Abstract
The cellular fate after infection with human coronaviruses (HCoVs) is typically death. Previous data suggest, however, that the transcriptional state of an individual cell may sometimes allow additional outcomes of infection. Here, to probe the range of interactions a permissive cell type can have with a HCoV, we perform a CRISPR activation screen with HCoV-229E. The screen identified the transcription factor ZBTB7A, which strongly promotes cell survival after infection. Rather than suppressing viral infection, ZBTB7A upregulation allows the virus to induce a persistent infection and homeostatic state with the cell. We also find that control of oxidative stress is a primary driver of cellular survival during HCoV-229E infection. These data illustrate that, in addition to the nature of the infecting virus and the type of cell that it encounters, the cellular gene expression profile prior to infection can affect the eventual fate.
Keywords: cellular fate, oxidative stress, persistent infection, CRISPRa
Graphical abstract
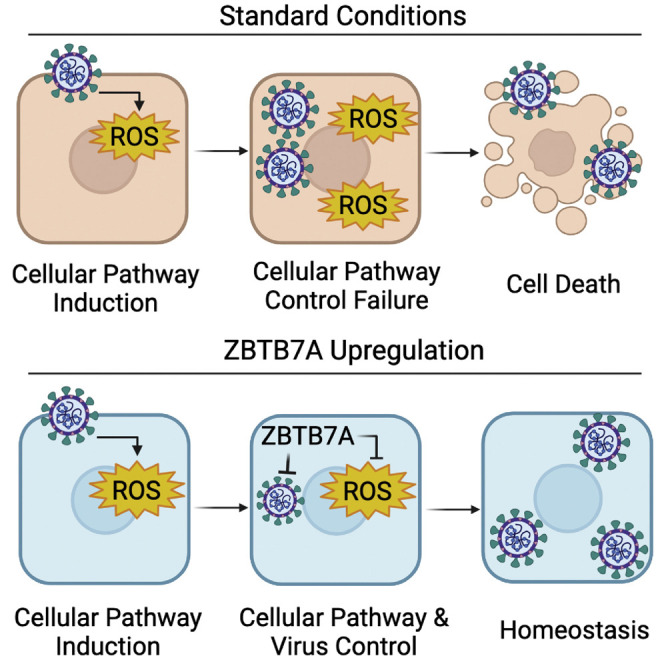
The fates of infected cells can affect viral pathogenesis. Zhu et al. show that the upregulation of ZBTB7A can convert a cytolytic human coronavirus infection to a persistent, homeostatic one. Continued work in this area may ultimately explain the broad range of clinical presentations associated with respiratory viral infections.
Introduction
Coronaviruses (CoVs) are a group of enveloped viruses with positive-stranded RNA genomes (V'Kovski et al., 2021). They are divided into four genera, α-, β-, γ-, and δ-CoV, which belong to order Nidovirales, family Coronaviridae, and subfamily Coronavirinae (The International Committee on Taxonomy of Viruses). Currently seven CoVs are known to infect humans. Four of the human CoVs, HCoV-229E, HCoV-OC43, HCoV-NL63, and HCoV-HKU1, are circulating seasonal pathogens and cause mild to moderate respiratory diseases. The three remaining HCoVs, severe acute respiratory syndrome (SARS)-CoV, Middle East respiratory syndrome (MERS)-CoV, and SARS-CoV-2, are highly pathogenic and pose a severe threat to public health (Fehr and Perlman, 2015; Zhou et al., 2020).
It has been reported that HCoV infections induce cell death, resulting in tissue damage (Lee et al., 2020; Leist et al., 2020; Mangalmurti and Hunter, 2020). These phenomena have been described in some detail for the more severe respiratory HCoVs that infect the respiratory tract in humans: SARS-CoV-2 infection triggers cell death in airway epithelial cells (Li et al., 2020; Zhu et al., 2020), and SARS-CoV infection results in denudation of lung epithelial cells (Nicholls et al., 2003). While the magnitude of tissue damage with the seasonal HCoVs is less, the same general cytopathic effects have been observed both during in vitro and in vivo infections (Jacomy et al., 2006; Mesel-Lemoine et al., 2012b; Milewska et al., 2014). There are, however, clinical manifestations of disease such as long-term asymptomatic shedders of virus (Agarwal et al., 2020; Avanzato et al., 2020) that suggest that not all infections induce rapid cell death, inflammation, and major tissue damage. In addition, at least with a murine coronavirus, some cells can resist virus-induced killing and intrinsically clear viral infection (Wheeler et al., 2017). These data together support the idea that the range of interactions between CoVs and the host cell may be broader than the exclusive killing of the infected cells. However, potential controllers of non-lytic death cell fates remain mostly undefined.
Genome-wide screening based on clustered regularly interspaced short palindromic repeats (CRISPR) is a powerful tool for the identification of host factors that affect viral infection (Shalem et al., 2015). Recently, multiple groups have performed both CRISPR gain-of-function and loss-of-function screens to identify essential genes and restriction factors for HCoV infections, with an emphasis on the host factors required for productive SARS-CoV-2 infection (Baggen et al., 2021; Biering et al., 2022; Daniloski et al., 2021; Danziger et al., 2022; Grodzki et al., 2022; Hoffmann et al., 2021; Israeli et al., 2022; Kratzel et al., 2021; Mac Kain et al., 2022; Rebendenne et al., 2021, 2022; Schneider et al., 2021; Sherman et al., 2022; Trimarco et al., 2021; Wang et al., 2021a, 2021b; Wei et al., 2021; Zhu et al., 2021, 2022). In order to define host factors that could alter infected cell fate, we decided to perform a CRISPR activation (CRISPRa) screen that would allow us to define host factors that by themselves are sufficient to alter CoV infection outcomes. CRISPRa is an approach based on a catalytically inactive Cas9 protein (dCas9) that recruits transcriptional activators to gene promoter regions by virtue of single guide RNA (sgRNA) targeting, leading transcriptional upregulation (Konermann et al., 2015). We hypothesized that, in addition to increasing our basic understanding of the range of interactions these viruses can have with their host cells, the identification of host factors regulating CoV-induced cell death might help predict disease susceptibilities or serve as potential therapeutic targets.
Using the normally highly cytopathic human seasonal HCoV-229E as a model contemporary CoV, we found that CRISPR activation screening almost exclusively enriched for the host transcription factor ZBTB7A. While upregulation of this factor dramatically limited HCoV-229E-induced cell death, further experiments revealed that ZBTB7A did not prevent infection but rather allowed the virus to induce homeostasis within the host cells in a quasi-persistent infection. To understand the mechanism by which ZBTB7A was allowing this phenotype, we performed RNA sequencing (RNA-seq) both before and during viral infection. While many cellular pathways were dysregulated by the upregulation of this transcription factor, we identified a number of oxidative stress-response genes induced by ZBTB7A. Further experimentation confirmed that control of oxidative damage during infection is a major contributor to cellular resistance of virus-induced killing. These data highlight the need for continued work understanding the true range of infection outcomes during authentic infection as well as how altering cellular fate may affect pathogenesis.
Results
A CRISPR activation screen identifies host factors that prevent HCoV-229E-induced cell killing
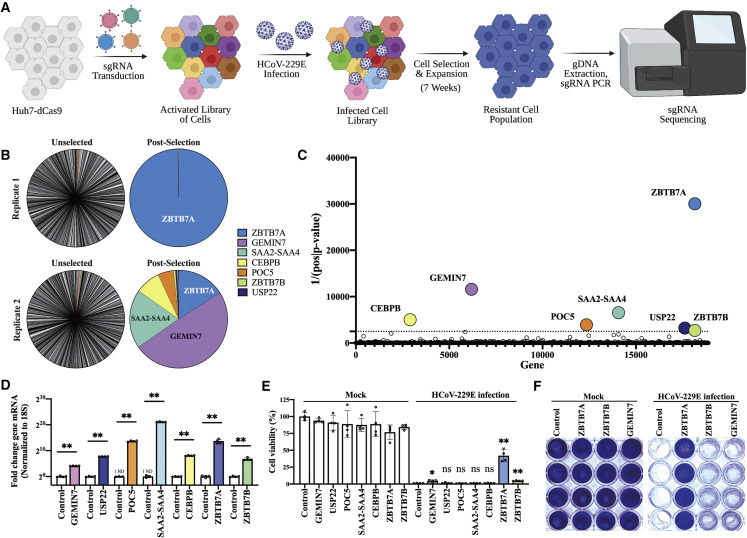
We were primarily interested in identifying the host factors that were sufficient for the avoidance of cellular death after HCoV infection. We therefore chose to perform a CRISPR activation screen with the expectation that we would enrich for general viral restriction factors as well as modulators of cell fate. For the virus, we chose the seasonal HCoV-229E as we have previously observed strong cytopathic effect in the human hepatoma Huh7 cell line (Trimarco et al., 2021); this allows us to apply a strong selection to the phenotypically altered population of cells. To perform the screen, catalytically inactive Cas9 (dCas9) was stably expressed in our Huh7 cell line and the Calabrese library (Sanson et al., 2018) of sgRNAs were introduced. After allowing sufficient time for gene upregulation, we infected the cells with HCoV-229E (Figure 1A).
Figure 1.
CRISPR activation screen identifies host factors that allow avoidance of HCoV-229E-induced cell death
(A) Overview of CRISPR activation screen.
(B) Distribution of sgRNA reads before (unselected) and after (post-selected) infection; the most abundant 500 sgRNAs after selection are shown.
(C) The top seven enriched genes based on the inverse of the p value.
(D) Quantification of mRNA by qRT-PCR from the indicated overexpression cell lines. n = 4 biological replicates. ND, not detected.
(E) Cell viability of transduced Huh7 cells after viral infection. Transduced cells were infected with HCoV-229E at 0.5 MOI. MTT assay was conducted 3 DPI. n = 4 biological replicates.
(F) Crystal violet staining of transduced Huh7 cells after infection. Control Huh7 cells expressing mCherry, ZBTB7A-Huh7 cells, ZBTB7B-Huh7 cells, and GEMIN7-Huh7 cells were infected with HCoV-229E at 0.5 MOI. The cells were stained at 5 DPI. n = 4 biological replicates.
All panels except (A)–(C) are representative of two independent experiments. For (D) and (E), p values were calculated by unpaired two-tailed Student’s t tests. ∗p < 0.05; ∗∗p < 0.001; ns, not significant. Data shown as mean ± SD.
Early after infection, we observed dramatic cellular killing and failed to enrich for resistant cell populations of any appreciable size; we therefore allowed the few remaining cells to co-incubate with the virus for approximately seven additional weeks. Toward the end of this time, resistant populations that were actively proliferating and expanding could be observed. These cells were then collected, the gDNA was extracted, and the sgRNAs present were identified by Illumina sequencing. Despite high diversity of sgRNAs present prior to infection, only a handful of sgRNAs were detected after viral selection, suggesting that the upregulation of most cellular genes is irrelevant for avoidance of virally induced cell killing. One sgRNA predicted to upregulate zinc finger and BTB domain containing 7A (ZBTB7A), however, was strongly enriched in both replicates and represented over 99% of the sgRNA reads in one of the samples (Figure 1B). After using the MAGeCK bioinformatic pipeline (Li et al., 2014) for statistical analysis, we selected the seven genes with the lowest p values for subsequent testing: ZBTB7A, GEMIN7, SAA2-SAA4, CEBPB, POC5, USP22, and ZBTB7B (Figure 1C; Table S1).
In order to avoid any potential CRISPR off-target effects during validation studies, we cloned the open reading frames (ORFs) for these seven genes and stably introduced them into Huh7 cells. Quantitative reverse transcription PCR (qRT-PCR) assays showed that the mRNA levels of all candidate genes were increased compared with the control Huh7 cells expressing mCherry (Figure 1D). These cell lines were then infected with HCoV-229E and cellular viability was observed and quantified via a 3-(4,5-dimethylthiazol-2-yl)-2,5-diphenyl tetrazolium bromide (MTT) assay or crystal violet staining. The overexpression of three genes, ZBTB7A, ZBTB7B, and GEMIN7, improved cell viability during infection; however, the effect was by far the most dramatic after ZBTB7A upregulation (Figures 1E, 1F, and S1).
ZBTB7A overexpression allows a quasi-persistent infection of Huh7 cells
As we were unable to find previous reports of a role for ZBTB7A during viral infection, we decided to further characterize how this protein was protecting cells from virus-induced killing. First, we expanded upon our qRT-PCR data and confirmed ZBTB7A overexpression at the protein level (Figure 2A). We next wanted to define whether the inhibition of cell killing was by restricting initial infection or preventing infected cell death. Immunofluorescence detection of the viral N protein after infection revealed that ZBTB7A upregulation only modestly reduced HCoV-229E infection (Figure 2B). Further, viral RNA levels and infectious viral titer from ZBTB7A-Huh7 cells were similarly only slightly, but still significantly, reduced (Figures 2C and 2D). Similar experiments at different temperatures and over a time course revealed similar trends, with a reduction in infection observable early after infection (Figures S2 and S3). To rule out cell-type-specific effects of ZBTB7A, human lung fibroblasts (MRC-5) and alveolar basal epithelial (A549) cells were transduced with lentivirus expressing ZBTB7A. HCoV-229E infection was again only modestly restricted, and ZBTB7A preserved most of the MRC-5 cells on the plate (Figure S4). Infection was similarly restricted in A549 cells, but, as these cells do not normally undergo appreciable lytic death after HCoV-229E infection, we were unable to measure that phenotype (Figure S5). These results together suggested that the effects of ZBTB7A upregulation are not restricted to Huh7 cells and that the cell-protective effects after HCoV-229E infection are not solely attributable to inhibition of viral infection.
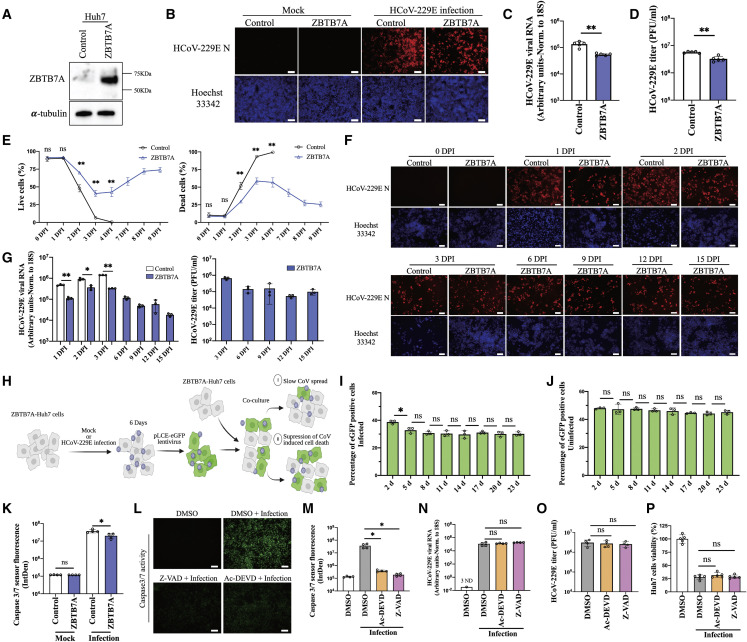
Figure 2.
ZBTB7A overexpression allows a quasi-persistent HCoV-229E infection
(A) ZBTB7A protein expression in transduced Huh7 cells.
(B) HCoV-229E N staining by immunofluorescence assay (IFA). Control-Huh7 cells and ZBTB7A-Huh7 cells were infected with HCoV-229E at 0.5 MOI and collected for immunofluorescence staining at 30 h post infection (hpi).
(C) Viral RNA detection by qRT-PCR. Control-Huh7 cells and ZBTB7A-Huh7 cells were infected as described in (B). n = 5 biological replicates.
(D) Viral titer by plaque assay. Control-Huh7 cells and ZBTB7A-Huh7 cells were infected as described in (B). n = 5 biological replicates.
(E) Proportion of live and dead cells during HCoV-229E infection. Control-Huh7 cells and ZBTB7A-Huh7 cells were infected at 1 MOI. Live and dead cells were stained and analyzed by flow cytometry at the indicated time points. n = 4 biological replicates. p values were calculated by two-way analysis of variance (ANOVA).
(F) HCoV-229E N staining during long-term infection. Control-Huh7 cells and ZBTB7A-Huh7 cells were infected at 1 MOI. HCoV-229E N was stained by IFA at the indicated time points.
(G) Viral RNA and titer during long-term infection. Left, Huh7 cells were infected at 1 MOI and viral RNA in cell lysate was detected by qRT-PCR. N = 3 biological replicates. Right, viral titer in supernatant from ZBTB7A-Huh7 cells was detected by plaque assay. n = 3 biological replicates.
(H) Overview of the co-culture of mock-infected and infected ZBTB7A-Huh7 cells. ZBTB7A-Huh7 cells were infected at 1 MOI. eGFP-encoding lentiviruses were then used to transduce the mock-infected cells or infected cells at 6 DPI. These cells were then co-cultured with normal ZBTB7A-Huh7 cells.
(I) Percentage of eGFP-positive cells with infection during co-culture. The ZBTB7A-Huh7 cells were treated as described in (H) and the percentage of eGFP-positive cells was analyzed by flow cytometry at the indicated time points. n = 3 biological replicates.
(J) Percentage of eGFP-positive cells without infection during co-culture. The ZBTB7A-Huh7 cells were treated as described in (H) and the percentage of eGFP-positive cells was analyzed by flow cytometry at the indicated time points. n = 3 biological replicates.
(K) Caspase 3/7 activity assay during infection. ZBTB7A-Huh7 cells and control-Huh7 cells were infected with HCoV-229E at 0.5 MOI and analyzed at 2 DPI. n = 4 biological replicates.
(L) Caspase 3/7 activity assay after apoptosis-related inhibitor treatment. Huh7 cells were infected with HCoV-229E at 0.5 MOI and treated with 30 μM Z-VAD or 30 μM Ac-DEVD at 30 hpi. Samples were analyzed at 2 DPI. Images representative of n = 4 biological replicates.
(M) Fluorescence intensity of caspase 3/7 activity calculated from the experiment shown in (L) via Image J. n = 4 biological replicates.
(N) Viral RNA detection after caspase inhibitor treatment. Huh7 cells were infected and treated as described for (L). Cells were collected at 2 DPI for qRT-PCR. n = 4 biological replicates. ND, not detected.
(O) Viral titer after caspase inhibitor treatment. Huh7 cells were infected and treated as described in (L). n = 4 biological replicates.
(P) Cell viability after caspase inhibitor treatment. Huh7 cells were infected and treated as described in (L). MTT assays were conducted at 3 DPI. N = 5 biological replicates. All panels are representative of two independent experiments. Unless otherwise noted, p values were calculated by unpaired two-tailed Student’s t tests.
For all panels, ∗p < 0.05; ∗∗p < 0.001; ns, not significant. Scale bars, 100 μm. Data shown as mean ± SD.
Due to the limited direct antiviral effects of ZBTB7A overexpression, we hypothesized that the protein may instead be preventing infected cell death. We therefore conducted a long-term culture of HCoV-229E-infected cells and monitored cell death via flow cytometry. Indeed, we found that, while there was an initial dip in the percentage of live cells with ZBTB7A overexpression, cellular viability returned almost to pre-infection levels within a week post infection in contrast to the control cells, which were all killed by 4 days post infection (DPI) (Figures 2E and S6). As these data could be explained by cell tolerance of infection or rapid cell intrinsic clearance of the virus, we next repeated the experiment but performed immunofluorescence microscopy for the viral N protein. Strikingly, we found that viral infection was continuously detected at similar levels from 1 to 15 DPI (Figure 2F). Intracelluar viral RNA and infectious virus in the supernatant of ZBTB7A-Huh7 cells also remained high across the entire time course (within ∼10-fold), although they were reduced relative to control at early time points (Figure 2G).
While the virus was clearly maintained in the ZBTB7A overexpression cultures, we wanted to distinguish between viral “persistence” in infected cells and consistent low-level spread of virus followed by cellular killing. We therefore designed a co-culture experiment where ZBTB7A-overexpressing cells infected with HCoV-229E were transduced with lentivirus expressing eGFP, a reporter protein that did not affect HCoV-229E-induced cytopathic effect (Figure S7). These infected fluorescent cells (or mock-infected fluorescent control cells) were then mixed with non-fluorescent, mock-infected ZBTB7A-overexpression cells at a 1:1 ratio and co-cultured for 23 days. If the virus was killing infected cells and spreading to new cells in the culture, we would expect the eGFP-positive cells to be eliminated from the cultures over time (Figure 2H). The results showed that, while eGFP-positive cell numbers were slightly reduced early after co-culture, 8 days later, the proportion of eGFP-positive cells then mostly stabilized (Figure 2I). The proportion of eGFP-positive cells in uninfected control similarly displayed no significant change over the time course (Figure 2J). Finally, to demonstrate that infected cells can actively divide, we performed an 5-ethynyl-2’-deoxyuridine (EdU)-based proliferation assay and could detect label in cells positive for viral protein (Figure S8). These data together support a model wherein ZBTB7A does not prevent or allow clearance of infection but at least partially allows the infected cells to better tolerate viral replication long term.
Our next question was whether this phenotype could simply be the avoidance of apoptosis, a cell death pathway known to be induced by CoV infection (Chu et al., 2016; Li et al., 2020). We first asked whether HCoV-229E infection activates the effector caspase proteins of the apoptosis pathway and whether ZBTB7A would affect that activation. Viral infection did indeed activate caspase 3/7, and ZBTB7A overexpression modestly but significantly reduced that activity (Figure 2K). To directly test whether apoptosis suppression could recapitulate the ZBTB7A phenotype, we treated HCoV-229E-infected cells with the caspase inhibitors Ac-DEVD or Z-VAD and infected with HCoV-229E. While caspase 3/7 activity was suppressed by drug treatment (Figures 2L and 2M), we did not detect an effect on either viral infection or cell survival in Huh7 cells (Figures 2N–2P). Thus, while we could not formally rule out a minor role for inhibition of apoptosis in the ZBTB7A-mediated avoidance of viral killing, it did not appear to be the major driver of the phenotype.
ZBTB7A modulates gene pathways related to control of oxidative stress
As a nuclear transcription factor, ZBTB7A has been reported to regulate a number of cellular pathways (Lunardi et al., 2013; Pessler and Hernandez, 2003; Pittol et al., 2018). To identify genes differentially regulated by ZBTB7A that may explain the persistent infection phenotype, we compared gene expression between control and ZBTB7A-overexpression cells with and without HCoV-229E infection using RNA-seq (Figure 3A and Table S2). We detected a slight reduction in viral RNA levels between control and ZBTB7A cells, consistent with our previous findings of a minor effect of ZBTB7A overexpression on viral replication (Figure 3B). Turning our attention to the host transcriptional profile, we compared mock or virally infected-control and ZBTB7A-overexpression cells and observed hundreds of genes differentially regulated by ZBTB7A (Tables S3 and S4). Gene Ontology analysis of the differentially regulated genes revealed that diverse metabolic processes were affected (Tables S5 and S6). When considering ZBTB7A-regulated transcriptional changes with an eye to understanding the viral homeostasis phenotype, we found that oxidative-stress-related genes and their associated pathways were upregulated in both mock-infected and infected samples (Figures 3C and 3D). Oxidative stress is frequently linked to inflammation and cell death processes (Imai et al., 2008; Xu et al., 2019), but not necessarily through the classical apoptosis pathway (Fiers et al., 1999; Gudipaty et al., 2018; Tait et al., 2014). As some of the oxidative-stress-response-related genes upregulated by ZBTB7A, such as AKR1C3 and APOA4, can act as general controllers of the oxidative stress response (Qin et al., 1998; Xiong et al., 2014), we decided to focus on the control of oxidative damage as a potential explanation for our HCoV-229E infection phenotype.
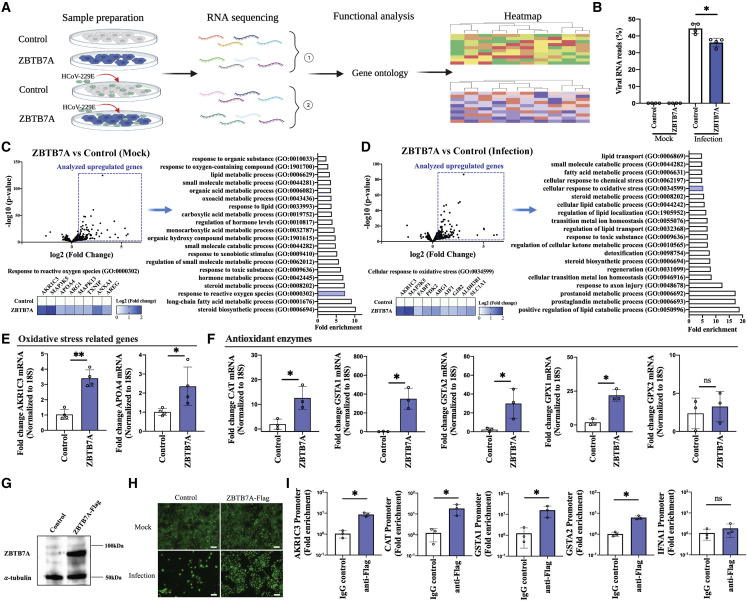
Figure 3.
RNA sequencing identifies oxidative-stress-related genes regulated by ZBTB7A
(A) Overview of RNA sequencing strategy. ZBTB7A-Huh7 cells and control-Huh7 cells were mock or infected with HCoV-229E (MOI = 1). Total RNA was collected at 1 DPI and prepared for sequencing. n = 4 biological replicates.
(B) Percentage of mapped viral RNA reads from the RNA sequencing data.
(C) Comparison of upregulated genes between mock control-Huh7 and mock ZBTB7A-Huh7 cells. Volcano plot of RNA sequencing from mock control-Huh7 cells and ZBTB7A-Huh7 cells. Gene Ontology (GO) of the upregulated genes (log2fold change >0.7 and −log10p value >3) was conducted and top 20 GO terms were plotted (−log10p value >4). The specific genes related to ROS are presented by heatmap.
(D) Comparison of upregulated genes between infected-control-Huh7 cells and infected-ZBTB7A-Huh7 cells. Volcano plot of RNA sequencing from infected-control-Huh7 cells and infected-ZBTB7A-Huh7 cells. GO of the upregulated genes (log2fold change >0.5 and −log10p value >3) was conducted and top 20 GO terms in upregulated genes were plotted (−log10p value >4). The specific genes related to oxidative stress are presented by heatmap.
(E) RNA levels of oxidative response inhibition-related genes were analyzed by qRT-PCR after infection. Huh7 cells were infected with HCoV-229E at 1 MOI and collected 1 DPI. n = 4 biological replicates.
(F) RNA levels of antioxidative enzymes were analyzed by qRT-PCR after infection. Huh7 cells were treated as described in (E). N = 3 biological replicates.
(G) ZBTB7A-FLAG expression (and tubulin loading control) from transduced Huh7 cells.
(H) Survival of cells during HCoV-229E infection. ZBTB7A-FLAG-Huh7 cells and control-Huh7 cells were infected with HCoV-229E (MOI = 1) and analyzed at 3 DPI. Scale bar, 100 μm.
(I) Promoter binding of ZBTB7A. ZBTB7A-FLAG-Huh7 cells and control-Huh7 cells were infected with HCoV-229E (MOI = 1). ChIP was conducted at 1 DPI and genomic DNA was analyzed by qPCR. (E–I) Representative of two independent experiments. For all panels, ∗p < 0.05; ∗∗p < 0.001; ns, not significant. For all panels, p values were calculated by unpaired two-tailed Student’s t tests. Data shown as mean ± SD.
First, we confirmed, via qRT-PCR, upregulation of the expected oxidative-response-related genes predicted by RNA-seq (Figure 3E). Next, we examined the expression of a panel of more canonical antioxidant enzymes and found that some, but not all, were also upregulated when ZBTB7A was overexpressed (Figure 3F). To verify whether ZBTB7A regulates oxidative-response-related genes by direct promoter binding, we constructed a ZBTB7A-FLAG overexpression Huh7 cell line (Figure 3G). As the FLAG-tag fusion at the N terminus did not affect the cell survival during the infection phenotype (Figure 3H), we used this cell line to perform chromatin immunoprecipitation (ChIP) experiments. Previous research has shown that ZBTB7A functions as a widespread promoter-associated protein and can bind to thousands of genomic sites (Han et al., 2019; Pittol et al., 2018). Perhaps unexpectedly, we could find our tagged and overexpressed ZBTB7A enriched at promoters of a number of the upregulated oxidative-response-related genes; however, we did observe some specificity as the promoter for IFNA1 did not show a similar enrichment (Figure 3I). Thus, while not ruling out indirect effects on gene transcription, these data are consistent with a direct role for ZBTB7A in promoting the transcription of antioxidant genes.
Control of oxidative stress contributes to cellular survival during HCoV-229E infection
We next tested whether the gene expression changes mediated by ZBTB7A overexpression would affect biological control of oxidative damage. Indeed, treatment of control or ZBTB7A-overexpression Huh7 cells with hydrogen peroxide (H2O2) revealed an enhanced tolerance to oxidative damage when ZBTB7A was present (Figures 4A and 4B). Next, we tested whether HCoV-229E infection induced appreciable reactive oxygen species (ROS) in our experimental system. Staining for ROS via fluorogenic probe in the presence or absence of ZBTB7A revealed ROS were clearly induced by viral infection and dramatically suppressed by ZBTB7A (Figures 4C, 4D, and S9). This induction of ROS is consistent with reports from other CoV infection systems; SARS-CoV-2 causes pulmonary diseases associated with oxidative stress (Sawalha et al., 2020), and an ROS imbalance has been reported in SARS-CoV-infected mice (Nicholls et al., 2003).
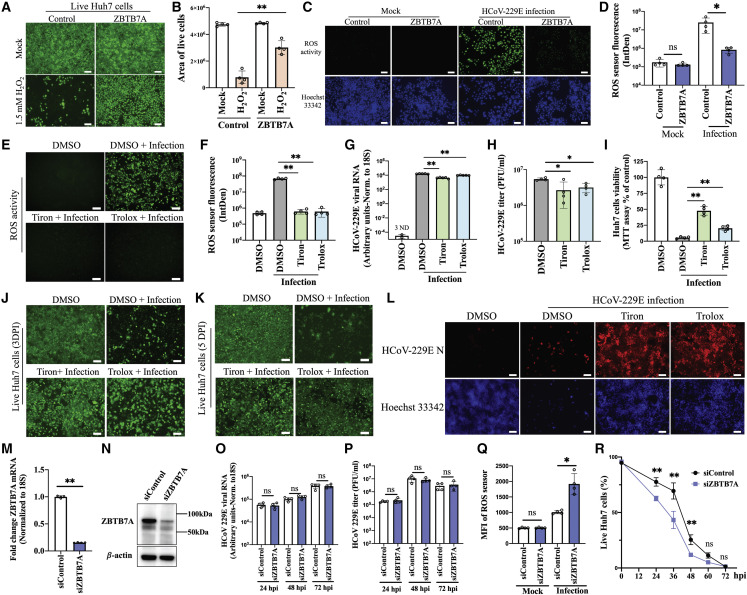
Figure 4.
ZBTB7A regulation of ROS-controlling factors contributes to cell tolerance of HCoV-229E
(A) Huh7 cell viability after H2O2 treatment. Control-Huh7 cells and ZBTB7A-Huh7 cells were incubated with 1.5 mM H2O2. After 24 h, live cells were stained.
(B) The area of live cells from (A) was calculated via Image J analysis. n = 4 images from two independent experiments.
(C) ROS activity during infection. ZBTB7A-Huh7 cells and control-Huh7 cells were infected with HCoV-229E at 0.5 MOI. ROS were detected via CellROX and imaged at 2 DPI.
(D) Fluorescence intensity of the ROS sensor was calculated from (C) by Image J. n = 4 biological replicates.
(E) ROS assay after superoxide scavenger treatment. Huh7 cells were infected with HCoV-229E at 0.5 MOI. 10 mM tiron or 200 μM trolox were added at 2 hpi and ROS sensor fluorescence was imaged at 2 DPI.
(F) Fluorescence intensity of the ROS sensor calculated from (E) by Image J. n = 4 biological replicates.
(G) Viral RNA detection after superoxide scavenger treatment. Huh7 cells were infected and treated as described in (E). Cells were collected at 2 DPI for qRT-PCR. n = 5 biological replicates. ND, not detected.
(H) Viral titer after superoxide scavenger treatment. Huh7 cells was treated as described in (E). Cells were collected at 2 DPI. n = 4 biological replicates.
(I) Cell viability after superoxide scavenger treatment. Huh7 cells were infected and treated as described in (E) and MTT assays were conducted at 3 DPI. n = 4 biological replicates.
(J and K) Survival of Huh7 cells after superoxide scavenger treatment. Huh7 cells were infected and treated as described in (E). The surviving cells were stained at 3 DPI (J) and 5 DPI (K).
(L) HCoV-229E N staining in surviving cells. Huh7 cells were infected and treated as described in (E). HCoV-229E N was stained in surviving cells at 5 DPI.
(M and N) ZBTB7A knockdown efficiency. Huh7 cells were transfected with ZBTB7A siRNA or negative control siRNA. After 48 h, samples were collected for qRT-PCR (n = 4 biological replicates) (M) and western blot (N).
(O) Viral RNA detection via qRT-PCR. Huh7 cells were transfected with ZBTB7A siRNA or control siRNA and infected with HCoV-229E at 0.5 MOI after 24 h. Viral RNA was measured via qRT-PCR at the indicated time points. n = 4 biological replicates.
(P) Viral titer assessed via plaque assay. Huh7 cells were treated as described in (O). n = 4 biological replicates.
(Q) ROS sensor fluorescence after ZBTB7A knockdown. Huh7 cells treated as described in (O). Samples were stained at 2 DPI and analyzed by flow cytometry. n = 4 biological replicates.
(R) Proportion of live cells after ZBTB7A knockdown. Huh7 cells treated as described in (O). Dead cells were stained at the indicated time points and analyzed by flow cytometry. n = 4 biological replicates. p values were calculated by two-way ANOVA.
Unless otherwise indicated, all panels are representative of two independent experiments. For all panels, p values were calculated by unpaired two-tailed Student’s t tests. ∗p < 0.05; ∗∗p < 0.001; ns, not significant. Scale bar, 100 μm. Data shown as mean ± SD.
To determine whether increased control of ROS was sufficient to prevent CoV-induced cell death, we utilized the superoxide scavengers tiron and trolox. As expected, tiron or trolox treatment both significantly decreased ROS activity during HCoV-229E infection (Figures 4E and 4F). While viral replication was only slightly affected by tiron or trolox treatment (Figures 4G and 4H), MTT assays and live cell imaging revealed that both treatments promoted cellular survival during infection at multiple time points (Figures 4I–4K). Staining for the HCoV-229E N protein revealed that, after tiron and trolox treatment, the live cells were also predominantly positive for viral antigen (Figure 4L). We repeated experiments in MRC-5 cells and again found good concordance with the Huh7 experiments (Figure S10). Thus, artificial suppression of ROS phenocopies ZBTB7A overexpression during HCoV-229E infection. Finally, as the previous experiments were done in the context of ZBTB7A overexpression, we tested the effects of endogenous levels of ZBTB7A. Huh7 cells were transfected with siRNA to reduce ZBTB7A transcript and protein levels (Figures 4M and 4N). While knockdown of endogenous ZBTB7A did not affect HCoV-229E RNA levels or release of infectious particles, it promoted ROS activity and cell death after infection (Figures 4O–4R). These data together suggest that, while ZBTB7A activity is important for many cellular pathways, its ability to facilitate control of ROS signaling drives the uncoupling of HCoV-229E infection and cell death.
Discussion
To fully understand the molecular basis of viral pathogenesis, a complete understanding of the potential outcomes of infection is necessary. In this study, we wanted to identify the host factors that could modulate CoV-induced cell death using a CRISPR activation screen. We identified that high levels of the host gene ZBTB7A could allow the establishment of a quasi-homeostasis between the virus and the infected cell, at least after HCoV-229E infection. Through RNA-seq and subsequent mechanistic analysis, we identified a number of ZBTB7A-regulated host pathways, with control of oxidative stress being particularly important for modulating host cell fate. While previous work with ZBTB7A has been primarily in the field of cancer biology (Jeon et al., 2008; Wang et al., 2013) and no links between ZBTB7A and viruses have previously been reported, it is clear that this host factor can alter the outcomes of infection.
While we have shown that control of ROS contributes to the ability of a cell to tolerate viral replication, how the full range of ZBTB7A-induced changes contributes to the viral/host homeostasis phenotypes remains incompletely answered. For example, ZBTB7A likely has differential effects on the various cell death pathways. We could not recapitulate the HCoV-229E cell survival phenotype with caspase inhibitors, suggesting that the virally/ROS-induced death pathway is at least partially caspase independent. This is consistent with previous reports showing a variety of cell death pathways are induced by HCoV infections (Lee et al., 2020; Mesel-Lemoine et al., 2012a); the relative contributions of different cell death pathways (and associated effects of ZBTB7A) during HCoV infections, however, remain an important area of future study. Additionally, while ROS inhibitor treatment increased cell survival, its effect did not fully recapitulate the ZBTB7A overexpression phenotype. We therefore believe it is likely that a number of alterations to cell physiology (in addition to the pathways that we tested) all contribute to the ability of the cell to harbor a normally lytic virus long term. By far, most of the pathways identified in our Gene Ontology analysis were related to metabolic alterations of the cell. We suspect that, in addition to avoiding cell death pathways and preventing oxidative damage, overall metabolic reprogramming of cells is likely required to balance the requirements of both the host and the virus for various metabolites.
Along those lines, we focused some efforts on understanding which ZBTB7A pathways may mediate the early suppression of infection. We found the alcohol dehydrogenase 1 (ADH1) gene family, including ADH1A, ADH1B, and ADH1C, were induced by ZBTB7A after viral infection (Table S2). It has been reported that expression of these genes is sufficient to complete the retinoic acid biosynthesis pathway in Huh7 cells and, interestingly, retinoic acid signaling has been reported to have antiviral activity (Cho et al., 2016; Liang et al., 2020; Trottier et al., 2008). Whether ZBTB7A does indeed mediate activation of retinoic acid metabolism and thereby contribute to the restriction of viral infection is an area that requires future study.
Perhaps the most important remaining question raised by this study is whether differences in physiological levels of ZBTB7A across different tissues or cell types truly modulate cell fate during a natural infection. ZBTB7A is expressed in virtually all tissues, with particularly high expression in the brain, lung, gastrointestinal (GI) tract, and lymphoid tissues (Thul et al., 2017; Uhlen et al., 2015), and therefore could be affecting cell fate during respiratory HCoV-229E infection. Whether or not there is any specific induction of the protein in cells after viral infection or innate immune activation remains an open question. It also remains unclear whether ZBTB7A levels mediate a dose-dependent effect on cellular survival, or if a threshold of expression is required for a binary survival versus the normal death fate. We suspect that the general activities of the protein normally establish a cellular environment that contributes to the amount of time that a cell can tolerate infection prior to death. However, it remains possible that cells that rapidly die or tolerate the virus for a disproportionately long time may do so in part because of fluctuations in the expression levels of ZBTB7A. The answers to these questions, as well as whether or not cellular fates after infection with other HCoVs are affected by ZBTB7A, will require additional investigation.
In summary, we have identified a previously unappreciated cellular fate after HCoV-229E infection, the establishment of a quasi-homeostatic state with the host cell. This fate alteration is mediated by ZBTB7A, a host transcription factor, which broadly alters the host cell physiology but likely prevents cell death in no small part due to control of oxidative damage during infection. Future work understanding this, and similar processes, may ultimately allow for a mechanistic understanding of certain clinical manifestations of respiratory virus disease, such as asymptomatic long-term viral shedders. Such insights will undoubtedly not only increase our understanding of viral pathogenesis but may also identify next-generation intervention strategies.
Limitations of the study
In the current study, we reveal a role for ZBTB7A in virus-host interactions. Although the control of ROS regulated by ZBTB7A was vital for cell survival during infection, the extent that other effects mediated by ZBTB7A expression (e.g., metabolic reprogramming) are required to establish a virus/host balance is still unclear. Additionally, our study was mainly focused on HCoV-229E infection and was conducted in immortalized cell culture systems. Further study will be required to understand whether other coronaviruses are similarly affected and whether or not the phenotypes extend in vivo.
STAR★Methods
Key resources table
| REAGENT or RESOURCE | SOURCE | IDENTIFIER |
|---|---|---|
| Antibodies | ||
| anti-ZBTB7A | Santa Cruz | Cat# sc-33683; RRID:AB_668999 |
| anti-ZBTB7A | R&D | Cat# mab3496; RRID: AB_10584995 |
| anti-alpha tubulin | Abcam | Cat# ab179484; RRID:AB_2890906 |
| anti-alpha actin | Santa Cruz | Cat# sc-47778; RRID:AB_626632 |
| anti-HCoV-229E N | Eurofins Technologies | Cat# M.30.HCo.I1E8 |
| anti-Flag | Sigma-Aldrich | Cat# F1804; RRID:AB_262044 |
| anti-GFP | Cell Signaling Technology | Cat# 2955; RRID:AB_1196614 |
| anti-rabbit-HRP | Thermo Fisher Scientific | Cat# A16104; RRID:AB_2534776 |
| anti-armenian hamster-HRP | Novus Biologicals | Cat# NB100-2066; RRID:AB_10000771 |
| Alexa Fluor 594 Goat Anti-Mouse | Thermo Fisher Scientific | Cat# A11005; RRID:AB_2534073 |
| Bacterial and virus strains | ||
| Human coronavirus 229E | ATCC | VR-740 strain |
| Chemicals, peptides, and recombinant proteins | ||
| Puromycin | Sigma-Aldrich | Cat# P5412 |
| Blasticidin S | Sigma-Aldrich | Cat# SBR00022 |
| Poly-L-lysine solution | Sigma-Aldrich | Cat# P8920 |
| Hoechst 33342 | Invitrogen | Cat# H3570 |
| Universal SYBR® Green Supermix | Bio-Rad | Cat# 1725274 |
| MTT solution | Abcam | Cat# ab211091 |
| Caspase 3/7 Green Detection Reagent | Thermo Fisher Scientific | Cat# C10423 |
| CellROX Green Reagent | Thermo Fisher Scientific | Cat# C10444 |
| Z-VAD-FMK | R&D | Cat# FMK001 |
| Ac-DEVD-CHO | Selleck | Cat# S7901 |
| Tiron | Abcam | Cat# ab146234 |
| Trolox | Sigma-Aldrich | Cat# 53188-07-1 |
| Hydrogen peroxide solution | Sigma-Aldrich | Cat# 7722-84-1 |
| Critical commercial assays | ||
| QIAamp DNA Blood Maxi Kit | QIAGEN | Cat# 51192 |
| NEB Monarch total RNA miniprep kit | NEB | Cat# T2010 |
| SuperScript™ III One-Step RT-PCR | Thermo Fisher Scientific | Cat# 12574026 |
| HiFi DNA assembly | NEB | Cat# M5520AA |
| One-Step Superscript™ qRT-PCR | Thermo Fisher Scientific | Cat# 11781200 |
| EdU Assay Kit | Abcam | Cat# ab219801 |
| LIVE/DEAD Fixable Green Dead Cell Stain Kit | Thermo Fisher Scientific | Cat# L34969 |
| LIVE/DEAD Cell Imaging Kit | Thermo Fisher Scientific | Cat# R37601 |
| Poly(A) messenger RNA Magnetic Isolation Module | NEB | Cat# E74905 |
| Ultra II RNA Library Prep Kit | NEB | Cat# E7770S |
| Pierce™ Immunoprecipitation, Magnetic ChIP Kit | Thermo Fisher Scientific | Cat# 26157 |
| Deposited data | ||
| CRISPR sequencing data | This paper | GEO: GSE197882 |
| RNA sequencing data | This paper | GEO: GSE197644 |
| Experimental models: Cell lines | ||
| Huh7 cells | Collaborating Laboratory, Duke University (US) | N/A |
| HEK-293T cells | ATCC | CRL-3216 |
| MRC5 cells | ATCC | CCL-171 |
| A549 cells | ATCC | CCL-185 |
| Oligonucleotides | ||
| Negative control siRNA | Thermo Fisher Scientific | Cat# AM4611 |
| siRNA targeting ZBTB7A | Thermo Fisher Scientific | Cat# AM16708 |
| NEBNext Multiplex Oligos for Illumina | NEB | Cat# E7335S |
| Primers and Probes targeting HCoV-229E RNA | Thermo Fisher Scientific | Vi06439671_s1 |
| Primers and Probes targeting 18S rRNA | Thermo Fisher Scientific | 4319413E |
| Recombinant DNA | ||
| CRISPR activation sgRNA library | Addgene | Cat# 92379 |
| pLEX-MCS | Collaborating Laboratory, Duke University (US) | N/A |
| pMD2.G | Collaborating Laboratory, Duke University (US) | N/A |
| pCMVR8.74 | Collaborating Laboratory, Duke University (US) | N/A |
| pLEX-GEMIN7 | This paper | N/A |
| pLEX-SAA2-SAA4 | This paper | N/A |
| pLEX-CEBPB | This paper | N/A |
| pLEX-POC5 | This paper | N/A |
| pLEX-USP22 | This paper | N/A |
| pLEX-ZBTB7A | This paper | N/A |
| pLEX-ZBTB7B | This paper | N/A |
| pLEX-ZBTB7A-Flag | This paper | N/A |
| Software and algorithms | ||
| ImageJ | NIH | https://imagej.nih.gov/ij/ |
| Illustrator | Adobe | N/A |
| FlowJo | BD | https://www.flowjo.com/solutions/flowjo |
| GraphPad Prism | GraphPad Software, Inc | N/A |
Resource availability
Lead contact
Further information and requests for resources and reagents should be directed to and will be fulfilled by the lead contact, Nicholas S. Heaton (nicholas.heaton@duke.edu).
Materials availability
All plasmids and cell lines generated in this study are available from the lead contact upon request.
Experimental model and subject details
The MRC-5, A549 and HEK-293T cell lines were obtained from American Type Culture Collection (ATCC). MRC-5 cells were cultured with minimum essential medium (MEM) containing 10% FBS, 1mM Pyruvate, 1×MEM NEAA and 1% Penicillin/Streptomycin. HEK-293T cell and A549 cells were both cultured with Dulbecco’s modified Eagle medium (DMEM) supplemented with 5% FBS, 1×Glutamax and 1% Penicillin/Streptomycin. Huh7 cells were a kind gift from Dr. Emily Derbyshire and cultured with DMEM containing 10% FBS, 1×Glutamax and 1% Penicillin/Streptomycin. All cells were grown at 37°C with 5% CO2.
Human coronavirus 229E (HCoV-229E), strain VR-740, was obtained from ATCC. Huh7 cells cultured in 15 cm dish were inoculated with 0.05 MOI HCoV-229E at 37°C. At 1-h post infection (hpi) the supernatant was replaced with 20 mL complete DMEM. The media supernatant was collected 72 hpi and stored in aliquots at −80°C. Plaque assay was conducted on Huh7 cells to calculate virus titer, as described previously (Trimarco et al., 2021).
Method details
CRISPR activation screen and next-generation sequencing
Huh7 cells were transduced with a lentivirus expressing dCAS9-VP64 fusion protein (Addgene #61425). Two days later the cells were selected with 3 μg/mL blasticidin until all control cells had died. The surviving Huh7-dCas9 cells were expanded and 3×107 Huh7-dCas9 cells were transduced with CRISPR activation sgRNA library (Calabrese set A, Addgene#92379) at 0.5 MOI in two replicates. Two days later, cells were split into 1 μg/mL puromycin selection. The cells were split and cultured under puromycin pressure for ten days. Next, half of the cells were collected as a control and the other half of the cells were infected with HCoV-229E (MOI = 0.02). The infected cells were cultured and expanded for seven weeks. Surviving cells were then harvested and genomic DNA from control and surviving cells were extracted with QIAamp DNA Blood Maxi Kit (QIAGEN, catalog no. 51192). The sgRNA sequences were amplified by ExTaq DNA polymerase (Takara, catalog no. RR001). Amplified libraries were further purified with GeneJet Gel Extraction Kit (Thermo, catalog no. K0692) and measured via Agilent Bioanlyzer. The PCR products were then pooled and sequenced on an Illumina MiSeq. sgRNA enrichment was compared between the post-infected and the transduced library input samples. FastQ files were analyzed by MAGeCK (Li et al., 2014); the normalized sgRNA read counts and raw data are available at NCBI GEO under accession number: GSE197882.
Generation of plasmids
Total RNA in Huh7 cells were extracted with NEB Monarch total RNA miniprep kit (NEB, T2010) and used as the template for RT-PCR. GEMIN7, SAA2-SAA4, CEBPB, POC5, USP22 and ZBTB7B were both successfully amplified via SuperScript™ III One-Step RT-PCR System (Thermo, 12574026). ZBTB7A was amplified from a codon-optimized gBlock (IDT). Flag-tags were further introduced on the N-terminus of ZBTB7A. All PCR products were cloned into the pLEX expression plasmid through HiFi DNA assembly (NEB, M5520AA). Plasmid DNA was propagated in DH5α (NEB, C2987P) and all plasmid clones were verified by Sanger sequencing.
Lentiviral production and transduction
The 6-well plates were treated with Poly-L-lysine solution (Sigma-Aldrich, P8920) and seeded with HEK-293T cells. 1 μg of the pLEX plasmid was transfected into HEK-293T cells along with 0.4 μg pMD2.G and 1 μg pCMVΔ8.74. 24 h later, the media in each well was replaced with 2 mL complete DMEM and cultured for another 48 h. Finally, lentiviral supernatant media was harvested and aliquoted at −80°C. To generate a stable expression cell line, Huh7 cells, MRC5 cells or A549 cells in 6-well plate were transduced with 2 mL lentivirus. After 48 h, the transduced cells were selected with 1 μg/mL puromycin. The surviving cells were expanded and maintained with 0.5 μg/mL puromycin.
qRT-PCR
Total RNA was extracted from experimental samples. To verify the expression of candidate genes in Huh7 cells, Taqman probes targeting GEMIN7 (Thermo, Hs01547773_m1), USP22 (Thermo, Hs00392751_m1), SAA2-SAA4 (Thermo, Hs00197854_m1), POC5 (Thermo, Hs00401506_m1), CEBPB (Thermo, Hs00270923_s1), ZBTB7B (Hs00757087_g1) were ordered. A custom primer-probe set (forward primer: GGTTCACTCGCCAGGATAAA, reverse primer: CCGCATGTGGTTCTTCAAATC, Probe:/56-FAM/ATGCGAAAG/ZEN/CACACTGGCGAAA/3IABkFQ) targeting ZBTB7A was synthesized by IDT. HCoV-229E viral RNA was detected by a commercially available probe (Thermo, Vi06439671_s1). AKR1C3 (Thermo, Hs00366267_m1) and APOA4 probes (Thermo, Hs00166636_m1) were used for oxidative response related genes detection. In addition, a GSTA1 probe (Thermo, Hs07292464_g1), GSTA2 probe (Thermo, Hs00747232_mH), CAT probe (Thermo, Hs00156308_m1), GPX1 probe (Thermo, Hs00829989_gH) and GPX2 probe (Thermo, Hs01591589_m1) were used to evaluate the mRNA levels of antioxidant genes. 18S rRNA (Thermo, 4319413E) was the endogenous control. EXPRESS One-Step Superscript™ qRT-PCR kit (Thermo, 11781200) was used to perform one-step qRT-PCR on the Applied Biosystems QuantStudio 3 Real-Time PCR System. In some cases, ΔΔCT analysis was conducted to determine the fold change of gene mRNA levels. In other cases, such as for viral RNA, a dilution curve (thus generating arbitrary RNA units) was used to calculate experimental values.
Western blotting
ZBTB7A stable overexpression cells were seeded into a 6-well plate. The next day, cells were trypsinized and collected through centrifugation. The cell pellets were resuspended with 100 μL PBS and lysed with 100 μL 2×Laemmli sample buffer (Bio-Rad, 1610737). The cell lysate was heated at 95°C for 5 min and quantified by Bradford assay (Pierce, 23246). 30 μg of cell lysate was loaded onto 4–20% Mini-PROTEAN TGX Stain-Free Protein gels (Bio-Rad, 4568094) and run at 100 V for 1 h. The gel was transferred to 0.45 μm nitrocellulose membrane at 60 V for 1 h and the membrane was further blocked with 5% milk diluted with PBST for 3 h. After three washes with PBST, recombinant anti-ZBTB7A antibody (R&D, mab3496) at a 1:1000 dilution, anti-alpha tubulin antibody (Abcam, ab179484) or anti-alpha actin antibody (Santa Cruz, sc-47778) at a 1:1000 dilution was incubated overnight at 4°C. After three washes with PBST, goat anti-rabbit-HRP antibody (Life Technologies, A16104) or goat anti-armenian hamster-HRP antibody (Novus, NB100-2066) was incubated with the membrane for 1 h. Finally, Clarity Western ECL Substrate (Bio-Rad, 1705060) was used for imaging with Amersham Hyperfilm ECL (GE Healthcare 28906839).
Immunofluorescence assays
Huh7 cells or MRC5 cells were seeded onto 24-well plates and then infected with HCoV-229E as described in the figure legends. Afterwards, cells were washed with PBS and then fixed with 4% paraformaldehyde for 15min. After washing three times, the cells were permeabilized with 0.1% saponin for 20 min and blocked with 5% BSA in 0.1% saponin for 3 h. To identify the HCoV-229E infected cells, anti-HCoV-229E N antibody (Eurofins, M.30.HCo.I1E8) was used at a 1:200 dilution was incubated overnight at 4°C. After washing three times, the cells were stained with Alexa Fluor 594 Goat Anti-Mouse antibody (Thermo, A-11005) at a 1:1000 dilution. Cell nuclei were stained with Hoechst 33342 (Invitrogen, H3570). All samples were imaged by ZOE Fluorescent Cell Imager (Bio-Rad, 1450031).
Proliferation assay
The infected ZBTB7A-Huh7 cells were plated in 24-well plate. EdU Assay Kit (Abcam, ab219801) was applied following the protocol. Briefly, EdU solution was diluted with DMEM media at 20μM and added into cells to be stained for 4 h. The cells were washed with PBS and then incubated with fixative solution and permeabilization buffer separately for 15 min. The reaction buffer containing iFluor 488 dye was added and incubated for 30 min. The samples were analyzed by fluorescence microscope.
Flow cytometry
Flow cytometry was performed according to standard methods. The cell samples were trypsinized and pelleted by centrifuge. GFP expressing samples were resuspended with 2% BSA in PBS and directly analyzed via FACSCanto II machine. To analyze live/dead cells, LIVE/DEAD Fixable Green Dead Cell Stain Kit (Thermo, L34969) was used. The cell pellets were washed once with 1 mL PBS and 1 μL fluorescent reactive dye was added to 1 mL cell suspension. The samples were protected from light and incubated at room temperature for 30 min. The samples were then washed once with 1 mL PBS and incubated with 4% formaldehyde for 15 min. Then, the samples were washed and suspended with 2% BSA in PBS and analyzed by flow cytometry. For the oxidative stress assay, CellROX Green Reagent (Thermo, C10444) was used to detect ROS. Cells cultured in the plate were incubated with the CellROX® Reagent at 5 μM concentration for 30 min. Then medium was removed and cells were washed three times with PBS. In some cases, the stained samples were trypsinized and fixed as previous described and analyzed by flow cytometry. The data was analyzed with FlowJo software.
Live cell staining
To evaluate HCoV-229E infection induced cytopathic effect, the live cells were stained with Crystal Violet. The infected samples were washed with PBS and fixed with 4% formaldehyde for 10 min. After three washes with PBS, the cells were stained with 0.1% Crystal Violet for 10 min before washing of residual stain. In addition, live cells were stained with LIVE/DEAD Cell Imaging Kit (Thermo, R37601). Live Green vial (A) was transferred into Dead Red vial (B) and added to cultured cells. After 30 min of incubation at 37°C, the live cells were imaged by ZOE Fluorescent Cell Imager.
RNA-seq
ZBTB7A-Huh7 cells and control cells were seeded into 6-well plates and mock-infected or infected with HCoV-229E at 1 MOI. Each group contained 4 replicates. The cells were collected 24 hpi and total RNA was extracted with Monarch Total RNA Miniprep Kit (NEB, T2010S). Samples were prepared with a NEBNext Poly(A) messenger RNA (mRNA) Magnetic Isolation Module (NEB #E74905). The NEBNext Ultra II RNA Library Prep Kit (E7770S) was used to prepare libraries with NEBNext Multiplex Oligos for Illumina (E7335S, E7500S). Samples were then sequenced on an Illumina NovaSeq 6000 and then raw reads were mapped to the human reference genome. After normalization, average read values were compared across samples. The data were further sorted based on the fold change and p-values. The upregulated and downregulated genes were categorized by gene ontology (pantherdb.org). The raw sequencing data are available at NCBI GEO under accession number: GSE197644.
ChIP and qPCR
Huh7 cells were transduced with a lentivirus expressing ZBTB7A-Flag. ZBTB7A-Flag-Huh7 cells were then infected with HCoV-229E at 1 MOI. After 24 h, cells were crosslinked and harvested with Pierce Magnetic ChIP Kit (Thermo, 26157). Following the ChIP Kit protocol, isolated chromatin from ZBTB7A-Flag-Huh7 cells was incubated with mouse anti-Flag antibody (Sigma, F1804) or mouse anti-GFP antibody (Cell Signaling Technology, 2955) as control for immunoprecipitation. After IP elution and DNA recovery, purified DNA binding by ZBTB7A and input DNA were collected. To identify candidate gene promoter binding by ZBTB7A, qPCR primers targeting genomic regions were designed: AKR1C3-F, TGAAACAGTTTGTGCCTTCA; AKR1C3-R, TGGACACATGGAATCCTCAA; CAT-F, CTGGGTATCTCCGGTCTTCA; CAT-R, GACTTCAGGCTCAGCCAATC; GSTA1-F, AGGCAGGGAAGGATTGTTCT; GSTA1-R, TGGTGGGAGTATGTGGGAGT; GSTA2-F, CTGGTGCAGGTCCTTGGTAT; GSTA2-R, CAGTGGCCCTCAGGTGTTAT; IFNA1-F, TGCATCCCAGGAATAAATCA; IFNA1-R, TGAAGCCAGCATTACCTTGA. Universal SYBR® Green Supermix (Bio-Rad, 1725274) was used to perform qPCR on the Applied Biosystems QuantStudio 3 Real-Time PCR System.
ZBTB7A knockdown
To knockdown ZBTB7A, we utilized siRNA targeting ZBTB7A (Thermo, siRNA ID 147596, AM16708) and negative control siRNA (Thermo, AM4611). Huh7 cells were transfected with 50nM siRNA using RNAiMAX Transfection Reagent (Thermo, 13778150) and infected with HCoV-229E as indicated. ZBTB7A mRNA and protein levels were detected with the following probe (Thermo, Hs00792219_m1) and ZBTB7A antibody (Santa Cruz, sc-33683).
MTT assay
MRC5 or Huh7 cells were cultured in 96-well plates. After infection and/or drug treatment, the supernatant was aspirated. 50 μL of serum-free media and 50 μL of 10 mg/mL MTT solution (Abcam, ab211091) were added to each well and incubated at 37°C for 3 h. Then 150 μL of 4 mM HCl, 0.1% NP40 in isopropanol was added. The plate was then agitated and the OD590 nm absorbance was measured.
Caspase 3/7 activity assay and apoptosis related inhibitor treatment
Huh7 cells were infected with HCoV-229E. Caspase 3/7 Green Detection Reagent (Invitrogen, C10423) was applied to measure caspase activity in live cells. After 30 min incubation, fluorescence intensity of caspase activity was imaged by ZOE Fluorescent Cell Imager (Bio-Rad, 1450031). In addition, the apoptosis related inhibitor 30 μM Z-VAD-FMK (R&D, FMK001) or 30 μM Ac-DEVD-CHO (Selleck, S7901) were used.
Superoxide scavenger and H2O2 treatment
The live Huh7 cells were analyzed at 3 DPI, and live MRC5 cells were further analyzed at 5 DPI. Tiron (Abcam, ab146234) and trolox (Sigma, 53188-07-1) were ordered and dissolved to a stock concentration of 500 mM and 100 mM. MRC5 cells and Huh7 cells were infected with HCoV-229E. After 2 h, the cells were treated with 10 mM tiron or 200 μM trolox. The live Huh7 cells were analyzed at 3 DPI, and live MRC5 cells were further analyzed at 5 DPI. Hydrogen peroxide solution (Sigma, 7722-84-1) was diluted with DMEM without FBS and used to treat Huh7 cells at the concentrations indicated in the figure legends.
Quantification and statistical analysis
Except for the CRISPR screen and RNA sequencing, all experiments were performed with at least three biological replicates and the full experiment was independently repeated at least twice as described in figure legend. Unless otherwise indicated, all data were analyzed based on the unpaired two-tailed, student’s t-test through Prism9 software (GraphPad). Data shown as mean ± SD. Error bars represent SD. p < 0.05, ∗; p < 0.001, ∗∗; not significant, ns.
Acknowledgments
We would like to thank Dr. Bryan Cullen for the sharing of key reagents. N.S.H. is funded in part by the Defense Advanced Research Projects Agency’s (DARPA) Preemptive Expression of Protective Alleles and Response Elements (PREPARE) program (cooperative agreement #HR00111920008). J.D.T. was partially funded by the following training grant: T32-CA00911. T.E.R. is funded in part by DARPA grant AWD-102840-G7. This work used a high-performance computing facility partially supported by grants 2016-IDG-1013 and 2020-IIG-2109 from the North Carolina Biotechnology Center. The views, opinions, and/or findings expressed are those of the authors and should not be interpreted as representing the official views or policies of the US Government.
Author contributions
X.Z., J.D.T., and C.A.W. performed experimentation. A.B., X.Z., and J.D.T. performed data analysis. T.E.R. and N.S.H. supervised research and acquired funding. X.Z. and N.S.H. wrote the manuscript.
Declaration of interests
The authors declare no competing interests.
Published: October 5, 2022
Footnotes
Supplemental information can be found online at https://doi.org/10.1016/j.celrep.2022.111540.
Supplemental information
Data and code availability
CRISPR sequencing data and RNA sequencing data have been deposited at NCBI GEO and are publicly available. Accession numbers are listed in the key resources table. This paper does not report original code and any additional information required to reanalyze the data reported in this work paper is available from the lead contact upon request.
References
- Agarwal V., Venkatakrishnan A.J., Puranik A., Kirkup C., Lopez-Marquez A., Challener D.W., Theel E.S., O'Horo J.C., Binnicker M.J., Kremers W.K., et al. Long-term SARS-CoV-2 RNA shedding and its temporal association to IgG seropositivity. Cell Death Discov. 2020;6:1–9. doi: 10.1038/s41420-020-00375-y. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Avanzato V.A., Matson M.J., Seifert S.N., Pryce R., Williamson B.N., Anzick S.L., Barbian K., Judson S.D., Fischer E.R., Martens C., et al. Case study: prolonged infectious SARS-CoV-2 shedding from an asymptomatic immunocompromised individual with cancer. Cell. 2020;183:1901–1912. doi: 10.1016/j.cell.2020.10.049. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Baggen J., Persoons L., Vanstreels E., Jansen S., Van Looveren D., Boeckx B., Geudens V., De Man J., Jochmans D., Wauters J., et al. Genome-wide CRISPR screening identifies TMEM106B as a proviral host factor for SARS-CoV-2. Nat. Genet. 2021;53:435–444. doi: 10.1038/s41588-021-00805-2. [DOI] [PubMed] [Google Scholar]
- Biering S.B., Sarnik S.A., Wang E., Zengel J.R., Leist S.R., Schäfer A., Sathyan V., Hawkins P., Okuda K., Tau C., et al. Genome-wide bidirectional CRISPR screens identify mucins as host factors modulating SARS-CoV-2 infection. Nat. Genet. 2022;54:1078–1089. doi: 10.1038/s41588-022-01131-x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cho N.E., Bang B.R., Gurung P., Li M., Clemens D.L., Underhill T.M., James L.P., Chase J.R., Saito T. Retinoid regulation of antiviral innate immunity in hepatocytes. Hepatology. 2016;63:1783–1795. doi: 10.1002/hep.28380. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chu H., Zhou J., Wong B.H.Y., Li C., Chan J.F.W., Cheng Z.S., Yang D., Wang D., Lee A.C.Y., Li C., et al. Middle East respiratory syndrome coronavirus efficiently infects human primary T lymphocytes and activates the extrinsic and intrinsic apoptosis pathways. J. Infect. Dis. 2016;213:904–914. doi: 10.1093/infdis/jiv380. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Daniloski Z., Jordan T.X., Wessels H.H., Hoagland D.A., Kasela S., Legut M., Maniatis S., Mimitou E.P., Lu L., Geller E., et al. Identification of required host factors for SARS-CoV-2 infection in human cells. Cell. 2021;184:92–105.e16. doi: 10.1016/j.cell.2020.10.030. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Danziger O., Patel R.S., DeGrace E.J., Rosen M.R., Rosenberg B.R. Inducible CRISPR activation screen for interferon-stimulated genes identifies OAS1 as a SARS-CoV-2 restriction factor. PLoS Pathog. 2022;18:e1010464. doi: 10.1371/journal.ppat.1010464. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fehr A.R., Perlman S. Coronaviruses: an Overview of their replication and pathogenesis. Methods Mol. Biol. 2015;1282:1–23. doi: 10.1007/978-1-4939-2438-7-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fiers W., Beyaert R., Declercq W., Vandenabeele P. More than one way to die: apoptosis, necrosis and reactive oxygen damage. Oncogene. 1999;18:7719–7730. doi: 10.1038/sj.onc.1203249. [DOI] [PubMed] [Google Scholar]
- Grodzki M., Bluhm A.P., Schaefer M., Tagmount A., Russo M., Sobh A., Rafiee R., Vulpe C.D., Karst S.M., Norris M.H. Genome-scale CRISPR screens identify host factors that promote human coronavirus infection. Genome Med. 2022;14:10. doi: 10.1186/s13073-022-01013-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gudipaty S.A., Conner C.M., Rosenblatt J., Montell D.J. Unconventional ways to live and die: cell death and survival in development, homeostasis, and disease. Annu. Rev. Cell Dev. Biol. 2018;34:311–332. doi: 10.1146/annurev-cellbio-100616-060748. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Han D., Chen S., Han W., Gao S., Owiredu J.N., Li M., Balk S.P., He H.H., Cai C. ZBTB7A mediates the transcriptional repression activity of the androgen receptor in prostate cancer. Cancer Res. 2019;79:5260–5271. doi: 10.1158/0008-5472.CAN-19-0815. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hoffmann H.H., Sánchez-Rivera F.J., Schneider W.M., Luna J.M., Soto-Feliciano Y.M., Ashbrook A.W., Le Pen J., Leal A.A., Ricardo-Lax I., Michailidis E., et al. Functional interrogation of a SARS-CoV-2 host protein interactome identifies unique and shared coronavirus host factors. Cell Host Microbe. 2021;29:267–280.e5. doi: 10.1016/j.chom.2020.12.009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Imai Y., Kuba K., Neely G.G., Yaghubian-Malhami R., Perkmann T., van Loo G., Ermolaeva M., Veldhuizen R., Leung Y.H.C., Wang H., et al. Identification of oxidative stress and toll-like receptor 4 signaling as a key pathway of acute lung injury. Cell. 2008;133:235–249. doi: 10.1016/j.cell.2008.02.043. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Israeli M., Finkel Y., Yahalom-Ronen Y., Paran N., Chitlaru T., Israeli O., Cohen-Gihon I., Aftalion M., Falach R., Rotem S., et al. Genome-wide CRISPR screens identify GATA6 as a proviral host factor for SARS-CoV-2 via modulation of ACE2. Nat. Commun. 2022;13:2237. doi: 10.1038/s41467-022-29896-z. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jacomy H., Fragoso G., Almazan G., Mushynski W.E., Talbot P.J. Human coronavirus OC43 infection induces chronic encephalitis leading to disabilities in BALB/C mice. Virology. 2006;349:335–346. doi: 10.1016/j.virol.2006.01.049. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jeon B.N., Yoo J.Y., Choi W.I., Lee C.E., Yoon H.G., Hur M.W. Proto-oncogene FBI-1 (Pokemon/ZBTB7A) represses transcription of the tumor suppressor Rb gene via binding competition with Sp1 and recruitment of co-repressors. J. Biol. Chem. 2008;283:33199–33210. doi: 10.1074/jbc.M802935200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Konermann S., Brigham M.D., Trevino A.E., Joung J., Abudayyeh O.O., Barcena C., Hsu P.D., Habib N., Gootenberg J.S., Nishimasu H., et al. Genome-scale transcriptional activation by an engineered CRISPR-Cas9 complex. Nature. 2015;517:583–588. doi: 10.1038/nature14136. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kratzel A., Kelly J.N., V'Kovski P., Portmann J., Brüggemann Y., Todt D., Ebert N., Shrestha N., Plattet P., Staab-Weijnitz C.A., et al. A genome-wide CRISPR screen identifies interactors of the autophagy pathway as conserved coronavirus targets. PLoS Biol. 2021;19:e3001490. doi: 10.1371/journal.pbio.3001490. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lee S., Channappanavar R., Kanneganti T.D. Coronaviruses: innate immunity, inflammasome activation, inflammatory cell death, and cytokines. Trends Immunol. 2020;41:1083–1099. doi: 10.1016/j.it.2020.10.005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Leist S.R., Dinnon K.H., 3rd, Schäfer A., Tse L.V., Okuda K., Hou Y.J., West A., Edwards C.E., Sanders W., Fritch E.J., et al. A mouse-adapted SARS-CoV-2 induces acute lung injury and mortality in standard laboratory mice. Cell. 2020;183:1070–1085.e12. doi: 10.1016/j.cell.2020.09.050. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li S., Zhang Y., Guan Z., Li H., Ye M., Chen X., Shen J., Zhou Y., Shi Z.L., Zhou P., Peng K. SARS-CoV-2 triggers inflammatory responses and cell death through caspase-8 activation. Signal Transduct. Target. Ther. 2020;5:235. doi: 10.1038/s41392-020-00334-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li W., Xu H., Xiao T., Cong L., Love M.I., Zhang F., Irizarry R.A., Liu J.S., Brown M., Liu X.S. MAGeCK enables robust identification of essential genes from genome-scale CRISPR/Cas9 knockout screens. Genome Biol. 2014;15:554. doi: 10.1186/s13059-014-0554-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liang Y., Yi P., Wang X., Zhang B., Jie Z., Soong L., Sun J. Retinoic acid modulates hyperactive T cell responses and protects vitamin A-deficient mice against persistent lymphocytic choriomeningitis virus infection. J. Immunol. 2020;204:2984–2994. doi: 10.4049/jimmunol.1901091. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lunardi A., Guarnerio J., Wang G., Maeda T., Pandolfi P.P. Role of LRF/Pokemon in lineage fate decisions. Blood. 2013;121:2845–2853. doi: 10.1182/blood-2012-11-292037. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mac Kain A., Maarifi G., Aicher S.M., Arhel N., Baidaliuk A., Munier S., Donati F., Vallet T., Tran Q.D., Hardy A., et al. Identification of DAXX as a restriction factor of SARS-CoV-2 through a CRISPR/Cas9 screen. Nat. Commun. 2022;13:2442. doi: 10.1038/s41467-022-30134-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mangalmurti N., Hunter C.A. Cytokine storms: understanding COVID-19. Immunity. 2020;53:19–25. doi: 10.1016/j.immuni.2020.06.017. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mesel-Lemoine M., Millet J., Vidalain P.O., Law H., Vabret A., Lorin V., Escriou N., Albert M.L., Nal B., Tangy F. A human coronavirus responsible for the common cold massively kills dendritic cells but not monocytes. J. Virol. 2012;86:7577–7587. doi: 10.1128/Jvi.00269-12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mesel-Lemoine M., Millet J., Vidalain P.O., Law H., Vabret A., Lorin V., Escriou N., Albert M.L., Nal B., Tangy F. A human coronavirus responsible for the common cold massively kills dendritic cells but not monocytes. J. Virol. 2012;86:7577–7587. doi: 10.1128/JVI.00269-12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Milewska A., Zarebski M., Nowak P., Stozek K., Potempa J., Pyrc K. Human coronavirus NL63 utilizes heparan sulfate proteoglycans for attachment to target cells. J. Virol. 2014;88:13221–13230. doi: 10.1128/JVI.02078-14. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nicholls J.M., Poon L.L.M., Lee K.C., Ng W.F., Lai S.T., Leung C.Y., Chu C.M., Hui P.K., Mak K.L., Lim W., et al. Lung pathology of fatal severe acute respiratory syndrome. Lancet. 2003;361:1773–1778. doi: 10.1016/S0140-6736(03)13413-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pessler F., Hernandez N. Flexible DNA binding of the BTB/POZ-domain protein FBI-1. J. Biol. Chem. 2003;278:29327–29335. doi: 10.1074/jbc.M302980200. [DOI] [PubMed] [Google Scholar]
- Ramos Pittol J.M., Oruba A., Mittler G., Saccani S., van Essen D. Zbtb7a is a transducer for the control of promoter accessibility by NF-kappa B and multiple other transcription factors. PLoS Biol. 2018;16:e2004526. doi: 10.1371/journal.pbio.2004526. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Qin X., Swertfeger D.K., Zheng S., Hui D.Y., Tso P. Apolipoprotein AIV: a potent endogenous inhibitor of lipid oxidation. Am. J. Physiol. 1998;274:1836–1840. doi: 10.1152/ajpheart.1998.274.5.H1836. [DOI] [PubMed] [Google Scholar]
- Rebendenne A., Roy P., Bonaventure B., Chaves Valadão A.L., Desmarets L., Arnaud-Arnould M., Rouillé Y., Tauziet M., Giovannini D., Touhami J., et al. Bidirectional genome-wide CRISPR screens reveal host factors regulating SARS-CoV-2, MERS-CoV and seasonal HCoVs. Nat. Genet. 2022;54:1090–1102. doi: 10.1038/s41588-022-01110-2. [DOI] [PubMed] [Google Scholar]
- Rebendenne A., Roy P., Bonaventure B., Chaves Valadão A.L., Desmarets L., Rouillé Y., Tauziet M., Arnaud-Arnould M., Giovannini D., Lee Y., et al. Bidirectional genome-wide CRISPR screens reveal host factors regulating SARS-CoV-2, MERS-CoV and seasonal coronaviruses. bioRxiv. 2021 doi: 10.1101/2021.05.19.444823. Preprint at. [DOI] [PubMed] [Google Scholar]
- Sanson K.R., Hanna R.E., Hegde M., Donovan K.F., Strand C., Sullender M.E., Vaimberg E.W., Goodale A., Root D.E., Piccioni F., Doench J.G. Optimized libraries for CRISPR-Cas9 genetic screens with multiple modalities. Nat. Commun. 2018;9:5416. doi: 10.1038/s41467-018-07901-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sawalha A.H., Zhao M., Coit P., Lu Q. Epigenetic dysregulation of ACE2 and interferon-regulated genes might suggest increased COVID-19 susceptibility and severity in lupus patients. Clin. Immunol. 2020;215:108410. doi: 10.1016/j.clim.2020.108410. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schneider W.M., Luna J.M., Hoffmann H.H., Sánchez-Rivera F.J., Leal A.A., Ashbrook A.W., Le Pen J., Ricardo-Lax I., Michailidis E., Peace A., et al. Genome-Scale identification of SARS-CoV-2 and pan-coronavirus host factor networks. Cell. 2021;184:120–132.e14. doi: 10.1016/j.cell.2020.12.006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shalem O., Sanjana N.E., Zhang F. High-throughput functional genomics using CRISPR-Cas9. Nat. Rev. Genet. 2015;16:299–311. doi: 10.1038/nrg3899. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sherman E.J., Mirabelli C., Tang V.T., Khan T.G., Leix K., Kennedy A.A., Graham S.E., Willer C.J., Tai A.W., Sexton J.Z., et al. Identification of cell type specific ACE2 modifiers by CRISPR screening. PLoS Pathog. 2022;18:e1010377. doi: 10.1371/journal.ppat.1010377. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tait S.W.G., Ichim G., Green D.R. Die another way - non-apoptotic mechanisms of cell death. J. Cell Sci. 2014;127:2135–2144. doi: 10.1242/jcs.093575. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Thul P.J., Åkesson L., Wiking M., Mahdessian D., Geladaki A., Ait Blal H., Alm T., Asplund A., Björk L., Breckels L.M., et al. A subcellular map of the human proteome. Science. 2017;356:eaal3321. doi: 10.1126/science.aal3321. [DOI] [PubMed] [Google Scholar]
- Trimarco J.D., Heaton B.E., Chaparian R.R., Burke K.N., Binder R.A., Gray G.C., Smith C.M., Menachery V.D., Heaton N.S. TMEM41B is a host factor required for the replication of diverse coronaviruses including SARS-CoV-2. PLoS Pathog. 2021;17:e1009599. doi: 10.1371/journal.ppat.1009599. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Trottier C., Chabot S., Mann K.K., Colombo M., Chatterjee A., Miller W.H., Jr., Ward B.J. Retinoids inhibit measles virus in vitro via nuclear retinoid receptor signaling pathways. Antiviral Res. 2008;80:45–53. doi: 10.1016/j.antiviral.2008.04.003. [DOI] [PubMed] [Google Scholar]
- Uhlén M., Fagerberg L., Hallström B.M., Lindskog C., Oksvold P., Mardinoglu A., Sivertsson Å., Kampf C., Sjöstedt E., Asplund A., et al. Proteomics. Tissue-based map of the human proteome. Science. 2015;347:1260419. doi: 10.1126/science.1260419. [DOI] [PubMed] [Google Scholar]
- V'Kovski P., Kratzel A., Steiner S., Stalder H., Thiel V. Coronavirus biology and replication: implications for SARS-CoV-2. Nat. Rev. Microbiol. 2021;19:155–170. doi: 10.1038/s41579-020-00468-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang C., Dinesh R.K., Qu Y., Rustagi A., Cousins H., Zengel J., Guo Y., Hall T., Beck A., Tso L., et al. CRISPRa screening with real world evidence identifies potassium channels as neuronal entry factors and druggable targets for SARS-CoV-2. bioRxiv. 2021 doi: 10.1101/2021.07.01.450475. Preprint at. [DOI] [Google Scholar]
- Wang G., Lunardi A., Zhang J., Chen Z., Ala U., Webster K.A., Tay Y., Gonzalez-Billalabeitia E., Egia A., Shaffer D.R., et al. Zbtb7a suppresses prostate cancer through repression of a Sox9-dependent pathway for cellular senescence bypass and tumor invasion. Nat. Genet. 2013;45:739–746. doi: 10.1038/ng.2654. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang R., Simoneau C.R., Kulsuptrakul J., Bouhaddou M., Travisano K.A., Hayashi J.M., Carlson-Stevermer J., Zengel J.R., Richards C.M., Fozouni P., et al. Genetic screens identify host factors for SARS-CoV-2 and common cold coronaviruses. Cell. 2021;184:106–119.e14. doi: 10.1016/j.cell.2020.12.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wei J., Alfajaro M.M., DeWeirdt P.C., Hanna R.E., Lu-Culligan W.J., Cai W.L., Strine M.S., Zhang S.M., Graziano V.R., Schmitz C.O., et al. Genome-wide CRISPR screens reveal host factors critical for SARS-CoV-2 infection. Cell. 2021;184:76–91.e13. doi: 10.1016/j.cell.2020.10.028. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wheeler D.L., Athmer J., Meyerholz D.K., Perlman S. Murine olfactory bulb interneurons survive infection with a neurotropic coronavirus. J. Virol. 2017;91 doi: 10.1128/JVI.01099-17. e01099-17. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Xiong W., Zhao J., Yu H., Li X., Sun S., Li Y., Xia Q., Zhang C., He Q., Gao X., et al. Elevated expression of AKR1C3 increases resistance of cancer cells to ionizing radiation via modulation of oxidative stress. PLoS One. 2014;9:e111911. doi: 10.1371/journal.pone.0111911. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Xu X., Xu Y., Zhang Q., Yang F., Yin Z., Wang L., Li Q. Porcine epidemic diarrhea virus infections induce apoptosis in Vero cells via a reactive oxygen species (ROS)/p53, but not p38 MAPK and SAPK/JNK signalling pathways. Vet. Microbiol. 2019;232:1–12. doi: 10.1016/j.vetmic.2019.03.028. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhou P., Yang X.L., Wang X.G., Hu B., Zhang L., Zhang W., Si H.R., Zhu Y., Li B., Huang C.L., et al. A pneumonia outbreak associated with a new coronavirus of probable bat origin. Nature. 2020;579:270–273. doi: 10.1038/s41586-020-2012-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhu N., Wang W., Liu Z., Liang C., Wang W., Ye F., Huang B., Zhao L., Wang H., Zhou W., et al. Morphogenesis and cytopathic effect of SARS-CoV-2 infection in human airway epithelial cells. Nat. Commun. 2020;11:3910. doi: 10.1038/s41467-020-17796-z. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhu S., Liu Y., Zhou Z., Zhang Z., Xiao X., Liu Z., Chen A., Dong X., Tian F., Chen S., et al. Genome-wide CRISPR activation screen identifies candidate receptors for SARS-CoV-2 entry. Sci. China Life Sci. 2022;65:701–717. doi: 10.1007/s11427-021-1990-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhu Y., Feng F., Hu G., Wang Y., Yu Y., Zhu Y., Xu W., Cai X., Sun Z., Han W., et al. A genome-wide CRISPR screen identifies host factors that regulate SARS-CoV-2 entry. Nat. Commun. 2021;12:961. doi: 10.1038/s41467-021-21213-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Data Availability Statement
CRISPR sequencing data and RNA sequencing data have been deposited at NCBI GEO and are publicly available. Accession numbers are listed in the key resources table. This paper does not report original code and any additional information required to reanalyze the data reported in this work paper is available from the lead contact upon request.