Abstract
Enteropathogenic Escherichia coli (EPEC) produces the bundle-forming pilus (BFP), a type IV fimbria that has been implicated in virulence, autoaggregation, and localized adherence to epithelial cells. The bfpE gene is one of a cluster of bfp genes previously shown to encode functions that direct BFP biosynthesis. Here, we show that an EPEC strain carrying a nonpolar mutation in bfpE fails to autoaggregate, adhere to HEp-2 cells, or form BFP, thereby demonstrating that BfpE is required for BFP biogenesis. BfpE is a cytoplasmic membrane protein of the GspF family. To determine the membrane topology of BfpE, we fused bfpE derivatives containing 3′ truncations and/or internal deletions to alkaline phosphatase and/or β-galactosidase reporter genes, whose products are active only when localized to the periplasm or cytoplasm, respectively. In addition, we constructed BfpE sandwich fusions using a dual alkaline phosphatase/β-galactosidase reporter cassette and analyzed BfpE deletion derivatives by sucrose density flotation gradient fractionation. The data from these analyses support a topology in which BfpE contains four hydrophobic transmembrane (TM) segments, a large cytoplasmic segment at its N terminus, and a large periplasmic segment near its C terminus. This topology is dramatically different from that of OutF, another member of the GspF family, which has three TM segments and is predominantly cytoplasmic. These findings provide a structural basis for predicting protein-protein interactions required for assembly of the BFP biogenesis machinery.
Bundle-forming pili (BFP) are type IV fimbriae expressed by enteropathogenic Escherichia coli (EPEC), a bacterium that causes infantile diarrhea (25). BFP are a demonstrated EPEC virulence factor (6) and can elicit an immune response in naturally occurring infections (40). The presence of BFP is required for two notable EPEC phenotypes. Localized adherence, the classical phenotype, describes the binding of EPEC to cultured epithelial cells in a characteristic clustered formation (56). A similar pattern has been noted in EPEC infections in vivo (52). Autoaggregation, the second phenotype, describes the formation of dynamic multicellular clusters of EPEC grown in tissue culture medium (6). The relevance of this phenomenon to EPEC infection of the human intestinal tract is unclear.
Few details are known about the molecular mechanisms of type IV fimbrial biogenesis. Our current concept of BFP synthesis is as follows (21). BFP fibers are polymers of a pilin protein known as bundlin. Coincident with or following their synthesis, bundlin precursors are probably anchored in the cytoplasmic membrane (via a stretch of hydrophobic residues located near their N termini), with the majority of the polypeptide being exported to the periplasm. On the cytoplasmic side of the membrane, the N-terminal leader peptide of prebundlin is removed by the BfpP prepilin peptidase (75). In the periplasm, the formation of a disulfide bond required for bundlin stability is catalyzed by the oxidant DsbA (74). Following these events, the modified bundlin monomers are somehow removed from the membrane and polymerized into fimbriae that are extruded through the outer membrane.
BFP biogenesis is dependent on a cluster of 14 bfp genes located on a large plasmid found in many EPEC strains (58, 60). Expression of these 14 genes in a K-12 laboratory strain of E. coli that is normally nonpiliated is sufficient to elicit BFP formation (60). The bfpA gene encodes prebundlin (17, 57), while the bfpP gene encodes the prepilin peptidase (4, 75). A third gene, bfpB, encodes an outer membrane lipoprotein that is a member of the secretin family and is required for BFP biogenesis (4, 49). The bfpF gene encodes a putative cytoplasmic membrane protein that is not essential for BFP biogenesis but is required for the dynamic behavior of EPEC autoaggregates and BFP bundles (3, 6, 33). The precise functions of the 10 remaining bfp gene products are unknown. At least four of them (bfpC, bfpD, bfpG, and bfpL) are required for BFP biogenesis, while bfpH is not (4, 6, 58, 60). The seventh gene in the bfp cluster, bfpE, encodes a putative polytopic cytoplasmic membrane protein, BfpE, which has been detected using inducible T7 RNA polymerase expression systems (58, 60). BfpE is a member of the GspF family of proteins, which includes components of other systems that transport macromolecules or macromolecular assemblies (including type IV pili) across bacterial envelopes (48, 53). In this report, we describe preliminary analyses of the function and structure of BfpE.
MATERIALS AND METHODS
Bacterial strains and growth conditions.
The E. coli strains used in this study are listed in Table 1. Strains were routinely cultured in Luria-Bertani (LB) broth or on LB agar at 37°C. Antibiotics and/or chromogenic substrates were added as necessary at the following concentrations: ampicillin, 200 μg/ml; chloramphenicol, 20 μg/ml; nalidixic acid, 50 μg/ml; kanamycin, 50 μg/ml; 5-bromo-4-chloro-3-indolylphosphate (BCIP), 40 μg/ml; 5-bromo-4-chloro-3-indolyl-β-d-galactopyranoside (X-Gal), 40 μg/ml. Dual-indicator plates were prepared as described elsewhere (1).
TABLE 1.
E. coli strains used in this study
| Strain | Reference or source | Description or genotype |
|---|---|---|
| CC118 | 38 | araD139 Δ(ara-leu)7697 argE(Am) galE galK ΔlacX74 ΔphoA20 recA1 rpoB rpsE thi |
| DH5α | 54 | deoR endA1 gyrA96 hsdR17 φ80ΔlacZM15 recA1 relA1 supE44 thi-1 Δ(lacIZYA-argF)U169 |
| DH5αλpir | 41 | DH5α carrying λpir function necessary for autonomous replication of pCVD422-based suicide vector |
| E2348/69 | 35 | Prototype O127:H6 EPEC strain carrying pMAR2; NaIr |
| HB101 | 54 | ara-14 galK2 Δ(gpt-proA)62 hsdS20 lacY1 leuB6 mtl-1 recA13 rpsL20 supE44 thi-1 xyl-5 Δ(mcrC-mrr) (F′) |
| TG1 | 54 | ΔhsdS Δ(lac-proAB) supE thi F′ [traD36 proAB+ laclq ΔlacZM15] |
| TOP10 | Invitrogen | araD139 Δ(ara-leu)7697 deoR endA1 galK galU ΔlacX74 φ80ΔlacZM15 mcrA Δ(mrr-hsdRMS-mcrBC) nupG recA1 rpsL (F−) |
| UMD901 | 74 | E2348/69 bfpA (C129S) |
| UMD932 | 4 | E2348/69 bfpP::aphA-3 |
| UMD934 | This study | E2348/69 bfpE::aphA-3 |
Autoaggregation assay.
EPEC strains were cultured overnight at 37°C in LB (plus ampicillin as appropriate). The resulting stationary-phase cultures were diluted 1:100 or 1:250 (making adjustments for the optical density at 600 nm [OD600]) into 20 ml of Dulbecco's modified Eagle medium plus nutrient mixture F-12 (DMEM/F-12) containing 15 mM HEPES buffer and lacking phenol red (Gibco-BRL Life Technologies no. 11039-021). The DMEM/F-12 cultures were shaken at 250 rpm for 5 h in 50-ml conical polypropylene tubes at 37°C. Autoaggregation was gauged by visually inspecting the cultures for bacterial aggregates and sedimentation, by examining 5-μl aliquots of the culture under phase-contrast microscopy, and by determining the aggregation index (AI) (4). To determine AI, two equivalent 1-ml samples were removed from each culture. The OD600 of the first sample was measured immediately using a spectrophotometer. The second sample was agitated for 1 min on a vortex mixer before the OD600 was measured. AI was calculated by subtracting the OD600 of the first sample from that of the second, dividing the result by the value of the first sample, and multiplying by 100.
Localized adherence assay.
Localized adherence to HEp-2 laryngeal carcinoma cells was assayed as described previously (19), using the eight-well chamber slide modification.
Transmission electron microscopy.
To prepare samples for electron microscopy, 1-ml aliquots were removed from EPEC cultures grown in DMEM for 6 h and the bacteria were pelleted by brief centrifugation. Most of the medium was decanted, and the bacterial pellet was gently resuspended in the remainder. A 10-μl aliquot was spotted onto electron microscopy grids, which were dried for 10 min and then stained and examined as described previously (3, 60).
Sequence analysis.
Eight computer programs available on the Internet were used to examine the BfpE protein sequence for the presence of hydrophobic regions having the potential to be transmembrane (TM) segments. These programs are DAS (13) (http://www.sbc.su.se/∼miklos/DAS/), HMMTOP (65) (www.enzim.hu/hmmtop/), PHDhtm/PHDtopology (50, 51) (www.embl-heidelberg.de/predictprotein/), PSORT (43) (http://psort.nibb.ac.jp/), SOSUI (29) (http://sosui.proteome.bio.tuat.ac.jp/sosuiframe0.html), TMHMM (59) (www.cbs.dtu.dk/services/TMHMM-1.0/), TMpred (30) (www.isrec.isb-sib.ch/software/TMPRED_form.html), and TopPred2 (12, 68) (http://www.sbc.su.se /∼erikw/toppred2/). Five of these programs (HMMTOP, PHDtopology, TMHMM, TMpred, and TopPred2) were also used to predict the membrane topology of BfpE.
Plasmids, primers, PCR, and sequencing.
Source plasmids used for this study are listed in Table 2. The plasmids constructed during the course of this work are described below. Oligonucleotide primers used for PCR and plasmid sequencing are listed in Table 3. The University of Maryland School of Medicine Biopolymer Laboratory performed all primer syntheses as well as plasmid sequencing. Standard techniques were used for DNA manipulation (54). Plasmids were maintained in strain DH5α or CC118 and were prepared using the Wizard Plus SV Minipreps DNA Purification System (Promega). PCR was carried out using Taq (Gibco-BRL) or Pfu (Stratagene) polymerase as specified by the manufacturer.
TABLE 2.
Source plasmids used in this study
| Plasmid | Reference or source | Description |
|---|---|---|
| pBAD/myc-His B | Invitrogen | Expression vector with araBAD promoter, araC regulator, and 3′ double-epitope tag (Ap) |
| pBAD/gIII A | Invitrogen | Expression vector with araBAD promoter, araC regulator, 5′ phage fd gene III signal sequence, and 3′ double-epitope tag (Ap) |
| pCVD442 | 18 | Positive-selection suicide vector carrying sacB, oriR6K, and mobRP4 (Ap) |
| pCVD433::31 TnphoA | 16 | MluI fragment from EPEC mutant 31-6-1(1) containing bfpA4::TnphoA cloned into the MluI site of pCVD433 (Cm) |
| pHZZ4B1 | 75 | pACYC184 carrying a 4.0-kb BamHI fragment of pMAR2 that contains a partial bfp gene cluster (′bfpD bfpE bfpF bfpP bfpH′) (Cm) |
| pKDS135 | 60 | pBR322 carrying the entire bfp gene cluster |
| pMA632 | 1 | pBluescript II SK (+) carrying ′phoA-lacZα cassette |
| pMAR2 | 5 | Enterocyte adherence factor plasmid present in EPEC strain E2348/69 |
| pMD103 | 31 | pBluescript derivative carrying lacZYA |
| pRK2073 | 10 | Helper plasmid used to mobilize suicide vector |
| pTrc99A | 2 | Low-copy trc promoter expression vector carrying the lacIq gene (Ap) |
| pUC18K | 41 | pUC18 carrying a nonpolar aphA-3 kanamycin resistance cassette (Ap Kn) |
TABLE 3.
Oligonucleotide primers used in this study
| Primer | Sequenceab | Annealing sitea | Restriction site(s)b |
|---|---|---|---|
| Donne-200 | 5′-GGAAGAACAGTATGTCGAGC-3′ | aphA-3 | |
| Donne-211 | 5′-GGGGATCCATTACCTCAGCCCCAGAGCGGCT-3′ | phoA | BamHI |
| Donne-222 | 5′-GATATTGCCGTGGTACGTT-3′ | phoA | |
| Donne-223 | 5′-AACGTACCACGGCAATATC-3′ | phoA | |
| Donne-224 | 5′-CGGGGTACCCCCGGGCCTGTTCTGGAAAACC-3′ | phoA | KpnI, AvaI-XmaI |
| Donne-227 | 5′-GGGGATCCGGCCTGCCCGGTTATTATTA-3′ | lacZ | BamHI |
| Donne-228 | 5′-ATAGGTACCCCCGGGCTGGCCGTCGTTTTACAA-3′ | lacZ | KpnI, AvaI-XmaI |
| Donne-243 | 5′-ATAGTCCATGGCGAAAGAGAAATTAAACAGACTGC-3′ | bfpE | NcoI |
| Donne-256 | 5′-TAATCCCGGGTGCTAGCTTACTCGAGGAATCTTGCTGCATCAC-3′ | bfpE | AvaI-XmaI, NheI, XhoI |
| Donne-272 | 5′-CCCATCGCCAATCAGCAAAATA-3′ | phoA | |
| Donne-273 | 5′-CGGGCCTCTTCGCTATTA-3′ | lacZ | |
| Donne-305 | 5′-TAATCCCGGGTGCGCCTTTTACTTTCT-3′ | bfpE | AvaI-XmaI |
| Donne-315 | 5′-TAACGCCGGCTCAGTTCAATGGCAGGATGAC-3′ | bfpE | Ngo-MIV |
| Donne-316 | 5′-TAAACCCGGGAGCAACATTTTTCAGAACAGTCAT-3′ | bfpE | AvaI-XmaI |
| Donne-317 | 5′-TAAACCCGGGAGCTATCGACCAGGGAATAAAT-3′ | bfpE | AvaI-XmaI |
| Donne-318 | 5′-TTAACCCGGGAGCAAAGAAGAGACGCAACCT-3′ | bfpE | AvaI-XmaI |
| Donne-319 | 5′-TAAACCCGGGAGCCCACCCCCTCATCGCTTCA-3′ | bfpE | AvaI-XmaI |
| Donne-343 | 5′-TCACTGCCGGCGAAGCCTTACGAATAAT-3′ | bfpE | NgoMIV |
| Donne-344 | 5′-CGGACATTTTTCACTGGTC-3′ | phoA | |
| Donne-383 | 5′-ACGCTGCGCGTAACCACCACA-3′ | f1 origin | |
| Donne-384 | 5′-ATATCCCGGGGCCCATTCGCCATTCAGG-3′ | lacZ | AvaI-XmaI |
| Donne-385 | 5′-TATCGGGCCCCTGTTCTGGAAAACCGGGCTGCTCAG-3′ | phoA | EcoO1091 |
| Donne-386 | 5′-AATTGGGGCCCATTCGCCATTCAGG-3′ | lacZ | EcoO1091 |
| Donne-486 | 5′-TCAGGACCATGGCGGAAGCCTTACGAATAAT-3′ | bfpE | NcoI |
| Donne-507 | 5′-CTGTTGCTCTAGAAATCTTGCTGCATCAC-3′ | bfpE | XbaI |
| Donne-508 | 5′-GGCGTGCTCTAGAACATTTTTCAGAACAGTCAT-3′ | bfpE | XbaI |
The underlined portion of the primer sequence anneals to the gene indicated.
The italicized portion of a primer sequence indicates a restriction site present in a PCR product prepared with that primer.
Construction of strain UMD934 containing a nonpolar bfpE mutation.
A previously described allelic replacement strategy (20) was modified to disrupt the bfpE gene in wild-type EPEC strain E2348/69. Plasmid pHZZ4B1 was linearized at a unique AflIII site specified by codons 130 and 131 of bfpE and then treated with Klenow fragment of DNA polymerase I. The blunted ends were ligated to the 850-bp SmaI fragment from pUC18K carrying the nonpolar aphA-3 cassette specifying kanamycin resistance (41), creating pTEB43-K1A. Restriction analysis followed by sequencing using the Donne-200 primer confirmed that the aphA-3 gene was inserted in the same orientation as bfpE and was in the proper reading frame to reinitiate translation from the 3′ end of the transcribed cassette. Plasmid pTEB43-K1A and suicide vector pCVD442 were both digested with SalI and then ligated to form pTEB44, in which the bfpE::aphA-3 and sacB genes are transcribed in opposite orientations. Plasmid pTEB44 was next digested with SacI to remove a 5.9-kb fragment containing the pACYC184 origin of replication, chloramphenicol resistance gene, and a portion of the bfp cluster. The remaining 9.3-kb SacI fragment was religated to form the bfpE::aphA-3 suicide plasmid pTEB45, which was introduced into DH5αλpir by electroporation. HB101 containing helper plasmid pRK2073 was used to mobilize pTEB45 from DH5αλpir into EPEC strain E2348/69. EPEC transconjugants were selected on LB plates containing nalidixic acid and kanamycin. From among these, an isolate was identified that was resistant to kanamycin and 5% sucrose but sensitive to ampicillin. Replacement of the wild-type bfpE allele by the bfpE::aphA-3 allele in this strain, UMD934, was confirmed by PCR analysis using primers Donne-243 and Donne-256.
Construction of plasmids for membrane topology analysis.
A matched set of plasmids for membrane topology analysis was derived from the low-copy expression vector pTrc99A (Amersham Pharmacia Biotech). One plasmid, pTrcphoA, carries the ′phoA gene (lacking codons Met 1 through Met 26 encoding the signal sequence), while the other, pTrclacZ, carries the ′lacZ gene (lacking codons Met 1 through Ser 7). To construct pTrcphoA, overlap extension PCR was first used to eliminate the NcoI site from ′phoA. Specifically, a portion of ′phoA upstream of and overlapping the NcoI site was amplified using primers Donne-222 and Donne-224, with plasmid pCVD433::31TnphoA as the template. A second portion of ′phoA overlapping and downstream of the NcoI site was prepared using primers Donne-211 and Donne-223. The two portions were annealed and amplified with primers Donne-224 and Donne-211, creating a ′phoA fragment carrying a silent mutation of CAT to CAC at histidine codon 272. The ends of the ′phoA fragment were digested with BamHI and KpnI and inserted into the multiple-cloning site of BamHI-KpnI-digested pTrc99A. To construct pTrclacZ, the ′lacZ gene was amplified by PCR using Donne-227 and Donne-228 as primers and pMD103 as template. The ends of the ′lacZ PCR product were digested with BamHI and KpnI and then ligated to BamHI-KpnI-digested pTrc99A. In both pTrclacZ and pTrcphoA, the reporter genes are fused in frame to a start codon directly downstream of the trc promoter. Fusion genes can be created in these plasmids by inserting gene segments between restriction sites present between the start codon and reporter gene.
Construction of random bfpE′::′phoA and bfpE′::′lacZ gene fusions.
PCR using primers Donn-243 and Donne-256 and pKDS135 as template was used to construct the bfpE355 allele, which differs slightly from wild-type bfpE at both ends. At the 5′ end of the PCR product, the initiator Met codon (Met-1) was replaced with a sequence creating an NcoI site and an extra Ala codon between Met-1 and Lys-2. At the 3′ end of PCR product, the TGA termination codon was replaced with a sequence encoding Leu-Glu-STOP and multiple restriction sites. The bfpE355 PCR product was digested with NcoI and XmaI and ligated to the large fragments of pTrcphoA and pTrclacZ resulting from digestion with the same enzymes, creating pTEB41 and pTEB42. In both plasmids, the bfpE355 gene is followed by a TAA termination codon and a 13-bp linker sequence, rendering the ′lacZ or ′phoA genes out of frame. Plasmid pTEB65 carrying bfpE but no reporter gene was constructed by deleting a 1.36-kb NheI-XbaI fragment containing ′phoA from pTEB41.
The Erase-a-Base system (Promega) for exonuclease III digestion (28) was used to generate 3′ deletion derivatives of the bfpE gene. Both pTEB41 and pTEB42 were linearized at the NheI site located between bfpE and ′lacZ or ′phoA. The NheI ends were filled in with α-phosphorothioates using Klenow DNA polymerase to protect the ′phoA or ′lacZ reporter genes from digestion. The linearized plasmids were next digested with XhoI to expose a 5′ overhang adjacent to bfpE355. Exonuclease III was added, and the digests were incubated at room temperature for 6 to 8 min. Samples were removed at 30-s or 1-min intervals; treated sequentially with S1 nuclease, Klenow, and T4 DNA ligase; and then used to transform strain DH5α, which was plated on media containing ampicillin and BCIP or X-Gal. Dark blue colonies were cultured, and plasmids were prepared from isolates that continued to exhibit the dark blue color after replating. To determine the approximate size of the partial bfpE segments, plasmids containing bfpE′::′lacZ fusions were analyzed by digestion with EcoRV, NcoI, XhoII, XmaI, and XmnI, and plasmids containing bfpE′::′phoA fusions were analyzed by PCR using primers Donne-243 and Donne-272. Sequencing of selected plasmids was carried out using primers Donne-272 (annealing to phoA) and Donne-273 (annealing to lacZ) to determine the precise location of the bfpE fusion junction. Most of the bfpE′ alleles are separated from the ′lacZ or ′phoA genes by a linker sequence (GCACCCGGG) that encodes Ala-Pro-Gly and contains an AvaI site allowing the bfpE′ allele to be subcloned into another context. The exceptions are bfpE352, which is followed by a linker sequence (CCACCCGGG) encoding Pro-Pro-Gly, and bfpE149′, which is fused directly to ′phoA.
Construction of specific bfpE′::′phoA and bfpE′::′lacZ fusions.
The randomly generated bfpE′ alleles described above were transferred as NcoI-AvaI fragments to the vector (pTrcphoA digested with NcoI-AvaI, or pTrclacZ digested with NcoI-XmaI) containing the complementary reporter gene. PCR was used to construct additional plasmids containing bfpE′ segments of specified sizes. Each of the PCR amplifications used pTEB65 as template and Donne-243 as the upstream primer. The downstream primers were Donne-305, Donne-316, Donne-317, Donne-318, and Donne-319. PCR products containing bfpE′ segments were digested with NcoI and AvaI and introduced into pTrcphoA digested with NcoI-AvaI or into pTrclacZ digested with NcoI-XmaI.
Construction of internal deletions in bfpE.
Three plasmids carrying bfpE::′phoA fusions containing internal deletions in bfpE were constructed by introducing downstream segments of bfpE into existing bfpE′::′phoA fusion plasmids. To construct the bfpE292′Δ(210–233)::′phoA plasmid, primers Donne-343 and Donne-344 were used to amplify a fragment from the bfpE292′::′phoA plasmid containing codons 234 through 292 of bfpE plus the proximal end of ′phoA. The resulting 722-bp PCR product was digested with NgoMIV and RsrII and then introduced into the bfpE78′::′phoA plasmid that had been digested with AvaI and RsrII. To construct the bfpE352Δ(116–301)::′phoA plasmid, primers Donne-315 and Donne-344 were used to amplify a fragment from the bfpE352::′phoA plasmid containing codons 302 through 352 of bfpE plus the proximal end of ′phoA. The resulting 697-bp PCR product was digested with NgoMIV and RsrII and then introduced into the bfpE115′::′phoA plasmid that had been digested with AvaI and RsrII. To construct the bfpE352Δ(210–233)::′phoA plasmid, primers Donne-222 and Donne-343 were used to amplify a fragment from the bfpE352::′phoA plasmid containing codons 234 through 352 of bfpE plus the proximal end of ′phoA. The resulting 1,202-bp PCR product was digested with NgoMIV and RsrII and then introduced into the bfpE209′::′phoA plasmid that had been digested with AvaI and RsrII. A linker sequence specifying Ala-Pro-Gly replaced the deleted bfpE codons in each of the new plasmids. A plasmid carrying bfpE with a deletion of sequences specifying HS3 was constructed by replacing an NcoI-EcoO109I fragment of pTEB65 with the corresponding fragment from the bfpE352Δ(210–233)::′phoA plasmid.
Construction of bfpE′::′phoA-lacZα::′bfpE sandwich fusions.
Sandwich fusion plasmids were constructed by in-frame insertion of a dual-reporter ′phoA-lacZα cassette from pMA632 into unique restriction sites within the bfpE gene of pTEB65. The cassette was inserted into the BtrI site as a SmaI-EcoRV fragment. The cassette was amplified from pMA632 by PCR using primers Donne-383 and Donne-384, cut with AvaI, and inserted into the BspEI site. Likewise, the cassette was amplified using primers Donne-385 and 386, cut with EcoO109I, and then inserted into the EcoO109I sites of both pTEB65 and its ΔHS3 derivative. A plasmid carrying a tandem repeat of the cassette at the EcoO109I site of the full-length bfpE gene was a by-product of this construction. The cassette was inserted into the filled-in AvaI site as a SmaI-NruI fragment. The NcoI site in the phoA gene of the AvaI insertion plasmid was eliminated by replacing a 331-bp EcoRI fragment with the corresponding fragment from pTrcphoA, generating pTEB120. Digestion of pTEB120 with NcoI and AvaI or XhoI will precisely remove the bfpE gene, allowing any gene fragments to be placed there to construct novel C-terminal dual-reporter fusions.
Construction of bfpE derivatives with a C-terminal epitope tag.
The full-length bfpE gene (codons 1 to 352) was amplified by PCR from pTEB65 using primers Donne-243 and Donne-507, digested with NcoI and XbaI, and inserted into the corresponding sites in the multiple-cloning site of pBAD/myc-His B. Partial bfpE genes were amplified using primers Donne-486 and Donne-507 (codons 234 to 352) or primers Donne-486 and Donne-508 (codons 234 to 323), digested with NcoI and XbaI, and inserted into the corresponding sites in the multiple-cloning site of pBAD/gIII A.
Immunoblotting.
For the bundlin immunoblot, overnight LB broth cultures of EPEC were diluted 1:100 into 20 ml of DMEM/F-12. After 6 h of growth at 37°C with shaking, the bacteria were pelleted by centrifugation at 2,500 × g for 10 min and then resuspended in 1.2 ml of cell lysis buffer (20 mM Tris-HCl [pH 8.0], 500 mM NaCl, 0.1 mM EDTA, 0.1% Triton X-100). The remaining procedures for sample preparation, protein concentration measurement, and electrophoresis were as previously described (4). The blots were blocked with phosphate-buffered saline containing 0.1% Tween-20 plus 5% nonfat dry milk and then incubated sequentially with a 1:10,000 dilution of ICA4, a monoclonal immunoglobulin G1 antibody raised against BFP (26), with an anti-mouse horseradish peroxidase (HRP) conjugate at a 1:30,000 dilution, and, finally, with enhanced chemiluminescence (ECL) Western blotting detection reagents (Amersham Pharmacia Biotech).
For immunoblots of BfpE-PhoA and BfpE-LacZ fusions, overnight cultures of CC118 plus the appropriate plasmids were diluted 1:20 into LB broth containing ampicillin and, for BfpE-LacZ fusions only, 0.2% glucose. Cultures were grown to an OD600 between 0.3 and 1.0. Aliquots (1 ml) were centrifuged in a microcentrifuge for 3 min, and the bacterial pellets were washed once with 1 ml of 1 M Tris-HCl (pH 8.0) and then resuspended in 1 ml of cell lysis buffer. The OD620s of the samples were determined using a Titertek Multiskan MCC microtiter plate reader. To prepare samples for electrophoresis, 25 μl of 4× sodium dodecyl sulfate (SDS) loading buffer (17) was added to 150 μl of bacterial suspensions, which had been diluted based on the OD620 readings. Samples of 25 μl (BfpE-PhoA fusions) or 10 μl (BfpE-LacZ fusions) were separated by SDS-polyacrylamide gel electrophoresis (PAGE), transferred to Immobilon-P polyvinylidene difluoride membranes (Millipore), and blocked as described above. To detect BfpE-PhoA fusions, blots were incubated sequentially with a 1:2,000 dilution of antibacterial alkaline phosphatase antibody (5 Prime→3 Prime Inc.), with a 1:30,000 dilution of anti-rabbit HRP conjugate (Amersham Pharmacia Biotech), and then with ECL detection reagents. To detect BfpE-LacZ fusions, blots were incubated sequentially with anti-β-galactosidase antibody (5 Prime→3 Prime Inc.) at a 1:4,000 dilution, with anti-rabbit HRP conjugate at a 1:40,000 dilution, and then with ECL Plus Western blotting detection reagents. To detect proteins carrying the myc-His epitope tag, blots were incubated with an anti-His(C-term)–HRP conjugate antibody (Invitrogen).
Enzyme assays.
To assay the enzyme activities of BfpE-PhoA fusions, cultures were grown as described above for immunoblotting. Samples were prepared and alkaline phosphatase assays were carried out by a microtiter plate procedure described previously (14), with the following modifications. The volume of diluted, permeabilized cultures added to the wells of the microtiter plate was 220 μl, and the volume of para-nitrophenyl phosphate solution was 20 μl. After a 15-min incubation at room temperature, the reactions were stopped by adding 20 μl of 1 M KH2PO4 plus 4 μl of 0.5 M EDTA (pH 8.0) to each well. OD readings were recorded at 620 nm (cell density), 414 nm (yellow color), and 540 nm (light scattering by cell debris). To assay the activities of BfpE-LacZ C-terminal fusions, β-galactosidase assays were carried out in a similar fashion, with the following modifications. Bacteria from 1-ml culture samples were pelleted, washed with 1 ml of 1 M Tris-HCl (pH 8.0), and resuspended in 1 ml of Z buffer (42). Aliquots of cultures that had been diluted 1:4 or 1:9 in Z buffer and then permeabilized were placed in the wells of a microtiter plate. The reaction was started by adding 37.5 μl of 10-mg/ml ortho-nitrophenyl-β-d-galactoside (ONPG) prepared in Z buffer and allowed to continue for 20 min at room temperature. The reactions were stopped by adding 75 μl of 1 M Na2CO3. Activity units were calculated by standard methods (37, 42), with appropriate adjustments for the dilution of the original cultures. The β-galactosidase activities of the BfpE–dual-reporter sandwich fusions could not be detected using ONPG. They were detectable using the more sensitive fluorescent substrate 4-methylumbelliferyl-β-d-galactoside (MUG). The MUG assay was carried out as described previously (42), except that a Shimadzu fluorescence spectrophotometer was utilized. A standard curve of 12.5 to 400 pM 4-methylumbelliferone was used as a reference to calculate units of specific β-galactosidase activity (42).
Sucrose density flotation gradient fractionation.
TOP10 strains containing plasmids with epitope-tagged bfpE derivatives were grown overnight in LB medium, diluted 1:100 into 30 ml of fresh LB medium, and shaken at 37°C for 4 h, after which arabinose was added to 0.2% to induce protein expression and shaking was continued for an additional 3 h. The remainder of the procedure was performed as previously described (4).
RESULTS
Phenotypes of a bfpE mutant.
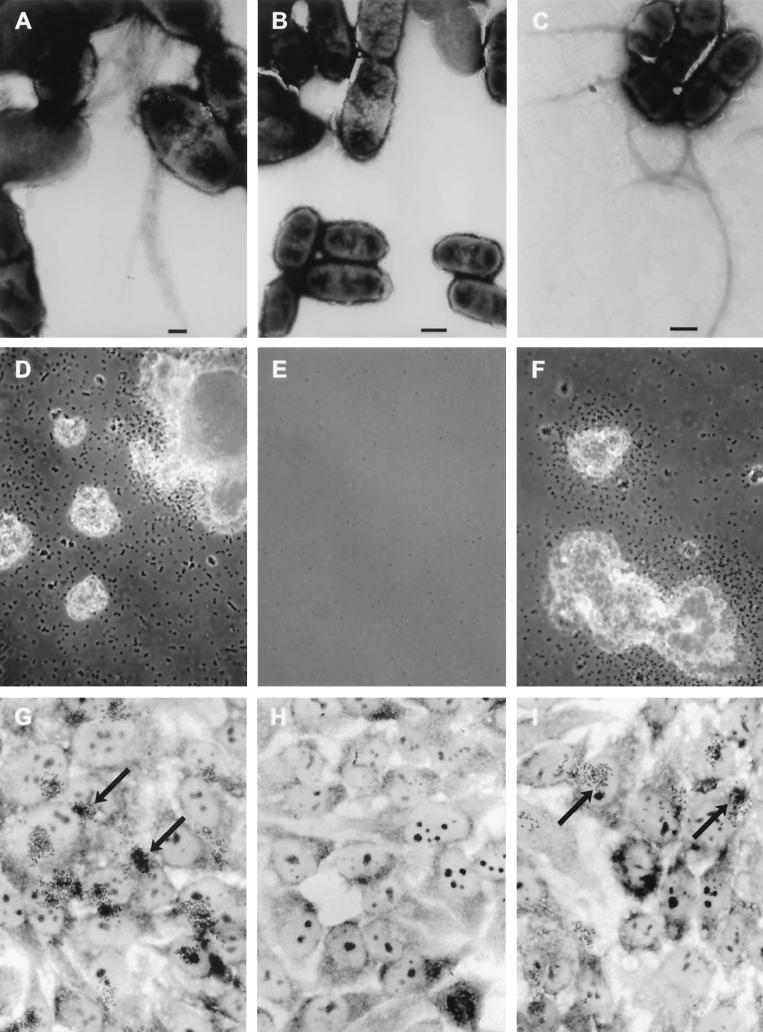
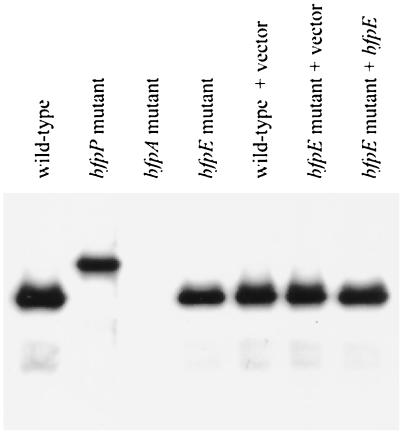
To determine whether the bfpE gene product plays a role in BFP biogenesis and BFP-associated phenotypes, we constructed UMD934, an EPEC strain containing a nonpolar insertion mutation in the bfpE gene. UMD934 was examined for BFP formation directly by transmission electron microscopy and indirectly by assaying for the BFP-dependent phenomena of autoaggregation and localized adherence. The isogenic wild-type EPEC strain E2348/69 and the bfpA mutant strain UMD901 were included in these experiments as positive and negative controls, respectively. BFP were readily detected emanating from cells of strain E2348/69 (Fig. 1A) but were not detected on cells of strain UMD901 or UMD934. When E2348/69 was grown in DMEM/F-12 tissue culture medium for at least 3 h, visible bacterial aggregates formed (Fig. 1D). In contrast, UMD901 and UMD934 cultures did not develop visible or even microscopic aggregates. To quantify the extent of autoaggregation in these strains, an AI was calculated. AI is a measure of the percent increase in OD that occurs after agitation of the culture to disperse bacterial aggregates (3, 4). E2348/69 exhibited a high AI value, while the bfpE mutant UMD934 exhibited a low AI value similar to that of UMD901 (Table 4). When tested for localized adherence to HEp-2 epithelial cells in tissue culture, E2348/69 exhibited the characteristic clustered pattern (Fig. 1G). UMD901 and UMD934 adhered poorly. To determine whether BfpE plays a role in bundlin expression and processing, extracts of EPEC strains were subjected to immunoblotting using an anti-bundlin antibody. UMD934 was found to produce completely processed bundlin at normal levels (Fig. 2).
FIG. 1.
Assay of BFP formation, autoaggregation, and localized adherence by EPEC. Shown are the wild-type EPEC strain E2348/69 (A, D, and G) and the isogenic bfpE mutant UMD934 bearing either the vector pTrcphoA (B, E, and H) or plasmid pTEB41 carrying the bfpE gene (C, F, and I). The top row (A to C) displays transmission electron micrographs of EPEC cultured in DMEM for 6 h. Magnifications, ×20,000 (A) and ×12,000 (B and C). Bar, 200 nm (A) or 500 nm (B and C). BFP can be seen in panels A and C. The center row (D to F) displays phase-contrast micrographs (magnifications, approximately ×460 for panels D and F and ×580 for panel E) of EPEC cultured in DMEM for 7 h and then examined in hanging drop slides. EPEC aggregates are seen in panels D and F. The bottom row (G to I) displays phase-contrast micrographs (magnification, ×630) of HEp-2 cells incubated for 3 hours with EPEC, washed, and fixed. Adherent EPEC microcolonies are seen in panels G and I. Arrows point to two of the microcolonies in each panel.
TABLE 4.
AI of EPEC strains
| Strain | Plasmid | AIa |
|---|---|---|
| E2348/69 (wild type) | None | 160 ± 28 |
| E2348/69 (wild type) | pTrcphoA (vector) | 77 ± 25 |
| UMD901 (bfpA mutant) | None | 3 ± 1 |
| UMD934 (bfpE mutant) | None | 7 ± 4 |
| UMD934 (bfpE mutant) | pTrcphoA (vector) | 5 ± 2 |
| UMD934 (bfpE mutant) | pTEB41 (bfpE) | 85 ± 26 |
Autoaggregation was quantitated as described in Materials and Methods. Values represent the mean and standard error of four independent cultures.
FIG. 2.
Examination of bundlin expression and processing by EPEC. Whole-cell extracts were prepared from EPEC strains (left to right) E2348/69, UMD932, UMD901, UMD934, E2348/69 (pTrcphoA), UMD934 (pTrcphoA), and UMD934 (pTEB41) after growth in DMEM/F-12 for 6 h. Extracts were separated by SDS-PAGE using a 15% polyacrylamide gel. A monoclonal antibody was used to detect bundlin.
Complementation of the bfpE mutation.
The low-copy plasmid pTEB41, which carries the cloned bfpE gene, was introduced into strain UMD934 to complement the bfpE mutation. UMD934 bearing pTEB41 exhibited BFP formation, autoaggregation, and localized adherence (Fig. 1C, G, and I; Table 4), while UMD934 bearing the control vector pTrcphoA did not (Fig. 1B, E, and H; Table 4). These results demonstrated conclusively that bfpE is required for BFP biogenesis. In pTEB41, a pTrc99A derivative, the bfpE gene is expressed under the control of the trc promoter. This promoter is supposed to be efficiently repressed by the product of the lacIq gene present on the plasmid and can be derepressed by the addition of isopropyl-β-d-thiogalactopyranoside (IPTG) (2). However, successful complementation of the bfpE mutant did not require the addition of IPTG. The repressed level of bfpE expression from pTrc99A derivatives was sufficient not only for complementation but also for the production of BfpE fusion proteins at a level that can be readily detected by immunoblotting and enzyme assays (see below).
Predictions of BfpE topology and TM segments.
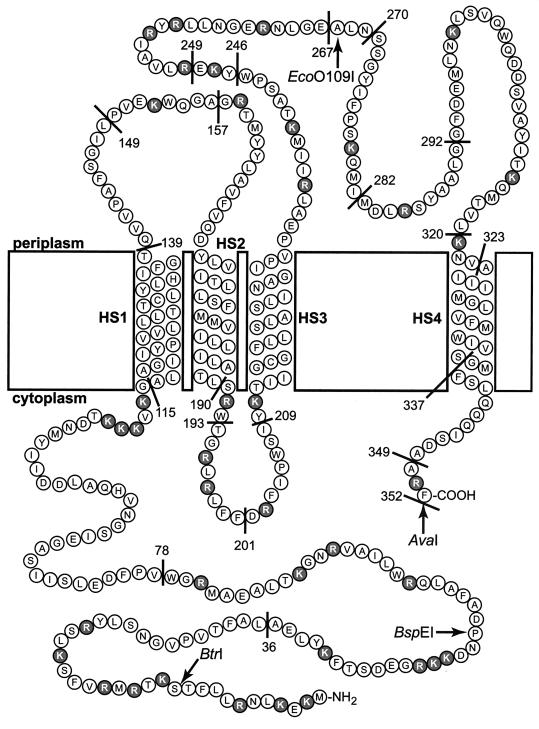
Hydropathy plots and computer programs were used to identify potential TM segments in BfpE and to suggest the most likely topology of the protein (see Materials and Methods). Four hydrophobic segments potentially long enough to span the cytoplasmic membrane were identified by all programs. These segments, designated HS1 through HS4, are located at residues 115 to 139, 171 to 191, 212 to 232, and 322 to 342, respectively, of BfpE (modal values from eight programs). In the predominant topology model (predicted by three of five programs), all four of the hydrophobic segments cross the membrane and both termini of the protein are found in the cytoplasm.
Construction of random and specific bfpE′::′phoA and bfpE′::′lacZ fusions.
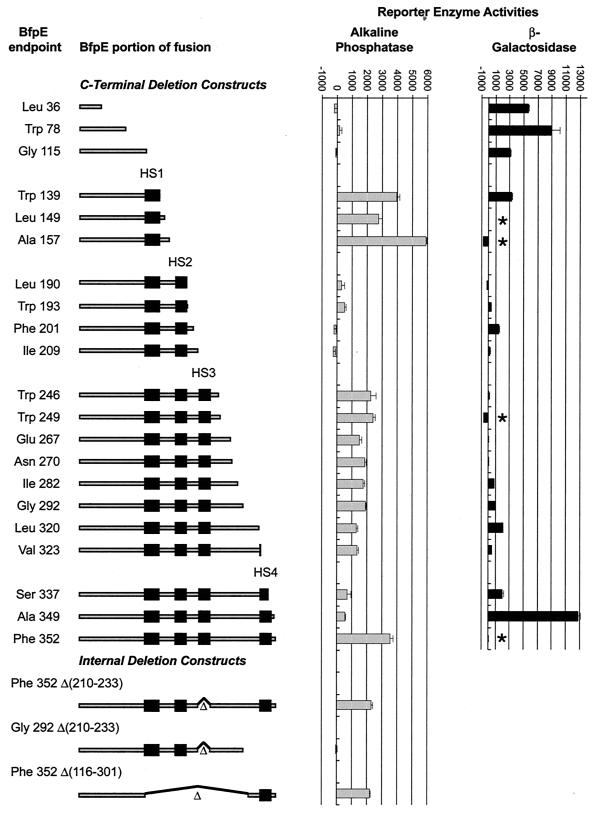
To determine the topology of BfpE experimentally, we turned to the construction and analysis of bfpE′::′lacZ and bfpE′::′phoA fusions. Such fusions are commonly used to study membrane protein topology (reviewed in references 37, 48, 64, and 66). Enzymatically active β-galactosidase (LacZ) fusions are expected to identify regions of BfpE that reside in the cytoplasm, while active alkaline phosphatase (PhoA) fusions are expected to identify regions of BfpE that reside in the periplasm. The bfpE gene was introduced into the expression vector pTrc99A upstream from and out of frame with ′lacZ or ′phoA reporter genes. Exonuclease III was used to degrade the bfpE gene from the 3′ end and create a nested set of bfpE′ C-terminal deletion fusions to ′phoA or ′lacZ. The plasmid pool carrying these gene fusions was introduced into E. coli DH5α. Colonies containing enzymatically active BfpE-LacZ and BfpE-PhoA fusions were identified on medium containing the chromogenic substrates X-Gal or BCIP.
To identify active in-frame fusions of bfpE′ to ′phoA, plasmids were isolated from 58 colonies that exhibited a dark blue color on medium containing BCIP. All plasmids were analyzed to determine the relative lengths of their partial bfpE genes. Twenty plasmids were selected for sequencing to determine the precise location of the fusion junction. All of these carried in-frame gene fusions, and 15 unique bfpE′ endpoints were identified among them. Most of the bfpE′::′phoA fusions specified proteins in which the BfpE endpoint was located in a relatively hydrophilic segment or at the margins of a putative TM segment. No endpoints were located in the N-terminal hydrophilic segment of BfpE. Three endpoints (Thr 139, Leu 149, and Ala 157) were located between candidate TM segments HS1 and HS2. Two endpoints (at Leu 190 and Trp193) were near the end of HS2, and seven (at Trp 246, Trp 249, Glu 267, Asn 270, Ile 282, Gly 292, and Leu 320) were between HS3 and HS4. One endpoint was near the end of HS4 (Ser 337), while two were C-terminal to HS4 (Ala 349 and Phe 352).
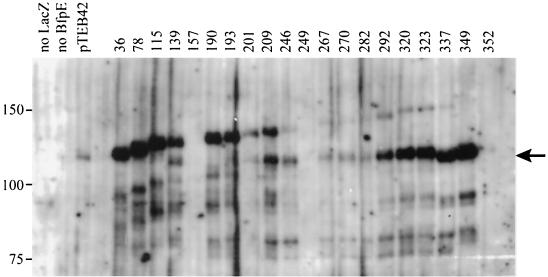
To identify active fusions of bfpE′ to ′lacZ, we prepared plasmids from 55 colonies that exhibited a dark blue color on medium containing X-Gal. This screen was complicated by the fact that colonies containing pTEB42 itself generally exhibited a light blue color on X-Gal. Despite the presence of a stop codon and frameshift and the absence of an initiation codon between the bfpE and lacZ genes on this plasmid, pTEB42 is capable of expressing a β-galactosidase-sized protein (Fig. 3) having detectable enzyme activity (1,780 ± 48 U). We do not know the mechanism by which such a protein is produced. Despite this complication, restriction analysis and sequencing of plasmids led to the identification of one in-frame bfpE′::′lacZ fusion gene, specifying a protein having a BfpE endpoint at Leu 36.
FIG. 3.
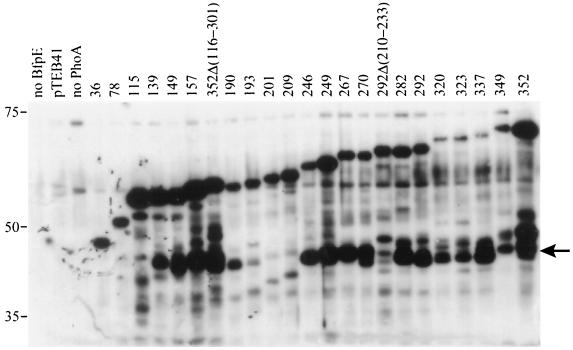
Expression of BfpE-LacZ fusion proteins. Whole-cell extracts were prepared from plasmid-bearing derivatives of E. coli CC118 and separated by SDS-PAGE on a 6% polyacrylamide gel. The fusion proteins were detected with an anti-β-galactosidase antibody. The positions of molecular mass markers are displayed to the left of the blot. The first three lanes display samples from strains carrying control plasmids pTEB65 (no LacZ), pTrclacZ (no BfpE), and pTEB42 (substrate for exonuclease III digestion). The remaining lanes display samples from strains carrying plasmids with bfpE′::′lacZ fusion genes. The number of the terminal amino acid in the BfpE portion of the fusion protein is noted above each lane. The arrow to the right of the blot indicates a prominent degradation product of many of the fusions that is similar in size to β-galactosidase (LacZ).
The bfpE′ segments from most of the random ′lacZ and ′phoA fusions described above were transferred by cloning into the alternate vector (pTrcphoA or pTrclacZ) so that they could be analyzed in the context of both reporter genes. It was expected that LacZ and PhoA fusion proteins having identical BfpE segments would exhibit complementary activities. Plasmids carrying additional bfpE′::′lacZ or bfpE′::′phoA fusions were constructed by PCR amplification of specific bfpE′ segments. These constructs, designed to supplement the randomly generated fusions at critical points, specified fusion proteins with BfpE endpoints at Trp 78, Gly 115, Phe 201, Ile 209, or Val 323. The entire set of constructs is depicted in Fig. 4.
FIG. 4.
Activities of BfpE-LacZ and BfpE-PhoA fusion proteins. In the diagrams to the left, the shaded bars represent the extent of BfpE included in each fusion protein and the black boxes represent potential transmembrane segments. Black lines and triangles indicate deleted portions of BfpE. Alkaline phosphatase or β-galactosidase enzyme assays were performed on permeabilized cultures of E. coli CC118 carrying fusion plasmids. The bar graph data represent the mean and standard error values (in units of enzyme activity) for four enzyme activity determinations from a single set of permeabilized cells per sample. Similar results were obtained in repeated experiments. Each datum point corresponds to the fusion depicted to the left of it. Four points are absent from the LacZ data (indicated by asterisks) because a plasmid expressing the fusion was not constructed (residue 149) or a fusion protein was not detected by immunoblotting (residues 157, 249, and 352).
Analyses of fusion protein enzyme activity and expression.
To indicate a cytoplasmic or periplasmic location for the reporter enzyme moieties of the BfpE fusion proteins, alkaline phosphatase or β-galactosidase activities were determined for permeabilized E. coli CC118 bearing bfpE′::′phoA or bfpE′::′lacZ plasmids (Fig. 4). To determine whether BfpE-LacZ and BfpE-PhoA fusion proteins were expressed, whole-cell extracts were prepared from these strains and subjected to immunoblotting using anti-LacZ and anti-PhoA antiserum (Fig. 3 and 5).
FIG. 5.
Expression of BfpE-PhoA fusion proteins. Whole-cell extracts were prepared from plasmid-bearing derivatives of E. coli CC118 and separated by SDS-PAGE on a 6.5% polyacrylamide gel. The fusion proteins were detected with an anti-PhoA antibody. The positions of molecular mass markers are displayed to the left of the blot. The first three lanes display samples from strains carrying control plasmids pTrcphoA (no BfpE), pTEB41, and pTEB65 (no PhoA). The remaining lanes display samples from strains carrying plasmids with bfpE′::′phoA fusion genes. The number of the terminal amino acid in the BfpE portion of the fusion protein is noted above each lane. The arrow to the right of the blot indicates a prominent degradation product of many of the fusions that is similar in size to alkaline phosphatase (PhoA).
The BfpE-PhoA fusions produced a reasonably clear picture of the topology of most of BfpE. All bfpE′::′phoA fusions produced readily detectable proteins (Fig. 5). As expected, the fusion protein size increased with the length of the BfpE segment included. The three PhoA fusions having BfpE endpoints located before HS1 (at residues 36, 78, and 115) had no or low alkaline phosphatase activities, indicating a cytoplasmic location (Fig. 4). PhoA fusions having BfpE endpoints between HS1 and HS2 (at residues 139, 149, and 157) exhibited particularly high activities, indicating a periplasmic location. Two randomly generated PhoA fusions located between HS2 and HS3 (at residues 190 and 193) exhibited low activities. These fusions contain none or only one of the five positively charged residues that are found between HS2 and HS3. Two other fusions constructed at residues 201 and 209 contain three or four positively charged residues downstream of HS2. These fusions had no alkaline phosphatase activity. These findings are in accord with previous data demonstrating that positive charges can promote cytoplasmic localization (7) and indicate that the region of BfpE between HS2 and HS3 is located in the cytoplasm. Fusions between HS3 and HS4 (at residues 246, 249, 267, 270, 282, 292, 320, and 323) had moderately high activities, suggesting a periplasmic location. The BfpE-PhoA fusion results up to HS4 are completely consistent with a BfpE topology in which the N terminus is located in the cytoplasm and HS1, HS2, and HS3 act as TM segments. It was expected that HS4 would also cross the membrane, leading to low PhoA activities in the fusions downstream. PhoA fusions at BfpE residues 337 and 349 had rather low activities. These data were difficult to interpret, however, because fusions at these points (and at residues 320 and 323) exhibit reduced amounts of fusion protein. In contrast, a fusion of full-length BfpE (residue 352) to PhoA had strikingly high activity and a high protein level. These results suggested that HS4, the final hydrophobic segment of BfpE, does not act as a TM segment. A second interpretation could be that while both HS3 and HS4 may be capable of spanning the membrane independently, HS3 is excluded from the membrane in the presence of HS4.
Many of the BfpE-PhoA fusion proteins appeared to be subject to degradation, producing a prominent protein having an electrophoretic mobility similar to that expected for processed alkaline phosphatase (∼47 kDa) (Fig. 5). Similar degradation products have been noted previously in other phoA-based topology studies (8, 24, 27, 39). They are thought to result from the action of a periplasmic protease that releases a properly folded PhoA moiety from the fusion protein. Therefore, the appearance of these degradation products may be a useful indicator of PhoA export. Accordingly, PhoA-sized degradation products were present in samples of all BfpE-PhoA fusions having moderate to high enzyme activity. Such degradation products were absent or scarce in samples of most of the low-activity fusions (especially those having BfpE endpoints at residues 36, 78, 115, 193, 201, and 209).
Four problems were encountered with the BfpE-LacZ fusions. First, four important BfpE-LacZ fusions having endpoints corresponding to those of the PhoA fusions either could not be properly constructed (residue 149) or did not produce a readily detectable protein on an immunoblot (residues 157, 249, and 352). Therefore, these could not be considered in the analysis. It was subsequently determined by sequencing that the 157 and 249 plasmids had a frameshift at the junction and lacked an insert, respectively. In contrast, no sequence defects could be detected in the entire bfpE gene or at the bfpE′::′lacZ junction of the 352 fusion plasmid. Second, CC118 containing particular bfpE′::′lacZ fusions grew poorly (data not shown). Third, the amount of BfpE-LacZ fusion protein was greatly reduced in fusions located after HS3 (Fig. 3). Fourth, many of the fusions, especially those located after residue 193, exhibited a conspicuous band with a mobility similar to that expected for native β-galactosidase (∼116 kDa). These bands may signify cytoplasmic β-galactosidase liberated by proteolysis of membrane-associated fusion proteins (23, 27). If so, they could result in spurious β-galactosidase activities.
LacZ fusions prior to HS1 (at BfpE residues 36, 78, and 115) exhibited high activities, indicating a cytoplasmic location, as would be expected from the predicted topology and alkaline phosphatase fusion results. The activities and expression levels of the remaining BfpE-LacZ fusions provided a picture of the topology of BfpE that was not completely consistent with the activities of the BfpE-PhoA fusions. A fusion at the end of HS1 (residue 139) also exhibited high activity, suggesting that the LacZ moiety was not transported to the periplasm as expected. In this fusion, LacZ may prevent the transport of HS1 across the membrane. LacZ fusions having BfpE endpoints between HS2 and HS3 had low or moderate activities despite their expected cytoplasmic location based on the PhoA fusion data. The BfpE-LacZ fusions between HS3 and HS4 likewise exhibited low activities or no activity. These also produced low levels of full-length fusion protein, making their activities difficult to evaluate. LacZ fusions within and following HS4 (residues 337 and 349) gave moderate and very high activities. The high activity level of the LacZ fusion at 349 appears to be in conflict with the high activity of the PhoA fusion at 352. We tentatively attribute the substantial β-galactosidase activity of the bfpE349′::′lacZ fusion to an active β-galactosidase degradation product (Fig. 3). However, we note that fusions at 292 though 337 also displayed large amounts of liberated β-galactosidase yet did not have correspondingly high activities. This may result from sequence differences in the degradation products. Overall, the β-galactosidase fusions provided little useful information to help us determine the topology of BfpE, while alkaline phosphatase fusions supported a topology similar to that predicted by sequence analysis, with the exception being the location of HS4 and the BfpE C terminus in the periplasm.
Analyses of dual-reporter sandwich fusions.
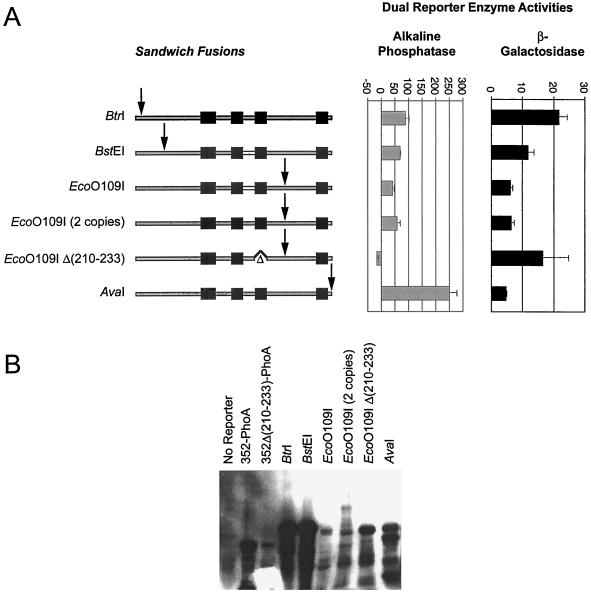
To further analyze the topology of BfpE, we constructed sandwich fusions, where the reporter moiety was placed at sites internal to the complete BfpE protein. This approach can provide a more reliable picture of a protein's topology than the C-terminal deletion fusion approach, especially when regions of the protein located both N- and C-terminal to the reporter enzyme must interact to establish the correct topology (22, 66). In particular, we wished to use such fusions to test the hypothesis, suggested by the BfpE-PhoA fusions, that HS3 of BfpE does not act as a TM domain when HS4 is also present. To make sandwich fusions in bfpE, we utilized a dual-reporter cassette containing the fused phoA gene and lacZα fragment, which allows both PhoA and LacZ enzyme activities to be analyzed in the context of a single protein (1). The cassette was inserted in frame into four restriction sites unique within the bfpE gene of plasmid pTEB65. As indicated in Fig. 6, these insertion sites correspond to regions in BfpE near the N terminus (BtrI and BspEI), between HS3 and HS4 (EcoO109I), and at the extreme C terminus (AvaI). Two variants of the EcoO109I construct were also prepared: one which carried two tandem copies of the dual reporter and one in which the dual reporter was inserted into a bfpE gene that lacked sequences encoding the putative TM domain HS3 (codons 210 to 233). The sandwich fusion plasmids were introduced into E. coli TG1. When plated on Red-Gal/BCIP dual indicator medium (1), TG1 colonies carrying the EcoO109I, EcoO109I×2, and AvaI insertions exhibited a blue color 1 day after plating, suggesting a periplasmic location for the dual reporter. In contrast, colonies carrying the BtrI, BstEI, and EcoO109IΔ(210–233) insertions exhibited a purplish red color that was not detectable the day after plating but was clearly seen after 1 week of storage at 4°C. This result suggests a cytoplasmic location for the dual reporter in these constructs. The sandwich fusion proteins were analyzed for enzyme activity and expression (Fig. 6). Their enzyme activities were extremely low but detectable, allowing certain conclusions. The relatively high alkaline phosphatase and low β-galactosidase activities produced by the AvaI insertion indicate that the C-terminal portion of the fusion is periplasmic. The striking reversal in the relative activities produced by the EcoO109I insertions that occurs on deletion of HS3 strongly supports a periplasmic location for the segment between HS3 and HS4 as well. These changes argue that HS3 acts as a TM segment even in the presence of HS4. The other activities were too low or inconsistent to yield firm conclusions.
FIG. 6.
Analyses of BfpE–dual-reporter sandwich fusion proteins. The labels indicate restriction sites in the bfpE gene into which a dual-reporter (′phoA-lacZα) cassette was inserted. (A) Activities of sandwich fusion proteins. The BfpE protein is depicted as in Fig. 4, with the arrows indicating the approximate point at which the dual reporter is inserted into the protein. Alkaline phosphatase or β-galactosidase enzyme assays were performed on cultures of E. coli TG1 carrying sandwich fusion plasmids. The bar graph data represent the mean and standard error values for three separate experiments, with four (alkaline phosphatase) or three (β-galactosidase) enzyme activity determinations per experiment. (B) Expression of sandwich fusion proteins. Whole-cell extracts were prepared from the strains described above and separated by SDS-PAGE on a 6% polyacrylamide gel. The fusion proteins were detected with an anti-PhoA antibody. The first three lanes display samples from strains carrying control plasmids, while the remaining lanes display samples from strains carrying sandwich fusions.
HS4 can act as a TM segment.
The substantial alkaline phosphatase activities produced by fusions of PhoA on either side of HS4 called into question the ability of this region to act as a TM segment in BfpE. To explore this issue further, we analyzed three bfpE′::′phoA constructs containing specific deletions within the bfpE gene (Fig. 4). In the first construct, sequences encoding HS3 (codons 210 to 233) were deleted from an otherwise full-length fusion of bfpE to phoA. This construct produced a weakly detectable protein (Fig. 6) that had moderately high activity (Fig. 4). As a control, a partial bfpE gene ending before the sequence encoding HS4 (at codon 292) and lacking sequences encoding HS3 was fused to phoA. The construct expressed a readily detectable protein (Fig. 5), but, in contrast to both the first construct and the C-terminal PhoA fusion at 292, it had no activity (Fig. 4). These data reconfirm the membrane-spanning capabilities of HS3 and, more importantly, suggest that HS4 can also act as a TM segment, exporting PhoA to the periplasm in the absence of HS3. To confirm this notion, a third phoA fusion construct was analyzed which contained a bfpE gene from which codons 116 through 301 had been deleted. The resulting fusion protein lacked hydrophobic segments HS1, HS2, and HS3, leaving HS4 as the only potential TM segment. This fusion construct produced a readily detectable protein with considerable PhoA activity (Fig. 4 and 5), clearly demonstrating that HS4 is capable of spanning the membrane. The limitation of these constructs was that HS4 was in an orientation opposite from the one it would be expected to hold in the native BfpE protein.
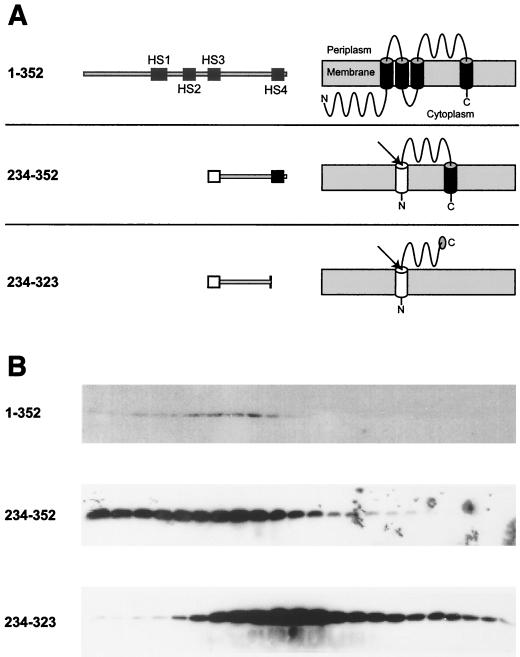
To further probe the ability of HS4 to act as a TM segment, we constructed three plasmids expressing epitope-tagged BfpE derivatives regulated by an arabinose-inducible promoter. One construct was a full-length epitope-tagged version of BfpE to be used as a control. A second construct contained only the region between HS3 and HS4 (residues 234 to 323), while a third contained the entire region of BfpE downstream of HS3 (residues 234 to 352). The last two constructs also carried an N-terminal phage fd gene III signal sequence to direct the expressed proteins to the periplasm, where the inter-HS3/HS4 region appears to be located in full-length BfpE. We used isopycnic sucrose density flotation gradient fractionation to indicate the location of the three constructs after expression in the E. coli strain TOP10. Whole-cell lysates were placed at the bottom of sucrose gradients and subjected to ultracentrifugation. Fractions were removed sequentially from the top of the gradient and subjected to electrophoresis followed by immunoblotting with an antibody recognizing the epitope tag. The protein composition of the fractions was shown to vary by Ponceau S staining of immunoblotting membranes, with an increasing number of proteins present near the bottom of the gradients, as expected for fractions containing the cytoplasmic and periplasmic contents of the cell (data not shown). We expected the full-length BfpE construct (residues 1 to 352) to localize to the inner membrane. Consistent with this notion, this protein appeared primarily in the less dense fractions of the gradient, the expected location for inner membrane vesicles (Fig. 7). We expected the BfpE segment from residues 234 to 323 to be soluble and periplasmic and to appear near the bottom of the gradient. In practice, this construct was present in samples from both the middle and bottom of the gradient but scarce in the lowest-density fractions. The location of this protein in the middle of the gradient, which characteristically contains outer membrane vesicles, was surprising. It may suggest that the protein sequence has some ability to associate with the outer membrane, a result that is consistent with localization predictions using the PSORT program (data not shown). The BfpE segment from residues 234 to 352 would be expected to localize in either the periplasm or inner membrane depending on the transmembrane properties of HS4. Experimentally, this protein was found primarily in the least dense fractions of the gradient, indicating an inner membrane location. The striking difference between the gradient distributions of the constructs containing residues 234 to 323 and 234 to 352 indicates that HS4 can mediate localization to the cytoplasmic membrane from the periplasmic side.
FIG. 7.
Sucrose density flotation gradient fractionation of three epitope-tagged BfpE derivatives. (A) Representations of epitope-tagged BfpE constructs. Each construct carries a C-terminal myc-His double epitope tag (not shown). For each construct, the number in the left column indicates the range of BfpE amino acids that are present. The middle column shows a linear representation of each protein. The right column displays the expected topology of each protein. Black boxes and cylinders indicate TM segments, while white boxes and cylinders indicate an exogenous N-terminal signal sequence, whose presumed removal is indicated by an arrow. (B) Lysates of TOP10 E. coli carrying plasmids producing the constructs shown in panel A were fractionated on sucrose density flotation gradients by centrifugation. Fractions were collected from the top (least dense portion) of the gradient, separated by SDS-PAGE on a 15% polyacrylamide gel, and subjected to immunoblotting using an antiserum recognizing the epitope tag. Fractions progress from the least dense on the left to the most dense on the right.
Complementation studies indicate the importance of HS3 and HS4 in the activity of BfpE.
To determine the extent of the BfpE C terminus that is required for activity, we tested the longest BfpE-PhoA fusions for the ability to restore autoaggregation to the bfpE mutant EPEC strain UMD934. Both the full-length bfpE352::′phoA fusion and the bfpE349′::′phoA fusion enabled UMD934 to form microscopic aggregates, with the aggregates being consistently larger and more numerous in the bfpE352::′phoA sample. In contrast, PhoA C-terminal fusions at BfpE residues 337, 323, 320, or 292 did not promote autoaggregation. These results indicate that an intact HS4 is required for the activity of BfpE. We note that it is unclear whether the intact BfpE-PhoA fusion or the BfpE portion remaining after the release of the PhoA moiety (see above) is the complementing protein in these experiments.
Plasmid pTEB65, which carries the full-length bfpE gene, was able to restore the ability of UMD934 to autoaggregate. The ΔHS3 derivative of pTEB65 did not do so, indicating the requirement of HS3 for the function of BfpE. Derivatives of pTEB65 carrying insertions of the phoA-lacZα dual reporter at the BtrI, BstEI, or EcoO109I site of bfpE were also unable to promote autoaggregation, indicating that the presence of the reporter moiety at these locations inhibits BfpE function. Of note, plasmids carrying dual-reporter insertions in the EcoO109I site of pTEB65, when present in EPEC strain UMD934, produced dark blue colonies on medium containing BCIP. Plasmids carrying reporter insertions in the BtrI or BspEI sites of pTEB65 or in the EcoO109I site of the ΔHS3 derivative of pTEB65 did not produce blue colonies. These observations support the notion that the topology of BfpE expressed in EPEC is the same as when expressed in laboratory strains of E. coli.
DISCUSSION
BfpE is required for BFP biogenesis.
The biogenesis of type IV fimbriae requires multiple protein components in addition to pilin, the fimbrial structural element. It is likely that in EPEC, many or all of the products of the bfp gene cluster constitute a molecular machine that assembles BFP fimbriae and extrudes them through the outer bacterial membrane. Other type IV fimbriae are presumably assembled by similar machines. In this study, we have demonstrated the requirement for BfpE, one component of the BFP assembly apparatus. We found that a bfpE mutant strain of EPEC fails to form detectable BFP and fails to carry out autoaggregation and localized adherence. The cloned bfpE gene, transcribed from a heterologous promoter on a low-copy plasmid, complements each of the defects of the mutant. These results indicate that the BfpE protein is essential for type IV fimbrial biogenesis in EPEC. The bfpE mutation does not alter the expression or leader peptide processing of prebundlin. Since catalysis of disulfide bond formation in bundlin by the periplasmic protein DsbA is required for pilin stability (74), BfpE probably participates in an aspect of BFP synthesis that takes place after the insertion of bundlin into the cytoplasmic membrane. For example, it may mediate the extraction of bundlin from the membrane, the polymerization of bundlin into BFP, the extrusion of BFP through the outer membrane, or the anchoring of BFP to the cell envelope. BfpE could associate with bundlin directly or could influence it indirectly through other components of the BFP synthesis machinery. Mutations introduced into many of the bfp genes, including bfpB, bfpC, bfpD, bfpG, and bfpL, elicit phenotypes identical to those found in the bfpE mutant (4, 6, 49, 58, 60). To understand the specific requirement for the products of each of these bfp genes in detail, it will be necessary to develop new means of examining BFP synthesis at the molecular level.
The phenotypes elicited by the bfpE mutation in EPEC are quite similar to those created by mutations in the corresponding genes of other type IV fimbrial systems. Insertion mutations in the pilC genes of Pseudomonas aeruginosa (34, 45) and Myxococcus xanthus (71), the pilG genes of Neisseria gonorrhoeae and Neisseria meningitidis (62), the pilR gene of the R64 plasmid (72), and the tcpE gene of Vibrio cholerae (9, 32) all generate strains that fail to elaborate functional type IV fimbriae. When tested, some of these mutants have been shown to exhibit normal levels of processed pilin, as does our bfpE mutant. Mutations in other genes of the gspF family disable protein secretion by the type II pathway present in particular gram-negative bacteria (15, 36, 47, 55) or reduce genetic transformation competence (i.e., binding and uptake of exogenous DNA) in some gram-negative and gram-positive bacteria (11, 62). It appears that intact GspF proteins are universally required for the operation of the molecular machines of which they are a component.
Topology of BfpE.
As a further step toward understanding the structure and function of BfpE and other GspF proteins, we have determined the arrangement of BfpE in the cytoplasmic membrane. For this purpose we isolated and constructed fusions of 3′-truncated bfpE derivatives to phoA (alkaline phosphatase) and lacZ (β-galactosidase) genes. These had a wide range of BfpE endpoints, covering each of the hydrophilic domains of the protein. We also constructed a small number of sandwich fusions. The ability to identify both active and inactive PhoA and LacZ fusions strongly supports the notion that BfpE is a cytoplasmic membrane protein, as do the results of sucrose density gradient centrifugation with an epitope-tagged version of BfpE. A study of the relative activities and levels of the fusion proteins allowed us to evaluate the locations and orientations of four putative TM segments, HS1 through HS4, identified through sequence analysis. The activities of the BfpE-PhoA fusion proteins, as well as the dual-reporter sandwich fusions, varied in a manner consistent with the topology shown in Fig. 8. This topology meets the criteria of the positive-inside rule (67), with the majority of the arginine and lysine residues being located in the cytoplasm. The region between HS3 and HS4 contains 12 such residues and is periplasmic but can be considered exempt from the rule due to its length of greater than 60 residues (69, 70). Overall, the BfpE protein can be thought of as being divided into thirds. The N-terminal third is located in the cytoplasm. The middle third serves as a membrane anchor, containing three membrane-spanning segments. The C-terminal third is located primarily in the periplasm, with its end anchored in the membrane. As discussed in more detail below, this organization has profound implications for the function of BfpE.
FIG. 8.
Proposed topology of the BfpE protein. The amino acids composing BfpE are represented by circles. These are displayed in a manner that specifies their arrangement in the E. coli cytoplasmic membrane. HS1 through HS4 denote TM segments. Positively charged amino acids (arginines and lysines) that may be topology determinants are shaded. C-terminal fusion protein junctions are indicated by a line and the number of the terminal BfpE residue in the fusion. The insertion sites for the dual reporter in sandwich fusions are indicated by a restriction enzyme name.
We obtained conflicting data on the location of HS4, the fourth putative TM segment in BfpE. When PhoA was fused to BfpE that had been truncated on either side of HS4, substantial alkaline phosphatase activities resulted, suggesting that HS4 remains in the periplasm instead of inserting into the membrane. This finding was unexpected, since all computer programs had predicted HS4 to be a TM domain. Furthermore, the activities of bfpE::phoA internal deletion constructs and the results of isopycnic sucrose gradient experiments using partial BfpE proteins indicated that HS4 is capable of spanning the membrane, at least as an isolated unit. Given the conflicting data, it remains somewhat unclear whether HS4 in the native BfpE protein crosses the membrane. We favor a model in which HS4 is a stop-transfer sequence that normally inserts into the membrane but is prevented from doing so in our fusions by the PhoA reporter, which prefers to reside in the periplasm. This model is most consistent with our data showing that HS4 is able to cross the membrane in either direction. The short (10-residue) segment following HS4 contains only one positive charge, which may be insufficient to retain the PhoA moiety in the cytoplasm. This situation would be analogous to that of the PhoA fusions at residues 190 and 193 of BfpE. These fusions are anchored by few positive charges and have alkaline phosphatase activity, although located in a region that is cytoplasmic in the full-length protein. We cannot entirely exclude the explanation that HS4 actually does not insert into the membrane, perhaps being prevented from doing so by other topological determinants in BfpE. A final possibility is that the topology of the HS4 region is variable, reflecting some ability of the protein to undergo conformation changes while in the membrane. Such a property has been demonstrated for SecG, a component of the E. coli preprotein translocase (44). Given the current data, the placement of HS4 in the membrane must be regarded as provisional.
BfpE has a different topology from another GspF protein.
The topology of OutF, a GspF protein from the type II protein secretion system of Erwinia carotovora, was previously determined using β-lactamase gene fusions (61). Like BfpE, OutF has a large N-terminal cytoplasmic segment plus TM domains that are analogs of HS1 and HS2. However, the C-terminal portion of the OutF topology is surprisingly divergent from that proposed here for BfpE. A TM domain corresponding to HS3 does not exist in OutF. As a result, most of the C-terminal third of OutF is located in the cytoplasm while the C-terminal third of BfpE is found mostly in the periplasm. The third TM segment in OutF is analogous to HS4 of BfpE in terms of its position in the protein sequence. However, in OutF this segment crosses the membrane from the cytoplasmic to the periplasmic side, while in BfpE it appears to have the opposite orientation. The topologies of the two proteins could be identical if HS3 of BfpE was excluded from crossing the membrane in the presence but not in the absence of HS4. However, this possibility was ruled out by BfpE sandwich fusion data, which indicate that the region downstream of HS3 is located in the periplasm even when HS4 is present. In summary, the presence of HS3 and disposition of HS4 define the differences between BfpE and OutF.
It is important to understand whether the differences in the experimentally determined membrane topologies of BfpE and OutF reflect the actual topologies of the native proteins or indicate artifacts introduced by the use of fusion proteins. In theory, the presence of the reporter protein could alter the topology of specific fusions. However, such effects are not been generally noted for PhoA, LacZ, or BlaM fusions (64). It is also possible that the BfpE fusion proteins could acquire an incorrect topology in the absence of other Bfp proteins in the E. coli K-12 strains used in this study. However, individual membrane proteins are thought to integrate into the membrane independently of one another (46, 48, 63, 73). Furthermore, the BfpE sandwich fusions appeared to have the same topology in EPEC and K-12 E. coli based on a chromogenic indicator assay, and the topology of OutF in the context of wild-type Erwinia did not differ from that deduced in an E. coli host that lacks a functional general secretion pathway (61). Thus, it appears that OutF and BfpE, although members of the same protein family, have dramatically different membrane topologies. These topologies are presumably adapted to the protein export machinery of Erwinia and the pilus biogenesis machinery of EPEC and may reflect interactions that occur with protein components unique to each respective system.
To suggest the extent to which the BfpE or OutF topologies prevail in the GspF family, we used TMHMM (59) to analyze the sequences of various GspF proteins (data not shown). This algorithm was chosen because its topology predictions for both OutF and BfpE correspond closely to the experimentally determined topologies. TMHMM identified three TM segments in each of the GspF proteins tested, located approximately at the same positions as those in OutF. Each of the predicted topologies was identical to that of OutF. A significantly hydrophobic segment corresponding to HS3 of BfpE was not identified in any of the other GspF proteins. Notably, such a segment was not present in the two proteins that are most similar in sequence to BfpE. These proteins are TcpE, encoded by the toxin-coregulated pilus gene cluster of V. cholerae (32), and PilR, encoded by the thin pilus gene cluster of the IncI plasmid R64 (72). Thus far, OutF appears to be structurally representative of the GspF family while the topology of BfpE appears to be novel.
An understanding of the membrane topology of BfpE provided by the results of this study should allow us to construct testable models of protein interactions integral to the BFP biogenesis machinery. With its large N-terminal cytoplasmic domain and large C-terminal periplasmic domain, BfpE appears poised to bridge the gap between components in multiple compartments. Because the N terminus of BfpE has a cytoplasmic location conserved with the other GspF proteins, it may retain a conserved function. The C terminus of BfpE, with its unique periplasmic location, may have a more specialized function.
In addition to the specific information provided regarding the topology of BfpE, several general points are emphasized by the results of this study. First, the results obtained by analyzing the enzymatic activity of a single type of reporter protein can convey misleading data. Second, the use of complementary systems can lead to conflicting data. To reconcile these data, it is important to analyze additional information such as the steady-state levels of the fusion proteins and the activities of fusion proteins generated by deletions of TM domains or sandwich fusions. Lastly, the topology of one member of a protein family (such as BfpE) cannot always be inferred from the analysis of another member of that family (such as OutF) but must be determined by experimentation.
ACKNOWLEDGMENTS
We thank Ravi Anantha for providing strain UMD932, Colin Manoil for providing strain CC118, Mikhail Alexeyev for providing plasmid pMA632 and strain TG1, John Albert and Jorge Girón for providing monoclonal antibody ICA4, David Silverman for the use of his fluorescence spectrophotometer, and Harry Mobley and Shanmuga Sozhamannan for helpful comments on the manuscript. We are grateful to members of the Donnenberg laboratory for helpful suggestions during the course of this work, especially Ravi Anantha, and Barry McNamara, who identified the optimal conditions for autoaggregation.
This investigation was supported by a Public Health Service grant (R01 AI-37606) to M.S.D. and a National Research Service Award postdoctoral training fellowship (F32 AI-10191) to T.E.B.
REFERENCES
- 1.Alexeyev M F, Winkler H H. Membrane topology of the Rickettsia prowazekii ATP/ADP translocase revealed by novel dual pho-lac reporters. J Mol Biol. 1999;285:1503–1513. doi: 10.1006/jmbi.1998.2412. [DOI] [PubMed] [Google Scholar]
- 2.Amann E, Ochs B, Abel K J. Tightly regulated tac promoter vectors useful for the expression of unfused and fused proteins in Escherichia coli. Gene. 1988;69:301–315. doi: 10.1016/0378-1119(88)90440-4. [DOI] [PubMed] [Google Scholar]
- 3.Anantha R P, Stone K D, Donnenberg M S. The role of BfpF, a member of the PilT family of putative nucleotide-binding proteins, in type IV pilus biogenesis and in interactions between enteropathogenic Escherichia coli and host cells. Infect Immun. 1998;66:122–131. doi: 10.1128/iai.66.1.122-131.1998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Anantha R P, Stone K D, Donnenberg M S. Effects of bfp mutations on biogenesis of functional enteropathogenic Escherichia coli type IV pili. J Bacteriol. 2000;182:2498–2506. doi: 10.1128/jb.182.9.2498-2506.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Baldini M M, Kaper J B, Levine M M, Candy D C, Moon H W. Plasmid-mediated adhesion in enteropathogenic Escherichia coli. J Pediatr Gastroenterol Nutr. 1983;2:534–538. doi: 10.1097/00005176-198302030-00023. [DOI] [PubMed] [Google Scholar]
- 6.Bieber D, Ramer S W, Wu C Y, Murray W J, Tobe T, Fernandez R, Schoolnik G K. Type IV pili, transient bacterial aggregates, and virulence of enteropathogenic Escherichia coli. Science. 1998;280:2114–2118. doi: 10.1126/science.280.5372.2114. [DOI] [PubMed] [Google Scholar]
- 7.Boyd D, Beckwith J. Positively charged amino acid residues can act as topogenic determinants in membrane proteins. Proc Natl Acad Sci USA. 1989;86:9446–9450. doi: 10.1073/pnas.86.23.9446. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Calamia J, Manoil C. lac permease of Escherichia coli: topology and sequence elements promoting membrane insertion. Proc Natl Acad Sci USA. 1990;87:4937–4941. doi: 10.1073/pnas.87.13.4937. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Chiang S L, Mekalanos J J. Use of signature-tagged transposon mutagenesis to identify Vibrio cholerae genes critical for colonization. Mol Microbiol. 1998;27:797–805. doi: 10.1046/j.1365-2958.1998.00726.x. [DOI] [PubMed] [Google Scholar]
- 10.Chikami G K, Fierer J, Guiney D G. Plasmid-mediated virulence in Salmonella dublin demonstrated by use of a Tn5-oriT construct. Infect Immun. 1985;50:420–424. doi: 10.1128/iai.50.2.420-424.1985. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Chung Y S, Dubnau D. All seven comG open reading frames are required for DNA binding during transformation of competent Bacillus subtilis. J Bacteriol. 1998;180:41–45. doi: 10.1128/jb.180.1.41-45.1998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Claros M G, Von Heijne G. TopPred II: an improved software for membrane protein structure predictions. Comput Appl Biosci. 1994;10:685–686. doi: 10.1093/bioinformatics/10.6.685. [DOI] [PubMed] [Google Scholar]
- 13.Cserzö M, Wallin E, Simon I, Von Heijne G, Elofsson A. Prediction of transmembrane α-helices in prokaryotic membrane proteins: the dense alignment surface method. Protein Eng. 1997;10:673–676. doi: 10.1093/protein/10.6.673. [DOI] [PubMed] [Google Scholar]
- 14.Daniels C, Vindurampulle C, Morona R. Overexpression and topology of the Shigella flexneri O-antigen polymerase (Rfc/Wzy) Mol Microbiol. 1998;28:1211–1222. doi: 10.1046/j.1365-2958.1998.00884.x. [DOI] [PubMed] [Google Scholar]
- 15.DeShazer D, Brett P J, Burtnick M N, Woods D E. Molecular characterization of genetic loci required for secretion of exoproducts in Burkholderia pseudomallei. J Bacteriol. 1999;181:4661–4664. doi: 10.1128/jb.181.15.4661-4664.1999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Donnenberg M S, Calderwood S B, Donohue-Rolfe A, Keusch G T, Kaper J B. Construction and analysis of TnphoA mutants of enteropathogenic Escherichia coli unable to invade HEp-2 cells. Infect Immun. 1990;58:1565–1571. doi: 10.1128/iai.58.6.1565-1571.1990. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Donnenberg M S, Girón J A, Nataro J P, Kaper J B. A plasmid-encoded type IV fimbrial gene of enteropathogenic Escherichia coli associated with localized adherence. Mol Microbiol. 1992;6:3427–3437. doi: 10.1111/j.1365-2958.1992.tb02210.x. [DOI] [PubMed] [Google Scholar]
- 18.Donnenberg M S, Kaper J B. Construction of an eae deletion mutant of enteropathogenic Escherichia coli by using a positive-selection suicide vector. Infect Immun. 1991;59:4310–4317. doi: 10.1128/iai.59.12.4310-4317.1991. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Donnenberg M S, Nataro J P. Methods for studying adhesion of diarrheagenic Escherichia coli. Methods Enzymol. 1995;253:324–336. doi: 10.1016/s0076-6879(95)53028-2. [DOI] [PubMed] [Google Scholar]
- 20.Donnenberg M S, Yu J, Kaper J B. A second chromosomal gene necessary for intimate attachment of enteropathogenic Escherichia coli to epithelial cells. J Bacteriol. 1993;175:4670–4680. doi: 10.1128/jb.175.15.4670-4680.1993. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Donnenberg M S, Zhang H-Z, Stone K D. Biogenesis of the bundle-forming pilus of enteropathogenic Escherichia coli: reconstitution of fimbriae in recombinant E. coli and role of DsbA in pilin stability—a review. Gene. 1997;192:33–38. doi: 10.1016/s0378-1119(96)00826-8. [DOI] [PubMed] [Google Scholar]
- 22.Ehrmann M, Boyd D, Beckwith J. Genetic analysis of membrane protein topology by a sandwich gene fusion approach. Proc Natl Acad Sci USA. 1990;87:7574–7578. doi: 10.1073/pnas.87.19.7574. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Georgiou C D, Dueweke T J, Gennis R B. β-Galactosidase gene fusions as probes for the cytoplasmic regions of subunits I and II of the membrane-bound cytochrome d terminal oxidase from Escherichia coli. J Biol Chem. 1988;263:13130–13137. [PubMed] [Google Scholar]
- 24.Ginn S L, Brown M H, Skurray R A. Membrane topology of the metal-tetracycline/H+ antiporter TetA(K) from Staphylococcus aureus. J Bacteriol. 1997;179:3786–3789. doi: 10.1128/jb.179.11.3786-3789.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Girón J A, Ho A S Y, Schoolnik G K. An inducible bundle-forming pilus of enteropathogenic Escherichia coli. Science. 1991;254:710–713. doi: 10.1126/science.1683004. [DOI] [PubMed] [Google Scholar]
- 26.Girón J A, Qadri F, Azim T, Jarvis K J, Kaper J B, Albert M J. Monoclonal antibodies specific for the bundle-forming pilus of enteropathogenic Escherichia coli. Infect Immun. 1995;63:4949–4952. doi: 10.1128/iai.63.12.4949-4952.1995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Gött P, Boos W. The transmembrane topology of the sn-glycerol-3-phosphate permease of Escherichia coli analysed by phoA and lacZ protein fusions. Mol Microbiol. 1988;2:655–663. doi: 10.1111/j.1365-2958.1988.tb00074.x. [DOI] [PubMed] [Google Scholar]
- 28.Henikoff S. Unidirectional digestion with exonuclease III creates targeted breakpoints for DNA sequencing. Gene. 1984;28:351–359. doi: 10.1016/0378-1119(84)90153-7. [DOI] [PubMed] [Google Scholar]
- 29.Hirokawa T, Boon-Chieng S, Mitaku S. SOSUI: classification and secondary structure prediction system for membrane proteins. Bioinformatics. 1998;14:378–379. doi: 10.1093/bioinformatics/14.4.378. [DOI] [PubMed] [Google Scholar]
- 30.Hofmann K, Stoffel W. A database of membrane spanning protein segments. Biol Chem Hoppe-Seyler. 1993;374:166. [Google Scholar]
- 31.Island M D, Mobley H L. Proteus mirabilis urease: operon fusion and linker insertion analysis of ure gene organization, regulation, and function. J Bacteriol. 1995;177:5653–5660. doi: 10.1128/jb.177.19.5653-5660.1995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Kaufman M R, Shaw C E, Jones I D, Taylor R K. Biogenesis and regulation of the Vibrio cholerae toxin-coregulated pilus: analogies to other virulence factor secretory systems. Gene. 1993;126:43–49. doi: 10.1016/0378-1119(93)90588-t. [DOI] [PubMed] [Google Scholar]
- 33.Knutton S, Shaw R K, Anantha R P, Donnenberg M S, Zorgani A A. The type IV bundle-forming pilus of enteropathogenic Escherichia coli undergoes dramatic alterations in structure associated with bacterial adherence, aggregation and dispersal. Mol Microbiol. 1999;33:499–509. doi: 10.1046/j.1365-2958.1999.01495.x. [DOI] [PubMed] [Google Scholar]
- 34.Koga T, Ishimoto K, Lory S. Genetic and functional characterization of the gene cluster specifying expression of Pseudomonas aeruginosa pili. Infect Immun. 1993;61:1371–1377. doi: 10.1128/iai.61.4.1371-1377.1993. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Levine M M, Bergquist E J, Nalin D R, Waterman D H, Hornick R B, Young C R, Sotman S, Rowe B. Escherichia coli strains that cause diarrhoea but do not produce heat-labile or heat-stable enterotoxins and are non-invasive. Lancet. 1978;i:1119–1122. doi: 10.1016/s0140-6736(78)90299-4. [DOI] [PubMed] [Google Scholar]
- 36.Lindeberg M, Salmond G P C, Collmer A. Complementation of deletion mutations in a cloned functional cluster of Erwinia chrysanthemi out genes with Erwinia carotovora out homologues reveals OutC and OutD as candidate gatekeepers of species-specific secretion of proteins via the type II pathway. Mol Microbiol. 1996;20:175–190. doi: 10.1111/j.1365-2958.1996.tb02499.x. [DOI] [PubMed] [Google Scholar]
- 37.Manoil C. Analysis of membrane protein topology using alkaline phosphatase and β-galactosidase gene fusions. Methods Cell Biol. 1991;34:61–75. doi: 10.1016/s0091-679x(08)61676-3. [DOI] [PubMed] [Google Scholar]
- 38.Manoil C, Beckwith J. TnphoA: A transposon probe for protein export signals. Proc Natl Acad Sci USA. 1985;82:8129–8133. doi: 10.1073/pnas.82.23.8129. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Manoil C, Beckwith J. A genetic approach to analyzing membrane protein topology. Science. 1986;233:1403–1408. doi: 10.1126/science.3529391. [DOI] [PubMed] [Google Scholar]
- 40.Martinez M B, Taddei C R, Ruiz-Tagle A, Trabulsi L R, Girón J A. Antibody response of children with enteropathogenic Escherichia coli infection to the bundle-forming pilus and locus of enterocyte effacement-encoded virulence determinants. J Infect Dis. 1999;179:269–274. doi: 10.1086/314549. [DOI] [PubMed] [Google Scholar]
- 41.Ménard R, Sansonetti P J, Parsot C. Nonpolar mutagenesis of the ipa genes defines IpaB, IpaC, and IpaD as effectors of Shigella flexneri entry into epithelial cells. J Bacteriol. 1993;175:5899–5906. doi: 10.1128/jb.175.18.5899-5906.1993. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Miller J H. A short course in bacterial genetics. Plainview, N.Y: Cold Spring Harbor Laboratory Press; 1992. [Google Scholar]
- 43.Nakai K, Kanehisa M. Expert system for predicting protein localization sites in gram-negative bacteria. Proteins. 1991;11:95–110. doi: 10.1002/prot.340110203. [DOI] [PubMed] [Google Scholar]
- 44.Nishiyama K, Suzuki T, Tokuda H. Inversion of the membrane topology of SecG coupled with SecA-dependent preprotein translocation. Cell. 1996;85:71–81. doi: 10.1016/s0092-8674(00)81083-1. [DOI] [PubMed] [Google Scholar]
- 45.Nunn D, Bergman S, Lory S. Products of three accessory genes, pilB, pilC, and pilD, are required for biogenesis of Pseudomonas aeruginosa pili. J Bacteriol. 1990;172:2911–2919. doi: 10.1128/jb.172.6.2911-2919.1990. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Popot J L, Engelman D M. Membrane protein folding and oligomerization: the two-stage model. Biochemistry. 1990;29:4031–4037. doi: 10.1021/bi00469a001. [DOI] [PubMed] [Google Scholar]
- 47.Possot O, d'Enfert C, Reyss I, Pugsley A P. Pullulanase secretion in Escherichia coli K-12 requires a cytoplasmic protein and a putative polytopic cytoplasmic membrane protein. Mol Microbiol. 1992;6:95–105. doi: 10.1111/j.1365-2958.1992.tb00841.x. [DOI] [PubMed] [Google Scholar]
- 48.Pugsley A P. The complete general secretary pathway in gram-negative bacteria. Microbiol Rev. 1993;57:50–108. doi: 10.1128/mr.57.1.50-108.1993. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Ramer S W, Bieber D, Schoolnik G K. BfpB, an outer membrane lipoprotein required for the biogenesis of bundle-forming pili in enteropathogenic Escherichia coli. J Bacteriol. 1996;178:6555–6563. doi: 10.1128/jb.178.22.6555-6563.1996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Rost B, Casadio R, Fariselli P, Sander C. Transmembrane helices predicted at 95% accuracy. Protein Sci. 1995;4:521–533. doi: 10.1002/pro.5560040318. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Rost B, Fariselli P, Casadio R. Topology prediction for helical transmembrane proteins at 86% accuracy. Protein Sci. 1996;5:1704–1718. doi: 10.1002/pro.5560050824. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Rothbaum R, McAdams A J, Giannella R, Partin J C. A clinicopathological study of enterocyte-adherent Escherichia coli: a cause of protracted diarrhea in infants. Gastroenterology. 1982;83:441–454. [PubMed] [Google Scholar]
- 53.Russel M. Macromolecular assembly and secretion across the bacterial cell envelope: type II protein secretion systems. J Mol Biol. 1998;279:485–499. doi: 10.1006/jmbi.1998.1791. [DOI] [PubMed] [Google Scholar]
- 54.Sambrook J, Fritsch E F, Maniatis T. Molecular cloning: a laboratory manual. 2nd ed. Cold Spring Harbor, N.Y: Cold Spring Harbor Laboratory Press; 1989. [Google Scholar]
- 55.Sandkvist M, Michel L O, Hough L P, Morales V M, Bagdasarian M, Koomey M, DiRita V J. General secretion pathway (eps) genes required for toxin secretion and outer membrane biogenesis in Vibrio cholerae. J Bacteriol. 1997;179:6994–7003. doi: 10.1128/jb.179.22.6994-7003.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Scaletsky I C A, Silva M L M, Trabulsi L R. Distinctive patterns of adherence of enteropathogenic Escherichia coli to HeLa cells. Infect Immun. 1984;45:534–536. doi: 10.1128/iai.45.2.534-536.1984. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Sohel I, Puente J L, Murray W J, Vuopio-Varkila J, Schoolnik G K. Cloning and characterization of the bundle-forming pilin gene of enteropathogenic Escherichia coli and its distribution in Salmonella serotypes. Mol Microbiol. 1993;7:563–575. doi: 10.1111/j.1365-2958.1993.tb01147.x. [DOI] [PubMed] [Google Scholar]
- 58.Sohel I, Puente J L, Ramer S W, Bieber D, Wu C-Y, Schoolnik G K. Enteropathogenic Escherichia coli: identification of a gene cluster coding for bundle-forming pilus morphogenesis. J Bacteriol. 1996;178:2613–2628. doi: 10.1128/jb.178.9.2613-2628.1996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Sonnhammer E L, Von Heijne G, Krogh A. A hidden Markov model for predicting transmembrane helices in protein sequences. Proc Int Conf Intell Syst Mol Biol. 1998;6:175–182. [PubMed] [Google Scholar]
- 60.Stone K D, Zhang H-Z, Carlson L K, Donnenberg M S. A cluster of fourteen genes from enteropathogenic Escherichia coli is sufficient for biogenesis of a type IV pilus. Mol Microbiol. 1996;20:325–337. doi: 10.1111/j.1365-2958.1996.tb02620.x. [DOI] [PubMed] [Google Scholar]
- 61.Thomas J D, Reeves P J, Salmond G P C. The general secretion pathway of Erwinia carotovora subsp. carotovora: analysis of the membrane topology of OutC and OutF. Microbiology. 1997;143:713–720. doi: 10.1099/00221287-143-3-713. [DOI] [PubMed] [Google Scholar]
- 62.Tønjum T, Freitag N E, Namork E, Koomey M. Identification and characterization of pilG, a highly conserved pilus-assembly gene in pathogenic Neisseria. Mol Microbiol. 1995;16:451–464. doi: 10.1111/j.1365-2958.1995.tb02410.x. [DOI] [PubMed] [Google Scholar]
- 63.Traxler B, Beckwith J. Assembly of a hetero-oligomeric membrane protein complex. Proc Natl Acad Sci USA. 1992;89:10852–10856. doi: 10.1073/pnas.89.22.10852. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Traxler B, Boyd D, Beckwith J. The topological analysis of integral cytoplasmic membrane proteins. J Membr Biol. 1993;132:1–11. doi: 10.1007/BF00233047. [DOI] [PubMed] [Google Scholar]
- 65.Tusnády G E, Simon I. Principles governing amino acid composition of integral membrane proteins: application to topology prediction. J Mol Biol. 1998;283:489–506. doi: 10.1006/jmbi.1998.2107. [DOI] [PubMed] [Google Scholar]
- 66.van Geest M, Lolkema J S. Membrane topology and insertion of membrane proteins: search for topogenic signals. Microbiol Mol Biol Rev. 2000;64:13–33. doi: 10.1128/mmbr.64.1.13-33.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.Von Heijne G. The distribution of positively charged residues in bacterial inner membrane proteins correlates with the trans-membrane topology. EMBO J. 1986;5:3021–3027. doi: 10.1002/j.1460-2075.1986.tb04601.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68.Von Heijne G. Membrane protein structure prediction. Hydrophobicity analysis and the positive-inside rule. J Mol Biol. 1992;225:487–494. doi: 10.1016/0022-2836(92)90934-c. [DOI] [PubMed] [Google Scholar]
- 69.Von Heijne G. Membrane proteins: from sequence to structure. Annu Rev Biophys Biomol Struct. 1994;23:167–192. doi: 10.1146/annurev.bb.23.060194.001123. [DOI] [PubMed] [Google Scholar]
- 70.Von Heijne G, Gavel Y. Topogenic signals in integral membrane proteins. Eur J Biochem. 1988;174:671–678. doi: 10.1111/j.1432-1033.1988.tb14150.x. [DOI] [PubMed] [Google Scholar]
- 71.Wu S S, Wu J, Kaiser D. The Myxococcus xanthus pilT locus is required for social gliding motility although pili are still produced. Mol Microbiol. 1997;23:109–121. doi: 10.1046/j.1365-2958.1997.1791550.x. [DOI] [PubMed] [Google Scholar]
- 72.Yoshida T, Kim S R, Komano T. Twelve pil genes are required for biogenesis of the R64 thin pilus. J Bacteriol. 1999;181:2038–2043. doi: 10.1128/jb.181.7.2038-2043.1999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 73.Yun C H, Van Doren S R, Crofts A R, Gennis R B. The use of gene fusions to examine the membrane topology of the L-subunit of the photosynthetic reaction center and of the cytochrome b subunit of the bc1 complex from Rhodobacter sphaeroides. J Biol Chem. 1991;266:10967–10973. [PubMed] [Google Scholar]
- 74.Zhang H-Z, Donnenberg M S. DsbA is required for stability of the type IV pilin of enteropathogenic Escherichia coli. Mol Microbiol. 1996;21:787–797. doi: 10.1046/j.1365-2958.1996.431403.x. [DOI] [PubMed] [Google Scholar]
- 75.Zhang H-Z, Lory S, Donnenberg M S. A plasmid-encoded prepilin peptidase gene from enteropathogenic Escherichia coli. J Bacteriol. 1994;176:6885–6891. doi: 10.1128/jb.176.22.6885-6891.1994. [DOI] [PMC free article] [PubMed] [Google Scholar]