Abstract
Background
Since early 2022, Omicron BA.1 has been eclipsed by BA.2, which was in turn outcompeted by BA.5, which displays enhanced antibody escape properties.
Methods
Here, we evaluated the duration of the neutralizing antibody (Nab) response, up to 18 months after Pfizer BNT162b2 vaccination, in individuals with or without BA.1/BA.2 breakthrough infection. We measured neutralization of the ancestral D614G lineage, Delta, and Omicron BA.1, BA.2, and BA.5 variants in 300 sera and 35 nasal swabs from 27 individuals.
Findings
Upon vaccination, serum Nab titers were decreased by 10-, 15-, and 25-fold for BA.1, BA.2, and BA.5, respectively, compared with D614G. We estimated that, after boosting, the duration of neutralization was markedly shortened from 11.5 months with D614G to 5.5 months with BA.5. After breakthrough, we observed a sharp increase of Nabs against Omicron subvariants, followed by a plateau and a slow decline after 5–6 months. In nasal swabs, infection, but not vaccination, triggered a strong immunoglobulin A (IgA) response and a detectable Omicron-neutralizing activity.
Conclusions
BA.5 spread is partly due to abbreviated vaccine efficacy, particularly in individuals who were not infected with previous Omicron variants.
Funding
Work in O.S.’s laboratory is funded by the Institut Pasteur, Urgence COVID-19 Fundraising Campaign of Institut Pasteur, Fondation pour la Recherche Médicale (FRM), ANRS, the Vaccine Research Institute (ANR-10-LABX-77), Labex IBEID (ANR-10-LABX-62-IBEID), ANR/FRM Flash Covid PROTEO-SARS-CoV-2, ANR Coronamito, and IDISCOVR, Laboratoire d’Excellence ‘Integrative Biology of Emerging Infectious Diseases’ (grant no. ANR-10-LABX-62-IBEID), HERA european funding and the NIH PICREID (grant no U01AI151758).
Keywords: SARS-Cov-2, variants, Omicron, BA.5, neutralization, vaccine, breakthrough, mucosal immunity, IgG, IgA
Graphical abstract
Context and significance
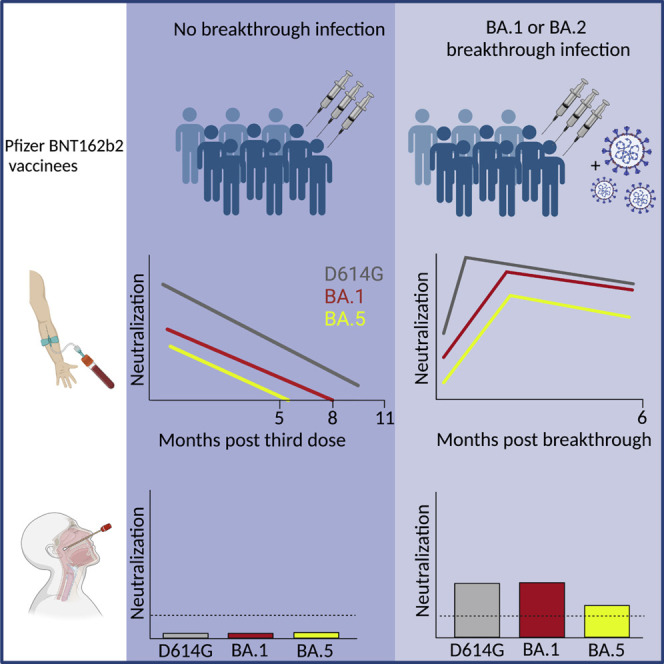
Since the emergence of the severe acute respiratory syndrome coronavirus 2 (SARS-CoV-2) Omicron variant in December 2022, approximately 50% of the world population has been infected, reflecting a poor protection against infection conferred by vaccination. Researchers from Paris, France, report that, upon vaccination, the duration of an efficient antibody response was significantly shorter for Omicron variants, including BA.5, compared with the ancestral strain. After breakthrough infection, antibody levels against Omicron subvariants increased, and remained elevated for at least 5–6 months. Breakthrough infection, but not vaccination, triggered detectable local response in the nasal mucosa against SARS-CoV-2. These results show that the longitudinal survey of antibody levels in blood and nasal samples may provide a reliable marker of the effectiveness of vaccination, natural, and hybrid immunity against acquisition of current and future SARS-CoV-2 variants.
Planas et al. analyze the extent and duration of the neutralizing antibody response after vaccination with Pfizer BNT162b2 mRNA in the sera and nasal swabs from individuals with or without Omicron breakthrough infection, finding a short duration of neutralization against BA.5 after boosting and strong immunoglobulin A (IgA) response upon breakthrough infection.
Introduction
The severe acute respiratory syndrome coronavirus 2 (SARS-CoV-2) Omicron BA.1 and BA.2 variants spread across the world and replaced the Delta variant in early 2022.1 It is estimated that more than 50% of the population were infected by BA.1 or BA.2 by March 20222 with little protection against infection conferred by vaccination.3 , 4 , 5 The incidence of breakthrough infections in vaccinated individuals has thus dramatically increased since Omicron emerged.6 BA.1 and BA.2 contain approximately 32 changes in the spike protein, promoting their high transmissibility and immune escape properties.7 , 8 , 9 , 10 , 11 , 12 , 13 , 14 , 15 , 16 , 17 The Omicron clade has rapidly evolved into sub-lineages, including BA.5, that outcompeted BA.1 and BA.2.18 The BA.5 spike shares multiple changes noted in BA.2 and bears a few additional modifications. BA.5 became predominant worldwide by mid-2022 and was responsible for a surge of infections in many countries.18 , 19 The neutralizing activity of sera from COVID-19 vaccine recipients is reduced against BA.5 by approximately three- to five-fold compared with BA.1 and BA.2.11 , 20 , 21 , 22 , 23 , 24 Here, we assessed the durability and magnitude of neutralizing antibody (Nab) responses against different Omicron variants, up to 18 months after Pfizer BNT162b2 vaccination. We also analyzed the evolution of Nabs in the sera and nasal swabs from vaccine recipients who experienced BA.1 or BA.2 breakthrough infections. We report a shortened duration of neutralization against BA.5 Nabs in the sera of vaccine recipients, and a presence of such antibodies in nasal swabs only after breakthrough infection.
Results
Cohort design
We longitudinally collected 300 sera and 35 nasal samples from a cohort of 27 health care workers in Orleans, France. We previously studied the ability of some of these sera to neutralize the Alpha, Beta, Delta, and Omicron BA.1 variants.10 The characteristics of each participant are indicated in Table S1. The participants, who were not previously infected at the time of inclusion, received two doses of Pfizer BNT162b2 vaccine within an interval of 21–28 days and a booster dose 154–361 days later. Of the 27 individuals, 11 experienced a pauci-symptomatic breakthrough infection 60–178 days after the third injection. Screening by polymerase chain reaction or whole-viral-genome sequencing confirmed an Omicron breakthrough infection. At the time of infection (between December 2021 and mid-February 2022), BA.1 and BA.2 represented 95% and 5% of the Omicron lineage in France, respectively.25 Each participant was sampled 3–23 times (mean, 11) during the 18 months of the survey. The days of vaccination, breakthrough infection, and sampling are displayed for each participant in Figure S1 and Table S1.
BA.5 serum neutralization in vaccine recipients, with or without omicron BA.1 or BA.2 infection
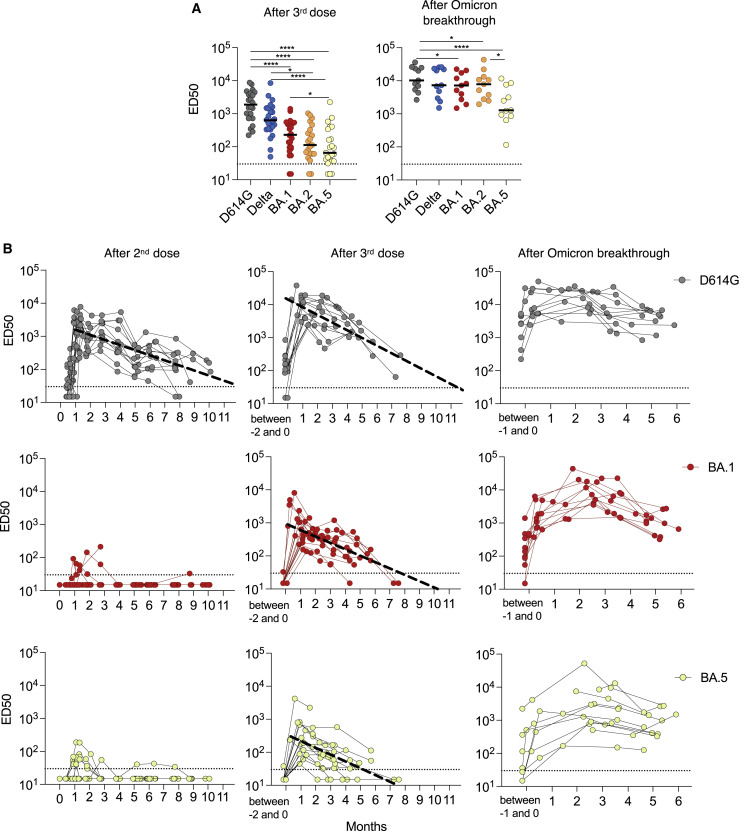
We first measured serum Nab titers against the D614G reference virus, the Delta variant, and the Omicron BA.1, BA.2, and BA.5 isolates in the 24 vaccine recipients that were sampled 1–5 months after the booster doses. We calculated the 50% effective dilution (ED50) for each combination of serum and virus. As previously reported, Omicron subvariants displayed considerable immune escape properties, compared with the D614G and Delta variants (Figure 1A). Among Omicron subvariants, BA.5 neutralization was extremely low, with a median ED50 of 60, decreased by a factor of 2.5 and 1.7 compared with BA.1 and BA.2 (ED50 of 148 and 100, respectively).
Figure 1.
Magnitude, cross-reactivity, and durability of antibodies in sera from Pfizer vaccinees, with or without breakthrough BA.1/BA.2 infection
(A) Nab titers against D614G, Delta and Omicron BA.1, BA.2 and BA.5 were quantified in sera from triple vaccinated individuals (n = 24; median = 120 days after the third dose) (left panel) and after Omicron BA.1 or BA.2 breakthrough infection (n = 11; median = 80 days after infection). Data are the mean from two independent experiments. The horizontal dotted line indicates the limit of detection (ED50 = 30). Black lines represent the median values. Two-sided Friedman test with Dunn’s test for multiple comparisons was performed between each viral strain at the different time points; ∗p < 0.05; ∗∗∗∗p < 0.0001.
(B) Temporal evolution of Nab titers (ED50) against D614G (black), BA.1 (red) and BA.5 (yellow) in 27 vaccine recipients and 11 participants who had an Omicron BA.1 or BA.2 breakthrough infection. The Nab titers were calculated at the indicated months after the second dose (left panels), third dose (middle panels), or breakthrough infection (right panels). The bold dotted line included in some panels represents a simple linear regression of Nab waning. In the other panels, the shape of the curves did not allow this analysis. Data are the mean from two independent experiments. The horizontal dotted line indicates the limit of detection (ED50 = 30).
One month after infection, the overall anti-SARS-CoV-2 IgG antibody levels increased by only 3.6-fold in the 11 individuals who experienced a breakthrough infection (Figure S2A). A strong augmentation of the cross-neutralization against Delta, BA.1, and BA.2 variants was observed, with ED50 ranging from 103 to 104 (Figure 1A). The Nab titer was lower against BA.5 (ED50 of 103). Therefore, post-vaccination infection by BA.1 or BA.2 led to an increase in Omicron-specific Nab titers, which was less marked against BA.5.
Temporal evolution of the neutralization profile
We next longitudinally analyzed the evolution of serum cross-neutralization in the 27 individuals, up to 18 months after initiation of the vaccination, in all available samples. We represented IgG levels and Nab titers at different time points after the second and third vaccine injections, as well as after Omicron BA.1 or BA.2 breakthrough infection. We performed modeling for the best fit curve of the data, using either continuous (simple linear regression), one-phase, or two-phase decays. We compared the three decays using the extra sum-of-squares F test, selecting the simpler model with a p value of less than 0.05. The best fitting statistical model for the decline of anti-S IgG and Nab was the simple linear regression. As previously reported,26 a peak level of anti-spike IgG was observed one month after the second dose, which subsequently decreased over the next 10 months (Figure S2B). A booster dose induced higher IgG peak levels than the second dose. The regression model indicated that IgGs should become undetectable about 1.91 and 1.95 years after the second and third doses, respectively. The breakthrough infection increased IgG levels, without an obvious decrease for up to 6 months, precluding the calculation of a time to undetectability.
The evolution of the neutralization profile showed disparities between variants. After the second dose, neutralization against Omicron BA.1, BA.2, and BA.5 was extremely low with undetectable titers is most samples, whereas D614G and to a lesser extent Delta were neutralized (Figures 1B and S2B). After boosting, the sera neutralized all variants, with differences not only in the peak level (Figure 1B), but also in the duration of neutralization. The time to undetectability was shortened from 11.5 months for D614G to 8 months for Delta and BA.1 and 5.5 months for BA.5 (Figures 1B and S2B). The kinetics of serum neutralization were different after breakthrough infection. With all variants, the peak was reached approximately 2 months after infection, followed by a slow decline not noticeable before 5–6 months.
To better visualize the influence of the lag between the booster and breakthrough infection on the evolution of cross-neutralization, we depicted the kinetics of Nab titers for six vaccine recipients with and three individuals without breakthrough infection (Figure S3). Breakthrough Omicron infections occurred 2–5 months after the third dose and caused a consistent increase of anti-S IgGs and Nabs against the different variants. In non-infected individuals, Nabs decreased progressively over the survey period.
Altogether, these results indicate a shorter neutralization efficacy of sera from triple vaccinated individuals against BA.5. A breakthrough Omicron infection triggered a longer lasting neutralizing response. There was no major impact of the timing of breakthrough infection relative to the vaccination on the extent of induction of cross-reactive Nabs.
Antibody responses at the nasal mucosae
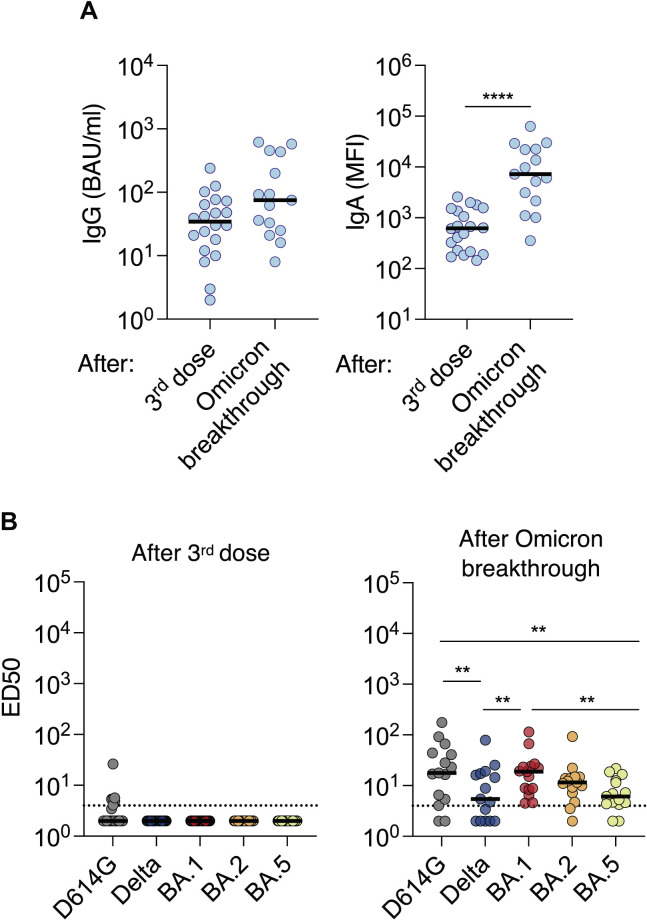
We then asked whether vaccination and breakthrough infections may trigger different antibody responses at the mucosal surfaces. We measured the levels of anti-spike IgG and cross-neutralizing activity in 35 nasal swabs. Twenty individuals were sampled 1 month after their third vaccine dose. Among them, 7 experienced a breakthrough infection and were sampled 1–3 times, up to 3 months after infection, yielding a total of 15 samples. Levels of anti-SARS-CoV-2 IgG and IgA were relatively low (Figure 2A), confirming that Pfizer vaccination does not induce a strong local immunity. The breakthrough infection triggered a moderate (2.2-fold) increase in IgGs, whereas IgAs were augmented by approximately 12-fold (Figure 2A). Accordingly, in triple vaccinated individuals, we did not detect any neutralizing activity against Delta or Omicron variants, and only one individual poorly neutralized the D614G ancestral strain. In sharp contrast, 10 of 15 nasal swabs collected after infection presented detectable neutralization against all variants. This neutralizing activity was higher against D614G, BA.1, and BA.2 than against Delta and BA.5. There was no obvious correlation between neutralization titers in the sera and nasal swabs (not shown). This is in agreement with a known compartmentalization of systemic and mucosal immune responses during acute SARS-CoV-2 infection.27
Figure 2.
Induction of cross-neutralizing antibodies at the nasal mucosae upon Omicron BA.1 or BA.2 breakthrough infection
Nasal swabs were collected at 1 month after the third dose for 20 participants (median = 38 days after the third dose) and 1–3 months after BA.1 or BA.2 breakthrough infection for 7 participants. In 5 participants who experienced a breakthrough infection, 2 or 3 time points after infection are shown, representing a total of 15 samples.
(A) Levels of anti-spike IgGs (left panel) and IgAs (right panel) measured by flow cytometry with the S-Flow assay. For IgGs, results are presented in BAU/mL. The dotted lines indicate the limit of detection (3 BAU/mL). For IgAs, the mean fluorescence intensity of binding is shown, since the lack of IgA reference samples precluded a calculation of BAU IgA/mL. Black lines represent the median values. A Mann-Whitney unpaired t-test was performed. ∗∗∗∗p < 0.0001.
(B) The Nab titers against D614G, Delta and Omicron BA.1, BA.2, and BA.5 were measured in the same samples from triple vaccinated individuals (left panel) and after breakthrough infection (right panel). The dotted lines indicate the limit of detection (ED50 = 4). Black lines represent the median values. A two-sided Friedman test with Dunn’s test for multiple comparisons was performed between each viral strain at the different time points; ∗∗p < 0.01.
Therefore, Omicron breakthrough infection, but not vaccination, triggers a local Nab response at the viral entry site in the host. Our results help us to understand why a hybrid immunity is more effective than vaccination alone to prevent infection by a novel SARS-CoV-2 variant.
Discussion
The Omicron lineage has evolved toward enhanced transmissibility and immune evasion properties. BA.5 surged in many countries and displays a three- to five-fold lower sensitivity to Nabs generated by vaccination, relative to prior Omicron variants.20 , 21 , 22 , 23 How this decrease impacts the duration of vaccine efficacy remains poorly characterized. Here, we longitudinally analyzed the levels of Nabs in a cohort of Pfizer-vaccinated recipients. A booster dose generated a neutralizing response against the ancestral strain D614G that lasted approximately 11.5 months. The duration of neutralization was shortened to 5.5 months against BA.5, suggesting an abbreviated efficacy of current vaccines against this variant. In contrast, a natural Omicron infection in vaccinated individuals induced a longer lasting neutralizing response, with no visible decrease up to 5–6 months after infection. We also report that vaccination did not generate a detectable local neutralizing immunity at the nasal mucosae, whereas a breakthrough infection in vaccine recipients induced such a response. The breakthrough infection triggered a moderate (2.2-fold) increase in IgGs, whereas IgAs were augmented by approximately 12-fold (Figure 2A). Accordingly, in triple vaccinated individuals, we did not detect any neutralizing activity against the Delta or Omicron variants, whereas only one individual poorly neutralized the D614G ancestral strain. In sharp contrast, 10 of 15 nasal swabs collected after infection presented detectable neutralization against all variants. This neutralizing activity was higher against D614G, BA.1, and BA.2 than against Delta and BA.5. Neutralization was in a large part likely owing to the increased levels of anti-SARS-CoV-2 IgAs, which are known to be more potent than IgGs.28 , 29
These findings are line with a recent national study in Portugal showing that a previous BA.1 or BA.2 infection had a protective effect against BA.5 infection.30 Our results help to explain why the BA.5 wave preferentially occurred in first-timer individuals who were not previously infected with Omicron, independent of their vaccination status.
Our results are in line with public health vaccine efficacy reports, analyzing tens of thousands of individuals in different countries.31 , 32 , 33 , 34 , 35 For instance, one study performed in England before the Omicron surge reported that two doses of the Pfizer vaccine were associated with high short-term protection against SARS-CoV-2 infection.32 This protection waned considerably after 6 months. Infection-acquired immunity boosted with vaccination remained high more than 1 year after infection.33 A study performed in Qatar reported that the effectiveness of Pfizer vaccination against symptomatic BA.2 infection with two doses of Pfizer vaccine was negligible,34 in agreement with our observation of a lack of detection Nabs against Omicron variants in sera from double vaccine recipients. The effectiveness of three doses of Pfizer, without or with previous infection, was 52% and 77%, respectively.34 Another report from the United States indicated that vaccine effectiveness against COVID-19-associated hospitalization was higher during the BA.1 period than during the BA.2/BA.2.12.1 period.35 A Danish nation-wide survey recently reported a high protection against BA.5 from prior omicron infection in triple-vaccinated individuals, and similar vaccine effectiveness for BA.5 and BA.2.31
Limitations of the study
The size of our cohort was relatively small, but the differences between variants were sufficiently marked to attain statistical significance. The size of our cohort was relatively small, but the differences between variants were sufficiently marked to obtain statistical significance. The sampling time points were not identical for all participants. Owing to the limited amounts of sera available in this cohort, we performed a longitudinal analysis with four different viral strains (D614G, Delta, BA.1, and BA.5), and not with BA.2. Age-related heterogeneity in the humoral response to SARS-CoV-2 vaccine has been previously described.36 , 37 , 38 In our study, 80% of the participants were above the age of 50. It is possible that stronger and extended antibody responses may be observed in a younger population. We focused our work on Pfizer vaccine recipients and did not assess the neutralization conferred by a fourth dose. We analyzed the temporal evolution of the neutralizing response only up to 6 months after a breakthrough infection. Future work will help to understand the efficacy of different vaccine regimen in various categories of individuals. It will be worth further examining the impact of a fourth vaccine dose on the extent and duration of the humoral immune response against BA.5 and other variants.
In summary, our data suggest that a longitudinal survey of the neutralizing humoral response in blood and nasal samples may provide a reliable marker of the duration of effectiveness of vaccination, natural and hybrid immunity against the acquisition of current and forthcoming SARS-CoV-2 variants.
STAR★Methods
Key resources table
| REAGENT or RESOURCE | SOURCE | IDENTIFIER |
|---|---|---|
| Antibodies | ||
| anti-IgG AlexaFluor647 | Jackson ImmunoResearch | Cat#A-21445 ; RRID:AB_2535862 |
| Anti-IgA AlexaFluor488 | Jackson ImmunoResearch | Cat#109-545-011 ; RRID:AB_2337834 |
| Bacterial and virus strains | ||
| D614G(hCoV-19/France/GE1973/2020) | National Reference Center for Respiratory Viruses (Institut Pasteur, Paris, France) | EPI_ISL_4146339 |
| Delta | Laboratory of Virology of Hopital Européen Georges Pompidou (Assistance Publique – Hopitaux de Paris) | EPI_ISL_202911340 |
| Omicron BA.1, BA.2 & BA.5 | NRC UZ/KU Leuven, Belgium | EPI_ISL_679490741 EPI_ISL_1065497942 |
| Chemicals, peptides, and recombinant proteins | ||
| Hoechst 33342 | Invitrogen | Cat#H3570 |
| Paraformaldehyde 4% | Alfa Aesar | Cat#J19943.K2 |
| Deposited data | ||
| Viral genome sequences | GISAID | EPI_ISL_41463 EPI_ISL_2029113 EPI_ISL_6794907 EPI_ISL_10654979 |
| Experimental models: Cell lines | ||
| 293T | ATCC | CRL-3216 ; RRID:CVCL_0063 |
| U2OS cells | ATCC | Cat#HTB-96 ; RRID:CVCL_0042 |
| Software and algorithms | ||
| Harmony High-Content Imaging and Analysis Software | PerkinElmer | Cat#HH17000012 |
| Excel 365 | Microsoft | https://www.microsoft.com/en-ca/microsoft-365/excel |
| Prism 8 | Graphpad | https://www.graphpad.com/ |
| FlowJo v10 | Tree Star | https://www.flowjo.com/ |
Resource availability
Lead contact
Further information and requests for resources and reagents should be directed to and will be fulfilled by the Lead Contact, Olivier Schwartz (Olivier.schwartz@pasteur.fr).
Materials availability
All unique/stable reagents generated in this study are available from the lead contact with a completed Materials Transfer Agreement.
Experimental models and subject details
We conducted a prospective, monocentric, longitudinal, interventional cohort clinical study (ABCOVID) since 27 August 2020 with the objective to study the kinetics of COVID-19 antibodies in patients with confirmed SARS-CoV-2 infection (NCT04750720). A sub-study aimed to describe the kinetic of neutralizing antibodies (in blood and nasal mucosae) after vaccination. The cohort was previously described.10 , 39 , 40 This study was approved by the Ile-de-France IV ethical committee. At enrollment, written informed consent was collected and participants completed a questionnaire covering sociodemographic characteristics, virological findings (SARS-CoV-2 RT–qPCR results, date of testing) and data related to SARS-CoV-2 vaccination (brand product, date of first, second and third vaccination). Participants information on sex and age was reported in Table S1. Information on socioeconomic status was not collected. Blood and nasal swabs were collected at several time points. Nasal swabs were collected in the two cavities and preserved in 1 or 3 mL of M4RT transport buffer (Standard Sigma Swabs MW910S, Sigma VCM). Study participants did not receive any compensation.
Method details
S-flow assay
The S-Flow assay is based on the recognition of the SARS-CoV-2 spike protein expressed on the surface of 293T cells. It was used to quantify SARS-CoV-2-specific IgG and IgA subtypes in sera, nasopharyngeal swab supernatants and saliva as previously described.41 , 42
Briefly, 293T cells were obtained from ATCC (ATCC Cat# CRL-3216, RRID:CVCL_0063) and tested negative for mycoplasma. 293T cells stably expressing Spike (293T S) or control (293T Empty) cells were transferred into U-bottom 96-well plates (105 cells/well). Cells were incubated at 4°C for 30 min with serum (1:300 dilution) or nasal swabs (1:50) in PBS containing 0.5% BSA and 2 mM EDTA. Then, cells were washed with PBS, and stained using anti-IgG Alexa Fluor 647 (Jackson ImmunoResearch cat# 109-605-170) and Anti-IgA Alexa Fluor 488 (Jackson ImmunoResearch cat# 109-545-011). Cells were washed with PBS and fixed for 10 min with 4% PFA. Data were acquired on an Attune Nxt instrument (Life Technologies). Stainings were also performed on control (293T Empty) cells. Results were analyzed with FlowJo 10.7.1 (Becton Dickinson). The gating strategy for IgGs and IgAs is shown in Figure S4. The specific binding was calculated as follows: 100 x (% binding 293T Spike - % binding 293T Empty)/(100 - % binding 293T Empty). The assay was standardized with whom international reference sera (20/136 and 20/130) and cross-validated with two commercially available ELISA (Abbott and Beckmann) using a Passing-Bablok linear regression model to allow calculation of BAU/mL.43
S-Fuse neutralization assay
U2OS-ACE2 GFP1-10 or GFP 11 cells, also termed S-Fuse cells, become GFP + when they are productively infected by SARS-CoV-2.40 , 44 Cells were tested negative for mycoplasma. Cells were mixed (ratio 1:1) and plated at 8 × 103 per well in a μClear 96-well plate (Greiner Bio-One). The indicated SARS-CoV-2 strains were incubated with serially diluted sera (first dilution 1:30) or nasal swabs (first dilution 1:4) for 15 min at room temperature and added to S-Fuse cells. The sera were heat-inactivated for 30 min at 56°C before use. 18 h later, cells were fixed with 2% PFA, washed and stained with Hoechst (dilution 1:1,000, Invitrogen). Images were acquired with an Opera Phenix high content confocal microscope (PerkinElmer). The GFP area and the number of nuclei were quantified using the Harmony software (PerkinElmer). The percentage of neutralization was calculated using the number of syncytia as value with the following formula: 100 x (1 – (value with serum – value in “non-infected”)/(value in “no serum” – value in “non-infected”)). Neutralizing activity of each serum was expressed as the half maximal effective dilution (ED50). ED50 values (dilution values for sera and nasal swabs) were calculated with a reconstructed curve using the percentage of the neutralization at the different concentrations.
Virus strains
The reference D614G strain (hCoV-19/France/GE1973/2020) was supplied by the National Reference Center for Respiratory Viruses hosted by Institut Pasteur (Paris, France) and headed by Pr. S. van der Werf.40 This viral strain was supplied through the European Virus Archive goes Global (Evag) platform, a project that has received funding from the European Union’s Horizon 2020 research and innovation program under grant agreement n° 653316. Delta was isolated from a nasopharyngeal swab of a hospitalized patient returning from India.39 The swab was provided and sequenced by the laboratory of Virology of Hopital Européen Georges Pompidou (Assistance Publique – Hopitaux de Paris). The Omicron BA.1 and BA.2 strains were supplied and sequenced by the NRC UZ/KU Leuven (Leuven, Belgium).45 The Omicron BA.5 was isolated from a nasopharyngeal swab of a woman hospitalized for rheumatological pain. The swab was sequenced by the National Reference Center for HIV-Associated laboratory of Tours, France. All patients provided informed consent for the use of the biological materials.
The variant strains were isolated from nasal swabs using Vero E6 cells and amplified by one or two passages. Viruses were sequenced directly on nasal swabs, and after one or two passages on Vero cells. Sequences of the swabs and amplified viruses were similar. Sequences were deposited on GISAID immediately after their generation, with the following IDs: D614G: EPI_ISL_41463; Delta ID: EPI_ISL_2029113; Omicron BA.1 ID: EPI_ISL_6794907; Omicron BA.2 GISAID ID: EPI_ISL_10654979; Omicron BA.5 ID: EPI_ISL_13660702. Titration of viral stocks was performed on Vero E6, with a limiting dilution technique allowing a calculation of TCID50, or on S-Fuse cells.
Quantification and statistical analysis
Flow cytometry data were analyzed with FlowJo v10 software (Tri-Star). Calculations were performed using Excel 365 (Microsoft). Figures were drawn on Prism 9 (GraphPad Software). Statistical analysis was conducted using GraphPad Prism 9. No methods were used to determine whether the data met assumptions of the statistical approach. Statistical significance between different groups was calculated using the tests indicated in each figure.
Additional resources
Clinical trial registration: NCT04750720.
Acknowledgments
Work in OS lab is funded by Institut Pasteur, Urgence COVID-19 Fundraising Campaign of Institut Pasteur, Fondation pour la Recherche Médicale (FRM), ANRS, the Vaccine Research Institute (ANR-10-LABX-77), Labex IBEID (ANR-10-LABX-62-IBEID), ANR/FRM Flash Covid PROTEO-SARS-CoV-2, ANR Coronamito, and IDISCOVR. DP is supported by the Vaccine Research Institute. Work in OS lab is funded by Institut Pasteur, the INCEPTION program (Investissements d’Avenir grant ANR-16-CONV-0005) and the French Government’s Investissement d’Avenir program, Laboratoire d’Excellence ‘Integrative Biology of Emerging Infectious Diseases’ (grant no. ANR-10-LABX-62-IBEID), HERA european funding and the NIH PICREID (grant no U01AI151758). The Opera system was co-funded by Institut Pasteur and the Région ile de France (DIM1Health). Work in UPBI is funded by grant ANR-10-INSB-04-01 and Région Ile-de-France program DIM1-Health.
The funders of this study had no role in study design, data collection, analysis and interpretation, or writing of the article.
The authors thank A. Fontanet for critical reading of the manuscript, the patients who participated to this study, members of the Virus and Immunity Unit and other teams for discussion and help, N. Aulner and the staff at the UtechS Photonic BioImaging (UPBI) core facility (Institut Pasteur), a member of the France BioImaging network, for image acquisition and analysis, and F. Peira, V. Legros, and L. Courtellemont for their help with the cohorts.
Author contributions
Experimental strategy design, experiments: D.P., I.S., F.P., F.G.-B., W.-H.B., T.B., M.P., E.S.-L., and O.S. Vital materials: L.H., J.P., H.P., D.V., A.S., T.P., K.S., and L.H. Statistical analyses: D.P. Manuscript writing, reviewing and editing D.P. and O.S. Manuscript editing D.P., W.-H. B., E. S-L., T.B., and O.S. D.P. and O.S. have unrestricted access to all data. All authors approved the final article and take responsibility for its content.
Declaration of interests
The authors declare no competing interests.
Published: October 5, 2022
Footnotes
Supplemental information can be found online at https://doi.org/10.1016/j.medj.2022.09.010.
Supplemental information
Data and code availability
SARS-CoV-2 variants genomes have been deposited at GISAID and are publicly available as of the date of publication. Accession numbers are listed in the key resources table.
This study did not generate any new codes.
Any additional information required to reanalyze the data reported in this paper is available from the lead contact upon request.
References
- 1.Viana R., Moyo S., Amoako D.G., Tegally H., Scheepers C., Althaus C.L., Anyaneji U.J., Bester P.A., Boni M.F., Chand M., et al. Rapid epidemic expansion of the SARS-CoV-2 Omicron variant in southern Africa. Nature. 2022;603:679–686. doi: 10.1038/s41586-022-04411-y. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Murray C.J.L. COVID-19 will continue but the end of the pandemic is near. Lancet. 2022;399:417–419. doi: 10.1016/S0140-6736(22)00100-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Andrews N., Stowe J., Kirsebom F., Toffa S., Rickeard T., Gallagher E., Gower C., Kall M., Groves N., O’Connell A.-M., et al. Covid-19 vaccine effectiveness against the omicron (B.1.1.529) variant. N. Engl. J. Med. 2022;386:1532–1546. doi: 10.1056/NEJMoa2119451. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Abu-Raddad L.J., Chemaitelly H., Ayoub H.H., AlMukdad S., Yassine H.M., Al-Khatib H.A., Smatti M.K., Tang P., Hasan M.R., Coyle P., et al. Effect of mRNA vaccine boosters against SARS-CoV-2 omicron infection in Qatar. N. Engl. J. Med. 2022;386:1804–1816. doi: 10.1056/NEJMoa2200797. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Accorsi E.K., Britton A., Fleming-Dutra K.E., Smith Z.R., Shang N., Derado G., Miller J., Schrag S.J., Verani J.R. Association between 3 doses of mRNA COVID-19 vaccine and symptomatic infection caused by the SARS-CoV-2 omicron and Delta variants. JAMA. 2022;327:639–651. doi: 10.1001/jama.2022.0470. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Kuhlmann C., Mayer C.K., Claassen M., Maponga T., Burgers W.A., Keeton R., Riou C., Sutherland A.D., Suliman T., Shaw M.L., Preiser W. Breakthrough infections with SARS-CoV-2 omicron despite mRNA vaccine booster dose. Lancet. 2022;399:625–626. doi: 10.1016/S0140-6736(22)00090-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Cameroni E., Bowen J.E., Rosen L.E., Saliba C., Zepeda S.K., Culap K., Pinto D., VanBlargan L.A., De Marco A., di Iulio J., et al. Broadly neutralizing antibodies overcome SARS-CoV-2 Omicron antigenic shift. Nature. 2022;602:664–670. doi: 10.1038/s41586-021-04386-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Cele S., Jackson L., Khoury D.S., Khan K., Moyo-Gwete T., Tegally H., San J.E., Cromer D., Scheepers C., Amoako D.G., et al. Omicron extensively but incompletely escapes Pfizer BNT162b2 neutralization. Nature. 2022;602:654–656. doi: 10.1038/s41586-021-04387-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Carreño J.M., Alshammary H., Tcheou J., Singh G., Raskin A.J., Kawabata H., Sominsky L.A., Clark J.J., Adelsberg D.C., Bielak D.A., et al. Activity of convalescent and vaccine serum against SARS-CoV-2 Omicron. Nature. 2022;602:682–688. doi: 10.1038/s41586-022-04399-5. [DOI] [PubMed] [Google Scholar]
- 10.Planas D., Saunders N., Maes P., Guivel-Benhassine F., Planchais C., Buchrieser J., Bolland W.-H., Porrot F., Staropoli I., Lemoine F., et al. Considerable escape of SARS-CoV-2 Omicron to antibody neutralization. Nature. 2022;602:671–675. doi: 10.1038/s41586-021-04389-z. [DOI] [PubMed] [Google Scholar]
- 11.Garcia-Beltran W.F., St Denis K.J., Hoelzemer A., Lam E.C., Nitido A.D., Sheehan M.L., Berrios C., Ofoman O., Chang C.C., Hauser B.M., et al. mRNA-based COVID-19 vaccine boosters induce neutralizing immunity against SARS-CoV-2 Omicron variant. Cell. 2022;185:457–466.e4. doi: 10.1016/j.cell.2021.12.033. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Hoffmann M., Krüger N., Schulz S., Cossmann A., Rocha C., Kempf A., Nehlmeier I., Graichen L., Moldenhauer A.-S., Winkler M.S., et al. The Omicron variant is highly resistant against antibody-mediated neutralization: implications for control of the COVID-19 pandemic. Cell. 2022;185:447–456.e11. doi: 10.1016/j.cell.2021.12.032. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Dejnirattisai W., Huo J., Zhou D., Zahradník J., Supasa P., Liu C., Duyvesteyn H.M.E., Ginn H.M., Mentzer A.J., Tuekprakhon A., et al. SARS-CoV-2 Omicron-B.1.1.529 leads to widespread escape from neutralizing antibody responses. Cell. 2022;185:467–484.e15. doi: 10.1016/j.cell.2021.12.046. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Gruell H., Vanshylla K., Tober-Lau P., Hillus D., Schommers P., Lehmann C., Kurth F., Sander L.E., Klein F. mRNA booster immunization elicits potent neutralizing serum activity against the SARS-CoV-2 Omicron variant. Nat. Med. 2022;28:477–480. doi: 10.1038/s41591-021-01676-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Pérez-Then E., Lucas C., Monteiro V.S., Miric M., Brache V., Cochon L., Vogels C.B.F., Malik A.A., De la Cruz E., Jorge A., et al. Neutralizing antibodies against the SARS-CoV-2 Delta and Omicron variants following heterologous CoronaVac plus BNT162b2 booster vaccination. Nat. Med. 2022;28:481–485. doi: 10.1038/s41591-022-01705-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Schmidt F., Muecksch F., Weisblum Y., da Silva J., Bednarski E., Cho A., Wang Z., Gaebler C., Caskey M., Nussenzweig M.C., et al. Plasma neutralization of the SARS-CoV-2 omicron variant. N. Engl. J. Med. 2022;386:599–601. doi: 10.1056/NEJMc2119641. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Muecksch F., Wang Z., Cho A., Gaebler C., ben Tanfous T., DaSilva J., Bednarski E., Ramos V., Zong S., Johnson B., et al. Increased memory B cell potency and breadth after a SARS-CoV-2 mRNA boost. Nature. 2022;607:128–134. doi: 10.1038/s41586-022-04778-y. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Parums D.V. Editorial: world health organization (WHO) variants of concern lineages under monitoring (VOC-LUM) in response to the global spread of lineages and sublineages of omicron, or B.1.1.529, SARS-CoV-2. Med. Sci. Monit. 2022;28:e937676. doi: 10.12659/MSM.937676. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Tegally H., Moir M., Everatt J., Giovanetti M., Scheepers C., Wilkinson E., Subramoney K., Makatini Z., Moyo S., Amoako D.G., et al. Emergence of SARS-CoV-2 omicron lineages BA.4 and BA.5 in South Africa. Nat. Med. 2022;28:1785–1790. doi: 10.1038/s41591-022-01911-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Qu P., Faraone J., Evans J.P., Zou X., Zheng Y.-M., Carlin C., Bednash J.S., Lozanski G., Mallampalli R.K., Saif L.J., et al. Neutralization of the SARS-CoV-2 omicron BA.4/5 and BA.2.12.1 subvariants. N. Engl. J. Med. 2022;386:2526–2528. doi: 10.1056/NEJMc2206725. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Cao Y., Yisimayi A., Jian F., Song W., Xiao T., Wang L., Du S., Wang J., Li Q., Chen X., et al. BA.2.12.1, BA.4 and BA.5 escape antibodies elicited by Omicron infection. Nature. 2022;608:593–602. doi: 10.1038/s41586-022-04980-y. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Tuekprakhon A., Nutalai R., Dijokaite-Guraliuc A., Zhou D., Ginn H.M., Selvaraj M., Liu C., Mentzer A.J., Supasa P., Duyvesteyn H.M.E., et al. Antibody escape of SARS-CoV-2 Omicron BA.4 and BA.5 from vaccine and BA.1 serum. Cell. 2022;185:2422–2433.e13. doi: 10.1016/j.cell.2022.06.005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Hachmann N.P., Miller J., Collier A.-R.Y., Ventura J.D., Yu J., Rowe M., Bondzie E.A., Powers O., Surve N., Hall K., Barouch D.H. Neutralization escape by SARS-CoV-2 omicron subvariants BA.2.12.1, BA.4, and BA.5. N. Engl. J. Med. 2022;387:86–88. doi: 10.1056/NEJMc2206576. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Wang Q., Guo Y., Iketani S., Nair M.S., Li Z., Mohri H., Wang M., Yu J., Bowen A.D., Chang J.Y., et al. Antibody evasion by SARS-CoV-2 Omicron subvariants BA.2.12.1, BA.4, & BA.5. Nature. 2022;608:603–608. doi: 10.1038/s41586-022-05053-w. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Santé publique France Point épidémiologique COVID-19 du 10 février 2022. 2022. https://www.santepubliquefrance.fr/presse/2022/point-epidemiologique-covid-19-du-10-fevrier-2022-le-ralentissement-de-la-circulation-du-sars-cov-2-se-confirme-et-s-accompagne-d-une-baisse-des
- 26.Zhang Z., Mateus J., Coelho C.H., Dan J.M., Moderbacher C.R., Gálvez R.I., Cortes F.H., Grifoni A., Tarke A., Chang J., et al. Humoral and cellular immune memory to four COVID-19 vaccines. bioRxiv. 2022 doi: 10.1101/2022.03.18.484953. Preprint at. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Smith N., Goncalves P., Charbit B., Grzelak L., Beretta M., Planchais C., Bruel T., Rouilly V., Bondet V., Hadjadj J., et al. Distinct systemic and mucosal immune responses during acute SARS-CoV-2 infection. Nat. Immunol. 2021;22:1428–1439. doi: 10.1038/s41590-021-01028-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Wang Z., Lorenzi J.C.C., Muecksch F., Finkin S., Viant C., Gaebler C., Cipolla M., Hoffman H.-H., Oliveira T.Y., Oren D.A., et al. Enhanced SARS-CoV-2 neutralization by dimeric IgA. Sci. Transl. Med. 2021;13:eabf1555. doi: 10.1126/scitranslmed.abf1555. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Sterlin D., Mathian A., Miyara M., Mohr A., Anna F., Claër L., Quentric P., Fadlallah J., Devilliers H., Ghillani P., et al. IgA dominates the early neutralizing antibody response to SARS-CoV-2. Sci. Transl. Med. 2021;13:eabd2223. doi: 10.1126/scitranslmed.abd2223. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Malato J., Ribeiro R.M., Leite P.P., Casaca P., Fernandes E., Antunes C., Fonseca V.R., Gomes M.C., Graca L. Risk of BA.5 infection among persons exposed to previous SARS-CoV-2 variants. N. Engl. J. Med. 2022;387:953–954. doi: 10.1056/NEJMc2209479. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Hansen C.H., Friis N.U., Bager P., Stegger M., Fonager J., Fomsgaard A., Gram M.A., Christiansen L.E., Ethelberg S., Rebecca Legarth T., et al. Risk of reinfection, vaccine protection, and severity of infection with the BA.5 omicron subvariant: a Danish nation-wide population-based study. 2022. https://ssrn.com/abstract=4165630 Available at SSRN. [DOI] [PMC free article] [PubMed]
- 32.Hall V., Foulkes S., Insalata F., Kirwan P., Saei A., Atti A., Wellington E., Khawam J., Munro K., Cole M., et al. Protection against SARS-CoV-2 after covid-19 vaccination and previous infection. N. Engl. J. Med. 2022;386:1207–1220. doi: 10.1056/NEJMoa2118691. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Altarawneh H.N., Chemaitelly H., Ayoub H.H., Tang P., Hasan M.R., Yassine H.M., Al-Khatib H.A., Smatti M.K., Coyle P., Al-Kanaani Z., et al. Effects of previous infection and vaccination on symptomatic omicron infections. N. Engl. J. Med. 2022;387:21–34. doi: 10.1056/NEJMoa2203965. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Chemaitelly H., Nagelkerke N., Ayoub H.H., Coyle P., Tang P., Yassine H.M., Al-Khatib H.A., Smatti M.K., Hasan M.R., Al-Kanaani Z., et al. Duration of immune protection of SARS-CoV-2 natural infection against reinfection in Qatar. medRxiv. 2022 doi: 10.1101/2022.07.06.22277306. Preprint at. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Link-Gelles R., Levy M.E., Gaglani M., Irving S.A., Stockwell M., Dascomb K., DeSilva M.B., Reese S.E., Liao I.C., Ong T.C., et al. Effectiveness of 2, 3, and 4 COVID-19 mRNA vaccine doses among immunocompetent adults during periods when SARS-CoV-2 omicron BA.1 and BA.2/BA.2.12.1 sublineages predominated, December 2021–June 2022. MMWR Morb. Mortal. Wkly. Rep. 2022;71:931–939. doi: 10.15585/mmwr.mm7129e1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Trigueros M., Pradenas E., Palacín D., Muñoz-López F., Ávila-Nieto C., Trinité B., Bonet-Simó J.M., Isnard M., Moreno N., Marfil S., et al. Reduced humoral response 3 months following BNT162b2 vaccination in SARS-CoV-2 uninfected residents of long-term care facilities. Age Ageing. 2022;51:afac101. doi: 10.1093/ageing/afac101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Müller L., Andrée M., Moskorz W., Drexler I., Walotka L., Grothmann R., Ptok J., Hillebrandt J., Ritchie A., Rabl D., et al. Clinical infectious Diseases age-dependent immune response to the biontech/Pfizer BNT162b2 coronavirus disease 2019 vaccination. Clin. Infect. Dis. 2021;73:2065–2072. doi: 10.1093/cid/ciab381. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Collier D.A., Ferreira I.A.T.M., Kotagiri P., Datir R.P., Lim E.Y., Touizer E., Meng B., Abdullahi A., CITIID-NIHR BioResource COVID-19 Collaboration. Kingston N., et al. Age-related immune response heterogeneity to SARS-CoV-2 vaccine BNT162b2 the CITIID-NIHR BioResource COVID-19 Collaboration. Nature. 2021;596:417–422. doi: 10.1038/s41586-021-03739-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Planas D., Veyer D., Baidaliuk A., Staropoli I., Guivel-Benhassine F., Rajah M.M., Planchais C., Porrot F., Robillard N., Puech J., et al. Reduced sensitivity of SARS-CoV-2 variant Delta to antibody neutralization. Nature. 2021;596:276–280. doi: 10.1038/s41586-021-03777-9. [DOI] [PubMed] [Google Scholar]
- 40.Planas D., Bruel T., Grzelak L., Guivel-Benhassine F., Staropoli I., Porrot F., Planchais C., Buchrieser J., Rajah M.M., Bishop E., et al. Sensitivity of infectious SARS-CoV-2 B.1.1.7 and B.1.351 variants to neutralizing antibodies. Nat. Med. 2021;27:917–924. doi: 10.1038/s41591-021-01318-5. [DOI] [PubMed] [Google Scholar]
- 41.Grzelak L., Temmam S., Planchais C., Demeret C., Tondeur L., Huon C., Guivel-Benhassine F., Staropoli I., Chazal M., Dufloo J., et al. A comparison of four serological assays for detecting anti–SARS-CoV-2 antibodies in human serum samples from different populations. Sci. Transl. Med. 2020;12:eabc3103. doi: 10.1126/SCITRANSLMED.ABC3103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Grzelak L., Velay A., Madec Y., Gallais F., Staropoli I., Schmidt-Mutter C., Wendling M.-J., Meyer N., Planchais C., Rey D., et al. Sex differences in the evolution of neutralizing antibodies to SARS-CoV-2. J. Infect. Dis. 2021;224:983–988. doi: 10.1093/infdis/jiab127. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Hadjadj J., Yatim N., Barnabei L., Corneau A., Boussier J., Smith N., Péré H., Charbit B., Bondet V., Chenevier-Gobeaux C., et al. Impaired type I interferon activity and inflammatory responses in severe COVID-19 patients. Science. 2020;369:718–724. doi: 10.1126/science.abc6027. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Buchrieser J., Dufloo J., Hubert M., Monel B., Planas D., Rajah M.M., Planchais C., Porrot F., Guivel-Benhassine F., Van der Werf S., et al. Syncytia formation by SARS-CoV-2-infected cells. EMBO J. 2020;39:e106267. doi: 10.15252/embj.2020106267. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Bruel T., Hadjadj J., Maes P., Planas D., Seve A., Staropoli I., Guivel-Benhassine F., Porrot F., Bolland W.-H., Nguyen Y., et al. Serum neutralization of SARS-CoV-2 Omicron sublineages BA.1 and BA.2 in patients receiving monoclonal antibodies. Nat. Med. 2022;28:1297–1302. doi: 10.1038/s41591-022-01792-5. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Data Availability Statement
SARS-CoV-2 variants genomes have been deposited at GISAID and are publicly available as of the date of publication. Accession numbers are listed in the key resources table.
This study did not generate any new codes.
Any additional information required to reanalyze the data reported in this paper is available from the lead contact upon request.