Abstract
Oral squamous cell carcinoma (OSCC) patients suffer from poor survival due to metastasis or locoregional recurrence, processes that are both facilitated by perineural invasion (PNI). OSCC has higher rates of PNI than other cancer subtypes, with PNI present in 80% of tumors. Despite the impact of PNI on oral cancer prognosis and pain we know little about the genes that drive PNI, which in turn drive pain, invasion and metastasis. In this study we leverage clinical data, preclinical and in vitro models to elucidate the role of neurotrophins in OSCC metastasis, PNI, and pain. Our expression data in OSCC patients with metastasis, PNI, or pain demonstrate dysregulation of neurotrophin genes. We focus on TrkA and NGFR, two receptors that are activated by nerve growth factor (NGF), a neurotrophin expressed at high levels in OSCC. We demonstrate that targeted knockdown of these two receptors inhibits proliferation and invasion in an in vitro and preclinical model of OSCC, and metastasis, PNI and pain. We further determine that TrkA knockdown alone inhibits thermal hyperalgesia, whereas NGFR knockdown alone inhibits mechanical allodynia. Collectively our results highlight the ability of OSCC to co-opt different components of the neurotrophin pathway in metastasis, PNI, and pain.
Graphical Abstract

In this study, we demonstrate that the neurotrophin gene pathway is dysregulated in oral squamous cell carcinoma (OSCC) patients with metastasis, perineural invasion (PNI), and pain. shRNA-mediated knockdown of the neurotrophin receptors, TrkA and NGFR, inhibits OSCC proliferation in vitro, and metastasis, PNI and pain in a preclinical OSCC model.
INTRODUCTION
Oral squamous cell carcinoma (OSCC) patients suffer from poor survival due to metastasis or locoregional recurrence, processes that are both facilitated by perineural invasion (PNI). OSCC has higher rates of PNI than many other cancer subtypes, with PNI present up to 80% of tumors [1]. PNI also correlates with cancer pain [2] and reduces quality of life in OSCC patients, especially in patients who will not survive their disease [3]. Despite the impact of PNI on oral cancer prognosis and pain we know little about the mediators and receptors that drive PNI, which in turn drive cancer pain, invasion and metastasis. The few available studies show that neurotrophins mediate crosstalk between cancer cells and neurons [1]. Neurotrophins bind to their respective receptors on cancer cells or neurons to activate downstream pathways that facilitate neuronal invasion of the cancer cells and tropism of neurons toward cancer [1]. Previous studies indicate that nerve growth factor (NGF) and its receptor, tropomyosin kinase receptor kinase type A (TrkA), are elevated in OSCCs that invade nerves [4]. Our own studies show that NGF blockade with a monocolonal antibody inhibits OSCC proliferation and pain in a cancer mouse model [5]. Furthermore, brain derived neurotrophic factor (BDNF) and its receptor TrkB are implicated in OSCC progression and pain [6, 7]. These preliminary findings form the basis of our current study, where we propose that neurotrophin-related gene pathways are dysregulated and mediate PNI and pain in OSCC. Aside from NGF and BDNF, other neurotrophin genes have not been mechanistically studied to clarify their role in OSCC PNI, metastasis, and pain. Oral cancers are genomically and phenotypically heterogeneous; therefore, we hypothesize that different oral cancers co-opt different members of the neurotrophin gene pathway. In this study, we leverage OSCC patient tissues to identify the different neurotrophins associated with OSCC PNI, metastasis, and pain, and test cause-and-effect of a subset of these genes in in vitro and in vivo models of PNI, metastasis, and pain. The proposed work bridges a gap in our understanding of the molecular mechanisms underlying PNI, metastasis, and pain in oral cancer patients. We select 87 candidate neurotrophin pathway gene features that are highly dysregulated in the cancer tissue using the results from a gene expression array. We then compare the expression of these candidate genes in OSCC tissues with and without PNI, with and without metastasis, and with and without pain. From the findings in patients, we focus on TrkA and nerve growth factor receptor (NGFR) as two high-priority genes for in vitro and in vivo mechanistic studies to determine their roles in OSCC PNI, metastasis and cancer pain. We choose these two genes as they both code for receptors that bind NGF, which has a defined role in OSCC pain and progression [5], as well as other neurotrophins, with varying affinity.
RESULTS
Neurotrophin pathway genes are dysregulated in oral cancer patients with perineural invasion, metastasis, and pain
We performed a gene expression on 22 OSCC patients (age range 30–92 years, mean 67 years; Table 1). Fourteen of the 22 patients had complete clinical information and gene expression data that passed QC. Batch correction was performed using Surrogate Variable Analysis (Supplemental Figure 1). Heat maps were generated from the 87 features of the neurotrophin pathway (Supplemental Figure 2) with the OSCC patients separated by each of the three categories: 1) PNI (5 present, 8 absent), 2) neck metastasis (5 present, 9 absent), and 3) pain levels (6 severe pain, 5 low pain). The 87 genes were shown to be part of the neurotrophin pathway in the Kyoto Encyclopedia of Genes and Genomes (KEGG) pathway analysis that we performed. The top 10 differentially expressed genes were shown between the two subgroups of patients within each of the three categories, resulting in 11 dysregulated probes spanning 10 genes for PNI vs no PNI, 13 dysregulated probes spanning 10 genes for neck metastasis vs. no metastasis, and 11 dysregulated probes spanning 10 genes for severe pain vs. low pain (Figure 1).
Table 1.
Patient Demographics
| Beadchip ID | Sex | Tumor Location | TNM Classification | Neck Metatasis | PNI | Pain |
|---|---|---|---|---|---|---|
| 9.806E+09 C | F | mandibular gingiva | T4aN0M0 | N | N | severe |
| 9.806E+09 G | F | mandibular gingiva | T1N0M0 | N | N | low |
| 9.965E+09 B | M | tongue | T1N0M0 | N | N | low |
| 9.806E+09 I | F | tongue | T1N2bM0 | Y | N | severe |
| 9.806E+09 K | F | mandibular gingiva | T2N2bM0 | Y | N | severe |
| 9.806E+09 A | F | tongue | T1N0M0 | N | Y | low |
| 9.806E+09 C | F | tongue | T1N1M0 | Y | N | severe |
| 9.806E+09 E | F | floor of mouth | T2N0M0 | N | Y | severe |
| 9.806E+09 I | M | lip | T1N0M0 | N | Y | unknown |
| 9.806E+09 K | F | tongue | T1N0M0 | N | Y | unknown |
| 9.965E+09 H | F | mandibular gingiva | T1N1M0 | Y | N | low |
| 9.965E+09 J | F | tongue | T1N0M0 | N | N | low |
| 9.965E+09 L | M | tongue | T3N0M0 | N | Y | severe |
| 9.806E+09 A | M | maxillary gingiva | T4N2bM0 | Y | unknown | unknown |
Figure 1.
The heat maps depict the top most differentially expressed genes between patients with and without PNI, with and without neck metastasis, or with and without pain. Neurotrophin pathway genes are differentially expressed in patients with neck metastasis, PNI and pain. Patients are dichotomized by “0” vs “1” to denote “absent” and “present” for PNI and metastasis, and “low” vs “severe” for pain.
Functional network analysis
We performed gene network analysis using KEGG and Gene Ontology (GO) to determine additional gene pathways that converge with the neurotrophin pathway to mediate metastasis, PNI and pain. Differential gene expression was calculated based on either metastasis, PNI or pain dichotomy in the cohort using all differentially expressed genes. Figures 2A–C detail the KEGG pathways that are linked to the differential genes based on metastasis status. Genes were also aggregated by gene ontology category (i.e., biological process, cellular compartment, molecular function). The complex associations between differentially expressed genes mapped to multiple related differentially expressed pathways were visualized using a geneset enrichment dotplot and a gene-concept network plot. Multiple significant pathways that already have established roles in metastasis were identified as being inter-related to the neurotrophin genes (p<0.05; Figures 2A–C). Gene expression was then analyzed based on PNI status (Figures 2D–E). The genes underwent gene concept network of ORA Reactome gene sets, revealing cytokine signaling and integrin signaling as inter-related pathways, which both have established roles in cancer pain [8, 9], metastasis and cancer progression through mediating immune cells and cancer associated fibroblasts, epithelial meschenymal transition (EMT), and extracellular matrix (ECM) cross-linking [10–13]. Additional significant and related pathways included ECM-receptor interaction and PI3K-Akt signaling, which have also been extensively examined in metastasis of different cancers, including oral cancer [14]. Previous studies have shown that NGF uses the PI3K-Akt signaling pathway to enhance oral cancer cell migration and PNI [14]. Functional network analysis of differential genes based on pain levels in the cohort shows pathways like opioid signaling and NMDA receptors, which control multiple pain conditions. Additionally, cell-cell communication, cell junction organization, and extracellular matrix organization pathways are mediated by genes that control metastasis (Figure 2F). These results illustrate the network of gene pathways that crosstalk with the neurotrophin genes; when dichotomized based on PNI status alone, multiple gene pathways that also regulate metastasis and pain are significantly expressed in the analysis.
Figure 2.
A) Functional network analysis was performed on genes differentially expressed based on metastasis status. Dotplot of over-representation analysis (ORA) GO Biological Processes (BP) pathways reveals additional pathways that converge with the neurotrophin pathway to mediate metastasis in the cohort of patients. B) The differential genes when sorted by metastasis status underwent enrichment mapping with ORA GO BP pathways. These results relate neuroptrophin genes to multiple pathways that are critical in metastasis. C) Similarly, functional network analysis of differential genes by methylation status was performed with ORA KEGG pathways. D) Differentially expressed genes based on PNI status underwent gene concept network of ORA Reactome gene sets, revealing cytokine signaling and integrin signaling as inter-related pathways with the neurotrophin pathway, with both of these pathways having established roles in cancer pain and metastasis. E) Differential genes based on PNI status analyzed by KEGG also reveals additional gene pathways (i.e., ECM-receptor interaction, PI3K-Akt signaling pathway) that have established roles in metastasis of different cancers. F) Functional network analysis of differential genes based on pain levels in the cohort shows pathways like opioid signaling and NMDA receptors, which control multiple pain conditions. Additionally, cell-cell communication, cell junction organization, and extracellular matrix organization pathways are mediated by genes that control metastasis.
shRNA treatment effectively silences NGFR and TrkA expression in a mouse oral cancer model
The GFP tag was used as a proxy for shRNA expression in all treatment groups (Figure 3A and 3B). In vitro transduction efficiency was comparable amongst all treatment groups (scrambled, NGFR shRNA, TrkA shRNA and combination). Following inoculation of the transduced cancer cells to form ex vivo OSCC in the mouse model, we determined the efficacy of shRNA-induced silencing by quantifying NGFR and TrkA mRNA after tissue harvesting. RT-PCR to quantify NGFR mRNA demonstrated that groups treated with shRNA to NGFR (i.e., the shRNA NGFR group and combination group) had significant silencing of the gene (Figure 3C). Similarly, groups treated with shRNA to TrkA (i.e., the shRNA TrKA group and the combination group) had significant lower TrkA mRNA compared to control scrambled shRNA (Figure 3D).
Figure 3.
(A and B) Transduction efficiency is comparable amongst all four treatment groups. Using GFP as a proxy for gene expression, the percentage of positive cells is similar amongst all four treatment groups. (C) NGFR expression is significantly lower in oral SCC tumors treated with shRNA to NGFR (*p<.05, compared to scrambled shRNA group, One way ANOVA, Holm Sidak). (D) TrkA expression is significantly lower in oral SCC tumors treated with shRNA to TrkA (***p<.001, compared to scrambled shRNA group, One way ANOVA, Holm Sidak). (E) NGFR and TrkA silencing inhibits oral SCC proliferation in vitro. The graph shows MTS assay absorbance as an index of cell proliferation for each treatment group. Silencing of NGFR or TrkA alone or in combination has a significant antiproliferative effect on oral SCC (Hela-O3) cells in vitro (**p<.01, ***p<.001, One way ANOVA and Tukey’s test.) (F) NGFR and TrkA silencing inhibits oral SCC proliferation in vivo. Tumor volume is quantified after tissue harvest of the in vivo model for the four treatment groups: scrambled RNA (n=10), combination treatment (n=9), shRNA NGFR (n=10), and shRNA TrkA (n=7). Combination silencing of NGFR and TrkA produces the most robust decrease in OSCC (Hela-O3) tumor growth (***p<.001, One way ANOVA, Holm Sidak).
Combination NGFR and TrkA knockdown had an antiproliferative effect in vitro and in vivo
We evaluated the effect of NGFR and TrkA knockdown alone or as a combination treatment on cancer proliferation in vitro. An MTS assay was performed on Hela-O3 cells transduced with adenovirus carrying control scrambled RNA, shRNA NGFR, shRNA TrkA, or the combination of shRNA NGFR and TrkA. Absorbance at 450nm, which was quantified as an index of proliferation, was significantly lower in the treatment groups with either shRNA NGFR and shRNA TrkA alone, or in combination, compared to absorbance of the control vehicle group (Figure 3E).
Cancer proliferation was quantified in the mouse model by quantifying tongue tumor volume after tissue harvest (see Supplemental Figure 3 for photomicrographs of tongue tumors and lymph node metastases). The treatment groups included 1) scrambled RNA (n=10), 2) combination treatment (n=9), 3) shRNA NGFR (n=10), and 4) shRNA TrkA (n=7). Tongue tumor volume was not different between control treatment and shRNA NGFR or shRNA TrkA groups. However, the combination treatment group in which both NGFR and TrkA were silenced had significantly smaller tumor volume compared to control scrambled RNA, demonstrating that this combination of NGFR and TrkA knockdown had an in vivo anti-tumor effect (Figure 3F).
Combination NGFR and TrkA knockdown inhibited invasion in vitro
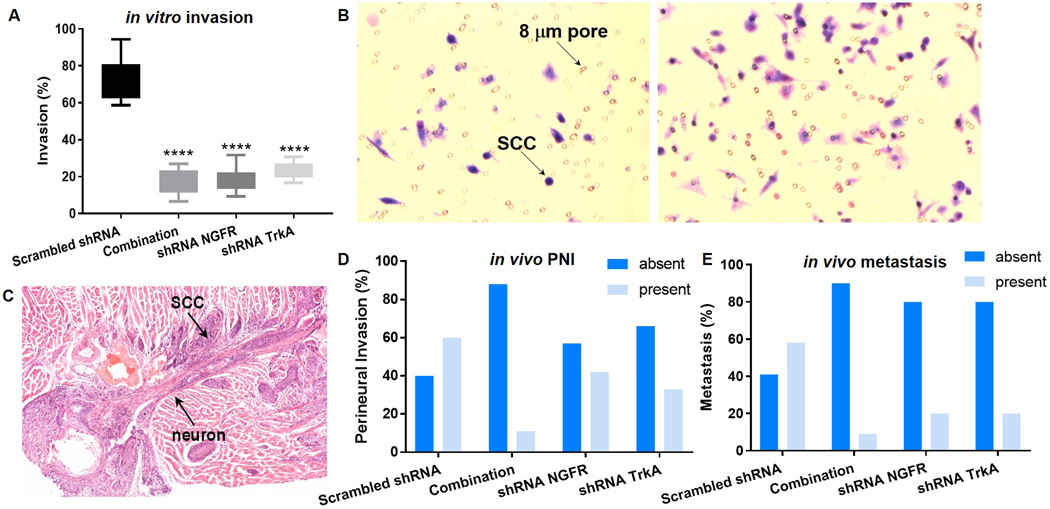
Using the invasion assay we determined whether NGFR and TrkA knockdown affected the ability of Hela-O3 migration toward 5% FBS chemoattractant. Figure 4A shows the percent invasion for each of the treatment group, with the values as follows for the treatment groups: 1) control scrambled shRNA 74.2%%, 2) combination shRNA NGFR and TrkA 17.8%, 3) shRNA NGFR alone 17.5%, and 4) shRNA TrkA alone 23.5%. All three treatment groups had significantly lower percent invasion than the control scrambled shRNA group (Figure 4B demonstrates a representative image of the invasion assay).
Figure 4.
(A) Box and whisker plot (showing minimum-maximum) of invasion assay results demonstrate inhibition of invasion with shRNA treatment to silence NGFR and TrkA. All three treatment groups have significantly lower percent invasion than control scrambled RNA (****p<.0001, One way ANOVA, Holm Sidak). (B) The two images show our co-culture model, where transwells are seeded with cancer cell, and the bottom well is loaded with serum media. SCC cells migrate through the pores at different frequency based on the treatment group (right greater than left). (C) Representative image of a mouse tongue SCC with PNI. (D) The contingency graph demonstrates the frequency of perineural invasion (PNI) in each treatment group in the preclinical model of oral SCC. The four treatment groups are significantly different from each other (p<0.0001, Chi square test, based on percentage of each group). (E) The contingency graph demonstrates the frequency of cervical metastasis in each treatment group in the preclinical model of oral SCC. The four treatment groups are significantly different from each other (p<.0001, Chi square test, based on percentage of each group).
Combination NGFR and TrkA knockdown resulted in reduced perineural invasion and cervical metastasis in a mouse model of oral cancer
The mouse tongue tumors were assessed for presence of PNI (Figure 4C). In each treatment group PNI was graded as negative (PNI absent) or positive (PNI present, or highly suspicious for PNI with extensive neurotropism and nerve entrapment). Percentage of PNI was significantly different between the four treatment groups (Table 2; Figure 4D).
Table 2.
Perineural invasion and metastasis rates
| in vivo treatment | perineural invasion | metastasis | ||
|---|---|---|---|---|
| absent | present | absent | present | |
| Scrambled shRNA | 40 | 60 | 42 | 58 |
| Combination | 88 | 11 | 92 | 9 |
| shRNA NGFR | 57 | 42 | 80 | 20 |
| shRNA TrkA | 66 | 33 | 80 | 20 |
Cervical metastasis was determined in the treatment groups after cervical lymph node harvest (Supplemental Figure 3). Percent metastasis is out lined in Table 2 and Figure 4E, with a statistically significant difference in metastasis rates among the four treatment groups.
NGFR and TrkA knockdown had an antinociceptive effect in a mouse oral cancer model
OSCC was established by inoculating transduced Hela-O3 cells in the tongue. Nociception to mechanical and thermal stimuli was determined in each of the treatment groups at baseline (day 0) and until PID 19. Nociceptive threshold change was calculated based on the pre-inoculation baseline values, demonstrated as a percent change, with a negative percent change signifying a decrease in threshold and therefore increase in nociception. Both mechanical and thermal nociceptive thresholds decreased with cancer inoculation (Figure 5), indicating that Hela-O3 cancers had a nociceptive effect. Gene knockdown was performed through adenovirus transduction at the time of cancer inoculation. TrkA knockdown significantly decreased thermal nociception, whereas NGFR knockdown alone significantly decreased mechanical nociception. Combined NGFR and TrkA knockdown in the tumor microenvironment produced a significant antinociceptive response to both mechanical and thermal stimuli (Figure 5; Table 3).
Figure 5.
The graphs demonstrate the change in (A) mechanical and (B) thermal threshold after cancer inoculation (day 0). Cancer cells are transduced with adenovirus expressing shRNA to the indicated genes. Combination knockdown of NGFR and TrkA produces antinociception to both mechanical and thermal stimuli, compared to the control scrambled shRNA group. NGFR knockdown alone produces significant mechanical antinociception, whereas TrkA knockdown alone produces significant thermal antinociception (*p<.05, **p<.01, ***p<.001, Two way ANOVA, Holm Sidak, see Table 3 for statistical analyses).
Table 3.
Statistical summary
| Two-way RM ANOVA | Holm-Sidak post hoc test | |||||
|---|---|---|---|---|---|---|
| Effects | DF | F | P | Groups | P | |
| Figure 5A | Treatment | 3 | 8.203 | 0.0003 | Combo v. Ctrl | <0.0001 |
| Time | 5 | 84.36 | <0.0001 | shRNA NGFR v. Ctrl | 0.0155 | |
| Interaction | 15 | 2.828 | 0.0006 | shRNA TrkA v. Ctrl | 0.084 | |
| Figure 5B | Treatment | 3 | 4.613 | 0.0079 | Combo v. Ctrl | 0.047 |
| Time | 5 | 199.8 | <0.0001 | shRNA NGFR v. Ctrl | 0.1087 | |
| Interaction | 15 | 4.309 | <0.0001 | shRNA TrkA v. Ctrl | 0.0025 | |
DISCUSSION
Perineural invasion and neck metastasis portend poor survival in oral cancer
Despite continued work toward targeted therapy for cancer patients, OSCC patients suffer from poor outcomes. Treatment failure is a consequence of invasion, metastasis, and recurrence. PNI drives local recurrence and metastasis, and is a worrisome pathologic feature that serves as an indication for post-operative adjuvant radiation due to its role in worsening prognosis [15–18]. The first report of PNI in 1862 is in oral cancer [19]. Since then, reports show that the incidence of PNI in OSCC is as high as 80% [1], and that PNI plays a critical role in treatment failure [20]. While the process of OSCC metastasis through lymphatic channels is well-characterized, we know little about the molecular mechanisms that facilitate PNI and cancer-nerve interactions. Previous work demonstrates that one of the mechanisms for cancer-nerve crosstalk is through the tumor suppressor gene TP53, with the loss of TP53 leading to adrenergic transdifferentiation of nerves and subsequent tumor growth [21]. The molecular relationship between PNI and metastasis has been explored, with NGF emerging as a molecule linking the two processes. High NGF expression is correlated with increased PNI and neck metastasis in a study of 101 OSCC patients. Furthermore, PNI was a significant independent predictor of neck metastasis and disease free survival [22]. In this study, we investigate the neurotrophin pathway in OSCC. Our recent publications and preliminary studies show that dysregulation of neurotrophin pathway genes in OSCC contributes to PNI and pain. Functional network analyses of our gene expression array data in OSCC patients reveal that the differentially expressed neurotrophin pathway genes have significant associations with multiple pathways involved in pain, cancer invasion and metastasis through mediating immune cell infiltration, cancer cell immune escape, EMT, and ECM cross-linking. While our gene expression results are derived from cancer tissue biopsies, which include heterogeneous cell types, making it difficult to draw conclusions about cell-cell interactions, specifically with regard to cancer cells, neurons, and immune cells, these dysregulated pathways present in the cancer microenvironment and otherwise absent in the normal microenvironment from matched normal biopsies, confirm the distinct role of neurotrophin genes in mediating the multitude of pathways.
The neurotrophin pathway mediates perineural invasion of different cancers
Neurotrophins are a family of proteins that are secreted by neuronal and cancer cells, and play a role in proliferation and differentiation. Studies in different cancers have shown that cancer cells secrete neurotrophins, which activate downstream pathways on neurons leading to PNI. Neurotrophins and their associated receptors mediate PNI and pain in pancreatic cancer, which has a high prevalence of PNI similar to OSCC [1]. Levels of NGF and TrkA in pancreatic cancer correlate with PNI [23]. BDNF mediates pancreatic cancer invasion and proliferation [24]. Similarly, PNI in adenoid cystic carcinoma, a rare salivary gland malignancy in which metastasis occurs through neuronal invasion, is mediated by NGF, BDNF, and the neurotrophin-3 receptor TrkC [20, 25]. In prostate cancer, NGF secretion induces angiogenesis and neurogenesis [26], which are linked to bone metastasis and pain [27].
The neurotrophin pathway in oral cancer perineural invasion and pain
In a clinical study of 21 PNI positive and 21 PNI negative OSCC samples, the expression of NGF and TrkA are significantly higher in PNI positive OSCC (Mann Whitney test, p=0.0001 for NGF and p=0.039 for TrkA) [4]. Our own publications have also demonstrated that OSCC cells secrete high levels of NGF relative to normal oral keratinocytes. NGF blockade with anti-NGF results in decreased OSCC proliferation as measured by in vitro assays and tumor growth in two oral cancer mouse models. Furthermore, anti-NGF decreases oral cancer-induced pain using two different nociceptive (i.e., pain) assays [5]. In one recent study using multiple oral cancer cell lines, NGF enhances oral cancer cell migration and potential PNI through a PI3K-Akt dependent manner [14]. Functional network analysis of differentially expressed genes in our patient cohort demonstrate that PI3K-Akt is one of the significant pathways that interacts with the neurotophin pathway. In addition to NGF, BDNF has been shown to play a pivotal role in oral cancer progression and pain. BDNF activation of its receptor TrkB regulates tumor growth, invasion, metastasis, and epithelial-to-mesenchymal transition (EMT) [28]. BDNF and TrkB overexpression are correlated with increased tumor invasion and worse survival in OSCC patients[6]. TrkB is critical in mediating cancer cell invasion and survival of cancer stem cells in mucoepidermoid carcinoma, another oral cancer subtype that is distinct from OSCC, but with similar PNI and metastasis risk in high grade cases [29]. BDNF-TrkB signaling also modulates interactions between oral SCC cells and Schwann cells, which support myelination of peripheral neurons, to induce de-differentiation of Schwann cells and intercalation of these cells between cancer cells, resulting in cancer cell dispersion [28]. These findings challenge the belief that PNI only results from cancer cell invasion into nerves. In addition to PNI, BDNF-TrkB signaling also promotes oral SCC pain [7, 30]. Oral SCC cells release BDNF, and TrkB is expressed in tumor-innervating lingual neurons [7, 30]. Pharmacologically blocking TrkB, or deleting TrkB in the trigeminal ganglion with siRNA administration reverses oral cancer pain [7, 30]. TrkB blockade also reverses tumor-induced mechanical sensitivity of putative thinly myelinated high threshold mechanoreceptors, captured using single-fiber tongue-nerve electrophysiology [7]. In contrast to the well-established roles of BDNF and TrkB in preclinical OSCC models, TrkA and NGFR, and their reciprocal interaction upon NGF activation, are not as well characterized. This study is a continuation of our previous work wherein we demonstrate that OSCC secretes high levels of NGF and that anti-NGF is a promising therapeutic approach to treat OSCC progression and pain [5].
Neurotrophin pathway mediation of different aspects of cancer pain: mechanical vs. thermal pain
Most of the research on the neurotrophin pathway to date has focused mainly on the role of NGF and BDNF and their respective high affinity receptors, TrkA and TrkB, in various pain conditions. Treatment strategies designed in preclinical models have included receptor blockade with antibodies or antagonists [31–33]. Due to previous studies and our own results demonstrating the important role of NGF in OSCC pain and PNI [4, 5], as well as our preliminary clinical data, this study focuses on the interplay between the high-affinity NGF receptor TrkA and low-affinity receptor NGFR. While TrkA has the highest affinity to NGF, the low affinity receptor NGFR not only binds neurotrophins [NGF, BDNF, neurotrophin (NT)-3 and NT-4/5], but also the precursor pro-neurotrophins (pro-NGF, pro-BDNF, pro-NT-3 and pro-NT-4/5). Rather than using antagonists or antibodies to block these two receptors, which only produce short-term effects, we perform gene silencing with shRNA. Previous studies in rats have shown that TrkA plays a predominant role in NGF-induced thermal hyperalgesia [34], whereas NGFR activation is essential for mechanical hypersensitivity [34]. Similarly, we demonstrate for the first time in a mouse OSCC model that TrkA gene silencing treats cancer-induced thermal nociception, whereas NGFR gene silencing treats cancer-induced mechanical nociception. Combined blockade of TrkA and NGFR produces a profound antinociceptive effect to both thermal and mechanical pain in our OSCC model. It is worth noting that while the tumor mass effect might theoretically contribute to nociceptive quantification in mouse models of oral cancer, we have demonstrated in both preclinical models and patients that tumor size does not correlate to nociception, that gene therapy of specific pain-mediating genes blocks nociception in mouse models without an effect on cancer growth, and that cancer pain is much more complex [35–39]. While the two receptors are both activated by NGF, which is found at high concentrations in the cancer microenvironment [5], their distinct roles in thermal and mechanical pain highlight the complexity of cancer pain and the need for targeted therapy. Studies in other models of pain have heterogeneous results with regard to the antinociceptive effect of TrkA or NGFR blockade. However, these differences might be explained by the fact that these studies use antagonists to block the receptors, whereas we use shRNA to silence receptor expression. For example, a mouse melanoma model shows no significant antinociceptive effect to thermal stimuli when the TrkA antagonist IPTRK3 is used [31]. On the other hand, in preclinical model of inflammatory pain, TrkA blockade with the antagonist IPTRK3 produces antinociception to both mechanical and thermal stimuli [40]. In a model of neuropathic pain, TrkA blockade by IPTRK3 suppresses both thermal hyperalgesia and mechanical allodynia [41]. TrkA-activation by NGF is associated with upregulation of transient receptor potential cation channel subfamily V member 1 (TRPV1), substance P, calcitonin gene-related peptide (CGRP), and the sodium channels (Nav1.8 and Nav1.9), all which are nociceptor proteins with defined roles in neuropathic pain as well as other pain conditions [32]. NGFR has also been shown to play an important role in mediating neuropathic pain [32]. NGFR activation is thought to induce neuropathic pain through NF-κB activation through the Jun pathway [33].
Conclusion
In this study, we elucidate the role of the neurotrophin pathway in OSCC progression and pain. We establish the clinical significance of targeted neurotrophin pathway blockade by demonstrating that candidate neurotrophin pathway genes are dysregulated in cancer tissues of OSCC patients with either PNI, metastasis, or pain. Using a targeted gene silencing approach with shRNA and focusing on the two main receptors of the pathway, we demonstrate that both TrkA and NGFR mediate PNI, metastasis and pain, and that TrkA and NGFR are each responsible for different facets of oral cancer pain. TrkA and NGFR blockade can serve as promising therapy for oral cancer, both as anti-cancer and analgesic strategies.
METHODS
Cell culture
Cancer cells:
The Hela-O3 cell line was obtained from Dr. Roberto Weigert at National Institutes of Health (NIH) after STR DNA authentication (Genetica DNA Laboratories), was confirmed to be free of Mycoplasma (Invivogen). We selected HeLa-O3 cells due to their highly metastatic potential when injected into the oral cavity of immunocompromised mice; this cell line is formerly known as OSCC3, and is a variant of HeLa adenocarcinoma cells, which has acquired the ability to metastasize to the neck if inoculated in the oral cavity. This cell line is commonly used to establish xenograft models of metastatic head and neck carcinoma, due to its metastatic ability and high EGFR expression, which is commonly upregulated in HNSCC [42–44]. The Hela-O3 was cultured in Dulbecco’s Modification of Eagle’s Medium (DMEM) with 4.5 g/L glucose, L-glutamine and sodium pyruvate, supplemented with 10% fetal bovine serum (FBS), and cultured at 37 °C in 5% CO2.
Transduction of shRNA
Short Hairpin RNA (shRNA) construction and viral particle purification were completed through Vector Biolabs. Sequences are as follows: NTRK1-shRNA 5′-CACC GCATGAGCAGGGATATCTACACTCGAGTGTAGATATCCCTGCTCATGC-TTTTTG-3′ and NGFR-shRNA 5′-CACC GGAACAGCTGCAAGCAGAACACTCGAGTGTTCTGCTTGCAGCTGTTCC-TTTTTG-3′. OSCC cells (Hela-O3) were transduced with recombinant adenovirus containing scrambled shRNA, shRNA NGFR or shRNA TrkA at increasing multiplicities of infection (MOI) to determine transduction efficiency. The vectors contained a GFP tag to allow for fluorescent imaging. Transduction was performed in DMEM with 2% fetal bovine serum (FBS) and the aforementioned supplements.
In vitro migration and invasion assay
The BD Biosciences invasion assay was used according to manufacturer’s recommendations. The assay consisted of an upper invasion chamber insert that fit into a 24-well cell culture plate. The upper invasion chamber was coated with BD Matrigel matrix to allow for assessment of invasive capacity. For each treatment, non-coated invasion chambers were used as the control. Equal numbers of Hela-O3 cells (2×105) in DMEM supplemented with 0.4% FBS were added to each upper chamber. The cells had either been transduced with adenovirus carrying scrambled shRNA, shRNA NGFR, shRNA TrkA, or both shRNA NGFR and TrkA. The bottom wells in the 24-well culture plate were filled with DMEM supplemented with 5% FBS, which served as a chemoattractant. The upper invasion chambers were placed in the cell culture plate and incubated at 37°C for 16 hours. At the end of the incubation period, cells from the upper surface of the filter were wiped off with a cotton swab. The lower surface of the filter was stained with DiffQuik (Dade Behring, Switzerland). The number of cells that migrated to the bottom of the chamber were counted in the light microscope on ten randomly selected fields at 10x magnification. Counting was performed by an investigator who was blinded to the treatment groups. The mean number of cells was calculated per field. Three sets of experiments were carried out, each in quadruplicate. The percent invasion was calculated for each well by dividing the mean number of cells in each well by the mean number of cells of all control wells for the given treatment.
MTS Assay
The MTS (3-(4,5-dimethylthiazol-2-yl)-5-(3-carboxymethoxyphenyl)-2-(4-sulfophenyl)-2H-tetrazolium) assay was used to quantify the effect of shRNA NGFR, TrkA or scrambled shRNA on Hela-O3 proliferation. 1×104 Hela-O3 cells that were transduced with adenovirues (at 20 MOI total) were seeded in each well in a 96-well plate. Eight wells were used for each treatment group. Cells were seeded with either macitentan 4.5μM or vehicle (DMSO) in 5% FBS supplemented DMEM. Cells were incubated at 37°C for 24 hours. 20μl of MTS (Promega BioSciences, San Luis Obispo, CA) was added to each well and incubated at 37°C for two hours. The absorbance of each well was read at 450nm. The absorbance value was averaged for each of the four treatment groups.
RT-PCR
Gene expression was quantified using RT-PCR with commercially available probes and primers to NTRK and TrkA (Taqman assays, Applied Biosystems), as previously described [35]. GUSB was used as the internal control gene.
Mouse cancer model
All animal experiments were approved by the Institutional Animal Care and Use Committee (IACUC) and all methods were carried out in accordance to relevant guidelines and regulations. Methods were reported in accordance with Animal Research: Reporting of In Vivo Experiments (ARRIVE) guidelines. A mouse tongue cancer model was produced by inoculating 106 Hela-O3 into the right tongue of athymic BALB/c mice [37]. The Hela-O3 cell line (formerly OSCC3) metastasizes to the cervical nodes when injected into oral cavity of athymic mice [42]. Inoculation was performed after adenoviruses containing shRNA were introduced into the cell media.
Mechanical allodynia measurement
We quantified facial allodynia using a graded series of von Frey filaments [45]. Testing was performed on day 0 prior to inoculation and twice a week after inoculation until post inoculation day (PID) 19. The graded von Frey assay is a valid assay for quantifying allodynia in mouse models of orofacial cancer. Three measurements were taken for each mouse.
Thermal hyperalgesia measurement
We determined thermal hyperalgesia using the facial cancer model. Thermal hyperalgesia was assessed as previously described [46] using a focused projection bulb to deliver a thermal stimulus to the right whisker pad with a cutoff of 20 seconds. Facial withdrawal to heat was calculated as a mean of three measurements. Testing was performed on day 0 prior to inoculation and then twice a week after inoculation, on the same day as mechanical allodynia testing.
Tissue harvesting, determination of perineural invasion and cervical metastasis
The biological samples were harvested as part of a larger study on oral cancer approved by the Institutional Animal Care and Use Committee (IACUC). All methods were carried out in accordance with the relevant guidelines and regulations. Mouse body weight was measured prior to behavioral experiments to ensure consistent body weight despite tongue tumors. Body weight loss of over 15% was not observed in any of the animals. Mouse tongue and cervical lymph nodes of the neck were harvested under vascular perfusion, which required surgical access to the thoracic and abdominal cavities and simultaneous examination for distant metastatic sites. Cancer volume of the tongue was calculated after tissue harvest by measurement of the length, width and depth of the cancer. Tissue were fixed in 4% paraformaldehyde and embedded in paraffin. 10μm sections were cut for hematoxylin and eosin staining. Three sections from the tongue or lymph node specimen were reviewed, across 60 μm of tissue. The tongue sections were analyzed for evidence of PNI and lymph node sections were analyzed for evidence of metastasis by a board-certified oral pathologist (Dr. Aditi Bhattacharya). PNI was defined by published criteria [47]. Tongue samples were classified as either absent for PNI, present for PNI, or highly suspicious for PNI if extensive neurotropism and nerve entrapment were seen. Presence of scattered neurotropism in a sample did not qualify the sample as highly suspicious for PNI. Cervical node metastasis was marked as absent if all three sections did not demonstrate presence of cancer cells. Cervical node metastasis was marked as present if at least one of the three sections demonstrated presence of cancer cells within the lymph node.
Patient tissue gene expression array analysis
All data and sample collection in patients were approved by the Institutional Review Board (IRB). Snap frozen tissue of the cancer and contralateral normal tissue were taken at the time of ablative surgery from 22 patients with oral cancer. Disease subsites included tongue, lip, maxillary and mandibular gingiva, and floor of mouth. RNA and DNA were extracted according to Qiagen AllPrep protocol (Qiagen). RNA and DNA were processed to prepare for the Illumina Gene Expression Array. Expression patterns were determined for the neurotrophin pathway genes, which included 40 genes listed in the Gene Ontology Browser. Patients were classified as either positive or negative for 1) PNI, 2) neck metastasis and 3) pain. Pain was graded as either severe or low after being quantified using the validated UCSF Oral Cancer Pain Questionnaire (UCSF-OPQ), an 8-question survey of spontaneous and function-induced pain in patients using a visual analog scale of 0–100mm [2]. A total score ≤400 placed the patient in the “low pain” group and a score of >400 placed the patient in the “severe pain” group. Gene expression data was interpreted using the limma package (version 3.48.3) for analysis and the results were represented as a Log fold change (FC) value, where “Log(FC)” = mean[log2(expression of the positive group)] – mean[log2(expression of the negative group)]. Quality control (QC) of the data was performed using the package arrayQualityMetrics (version 3.48.0) and samples that did not pass all 3 QC metrics were removed. Samples that passed QC underwent data normalization. We then performed subset analysis of the normalized expression values for the 87 genes that are of interest. Two heat maps were generated: 1) for all 87 features within the neurotrophin pathway and 2) the top 10 most differentially expressed genes form among all genes measured by the HT12 array that passed all QC metrics. To evaluate for enrichment of differentially expressed genes among pathways, pathway analysis was conducted using three complementary and overlapping annotations: gene ontology (GO[48]), Kyoto Encyclopedia of Genes and Genomes (KEGG[49]), and Reactome. Over-representation analysis (ORA) on the differentially expressed genes was performed using the clusterprofiler package (version 3.16.1) using GO, KEGG, and Reactome databases. Multiple testing was accounted for using the Benjamini Hocheberg correction. For ORA employing GO annotations, pathways were categorized further into biological process, molecular function, and cellular compartment. Differentially expressed pathways were evaluated in relation to each other and contributing differentially expressed sites by two visualizations of functional enrichment (i.e., dot plot and gene-concept networks) using the enrichplot package (version 1.12.3) in R [50].
Statistical analysis
Results were analyzed using Sigma Plot version 13.0 or GraphPad Prism version 7.01. ANOVA and post hoc analysis were performed as appropriate, and p-values were reported.
Supplementary Material
Funding:
American Society of Clinical Oncology - Conquer Cancer Foundation, NIDCR K23DE030250, NIDA R01DA047063 and R01DA047820
Footnotes
DECLARATIONS
Ethics approval and consent to participate: Institutional Review Board and Institutional Animal Care and Use Committee approval were obtained.
Consent for publication: Not applicable.
Competing interests: Not applicable.
Availability of data and materials:
All data generated for this study are included in this article.
References
- 1.Bapat AA, Hostetter G, Von Hoff DD, and Han H, Perineural invasion and associated pain in pancreatic cancer. Nat Rev Cancer, 2011. 11(10): p. 695–707. [DOI] [PubMed] [Google Scholar]
- 2.Connelly ST and Schmidt BL, Evaluation of pain in patients with oral squamous cell carcinoma. J Pain, 2004. 5(9): p. 505–10. [DOI] [PubMed] [Google Scholar]
- 3.Hammerlid E, Bjordal K, Ahlner-Elmqvist M, Boysen M, Evensen JF, Biorklund A, Jannert M, Kaasa S, Sullivan M, and Westin T, A prospective study of quality of life in head and neck cancer patients. Part I: at diagnosis. Laryngoscope, 2001. 111(4 Pt 1): p. 669–80. [DOI] [PubMed] [Google Scholar]
- 4.Kolokythas A, Cox DP, Dekker N, and Schmidt BL, Nerve Growth Factor and Tyrosine Kinase A Receptor in Oral Squamous Cell Carcinoma: Is There an Association With Perineural Invasion? Journal of Oral and Maxillofacial Surgery, 2010. 68(6): p. 1290–1295. [DOI] [PubMed] [Google Scholar]
- 5.Ye Y, Dang D, Zhang J, Viet CT, Lam DK, Dolan JC, Gibbs JL, and Schmidt BL, Nerve growth factor links oral cancer progression, pain, and cachexia. Mol Cancer Ther, 2011. 10(9): p. 1667–76. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Moriwaki K, Ayani Y, Kuwabara H, Terada T, Kawata R, and Asahi M, TRKB tyrosine kinase receptor is a potential therapeutic target for poorly differentiated oral squamous cell carcinoma. Oncotarget, 2018. 9(38): p. 25225–25243. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Grayson M, Arris D, Wu P, Merlo J, Ibrahim T, Fang-Mei C, Valenzuela V, Ganatra S, and Ruparel S, Oral squamous cell carcinoma-released brain-derived neurotrophic factor contributes to oral cancer pain by peripheral tropomyosin receptor kinase B activation. Pain, 2021. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Donnelly CR, Jiang C, Andriessen AS, Wang K, Wang Z, Ding H, Zhao J, Luo X, Lee MS, Lei YL, Maixner W, Ko MC, and Ji RR, STING controls nociception via type I interferon signalling in sensory neurons. Nature, 2021. 591(7849): p. 275–280. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Heussner MJ, Folger JK, Dias C, Massri N, Dahdah A, Vermeer PD, and Laumet G, A Novel Syngeneic Immunocompetent Mouse Model of Head and Neck Cancer Pain Independent of Interleukin-1 Signaling. Anesth Analg, 2021. 132(4): p. 1156–1163. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Syed V, TGF-beta Signaling in Cancer. J Cell Biochem, 2016. 117(6): p. 1279–87. [DOI] [PubMed] [Google Scholar]
- 11.Li J, Duran MA, Dhanota N, Chatila WK, Bettigole SE, Kwon J, Sriram RK, Humphries MP, Salto-Tellez M, James JA, Hanna MG, Melms JC, Vallabhaneni S, Litchfield K, Usaite I, Biswas D, Bareja R, Li HW, Martin ML, Dorsaint P, Cavallo JA, Li P, Pauli C, Gottesdiener L, DiPardo BJ, Hollmann TJ, Merghoub T, Wen HY, Reis-Filho JS, Riaz N, Su SM, Kalbasi A, Vasan N, Powell SN, Wolchok JD, Elemento O, Swanton C, Shoushtari AN, Parkes EE, Izar B, and Bakhoum SF, Metastasis and Immune Evasion from Extracellular cGAMP Hydrolysis. Cancer Discov, 2021. 11(5): p. 1212–1227. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Gonzalez DM and Medici D, Signaling mechanisms of the epithelial-mesenchymal transition. Sci Signal, 2014. 7(344): p. re8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Yang L, Pang Y, and Moses HL, TGF-beta and immune cells: an important regulatory axis in the tumor microenvironment and progression. Trends Immunol, 2010. 31(6): p. 220–7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Alkhadar H, Macluskey M, White S, and Ellis I, Nerve growth factor-induced migration in oral and salivary gland tumour cells utilises the PI3K/Akt signalling pathway: Is there a link to perineural invasion? J Oral Pathol Med, 2020. 49(3): p. 227–234. [DOI] [PubMed] [Google Scholar]
- 15.Kurtz KA, Hoffman HT, Zimmerman MB, and Robinson RA, Perineural and vascular invasion in oral cavity squamous carcinoma: increased incidence on re-review of slides and by using immunohistochemical enhancement. Arch Pathol Lab Med, 2005. 129(3): p. 354–9. [DOI] [PubMed] [Google Scholar]
- 16.Bernier J, Domenge C, Ozsahin M, Matuszewska K, Lefebvre JL, Greiner RH, Giralt J, Maingon P, Rolland F, Bolla M, Cognetti F, Bourhis J, Kirkpatrick A, van Glabbeke M, European Organization for R, and Treatment of Cancer T, Postoperative irradiation with or without concomitant chemotherapy for locally advanced head and neck cancer. N Engl J Med, 2004. 350(19): p. 1945–52. [DOI] [PubMed] [Google Scholar]
- 17.Bernier J, Cooper JS, Pajak TF, van Glabbeke M, Bourhis J, Forastiere A, Ozsahin EM, Jacobs JR, Jassem J, Ang KK, and Lefebvre JL, Defining risk levels in locally advanced head and neck cancers: a comparative analysis of concurrent postoperative radiation plus chemotherapy trials of the EORTC (#22931) and RTOG (# 9501). Head Neck, 2005. 27(10): p. 843–50. [DOI] [PubMed] [Google Scholar]
- 18.Cooper JS, Pajak TF, Forastiere AA, Jacobs J, Campbell BH, Saxman SB, Kish JA, Kim HE, Cmelak AJ, Rotman M, Machtay M, Ensley JF, Chao KS, Schultz CJ, Lee N, and Fu KK, Postoperative concurrent radiotherapy and chemotherapy for high-risk squamous-cell carcinoma of the head and neck. The New England journal of medicine, 2004. 350(19): p. 1937–44. [DOI] [PubMed] [Google Scholar]
- 19.Neumann E, Secondare cancroid infiltration des nervus mentalis bei einem. Arch Pathol Anat, 1862. 24: p. 201. [Google Scholar]
- 20.Liu J, Shao C, Tan ML, Mu D, Ferris RL, and Ha PK, Molecular biology of adenoid cystic carcinoma. Head Neck, 2012. 34(11): p. 1665–77. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Amit M, Takahashi H, Dragomir MP, Lindemann A, Gleber-Netto FO, Pickering CR, Anfossi S, Osman AA, Cai Y, Wang R, Knutsen E, Shimizu M, Ivan C, Rao X, Wang J, Silverman DA, Tam S, Zhao M, Caulin C, Zinger A, Tasciotti E, Dougherty PM, El-Naggar A, Calin GA, and Myers JN, Loss of p53 drives neuron reprogramming in head and neck cancer. Nature, 2020. 578(7795): p. 449–454. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Yu EH, Lui MT, Tu HF, Wu CH, Lo WL, Yang CC, Chang KW, and Kao SY, Oral carcinoma with perineural invasion has higher nerve growth factor expression and worse prognosis. Oral Dis, 2014. 20(3): p. 268–74. [DOI] [PubMed] [Google Scholar]
- 23.Zhu Z, Friess H, diMola FF, Zimmermann A, Graber HU, Korc M, and Buchler MW, Nerve growth factor expression correlates with perineural invasion and pain in human pancreatic cancer. J Clin Oncol, 1999. 17(8): p. 2419–28. [DOI] [PubMed] [Google Scholar]
- 24.Miknyoczki SJ, Lang D, Huang L, Klein-Szanto AJ, Dionne CA, and Ruggeri BA, Neurotrophins and Trk receptors in human pancreatic ductal adenocarcinoma: expression patterns and effects on in vitro invasive behavior. Int J Cancer, 1999. 81(3): p. 417–27. [DOI] [PubMed] [Google Scholar]
- 25.Ivanov SV, Panaccione A, Brown B, Guo Y, Moskaluk CA, Wick MJ, Brown JL, Ivanova AV, Issaeva N, El-Naggar AK, and Yarbrough WG, TrkC signaling is activated in adenoid cystic carcinoma and requires NT-3 to stimulate invasive behavior. Oncogene, 2013. 32(32): p. 3698–710. [DOI] [PubMed] [Google Scholar]
- 26.Mapp PI and Walsh DA, Mechanisms and targets of angiogenesis and nerve growth in osteoarthritis. Nature reviews. Rheumatology, 2012. 8(7): p. 390–8. [DOI] [PubMed] [Google Scholar]
- 27.Jimenez-Andrade JM, Ghilardi JR, Castaneda-Corral G, Kuskowski MA, and Mantyh PW, Preventive or late administration of anti-NGF therapy attenuates tumor-induced nerve sprouting, neuroma formation, and cancer pain. Pain, 2011. 152(11): p. 2564–74. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Ein L, Mei C, Bracho O, Bas E, Monje P, Weed D, Sargi Z, Thomas G, and Dinh C, Modulation of BDNF-TRKB Interactions on Schwann Cell-induced Oral Squamous Cell Carcinoma Dispersion In Vitro. Anticancer Res, 2019. 39(11): p. 5933–5942. [DOI] [PubMed] [Google Scholar]
- 29.Wagner VP, Martins MD, Amoura E, Zanella VG, Roesler R, de Farias CB, Bingle CD, Vargas PA, and Bingle L, TrkB-Targeted Therapy for Mucoepidermoid Carcinoma. Biomedicines, 2020. 8(12). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Chodroff L, Bendele M, Valenzuela V, Henry M, and Ruparel S, EXPRESS: BDNF Signaling Contributes to Oral Cancer Pain in a Preclinical Orthotopic Rodent Model. Mol Pain, 2016. 12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Tabata M, Murata E, Ueda K, Kato-Kogoe N, Kuroda Y, and Hirose M, Effects of TrkA inhibitory peptide on cancer-induced pain in a mouse melanoma model. J Anesth, 2012. 26(4): p. 545–51. [DOI] [PubMed] [Google Scholar]
- 32.Khan N. and Smith MT, Neurotrophins and Neuropathic Pain: Role in Pathobiology. Molecules, 2015. 20(6): p. 10657–88. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Reichardt LF, Neurotrophin-regulated signalling pathways. Philos Trans R Soc Lond B Biol Sci, 2006. 361(1473): p. 1545–64. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Khodorova A, Nicol GD, and Strichartz G, The TrkA receptor mediates experimental thermal hyperalgesia produced by nerve growth factor: Modulation by the p75 neurotrophin receptor. Neuroscience, 2017. 340: p. 384–397. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Viet CT, Dang D, Ye Y, Ono K, Campbell RR, and Schmidt BL, Demethylating Drugs as Novel Analgesics for Cancer Pain. Clin Cancer Res, 2014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Viet CT and Schmidt BL, Biologic mechanisms of oral cancer pain and implications for clinical therapy. Journal of dental research, 2012. 91(5): p. 447–53. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Viet CT, Ye Y, Dang D, Lam DK, Achdjian S, Zhang J, and Schmidt BL, Re-expression of the methylated EDNRB gene in oral squamous cell carcinoma attenuates cancer-induced pain. Pain, 2011. 152(10): p. 2323–32. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Lam DK and Schmidt BL, Orofacial pain onset predicts transition to head and neck cancer. Pain, 2011. 152(5): p. 1206–9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Viet CT, Corby PM, Akinwande A, and Schmidt BL, Review of preclinical studies on treatment of mucositis and associated pain. J Dent Res, 2014. 93(9): p. 868–75. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Ueda K, Hirose M, Murata E, Takatori M, Ueda M, Ikeda H, and Shigemi K, Local administration of a synthetic cell-penetrating peptide antagonizing TrkA function suppresses inflammatory pain in rats. J Pharmacol Sci, 2010. 112(4): p. 438–43. [DOI] [PubMed] [Google Scholar]
- 41.Ma WY, Murata E, Ueda K, Kuroda Y, Cao MH, Abe M, Shigemi K, and Hirose M, A synthetic cell-penetrating peptide antagonizing TrkA function suppresses neuropathic pain in mice. J Pharmacol Sci, 2010. 114(1): p. 79–84. [DOI] [PubMed] [Google Scholar]
- 42.Amornphimoltham P, Rechache K, Thompson J, Masedunskas A, Leelahavanichkul K, Patel V, Molinolo A, Gutkind JS, and Weigert R, Rab25 regulates invasion and metastasis in head and neck cancer. Clin Cancer Res, 2013. 19(6): p. 1375–88. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Kamarajan P, Alhazzazi TY, Danciu T, D’Silva N J, Verdin E, and Kapila YL, Receptor-interacting protein (RIP) and Sirtuin-3 (SIRT3) are on opposite sides of anoikis and tumorigenesis. Cancer, 2012. 118(23): p. 5800–10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Henson B, Li F, Coatney DD, Carey TE, Mitra RS, Kirkwood KL, and D’Silva NJ, An orthotopic floor-of-mouth model for locoregional growth and spread of human squamous cell carcinoma. J Oral Pathol Med, 2007. 36(6): p. 363–70. [DOI] [PubMed] [Google Scholar]
- 45.Vos BP, Strassman AM, and Maciewicz RJ, Behavioral evidence of trigeminal neuropathic pain following chronic constriction injury to the rat’s infraorbital nerve. J Neurosci, 1994. 14(5 Pt 1): p. 2708–23. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Hargreaves K, Dubner R, Brown F, Flores C, and Joris J, A new and sensitive method for measuring thermal nociception in cutaneous hyperalgesia. Pain, 1988. 32(1): p. 77–88. [DOI] [PubMed] [Google Scholar]
- 47.Chi AC, Katabi N, Chen HS, and Cheng YL, Interobserver Variation Among Pathologists in Evaluating Perineural Invasion for Oral Squamous Cell Carcinoma. Head Neck Pathol, 2016. 10(4): p. 451–464. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Ashburner M, Ball CA, Blake JA, Botstein D, Butler H, Cherry JM, Davis AP, Dolinski K, Dwight SS, Eppig JT, Harris MA, Hill DP, Issel-Tarver L, Kasarskis A, Lewis S, Matese JC, Richardson JE, Ringwald M, Rubin GM, and Sherlock G, Gene ontology: tool for the unification of biology. The Gene Ontology Consortium. Nat Genet, 2000. 25(1): p. 25–9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Kanehisa M, The KEGG database. Novartis Found Symp, 2002. 247: p. 91–101; discussion 101–3, 119–28, 244–52. [PubMed] [Google Scholar]
- 50.Viet CT, Yu G, Asam K, Thomas CM, Yoon AJ, Wongworawat YC, Haghighiabyaneh M, Kilkuts CA, McGue CM, Couey MA, Callahan NF, Doan C, Walker PC, Nguyen K, Kidd SC, Lee SC, Grandhi A, Cheng AC, Patel AA, Philipone E, Ricks OL, Allen CT, and Aouizerat BE The REASON Score: An Epigenetic and Clinicopathologic Score to Predict Risk of Poor Survival in Patients with Early Stage Oral Squamous Cell Carcinoma Biomarker Research, 2021. 9(1). [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Data Availability Statement
All data generated for this study are included in this article.