Abstract
Vero cells are widely used for antiviral tests and virology research for SARS-CoV-2 as well as viruses from various other families. However, Vero cells generally express high levels of multi-drug resistance 1 (MDR1) or Pgp protein, the efflux transporter of foreign substances including many antiviral compounds, affecting the antiviral activity as well as interpretation of data. To address this, a Pgp gene knockout VeroE6 cell line (VeroE6-Pgp-KO) was generated using CRISPR-CAS9 technology. These cells no longer expressed the Pgp protein as indicated by flow cytometry analysis following staining with a Pgp-specific monoclonal antibody. They also showed significantly reduced efflux transporter activity in the calcein acetoxymethyl ester (calcein AM) assay. The VeroE6-Pgp-KO cells and the parental VeroE6 cells were each infected with SARS-CoV-2 to test antiviral activities of remdesivir and nirmatrelvir, two known Pgp substrates, in the presence or absence of a Pgp inhibitor. The compounds showed antiviral activities in VeroE6-Pgp-KO cells similar to that observed in the presence of the Pgp inhibitor. Thus, the newly established VeroE6-Pgp-KO cell line adds a new in vitro virus infection system for SARS-CoV-2 and possibly other viruses to test antiviral therapies without a need to control the Pgp activity. Removal of the Pgp inhibitor for antiviral assays will lead to less data variation and prevent failed assays.
Keywords: Nirmatrelvir, SARS-CoV-2, Vero cells, Pgp, Knockout
1. Introduction
The COVID-19 pandemic caused by SARS-CoV-2 (severe acute respiratory syndrome coronavirus 2) infection has led to 505 million confirmed cases and over 6.2 million deaths worldwide (World Health Organization, 2022). A range of prophylactic and therapeutic agents against COVID-19 have been developed at unprecedented speed to combat this deadly pandemic, including vaccines (Food and Drug Administration, 2021a, 2022), monoclonal antibodies (Food and Drug Administration, 2020a, b), and small molecule antiviral drugs including remdesivir for IV infusion (Food and Drug Administration, 2020c) and more recently oral compounds nirmatrelvir and molnupiravir (Food and Drug Administration, 2021b, c). However, the newly emerging SARS-CoV-2 variants such as Omicron are showing a potential to escape from vaccine and monoclonal antibody protection, highlighting the need for additional prophylactic and therapeutic options. Therefore, additional research into SARS-CoV-2 virology and different antiviral strategies are needed to prepare for the evolving nature of this pandemic.
Cell culture virus replication systems are essential for the study of virus replication and pathogenesis, antiviral drug testing, and viral research. SARS-CoV-2, a beta-coronavirus, enters host cells via the angiotensin-converting enzyme 2 (ACE2) receptor, but also requires the transmembrane serine proteinase 2 (TMPRSS2) activity to prime the virus Spike protein (Hoffmann et al., 2020). The most physiologically relevant primary human airway epithelial cells support the SARS-CoV-2 replication cycle but cannot proliferate indefinitely and vary in infectability between different donors. Several other cell lines also support SARS-CoV-2 replication, and have been used in SARS-CoV-2 infection studies. These include Vero cells, HEK293, Calu-3, Caco2, Huh-7, and A549 cells with stable expression of exogenously introduced ACE2 (Takayama, 2020). Vero cells, derived from African Green monkey kidney cells (Yasumura and Kawakita, 1963), are susceptible to SARS-CoV-2 infection, as well as SARS-CoV-1 and MERS viruses (Chu et al., 2020; Lau et al., 2018). Vero cells, along with their various derivatives such as VeroE6 and VeroE6/TMPRSS2 cells, are easy to maintain, offer high levels of infectivity and support efficient SARS-CoV-2 replication and virus production. As a result, they are the most widely used cell culture for SARS-CoV-2 replication, antiviral screening, and virology studies (Kumar et al., 2021).
Despite the multiple benefits, Vero cells express high levels of multi-drug resistance protein 1 (MDR1), also named Pgp (P-glycoprotein) or ATP-binding cassette sub-family B member 1 (ABCB1) (De Rosa et al., 2004; Gottesman and Ling, 2006). Pgp is an ATP-dependent efflux pump with a broad substrate specificity (Gottesman and Ling, 2006). Because of this, it is necessary to include a Pgp inhibitor in the cell culture to prevent tested compounds from being exported out of cells (Boras et al., 2021; He et al., 2021; Owen et al., 2021). Consequently, the observed antiviral activities could be affected by co-administered Pgp inhibitors, resulting in a synergistic or antagonistic effect of a Pgp inhibitor or an added toxicity effect with the test compound. The co-administration of a Pgp inhibitor would also limit the use of the otherwise highly permissive VeroE6 or VeroE6/TMPRSS2 cells in studies such as virus drug resistance selections. We, therefore, attempted to abolish the Pgp expression in VeroE6 cells by introducing a genomic mutation using the CRISPR-CAS9 technology. The resulting VeroE6 cells with the Pgp gene knockout were characterized and used in antiviral studies. The antiviral activities of nirmatrelvir and remdesivir against SARS-CoV-2 in this newly established cell culture are evaluated.
2. Materials and Methods
2.1. Virus and cells
The SARS-CoV-2 USA-WA1/2020 strain (Cat. No. NR-52281) was purchased from BEI Resources (Manassas, VA), propagated, and titrated, and the genome sequence was confirmed by the NGS analysis as described previously (Rai et al., 2022). VeroE6 cells were obtained from Pfizer's in-house cell culture repository. VeroE6-Pgp-KO cells were generated by targeting exon 4 of Pgp gene (ENSCSAG00000012391.1) using the CRISPR-CAS9 method through a fee-for-service contract by Synthego, Inc. (Menlo Park, CA). Both VeroE6 and VeroE6-Pgp-KO cells were maintained in a complete growth medium (DMEM supplemented with 1% antibiotic – antimycotic and 10% FBS). NIH-MDCKI-MDR1 and Borst-MDCKII-MDR1 cell lines were each obtained from the National Institutes of Health (Bethesda, MD, USA) and Dr. Piet Borst (Netherlands Cancer Institute, Amsterdam, Netherlands), respectively.
2.2. Cell staining and flow cytometry analysis
Vero E6 and VeroE6-Pgp-KO cells grown to approximately 90–100% confluency were each trypsinized, seeded into a 96-well V-bottom plate at 400,000 cells/well, pelleted, and washed with 1X DPBS. Live/dead cell staining with Aqua 405 fixable viability stain (Invitrogen, Carlsbad, CA) was followed by fixation and permeabilization using the BD Bioscience's Cytofix/Cytoperm (fix/perm) Kit (BD, Franklin Lakes, NJ). Pgp staining was conducted or performed with an anti-Pgp monoclonal primary antibody (Invitrogen, Carlsbad, CA) diluted to 400 ng/mL followed by an anti-mouse IgG antibody conjugated to FITC (Invitrogen, Carlsbad, CA) secondary antibody solution diluted to 1.6 μg/mL.
Cellular Pgp data collection and quantitation were carried out using a BD Biosciences Fortessa X-20 flow cytometer. Aqua 405 viability stain was excited with a 405 nm laser and detected in a violet laser channel with a 450/50 bandpass filter. FITC was excited with a 488 nm laser and emission was detected in a blue laser channel with a 505LP dichroic filter and 530/30 bandpass filter. Single color controls were run for both the Aqua 405 and FITC fluorophores, and compensation was calculated and applied. For each sample, 25,000 events were recorded and gating was performed to eliminate dead cells and debris. Doublet discrimination was performed in both the forward and side scatter directions. Live single cells were then displayed on a FITC histogram and positive and negative population percentages and mean fluorescence intensity (MFI) were measured for each sample. Raw data FCS files were exported and analyzed using the FlowJo 10.7.1 software to generate histograms, and MFI values were analyzed with GraphPad Prism 9.0.0 to generate bar charts comparing cellular PgP levels.
2.3. Calcein AM (CAM) based Pgp activity assay
Calcein AM (CAM) is a cell permeable and non-fluorescent Pgp substrate that is hydrolyzed by intracellular esterases to form a poorly permeable, fluorescent compound, calcein. VeroE6 and pooled VeroE6-Pgp-KO cells were plated in 96-well poly-D-lysine coated plates at the density of 3.7 × 104 cells/well for 48 h. To evaluate the Pgp activity, wild-type and KO cells were pre-incubated for 30 min at 37 °C in Hanks Balanced Salt Solution (HBSS), pH 7.4, in the presence of a Pgp inhibitor, PSC833 (10 μM), or a DMSO control. Cells were incubated with 0.5 μM CAM and PSC833 or DMSO for 30 min at 37 °C and washed with ice-cold HBSS buffer four times. Intracellular calcein levels were quantified using a Spectramax Gemini EM (San Jose, CA, USA) at excitation and emission wavelengths of 486 and 535 nm, respectively, and expressed as background-subtracted relative fluorescent units (RFU) (i.e., relative fluorescence in CAM-treated cells – cells only). Statistical significance was assessed using a two-way analysis of variance followed by Sidak's multiple comparison test. Two well-established Pgp expressing cell lines, NIH-MDCKI-MDR1 and Borst-MDCKII-MDR1, were also evaluated as additional assay controls. Uptake ratios were calculated as the RFU ratio between PSC833-treated:DMSO-treated cells, and statistical significance was assessed using a one-way analysis of variance followed by Dunnett's multiple comparison test.
2.4. Antiviral activity assay
The antiviral assay with a readout of percent inhibition of virus RNA levels was described previously (Rai et al., 2022). Briefly, two-fold, ten-point serial dilutions of remdesivir and PF-07321322 were each prepared in virus growth medium (VGM) containing DMEM, 2% FBS, 10 mM HEPES, 1X antimycotic-antibiotic, and 2% DMSO or in VGM containing 8 μM Pgp inhibitor drug CP-100356. Next, 20,000 cell/well were mixed at 1:1 ratio with the remdesivir or PF-07321332 dilutions. Finally, the cell-drug mixture was mixed at a 1:1 (V/V) ratio with SARS-CoV-2 USA-WA1 strain to get 0.041 multiplicity of infection (MOI), as well as 2 μM CP-100356 final concentration for test conditions including a Pgp inhibitor. Final starting concentration of remdesivir was 2 μM in Vero E6 and 0.25 μM in Vero E6-Pgp-KO cells. Nirmatrelvir starting concentration was 2 μM in Vero E6 and 1.25 μM in Vero E6-Pgp-KO cells. After a 48-h incubation at 37 °C, an in-plate cell lysis and a viral genome copy number were quantified by qRT-PCR of the NSP10 region using the same primer/probe, PCR conditions, and RNA standards described in detail before (Rai et al., 2022). The percent inhibition of virus replication (genome copy number) was calculated against a no-drug virus-infected cell control. EC50 and EC90 values were calculated in GraphPad Prism using the log(inhibitor) vs. response—Variable slope (four parameters) and [Agonist] vs. response-Find EC anything parameters, respectively, with Hill Slope set to “must be less than 3.”
3. Results
3.1. Generation of VeroE6-Pgp-KO cell line
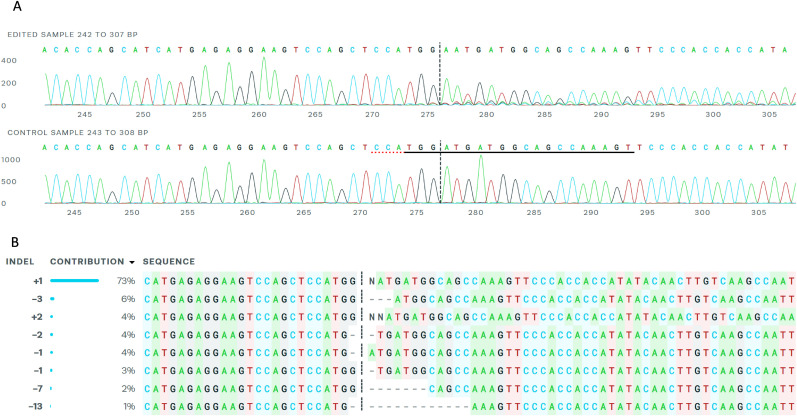
The VeroE6 cells were a clone of Vero cells originally derived from African green monkey kidney cells (Yasumura and Kawakita, 1963). Vero cells express high levels of MDR1 or Pgp activities (De Rosa et al., 2004), making it necessary to include an efflux inhibitor in the cell culture antiviral testing to prevent the export of compounds from inside the cells (Boras et al., 2021; He et al., 2021; Owen et al., 2021). To generate the Pgp gene knockout in VeroE6 cells, the CRISPR-CAS9 technology was employed to introduce mutations through non-homologous end joining into exon 4 of the MDR1 gene as described in Materials and Methods. The resulting mutant VeroE6 Pgp gene knockout cell population contained 97% cells with indel mutations in the Pgp gene exon 4 (Fig. 1 A). About 73% contained a +1 indel insertion at the CRISPR cut site in exon 4 (Fig. 1B), causing a frame shift mutation. Cell cloning of the mutant population led to the identification of 2 cell clones, clone B3 containing a +1 indel insertion and clone D1 containing a −2/+1 mutation in Pgp gene exon 4. Clone B3 was selected for further characterization using the FACS analysis and antiviral assay and herein was referred to as VeroE6-Pgp-KO cell line. Both generated VeroE6-Pgp-KO cell clones replicated similar to the parental VeroE6 cells and no differences in cell culture properties were detected, eg, passage number, cell size, etc (data not shown).
Fig. 1.
Genomic sequences of Pgp gene exon 4 region of VeroE6-Pgp-KO cells. A, Sequences around the CRISPR edited Pgp exon 4 site. Upper panel indicates VeroE6-Pgp-KO genomic sequence, while lower panel VeroE6 parent sequence. B, Frequency of CRISPR induced indel mutations in Pgp gene exon 4 of VeroE6-Pgp-KO cell population.
3.2. Lack of Pgp protein expression in VeroE6-Pgp-KO cell line
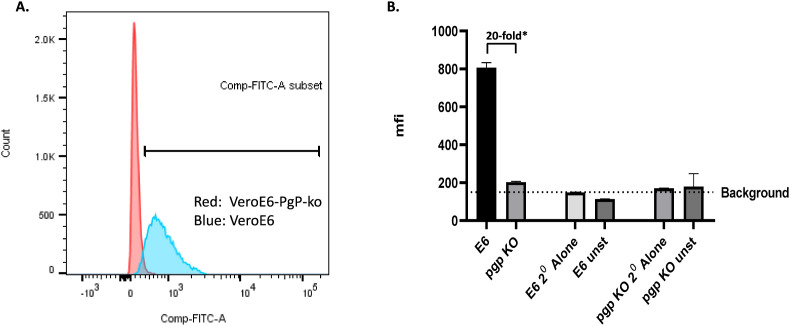
To evaluate whether the introduced frame shift mutation in VeroE6-Pgp-KO cells abrogated the Pgp protein expression, staining with an anti-Pgp monoclonal antibody followed by a secondary antibody conjugated to FITC was performed to detect the total cellular Pgp protein expression levels. VeroE6 was analyzed in parallel as a control. FACS analysis was used to count cells at different MFI levels from a total of about 15,000 cells, as described in Materials and Methods. VeroE6-Pgp-KO cells peaked around the background MFI (∼200) levels (Fig. 2 A, red peak, and Fig. 2B), while that of VeroE6 had a wide range of MFI levels with a mean of 800 MFI (Fig. 2A, blue peak, and Fig. 2B), representing about 20-fold signal/noise ratio after subtracting the background MFI. VeroE6 and VeroE6-Pgp-KO cells stained with a non-specific secondary antibody alone served as background (Fig. 2B). These results indicated that the VeroE6-Pgp-KO cell population lacked an antibody-detectible Pgp expression as shown by the red narrow peak (Fig. 1A), and that there was a range of Pgp expression levels among VeroE6 cells as shown by the wide blue peak (Fig. 1A).
Fig. 2.
Detection of Pgp protein expression in VeroE6 and VeroE6-Pgp-ko Cells by FACS Analysis. A. Cell count and overlay chromatogram depicting Pgp expression in VeroE6 and VeroE6-Pgp-ko cells detected using FITC. Vero E6 cells are represented by the blue peak and VeroE6-PgP-ko cells are represented by the red peak. A right shift in the peak can be observed in the VeroE6 (blue) cells, representing detection of Pgp. B. Mean Fluorescence Intensity (MFI) of VeroE6 (E6) and VeroE6-Pgp-KO (Pgp KO) cells. Cells fixed and permeabilized, but unstained or stained only with secondary antibody served as background controls. Background Mean Fluorescence Intensity (MFI) was calculated by taking a geometric mean of the MFI of the above controls. *After background subtraction, the MFI of VeroE6 was greater than 20-fold that of VeroE6-Pgp-KO cells.
3.3. Reduction of Pgp activity in VeroE6-Pgp-KO cells in CAM uptake assay
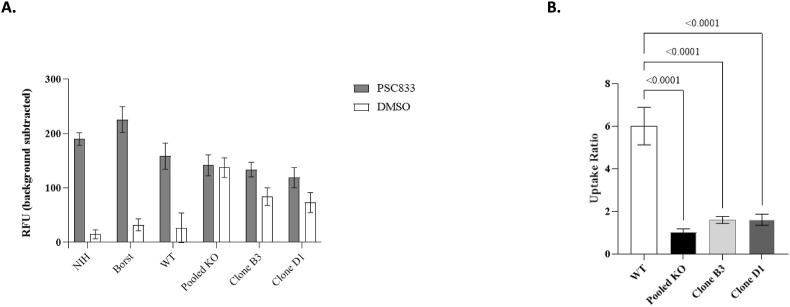
To evaluate if the Pgp gene knockout led to a change in the Pgp function, the efflux activity in the parental VeroE6 cells and VeroE6-Pgp-KO cells was assessed using the CAM assay (Glavinas et al., 2011; Rautio et al., 2006). Assay controls, NIH-MDCKI-MDR1 and Borst-MDCKII-MDR1 cells, that express Pgp were also tested in parallel. In the presence of 10 μM Pgp inhibitor PSC833, background-subtracted calcein levels were approximately 6-fold higher than in the DMSO control cells for the parental VeroE6 cells (Fig. 3 A), similar to that observed with NIH and Borst cells. On the contrary, calcein levels in the pooled VeroE6-Pgp-KO cells were similar in the DMSO vs PSC833 treated groups, suggesting minimal Pgp-mediated efflux activity (Fig. 3A). The uptake ratio (ratio between PSC833:DMSO) was calculated for all cell lines (Fig. 3B). The uptake ratio decreased to 1.6-fold in VeroE6-Pgp-KO cell clones B3 and D1 compared to 5.7-fold in the parental VeroE6 cells. The uptake ratios in pooled cells as well as B3 and D1 VeroE6-Pgp-KO cells were <1.6 fold and were significantly different (P < 0.001) from that of the 5.7-fold in wild-type VeroE6 (Fig. 3B). These results are consistent with a lack of Pgp efflux activity in VeroE6-Pgp-KO cells.
Fig. 3.
Calcein AM assay in different cells in the presence and absence of a Pgp inhibitor. A. Calcein AM uptake in DMSO or PSC833 treated cells. Calcein AM uptake across cell lines in DMSO treated (openbar) and PSC833 inhibited cells (grey bar) expressed as background-subtracted relative fluorescent units (RFU). Data represent mean ± SD (N ≥ 6). B. Calcein AM uptake ratios in WT (VeroE6) and KO (VeroE6-Pgp-KO) cell lines. Uptake ratios were calculated in WT (open bar), pooled KO (black), KO Clone B3 (light grey), and KO Clone D1 (dark grey) by determining the ratio of calcein levels in PSC833-treated cells to DMSO treated cells. Data represent mean uptake ratios ± SD (N ≥ 12).
3.4. Antiviral activities of remdesivir and nirmatrelvir in VeroE6-Pgp-KO cell line
To evaluate the effect of the Pgp gene knockout in VeroE6-Pgp-KO cells on observed antiviral activities, VeroE6-Pgp-KO cells were infected with SARS-CoV-2 WA-1 strain and the antiviral activities of nirmatrelvir and remdesivir were each assessed as described in Materials and Methods. Nirmatrelvir is a SARS-CoV-2 main protease (Mpro) inhibitor recently authorized by the FDA under an EUA (PAXLOVID™) as an oral treatment for COVID-19 (Food and Drug Administration, 2021c), while remdesivir is a SARS-CoV-2 RNA dependent RNA polymerase (RdRp) inhibitor approved for COVID-19 (VEKLURY™) as an IV infusion treatment (Food and Drug Administration, 2020c). The compounds were each tested in the presence or absence of 2 μM CP-100356, a well characterized Pgp inhibitor, which at 2 μM had a strong inhibition of Pgp but showed no antiviral activity against SARS-CoV-2 (Boras et al., 2021). The parental VeroE6 cells were infected and tested as controls under the same conditions with or without 2 μM Pgp inhibitor CP-100356.
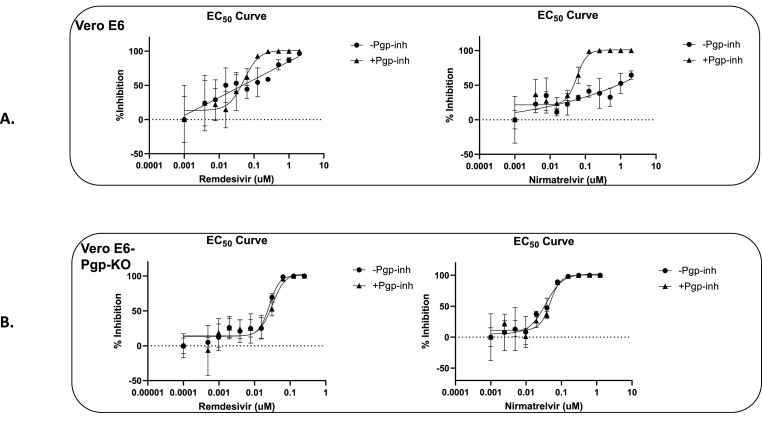
As summarized in Table 1 , in the parental VeroE6 cells, remdesivir inhibited SARS-CoV-2 with an average EC50 of 60 nM in the presence of 2 μM CP-100356, but with reduced and variable efficacies in the absence of Pgp inhibitor, as indicated also by the right shift of dose response curve in Fig. 4 A. Such a change in remdesivir potency was not observed in VeroE6-Pgp-KO cells, as shown by the EC50 values (Table 1) and a lack of shift in dose response curves (Fig. 4B). Similar phenomena were observed for nirmatrelvir's anti-SARS-CoV-2 activities with and without 2 μM CP-100356, in VeroE6 vs VeroE6-Pgp-KO cells (Fig. 4A and B). In VeroE6 cells, nirmatrelvir showed an EC50 of 45 nM in the presence of 2 μM CP-100356 (Table 1), but reduced and variable efficacies in the absence of the Pgp inhibitor (Fig. 4A). On the contrary, in VeroE6-Pgp-KO cells, nirmatrelvir's EC50 values in the presence or absence of 2 μM Pgp inhibitor CP-100356 were comparable and within the assay ranges (Table 1). Together, these results indicated that in VeroE6-Pgp-KO cells, the two evaluated drugs (Mpro and RdRp inhibitors) displayed antiviral activities as they would in the presence of a strong inhibitor of Pgp, 2 μM CP-100356, thereby negating the requirement for inclusion of a Pgp inhibitor in antiviral drug tests when using these cells.
Table 1.
Remdesivir and nirmatrelvir anti-SARS-CoV-2 activities in VeroE6 and VeroE6-Pgp-KO cells.
| Cell Line | Remdesivir |
Nirmatrelvir |
||||||
|---|---|---|---|---|---|---|---|---|
| EC50 (nM) Average (2 individual values)a |
EC90 (nM) Average (2 individual values) |
EC50 (nM) Average (2 individual values) |
EC90 (nM) Average (2 individual values) |
|||||
| -Pgp-inh | +Pgp-inh | -Pgp-inh | +Pgp-inh | -Pgp-inh | +Pgp-inh | -Pgp-inh | +Pgp-inh | |
| VeroE6 | Variable | 60 (50, 70) | Variable | 160 (150, 170) | Variable | 45 (30, 60) | Variable | 85 (50, 120) |
| VeroE6-Pgp-KO | 75 (30, 120) | 115 (30, 200) | 150 (50, 250) | 240 (80, 400) | 35 (30, 40) | 70 (20, 120) | 95 (50, 140) | 185 (100, 270) |
Individual EC50 and EC90 values were from independent experiments.
Fig. 4.
Effect of Pgp inhibitor (CP-100356) on in vitro anti-SARS-CoV-2 efficacy of nirmatrelvir and remdesivir in VeroE6 and VeroE6-Pgp-KO cells. The antiviral assays were carried out as described in materials & methods, in a 96 well plate with quadruplicate wells at each tested drug concentrations. Two days after infection and drug treatment, virus RNA were extracted and quantified using qPCR, and normalized to and subtracted from that of no drug infection control (% inhibition). The averages of the quadruplicate wells were plotted against log10 of drug concentration. Fifty percent effective concentration, EC50 was determined for remdesivir and Nirmatrelvir using four-parameter IC50 equation using GraphPad Prism 9. Circle symbols represent the % virus inhibition in the absence of Pgp inhibitor drug (-Pgp-inh) and triangle symbols represent % virus inhibition in the presence of 2 μM Pgp inhibitor drug (+Pgp-inh). A. Representative dose response curves of remdesivir and nirmatrelvir in VeroE6 cells, with and without 2 μM Pgp inhibitor CP-100356. B. Representative dose response curves of remdesivir and nirmatrelvir in VeroE6-Pgp-KO cells.
4. Discussion
African green monkey-derived Vero cells are susceptible to a range of different viruses, including simian polyoma virus SV-40, measles virus, rubella virus, arboviruses, adenoviruses, influenza A and B viruses, Ebola virus, and several human beta-coronaviruses including SARS-CoV-1, MERS and SARS-CoV-2 (Govorkova et al., 1996; Osada et al., 2014). Owing to their ease of propagation and maintenance, and the lack of tumorigenicity, Vero cells have been widely used for studying virus replication, antiviral drug testing, and virus vaccine production (Montomoli et al., 2012; Osada et al., 2014). In the campaign for discovering new drug or drug repurposing to battle the ongoing COVID-19 pandemic, Vero cells (e.g. Vero76, VeroE6, VeroE6/TMPRSS2, VeroE6/TMPRSS2/ACE2) are the most widely used in vitro cell culture system for SARS-CoV-2 infection and antiviral assays (Kumar et al., 2021). However, Vero cell usage in antiviral drug screening and testing is affected and confounded by their expression of Pgp or MDR1 gene, the major export transporter that pumps out foreign substance, necessitating the inclusion of Pgp inhibitors to prevent the export of compounds from host cells (Boras et al., 2021; He et al., 2021; Owen et al., 2021). On the other hand, the addition of a Pgp inhibitor would introduce an uncertain variable in the interpretation of antiviral results and cytotoxicity data, as well as limit the use of Vero cells in drug resistance and drug combination studies. We, therefore, created a Vero cell line with the Pgp gene knocked out and used it for an antiviral activity assessment in the current study.
Employing the CRISPR-CAS9 methodology, VeroE6-Pgp-KO cell with the Pgp gene knock out was generated. FACS analysis using a monoclonal antibody against Pgp indicated that the fluorescence signals from the Pgp KO cells were uniform and at the background levels (Fig. 2A), indicating a lack of Pgp expression in these cells. Interestingly, the parental VeroE6 cells expressed Pgp over a range of levels (Fig. 2A). Such an observation could explain the wide variation in some of the antiviral activities in Vero cells, for example the EC50 values of remdesivir varied between 0.77 and 26.9 μM in VeroE6 cells as shown in an RT-PCR based assay (Simonis et al., 2021; Wang et al., 2020), further emphasizing the importance of inhibition/elimination of Pgp efflux activity in antiviral tests to yield consistent and comparable antiviral activity readouts across different groups.
The abrogation of Pgp expression in VeroE6-Pgp-KO cells was reflected by a lack of Pgp-mediated transporter activity demonstrated in the CAM assay. In the presence vs absence of 10 μM Pgp inhibitor PSC833, calcein uptake ratio was about the same for the pooled VeroE6-Pgp-KO cells and was <1.6-fold for the two cell clones, as opposed to a 5.7-fold difference in the parental VeroE6 cells. The residual transport activity in the B3 and D1 VeroE6-Pgp-KO clones could be due to upregulation of other calcein efflux transporters (Legrand et al., 1998). The phenotypic functional difference in Pgp activity between VeroE6-Pgp-KO and VeroE6 cells was then assessed in the antiviral activities of two anti-SARS-CoV-2 compounds, nirmatrelvir and remdesivir, that are known substrates of Pgp (Boras et al., 2021; Owen et al., 2021). As expected, the Pgp gene knockout rendered the consistency of the antiviral activities of nirmatrelvir and remdesivir each in VeroE6-Pgp-KO cells with and without the addition of Pgp inhibitor CP-100356, which otherwise would have required the inhibition of Pgp function as in the parental VeroE6 cells. These data also demonstrated the potential for use of the VeroE6-Pgp-KO cells in SARS-CoV-2 antiviral testing without the need to control Pgp activity through addition of a Pgp inhibitor.
Because Pgp is the major efflux transporter of foreign substances and is highly expressed in Vero cell lines, and Vero cells are widely used in SARS-CoV-2 new drug screening and drug repurpose tests in which most of the time the effect of Pgp activity was not accounted for (Kumar et al., 2021), the reported antiviral activities could have been dramatically affected. The varying Pgp expression in Vero cells could also have been the cause for the vast variation of antiviral activity data of the same compounds reported by different groups (Simonis et al., 2021). The usage of VeroE6-Pgp-KO cells could eliminate these confounding factors and variations while retaining the high infectibility and ease of use of Vero cells. Because of the wide range permissibility of Vero cells to many other viruses, e.g. SARS-CoV-1, MERS, H5N1 influenza virus, and Ebola virus, the established VeroE6-Pgp-KO cell line also has a potential to be used for antiviral drug screening and tests against other viruses for future virus epidemics.
Funding
The study was supported by Pfizer Inc.
Disclaimer
The content of this publication reflects the views of the authors and neither the IMI nor the European Union, EFPIA or any other partners are liable for any use that may be made of the information it contains.
Declaration of competing interest
The authors declare the following financial interests/personal relationships which may be considered as potential competing interests: All authors are employees of Pfizer, Inc.
Acknowledgments
The CARE project has received funding from the Innovative Medicines Initiative 2 Joint Undertaking (JU) under grant agreement No 101005077. The JU receives support from the European Union's Horizon 2020 research and innovation programme and EFPIA and BILL & MELINDA GATES FOUNDATION, GLOBAL HEALTH DRUG DISCOVERY INSTITUTE, UNIVERSITY OF DUNDEE.
The authors thank Nataliya Kushnir (Pfizer Inc) for the editorial assistance.
Data availability
Data will be made available on request.
References
- Boras B., Jones R.M., Anson B.J., Arenson D., Aschenbrenner L., Bakowski M.A., Beutler N., Binder J., Chen E., Eng H., Hammond H., Hammond J., Haupt R.E., Hoffman R., Kadar E.P., Kania R., Kimoto E., Kirkpatrick M.G., Lanyon L., Lendy E.K., Lillis J.R., Logue J., Luthra S.A., Ma C., Mason S.W., McGrath M.E., Noell S., Obach R.S., Mn O.B., O'Connor R., Ogilvie K., Owen D., Pettersson M., Reese M.R., Rogers T.F., Rosales R., Rossulek M.I., Sathish J.G., Shirai N., Steppan C., Ticehurst M., Updyke L.W., Weston S., Zhu Y., White K.M., Garcia-Sastre A., Wang J., Chatterjee A.K., Mesecar A.D., Frieman M.B., Anderson A.S., Allerton C. Preclinical characterization of an intravenous coronavirus 3CL protease inhibitor for the potential treatment of COVID19. Nat. Commun. 2021;12:6055. doi: 10.1038/s41467-021-26239-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chu H., Chan J.F., Yuen T.T., Shuai H., Yuan S., Wang Y., Hu B., Yip C.C., Tsang J.O., Huang X., Chai Y., Yang D., Hou Y., Chik K.K., Zhang X., Fung A.Y., Tsoi H.W., Cai J.P., Chan W.M., Ip J.D., Chu A.W., Zhou J., Lung D.C., Kok K.H., To K.K., Tsang O.T., Chan K.H., Yuen K.Y. Comparative tropism, replication kinetics, and cell damage profiling of SARS-CoV-2 and SARS-CoV with implications for clinical manifestations, transmissibility, and laboratory studies of COVID-19: an observational study. Lancet Microbe. 2020;1:e14–e23. doi: 10.1016/S2666-5247(20)30004-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- De Rosa M.F., Sillence D., Ackerley C., Lingwood C. Role of multiple drug resistance protein 1 in neutral but not acidic glycosphingolipid biosynthesis. J. Biol. Chem. 2004;279:7867–7876. doi: 10.1074/jbc.M305645200. [DOI] [PubMed] [Google Scholar]
- Food and Drug Administration Coronavirus (COVID-19) update: FDA authorizes monoclonal antibodies for treatment of COVID-19. 2020. https://www.fda.gov/news-events/press-announcements/coronavirus-covid-19-update-fda-authorizes-monoclonal-antibodies-treatment-covid-19 Available from:
- Food and Drug Administration Coronavirus (COVID-19) update: FDA authorizes monoclonal antibody for treatment of COVID-19. 2020. https://www.fda.gov/news-events/press-announcements/coronavirus-covid-19-update-fda-authorizes-monoclonal-antibody-treatment-covid-19 Available from:
- Food and Drug Administration FDA approves first treatment for COVID-19. News release. 2020. https://www.fda.gov/news-events/press-announcements/fda-approves-first-treatment-covid-19 Available from:
- Food and Drug Administration COMIRNATY® (COVID-19 Vaccine, mRNA) suspension for injection, for intramuscular use. 2021. https://www.fda.gov/media/151707/download Available from:
- Food and Drug Administration Coronavirus (COVID-19) update: FDA authorizes additional oral antiviral for treatment of COVID-19 in certain adults. 2021. https://www.fda.gov/news-events/press-announcements/coronavirus-covid-19-update-fda-authorizes-additional-oral-antiviral-treatment-covid-19-certain Available at:
- Food and Drug Administration Coronavirus (COVID-19) update: FDA authorizes first oral antiviral for treatment of COVID-19. 2021. https://www.fda.gov/news-events/press-announcements/coronavirus-covid-19-update-fda-authorizes-first-oral-antiviral-treatment-covid-19 Available at:
- Food and Drug Administration Emergency use authorization (EUA) of the moderna COVID-19 vaccine to prevent coronavirus disease 2019 (COVID-19) 2022. https://www.fda.gov/media/144637/download Available from:
- Glavinas H., von Richter O., Vojnits K., Mehn D., Wilhelm I., Nagy T., Janossy J., Krizbai I., Couraud P., Krajcsi P. Calcein assay: a high-throughput method to assess P-gp inhibition. Xenobiotica. 2011;41:712–719. doi: 10.3109/00498254.2011.587033. [DOI] [PubMed] [Google Scholar]
- Gottesman M.M., Ling V. The molecular basis of multidrug resistance in cancer: the early years of P-glycoprotein research. FEBS Lett. 2006;580:998–1009. doi: 10.1016/j.febslet.2005.12.060. [DOI] [PubMed] [Google Scholar]
- Govorkova E.A., Murti G., Meignier B., de Taisne C., Webster R.G. African green monkey kidney (Vero) cells provide an alternative host cell system for influenza A and B viruses. J. Virol. 1996;70:5519–5524. doi: 10.1128/jvi.70.8.5519-5524.1996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- He X., Quan S., Xu M., Rodriguez S., Goh S.L., Wei J., Fridman A., Koeplinger K.A., Carroll S.S., Grobler J.A., Espeseth A.S., Olsen D.B., Hazuda D.J., Wang D. vol. 118. Proc Natl Acad Sci U S A; 2021. (Generation of SARS-CoV-2 Reporter Replicon for High-Throughput Antiviral Screening and Testing). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hoffmann M., Kleine-Weber H., Schroeder S., Kruger N., Herrler T., Erichsen S., Schiergens T.S., Herrler G., Wu N.H., Nitsche A., Muller M.A., Drosten C., Pohlmann S. SARS-CoV-2 cell entry depends on ACE2 and TMPRSS2 and is blocked by a clinically proven protease inhibitor. Cell. 2020;181:271–280 e278. doi: 10.1016/j.cell.2020.02.052. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kumar S., Sarma P., Kaur H., Prajapat M., Bhattacharyya A., Avti P., Sehkhar N., Kaur H., Bansal S., Mahendiratta S., Mahalmani V.M., Singh H., Prakash A., Kuhad A., Medhi B. Clinically relevant cell culture models and their significance in isolation, pathogenesis, vaccine development, repurposing and screening of new drugs for SARS-CoV-2: a systematic review. Tissue Cell. 2021;70 doi: 10.1016/j.tice.2021.101497. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lau S.K.P., Fan R.Y.Y., Luk H.K.H., Zhu L., Fung J., Li K.S.M., Wong E.Y.M., Ahmed S.S., Chan J.F.W., Kok R.K.H., Chan K.H., Wernery U., Yuen K.Y., Woo P.C.Y. Replication of MERS and SARS coronaviruses in bat cells offers insights to their ancestral origins. Emerg. Microb. Infect. 2018;7:209. doi: 10.1038/s41426-018-0208-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Legrand O., Simonin G., Perrot J.Y., Zittoun R., Marie J.P. Pgp and MRP activities using calcein-AM are prognostic factors in adult acute myeloid leukemia patients. Blood. 1998;91:4480–4488. [PubMed] [Google Scholar]
- Montomoli E., Khadang B., Piccirella S., Trombetta C., Mennitto E., Manini I., Stanzani V., Lapini G. Cell culture-derived influenza vaccines from Vero cells: a new horizon for vaccine production. Expert Rev. Vaccines. 2012;11:587–594. doi: 10.1586/erv.12.24. [DOI] [PubMed] [Google Scholar]
- Osada N., Kohara A., Yamaji T., Hirayama N., Kasai F., Sekizuka T., Kuroda M., Hanada K. The genome landscape of the african green monkey kidney-derived vero cell line. DNA Res. 2014;21:673–683. doi: 10.1093/dnares/dsu029. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Owen D.R., Allerton C.M.N., Anderson A.S., Aschenbrenner L., Avery M., Berritt S., Boras B., Cardin R.D., Carlo A., Coffman K.J., Dantonio A., Di L., Eng H., Ferre R., Gajiwala K.S., Gibson S.A., Greasley S.E., Hurst B.L., Kadar E.P., Kalgutkar A.S., Lee J.C., Lee J., Liu W., Mason S.W., Noell S., Novak J.J., Obach R.S., Ogilvie K., Patel N.C., Pettersson M., Rai D.K., Reese M.R., Sammons M.F., Sathish J.G., Singh R.S.P., Steppan C.M., Stewart A.E., Tuttle J.B., Updyke L., Verhoest P.R., Wei L., Yang Q., Zhu Y. An oral SARS-CoV-2 M(pro) inhibitor clinical candidate for the treatment of COVID-19. Science. 2021;374:1586–1593. doi: 10.1126/science.abl4784. [DOI] [PubMed] [Google Scholar]
- Rai D.K., Yurgelonis I., McMonagle P., Rothan H.A., Hao L., Gribenko A., Titova E., Kreiswirth B., White K.M., Zhu Y., Anderson A.S., Cardin R.D. Nirmatrelvir, an orally active Mpro inhibitor, is a potent inhibitor of SARS-CoV-2 Variants of Concern. 2022. BioRxiv. Available from: [DOI]
- Rautio J., Humphreys J.E., Webster L.O., Balakrishnan A., Keogh J.P., Kunta J.R., Serabjit-Singh C.J., Polli J.W. In vitro p-glycoprotein inhibition assays for assessment of clinical drug interaction potential of new drug candidates: a recommendation for probe substrates. Drug Metab. Dispos. 2006;34:786–792. doi: 10.1124/dmd.105.008615. [DOI] [PubMed] [Google Scholar]
- Simonis A., Theobald S.J., Fatkenheuer G., Rybniker J., Malin J.J. A comparative analysis of remdesivir and other repurposed antivirals against SARS-CoV-2. EMBO Mol. Med. 2021;13 doi: 10.15252/emmm.202013105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Takayama K. In vitro and animal models for SARS-CoV-2 research. Trends Pharmacol. Sci. 2020;41:513–517. doi: 10.1016/j.tips.2020.05.005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang M., Cao R., Zhang L., Yang X., Liu J., Xu M., Shi Z., Hu Z., Zhong W., Xiao G. Remdesivir and chloroquine effectively inhibit the recently emerged novel coronavirus (2019-nCoV) in vitro. Cell Res. 2020;30:269–271. doi: 10.1038/s41422-020-0282-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- World Health Organization WHO COVID-19 dashboard. 2022. https://covid19.who.int/ Available from:
- Yasumura Y., Kawakita Y. Studies on SV40 in tissue culture: preliminary step for cancer research in vitro. Nihon Rinsho. 1963;21:1201–1215. (in Japanese) [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Data Availability Statement
Data will be made available on request.