Abstract
Due to the recrudescence of severe acute respiratory syndrome coronavirus 2 (SARS-CoV-2) infections worldwide, mainly caused by the Omicron variant of concern (VOC) and its sub-lineages, several jurisdictions are administering an mRNA vaccine boost. Here, we analyze humoral responses induced after the second and third doses of an mRNA vaccine in naive and previously infected donors who received their second dose with an extended 16-week interval. We observe that the extended interval elicits robust humoral responses against VOCs, but this response is significantly diminished 4 months after the second dose. Administering a boost to these individuals brings back the humoral responses to the same levels obtained after the extended second dose. Interestingly, we observe that administering a boost to individuals that initially received a short 3- to 4-week regimen elicits humoral responses similar to those observed in the long interval regimen. Nevertheless, humoral responses elicited by the boost in naive individuals do not reach those present in previously infected vaccinated individuals.
Keywords: coronavirus, COVID-19, SARS-CoV-2, spike glycoproteins, long interval, third mRNA vaccine dose, humoral responses, variants of concern
Graphical abstract
In this study, Tauzin et al. report that the third dose of SARS-CoV-2 mRNA vaccine elicits strong humoral responses against VOCs in naive individuals, regardless of the interval between the first two doses. However, these responses remain lower than those induced by hybrid immunity.
Introduction
Two years after the coronavirus disease 2019 (COVID-19) was declared pandemic by the WHO, the severe acute respiratory syndrome coronavirus 2 (SARS-CoV-2) continues to circulate worldwide and has evolved into several variants. The variants of concern (VOCs), defined as variants with increased transmissibility, virulence, and/or against which vaccines and monoclonal antibody treatments are less effective (WHO, 2022a), are now the main source of concern about the evolving pandemic. Currently, the Delta and Omicron variants are the main circulating VOCs. The Delta (B.1.617.2) variant was declared as a VOC in May 2021 and the Omicron (B.1.1.529) variant in November 2021 (Choi and Smith, 2021; WHO, 2022a). Delta became the dominant strain in the summer/autumn of 2021. Omicron is divided into several sub-lineages, including BA.1 (the main variant, named Omicron hereafter), BA.1.1, BA.2, and BA.4 and BA.5, which emerged more recently (Kumar et al., 2022; Mohapatra et al., 2022; Viana et al., 2022). Due to their relatively high number of mutations, notably in the spike (S) glycoprotein, Omicron variants are more resistant to humoral responses elicited by vaccination or natural infection. This phenotype, in combination with a higher transmissibility rate compared with Delta, likely explains why they have become the dominant strains worldwide since January 2022 (Chen et al., 2022; Dhar et al., 2021).
Vaccination campaigns began over a year ago, and in several parts of the world, public health authorities are administering a third dose of vaccine (boost). Vaccine scarcity at the beginning of the vaccination campaign led some public health authorities to increase the interval between the first two doses, notably in the province of Quebec, Canada, where this interval was delayed to 16 weeks instead of 3 to 4 weeks. Several studies have now shown that this strategy leads to improved humoral and T and B cell responses after the second dose compared with the short vaccine regimen, in particular against VOCs including Delta and Omicron variants (Chatterjee et al., 2021; Nayrac et al., 2022; Payne et al., 2021; Tauzin et al., 2022a).
A vaccine boost is now recommended in several jurisdictions worldwide in response to the Omicron wave (Ferdinands, 2022). Recent studies have shown that this boost, following the 3- to 4-week dose interval regimen, strongly improves humoral responses against VOCs, for which poor responses were observed after the second dose (Doria-Rose et al., 2021; Gruell et al., 2022; Nemet et al., 2022; Schmidt et al., 2022). Here, we analyzed humoral responses elicited after the second and third doses of mRNA vaccine in a cohort of SARS-CoV-2-naive and previously infected donors who received their first two doses of Pfizer BioNTech mRNA vaccine with a 16-week interval and compared them with individuals receiving a short interval.
Results
We analyzed humoral responses induced after the second and third doses of BNT162b2 mRNA vaccine in a cohort of donors who received their first two doses with an extended interval of 16 weeks (median [range]: 111 days [76–134 days]). These donors received their third dose around 7 months after the second dose (median [range]: 219 days [167–235 days]). The cohort included 20 SARS-CoV-2-naive and 11 previously infected (PI) individuals who were infected during the first wave of COVID-19 (early 2020) and tested SARS-CoV-2 positive by nasopharyngeal swab PCR around 10 months before their first dose (median [range]: 300 days [247–321 days]). Blood samples were analyzed 3 weeks (V3, median [range]: 21 days [13–42 days]) and 4 months (V4, median [range]: 112 days [90–156 days]) after the second dose and 4 weeks (V5, median [range]: 27 days [19–38 days]) and 4 months (V6, median [range]: 119 days [111–131 days]) after the third dose of mRNA vaccine. Basic demographic characteristics of the cohorts and detailed vaccination time points are summarized in Table 1 and Figure 1A.
Table 1.
Characteristics of the vaccinated SARS-CoV-2 cohorts
| SARS-CoV-2 naive | SARS-CoV-2 previously infected | ||
|---|---|---|---|
| Number | 20 | 11 | |
| Age | 52 (33–64) | 48 (39–65) | |
| Gender | male (n) | 8 | 8 |
| female (n) | 12 | 3 | |
| Days between symptom onset and the first dosea | N/A | 300 (247–321) | |
| Days between the first and second dosesa | 111 (76–120) | 110 (90–134) | |
| Days between the second and third dosesa | 219 (167–230) | 219 (187–235) | |
| Days between the second dose and V3a | 21 (17–34) | 22 (13–42) | |
| Days between the second dose and V4 a | 112 (96–156) | 113 (90–127) | |
| Days between the third dose and V5a | 27 (20–38) | 27 (19–37) | |
| Days between the third dose and V6a | 119 (113–131) | 117 (111–127) | |
Values displayed are medians, with ranges in parentheses.
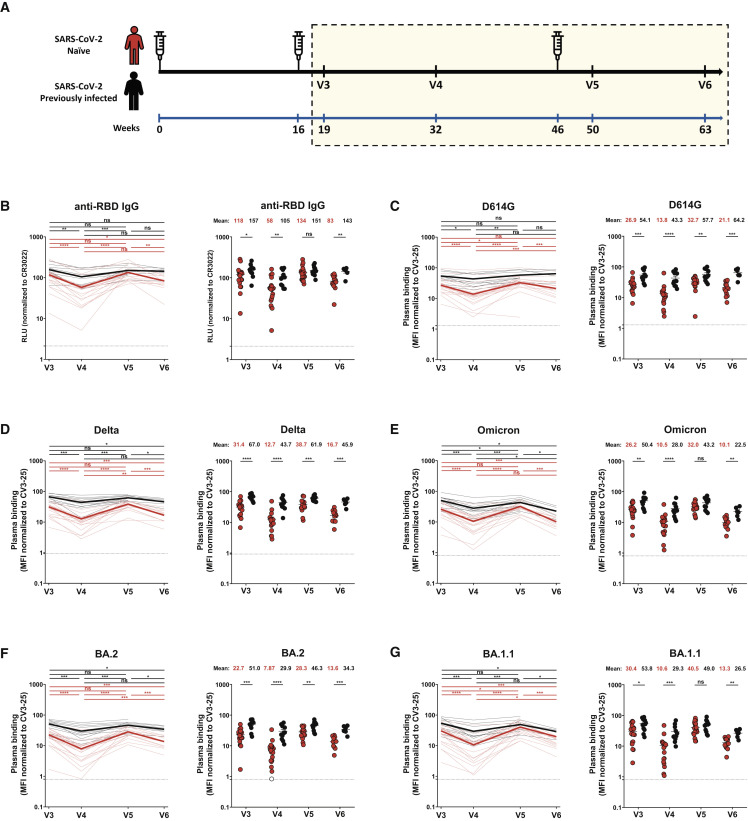
Figure 1.
RBD-specific IgG and recognition of SARS-CoV-2 spike variants by vaccine-elicited antibodies in SARS-CoV-2-naive and previously infected individuals after the second and third doses of mRNA vaccine
(A) SARS-CoV-2 vaccine cohort design. The yellow box represents the period under study.
(B) Indirect ELISA was performed by incubating plasma samples from naive and previously infected (PI) donors collected at V3, V4, V5, and V6 with recombinant SARS-CoV-2 RBD protein. Anti-RBD Ab binding was detected using HRP-conjugated anti-human IgG. Relative light unit (RLU) values obtained with BSA (negative control) were subtracted and further normalized to the signal obtained with the anti-RBD CR3022 monoclonal antibodies (mAbs) present in each plate.
(C–G) 293T cells were transfected with the indicated full-length spike (S) from different SARS-CoV-2 variants S and stained with the CV3-25 Ab or with plasma from naive or PI donors collected at V3, V4, V5, and V6 and analyzed by flow cytometry. The values represent the median fluorescence intensities (MFI) normalized by CV3-25 Ab binding.
(B–G) (Left panels) Each curve represents the values obtained with the plasma of one donor at every time point. Mean of each group is represented by a bold line. (Right panels) Plasma samples were grouped in different time points (V3, V4, V5, and V6). Naive and PI donors are represented by red and black points, respectively. Undetectable measures are represented as white symbols, and limits of detection are plotted. Error bars indicate means ± SEM (∗p < 0.05; ∗∗p < 0.01; ∗∗∗p < 0.001; ∗∗∗∗p < 0.0001; ns, non-significant). For naive donors, n (number of individuals) = 20 at V3, V4, and V5 and n = 13 at V6, and for PI donors, n = 11 at V3, V4, and V5 and n = 6 at V6.
Anti-RBD IgG levels of vaccine-elicited antibodies
We first measured the level of anti-receptor-binding domain (RBD) immunoglobulin G (IgG) induced after the second and third doses of mRNA vaccine by ELISA assay (Anand et al., 2021; Beaudoin-Bussières et al., 2020; Prévost et al., 2020; Tauzin et al., 2021). Three weeks after the second dose of mRNA vaccine (V3), both naive and PI individuals presented high levels of anti-RBD IgG (Figure 1B). Four months after the second dose (V4), the level of antibodies (Abs) decreased for both groups, reaching significantly lower levels for naive individuals, in agreement with previous observations (Tauzin et al., 2022a). The third dose (V5) led to an increase of the anti-RBD IgG level, similar in both groups, that reached the same levels as after the second dose (V3). Four months after the third dose (V6), this level decreased again for naive donors but remained stable for PI individuals (Figure 1B).
Recognition of SARS-CoV-2 S variants by plasma from vaccinated individuals
We evaluated the ability of plasma IgG to recognize SARS-CoV-2 full-length S variants after the second and third doses of vaccine (Figures 1C–1G). After the second dose, plasma from naive donors recognized the D614G S less efficiently than plasma from PI individuals (Figure 1C). Four months after the second dose (V4), we observed a decreased recognition for both groups, but it was more pronounced in the naive group. The third dose (V5) increased D614G S recognition by the naive group, reaching levels similar to those achieved after the second dose (V3) (Figure 1C). However, even after the boost, the level of recognition in naive donors did not reach the same level as in the PI group and significantly decreased at V6, while it remained stable for PI donors.
The original Wuhan strain was used to develop mRNA SARS-CoV-2 vaccines. Numerous mutations, particularly in the S glycoprotein, reduced the ability of vaccine-induced Abs to recognize currently circulating strains. We tested the S recognition of several VOCs in circulation after mRNA vaccination (Figures 1D–1G and S1C–S1E). For all the VOCs S tested, a similar pattern of response as for D614G S was observed. Except for Omicron and BA.1.1 S at V5, plasma from PI donors more efficiently recognized S variants than naive donors at all time points. Again, booster-elicited Abs able to recognize the different S glycoproteins reached levels similar to those obtained after the second dose. Four months after the third dose, the level of S variants recognition decreased in both groups but to a higher extent for naive individuals (Figures 1D–1G). Comparable responses were observed when we measured the capacity of plasma to recognize the S2 subunit (Figure S1A).
We also evaluated whether the booster impacted the capacity of plasma to recognize the S glycoprotein of the endemic HKU1 human Betacoronaviruses (Figure S1B). We did not observe major changes in recognition after the second and third doses. However, for all time points, plasma from PI donors always recognized the HCoV-HKU1 S better than the naive group. This suggests that natural infection elicits more cross-reactive Abs.
Functional activities of vaccine-elicited Abs
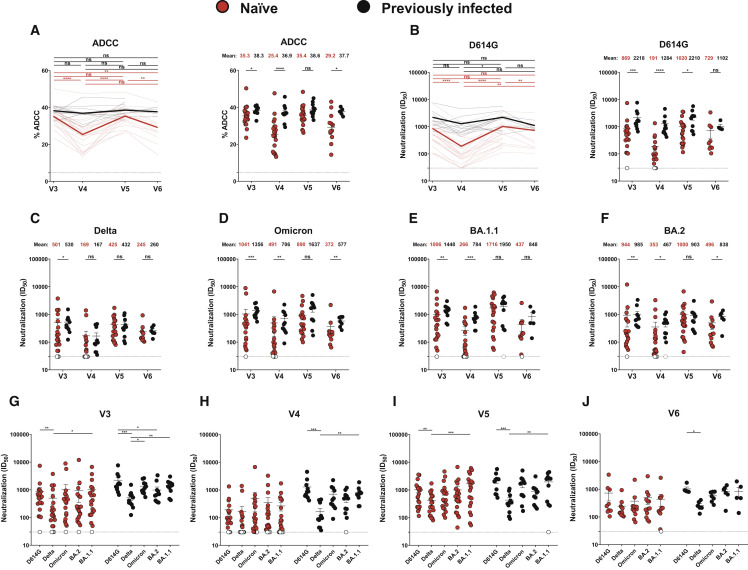
We evaluated functional activities of vaccine-elicited Abs after the second and third doses (Figure 2 ). We measured Fc-effector functions using a well-described antibody-dependent cellular cytotoxicity (ADCC) assay (Anand et al., 2021; Beaudoin-Bussières et al., 2020, 2021; Ullah et al., 2021). Plasma from PI individuals presented significantly higher ADCC activity after the second dose (V3 and V4), with naive individuals reaching similar levels after the third dose (V5) (Figure 2A). However, 4 months after the third dose (V6), PI donors again had a significantly higher ADCC activity than naive individuals. We noted that while ADCC remained relatively stable over time for PI individuals, it significantly decreased 4 months after the second and third doses for naive individuals (V4 and V6). A similar pattern of responses was observed for the neutralizing activity against pseudoviruses carrying the D614G S (Figure 2B). At the four time points, the neutralizing activity was better in the PI group compared with the naive group, but this difference was not significant at V6. The level of neutralizing Abs remained stable in PI individuals. On the contrary, the level of neutralizing Abs was significantly increased by the boost in naive donors but significantly decreased 4 months after the second and third doses at V4 and V6.
Figure 2.
Fc-effector functions and neutralization activities induced by mRNA vaccination in SARS-CoV-2-naive and PI individuals
(A) CEM.NKr parental cells were mixed at a 1:1 ratio with CEM.NKr-S cells and were used as target cells. Peripheral blood mononuclear cells (PBMCs) from uninfected donors were used as effector cells in a fluorescence-activated cell sorting (FACS)-based ADCC assay.
(B–J) Neutralizing activity was measured by incubating pseudoviruses bearing SARS-CoV-2 S glycoproteins, with serial dilutions of plasma for 1 h at 37°C before infecting 293T-ACE2 cells. Neutralization half-maximal inhibitory serum dilution (ID50) values were determined using a normalized non-linear regression using GraphPad Prism software.
(A and B, left) Each curve represents the values obtained with the plasma of one donor at every time point. Mean of each group is represented by a bold line.
(A and B, right; C–F) Plasma samples were grouped in different time points (V3, V4, V5, and V6).
(G–J) Neutralization activities against several SARS-CoV-2 variants S were analyzed at the different time points (V3, G; V4, H; V5, I; and V6, J). Naive and PI donors are represented by red and black points, respectively. Undetectable measures are represented as white symbols, and limits of detection are plotted. Error bars indicate means ± SEM. (∗p < 0.05; ∗∗p < 0.01; ∗∗∗p < 0.001; ∗∗∗∗p < 0.0001; ns, non-significant). For naive donors, n = 20 at V3, V4, and V5 and n = 13 at V6, and for PI donors, n = 11 at V3, V4, and V5 and n = 6 at V6.
When looking at the neutralizing activity against VOCs, we observed that plasma from the PI group neutralized more efficiently all pseudoviruses than the naive group after the second dose (Figures 2C–2H). Interestingly, this difference disappeared 4 weeks after the boost (V5), but the level of neutralizing Abs declined to a lower level in naive donors than in PI individuals at V6 for the Omicron and BA.2 variants (Figures 2C–2F, 2I, and 2J).
Integrated analysis of vaccine responses elicited by the second and third doses
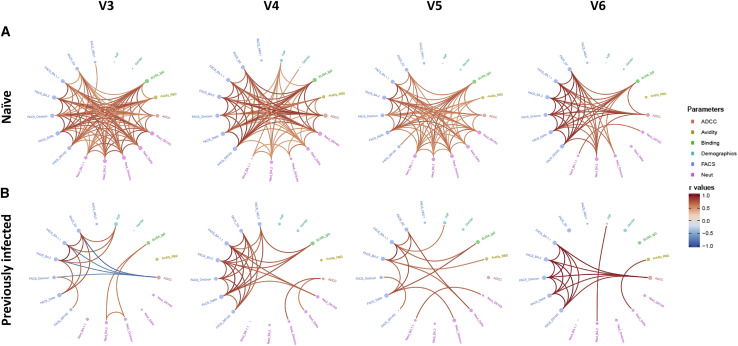
We evaluated the network of pairwise correlations among all studied immune variables on 11 randomly selected naive donors and the 11 PI individuals. For SARS-CoV-2-naive individuals (Figure 3A), we observed a dense network of positive correlations after the second dose (V3) involving all immune variables tested, except for HCoV-HKU1 S binding. Four months after the second dose, this network became less dense, and the third dose did not substantially alter the network of correlations except for improved correlations of neutralization responses against Omicron and BA.1.1 with other anti-SARS-CoV-2 immune responses. Four months after the third dose, the network of correlations became less dense. Interestingly, in PI individuals, the integrated network was less dense after the second dose compared with naive donors (Figure 3B), suggesting a less-focused immune response possibly due to the heterogenous immune stimulations by natural infection and vaccination. Whereas the network became slightly denser among the S binding responses 4 months after the second dose, it remained sparsely connected overall without major changes after the third dose of the mRNA vaccine.
Figure 3.
Mesh correlations of humoral response variables after the second and third doses of the mRNA vaccine
Edge-bundling correlation plots where red and blue edges represent positive and negative correlations between connected variables, respectively. Only significant correlations (p < 0.05, Spearman rank test) are displayed. Nodes are color coded based on the grouping of variables according to the legend. Node size corresponds to the degree of relatedness of correlations. Edge-bundling plots are shown for correlation analyses using eight different datasets, i.e., SARS-CoV-2-naive (A) or PI (B) individuals at V3, V4, V5, and V6, respectively. For naive and PI donors, n = 11.
Evolution of anti-RBD avidity induced after a short or a long interval between mRNA vaccine doses
We and others previously described that an extended interval between the first two doses of mRNA vaccine led to better humoral and cellular responses than the 3- to 4-week standard regimen, especially against VOCs (Nayrac et al., 2022; Payne et al., 2021; Tauzin et al., 2022a). However, whether the humoral advantages observed with the long interval persist after the boost remains unclear. To address this important question, we measured longitudinally the level of anti-RBD IgG in cohorts of naive and PI individuals that received their first two doses with the standard (short interval [SI]) or the extended regimen (long interval [LI]). Basic demographic characteristics of the cohorts and detailed vaccination time points are summarized in Table 2 and Figure 4A. We therefore decided to measure the avidity for the RBD of the induced IgG using a previously described assay (Björkman et al., 1999; Fialová et al., 2017; Tauzin et al., 2022b). This assay consists of parallel ELISAs with washing buffer having or not a chaotropic agent (8M urea). The RBD-avidity index therefore becomes a surrogate of Ab maturation, since only Abs with the highest avidity remain attached to the RBD after 8M urea washing (Figure S2).
Table 2.
Characteristics of the longitudinal SARS-CoV-2 cohorts vaccinated with a short or a long interval
| Short interval | Long interval | ||||
|---|---|---|---|---|---|
| SARS-CoV-2 naive | SARS-CoV-2 previously infected | SARS-CoV-2 naive | SARS-CoV-2 previously infected | ||
| Number | 45 | 16 | 30 | 15 | |
| Age | 35 (22–67) | 35 (23–59) | 51 (21–64) | 47 (29–65) | |
| Gender | male (n) | 21 | 10 | 12 | 10 |
| female (n) | 24 | 6 | 18 | 5 | |
| Days between symptom onset and the first dosea | N/A | 102 (50–275) | N/A | 274 (166–321) | |
| Days between the first and second dosesa | 21 (21–35) | 21 (21–30) | 111 (76–120) | 110 (90–134) | |
| Days between the second and third dosesa | 258 (183–342) | 276 (244–308) | 219 (167–230) | 219 (187–235) | |
Values displayed are medians, with ranges in parentheses.
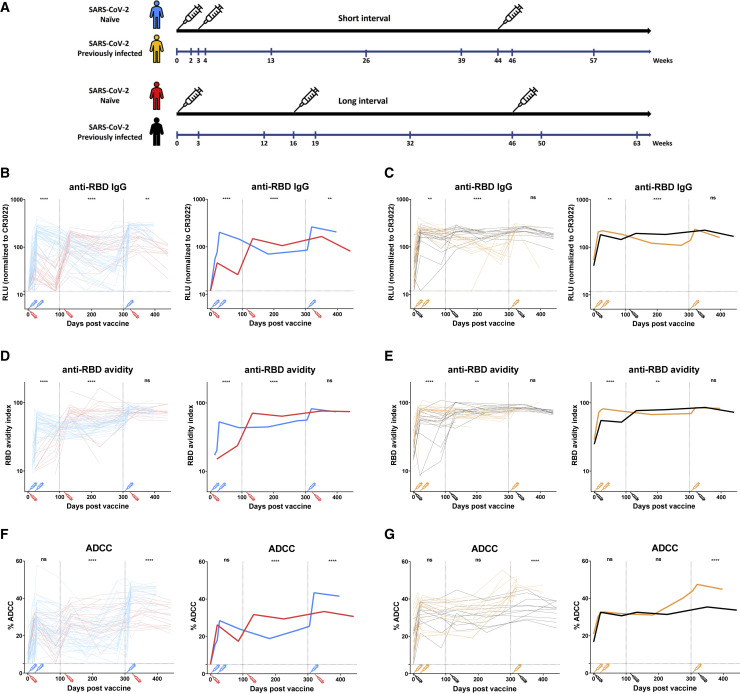
Figure 4.
Evolution of the RBD-specific IgG associated anti-RBD avidity and ADCC activity in SARS-CoV-2-naive and PI individuals vaccinated with a short or a long interval
(A) SARS-CoV-2 vaccine cohorts design.
(B–E) Indirect ELISAs were performed by incubating plasma samples from naive (B and D) and PI (C and E) donors vaccinated with a short interval (SI) or a long interval (LI) with recombinant SARS-CoV-2 RBD protein. The plasmas were collected at different time points from prior vaccination to after the third dose of mRNA vaccine. Anti-RBD Ab binding was detected using HRP-conjugated anti-human IgG.
(B and C) RLU values obtained were normalized to the signal obtained with the anti-RBD CR3022 mAb present in each plate.
(D and E) The RBD avidity index corresponded to the value obtained with the stringent (8M urea) ELISA divided by that obtained without urea.
(F and G) CEM.NKr parental cells were mixed at a 1:1 ratio with CEM.NKr-S cells and were used as target cells. PBMCs from uninfected donors were used as effector cells in a FACS-based ADCC assay.
(B–G) (Left panels) Each curve represents the values obtained with the plasma of one donor at every time point. (Right panels) The bold lines represent the mean of each group. Naive and PI donors vaccinated with the SI are represented by blue and yellow lines, respectively, and naive and PI donors vaccinated with the LI are represented by red and black lines, respectively. The time of vaccine dose injections is indicated by an associated color syringe. Limits of detection are plotted. Statistical analyses indicate the differences measured between plasma samples from naive donors vaccinated with an SI or an LI (B, D, and F) or PI donors vaccinated with an SI or an LI (C, E, and G), collected between 0 and 100 days post-vaccination (dpv), 101 and 300 dpv, or 300 + dpv (for more details, see Figure S3) (∗∗p < 0.01; ∗∗∗∗p < 0.0001; ns, non-significant). For naive donors vaccinated with the SI, n = 44 at weeks 0, 3, and 26; n = 45 at week 2; n = 46 at weeks 4 and 13; n = 30 at week 39; n = 22 at week 44; n = 35 at week 46; and n = 20 at week 57. For naive donors vaccinated with the LI, n = 25 at weeks 0, 3, and 12; n = 29 at week 19; n = 28 at week 32; n = 20 at week 50; and n = 14 at week 63. For PI donors vaccinated with the SI, n = 13 at weeks 0 and 26; n = 16 at week 2; n = 14 at week 3; n = 15 at weeks 4 and 13; n = 10 at week 39; n = 5 at week 44; n = 11 at week 46; and n = 3 at week 57. For PI donors vaccinated with the LI, n = 10 at week 0; n = 14 at weeks 3 and 32; n = 15 at weeks 12 and 19; and n = 12 at weeks 50 and 63.
Anti-RBD Abs reached their peak level faster with the SI compared with the LI in naive individuals (Figures 4B and S3A). While Ab levels rapidly decreased after the second dose in the SI regimen, the decay in the LI group was slower. In both groups, a booster elicited the highest levels of Abs (Figures 4B and S4B). Consistent with the proposed use of the RBD-avidity index as a surrogate for Ab maturation, the kinetics differed from those of the regular ELISA that only measure total levels of anti-RBD Abs. For example, 12 weeks after the first dose in the LI regimen, there was a significant decline in anti-RBD IgG levels, but the affinity of anti-RBD Abs likely improved, as shown by an increase in their avidity (Figures 4B and 4D), consistent with recent results (Tauzin et al., 2022b). We previously reported that several humoral responses, including RBD avidity, were lower in individuals receiving an SI (Tauzin et al., 2022a). However, whether this difference remained after the boost was unknown. Here, we report that the boost brought the RBD-avidity index to the same level, independent of the vaccine regimen, suggesting that the boost is required to further improve Ab responses in naive individuals that received the SI regimen (Figures 4D, S3C, and S4C).
For the PI groups, the SI and LI led rapidly to high levels of IgG (Figures 4C and S3B). These levels slightly decreased over time; however, they remained more stable in the LI group, likely due to the delayed dose. After the boost, we observed similar levels of IgG in both groups. Regarding the avidity, the SI rapidly led to IgG with strong avidity, which remained stable over time with only a minor effect induced by the boost (Figures 4E and S3D). For the LI group, the first dose increased the avidity but to a lower extent than in the SI group. The second dose boosted the avidity to the same level as in the SI group. As observed in the SI group, the boost did not improve the avidity, indicating that individuals that developed hybrid immunity due to natural infection already had a higher Ab avidity than naive, vaccinated individuals.
The third dose strongly boosted functional activities of the Abs in donors vaccinated with a short interval between mRNA vaccine doses
To assess functional activities of the Abs, we also measured longitudinally the ADCC activity in cohorts vaccinated with an SI or an LI (Figures 4F and 4G). In naive donors, we observed that donors vaccinated with an SI reached similar levels of ADCC activity after the second dose than after just one dose for donors vaccinated with the LI, indicating that administering the second dose 3 weeks after the first did not importantly improve ADCC activity (Figures 4F and S3E). However, the decay was slower in donors who received two doses. In the LI group, the second dose increased the level of ADCC compared with after the first dose, and the third dose allowed it to reach the same level of ADCC as after the second dose (Figures 2A and 4F). In contrast, in the SI group, the boost strongly increased the ADCC activity to a higher extent than observed in the LI group (Figures 4F, S3E, and S4D).
For PI groups, we observed that an SI or an LI led to the same level of ADCC after the first and the second doses of vaccine (Figures 4G and S3F). Surprisingly, the third dose highly increased the ADCC activity for the SI group. This level decreased after the boost, whereas it remained stable for the LI group (Figure 4G).
For naive donors vaccinated with the SI, we also observed that the third dose strongly increased the level of recognition of the D614G and several VOC spikes (Figure S4E). When looking at the network of correlations, we saw that it was less dense after the second dose in the SI group compared with the LI group and that the different parameters became more connected after the third dose, as observed for the LI group (Figures 3A and S5).
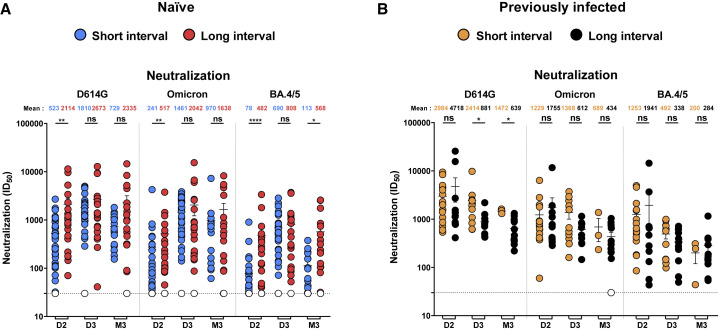
We then measured neutralizing responses. We observed significantly higher neutralizing activity against D614G, Omicron, and BA.4/5 pseudoviral particles after the second dose (D2) in naive donors vaccinated with the LI, compared with donors vaccinated with the SI (Figures 5A and S6A), consistent with previous studies showing improved neutralization for donors receiving the first two doses with an LI compared to an SI (Chatterjee et al., 2022; Tauzin et al., 2022a). The third dose of vaccine strongly boosted the level of neutralizing activity in the SI group (Figures 5A, S4F, and S6A). Similar levels of neutralization were observed between SI and LI after the third dose (D3). Interestingly, 3 to 4 months after the third dose (M3), the LI group presented significantly better neutralization against BA.4/5 than the LI group. Of note, we observed no major differences in neutralization for PI donors vaccinated with the SI and LI (Figures 5B and S6B).
Figure 5.
Neutralization activity induced by mRNA vaccination in SARS-CoV-2 naive and PI individuals vaccinated with a short or a long interval
(A and B) Neutralizing activity was measured by incubating pseudoviruses bearing SARS-CoV-2 S glycoproteins (D614G, Omicron, or BA.4/5), with serial dilutions of plasma for 1 h at 37°C before infecting 293T-ACE2 cells. Neutralization ID50 values were determined using a normalized non-linear regression using GraphPad Prism software. D2: 1 (SI) or 3 (LI) weeks after the second dose, D3: 2 (SI) or 4 (LI) weeks after the third dose, and M3: 3 (SI) or 4 (LI) months after the third dose. Naive (A) and PI (B) donors vaccinated with the SI are represented by blue and yellow points, respectively, and naive (A) and PI (B) donors vaccinated with the LI are represented by red and black points, respectively. Undetectable measures are represented as white symbols, and limits of detection are plotted. Error bars indicate means ± SEM (∗p < 0.05; ∗∗p < 0.01; ∗∗∗p < 0.001; ns, non-significant). For naive donors vaccinated with the SI, n = 45 at D2, n = 33 at D3, and n = 18 at M3. For naive donors vaccinated with the LI, n = 20 at D2, D3, and M3. For PI donors vaccinated with the SI, n = 16 at D2, n = 11 at D3, and n = 3 at M3. For PI donors vaccinated with the LI, n = 11 at D2, D3, and M3.
Discussion
Currently, a large part of the world’s population has received two or three doses of SARS-CoV-2 mRNA vaccines (WHO, 2022b). These vaccines were based on the ancestral Wuhan strain. One of the major challenges of the pandemic is the frequent emergence of variants that are more resistant to vaccine-elicited humoral responses, thus fueling new waves. In this study, we found that individuals who received their first two doses of mRNA vaccine with a 16-week extended interval had strong humoral responses against VOCs induced after the second dose. These responses significantly decreased 4 months later but came back to peak levels after the boost. However, they also rapidly declined after the boost, suggesting that although the vaccine induces good humoral responses, these are not sustained over time, particularly in naive donors.
Many studies have shown that an extended interval between doses elicits humoral responses that outperform those elicited by a short interval in SARS-CoV-2-naive individuals. Accordingly, we observed a network of correlation less dense after the second dose in individuals vaccinated with an SI compared with an LI (Figures 3A and S5). However, the third dose of the mRNA vaccine, administered several months after the second dose in the SI regimen, strongly improved the humoral responses against VOCs (Figure S4), and the network of correlations became denser, as observed after the LI regimen (Figures 3A and S5), emphasizing the importance of administering the boost.
Antibody maturation is a complex process that takes place in the germinal center (Young and Brink, 2021). This process is important for vaccine efficacy, since it allows the immune system to generate Abs with greater potency for viral neutralization and Fc-effector functions. We recently developed a high-throughput assay that could be used as a surrogate for Ab maturation (Tauzin et al., 2022b). Using this assay, we observed that the avidity of anti-RBD Abs was significantly higher in the LI after the second dose compared with the SI, suggesting that increasing the time between exposure to the antigen led to a better maturation of the B cells and so to Abs with higher avidity (Tauzin et al., 2022a). Here, we report that a boost allows the SI group to elicit Abs with the same avidity as in the LI group. In agreement with this, we observed that ADCC and neutralizing activities, which play an important role in protection against severe outcomes of SARS-CoV-2, were strongly improved by the boost in donors vaccinated with an SI. Since the RBD is highly immunogenic, it is possible that humoral responses against this domain compromise responses against less immunogenic domains of the spike or against RBD of variants that carry many mutations. Thus, in future studies, it would be interesting to measure the evolution of the avidity against other spike domains.
Vaccinated PI individuals presented a better avidity than naive individuals at all time points. These results are consistent with previous observations indicating that hybrid immunity led to broad and stronger humoral responses, but the mechanisms remain unclear (Andreano et al., 2021; Crotty, 2021; Goel et al., 2021). In correlation networks after the second and third doses of the mRNA vaccine, we observed strikingly different profiles of correlations in naive compared with PI donors, suggesting that infection primes the immune system in a different way than vaccination does. Whether this is linked to the immune stimulation with all components of the entire virus or the transmission mode of SARS-CoV-2, which infects host by the mucosa, therefore activating resident immune cells, remains poorly understood.
While vaccination confers good protection against severe COVID-19, it is less efficient against viral transmission. Thus, breakthrough infection in vaccinated individuals appears frequently. It was recently shown that breakthrough infection in vaccinated individuals induced strong neutralizing Abs efficient against VOCs, including Omicron (Miyamoto et al., 2022; Tauzin et al., 2022c). Interestingly, an LI between vaccination and breakthrough infection also induced better humoral responses against VOCs than an SI, as observed for vaccination. It is possible that breakthrough infection in fully vaccinated individuals led to hybrid immunity with humoral responses as strong as those observed in infected-then-vaccinated individuals. Moreover, it will be interesting to see the impact of breakthrough infection on the avidity of the Abs.
The third dose of the SARS-CoV-2 mRNA vaccine led to high levels of humoral responses against VOCs, irrespective of the interval between the first two doses. We do not yet know about the durability of this immunity, but epidemiological studies have shown a decline in vaccine effectiveness against Omicron within a few months of the third dose (Andrews et al., 2022). We observed a rapid decrease of Ab levels after the three doses with both intervals and to a greater extent in SARS-CoV-2-naive donors. However, it is possible that while humoral responses rapidly decreased after vaccination, cellular responses remain stable. Monitoring the evolution of humoral and cellular responses after the third dose of vaccine, and in particular in the numerous individuals who had Omicron subvariants breakthrough infection after their third dose, will be necessary to determine the need for additional boosts, the best interval time between dose injections, and the populations to be targeted (general population or only population at risk).
Limitations of the study
One limitation of this study is the relatively low number of individuals per group. Our results are nevertheless in agreement with recent epidemiological studies supporting the efficacy of a third dose to protect from Omicron subvariants (Accorsi et al., 2022; Ionescu et al., 2022; Thompson et al., 2022). Another limitation is that the time of blood collection was not exactly the same between the cohorts of vaccinated individuals with the SI and the LI, making side-by-side comparisons more difficult to visualize. Nevertheless, we believe that this is mitigated by presenting the results longitudinally.
STAR★Methods
Key resources table
| REAGENT or RESOURCE | SOURCE | IDENTIFIER |
|---|---|---|
| Antibodies | ||
| LIVE-DEAD Fixable AquaVivid Cell Stain | Thermo Fischer Scientific | Cat# P34957 |
| Mouse monoclonal anti-SARS-CoV-2 Spike (CR3022) | Dr M. Gordon Joyce (Meulen et al., 2006) | RRID: AB_2848080 |
| CV3-25 | (Jennewein et al., 2021) | N/A |
| Peroxidase AffiniPure Goat Anti-Human IgA + IgG + IgM (H + L) | Jackson ImmunoResearch | Cat # 109-035-064; RRID: AB_2337583 |
| Goat anti-Human IgG Fc Cross-Adsorbed Secondary Antibody, HRP | Invitrogen | Cat # 31413; RRID: AB_429693 |
| Alexa Fluor 647 AffiniPure Goat Anti-Human IgA + IgG + IgM (H + L) | Jackson ImmunoResearch | Cat # 109-605-064; RRID: AB_2337886 |
| Cell Proliferation Dye eFluor 670 | Thermo Fisher Scientific | Cat # 65-0840-85 |
| Cell Proliferation Dye eFluor450 | Thermo Fisher Scientific | Cat # 65-0842-85 |
| Biological samples | ||
| SARS-CoV-2 naïve donor blood samples | This paper | N/A |
| SARS-CoV-2 previously infected donor blood samples | This paper | N/A |
| Chemicals, peptides, and recombinant proteins | ||
| Dulbecco’s Modified Eagle’s medium (DMEM) | Wisent | Cat# 319-005-CL |
| Roswell Park Memorial Institute (RPMI) | Thermo Fischer Scientific | Cat# 61870036 |
| Penicillin/Streptomycin | Wisent | Cat# 450-201-EL |
| Fetal Bovine Serum (FBS) | VWR | Cat# 97068–085 |
| Bovine Serum Albumin (BSA) | Sigma | Cat# A7638 |
| Phosphate Buffered Saline (PBS) | ThermoFischer Scientific | Cat# 10010023 |
| Tween 20 | Sigma | Cat# P9416-100ML |
| Puromycin Dihydrochloride | Millipore Sigma | Cat# P8833 |
| Passive Lysis Buffer | Promega | Cat# E1941 |
| Freestyle 293F expression medium | Thermo Fischer Scientific | Cat# A14525 |
| D-Luciferin Potassium Salt | Thermo Fischer Scientific | Cat# L2916 |
| Formaldehyde 37% | Thermo Fischer Scientific | Cat# F79-500 |
| ExpiFectamine 293 transfection reagent | ThermoFisher Scientific | Cat# A14525 |
| Western Lightning Plus-ECL, Enhanced Chemiluminescence Substrate |
Perkin Elmer Life Sciences | Cat# NEL105001EA |
| Ni-NTA agarose | Invitrogen | Invitrogen |
| Experimental models: Cell lines | ||
| HEK293T cells | ATCC | Cat# CRL-3216; RRID: CVCL_0063 |
| 293T-ACE2 cells | (Prévost et al., 2020) | N/A |
| FreeStyle 293F cells | ThermoFischer Scientific | Cat# R79007; RRID: CVCL_D603 |
| CEM.NKr CCR5+ cells | NIH AIDS reagent program | Cat# ARP-4376; RRID: CVCL_X623 |
| CEM.NKr CCR5+.S cells | (Anand et al., 2021) | N/A |
| Recombinant DNA | ||
| pNL4.3 R-E− Luc | NIH AIDS reagent program | Cat# 3418 |
| pIRES2-EGFP | Clontech | Cat# 6029–1 |
| pCG1-SARS-CoV-2 D614G-Spike | (Beaudoin-Bussières et al., 2020) | N/A |
| pCMV3-HCoV-HKU1-Spike | Sino Biological | Cat# VG40021-UT |
| pCAGGS-SARS-CoV-2-B.1.617.2-Spike (Delta) | (Gong et al., 2021) | N/A |
| pCAGGS-SARS-CoV-2-B.1.1.529-Spike (Omicron) | (Chatterjee et al., 2021) | N/A |
| pCAGGS-SARS-CoV-2-BA.1.1 Spike | This paper | N/A |
| pCAGGS-SARS-CoV-2-BA.2 Spike | This paper | N/A |
| pCAGGS-SARS-CoV-2-BA.4/5 Spike | (Tauzin et al., 2022c) | N/A |
| pCMV3-SARS-CoV-2 S2 N-His tag | Sino Biological | Cat # VG40590-NH |
| Software and algorithms | ||
| Flow Jo v10.7.1 | Flow Jo | https://www.flowjo.com |
| GraphPad Prism v8.4.3 | GraphPad | https://www.graphpad.com |
| R | R Core Team | http://www.R-project.org |
| Microsoft Excel v16 | Microsoft Office | https://www.microsoft.com/en-ca/microsoft-365/excel |
| Other | ||
| BD LSRII Flow Cytometer | BD Biosciences | N/A |
| TriStar LB942 Microplate Reader | Berthold Technologies | N/A |
Resource availability
Lead contact
Further information and requests for resources and reagents should be directed to and will be fulfilled by the lead contact, Andrés Finzi (andres.finzi@umontreal.ca).
Materials availability
All unique reagents generated during this study are available from the lead contact without restriction.
Experimental model and subject details
Ethics statement
All work was conducted in accordance with the Declaration of Helsinki in terms of informed consent and approval by an appropriate institutional board. Blood samples were obtained from donors who consented to participate in this research project at CHUM (19.381) and University of Pennsylvania (University of Pennsylvania Institutional Review Board, IRB no. 845061). Plasmas were isolated by centrifugation and Ficoll gradient, and samples stored at −80°C until use.
Human subjects
The study was conducted in 20 SARS-CoV-2 naïve individuals (8 males and 12 females; age range: 33–64 years) and 11 SARS-CoV-2 previously infected individuals (8 males and 3 females; age range: 39–65 years). PI donors were infected during the first wave of COVID-19 in early 2020. All this information is summarized in Table 1. For the comparison between the SI and LI, the study was conducted in 45 SARS-CoV-2 naïve individuals (21 males and 24 females; age range: 22–67 years) and 16 SARS-CoV-2 previously-infected individuals (10 males and 6 females; age range: 23–59 years) for the SI and in 30 SARS-CoV-2 naïve individuals (12 males and 18 females; age range: 21–64 years) and 15 SARS-CoV-2 previously-infected individuals (10 males and 5 females; age range: 29–65 years) for the LI. All this information is summarized in Table 2. No specific criteria such as number of patients (sample size), gender, clinical or demographic were used for inclusion, beyond PCR confirmed SARS-CoV-2 infection in adults and no detection of Abs recognizing the N protein for naïve donors. Some naïve donors (n = 5) were tested positive at V6 and were removed from the analysis at this time point.
Plasma and antibodies
Plasma from SARS-CoV-2 naïve and PI donors were collected, heat-inactivated for 1 h at 56°C and stored at −80°C until ready to use in subsequent experiments. Plasma from uninfected donors collected before the pandemic were used as negative controls and used to calculate the seropositivity threshold in our ELISA, ADCC and flow cytometry assays (see below). The RBD-specific monoclonal antibody CR3022 was used as a positive control in ELISA assays, and the CV3-25 antibody in flow cytometry assays and were previously described (Anand et al., 2020; Beaudoin-Bussières et al., 2020; Jennewein et al., 2021; Meulen et al., 2006; Prévost et al., 2020). Horseradish peroxidase (HRP)-conjugated Abs able to detect the Fc region of human IgG (Invitrogen) was used as secondary Abs to detect Ab binding in ELISA experiments. Alexa Fluor-647-conjugated goat anti-human Abs able to detect all Ig isotypes (anti-human IgM+IgG+IgA; Jackson ImmunoResearch Laboratories) were used as secondary Ab to detect plasma binding in flow cytometry experiments.
Cell lines
293T human embryonic kidney cells (obtained from ATCC) were maintained at 37°C under 5% CO2 in Dulbecco’s modified Eagle’s medium (DMEM) (Wisent) containing 5% fetal bovine serum (FBS) (VWR) and 100 μg/mL of penicillin-streptomycin (Wisent). CEM.NKr CCR5+ cells (NIH AIDS reagent program) were maintained at 37°C under 5% CO2 in Roswell Park Memorial Institute (RPMI) 1640 medium (Gibco) containing 10% FBS and 100 μg/mL of penicillin-streptomycin. 293T-ACE2 cell line was previously reported (Prévost et al., 2020). CEM.NKr CCR5+ cells stably expressing the SARS-CoV-2 S glycoproteins were previously reported (Anand et al., 2021; Beaudoin-Bussières et al., 2021).
Method details
Plasmids
The plasmids encoding the SARS-CoV-2 S variants D614G, Delta (B.1.617.2) and Omicron (B.1.1.529) and the S2 subunit were previously reported (Beaudoin-Bussières et al., 2020; Chatterjee et al., 2021; Gong et al., 2021; Tauzin et al., 2022a). The HCoV-HKU1 S was purchased from Sino Biological. The plasmids encoding the BA.1.1, BA.2 and BA.4/5 S were generated by overlapping PCR for mutagenesis of a codon-optimized wild-type SARS-CoV-2 S gene (GeneArt, ThermoFisher) that was synthesized (Biobasic) and cloned in pCAGGS as a template. All constructs were verified by Sanger sequencing.
Protein expression and purification
FreeStyle 293F cells (Invitrogen) were grown in FreeStyle 293F medium (Invitrogen) to a density of 1 × 106 cells/mL at 37°C with 8% CO2 with regular agitation (150 rpm). Cells were transfected with a plasmid coding for SARS-CoV-2 S WT RBD (Beaudoin-Bussières et al., 2020) using ExpiFectamine 293 transfection reagent, as directed by the manufacturer (Invitrogen). One week later, cells were pelleted and discarded. Supernatants were filtered using a 0.22 μm filter (Thermo Fisher Scientific). The recombinant RBD proteins were purified by nickel affinity columns, as directed by the manufacturer (Invitrogen). The RBD preparations were dialyzed against phosphate-buffered saline (PBS) and stored in aliquots at −80°C until further use. To assess purity, recombinant proteins were loaded on SDS-PAGE gels and stained with Coomassie Blue.
Enzyme-linked immunosorbent assay (ELISA) and RBD avidity index
The SARS-CoV-2 WT RBD ELISA assay used was previously described (Beaudoin-Bussières et al., 2020; Prévost et al., 2020). Briefly, recombinant SARS-CoV-2 WT RBD proteins (2.5 μg/mL), or bovine serum albumin (BSA) (2.5 μg/mL) as a negative control, were prepared in PBS and were adsorbed to plates (MaxiSorp Nunc) overnight at 4°C. Coated wells were subsequently blocked with blocking buffer (Tris-buffered saline [TBS] containing 0.1% Tween20 and 2% BSA) for 1h at room temperature. Wells were then washed four times with washing buffer (Tris-buffered saline [TBS] containing 0.1% Tween20). CR3022 mAb (50 ng/mL) or a 1/500 dilution of plasma were prepared in a diluted solution of blocking buffer (0.1% BSA) and incubated with the RBD-coated wells for 90 min at room temperature. Plates were washed four times with washing buffer followed by incubation with secondary Abs (diluted in a diluted solution of blocking buffer (0.4% BSA)) for 1h at room temperature, followed by four washes. To calculate the RBD-avidity index, we performed in parallel a stringent ELISA, where the plates were washed with a chaotropic agent, 8M of urea, added of the washing buffer. This assay was previously described (Tauzin et al., 2022b). HRP enzyme activity was determined after the addition of a 1:1 mix of Western Lightning oxidizing and luminol reagents (Perkin Elmer Life Sciences). Light emission was measured with a LB942 TriStar luminometer (Berthold Technologies). Signal obtained with BSA was subtracted for each plasma and was then normalized to the signal obtained with CR3022 present in each plate. The seropositivity threshold was established using the following formula: mean of pre-pandemic SARS-CoV-2 negative plasma + (3 standard deviation of the mean of pre-pandemic SARS-CoV-2 negative plasma).
Cell surface staining and flow cytometry analysis
293T cells were co-transfected with a GFP expressor (pIRES2-GFP, Clontech) in combination with plasmids encoding the full-length S of SARS-CoV-2 variants (D614G, Delta and Omicron, BA.1.1 and BA.2), the S2 subunit or the HCoV-HKU1 S. 48h post-transfection, S-expressing cells were stained with the CV3-25 Ab (Jennewein et al., 2021) or plasma (1/250 dilution). AlexaFluor-647-conjugated goat anti-human IgM+IgG+IgA Abs (1/800 dilution) were used as secondary Abs. The percentage of transfected cells (GFP + cells) was determined by gating the living cell population based on viability dye staining (Aqua Vivid, Invitrogen). Samples were acquired on a LSRII cytometer (BD Biosciences) and data analysis was performed using FlowJo v10.7.1 (Tree Star). The seropositivity threshold was established using the following formula: mean of pre-pandemic SARS-CoV-2 negative plasma + (3 standard deviation of the mean of pre-pandemic SARS-CoV-2 negative plasma). The conformational-independent S2-targeting mAb CV3-25 was used to normalize S expression. CV3-25 was shown to effectively recognize all SARS-CoV-2 S variants (Chatterjee et al., 2021; Li et al., 2022).
ADCC assay
This assay was previously described (Anand et al., 2021; Beaudoin-Bussières et al., 2021). For evaluation of anti-SARS-CoV-2 antibody-dependent cellular cytotoxicity (ADCC), parental CEM.NKr CCR5+ cells were mixed at a 1:1 ratio with CEM.NKr cells stably expressing a GFP-tagged full length SARS-CoV-2 S (CEM.NKr.SARS-CoV-2.Spike cells). These cells were stained for viability (AquaVivid; Thermo Fisher Scientific, Waltham, MA, USA) and cellular dyes (cell proliferation dye eFluor670; Thermo Fisher Scientific) to be used as target cells. Overnight rested PBMCs were stained with another cellular marker (cell proliferation dye eFluor450; Thermo Fisher Scientific) and used as effector cells. Stained target and effector cells were mixed at a ratio of 1:10 in 96-well V-bottom plates. Plasma (1/500 dilution) or monoclonal antibody CR3022 (1 μg/mL) were added to the appropriate wells. The plates were subsequently centrifuged for 1 min at 300g, and incubated at 37°C, 5% CO2 for 5 h before being fixed in a 2% PBS-formaldehyde solution. ADCC activity was calculated using the formula: [(% of GFP + cells in Targets plus Effectors) - (% of GFP + cells in Targets plus Effectors plus plasma/antibody)]/(% of GFP + cells in Targets) x 100 by gating on transduced live target cells. All samples were acquired on an LSRII cytometer (BD Biosciences) and data analysis was performed using FlowJo v10.7.1 (Tree Star). The specificity threshold was established using the following formula: mean of pre-pandemic SARS-CoV-2 negative plasma + (3 standard deviation of the mean of pre-pandemic SARS-CoV-2 negative plasma).
Virus neutralization assay
To produce the pseudoviruses, 293T cells were transfected with the lentiviral vector pNL4.3 R-E− Luc (NIH AIDS Reagent Program) and a plasmid encoding for the indicated S glycoprotein (D614G, Delta and Omicron, BA.1.1, BA.2 and BA.4/5) at a ratio of 10:1. Two days post-transfection, cell supernatants were harvested and stored at −80°C until use. For the neutralization assay, 293T-ACE2 target cells were seeded at a density of 1×104 cells/well in 96-well luminometer-compatible tissue culture plates (Perkin Elmer) 24h before infection. Pseudoviral particles were incubated with several plasma dilutions (1/50; 1/250; 1/1250; 1/6250; 1/31250) for 1h at 37°C and were then added to the target cells followed by incubation for 48h at 37°C. Then, cells were lysed by the addition of 30 μL of passive lysis buffer (Promega) followed by one freeze-thaw cycle. An LB942 TriStar luminometer (Berthold Technologies) was used to measure the luciferase activity of each well after the addition of 100 μL of luciferin buffer (15mM MgSO4, 15mM KPO4 [pH 7.8], 1mM ATP, and 1mM dithiothreitol) and 50 μL of 1mM d-luciferin potassium salt (Prolume). The neutralization half-maximal inhibitory dilution (ID50) represents the plasma dilution to inhibit 50% of the infection of 293T-ACE2 cells by pseudoviruses.
Quantification and statistical analysis
Statistical analysis
Symbols represent biologically independent samples from SARS-CoV-2 naïve or SARS-CoV-2 PI individuals. Lines connect data from the same donor. Statistics were analyzed using GraphPad Prism version 8.0.1 (GraphPad, San Diego, CA). Every dataset was tested for statistical normality and this information was used to apply the appropriate (parametric or nonparametric) statistical test. Differences in responses for the same patient before and after vaccination were performed using Wilcoxon tests. Differences in responses between naïve and PI individuals at each time point (V3, V4, V5 and V6) were measured by Mann-Whitney tests. Differences in responses between naïve vaccinated with a SI or a LI, and PI individuals vaccinated with a SI or a LI were measured by Mann-Whitney tests or unpaired t-tests. Differences in responses against the different S for the same patient were measured by Friedman tests. p values <0.05 were considered significant; significance values are indicated as ∗ p < 0.05, ∗∗ p < 0.01, ∗∗∗ p < 0.001, ∗∗∗∗ p < 0.0001. Spearman’s R correlation coefficient was applied for correlations. Statistical tests were two-sided and p < 0.05 was considered significant.
Software scripts and visualization
Edge bundling graphs were generated in undirected mode in R and RStudio using ggraph, igraph, tidyverse, and RColorBrewer packages (R Core Team, 2014). Edges are only shown if p < 0.05, and nodes are sized according to the connecting edges’ r values. Nodes are color-coded according to groups of parameters.
Acknowledgments
The authors are grateful to the donors who participated in this study. The authors thank the CRCHUM BSL3 and Flow Cytometry Platforms for technical assistance. We thank Dr. Stefan Pöhlmann (Georg-August University, Germany) for the plasmid coding for SARS-CoV-2 S glycoproteins and Dr. M. Gordon Joyce (U.S. MHRP) for the monoclonal Ab CR3022. This work was supported by le Ministère de l’Économie et de l’Innovation du Québec, Programme de soutien aux organismes de recherche et d’innovation to A.F. and by the Fondation du CHUM. This work was also supported by CIHR Foundation grant 352417, by CIHR Operating Pandemic and Health Emergencies Research grant 177958, a CIHR stream 1 and 2 for SARS-CoV-2 Variant Research to A.F., and by Exceptional Fund COVID-19 from the Canada Foundation for Innovation (CFI) 41027 to A.F. and D.E.K. Work on variants presented was also supported by the Sentinelle COVID Quebec network led by the LSPQ in collaboration with Fonds de Recherche du Québec - Santé (FRQS) to A.F. This work was also partially supported by a CIHR COVID-19 Rapid Response grant (OV3 170632) and CIHR stream 1 SARS-CoV-2 Variant Research to M.C. and CITF to R.B. and A.F. A.F. is the recipient of Canada Research Chair on Retroviral Entry no. RCHS0235 950-232424. M.C. is a Tier II Canada Research Chair in Molecular Virology and Antiviral Therapeutics. V.M.-L. is supported by a FRQS Junior 1 salary award. D.E.K. is a FRQS Merit Research Scholar. G.B.-B. is the recipient of a FRQS PhD fellowship. A.T. and A.L. were supported by MITACS Accélération postdoctoral fellowships. This work was also supported by NIH grants AI108545, AI155577, AI149680, and U19AI082630 (to E.J.W.); the University of Pennsylvania Perelman School of Medicine COVID Fund (to R.R.G. and E.J.W.); the University of Pennsylvania Perelman School of Medicine 21st Century Scholar Fund (to R.R.G.); and the Paul and Daisy Soros Fellowship for New Americans (to R.R.G.). The funders had no role in study design, data collection and analysis, decision to publish, or preparation of the manuscript.
Author contributions
A.T. and A.F. conceived the study. A.T., S.Y.G., D.C., G.B.-B., L.M., M.B., A.L., C.B., G.G.-L., Y.B., S.D., J.R., and A.F. performed, analyzed, and interpreted the experiments. A.T., J.R., and R.D. performed statistical analysis. S.Y.G., M.M.P., R.R.G., G.B.-B., A.L., J.O., G.G.-L., H.M., G.G., Y.B., M.C., A.R.G., E.J.W., and A.F. contributed unique reagents. J.C.W., L.G., C.M., P.A., C.T., D.E.K., and V.M.-L. collected and provided clinical samples. G.D.S., J.F., D.E.K., E.J.W., and R.B. provided scientific input related to VOCs and vaccine efficacy. A.T. and A.F. wrote the manuscript with input from others. Every author has read, edited, and approved the final manuscript.
Declaration of interests
A.R.G. is a consultant for Relation Therapeutics. E.J.W. is consulting for or is an advisor for Merck, Marengo, Janssen, Related Sciences, Synthekine, and Surface Oncology. E.J.W. is a founder of Surface Oncology, Danger Bio, and Arsenal Biosciences.
Published: October 25, 2022
Footnotes
Supplemental information can be found online at https://doi.org/10.1016/j.celrep.2022.111554.
Supplemental information
Data and code availability
-
•
All data reported in this paper will be shared by the lead contact (andres.finzi@umontreal.ca) upon request.
-
•
This paper does not report original code.
-
•
Any additional information required to reanalyze the data reported in this paper is available from the lead contact (andres.finzi@umontreal.ca) upon request.
References
- Accorsi E.K., Britton A., Fleming-Dutra K.E., Smith Z.R., Shang N., Derado G., Miller J., Schrag S.J., Verani J.R. Association between 3 doses of mRNA COVID-19 vaccine and symptomatic infection caused by the SARS-CoV-2 Omicron and Delta variants. JAMA. 2022;327:639–651. doi: 10.1001/jama.2022.0470. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Anand S.P., Prévost J., Nayrac M., Beaudoin-Bussières G., Benlarbi M., Gasser R., Brassard N., Laumaea A., Gong S.Y., Bourassa C., et al. Longitudinal analysis of humoral immunity against SARS-CoV-2 Spike in convalescent individuals up to eight months post-symptom onset. Cell Rep. Med. 2021;2:100290. doi: 10.1016/j.xcrm.2021.100290. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Anand S.P., Prévost J., Richard J., Perreault J., Tremblay T., Drouin M., Fournier M.-J., Lewin A., Bazin R., Finzi A. 2020. High-throughput Detection of Antibodies Targeting the SARS-CoV-2 Spike in Longitudinal Convalescent Plasma Samples (Microbiology) [DOI] [PMC free article] [PubMed] [Google Scholar]
- Andreano E., Paciello I., Piccini G., Manganaro N., Pileri P., Hyseni I., Leonardi M., Pantano E., Abbiento V., Benincasa L., et al. Nature. 2021;600:530–535. doi: 10.1038/s41586-021-04117-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Andrews N., Stowe J., Kirsebom F., Toffa S., Rickeard T., Gallagher E., Gower C., Kall M., Groves N., O’Connell A.-M., et al. Covid-19 vaccine effectiveness against the Omicron (B.1.1.529) variant. N. Engl. J. Med. 2022;386:1532–1546. doi: 10.1056/NEJMoa2119451. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Beaudoin-Bussières G., Laumaea A., Anand S.P., Prévost J., Gasser R., Goyette G., Medjahed H., Perreault J., Tremblay T., Lewin A., et al. Decline of humoral responses against SARS-CoV-2 spike in convalescent individuals. mBio. 2020;11 doi: 10.1128/mBio.02590-20. 025900-e2620. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Beaudoin-Bussières G., Richard J., Prévost J., Goyette G., Finzi A. A new flow cytometry assay to measure antibody-dependent cellular cytotoxicity against SARS-CoV-2 Spike-expressing cells. STAR Protoc. 2021;2:100851. doi: 10.1016/j.xpro.2021.100851. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Björkman C., Näslund K., Stenlund S., Maley S.W., Buxton D., Uggla A. An IgG avidity ELISA to discriminate between recent and chronic neospora caninum infection. J. Vet. Diagn. Invest. 1999;11:41–44. doi: 10.1177/104063879901100106. [DOI] [PubMed] [Google Scholar]
- Chatterjee D., Tauzin A., Marchitto L., Gong S.Y., Boutin M., Bourassa C., Beaudoin-Bussières G., Bo Y., Ding S., Laumaea A., et al. SARS-CoV-2 Omicron Spike recognition by plasma from individuals receiving BNT162b2 mRNA vaccination with a 16-weeks interval between doses. Cell Rep. 2021;38:110429. doi: 10.1016/j.celrep.2022.110429. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chatterjee D., Tauzin A., Marchitto L., Gong S.Y., Boutin M., Bourassa C., Beaudoin-Bussières G., Bo Y., Ding S., Laumaea A., et al. SARS-CoV-2 Omicron Spike recognition by plasma from individuals receiving BNT162b2 mRNA vaccination with a 16-week interval between doses. Cell Rep. 2022;38:110429. doi: 10.1016/j.celrep.2022.110429. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chen J., Wang R., Gilby N.B., Wei G.-W. Omicron variant (B.1.1.529): infectivity, vaccine breakthrough, and antibody resistance. J. Chem. Inf. Model. 2022;62:412–422. doi: 10.1021/acs.jcim.1c01451. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Choi J.Y., Smith D.M. SARS-CoV-2 variants of concern. Yonsei Med. J. 2021;62:961–968. doi: 10.3349/ymj.2021.62.11.961. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Crotty S. Hybrid immunity. Science. 2021;372:1392–1393. doi: 10.1126/science.abj2258. [DOI] [Google Scholar]
- Dhar M.S., Marwal R., Vs R., Ponnusamy K., Jolly B., Bhoyar R.C., Sardana V., Naushin S., Rophina M., Mellan T.A., et al. Genomic characterization and epidemiology of an emerging SARS-CoV-2 variant in Delhi, India. Science. 2021;374:995–999. doi: 10.1126/science.abj9932. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Doria-Rose N.A., Shen X., Schmidt S.D., O’Dell S., McDanal C., Feng W., Tong J., Eaton A., Maglinao M., Tang H., et al. Booster of mRNA-1273 strengthens SARS-CoV-2 Omicron neutralization. medRxiv. 2021 doi: 10.1101/2021.12.15.21267805. Preprint at. [DOI] [Google Scholar]
- Ferdinands J.M., Rao S., Dixon B.E., Mitchell P.K., DeSilva M.B., Irving S.A., Lewis N., Natarajan K., Stenehjem E., Grannis S.J., et al. Waning 2-dose and 3-dose effectiveness of mRNA vaccines against COVID-19–associated emergency department and urgent care encounters and hospitalizations among adults during periods of Delta and Omicron variant predominance — VISION network, 10 states, august 2021–january 2022. MMWR Morb. Mortal. Wkly. Rep. 2022;71:255–263. doi: 10.15585/mmwr.mm7107e2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fialová L., Petráčková M., Kuchař O. Comparison of different enzyme-linked immunosorbent assay methods for avidity determination of antiphospholipid antibodies. J. Clin. Lab. Anal. 2017;31:e22121. doi: 10.1002/jcla.22121. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Goel R.R., Apostolidis S.A., Painter M.M., Mathew D., Pattekar A., Kuthuru O., Gouma S., Hicks P., Meng W., Rosenfeld A.M., et al. Distinct antibody and memory B cell responses in SARS-CoV-2 naïve and recovered individuals following mRNA vaccination. Sci. Immunol. 2021;6:eabi6950. doi: 10.1126/sciimmunol.abi6950. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gong S.Y., Chatterjee D., Richard J., Prévost J., Tauzin A., Gasser R., Bo Y., Vézina D., Goyette G., Gendron-Lepage G., et al. Contribution of single mutations to selected SARS-CoV-2 emerging variants spike antigenicity. Virology. 2021;563:134–145. doi: 10.1016/j.virol.2021.09.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gruell H., Vanshylla K., Tober-Lau P., Hillus D., Schommers P., Lehmann C., Kurth F., Sander L.E., Klein F. mRNA booster immunization elicits potent neutralizing serum activity against the SARS-CoV-2 Omicron variant. Nat. Med. 2022;28:477–480. doi: 10.1038/s41591-021-01676-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ionescu I.G., Skowronski D.M., Sauvageau C., Chuang E., Ouakki M., Kim S., Serres G.D. BNT162b2 Effectiveness against Delta & Omicron Variants in Teens by Dosing Interval and Duration. 2022. [DOI] [PMC free article] [PubMed]
- Jennewein M.F., MacCamy A.J., Akins N.R., Feng J., Homad L.J., Hurlburt N.K., Seydoux E., Wan Y.-H., Stuart A.B., Edara V.V., et al. Isolation and characterization of cross-neutralizing coronavirus antibodies from COVID-19+ subjects. Cell Rep. 2021;36:109353. doi: 10.1016/j.celrep.2021.109353. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kumar S., Karuppanan K., Subramaniam G. Omicron (BA.1) and sub-variants (BA.1, BA.2 and BA.3) of SARS-CoV-2 spike infectivity and pathogenicity: a comparative sequence and structural-based computational assessment. bioRxiv. 2022 doi: 10.1101/2022.02.11.480029. Preprint at. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li W., Chen Y., Prévost J., Ullah I., Lu M., Gong S.Y., Tauzin A., Gasser R., Vézina D., Anand S.P., et al. Structural basis and mode of action for two broadly neutralizing antibodies against SARS-CoV-2 emerging variants of concern. Cell Rep. 2022;38:110210. doi: 10.1016/j.celrep.2021.110210. [DOI] [PMC free article] [PubMed] [Google Scholar]
- ter Meulen J., van den Brink E.N., Poon L.L.M., Marissen W.E., Leung C.S.W., Cox F., Cheung C.Y., Bakker A.Q., Bogaards J.A., van Deventer E., et al. Human monoclonal antibody combination against SARS coronavirus: synergy and coverage of escape mutants. PLoS Med. 2006;3:e237. doi: 10.1371/journal.pmed.0030237. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Miyamoto S., Arashiro T., Adachi Y., Moriyama S., Kinoshita H., Kanno T., Saito S., Katano H., Iida S., Ainai A., et al. Vaccination-infection interval determines cross-neutralization potency to SARS-CoV-2 Omicron after breakthrough infection by other variants. Med. 2022;3:249–261.e4. doi: 10.1016/j.medj.2022.02.006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mohapatra R.K., Kandi V., Sarangi A.K., Verma S., Tuli H.S., Chakraborty S., Chakraborty C., Dhama K. The recently emerged BA.4 and BA.5 lineages of Omicron and their global health concerns amid the ongoing wave of COVID-19 pandemic – Correspondence. Int. J. Surg. 2022;103:106698. doi: 10.1016/j.ijsu.2022.106698. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nayrac M., Dubé M., Sannier G., Nicolas A., Marchitto L., Tastet O., Tauzin A., Brassard N., Lima-Barbosa R., Beaudoin-Bussières G., et al. Temporal associations of B and T cell immunity with robust vaccine responsiveness in a 16-week interval BNT162b2 regimen. Cell Rep. 2022;39:111013. doi: 10.1016/j.celrep.2022.111013. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nemet I., Kliker L., Lustig Y., Zuckerman N., Erster O., Cohen C., Kreiss Y., Alroy-Preis S., Regev-Yochay G., Mendelson E., Mandelboim M. Third BNT162b2 vaccination neutralization of SARS-CoV-2 Omicron infection. N. Engl. J. Med. 2022;386:492–494. doi: 10.1056/NEJMc2119358. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Payne R.P., Longet S., Austin J.A., Skelly D.T., Dejnirattisai W., Adele S., Meardon N., Faustini S., Al-Taei S., Moore S.C., et al. Immunogenicity of standard and extended dosing intervals of BNT162b2 mRNA vaccine. Cell. 2021;184:5699–5714.e11. doi: 10.1016/j.cell.2021.10.011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Prévost J., Gasser R., Beaudoin-Bussières G., Richard J., Duerr R., Laumaea A., Anand S.P., Goyette G., Benlarbi M., Ding S., et al. Cross-sectional evaluation of humoral responses against SARS-CoV-2 spike. Cell Rep. Med. 2020;1:100126. doi: 10.1016/j.xcrm.2020.100126. [DOI] [PMC free article] [PubMed] [Google Scholar]
- R Core Team . R Foundation for Statistical Computing; 2014. R: A Language and Environment for Statistical Computing.http://www.R-project.org/ [Google Scholar]
- Schmidt F., Muecksch F., Weisblum Y., Da Silva J., Bednarski E., Cho A., Wang Z., Gaebler C., Caskey M., Nussenzweig M.C., et al. Plasma neutralization of the SARS-CoV-2 Omicron variant. N. Engl. J. Med. 2022;386:599–601. doi: 10.1056/NEJMc2119641. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tauzin A., Chatterjee D., Dionne K., Gendron-Lepage G., Medjahed H., Bo Y., Perreault J., Goyette G., Gokool L., Arlotto P., et al. SARS-CoV-2 BA.4/5 Spike recognition and neutralization elicited after the third dose of mRNA vaccine. 2022. [DOI] [PMC free article] [PubMed]
- Tauzin A., Gendron-Lepage G., Nayrac M., Anand S.P., Bourassa C., Medjahed H., Goyette G., Dubé M., Bazin R., Kaufmann D.E., Finzi A. Evolution of anti-RBD IgG avidity following SARS-CoV-2 infection. Viruses. 2022;14:532. doi: 10.3390/v14030532. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tauzin A., Gong S.Y., Beaudoin-Bussières G., Vézina D., Gasser R., Nault L., Marchitto L., Benlarbi M., Chatterjee D., Nayrac M., et al. Strong humoral immune responses against SARS-CoV-2 Spike after BNT162b2 mRNA vaccination with a 16-week interval between doses. Cell Host Microbe. 2022;30:97–109.e5. doi: 10.1016/j.chom.2021.12.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tauzin A., Nayrac M., Benlarbi M., Gong S.Y., Gasser R., Beaudoin-Bussières G., Brassard N., Laumaea A., Vézina D., Prévost J., et al. A single dose of the SARS-CoV-2 vaccine BNT162b2 elicits Fc-mediated antibody effector functions and T cell responses. Cell Host Microbe. 2021;29:1137–1150.e6. doi: 10.1016/j.chom.2021.06.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Thompson M.G., Natarajan K., Irving S.A., Rowley E.A., Griggs E.P., Gaglani M., Klein N.P., Grannis S.J., DeSilva M.B., Stenehjem E., et al. Effectiveness of a third dose of mRNA vaccines against COVID-19-associated emergency department and urgent care encounters and hospitalizations among adults during periods of Delta and Omicron variant predominance - VISION network, 10 states, august 2021-january 2022. MMWR Morb. Mortal. Wkly. Rep. 2022;71:139–145. doi: 10.15585/mmwr.mm7104e3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ullah I., Prévost J., Ladinsky M.S., Stone H., Lu M., Anand S.P., Beaudoin-Bussières G., Symmes K., Benlarbi M., Ding S., et al. Live imaging of SARS-CoV-2 infection in mice reveals that neutralizing antibodies require Fc function for optimal efficacy. Immunity. 2021;54:2143–2158.e15. doi: 10.1016/j.immuni.2021.08.015. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Viana R., Moyo S., Amoako D.G., Tegally H., Scheepers C., Althaus C.L., Anyaneji U.J., Bester P.A., Boni M.F., Chand M., et al. Rapid epidemic expansion of the SARS-CoV-2 Omicron variant in southern Africa. Nature. 2022;603:679–686. doi: 10.1038/s41586-022-04411-y. [DOI] [PMC free article] [PubMed] [Google Scholar]
- WHO . 2022. Tracking SARS-CoV-2 Variants.https://www.who.int/en/activities/tracking-SARS-CoV-2-variants/ [Google Scholar]
- WHO . 2022. Coronavirus (COVID-19) Dashboard | WHO Coronavirus (COVID-19) Dashboard with Vaccination Data.https://covid19.who.int/table [Google Scholar]
- Young C., Brink R. The unique biology of germinal center B cells. Immunity. 2021;54:1652–1664. doi: 10.1016/j.immuni.2021.07.015. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Data Availability Statement
-
•
All data reported in this paper will be shared by the lead contact (andres.finzi@umontreal.ca) upon request.
-
•
This paper does not report original code.
-
•
Any additional information required to reanalyze the data reported in this paper is available from the lead contact (andres.finzi@umontreal.ca) upon request.