substitutions and appears to be highly divergent from the existing members of the Ia subfamily. In addition, the P. falciparum enzyme is the first aminotransferase found to lack the invariant residue G197 (P. K. Mehta, T. I. Hale, and P. Christen, Eur. J. Biochem. 214:549–561, 1993), a feature shared by sequences discovered in P. vivax and P. berghei. All five enzymes were able to catalyze aspartate-ketoglutarate, tyrosine-ketoglutarate, and amino acid-ketomethiobutyrate aminotransfer reactions. In the latter, glutamate, phenylalanine, tyrosine, tryptophan, and histidine were all found to be effective amino donors. The crithidial and trypanosomal cytosolic aminotransferases were also able to catalyze alanine-ketoglutarate and glutamine-ketoglutarate aminotransfer reactions and, in common with the giardial aminotransferase, were able to catalyze the leucine-ketomethiobutyrate aminotransfer reaction. In all cases, the kinetic constants were broadly similar, with the exception of that of the plasmodial enzyme, which catalyzed the transamination of ketomethiobutyrate significantly more slowly than aspartate-ketoglutarate aminotransfer. This result obtained with the recombinant P. falciparum aminotransferase parallels the results seen for total ketomethiobutyrate transamination in malarial homogenates; activity in the latter was much lower than that in homogenates from other organisms. Total ketomethiobutyrate transamination in Trichomonas vaginalis and G. intestinalis homogenates was extensive and involved lysine-ketomethiobutyrate enzyme activity in addition to the aspartate aminotransferase activity. The methionine production in these two species could be inhibited by the amino-oxy compounds canaline and carboxymethoxylamine. Canaline was also found to be an uncompetitive inhibitor of the plasmodial aspartate aminotransferase, with a Ki of 27 μM.
The amino acid methionine (Met) is required for a number of vital cellular functions, including the initiation of protein synthesis, the methylation of rRNA and xenobiotics, and the biosynthesis of cysteine, phospholipids, and polyamines. This latter function is particularly important in rapidly growing cells, such as most parasites, bacteria, and cancer cells, which synthesize large amounts of polyamines immediately prior to DNA replication (31). The formation of spermidine from putrescine and of spermine from spermidine consumes Met (in the form of decarboxylated S-adenosylmethionine) as a source of aminopropyl groups, yielding methylthioadenosine as a by-product. As de novo biosynthesis of Met is energetically expensive (from aspartate, it requires one ATP molecule, two NADPH molecules, succinyl coenzyme A [CoA], cysteine or H2S, and 5-methyltetrahydrofolate) and many organisms lack the ability to synthesize the amino acid, Met tends to be present in limiting amounts. In order to prevent depletion of free Met, there exists a unique pathway which regenerates Met from methylthioadenosine in seven or eight steps (see reference 19 for a diagram of the pathway and the enzymes involved); the final step is the transamination of α-ketomethiobutyrate (KMTB) to yield Met.
The Met regeneration pathway, sometimes referred to as the methylthioadenosine cycle, has been partially characterized for a number of organisms, including rat liver (4, 5, 50), plants (47), yeasts (30), and protozoal parasites (16, 37, 41). However, the complete pathway has only been fully delineated for the gram-negative bacterium Klebsiella pneumoniae, where a series of unusual enzymes have been found to be responsible for the production of KMTB and tyrosine aminotransferase (TyrAT) has been found to catalyze the final step (14, 19, 34, 44, 49, 50). Aside from our recent studies with K. pneumoniae, very little is known about the identity of the aminotransferase(s) responsible for Met recycling. In the original studies which discovered KMTB conversion to Met in rat liver, Backlund et al. (4, 5) demonstrated that Met could be produced in tissue homogenates using glutamine or asparagine, but not glutamate or aspartate, as the amino donor. No other amino acids were examined as potential amino donors, and no purification of the aminotransferase(s) involved was undertaken. These results were consistent with the findings of Cooper and Meister (10), who found that purified rat liver or kidney glutamine aminotransferase (GlnAT) could use the substrate pairs KMTB-glutamine and hydroxyphenylpyruvate-Met. Again, a wider examination of KMTB amino donor specificity was not conducted. These early studies led to the unwarranted assumption that GlnAT was responsible for Met recycling in all organisms and that glutamine and asparagine were the primary amino donors.
In a previous study of Met regeneration with the trypanosomatids Crithidia fasciculata and Trypanosoma brucei brucei, it was found that aromatic amino acids were the preferred amino donors for KMTB and that glutamine and asparagine were very poor amino donors (6). Subsequent purification of this activity from C. fasciculata yielded an aminotransferase that actively catalyzed KMTB-tryptophan, KMTB-phenylalanine, KMTB-tyrosine, KMTB-glutamate, α-ketoglutarate (KG)– tyrosine, and KG-aspartate aminotransfer (7). Amino acid sequencing of an internal peptide from this enzyme gave a sequence with very high identity to sequences of eukaryotic cytosolic aspartate aminotransferases (AspATs). Purified, commercial pig heart AspAT was found to be a very poor catalyst of Met production from KMTB (7). A similar study with K. pneumoniae found that TyrAT (tyrB gene product) was responsible for Met regeneration in this organism (19). Again, aromatic amino acids and glutamate were the preferred amino donors, and the enzyme was found to catalyze the KMTB-tyrosine aminotransfer reaction equally as well as the KG-tyrosine reaction normally associated with TyrAT.
In the present study, we have cloned and expressed the C. fasciculata aminotransferase identified in the previous experiments and have examined its substrate specificity and apparent kinetics. In addition, the aminotransferases involved in Met formation in Plasmodium falciparum, Giardia intestinalis, and Trichomonas vaginalis have been studied with cell homogenates, and the enzymes have been cloned and expressed from P. falciparum, G. intestinalis, and T. brucei brucei. The amino acid sequences, substrate specificities, and kinetic parameters of all of the recombinant enzymes identify them as AspATs.
MATERIALS AND METHODS
Parasites and tissues.
C. fasciculata clone HS6 was grown in undefined yeast extract-tryptone medium as described previously (24) and was harvested by centrifugation for 10 min at 3,500 × g when the cells were in early to middle exponential growth. P. falciparum clone 3D7 was cultured in A-positive human erythrocytes grown at pH 7.4 in RPMI 1640 medium (Life Technologies, Paisley, United Kingdom) supplemented (per liter) with 1 g of bicarbonate, 2 g of glucose, 26 mg of hypoxanthine (Sigma, Poole, United Kingdom), and 5 g of Albumax II (Life Technologies). The malaria parasites were allowed to grow asynchronously and were harvested by saponin lysis followed by sequential washes in phosphate-buffered saline. T. brucei brucei clone S427-117 procyclics were cultured in SDM-79 medium and were harvested by centrifugation for 10 min at 3,500 × g. G. intestinalis and T. vaginalis were obtained as frozen pellets of trophozoites. Pig kidney was obtained fresh from the Dundee City Abatoir (Dundee, United Kingdom) and kept on ice before being cut into small pieces and frozen at −20°C.
To make subcellular homogenates for determining aminotransferase activities and for subsequent purification, parasite cell pellets or minced pig kidney samples were resuspended in 25 mM potassium phosphate (pH 7.8)–120 mM KCl–1 mM dithiothreitol (DTT)–2.5 mM KG (Sigma)–0.2 mM pyridoxal-5-phosphate (PLP; Sigma)–1 mM phenylmethylsulfonyl fluoride–5 μM leupeptin–2 μM pepstatin–0.5 mM Nα-p-tosyl-l-lysine chloromethyl ketone (TLCK) and then sonicated on ice (parasites) or ground with a motorized Ystral (Dottingen, Germany) homogenizer (kidney). The resulting homogenate was then centrifuged at 14,000 × g for 20 min, and the supernatant was dialyzed against two changes of 10 mM phosphate buffer–1 mM EDTA–1 mM DTT (buffer A) (pH 7.4).
DNA was isolated from each parasite by resuspending the pelleted cells in an equal volume of 100 mM NaCl–10 mM Tris-HCl (pH 8.0)–25 mM EDTA–0.5% (wt/vol) sodium dodecyl sulfate–0.1 mg of proteinase K (Bioline, London, United Kingdom)/ml and incubating the suspension at 37°C for 1 h prior to phenol-chloroform-isoamyl alcohol extraction. The isolated DNA was precipitated in 300 mM sodium acetate (pH 5.2)–95% ethanol, vacuum dried, and resuspended in distilled water. P. falciparum total RNA was isolated from saponin-freed asynchronous parasites by resuspension of a 100-μl cell pellet in 500 μl of RNAce lysis buffer (Bioline) and subsequent phenol-chloroform extraction as outlined in the RNAce kit instructions. A 100-ng quantity of total RNA was used for reverse transcription at 37°C for 30 min with Moloney murine leukemia virus reverse transcriptase (Promega, Southampton, United Kingdom) and antisense primers specific for the 3′ end of the plasmodial AspAT gene (see below) or the plasmodial lactate dehydrogenase gene (positive control for expression ). Incubations without primers were used to control for any DNA contamination. After reverse transcription, the products were diluted fourfold and an aliquot was used for PCR with 35 cycles of 95°C for 1 min, 55°C for 1 min, and 72°C for 1 min.
Native aminotransferases
The number of aminotransferases catalyzing Met formation was determined by loading 2 ml of dialyzed supernatant into a 10-cm DEAE-Sepharose FF (Pharmacia, St. Albans, United Kingdom) column equilibrated with buffer A (pH 7.8) and eluting the sample with a linear gradient of 0 to 250 mM KCl in buffer A. The column was connected to a Biosys (Beckman, High Wycombe, United Kingdom)-biocompatible high-pressure liquid chromatograph run at 1 ml/min and 4°C, with UV detection at 280 nm. One-milliliter fractions were kept from each run and analyzed for general KMTB-amino acid aminotransfer (KMAT activity) as outlined below. Peaks of activity within a given run were then reassayed for the amino donor specificity of the KMAT reaction as outlined below. Total KMAT activity in T. vaginalis and G. intestinalis homogenates was inhibited by incubating 10 μl of the enzyme preparation as described below for general KMAT activity in the presence of 100 μM or 1.0 mM malic acid, serine-O-sulfate, canaline, carboxymethoxylamine, or nitrophenylalanine (all from Sigma). The amount of Met produced was then quantified by high-pressure liquid chromatography (HPLC) and compared to that of control incubations.
Enzyme assays.
In order to assay for general KMAT activity, 10 μl of test fraction was added to 100 μl of 100 mM potassium phosphate (pH 7.4)–2 mM ADEFGHIKNQRSTWY–1 mM KMTB–50 μM PLP, and the mixture was incubated at 37°C for 30 min. At the end of the incubation, samples were frozen at −20°C until analyzed further. After thawing of the samples, 20 μl of sample was mixed with 100 μl of 0.4 M borate (pH 10.5) and then with 20 μl of o-phthalaldehyde (10 mg/ml)–3-mercaptopropionate (12 μl/ml)–0.4 M borate (pH 10.5). Seven microliters of this mixture was then immediately injected into an ODS-AA column (2.1 by 200 mm; Hewlett-Packard, Stockport, United Kingdom) run on a Beckman HPLC system consisting of a model 126 binary pump, a 166 photodiode array detector, a 507e autosampler, and System Gold operating software. The column was run at ambient temperature with an initial flow rate of 0.45 ml/min and with 2.72 mg of sodium acetate (pH 7.2)/ml–0.018% (vol/vol) triethylamine–0.3% tetrahydrofuran as solvent A and 2.72 mg of sodium acetate (pH 7.2)/ml–40% (vol/vol) methanol–40% (vol/vol) acetonitrile as solvent B. Elution was accomplished with a linear gradient of 0 to 60% solvent B over 17 min, 60 to 100% solvent B over 1 min, and 100% solvent B for 6 min. A flow rate of 0.45 ml/min was used over the first 18 min, and a flow rate of 0.8 ml/min was used over the final 6 min. The o-phthalaldehyde-derivatized amino acids were detected by UV spectrophotometry at 338 nm.
To measure the capacity of a single amino acid to act as an amino donor for KMTB, the reaction mixture consisted of a 2 mM concentration of the test amino acid in place of ADEFGHIKNQRSTWY. The HPLC assay was also capable of measuring the aminotransfer of any amino acid-keto acid pairing by replacement of ADEFGHIKNQRSTWY with the appropriate amino acid and KMTB with the desired keto acid. In this manner, AspAT activity was assayed with 2 mM aspartate–1 mM KG or 2 mM glutamate–1 mM oxaloacetate, alanine aminotransferase (AlaAT) activity was assayed with 2 mM alanine–1 mM KG or 2 mM glutamate–1 mM pyruvate, and TyrAT activity was assayed with 2 mM tyrosine–1 mM KG.
To determine the kinetic parameters of the recombinant enzymes, samples for HPLC analysis were constructed using 0.5 mM PLP, 10 mM cosubstrate, and 0.1, 0.5, 1.0, 2.5, 5.0, or 10 mM substrate. These samples were incubated at 37°C for 15 min and were then stored at −20°C until analysis. Following conversion of HPLC peak areas to nanomoles per minute per milligram of protein, kinetic constants were determined via the Scientist program (MicroMath, Salt Lake City, Utah) with the Michaelis-Menten equation and nonlinear least-squares fitting. For inhibition studies, recombinant P. falciparum AspAT was added to 1.0, 2.0, or 3.0 mM Tyr, 5.0 mM KMTB, 0.5 mM PLP, and 10 mM potassium phosphate (pH 7.4) containing 0, 100, 200, 300, 400, or 500 μM canaline (Sigma) before incubation at 37°C for 30 min.
Cloning of aminotransferases.
For the C. fasciculata enzyme, selected cytosolic AspATs from lower and higher eukaryotes were aligned using the Megalign program (DNAStar, Madison, Wis.) and the Clustal algorithm (42). Two regions of very high conservation and relatively low redundancy were chosen for the design of degenerate oligonucleotide primers (5′-CTNCACGCNTGCGCNCACAACCCNACNGG-3′ [sense] and 5′-CGCATSGWSACGATNCGGTCNGCCAT-3′ [antisense]). After PCR amplification using 5 μg of C. fasciculata genomic DNA, BioTaq DNA polymerase (Bioline), 1.5 mM MgCl2, and 30 cycles of denaturation at 95°C for 1 min, annealing at 60°C for 1.5 min, and extension at 72°C for 1.5 min, the anticipated product of approximately 480 bp was isolated from an agarose gel using Qiaex II resin (Qiagen, Crawley, United Kingdom) and ligated into PCRScript according to the manufacturer's instructions (Stratagene, Amsterdam, The Netherlands). The plasmid containing the PCR product was isolated from cultures of Escherichia coli using a Qiaspin Mini kit (Qiagen), and the insert was sequenced in both directions by automated, dye-labeled DNA sequencing (ABI, Warrington, United Kingdom) at the Department of Biochemistry, University of Dundee. The translated amino acid sequence of the PCR product was then used in multiple alignments to confirm the identity of the C. fasciculata gene fragment. The nucleotide sequence of the insert was then used to design exact primers for the amplification of a 380-bp portion of the gene (CfAT primers): 5′-ACAGGCGTCGACCCCTCGCACGCGCA-3′ (sense) and 5′-TTCCGCAGCTCCTTATCGCTCAGCA-3′ (antisense).
One hundred micrograms of C. fasciculata genomic DNA was digested with 0.248 U of Sau3A (Promega) for 30 min at 37°C and then for 10 min at 70°C, treated with alkaline phosphatase (Promega) for 30 min at 37°C and then for 10 min at 70°C, and ligated to BamHI (Promega)-digested λ-Dash at a 1:0.2 ratio of arms to insert. After Sau3A digestion, alkaline phosphatase treatment, and ligation, a sample was subjected to PCR with the CfAT primers to confirm the presence of the 380-bp sequence. The ligated DNA was then packaged according to the Packagene protocol (Promega), and the resulting library was titered, amplified, and stored at 4°C with 0.3% chloroform in SM broth. A 5-μl aliquot of the amplified library was heated at 99°C for 5 min and then used as the template for PCR with the CfAT primers to confirm the presence of the target gene in the library. An aliquot (0.1 μl) of the amplified library was then plated, and plaque lifting was performed in duplicate using Hybond NX filters (Amersham, St. Albans, United Kingdom). One of the filters was rinsed with 2.0 ml of 100 mM NaCl–8 mM MgSO4–50 mM Tris-HCl (pH 7.5)–0.1% (wt/vol) gelatin, and then 20 μl of this liquid was heated to 99°C for 5 min and used as the template for PCR with the CfAT primers to confirm the presence of the target gene on the plate. The second filter was dried, denatured in 0.2 M NaOH–1.5 M NaCl, and subjected to UV cross-linking. The filter was then probed with a fluorescein-labeled oligonucleotide prepared from the PCR product of the CfAT primers and C. fasciculata genomic DNA. Labeling of the probe, hybridization, and detection were performed according to the manufacturer's instructions for the Gene Images kit (Amersham). Positive plaques were picked from the agar plate into 1 ml of SM broth, and 5 μl of this liquid was used for PCR with the CfAT primers to confirm the presence of the target gene. A large-scale amplification of a single positive plaque was then performed, and the DNA was isolated and subjected to nucleotide sequencing in both directions. The complete open reading frame was amplified from genomic DNA using 5′-GACGACGACAAGATGTCTGGATCTCCGACCGA-3′ (sense) and 5′-GGAACAAGACCCGTTTACACCGTGCGAACCGCCTC-3′ (antisense) and cloned into pCALnEK (Stratagene).
The P. falciparum AspAT was identified by similarity searching of the malaria genome project (www.ncbi.nlm.gov/Malaria/blastindex.html) using the BLAST program (1) and the translated sequence of the 486-bp fragment of the C. fasciculata AspAT as the query sequence. The complete open reading frame was amplified from genomic DNA using 5′-GACGACGACAAGATGGATAAGTTATTAAGCAGCTTAGA-3′ (sense) and 5′-GGAACAAGACCCGTTCATATTTGACTTAGCGAAAGACAA-3′ (antisense) and cloned into pCALnEK. The G. intestinalis enzyme was similarly identified by similarity searching of the Giardia genome project (www.mbl.edu/Giardia) (32) and assembly of the nucleotide fragments from high-identity matches using the Seqman program (DNAStar). The complete open reading frame was amplified from genomic DNA using 5′-GACGACGACAAGATGTCTGTCTTCTCAGGGTTTCCTG-3′ (sense) and 5′-GGAACAAGACCCGTTTCATTTCTTGAACGGGAGCTTCG-3′ (antisense) and cloned into pCALnEK. Two AspATs were identified in T. brucei brucei by similarity searching of the Trypanosoma brucei genome project (www.tigr/org/tdb/mdb/tbdb) and assembly of the nucleotide fragments from high-identity matches using Seqman. The complete open reading frame of the mitochondrial AspAT was amplified from genomic DNA using 5′-GACGACGACAAGATGGGGAAACCGGATCCCA-3′ (sense) and 5′-GGAACAAGACCCGTTCATTTAGTAACGTTGTGA-3′ (antisense), and that of the cytosolic AspAT was amplified using 5′-GACGACGACAAGATGTCCAGGCCCTTTAAGGACT-3′ (sense) and 5′-GGAACAAGACCCGTCTACTTGTTACGCACGTGTCGGACAACATCGTCAATC-3′ (antisense). Both trypanosomal AspATs were cloned into pCALnEK.
Amino acid sequences were compared by multiple alignments using ClustalW (42) and Clustal analysis with the PAM250 sequence substitution table (12). The aligned sequences were then subjected to distance analysis using the ProtDist program in the Phylip package (13). The resulting distance matrix was used in the Neighbor program of Phylip for tree generation by the method of neighbor joining (39), and all trees were visualized using the TreeView program (http://taxonomy.zoology.gla.ac.uk/rod/rod.html).
Expression of recombinant enzymes.
The pCALnEK constructs were transformed into E. coli BL21-CodonPlus cells (Stratagene), which were then grown in Luria-Bertani medium at 37°C until the A600 reached 0.6 to 0.8. Isopropyl-β-d-thiogalactopyranoside (IPTG) was added to a final concentration of 1 mM, and the cultures were grown for an additional 5 to 7 h at 27°C before the cells were harvested by centrifugation. The cell pellet was resuspended in 10 mM HEPES (pH 7.8)–150 mM NaCl–1 mM DTT–1 mM imidazole–1 mM magnesium acetate–2 mM CaCl2 (buffer B) and sonicated on ice before centrifugation at 4,000 × g for 20 min. The cell supernatant was loaded into a calmodulin-agarose (Stratagene) column (1.0 by 10 cm) equilibrated and washed with buffer B and then was eluted with 10 mM HEPES (pH 7.8)–1.2 M NaCl–1 mM DTT–3 mM EGTA. Fractions containing the recombinant protein were dialyzed against 10 mM HEPES (pH 7.4)–1 mM DTT–1 mM EDTA and concentrated to less than 5 ml using a 30-kDa molecular mass cutoff filter.
Nucleotide sequence accession numbers.
The sequences reported in this paper have been submitted to GenBank under accession numbers AF326988, AF326989, AF326990, and AF326991.
RESULTS
C. fasciculata cytoplasmic AspAT
Previously, it had been demonstrated that C. fasciculata and T. brucei brucei were capable of generating Met from methylthioadenosine (6) and that C. fasciculata contained three different aminotransferases capable of catalyzing the final step in this pathway. An internal peptide sequence with high identity to the sequence of eukaryotic cytosolic AspAT was obtained from the most active of these enzymes (7), and degenerate oligonucleotide primers were designed for amplification of the middle third of the gene. The resulting 480-bp product was sequenced and, upon translation, was found to have a very high identity to eukaryotic cytosolic AspAT (51% to the chicken cytoplasmic AspAT). The gene fragment contained a unique SalI site, and Southern analysis of restriction enzyme-digested C. fasciculata DNA yielded two bands (of 4.7 and 1.8 kbp) with SalI and single bands with other endonucleases when probed with the 480-bp fragment. The AspAT gene therefore appears to exist as a single gene copy in the parasite genome.
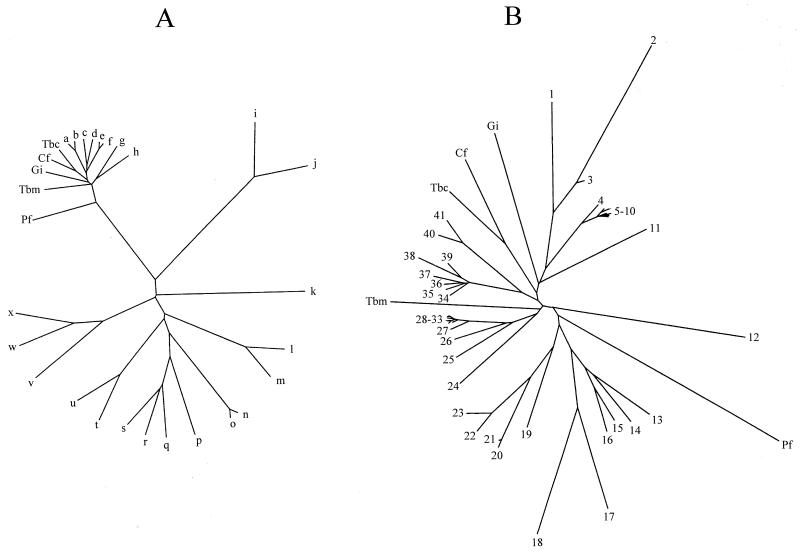
The complete coding sequence for the AspAT was cloned from a genomic library and was found to be 1,218 bp long (GenBank accession number AF326988). The gene codes for a polypeptide of 405 amino acids with high similarity to eukaryotic cytosolic AspAT. Figure 1 shows an alignment of the crithidial AspAT with selected AspATs for clarity. A larger alignment, consisting of the sequences discussed in this paper and the sequences of the currently known members of the aminotransferase Ia subfamily or the members of the aminotransferase I family, is available from B. J. Berger; see references 20 and 23 for information on the classification of enzymes within the aminotransferase I family and reference 19 for a more recent dendrogram. As expected, the crithidial AspAT shares a relatively low number of completely conserved residues across even the limited number of sequences shown in Fig. 2 and is most closely related to the T. brucei brucei cytoplasmic AspAT (see below), with an identity of 56% and a similarity of 75%. Of sequences outside those shown in this article, the crithidial AspAT most closely resembles the chicken cytoplasmic AspAT, with an identity of 42% and a similarity of 65%. With conservation of the motifs LLHXCXHNPTGXDX5W, DXAYQGX3GX4D, and SKX3 LYXERXG around key residues, the crithidial AspAT is clearly a member of the Ia subfamily of aminotransferases (23). The residues G192(G197), D217(D222), K253(K258), and R380(R386) are the ones identified by Mehta et al. (33) as being essential for the binding and stabilization of the PLP cofactor; the residues given in parentheses are those for the pig cytosolic AspAT, which Mehta et al. (33) proposed as the standard for comparative nomenclature. These residues are purportedly the only ones completely conserved across all four families of aminotransferases (33). Phylogenetic analysis of selected family I aminotransferases demonstrated that the enzyme clustered within the Ia subfamily, which consists of eukaryotic AspATs, gram-negative bacterial AspATs, and prokaryotic TyrATs (Fig. 2A). A similar analysis of the subfamily Ia aminotransferases showed that the crithidial AspAT clustered with the other known eukaryotic cytosolic AspATs (Fig. 2B).
FIG. 1.
Alignment of parasite AspATs with selected eukaryotic enzymes. The enzymes are as follows: CfC, C. fasciculata cytoplasmic AspAT; Pf, P. falciparum AspAT; GiC, G. intestinalis cytoplasmic AspAT; TbC, T. brucei brucei cytoplasmic AspAT; TbM, T. brucei brucei mitochondrial AspAT, HuC, Homo sapiens cytoplasmic AspAT; ChC, Gallus gallus cytoplasmic AspAT; YeC, S. cerevisiae cytoplasmic AspAT; HuM, H. sapiens mitochondrial AspAT; ChM, G. gallus mitochondrial AspAT; and YeM, S. cerevisiae mitochondrial AspAT. Boxes surround residues which are conserved across all 11 sequences, while the underlined residues in the C. fasciculata enzyme represent the sequence determined previously by amino acid sequencing of a purified aminotransferase (7). The residues marked with asterisks are those reported by Jensen and Gu (23) as being conserved in all members of the Ia subfamily of aminotransferases, while those marked with number signs are those reported by Mehta et al. (33) as being conserved in all aminotransferase families.
FIG. 2.
Phylogenetic analysis of parasite AspATs. In both trees, constructed by neighbor joining with the Phylip package, parasite AspATs are abbreviated as follows: Cf, C. fasciculata cytoplasmic AspAT; Tbc, T. brucei brucei cytoplasmic AspAT; Tbm, T. brucei brucei mitochondrial AspAT; Gi, G. intestinalis cytoplasmic AspAT; and Pf, P. falciparum AspAT. Tree A was formed using the following additional sequences: a, H. sapiens cytoplasmic AspAT; b, G. gallus cytoplasmic AspAT; c, Lupinus angustifolicus mitochondrial AspAT; d, Oryza sativa cytoplasmic AspAT; e, H. sapiens mitochondrial AspAT; f, G. gallus mitochondrial AspAT; g, E. coli AspAT; h, E. coli TyrAT; i, Schizosaccharomyces pombe TyrAT; j, S. cerevisiae TyrAT; k, E. coli alanine-valine aminotransferase; l, H. sapiens AlaAT; m, S. cerevisiae AlaAT; n, H. sapiens kynurenine aminotransferase; o, Rattus norvegicus kynurenine aminotransferase; p, B. subtilis YHDR gene product; q, B. subtilis AspAT; r, B. subtilis PATA gene product; s, B. subtilis YUGH gene product; t, H. sapiens TyrAT; u, Trypanosoma cruzi TyrAT; v, Halobacter sp. histidinol-phosphate aminotransferase; w, E. coli histidinol-phosphate aminotransferase; and x, S. pombe histidinol-phosphate aminotransferase. Tree B was formed using the following additional sequences from members of the aminotransferase Ia subfamily: 1, C. elegans CE07462 gene product; 2, C. elegans CE06829 gene product; 3, C. elegans CE07461 gene product; 4, G. gallus cytoplasmic AspAT; 5, Mus muris cytoplasmic AspAT; 6, R. norvegicus cytoplasmic AspAT; 7, H. sapiens cytoplasmic AspAT; 8, Equus caballus cytoplasmic AspAT; 9, Bos taurus cytoplasmic AspAT; 10, Sus scrofus cytoplasmic AspAT; 11, S. cerevisiae cytoplasmic AspAT; 12, S. cerevisiae mitochondrial AspAT; 13, Vibrio cholerae AspAT; 14, E. coli AspAT; 15, Haemophilus influenzae AspAT; 16, Neisseria gonorrhoeae AspAT; 17, Paracoccus denitrificans TyrAT; 18, Rhizobium meliloti TyrAT; 19, Pseudomonas aeruginosa TyrAT; 20, N. gonorrhoeae TyrAT; 21, N. meningitidis TyrAT; 22, E. coli TyrAT; 23, K. pneumoniae TyrAT; 24, A. thaliana AspAT1; 25, Drosophila melanogaster CT10757 gene product; 26, C. elegans CE02477 gene product; 27, G. gallus mitochondrial AspAT; 28, H. sapiens mitochondrial AspAT; 29, E. caballus mitochondrial AspAT; 30, B. taurus mitochondrial AspAT; 31, S. scrofus mitochondrial AspAT; 32, R. norvegicus mitochondrial AspAT; 33, M. muris mitochondrial AspAT; 34, Medicago sativa cytoplasmic AspAT; 35, Daucus caroti cytoplasmic AspAT; 36, O. sativa cytoplasmic AspAT; 37, A. thaliana AspAT3; 38, A. thaliana AspAT4; 39, A. thaliana AspAT2; 40, L. angustifolicus mitochondrial AspAT; and 41, A. thaliana mitochondrial AspAT.
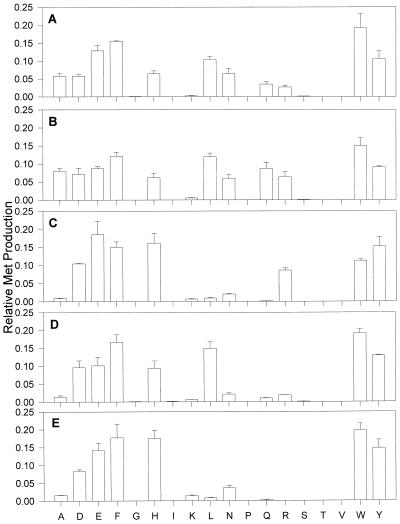
The crithidial AspAT was expressed in E. coli containing an extra plasmid expressing the argU, ileY, and leuW genes. These extra tRNAs were not essential for expression of the enzyme but greatly enhanced the amount of recombinant protein obtained (data not shown). The enzyme was found to be active and readily utilized glutamate, phenylalanine, tyrosine, tryptophan and, to a lesser degree, alanine, aspartate, histidine, leucine, asparagine, and glutamine as amino donors for Met production from KMTB (Fig. 3A). The crithidial AspAT was also fully capable of catalyzing AspAT and TyrAT activities. These results are in full agreement with those obtained previously with the native, purified enzyme (7). Selected substrate pairings were examined further to determine the apparent kinetic constants (Table 1). As was found for the K. pneumoniae TyrAT (19), the crithidial AspAT was able to catalyze Met production equally as well as aspartate-KG and tyrosine-KG aminotransfer, with Vmax values of 1.97 to 4.00 μmol/min/mg of protein. Surprisingly, the AspAT also catalyzed AlaAT and GlnAT reactions at equal or better rates. To date, there have been no reports of an AspAT capable of catalyzing such a broad range of activities, and the biochemical significance of such a broad substrate specificity is unclear. However, based on previous studies, this aminotransferase is the main, but not sole, mechanism of KMTB transamination in C. fasciculata (7).
FIG. 3.
Amino donor spectrum for recombinant parasite AspATs. Purified recombinant aminotransferases were incubated with a 2 mM concentration of a single amino acid and 1 mM KMTB as described in Materials and Methods and analyzed for Met production by HPLC. Met production for each amino acid is shown as the fraction of total Met production. (A) C. fasciculata cytoplasmic AspAT. (B) T. brucei brucei cytoplasmic AspAT. (C) T. brucei brucei mitochondrial AspAT. (D) G. intestinalis cytoplasmic AspAT. (E) P. falciparum AspAT.
TABLE 1.
Kinetic parameters for selected substratesa
| AspAT | Substrate | Cosubstrate | Apparent Vmax (μmol/min/mg) | Apparent Km (mM) |
|---|---|---|---|---|
| C. fasciculata cytoplasmic | Asp | KG | 3.37 ± 0.31 | 5.00 ± 0.98 |
| Tyr | KG | 3.30 ± 0.08 | 4.77 ± 0.20 | |
| Gln | KG | 23.82 ± 2.52 | 32.02 ± 4.23 | |
| Ala | KG | 2.76 ± 0.14 | 3.10 ± 0.38 | |
| KG | Tyr | 3.30 ± 0.11 | 6.78 ± 0.41 | |
| KMTB | Glu | 1.87 ± 0.13 | 6.95 ± 0.89 | |
| Glu | KMTB | 2.27 ± 0.06 | 11.34 ± 0.50 | |
| Tyr | KMTB | 1.97 ± 0.24 | 10.12 ± 1.72 | |
| Phe | KMTB | 2.50 ± 0.08 | 25.99 ± 1.13 | |
| Trp | KMTB | 2.54 ± 0.09 | 11.94 ± 0.69 | |
| His | KMTB | 4.00 ± 0.65 | 16.38 ± 3.90 | |
| Leu | KMTB | 2.97 ± 0.03 | 12.77 ± 0.20 | |
| T. brucei brucei cytoplasmic | Asp | KG | 4.59 ± 0.33 | 10.38 ± 1.22 |
| Tyr | KG | 8.68 ± 1.24 | 17.10 ± 3.04 | |
| Gln | KG | 16.68 ± 1.53 | 21.47 ± 2.67 | |
| Ala | KG | 3.50 ± 0.26 | 7.83 ± 0.88 | |
| KG | Tyr | 2.83 ± 0.16 | 6.96 ± 0.75 | |
| KMTB | Glu | 2.05 ± 0.13 | 8.69 ± 0.24 | |
| Glu | KMTB | 1.97 ± 0.11 | 8.93 ± 0.84 | |
| Tyr | KMTB | 1.47 ± 0.21 | 7.25 ± 1.53 | |
| Phe | KMTB | 1.69 ± 0.08 | 16.12 ± 1.07 | |
| Trp | KMTB | 1.63 ± 0.16 | 6.85 ± 1.29 | |
| His | KMTB | 4.63 ± 0.37 | 19.53 ± 2.20 | |
| Leu | KMTB | 2.09 ± 0.06 | 8.76 ± 0.47 | |
| T. brucei brucei mitochondrial | Asp | KG | 8.40 ± 0.44 | 9.80 ± 0.87 |
| Tyr | KG | 3.14 ± 0.27 | 1.94 ± 0.39 | |
| Gln | KG | 0 | ||
| Ala | KG | 0 | ||
| KG | Tyr | 2.88 ± 0.88 | 2.13 ± 0.17 | |
| KMTB | Glu | 2.62 ± 0.07 | 4.23 ± 0.26 | |
| Glu | KMTB | 4.43 ± 0.25 | 10.39 ± 0.96 | |
| Tyr | KMTB | 0.42 ± 0.02 | 0.79 ± 0.09 | |
| Phe | KMTB | 0.92 ± 0.06 | 4.28 ± 0.65 | |
| Trp | KMTB | 0.31 ± 0.01 | 0.44 ± 0.08 | |
| His | KMTB | 4.62 ± 1.17 | 27.42 ± 8.97 | |
| Leu | KMTB | 0.06 ± 0.01 | 0.38 ± 0.26 | |
| G. intestinalis cytoplasmic | Asp | KG | 7.29 ± 1.39 | 11.30 ± 3.47 |
| Tyr | KG | 5.88 ± 0.62 | 6.90 ± 1.13 | |
| Gln | KG | 0 | ||
| Ala | KG | 0 | ||
| KG | Tyr | 3.75 ± 0.24 | 5.44 ± 0.72 | |
| KMTB | Glu | 3.67 ± 0.19 | 10.64 ± 0.92 | |
| Glu | KMTB | 3.63 ± 0.18 | 10.94 ± 0.91 | |
| Tyr | KMTB | 1.84 ± 0.15 | 5.61 ± 0.72 | |
| Phe | KMTB | 3.34 ± 0.15 | 23.95 ± 1.41 | |
| Trp | KMTB | 4.85 ± 0.32 | 16.09 ± 1.54 | |
| His | KMTB | 6.60 ± 0.41 | 17.47 ± 1.54 | |
| Leu | KMTB | 2.88 ± 0.13 | 8.11 ± 0.64 | |
| P. falciparum | Asp | KG | 3.69 ± 0.41 | 5.48 ± 1.24 |
| Tyr | KG | 1.35 ± 0.08 | 1.47 ± 0.23 | |
| Gln | KG | 0 | ||
| Ala | KG | 0 | ||
| KG | Tyr | 1.03 ± 0.04 | 1.26 ± 0.15 | |
| KMTB | Glu | 0.45 ± 0.03 | 1.06 ± 0.28 | |
| Glu | KMTB | 0.96 ± 0.06 | 8.90 ± 1.00 | |
| Tyr | KMTB | 0.14 ± 0.01 | 0.44 ± 0.13 | |
| Phe | KMTB | 0.16 ± 0.01 | 0.79 ± 0.10 | |
| Trp | KMTB | 0.12 ± 0.01 | 0.72 ± 0.06 | |
| His | KMTB | 1.06 ± 0.16 | 16.34 ± 3.69 | |
| Leu | KMTB | 0.02 ± 0.01 | 0.77 ± 0.28 |
The recombinant enzymes were incubated with 10 mM cosubstrate (5 mM in the case of Tyr) and various amounts of substrate as outlined in Materials and Methods. Met production was quantified by HPLC, and kinetic constants were determined using nonlinear least-squares fitting to the Michaelis-Menten equation. Vmax and Km values are reported as the mean and standard deviation.
T. brucei brucei cytoplasmic and mitochondrial AspATs
Screening of the T. brucei brucei GuTat10.1 genome project database with the 480-bp gene fragment sequence from C. fasciculata yielded several shotgun gene fragments which could be assembled into two unique aminotransferase genes. The complete coding sequences of both of these genes were cloned from genomic T. brucei brucei S427/117 DNA. The first sequence (GenBank accession number AF326989) was 1,212 bp long and encoded a polypeptide of 403 amino acids, while the second (GenBank accession number AF326990) was 1,173 bp long and encoded a protein of 390 amino acids. The presence of the motifs HNPTGXDX5W, PXXDXAYqgX3GX4D, and SXXKXXGLYXXRXG in the deduced amino acid sequences of both enzymes suggested that both AspATs were members of the aminotransferase Ia subfamily (Fig. 1); lowercase letters indicate substitutions relative to the consensus sequence. While Q226 and G227 have been reported to be conserved across the Ia subfamily (25), the second trypanosomal AspAT had P207(P226) and S208(S227) in these positions.
Phylogenetic analysis of selected family I aminotransferases supported the placement of both enzymes in the Ia subfamily (Fig. 2A). A similar analysis of the Ia subfamily showed that the first trypanosomal AspAT clustered with known eukaryotic, nonplant cytoplasmic AspATs, while the second one clustered with known eukaryotic mitochondrial AspATs (with the notable exception of the Saccharomyces cerevisiae mitochondrial AspAT) (Fig. 2B). After multiple sequence alignment or sequential pairwise alignment, the trypanosomal cytoplasmic AspAT was found to be most similar to the crithidial AspAT (see above) and was equally distant from mammalian, plant, and yeast cytoplasmic AspATs (with identities ranging from 39 to 45% and similarities of 63 to 66%). The trypanosomal mitochondrial AspAT was found to be most similar to a mitochondrial AspAT from Arabidopsis thaliana (GenBank accession number P46248), with an identity of 38% and a similarity of 60%, and a mitochondrial AspAT from Caenorhabditis elegans (GenBank accession number U39645), at 39 and 63%, respectively.
The trypanosomal enzymes were individually expressed in E. coli and purified by affinity chromatography. As with the C. fasciculata enzyme, coexpression with the argU, ileY, and leuW tRNA genes greatly increased the amounts of trypanosomal aminotransferases obtained. Both enzymes were screened for aminotransferase activity using KMTB as the amino acceptor and each amino acid as the donor (Fig. 3B and C). Both enzymes effectively catalyzed reactions with tryptophan, tyrosine, phenylalanine, glutamate, aspartate, histidine, and arginine. In addition, the trypanosomal cytoplasmic AspAT also utilized alanine, leucine, and glutamine as amino donors. The pattern seen for the trypanosomal cytoplasmic AspAT closely follows that seen previously with homogenates prepared from bloodstream T. brucei brucei (6). In detailed kinetic analyses, the cytoplasmic and mitochondrial AspATs were similar (Table 1), with the exception that glutamate and the aromatic amino acids were equally as effective as amino donors for KMTB with the cytoplasmic AspAT (Vmax values of 1.63 to 1.97 nmol/min/mg of protein), whereas glutamate was 5- to 10-fold more effective than the aromatic amino acids with the mitochondrial AspAT (4.43 versus 0.31 to 0.92 nmol/min/mg of protein). In addition, the cytoplasmic AspAT readily catalyzed GlnAT, AlaAT, and leucine-KMTB aminotransfer reactions, while the mitochondrial AspAT did not catalyze these reactions. Both enzymes catalyzed AspAT and TyrAT activities.
Methionine formation in T. vaginalis and G. intestinalis.
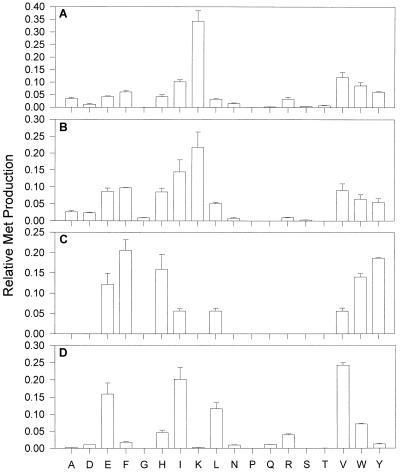
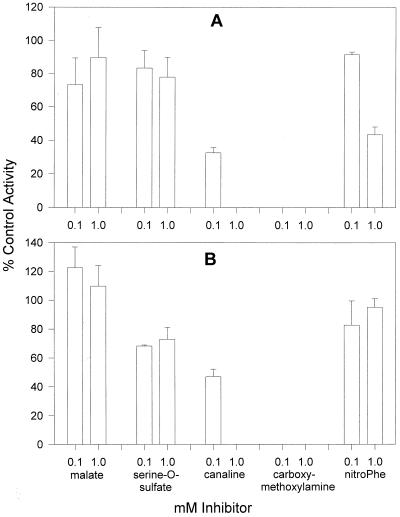
Cellular homogenates of T. vaginalis and G. intestinalis were examined for their ability to catalyze the formation of Met from KMTB (Fig. 4A and B). Both organisms were found to have similar patterns for the amino donor preference for the KMAT reaction, with lysine, glutamate, phenylalanine, histidine, isoleucine, valine, tryptophan, and tyrosine all being effective amino donors. The strong preference for lysine as an amino donor was very unusual, as this amino acid is a very ineffective amino donor for KMTB in all the other organisms studied here and previously (6, 7, 19). In addition to the unusual amino donor preference, both anaerobic organisms catalyzed the KMAT reaction much more readily than the other parasites and bacteria examined. An example of this phenomenon can be seen by comparing the rates of 14.07 nmol/min/mg of protein (T. vaginalis) and 14.96 nmol/min/mg of protein (G. intestinalis) for lysine-KMTB aminotransfer with the rate of 0.64 nmol/min/mg of protein for phenylalanine-KMTB aminotransfer in C. fasciculata cellular homogenates or 1.92 nmol/min/mg of protein for glutamate-KMTB aminotransfer in K. pneumoniae homogenates. KMAT activity in T. vaginalis and G. intestinalis homogenates was also examined in the presence of selected aminotransferase inhibitors (Fig. 5). Only the amino-oxy compounds canaline and carboxymethoxylamine showed appreciable inhibition, with both compounds completely abolishing KMAT activity at 1.0 mM. At 100 μM, carboxymethoxylamine also completely inhibited KMAT activity, while canaline reduced KMAT activity to 35 to 45% that in control incubations. These results are similar to the inhibition seen in homogenates of C. fasciculata and K. pneumoniae (7, 19).
FIG. 4.
Amino donor spectrum for parasite or tissue homogenates. Aliquots of subcellular homogenates were incubated with a 2 mM concentration of a single amino acid and 1 mM KMTB as described in Materials and Methods and analyzed for Met production by HPLC. Met produced by each amino acid is shown as the fraction of total Met production. (A) T. vaginalis homogenates. (B) G. intestinalis homogenates. (C) P. falciparum homogenates. (D) Pig kidney homogenates.
FIG. 5.
Inhibition of Met production in homogenates of T. vaginalis (A) and G. intestinalis (B). Homogenates of each parasite were incubated with 2 mM ADEFGHIKLNQRSTWY, 1 mM KMTB, and 0.1 or 1.0 mM individual inhibitors as described in Materials and Methods and analyzed for Met production. Inhibition of Met production relative to the results for control incubations is shown. nitroPhe, nitrophenylalanine.
The T. vaginalis and G. intestinalis homogenates were separated with a DEAE column, yielding two active fractions for each organism which eluted in the voided volume and at 800 mM KCl. The first fraction was found to catalyze KMAT activity with isoleucine, valine, histidine, phenylalanine, tryptophan, tyrosine, and glutamate, whereas the second fraction used only lysine as an amino donor. With the exception of isoleucine and valine utilization, the amino donor preference of the first fraction closely resembled that of parasite AspATs or bacterial TyrATs (see above and references 6, 7, and 19).
G. intestinalis cytoplasmic AspAT
By similarity searching of the G. intestinalis genome project shotgun sequence data, it was possible to identify the two ends of the cytoplasmic AspAT. The complete gene was cloned and was found to be 1,284 bp long and to encode a protein of 427 amino acids (GenBank accession number AF326991). As with the other parasite AspATs, the presence of the motifs HXCXHNPsGXDX5W, DXAYQGX3GX4D, and SXXKXXGLYXERXG around the anchor residues identified the enzyme as a member of the aminotransferase Ia subfamily (Fig. 1). While T196 has been reported to be conserved across the Ia subfamily (23), the giardial AspAT had S194(S196) in this position. In addition, phylogenetic analysis of the protein sequence placed the enzyme with other members of the Ia subfamily (Fig. 2A and B), where it clustered with other eukaryotic cytoplasmic AspATs. The G. intestinalis AspAT showed almost equal sequence similarities to the C. fasciculata AspAT, the T. brucei brucei cytoplasmic AspAT, human cytoplasmic AspAT (GenBank accession number NM002079), chicken cytoplasmic AspAT (GenBank accession number X15636), and S. cerevisiae cytoplasmic AspAT (GenBank accession number NC001144), with identities of 38 to 44% and similarities of 59 to 64%.
The enzyme was expressed in E. coli, where coexpression with a vector containing the argU, ileY, and leuW genes was necessary to obtain a good yield of active recombinant material. After purification by affinity chromatography, the enzyme was examined for amino donor specificity in the KMAT reaction (Fig. 3D); glutamate, aspartate, phenylalanine, histidine, leucine, tryptophan, and tyrosine were found to be effective donors. In this respect, the G. intestinalis AspAT is very similar to the C. fasciculata and T. brucei brucei cytoplasmic AspATs. However, the G. intestinalis enzyme demonstrates reduced utilization of alanine, asparagine, glutamine, and arginine as amino donors relative to the two trypanosomal cytoplasmic AspATs. With the exception of an inability to use valine and isoleucine as amino donors, the purified G. intestinalis AspAT catalyzes all the KMAT activity seen in the DEAE voided fraction isolated from cellular homogenates. Therefore, with regard to global Met regeneration from KMTB, G. intestinalis utilizes AspAT, an aminotransferase with a strict dependence on lysine as the amino donor, and an additional aminotransferase activity which can utilize valine and isoleucine as amino donors.
Selected substrate pairs were further examined to characterize the kinetics of the enzyme (Table 1). The Km and Vmax values for KMTB, KG, glutamate, tyrosine, phenylalanine, tryptophan, histidine, and leucine were very similar to those seen for the C. fasciculata and T. brucei brucei cytoplasmic AspATs, with Km values ranging from 5.44 to 23.95 mM and Vmax values of 1.84 to 6.60 μmol/min/mg of protein. Like the other enzymes, the G. intestinalis AspAT catalyzed AspAT and TyrAT reactions but, in contrast to the trypanosomal cytoplasmic AspATs, did not perform any glutamine-KG or alanine-KG aminotransfer. Therefore, while the giardial cytoplasmic AspAT is very similar to the trypanosomal homologues, it has a narrower substrate specificity.
Methionine regeneration in P. falciparum
Cellular homogenates of malarial parasites isolated from human erythrocytes were examined for their ability to catalyze the KMAT reaction (Fig. 4C). Glutamate, phenylalanine, histidine, isoleucine, leucine, valine, tryptophan, and tyrosine were the only amino acids capable of acting as amino donors, with the branched-chain amino acids being less active as substrates. The amount of KMAT activity detected in the plasmodial homogenates was lower than that found in other organisms screened to date. When the most active amino donor, glutamate, was used, only 0.22 nmol of Met/min/mg of protein could be produced from KMTB, as opposed to 0.64 nmol/min/mg of protein for phenylalanine-KMTB in C. fasciculata homogenates and 1.92 nmol/min/mg of protein for glutamate-KMTB in K. pneumoniae homogenates. The plasmodial homogenates were separated with a DEAE column, where a single active fraction, eluting at 700 mM KCl, was discovered. This fraction was able to catalyze Met formation from KMTB using the same amino donors as those used by the original homogenates, suggesting that only one aminotransferase catalyzes Met regeneration in P. falciparum.
P. falciparum AspAT.
The published sequence for P. falciparum chromosome 2 contains one region with significant similarity to AspATs (15). In addition, BLAST searching of all the sequence data available for the remaining chromosomes yielded no further AspAT homologues. The putative chromosome 2 AspAT was cloned, and the published sequence was confirmed to be 1,218 bp long and to code for a protein of 405 amino acids. The presence of the motifs qX3yNPcsXyX5y, DXAYQGX3tX4D, and SXXKXXsLYXERXG (Fig. 1) around the anchor residues suggests that the plasmodial enzyme is a member of the aminotransferase Ia subfamily. However, it is clear from the number of residues in these motifs shown as lowercase letters that the plasmodial AspAT has a number of substitutions relative to the reported Ia consensus sequence (23). In a most striking variation, it was noted that the normal anchor residue G197, which is postulated to be an essential requirement of all aminotransferases (33), is S188(S197) in the P. falciparum AspAT. As the published DNA sequence (GenBank accession number AE001380) and the sequence obtained in this manuscript are identical, it is highly unlikely that S188(S197) is due to a sequencing error or a PCR-induced mutation. In addition, similarity searching of the P. vivax and P. berghei genome surveys (parasite.vetmed.ufl.edu) (9) has uncovered partial sequences for the homologous AspATs (Fig. 6A). S188(S197) is present in all three sequences, indicating that the postulated requirement of G197 for aminotransferase activity is not absolute and that plasmodial (and perhaps apicomplexan) AspATs contain unusual sequence variations compared to enzymes from other phyla and kingdoms.
FIG. 6.
Alignment of plasmodial and trypanosomatid AspATs. (A) Clustal alignment of a portion of the P. falciparum AspAT with the deduced amino acid sequences of gene fragments obtained from the P. vivax and P. berghei sequence tag projects (9). (B) Clustal alignment of portions of the C. fasciculata cytoplasmic AspAT and T. brucei brucei cytoplasmic (c) AspAT with the deduced amino acid sequence of a gene fragment obtained from the L. major genome project (35). For both sets of sequences, the boxed residues were conserved by all three sequences, the residues above the sequences are those reported to be conserved in all aminotransferases in the Ia subfamily (23), and the residues marked with asterisks are those reported to be conserved in all aminotransferase families (33). The residues underlined in the L. major fragment represent the amino acid sequence determined by Vernal et al. (45) from a purified L. mexicana enzyme. The numbers in parentheses represent the total length of the amino acid sequence obtained for each enzyme.
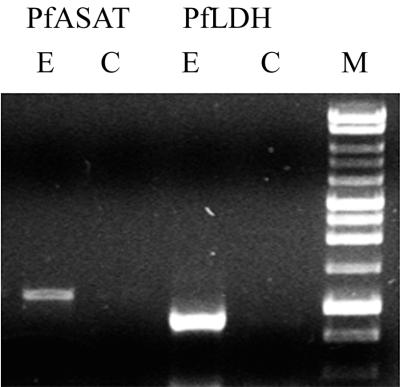
Phylogenetic analysis of the plasmodial AspAT and the family I aminotransferases unambiguously placed the enzyme in the Ia subfamily (Fig. 2A). However, when only the Ia subfamily is examined, the malarial enzyme is the most highly divergent sequence available, clustering with neither the eukaryotic cytoplasmic AspATs, the eukaryotic mitochondrial AspATs, the bacterial AspATs, nor the bacterial TyrATs (Fig. 2B). In terms of sequence similarity, the P. falciparum AspAT has only 24 to 31% identity and 49 to 55% similarity with any of the other Ia subfamily members. With these terms of reference, it is impossible to unambiguously identify the enzyme as cytoplasmic, mitochondrial, or apicoplastic. At a point in the primary amino acid sequence where the various AspATs reproducibly differ, around D222 (DS/TAY for cytoplasmic AspATs, DMAY for mitochondrial AspATs, DVAY for chloroplast AspATs, and DFAY for gram-negative bacterial AspATs), the malarial enzyme has a different sequence (DIAY). Nevertheless, the AspAT is expressed in the erythrocytic stages of the life cycle, as determined by reverse transcription-PCR (Fig. 7).
FIG. 7.
Reverse transcription-PCR of P. falciparum AspAT. RNA isolated from asynchronous P. falciparum was incubated in the presence (lanes E) or absence (lanes C) of reverse transcriptase and then subjected to PCR with primers specific for the full-length P. falciparum AspAT gene (PfASAT) or the P. falciparum lactate dehydrogenase gene (PfLDH) as a positive control. The products were then analyzed on an agarose gel together with DNA markers (lane M). The lengths of the markers are (from the gel bottom) 0.5, 0.75, 1.0, 1.5, 2.0, 2.5, 3.0, 4.0, 5.0, 6.0, 8.0, and 10.0 kbp. The expected length of AspAT was 1,218 bp, and that of lactate dehydrogenase was 951 bp.
Expression of the P. falciparum AspAT was difficult, as a series of expression vectors yielded a truncated product or no product in E. coli or S. cerevisiae (data not shown). Only the use of an E. coli strain coexpressing extra argU, ileY, and leuW genes gave rise to any active, full-length product. Even in this case, the yield was low and the bacterial biomass greatly decreased following induction, suggesting that expression of the malarial AspAT was in some manner toxic to the host cell (a phenomenon which did not occur with any of the other parasitic AspATs). The purified recombinant enzyme was examined for KMAT activity and was found to produce Met from KMTB with aspartate, glutamate, histidine, tryptophan, phenylalanine, and tyrosine as effective amino donors (Fig. 3E). Therefore, this AspAT is responsible for the majority of KMAT activity seen in plasmodial homogenates, with the exception of that related to the use of branched-chain amino acids as amino donors. The plasmodial AspAT was found to catalyze AspAT and TyrAT activities but not Gln-KG or Ala-KG aminotransfer (Table 1). The Km and Vmax values for most of the substrates were lower than those for the other parasitic AspATs, and this phenomenon was particularly striking for the KMAT reaction. It would appear that the plasmodial AspAT catalyzes Met regeneration rather poorly and that this phenomenon is responsible for the low level of KMAT activity detected in the cellular homogenates.
In previous work, it was shown that the aminooxy compound canaline can inhibit KMAT activity in homogenates and has antimalarial effects in vitro (7, 8, 19). Against the purified recombinant plasmodial AspAT, canaline was found to inhibit uncompetitively, with a Ki of 27.6 ± 3.1 μM.
Methionine regeneration in pig kidney.
In previous studies (7), it has been demonstrated that pig heart AspAT and AlaAT poorly catalyze the formation of Met from KMTB. In order to better compare mammalian and parasite Met regeneration, pig kidney was chosen as a model. Homogenates of freshly acquired tissue were found to readily catalyze the KMAT reaction, with glutamate, isoleucine, leucine, and valine being the major amino donors and aspartate, phenylalanine, histidine, asparagine, glutamine, arginine, tryptophan, and tyrosine also being able to act as amino donors (Fig. 4D). The homogenates were applied to a DEAE column, and two fractions catalyzing the KMAT reaction were discovered. The first, eluting in the column voided volume, utilized branched-chain amino acids and, to a lesser extent, glutamate, aromatic amino acids, and histidine. The second, eluting at 400 mM KCl, used histidine and, to a lesser extent, asparagine. Neither of these fractions had AspAT or TyrAT activity.
DISCUSSION
The international situation regarding the chemotherapy of bacterial and parasitic diseases continues to worsen with the spread of drug-resistant pathogenic strains. In particular, P. falciparum malaria currently threatens half of the world's population, with at least 200 million infections and 2 to 3 million deaths per year (48). Resistance to aminoquinolines and antifolates has become commonplace, and few effective or novel agents are clinically available. In a similar manner, trypanosomal diseases are an increasing concern, as evidenced by the recent epidemic of African sleeping sickness in the Congo, Uganda, and southern Sudan (40) and the reports of arsenical-resistant trypanosomiasis and antimony-resistant leishmaniasis (25, 26). Cases of trichomoniasis and giardiasis have become commonplace, and reports of metronidazole resistance have occurred (43). These disease resurgences require the discovery and development of novel drug targets. In past studies, it has been demonstrated that interference with enzymes involved in the regeneration of Met from methylthioadenosine can lead to cell death in K. pneumoniae, P. falciparum, and T. brucei (3, 17, 36, 41). These studies have focused on the enzymes methylthioadenosine phosphorylase and methylthioribose kinase, which convert methylthioadenosine to methylthiophosphoribose in eukaryotes and prokaryotes, respectively. It has been shown that the final step of Met regeneration from methylthioadenosine, the transamination of KMTB to Met, can be inhibited in homogenates of C. fasciculata and K. pneumoniae (7, 19). In addition, selected inhibitors which are active for this process have a cytocidal effect in vitro against C. fasciculata and P. falciparum (7, 8).
Identity and relationship of aminotransferases catalyzing Met formation.
In the present study, we have continued the characterization of the final step of Met recycling in parasitic organisms and have found that AspAT plays an important role in all of the organisms examined. In C. fasciculata, T. brucei brucei, and P. falciparum, AspAT represents the major source of activity for Met regeneration, and in G. intestinalis, the enzyme is second only to an as-yet-uncharacterized source of activity utilizing lysine as the amino donor. The consistency of these results across diverse phyla suggests that AspAT is an essential component of Met recycling in lower eukaryotes. However, aside from our work on the same reaction in K. pneumoniae, nothing is known about the exact aminotransferases active in other organisms. In particular, higher mammals, fungi, gram-positive bacteria, and archaea have not been fully examined. Nevertheless, it is clear that different aminotransferases are responsible for Met regeneration in different groups of organisms. The activity in K. pneumoniae has been unequivocally identified as TyrAT, whereas the protozoa examined here rely on AspAT. In mammals, using pig tissues as a model, we have been able to discount AspAT, TyrAT, and AlaAT as being responsible for the activity (7) but have been unable to fully purify the exact aminotransferase responsible. In studies with purified rat GlnAT (kynurenine aminotransferase), Cooper and Meister have shown that the enzyme actively catalyzes phenylalanine-KMTB aminotransfer (10). In addition, Davoodi et al. (11) and Hall et al. (18) have found that purified and recombinant human and rat branched-chain aminotransferases can catalyze branched-chain amino acid–KMTB aminotransfer. Given the amino donor preference for the KMAT reaction in pig kidney homogenates, it is entirely possible that the branched-chain aminotransferase plays a large role in the reaction in vivo, while a role for the kynurenine aminotransferase is less clear. Full purification of the mammalian KMAT aminotransferase(s) and a broader characterization of recombinant mammalian branched-chain and kynurenine aminotransferases is required to completely solve this problem.
It is interesting that the parasitic protozoa and K. pneumoniae, while using different types of aminotransferases to catalyze the KMAT reaction, all use enzymes from the Ia subfamily. However, from the numerous genome projects completed, we have been unable to find any members of the Ia subfamily in gram-positive bacteria or archaea. The AspATs in these organisms are members of the If subfamily (see the phylogenic tree in reference 19); thus, they are relatives of eukaryotic TyrATs and kynurenine aminotransferases. Branched-chain aminotransferases are all members of the III family of aminotransferases (33). We are currently characterizing KMAT activity in several gram-positive models, with an emphasis on Bacillus subtilis, where there is a strong preference for branched-chain amino acids as amino donors (data not shown). As mentioned above, we have also been able to discount mammalian AspAT as being responsible for KMAT activity, thus eliminating the only higher eukaryotic members of the Ia subfamily. Therefore, diverse subfamilies and, potentially, different families of aminotransferases may have evolved specificity for KMTB.
In terms of substrate specificity, the crithidial and trypanosomal cytosolic AspATs are unusually broad. In addition to the expected aspartate-KG and tyrosine-KG activities, these enzymes also catalyzed glutamine-KG and alanine-KG aminotransfer. In previous studies (19), the K. pneumoniae TyrAT was unable to perform these latter two reactions, and there are no other literature reports of AspATs catalyzing AlaAT or GlnAT activity. When the primary amino acid sequences are examined, there are no obvious differences between the sequences for the two kinetoplastid AspATs and the sequence for the giardial enzyme, which lacks AlaAT and GlnAT activities. Crystallization and structural analyses of the crithidial and/or trypanosomal enzymes may assist in further understanding the basis for the broad substrate specificity compared to the known structures for the E. coli, S. cerevisiae, and chicken AspATs (21, 22, 29). As both C. fasciculata and T. brucei brucei cytoplasmic AspATs have this wider range of activity, it is likely that the homologous enzyme from other trypanosomatids would share this feature. Vernal et al. purified from Leishmania mexicana promastigotes an aminotransferase which displayed an unusually broad range of activity and yielded an internal peptide sequence consistent with the sequence of a cytoplasmic AspAT (45). By similarity searching of shotgun sequence data from the L. major genome project, we have been able to identify approximately half of the leishmanial cytoplasmic AspAT sequence (Fig. 6B). It is clear, from the presence of the peptide sequence obtained by Vernal et al. (45), that this cytoplasmic AspAT represents the enzyme previously purified in that study. It is also apparent that the leishmanial sequence has a high identity to the crithidial and trypanosomal sequences. While we have not examined the biochemical properties of the leishmanial enzyme, the present work suggests that this AspAT should be active in Met regeneration in Leishmania spp.
Sequence of the plasmodial AspAT.
The AspAT from P. falciparum is highly unusual in that it contains sequence substitutions not found in aminotransferases from any of the four families (33). While, overall, the plasmodial AspAT has sufficient homology to subfamily Ia aminotransferases to unequivocally be grouped with these enzymes, the sequence displays a very high level of divergence from cytoplasmic AspATs, mitochondrial AspATs, plastid AspATs, gram-negative bacterial AspATs, and bacterial TyrATs. The partial sequences obtained from P. vivax and P. berghei clearly demonstrate that the unusual AspAT sequence is conserved within the plasmodia. Unfortunately, there is a paucity of aminotransferase sequences from nonfungal lower eukaryotes, so it is impossible to state whether the P. falciparum AspAT is unique to plasmodia or is indicative of a new subtype of Ia aminotransferases localized to apicomplexans or other phyla of protists. As dinoflagellates are often hypothesized as representing the phylum closest to the apicomplexans (2), AspAT sequences from these organisms would be particularly enlightening. Structural analysis of the P. falciparum AspAT would be helpful in determining whether the unusual primary sequence is the source for the relatively low activity of this enzyme for KMTB transamination.
KMTB transamination in G. intestinalis and T. vaginalis.
The finding that both G. intestinalis and T. vaginalis rely on lysine as a central amino donor for KMTB transamination is unusual. No other organism examined to date makes any use of lysine in this context. In addition, while in the other parasites and K. pneumoniae aminotransferases other than AspAT or TyrAT clearly play some role in Met regeneration, these additional enzymes are a minor component of total activity. In G. intestinalis and T. vaginalis, the unknown aminotransferase catalyzing the KMTB-lysine reaction appears to be quantitatively more important than AspAT for transaminating KMTB. In a previous study, Lowe and Rowe examined aminotransferase activities in T. vaginalis (27). Among their discoveries was an aminotransferase which readily catalyzed KG-lysine and phenylpyruvate-lysine aminotransfer. This enzyme was partially purified and found to copurify with an ornithine aminotransferase activity. While the enzyme was found to utilize ornithine, lysine, KG, and phenylpyruvate, neither KMTB nor other substrates were examined. It is quite possible that this enzyme is responsible for the KMTB-lysine activity seen in T. vaginalis, but it should be pointed out that Lowe and Rowe (27) also detected lysine-oxaloacetate activity in T. vaginalis homogenates that did not copurify with an ornithine or lysine aminotransferase. In related work, Lowe and Rowe also purified an AspAT from T. vaginalis (28) which had both AspAT and TyrAT activities but did not catalyze GlnAT or AlaAT reactions. While this enzyme was not examined for KMTB transamination, the kinetic properties for aspartate-KG and tyrosine-KG suggest that their T. vaginalis AspAT is the homologue of the G. intestinalis AspAT presented here.
Subsequent to the studies outlined in this paper, a report demonstrating the lack of any putrescine or spermidine aminopropyltransferase activity in T. vaginalis was published (51). Clearly, the KMAT activity measured in this organism cannot be related to Met regeneration from polyamine biosynthesis. The biochemical source of any cellular KMTB utilized in the KMAT reaction in T. vaginalis is presently a mystery. However, several alternatives to polyamine synthesis exist, such as S-adenosylmethionine conversion to methylthioadenosine and 1-aminocyclopropane-1-carboxylate (47), the action of amino acid oxidase on d-methionine, the conversion of hydroxymethiobutyrate via an α-hydroxy acid dehydrogenase, and the possibility of compartmental Met-KMTB cycling within the cell (46). The presence or absence of these alternative sources of KMTB has not been investigated.
Inhibition of Met formation.
We have shown that the amino-oxy compounds canaline and carboxymethoxylamine can inhibit total KMAT activity in homogenates of T. vaginalis and G. intestinalis. At 100 μM concentrations, canaline was as potent as in corresponding experiments with C. fasciculata or K. pneumoniae homogenates (7, 19). At present, neither or these compounds has been tested by us against T. vaginalis or G. intestinalis in vitro. However, Rowe and Lowe found that carboxymethoxylamine can completely inhibit T. vaginalis cell growth in vitro, albeit at a high concentration (5 mM) (38).
In previous work, canaline was demonstrated to be an effective antimalarial agent in vitro (8). The compound, an aminooxy analogue of ornithine, has been shown here to be an uncompetitive inhibitor of the plasmodial AspAT, with a Ki of 27 μM. As Met regeneration in malaria has been shown to be necessary for cellular survival (41) and our present work has demonstrated that P. falciparum catalyzes the KMAT reaction much less readily than the other protozoa examined, the malaria parasite may be uniquely susceptible to interference with the final step in Met recycling. It should be pointed out, however, that aminooxy inhibitors are not necessarily specific to a single aminotransferase. We have found that canaline also inhibits the plasmodial ornithine aminotransferase (data not shown), suggesting that the compound may exert an effect on multiple targets and pathways.
ACKNOWLEDGMENTS
We thank Alan H. Fairlamb for helpful discussions during the course of these investigations and Graham Coombs (Institute of Biomedical and Life Sciences, University of Glasgow) for providing the G. intestinalis and T. vaginalis cell pellets.
This work was funded by the Wellcome Trust. The sequences in this study were obtained from preliminary data made available by a number of ongoing sequence projects. Sequencing of the P. falciparum genome was performed at The Institute for Genomic Research, the Naval Medical Research Center, Stanford University, and the Sanger Centre, with financial support provided by the Burroughs Wellcome Fund, the Wellcome Trust, the National Institutes of Health (NIAID), and the U.S. Department of Defense Military Infectious Diseases Research Program. The giardial genome was sequenced at the Marine Biological Laboratory in Woods Hole, Mass., the University of Texas at El Paso, the University of Arizona at Tucson, and the University of Illinois at Urbana-Champaign, with funding provided by the National Institutes of Health (NIAID), the LI-COR Biotechnology Division, and the G. Unger Vetlesen Foundation. Sequencing of the T. brucei brucei genome was performed at The Institute for Genomic Research and the Sanger Centre and was funded by the National Institutes of Health (NIAID), the Wellcome Trust, and Beowulf Genomics. Sequencing of the L. major genome was performed at the Seattle Biomedical Research Institute, Fiocruz, EULeish, and the Sanger Centre, with funding from the Burroughs Wellcome Fund, the National Institutes of Health (NIAID), the European Union, Beowulf Genomics, and the World Health Organization (TDR). The P. vivax and P. berghei genome sequence tag projects were undertaken at the University of Florida, with funding from the National Institutes of Health (NIAID).
REFERENCES
- 1.Altschul S F, Madden T L, Schaffer A A, Zhang J, Zhang Z, Miller W, Lipman D J. Gapped BLAST and PSI-BLAST: a new generation of protein database search programs. Nucleic Acids Res. 1997;25:3389–3402. doi: 10.1093/nar/25.17.3389. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Ayala F J, Escalante A A, Lal A A, Rich S M. Evolutionary relationships of human malaria parasites. In: Sherman I, editor. Malaria: parasite biology, pathogenesis, and protection. Washington, D.C.: ASM Press; 1998. pp. 285–300. [Google Scholar]
- 3.Bacchi C J, Sufrin J R, Nathan H C, Spiess A J, Hannon T, Garafolo J, Alecia K, Katz L, Yarlett N. 5′-Alkyl-substituted analogs of 5′-methylthioadenosine as trypanocides. Antimicrob Agents Chemother. 1991;35:1904–1906. doi: 10.1128/aac.35.7.1315. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Backlund P S, Chang C P, Smith R A. Identification of 2-keto-4-methylthiobutyrate as an intermediate compound in methionine synthesis from 5′-methylthioadenosine. J Biol Chem. 1982;257:4196–4202. [PubMed] [Google Scholar]
- 5.Backlund P S, Smith R A. Methionine synthesis from 5′-methylthioadenosine in rat liver. J Biol Chem. 1980;256:1533–1535. [PubMed] [Google Scholar]
- 6.Berger B J, Dai W W, Wang H, Stark R E, Cerami A. Aromatic amino acid transamination and methionine recycling in trypanosomatids. Proc Natl Acad Sci USA. 1996;93:4126–4130. doi: 10.1073/pnas.93.9.4126. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Berger B J, Dai W W, Wilson J. Methionine formation from α-ketomethiobutyrate in the trypanosomatid Crithidia fasciculata. FEMS Microbiol Lett. 1998;165:305–312. doi: 10.1111/j.1574-6968.1998.tb13162.x. [DOI] [PubMed] [Google Scholar]
- 8.Berger B J. Antimalarial activities of aminooxy compounds. Antimicrob Agents Chemother. 2000;44:2540–2542. doi: 10.1128/aac.44.9.2540-2542.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8a.Brun R, Schonenberger M. Cultivation and in vitro cloning of procyclic culture forms of Trypanosoma brucei in a semi-defined medium. Acta Trop. 1979;36:289–292. [PubMed] [Google Scholar]
- 9.Carlton J M, Dame J B. The Plasmodium vivax and P. berghei gene sequence tag projects. Parasitol Today. 2000;16:409. doi: 10.1016/s0169-4758(00)01781-6. [DOI] [PubMed] [Google Scholar]
- 10.Cooper A J L, Meister A. Isolation and properties of highly purified glutamine aminotransferase. Biochemistry. 1972;11:661–671. doi: 10.1021/bi00755a001. [DOI] [PubMed] [Google Scholar]
- 11.Davoodi J, Drown P M, Bledsoe R K, Wallin R, Reinhart G D, Hutson S M. Overexpression and characterization of the human mitochondrial and cytosolic branched-chain aminotransferases. J Biol Chem. 1998;273:4982–4989. doi: 10.1074/jbc.273.9.4982. [DOI] [PubMed] [Google Scholar]
- 12.Dayhoff M O, Schwartz R M, Orcutt B C. A model of evolutionary change in proteins. In: Dayhoff M O, editor. Atlas of protein sequence and structure. Washington, D.C.: NBRF; 1978. pp. 345–352. [Google Scholar]
- 13.Felsenstein J. PHYLIP—Phylogeny Inference Package (version 3.2) Cladistics. 1989;5:164–166. [Google Scholar]
- 14.Furfine E S, Abeles R H. Intermediates in the conversion of 5′-methylthioadenosine to methionine in Klebsiella pneumoniae. J Biol Chem. 1988;263:9596–9606. [PubMed] [Google Scholar]
- 15.Gardner M J, Tettelin H, Carucci D J, Cummings L M, Aravind L, Koonin E V, Shallom S, Mason T, Yu K, Fujii C, Pederson J, Shen K, Jing J, Aston C, Lai Z, Schwartz D C, Pertea M, Salzberg S, Zhou L, Sutton G G, Clayton R, White O, Smith H O, Fraser C M, Adams M D, Venter J C, Hoffman S L. Chromosome 2 sequence of the human malaria parasite Plasmodium falciparum. Science. 1998;282:1126–1132. doi: 10.1126/science.282.5391.1126. [DOI] [PubMed] [Google Scholar]
- 16.Ghoda L Y, Savarese T M, Northrup C H, Parks R E, Garafolo J, Katz L, Ellenbogen B B, Bacchi C J. Substrate specificities of 5′-deoxy-5′methylthioadenosine phosphorylase from Trypanosoma brucei brucei and mammalian cells. Mol Biochem Parasitol. 1988;27:109–118. doi: 10.1016/0166-6851(88)90030-8. [DOI] [PubMed] [Google Scholar]
- 17.Gianotti A J, Tower P A, Sheley J H, Conte P A, Spiro C, Ferro A J, Fitchen J H, Riscoe M K. Selective killing of Klebsiella pneumoniae by 5-trifluoromethylthioribose. Chemotherapeutic exploitation of the enzyme 5-methylthioribose kinase. J Biol Chem. 1990;265:831–837. [PubMed] [Google Scholar]
- 18.Hall T R, Wallin R, Reinhart G D, Hutson S M. Branched chain aminotransferase isoenzymes. Purification and characterization of the rat brain isoenzyme. J Biol Chem. 1993;268:3092–3098. [PubMed] [Google Scholar]
- 19.Heilbronn J, Wilson J, Berger B J. Tyrosine aminotransferase catalyzes the final step of methionine recycling in Klebsiella pneumoniae. J Bacteriol. 1999;181:1739–1747. doi: 10.1128/jb.181.6.1739-1747.1999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Iraqui I, Vissers S, Cartiaux M, Urrestarazu A. Characterisation of Saccharomyces cerevisiae ARO8 and ARO9 genes encoding aromatic aminotransferase I and II reveals a new aminotransferase subfamily. Mol Gen Genet. 1998;257:238–248. doi: 10.1007/s004380050644. [DOI] [PubMed] [Google Scholar]
- 21.Jager J, Pauptit R A, Sauder U, Jansonius J N. Three-dimensional structure of a mutant E. coli aspartate aminotransferase with increased enzymic activity. Protein Eng. 1994;7:605–612. doi: 10.1093/protein/7.5.605. [DOI] [PubMed] [Google Scholar]
- 22.Jeffery C J, Barry T, Doonan S, Petsko G A, Ringe D. Crystallization and preliminary X-ray diffraction analysis of aspartate aminotransferase from Saccharomyces cerevisiae. Protein Sci. 1998;7:1380–1387. doi: 10.1107/s0907444997016235. [DOI] [PubMed] [Google Scholar]
- 23.Jensen R A, Gu W. Evolutionary recruitment of biochemically specialized subdivisions of family I within the protein superfamily of aminotransferases. J Bacteriol. 1996;178:2161–2171. doi: 10.1128/jb.178.8.2161-2171.1996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Kidder G W, Dutta B N. The growth and nutrition of Crithidia fasciculata. J Gen Microbiol. 1958;18:621–638. doi: 10.1099/00221287-18-3-621. [DOI] [PubMed] [Google Scholar]
- 25.Legros D, Fournier C, Gastellu Etchegorry M, Maiso F, Szumilin E. Therapeutic failure of melarsoprol among patients treated for late stage T.b. gambiense human African trypanosomiasis in Uganda. Bull Soc Pathol Exot. 1999;92:171–172. [PubMed] [Google Scholar]
- 26.Lira R, Sundar S, Makharia A, Kenney R, Gam A, Saraiva E, Sacks D. Evidence that the high incidence of treatment failures in Indian kala-azar is due to the emergence of antimony-resistant strains of Leishmania donovani. J Infect Dis. 1999;180:564–567. doi: 10.1086/314896. [DOI] [PubMed] [Google Scholar]
- 27.Lowe P N, Rowe A F. Aminotransferase activities in Trichomonas vaginalis. Mol Biochem Parasitol. 1986;21:65–74. doi: 10.1016/0166-6851(86)90080-0. [DOI] [PubMed] [Google Scholar]
- 28.Lowe P N, Rowe A F. Aspartate:2-oxoglutarate aminotransferase from Trichomonas vaginalis. Biochem J. 1985;232:689–695. doi: 10.1042/bj2320689. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Malashkevich V N, Stokopytor B V, Borisov V V, Dauter Z, Wilson K S, Torchinsky Y M. Crystal structure of the closed form of chicken cytosolic aspartate aminotransferase at 1.9 A resolution. J Mol Biol. 1995;247:111–124. doi: 10.1006/jmbi.1994.0126. [DOI] [PubMed] [Google Scholar]
- 30.Marchitto K S, Ferro A J. The metabolism of 5-methylthioadenosine and 5-methyl-1-phosphate in Saccharomyces cerevisiae. J Gen Microbiol. 1985;131:2153–2164. doi: 10.1099/00221287-131-9-2153. [DOI] [PubMed] [Google Scholar]
- 31.Marton L J, Pegg A E. Polyamines as targets for therapeutic intervention. Annu Rev Pharmacol Toxicol. 1995;35:55–91. doi: 10.1146/annurev.pa.35.040195.000415. [DOI] [PubMed] [Google Scholar]
- 32.McArthur A G, Morrison H G, Nixon J E, Passamaneck N Q, Kim U, Hinkle G, Crocker M K, Holder M E, Farr R, Reich C I, Olsen G E, Aley S B, Adam R D, Gillin F D, Sogin M L. The Giardia genome project database. FEMS Microbiol Lett. 2000;189:271–273. doi: 10.1111/j.1574-6968.2000.tb09242.x. [DOI] [PubMed] [Google Scholar]
- 33.Mehta P K, Hale T I, Christen P. Aminotransferases: demonstration of homology and division into evolutionary subgroups. Eur J Biochem. 1993;214:549–561. doi: 10.1111/j.1432-1033.1993.tb17953.x. [DOI] [PubMed] [Google Scholar]
- 34.Myers R W, Abeles R H. Purification and characterization of an enzyme involved in oxidative carbon-carbon cleavage reactions in the methionine salvage pathway of Klebsiella pneumoniae. J Biol Chem. 1993;268:24785–24791. [PubMed] [Google Scholar]
- 35.Myler P J, Stuart K D. Recent developments from the Leishmania genome project. Curr Opin Microbiol. 2000;3:412–416. doi: 10.1016/s1369-5274(00)00113-2. [DOI] [PubMed] [Google Scholar]
- 36.Riscoe M K, Ferro A J, Fitchen J H. Analogs of 5-methylthioribose, a novel class of antiprotozoal agents. Antimicrob Agents Chemother. 1988;32:1904–1906. doi: 10.1128/aac.32.12.1904. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Riscoe M K, Tower P A, Peyton D H, Ferro A J, Fitchen J H. Methionine recycling as a target for antiprotozoal drug development. In: Coombs G, North M, editors. Biochemical protozoology. London, United Kingdom: Taylor and Francis; 1991. pp. 450–457. [Google Scholar]
- 38.Rowe A F, Lowe P N. Modulation of amino acid and 2-oxo acid pools in Trichomonas vaginalis by aspartate aminotransferase. Mol Biochem Parasitol. 1986;21:17–24. doi: 10.1016/0166-6851(86)90074-5. [DOI] [PubMed] [Google Scholar]
- 39.Saitou N, Nai M. The neighbor-joining method: a new method for reconstructing phylogenetic trees. Mol Biol Evol. 1987;4:406–425. doi: 10.1093/oxfordjournals.molbev.a040454. [DOI] [PubMed] [Google Scholar]
- 40.Smith D H, Pepin J, Stich A H. Human African trypanosomiasis: an emerging public health crisis. Br Med Bull. 1998;54:341–355. doi: 10.1093/oxfordjournals.bmb.a011692. [DOI] [PubMed] [Google Scholar]
- 41.Sufrin J R, Meshnik S R, Spiess A J, Garafolo-Hannon J, Pan X Q, Bacchi C J. Methionine recycling pathways and antimalarial drug design. Antimicrob Agents Chemother. 1995;39:2511–2515. doi: 10.1128/aac.39.11.2511. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Thompson J D, Higgins D G, Gibson T J. CLUSTAL W: improving the sensitivity of progressive multiple sequence alignment through sequence weighting, position-specific gap penalties and weight matrix choice. Nucleic Acids Res. 1994;22:4673–4680. doi: 10.1093/nar/22.22.4673. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Townson S M, Boreham P F, Upcroft P, Upcroft J A. Resistance to the nitroheterocyclic drugs. Acta Trop. 1994;56:173–194. doi: 10.1016/0001-706x(94)90062-0. [DOI] [PubMed] [Google Scholar]
- 44.Trackman P C, Abeles R H. Methionine synthesis from 5′-S-1-phospho-5-S-methylthioribulose. J Biol Chem. 1983;258:6717–6720. [PubMed] [Google Scholar]
- 45.Vernal J, Cazzulo J J, Nowicki C. Isolation and partial characterization of a broad specificity aminotransferase from Leishmania mexicana promastigotes. Mol Biochem Parasitol. 1998;96:83–92. doi: 10.1016/s0166-6851(98)00117-0. [DOI] [PubMed] [Google Scholar]
- 46.Walker J, Barrett J. Parasite sulphur amino acid metabolism. Int J Parasitol. 1997;27:883–897. doi: 10.1016/s0020-7519(97)00039-8. [DOI] [PubMed] [Google Scholar]
- 47.Wang S Y, Adams D O, Lieberman M. Recycling of 5′-methylthioadenosine-ribose carbon atoms into methionine in tomato tissue in relation to ethylene production. Plant Physiol. 1982;70:117–121. doi: 10.1104/pp.70.1.117. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.World Health Organization. Tropical disease research: progress 1997–1998. Geneva, Switzerland: World Health Organization; 1999. [Google Scholar]
- 49.Wray J W, Abeles R H. A bacterial enzyme that catalyzes formation of carbon monoxide. J Biol Chem. 1993;268:21466–21469. [PubMed] [Google Scholar]
- 50.Wray J W, Abeles R H. The methionine salvage pathway in Klebsiella pneumoniae and rat liver. Identification and characterization of two novel dioxygenases. J Biol Chem. 1995;270:3147–3152. doi: 10.1074/jbc.270.7.3147. [DOI] [PubMed] [Google Scholar]
- 51.Yarlett N, Martinez M P, Goldberg B, Kramer D L, Porter C W. Dependence of Trichomonas vaginalis upon polyamine backconversion. Microbiology. 2000;146:2715–2722. doi: 10.1099/00221287-146-10-2715. [DOI] [PubMed] [Google Scholar]